The members of the MYC family of oncogenes, in particular c-Myc, N-Myc, and L-Myc, are activated either directly or indirectly in many, if not most, human tumors. These structurally and functionally related nuclear phosphoproteins (Fig. 1A) appear to promote cell growth and transformation by regulating the transcription of target genes required for proliferation (33, 44, 57). Myc's transcription functions require a C-terminal basic helix-loop-helix–leucine zipper (bHLHZip) DNA-protein interaction motif and conserved regulatory domains in the N terminus (Myc boxes, MB1 and MB2), which regulate Myc's transactivation and/or transrepression functions (Fig. 1A). Normally, the expression of Myc family genes is cell context specific, mutually exclusive, and tightly dependent upon mitogens (8, 47, 71, 88). Furthermore, the life span of Myc proteins is brief, and they are rapidly degraded by the ubiquitin-linked proteasome machinery (45). However, these controls are lost in cancer cells, resulting in aberrantly high levels of Myc proteins (2, 57).
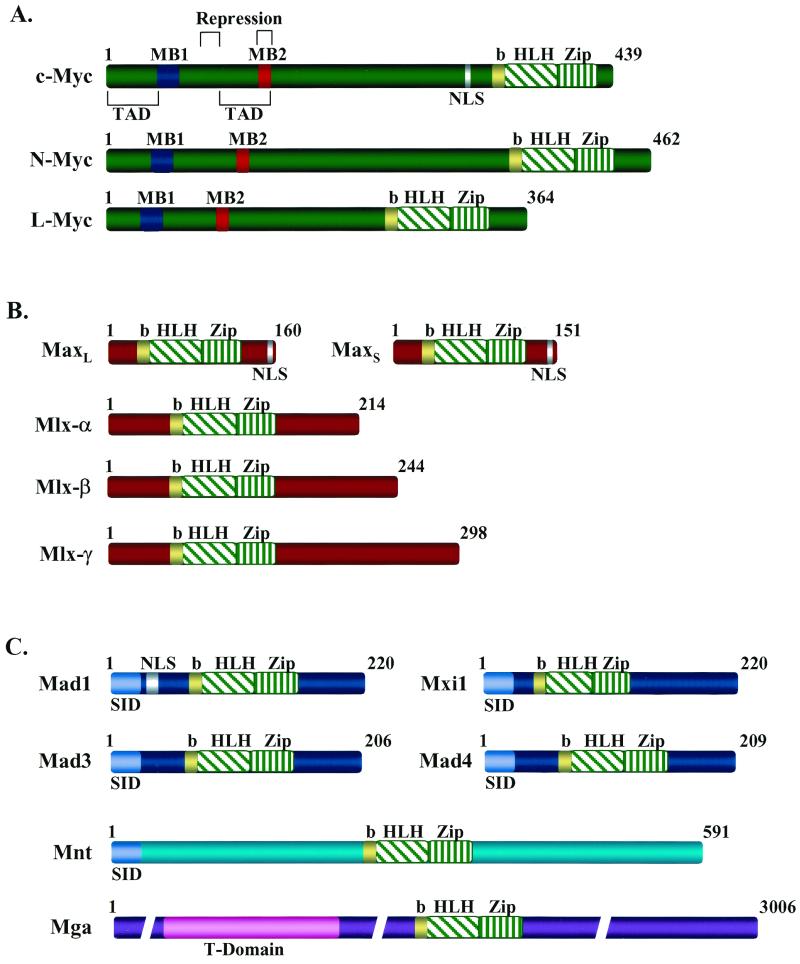
FIG. 1.
Domain structure of Myc, Max, and Mad family members and of Mnt and Mga. The structures of the bHLHZip factors of the Myc-Max-Mad network are shown. (A) The Myc family members activated in human cancer (c-Myc, N-Myc, and L-Myc). MB1 and MB2 are conserved Myc homology boxes found in all Myc family members that are required for Myc's transactivation and/or transrepression functions (113). MB2 is also required for interaction with the TRRAP transcriptional coactivator. TAD, c-Myc transactivation domain. (B) Max and its relative Mlx, which is expressed as three isoforms (18, 21, 85, 91). Max can homodimerize or form heterodimers with all members of the network through the HLHZip interactions, and the basic region confers specific DNA binding to CACGTG elements. Mlx selectively dimerizes with Mad1, Mad4, and Mnt but fails to dimerize with either Myc or Max (18, 91). (C) The Myc antagonists Mad1, Mxi1, Mad3, Mad4, and the related proteins Mnt (Rox) and Mga. Mad family members and Mnt (Rox) contain an alpha-helical amino-terminal domain (SID) that is required for interactions with the transcriptional corepressors Sin3a and Sin3b. By contrast, Mga contains a T domain near its amino terminus which is a conserved DNA- and protein-binding motif found in the Brachyury family of cell specification transcription factors (64). NLS indicates the location of identified nuclear localization signals. b, basic.
The strong selection for Myc overexpression in tumors appears to reflect its ability to provide constitutive signals that promote proliferation and angiogenesis within the growth-restrictive conditions of the tumor microenvironment (9, 25, 40, 108, 116). For example, c-Myc expression is necessary and sufficient for the entry of most cells into the DNA-synthetic (S) phase of the cell cycle (6, 9, 40, 42, 43, 50) and activates an angiogenic switch (108), whereas inhibition of Myc promotes cell cycle withdrawal and terminal differentiation (31, 50, 54, 116). Furthermore, programmed expression of Myc in transgenic animals is capable of promoting a diverse array of tumor types, and yet this requires cooperating mutations that disable Myc's penchant for activating apoptosis (9, 41, 43), an endogenous form of cell suicide (93).
In a yin-yang scenario typical in biology, Myc's ability to promote growth and transformation has been suggested to be opposed by a number of structurally related Myc antagonists. Their discovery followed the landmark findings of Blackwood and Eisenman (21) and others (114) that Myc's ability to bind to its cognate DNA recognition E-box site (CACGTG), activate transcription, and promote cell proliferation, transformation, or apoptosis requires its dimerization with an apparently obligate bHLHZip binding partner dubbed Max (4–7, 21, 22, 114). Unlike that of Myc genes, Max expression is ubiquitous and constitutive (16), and this small protein (two alternatively spliced forms [Fig. 1B]) is stable, resulting in Max levels that far exceed those of Myc (21, 69). However, Max, but not Myc, is also capable of forming homodimers, and at least in nuclear extracts these complexes appear to exist in cells (84), although the role that they may play remains obscure (16, 75, 114, 141).
Max interactions are rather promiscuous, as Max also dimerizes to other short-lived factors sharing a similar bHLHZip domain, including the Mad family members (Mad1, Mxi1, Mad3, and Mad4 [Fig. 1C]) (10, 11, 63, 143) as well as the much larger and more distantly related bHLHZip factors Mnt (Rox) and Mga (Fig. 1C) (62, 64, 92). Based upon overexpression studies, these six additional Max partners have been proposed to function as natural antagonists of Myc, as they can effectively compete for interactions with Max and generally repress, rather than activate, transcription by binding to the same CACGTG elements (11, 62–64, 92, 143). Transcriptional repression by the Mad proteins, and by Mnt, occurs through the assembly of a multisubunit complex that includes the transcriptional corepressors N-CoR and Sin3a/Sin3b, the histone deacetylases 1 and 2, and the oncoproteins Ski and Sno (3, 12, 51, 52, 55, 77, 78, 99, 125, 131). Assembly of this complex requires a small alpha-helical domain present in the amino terminus of Mnt and all Mads, which is required for binding to Sin3a/Sin3b (the Sin3 interaction domain [SID] [Fig. 1C]) (39, 51, 125). The net result is the formation of Max–Mad–Sin3–N-CoR–histone deacetylase–Ski-Sno complexes, which repress transcription of target genes through deacetylation of histone residues, resulting in the remodeling of chromatin into a closed conformation (3, 52, 55, 78, 99). The exception is Mga (64), which does not contain a SID motif but rather has a T domain within its amino terminus, which is a highly conserved DNA-binding and dimerization motif originally identified in the protein Brachyury (133) (Fig. 1C). Mga has been suggested to repress Myc-mediated transactivation simply by competing for Max, but Mga-Max interactions are complex and lead to unexpected transcriptional effects (see below).
Mad proteins, and Mnt and Mga, can effectively compete with c-Myc for binding to Max, at least in vitro. Mad1 and Mad4 are generally expressed in terminally differentiated cells, whereas Mxi1, Mad3, Mnt, and Mga, like all Myc genes, are also expressed in proliferating cells (61–64, 118, 143). These findings have prompted the hypothesis that collectively these proteins form a network centered around Max, whereby Myc activity is harnessed by competition for Max by Mad or Mnt (Fig. 2A). Why so many antagonists would exist to modulate Myc functions is unclear, and under such constraints one wonders how Myc could ever work. More parsimonious interpretations are clearly possible. In particular, recent biochemical and genetic studies have revealed additional layers of complexity, and gene targeting studies of mice have failed to make the expected links, promoting a fresh look at the network.
