Abstract
Brazil has become one of the epicentres of the COVID-19 pandemic, with cases heavily concentrated in large cities. Testing data is extremely limited and unreliable, which restricts health authorities’ ability to deal with the pandemic. Given the stark demographic, social and economic heterogeneities within Brazilian cities, it is important to identify hotspots so that the limited resources available can have the greatest impact. This study shows that decentralised monitoring of SARS-CoV-2 RNA in sewage can be used to assess the distribution of COVID-19 prevalence in the city. The methodology developed in this study allowed the identification of hotspots by comprehensively monitoring sewers distributed through Belo Horizonte, Brazil's third largest city. Our results show that the most vulnerable neighbourhoods in the city were the hardest hit by the pandemic, indicating that, for many Brazilians, the situation is much worse than reported by official figures.
Keywords: Wastewater-based epidemiology, Covid-19, prevalence, Hotspots, Decentralised sewage monitoring, Health vulnerability
Graphical abstract
1. Introduction
On March 11, 2020, the World Health Organization (WHO) declared COVID-19 a pandemic. COVID-19 is a respiratory syndrome caused by the SARS-CoV-2 virus, with over 130 million confirmed cases and nearly 3 million confirmed deaths worldwide as of April 12, 2021. Official data suggest that over 13 million Brazilians have been infected, with a death toll second only to the USA (Worldometers, 2021). Cases have been heavily concentrated in big cities but have spread to the countryside, affecting isolated indigenous communities. These statistics are fraught with uncertainties, because the tests are notoriously unreliable, and the sample size (less than 0.1% of the population) is very small (Worldometers, 2021). In large Brazilian cities, testing is mainly occurring among people who suffer from symptoms severe enough to seek health care or have private health insurance. Brazil is one of the most unequal countries in the world (OECD et a. 2019), and its large cities are starkly heterogeneous in demographic, social and economic terms. During the pandemic, it is imperative that limited available resources are directed to neighbourhoods where they will have the greatest impact, which requires the spatial distribution of infections to be characterized.
Following the detection of SARS-CoV-2 RNA in faeces of COVID-19 patients (Wang et al. 2020) and later in sewage (Medema et al. 2020) and sewage sludge (Peccia et al. 2020), an increasing number of studies have reported the use of sewage monitoring as a tool for epidemiological surveillance. Sewage-based surveillance of other viruses has been well documented in the literature (Wigginton et al., 2015; Sims and Kasprzyk-Hordern 2020) and has been successfully applied to assess the spread of polio in populations since 1980s (Hovi et al. 2012). By sampling raw sewage from the inlet of treatment plants and determining SARS-CoV-2 RNA concentrations, it is possible to extract valuable information on infection levels for the entire population that contributes sewage to the treatment plant. This approach has shown that SARS-CoV-2 RNA concentrations in sewage correlated with the local numbers of infected individuals (Ahmed et al. 2020; Randazzo et al. 2020). Extending this principle by sampling wastewater directly from the sewers at multiple points upstream of the treatment plant could reveal the spatial distribution in prevalence of the virus in a large city. This can be even more critical in cities where not all collected sewage reaches the treatment plant but is instead discharged in water bodies, a common limitation in sanitation infrastructure in the developing world. In places with severely scarce testing data during the current pandemic, such as Brazil, information obtained from decentralised sewage monitoring could be vital in devising potentially life-saving public health interventions.
We have carried out a comprehensive sewage monitoring programme for SARS-CoV-2 RNA in Belo Horizonte, Minas Gerais, Brazil since April 2020, early on its epidemic curve. Our objective was to assess temporal and spatial information on the virus load in different neighbourhoods to assist local health authorities in decision-making through weekly bulletins. Here, we present the results for the period between epidemiological weeks 20 and 32 (from May 11 to August 07). To the best of our knowledge, this is the first study to use decentralised sewage monitoring to assess spatial distribution of COVID-19 prevalence in a large city.
2. Material and methods
2.1. Selection of sewage sampling points
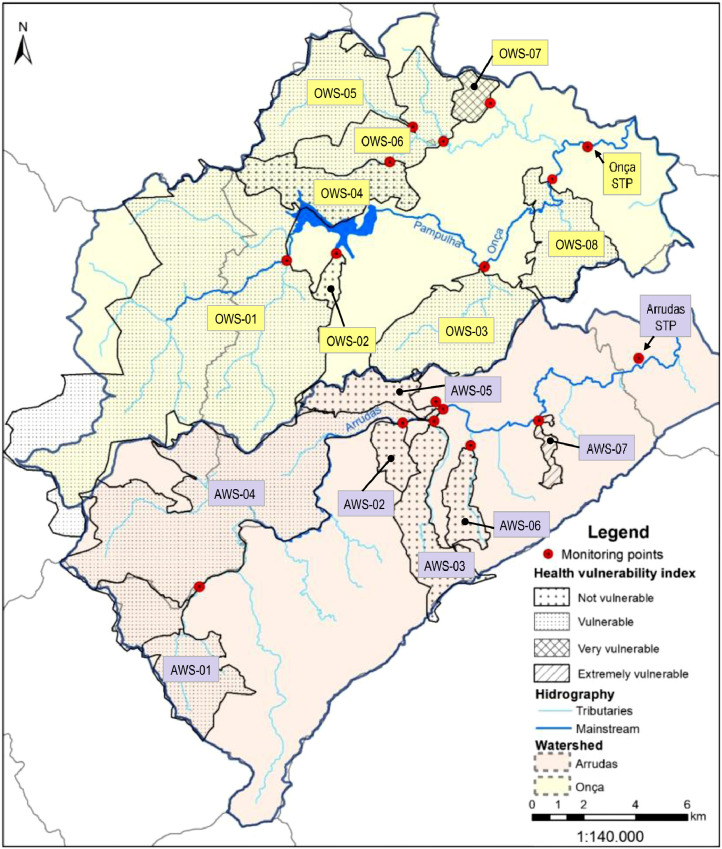
