Abstract
Hematopoietic stem and progenitor cell (HSPC)-based ex vivo gene therapy has demonstrated clinical success for X-linked severe combined immunodeficiency (SCID-X1) patients who lack a suitable donor for HSPC transplantation. Nevertheless, this form of treatment is associated with an increased risk of infectious disease complications and genotoxicity mainly due to the conditioning regimen. In addition, ex vivo gene therapy approaches require sophisticated facilities to manufacture gene-modified cells and to care for the patients after chemotherapy. Considering these impediments, we have developed an in vivo gene therapy approach to treat canine SCID-X1 after HSPC mobilization and systemic delivery of the therapeutic vector. Here, we investigated the use of the cocal envelope to pseudotype a lentiviral (LV) vector expressing a functional gammaC gene. The cocal envelope is resistant to serum inactivation compared with the commonly used vesicular stomatitis virus envelope glycoprotein (VSV-G) envelope and thus well suited for systemic delivery. Two SCID-X1 neonatal canines treated with this approach achieved long-term therapeutic immune reconstitution with no prior conditioning. Therapeutic levels of gene-corrected CD3+ T cells were demonstrated for at least 16 months, and all other correlates of T cell functionality were within normal range. Retroviral integration-site analysis demonstrated polyclonal T cell reconstitution. Comparative analysis of integration profiles of foamy viral (FV) vector and cocal LV vector after in vivo gene therapy found distinct integration-site patterns. These data demonstrate that clinically relevant and durable correction of canine SCID-X1 can be achieved with in vivo delivery of cocal LV. Since manufacturing of cocal LV is similar to VSV-G LV, this approach is easily translatable to a clinical setting, thus providing for a highly portable and accessible gene therapy platform for SCID-X1.
Keywords: hematopoietic stem cells, severe combined immunodeficiency, SCID-X1, in vivo gene therapy, lentiviral vector, stem cell mobilization, canine animal model
Introduction
X-linked severe combined immunodeficiency (SCID-X1) is a genetically inherited life-threatening disease associated with mutations in the interleukin-2 receptor γ chain (IL-2RG or γc) gene.1 The γ chain is essential in the development and function of lymphocytes because the receptor is shared by diverse cytokines critical for the biology of lymphocytes.2 Mutations in the γ chain gene lead to a lack of γc protein, which in turn leads to an absence of T, natural killer, and functional B cells.3 SCID-X1 is universally fatal within the first year of life in infants due to severe opportunistic infections owing to a defect in the cellular and humoral immune system. Implementation of newborn screening (NBS) has aided in early diagnosis of the disease and designing a treatment strategy.4 The current mainstay to restore the immunity includes either allogeneic hematopoietic stem cell transplantation (allo-HSCT) or autologous ex vivo stem cell gene therapy (auto-SCGT).5 Allo-HSCT with a matched sibling donor is curative but available for <20% of patients. Transplants from unrelated donors yield to increased morbidity and mortality due to transplant-associated risks of graft-versus-host-disease, genotoxicity of conditioning regimen, and suboptimal restoration in immunity.6,7
Ex vivo gene therapy with hematopoietic stem and progenitor cells (HSPCs) utilizing viral vectors has been used in multiple clinical trials5,8 as a surrogate measure to circumvent the complications associated with allo-HSCT. In the ex vivo auto-SCGT, HSPCs are collected from the patient's bone marrow (BM) or from mobilized peripheral blood (PB), and modified with a functional copy of the coding region of γc using gamma-retroviral or lentiviral (LV) vectors.9 Despite the undeniable therapeutic benefits offered by auto-SCGT for SCID-X1, this approach poses several limitations. The first and foremost concern is the requirement of chemotherapy conditioning, which could cause severe genotoxicity.10 Second, manipulation of HSPCs outside the patient's body may compromise “stemness,” which could lead to reduced engraftment after transplantation.11 Third, due to suboptimal immune reconstitution, gene therapy patients still require lifelong administration of intravenous immunoglobulins.12 Furthermore, even though the diagnosis of infants is confirmed in very first week of birth with NBS, the treatment is not administered until 2–6 months postdiagnosis, in part, due to the delays in the manufacture of the genetically modified cells.4 Finally, the limited availability of sophisticated transplant centers and elegant good manufacturing product cell manufacturing facilities became evident in the most recent human SCID-X1 clinical trials.5
Considering these impediments, we previously developed a novel and accessible in vivo gene therapy approach using foamy viral (FV) vectors without prior conditioning in a canine model of SCID-X1.13,14 In vivo gene therapy consists of administration of viral vector carrying the functional copy of γC cDNA directly into the patient's bloodstream, thereby circumventing the many limitations of ex vivo auto-SCGT, including the need for manipulation of HSPCs. We utilized the SCID-X1 canine model, which exhibits clinical and immunologic characteristics representative of human SCID-X1, thereby making it an ideal preclinical model to perform exploratory gene therapy strategies for human SCID-X1.15 We previously demonstrated that in vivo gene therapy combined with mobilization and FV expressing mCherry and γC under human phosphoglycerate kinase promoter (FV-PGK-mCherry-γC) not only therapeutically corrected the disease phenotype but also outperformed the clinically used elongation factor 1 alpha (EF1α) promoter in SCID-X1 dogs.14 Furthermore, dogs mobilized with a combination of AMD3100 plus recombinant canine granulocyte colony stimulating factor (rc-G-CSF) and intravenous injection of FV-PGK-mCherry-γC resulted in rapid immune reconstitution in the CD3+T cell compartment. Moreover, the treatment provided safe and effective long-term immunity, with overall survival spanning almost 4 years and one animal continues to be monitored. Although animals treated with FV-PGK-mCherry-γC exhibited enhanced kinetics of immune reconstitution and normal T cell functionality, the gene marking of B cells and myeloid cells measured very low.14 This low marking could be due to inefficient targeting of stem cells or due to lack of conditioning in the treated dogs as similar results were seen in human clinical trials. An additional challenge with the use of FV was the lack of any clinical-grade production pipeline to generate vector with adequate titers. Together, these findings in dogs prompted us to investigate the use of an LV vector pseudotype with our previously described cocal envelope as in vivo gene therapy platform.
Materials and Methods
Dog colony, animal experiments, and study approval
The SCID-X1 dog colony is maintained at the Fred Hutchinson Cancer Research Center (Fred Hutch) animal facility, and the Fred Hutch Institutional Animal Care and Use Committee (IACUC no. 50855) approved all animal experiments.
Production of cocal-pseudotyped LV vector and determination of vector titer
To generate LV expressing green fluorescent protein (GFP) and γC under the PGK promoter (PGK-GFP-2A-γC), FV transfer plasmid PGK-GFP-2A-γC-FV14 was digested with SpeI to extract the transgene and part of the promoter. The SpeI fragment was ligated into the LV transfer plasmid pRRL-cPPT-hPGK-MGMT P140K-WPRE16 digested with the same enzyme. Orientation of the insert was screened by digestion of potential clones with the NotI enzyme. The resulting LV vector was pseudotyped with cocal envelope, produced by transient transfection of 293T cells, and concentrated 100-fold as previously described.17
The titer of vector preparations was determined by adding vector preparations to human HT1080 fibrosarcoma cells plated at 1 × 105 cells/mL the day before vector addition. Protamine sulfate was added immediately before addition of vector at a final concentration of 8 μg/mL. Transduced cells were assayed by flow cytometry 3–4 days after vector exposure, and the percentage of fluorophore-expressing cells was used to calculate the number of transduction units per milliliter of vector preparation. In addition, endotoxin levels were measured in each preparation that was subsequently administered intravenously.
SCID-X1 dog treatment plan
SCID-X1 canines were treated as per the previously established protocol.14 Briefly, SCID-X1-affected neonatal pups at ∼1 kg (age ∼3 weeks) were injected subcutaneously (SQ) with 5 μg/kg of rc-G-CSF twice per day for 4 days and a last single dose of rc-G-CSF (5 μg/kg SQ) with AMD3100 (4 mg/kg SQ) administered 6–8 h before cocal LV injection (Supplementary Fig. S1 and Supplementary Table S1). Thirty minutes before vector administration, prophylactic diphenhydramine (antihistamine) was administered. For vector administration, the injection site was sterilely prepared, and a 23G catheter was placed in the peripheral vein. Vector was administered slowly over 2–5 min, and animals were monitored for injection-related reactions. Animals were monitored for temperature, pulse, and respiration rates every 15 min for the next 2 h, and hourly thereafter for the following 24 h.
In vivo gene marking and phenotypic analysis (flow cytometry)
Six hours after AMD3100 administration and immediately before cocal LV injection, 0.5 mL of PB was collected to measure CD34+ cell frequency in PB by staining with the anti-canine CD34 monoclonal antibody (clone 1H6; Serotec, Raleigh, NC). Gene marking and phenotype analysis of PB leukocytes were determined by flow cytometry using antibodies described in our previous study.13,14 Surface IL-2 receptor γC expression was determined by staining with the APC anti-human CD132 antibody clone TUGh4 (BioLegend, San Diego, CA). Blood was collected in ethylenediaminetetraacetic acid or heparin tubes, subjected to hemolysis, and washed in phosphate-buffered saline plus 2% fetal bovine serum. Flow cytometry sorting was performed on a BD FACS ARIA cell sorter, and gene marking analysis was conducted on a Symphony flow cytometer (Becton Dickinson, San Jose, CA) to measure fluorescent gene marking or fluorescent antibody cell surface receptor expression. Data containing flowcytometric files (fcs) were analyzed by FlowJo (FlowJo LLC, Ashland, OR) analysis software.
