Abstract
Objective
Even a small fragment from the body of planarian can regenerate an entire animal, implying that the different fragments from this flatworm eventually reach the same solution. In this study, our aim was to reveal the differences and similarities in mechanisms between different regenerating fragments from this worm.
Materials and Methods
In this experimental study, we profiled the dynamic proteome of regenerating head and tail to reveal the differences and similarities between different regenerating fragments using 2-DE combined with MALDI- TOF/TOF MS.
Results
Proteomic profiles of head and tail regeneration identified a total of 516 differential expressed proteins (DEPs) and showed a great difference in quantity and fold changes of proteome profiles between the two scenarios. Briefly, out of the 516 DEPs, 314 were identified to be specific for anterior regeneration, while 165 were specific for posterior regeneration. Bioinformatics analysis showed a wide discrepancy in biological activities between two regenerative processes; especially, differentiation & development and signal transduction in head regeneration were much more complex than that in tail regeneration. Protein functional analysis combined with protein-protein interaction (PPI) analysis showed a significant contribution of both Wnt and BMP signaling pathways to head regeneration not but tail regeneration. Additionally, several novel proteins showed completely opposite expression between head and tail regeneration.
Conclusion
Proteomic profiles of head and tail regeneration identified a total of 516 differential expressed proteins (DEPs) and showed a great difference in quantity and fold changes of proteome profiles between the two scenarios. Briefly, out of the 516 DEPs, 314 were identified to be specific for anterior regeneration, while 165 were specific for posterior regeneration. Bioinformatics analysis showed a wide discrepancy in biological activities between two regenerative processes; especially, differentiation & development and signal transduction in head regeneration were much more complex than that in tail regeneration. Protein functional analysis combined with protein-protein interaction (PPI) analysis showed a significant contribution of both Wnt and BMP signaling pathways to head regeneration not but tail regeneration. Additionally, several novel proteins showed completely opposite expression between head and tail regeneration.
Keywords: Planarian, Proteomics, Regeneration, Signaling Pathway
Introduction
When damaged or wounded, some animals can restore the damaged structures, while some respond by undergoing wound healing and scarring (1). It has been widely recognized that many animals like nematode worm, snails, salamanders, frog tadpoles and planarian, have different degrees of regenerative capabilities. However, as far as we know, no other animals have yet been found to have the regenerative ability as powerful as planarian. Planarian is one of dorsoventrally flattened, free-living freshwater members of Phylum Platyhelminthes with amazing feats of restorative and physiological regeneration (2). This remarkable morphological plasticity has long since attracted the interest of many researchers (3, 4). In addition, this freshwater species possess the unique advantages of small body size, easy maintenance and low cost; accordingly, it has become an ideal model system for studying the regeneration phenomena, like morphogenesis, restoration of pattern and polarity, the underlying mechanism of stem cell proliferation and differentiation (5, 6).
Once wounding or amputation, the activated adult stem cells, collectively referred to as “neoblasts” abundant in the flatworms, are enter the cell cycle for proliferation and then differentiate to regenerate or reconstruct the damaged or missing tissue via a series of regulation mechanisms (7). Surprisingly, even one small fragment of a whole animal can rebuild an entire body within 1-2 weeks as quick (8). For instance, after amputating the head of planarian, the tail stump pieces will regenerate a new head structure accompanied with the formation of many organs like nerve system, brain, eyespot, epidermis and muscle, which is called planarian head regeneration (PHR); following amputation of tail, the remaining anterior fragment re-grows a new tail with the development of epidermis, muscle, nerve system and so on, which is named planarian tail regeneration (PTR); when amputating both head and tail, the remaining worm trunk fragment reformed the new anterior and posterior blastema, and the latter further differentiates to reform the missing structure of the flatworm (9).
Nowadays, it has been realized that polarity establishment and patterning programs play vital roles in deciding whether a head regenerates at anterior-facing wound or a tail at posterior-facing wound (10). This decision is made through the coordination among various signaling pathways like Hedgehog (Hh), Wnt/β-catenin and BMP pathways whose role in the reconstitution of anterior-posterior (A/P) and dorsal-ventral (D/V) polarity has been broadly approved (11- 14). In detail, studies have shown that Wnt/β-catenin pathway is extremely necessary for reestablishment of posterior polarity in planarian, and blocking this signaling pathway can lead to anterior regeneration; furthermore, Wnt pathway can be also accelerated by Hh pathway which is required for posterior polarity during planarian regeneration (15). Orii and Watanabe (16) reported that BMP4 gene silencing by RNA interference caused the transformation of dorsal side into ventral side in planarians. However, the knockdown of BMP pathway inhibitor Noggin, in turn caused a complete dorsal phenotype in planarians. From above description, it could be concluded that, in spite of the convergence of some biological processes (like muscle formation) in both scenarios, great divergence exists in many events (including above signaling pathways) which are involved in PHR and PTR. In recent years, the relevant research studies have been done on the similarity and difference between the two different regenerative scenarios. A representative study was that Kao et al. (17) performed a time-course RNA-seq on regenerating head and tail fragments was, compared differentially expressed transcripts at various time points between these two regenerative events, and found a huge difference in transcriptome profiles between the beginning of head and tail regeneration, whereas a similar transcriptional profile at 48 hours post amputation (hpa). Roberts-Galbraith et al. (18) analyzed the dynamic gene expression during the first 3 days of head regeneration in planarian and identified some genes specifically induced at the early phase, like soxP-1 acting as a transcriptional regulator of brain regeneration. Whereas, Tewari et al. (19) pointed out that Hox gene Post-2d was required for tail regeneration in planarian after activated by Wnt signaling pathway. Above studies suggested the remarkable difference in the transcripts involved in head and tail regeneration. However, above conclusion was just only obtained from the in vivo and in vitro transcriptional studies, and very little research at the proteome level has been done on the comparative analysis between PHR and PTR (20).