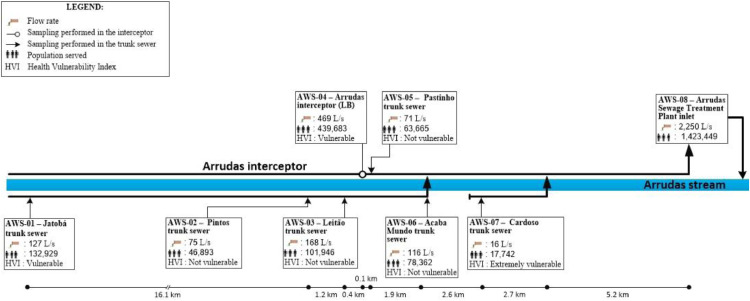
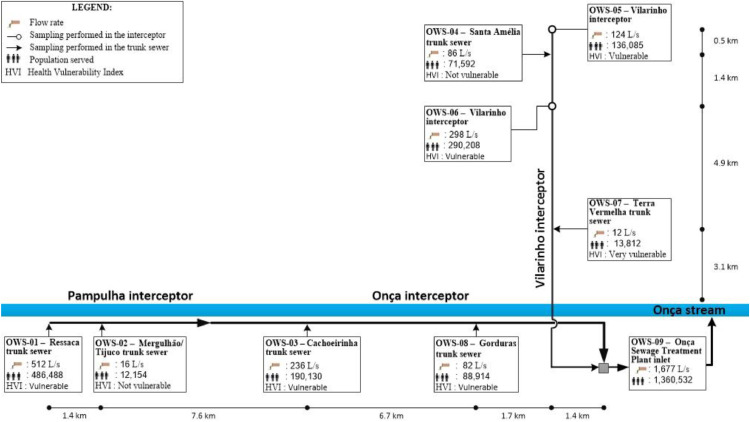
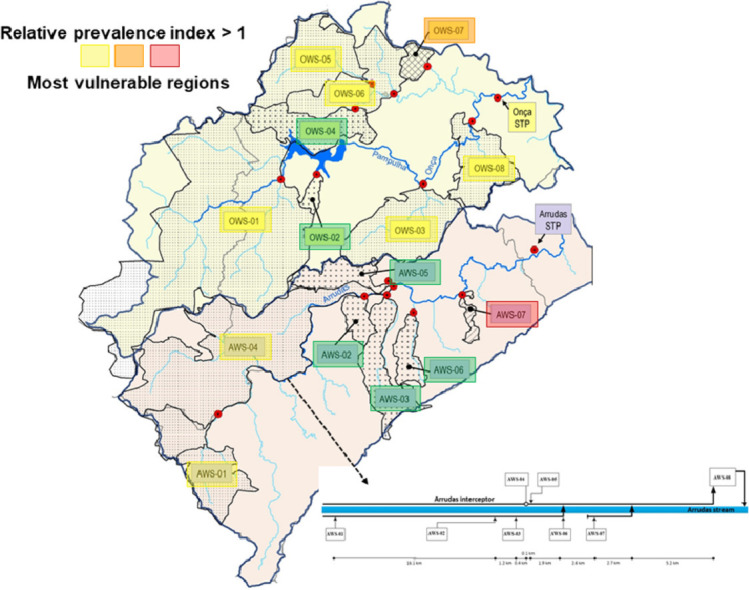
Belo Horizonte has a population of approximately 2 million people and is located in southeast Brazil. It has two major sewage treatment plants (STPs), each with a dedicated sewer network (Arrudas and Onça sewersheds). Weekly samples were collected from the inlet of both treatment plants and from a total of 15 points in the sewer networks (Chernicharo et al. 2020), purposely chosen to collect sewage from regions with starkly different socio-economic and health vulnerability indicators. Therefore, a total of 17 points were monitored: i) two points at the inlet of sewage treatment plants Arrudas and Onça; ii) 15 points in the sewer system (7 in the Arrudas sewershed and 8 in the Onça sewershed). A total of 202 samples were collected from May 11 to August 07. Fig. 1 shows a map with the sampling points and health vulnerability indexes for each region sampled. Fig. 2 shows the single-line diagram of both sewer networks, including the population served, flow rate, and distance between sampling points. Composite samples (24hr-composites for both treatment plants, and 4-hr composites for all other points) were collected on fixed days (Mondays for points on the Arrudas sewershed and Wednesdays for points on the Onça sewershed), from 8:30am to 12:30pm, using autosamplers. The sewers were designed as separate sewer systems. It did not rain during any of the sampling days.
Fig. 1.
Map showing sampling points and health vulnerability indexes for each region sampled.
Fig. 2.
a Single-line diagram of sewer network Arrudas, showing the population served, flow rates, and distances between sampling points. b Single-line diagram of sewer network Onça, showing the population served, flow rates, and distances between sampling points.
2.2. Health Vulnerability Index (HVI)
The Health Vulnerability Index was formulated by Belo Horizonte City Hall (PBH - Belo Horizonte City Hall 2018) considering socioeconomic (residents per household, illiteracy rates, income, skin color and ethnicity) and sanitation indicators (access to adequate water supply, sanitation and municipal solid waste collection and disposal). The HVI is divided into four classes (extremely vulnerable, very vulnerable, vulnerable and not vulnerable) and published in a shapefile format. For each region, the layers correspondent to the same class were dissolved and then the area of each class was calculated. The final class of each sampled region (presented in this study) corresponds to the dominant class in terms of area in each region. All data used in HVI calculations were derived from the last Brazilian Census (2010), and the relative weights used in the calculation were defined using the Analytic Hierarchy Process (as shown in Table 1 ).
Table 1.
Weight of each indicator used in the calculation of the health vulnerability index (HVI)
| Indicator | Weight |
| Improper water supply | 0.424 |
| Improper sanitation | 0.375 |
| Improper final disposal of solid waste | 0.201 |
| Residents per household | 0.073 |
| Illiteracy | 0.283 |
| Per capita income up to 0.5 minimal wage | 0.288 |
| Average income of heads of household | 0.173 |
| Percentage of black and indigenous population | 0.185 |
2.3. Collection, preservation and transport of sewage samples
The following procedures were adopted for sampling, storage and transport of sewage samples:
-
•
Isolation of the area near the access of the manhole using signal cones and easels.
-
•
Installation of automatic sampling devices (Fig. S1) equipped with battery, sampling container and reusable ice packs, keeping the samples at 4°C.
-
•
Collection of composite samples representing the morning period, usually between 8:30am and 12:30pm. The sample container (10 liters) was kept surrounded by ice packs throughout the sampling duration. The automatic sampling device was set to collect one portion of sample of approximate 400 mL every 10 minutes, resulting in 10 liters after 4 hours sampling.
-
•
Following sampling, composite samples were homogenized by vigorously shaking the container and transferred to two 1 liter bottles. One bottle was taken to the laboratory in the Department of Environmental and Sanitary Engineering (DESA) of the Federal University of Minas Gerais for the quantification of the SARS-CoV-2 RNA. The second bottle was taken to the central laboratory of the Sanitation Company for Minas Gerais for the determination of chemical oxygen demand (COD), total suspended solids (TSS), surfactants and thermotolerant coliforms (or E. coli), according to APHA (Baird and Bridgewater 2017). Both bottles were kept on ice during transport.
