Abstract
Background and aim
The ongoing global pandemic due to SARS-CoV-2 caused a medical emergency. Since December 2019, the COVID-19 disease is spread across the globe through physical contact and respiratory droplets. Coronavirus caused a severe effect on the human immune system where some of the non-structural proteins (nsp) are involved in virus-mediated immune response and pathogenesis. To suppress the viral RNA replication mechanism and immune-mediated responses, we aimed to identify limonoids and triterpenoids as antagonists by targeting helicases (nsp13), exonuclease (nsp14), and endoribonuclease (nsp15) of SARS-CoV-2 as therapeutic proteins.
Experimental procedure
In silico molecular docking and drug-likeness of a library of 369 phytochemicals from limonoids and triterpenoids were performed to screen the potential hits that binds effectively at the active site of the proteins target. In addition, the molecular dynamics simulations of the proteins and their complexes with the potential hits were performed for 100 ns by using GROMACS.
Results and conclusion
The potential compounds 26-deoxyactein and 25-O-anhydrocimigenol 3-O-beta-d-xylopyranoside posing strong interactions with a minimum binding energy of −10.1 and −9.5 kcal/mol, respectively and sustained close contact with nsp13 for 100 ns. The nsp14 replication fork activity was hindered by the tomentosolic acid, timosaponin A-I, and shizukaol A with the binding affinity score of −9.2, −9.2, and −9.0 kcal/mol, respectively. The nsp15 endoribonuclease catalytic residues were inhibited potentially by limonin, 25-O-anhydrocimigenol 3-O-alpha-l-arabinopyranoside, and asperagenin posing strong binding affinity scores of −9.0, −8.8, and −8.7 kcal/mol, respectively. Computationally predicted potential phytochemicals for SARS-CoV-2 are known to possess various medicinal properties.
Keywords: COVID-19, Limonoids, Triterpenoids, Molecular docking, Dynamics simulation
Graphical abstract
Highlights
-
•
Protein-ligand docking was performed to search potential phytochemicals against SARS-CoV-2.
-
•
A library of 369 limonoids and triterpenoids were screened against the protein targets nsp13, nsp14, and nsp15.
-
•
Roles of the proteins nsp13, nsp14, and nsp15 of SARS-CoV-2 are discussed.
-
•
Molecular dynamics simulations of the proteins and their complexes with some potential hits were performed.
1. Introduction
After the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in December 2019, the fight against COVID-19 pandemic is still underway globally due to the continuous increase in infected cases and deaths.1,2 Structurally, SARS-CoV-2 has a positive sense single-stranded viral RNA genome of length ∼30 kb with 16 non-structural proteins (nsp1 to nsp16), 3 structural proteins, and 8 accessory proteins.3 Therapeutic targets such as papain-like protease (PLpro), 3C-like protease (3CLpro), RNA-dependent RNA polymerase (RdRp), and spike glycoprotein (SGp) are targeted primarily for repositioning approved drugs and developing adjuvant therapies with phytochemicals and vaccines.4, 5, 6, 7
The three less focused targets, i.e., nsp13, nsp14, and nsp15 are also the critical components of the replication-transcription process to promote the life cycle of SARS-CoV-2. The nsp13 belongs to the helicase superfamily-1 driven by the nucleoside triphosphate hydrolase (NTPase), and the RNA helicase activity is pivotal in SARS-CoV-2 RNA replication.8,9 In nsp13, the zinc-binding sites that function with the involvement of divalent metallic ions, i.e., Mg2+, Mn2+, Ca2+, or Zn2+ critical in NTPase and ATPase activity.10 Some studies have highlighted the importance of nsp13 for its inhibitory role in regulating type I interferon production, as well as the suppression of interferons (IFNs) levels due to nsp13 overexpression via the host USP13.11 Some recent in silico studies have proposed cepharanthine, idarubicin, and nilotinib as potential inhibitory agents against nsp13.12 The uridine-specific endoribonuclease nsp15 is highly conserved in the coronavirus family at the C-terminal catalytic domain to recognize uridine moiety, and it is also responsible for the interference of the host immune system.13 There is a scope for experimenting and validating the key functions of nsp15 endoribonuclease with the host system, the uridine vanadate interaction with the catalytic residues including (HIS235, HIS230, LYS290, THR341, and GLY247) proposed to be the therapeutic target site to screen potential inhibitors. Along with nsp13, nsp15 also requires Mn2+ metal ions for various key functions.14 Tiparacil and uridine 2′,3′-vanadate have been reported to inhibit the putative active and catalytic sites of nsp15.13 The nsp14 N-terminal domain possesses exoribonucleolytic activity that belongs to the DEDDh exonucleases super family. An exonucleolytic activity is crucial for the RNA proofreading activity found in coronavirus replication is an exceptional feature not found in other viruses, which is critical for nidoviral evolution and genome expansion. It involves catalysis for the excision of endonucleoside monophosphates from nucleic acid.15 The nsp14 requires two divalent metal ions and reactive water molecules. The nsp14 exonuclease requires divalent cations like Mg2+, Mn2+, Ca2+, Ni2+, Cu2+, Co2+, or Zn2+ for inducing structural changes and reaction activity.16,17 The proofreading activity of nsp14 was associated with nsp10, and this activity could be hindered by sofosbuvir.18
