Abstract
Cancer will directly affect the lives of over one-third of the population. The DNA Damage Response (DDR) is an intricate system involving damage recognition, cell cycle regulation, DNA repair, and ultimately cell fate determination, playing a central role in cancer etiology and therapy. Two primary therapeutic approaches involving DDR targeting include: combinatorial treatments employing anticancer genotoxic agents; and synthetic lethality, exploiting a sporadic DDR defect as a mechanism for cancer-specific therapy. Whereas, many DDR proteins have proven “undruggable”, Fragment- and Structure-Based Drug Discovery (FBDD, SBDD) have advanced therapeutic agent identification and development. FBDD has led to 4 (with ~50 more drugs under preclinical and clinical development), while SBDD is estimated to have contributed to the development of >200, FDA-approved medicines. Protein X-ray crystallography-based fragment library screening, especially for elusive or “undruggable” targets, allows for simultaneous generation of hits plus details of protein-ligand interactions and binding sites (orthosteric or allosteric) that inform chemical tractability, downstream biology, and intellectual property. Using a novel high-throughput crystallography-based fragment library screening platform, we screened five diverse proteins, yielding hit rates of ~2–8% and crystal structures from ~1.8 to 3.2 Å. We consider current FBDD/SBDD methods and some exemplary results of efforts to design inhibitors against the DDR nucleases meiotic recombination 11 (MRE11, a.k.a., MRE11A), apurinic/apyrimidinic endonuclease 1 (APE1, a.k.a., APEX1), and flap endonuclease 1 (FEN1).
Keywords: Fragment-based drug discovery, Structure-based drug discovery, X-ray crystallography, Cancer therapeutics, DNA damage Response, DNA repair, MRE11, APE1, FEN1
1. Introduction
1.1. History of and milestone advances in fragment-based screening technologies
Fragment-Based Drug Discovery (FBDD), as it is commonly known now, started about 25 years ago. In its earliest years, it was often referred to as “Fragment-Based Lead Design” and later on (and even now sometimes) as “Fragment-Based Drug Design”. The FBDD approach relies on hit generation by starting with stable molecular fragments that share common features: typically, a molecular weight of <300 Da, cLog P ≤ 3 (a measure of hydrophilicity, with low values enhancing absorption), and number of hydrogen bond donors, or acceptors, of 3 or less. The number of rotatable bonds of ≤3 and polar surface area ≤60 Å2 may also be useful criteria for FBDD. Such fragments are smaller than regular lead-like molecules in drug discovery and are said to be Rule of Three (Ro3) compliant based on their chemical size and composition (Congreve et al., 2003). The first example of FBDD can be traced back to seminal work published from Abbott Laboratories, IL, USA on structure-activity-relationships “SAR by NMR” in 1996 (Shuker et al., 1996). Since then and until ~2010, FBDD played a prominent role in the early drug discovery programs of many pharmaceutical companies, including Abbott, Astex, Plexxikon, Syrxx, Genentech, SGX Pharma, Vernalis, Vertex, and AstraZeneca, among others. Since then, FBDD has grown to be an integral part of many drug discovery efforts in both industry and academia.
Multiple biophysical techniques are used for the initial screening of fragment libraries for hit generation in FBDD, with two of the most popular methods being nuclear magnetic resonance (NMR) (Gossert and Jahnke 2016) and surface plasmon resonance (SPR) (Navratilova and Hopkins 2010). Other biophysical methods include thermal shift assay (TSA), isothermal titration calorimetry (ITC) (Ciulli 2013), capillary electrophoresis (CE) (Austin et al., 2012), microscale thermophoresis (MST) (Linke et al., 2016), biolayer interferometry (BLI) (Wartchow et al., 2011), weak affinity chromatography (WAC) (Duong-Thi et al., 2011), photoaffinity probes (Grant et al., 2020; Parker et al., 2017), plus new approaches for fragment screening by grating-coupled interferometry (GCI) and Cryo-EM (Saur et al., 2020). Some FBDD strategies also utilize computational/virtual screening and biochemical screening. Computational screening methods have made significant advances and can be used to virtually screen millions of compounds and analyze fragment hits to narrow down on a few hundred to a few thousand fragments. However, results may exhibit varying degrees of reliability, and many screening campaigns additionally employ orthogonal screening strategies with the goal of advancing overlapping hits. Experimental structure determination is still required to determine the precise atomic details and to evaluate protein-ligand interactions to guide medicinal chemistry efforts (Kumar et al. 2012; Lolli and Caflisch 2016).
Among the potential drug discovery methods, the only one that can best provide detailed atomic level 3D protein-ligand interactions is protein X-ray crystallography (PX). PX is an essential component of FBDD and structure-based drug discovery (SBDD), since it is the gold standard for determining the exact orientation of a fragment bound to a target protein (Murray and Blundell 2010). PX probes the widest range of molecular affinities, from millimolar to nanomolar (Price et al. 2017), a key benefit in FBDD as initial fragment hits will likely bind weakly. Unlike other structure determination techniques, PX is independent of protein size, and is free from false positive issues that plague biophysical methods (Silvestre et al., 2013; Schiebel et al., 2015), as the electron density of bound fragments will be visible in the final electron density maps. Despite these advantages, PX can be expensive and inefficient as a primary screening process, and thus other methods are often used to pre-filter fragments prior to pursuing co-crystal structures. In typical drug discovery campaigns, only a relatively small number of the best fragment hits (and their analogs) from biophysical, computational and biochemical screening efforts are advanced to crystallographic studies. The need to limit the number of hits stems from the challenges and complexities associated with generating co-crystal structures. For example, several steps are needed to transfer crystals into fragment-soaks followed by crystal harvesting and freezing for X-ray diffraction experiments. These processes are typically involved and error-prone, particularly when dealing with hundreds of fragments and crystals, and need extensive optimization. Fragment cocktails (of 3e10 molecules) have therefore been used to improve the efficiency of fragment soaking for crystallography (Verlinde et al., 2018; Nienaber et al., 2000), but other challenges remain.
A strong interest has remained in enabling primary fragment screening using PX (Davies and Tickle 2012), particularly when used in parallel with complementary biophysical and computational screens. With the advent and development of high-throughput PX in structural genomics (Zarembinski et al., 1998), especially programs from 2000 to 2015 under the NIH/NIGMS Protein Structure Initiative (Montelione 2012; Burley et al., 2008), like the Joint Center for Structural Genomics (JCSG) (Elsliger et al., 2010) and Berkeley Structural Genomics Center (BSGC) (Shin et al., 2007), as well as similar global efforts like The Structural Genomics Consortium (SGC) (O. Gileadi et al., 2005; Opher Gileadi et al., 2007), it has become feasible to consider PX as a primary screening method in FBDD. Notably, this advance is paradigm-shifting for hit generation in early discovery, as it immediately provides hit location and information on critical protein-ligand interactions. In addition, the methodology permits biologists and biochemists to assess the potential biological and biochemical value of hits at a particular site, and chemists to determine chemical tractability and synthesis strategies very early in the process. The progress since 2015 has culminated in the setup of the XChem fragment screening facility at the Diamond Light Source, Oxford, UK (Collins et al., 2018) and the ABS-OneStep™ fragment screening platform at Accelero Biostructures, San Francisco, USA, thereby allowing a path for the transition and translation of learning from structural genomics and high-throughput PX into PX-based primary screening of fragment libraries for FBDD. Based on the success of XChem, a couple of other synchrotrons in Europe are working to set up fragment-based screening facilities. Another important contribution related to XChem has been the development of the PanDDA method (Pearce et al., 2017) that is powerful in extracting information about the binding of fragments by using data sets from a large number of crystals, which allows the detection of even weak binders to be picked up from what may otherwise be challenging interpretation, by conventional methods, of electron density maps.
1.2. Drug discovery successes using fragment-based screening approaches
FBDD is an important strategy for developing high-affinity lead compounds and has been widely adopted in the pharmaceutical industry over the last 10e15 years with proven success. Small molecule fragments (<300 Da MW), which typically bind with low millimolar affinity, are used as building blocks to develop potent drug-like molecules (Erlanson 2011). FBDD allows efficient sampling of chemical space (Hall et al. 2014; Boyd and de Kloe 2010) and has found success in generating leads for targets that have failed in high-throughput screening (Hajduk and Greer 2007).
Once fragment hits against a target are obtained, the hits are ranked by priority score. This ranking depends on the method used to obtain the hits. Where hits are obtained by crystallography, rankings can be based on the electron density of the bound fragment. When hits are obtained by biophysical screening, the ranking can be based on estimated affinities. Hits are then advanced into iterative molecular optimizations combining computational chemistry, medicinal chemistry, synthetic chemistry, and biophysical and biochemical assays. These are used to grow the size of the fragments by adding functionalities through, for example, amination, arylation and alkylation. In cases where there is evidence of multiple fragments bound in proximity, fragment linking can be another way to increase compound size. The number of hits that are advanced to fragment growth or linking strategies depends on chemistry and other resources available for downstream work. Where crystal structures of protein-ligand complexes are available, these can be used directly for adding functional groups by computational chemistry. In cases where crystals structures are unavailable, compounds can be docked at the binding sites by computational chemistry prior to functional group elaborations. This iterative process to grow the fragment hits, which are usually weak binders, into regular small molecule compounds, increases the potencies of the compounds as the molecular weight increases to ultimately lead to a useful inhibitor, as for example in the “SAR by Space” approach used in FBDD of bromodomain containing protein 4 (BRD4) (Borysko et al., 2018) and the “anchored plasticity” approach used in development of nitric oxide synthase inhibitors (Garcin et al., 2008).
FBDD programs have led to four approved drugs (Plexxikon’s Vemurafenib, a B-Raf kinase inhibitor, FDA approved in 2011; Plexxikon’s Pexidartinib, a CSF1R and c-Kit inhibitor, approved in 2019; Astex/J&J’s Erdafitinib, a pan FGFR1–4 inhibitor, approved in 2019, and AbbVie/Genentech’s Venetoclax, a Bcl-2 inhibitor, approved in 2016). In addition, ~50 compounds have entered clinical trials (see list at http://practicalfragments.blogspot.com/2020/03/fragments-in-clinic-2020-edition.html). As it is still early in its implementation phase, expanded use of FBDD presents many opportunities to identify novel chemical matter for a range of therapeutically relevant proteins, including those that function as part of the DNA Damage Response (DDR). As will be discussed next, the DDR is a critical system in not only the prevention of disease, but in several treatment paradigms, most notably the eradication of cancer; and understanding DDR network complexity makes efficient identification of inhibitors for multiple targets key to advance both biology and medicine.
1.3. DNA damage formation and consequences
DNA damage is generated by intrinsic and extrinsic mechanisms (Chatterjee and Walker 2017). The intrinsic processes include (i) spontaneous hydrolysis, (ii) attack by intracellular reactive chemicals [most notably reactive oxygen species (e.g., the hydroxyl radical)], (iii) misincorporation errors during DNA replication, and (iv) replicative stress that arises when a progressing replication fork is interrupted by an obstructive lesion, difficult to copy sequence (e.g., repetitive elements) or active transcription. Some of the most common extrinsic sources include sunlight, ionizing radiation, and many chemicals found in the environment, food and water (e.g., aflatoxin, heavy metals, tobacco components, certain emissions, etc.). Extrinsic DNA-damaging agents also consist of the numerous genotoxins used as front-line therapeutics in the eradication of rapidly dividing cells, such as observed in cancer (discussed below).
The types of DNA damage generated are vast but are comprised of primarily (i) simple base modifications that might alter the coding properties of the nucleotide without disrupting the helical structure and (ii) more complex lesions that involve bulky chemical additions, strand breakage, or crosslinking of neighboring bases (intrastrand crosslinks) or the complementary strands (interstrand crosslinks), often promoting gross conformational changes to the duplex. Depending on the nature of the modification, persistent DNA damage can lead to (i) altered genomic landscapes that disrupt gene regulatory networks, (ii) inaccurate copying during chromosome duplication, (iii) replication fork collapse and the formation of one-ended double-strand breaks or complex DNA intermediates, or (iv) transcription stalling or errors. These molecular events trigger cellular outcomes such as transformation, senescence or apoptosis, endpoints that underlie cancer development, degenerative disease and premature aging (Tiwari and Wilson 2019).