2.4. Quantification of SARS-CoV-2 RNA
The virus concentration step was performed according to methods previously described (Symonds et al., 2014; Ahmed et al. 2015). This method is based on the adsorption-extraction method using electronegative membranes (0.45 µm-HAWP04700-Millipore). Before the sewage filtration (average volume of 50 mL) through the membrane, the method began with the addition of MgCl2 (2.5 M) followed by adjustment of the sample pH to 3 to 3.5 using acetic acid (1M) (Symonds et al., 2014). Following sample filtration, the membrane was folded, put into a 2 mL tube with 600 µL of PM1 solution (provided in the DNA/RNA extraction kit), glass beads and 6 µL of beta-mercaptoethanol and stored at -80 °C until RNA extraction. RNA was extracted using a commercial kit (AllPrep PowerViral DNA/RNA, Qiagen®, Hilden, Germany), according to instructions provided by the manufacturer. A bead beater was used (instead of vortex) in the first step of RNA extraction to remove viral particles from the membrane. Following extraction, RNA was resuspended in 100 µL of ultrapure water (free of RNAses) and stored at – 80°C. The detection of SARS-CoV-2 was performed using available RT-qPCR assays using different primer-probe sets recommended by the US Center for Disease Control and Prevention (Center for Disease Control and Prevention 2020), targeting the nucleocapsid protein (N) gene-region N1 (Lu et al. 2020). All RT-qPCR amplifications were performed in 20 µL reaction mixtures using iTaqTM Universal probes One Step reaction mix and reverse transcriptase. For both RT-qPCR assays, plasmids were purchased from Integrated DNA Technologies (Coralville, IA, USA) and used as standards or positive controls. The RT-qPCR assays were performed using an Applied Biosystems 7500 Real Time PCR System. All RT-qPCR reactions were performed in triplicate. For each RT-qPCR run, a series of three positive and negative controls were included. Standard curves were built using serial dilutions (Table S1) of the standard plasmid SARS-CoV-2 obtained from IDT (2019-nCoV_N_Positive Control). An exponential model (Verbyla et al. 2016) was used to estimate the limit of detection (LOD), using 95% probability of amplification. The exponential model assumes that DNA/cDNA in samples are randomly distributed in reaction wells; DNA/cDNA copy has an independent and identical probability of “surviving” the conditions to complete the qPCR, and at least one target DNA/cDNA copy has to be physically present in the qPCR in order to cause amplification within 40 cycles. Based on these assumptions, the LOD with a 95% probability of amplification was determined according to Eq.1:
| (1) |
where x represents the mean number of target genome copies added to each qPCR well (mean concentration per reaction), and k represents the probability of “survival”.
The model determined an LOD of 15 copies per reaction (or 6 copies/mL of sewage) (Fig. S2). The limit of quantification (i.e. the lowest concentration where the dilution curve stops being linear) was 50 copies/reaction (or 20 gene copies/mL of sewage) (Table S1). The master standard curves used in this study were within the prescribed range of Minimum Information for Publication of Quantitative (MIQE) real-time PCR guidelines (Bustin et al., 2009). The amplification efficiencies for N1 assays ranged from 94.7% to 109%; R2 ranged from 0.993 to 0.997; slope ranged from -3.118 to -3.454 and Y-inter ranged from 35.726 to 38.667. All qPCR assays were performed in triplicate according to the MIQE guidelines (Bustin et al., 2009).
In order to check PCR inhibition, internal control HsRPP 30 was used, as recommended by the CDC (Center for Disease Control and Prevention 2020). Whenever the sample did not amplify with the internal control (negative for RNAse P), the sample was diluted 10x and 100x to check for inhibition and confirm that the result was negative. Since the RT-qPCR standard curves were built using double stranded plasmid and the target molecule in the environmental samples (SARS-CoV-2 RNA) is single stranded, all concentrations determined for the environmental samples were multiplied by a factor of 2. This was done because two viral N1 gene fragments (N1 gene copies, single-stranded) are required to give the equivalent fluorescence signal (Ct value on the RT-qPCR assay) of one molecule (N1 gene copy, double-stranded) of the plasmid standard (Fig. S3). In other words, if a sample that has the same Ct of 1000 copies of the plasmid standard, that sample has 1000 copies of double-stranded target. However, 1000 copies of double-stranded target would actually correspond to 2000 viral particles, since the virus is single-stranded. Alternatively, the nominal gene concentrations used to build the standard curve with double-stranded plasmid could be divided by a factor of 2, while maintaining unaltered the concentration values determined for the environmental samples.
2.5. Estimation ofin-sewerdecay ofSARS-CoV-2 RNA
In-sewer decay of the RT-qPCR signal for SARS-CoV-2 RNA was estimated for each of the 15 monitored regions based on the first-order decay rate constant determined by Ahmed et al., 2020. Design characteristics of all sampled trunk sewers and interceptors were provided by the Sanitation Company for Minas Gerais including: pipe diameter and total length, average slope and average flow rate at the sampling point. Sewage flow velocities and in-sewer travel time were calculated based on design characteristics (Table 2 ).
Table 2.
Design characteristics of sampled trunk sewers and interceptors and results of in-sewer travel time calculations.
| Sewershed | Diameter (mm) | Average slope (m.m−1) | Average flow rate (L.s−1) | Length (m) | Flow velocity (m.s−1) | In-sewer travel time (h) |
| OWS-1 | 1500 | 0.0059 | 512.83 | 4000 | 1.9 | 0.58 |
| OWS-2 | 400 | 0.0053 | 16.06 | 1500 | 0.8 | 0.53 |
| OWS-3 | 1200 | 0.0433 | 236.15 | 6000 | 3.1 | 0.54 |
| OWS-4 | 400 | 0.0117 | 86.82 | 4000 | 1.6 | 0.68 |
| OWS-5 | 600 | 0.0050 | 124.52 | 6200 | 1.3 | 1.31 |
| OWS-6 | 1000 | 0.0050 | 298.06 | 8200 | 1.6 | 1.44 |
| OWS-7 | 300 | 0.0278 | 12.40 | 1000 | 1.3 | 0.21 |
| OWS-8 | 500 | 0.0137 | 82.28 | 4500 | 1.7 | 0.75 |
| AWS-1 | 700 | 0.0090 | 127.44 | 5400 | 1.6 | 0.94 |
| AWS-2 | 600 | 0.0475 | 75.29 | 2000 | 2.6 | 0.22 |
| AWS-3 | 800 | 0.0193 | 168.31 | 7500 | 2.2 | 0.95 |
| AWS-4 | 1500 | 0.0038 | 460.68 | 16200 | 1.6 | 2.81 |
| AWS-5 | 500 | 0.0375 | 71.05 | 4000 | 2.4 | 0.47 |
| AWS-6 | 1600 | 0.0281 | 116.52 | 4000 | 2.4 | 0.47 |
| AWS-7 | 400 | 0.0094 | 16.20 | 2700 | 1.0 | 0.77 |
| STP-A | 1600 | 0.0050 | 1904.00 | 28400 | 2.5 | 3.11 |
| STP-O | 1600 | 0.0050 | 1691.00 | 27200 | 2.5 | 3.02 |
The degradation rate of SARS-CoV-2 RNA in sewage was assumed to follow exponential decay. Therefore, the RNA concentration that was excreted and discharged into the sewer collection system (N0) is described by the following equation (Eq. 1):
| (1a) |
Where N(t) is the RNA concentration measured by the sampling campaign (i.e. the amount of RNA that resists decay after a time t; t is the time elapsed between the time of excretion (time= 0) and time of sample collection (time= t) (i.e. in-sewer travel time), and k (0.183.d−1 – ref.14) is the first-order decay rate constant of the SARS-CoV-2 RNA at 25°C (average measured sewage temperature).