Phytochemicals have a profound role in inhibiting the viral targets of SARS-CoV-2. The validated virtual screening of natural compounds was found to deliver antagonist activity against the therapeutic target in vitro and in vivo experiments. Some examples justify the antagonism, i.e., betulonic acid, savinin, bavachinin, neobavaisoflavone, kaempferol, quercetin, catechin and curcumin could potentially inhibit SARS-CoV-2 3CLpro. Some triterpenoids like limonin, ursolic acid, glycyrrhizin, and 7-deacetyl-7-benzoylepoxyazadiradione possess activity against Mpro, RdRp, and SGp-RBD.19, 20, 21, 22 The active ingredient glycyrrhizin from Glycyrrhiza radix can inhibit the replication of SARS-CoV related viruses with minimal toxic effects.19 The natural products such as berberine, baicalin, resveratrol, catechin, procyanidins are reported to interfere with SARS-CoV-2 cellular infection and replication through their action on autophagy.22 The molecular docking studies of raddeanine, tomentodiplacone B, osajin, sesquiterpene glycoside, rhamnetin, silydianin, and eryvarinM showed higher binding affinity at the active site of nsp13.23 Similarly, guanosine-P3-adenosine-5′,5′-triphosphate (G3A), and TCM 57025 pose strong binding affinity to guanine-N7 methyltransferase of nsp14.24 Sarsasapogenin, ursolic acid, ajmalicine, silymarin, curcumin, gingerol and rosmarinic acid were found to be effective inhibitors against nsp15.25 In a recent experimental study, glycyrrhizic acid nanoparticles were developed and applied against SARS-CoV-2.26 Betulonic acid derivatives interfere with hCoV229E nsp15 mutant active site replication with an inhibitory concentration EC50 of 0.6 μM.27
Considering the role of nsp13, nsp14, and nsp15 in the SARS-CoV-2 life cycle including the importance of phytochemicals in formulating adjuvant therapies against COVID-19, as a part of our ongoing research,20,28 we are interested in finding promising lead compounds from limonoids and triterpenoids. We performed a computational screening for 369 limonoids and triterpenoids using molecular docking and pharmacokinetic properties, and proposed potential lead phytochemicals that could inhibit therapeutic targets and suppress the SARS-CoV-2 life cycle. In addition, the molecular dynamics simulations of docked complexes were performed for 100 ns to study the stability and flexibility of ligands at active site of target protein.
2. Computational methods
2.1. Preparation of macromolecules
The FASTA sequences of helicases (nsp13), endonucleases (nsp14), and exonucleases (nsp15) were retrieved from NCBI databases with gene ID: QHD43415. The secondary structures were modelled using Modeller 10.0 protein structure modelling tool.29 For nsp13 modelling, we chose X-ray crystallography structures, i.e., 5RL6, 6ZSL, 6XEZ, 6JYT retrieved from the RCSB protein data bank. The protein templates 5C8T, 7MC5, 7MC6 and 7D1Y were used for nsp14, whereas for nsp15 6WLC, 6VWW, 7KEG, and 7MED. The modelled protein structures were validated through Ramachandran plot, ERRAT plot, and Verify 3D structure validation by online tool (https://saves.mbi.ucla.edu/).30
2.2. Preparation of ligands library
The 3D structure of 369 phytochemicals (limonoids and triterpenoids) were collected from the Chemical Entities of Biological Interest (ChEBI) database (Table S1). The structure data files in ‘.sdf’ were converted to ‘.pdb’ format using the Open Babel 2.3.2 tool. Screening of 369 phytochemicals for ADME properties was performed by using the Swiss ADME online tool (http://www.swissadme.ch/index.php).31
2.3. Molecular docking
The Autodock4 tools was used to generate the ‘pdbqt’ file of the proteins and ligands.32 The grid coordinates for the site-specific and random docking are given in Table S2. The molecular docking is performed using the AutoDock Vina 1.1.2 scoring function, whereas the strawberry-pearl 5.32.1.1 script was used to perform virtual screening.33 The Lamarckian genetic algorithm (LGA) was used for the molecular docking studies. The docked structures were analysed by using Biovia Discovery studio 2021.