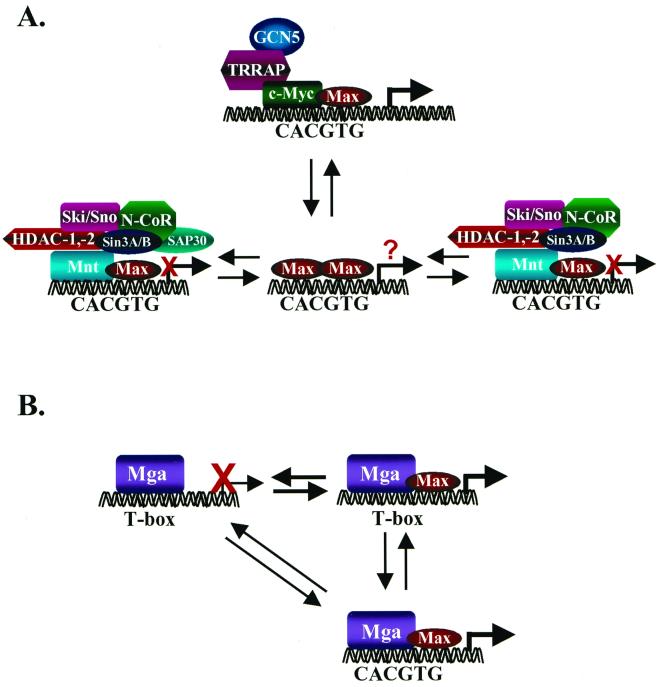
FIG. 2.
The conventional model of transcriptional regulation by the Myc-Max-Mad network (A) and transcriptional regulation by Mga (B). (A) As proposed by Eisenman and colleagues (11, 75), transcriptional activation is mediated exclusively by Myc-Max complexes which bind to CACGTG elements (and also to CACATG) in target genes. Myc's ability to activate transcription appears to require its association with the transcriptional coactivator TRRAP (89), which tethers the histone acetylase GCN5 (90). Transcriptional repression is mediated through the binding of Mad-Max or Mnt-Max complexes to identical sites and to the transcriptional corepressors Sin3a and Sin3b, through the SID. Sin3a/Sin3b interactions tether Mad/Mnt-Max complexes to a large transcriptional repressor complex that appears to contain the corepressors N-CoR, Ski, and Sno; an adapter protein, SAP30 (77, 145); and the histone deacetylases 1 and 2. An equilibrium among the various components of this network, regulated by changes in their expression and/or signaling events, dictates whether there is transcriptional activation or repression of target genes. The precise function of Max homodimers is not resolved, although they clearly exist in cells (142) and have been suggested to passively repress transcription by competition for DNA-binding elements (5). (B) Functions of Mga. Mga contains two DNA-binding protein interaction motifs, a bHLHZip domain related to that present in Myc, Max, Mad, and Mnt, and a T domain, which facilitates binding to large recognition elements that are bound by the Brachyury family of transcription factors, which play an essential role in cell specification (64). When bound to T boxes, Mga functions as a transcriptional repressor, but this is relieved by overexpression of Max. Binding of Mga to the consensus Myc-Max binding site CACGTG requires Max interactions, and at least when both are overexpressed, this complex activates gene expression (64). Thus, in theory, Mga can compete with Myc by sequestering Max, but the net result may be that Mga induces the same targets as does Myc.
CHALLENGES TO THE MYC-MAX-MAD NETWORK
The Myc-Max-Mad network model proposed by Eisenman and colleagues pins a central role on Max (11), which forms transactivating complexes when associated with Myc but repressive complexes when bound to Mads or Mnt. This model implies that Myc-Max, Max-Max, and Mad/Mnt-Max complexes exist in an equilibrium and that shifts in this equilibrium dictate whether transcription of target genes containing CACGTG elements is activated or repressed (Fig. 2A) (47). Changes in the balance could occur through well-documented fluctuations in the expression of Max-interacting proteins, for example, the down-regulation of Myc and concomitant up-regulation of some Mad genes when cells receive signals to undergo growth arrest or differentiation (10, 11, 23, 31, 143). In principle, signaling events that regulate their subcellular localization, degradation, protein-protein interactions, and/or DNA-binding or transcriptional activity could also skew the balance of Myc-Max versus Mad/Mnt-Max complexes. For example, in vitro phosphorylation of serines 2 and 11 of Max by casein kinase II impairs the DNA binding of Max-Max complexes (15, 16), and the phosphorylation of c-Myc by casein kinase II and glycogen synthase kinase 3β has been implicated in regulating its transcriptional and transforming activity and its ubiquitin-mediated degradation (48, 56, 98, 117). Finally, the model implies that all of the dimer complexes in the Myc-Mad-Max network compete for common DNA target sites (47). However, Myc-Max heterodimers bind only a subset of the sites bound by Max homodimers, due to a differential recognition of the flanking sequences (20, 72, 101, 134), and different residues within the bHLH region of Mad family members have been suggested to play a role in the recognition of different E-box sites (101). Furthermore, Myc has also been shown to bind to noncanonical (non-E-box) sites (20). Thus, although this model has been useful for understanding the opposing effects of Mads, Mnt, or Mga on targets that are activated by c-Myc, several issues challenge this paradigm.
Myc antagonists are often coexpressed with Myc.
The concept of the switch from Myc-activating to Mad/Mnt-repressing complexes is simple enough, but a paradox is that Mxi1, Mad3, Mnt, and Mga are often expressed in proliferating cells that also express Myc. Myc protein levels are never exceedingly high in proliferating cells (42, 88), and when they are, cells undergo apoptotic suicide (9, 19, 43, 102, 104). If it is simply a question of levels, then one would expect that the sum of the levels of these antagonists should far exceed the level of Myc, and if these complexes truly share targets, the equilibrium should always be in favor of transcriptional repression. In primary mouse embryo fibroblasts the creation of a conditional knockout of c-myc has shown that c-Myc functions are essential for entry into S phase (A. Trumpp, personal communication). So how then can cells grow if they express these antagonists? Two possibilities are that their functions are inactivated by signaling events and that they are active at specific phases of the cell cycle, as at least Mad3 appears to be expressed in an S-phase-specific manner (119). However, at this juncture there are no data directly supporting either of these prospects.
Yet another dilemma is that Mga-Max interactions extend the network into a new arena. The T domain present in Mga places it in the Tbx family of transcription factors, which play a critical role in the specification of embryonic mesoderm (59). Typically, these factors activate transcription from large palindromic response elements dubbed T boxes (59, 105). Curiously, Mga normally acts to repress promoters harboring T boxes but activates transcription from CACGTG elements, at least when overexpressed in conjunction with Max. Furthermore, Max interactions with Mga somehow convert Mga from a repressor to an activator of promoters bearing T boxes (64) (Fig. 2B). A review of Myc target genes reveals that most lack clearly identifiable T-box elements. Since Mga is coexpressed with Myc in proliferating cells (64), it is thus possible that many Myc-regulated targets are in fact activated by Mga-Max complexes.
Myc is promiscuous.
Myc has been shown to physically associate with a variety of regulatory proteins, and at last reckoning, there are 14 partners in addition to Max (for a recent review, see reference 123). At least half of these interacting proteins bind to Myc's HLHZip domain; these interactions are therefore mutually exclusive with Max and should in principle effect the equilibrium of Myc-Max and Mad/Mnt-Max complexes. Two examples underscore this notion. First, transactivation by c-Myc appears to require interactions of Myc's MB2 domain with an essential cofactor dubbed TRRAP (89), which tethers the histone acetylase GCN5 (Fig. 2A) (90), and Myc's HLHZip interaction with INI1/Snf5 (27), a factor that recruits the Swi-Snf complex that activates transcription through chromatin remodeling (141). At this juncture, it is not clear whether INI1/Snf5-Myc interactions are mutually exclusive with Max (Fig. 3A). Second, the HLH domain of Myc can also interact with Miz1 (Myc-interacting Zn-finger protein 1), a POZ domain zinc finger protein that activates transcription through initiator (Inr) elements and induces G1 cell cycle arrest (111). Myc binds to and inhibits Miz 1 DNA binding (111), suggesting that Myc may repress transcription of genes vis-à-vis its effects on Miz1 or Miz1-like proteins (Fig. 3A). These results and others (see below) raise the interesting notion that we may be thinking about Myc the wrong way round. Perhaps Myc is the real antagonist, and its primary function is to relieve transcriptional repression of its targets by effectively competing for Max, which is required for Mad- and Mnt-mediated repression of gene expression (Fig. 3B).
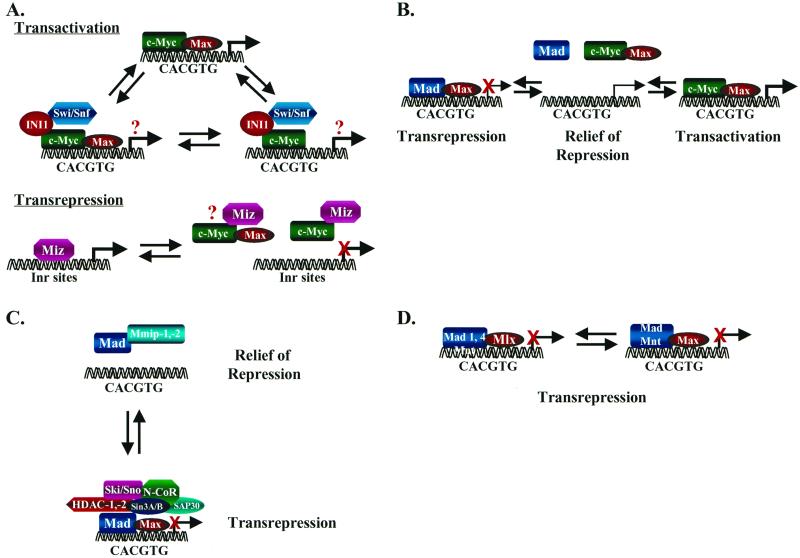
FIG. 3.