In vitro T lymphocyte functional assay and estimation of immunoglobulins
In the mitogen-induced proliferation assay, CD3+ lymphocytes were enriched using MACS® Columns and anti-mouse IgG microbeads (cat no. 130-048-401) by magnetic separation (Miltenyi Biotech, Germany), and 1 × 106 to 2 × 106 cells were stimulated with 5 μg/mL of phytohemagglutinin (Sigma, St. Louis, MO) for 48 h in complete medium at 37°C and 5% carbon dioxide. Cell proliferation was assessed using flow cytometric CellTracker™ dye assay (Thermo Fisher Scientific) as per the manufacturer's instructions. For phosphorylated STAT5 (pSTAT5) and phosphorylated STAT3 (pSTAT3) analyses, white blood cells (WBC) were incubated for 4–6 h at 37°C and 5% carbon dioxide in complete medium (Roswell Park Memorial Institute with 10% fetal calf serum, 1% l-glutamine, and 0.5% pen/strep), after which they were stimulated with IL-2 or IL-21 for 20–25 min as described previously.13,14 pSTAT3 and pSTA5 phosphorylation was subsequently monitored by intracellular staining with pSTAT3 antibody (BD Phosflow cat no. 557815) and pSTAT5 y694 antibody (BD Phosflow cat no. 612599), respectively, and analyzed by flow cytometry. Levels of polyclonal antibodies (pan immunoglobulin G [IgG], immunoglobulin A [IgA], and immunoglobulin M [IgM]) were measured from serum collected from FV- and cocal LV-treated animals from a commercially available source (test code SO633; Phoenix Laboratories, Seattle, WA).
T cell receptor excision circles and T cell receptor spectratyping analysis
For T cell receptor (TCR) excision circle (TCR excision circle [TREC]) analysis, PB was lysed and DNA was extracted from 5 × 106 cells using the Qiagen QIAamp DNA Blood Mini Kit (cat no. 51106). A real-time quantitative PCR method was used as previously described to detect signal joint TRECs.14 For spectratyping analysis, PB was hemolyzed and RNA was extracted from 5 × 106 WBC using the RNeasy Mini Kit (cat no. 74104; Qiagen, Valencia, CA). Complementary DNA was generated from 100 to 400 ng of RNA using 200 U of SuperScript II Reverse Transcriptase (cat no. 18064-022; Invitrogen, Grand Island, NY) and oligo dT following the manufacturer's instructions. Complementary DNA was amplified using 17 specific forward TCR vector β primers and a common 6-FAM-conjugated reverse primer, as previously published.14,18 The products were analyzed on an Applied Biosystems ABI 3730xl DNA Analyzer, and GeneMapper software (version 4.0) was used for the analysis of peak sizes (Life Technologies, Grand Island, NY).
Integration-site and bioinformatic analyses
Retroviral integration-site (RIS) analysis was performed on WBC collected longitudinally from PB and BM, and flow sorted T cells for H983 on day 273 after viral vector injection, as described previously.19 Processing of gDNA to amplify integration loci included modified genomic sequencing-PCR methods.20 Briefly, amplification of integration loci occurs through two PCRs. In the first PCR, the forward primer is aligned to the lentivirus long terminal repeat (LTR) and the reverse primer is aligned to a synthetic linker oligonucleotide that was assembled and ligated to fragmented genomic DNA. The forward primer in the second PCR is nested and aligns to the 3′-end of the sequence amplified by the LTR primer used in the first PCR. The reverse primer for the linker cassette is the same for both reactions.
RIS libraries were sequenced using the Illumina MiSeq next-generation sequencing platform for paired-end reads. For the Illumina data, the forward and reverse reads were stitched using PEAR with the -q 30 option to trim sequence reads after two bases, with a quality score below 30 being observed.21 Stitched FASTQ files and raw fast adaptive shrinkage thresholding algorithm (FASTA) files for all sequencing data were filtered using a custom C++ program. Each read was compared to the reference provirus LTR sequence. Reads with a <90% match to the LTR sequence were discarded. The LTR sequence was trimmed off remaining reads. Reads were then compared with vector sequence. Reads with ≥80% matches to the vector sequence were discarded. Remaining reads were output in FASTA format for alignment.
Identified genomic fragments were aligned to the dog genome assembly (CanFam3.1) (GenBank Assembly: GCA_000002285.2) to determine the chromosome, locus, and orientation of integration (e.g., Chr14_8020175_+). CanFam3.1 was downloaded from the Ensembl Genome Browser (https://www.ensembl.org/).22 Filtered and trimmed sequence reads were aligned to the reference genome using BLAST-like alignment tool (BLAT) with options -out = blast8, -tileSize = 11, -stepSize = 5, and -ooc = canFam3.11.ooc.23
The canFam3.11.ooc file contains a list of 11-mers occurring at least 2,253 times in the genome to be masked by BLAT and was generated using the following command:
$blat canFam3.2bit/dev/null/dev/null -tileSize = 11 -stepSize = 5 -makeOoc = canFam3.11.ooc -repMatch = 2253 as recommended by UCSC (http://genome.ucsc.edu/goldenpath/help/blatSpec.html23 and http://genome.ucsc.edu/FAQ/FAQblat.html#blat6). Resulting blast8 files were parsed using a custom python script. Any read with BLAT alignment length <30 or an alignment start >10 is discarded. The top scoring alignment and all alignments within 95% of the top score were saved for each read. The ratio of the best alignment to the second-best alignment is the degree with which the insert can be mapped to one location in the genome (multi-align-ratio). Starting with the highest count reads, reads with matching alignments were combined. Reads with multiple possible alignments were not discarded at this point but were grouped together with other reads with the same alignment(s). For each group of reads with matching alignments, the original FASTA sequence files were read, and then, the sequences were aligned with Clustal Omega.24 This alignment was used to build a single consensus sequence for the alignment group. The consensus sequence was then used to search the pool of all sequences that could not be aligned by BLAT, and any sequence with ≥90% identity was merged into the group. Starting with the highest count groups and comparing with the lowest count groups, groups with ≥90% sequence similarity were merged. Finally, when comparing all sequence files for one test subject, all groups with exact alignment matches were merged into one clone ID. Clone IDs with exact consensus sequence matches were also merged. Nonuniquely aligned groups (multi-align-ratio ≥0.9) that had ≥90% similar reference sequences were also merged into a single clone ID.25
Clone contribution graphs
RIS clone contributions were calculated by dividing the number of sequence reads for each clone by the total number of sequence reads associated with any clone ID for the sample. The mean maximum frequency of clones was calculated. A data frame was constructed including three columns: (1) days after treatment (or infusion product); (2) clone frequency in the sample; and (3) clone identifier (genomic locus). The data frame was visualized using the R package ggplot2 (http://ggplot2.org/).
We then used normalization to convert the number of sequence reads associated with each clone to frequencies for each sample. At this point, we determined the maximum contribution frequency of each clone observed. The contribution frequencies for each of these clones at each sampling time point are recorded. Each clone has two entries in the data frame, one for each sampling time point, with 0% values entered where sequence reads were not observed. This is repeated for every clone identified in any sample from the same subject. Visualization is then done using ggplot2 (http://ggplot2.org/) using geom_area/geom_bar. Clones contributing ≥1% of the detected pool in any sample from the same subject are denoted by a color ribbon/bar. All remaining identified clones are grouped into the gray ribbon/bar at the top of each plot.
The COSMIC database26 was used to analyze the genes implicated in cancer, and accordingly defined the tier 1 and tier 2 genes. To be classified into tier 1, a gene must possess a documented activity relevant to cancer, along with evidence of mutations in cancer, which change the activity of the gene product in a way that promotes oncogenic transformation. Tier 2 genes consist of genes with strong indications of a role in cancer but with less extensive available evidence.