The free energy of the protein-ligand interaction was computed by AutoDock 4.2. The free energy of binding is calculated as ΔG (bind) = ΔH - TΔS, where the ΔH represents the enthalpic, and TΔS the entropic contribution (only a negative ΔG value is energetically favourable). The energy of ligand and protein in the unbound state was estimated first, and then the energy of the protein-ligand complex was computed. The difference was used to estimate ΔG by following equation.34,35
| ΔG= (VboundL−L - VunboundL−L) + (VboundP−P - VunboundP−P) + (VboundP−L - VunboundP−L +ΔSconf) |
2.4. Molecular dynamics simulations
The molecular dynamics simulation was performed by using the Groningen machine chemical simulations (Gromacs 2019.2).36 The best docked confirmation was chosen for molecular dynamics simulations. The ligand topologies were built by using the PRODRG server. Protein parameters were generated by using the GROMOS96 43a1 force fields. For the solvation system, we have generated a triclinic simulation box fitting the complex provided by the SPC water model. In addition, 0.15 M sodium chloride has been added to the system. The total system is set up for energy minimization and neutralisation of 5000 steps for structure optimizations. The system had been equilibrated for 300 K temperature and 1.0 bar pressure, and these parameters were set for 150000 steps to perform simulations. The equilibrated system is set for MD simulations run up to 50000000 steps per 100 ns on an Intel (R) Xeon (R) CPU E5-2680v4@2.40GHz computer. We analysed the molecular dynamics simulation data and position restraints of the docking complex at 100 ns trajectories by using Graphpad Prism 6.0 software. The simulation data was analysed for RMSD (root mean square deviation) of the given structure over time, RMSF (root mean square fluctuation) of each residue in the given structure, radius of gyration, solvent-accessible surface area, and average number of H-bonds in each frame over time. Thousands of frames of simulation structures were interpreted using the VMD and Pymol visualization tools.
3. Results and discussion
3.1. Proteins validation and phytochemicals
The ion-dependent enzymes helicases (nsp13), exonucleases (nsp14), and endoribonuclease (nsp15) are involved in viral replication and proliferation activities of SARS-CoV-2. The protein structure was first prepared and validated. The DOPE scores of the best models of nsp13, nsp14, and nsp15 are given in Table S3. Protein structure validations of nsp13, nsp14, and nsp15 by Ramachandran plot, Errat plot, and Verify 3D are shown in Fig. S1-S9. The overall quality of the target proteins nsp13, nsp14, and nsp15 was 94.49, 98, and 96.99%, whereas the verified 3D structure quality was 95.78, 87.81, and 88.4%, respectively.
The tremendous research scope and therapeutic activities of limonoids and triterpenoids have been proven and reported in studies against SARS-CoV and SARS-CoV-2 via in silico, in vitro, and in vivo studies. A library of 369 limonoids and triterpenoids were created and screened for ADME properties (Table S4) and molecular docking studies with nsp13, nsp14 and nsp15 (Table S1). The triterpenoid, i.e., glycyrrhizin found in licorice roots effectively blocks the main protease activity of SARS-CoV and SARS-CoV-2.66,67 Betunolic acid and betulinic acid showed IC50 values ranging between 0.63 and 2.4 μM against SARS-CoV.68 Ursonic and ursolic acid potentially interact with SARS-CoV-2 endoribonuclease, which is essential for the lifecycle of coronaviruses.69 Experimental evidence highlighted limonin is useful against HIV-I and HTLV-I, whereas the theoretical studies supported inhibitory properties against SARS-CoV-2 RdRp.20 Limonin and limonin glycoside pose strong affinity against TMPRSS2 and furin by blocking SARS-CoV-2 spike glycoprotein interacting with proteolytic cleavage sites.28,70 Some of the phytochemicals reported for anti-cancerous properties, like lucidenic acid A, are a tetracyclic triterpenoid found in ganoderma lucidum showed anti-cancerous and antioxidant activities.38 Also, lucidenic A acid possesses cytotoxicity against P388 cancer cells at 17 nM. It is also considered as an active promoter of MMP-2 (matrix metallo proteases-2).55,56 These research outcomes encouraged us to develop a library of limonoids and triterpenoids for screening potential phytochemicals against the three key therapeutic proteins targets nsp13, nsp14 and nsp15 of SARS-CoV-2.
3.2. Screening of inhibitors for nsp13
Helicases, the NTP-dependent enzymes are widely spread in various kingdoms including the (+)RNA viruses with a genome greater than 7 kb.48 They play a vital role in unwinding the dsDNA/dsRNA substrates, displacing the proteins that are bound to nucleic acid, remodeling the DNA or RNA secondary structures, and translocating along with the double-stranded nucleic acid without unwinding.49 The nsp13 of SARS-CoV-2 has 596 amino acids. These amino acids are organised in a triangular pyramid shape with two RecA-like domains IA and IIA, and the domain IB forms a triangular base. The N-terminal zinc-binding domain is connected to the IB domain, and the stalk domain forms the apex of the pyramid and belongs to the SF1 super family of helicases (Fig. 1a).50 Nsp13 also shares similar structural features with eukaryotic Upf1 helicases. This also exhibits various enzymatic processes such as the hydrolysis of NTP during the capping mechanism, unwinding of RNA duplexes with 50-30 directionality, and the RNA triphosphatase activity.51 Additionally, RNA unwinding activity stimulated by the interaction with the RdRp, nsp12 and nsp13 is highly conserved in all coronaviruses and acts as a key enzyme in viral replication.52 In this context, a potent non-competitive inhibitor (SSYA10-001) blocks viral replication by inhibiting the unwinding activity of the helicases nsp13 in SARS-CoV, MHV and MERS.53,54
Fig. 1.