1.4. Responses to DNA damage and DNA repair mechanisms
To prevent the deleterious consequences of persistent DNA damage, organisms have evolved coping mechanisms that entail damage recognition, cell cycle checkpoint modulation, and activation of the appropriate response. In situations of manageable damage, DNA repair mechanisms are called into action to remove substrate lesions or intermediates, preserving genome integrity and restoring normal biochemical operations. However, when DNA damage is excessive, the cell will activate pathways that promote death (typically apoptosis) or arrested growth, such as senescence, preventing almost certain genomic instability and associated dysfunction. Collectively, the protective mechanisms are known as the DDR, a global system designed to choose shrewdly between cell survival or elimination (Pilie et al., 2019).
The major DNA repair mechanisms include the following, with many of them involving specialized sub-pathways: (i) direct reversal (DR), (ii) mismatch repair (MMR), (iii) base excision repair (BER), (iv) single-strand break repair (SSBR), (v) ribonucleotide excision repair (RER), (vi) nucleotide excision repair (NER), and (vii) double-strand break repair (DSBR), which includes non-homologous end-joining (NHEJ) and homologous recombination (HR). It is not our intent to provide a comprehensive discussion of the different DNA repair mechanisms, but instead, we have provided Table 1 to summarize the major repair pathways, DNA substrates, and protein participants, and direct the reader to reviews for further information (Soll et al. 2017; Fedeles et al., 2015; Modrich 2016; Krokan and Bjørås 2013; Andres et al., 2015; Mullenders 2018; Scully et al., 2019; Sun et al., 2020; Chaplin and Blundell 2020).
Table 1. Major DNA Repair Pathways, Main Substrates and Biochemical Steps, and Participating Proteins.
For further information and a more expansive list of the DDR proteins, please visit: https://www.mdanderson.org/documents/Labs/Wood-Laboratory/human-dna-repair-genes.html.
| DNA Repair Pathway Specific Biochemical Step |
Main Substrate(s) | Primary Protein(s) (Sub-Pathway) |
|---|---|---|
| Direct Reversal (DR) | ||
| Direct Transfer (to active site cysteine) | Methylated bases (e.g., O6-MeG) | MGMT |
| Direct Removal (via a-ketoglutarate-dependent reaction) | Alkylated bases (e.g., 1-MeA, 3-EtA, 3-MeC), certain ethenoadducts | ALKBH2, ALKBH3 |
| Mismatch Repair (MMR) | ||
| Recognition | Base-base mismatch, small insertion/deletion (INDEL) | MSH2/MSH6 (a.k.a., MUTSα) |
| Small and larger (~10 nt) INDEL | MSH2/MSH3 (a.k.a., MUTSβ) | |
| Incision | Leading Strand | MLH1/PMS2 (a.k.a., MUTLα) |
| Excision | 5′-Nick | EXO1 (EXO1-dependent) |
| 3′-Nick | MUTLα, POLδ (EXO1-independent) | |
| DNA Synthesis | Large Gap | POLδ |
| Ligation | Nick | LIG1 |
| Base Excision Repair (BER) | ||
| Base Excision | Uracil | UDG, SMUG1 |
| Oxidized Base | OGG1, MUTYH, NTH1, NEIL1, NEIL2, NEIL3 | |
| Alkylated Base | MPG | |
| Mismatched Base | TDG, MBD4 | |
| AP Site Incision | AP Site | APE1, AP Lyase |
| Termini Clean-Up (also part of single-strand break repair) | 5′-deoxyribose phosphate | POLβ |
| 3′-α,β-polyunsaturated aldehyde | APE1 | |
| 3′-phosphate | PNKP (APE1-independent) | |
| 3′-topoisomerase 1 adduct | TDP1 | |
| 5′-adenylate | APTX | |
| 5′-Flap (product of Long-patch BER) | FEN1 | |
| Non-enzymatic scaffold protein | XRCC1 | |
| DNA Synthesis | Single Nucleotide Gap | Short-patch (SP) BER: POLβ |
| Gap with Obstructive 5′-Terminus | Long-patch (LP) BER: POLε or POLδ | |
| Ligation | Nick | XRCC1/LIG3α or LIG1 (mainly LP-BER) |
| Nucleotide Excision Repair (NER) | ||
| Recognition & Repair Initiation | Helix-distorting Lesion | Global Genome (GG-NER): XPC/RAD23B, XPE/DDB2 |
| Transcription-blocking Lesion; Stalled RNAPII | Transcription-Coupled (TC-NER): CSB, CSA | |
| Damage Verification & Helix Unwinding | Initiating NER Complex | XPA, RPA, TFIIH |
| Damage Excision | Bubble Structure | XPG (3′ incision), ERCC1/XPF (5′ incision) |
| DNA Synthesis | Gap | POLδ |
| Ligation | Nick | LIG1 (replicating cells), LIG3 (non-replicating cells) |
| Non-Homologous End-Joining (NHEJ) | ||
| Recognition & Signaling | Two-Ended Double-strand Break (DSB) | KU70/KU80, DNA-PKcs |
| End Processing | DSB End | Artemis, POLλ, POLμ, TdT (see Termini Clean-Up as well) |
| End-Joining Ligation | Compatible Ends | XLF, XRCC4, LIG4 |
| Homologous Recombination (HR) | ||
| Recognition & Signaling | Two-Ended or One-Ended (collapsed replication fork) DSB | MRN (MRE11/RAD50/NBS1) complex, ATM, RBBP8 |
| Long-Range Resection | DSB End | EXO1, DNA2, BLM |
| Strand Exchange | Single-stranded Resection Products | RAD51, RAD51 paralogs, BRCA2, RAD54 |
| DNA Synthesis | Exchange Primer-Template | POLε, POLδ, POLκ, POLη |
| Holliday Junction Resolution | Cross-Shape Recombination Intermediate | TOPO3α/BLM, GEN1, MUS81/EME1, SLX1/SLX4 |
| Ligation | Nick | LIG1 |
1.5. Overview of the DDR in cancer etiology and treatment
Individuals that harbor a germline defect in a key component of the DDR commonly exhibit cancer predisposition as a primary clinical phenotype (Torgovnick and Schumacher 2015; Romero-Laorden and Castro 2017). Given that DNA damage can drive mutagenic events and genomic instability, outcomes known to underlie transformation and consequent carcinogenesis, the association between ineffective DNA repair and cancer development is obvious. Indeed, the connection between DDR defects and neoplasia is seen in genetic syndromes involving inefficient MMR (e.g., Lynch syndrome), BER (MUTYH- or NTHL1-related colorectal cancer syndromes), NER (e.g., xeroderma pigmentosum), and replicative stress or DSBR (e.g., Fanconi anemia, Bloom syndrome, Nijmegen breakage syndrome, ataxia telangiectasia). In addition to the inherited mutations, sporadic, deleterious genetic changes can arise in DDR genes. Such events can give rise to DNA repair deficiency and a consequent mutator phenotype driving the multi-hit theory of carcinogenesis that involves several genetic and biochemical changes that culminate in unregulated cell growth (Ma et al., 2018).
Beyond their function as tumor suppressor mechanisms, the DDR and discrete DNA repair pathways operate prominently in dictating the efficacy of many anti-cancer therapies. As alluded to earlier, several of the agents used to destroy cancer cells act via the introduction of toxic DNA damage or intermediates (Table 2) (Helleday et al., 2008). The concept is that complex DNA alterations will induce lethal stress responses (mainly due to collapsed replication events) in rapidly dividing cells, such as found in cancer. Consistently, defects in the DDR are associated with increased sensitivity to many of the DNA-damaging agents employed in anticancer chemo or radiotherapy. Furthermore, increased DNA repair capacity, a phenotype often seen in cancer cells, is associated with genotoxin agent resistance.
Table 2.
Anti-cancer genotoxic agents.
| Agent Classification | Examples | Mechanism of Toxicity (Cellular Target) |
|---|---|---|
| Alkylators | Nitrogen mustards, nitrosoureas, alkyl sulfonates, triazines, ethylenimines, platins | DNA damage: alkylation, monoadducts, intra- and inter-strand crosslinks |
| Antimetabolites | Folate antagonists, purine and pyrimidine analogs | Inhibition of DNA synthesis: disruption of nucleotide pools, chain termination |
| Plant alkaloids | Taxanes, vinca alkaloids | Disruption of chromosome architecture by spindle dissolution |
| Antibiotics | Anthracyclines, bleomycin, mitomycin | DNA damage: trapped protein-DNA intermediates, free radical-induced damage, crosslinks |
| Topoisomerase inhibitors | Topotecan, irinotecan, etoposide | Inhibition of essential DNA metabolic processes |
| Photosensitizers | Aminolevulinic acid, porfimer sodium | Macromolecular damage (e.g., to DNA) through photoexcitation and energy transfer |
1.6. DDR as a target in combinatorial and synthetic lethality treatment paradigms
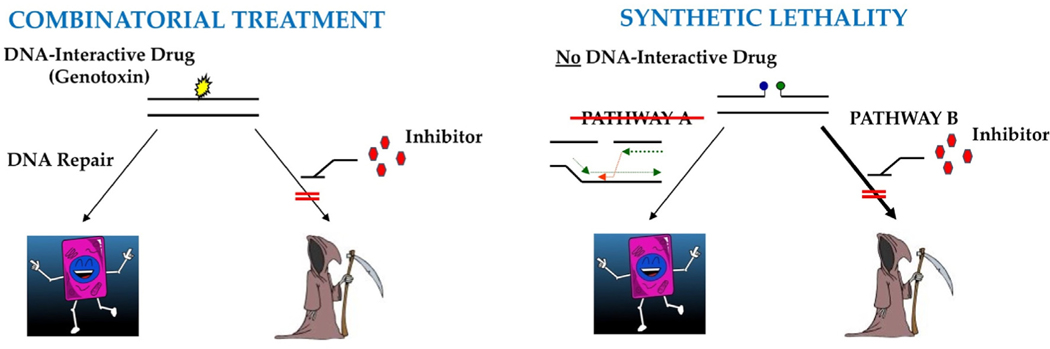
A current emphasis in the effort to improve anti-cancer therapies is to (i) selectively regulate the DDR in cancer cells, thereby increasing sensitivity to applied genotoxins or (ii) exploit distinctive genetic alterations in the DDR that give rise to cancer, particularly those originating sporadically (Fig. 1) (Gavande et al., 2016). The former strategy, while typically more complicated in terms of specificity, entails co-administration of a specific DNA repair inhibitor with a relevant genotoxin, aiming to augment the killing effect of the DNA-damaging agent in highly replicative cells. The latter approach, commonly known as “synthetic lethality”, involves disabling an essential compensatory pathway that takes on greater importance in the absence of a specific DDR gene/function.
Fig. 1. Major Anti-Cancer Therapeutic Approaches Involving a DNA Repair Inhibitor. (Left) Combinatorial Treatment.
Many anticancer treatments involve the use of a genotoxic agent (or DNA interactive drug) to kill rapidly dividing cancer cells. Thus, DNA repair plays an important role in dictating anticancer therapy effectiveness, by resolving toxic DNA damage and preserving viability. Selective inhibition of DNA repair has therefore been considered a logical mechanism for enhancing therapeutic efficacy of genotoxic agents. (Right) Synthetic Lethality. Many cancers arise from inherited or sporadic defects in the DDR. For example, mutations in pathway A (e.g., HR, as shown), which result in repair deficiency, give rise to or promote cancer development. While these cells may exhibit genomic instability, they are still viable, largely due to a compensatory mechanism, i.e., pathway B (e.g., SSBR). However, a selective inhibitor against pathway B (e.g., olaparib, a PARP inhibitor) interferes with or blocks the compensatory response, leading to an overwhelming accumulation of DNA damage and ultimately cell death.
1.6.1. Combinatorial therapies
The use of cytotoxins in the treatment of cancer stems largely from observations made following the employment of nitrogen mustards during the first and second world wars (Chabner and Roberts 2005). In particular, soldiers exposed to these chemical warfare agents suffered from depleted bone marrow and reduced lymph nodes, suggesting possible benefits of the compounds in the eradication of lymphatic tumors. After the advent of more stable chemical forms of nitrogen mustards, pre-clinical and clinical application of the drugs was shown to regress the growth of (i) a transplanted lymphoid tumor in mice and (ii) the mediastinal and lymphatic masses of a non-Hodgkin lymphoma patient, albeit only temporarily. This work was followed by the tactical administration of antifolates, such as aminopterin, in the treatment of leukemia, an approach similarly revealed to be effective in inducing disease remission, although again, only briefly. Some years later, it was discovered that both nitrogen mustards and antifolates elicit their lethal effects via the induction of DNA damage and replicative stress. The success of targeting DNA to selectively kill rapidly dividing cancer cells prompted the development and administration of several genotoxins (broadly defined in Table 2), including temozolomide (TMZ), cisplatin, doxorubicin, 5-fluorouracil, etoposide, and gemcitabine.