Therefore, in this study we applied 2-dimensional electrophoresis (2-DE) combined with matrix-assisted laser desorption/ionization time-of-flight/time-of-flight mass spectrometry (MALDI-TOF/TOF MS) techniques to separately measure the dynamic proteome of the regenerating head and tail from 6 to 168 hpa and construct a detailed protein database for PHR and PTR. By comparing proteomic profiles between PHR and PTR, we revealed the functional differences between these two regenerative scenarios, which maybe provided some valuable data for understanding the mechanisms undergoing planarian regeneration. For instance, the most significant expressed proteins in two processes might be extremely important to the corresponding regenerative process. The commonly-expressed proteins identified in this study show the differential regulation on head regeneration and tail regeneration because of its different dynamic expressions in two regenerative processes. Furthermore, some signal pathways, like BMP signaling, were more active in regenerating head, and how this signal pathway affects the amputated planarians needs an in-deep study.
Materials and Methods
Animal culture and treatments
In this experimental study, we used a clonal line of the planarian Dugesia japonica as experimental animals. The animals were obtained from Tagang Reservoir (Xinxiang, China) and reared in filtered tap water at 20°C in the dark. Planarians were fed with fresh fish spleen once every two weeks. All worms were 8-10 mm in length. Before amputation, planarians were starved for 7-10 days. The procedure of this research complied with national and international research ethics standards and was approved by the medical Ethics committee of the First Affiliated Hospital of Henan University of Science and Technology (No. 410305004330).
Preparation of the model for head regeneration and tail regeneration in planarian
Planarians were starved for at least one week prior to amputation. When surgery, the animals were placed on a pre-cooled block and amputated with a razor blade. For head regeneration, a cut at the anterior end of the pharynx was done to remove the anterior portion of the body, and the resting posterior part was allowed for continual growth for 6, 12, 24, 72, 120 and 168 hpa. The regenerating head pieces of 150 worms for each time point were pooled according to above-mentioned method, and immediately deep-frozen. 150 non-regenerating pieces (0-hour time point) served as the control. For tail regeneration, the animals were subjected to the same surgery as head regeneration, except for only one difference that regenerating tail fragments were pooled.
Extraction and quantitation of total proteins
The samples from the same group were harvested, snap frozen and grounded to fine powder in liquid nitrogen after adding a 2 mL of lysis buffer containing 7 M urea, 2 M thiourea, 18 mM DTT and 1% CHAPS (pH=7), (all of Yeasen Biotech Co., Ltd., Shanghai, China) followed by homogenization for 1 hour at 4°C. The homogenates were centrifuged at 4°C, 20000 g for 45 minutes. The supernatant was collected and immediately stored at -80°C for use. For the remaining precipitates, the same procedure as mentioned above was performed for further collecting the supernatant. And the concentration of protein in the supernatant was measured using a 2-D Quant kit according to the manufacturer’s protocol (GE Healthcare, USA).
Two-dimensional gel electrophoresis
One thousand μg of protein extract was loaded onto a 24 cm non-linear pH=3-10 immobilized pH gradient (IPG) strips (GE Healthcare) by rehydration. First-dimension isoelectric focusing was done using an Ettan IPGphor III (Bio-Rad, USA) under the following conditions: 30 V for 6 hours, 40 V for 7 hours, 100 V for 1 hour, 250 V for 2 hours, 500 V for 2 hours, 1000 V for 3 hours, gradient to 10 kV within 3 hours, finally keeping 10 kV for 12 hours. Following isoelectric focusing, the strips were washed with buffer solution I [stock solution (6 M Urea, 75 mM Tris-HCl pH=8.8, 29.3% glycerol, 2% sodium dodecyl sulfate (SDS), 0.002% bromophenol blue] plus 1% w/v DTT) for 15 minutes and then with buffer solution II (stock solution plus 2.5% w/v iodoacetamide) for 15 minutes at room temperature. Then, second-dimension separation based on molecular weights was carried out on a 12.5% SDS-PAGE. Gels were fixed with an aqueous solution consisting of 40% ethanol and 10% acetic acid, then stained in Coomassie Blue G-250 and visualized using QuantityOne software.
Analysis of 2-DE gel images
The Coomassie-stained 2-DE gels were scanned with a Typhoon FLA 9500 (GE Healthcare) and analyzed using the Imagemaster™ 2D Platinum software (v7.0, GE Healthcare) which is always used for spot intensity calibration, spot detection and background subtraction. For each group, three independent gels were run to minimize the experimental error. The stained spots were filtered and the quantity of each spot was normalized by total spot intensity. Student’s t test was used to evaluate the statistical significance of each spot. And P<0.05 was considered significant . Those spots with ≥2-fold change in expression level were chosen for subsequent mass spectrum (MS) analysis.
In-gel tryptic digestion and MALDI-TOF/TOF MS analysis
Those gel blocks stained with coomassie were manually excised, transferred into a 1.5 ml Eppendorf tube, destained using 2009L Milli-Q for 6 hours, and finally digested with sequencing-grade modified 0.01 μg/μL trypsin (2-3 μL) at 37°C overnight. 1 μL of the digested samples was eluted with an equal volume of matrix solution α-Cyano-4-Hydroxycinnamic Acid (HCCA, Sigma. USA) containing in 0.1% trifluoroacetic acid (TFA) and 50% acetonitrile (ACN), were dotted onto an AnchorChip™ MALDI target plate (Bruker Daltonics, Billerica, MA, USA). Peptide sequencing and protein identification were performed by MALDI-TOF/TOF MS method on an AutoFlex III mass spectrometer (BrukerDalton, Bremen, Germany) working in reflection mode as previously described (21). Polypeptide calibrator was used as an internal reference.
Mass spectrometry data processing and analysis
For peptide and protein identification of MALDI-TOF/TOF MS data, the resulting peptide peak lists were submitted to the MASCOT database search engine. Search parameters were selected as follows: trypsin digestion, one missed cleavage, carbamidomethylation as fixed modification, MetOxidation as the variable modifications, ± 100 ppm as precursor ion mass error tolerance, ± 0.5 Da as MS/MS fragment ion mass error tolerance, and 30:1 and 20:1 of signal-to-noise ratio of first order MS and secondary order MS, respectively. Confident protein identifications were defined as the highest protein score (at least 91% confidence level) on the database searching report, and a minimum of two matched peptides.