Results of molecular docking: a. helicases nsp13 protein structure domains and active sites represented in color and labeled, and best dock pose of 5 compounds (26-deoxyactein, 25-O-anhydrocimigenol 3-O-beta-d-xylopyranoside, 21-hydroxyisoglabrolide, 25-O-anhydrocimigenol 3-O-alpha-l-arabinopyranoside, timosaponin A-I represented in cyan, metallic grey, violet, brown and thick grey, respectively). b. The surface structure of nsp13 with compounds binding at the active site. c. Ligand position (RMSD) and binding affinity score dot plot. d. 2D structure of 26-deoxyactein interacting with nsp13 active site residues.
The study of ADME properties of 369 phytochemicals and their dock score at the active site of nsp13 showing more than SSYA10 resulted several lead compounds are tabulated in Table S5. The dock score and inhibition constant of the five best compounds (26-deoxyactein, 21-hydroxyisoglabrolide, 3-O-alpha-l-arabinopyranoside, 25-O-anhydrocimigenol 3-O-beta-d-xylopyranoside and timosaponin A-I) along with their interaction with the residues of nsp13 are summarized in Table 1. As shown in Fig. 1, 26-deoxyactein is covalently bound to LYS290, GLY289, THR288, GLY287, GLY540 and ARG445 residues. The hydrophobic surface of the protein attracts the ligand carbon backbone by Van der Waal forces with GLU321, SER291, ALA318, PRO285 and PRO286 residues. The strong engagement of the complex possesses binding affinity of −10.1 kcal/mol and a predicted inhibition concentration of 223.38 nM (Table 1). 25-O-anhydrocimigenol 3-O-beta-d-xylopyranoside was found to be a potent inhibitor of helicases by suppressing catalytic activity with covalent binding to GLU321, GLY289, THR288, LYS290, GLY287, ARG445, GLY540 and strong Van der Waal forces interactions with ALA318, SER291, PRO285, PRO286, LYS322, LYS325 residues with −9.7 kca/mol binding affinity and 4.04 μM inhibitory concentration (Table 1). From the top five compounds, 21-hydroxyisoglabrolide, a glycyrrhetol, has been suggested as a promising drug compound. It is a triterpenoid with six isoprene units and a weak basic nature, posing strong interactions at the active sites with a minimum binding energy of −9.7 kcal/mol and a predicted inhibitory concentration of 965.44 nM. The possible interaction of the protein with ligand bound to catalytic sites confirms the inhibitory nature and it is binding with LYS290, THR288, GLY289, ARG445 residues. The lead compound 25-O-anhydrocimigenol 3-O-alpha-l-arabinopyranoside is showing efficient binding affinity by forming Pi-alkyl and hydrophobic interactions with ALA315, ALA314, ALA318, HIS292 residues at the active sites of nsp13. It exhibits inhibitory charectersitics with predicted concentrations of 2.26 μM and interacts with the catalytic sites with a minimum binding energy of −9.7 kcal/mol. Timosaponin also binds to the active pocket residues GLU377 and LYS290 with a minimum binding energy and inhibitory concentrations of −9.7 kcal/mol and 4.44 μM, respectively.
Table 1.
Best five compounds pose at the nsp13 catalytic sites with predicted inhibition constant (Ki).
| Sr. No. | Compounds | Autodock Vina Binding affinity score | Autodock Free energy of binding kcal/mol | Inhibition constant (Ki) | Hydrogen bond interactions | Van der Waal interactions |
|---|---|---|---|---|---|---|
| 1 | 26-Deoxyactein | −10.1 | −9.07 | 223.38 nM | GLY289, THR288, LYS290, GLY287, GLY540, ARG445, LYS325, LEU324 | GLU321, SER291, ALA318, PRO285, PRO286, LYS322, TYR326 |
| 2 | 21-Hydroxyisoglabrolide | −9.7 | −8.21 | 965.44 nM | LYS290, THR288, GLY289, ARG445 | GLU321, ALA318, SER291, GLY540, LYS322, GLU542, GLY287 |
| 3 | 25-O-Anhydrocimigenol 3-O-alpha-l-arabinopyranoside | −9.7 | −7.7 | 2.26 uM | ARG444, Pi-Alkyl: ALA315, ALA314, ALA318, HIS292 | GLU263, LYS322, SER291, GLY289, GLY287, ARG445, LYS290ASP376, GLY540, GLU377, SER312 |
| 4 | Timosaponin A-I | −9.7 | −7.36 | 4.04 uM | GLU377, LYS290 | ASP376, ALA315, ALA318, SER291, LYS322, GLY289, GLY287, GLY287, ARG444, LYS467, GLU542, ARG445, GLN406, GLY540, GLN539 |