SARS-CoV-2 RNA concentrations determined for each monitored region over epidemiological weeks 20 to 32 (referred to as N (t) – Eq. 1), as well as the estimated initial RNA concentrations taking into account in-sewer RNA degradation (referred as N0 – Eq. 1) are shown in Table 3 and Table 4 , respectively.
Table 3.
SARS-CoV-2 RNA concentrations determined for each monitored region over epidemiological weeks 20 to 32 (N (t))
| N(t) – SARS-CoV-2 genomic copies.L−1 | |||||||||||||
| Ep. Week → Sewershed↓ | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 |
| OWS-1 | 314 | 220 | 2750 | 37142 | 9786 | 59456 | 85204 | 22742 | 31042 | 25120 | 59586 | 45822 | 20206 |
| OWS-2 | 392 | 338 | 460 | 322 | 2498 | 8250 | 4786 | 21058 | 24206 | 3674 | 15750 | 20460 | |
| OWS-3 | 1282 | 306 | 2172 | 2134 | 51690 | 17042 | 43064 | 35058 | 79224 | 25150 | 40166 | ||
| OWS-4 | 198 | 570 | 1028 | 3070 | 10894 | 23866 | 35034 | 46074 | 36286 | 67490 | 102382 | 45284 | |
| OWS-5 | 1624 | 224 | 1532 | 2820 | 5762 | 54174 | 102474 | 59770 | 83102 | 54828 | 114420 | 26154 | |
| OWS-6 | 312 | 606 | 870 | 5714 | 5594 | 28520 | 62258 | 122502 | 39862 | 205642 | 72628 | 69052 | 85792 |
| OWS-7 | 3082 | 714 | 8068 | 5678 | 12456 | 60512 | 51970 | 102360 | 102594 | 254594 | 66566 | 22302 | |
| OWS-8 | 726 | 1040 | 1464 | 12016 | 20286 | 51112 | 123050 | 1816 | 56158 | 101018 | 56882 | 141910 | 50580 |
| AWS-1 | 606 | 1982 | 1160 | 3226 | 40138 | 21606 | 82486 | 65614 | 59304 | 94704 | 58594 | ||
| AWS-2 | 540 | 400 | 632 | 312 | 7112 | 16370 | 48860 | 9316 | 23892 | 41970 | 55726 | 50058 | |
| AWS-3 | 226 | 856 | 1118 | 710 | 5498 | 16210 | 38474 | 19282 | 57130 | 34374 | 28594 | 28938 | |
| AWS-4 | 326 | 4028 | 1436 | 8112 | 42214 | 28126 | 39410 | 42758 | 97222 | 59674 | 32928 | 53226 | |
| AWS-5 | 1762 | 66828 | 24724 | 32842 | 35282 | 5044 | 14710 | 46454 | 15768 | ||||
| AWS-6 | 380 | 274 | 236 | 9990 | 30896 | 9410 | 25812 | 10486 | 29954 | 19198 | 4544 | ||
| AWS-7 | 1788 | 1660 | 748 | 137030 | 157298 | 33626 | 214430 | 171954 | 37802 | 47066 | 114942 | ||
| STP-A | 578 | 404 | 2268 | 730 | 3102 | 22242 | 33578 | 50692 | 39556 | 65578 | 167218 | 47430 | 31740 |
| STP-O | 450 | 56 | 972 | 3586 | 6306 | 38490 | 22416 | 78416 | 51548 | 65210 | 47754 | 80186 | 19234 |
Table 4.
Estimated SARS-CoV-2 RNA concentrations determined for each monitored region over the epidemiological weeks 20 to 32 (N (0))
| N(0) – SARS-CoV-2 genomic copies.L−1 | |||||||||||||
| Ep. Week → Sewershed↓ | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 |
| OWS-1 | 314 | 220 | 2762 | 37308 | 9830 | 59722 | 85584 | 22844 | 31182 | 25232 | 59852 | 46026 | 20296 |
| OWS-2 | 392 | 340 | 462 | 322 | 2508 | 8284 | 4806 | 21144 | 24306 | 3690 | 15814 | 20544 | |
| OWS-3 | 1286 | 306 | 2182 | 2142 | 51904 | 17114 | 43242 | 35204 | 79552 | 25254 | 40334 | ||
| OWS-4 | 198 | 574 | 1034 | 3086 | 10950 | 23990 | 35216 | 46314 | 36474 | 67842 | 102914 | 45520 | |
| OWS-5 | 1640 | 226 | 1546 | 2848 | 5820 | 54720 | 103506 | 60372 | 83940 | 55380 | 115572 | 26416 | |
| OWS-6 | 316 | 614 | 878 | 5778 | 5656 | 28836 | 62946 | 123856 | 40302 | 207916 | 73430 | 69816 | 86740 |
| OWS-7 | 3088 | 714 | 8080 | 5688 | 12476 | 60608 | 52052 | 102524 | 102758 | 255000 | 66672 | 22336 | |
| OWS-8 | 730 | 1046 | 1472 | 12086 | 20402 | 51406 | 123758 | 1828 | 56480 | 101600 | 57208 | 142728 | 50872 |
| AWS-1 | 610 | 1996 | 1168 | 3248 | 40426 | 21760 | 83078 | 66086 | 59730 | 95384 | 59016 | ||
| AWS-2 | 540 | 400 | 634 | 312 | 7124 | 16396 | 48940 | 9332 | 23932 | 42038 | 55816 | 50140 | |
| AWS-3 | 226 | 862 | 1126 | 716 | 5538 | 16328 | 38754 | 19424 | 57546 | 34624 | 28802 | 29148 | |
| AWS-4 | 332 | 4116 | 1468 | 8288 | 43130 | 28736 | 40264 | 43686 | 99328 | 60966 | 33642 | 54380 | |
| AWS-5 | 1768 | 67068 | 24812 | 32960 | 35410 | 5062 | 14762 | 46622 | 15824 | ||||
| AWS-6 | 382 | 274 | 236 | 10026 | 31006 | 9444 | 25904 | 10524 | 30062 | 19266 | 4560 | ||
| AWS-7 | 1670 | 752 | 137840 | 158228 | 33824 | 215698 | 172970 | 38024 | 47344 | 115620 | |||
| STP-A | 592 | 412 | 2322 | 746 | 3176 | 22774 | 34382 | 51906 | 40504 | 67148 | 171226 | 48566 | 32500 |
| STP-O | 462 | 58 | 994 | 3668 | 6454 | 39386 | 22938 | 80244 | 52750 | 66730 | 48866 | 82054 | 19682 |
Table 5 shows the percentage degradation of RNA at each sampling point, calculated based on data shown in Tables 2 and 3.