Results
FV maintains long-term immunity in the SCID-X1 canines
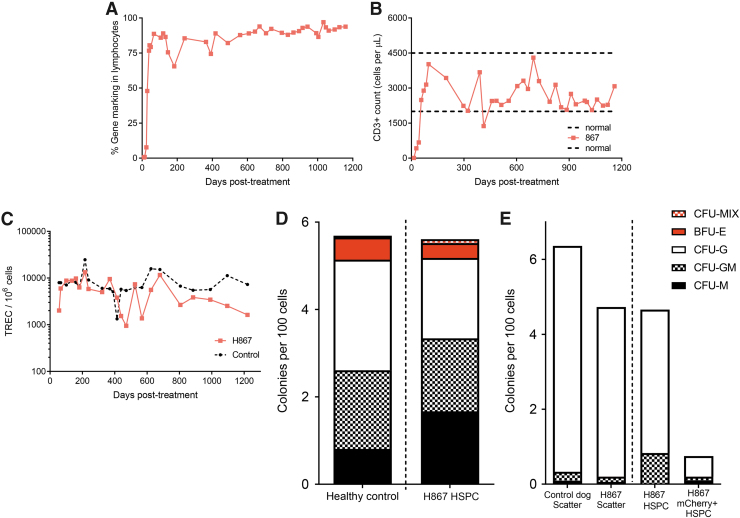
In our previous in vivo gene therapy studies to treat canine SCID-X1, we demonstrated that FV-PGK-mCherry-γC combined with the use of a mobilization regimen (rc-G-CSF and AMD3100) enhanced the kinetics of T lymphocyte reconstitution.14 Long-term follow-up of one animal treated with this approach (H867) confirmed the persistence of a functional adaptive immune response as indicated by normal number and functionality of T cells (Fig. 1A–C) and production of IgG and IgM (Supplementary Table S2). More importantly, H867 was able to mount normal immunoglobulin titers to common dog vaccines (canine distemper and canine parvovirus) (data not shown). Moreover, this animal maintained normal levels of TRECs (Fig. 1C) and CD45ra+CD3+ naive T cells (data not shown) in the periphery for over 45 months post-treatment that reflect sustained thymic output of naive T cells. Histopathological evaluation of tissues of the other treated animal at time of necropsy (H864, 16 months post-treatment) demonstrated the presence of plasma cells in the spleen and liver (data not shown). Long-term immune reconstitution in these dogs suggested that we were able to target either primitive HSPCs that reside in BM or early lymphoid progenitors that include extra- and intrathymic T cell progenitors, which subsequently gained a survival advantage upon correction.27 Upon BM aspiration ∼2.5–3 years post-treatment, CD34+ HSPCs obtained from treated animal H867 showed normal levels of colony forming potential when compared with healthy dogs (Fig. 1D). A small but reproducible frequency (0.02–0.08%) of these HSPCs was mCherry+CD34+ cells at 33 and 42 months post-treatment, suggesting that BM-derived HSPCs were transduced by FV-PGK-mCherry-γC. Flow cytometric sorting of these very rare CD45+CD34+mCherry+ cells showed clonogenic potential in colony forming assay (Fig. 1E), indicative of multilineage differentiation potential. These colonies were further confirmed to contain the FV provirus by mCherry-specific PCR amplification from genomic DNA isolated from single colonies (data not shown). These data indicate that we were able to successfully transduce a small number of HSPCs or their progenitors in situ with FV-PGK-mCherry-γC that maintained long-term differentiation potential, which might be sufficient to maintain long-term cellular (T cells) and humoral immunity (B cells). Although the FV-PGK-mCherry-γC treatment successfully corrected T cells and brought IgG and IgM to normal levels, gene marking in B cells, myeloid cells, and long-term HSPCs was low. Treated dogs remained susceptible to opportunistic infectious disease complications, resulting in the death of one of the FV-PGK-mCherry-γC-treated dogs due to a bacterial (Bordetella bronchiseptica) infection (Dog ID-H86428). Overall, intravenous delivery of FV to SCID-X1 dogs corrected immunological phenotype, maintained long-term immunity, and clinical outcome was substantially improved but partially unable to fight infections.
Figure 1.
Long-term immune restoration in T cell immunity and in situ transduction of BM CD34+ HSPCs. (A) Kinetics of gene marking based on fluorophore expression in PB lymphocytes from H867 treated with FV-PGK-mCherry-γC. Lymphocyte population was defined based on forward and side scatter. (B) Kinetics of CD3+ cell reconstitution (based on CD3+ cells per microliter of blood) in H867. (C) TRECs in PB of H867 (D) colony forming potential of BM CD34+ HSPCs in comparison with healthy dog 3.5 years postviral vector injection. (E) Colony forming potential of mCherry+ HSPCs, which shows transduction of stem cells by FV vector. Scatter was defined based on forward scatter (FSC) and side scatter (SSC) profile. HSPCs were defined as CD45+CD34+ cells from BM. BM, bone marrow; FSC, forward scatter; FV, foamy viral; FV-PGK-mCherry-γC, FV expressing mCherry and γC under human phosphoglycerate kinase promoter; HSPC, hematopoietic stem and progenitor cell; PB, peripheral blood; SSC, side scatter; TREC, T cell receptor excision circle.
Cocal-pseudotyped LV resists serum inactivation and transduces dog CD34+ HSPCs efficiently
In an effort to achieve a comprehensive immune reconstitution and to establish clinical utility, we investigated the use of LV pseudotyped with cocal envelope. The cocal-pseudotyped LV used in this study is self-inactivating (SIN) and uses a human PGK promoter to drive a codon-optimized human γC cDNA with enhanced green fluorescent protein (EGFP) expression for ease of in vivo gene marking monitoring. Using a transient transfection method and downstream processing to concentrate viral particles, we produced up to 3 × 108 TU/mL of cocal LV-PGK-EGFP-γC as previously published.17
To assess the feasibility of cocal-pseudotyped LVs for in vivo injection, we first examined how this vector resisted neutralizing by serum obtained from newborn SCID-X1 pups. Incubation of cocal LV with serum from SCID-X1 resulted in a drop in canine CD34+ cell transduction, but inactivation was two- to fourfold less than vesicular stomatitis virus envelope glycoprotein (VSV-G)-pseudotyped LV (Supplementary Fig. S2A). However, FV showed the maximum resistance from serum inactivation compared with cocal LV and VSV-G LV, consistent with our previous finding.29 Second, we investigated the transduction efficiency of cocal LV and FV on canine CD34+ HSPcs. In comparison with FV, cocal LV consistently showed higher transduction efficiency in canine CD34+ HSPCs ex vivo (Supplementary Fig. S2B). Based on these findings, we decided to pursue cocal LV for in vivo gene therapy.
Mobilization regimen with rc-G-CSF and AMD3100 increases peripheral WBC counts and efficiently mobilizes CD34+ HSPCs
HSPC mobilization is a process in which stem cells are forced to egress from BM into the blood.30 In the context of HSCT, these cells are used to repopulate the BM during a stem cell transplant. For auto-SCGT, mobilization is used to increase the yield of stem cells. In the context of in vivo gene therapy, HSPCs in the BM may not be accessible to intravenously injected gene transfer vectors. Mobilization is thus used to increase the number of circulating HSPCs for efficient targeting by systemic administration of viral vector. Previously, we have shown that the combination of rc-G-CSF and AMD3100 efficiently mobilizes canine CD34+ HSPCs.14
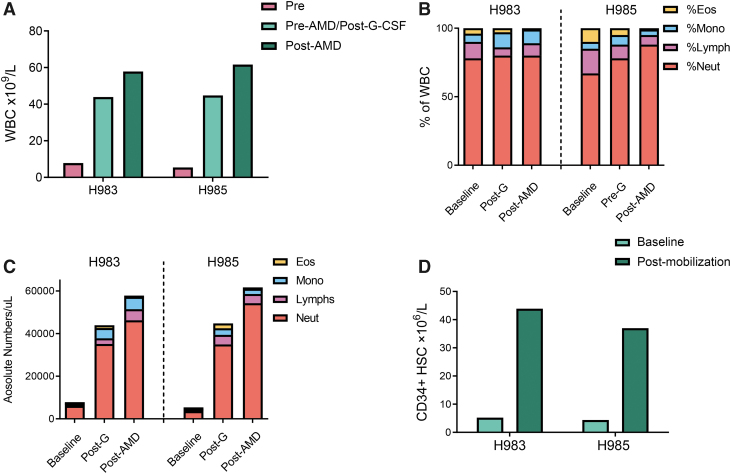
Treatment of two dogs (H983 and H985) with a mobilization regimen consisting of rc-G-CSF (5 μg/kg/bid SQ for 5 days) and a single administration of AMD3100 (4 mg/kg, SQ, 6 h before virus injection) uniformly resulted in leukocytosis. A significant increase in total WBC (7.4–11.4-fold from baseline, Fig. 2A) and the absolute neutrophil counts (7.50–14.90-fold from baseline, Fig. 2B, C) was observed at the time of vector administration. This finding is consistent with previously published data from adult healthy wild-type or SCID-X1 neonate dogs.14,31 Absolute lymphocyte and monocyte counts also increased by 4.4–5.5-fold and 9.0–12.0-fold, respectively (Fig. 2B, C). Interestingly, platelet counts in PB decreased in both pups postmobilization (Supplementary Fig. S8). There was no significant change in the hematocrits (data not shown). In addition, we found an 8.3–8.4-fold increase in the numbers of circulating CD34+ cells, respectively, in H983 and H985 before virus administration (Fig. 2D), which is in agreement with our previous study.14 Both pups tolerated the mobilization regimen without any appreciable short-term or long-term adverse effects. Based on these results, the combination of rc-G-CSF and AMD3100 efficiently mobilized CD34+cells in ∼3-week-old SCID-X1 puppies.
Figure 2.