| 5 | 25-O-Anhydrocimigenol 3-O-beta-d-xylopyranoside | −9.5 | −8.28 | 852.68 nM | GLU321, GLY289, THR288, LYS290, GLY287, ARG445, GLY540 | ALA318, SER291, PRO285, PRO286, LYS322, LYS325 |
The literature revealed that 26-deoxyactein is a tetracyclic triterpenoid isolated from the rhizomes of cimicifuga racemose herbs and showed the therapeutic activities against HL60 (human promyelocytic leukemia), A549 (human epithelial cell line), AGZY (human lung adenocarcinoma cell lines), and U937 (human histiocytic lymphoma) via cell cycle arrest and angiogenesis inhibition.57 It is also found in Actaea racemose and showed anti-multiple myeloma activity.37 The rhizomes of actaea recemosa contain 25-O-anhydrocimigenol 3-O-beta-d-xylopyranoside and 25-O-anhydrocimigenol 3-O-alpha-l-arabinopyranoside, which have been found to be cytotoxic against MCF-7 and HepG2 cancer cells.39,40 Timosaponin A-I is found in the rhizomes of Anemarrhena asphodeloides Bunge is reported for the treatment of diabetes mellitus, inhibiting dipeptidyl peptidase-4 DPP-4 activity at a concentration of 5–50 μM in a dose-dependent manner.41
Molecular dynamics properties of nsp13 and the complexes with two lead compounds 26-deoxyactein and 25-O-anhydrocimigenol 3-O-beta-d-xylopyranoside were investigated by simulating the structures for 100 ns. The solvated system was wrapped into the triclinic cell for equilibration and energy optimization of structure set for 5000 steps by steepest descent integrator and NVT/NPT set for 50 million steps for calculating the dynamics properties, i.e., temperature, pressure, density and total energy. The total system was found to have 864 donors and 1656 acceptors, including the ligand hetero atoms. The molecular dynamics calculations are ensured by the average RMSD calculations plot shown in Fig. 2a. The average root mean square deviation in 100 ns of nsp13 and its complexes with 26-deoxyactein and 25-O-anhydrocimigenol 3-O-beta-d-xylopyranoside is 0.33 nm and 0.34 nm, respectively. Nsp13 is regulated after very close binding with lead compounds at 20–100 ns simulation time. The equilibrated complex simulation trajectories revealed that the ligands bound to nsp13 protein were stable. The residual fluctuations during the simulation of ligand-bound protein dynamics were calculated as RMSF was shown in Fig. 2b. At the ligand-protein active site interactions, the residual fluctuations were observed. The peaks in the figure depict the behaviour of nsp13 in relation to ligands (26-deoxyactein and 25-O-anhydrocimigenol 3-O-beta-d-xylopyranoside) bound to the residues 16–75, 280–290, 404–450, and 535–570 (Fig. 2b). The helicases protein's zinc-binding sites, nucleic acid binding sites, and ATP binding sites have maximum RMSD peaks between 0.4 nm and 0.8 nm (Fig. 2a). The free energy calculations for 26-deoxyactein and 25-O-anhydrocimigenol 3-O-beta-d-xylopyranoside with nsp13 are −1372.44 kcal/mol and −1372.46 kcal/mol, respectively. By using the AutoDock scoring function, we estimated the inhibitory concentration (Ki) of 223.38 nM and 852.68 nM (Table 1), respectively. The quantified radius of gyration, solvent-accessible surface area, and average number of H-bonds in each frame over time of the ligand and receptor, notably 26-deoxyactein and 25-O-anhydrocimigenol 3-O-beta-d-xylopyranoside are shown in Fig S10 and Fig S11, respectively.
Fig. 2.
Results of MD simulation: (a) RMSD of solvated nsp13 protein, nsp13 complexed with 26-deoxyactein and 25-O-anhydrocimigenol 3-O-beta-d-xylopyranoside during 100 ns MD simulation represented in green, red and blue colour, respectively. (b) RMSF values of nsp13 complexes with 26-deoxyactein and 25-O-anhydrocimigenol 3-O-beta-d-xylopyranoside shown in green, red and blue colour, respectively.
3.3. Screening of inhibitors for nsp14
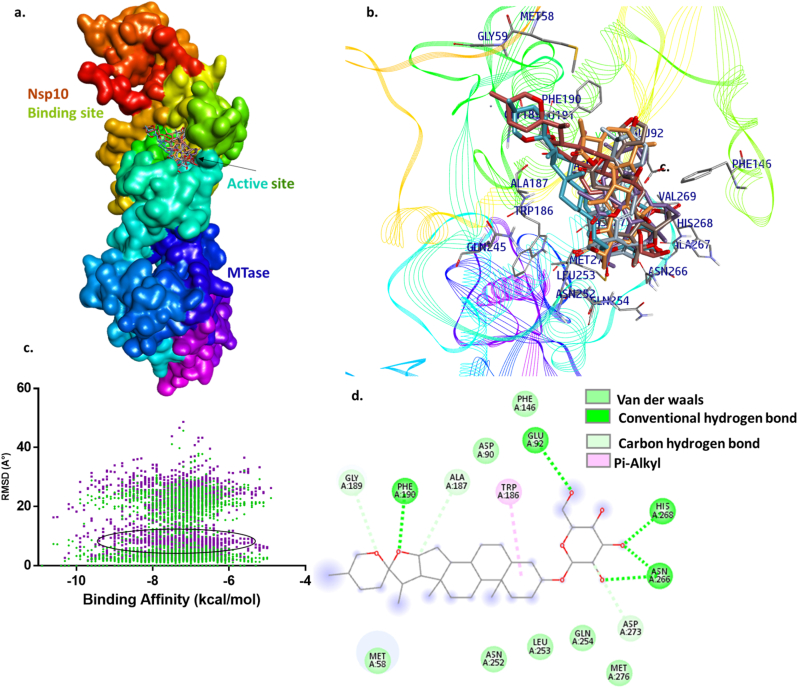