Table 5.
In-sewer percentage degradation of SARS-CoV-2 RNA for each of the 17 sampling points.
| Sewershed | In-sewer degradation (%) |
| OWS-1 | 0.4% |
| OWS-2 | 0.4% |
| OWS-3 | 0.4% |
| OWS-4 | 0.5% |
| OWS-5 | 1.0% |
| OWS-6 | 1.1% |
| OWS-7 | 0.2% |
| OWS-8 | 0.6% |
| AWS-1 | 0.7% |
| AWS-2 | 0.2% |
| AWS-3 | 0.7% |
| AWS-4 | 2.1% |
| AWS-5 | 0.4% |
| AWS-6 | 0.4% |
| AWS-7 | 0.6% |
| STP-A | 2.3% |
| STP-O | 2.3% |
2.6. Relative Prevalence Index (RPI) determination
SARS-CoV-2 RNA concentrations in sewage determined in the laboratory are expressed as the number of viral genome copies per litre of sewage. This can be determined based on the population of infected individuals that excrete viral particles and the population that is served by the sewer network (Eq. 2):
| (2) |
Where C is the viral genome concentration (RNA copies.L−1), PIN is the number of infected individuals (inhabitants), F is the faecal mass excreted per inhabitant per day (g faeces.inhab−1.day−1), NF is the number of viral genome copies per gram of faeces (RNA copies. g faeces−1), P is the population that contributes to the sewershed (inhabitants), and V is the per capita sewage volume generation (L.inhab−1.day−1).
In the previous equation, the term PIN/P can be interpreted as Prevalence Index (PI). Therefore, Eq. 2 can be rewritten to highlight the contribution of the PI to the viral genome concentration, as follows:
| (3) |
The per capita mass of faeces excreted per day ranges from 150 g to 300 g (Brown et al. 1996). The number of viral genome copies per gram of faeces varies by several orders of magnitude, between 103 to 107 copies per gram of faeces (La Rosa et al. 2020; Zheng et al. 2020). The volume of sewage generated per inhabitant per day varies between 120 and 200 litters in Brazilian urban areas (SNIS. Diagnóstico dos Serviços de Água e Esgotos –2018 2019). Therefore, PI determination based on the viral concentration in sewage is marred by uncertainties, due to the large intrinsic variations of the parameters required for its calculation. However, such uncertainties can be reduced if the PI in each region is evaluated relative to the overall PI in the sewershed, determined based on the viral genome concentration at the inlet of the sewage treatment plant (STP).
It is reasonable to consider that the ratio between the viral genome load in faeces (F x NF) and the volume of sewage generated per inhabitant per day is similar for each monitored region and the entire sewershed (monitored at the inlet of the STP). Therefore, these parameters can be grouped and expressed as a constant (K) throughout different regions and the entire sewershed (Eq. 4).
| (4) |
COD was used as a normalisation parameter to account for possible variations in water consumption amongst regions and differences in sewage composition due to distinct sample composite times. The measured COD in each sampling point was compared to the COD measured at the respective sewage treatment plant (Arrudas or Onça STP). Paired Kolmogorov Smirnov statistical tests (using SPSS Statistics software – IBM Corp., v27) were performed on COD data (Table S2). When significant differences were detected, a correction factor was applied to V, based on the ratio between the median COD concentration measured at the sampling point and the median COD concentration measured at the respective STP.
Eq. 3 can then be rewritten as follows (Eq. 5):
| (5) |
The Relative Prevalence Index of each monitored region (RPIi) is calculated as the ratio of the PI of each monitored region (PIi) and the PI of the entire sewershed (PISTP; monitored at the inlet of the STP), and is described by the following equation (Eq. 6):
| (6) |
Where Ci is the viral genome concentration determined at each monitored region (RNA copies.L−1) adjusted for decay and CSTP is the viral genome concentration determined for the entire sewershed (inlet of the STP; RNA copies.L−1), adjusted for decay. Regions with RPI higher than 1 have COVID-19 prevalence higher than the average prevalence in the sewershed and could hence be considered hotspots.
3. Results and discussion
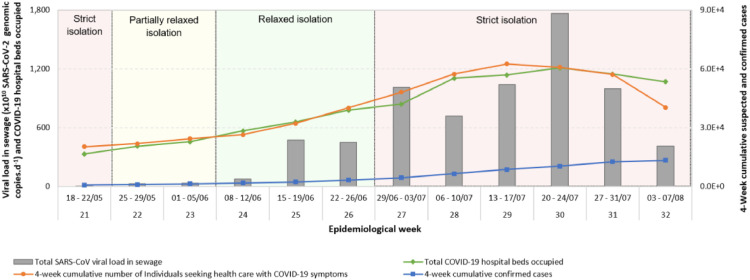
Due to limitations on COVID-19 testing data, authorities in Belo Horizonte have planned their pandemic control actions based on COVID-19 bed occupation data collected from local hospitals and municipal health centres and on the effective transmission rate (Rt). Likewise, we have collected and evaluated data that do not depend on COVID-19 testing, including the number of people seeking health care with COVID-19 symptoms. Partial lockdown measures were pre-emptively introduced to contain the spread of the virus from March 18, 2020 (epidemiological week 12), when a municipal decree ordered the closure of all non-essential services. These restrictions were partially lifted on May 25, 2020 (epidemiological week 22), when virtually the entire retail sector was allowed to open. Further relaxation of restrictions took place on June 8, 2020 (epidemiological week 24), when mainly agglomeration-prone establishments remained closed, including schools, bars, restaurants, and cinemas. The lifting of restrictions resulted in the aggravation of the pandemic from week 25, as shown by increases in the number of local COVID-19 hospital bed occupations, the number of individuals seeking health care with COVID-19 symptoms, and total SARS-CoV-2 load in sewage (Fig. 3 ). By week 26, the total number of intensive care and infirmary hospital beds occupied by COVID-19 patients reached, respectively, 85% and 69% of locally available COVID-19 beds. As a result, strict isolation measures were reinstated on week 27 (June 29, 2020).