Identification of differential expressed proteins
As described above, in order to ensure the reliability of the results, MALDI-TOF/TOF MS identification was performed three times. The triplicate data were analyzed separately, and one identity observed in at least two replicates was considered as valid. For one identified protein, the average of three biological replicates was calculated as its expression level. The ratio of average value of regenerative group to that of the control group was defined as the fold change of one protein in expression. Briefly, the proteins with more than 2-fold, and less than 0.5-fold were considered as up-regulation, and down-regulation, respectively. Then the Student’s t test was used to evaluate the significance of DEPs, and a value of P<0.05 was considered significant.
Bioinformatics analysis
Biological themes for DEPs in planarian regeneration were annotated by using the the National Center for Biotechnology Information (NCBI) database platform and retrieving the relevant documents. To assess the similarities and differences of proteome profile among two different scenarios, and to obtain a visual understanding of the relationship between two regenerative scenarios, hierarchical clustering and cluster visualization was performed using cluster 3.0 data analysis tool combined with Eisen Treeview v1.6 based on the clusters of protein expression profiles. STRING analysis (https://string-db.org) was carried out for exploring the interaction relationship between DEPs in the regenerative process. The parameters for STRING search included interaction score of medium confidence, active interaction sources of text mining, experiments, databases, co-expression, neighborhood, gene fusion, and co-occurrence. The hits considered in this study had false discovery rates (FDR) lower than 0.01.
Statistical analysis
In this study, three biological replicates were analyzed for each pooled samples. And significant differences between the control and regenerative samples for all measurements were estimated by Student’s t test. A value of P<0.05 was taken as indicative of a statistically significant difference.
Results
Comparative proteome analyses between head regeneration and tail regeneration
To explore the difference of molecular mechanism undergoing head and tail regeneration in planarian, this study measured the dynamic protein expressions in the regenerating planarian at different recovery time (from 6 hpa to 168 hpa) on a proteome-wide scale. Three independent 2-D gel electrophoresis analyses were performed to avoid the experimental variation as possible. Then, triplicate gel images were integrated into a master gel for each pooled group. Totally 14 different groups (7 for each regenerative scenario) were established for identifying DEPs. On average, for head regeneration, the distinctive changes in 1595, 1591, 1506, 1300, 1448 and 1141 protein spots were detected in 2-DE gels at 6, 12, 24, 72, 120 and 168 hours after decapitation, respectively; while 1722, 1562, 1324, 1363, 1616 and 1401 spots were differently expressed at the corresponding time points in the regenerating tail. A further screening was performed on these protein spots according to the following stringent criteria: ≥2-fold change in expression when compared to the control, and consistency in expression trends in three replicates. Results showed that, out of the above spots, 1635 spots in PHR and 1641 spots in PTR met these criteria (data not shown).
As for those protein spots meeting above criteria, student’s t test was used for difference analysis on abundance changes between the regenerative group and the control group. The result was shown in Figure 1, in which the numbers of protein spots related to PHR and PTR were 1146 and 1053, respectively. The representative 2-DE gel maps are displayed in Figure S1 (See Supplementary Online Information at www.celljournal.org).
Fig.1.
A heatmap indicating DEPs spots related to planarian regeneration. A. Proteome profiles for PHR and B. Proteome profiles for PTR. Red-colored bin; Protein spots with ≥2-fold up-regulation, Green-colored bin; Protein spots with ≤0.5-fold down-regulation, Black-colored bin; The spots with insignificant difference in expression level, DEP; Differential expressed proteins, PHR; Planarian head regeneration, PTR; Planarian tail regeneration, and h; Hour.
Planarian regeneration-associated protein identification
Above identified protein spots were further sequenced by MALDI-TOF/TOF MS and calibrated, annotated and filtrated by MASCOT 2.2 database search engine. Results revealed that 1146 differentially expressed protein spots related to PHR were matched to 798 DEPs including 361 identified and 437 uncertain (or undefined) ones, and 1053 protein spots related to PTR were matched to 531 DEPs including 212 identified and 319 uncertain (or undefined) ones (Tables S1, S2, See Supplementary Online Information at www.celljournal.org).
Among above identified proteins, 361 PHR-related and 212 PTR-related DEPs were selected for heatmap clustering analysis (Fig .S2, See Supplementary Online Information at www.celljournal.org). In detail, among 361 PHR-related proteins, up-regulated DEPs were the most abundant (205 DEPs) accounting for 56.2% of the total, then down-regulated ones (107 DEPs), and up/down-regulated DEPs were least in number (49 DEPs); among 212 PTR-related proteins, the amount of up-regulated ones (98 DEPs) was predominant, down-regulated ones (76 DEPs) were the next, and the up/down-regulated ones were least. The detailed data was listed in Tables S3 and S4 (See Supplementary Online Information at www.celljournal.org), respectively. Notably, 34 proteins in regenerating head and 11 proteins in regenerating tail were interpreted as uncharacterized or hypothetical proteins, or proteins without specific function in the database, indicating that many planarians regeneration-related proteins still remain unknown in terms of their biological functions.
Fig.2.

Comparison of distribution of DEPs between two different regenerative processes. A. PHR and B. PTR. Overlapping part represents the DEPs commonly expressing in two regeneration scenarios. DEP; Differential expressed proteins, PHR; Planarian head regeneration, and PTR; Planarian tail regeneration.