Nsp14 is an N-terminal exonuclease, a C-terminal N7-methyltransferase and a bifunctional enzyme for mRNA capping that is involved in replication.58 During the virus life cycle, nsp14 exhibits specific processes including pathogenicity, genome recombination, and innate immune responses. From the 3’ end of the growing RNA strand, the mismatched nucleotides were removed by the exoN functional domain, which corrects the strand for errors created by RdRp.8 ExoN has a distance relationship with the N-terminal DeDD protein superfamily. The zinc finger 1 is present and involved in nsp10 binding with nsp14, which reduces the catalytic activity of the exonuclease. The zinc finger 2 is adjacent to the catalytic sites i.e. ASP90, GLU92, GLU191, HIS268, and ASP273.59 The five catalytic residues are predicted to coordinate with Mg2+ ion and that is adjacent to the removal of incorporated nucleotide errors. ExoN activity is inhibited by blocking the amino acids DeDD, ASP and GLU.60
The potential lead compounds were identified through molecular docking where they bound at the active site of nsp14 and also obeyed the Lipinski rule for ADME properties (Table S6). The top 6 compounds and their interactions with the residues of nsp14 at the active site are summarized in Table 2. Molecular docking of the exonuclease enzyme with tomentosolic acid pose at the active site through multiple non-covalent interactions with a dock score of −9.2 kcal/mol (Fig. 3). The residues like VAL91, GLU191, HIS268, PHE146, GLU92, LEU149, GLY93, PRO141, ASN104, PHE190, ASP90, ASP273, MET276 and ASN252 of nsp14 are interacted with the tomentosolic acid (Table 2). Tomentosolic acid is known to show some medicinal properties, such as FPR1 inhibitor in human neutrophils with the ability to suppress psoriatic symptoms in mice and anti-inflammatory properties.43 The lead compound timosaponin A-I is also found to be potent against the nsp14. Timosaponin A-I exhibits strong affinity to the catalytic site by posing hydrogen bonds and Pi-alkyl to PHE190, GLU92, HIS268, ASN266, TRP186, and carbon hydrogen bonds with GLY189, ALA187 and ASP273 residues. Shizukaol A forms hydrogen bonds with HIS268 catalytic residue and also associates with GLN145, PHE146, VAL91, ASN104, PHE190, GLU191, ALA187, TRP186, LEU253, ASN252, GLN254, ASN266 by Van der Waal interactions (Table 2). Shizukaol A is isolated from chloranthus japonicus and inhibits inflammation by targeting HMGB1 to regulate the Nrf2/HO-1 signalling pathway, and a similar compound, shizukaol B, is reported as an inhibitor of HIV-1 reverse transcriptase RNase H.44,45 In addition, the triterpene lactone phytochemical absinthin listed in Table 2 found in artemisia absinthium showed anti-inflammatory activity.42 The proposed lead compounds bind at the active site of nsp14, and their various medicinal properties were also reported.
Table 2.
Best 6 compounds pose at the nsp14 catalytic sites and with predicted inhibition constant (Ki).
| Sr. No. | Compounds | Autodock Vina binding affinity score | Autodock Free energy of binding kcal/mol | Inhibition constant (Ki) | Hydrogen bond interactions | Van der Waal interactions |
|---|---|---|---|---|---|---|
| 1 | Timosaponin A-I | −9.2 | −7.31 | 2.56 uM | PHE190, GLU92, HIS268, ASN266, Pi-Alkyl: TRP186, Carbon hydrogen bond: GLY189, ALA187, ASP273 | PHE146, ASN252, LEU253, SLN254, MET276, MET58 |
| 2 | Absinthin | −9.2 | −7.24 | 21.17 uM | HIS268, GLY93, ASP273, Carbon hydrogen bond: GLU92 | GLN145, PHE146, VAL91, ASN104, PHE190, GLU191, ALA187, TRP186, LEU253, ASN252, GLN254, ASN266 |
| 3 | Tomentosolic acid | −9.2 | −8.49 | 602.47 nM | GLN254, ASN266, VAL91, GLU191 | HIS268, PHE146, GLU92, LEU149, GLY93, PRO141, ASN104, PHE190, ASP90, ASP273, MET276, ASN252 |
| 4 | Shizukaol A | −9 | −7.54 | 19.33 uM | HIS268, ASN266, GLN254, Pi-Alkyl: PHE146, Carbon hydrogen bond: ASP273 | GLN145, HIS148, GLU92, PRO141, LEU149, GLY93, PHE190, VAL91, ALA187, LEU253, MET276, ASN252 |
| 5 | Convallagenin A 3-O-alpha-l-arabinopyranoside | −8.9 | −7.26 | 22.03 uM | ASP273, ASN266, HIS268 | ALA267, GLU92, ASP90, PHE190, ALA187, GLY189, PHE146, VAL91, GLY93, ASN104, MET58, GLY59 |
| 6 | Limonin | −8.6 | −7.29 | 26.5 uM | HIS268, ASN252 Pi-Sigma: ALA187, Carbon hydrogen bond: GLU191 | GLN254, LEU253, TRP186, ASP90, ASN266, PHE146, GLU92, ASP273 |
Fig. 3.