Alternative mechanisms regulating Myc transcriptional activity. (A) Myc interacts through its HLH domain, perhaps in a mutually exclusive fashion, with the transcriptional regulatory proteins Max (top), INI1/Snf5 (middle), and Miz1 (bottom) to activate (Max and INI1) or repress (Miz1) transcription. It is unknown whether ternary complexes form. (B) Target genes bearing CACGTG elements are actively repressed by Mad/Mnt-Max complexes. Myc may then relieve active repression by effectively sequestering Max into solution and/or may bind CACGTG elements in conjunction with Max to activate transcription. (C) The Mad-specific interacting proteins Mmip-1 and Mmip-2 relieve Mad-mediated active repression by sequestering Mad into solution, which may lead to transcriptional activation. Mad–Mmip-1 or Mad–Mmip-2 complexes apparently do not bind DNA (49, 143). (D) The Max-related protein Mlx selectively binds to Mad1, Mad4, and Mnt, and these complexes actively repress transcription through binding to CACGTG elements (18, 91). Thus, Mad-mediated repression of transcription can occur independently of Max.
Mad also has other partners.
A tenet of the Myc-Max-Mad network is that Mad functions antagonize those of Myc by competing for Max (47), but recently, two additional Mad-specific interacting proteins have called this into question. First, Mad-member-interacting protein 1 (Mmip-1), a protein that contains a RING finger and a Zip domain, dimerizes with the Zip domains of all Mad family members but not with those of c-Myc or Max, and this interaction blocks Mad's suppressive effects on Myc functions (49). Mmip-2, yet another RING finger protein, also blocks Mad functions through interactions with the Mad Zip domain and, when overexpressed, can sequester Mad1 into the cytoplasm (142). A caveat for the Mmip-1 and Mmip-2 studies is that their interactions with Mad have been shown only in overexpression experiments, a problem common to many studies in the field. However, if these interactions are physiological, the function of Mad-Max complexes can be inactivated by mutually exclusive interactions of Mad with Mmips. The net result of this scenario is that more Max would be available to bind to Myc (Fig. 3C).
Mlx, a relative of Max.
To add even more complexity to the mix, two groups have independently cloned a structurally and functionally Max-related protein, termed Mlx (Fig. 1B) (18, 91). Like Max, Mlx is a long-lived and ubiquitously expressed bHLHZip protein, which can form homodimers and bind to the CACGTG element. However, there are three distinct isoforms of Mlx (α, β, and γ), and only one form, Mlxγ, is nuclear in its localization. Curiously, there appears to be a level of interplay between these isoforms, as Mlxγ can sequester Mlxα into the nucleus (91). Moreover, unlike Max, Mlx selectively forms heterodimers with Mad1, Mad4, and Mnt but fails to dimerize with Mxi1, Mad3, Myc, or Max (18, 91). This specificity, coupled with the ability of Mad1-Mlx and Mnt-Mlx dimers to repress transcription, again demonstrates that there are parallel pathways that regulate Mad activity independent of Max (Fig. 3D).
Network models.
The conventional model states that Myc-Max complexes specifically bind to DNA at CACGTG E-box elements in target genes such as ODC and cad (14, 94) and activate transcription through Myc's interactions with TRRAP and histone acetylases such as GCN5 (89, 90). Indeed, chromatin immunoprecipitation experiments have suggested that c-Myc–Max complexes can bind to the CACGTG response elements (24, 37). If this is indeed reflective of Myc-Max activity in vivo, the regulation of the Myc-Max-Mad network may have parallels with transcriptional regulation by the nuclear hormone receptors (87) (Fig. 4A). In both scenarios, active short-term repression is mediated by the association of DNA-binding complexes with the transcriptional corepressors such as N-CoR and Sin3a or Sin3b and the activation of associated histone deacetylases. Relief of repression by the Mad complex may be mediated by signaling pathways that disrupt the complex and/or the binding of a factor (like ligand for the hormone receptors) that inactivates the complex. Activation of the Myc target gene is then achieved by dissolution of this complex and binding displacement by the activating Myc-Max complex, which tethers the coactivator TRRAP and the histone acetylase GCN5 (Fig. 4A).
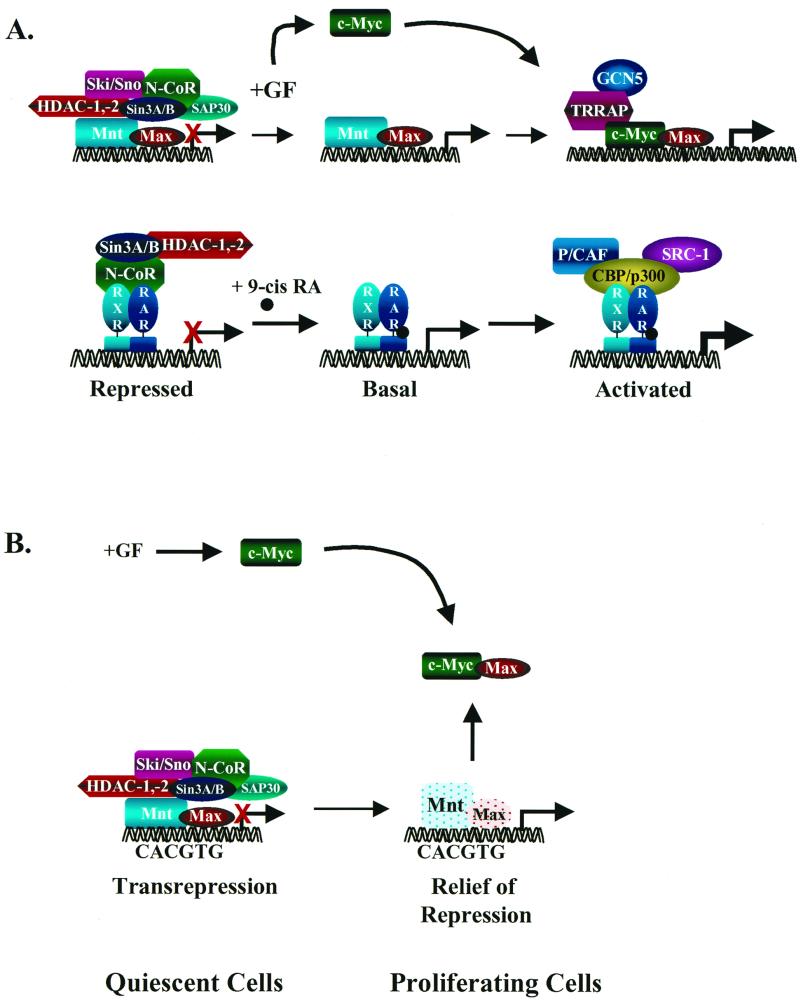
FIG. 4.
Transcriptional regulation by displacement-recruitment versus sequestration. (A) At top is shown a model of transcriptional regulation of Myc target genes by displacement and recruitment. In the repressed state, Mad/Mnt-Max DNA-binding complexes tether the transcription repressor complex. Following the addition of mitogens, c-Myc expression is up-regulated and the corepressor complex dissociates from Mad/Mnt-Max complexes, relieving active repression. c-Myc then complexes with Max, binds DNA, and recruits the transcriptional coactivator TRRAP, which tethers histone acetyltransferase GCN5. This complex then activates transcription through chromatin remodeling. This model has many parallels to the regulation of transcription by nuclear receptors (bottom panel) such as the retinoic acid receptor and retinoid X receptor (87). (B) Myc target genes are activated indirectly by the sequestration of Max away from Mad or Mnt. In this model, Myc-Max complexes do not bind DNA and the activation of target genes is due to relief of repression. Targets are then activated in proliferating cells simply by virtue of inducing c-Myc expression.
An alternative to this model is that Myc functions as the central antagonist in the pathway and blocks Mad- or Mnt-mediated repression by simply and effectively sequestering Max (Fig. 4B). A long-standing paradox for the field has been that Myc-Max complexes are exceedingly difficult to detect in whole-cell or nuclear extracts using gel shift analyses (84, 135). By contrast, the DNA binding of other complexes in cell extracts, particularly Mnt-Max and Max-Max homodimers, is easily detected (22, 84, 135; J. L. Cleveland, unpublished data). If these binding activities are reflective of the state of affairs in vivo, then Myc may activate its targets by virtue of relieving repression by Mad or Mnt by sequestering Max (Fig. 4B). In support of this notion, some of the transcriptional targets ascribed to Myc are in fact inducible by treating quiescent or growth factor-starved cells with cycloheximide (103), which would decrease levels of Mads or Mnt, which are all short-lived proteins (6).
LESSONS FROM GENE TARGETING
The ultimate genetic test for any pathway is an evaluation of the consequences of gene deletion or overexpression in vivo, and in this respect there are both pros and cons supporting the tenets of the Myc-Max-Mad network. The common denominator of Myc activation in cancers is overexpression, and among the first animal models created were the c-myc transgenic mice created by the Leder and Adams laboratories, who established that c-Myc was indeed capable of promoting transformation (1, 79, 80). Subsequent work established that N-Myc is functionally equivalent in this regard (122, 139). Thus, it was postulated that the Myc antagonists should function as tumor suppressors. Indeed, overexpression studies have demonstrated that Mad1, Mxi1, or Mnt overexpression blocks transformation of primary fibroblasts induced by Myc plus activated Ras (63, 74, 125), and transgenic animal studies have established that the programmed overexpression of Max, Mad1, and Mnt compromises lymphoid or embryonic development (62, 83, 120). However, a hallmark of tumor suppressors is their loss of function in human cancers, and here all of the Myc antagonists have fallen well short of the mark. To date, none of the loci encoding the Mad proteins, or Mnt or Mga, have been shown to be convincingly altered in human malignancies or to be lost in animal tumor models (38, 46, 70, 76, 136).
c-Myc or N-Myc deficiency arrests growth and development.