With the addition of DNA-damaging chemicals to the anticancer arsenal, which already included ionizing radiation, a physical agent that directly and indirectly induces DNA modifications (mainly toxic DSBs and multiply-damaged sites) to kill cells, the interest of basic and clinical researchers in the development of DNA repair inhibitors for combinatorial treatments amplified around 1980. Again, the concept was that inhibition of the DDR would improve the efficacy of a relevant genotoxin, although specificity to cancer cells was an obvious hurdle, particularly for therapies involving systemic administration of chemical agents. Some of the earliest DNA repair targeting work focused on poly(ADP)ribose polymerase (now known as PARP1), an enzyme shown to be specifically activated in response to DNA strand breaks (Alemasova and Lavrik 2019). Association of PARP1 at either a single- or double-strand break results in an enzyme-catalyzed consumption of NAD+ that facilitates both auto-modification of the protein and ribosylation of other DDR factors, such as X-ray cross complementing 1 (XRCC1). Auto-modification promotes consequent release of PARP1 from DNA, permitting subsequent repair and response measures. Given that many DNA-damaging agents, including alkylators and ionizing radiation, promote PARP1-driven repair reactions, the idea of selectively inhibiting the enzyme was an attractive approach for therapeutic agent sensitization. While the combinatorial strategy with PARP inhibitors and a relevant anti-cancer genotoxin has passed many preclinical tests, the application of DNA repair inhibitors as part of standard genotoxin-based therapy often suffers from off-target toxicity and thus remains unsatisfactorily proven (Lupo and Trusolino 2014). It was not in fact until the observation of synthetic lethality between PARP inhibitors and defects in HR (see later) that such inhibitors received clinical traction.
Around the same time investigators were pursuing PARP inhibitors, studies revealed a correlation between the expression of O6-methylguanine-DNA-methyltransferase (MGMT) and cellular sensitivity to clinical alkylators, such as nitrosourea (Pegg 2011). Human MGMT, and its conserved homologues in many other organisms, functions as a DNA repair protein by directly transferring a methyl group from the O6 position of guanine to an active site cysteine residue, leading to ‘suicide’ inactivation and targeting of the protein for degradation. Persistent O6-methylguanine lesions can cause mutagenic or inappropriate DNA transactions, ultimately leading to cell death. Like PARP inhibitors, small molecules generated to inactivate and consequently deplete cells of MGMT are effective at increasing relevant genotoxin sensitivity in pre-clinical paradigms, both cell culture and xenograft models. However, clinical trials using an MGMT-inactivating pseudosubstrate (or guanine analogue), such as O6-benzylguanine, in combination with an alkylator have revealed that many of the intended synergistic drug effects occur against non-target tissue, e.g., the bone marrow, leading to myelosuppression and general toxicity. Thus, while combinatorial strategies involving a systemic genotoxin remain of great therapeutic interest, improved cancer-specific targeting is necessary when it comes to DDR inhibitor use in the clinic. Some success has been attained for brain cancers, where local administration of drugs can be achieved via a slow-release polymer (e.g., the Gliadel wafer) that is precisely implanted during surgical resection of the primary tumor (Wait et al., 2015). In addition, since radiotherapy can be directed locally to eradicate cancer cells, there remains a push to apply DDR inhibitors, mainly against components of the DSBR response, as radiosensitizers (Gavande et al., 2016). Still, outside of the above example, to our knowledge, no combinatorial therapy involving a DNA repair inhibitor and genotoxic agent is being employed as front-line care in the treatment of cancer, indicating a need for better agents, treatment designs (namely targeting) or both.
1.6.2. Synthetic lethality therapies
While around for many decades, the application of synthetic lethality in situations involving the DDR gained widespread attention as a strategy in cancer treatment when the Helleday and Ashworth laboratories reported that chemical inhibitors of PARP1 induce killing of cells deficient in either breast cancer 1 (BRCA1) or BRCA2, proteins that operate in recombinational repair (Bryant et al., 2005; Farmer et al., 2005). The mechanism underlying cell death appears to involve inhibition of PARP1-mediated SSBR, resulting in the accumulation of strand breaks that are tightly bound by trapped PARP1, replication fork collapse, and the formation of one-ended DSBs, an outcome that HR-deficient cells are unable to handle, leading to replicative stress and the activation of apoptotic cell death. Similarly, inhibitors of the PARG glycohydrolase that removes poly-ADP ribose and releases PARP1 are also under active preclinical cancer investigation (Houl et al., 2019). Since the identification of the synthetic lethal relationship between SSBR and HR (overviewed in Fig. 1, right), albeit one that involves a dominant effect of PARP1 inhibition (Zimmermann et al., 2018; Murai et al., 2012; D’Andrea 2018), numerous other compensatory relationships have been reported involving DDR mechanisms, including a situation in which cancers that exhibit microsatellite instability (commonly associated with MMR defects) require WRN helicase function for survival (Chan et al., 2019; Lieb et al., 2019; Kategaya et al., 2019). Given that large-scale sequencing efforts have revealed that many cancers arise due to sporadic genetic defects in components of the DDR, a molecular outcome that likely provides a survival and adaptability advantage, there has been growing interest in exploiting DNA repair alterations in the treatment of neoplasia (Yar et al., 2020; Klinakis et al. 2020; Yap et al., 2019; Motegi et al., 2019).
1.7. FBDD approach for the development of inhibitors against selected DNA repair targets
Many DDR targets have been considered “undruggable”, primarily due to the challenge of developing compounds that can interfere with the typically large interface of protein-nucleic acid interactions, as compared to the discrete pockets or cavities found in many other classically druggable targets, such as protein kinases. In some cases, as for example with apurinic/apyrimidinic endonuclease 1 (APE1; see below), hits have been identified by conventional screening methods, but have failed to advance to effective clinical inhibitors. It is reasonable to hypothesize that the lack of experimental structural information to verify ligand poses and drive compound optimization hindered the progress of these programs. The approach with ABS-OneStep™ provides a significant value proposition for DDR drug discovery, offering a multi-pronged approach to inhibitor discovery and design: target-specific fragment hit generation with target-specific, structure-based hit expansion and linking.
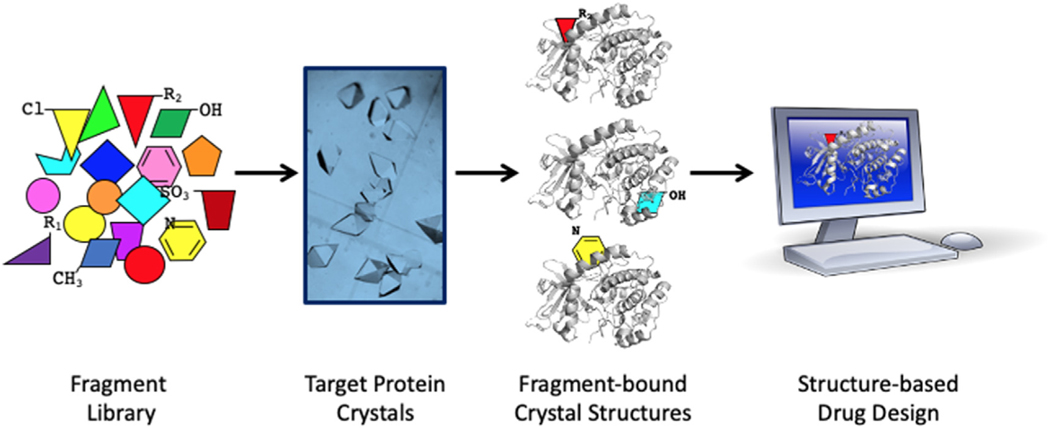
Results from ABS-OneStep™ for hit generation on 5 diverse proteins are shown in Table 3, and include DDR targets, meiotic recombination 11 (MRE11) and APE1, chosen for their therapeutic potential in cancer, as well as our extensive experience in crystallography, structural biochemistry, and biology of DNA-binding and DNA repair proteins, including polymerases, transcription factors and nucleases (Das and Georgiadis 2004; Hayashi et al., 2015; Das et al., 2010; H. He, Chen, and Georgiadis 2014; Trilles et al., 2019; Singh et al., 2018; Goodwin et al., 2010; Crowther et al., 2004; M. Li and Wilson 2014; Mol et al., 2000). The ABS-OneStep™ platform (Fig. 2) involves: (i) the generation of hundreds of crystals, (ii) efficient fragment soaking of crystals optimized for each target of interest, and (iii) high-volume X-ray diffraction data collection, processing, structure determination and analysis. The platform can currently handle screening of ~500 fragments in ~2 days, permitting scaling to 1000–2000 in approximately 1 week. We next discuss advances thus far on three DDR nucleases. These DDR nucleases are DNA damage structure-specific and offer the advantage that their specificity for damaged DNA enables precise targeting of DDR features to achieve inhibition (Tsutakawa et al. 2014).
Table 3.
Summary of screening campaigns.
| Protein | Fragments screened | Unique hits in electron density maps | Hit rate |
|---|---|---|---|
| Lysozyme | 100 | 4 | 4% |
| Trypsin | 50 | 1 | 2% |
| TmMRE11 | 30 | 2 | 6.7% |
| hAPE1 | 300 | 25 | 8.3% |
| hKDM4D | 100 | 3 | 3% |
Fig. 2. ABS-OneStep™ schematic.
Simplified schematic of the ABS-OneStep™ platform that includes generation of hundreds of crystals, efficient fragment soaking of crystals optimized for each target, and high-volume and high-throughput X-ray diffraction data collection, processing, structure determination and analysis. The platform can currently handle screening of ~500 fragments in ~2 days, permitting scaling to 1000–2000 in approximately 1 week.
1.7.1. MRE11
The MRE11 (a.k.a., MRE11A) nuclease, which forms a complex with the RAD50 ABC ATPase and Nijmegen breakage syndrome 1 (NBS1), plays a central role in the repair of cytotoxic and mutagenic DNA DSBs. MRE11 enzymes are present in all kingdoms of life and share significant sequence similarities in their nuclease domain. The MRN complex mediates DSB detection and facilitates resolution via HR or NHEJ (or non-canonical NHEJ such as microhomology-mediated end joining (MMEJ)) by processing and tethering DNA ends during the repair process (Syed and Tainer 2018; Dutta et al., 2017). DSBs can occur as intermediates during normal cellular processes, such as meiosis, mating type switching or V(D)J recombination, as well as through the action of environmental or therapeutic agents, such as ionizing radiation and genotoxic chemicals. MRE11 is the central player in the eukaryotic triad of MRN proteins (MRE11-RAD50-NBS1/XSR2) (Lafrance-Vanasse et al. 2015) and in the prokaryotic MR (MRE11-RAD50) assembly that is crucial to the DSBR process. MRE11 has broad substrate recognition and can act on single-stranded DNA (ssDNA), double-stranded DNA (dsDNA), and DNA hairpin structures. Its functionality is characterized by ssDNA endonuclease activity and dsDNA 3′–5′ exonuclease activity (Paull and Gellert 1998).