Fig 3.
Evolution of SARS-CoV-2 RNA loads in sewage (sum of viral loads in the influent of sewage treatment plants Arrudas and Onça), occupation of COVID-19 hospital beds, 4-week cumulative confirmed cases, and 4-week cumulative number of individuals seeking health care with COVID-19 symptoms in Belo Horizonte.
The four-week cumulative number of individuals seeking health care with COVID-19 symptoms and the load of SARS-CoV-2 RNA in sewage followed similar trends (Fig. 3), with sharp increases in weeks 24 to 26, followed by surges and later decreases in weeks 27 to 32. The comparison based on a four-week period was carried out because virus shedding in stool may occur for up to 4 weeks after the onset of respiratory symptoms in mild to severe cases (Zheng et al. 2020). The observed low number of confirmed cases in the same period highlights the scarcity and inadequacy of local clinical testing data. Sewage monitoring can detect pre- and asymptomatic individuals (La Rosa et al. 2020) who may excrete the virus in faeces but are not tested and hence are not included in official statistics.
Accurately estimating the number of COVID-19 cases based solely on sewage data may be problematic due to the intrinsic wide variability of some parameters necessary to convert SARS-CoV-2 RNA concentrations in sewage to the number of cases. For example, the number of viral genome copies excreted per gram of faeces can vary from 103 to 107 (Woelfel et al. 2020). To circumvent that limitation and to identify hotspots within the city, we propose data evaluation based on the Relative Prevalence Index (RPI, Section 2.6, Material and methods) and the concentration of SARS-CoV-2 RNA in sewage for each of the sampled regions, as both parameters do not depend on conversion factors. We define the RPI as the ratio of the regional prevalence (based on SARS-CoV-2 RNA concentrations determined at each monitored region) over the prevalence in the sewershed (based on SARS-CoV-2 RNA concentrations determined at the treatment plant inlet). RPI calculations take into consideration the estimated decay of SARS-CoV-2 RNA (Section 2.5, Material and methods) as it travels through sewers (Ahmed et al. 2020). A region with RPI higher than 1 has a COVID-19 prevalence higher than the average prevalence in the sewershed and could hence be considered a hotspot. Depending on the uncertainties related to the determination of some of the factors that may affect the RPI (e.g., sampling regime and storage, RNA quantification protocol, etc.), the alert level for regions of concern could be set at values higher than 1 in order to avoid overly alarmist prognostics based solely on sewage.
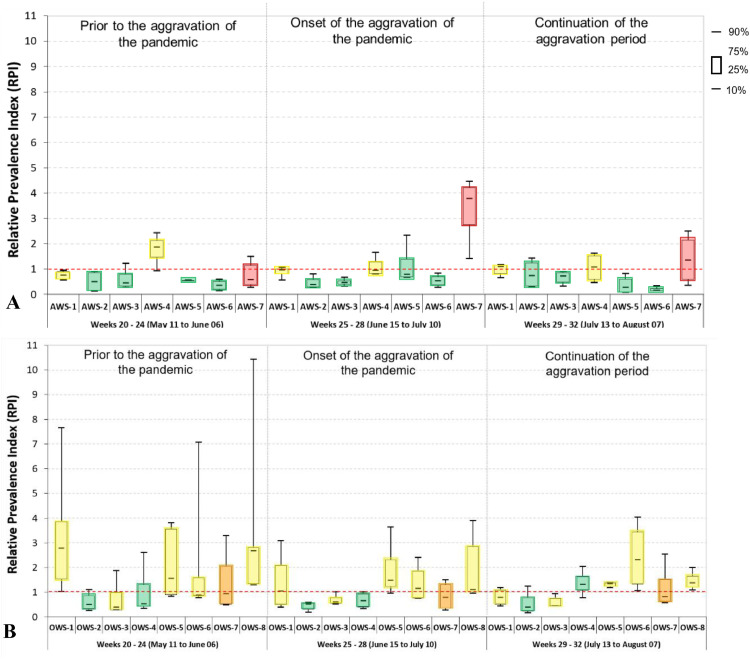
Fig. 4 shows RPI values determined for each of the 15 monitored regions in three distinct periods during the pandemic: epidemiological weeks 20 and 24 (prior to the aggravation of the pandemic); 25 and 28 (includes the onset of the aggravation period); and 29 and 32 (continuation of the aggravation period and beginning of the attenuation period). In addition, Fig. 4 shows the Health Vulnerability Index - HVI (Section 2.2, Material and methods) for each of the sampled regions. Regions with high HVI are considered vulnerable, as they are dominated by unplanned occupations (also known as favelas or shanty towns), with poor living conditions characterised by densely populated households with poor access to sanitation, health care, and other basic public services (Scalzaretto 2020). Our data shows that the most vulnerable regions were the hardest hit by the pandemic. For instance, the most vulnerable regions in the Arrudas (AWS-1, AWS-4 and AWS-7) and Onça (OWS-1, OWS-5, OWS-6, OWS-7 and OWS-8) sewersheds showed persistently high RPIs. In the period prior to the aggravation of the pandemic, the median COVID-19 prevalence in region OWS-8 was nearly three times the average prevalence in the Onça sewershed. In the period comprising weeks 25 to 28, the median COVID-19 prevalence in region AWS-7 was nearly four times the average prevalence in the Arrudas sewershed. Both these regions are classified as medium to extremely vulnerable. Regions with low HVI were less affected by the pandemic (e.g., OWS-2, AWS-2, AWS-3, AWS-5 and AWS-6), with RPI values below one during most of the monitoring period. RPI data proved to be particularly relevant as an early warning tool, as it helped to identify regions with relatively high COVID-19 prevalence before the aggravation of the pandemic that affected the entire city.
Fig 4.
Relative Prevalence Index (RPI) for different regions in Belo Horizonte in three distinct periods during the pandemic. RPI is the prevalence of COVID-19 in each region relative to the prevalence in the sewershed: (A) Arrudas sewershed; (B) Onça sewershed. A region with RPI of 1 has a prevalence similar to that in the sewershed. The Health Vulnerability Index (HVI) of each region is indicated in colours: red: extremely vulnerable; orange: very vulnerable; yellow: vulnerable; and green: not vulnerable.