Gene targeting approaches have demonstrated that c-Myc and N-Myc functions are critical for cell proliferation and development. Gene targeting has established that both c-Myc and N-Myc are essential for murine development (26, 34, 95, 124, 137). By contrast, mice lacking L-Myc are viable and lack any defects (53). c-Myc-deficient mice die at embryonic days E9.5 to 10.5 and are developmentally retarded, with embryos having a smaller size and defects in the pericardium and neural tube closure. Further, developmental scoring suggested defects in yolk sac circulation and hind limb formation. Indeed, rederivation and analyses of these mice have revealed that the severe developmental delay in these mice is associated with marked defects in primitive hematopoiesis and vasculogenesis (C. McKay, H. Pendeville, T. A. Baudino, A. C. Davis, J. N. Ihle, and J. L. Cleveland, submitted for publication). By contrast, the N-Myc knockout is lethal at day E11.5 and is associated with multiple defects in the heart, lung, mesonephros, gut, and central and peripheral nervous system (95, 124, 137). Analyses of both of these types of mice have failed to reveal any signs of inappropriate apoptosis. Rather, on the basis of staining with S-phase markers, these defects are uniformly associated with the failure of Myc-expressing cells to proliferate (67). In support of this concept, in primary mouse embryonic fibroblasts the conditional loss of c-myc results in an absolute block in cell proliferation in the G1 phase of the cell cycle (Trumpp, personal communication).
While mice deficient in N-Myc or c-Myc die during midgestation, their cells are able to divide extensively beforehand. This is likely due to redundancy of the Myc proteins, as early in development their expression is overlapping (35, 60, 146). Indeed, formal genetic proof of this concept has come from the recent studies demonstrating that the knock-in of N-myc into the c-myc gene is compatible with development (86). Of course, one unresolved issue is why cells lacking functional Myc fail to divide. The conventional model is that c-Myc interfaces with regulators of the cell cycle. This could occur by inducing the expression of cyclins (32, 65) or the ubiquitin degradation of the cyclin-dependent kinase inhibitor p27Kip1 (139), which leads to activation of cyclinE-cdk2 complexes that are necessary and sufficient for entry and progression through S phase (96, 130). Interestingly, the link to this pathway appears to be Cul1, a critical component of the ubiquitin ligase SKP1–CDC53 (cullin)–F-box protein complex and a direct transcriptional target induced by c-Myc (100). However, another intriguing possibility is that Myc regulates the accumulation of cell mass, rather than cell division per se, by regulating targets involved in cellular growth and metabolism. For cells to divide, they must attain a certain mass and volume. The creation of the Drosophila melanogaster dMyc knockout revealed a minute phenotype, and mosaic analyses suggested that loss of dMyc reduces cell size (127). However, in the murine c-Myc knockouts cell size is apparently not diminished (132; T. A. Baudino and J. L. Cleveland, unpublished data), and thus exactly how Myc promotes growth is unresolved.
Max is also essential.
Max is ubiquitously expressed, and by creating the Max knockout mouse, DePinho and colleagues established that Max provides essential, nonredundant functions in growth and development. Max−/− mice die of an early embryonic lethality at days E5.5 to 6.5 (129), which is associated with a generalized developmental arrest of both embryonic and extraembryonic tissues. These defects are not associated with inappropriate apoptosis, but rather with a failure of cells to divide. Furthermore, the timing of the lethality appears to reflect the loss of stores of maternal Max, which is expressed at high levels in oocytes (129). The essential nature of Max in growth and development underscores the central role for this factor in regulating Myc functions, and one prediction is that the creation of a Myc-less mouse should result in a comparable phenotype. Furthermore, although Mlx is structurally related to Max, it appears that Mlx cannot compensate for the loss of Max functions, suggesting that Mlx functions may indeed be limited to regulating those of Mad and/or Mnt (18, 91), which are, at least to date, not required for growth and development.
Mad1, Mxi1, and Mad3 are dispensable.
Mad1 and Mad4 are generally expressed during terminal stages of differentiation (11, 63, 118), whereas Mxi1 and Mad3 are apparently expressed early during differentiation programs or in proliferating cells, where their expression can overlap with that of c-Myc and N-Myc (63, 118, 119, 144). Given the purported role of Mxi1 and Mad3 as antagonists of Myc, these knockout mice were expected to be tumor prone and cells from these mice should display augmented levels of Myc activity. Furthermore, all four Mad genes were expected to play an important role in development, as all are expressed during gestation, particularly in the embryonic nervous system, heart, and lung (118). These predictions have not borne fruit. First, Mad1-deficient mice displayed a modest delay in granulocyte differentiation, and this phenotype was manifest only using in vitro culture (11). Second, despite the interesting pattern of expression of Mad3 in proliferating and committed progenitors in the neural tube, loss of Mad3 also results in a very modest phenotype. As reported in this issue (119), there are subtle effects of Mad3 loss on the sensitivity of some cells to gamma irradiation. One important caveat is that the relatively uninformative phenotypes of these three Mad knockouts could be due to functional redundancy, as several members are expressed in overlapping patterns. Indeed, the expression of Mxi1 and Mad3 is up-regulated in an ectopic fashion in Mad1-knockout mice (11). However, this is apparently not the case in mice lacking Mad3 (119) or Mxi1 (144), and thus one is left to wonder if the generation of mice lacking multiple Mad family members will be more informative.
Of mice deficient in any of the three Mad family members so far analyzed, only Mxi1-deficient mice appear to have a modicum of the phenotypes typical of classical tumor suppressors (126). First, cells from Mxi1-deficient mice appear to have an accelerated rate of growth and the mice exhibit hyperplasia within some tissues. Second, although spontaneous tumors do not develop in these mice, a phenotype is manifest when they are treated with carcinogens or when they are crossed to mice lacking the bona fide tumor suppressors p16Ink4a and p19ARF (126), which target the retinoblastoma and p53 tumor suppressors, respectively (28, 68, 112, 128, 130). However, loss of Mxi1 function in the context of this double knockout does not alter the tumor spectrum from that observed for mice lacking p16Ink4a-p19ARF alone (126). There have also been suggestions that loss of MXI1 may also occur in human prostate cancer (36, 115), and missense mutations and MXI1 loss of heterozygosity have been reported for neurofibrosarcomas and desmoplastic melanoma, respectively (81, 121). However, many of these findings have been challenged (13, 38), and overall, mutations appear to be very rare (58, 70, 73, 132).
A case for Mnt?
Collectively, the data indicate that, if there indeed exists a true antagonist for Myc, perhaps it is Mnt. First, Mnt is expressed in an overlapping pattern with c-Myc and N-Myc during development. Second, like Myc proteins, Mnt is expressed in proliferating cells and Mnt-Max DNA-binding complexes are readily detected in cell extracts (92, 135). Third, in overexpression studies Mnt is a potent antagonist of Myc transcriptional and transforming activity (62). Fourth, deregulated Mnt expression in mice results in an embryonic lethality at E9.5 to 10.5 with a phenotype that, at least on the surface, appears identical to that of c-Myc-null embryos, including defects in the pericardium and a general delay in growth (62). Finally, Mnt is localized at 17p13.3, a region commonly deleted in many cancers. However, Mnt is not inactivated in any cancers having alterations in this region (136). Thus, the jury is still out as to whether Mnt is a tumor suppressor.
The Ski-Sno components of the Mad transcriptional repressor complex behave as tumor suppressors.
Of the five components of the Mad transcriptional complex (Fig. 2), knockout mice have been created for the nuclear corepressor N-CoR (66), Ski (17), and Sno (131). Sin3 has been deleted in Drosophila and results in an embryonic lethality (110). Loss of N-CoR in mice also leads to embryonic lethality that is associated with profound defects in definitive erythropoiesis and central nervous system development. Furthermore, Mad-me- diated repression is ablated in N-CoR-deficient cells (66). However, loss of N-CoR has not been linked to inappropriate activation of Myc.
The Ski transcription factor was originally identified as a truncated avian retrovirus oncogene (82) and, like N-CoR (66), can function as either a transcriptional coactivator or a corepressor. Thus, depending on the cell context, Ski overexpression can result in either transformation or differentiation (30, 97). When associated with histone deacetylase 1 (as in Mad complexes), Ski functions as a corepressor through its ability to directly bind to both N-CoR and Sin3 (99). However, Ski can also directly bind to DNA (to GTCTAGAC elements), by forming homodimers or by heterodimerizing with a structurally related transcription factor termed Sno (29, 97). Expression of Ski occurs in both proliferating and differentiating cells during development, and although some Ski−/− pups are born, nullizygous embryos generally display excessive apoptosis in cranial neuroepithelium and retarded development of skeletal muscle and the central nervous system (17). Interestingly, loss of Ski in some cell contexts leads to the ectopic expression of the Myc target ornithine decarboxylase (99).