Mobilization regimen of rc-G-CSF and AMD3100 efficiently mobilizes WBC and HSPCs. (A) Baseline and postmobilization WBC counts. (B) Pre- and postmobilization differential counts from complete blood count. (C) Absolute number of WBC pre- and postmobilization, G = rc-G-CSF; AMD = AMD3100. (D) Flowcytometric quantitation of canine HSPCs pre- and postmobilization in H983 and H985. HSPCs were defined as CD45int+CD34+ cells in PB. rc-G-CSF, recombinant canine granulocyte colony stimulating factor; WBC, white blood cells.
Intravenous injection of cocal LV expressing γC restores humoral immunity
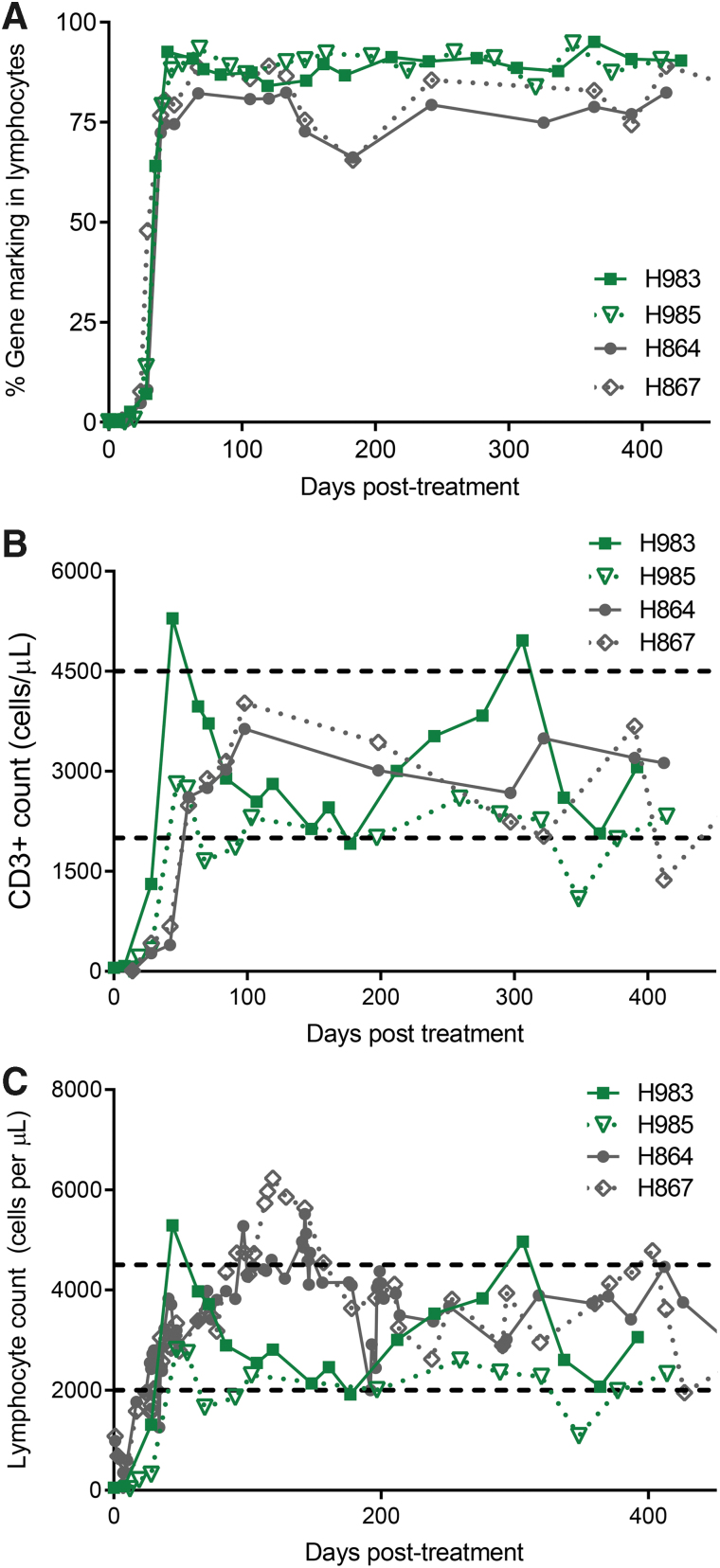
At birth, genomic analysis of canine IL2RG locus established genetic mutation, and analysis of WBC from H983 and H985 confirmed the absence of CD3+ T cells. Cocal LV-PGK-EGFP-γC vector, 4 × 108 IU, was injected in each pup (Supplementary Table S1) at 6 h post-AMD3100 administration, resulting in a rapid increase in gene-corrected T lymphocytes. Within 6 weeks, after vector injection, gene-corrected (GFP+) lymphocytes in PB reached 88–92% (Fig. 3A) of CD3+T lymphocytes, a level slightly higher than that reached in our previous study with FV (74–80%)14 at a similar time point. H983 showed 92.6% GFP+ CD3+ lymphocytes on day 44, and H985 showed 88% GFP+ CD3+ lymphocytes on day 47. In addition, both dogs showed normal levels of CD3+T lymphocytes in PB; accordingly, absolute CD3+ counts reached 2.3 × 109/L on day 35 in H983 and 2.4 × 109/L on day 47 postvirus injection (Fig. 3B). Correspondingly, various blood cell lineages remained within normal range in both dogs (Fig. 3C and Supplementary Fig. S3). H983 showed slightly faster reconstitution kinetics compared with H985, and both H983 and H985 showed slightly faster T cell kinetics compared with FV-PGK-mCherry-γC-treated dogs (H864 and H867, Fig. 4A, B). Vector copy number measured longitudinally from PB of cocal LV-treated dogs was comparable with FV-treated dogs14 (data not shown). We also verified by surface antibody staining that corrected lymphocytes expressed normal levels of γC as indicated by equivalent proportions and mean fluorescence intensity with the littermate control (H980) (Supplementary Fig. S7C). Despite very efficient HSPC mobilization and improvement in T lymphocyte reconstitution compared with FV-treated animals, cocal LV-treated dogs did not demonstrate correction of myeloid cells and B lymphocytes in an appreciable manner, with gene marking remaining minimal in both lineages similar to FV-treated dogs (Supplementary Fig. S5).
Figure 3.
T lymphocyte reconstitution with cocal-LV vector injection. (A) Kinetics of gene marking in PB lymphocytes from dogs treated with cocal-LV (H983 and H985) and compared with FV vector (H864 and H867) based on fluorophore expression. Lymphocyte population was defined based on forward and side scatter. (B) Kinetics of CD3+ cell reconstitution (based on CD3+ cells per microliter of blood) in the same animals as described in (A). Normal range of lymphocyte counts is shown by horizontal dashed lines. (C) Kinetics of lymphocyte reconstitution (based on lymphocyte per microliter count) in the same animals as described in (A). Horizontal dashed lines show normal range of lymphocyte counts. LV, lentiviral vector.
Figure 4.
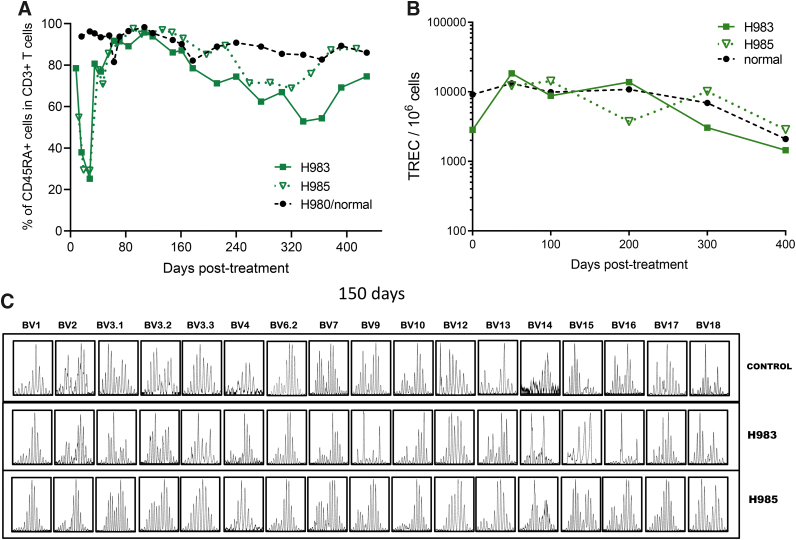
Thymopoiesis and TCR diversity in cocal-LV-treated dogs. (A) Fraction of CD45RA+ cells within the CD3+ population in PB of animals H983, H985 treated with cocal-LV-PGK-EGFP-γC vector. Data were compared with healthy control animal H980. (B) TRECs in PB of animals described in (A). (C) Rearrangement of the TCR beta chain was assessed by PCR amplification of complementary DNA using 17 different primer pairs (annotated on top) at 5 months post-treatment in an age-matched littermate control H980 and in treated SCID-X1 dogs H983 and H985. SCID-X1, X-linked severe combined immunodeficiency; TCR, T cell receptor.