According to above results, a total of 529 DEPs were identified as regeneration-related proteins. By comparing proteomic profile between PHR and PTR, it was found that i. The number of PHR-related DEPs was 361, obviously much more than that in tail regeneration (totally 212), ii. 314 DEPs were specific for head regeneration, while 165 for tail regeneration, iii. 47 DEPs were common in these two regeneration scenarios (Fig .2). Among 361 PHR-related DEPs, 15 proteins were upregulated by more than 4-fold. More specifically, CCD27, GI:223556, UNC4, D1YY19 and SRE2 proteins were predominantly up-regulated in the early stage (6-12 hpa). The resting 9 DEPs were increased in expression mainly between 24 hours-168 hours. Among the 15 DEPs, the upregulation of RENR was found to be the most significant, reaching a peak of 7.22-fold higher than the control at 24 hours of PHR. Of 212 PTR-related DEPs, there were just only 6 DEPs with >4-fold changes that were upregulated mainly at middle phase (72 pha) of tail regeneration. Among the 6 DEPs, the largest increase in expression was ALF2 taht showed a peak 6.43-fold upregulation at 72 hours of PTR. In addition, 47 DEPs were detected to be commonly expressed in two regenerative processes; including 15 up- and 9 down-regulated in both scenarios; and 23 DEPs with complicated expression patterns.
Of 47 common DEPs, 15 ones were up-regulated and 9 ones were down-regulated. Interestingly, the remaining 22 DEPs exerted different expression trends in these two scenarios. And the most upregulated DEPs (fold change>5) in head regeneration showed the opposite patterns in the regenerating tail. For instance, CA163 (Hcp beta-lactamase-like protein C1orf163 homolog) were 31.7-fold upregulated in PHR, while 0.22-fold downregulated in PTR; and DRE2 (Anamorsin homolog) were increased by 26.4-fold in PHR, while decreased by 0.07-fold in PTR. The most decreased DEPs during head regeneration, like D0VYP9 (Glyceraldehyde-3-phosphate dehydrogenase, 0.1-fold) and ENTP5 (Ectonucleoside triphosphate diphosphohydrolase 5, 0.08-fold), showed an extremely significant increase in the regenerating tail.
In addition, from the expression dynamics of planarian regeneration, the largest number of DEPs during head and tail regeneration were 133 DEPs up-regulated between 24-72 hours in regenerating heads and 88 DEPs up-regulated between 24-72 hours in tails, during which there was also obvious increase in the up-regulated DEPs in the regenerating tail, but not in the regenerating head. This implied the more complicated expression changes in the regenerating tail. However, the number of DEPs in regenerating head were more than that in the tail fragments in any phase of regeneration, suggesting that the regenerating head underwent the more drastic expression regulation than the tail fragments, which perhaps can be explained by the fact that a brain regulated by more rich population of proteins or homolog regenerates during head regeneration.
Functional classification of differentially expressed proteins
To gain insight into the potential difference in biological functions between PHR and PTR, the above-mentioned 361 head regeneration-related and 212 tail regeneration-related DEPs were analyzed by a DAVID web tool. As shown in Figure 3A, during head regeneration361 DEPs were mainly classified into 18 gene ontology (GO) categories based on the major category of “biological process”, out of which the largest group was differentiation and development (67 DEPs, 16.4%), followed by signal transduction (46 DEPs, 11.2%), metabolic process (11%), immunity and inflammation (8.0%) and cell proliferation (7.0%). Similarly, 212 tail regeneration-related DEPs were also classified into about 18 functional groups in “biological process” category (Fig .3B), and the top five groups were differentiation and development (30 DEPs, 16.0%), signal transduction (24 DEPs, 13.3%), metabolic process (11.0%), cell proliferation (8.3%) and cell biogenesis (6.1%). As shown in Figure 4, the two regeneration processes shared the same enriched functional groups, i.e., differentiation and development, and signal transduction, seeming no obvious difference between these two scenarios.
Fig.3.
The functional category distribution of differentially expressed proteins in two different regenerative events based on GO biological process. A. PHR and B. PTR. PHR; Planarian head regeneration, PTR; Planarian tail regeneration, and GO; gene ontology.
Function divergence of differentiation and development-related proteins between head regeneration and tail regeneration
The percentage of differentiation and development-involved proteins was the highest in both PHR and PTR, and there seemed to be no obvious difference between the two scenarios in this functional category. To unravel whether there was the distinction existing between two scenarios, we further performed sub-categorization analysis of these DEPs. The result showed that 67 development-related DEPs involved in PHR were mainly implicated in anterior/posterior pattern specification (e.g., homeobox protein Hox-B3, 14-3-3-like protein 1), eyespot formation (e.g., one transducing alpha subunit, retinoic acid receptor gamma 2I), nervous system development (e.g., protein Wnt-8b, alpha-tubulin N-acetyltransferase 2, ABC transporter ATP binding protein) and so on; for 30 differentiation and development-related proteinsin PTR, they were predominantly subgrouped into the following categories: brain development (e.g., arrestin, tubulin T beta15), epithelium morphogenesis (e.g., proteasome subunit beta type-5), and nervous system development (e.g., cullin-2, fasciculation and elongation protein zeta-1, kallikrein) as well (Fig .4).
Fig.4.
The expression profiling of development and differentiation-related proteins during two different regeneration processes. A. The expression profiles of related proteins during PHR and B. The expression profiles of related proteins during PTR. Expression changes of each protein were indicated by different color bars. Up-regulated, down-regulated and invariant proteins were highlighted in red, green and black -colored bins, respectively. PHR; Planarian head regeneration and PTR; Planarian tail regeneration.
Comparison analysis of signal transduction-related proteins between head regeneration and tail regeneration
It also can be seen from above results that the proportion of signal transduction-related DEPs was only second to those involved in differentiation development. To figure out whether there were the differences in signaling pathways involving PHR and PTR, this study carried out a further analysis on the “signal transduction” category. And the results were displayed in Figure 5. Briefly, during head regeneration, the “signal transduction” category was sub-grouped into Wnt signaling pathway (DEPs Wnt-8b, oxidoreductase domain-containing protein etc.), BMP signaling pathway (DEPs transforming growth factor beta-2, EIF4A isoform 1A etc.), Notch signaling pathway (Y-Box factor etc.) etc.; during regenerating tail, this category was further divided into the following signaling pathways: Rab protein signal transduction (Ras-related protein Rab-27A etc.), Wnt signaling pathway (DEPs V-type proton ATPase subunit C etc.) and others.
Fig.5.