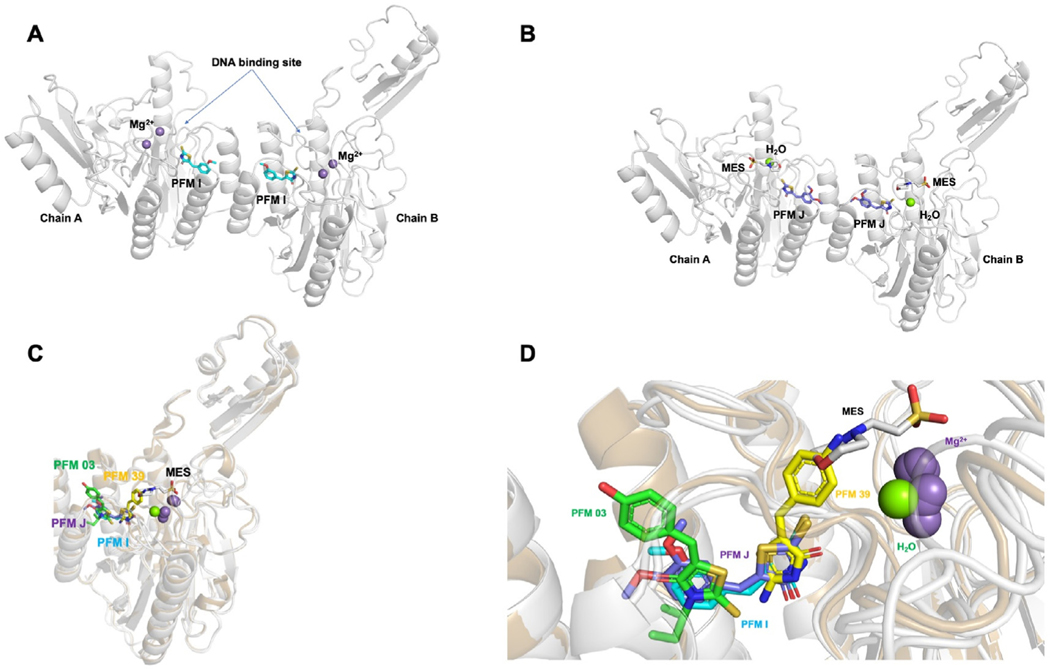
Recent interest and investigations have focused on discovery and development of inhibitors for MRE11. This ambition is driven by the role of the protein in genotoxin agent resistance, particularly agents that generated DSBs, such as ionizing radiation; as well as the synthetic lethality relationship with FEN1 described below. Since we elucidated the first crystal structure of a bacterial MRE11 from Thermotoga maritima, TmMRE11 (Das et al., 2010) (<20% sequence identity to human MRE11), we (J.A.T) have pursued different inhibitors against TmMRE11, which we hypothesize could be employed as chemical probes to specifically interrogate its endonuclease or exonuclease activities (Shibata et al., 2014; Moiani et al., 2018). Toward this end, the following compounds were indentified and used: PFM39, which is an aryl-pseudothiohydantoin, has exonuclease inhibitor activity and binds close to the active site; PFM03, which is an aryl-rhodanine, has endonuclease inhibitor activity and binds closer to the dimerization site; PFMI ((5Z)-5-[(3-methoxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one) and PFMJ ((5Z)-5-[(3,4-dimethoxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one), which are also aryl-rhodanine, but without a polar group substitute to the phenyl ring. The crystal structures of the modulators PFMI and PFMJ bound to TmMRE11 (PDB IDs 6X1Y and 6X1Z) have recently been determined and compared with PFM03 and PFM39 that were previously reported (PDB IDs 4O43 and 4O5G, respectively) (Moiani et al., 2018), leading to a tentative pharmacophore (a pharmacophore is a description of molecular features, which define the molecular interactions between a drug target and its ligand, that are necessary to trigger or block the biological response of the target). PFMI and PFMJ, which are aryl-rhodanine, span both endo- and exonuclease inhibitor sites because the aryl moiety overlaps the endonuclease and the rhodanine moiety overlaps the exonuclease site; but PFMI and PFMJ, with their substitution, bind in a perpendicular orientation compared to PFM39 and PFM03 as can be seen in the pharmacophore overlap (Fig. 3).
Fig. 3. TmMRE11 crystal structures.
Crystal structures of TmMRE11-PFMI, TmMRE11-PFMJ, and their superposition with other MRE11 inhibitor structures. Panels (A) and (B) depict, respectively, metal ions, buffer and water from crystallization conditions illustrating separately, variations in active site ligands (inhibitors and crystallization reagents) in different crystals structures determined; panels (C) and (D) depict metals, water, buffer and inhibitors all together to illustrate their relative juxtapositions. (A) TmMRE11 dimer in complex with small molecule modulator PFMI (PDB ID 6X1Y). One PFMI bound to each MRE11 subunit, with the rhodanine ring aimed toward the metal active site and methoxyphenyl moiety aimed toward dimerization site. The site of DNA binding has been indicated. (B) TmMRE11 dimer in complex with small molecule modulator PFMJ (PDB ID 6X1Z). One PFMJ bound to each MRE11 subunit, with the rhodanine ring aimed toward the active site, one molecule of MES (2-(N-morpholino)ethanesulfonic acid) buffer coordinated and with methoxy-phenyl moiety aimed toward the dimerization site. (C) Superposition of both complexes TmMRE11-PFMI and TmMRE11-PFMJ in comparison with complex TmMRE11-PFM03 (endo-inhibitor, PDB ID 4O43) and TmMRE11-PFM39 (exo-inhibitor, PDB ID 4O5G), revealing the different binding of the three types of small molecules. The compounds are color coded: PFM03-green; PFMJ-purple; PFMI-cyan; and PFM39-yellow (D) Detail of the superpositions described in (C) in a similar view, but with a slight rotation for a better view, where only one TmMRE11 subunit is visualized to maximize the description of superposed molecular moieties aimed to generate a preliminary pharmacophore.
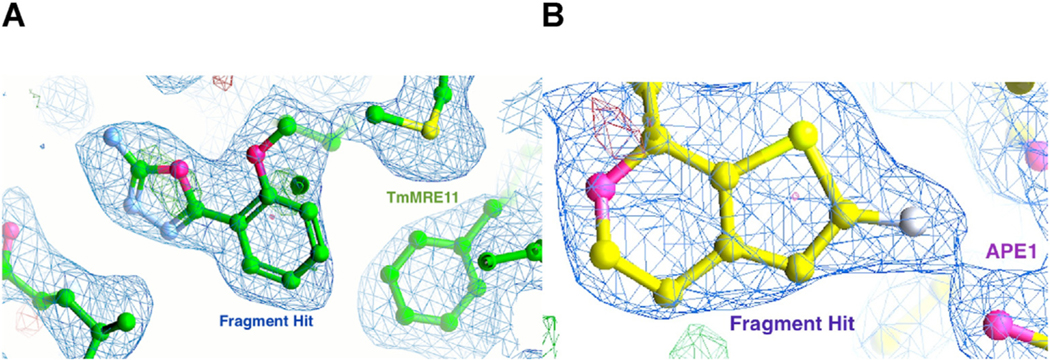
To find additional chemical moieties to probe nuances in TmMRE11 function, and demonstrate the power of our approach for hit generation, we performed a small proof-of-concept fragment screen. TmMRE11 crystals were reproduced and optimized for this screening with 30 fragments, resulting in 2 hits as assessed by calculating electron density maps with PanDDA (Pearce et al., 2017) (Fig. 4), including a hit with a positive control inhibitor (PFM01). These structures result in new hypotheses that can be tested, for example: investigating if and how the new compounds can modulate endonuclease or exonuclease activity; combining compounds to generate a new scaffold that can modulate MRE11 function; deploying the platform on a larger fragment library to generate additional hits; and using the new scaffold to test human MRE11 inhibitor design. MRE11 is exemplary for the power of PX with fragments to provide initial inhibitors that can then be enlarged to gain added specificity by employing the anchored plasticity approach that yielded high affinity and isotype specific nitric oxide synthase inhibitors (Garcin et al., 2008).
Fig. 4. Fragment hits by crystallography.
(A) Electron density of a fragment hit obtained with TmMRE11. (B) Electron density of a fragment hit obtained with APE1.
1.7.2. APE1
APE1 (a.k.a., APEX1) is a multifunctional protein with a primary function as a DNA repair nuclease and a separate function as a regulator of transcription factor DNA binding via a redox mechanism (McNeill et al., 2020; Wallace 2014). As an apurinic/apyrimidinic (AP) endonuclease, APE1 is a central player in the BER pathway, a process involved in resolving spontaneous, alkylative and oxidative DNA damage (M. Li and Wilson 2014). Specifically, APE1 cleaves at AP sites generated spontaneously, by damage-induction, or through the action of DNA glycosylases, which initiate classic BER by excising a substrate (often damaged) base moiety. Following the APE1-directed incision event, the remaining abasic fragment is removed, the gap filled, and the nick sealed (McNeill et al., 2020; Wallace 2014). Due to its prominent role in BER, and since rapidly proliferating cancer cells often upregulate DNA repair enzymes, such as APE1, the protein has emerged as a promising anticancer target.
Every day more than 10,000 AP sites are created in each cell under normal metabolic conditions (Lindahl and Nyberg 1972), and treatment with some chemotherapeutic agents, as well as ionizing radiation, increases the total number of genomic abasic lesions. If left unrepaired, non-coding AP sites can lead to mutations or replication fork collapse and DSBs that are highly cytotoxic (Boiteux and Guillet 2004). The importance of APE1 in BER is highlighted by the fact that the enzyme is responsible for greater than 95% of the total AP endonuclease activity in human cells (D. M. Wilson 3rd and Barsky 2001) and that depletion of the protein leads to mammalian cell inviability (Fung and Demple 2005; Izumi et al., 2005). In addition, siRNA knockdown of APE1 increases the sensitivity of cells to several DNA-damaging agents, most notably alkylators, such as methylmethane sulfonate (MMS) (Walker et al.,1994; Silber et al., 2002; Wang et al. 2004; Liu et al., 2003). Thus, APE1 is potentially a relevant therapeutic target for a number of cancers and has been validated in a xenograft model for ovarian cancer, with knockdown of APE1 greatly reducing tumor growth (Fishel et al., 2008). APE1 may be particularly relevant in glioblastoma for which the alkylator TMZ is the preferred chemotherapeutic, as resistance to TMZ is in part mediated by elevated levels of APE1 (Silber et al., 2002; M. S. Bobola et al., 2001; Michael S. Bobola, Finn, et al., 2005a; Michael S. Bobola, Silber, et al., 2005b). The strategy of combining a DNA-damaging agent with a BER inhibitor, i.e., TMZ with the PARP inhibitor, Veliparib, is currently being explored in a phase I trial for the treatment of acute myeloid leukemia (Gojo et al., 2017). An even more effective strategy might be to employ an APE1 selective inhibitor with TMZ.
In addition to the promise of targeting APE1 in combinatorial therapies, recent work has demonstrated that APE1 nuclease inhibitors induce synthetic lethality of cancer cells deficient in DNA DSBR, i.e., BRCA or ataxia telangiectasia mutated (ATM) defective cell models (Sultana et al., 2012). Inhibition of APE1 results in the accumulation of DSBs and G2/M cell cycle arrest, presumably due to AP site accumulation and replication fork collapse, ultimately leading to genomic instability and cell death. A similar synthetic lethal relationship has been observed with APE1 inactivation in PTEN-deficient melanoma cells (Abbotts et al., 2014), which likely suffer from analogous DSBR defects. Thus, clinically effective inhibitors against APE1 nuclease activity have the potential to be used in combination with or as an alternative to PARP inhibitors in the treatment of HR deficient cancers.
Efforts to date to identify high affinity, selective small molecule APE1 inhibitors have met with limited success as evidenced by the few follow-up studies to improve the compounds. The published approaches have relied primarily on (i) screening commercially available compounds that were synthesized for other molecular targets, (ii) computational screening, and (iii) pharmacophore modeling (Madhusudan et al., 2005; Abbotts et al., 2014; Aiello et al., 2012; Zawahir et al., 2009; Seiple et al., 2008; Dorjsuren et al., 2012; Rai et al., 2013; 2012; Simeonov et al., 2009; Sultana et al., 2012; David M. Wilson 3rd and Simeonov 2010). The limited success of these efforts stems in part from the inherent difficulty in targeting a protein-nucleic acid interaction. In the case of APE1, the DNA binding site is large, accommodating 9 base pairs of duplex DNA, and is polar (Mol et al., 2000; Tsutakawa et al., 2013). Thus, a new approach is warranted for the discovery of novel and selective APE1 inhibitors, and this has been addressed by applying the ABS-OneStep™ platform for hit generation to APE1.
Briefly, we first identified a crystallization condition for APE1 that was compatible with fragment screening, starting with crystals that were optimized to yield the high resolution crystal structure of apo-APE1 to 1.4 Å (PDB ID 4QHE) (H. He, Chen, and Georgiadis 2014). Based on this methodology, we then developed a scalable crystal system for obtaining hundreds of protein crystals of APE1 with ligands and subsequently screened a 300-fragment library. We obtained ~25 high quality crystal structures showing unique and diverse fragment hits bound at the endonuclease site, but also at a previously unidentified secondary site, thus presenting multiple novel inhibition strategies (for future publication due to intellectual property considerations at this time). Starting with fragment hits with low affinity, but for which structural information is available, will allow successful evolution to high affinity APE1-specific inhibitors by fragment elaboration using reiterative crystallography, computational modeling, and synthetic chemistry. The simultaneous discovery of fragment hits, binding sites (orthosteric and potentially allosteric), and the visualization of protein-ligand interactions at high resolution, will propel the development of new APE1 lead inhibitors entering the path to clinical development.