The functional correction of T cells from cocal LV-treated dogs was validated by subjecting the CD3+ cells to T cell mitogen phytohemagglutinin (PHA). CD3+T cells from both treated animals did respond and proliferate in response to PHA stimulation as indicated by the dilution of cell tracer dye (Supplementary Fig. S7A). The functionality of the γC-dependent signaling pathways in corrected lymphocytes obtained from both cocal LV-treated animals was verified by measuring tyrosine phosphorylation of the effector molecules STAT3 and STAT5 following in vitro stimulation with IL-21 and IL-2, respectively. Compared with cells obtained from the normal littermate control, equivalent levels of STAT3 and STAT5 phosphorylation were detected in CD3+ lymphocytes from mobilized cocal LV-treated animals, H983 and H985 (Supplementary Fig. S7B). Recovery in B lymphocyte function was indirectly assessed with the quantitative measurement of various immunoglobulins (IgG, IgM, and IgA). By 13 months, levels of IgM were within normal range, and IgG levels were close to normal or subnormal range (Supplementary Table S3), indicating that both treated pups were able to class-switch immunoglobulins and generate a mature humoral immune response. In conclusion, these results demonstrated restoration of T cell-specific signaling pathways and B cell-specific isotype switching in cocal LV-treated SCID-X1 dogs.
Sustained thymic output, broad and diverse TCR repertoire generated in the cocal LV-treated animals
T cell reconstitution can involve both a thymus-dependent pathway (the generation of new naive T cells from HSPCs) and a thymus-independent pathway (peripheral expansion of preexisting memory T cells).27 In the newborn SCID-X1 pups, which lack T cells, thymus-dependent pathways have a major significance in the development of T cell immunity. In addition, the thymus-dependent pathway has the potential advantage of reconstituting antigen-specific responses. Thymus-dependent T lymphopoiesis studies have relied on cell surface molecules such as isoforms of CD45 (CD45RA) to differentiate naive recent thymic emigrants from memory or effector T cells. The two mobilized cocal LV-treated animals (H983 and H985) showed normal frequency (∼90%) of CD3+CD45RA+ T cells in PB starting at 6 weeks post-treatment (Fig. 4A) and stabilizing at 100 days postinjection. Importantly, naive CD4 (CD45RA+CD3+CD4+) and CD8 (CD45RA+CD3+CD8+) T cell subpopulations were also found to be within normal range compared with the normal healthy littermates (data not shown). Levels of CD3+CD45RA+ cells remained stable in H983 and H985 for at least 14 months post-treatment and continue to be monitored (Fig. 4A).
Although CD45RA+ naive T cells measure thymic output, T cells expressing a naive phenotype are not necessarily accurate surrogate markers of thymic function.32 T cell receptor gene rearrangement excisional circles are markers of developmental proximity to the thymus, and their concentrations in PB would allow for direct measurement of thymic output. Therefore, we measured TRECs in peripheral WBC in the cocal LV-treated animals. Normal TREC levels were seen as early as 3 months post-treatment and remained identical to the littermate control more than 400 days post-treatment (Fig. 4B), demonstrating that treatment of SCID-X1 canines increased the thymic output to levels comparable with a healthy control.
Naive T cells are continuously generated in the thymus where each cell undergoes DNA rearrangement to generate a unique T cell receptor.33 We assessed TCR diversity in each treated animal using TCRVbeta spectratyping, which analyzes genetic rearrangement of the variable region of the TCR beta gene. At 2 months of age, the TCR diversity between the control littermate (H980) and cocal LV-treated animals was limited as shown by a weak spectratyping profile (Supplementary Fig. S6A). However, by 5 months post-treatment, the two animals H983/H985 showed robust spectratype profiles, characterized by a Gaussian distribution of fragments sized across 17 families of TCRVbeta segments up to at 400 days post-treatment, similar to that of an age-matched normal littermate (Fig. 4C). These results demonstrate normal T cell maturation and TCR diversity in the cocal LV-treated SCID-X1 animals, representative of healthy animals.
Distinct CD4+ and CD8+ T cell dynamics within the CD3+ T cell population is a prognostic indicator of a healthy immune system and usually the CD4/CD8 ratio stays close to 2:1 in healthy adult dogs. Immunodeficiency disorders and infectious diseases are implicated in the deviation from this ratio. Upon examination of CD4+ and CD8+ T cell distribution within CD3+ T cells in PB during the early phase (until 3–4 weeks post-treatment) of reconstitution, the cells were either made of double-negative (CD4-CD8-) phenotypes or of CD4+ single-positive cells. By days 28–35, the majority of expanded CD3+ lymphocytes were mature, expressing the coreceptors CD4 or CD8, with a small fraction of cells being CD4/CD8 double positive or double negative (Supplementary Fig. S4). H985 showed normal CD4 to CD8 cell ratios averaging 2, which is similar to the control animal (H980; Supplementary Fig. S4). Interestingly, in dog H983, the ratio of CD4 to CD8 was reversed after day 175 post-treatment (Supplementary Fig. S4). The majority of circulating T lymphocytes in H983/H985 stained positive for TCR αβ starting at 2 months post-treatment (Supplementary Fig. S6b), while a small percentage (1–2%) showed the expression of TCR-gamma delta (data not shown), consistent with observations from healthy canines. Altogether, cocal LV-treated animals comprehensively exhibited all the attributes of a normal T cell immune repertoire.
RIS analysis reveals diverse and polyclonal integration pattern
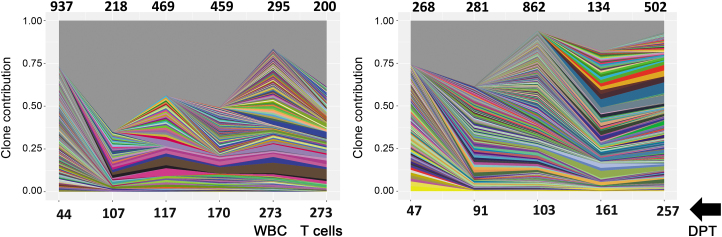
In our previous study with FV-PGK-mCherry-γC-treated dogs, we observed a polyclonal integration pattern with higher numbers of unique integrations in mobilized animals compared with nonmobilized animals.14 In the cocal LV-treated animals, RIS analysis showed a polyclonal integration pattern similar to FV-treated animals, although the total number of unique integrations at a given time point in the blood was consistently lower14 (Fig. 5), a result that is also confirmed by a previous study involving ex vivo modified human SCID repopulating cells.34 No clonal dominance was observed in cocal LV animals for nearly ∼270 days postvector injection, with the most abundant clones consistently detected over time (Fig. 5).
Figure 5.
ISA shows polyclonal diversity in cocal-LV-treated dogs. Clonal diversity in mobilized canines H983 (left) and H985 (right) at the indicated time points post-treatment with G-CSF/AMD3100 mobilization and vector cocal-LV-hPGK-EGFP-γC. In the dog H983, a comparative analysis was done on flow-sorted CD3+T cells and WBC on day 273 DPT. In all graphs, unique IS is plotted based on the number of times the IS was sequenced and normalized to the percentage of total IS captured at each time point for each animal. Each unique IS appearing at a frequency >1% in each sample is represented by a colored ribbon. The gray portion of the graph depicts all retroviral integration sites with a frequency <1% at each time point. Numbers on top of the graph represents unique integrations at each time point depicted in the bottom of chart. DPT, day post-treatment; ISA, integration-site analysis.
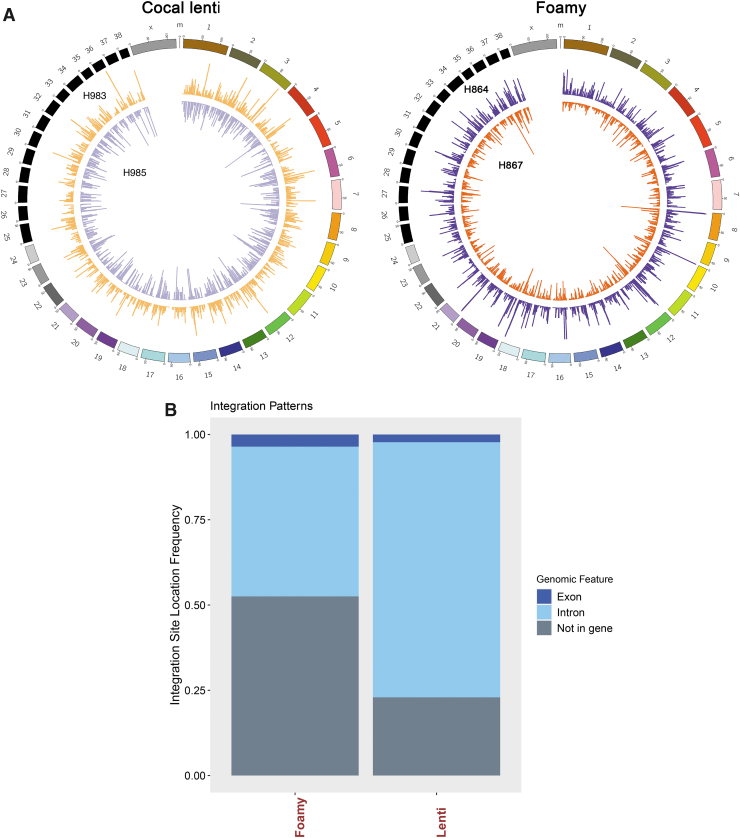
To compare the safety profile of each viral vector, we performed a comprehensive side-by-side analysis in the integration profile in all four dogs (Supplementary Table S4). The pattern of intragenic integration in cocal LV-treated dogs was distinct compared with FV-PGK-mCherry-γC-treated dogs (Fig. 6A, circos plots). In cocal LV-treated dogs, 77% and 47% of all integrations were found within the genes for cocal LV- and FV-treated dogs, respectively (exons and introns combined, Fig. 6B). In all dogs, integrations in introns were more frequent than exons.