Of all of the proteins involved in the transcriptional repression arm of the network, perhaps the physiological role of the Sno transcriptional corepressor is most compelling. Sno is expressed as several isoforms (106, 107, 109) and appears required for transcriptional repression by Mad, Rb, and nuclear hormone receptors (131). The creation of Sno−/− mice revealed that Sno is required for proper formation of the blastocyst and is thus lethal at E3.5 (131). However, Sno+/− mice develop spontaneous B- and T-cell lymphoma at a low frequency, and this is dramatically augmented when they are challenged with tumor promoters. Furthermore, the tumors that do arise in these animals display loss of the wild-type Sno allele, proving that Sno behaves as a tumor suppressor (131). However, although the biological effects of Ski and Sno loss are compelling, an important consideration is whether their effects are truly selective for Mad, particularly since these factors appear to be components of a wide array of transcriptional repressor complexes.
THE MAX MATRIX
Myc's regulation of various cellular processes appears to be a function of its interactions with a wide array of regulatory and effector molecules (123). Several Myc-interacting proteins bind to Myc through HLHZip interactions that are also required for binding to Max (123). Thus, it seems unlikely that all of the Myc functions can be attributed to its dimerization with Max. The precise contributions of other Myc partners await genetic studies with mice and elsewhere, where they can be put to the test. Indeed, Max makes the grade in this scenario, as its deletion, like that of Myc, compromises cell growth. This is not so for the other factors that have been proposed to regulate Myc activity by dancing with Max. In particular, the deletion of three of the Mad family members has essentially no impact on development; cell growth, survival, and differentiation; or tumor susceptibility. Thus, while one can still invoke redundancy, it remains to be determined whether Mad, Mnt, Mga, and possibly other yet unidentified factors play an important physiological role in regulating Myc activity.
At this juncture, it is difficult to reconcile all of the available data into a linear Myc-Max-Mad network. Rather, the constellation of data fits better with the concept of a matrix, in which several parallel yet sometimes intersecting pathways are involved in decisions that control cell fate. Max certainly plays a central role in this matrix by targeting multiple transcription factors, including those of the Brachyury family. Moreover, Max does not function in this matrix alone, as Mlx also appears to contribute to Mad and Mnt protein functions. Furthermore, it remains possible that Myc functions as an effective antagonist of this collective group of transcriptional repressors by effectively sequestering Max or other factors from their binding partners. The bottom line is that many more genetic studies need to be performed before we can hope to understand the multiplicity of interactions, and the generation of double or perhaps triple knockouts will hopefully address the relevance, if any, of the Mad family of transcription factors to regulation of Myc activity.
ACKNOWLEDGMENTS
We thank Gerard Zambetti for critical review.
In part, this review is supported by NIH grants DK44158 and CA76379 (J.L.C.), an NIH postdoctoral training grant (T.A.B.), and Cancer Center core grant CA21765 and by the American Lebanese Syrian Associated Charities (ALSAC) of St. Jude Children's Research Hospital.
REFERENCES
- 1.Adams J M, Harris A W, Pinkert C A, Corcoran L M, Alexander W S, Palmiter R D, Brinster R L. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 2.Alitalo K, Koskinen P, Makela T P, Saksela K, Sistonen L, Winqvist R. myc oncogenes: activation and amplification. Biochim Biophys Acta. 1987;907:1–32. doi: 10.1016/0304-419x(87)90016-3. [DOI] [PubMed] [Google Scholar]
- 3.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 4.Amati B, Brooks M W, Levy N, Littlewood T D, Evan G I, Land H. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell. 1993;72:233–245. doi: 10.1016/0092-8674(93)90663-b. [DOI] [PubMed] [Google Scholar]
- 5.Amati B, Dalton S, Brooks M W, Littlewood T D, Evan G I, Land H. Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature. 1992;359:423–426. doi: 10.1038/359423a0. [DOI] [PubMed] [Google Scholar]
- 6.Amati B, Land H. Myc-Max-Mad: a transcription factor network controlling cell cycle progression, differentiation, and death. Curr Opin Genet Dev. 1994;4:102–108. doi: 10.1016/0959-437x(94)90098-1. [DOI] [PubMed] [Google Scholar]
- 7.Amati B, Littlewood T D, Evan G I, Land H. The c-Myc protein induces cell cycle progression and apoptosis through dimerization with Max. EMBO J. 1993;12:5083–5087. doi: 10.1002/j.1460-2075.1993.tb06202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armelin H A, Armelin M C S, Kelly K, Stewart T, Leder P, Cochran B H, Stiles C D. Functional role of c-myc in mitogenic response to platelet-derived growth factor. Nature. 1984;310:655–660. doi: 10.1038/310655a0. [DOI] [PubMed] [Google Scholar]
- 9.Askew D S, Ashmun R A, Simmons B C, Cleveland J L. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene. 1991;6:1915–1922. [PubMed] [Google Scholar]
- 10.Ayer D E, Eisenman R N. A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation. Genes Dev. 1993;7:2110–2119. doi: 10.1101/gad.7.11.2110. [DOI] [PubMed] [Google Scholar]
- 11.Ayer D E, Kretzner L, Eisenman R N. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- 12.Ayer D E, Lawrence Q A, Eisenman R N. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 13.Bartsch D, Peiffer S L, Kaleem Z, Wells S A, Jr, Goodfellow P J. Mxi1 tumor suppressor gene is not mutated in primary pancreatic adenocarcinoma. Cancer Lett. 1996;102:73–76. doi: 10.1016/0304-3835(96)04167-5. [DOI] [PubMed] [Google Scholar]
- 14.Bello-Fernandez C, Packham G, Cleveland J L. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci USA. 1993;90:7804–7808. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berberich S, Cole M D. Casein kinase II inhibits the DNA-binding activity of Max homodimers but not Myc/Max heterodimers. Genes Dev. 1992;6:166–176. doi: 10.1101/gad.6.2.166. [DOI] [PubMed] [Google Scholar]
- 16.Berberich S, Hyde-DeRuyscher N, Espenshade P, Cole M. max encodes a sequence-specific DNA-binding protein and is not regulated by serum growth factors. Oncogene. 1992;7:775–779. [PubMed] [Google Scholar]
- 17.Berk M, Desai S Y, Heyman H C, Colmenares C. Mice lacking the ski proto-oncogene have defects in neurulation, craniofacial patterning, and skeletal muscle development. Genes Dev. 1997;11:2029–2039. doi: 10.1101/gad.11.16.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Billin A N, Eilers A L, Queva C, Ayer D E. Mlx, a novel Max-like BHLHZip protein that interacts with the Max network of transcription factors. J Biol Chem. 1999;274:36344–36350. doi: 10.1074/jbc.274.51.36344. [DOI] [PubMed] [Google Scholar]
- 19.Bissonnette R P, Echeverri F, Mahboubi A, Green D R. Apoptotic cell death induced by c-myc is inhibited by bcl-2. Nature. 1992;359:552–556. doi: 10.1038/359552a0. [DOI] [PubMed] [Google Scholar]
- 20.Blackwell T K, Huang J, Ma A, Kretzner L, Alt F W, Eisenman R N, Weintraub H. Binding of myc proteins to canonical and noncanonical DNA sequences. Mol Cell Biol. 1993;13:5216–5224. doi: 10.1128/mcb.13.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blackwood E M, Eisenman R N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 22.Blackwood E M, Luscher B, Eisenman R N. Myc and Max associate in vivo. Genes Dev. 1992;6:71–80. doi: 10.1101/gad.6.1.71. [DOI] [PubMed] [Google Scholar]
- 23.Bouchard C, Staller P, Eilers M. Control of cell proliferation by Myc. Trends Cell Biol. 1998;8:202–206. doi: 10.1016/s0962-8924(98)01251-3. [DOI] [PubMed] [Google Scholar]
- 24.Boyd K E, Farnham P J. Coexamination of site-specific transcription factor binding and promoter activity in living cells. Mol Cell Biol. 1999;19:8393–8399. doi: 10.1128/mcb.19.12.8393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brandvold K A, Neiman P, Ruddell A. Angiogenesis is an early event in the generation of myc-induced lymphomas. Oncogene. 2000;19:2780–2785. doi: 10.1038/sj.onc.1203589. [DOI] [PubMed] [Google Scholar]
- 26.Charron J, Malynn B A, Fisher P, Stewart V, Jeannotte L, Goff S P, Robertson E J, Alt F W. Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Genes Dev. 1992;6:2248–2257. doi: 10.1101/gad.6.12a.2248. [DOI] [PubMed] [Google Scholar]
- 27.Cheng S W, Davies K P, Yung E, Beltran R J, Yu J, Kalpana G V. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat Genet. 1999;22:102–105. doi: 10.1038/8811. [DOI] [PubMed] [Google Scholar]