These results are hardly surprising, as it has been fairly well documented that socioeconomically disadvantaged communities are being disproportionately affected by COVID-19 (Adhikari et al. 2020; Wadhera et al. 2020; Bilal et al. 2021; Ogedegbe et al. 2021; Passos et al. 2021). However, the novelty of the current work is that these findings were achieved using decentralized sewage monitoring, rather than based on clinical data.
A recent study by Passos et al. (Passos et al. 2021) based on census and calculated excess deaths data also showed that people living in vulnerable areas in Belo Horizonte were more affected by COVID-19 (epidemiological weeks 10 to 43). This was especially true for the elderly, as the mortality rate per 100,000 for 60-year-old and over was 179.2, 353.6 and 472.6 in low, medium and high/very high vulnerability sectors, respectively.
Bilal et al. (2021) evaluated the correlation between the Social Vulnerability Index (SVI) and the number of COVID-19 tests done, confirmed COVID-19 cases, and COVID-19 deaths for hundreds of zones in Chicago, Philadelphia, and New York City. The SVI used included 15 variables from the American Community Survey, comprising 4 domains: socioeconomic status, household composition, race and language barriers, and housing and transportation. Sectors with higher SVI (more vulnerable) had lower testing rates, higher positivity rates, more confirmed cases, and greater COVID-19–related mortality.
Vulnerable communities are more affected by the pandemic likely due to a number of factors or a combination of them, including higher risk of exposure to the virus (e.g., due to their stronger reliance on public transport, which can be overcrowded, even during the pandemic), lower access to safe water and sanitation, higher number of people per household, cohabitation with relatives who work in essential services, higher comorbidity burden, decreased access to health care, barriers to testing, and other factors.
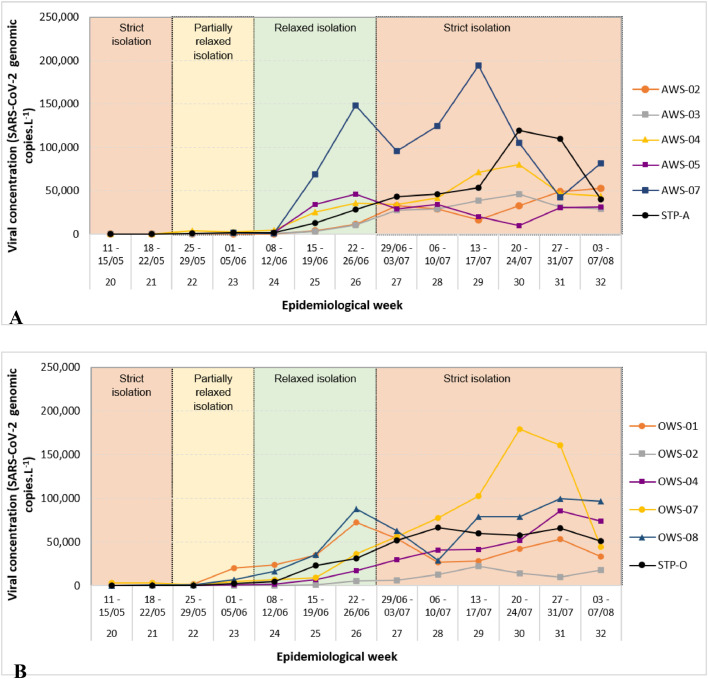
Our data on SARS-CoV-2 RNA concentrations in sewage (Fig. 5 ) indicate that the monitored regions had distinct epidemiologic dynamics, with clear differences in peak duration, week of peak occurrence and number of peaks among regions. The factors that govern these differences have yet to be determined.
Fig 5.
Two-week moving average of SARS-CoV-2 concentrations (N1 copies per L) in sewage for selected regions in sewersheds Arrudas (A) and Onça (B). The values shown in these graphs have been adjusted to account for the decay of RNA in the sewers (Section 2.5, Material and methods).
The current study has important limitations that should be acknowledged. For instance, RNA recovery rates during sample concentration and RNA extraction were not systematically assessed. Therefore, any possible extraction biases were assumed constant throughout the study, which might not necessarily be accurate. SARS-CoV-2 faecal shedding patterns were assumed to be similar among all sampled regions. Considering that the smallest population-equivalent of the monitored regions was fairly large (approximately 12 thousand people), this basic assumption might be reasonable, as viral shedding patterns in large populations tend to be less affected by groups of individuals with highly different shedding rates. Finally, the volume and composition of sewage might differ among monitored regions, especially due to socioeconomic factors. To address this, chemical oxygen demand (COD) was used to calculate a correction factor for RPI calculations. COD is frequently used as a normalizing parameter in wastewater-based epidemiology (WBE) studies (Rico et al. 2017).
4. Conclusion
Our data suggested that local restrictions had a strong effect on hospital bed occupation, as well as suspected COVID-19 cases and SARS-CoV-2 RNA loads in sewage. The monitoring of wastewater closer to the target population, in several neighbourhoods and not only at the treatment plant, revealed the spatial distribution in prevalence of the virus in a large, heterogeneous Brazilian city. The relative prevalence index (RPI) was proposed to help identify hotspots during different periods in the pandemic. Since the RPI assesses the regional prevalence relative to the prevalence in the entire sewershed, several parameters are normalized in its calculation and are therefore not needed, including shedding rates per infected person and sewage flow rates. This can be greatly advantageous, as shedding rates per infected person can vary by several orders of magnitude among individuals, and accurately measuring sewage flowrates during sampling can be very challenging. In-sewer SARS-CoV-2 RNA decay was modelled, and the results showed negligible decay estimates in the local sewer network.
The novelty of the current work is that hotspots could be identified in the city based on data generated by decentralized sewage monitoring, rather than based on clinical data. This could help major cities deal with the current pandemic, especially in places with utterly inadequate clinical testing data, such as Brazil and most developing countries.