1.7.3. FEN1
Flap endonuclease 1 (FEN1) was first isolated from rabbit tissue as a protein able to cleave 5′ ends of double-stranded DNA, thereby liberating 5′-mononucleotides, and at the time was termed DNase IV (Lindahl et al. 1969). The homologous protein was purified over two decades later from murine cells based on its ability to cleave 5′-flap structures in DNA, and was coined FEN1, its current designation (Harrington and Lieber 1994). As a structure-specific endonuclease, the enzyme recognizes and releases 5′-unannealed single-stranded flap regions, creating a nicked duplex product that is eventually sealed by DNA ligase. 5′-flap removal by FEN1 is critical for Okazaki fragment processing during lagging strand DNA synthesis or long-patch BER, and in regulating or facilitating recombination events (Zheng et al., 2011; Balakrishnan and Bambara 2013). FEN1 has more recently been found to possess gap endonuclease (i.e. GEN) activity, whereby the protein cleaves single-stranded regions with gaps of four or more nucleotides to aid in the resolution of stalled replication forks or to promote apoptotic DNA fragmentation (Zheng et al., 2005).
Studies from Kucherlapati et al., (2002) and Larsen et al., (2003) have demonstrated that mice carrying a homozygous null genotype (fen1−/−) exhibit early embryonic lethality (E4.5), indicating that FEN1 is essential for animal development. Cells from fen1−/− blastocysts show increased apoptotic cell death after ionizing radiation, and chicken cells lacking fen1 display hypersensitivity to DNA alkylating agents, e.g. MMS and N-methyl-N′-nitro-N-nitrosoguanidine (MNNG), as well as hydrogen peroxide (Matsuzaki et al. 2002). Furthermore, down-regulation of FEN1 has been shown to sensitize human glioblastoma cells to MMS, and the clinical drugs TMZ and cisplatin (Nikolova et al. 2009). These observations indicate that FEN1 deficiency results in failure to repair DNA lesions generated by DNA alkylating and oxidizing compounds, and possibly crosslinking agents, and provide evidence for an essential role of the protein in dividing cells and in preserving genome integrity via its contributions to replication and repair.
FEN1 gene expression has been reported to be increased in many human cancers, including lung, gastric prostate, pancreatic, brain, and breast cancer (Zheng et al., 2011; Illuzzi and Wilson 2012). Moreover, FEN1 expression is induced during cell proliferation and down-regulated during cell differentiation. These findings suggest that the level of FEN1 may influence tumor development and/or progression. Additionally, over-expression of FEN1 associates with a high Gleason score, which is reflective of aggressive and malignant tumors, increased serum prostate-specific antigen (PSA) levels, and seminal vesicle invasion in prostate cancer specimens (Lam et al., 2006). Elevated FEN1 expression also leads to improved cancer cell survival in response to anticancer therapy, where, for example, high FEN1 levels in hormone refractory human prostate cancer cells correlate with resistant to various anticancer agents, including ionizing radiation, doxorubicin, paclitaxel and vinblastine (Freedland et al., 2003). As noted earlier, studies indicate that down-regulation or inhibition of FEN1 sensitizes cells to a number of chemotherapeutic agents, such as TMZ and cisplatin, as well as radiotherapy (Xie et al. 2016; Deshmukh et al., 2017; L. He et al., 2017; J.-L. Li et al., 2019). The studies collectively suggest that the levels of FEN1 expression influence cancer cell function as it relates to proliferation potential, survival and apoptosis.
McManus et al., (2009) reported on one of the early efforts to identify a synthetic lethal relationship involving human FEN1. Specifically, they observed that RAD54B-deficient colorectal cancer cells, which are ineffective in HR, exhibit a proliferation defect and increased cellular cytotoxicity when FEN1 expression is reduced. Subsequent work revealed that inhibitors of FEN1 nuclease activity or siRNA knockdown of the protein resulted in synthetic lethality of cells harboring disrupted MRE11 or ATM activity, two factors central to the DDR (van Pel et al., 2013; Ward et al. 2017). More recent studies have found that FEN1 is synthetically lethal with BRCA1 or BRCA2 loss of function (Guo et al., 2020), substantiating a critical compensatory role for the nuclease in cells defective in resolving DNA intermediates (most likely DSBs) that arise from replicative stress (Mengwasser et al., 2019). Additionally, genome wide-based analyses identified FEN1 as the strongest determining factor in ERα-positive breast cancer patient prognostication, with high levels of the endonuclease correlating with poor survival, while complementary preclinical work provided evidence that pharmacological inhibition of FEN1 is a treatment option for tamoxifen-resistant cancer cells (Flach et al., 2020).
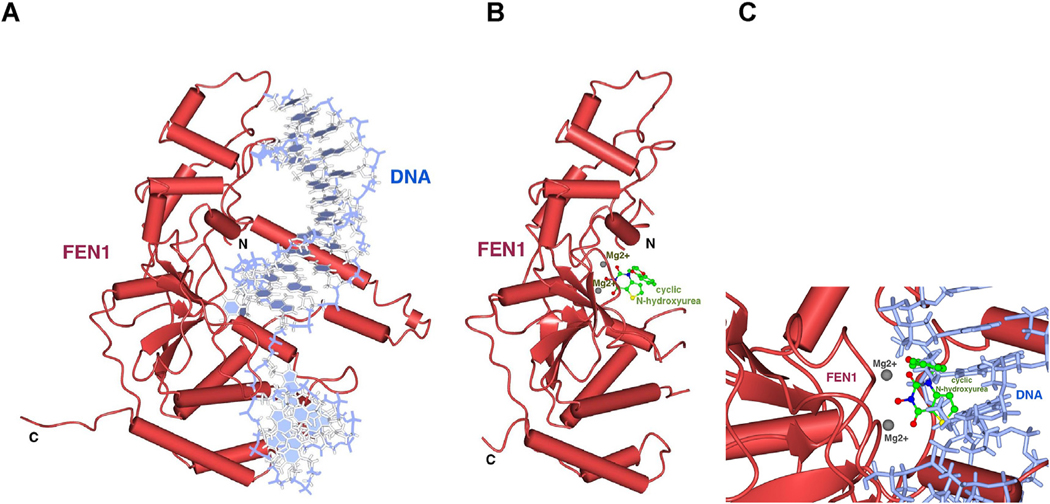
In light of the findings above, FEN1 serves as an attractive target for the treatment of many cancer types. Towards the development of FEN1-specific inhibitors, several related compounds have been developed with slightly different mechanisms of action, including the elucidation of the crystal structure of FEN1 in complex with N-hydroxyurea (Exell et al., 2016) (PDB 5FV7) as well as the crystal structure of FEN1 in complex with 5′-flap DNA (Tsutakawa et al., 2017) (PDB ID 5UM9) (Fig. 5). The crystal structure revealed coordination of the compound to the catalytic magnesium ions, which assist in the hydrolysis of phosphodiester bonds in the DNA substrate. We are currently in the process of interrogating current crystallographic data to optimize inhibitor design as well as generating FEN1 protein for crystallization and fragment library screening using the ABS-OneStep™ approach. These FEN1 studies and results may provide foundational insights and compounds to strategically inhibit other FEN1 family nucleases, including the XPG nuclease needed for bulky lesion removal in NER, whose structure was recently determined (Tsutakawa et al., 2020).
Fig. 5. Crystal structures of FEN1.
(A) Crystal structure of FEN1 bound to 5′-flap DNA (PDB ID 5UM9) (B) Crystal structure of FEN1 in complex with N-hydroxyurea (PDB ID 5FV7) revealing interaction of the N-hydroxyurea (green sticks) with the catalytic magnesium ions (grey balls), which help to hydrolyze phosphodiester bonds in the DNA substrate, in the same view as (A). The N- and C-terminal ends of FEN are labeled. Several secondary structure elements that are ordered in the presence of bound DNA (A) are disordered in the crystal structure without DNA (B). (C) Expanded view of the active site with DNA and the N-hydroxyurea compound coordinating the active site metals, obtained by superimposing the individual crystal structures in (A) and (B).
2. Closing remarks
Due to the development and implementation of high-throughput “omics” approaches, we are in the midst of a massive wave of scientific advances in our understanding of molecular processes that underlie disease risk and etiology. With this huge amount of mechanistic insight, novel therapeutic targets are being revealed regularly. Yet, despite our capacity to identify rational drug targets, our ability to develop clinical-grade therapeutic agents against these targets has been handcuffed for a variety of reasons. With the advent and continued optimization of FBDD/SBDD, and the development of a hit generation platform like ABS-OneStep™, we are poised at the cutting edge of advancing early drug discovery efforts on novel therapeutic targets. Although we focus here on three nucleases, there are many other DNA metabolizing enzymes with known crystallographic structures that are suitable targets for cancer therapy, such as WRN that is selectively essential in the context of microsatellite instability in multiple cancers (Perry et al., 2006; Lou et al. 2019). In light of their role in both combinatorial and synthetic lethality therapeutic paradigms, and given its track record as an undruggable space, proteins of the DDR offer prime ground for the application of FBDD/SBDD methods in the quest to create new clinical agents.
Acknowledgements
Research included in this publication was supported by the National Center For Advancing Translational Sciences of the NIH under Award Number R43 TR001736, and the National Institute Of General Medical Sciences of the NIH under Award Number R44 GM132796, to Accelero Biostructures Inc.; National Cancer Institute of the NIH under Award Number R43 CA254552 to XPose Therapeutics, Inc.; and NIH/NCI grants to John A. Tainer (P01 CA092584, R01 CA117638, R01 CA200231, and R35 CA220430). J.A.T. also acknowledges support by a Robert A. Welch Chemistry Chair, and the Cancer Prevention Research Institute of Texas (CPRIT) grants RP180813 and RP130397. X-ray diffraction data encompassing the work in this manuscript were collected by Accelero Biostructures Inc. at Stanford Synchrotron Radiation Lightsource (SSRL), SLAC National Accelerator Laboratory, Menlo Park, California; and at the Berkeley Center for Structural Biology (BCSB) synchrotron beamlines at the Advanced Light Source (ALS), Lawrence Berkeley National Laboratory, Berkeley, California. Use of the Stanford Synchrotron Radiation Lightsource (SSRL), SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (P41 GM103393). The BCSB is supported in part by the Howard Hughes Medical Institute. The ALS is a Department of Energy Office of Science User Facility under Contract No. DE-AC02-05CH11231. The Pilatus detector on 5.0.1. was funded under NIH grant S10 OD021832. The ALS-ENABLE beamlines are supported in part by the National Institutes of Health, National Institute of General Medical Sciences (P30 GM124169). Portions of this work (J.A.T) were also conducted at ALS through the Integrated Diffraction Analysis Technologies (IDAT) program, supported by DOE Office of Biological and Environmental Research with additional support from a High-End Instrumentation Grant (S10 OD018483). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
References
- Abbotts Rachel, Jewell Rosalyn, Nsengimana Jeremie, Maloney David J., Simeonov Anton, Seedhouse Claire, Elliott Faye, et al. , 2014. Targeting human apurinic/apyrimidinic endonuclease 1 (APE1) in phosphatase and tensin homolog (PTEN) deficient melanoma cells for personalized therapy. Oncotarget 5 (10), 3273–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiello Francesca, Shabaik Yumna, Esqueda Adrian, Sanchez Tino W., Grande Fedora, Garofalo Antonio, Neamati Nouri, 2012. Design and synthesis of 3-carbamoylbenzoic acid derivatives as inhibitors of human apurinic/apyrimidinic endonuclease 1 (APE1). ChemMedChem 7 (10), 1825–1839. [DOI] [PubMed] [Google Scholar]