- 28.Chin L, Pomerantz J, DePinho R A. The INK4a/ARF tumor suppressor: one gene—two products—two pathways. Trends Biochem Sci. 1998;23:291–296. doi: 10.1016/s0968-0004(98)01236-5. [DOI] [PubMed] [Google Scholar]
- 29.Cohen S B, Zheng G, Heyman H C, Stavnezer E. Heterodimers of the SnoN and Ski oncoproteins form preferentially over homodimers and are more potent transforming agents. Nucleic Acids Res. 1999;27:1006–1014. doi: 10.1093/nar/27.4.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colmenares C, Stavnezer E. The ski oncogene induces muscle differentiation in quail embryo cells. Cell. 1989;59:293–303. doi: 10.1016/0092-8674(89)90291-2. [DOI] [PubMed] [Google Scholar]
- 31.Coppola J A, Cole M D. Constitutive c-myc oncogene expression blocks mouse erythroleukaemia cell differentiation but not commitment. Nature. 1986;320:760–763. doi: 10.1038/320760a0. [DOI] [PubMed] [Google Scholar]
- 32.Daksis J I, Lu R Y, Facchini L M, Marhin W W, Penn L J Z. Myc induces cyclin D1 expression in the absence of de novo protein synthesis and links mitogen-stimulated signal transduction to the cell cycle. Oncogene. 1994;9:3635–3645. [PubMed] [Google Scholar]
- 33.Dang C V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis A C, Wims M, Spotts G D, Hann S R, Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 1993;7:671–682. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- 35.Downs K M, Martin G R, Bishop J M. Contrasting patterns of myc and N-myc expression during gastrulation of the mouse embryo. Genes Dev. 1989;3:860–869. doi: 10.1101/gad.3.6.860. [DOI] [PubMed] [Google Scholar]
- 36.Eagle L R, Yin X, Brothman A R, Williams B J, Atkin N B, Prochownik E V. Mutation of the MXI1 gene in prostate cancer. Nat Genet. 1995;9:249–255. doi: 10.1038/ng0395-249. [DOI] [PubMed] [Google Scholar]
- 37.Eberhardy S R, D'Cunha C A, Farnham P J. Direct examination of histone acetylation on Myc target genes using chromatin immunoprecipitation. J Biol Chem. 2000;275:33798–33805. doi: 10.1074/jbc.M005154200. [DOI] [PubMed] [Google Scholar]
- 38.Edwards S M, Dearnaley D P, Ardern-Jones A, Hamoudi R A, Easton D F, Ford D, Shearer R, Dowe A, Eeles R A. No germline mutations in the dimerization domain of MXI1 in prostate cancer clusters. Br J Cancer. 1997;76:992–1000. doi: 10.1038/bjc.1997.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eilers A L, Billin A N, Liu J, Ayer D E. A 13-amino acid amphipathic alpha-helix is required for the functional interaction between the transcriptional repressor Mad1 and mSin3A. J Biol Chem. 1999;274:32750–32756. doi: 10.1074/jbc.274.46.32750. [DOI] [PubMed] [Google Scholar]
- 40.Eilers M, Schirm S, Bishop J M. The MYC protein activates transcription of the a-prothymosin gene. EMBO J. 1991;10:133–141. doi: 10.1002/j.1460-2075.1991.tb07929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eischen C M, Weber J D, Roussel M F, Sherr C J, Cleveland J L. Disruption of the ARF-Mdm2–p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Evan G I, Littlewood T D. The role of c-myc in cell growth. Curr Opin Genet Dev. 1993;3:44–49. doi: 10.1016/s0959-437x(05)80339-9. [DOI] [PubMed] [Google Scholar]
- 43.Evan G I, Wyllie A H, Gilbert C S, Littlewood T D, Land H, Brooks M, Waters C M, Penn L Z, Hancock D C. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 44.Facchini L M, Penn L Z. The molecular role of Myc in growth and transformation: recent discoveries lead to new insights. FASEB J. 1998;12:633–651. [PubMed] [Google Scholar]
- 45.Flinn E M, Busch C M C, Wright A P H. Myc boxes, which are conserved in Myc family proteins, are signals for protein degradation via the proteasome. Mol Cell Biol. 1998;18:5961–5969. doi: 10.1128/mcb.18.10.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gesk S, Siebert R, Wacker H H, Nurnberg N, Harder L, Lehman J, Kloppel G, Grote W, Stockle M, Schlegelberger B. Lack of deletions of PTEN/MMAC1 and MXI1 loci in renal cell carcinoma by interphase cytogenetics. Cancer Genet Cytogenet. 2000;118:87–88. doi: 10.1016/s0165-4608(99)00190-9. [DOI] [PubMed] [Google Scholar]
- 47.Grandori C, Cowley S M, James L P, Eisenman R N. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 48.Gregory M A, Hann S R. c-Myc proteolysis by the ubiquitin-proteosome pathway: stabilization of c-Myc in Burkitt's lymphoma cells. Mol Cell Biol. 2000;20:2423–2435. doi: 10.1128/mcb.20.7.2423-2435.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gupta K, Anand G, Yin X, Grove L, Prochownik E V. Mmip1: a novel leucine zipper protein that reverses the suppressive effects of Mad family members on c-myc. Oncogene. 1998;16:1149–1159. doi: 10.1038/sj.onc.1201634. [DOI] [PubMed] [Google Scholar]
- 50.Hanson K D, Shichiri M, Follansbee M R, Sedivy J M. Effects of c-myc expression on cell cycle progression. Mol Cell Biol. 1994;14:5748–5755. doi: 10.1128/mcb.14.9.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harper S E, Qiu Y, Sharp P A. Sin3 corepressor function in Myc-induced transcription and transformation. Proc Natl Acad Sci USA. 1996;93:8536–8540. doi: 10.1073/pnas.93.16.8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 53.Hatton K S, Mahon K, Chin L, Chiu F C, Lee H W, Peng D, Morgenbesser S D, Horner J, DePinho R A. Expression and activity of L-Myc in normal mouse development. Mol Cell Biol. 1996;16:1794–1804. doi: 10.1128/mcb.16.4.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heikkila R, Schwab G, Wickstrom E, Loke S L, Pluznik D H, Watt R, Neckers L M. A c-myc antisense oligodeoxynucleotide inhibits entry into S phase but not progress from G0 to G1. Nature. 1987;328:445–449. doi: 10.1038/328445a0. [DOI] [PubMed] [Google Scholar]
- 55.Heinzel T, Lavinsky R M, Mullen T-M, Soderstrom M, Laherty C D, Torchia J, Yang W-M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3, and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–47. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 56.Henriksson M, Barkardjiev A, Klein G, Luscher B. Phosphorylation sites mapping in the N-terminal domain of c-myc modulate its transforming potential. Oncogene. 1993;8:3199–3209. [PubMed] [Google Scholar]
- 57.Henriksson M, Luscher B. Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv Cancer Res. 1996;68:109–182. doi: 10.1016/s0065-230x(08)60353-x. [DOI] [PubMed] [Google Scholar]
- 58.Herbst R A, Podewski E K, Mommert S, Kapp A, Weiss J. PTEN and MXI1 allelic loss on chromosome 10q is rare in melanoma in vivo. Arch Dermatol Res. 1999;291:567–569. doi: 10.1007/s004030050456. [DOI] [PubMed] [Google Scholar]
- 59.Herrmann B G, Kispert A. The T genes in embryogenesis. Trends Genet. 1994;10:280–286. doi: 10.1016/0168-9525(90)90011-t. [DOI] [PubMed] [Google Scholar]
- 60.Hirning U, Schmid P, Schulz W A, Retternberger G, Hameister H. A comparative analysis of N-myc and c-myc expression and cellular proliferation in mouse organogenesis. Mech Dev. 1991;33:119–126. doi: 10.1016/0925-4773(91)90078-k. [DOI] [PubMed] [Google Scholar]
- 61.Hurlin P J, Foley K P, Ayer D E, Eisenman R N, Hanahan D, Arbeit J M. Regulation of Myc and Mad during epidermal differentiation and HPV-associated tumorigenesis. Oncogene. 1995;11:2487–2501. [PubMed] [Google Scholar]
- 62.Hurlin P J, Queva C, Eisenman R N. Mnt, a novel Max-interacting protein, is coexpressed with Myc in proliferating cells and mediates repression at Myc binding sites. Genes Dev. 1997;11:44–58. doi: 10.1101/gad.11.1.44. [DOI] [PubMed] [Google Scholar]
- 63.Hurlin P J, Queva C, Koskinen P J, Steingrimsson E, Ayer D E, Copeland N G, Jenkins N A, Eisenman R N. Mad3 and Mad4: novel Max-interacting transcriptional repressors that suppress c-myc dependent transformation and are expressed during neural and epidermal differentiation. EMBO J. 1995;14:5646–5659. doi: 10.1002/j.1460-2075.1995.tb00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hurlin P J, Steingrimsson E, Copeland N G, Jenkins N A, Eisenman R N. Mga, a dual specificity transcription factor that interacts with Max and contains a T-domain DNA-binding motif. EMBO J. 1999;18:7019–7028. doi: 10.1093/emboj/18.24.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jansen-Durr P, Meichle A, Steiner P, Pagano M, Finke K, Botz J, Wessbecher J, Draetta G, Eilers M. Differential modulation of cyclin gene expression by MYC. Proc Natl Acad Sci USA. 1993;90:3685–3689. doi: 10.1073/pnas.90.8.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jepsen K, Hermanson O, Onami T M, Gleiberman A S, Lunyak V, McEvilly R J, Kurokawa R, Kumar V, Liu F, Seto E, Hedrick S M, Mandel G, Glass C K, Rose D W, Rosenfeld M G. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102:753–763. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- 67.Johnston L A, Prober D A, Edgar B A, Eisenman R N, Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98:779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kamijo T, Zindy F, Roussel M F, Quelle D E, Downing J R, Ashmun R A, Grosveld G, Sherr C J. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 69.Kato G J, Lee W M F, Chen L, Dang C V. Max: functional domains and interaction with c-Myc. Genes Dev. 1992;6:81–92. doi: 10.1101/gad.6.1.81. [DOI] [PubMed] [Google Scholar]