Molecular docking of phytochemicals with exonuclease nsp14: a. nsp14 protein structure domains and active sites represented in color and labeled. b. The line ribbon structure of nsp14 with compounds binding at the active site (timosaponin A-I, absinthin, tomentosolic acid, shizukaol A, convallagenin A 3-O-alpha-l-arabinopyranoside, limonin represented in cyan, orange, metallic grey, violet, brown, dark grey, respectively). c. Dot plot of ligands RMSD with nsp14 representing the binding affinity scores. d. 2D structure of timosaponin A-I interacting with nsp14 active site residues.
The dynamics properties of the nsp14 and its protein-ligand complexes with tomentosolic acid, timosaponin A-I and shizukaol A were obtained by the Gromacs MD simulations for 100 ns. The solvated system was wrapped into a triclinic cell for equilibration and energy optimization of the structure set for 5000 steps by the steepest descent integrator and the NVT/NPT set for 50 million steps for calculating the dynamics properties, i.e. temperature, pressure, density, and total energy. The total system found 757 donors and 1476 acceptors, including the ligand hetero atoms. System molecular dynamics calculations are ensured by the average RMSD calculations plot shown in Fig. 4a. The average root mean square deviation in 100 ns of nsp14, tomentosolic acid, timosaponin A-I and shizukaol A is 0.64 nm, 0.63 nm, 0.63 nm and 0.56 nm, respectively. Nsp14 is stabilised after the binding of lead compounds was very close to the protein at 100 ns simulation time. The equilibrated complex simulation trajectories showed that the ligands, i.e. tomentosolic acid, timosaponin A-I, and shizukaol A bound to the nsp13 protein were more stable. The residual fluctuations during the ligand bound to protein dynamics simulation were calculated as RMSF (Fig. 4b). The fluctuations were noticed in the ligand interactions with protein active site residues. The peaks in Fig. 4b showing the nsp14 behavior concerning ligands (tomentosolic acid, timosaponin A-I, and shizukaol A) bound to residues, i.e., 10–70, 100–120, 210–215, and 450–475. The maximum peaks between 0.5 and 1.3 nm are of Mg2+ ion binding sites, MTase sites, and catalytic sites of the nsp14 protein. The free energy calculation of tomentosolic acid, timosaponin A-I, and shizukaol A with nsp13 is −1372.72, −1370.55 and −1370.68 kcal/mol, whereas the AutoDock scoring function calculates the estimated inhibition constant (Ki) of 602.47 nM, 2.56 μM and 19.33 μM, respectively. The computed radius of gyration, solvent-accessible surface area, and average number of H-bonds in each frame over time of the ligand and receptor, namely tomentosolic acid, timosaponin A-I, and shizukaol A are shown in Fig. S12, Fig. S13 and Fig. S14, respectively.
Fig. 4.
MD simulation of nsp14 protein and its protein-ligand complxes with tomentosolic acid, timosaponin A-I and shizukaol A: (a) RMSD of solvated nsp14 protein shown in light green, nsp14 complexes with tomentosolic acid, timosaponin A-I and shizukaol A during 100 ns MD simulation represented in blue, red and pink respectively. (b) RMSF values of nsp14 protein and its complexes with tomentosolic acid, timosaponin A-I, and shizukaol A.
3.4. Screening of inhibitors for nsp15
Nsp15 is an endoribonuclease (endoU) found in all coronavirus families. It cleaves RNA substrates 3′ of uredines.61 Nsp15 cleavage target regulates the RNA activation and accumulation of viral RNA. This eludes the active viral RNA from getting recognised by the host's antibodies and defenders. Initially, it is thought that nsp15 is involved directly in the replication of the viral genome.62 Later, it was found that the coronaviruses with inadequate nsp15 were more operational and replicating. Nsp15 is known for implying its cleavage specificity. It is vital for the life cycle of the coronaviridae. The endonuclease function of nsp15 plays an important role in the viral life cycle. Nsp15 preferentially cleaves uredine-containing RNA substrates.63 So, this is generally known as endoU for its particularity in partitioning. Rather than its function in RNA replication and synthesis, this nuclease is quite important for dodging host immune responses. The crystal structure of nsp15 is composed of a common hexameric assembly that consists of dimers made up of nsp15 trimers. The nsp15 protomer consists of three domains which include the N-terminal domain, the middle domain, and the C-terminal endonuclease domain (Fig. 5a).64 Each domain performs their specific functions. The N-terminal domain performs a vital function in the process of oligomerization, whereas the middle domain and the C-terminal endonuclease are profoundly homologous with the other endoU enzymes. Moreover, the nsp15 is said to be only active in its hexamer form. Recent studies stated that the molecular requirements of nsp15 to participate in the oligomerization are still unknown.65 The active site of nsp15 consists of two histidine residues. These residues facilitate the catalysis and also resemble the RNase A active site. The nsp15 consists of conserved histidine residues at the active site that are involved in catalysis and reminiscent of the postulated RNase A. Catalysis of RNase A followed by a two-step reaction in which, initially a 2′-3′-cyclic phosphate is hydrolyzed to form a 3′-phosphate. HIS235, HIS250, LYS290, SER294 and TYR343 are 5′-UMP-bound nsp15 endoU active residues.13
Fig. 5.