The distribution of sub-categories of signal transduction-related proteins in two different regeneration processes. A. Sub-categories of the related proteins during PHR and B. Sub-categories of the related proteins during PTR. Red and green represent up-regulated and down-regulated proteins, respectively. PHR; Planarian head regeneration and PTR; Planarian tail regeneration.
PPI network analysis of DEPs involved in signal pathways
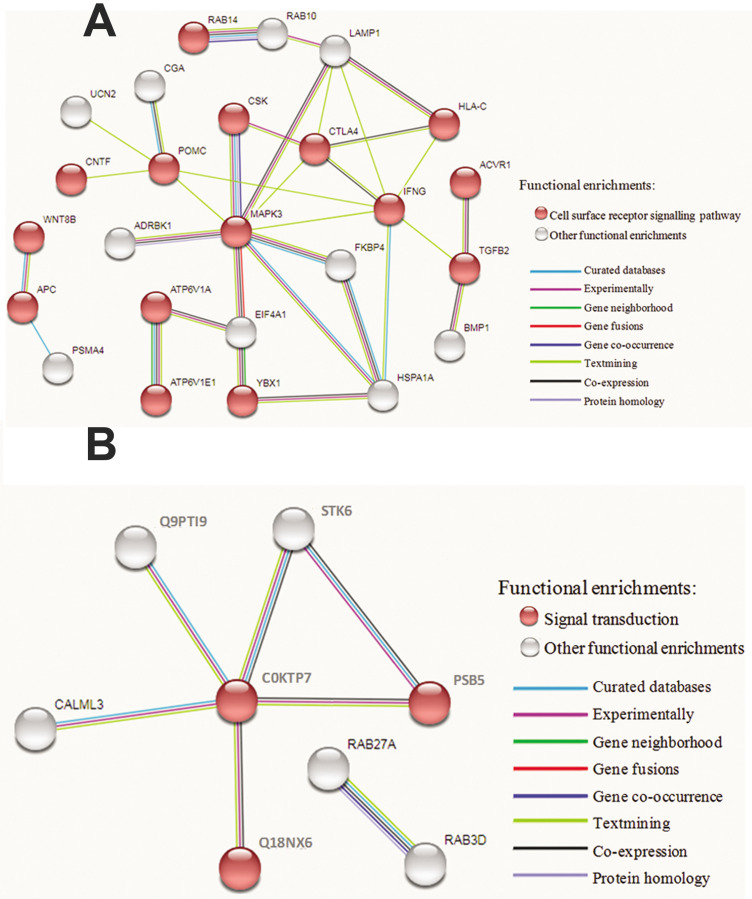
To find out what signaling pathway play a significant role in two different regenerative scenarios, respectively, the PPI network of DEPs involved in signal transduction was constructed according to the data in STRING. The results were shown in Figure 6. Briefly, for head regeneration PPI network comprised a total of 28 nodes and 33 edges with average node degree of 2.36 and average local clustering coefficient of 0.538, and the statistical analysis showed a significant change in "cell surface receptor signaling pathway" (FDR of 1.79e-08). Meanwhile, for tail regeneration, the PPI network obtained from STRING database contained 18 nodes and 7 edges with average node degree of 0.778 and local clustering coefficient of 0.394, and an obvious alteration with 0.0074 FDR in "signal transduction" was found through STRING analysis.
Fig.6.
STRING analyses of differentially expressed proteins (DEPs) involved in signaling pathways during two different regeneration processes. A. Analysis result of signaling pathways-involved DEPs during head restoration and B. Analysis result of signaling pathways-involved DEPs during tail regeneration.
Discussion
It is well-known that planarians have a remarkable regenerative ability and can regenerate whole animal body from small tissue fragments within an extremely short time (22). For instance, after cutting this flatworm into pieces along the front end of pharynx, the anterior segment can regenerate the posterior part from the wound, and vice versa, which implies that both of these two fragments from the animal contain the information required for reconstituting an entire body (23, 24). However, we lack an understanding of the discrepancies of the regeneration of different fragments especially at the proteomic level.
For this reason, we profiled the dynamic proteome of these two regeneration scenarios separately at 6, 12, 24, 72, 120, 168 hpa. According to the data, the protein expression pattern showed the temporal changes in both PHR and PTR. By comparing the proteomic profiles between the two scenarios, we identified many biologically meaningful DEPs. In brief, 361 proteins and 212 proteins were significantly differentially expressed during PHR and PTR, respectively, which was obvious that the number of DEPs in PHR was more than that in the latter. A possible reason for this discrepancy may be that a brain involving a rich battery of proteins regenerates during PHR. During head regeneration, 15 proteins showed a more than 4-fold increase in expression and were considered to play the critical role in this process. More interestingly, all of them were regulated in the whole regeneration process. Among these DEPs, RENR (renin receptor) showed the most significant upregulated expression with up to 7.22-fold higher than the control at 24 hpa. As a receptor for rennin that is widely expressed in brain, RENR is can strengthen the activity of angiotensin converting enzyme (ACE), furthermore influence the cardiovascular activity. Also, this receptor activates intracellular signal transductions system, such as MAPK pathway and Wnt pathway. Recently, a body of study proves that RENR plays an important role in embryonic development (25). In addition, it has been documented that, within 24 h following head amputation, brain primordium is formed and continues to develop into brain (26). Based on above description, it was speculated that RENR is possibly implicated in brain regeneration during PHR.
Compared to head regeneration, the DEPs upregulated by >4-fold obviously decreased in number (just only 6 DEPs) during tail regeneration, out of which the expression of ALF2 (Fructose-bisphosphate aldolase 2) began to increase at 6 h and reached a peak of 6.43-fold at 72 hours post amputation. The function of this enzyme is to catalyze the condensation of dihydroxyacetone phosphate with glyceraldehyde-3-phosphate into fructose-1,6-bisphosphate, which is closely related to glucose and fructose metabolism (27). Considered together, the upregulation of this protein in early phase of tail regeneration maybe supplies the sufficient energy and the carbon sources for macromolecular synthesis required for completion of the regeneration. Whereas, there were 47 DEPs commonly expressed in two regenerative processes; among them, 15 were up-regulated and 9 were down-regulated in both processes, which might be involved in the biological activities occurring on both scenarios; while the remaining 23 DEPs showed the different expression patterns in the two processes, which may be involved in the occurrence or regulation of the activities specific to PHR or PTR.