- 70.Kawamata N, Park D, Wilczynski S, Yokota J, Koeffler H P. Point mutations of the Mxi1 gene are rare in prostate cancers. Prostate. 1996;29:191–193. doi: 10.1002/1097-0045(199609)29:3<191::aid-pros2990290305>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 71.Kelly K, Cochran B H, Stiles C D, Leder P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platlet-derived growth factor. Cell. 1982;35:603–610. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- 72.Kerkhoff E, Bister K, Klempnauer K H. Sequence-specific DNA binding by Myc proteins. Proc Natl Acad Sci USA. 1991;88:4323–4327. doi: 10.1073/pnas.88.10.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim S K, Ro J Y, Kemp B L, Lee J S, Kwon T J, Hong W K, Mao L. Identifications of two distinct tumor-suppressor loci on the long arm of chromosome 10 in small cell lung cancer. Oncogene. 1998;17:1749–1753. doi: 10.1038/sj.onc.1202073. [DOI] [PubMed] [Google Scholar]
- 74.Koskinen P J, Ayer D E, Eisenman R N. Repression of Myc-Ras cotransformation by Mad is mediated by multiple protein-protein interactions. Cell Growth Differ. 1995;6:623–629. [PubMed] [Google Scholar]
- 75.Kretzner L, Blackwood E M, Eisenman R N. Myc and Max proteins possess distinct transcriptional activities. Nature. 1992;359:426–429. doi: 10.1038/359426a0. [DOI] [PubMed] [Google Scholar]
- 76.Kuczyk M A, Serth J, Bokemeyer C, Schwede J, Herrmann R, Machtens S, Gruneweld V, Hofner K, Jonas U. The Mxi1 tumor supressor gene is not mutated in primary prostate cancer. Oncol Rep. 1998;5:213–216. [PubMed] [Google Scholar]
- 77.Laherty C D, Billin A N, Lavinsky R M, Yochum G S, Bush A C, Sun J-M, Mullen T-M, Davie J R, Rose D W, Glass C K, Rosenfeld M G, Ayer D E, Eisenman R N. SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol Cell. 1998;2:33–42. doi: 10.1016/s1097-2765(00)80111-2. [DOI] [PubMed] [Google Scholar]
- 78.Laherty C D, Yang W-M, Sun J-M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 79.Langdon W Y, Harris A W, Cory S, Adams J M. The c-myc oncogene perturbs B lymphocyte development in E m-myc transgenic mice. Cell. 1986;47:11–18. doi: 10.1016/0092-8674(86)90361-2. [DOI] [PubMed] [Google Scholar]
- 80.Leder A, Pattengale P K, Kuo A, Stewart T A, Leder P. Consequences of widespread deregulation of the c-myc gene in transgenic mice: multiple neoplasms and normal development. Cell. 1986;45:485–495. doi: 10.1016/0092-8674(86)90280-1. [DOI] [PubMed] [Google Scholar]
- 81.Li X J, Wang D Y, Zhu Y, Guo R J, Wang X D, Lubomir K, Mukai K, Sasaki H, Yoshida H, Oka T, Machinami R, Shinmura K, Tanaka M, Sugimara H. Mxi1 mutations in human neurofibrosarcomas. Jpn J Cancer Res. 1999;90:740–746. doi: 10.1111/j.1349-7006.1999.tb00809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li Y, Turck C M, Teumer J K, Stavnezer E. Unique sequence, ski, in Sloan-Kettering avian retroviruses with properties of a new cell-derived oncogene. J Virol. 1986;57:1065–1072. doi: 10.1128/jvi.57.3.1065-1072.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lindeman G J, Harris A W, Bath M L, Eisenman R N, Adams J M. Overexpressed max is not oncogenic and attenuates myc-induced lymphoproliferation and lymphomagenesis in transgenic mice. Oncogene. 1995;10:1013–1017. [PubMed] [Google Scholar]
- 84.Littlewood T D, Amati B, Land H, Evan G I. Max and c-Myc/Max DNA-binding activities in cell extracts. Oncogene. 1992;7:1783–1792. [PubMed] [Google Scholar]
- 85.Makela T P, Koskinen P J, Vastrik I, Aitalo K. Alternative forms of Max as enhancers or suppressors of Myc-Ras cotransformation. Science. 1992;256:373–377. doi: 10.1126/science.256.5055.373. [DOI] [PubMed] [Google Scholar]
- 86.Malynn B A, Moreno de Alboran I, O'Hagan R C, Bronson R, Davidson L, DePinho R A, Alt F W. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev. 2000;14:1390–1399. [PMC free article] [PubMed] [Google Scholar]
- 87.Mangelsdorf D J, Evans R M. The RXR heterodimers and orphan receptors. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 88.Marcu K B, Bossone S A, Patel A J. Myc function and regulation. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- 89.McMahon S B, Van Buskirk H A, Dugan K A, Copeland T D, Cole M D. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 90.McMahon S B, Wood M A, Cole M D. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Meroni G, Cairo S, Merla G, Messali S, Brent R, Ballabio A, Reymond A. Mlx, a new Max-like bHLHZip family member: the center stage of a novel transcription factor regulatory pathway? Oncogene. 2000;19:3266–3277. doi: 10.1038/sj.onc.1203634. [DOI] [PubMed] [Google Scholar]
- 92.Meroni G, Reymond A, Alcalay M, Borsani G, Tanigami A, Tonlorenzi R, Lo Nigro C, Messali S, Zollo M, Ledbetter D H, Brent R, Ballabio A, Carrozzo R. Rox, a novel bHLHZip protein expressed in quiescent cells that heterodimerizes with Max, binds a non-canonical E box, and acts as a transcriptional repressor. EMBO J. 1997;16:2892–2906. doi: 10.1093/emboj/16.10.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Metzstein M M, Stanfield G M, Horvitz H R. Genetics of programmed cell death in C. elegans: past, present, and future. Trends Genet. 1998;14:410–416. doi: 10.1016/s0168-9525(98)01573-x. [DOI] [PubMed] [Google Scholar]
- 94.Miltenberger R J, Sukow K A, Farnham P J. An E-box-mediated increase in cad transcription at the G1/S-phase boundary is suppressed by inhibitory c-Myc mutants. Mol Cell Biol. 1995;15:2527–2535. doi: 10.1128/mcb.15.5.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Moens C B, Stanton B R, Parada L F, Rossant J. Defects in heart and lung development in compound heterozygotes for two different targeted mutations at the N-myc locus. Development. 1993;119:485–499. doi: 10.1242/dev.119.2.485. [DOI] [PubMed] [Google Scholar]
- 96.Muller D, Bouchard C, Rudolph B, Steiner P, Stuckmann I, Saffrich R, Ansorge W, Huttner W, Eilers M. Cdk2-dependent phosphorylation of p27 facilitates its Myc-induced release from cyclin E/cdk2 complexes. Oncogene. 1997;15:2561–2576. doi: 10.1038/sj.onc.1201440. [DOI] [PubMed] [Google Scholar]
- 97.Nicol R, Zheng G, Sutrave P, Foster D N, Stavnezer E. Association of specific DNA binding and transcriptional repression with the transforming and myogenic activities of c-Ski. Cell Death Differ. 1999;10:243–254. [PubMed] [Google Scholar]
- 98.Niklinski J, Claassen G, Meyers C, Gregory M A, Allegra C J, Kaye F J, Hann S R, Zajac-Kaye M. Disruption of Myc-tubulin interaction by hyperphosphorylation of c-Myc during mitosis or by constitutive hyperphosphorylation of mutant c-Myc in Burkitt's lymphoma. Mol Cell Biol. 2000;20:5276–5284. doi: 10.1128/mcb.20.14.5276-5284.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nomura T, Khan M M, Kaul S C, Dong H-D, Wadhwa R, Colmenares C, Kohno I, Ishii S. Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes Dev. 1999;13:412–423. doi: 10.1101/gad.13.4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.O'Hagen R C, Ohh M, David G, de Alboran I M, Alt F W, Kaelin W G, Jr, DePinho R A. Myc-enhanced expression of Cul1 promotes ubiquitin-dependent proteolysis and cell cycle progression. Genes Dev. 2000;14:2185–2191. doi: 10.1101/gad.827200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.O'Hagen R C, Schreiber-Agus N, Chen K, David G, Engelman J A, Schwab R, Alland L, Thomson C, Ronning D R, Sacchettini J C, Meltzer P, DePinho R A. Gene-target recognition among members of the Myc superfamily and implications for oncogenesis. Nat Genet. 2000;24:113–119. doi: 10.1038/72761. [DOI] [PubMed] [Google Scholar]
- 102.Packham G, Cleveland J L. c-Myc and apoptosis. Biochim Biophys Acta. 1995;1242:11–28. doi: 10.1016/0304-419x(94)00015-t. [DOI] [PubMed] [Google Scholar]
- 103.Packham G, Cleveland J L. Induction of ornithine decarboxylase by IL-3 is mediated by sequential c-Myc-independent and c-Myc-dependent pathways. Oncogene. 1997;15:1219–1232. doi: 10.1038/sj.onc.1201273. [DOI] [PubMed] [Google Scholar]
- 104.Packham G, Porter C W, Cleveland J L. c-Myc induces apoptosis and cell cycle progression by seperable, yet overlapping, pathways. Oncogene. 1996;13:461–469. [PubMed] [Google Scholar]