- Alemasova Elizaveta E., Lavrik Olga I., 2019. Poly(ADP-Ribosyl)ation by PARP1: reaction mechanism and regulatory proteins. Nucleic Acids Res. 47 (8), 3811–3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres Sara N., Schellenberg Matthew J., Wallace Bret D., Tumbale Percy, Williams Scott R., 2015. Recognition and repair of chemically heterogeneous structures at DNA ends. Environ. Mol. Mutagen 56 (1), 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin Carol, Pettit Simon N., Magnolo Sharon K., Sanvoisin Jonathan, Chen Wenjie, Wood Stephen P., Freeman Lauren D., Pengelly Reuben J., Hughes Dallas E., 2012. Fragment screening using capillary electrophoresis (CEfrag) for hit identification of heat shock protein 90 ATPase inhibsitors. J. Biomol. Screen 17 (7), 868–876. [DOI] [PubMed] [Google Scholar]
- Balakrishnan Lata, Bambara Robert A., 2013. Flap endonuclease 1. Annu. Rev. Biochem 82, 119–138. February. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobola Michael S., Finn Laura S., Ellenbogen Richard G., Geyer Russell, Berger Mitchel S., Braga Justin M., Meade Elizabeth H., Gross Mary E., Silber John R., 2005a. Apurinic/apyrimidinic endonuclease activity is associated with response to radiation and chemotherapy in medulloblastoma and primitive neuroectodermal tumors. Clin. Canc. Res.: Off. J. Am. Assoc. Canc. Res 11 (20), 7405–7414. [DOI] [PubMed] [Google Scholar]
- Bobola Michael S., Silber John R., Ellenbogen Richard G., Geyer J. Russell, Blank A, Goff Ryan D., 2005b. O6-Methylguanine-DNA methyltransferase, O6-benzylguanine, and resistance to clinical alkylators in pediatric primary brain tumor cell lines. Clin. Canc. Res.: Off. J. Am. Assoc. Canc. Res 11 (7), 2747–2755. [DOI] [PubMed] [Google Scholar]
- Bobola MS, Blank A, Berger MS, Stevens BA, Silber JR, 2001. Apurinic/apyrimidinic endonuclease activity is elevated in human adult gliomas. Clin. Canc. Res.: Off. J. Am. Assoc. Canc. Res 7 (11), 3510–3518. [PubMed] [Google Scholar]
- Boiteux Serge, Guillet Marie, 2004. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair 3 (1), 1–12. [DOI] [PubMed] [Google Scholar]
- Borysko Petro, Moroz Yurii S., Vasylchenko Oleksandr V., Hurmach Vasyl V., Starodubtseva Anastasia, Stefanishena Natalia, Nesteruk Kateryna, Zozulya Sergey, Kondratov Ivan S., Grygorenko Oleksandr O., 2018. Straightforward hit identification approach in fragment-based discovery of bromodomain-containing protein 4 (BRD4) inhibitors. Bioorg. Med. Chem 26 (12), 3399–3405. [DOI] [PubMed] [Google Scholar]
- Boyd Susan M., de Kloe Gerdien E., 2010. Fragment library design: efficiently hunting drugs in chemical space. Drug Discov. Today Technol 10.1016/j.ddtec.2010.11.010. [DOI] [PubMed]
- Bryant Helen E., Schultz Niklas, Thomas Huw D., Parker Kayan M., Flower Dan, Lopez Elena, Kyle Suzanne, Meuth Mark, Curtin Nicola J., Helleday Thomas, 2005. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434 (7035), 913–917. [DOI] [PubMed] [Google Scholar]
- Burley Stephen K., Joachimiak Andrzej, Montelione Gaetano T., Wilson Ian A., 2008. Contributions to the NIH-NIGMS protein structure initiative from the PSI production centers. Structure 16 (1), 5–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabner Bruce A., Roberts Jr., Thomas G, 2005. Timeline: chemotherapy and the war on cancer. Nat. Rev. Canc 5 (1), 65–72. [DOI] [PubMed] [Google Scholar]
- Chan Edmond M., Shibue Tsukasa, McFarland James M., Gaeta Benjamin, Ghandi Mahmoud, Dumont Nancy, Gonzalez Alfredo, et al. , 2019. WRN helicase is a synthetic lethal target in microsatellite unstable cancers. Nature 568 (7753), 551–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaplin Amanda K., Blundell Tom L., 2020. Structural biology of multicomponent assemblies in DNA double-strand-break repair through non-homologous end joining. Curr. Opin. Struct. Biol 61, 9–16. April. [DOI] [PubMed] [Google Scholar]
- Chatterjee Nimrat, Walker Graham C., 2017. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen 58 (5), 235–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciulli Alessio, 2013. Biophysical screening for the discovery of small-molecule ligands. In: Williams Mark A., Daviter Tina (Eds.), In Protein-Ligand Interactions: Methods and Applications. Humana Press, Totowa, NJ, 357–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins Patrick M., Douangamath Alice, Talon Romain, Dias Alexandre, Brandão-Neto José, Krojer Tobias, von Delft Frank, 2018. Achieving a good crystal system for crystallographic X-ray fragment screening. Methods Enzymol. 610, 251–264. [DOI] [PubMed] [Google Scholar]
- Congreve Miles, Carr Robin, Murray Chris, Jhoti Harren, 2003.A ‘Rule of three’ for fragment-based lead discovery? Drug Discov. Today [DOI] [PubMed]
- Crowther Robert L., Remeta David P., Minetti Conceição A.S. A., Das Debanu, Montano Sherwin P., Georgiadis Millie M., 2004. Structural and energetic characterization of nucleic acid-binding to thefingers domain of moloney murine leukemia virus reverse transcriptase. Proteins 57 (1), 15–26. [DOI] [PubMed] [Google Scholar]
- D’Andrea Alan D., 2018. Mechanisms of PARP inhibitor sensitivity and resistance. DNA Repair 71, 172e176. November. [DOI] [PubMed] [Google Scholar]
- Das Debanu, Georgiadis Millie M., 2004. The crystal structure of the monomeric reverse transcriptase from moloney murine leukemia virus. Structure 12 (5), 819–829. [DOI] [PubMed] [Google Scholar]
- Das Debanu, Moiani Davide, Axelrod Herbert L., Miller Mitchell D., McMullan Daniel, Jin Kevin K., Abdubek Polat, et al. , 2010. Crystal structure of the first eubacterial Mre11 nuclease reveals novel features that may discriminate substrates during DNA repair. J. Mol. Biol 397 (3), 647–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies Thomas G., Tickle Ian J., 2012. Fragment screening using X-ray crystallography. Top. Curr. Chem 317, 33–59. [DOI] [PubMed] [Google Scholar]
- Deshmukh Amit Laxmikant, Chandra Sharat, Singh Deependra Kumar, Siddiqi Imran, Mohammad Banerjee, Dibyendu, 2017. Identification of human flap endonuclease 1 (FEN1) inhibitors using a machine learning based consensus virtual screening. Mol. Biosyst 13 (8), 1630–1639. [DOI] [PubMed] [Google Scholar]
- Dorjsuren Dorjbal, Kim Daemyung, Vyjayanti Vaddadi N., Maloney David J., Jadhav Ajit, David M, Wilson Simeonov 3rd, Anton, 2012. Diverse small molecule inhibitors of human apurinic/apyrimidinic endonuclease APE1 identified from a screen of a large public collection. PloS One 7 (10), e47974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong-Thi Minh-Dao, Meiby Elinor, Bergström Maria, Tomas Fex, Roland Isaksson, Ohlson Sten, 2011. Weak affinity chromatography as a new approach for fragment screening in drug discovery. Anal. Biochem 414 (1), 138–146. [DOI] [PubMed] [Google Scholar]
- Dutta Arijit, Bradley Eckelmann, Adhikari Sanjay, Ahmed Kazi Mokim, Sengupta Shiladitya, Pandey Arvind, Hegde Pavana M., et al. , 2017. Microhomology-mediated end joining is activated in irradiated human cells due to phosphorylation-dependent formation of the XRCC1 repair complex. Nucleic Acids Res. 45 (5), 2585–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsliger Marc Andre, Deacon Ashley M., Adam Godzik, Lesley Scott A., Wooley John, Kurt Wüthrich, Wilson Ian A., 2010. The JCSG high-throughput structural biology pipeline. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun 66 (Pt 10), 1137–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlanson Daniel A., 2011. Introduction to fragment-based drug discovery. Top. Curr. Chem 10.1007/128_2011_180. [DOI] [PubMed]
- Exell Jack C., Thompson Mark J., Finger L. David, Shaw Steven J., Debreczeni Judit, Ward Thomas A., McWhirter Claire, et al. , 2016. Cellularly active N-hydroxyurea FEN1 inhibitors block substrate entry to the active site. Nat. Chem. Biol 12 (10), 815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer Hannah, McCabe Nuala, Lord Christopher J., Tutt Andrew N.J., Johnson Damian A., Richardson Tobias B., Santarosa Manuela, et al. , 2005. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434 (7035), 917–921. [DOI] [PubMed] [Google Scholar]
- Fedeles Bogdan I., Singh Vipender, Delaney James C., Li Deyu, Essigmann John M., 2015. The AlkB family of Fe(II)/α-Ketoglutarate-Dependent dioxygenases: repairing nucleic acid alkylation damage and beyond. J. Biol. Chem 290 (34), 20734–20742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel Melissa L., He Ying, Reed April M., Chin-Sinex Helen, Hutchins Gary D., Mendonca Marc S., Kelley Mark R., 2008. Knockdown of the DNA repair and redox signaling protein ape1/ref-1 blocks ovarian cancer cell and tumor growth. DNA Repair 7 (2), 177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flach Koen D., Periyasamy Manikandan, Jadhav Ajit, Dorjsuren Dorjbal, Siefert Joseph C., Hickey Theresa E., Opdam Mark, et al. , 2020. Endonuclease FEN1 coregulates ERα activity and provides a novel drug interface in tamoxifen-resistant breast cancer. Canc. Res 80 (10), 1914–1926. [DOI] [PubMed] [Google Scholar]
- Freedland Stephen J., Pantuck Allan J., Sun H. Paik, Zisman Amnon, Graeber Thomas G., David Eisenberg, McBride William H., Nguyen David, Tso Cho-Lea, Belldegrun Arie S., 2003. Heterogeneity of molecular targets on clonal cancer lines derived from a novel hormone-refractory prostate cancer tumor system. Prostate 55 (4), 299–307. [DOI] [PubMed] [Google Scholar]
- Fung Hua, Bruce Demple, 2005. A vital role for ape1/ref1 protein in repairing spontaneous DNA damage in human cells. Mol. Cell 17 (3), 463–470. [DOI] [PubMed] [Google Scholar]
- Garcin Elsa D., Arvai Andrew S., Rosenfeld Robin J., Kroeger Matt D., Crane Brian R., Andersson Gunilla, Andrews Glen, et al. , 2008. Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase. Nat. Chem. Biol 4 (11), 700–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavande Navnath S., VanderVere-Carozza Pamela S., Hinshaw Hilary D., Jalal Shadia I., Sears Catherine R., Pawelczak Katherine S., Turchi John J., 2016. DNA repair targeted therapy: the past or future of cancer treatment? Pharmacol. Ther 160, 65–83. April. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gileadi O, Colebrook S, Burgess N, Berridge G, Haroniti A, Shrestha B, Smee C, Sundstrom M, 2005. The structural genomics Consortium: high-output structure determination. https://ora.ox.ac.uk/objects/uuid:65ae0be5-8cce-4484-9f75-9e38b7e473c4.