Furthermore, bioinformatics analysis revealed the similar biological activities occurring during both regeneration processes at the protein level, such as differentiation and development, signal transduction, metabolic process, immunity and inflammation, cell apoptosis and cell proliferation etc. Notably, DEPs in the categories "differentiation and development" and "signal transduction" shared the highest proportion of the total DEPs in head regeneration and tail regeneration, appearing to be no obvious difference between the two scenarios. To find out the difference in biological activities between the two scenarios, sub-categorization was especially performed on the groups "differentiation and development" and "signal transduction", and the result revealed the significant difference in biological processes among these two scenarios.
Differentiation and development
There is compelling evidence that extraordinary regenerative ability of planarian is supported by a population of neoblasts (28). Upon damage, neoblasts rapidly enter the mitotic cycle and form a regenerative blastema at the wound site, which generates the missing part of the body (29). More specifically, after decapitation the neoblast-derived progenitors regenerate the missing anterior tissue, such as muscle, epidermis, brain, eyespots etc (30, 31). After tail cutting, blastema directs the development of posterior fragment, like muscle, epidermis and nervous system (32). Our result found that, among 361 head regeneration-related DEPs, 67 were involved in differentiation and development, accounting for 16.4% of total DEPs; and were functionally subcategorized into anterior/posterior pattern specification, eyespot formation, nervous system development, muscle and epidermis development; while in the regenerating tail, 30 development and differentiation-involved DEPs were further functionally sorted into brain development, epithelium morphogenesis and nervous system formation. It can be concluded that biological activities in PHR were much more sophisticate than that in PTR. Notably, a majority of DEPs involved in anterior/posterior pattern specification and eyespot formation were upregulated during head regeneration, while brain development-involved DEPs in tail regeneration were just opposite. For example, proteins RSMB, APC and LEC3, involved in brain development in anterior regeneration, showed a significant increase in the expression; whereas it was just on the contrary for the proteins BB2B, DC1L1, H6V057 and WWOX showing downregulation in posterior regeneration. Inoue et al. (33) found that blastema at the wound site was triggered to differentiate into neurons within 24 hours after amputation and develop into nervous system after 5 days. Consistently, our study detected the up-regulation of almost all the brain development-related proteins at early phase (6-12 hpa) of PHR. Meanwhile, the inhibition of brain and eyespots formation in PTR was observed in the study of Umesono et al. (34), also supported by our finding that almost all the brain development-related proteins were downregulated during PTR.
Meanwhile, we also found some novel planarian regeneration-related proteins, like the down-regulated GI: 8895467 (Accession ID) homologous to retinoic acid receptor gamma 2 (RARG2). Studies have reported that RARG2 impacted axial patterning and eyespot formation by interacting with transcription factor Cdx1, and the mutation of Rarg gene caused an abnormality of ocular phenotype in animals (35). Whether GI:8895467 has the similar functions needs to be experimentally verified. In addition, another protein GI:3789980, a homolog to cone transducin alpha subunit, was also significantly increased in expression. It has been documented that above subunit forms G protein with beta and gamma subunits, and transduces visual signals in optic system (36). Similarly, the exact role of GI:3789980 in visual development needs to be tested.
Signal transduction
Studies have confirmed the involvement of many activities in both PHR and PTR, such as cell-fate determination, cell proliferation, polarity, adhesion, motility, apoptosis, differentiation, patterning and morphogenesis. These biological activities cannot be completed without the specific signal pathway. For instance, Adell et al. (37) and Reuter et al. (38) reported that inactivation of Wnt signaling resulted in the formation of anterior determinants-expressing cells in tail regions; on the contrary, overexpression of Wnts caused the lack of head regenerative ability of tail fragments, proving the necessity of Wnt signaling in A/P polarity establishment of planarian. Our study found that most components in Wnt signaling were downregulated in the regenerating head, whereas one exception was Wnt8B upregulated at 120 hpa. Consistently, studies have found the highly brain-restricted expression of Wnt8B in early embryogenesis (39). In view of this, we hypothesized that Wnt8B maybe regulates brain formation after decapitation via some alternative pathway.
There are accumulating evidence that bone morphogenetic proteins (BMPs) are essential for D/V patterning of many organisms. For instance, BMP knockdown caused the formation of double dorsal and the failure to regenerate dorsal root axonal in planarian. Gaviño and Reddien (13), found that RNAi-directed downregulation of components (SMAD1, BMP4, SMAD4) of BMP pathway resulted in the abnormality of D/V patterning, suggesting the requirement of BMP signaling in the maintenance of body plan. Also, our data showed the obvious increase in components (TGFB2, MAPK3, BMP1 homologue O76147) of BMP pathway in PHR, but insignificant alteration in PTR. It can be speculated that BMP pathway plays key role in D/V patterning in PHR, while other pathways for regenerating tail.
Above conclusion was only drawn from the proteome data of regenerating planarian. To evaluate the difference in signaling pathways between PHR and PTR, we constructed the PPI network of signal transduction-involved DEPs. For head regeneration, STRING analysis suggested the significant change in cell surface receptor signaling pathway in which the components were subgrouped into BMP signaling, Wnt signaling and Notch signaling. For tail regeneration, the enrichment in the Reactome "signal transduction" was observed. Unlike head regeneration, the components in category "signal transduction" were subgrouped into Wnt signaling.