Figure 6.
Comparative ISA (ISA in cocal LV and foamy vector-treated dogs) indicates random integration pattern. (A) Circos plot depicting integration sites across the dog genome and comparison of cocal-LV-hPGK-EGFP-γC-treated animals (H983 and H985, left circos plot) with that of FV-hPGK-mCherry-γC-treated (H864 and H867, right circos plot) animals. Each chromosome is represented on the exterior of the ring and is broken down into sequential bins, each 100 kB in size. The total number of unique integrations occurring within each bin is represented by the height of histogram bars. (B) Bar chart depicts the percentage of integration sites found within an exon, intron, or not in a gene for foamy- and cocal LV vector-treated dog. The total integrations plotted for foamy are 12,471 and for cocal LV dogs are 5,244.
We next identified vector-specific as well as common integration sites. Performing initially an unrestricted analysis, we found 3,158 genes with at least one read in at least one out of four dogs (Supplementary Table S11). Approximately 11% of these genes showed more than one read (365 genes, Supplementary Table S12). From this gene list, 239 (cocal LV) and 269 (FV) treatment-specific/unique integration sites were found with at least one read in both dogs (Supplementary Tables S5 and S6). Only 19 common genes were identified in both FV and cocal LV dogs (at least one dog in each group) (Supplementary Table S13). Among these 19 genes, 1 gene (MAML2) showed integration in all 4 dogs, and 2 genes (BCL2 and ADK2) showed integration in 3 dogs. These data indicate that both FV and cocal LV vectors show distinct and nonrandom integration patterns with overlap in only few loci.
For the distinct patterns, we next determined the vector-specific hot spots for viral integration. For this analysis only integrations with greater than five reads in at least one dog and present at any count in the second dog were considered (genes summarized in Supplementary Tables 7–10). Overall, virus-specific hot spots were found in cocal LV-treated dogs (164 total; H983:79 and H985: 85) in comparison with FV-treated dogs (total: 271; H864:68 and H867: 203). To evaluate whether these integration hot spots are associated with known oncogenes, we analyzed our data with that of the COSMIC database, a catalog for somatically acquired mutations found in various human cancers.26 Vector-specific integrations were present in 17 tier-1 genes in cocal LV dogs (Supplementary Table S14) and 17 tier-1 and 2 tier-2 genes in FV dogs (see the Materials and Methods section for tier 1 and 2 description, Supplementary Table S15).
In summary, we confirmed that both FV and cocal LV vectors could safely be utilized for SCID-X1 in vivo gene therapy. Integration-site analysis revealed a diverse polyclonal integration pattern, vector-specific integrations and hot spots, as well as the absence of clonal expansion.
Discussion
This study demonstrates that in vivo gene therapy with FV and cocal LV offers a portable and accessible treatment for canine SCID-X1 without the need of a conditioning regimen. Here, we present data obtained longitudinally for over 45 months that demonstrate a long-term therapeutic benefit of FV to rescue dogs with the SCID-X1 disease phenotype. Furthermore, we improved this platform by using clinically advanced LV pseudotyped with a cocal envelope to obtain immune correction in the SCID-X1 dogs. We also show that a regimen of G-CSF and AMD3100 efficiently mobilized dog HSPCs as indicated by an approximately eightfold increase of CD34+ HSPCs in PB. Rapid reconstitution of T cells was achieved, and treated dogs showed a naive (CD45RA+) phenotype and a rearranged TCRVβ locus. Importantly, the overall safety profile of the vector was favorable as shown by RIS analysis and we did not observe any genotoxicity.
LVs developed in the past two decades have demonstrated remarkable safety and efficacy in various clinical trials, including a recent human SCID-X1 clinical trial.5 Owing to their ability to transduce quiescent cells, LVs have been optimized to efficiently target CD34+ HSPCs in humans and a variety of preclinical animal models, including canine HSPCs.35–37 LVs are commonly pseudotyped with VSV-G, which confers a broad tropism and efficiently transduces HSCs in large animal models.36,38 However, there are several limitations of using VSV-G-pseudotyped LV, including a general scarcity in stable packaging cell lines to produce consistent and clinically relevant titers,39 inactivation by human serum complement,40 and potent immune responses against this envelope that may result in the elimination of gene-modified cells and prevent the repeated administration of VSV-G-pseudotyped vectors.41,42 Thus, many alternative pseudotypes43 have been evaluated, including the cocal vesiculovirus envelope glycoprotein,29 feline endogenous virus (RD11444), gibbon ape leukemia virus,45 baboon retroviral envelope glycoprotein,46 and, very recently, measles virus envelope.47 Although the cocal envelope is closely related to the VSV-G Indiana strain, it is serologically distinct, which is advantageous in terms of mounting an immune response and inactivation by serum. Indeed, cocal LVs were found to have broad tropism by efficiently transducing human, nonhuman primate, and canine HSCs, and to be more resistant to inactivation by human and canine serum than VSV-G LV.29 In addition, cocal LV can be produced at high titers due to availability of third-generation SIN LV producer cell lines.17 Therefore, cocal LVs may be ideally suited for in vivo injection to correct SCID-X1 canines.
In the current study, cocal LV showed greater resistance to inhibition by canine pup serum compared with VSV-G LV, which demonstrated the greatest reduction in the viral titers and transduction ability relative to cocal LV and FV. The observed reduction in titers could be due to potential neutralization by maternal antibodies, which are viable up to 12 weeks of age in pups48 or due to complement-mediated inactivation.49 Differences in the transduction performance of LV pseudotyped with a variety of envelopes may be attributed to quiescent versus activated HSPCs and different receptor usages,50,51 although cocal LV and VSV-G likely share the same receptor.17 In the current study, we found that cocal LV consistently outperformed FV in transducing canine CD34+ cells ex vivo. Therefore, in using cocal LV, we can significantly decrease the required vector doses (higher transduction efficiency and lower serum inactivation tendency) without a significant loss of gene transfer efficiency for in vivo applications, thereby making this treatment accessible to a higher number of patients.
Hallmarks of immune reconstitution after cocal LV treatment reflect the general observations made previously in FV-treated dogs14 but with a slightly faster recovery of CD3+ T cells in cocal LV-treated dogs, presumably due to transduction of a larger pool of primitive HSPC and T cell progenitors in situ or more efficient targeting of these cells by cocal LV relative to FV. Although in the cocal LV-treated dogs, gene marking levels in myeloid and B cells remained low, akin to FV-treated dogs, notably, we document the serum IgG and IgA production and normal levels of IgM, which is suggestive of antibody isotype switching in a similar time frame to FV dogs. In FV-treated dogs, the antibody levels were not in the normal range until 18–24 months postinjection, a circumstance that we have now (∼16 months post-treatment) begun to observe in cocal LV-treated dogs. Furthermore, on histopathology, the presence of plasma cells in FV-treated dog H864 could explain the normal levels of antibody isotypes. While we found low levels of gene marking in CD20+ B cells, the mechanism behind the generation of antibody-secreting plasma cells will need to be explored in detail in future studies. Interestingly, γc is required for biologic responses by B cells to IL-2 and IL-15, but expression of functional γc is not required for B cell activation, proliferation, or IgE secretion in response to IL-4 or IL-13.52 In addition, the ability of SCID-X1 B cells to respond to both IL-4 and IL-13 suggests that B cell immunodeficiency in these patients may be caused by defective responses to other cytokines or simply by an absence of T cells. On a similar note, IL-21 is considered to be the primary cytokine required for human B cell differentiation in vivo, for the generation of plasma cells and cross talk between CD4+T cell and B cells.53,54 Therefore, the restoration of T cell functions and IL-2, IL-15, and IL-21 cytokine signaling may have helped to generate at least some of the B cell's normal functions, such as the ability to undergo isotype switching. In addition, observations made from past HSCT indicate that it is possible to have normal B cell function in the absence of B cell engraftment, which may occur both in the absence and presence of conditioning regimen.55 It may be true in the case of in vivo gene therapy where we have seen low gene marking in B cells, but normal levels of antibody isotypes with no conditioning. In addition, the presence and long-term maintenance of rare populations of gene-marked CD34+ HSPCs in FV-treated dogs more than 3.5 years after treatment may have contributed toward the generation of plasma cells. Low levels of gene marking in B and myeloid cells could also be explained by the fact that B and myeloid progenitors do not have selective survival advantage compared with T cell progenitors. Overall, the presence of various antibody isotypes in both cocal LV- and FV-treated dogs, and the presence of plasma cells in an FV-treated dog (H864) indicate that in vivo gene therapy has the ability to correct at least some of the deficiencies in the B cell compartment.