- 105.Papaioannou V E, Silver L M. The T-box gene family. Bioessays. 1998;20:9–19. doi: 10.1002/(SICI)1521-1878(199801)20:1<9::AID-BIES4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 106.Pearson-White S. SnoI, a novel alternatively spliced isoform of the ski protooncogene homolog, sno. Nucleic Acids Res. 1993;21:4632–4638. doi: 10.1093/nar/21.19.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pearson-White S, Crittenden R. Proto-oncogene Sno expression, alternative isoforms and immediate early serum response. Nucleic Acids Res. 1997;25:2930–2937. doi: 10.1093/nar/25.14.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pelengaris S, Littlewood T, Khan M, Elia G, Evan G. Reversible activation of c-Myc in skin: induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol Cell. 1999;3:565–577. doi: 10.1016/s1097-2765(00)80350-0. [DOI] [PubMed] [Google Scholar]
- 109.Pelzer T, Lyons G E, Kim S, Moreadith R W. Cloning and characterization of the murine homolog of the sno proto-oncogene reveals a novel splice variant. Dev Dyn. 1996;205:114–125. doi: 10.1002/(SICI)1097-0177(199602)205:2<114::AID-AJA3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 110.Pennetta G, Pauli D. The Drosophila Sin3 gene encodes a widely distributed transcription factor essential for embryonic viability. Dev Genes Evol. 1998;208:531–536. doi: 10.1007/s004270050212. [DOI] [PubMed] [Google Scholar]
- 111.Peukert K, Staller P, Schneider A, Carmichael G, Hanel F, Eilers M. An alternative pathway for gene regulation by Myc. EMBO J. 1997;16:5672–5682. doi: 10.1093/emboj/16.18.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pomerantz J, Schreiber-Agus N, Liegeois N J, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H-W, Cordon-Cardo C, DePinho R A. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 113.Prendergast G C. Myc structure and function. In: Yaniv M, Ghysdael J, editors. Oncogenes as transcriptional regulators. 1. Retroviral oncogenes. Basel, Switzerland: Birkhauser Verlag; 1997. pp. 1–28. [Google Scholar]
- 114.Prendergast G C, Lawe D, Ziff E B. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991;65:395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- 115.Prochownik E V, Eagle G L, Deubler D, Zhu X L, Stephenson R A, Rohr L R, Yin X, Brothman A R. Commonly occurring loss and mutation of the MXI1 gene in prostate cancer. Genes Chromosomes Cancer. 1998;22:295–304. [PubMed] [Google Scholar]
- 116.Prochownik E V, Kukowska J, Rodgers C. c-myc antisense transcripts accelerate differentiation and inhibit G1 progression in murine erythroleukemia cells. Mol Cell Biol. 1988;8:3683–3695. doi: 10.1128/mcb.8.9.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pulverer B J, Fisher C, Vousden K, Littlewood T, Evan G, Woodjett J R. Site-specific modulation of c-Myc cotransformation by residues phosphorylated in vivo. Oncogene. 1994;9:59–70. [PubMed] [Google Scholar]
- 118.Queva C, Hurlin P J, Foley K P, Eisenman R N. Sequential expression of the MAD family of transcriptional repressors during differentiation and development. Oncogene. 1998;16:967–977. doi: 10.1038/sj.onc.1201611. [DOI] [PubMed] [Google Scholar]
- 119.Queva C, McArthur G A, Iritani B M, Eisenman R N. Targeted deletion of the S-phase specific Myc antagonist Mad3 sensitizes neuronal and lymphoid cells to radiation-induced apoptosis. Mol Cell Biol. 2001;21:703–712. doi: 10.1128/MCB.21.3.703-712.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Queva C, McArthur G A, Ramos L S, Eisenman R N. Dwarfism and dysregulated proliferation in mice overexpressing the MYC antagonist MAD1. Cell Growth Differ. 1999;10:785–796. [PubMed] [Google Scholar]
- 121.Rao U N, Bakker A, Swalsky P A, Finkelstein S D. Max interacting protein 1: loss of heterozygosity is frequent in desmoplastic melanoma. Mod Pathol. 1999;12:344–350. [PubMed] [Google Scholar]
- 122.Rosenbaum H, Webb E, Adams J M, Cory S, Harris A W. N-myc transgene promotes B lymphoid proliferation, elicts lymphomas and reveals cross-regulation with c-myc. EMBO J. 1989;8:749–755. doi: 10.1002/j.1460-2075.1989.tb03435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sakamuro D, Prendergast G C. New Myc-interacting proteins: a second Myc network emerges. Oncogene. 1999;18:2942–2954. doi: 10.1038/sj.onc.1202725. [DOI] [PubMed] [Google Scholar]
- 124.Sawai A, Shimono A, Wakamatsu Y, Palmes C, Hanaoka K, Kondoh H. Defects of embryonic organogenesis resulting from targeted disruption of the N-myc gene in the mouse. Development. 1993;117:1445–1455. doi: 10.1242/dev.117.4.1445. [DOI] [PubMed] [Google Scholar]
- 125.Schreiber-Agus N, Chin L, Chen K, Torres R, Rao G, Guida P, Skoultchi A I, DePinho R A. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- 126.Schreiber-Agus N, Meng Y, Hoang T, Hou H, Jr, Chen K, Greenberg R, Cordon-Cardo C, Lee H-W, DePinho R A. Role of Mxi1 in ageing organ systems and the regulation of normal and neoplastic growth. Nature. 1998;393:483–487. doi: 10.1038/31008. [DOI] [PubMed] [Google Scholar]
- 127.Schreiber-Agus N, Stein D, Chen K, Goltz J S, Stevens L, DePinho R A. Drosophila Myc is oncogenic in mammalian cells and plays a role in the diminutive phenotype. Proc Natl Acad Sci USA. 1997;94:1235–1240. doi: 10.1073/pnas.94.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Serrano M. The tumor suppressor protein p16INK4a. Exp Cell Res. 1997;237:7–13. doi: 10.1006/excr.1997.3824. [DOI] [PubMed] [Google Scholar]
- 129.Shen-Li H, O'Hagan R C, Hou H, Horner II J W, Lee H-W, DePinho R A. Essential role for Max in early embryonic growth and development. Genes Dev. 2000;14:17–22. [PMC free article] [PubMed] [Google Scholar]
- 130.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 131.Shinagawa T, Dong H-D, Xu M, Maekawa T, Ishii S. The sno gene, which encodes a component of the histone deacetylase complex, acts as a tumor suppressor in mice. EMBO J. 2000;19:2280–2291. doi: 10.1093/emboj/19.10.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Siebert R, Gesk S, Harder S, Plotz S, Matthiesen P, Grote W, Schlegelberger B, Jandrig B, Grasmo-Wendler U H, Scherneck S, Rosenwald A, Ott G. Deletions in the long arm of chromosome 10 in lymphomas with t(14;18): a pathogenetic role of the tumor suppressor genes PTEN/MMAC and MXI1. Blood. 1998;92:4487–4489. [PubMed] [Google Scholar]
- 133.Smith J. T-box genes: what they do and how they do it. Trends Genet. 1999;15:154–158. doi: 10.1016/s0168-9525(99)01693-5. [DOI] [PubMed] [Google Scholar]
- 134.Solomon D L C, Amati B, Land H. Distinct DNA binding preferences for the c-Myc/Max and Max/Max dimers. Nucleic Acids Res. 1993;21:5372–5376. doi: 10.1093/nar/21.23.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sommer A, Bousset K, Kremmer E, Austen M, Luscher B. Identification and characterization of specific DNA-binding complexes containing members of the Myc/Max/Mad network of transcriptional regulators. J Biol Chem. 1998;273:6632–6642. doi: 10.1074/jbc.273.12.6632. [DOI] [PubMed] [Google Scholar]
- 136.Sommer A, Waha A, Tonn J, Sorensen N, Hurlin P J, Eisenman R N, Luscher B, Pietsch T. Analysis of the Max-binding protein MNT in human medulloblastomas. Int J Cancer. 1999;82:810–816. doi: 10.1002/(sici)1097-0215(19990909)82:6<810::aid-ijc7>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 137.Stanton B R, Perkins A S, Tessarollo L, Sassoon D A, Parada L F. Loss of N-myc function results in embryonic lethality and failure of the epithelial component of the embryo to develop. Genes Dev. 1992;6:2235–2247. doi: 10.1101/gad.6.12a.2235. [DOI] [PubMed] [Google Scholar]
- 138.Vlach J, Hennecke S, Alevizopoulos K, Conti D, Amati B. Growth arrest by the cyclin-dependent kinase inhibitor p27Kip is abrogated by c-Myc. EMBO J. 1996;15:6595–6604. [PMC free article] [PubMed] [Google Scholar]
- 139.Weiss W A, Aldape K, Mohapatra G, Feuerstein B G, Bishop J M. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 1997;16:2985–2995. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Winston F, Carlson M. Yeast SNFISWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 141.Yin X, Grove L, Prochownik E V. Lack of transcriptional repression by Max homodimers. Oncogene. 1998;16:2629–2637. doi: 10.1038/sj.onc.1201777. [DOI] [PubMed] [Google Scholar]
- 142.Yin X-Y, Gupta K, Han W P, Levitan E S, Prochownik E V. Mmip-2, a novel RING finger protein that interacts with Mad members of the Myc oncoprotein network. Oncogene. 1999;18:6621–6634. doi: 10.1038/sj.onc.1203097. [DOI] [PubMed] [Google Scholar]
- 143.Zervos A S, Gyruis J, Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
- 144.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Cell. 1997;89:357–364. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]
- 145.Zimmerman K A, Yancopoulos G D, Collum R G, Smith R K, Kohl N E, Denis K A, Nau M M, Witte O N, Toran-Allerand D, Gee C E. Differential expression of myc family genes durning murine development. Nature. 1986;319:780–783. doi: 10.1038/319780a0. [DOI] [PubMed] [Google Scholar]