Conclusion
In this study, we established a relatively comprehensive proteome database using MALDI-TOF/TOF MS, and observed totally 529 planarian regeneration-related DEPs. By comparing the proteome between PHR and PTR, 314 out of 529 DEPs were identified to be specific for head regeneration, obviously more than number of DEPs (totally 165) in tail regeneration, indicating the more complicated regulation in PHR compared to PTR. Bioinformatics analysis showed the difference between two scenarios in some biological processes such as signaling pathways, and further analysis showed a more significant contribution of both Wnt and BMP signaling to PHR. As mentioned above, the conclusion were only drawn from high-throughput proteome analyses, there are many questions to be answered with respect to whether the most significantly upregulated proteins in two scenarios are key to the corresponding regenerative processes? How do the commonly-expressed proteins identified in this study function during head and tail regeneration, or differentially regulate these two processes? How the signal pathways like Wnt signaling affects the amputated planarians? What is the exact function of the novel proteins confirmed in this study (Fig .S3, See Supplementary Online Information at www. celljournal.org)? These issues need to be solved by the in-depth researches in future.
Supplementary PDF
Supplementary Exels
Acknowledgements
This study was approved by Henan University of Science and Technology, and funded by the National Natural Science Foundation of China (No. 31401209) from Ministry of Science and Technology of the People’s Republic of China (MOST) and Student Research Training Program (SRTP) from Henan University of Science and Technology (No. 2020355). The authors declare that there are no conflicts of interest.
Authors’ Contributions
X.C., Y.L.; Participated in study design, data collection, drafting and statistical analysis. X.Z.; Contributed in total protein extraction, 2-DE gel analysis, in-gel digestion, and MALDI-TOF/TOF MS analysis and data processing. Q.L.; Contributed in sample collection used in this study, differential expressed protein identification, bioinformatics analysis and interpretation of the data. All authors read and approved the final manuscript.
References
- 1.Imperadore P, Fiorito G. Cephalopod tissue regeneration: consolidating over a century of knowledge. Front Physiol. 2018;9:593–593. doi: 10.3389/fphys.2018.00593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu JP, Li MH. The use of freshwater planarians in environmental toxicology studies: Advantages and potential. Ecotoxicol Environ Saf. 2018;161:45–56. doi: 10.1016/j.ecoenv.2018.05.057. [DOI] [PubMed] [Google Scholar]
- 3.Jones S, Osman S, Howl J. The planarian Schmidtea mediterranea as a model system for the discovery and characterization of cellpenetrating peptides and bioportides. Chem Biol Drug Des. 2019;93(6):1036–1049. doi: 10.1111/cbdd.13483. [DOI] [PubMed] [Google Scholar]
- 4.Li Y, Zeng A, Han XS, Li G, Li YQ, Shen B, et al. Small RNAome sequencing delineates the small RNA landscape of pluripotent adult stem cells in the planarian Schmidtea mediterranea. Genom Data. 2017;14:114–125. doi: 10.1016/j.gdata.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong Z, Yuwen Y, Sima Y, Dong Y, Zhan H, Chen G, et al. Photokinesis and Djopsin gene expression analysis during the regeneration of planarian eyes. Integr Zool. 2017;12(2):157–164. doi: 10.1111/1749-4877.12234. [DOI] [PubMed] [Google Scholar]
- 6.Durant F, Lobo D, Hammelman J, Levin M. Physiological controls of large-scale patterning in planarian regeneration: a molecular and computational perspective on growth and form. Regeneration (Oxf) 2016;3(2):78–102. doi: 10.1002/reg2.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alessandra S, Rossi L. Planarian stem cell heterogeneity. Adv Exp Med Biol. 2019;1123:39–54. doi: 10.1007/978-3-030-11096-3_4. [DOI] [PubMed] [Google Scholar]
- 8.Kashima M, Agata K, Shibata N. Searching for non-transposable targets of planarian nuclear PIWI in pluripotent stem cells and differentiated cells. Dev Growth Differ. 2018;60(5):260–277. doi: 10.1111/dgd.12536. [DOI] [PubMed] [Google Scholar]
- 9.Rojo-Laguna JI, Garcia-Cabot S, Saló E. Tissue transplantation in planarians: A useful tool for molecular analysis of pattern formation. Semin Cell Dev Biol. 2019;87:116–124. doi: 10.1016/j.semcdb.2018.05.022. [DOI] [PubMed] [Google Scholar]
- 10.Durant F, Bischof J, Fields C, Morokuma J, LaPalme J, Hoi A, et al. The role of early bioelectric signals in the regeneration of planarian anterior/posterior polarity. Biophys J. 2019;116(5):948–961. doi: 10.1016/j.bpj.2019.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kijimoto T, Moczek AP. Hedgehog signaling enables nutrition-responsive inhibition of an alternative morph in a polyphenic beetle. Proc Natl Acad Sci USA. 2016;113(21):5982–5987. doi: 10.1073/pnas.1601505113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Windsor Reid PJ, Matveev E, McClymont A, Posfai D, Hill AL, Leys SP. Wnt signaling and polarity in freshwater sponges. BMC Evol Biol. 2018;18(1):12–12. doi: 10.1186/s12862-018-1118-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaviño MA, Reddien PW. A Bmp/Admp regulatory circuit controls maintenance and regeneration of dorsal-ventral polarity in planarians. Curr Biol. 2011;21(4):294–299. doi: 10.1016/j.cub.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darras S, Fritzenwanker JH, Uhlinger KR, Farrelly E, Pani AM, Hurley IA, et al. Anteroposterior axis patterning by early canonical Wnt signaling during hemichordate development. PLoS Biol. 2018;16(1):e2003698–e2003698. doi: 10.1371/journal.pbio.2003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pelullo M, Zema S, Nardozza F, Checquolo S, Screpanti I, Bellavia D. Wnt, Notch, and TGF-β pathways impinge on hedgehog signaling complexity: an open window on cancer. Front Genet. 2019;10:711–711. doi: 10.3389/fgene.2019.00711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orii H, Watanabe K. Bone morphogenetic protein is required for dorso-ventral patterning in the planarian Dugesia japonica. Dev Growth Differ. 2007;49(4):345–349. doi: 10.1111/j.1440-169X.2007.00931.x. [DOI] [PubMed] [Google Scholar]