- Gileadi Opher, Knapp Stefan, Lee Wen Hwa, Marsden Brian D., Müller Susanne, Niesen Frank H., Kavanagh Kathryn L., et al. , 2007. The scientific impact of the structural genomics Consortium: a protein family and ligand-centered approach to medically-relevant human proteins. J. Struct. Funct. Genom 10.1007/s10969-007-9027-2. [DOI] [PMC free article] [PubMed]
- Gojo Ivana, Beumer Jan H., Pratz Keith W., McDevitt Michael A., Baer Maria R., Blackford Amanda L., Smith Douglas B., et al. , 2017. A phase 1 study of the PARP inhibitor Veliparib in combination with temozolomide in acute myeloid leukemia. Clin. Canc. Res 10.1158/1078-0432.ccr-16-0984. [DOI] [PMC free article] [PubMed]
- Goodwin Kristie D., He Hongzhen, Imasaki Tsuyoshi, Lee Suk-Hee, Georgiadis Millie M., 2010. Crystal structure of the human hsmar1-derived transposase domain in the DNA repair enzyme metnase. Biochemistry 49 (27), 5705–5713. [DOI] [PubMed] [Google Scholar]
- Gossert Alvar D., Jahnke Wolfgang, 2016. NMR in drug discovery: a practical guide to identification and validation of ligands interacting with biological macromolecules. Prog. Nucl. Magn. Reson. Spectrosc 97, 82–125. November. [DOI] [PubMed] [Google Scholar]
- Grant Emma K., Fallon David J., Hann Michael M., Fantom Ken G.M., Quinn Chad, Zappacosta Francesca, Annan Roland S., et al. , 2020. PhotoAffinity bits: a photoaffinity-based fragment screening platform for efficient identification of protein ligands. http://itempdf74155353254prod.s3.amazonaws.com/12053445/PhotoAffinity_Bits__A_Photoaffinity-Based_Fragment_Screening_Platform_for_Efficient_Identification_of_Protein_Ligands_v1.pdf. [DOI] [PubMed]
- Guo Elaine, Ishii Yuki, Mueller James, Srivatsan Anjana, Gahman Timothy, Putnam Christopher D., Jean Y, Wang J, Kolodner Richard D., 2020. FEN1 endonuclease as a therapeutic target for human cancers with defects in homologous recombination. Proc. Natl. Acad. Sci. U.S.A 117 (32), 19415–19424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajduk Philip J., Greer Jonathan, 2007. A decade of fragment-based drug design: strategic advances and lessons learned. Nat. Rev. Drug Discov 10.1038/nrd2220. [DOI] [PubMed]
- Hall Richard J., Mortenson Paul N., Murray Christopher W., 2014. Efficient exploration of chemical space by fragment-based screening. Prog. Biophys. Mol. Biol 116 (2e3), 82–91. [DOI] [PubMed] [Google Scholar]
- Harrington JJ, Lieber MR, 1994. The characterization of a mammalian DNA structure-specific endonuclease. EMBO J. 13 (5), 1235–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Yohei, Caboni Laura, Das Debanu, Yumoto Fumiaki, Clayton Thomas, Deller Marc C., Nguyen Phuong, et al. , 2015. Structure-based discovery of NANOG variant with enhanced properties to promote self-renewal and reprogramming of pluripotent stem cells. Proc. Natl. Acad. Sci. U.S.A 112 (15), 4666–4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Hongzhen, Chen Qiujia, Georgiadis Millie M., 2014. High-resolution crystal structures reveal plasticity in the metal binding site of apurinic/apyrimidinic endonuclease I. Biochemistry 53 (41), 6520–6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Lingfeng, Luo Libo, Zhu Hong, Yang Huan, Zhang Yilan, Wu Huan, Sun Hongfang, et al. , 2017. FEN1 promotes tumor progression and confers cisplatin resistance in non-small-cell lung cancer. Mol. Oncol 11 (9), 1302–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday Thomas, Petermann Eva, Lundin Cecilia, Hodgson Ben, Ricky A, Sharma, 2008. DNA repair pathways as targets for cancer therapy. Nat. Rev. Canc 8 (3), 193–204. [DOI] [PubMed] [Google Scholar]
- Houl Jerry H., Ye Zu, Brosey Chris A., Balapiti-Modarage Lakshitha P.F., Namjoshi Sarita, Bacolla Albino, Laverty Daniel, et al. , 2019. Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death. Nat. Commun 10 (1), 5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illuzzi JL, Wilson DM 3rd, 2012. Base excision repair: contribution to tumorigenesis and target in anticancer treatment paradigms. Curr. Med. Chem 19 (23), 3922–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi Tadahide, Brown David B., Naidu, Bhakat Kishor K., Macinnes Mark A., Saito Hiroshi, Chen David J., Mitra Sankar, 2005. Two essential but distinct functions of the mammalian abasic endonuclease. Proc. Natl. Acad. Sci. U.S.A 102 (16), 5739–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kategaya Lorn, Perumal Senthil K., Hager Jeffrey H., Belmont Lisa D., 2019. Werner syndrome helicase is required for the survival of cancer cells with microsatellite instability. iScience 13, 488–497. March. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinakis Apostolos, Karagiannis Dimitris, Rampias Theodoros, 2020. Targeting DNA repair in cancer: current state and novel approaches. Cell. Mol. Life Sci.: CMLS 77 (4), 677–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokan Hans E., Bjørås Magnar, 2013. Base excision repair. Cold Spring Harb. Perspect. Biol 5 (4), a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucherlapati Melanie, Yang Kan, Kuraguchi Mari, Zhao Jie, Lia Maria, Heyer Joerg, Kane Michael F., et al. , 2002. Haploinsufficiency of flap endonuclease (Fen1) leads to rapid tumor progression. Proc. Natl. Acad. Sci. U.S.A 99 (15), 9924–9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Voet A, Zhang KYJ, 2012. Fragment based drug design: from experimental to computational approaches. Curr. Med. Chem 19 (30), 5128–5147. [DOI] [PubMed] [Google Scholar]
- Lafrance-Vanasse Julien, Williams Gareth J., Tainer John A., 2015. Envisioning the dynamics and flexibility of mre11-rad50-nbs1 complex to decipher its roles in DNA replication and repair. Prog. Biophys. Mol. Biol 117 (2–3), 182–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam John S., Seligson David B., Yu Hong, Li Ai, Eeva Mervi, Pantuck Allan J., Zeng Gang, Horvath Steve, Arie S, Belldegrun, 2006. Flap endonuclease 1 is overexpressed in prostate cancer and is associated with a high Gleason score. BJU Int. 98 (2), 445–451. [DOI] [PubMed] [Google Scholar]
- Larsen Elisabeth, Gran Christine, Saether Barbro Elisabet, Seeberg Erling, Klungland Arne, 2003. Proliferation failure and gamma radiation sensitivity of Fen1 null mutant mice at the blastocyst stage. Mol. Cell Biol 23 (15), 5346–5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb Simone, Blaha-Ostermann Silvia, Kamper Elisabeth, Rippka Janine, Schwarz Cornelia, Ehrenhöfer-Wölfer Katharina, Schlattl Andreas, et al. , 2019. Werner syndrome helicase is a selective vulnerability of microsatellite instability-high tumor cells. eLife 8. 10.7554/eLife.43333. March. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Jin-Li, Wang Jian-Ping, Chang Hong, Deng Sheng-Ming, Du Jia-Hui, Wang Xiao-Xiao, Hu He-Juan, et al. , 2019. FEN1 inhibitor increases sensitivity of radiotherapy in cervical cancer cells. Canc. Med 8 (18), 7774–7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Mengxia, Wilson David M. 3rd, 2014. Human apurinic/apyrimidinic endonuclease 1. Antioxidants Redox Signal. 20 (4), 678–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T, Gally JA, Edelman GM, 1969. Deoxyribonuclease IV: a new exonuclease from mammalian tissues. Proc. Natl. Acad. Sci. U.S.A 62 (2), 597–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T, Nyberg B, 1972. Rate of depurination of native deoxyribonucleic acid. Biochemistry 11 (19), 3610–3618. [DOI] [PubMed] [Google Scholar]
- Linke Pawel, Kwame Amaning, Maschberger Melanie, Vallee Francois, Steier Valerie, Baaske Philipp, Duhr Stefan, Breitsprecher Dennis, Rak Alexey, 2016. An automated microscale thermophoresis screening approach for fragment-based lead discovery. J. Biomol. Screen 21 (4), 414–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Lili, Yan Ling, Donze Jon R., Gerson Stanton L., 2003. Blockage of abasic site repair enhances antitumor efficacy of 1,3-bis-(2-chloroethyl)-1-nitrosourea in colon tumor xenografts. Mol. Canc. Therapeut 2 (10), 1061–1066. [PubMed] [Google Scholar]
- Lolli Graziano, Caflisch Amedeo, 2016. High-throughput fragment docking into the BAZ2B bromodomain: efficient in silico screening for X-ray crystallography. ACS Chem. Biol 11 (3), 800–807. [DOI] [PubMed] [Google Scholar]
- Lou Kevin, Gilbert Luke A., Shokat Kevan M., 2019. A bounty of new challenging targets in oncology for chemical discovery. Biochemistry 58 (31), 3328–3330. [DOI] [PubMed] [Google Scholar]
- Lupo Barbara, Trusolino Livio, 2014. Inhibition of poly(ADP-ribosyl)ation in cancer: old and new paradigms revisited. Biochim. Biophys. Acta 1846 (1), 201–215. [DOI] [PubMed] [Google Scholar]
- Madhusudan Srinivasan, Smart Fiona, Paul Shrimpton, Parsons Jason L., Gardiner Laurence, Houlbrook Sue, Talbot Denis C., et al. , 2005. Isolation ofa small molecule inhibitor of DNA base excision repair. Nucleic Acids Res. 33 (15), 4711–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Jennifer, Setton Jeremy, Lee Nancy Y., Riaz Nadeem, Powell Simon N., 2018. The therapeutic significance of mutational signatures from DNA repair deficiency in cancer. Nat. Commun 9 (1), 3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Yasuo, Adachi Noritaka, Koyama Hideki, 2002. Vertebrate cells lacking FEN-1 endonuclease are viable but hypersensitive to methylating agents and H2O2. Nucleic Acids Res. 30 (14), 3273–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus Kirk J., Barrett Irene J., Nouhi Yasaman, Hieter Philip, 2009. Specific synthetic lethal killing of RAD54B-deficient human colorectal cancer cells by FEN1 silencing. Proc. Natl. Acad. Sci. U.S.A 106 (9), 3276–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill Daniel R., Whitaker Amy M., Stark Wesley J., Illuzzi Jennifer L., McKinnon Peter J., Freudenthal Bret D., Wilson David M. 3rd, 2020. Functions of the major abasic endonuclease (APE1) in cell viability and genotoxin resistance. Mutagenesis 35 (1), 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengwasser Kristen E., Adeyemi Richard O., Leng Yumei, Choi Mei Yuk, Clairmont Connor, D’Andrea Alan D., Elledge Stephen J., 2019. Genetic screens reveal FEN1 and APEX2 as BRCA2 synthetic lethal targets. Mol. Cell 73 (5), 885–899e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich Paul, 2016. Mechanisms in E. Coli and human mismatch repair (nobel lecture). Angew. Chem 55 (30), 8490–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiani Davide, Ronato Daryl A., Brosey Chris A., Arvai Andrew S., Syed Aleem, Masson Jean-Yves, Elena Petricci, Tainer John A., 2018. Targeting allostery with avatars to design inhibitors assessed by cell activity: dissecting MRE11 endo- and exonuclease activities. Methods Enzymol. 601, 205–241. February. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol CD, Izumi T, Mitra S, Tainer JA, 2000. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected]. Nature 403 (6768), 451–456. [DOI] [PubMed] [Google Scholar]
- Montelione Gaetano T., 2012. The protein structure initiative: achievements and visions for the future. F1000 Biol. Rep 4, 7. April. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi Akira, Masutani Mitsuko, Yoshioka Ken-Ichi, Bessho Tadayoshi, 2019. Aberrations in DNA repair pathways in cancer and therapeutic significances. Semin. Canc. Biol 58, 29–46. October. [DOI] [PubMed] [Google Scholar]
- Mullenders Leon H.F., 2018. Solar UV damage to cellular DNA: from mechanisms to biological effects. Photochem. Photobiol. Sci.: Off. J. Eur. Photochem. Assoc. Eur. Soc. Photobiol 17 (12), 1842–1852. [DOI] [PubMed] [Google Scholar]
- Murai Junko, Huang Shar-Yin N., Das Benu Brata, Renaud Amelie, Zhang Yiping, Doroshow James H., Ji Jiuping, Takeda Shunichi, Pommier Yves, 2012. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Canc. Res 72 (21), 5588–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray Christopher W., Blundell Tom L., 2010. Structural biology in fragment-based drug design. Curr. Opin. Struct. Biol 20 (4), 497–507. [DOI] [PubMed] [Google Scholar]
- Navratilova Iva, Hopkins Andrew L., 2010. Fragment screening by surface plasmon resonance. ACS Med. Chem. Lett 1 (1), 44–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nienaber VL, Richardson PL, Klighofer V, Bouska JJ, Giranda VL, Greer J, 2000. Discovering novel ligands for macromolecules using X-ray crystallographic screening. Nat. Biotechnol 18 (10), 1105–1108. [DOI] [PubMed] [Google Scholar]
- Nikolova Teodora, Christmann Markus, Kaina Bernd, 2009. FEN1 is overexpressed in testis, lung and brain tumors. Anticancer Res. 29 (7), 2453–2459. [PubMed] [Google Scholar]
- Parker Christopher G., Galmozzi Andrea, Wang Yujia, Correia Bruno E., Sasaki Kenji, Joslyn Christopher M., Kim Arthur S., et al. , 2017. Ligand and target discovery by fragment-based screening in human cells. Cell 168 (3), 527–541 e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull Tanya T., Martin Gellert, 1998. The30 to50exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol. Cell 1 (7), 969–979. [DOI] [PubMed] [Google Scholar]