Ex vivo gene therapy with next-generation SIN gammaretroviral and LV vectors in human clinical trials has shown improved safety profiles so far relative to conventional gammaretroviral vectors. While no serious adverse events have been reported, clonal expansion has been observed in patients treated with LV vectors56 and therefore is a cause for concern. Genome-wide RIS analysis can help define the spectrum of insertional mutagenesis expected of a viral vector and thereby its potential of genotoxicity. In our study, RIS showed a diverse polyclonal contribution, with a similar or greater number of unique clones in total nucleated cells from PB compared with the patients treated in the most recent ex vivo gene therapy clinical trial after 9–12 months of treatment.5 Comparative analysis of FV and cocal LV vector RIS displayed integration profiles consistent to what has previously been reported in other studies,19,34,57,58 but with the important distinction that in the current study, these viral vectors were directly injected into the bloodstream of the dogs. FV showed a higher number of unique clones, the cocal LV has an integration preference within genes, and both FV and cocal LV have a higher tendency of integration in introns over exons. Based on these observations, we can conclude that the genome-wide distribution of both FV and cocal LV proviruses is nonrandom in the SCID-X1dogs. Whether there is an FV- or cocal LV-specific tendency of integrations in a genomic locus requires further assessment and validation. While we see genes with a higher integration frequency in all four dogs, whether these genes can be classified, as “hot spots” still needs to be investigated. However, none of the genes with the highest integration frequencies (putative hotspots) was associated with clonal dominance in our study. In addition, it is important to stress that absence of integrations at any given time point could be affected by sampling depth (not having enough samples), capture efficiency, and sensitivity of the RIS pipeline.59 Therefore, it will be necessary to continue to monitor the treated animals long term. Nevertheless, none of the dogs (H867 nearly 45 months; H983 and H985 nearly 18 months) has shown any adverse clinical outcome due to clonal expansion, reflecting the safety of these vectors as in vivo gene therapy platforms to treat SCID-X1.
Altogether, in vivo gene therapy offers a great strategy particularly for SCID-X1 and Fanconi anemia (FA) patients owing to the ability to transduce CD34+ HSPCs in situ in their natural niche. In the case of SCID-X1, a small percentage of corrected stem cells will have a selective survival advantage and will be enough to correct the disease phenotype. In the case of FA, the genetic defect causes fragility of the HSPCs, which are strongly reduced in number in these patients.60 Moreover, cytokine stimulation induces apoptosis in HSPCs in FA patients. Therefore, abolishing HSPC manipulation combined with efficient gene transfer in resting HSPCs in vivo without a genotoxic-conditioning regimen would be invaluable for FA gene therapy.61,62
In summary, the cocal LV allowed for the correction of SCID-X1 dogs' clinical phenotype and achieved long-term immune reconstitution. We believe our encouraging findings in two dogs with cocal LV merit additional studies with this approach to further consolidate these findings and evaluate potential long-term toxicity. Therefore, we propose to expand our in vivo gene therapy toolbox by adding cocal LV to the already established FV-based platform. We further propose that in vivo gene therapy with FV or cocal LV could serve as a bridging therapy for patients waiting for bone marrow transplantation (BMT) or auto-SCGT. A good starting point could be to utilize in vivo gene therapy in patients where BMT and ex vivo gene therapy have only resulted in partial immune reconstitution. Thus, the development and validation of in vivo gene therapy in clinical trials with human patients are warranted as a potential alternative to current hematopoietic stem and progenitor cell transplantation and ex vivo approaches. In conclusion, cocal LV- and FV-based vectors can be used for HSPC-based in vivo gene therapy to treat SCID-X1 and hold promise to treat other nonmalignant monogenetic blood disorders.
Supplementary Material
Authors' Contributions
H.-P.K. is the principal investigator of the study and coordinated the overall execution of the projects. H.-P.K., Y.S.R, and O.H. conceived the project; Y.S.R. designed the experiments; Y.S.R. and S.M.C. performed the experiments; Y.S.R. analyzed the data; S.R. helped in flow cytometric and CFU assays. D.P. and M.E. performed all bioinformatic analysis and generated all RIS figures; Y.S.R. wrote the article, and OH, H.-P.K., S.R., D.P., and M.E. edited the article. Y.S.R. assembled the figures.
Acknowledgments
We thank Helen Crawford for help preparing and formatting this article and the figures. We thank the veterinary and animal care staff of the Comparative Medicine Department, at the Fred Hutchinson Cancer Research Center, for taking care of the canine colony; H.-P.K. is a Markey Molecular Medicine Investigator and received support as the inaugural recipient of the Jose Carreras/E. Donnall Thomas Endowed Chair for Cancer Research and the Endowed Chair for Stem Cell and Gene Therapy.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health (NIH), which had no involvement in the in-study design; the collection, analysis, and interpretation of data; the writing of the report; nor in the decision to submit the article for publication.
Author Disclosure
H.-P.K is consulting for Rocket Pharma, Homology Medicines, CSL Behring, Vor Biopharma, and Magenta Therapeutics. Other authors declare that they have no competing financial interests.
Funding Information
This work has been supported, in part, by grants from the NIH, Bethesda, MD: HL122173 (H.-P.K.).
Supplementary Material
References
- 1. Noguchi M, Yi H, Rosenblatt HM, et al. . Interleukin-2 receptor gamma chain mutation results in X-linked severe combined immunodeficiency in humans. Cell 1993;73:147–157. [DOI] [PubMed] [Google Scholar]
- 2. Leonard WJ, Lin JX, O'Shea JJ. The gammac family of cytokines: basic biology to therapeutic ramifications. Immunity 2019;50:832–850. [DOI] [PubMed] [Google Scholar]
- 3. Buckley RH. Molecular defects in human severe combined immunodeficiency and approaches to immune reconstitution (Review). Annu Rev Immunol 2004;22:625–655. [DOI] [PubMed] [Google Scholar]
- 4. Dorsey MJ, Dvorak CC, Cowan MJ, et al. . Treatment of infants identified as having severe combined immunodeficiency by means of newborn screening. J Allergy Clin Immunol 2017;139:733–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mamcarz E, Zhou S, Lockey T, et al. . Lentiviral gene therapy combined with low-dose busulfan in infants with SCID-X1. N Engl J Med 2019;380:1525–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pai SY, Logan BR, Griffith LM, et al. . Transplantation outcomes for severe combined immunodeficiency, 2000–2009. N Engl J Med 2014;371:434–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wilhelmsson M, Vatanen A, Borgstrom B, et al. . Adverse health events and late mortality after pediatric allogeneic hematopoietic SCT-two decades of longitudinal follow-up. Bone Marrow Transplant 2015;50:850–857. [DOI] [PubMed] [Google Scholar]
- 8. De Ravin SS, Wu X, Moir S, et al. . Lentiviral hematopoietic stem cell gene therapy for X-linked severe combined immunodeficiency. Sci Transl Med 2016;8:335ra357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Staal FJT, Aiuti A, Cavazzana M. Autologous stem-cell-based gene therapy for inherited disorders: state of the art and perspectives. Front Pediatr 2019;7:443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thompson AA, Walters MC, Kwiatkowski J, et al. . Gene therapy in patients with transfusion-dependent beta-thalassemia. N Engl J Med 2018;378:1479–1493. [DOI] [PubMed] [Google Scholar]
- 11. Glimm H, Oh IH, Eaves CJ. Human hematopoietic stem cells stimulated to proliferate in vitro lose engraftment potential during their S/G(2)/M transit and do not reenter G(0). Blood 2000;96:4185–4193. [PubMed] [Google Scholar]
- 12. Hacein-Bey-Abina S, Hauer J, Lim A, et al. . Efficacy of gene therapy for X-linked severe combined immunodeficiency. N Engl J Med 2010;363:355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Burtner CR, Beard BC, Kennedy DR, et al. . Intravenous injection of a foamy virus vector to correct canine SCID-X1. Blood 2014;123:3578–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Humbert O, Chan F, Rajawat YS, et al. . Rapid immune reconstitution of SCID-X1 canines after G-CSF/AMD3100 mobilization and in vivo gene therapy. Blood Adv 2018;2:987–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Felsburg PJ, De Ravin SS, Malech HL, et al. . Gene therapy studies in a canine model of X-linked severe combined immunodeficiency. Hum Gene Ther Clin Dev 2015;26:50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gori JL, Beard BC, Ironside C, et al. . In vivo selection of autologous MGMT gene-modified cells following-reduced intensity conditioning with BCNU and temozolomide in the dog model. Cancer Gene Ther 2012;19:523–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Humbert O, Gisch DW, Wohlfahrt ME, et al. . Development of third-generation cocal envelope producer cell lines for robust lentiviral gene transfer into hematopoietic stem cells and T-cells. Mol Ther 2016;24:1237–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vernau W, Hartnett BJ, Kennedy DR, et al. . T cell repertoire development in XSCID dogs following nonconditioned allogeneic bone marrow transplantation. Biol Blood Marrow Transplant 2007;13:1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sheih A, Voillet V, Hanafi LA, et al. . Clonal kinetics and single-cell transcriptional profiling of CAR-T cells in patients undergoing CD19 CAR-T immunotherapy. Nat Commun 2020;11:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Beard BC, Adair JE, Trobridge GD, et al. . High-throughput genomic mapping of vector integration sites in gene therapy studies. Methods Mol Biol 2014;1185:321–344. [DOI] [PubMed] [Google Scholar]