- 17.Kao D, Felix D, Aboobaker A. The planarian regeneration transcriptome reveals a shared but temporally shifted regulatory program between opposing head and tail scenarios. BMC Genomics. 2013;14(1):797–797. doi: 10.1186/1471-2164-14-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts-Galbraith RH, Brubacher JL, Newmark PA. A functional genomics screen in planarians reveals regulators of whole-brain regeneration. Elife. 2016;5:e17002–e17002. doi: 10.7554/eLife.17002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tewari AG, Owen JH, Petersen CP, Wagner DE, Reddien PW. A small set of conserved genes, including sp5 and Hox, are activated by Wnt signaling in the posterior of planarians and acoels. PLoS Genet. 2019;15(10):e1008401–e1008401. doi: 10.1371/journal.pgen.1008401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubert A, Henderson JM, Cowles MW, Ross KG, Hagen M, Anderson C, et al. A functional genomics screen identifies an importin-α homolog as a regulator of stem cell function and tissue patterning during planarian regeneration. BMC Genomics. 2015;16:769–769. doi: 10.1186/s12864-015-1979-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H, Deery MJ, Gannon L, Powers SJ, Lilley KS, Theodoulou FL. Quantitative proteomics analysis of the Arg/N-end rule pathway of targeted degradation in Arabidopsis roots. Proteomics. 2015;15(14):2447–2457. doi: 10.1002/pmic.201400530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tewari AG, Stern SR, Oderberg IM, Reddien PW. Cellular and molecular responses unique to major injury are dispensable for planarian regeneration. Cell Rep. 2018;25(9):2577–2590. doi: 10.1016/j.celrep.2018.11.004. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karge A, Bonar NA, Wood S, Petersen CP. tec-1 kinase negatively regulates regenerative neurogenesis in planarians. Elife. 2020;9:e47293–e47293. doi: 10.7554/eLife.47293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ermakov AM, Ermakova ON, Popov AL, Manokhin AA, Ivanov VK. Opposite effects of low intensity light of different wavelengths on the planarian regeneration rate. J Photochem Photobiol B. 2020;202:111714–111714. doi: 10.1016/j.jphotobiol.2019.111714. [DOI] [PubMed] [Google Scholar]
- 25.Ramkumar N, Kohan DE. The (pro)renin receptor: an emerging player in hypertension and metabolic syndrome. Kidney Int. 2019;95(5):1041–1052. doi: 10.1016/j.kint.2018.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wenemoser D, Reddien PW. Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev Biol. 2010;344(2):979–991. doi: 10.1016/j.ydbio.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacques B, Coinçon M, Sygusch J. Active site remodeling during the catalytic cycle in metal-dependent fructose-1,6-bisphosphate aldolases. J Biol Chem. 2018;293(20):7737–7753. doi: 10.1074/jbc.RA117.001098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dattani A, Kao D, Mihaylova Y, Abnave P, Hughes S, Lai A, et al. Epigenetic analyses of planarian stem cells demonstrate conservation of bivalent histone modifications in animal stem cells. Genome Res. 2018;28(10):1543–1554. doi: 10.1101/gr.239848.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Issigonis M, Newmark PA. From worm to germ: Germ cell development and regeneration in planarians. Curr Top Dev Biol. 2019;135:127–153. doi: 10.1016/bs.ctdb.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Levin M, Pietak AM, Bischof J. Planarian regeneration as a model of anatomical homeostasis: recent progress in biophysical and computational approaches. Semin Cell Dev Biol. 2019;87:125–144. doi: 10.1016/j.semcdb.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petersen CP. Regeneration: organizing the blastema in planarians. Curr Biol. 2017;27(5):R181–R183. doi: 10.1016/j.cub.2017.01.057. [DOI] [PubMed] [Google Scholar]
- 32.Cervera J, Meseguer S, Levin M, Mafe S. Bioelectrical model of head-tail patterning based on cell ion channels and intercellular gap junctions. Bioelectrochemistry. 2020;132:107410–107410. doi: 10.1016/j.bioelechem.2019.107410. [DOI] [PubMed] [Google Scholar]
- 33.Inoue T, Hoshino H, Yamashita T, Shimoyama S, Agata K. Planarian shows decision-making behavior in response to multiple stimuli by integrative brain function. Zoological Lett. 2015;1:7–7. doi: 10.1186/s40851-014-0010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Umesono Y, Tasaki J, Nishimura Y, Hrouda M, Kawaguchi E, Yazawa S, et al. The molecular logic for planarian regeneration along the anterior-posterior axis. Nature. 2013;500(7460):73–76. doi: 10.1038/nature12359. [DOI] [PubMed] [Google Scholar]
- 35.Thompson B, Katsanis N, Apostolopoulos N, Thompson DC, Nebert DW, Vasiliou V. Genetics and functions of the retinoic acid pathway, with special emphasis on the eye. Hum Genomics. 2019;13(1):61–61. doi: 10.1186/s40246-019-0248-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dexter PM, Lobanova ES, Finkelstein S, Spencer WJ, Skiba NP, Arshavsky VY. Transducin β-subunit can interact with multiple gprotein γ-subunits to enable light detection by rod photoreceptors. eNeuro. 2018;5(3):ENEURO-0144-18–ENEURO-0144-18. doi: 10.1523/ENEURO.0144-18.2018. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adell T, Cebrià F, Saló E. Gradients in planarian regeneration and homeostasis. Cold Spring Harb Perspect Biol. 2010;2(1):a000505–a000505. doi: 10.1101/cshperspect.a000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reuter H, März M, Vogg MC, Eccles D, Grífol-Boldú L, Wehner D, et al. Β-catenin-dependent control of positional information along the AP body axis in planarians involves a teashirt family member. Cell Rep. 2015;10(2):253–265. doi: 10.1016/j.celrep.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 39.Liu W, Lagutin O, Swindell E, Jamrich M, Oliver G. Neuroretina specification in mouse embryos requires Six3-mediated suppression of Wnt8b in the anterior neural plate. J Clin Invest. 2010;120(10):3568–3577. doi: 10.1172/JCI43219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.