- Pearce Nicholas M., Krojer Tobias, Bradley Anthony R., Collins Patrick, Nowak Radosław P., Talon Romain, Marsden Brian D., et al. , 2017. A multicrystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density. Nat. Commun 8, 15123. April. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg Anthony E., 2011. Multifaceted roles of alkyltransferase and related proteins in DNA repair, DNA damage, resistance to chemotherapy, and Research tools. Chem. Res. Toxicol 24 (5), 618–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Pel Derek M., Barrett Irene J., Shimizu Yoko, Sajesh Babu V., Guppy Brent J., Pfeifer Tom, McManus Kirk J., Hieter Philip, 2013. An evolutionarily conserved synthetic lethal interaction network identifies FEN1 as a broad-spectrum target for anticancer therapeutic development. PLoS Genet. 9 (1), e1003254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J, Jefferson P, Yannone Steven M., Holden Lauren G., Hitomi Chiharu, Asaithamby Aroumougame, Han Seungil, Priscilla K, Cooper David J. Chen Tainer, John A, 2006. WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing. Nat. Struct. Mol. Biol 13 (5), 414–422. [DOI] [PubMed] [Google Scholar]
- Pilié Patrick G., Tang Chad, Mills Gordon B., Yap Timothy A., 2019. State-of-the-Art strategies for targeting the DNA damage response in cancer. Nat. Rev. Clin. Oncol 16 (2), 81–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price Amanda J., Howard Steven, Cons Benjamin D., 2017. Fragment-based drug discovery and its application to challenging drug targets. Essays Biochem. 61 (5), 475–484. [DOI] [PubMed] [Google Scholar]
- Rai Ganesha, Vyjayanti Vaddadi N., Dorjsuren Dorjbal, Simeonov Anton, Jadhav Ajit, Wilson David M. 3rd, Maloney David J., 2012. Synthesis, biological evaluation, and structure-activity relationships of a novel class of apurinic/apyrimidinic endonuclease 1 inhibitors. J. Med. Chem 55 (7), 3101–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai Ganesha, Vyjayanti Vaddadi N., Dorjsuren Dorjbal, Simeonov Anton, Jadhav Ajit, Wilson David M., Maloney David J., 2013. Small molecule inhibitors of the human apurinic/apyrimidinic endonuclease 1 (APE1). In: Probe Reports from the NIH Molecular Libraries Program [Internet]. National Center for Biotechnology Information (US). [PubMed] [Google Scholar]
- Romero-Laorden Nuria, Castro Elena, 2017. Inherited mutations in DNA repair genes and cancer risk. Curr. Probl. Canc 41 (4), 251–264. [DOI] [PubMed] [Google Scholar]
- Saur Michael, Hartshorn Michael J., Dong Jing, Reeks Judith, Bunkoczi Gabor, Jhoti Harren, Pamela A, Williams, 2020. Fragment-based drug discovery using cryo-EM. Drug Discov. Today 25 (3), 485–490. [DOI] [PubMed] [Google Scholar]
- Schiebel Johannes, Radeva Nedyalka, Köster Helene, Metz Alexander, Krotzky Timo, Kuhnert Maren, Diederich Wibke E., et al. , 2015. One question, multiple answers: biochemical and biophysical screening methods retrieve deviating fragment hit lists. ChemMedChem 10 (9), 1511–1521. [DOI] [PubMed] [Google Scholar]
- Scully Ralph, Panday Arvind, Elango Rajula, Willis Nicholas A., 2019. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol 20 (11), 698–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiple Lauren A., Cardellina John H. 2nd, Akee Rhone, Stivers James T., 2008. Potent inhibition of human apurinic/apyrimidinic endonuclease 1 by arylstibonic acids. Mol. Pharmacol 73 (3), 669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Atsushi, Moiani Davide, Arvai Andrew S., Perry Jefferson, Harding Shane M., Genois Marie-Michelle, Maity Ranjan, et al. , 2014. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell 53 (1), 7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Dong Hae, Hou Jingtong, Chandonia John-Marc, Das Debanu, Choi In-Geol, Kim Rosalind, Kim Sung-Hou, 2007. Structure-based inference of molecular functions of proteins of unknown function from Berkeley structural genomics center. J. Struct. Funct. Genom 8 (2–3), 99–105. [DOI] [PubMed] [Google Scholar]
- Shuker SB, Hajduk PJ, Meadows RP, Fesik SW, 1996. Discovering high-affinity ligands for proteins: SAR by NMR. Science 274 (5292), 1531–1534. [DOI] [PubMed] [Google Scholar]
- Silber John R., Bobola Michael S., Blank A, Schoeler Kathryn D., Haroldson Peter D., Huynh Mary B., Kolstoe Douglas D., 2002. The apurinic/apyrimidinic endonuclease activity of ape1/ref-1 contributes to human glioma cell resistance to alkylating agents and is elevated by oxidative stress. Clin. Canc. Res.: Off. J. Am. Assoc. Canc. Res 8 (9), 3008–3018. [PubMed] [Google Scholar]
- Silvestre Hernani Leonardo, Blundell Thomas L., Abell Chris, Ciulli Alessio, 2013. Integrated biophysical approach to fragment screening and validation for fragment-based lead discovery. Proc. Natl. Acad. Sci. U.S.A 110 (32), 12984–12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeonov Anton, Kulkarni Avanti, Dorjsuren Dorjbal, Jadhav Ajit, Shen Min, McNeill Daniel R., Austin Christopher P., Wilson David M. 3rd, 2009. Identification and characterization of inhibitors of human apurinic/apyrimidinic endonuclease APE1. PloS One 4 (6), e5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh Isha, Laos Roberto, Hoshika Shuichi, Benner Steven A., Georgiadis Millie M., 2018. Snapshots of an evolved DNA polymerase pre- and post-incorporation of an unnatural nucleotide. Nucleic Acids Res. 46 (15), 7977–7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll Jennifer M., Sobol Robert W., Mosammaparast Nima, 2017. Regulation of DNA alkylation damage repair: lessons and therapeutic opportunities. Trends Biochem. Sci 42 (3), 206–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana Rebeka, McNeill Daniel R., Abbotts Rachel, Mohammed Mohammed Z., Zdzienicka Malgorzata Z., Qutob Haitham, Seedhouse Claire, et al. , 2012. Synthetic lethal targeting of DNA double-strand break repair deficient cells by human apurinic/apyrimidinic endonuclease inhibitors. J. Int. Canc 131 (10), 2433–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Yueru, McCorvie Thomas J., Yates Luke A., Zhang Xiaodong, 2020. Structural basis of homologous recombination. Cell. Mol. Life Sci.: CMLS 77 (1), 3–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed Aleem, Tainer John A., 2018. The MRE11-RAD50-NBS1 complex conducts the orchestration of damage signaling and outcomes to stress in DNA replication and repair. Annu. Rev. Biochem 87, 263–294. June. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari Vinod, Wilson David M. 3rd, 2019. DNA damage and associated DNA repair defects in disease and premature aging. Am. J. Hum. Genet 105 (2), 237–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgovnick Alessandro, Schumacher Bjo€rn, 2015. DNA repair mechanisms in cancer development and therapy. Front. Genet 6, 157. April. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trilles Richard, Beglov Dmitri, Chen Qiujia, He Hongzhen, Randall Wireman, Reed April, Chennamadhavuni Spandan, et al. , 2019. Discovery of macrocyclic inhibitors of apurinic/apyrimidinic endonuclease 1. J. Med. Chem 62 (4), 1971–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa Susan E., Lafrance-Vanasse Julien, Tainer John A., 2014. The cutting edges in DNA repair, licensing, and fidelity: DNA and RNA repair nucleases sculpt DNA to measure twice, cut once. DNA Repair 19, 95–107. July. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa Susan E., Sarker Altaf H., Ng Clifford, Arvai Andrew S., Shin David S., Shih Brian, Jiang Shuai, et al. , 2020. Human XPG nuclease structure, assembly, and activities with insights for neurodegeneration and cancer from pathogenic mutations. Proc. Natl. Acad. Sci. U.S.A 117 (25), 14127–14138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa Susan E., Shin David S., Mol Clifford D., Izumi Tadahide, Arvai Andrew S., Mantha Anil K., Szczesny Bartosz, et al. , 2013. Conserved structural chemistry for incision activity in structurally non-homologous apurinic/apyrimidinic endonuclease APE1 and endonuclease IV DNA repair enzymes. J. Biol. Chem 10.1074/jbc.m112.422774. [DOI] [PMC free article] [PubMed]
- Tsutakawa Susan E., Thompson Mark J., Arvai Andrew S., Neil Alexander J., Shaw Steven J., Algasaier Sana I., Kim Jane C., et al. , 2017. Phosphate steering by flap endonuclease 1 promotes 50-flap specificity and incision to prevent genome instability. Nat. Commun 8 (1), 15855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlinde Christophe L.M. J., Christophe LM, Kim Hidong, Bernstein Bradley E., Mande Shekhar C., Hol Wim G.J., 2018. Antitrypanosomiasis drug development based on structures of glycolytic enzymes. Struct. Based Drug Des 10.1201/9780203738023-15. [DOI]
- Wait Scott D., Prabhu Roshan S., Burri Stuart H., Atkins Tyler G., Asher Anthony L., 2015. Polymeric drug delivery for the treatment of glioblastoma. Neuro Oncol. 17 (Suppl. 2) (March): ii9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LJ, Craig RB, Harris AL, Hickson ID, 1994. A role for the human DNA repair enzyme HAP1 in cellular protection against DNA damaging agents and hypoxic stress. Nucleic Acids Res. 22 (23), 4884–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace Susan S., 2014. Base excision repair: a critical player in many games. DNA Repair. 10.1016/j.dnarep.2014.03.030. [DOI] [PMC free article] [PubMed]
- Wang Dong, Luo Meihua, Mark R, Kelley, 2004. Human apurinic endonuclease1(APE1) expression and prognostic significance in osteosarcoma: enhanced sensitivity of osteosarcoma to DNA damaging agents using silencing RNA APE1 expression inhibition. Mol. Canc. Therapeut 3 (6), 679–686. [PubMed] [Google Scholar]
- Ward Thomas A., McHugh Peter J., Durant Stephen T., 2017. Small molecule inhibitors uncover synthetic genetic interactions of human flap endonuclease1 (FEN1) with DNA damage response genes. PloS One 12 (6), e0179278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wartchow Charles A., Frank Podlaski, Li Shirley, Rowan Karen, Zhang Xiaolei, Mark David, Huang Kuo-Sen, 2011. Biosensor-based small molecule fragment screening with biolayer interferometry. J. Comput. Aided Mol. Des 25 (7), 669–676. [DOI] [PubMed] [Google Scholar]
- Wilson David M. 3rd, Simeonov Anton, 2010. Small molecule inhibitors of DNA repair nuclease activities of APE1. Cell. Mol. Life Sci.: CMLS 67 (21), 3621–3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DM 3rd, Barsky D, 2001. The major human abasic endonuclease: formation, consequences and repair of abasic lesions in DNA. Mutat. Res 485 (4), 283–307. [DOI] [PubMed] [Google Scholar]
- Xie Chunhong, Wang Kejia, Chen Daorong, 2016. Flap endonuclease 1 silencing is associated with increasing the cisplatin sensitivity of SGC-7901 gastric cancer cells. Mol. Med. Rep 13 (1), 386–392. [DOI] [PubMed] [Google Scholar]
- Yap Timothy A., Plummer Ruth, Azad Nilofer S., Thomas Helleday, 2019. The DNA damaging revolution: PARP inhibitors and beyond. In: American Society of Clinical Oncology Educational Book. American Society of Clinical Oncology. Annual Meeting, vol. 39, pp. 185–195. January. [DOI] [PubMed] [Google Scholar]
- Yar M. Shahar, Haider, Kashif, Gohel, Vivek, Siddiqui Ali, Nasir, Ahmed, Kamal, 2020. Synthetic lethality on drug discovery: an update on cancer therapy. Expet Opin. Drug Discov 1–10. March. [DOI] [PubMed]
- Zarembinski TI, Hung LW, Mueller-Dieckmann HJ, Kim KK, Yokota H, Kim R, Kim SH, 1998. Structure-based assignment of the biochemical function of a hypothetical protein: a test case of structural genomics. Proc. Natl. Acad. Sci. U.S.A 95 (26), 15189–15193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawahir Zahrah, Dayam Raveendra, Deng Jinxia, Pereira Cherelene, Neamati Nouri, 2009. Pharmacophore guided discovery of small-molecule human apurinic/apyrimidinic endonuclease 1 inhibitors. J. Med. Chem 52 (1), 20–32. [DOI] [PubMed] [Google Scholar]
- Zheng Li, Jia Jia, David Finger L, Guo Zhigang, Zer Cindy, Shen Binghui, 2011. Functional regulation of FEN1 nuclease and its link to cancer. Nucleic Acids Res. 39 (3), 781–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Li, Zhou Mian, Chai Qing, Parrish Jay, Ding Xue, Patrick Steve M., Turchi John J., Yannone Steven M., Chen David, Shen Binghui, 2005. Novel function of the flap endonuclease 1 complex in processing stalled DNA replication forks. EMBO Rep. 6 (1), 83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann Michal, Murina Olga, Reijns Martin A.M., Angelo Agathanggelou, Challis Rachel, Tarnauskaite Žygimantė, Muir Morwenna, et al. , 2018. CRISPR screens identify genomic ribonucleotides as a source of PARP-trapping lesions. Nature 559 (7713), 285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]