- 21. Zhang J, Kobert K, Flouri T, et al. . PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014;30:614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cunningham F, Achuthan P, Akanni W, et al. . Ensembl 2019. Nucleic Acids Res 2019;47:D745–D751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res 2002;12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sievers F, Higgins DG. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci 2018;27:135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hocum JD, Battrell LR, Maynard R, et al. . VISA—Vector Integration Site Analysis server: a web-based server to rapidly identify retroviral integration sites from next-generation sequencing. BMC Bioinform 2015;16:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tate JG, Bamford S, Jubb HC, et al. . COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res 2019;47:D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bhandoola A, von Boehmer H, Petrie HT, et al. . Commitment and developmental potential of extrathymic and intrathymic T cell precursors: plenty to choose from. Immunity 2007;26:678–689. [DOI] [PubMed] [Google Scholar]
- 28. Rajawat YS, Humbert O, Kiem HP. In-vivo gene therapy with foamy virus vectors. Viruses 2019;11:E1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Trobridge GD, Wu RA, Hansen M, et al. . Cocal-pseudotyped lentiviral vectors resist inactivation by human serum and efficiently transduce primate hematopoietic repopulating cells. Mol Ther 2010;18:725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hopman RK, DiPersio JF. Advances in stem cell mobilization. Blood Rev 2014;28:31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Burroughs L, Mielcarek M, Little MT, et al. . Durable engraftment of AMD3100-mobilized autologous and allogeneic peripheral blood mononuclear cells in a canine transplantation model. Blood 2005;106:4002–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Douek DC, Vescio RA, Betts MR, et al. . Assessment of thymic output in adults after haematopoietic stem-cell transplantation and prediction of T-cell reconstitution. Lancet 2000;355:1875–1881. [DOI] [PubMed] [Google Scholar]
- 33. Thome JJ, Grinshpun B, Kumar BV, et al. . Longterm maintenance of human naive T cells through in situ homeostasis in lymphoid tissue sites. Sci Immunol 2016;1:eaah6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Everson EM, Olzsko ME, Leap DJ, et al. . A comparison of foamy and lentiviral vector genotoxicity in SCID-repopulating cells shows foamy vectors are less prone to clonal dominance. Mol Ther Methods Clin Dev 2016;3:16048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Case SS, Price MA, Jordan CT, et al. . Stable transduction of quiescent CD34(+)CD38(-) human hematopoietic cells by HIV-1-based lentiviral vectors. Proc Natl Acad Sci U S A 1999;96:2988–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Horn PA, Keyser KA, Peterson LJ, et al. . Efficient lentiviral gene transfer to canine repopulating cells using an overnight transduction protocol. Blood 2004;103:3710–3716. [DOI] [PubMed] [Google Scholar]
- 37. Jang Y, Kim YS, Wielgosz MM, et al. . Optimizing lentiviral vector transduction of hematopoietic stem cells for gene therapy. Gene Ther 2020. [E-pub]; DOI: 10.1038/s41434-41020-40150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Trobridge GD, Beard BC, Gooch C, et al. . Efficient transduction of pigtailed macaque hematopoietic repopulating cells with HIV-based lentiviral vectors. Blood 2008;111:5537–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ory DS, Neugeboren BA, Mulligan RC. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci U S A 1996;93:11400–11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. DePolo NJ, Reed JD, Sheridan PL, et al. . VSV-G pseudotyped lentiviral vector particles produced in human cells are inactivated by human serum. Mol Ther 2000;2:218–222. [DOI] [PubMed] [Google Scholar]
- 41. Carbonaro-Sarracino DA, Tarantal AF, Lee CCI, et al. . Dosing and re-administration of lentiviral vector for in vivo gene therapy in rhesus monkeys and ADA-deficient mice. Mol Ther Methods Clin Dev 2020;16:78–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shirley JL, de Jong YP, Terhorst C, et al. . Immune responses to viral gene therapy vectors. Mol Ther 2020;28:709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Levy C, Verhoeyen E, Cosset FL. Surface engineering of lentiviral vectors for gene transfer into gene therapy target cells. Curr Opin Pharmacol 2015;24:79–85. [DOI] [PubMed] [Google Scholar]
- 44. Kelly PF, Vandergriff J, Nathwani A, et al. . Highly efficient gene transfer into cord blood nonobese diabetic/severe combined immunodeficiency repopulating cells by oncoretroviral vector particles pseudotyped with the feline endogenous retrovirus (RD114) envelope protein. Blood 2000;96:1206–1214. [PubMed] [Google Scholar]
- 45. Glimm H, Kiem HP, Darovsky B, et al. . Efficient gene transfer in primitive CD34 +/CD38 lo human bone marrow cells reselected after long-term exposure to GALV-pseudotyped retroviral vector. Hum Gene Ther 1997;8:2079–2086. [DOI] [PubMed] [Google Scholar]
- 46. Girard-Gagnepain A, Amirache F, Costa C, et al. . Baboon envelope pseudotyped LVs outperform VSV-G-LVs for gene transfer into early-cytokine-stimulated and resting HSCs. Blood 2014;124:1221–1231. [DOI] [PubMed] [Google Scholar]
- 47. Levy C, Amirache F, Girard-Gagnepain A, et al. . Measles virus envelope pseudotyped lentiviral vectors transduce quiescent human HSCs at an efficiency without precedent. Blood Adv 2017;1:2088–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hartnett BJ, Somberg RL, Krakowka S, et al. . B-cell function in canine X-linked severe combined immunodeficiency. Vet Immunol Immunopathol 2000;75:121–134. [DOI] [PubMed] [Google Scholar]
- 49. Guibinga GH, Friedmann T.. Preparation of pseudotyped lentiviral vectors resistant to inactivation by serum complement. Cold Spring Harb Protoc 2010;2010; DOI: 10.1101/pdb.prot5420. [DOI] [PubMed] [Google Scholar]
- 50. Finkelshtein D, Werman A, Novick D, et al. . LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc Natl Acad Sci U S A 2013;110:7306–7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Marin M, Lavillette D, Kelly SM, et al. . N-linked glycosylation and sequence changes in a critical negative control region of the ASCT1 and ASCT2 neutral amino acid transporters determine their retroviral receptor functions. J Virol 2003;77:2936–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Matthews DJ, Clark PA, Herbert J, et al. . Function of the interleukin-2 (IL-2) receptor gamma-chain in biologic responses of X-linked severe combined immunodeficient B cells to IL-2, IL-4, IL-13, and IL-15. Blood 1995;85:38–42. [PubMed] [Google Scholar]
- 53. Moens L, Tangye SG. Cytokine-mediated regulation of plasma cell generation: IL-21 takes center stage. Front Immunol 2014;5:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Recher M, Berglund LJ, Avery DT, et al. . IL-21 is the primary common gamma chain-binding cytokine required for human B-cell differentiation in vivo. Blood 2011;118:6824–6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Haddad E, Leroy S, Buckley RH. B-cell reconstitution for SCID: should a conditioning regimen be used in SCID treatment? (Review). J Allergy Clin Immunol 2013;131:994–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cavazzana-Calvo M, Payen E, Negre O, et al. . Transfusion independence and HMGA2 activation after gene therapy of human -thalassaemia. Nature 2010;467:318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Beard BC, Keyser KA, Trobridge GD, et al. . Unique integration profiles in a canine model of long-term repopulating cells transduced with gammaretrovirus, lentivirus, and foamy virus. Human Gene Ther 2007;18:423–434. [DOI] [PubMed] [Google Scholar]
- 58. Trobridge GD, Miller DG, Jacobs MA, et al. . Foamy virus vector integration sites in normal human cells. Proc Natl Acad Sci U S A 2006;103:1498–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Adair JE, Enstrom MR, Haworth KG, et al. . DNA barcoding in nonhuman primates reveals important limitations in retrovirus integration site analysis. Mol Ther Methods Clin Dev 2020;17:796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tolar J, Adair JE, Antoniou M, et al. . Stem cell gene therapy for Fanconi anemia: report from the 1st International Fanconi Anemia Gene Therapy Working Group meeting. Mol Ther 2011;19:1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Srikanthan MA, Humbert O, Haworth KG, et al. . Effective multi-lineage engraftment in a mouse model of Fanconi anemia using non-genotoxic antibody-based conditioning. Mol Ther Methods Clin Dev 2020;17:455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rio P, Navarro S, Wang W, et al. . Successful engraftment of gene-corrected hematopoietic stem cells in non-conditioned patients with Fanconi anemia. Nat Med 2019;25:1396–1401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.