Our results highlight the importance of planning and carrying out control measures at targeted areas. This is especially true for favelas, which tend to be densely populated, with limited sanitation infrastructure, and where it may be virtually impossible for residents to effectively isolate and follow basic prevention measures such as hand washing. Our results show that the most vulnerable neighbourhoods in the city were the hardest hit by the pandemic, indicating that, for many Brazilians, the situation is a lot worse than reported by official figures. Therefore, specific pandemic control actions should be prioritised in the identified hotspots, including carrying out targeted epidemiologic surveys, intensification of contact tracing of patients, implementation of full, mandatory local lockdowns, and possibly prioritizing vaccination of people living under such conditions.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The project was led by the National Institute of Science and Technology on Sustainable Sewage Treatment Plants (coordinated by the Federal University of Minas Gerais – UFMG) and the National Water and Sanitation Agency, in partnership with the Minas Gerais State Health Authority, the Sanitation Company for Minas Gerais and the Minas Gerais Institute for Water Management. The authors acknowledge the financial support obtained from the following institutions: Agência Nacional de Água e Saneamento Básico – ANA; Conselho Nacional de Desenvolvimento Científico e Tecnológico – CNPq; Fundação de Amparo à Pesquisa do Estado de Minas Gerais – FAPEMIG; Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – CAPES; Instituto Nacional de Ciência e Tecnologia em Estações Sustentáveis de Tratamento de Esgoto – INCT ETEs Sustentáveis (INCT Sustainable Sewage Treatment Plants). The authors would like to thank Professors Tom Curtis (Newcastle University) and William T. Sloan (University of Glasgow) for their review of the paper and valuable comments and feedback.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.watres.2021.117388.
Appendix. Supplementary materials
References
- Adhikari S., Pantaleo N.P., Feldman J.M., et al. Assessment of community-level disparities in coronavirus disease 2019 (COVID-19) infections and deaths in large US metropolitan areas. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.1693. [PMID: 32721027](2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728(138764):1–8. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsh P.M., Bibby H., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.T., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;81 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Harwood V.J., Gyawali P., Sidhu J.P.S., Toze S. Comparison of concentration methods for quantitative detection of sewage-associated viral markers in environmental waters. Appl. Environ. Microbiol. 2015;81(6):2042–2049. doi: 10.1128/AEM.03851-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird R., Bridgewater L. 23rd ed. American Public Health Association; Washington, D.C: 2017. Standard methods for the examination of water and wastewater. [Google Scholar]
- Bilal U., Tabb L.P., Barber S., et al. Spatial inequities in COVID-19 testing, positivity, confirmed cases, and mortality in 3 U.S. cities. an ecological study. Ann. Intern. Med. 2021 doi: 10.7326/M20-3936. 30 March 2021[Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D.M., Butler D., Orman N.R., Davies J.W. Gross solids transport in small diameter sewers. Water Sci. Technol. 1996 doi: 10.2166/wst.1996.0168. [DOI] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Center for Disease Control and Prevention. 2019-Novel coronavirus (2019-nCoV) Real-time rRT-PCR panel primers and probes. https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html (accessed 12 May, 2020).
- Chernicharo C.A.L., Araújo J.C., Mota Filho C.R., et al. Engenharia Sanitária e Ambiental; 2020. Sewage monitoring as an epidemiological surveillance tool to control covid-19: a case study in the city of Belo Horizonte. (online), http://abes-dn.org.br/?page_id=35681. [Google Scholar]
- Hovi, et al. Role of environmental poliovirus surveillance in global polio eradication and beyond. Epidemiol. Infect. 2012;140:1–13. doi: 10.1017/S095026881000316X. [DOI] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Ferraro G.B., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, et al. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26:2020. doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020 doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- OECD . OECD Publishing; Paris: 2019. Latin American Economic Outlook 2019: Development in Transition. [DOI] [Google Scholar]
- Ogedegbe G., Inouye S.K. 2021. Injustice in Health: Now Is the Time to Change the Story. Annals of Internal Medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passos V.M.A., Brant L.C.C., Pinheiro P.C., et al. Mortality and social inequality during COVID-19 pandemic in Belo Horizonte. Brazilian J. Epidemiol. 2021 doi: 10.1590/1980-549720210025. [DOI] [Google Scholar]
- PBH - Belo Horizonte City Hall . 2018. Health Vulnerability Index. Índice de vulnerabilidade da saúde.http://bhmap.pbh.gov.br/v2/mapa/idebhgeo (accessed April 14, 2020) [Google Scholar]
- Peccia J., et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020 doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Fernando E., Simon P., Allende A., Sanchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Wat. Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico. M., Andrés-Costa M.J., Picó Y. Estimating population size in wastewater-based epidemiology. Valencia metropolitan area as a case study. J. Hazard. Mater. 2017 doi: 10.1016/j.jhazmat.2016.05.079. [DOI] [PubMed] [Google Scholar]
- Scalzaretto N. 2020. Mapping the Covid-19 spread in Brazilian favelas.https://brazilian.report/coronavirus-brazil-live-blog/2020/04/20/mapping-covid-19-spread-brazilian-favelas/ [Google Scholar]
- Sims N., Kasprzyk-Hordern B. Future perspectives of wastewater-based epidemiology: Monitoring infectious disease spread and resistance to the community level. Environ. Int. 2020;139 doi: 10.1016/j.envint.2020.105689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SNIS. Diagnóstico dos Serviços de Água e Esgotos –2018. Brasília. Disponível em: <http://www.snis.gov.br/diagnostico-anual-agua-e-esgotos/diagnostico-dos-servicos-de-agua-e-esgotos-2018>. (Accessed in: jul.2020). In Portuguese (2019).
- Symonds E.M., et al. A case study of enteric virus removal and insights into the associated risk of water reuse for two wastewater treatment pond systems in Bolivia. Water Res. 2014;65:257–270. doi: 10.1016/j.watres.2014.07.032. [DOI] [PubMed] [Google Scholar]
- Verbyla M.E., Symonds E.M., Kafle R.C., et al. Managing microbial risks from indirect wastewater reuse for irrigation in urbanizing watersheds. Environ. Sci. Technol. 2016;50:6803–6813. doi: 10.1021/acs.est.5b05398. [DOI] [PubMed] [Google Scholar]
- Wadhera R.K., Wadhera P., Gaba P., et al. Variation in COVID-19 hospitalizations and deaths across New York City boroughs. JAMA. 2020;323(2020):2192–2195. doi: 10.1001/jama.2020.7197. [PMID: 32347898] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigginton K.R., Ye Y., Ellenberg R.M. Emerging investigators series: the source and fate of pandemic viruses in the urban water cycle. Environ. Sci. Water Res. Technol. 2015;1:735–746. doi:0.1039/C5EW00125K. [Google Scholar]
- Woelfel R., Corman V.M., Guggemos W., Seilmaier M., et al. Nature; 2020. Virological assessment of hospitalized cases of coronavirus disease 2019. [DOI] [PubMed] [Google Scholar]
- Worldometers, https://www.worldometers.info/coronavirus/(accessed 3 12 DecemberApril, 20202021) (2020 2021).
- Zheng S., Fan J., Yu F., Feng B., et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;2020(369):m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.