Molecular docking of phytochemicals with nsp15: a. edoribonuclease nsp15 protein structure domains and active site represented in color and labeled, and best dock pose of 6 compounds (hecogenin, limonin, picrasin A, 25-O-anhydrocimigenol 3-O-alpha-l-arabinopyranoside, 23-trans-p-coumaroyloxytormentic acid, asperagenin). b. The surface structure of nsp15 with compounds binding at the active site. c. Ligand position (RMSD) and binding affinity scores plotted using dot plot. d. 2D structure of limonin interacting with nsp15 active site residues.
The best compounds with drug-like properties and strong binding affinity to endoribonuclease catalytic site were screened (Table S7). The best six lead compounds such as hecogenin, limonin, picrasin A, 25-O-anhydrocimigenol, 3-O-alpha-l-arabinopyranoside, 23-trans-p-coumaryloxytormentic acid, and asperagenin pose various non-covalent interactions and binds effectively with nsp15, and their minimum inhibition concentrations are summarized in Table 3. Further, the MD simulations of protein-ligand complexes of limonin, 25-O-anhydrocimigenol 3-O-alpha-l-arabinopyranoside and aspergenin with nsp15 were performed for 100 ns.
Table 3.
Best six compounds pose at the nsp15 catalytic sites and with predicted inhibition constant (Ki).
| Sr. No. | Compounds | Autodock Vina Binding affinity score | Autodock Free energy of binding kcal/mol | Inhibition constant (Ki) | Hydrogen bond interactions | Van der Waal interactions |
|---|---|---|---|---|---|---|
| 1 | 23-trans-p-Coumaroyloxytormentic acid | −9 | −7.59 | 2.72 uM | LYS290, GLY248, HIS250, THR341, Pi-Alkyl: MET331, Pi-Pi: TYR343, Pi-Donor hydrogen bond: TRP333 | GLY247, HIS235, GLN245, CYS291, VAL292 |
| 2 | Limonin | −8.8 | −7.67 | 1.98 uM | LYS290, HIS250, THR341, HIS235, Carbon hydrogen bond: HIS250 | VAL292, SER294, CYS293, GLN245, TYR343, GLY248 |
| 3 | Asperagenin | −8.7 | −8.21 | 965.14 nM | HIS235, SER294, Pi-Alkyl: VAL292, TYR343, Carbon hydrogen bond: HIS250 | GLU340, GLN245, THR341, GLY248, LYS290, CYS293 |
| 4 | Hecogenin | −8.7 | −7.76 | 2.03 uM | HIS235, Pi-Alkyl: lys335, ALA232, Carbon hydrogen bond: GLU340 | HIS338, VAL339, GLU234, MET219, ASP240, GLY239, PHE241 |
| 5 | 25-O-Anhydrocimigenol 3-O-alpha-l-arabinopyranoside | −8.7 | −8.98 | 260.39 nM | HIS235, ARG258, Carbon hydrogen bond: ASP240. | PHE241, SER242, VAL339, ALA232, GLU340, GLU234, GLY239, MET219, TYR238 |
| 6 | Picrasin A | −8.6 | −7.48 | 3.30 uM | GLY248, THR341, HIS235, HIS250, SER294, GLN255 | GLU340, LYS345, PRO344, CYS293, LYS290, VAL292 |
Limonin is a potent ligand having versatile medicinal properties posing at the nsp15 active site with strong interaction with the residues LYS290, HIS250, THR341, HIS235 and HIS250 (Fig. 5b). It exhibits strong binding affinity with a minimum binding energy of −8.8 kcal/mol and a predicted inhibitory concentration (Ki) of 1.98 μM (Table 3). Limonin is reported to have enormous medicinal properties, i.e. anti-proliferative, proapoptotic, antiviral (HIV and HTLV-I), neuroprotective, and anti-obesity in mice.46,47 Asperagenin, a rare type of steroidal sapogenin with the 25-hydroxyl group posing in the nsp15 active pocket with a minimum binding energy of −8.7 kcal/mol and a Ki of 965.14 nM. The compound 25-O-anhydrocimigenol 3-O-alpha-l-arabinopyranoside binds covalently with HIS235, ARG258 and carbon hydrogen bonds with ASP240. It is found to be a highly potent compound with a minimum inhibitory concentration of 260.39 nM against nsp15 and showed an affinity score of −8.7 kcal/mol. It also poses hydrophobic interactions with PHE241, SER242, VAL339, ALA232, GLU340, GLU234, GLY239, MET219, TYR238 residues by Van der Waal forces. The compound hecogenin is posing strong binding affinity to the catalytic residues HIS235, HIS250, LYS290, SER294 with a binding energy of −8.7 kcal/mol. Hecogenin, the natural steroid sapogenin has been shown to have therapeutic properties against gastric ulcers in mice. It is also reported as a potential lead compound with medicinal properties, i.e., antiarthritis properties, and down-regulates the pro-inflammatory cytokines. Picrasin A is effectively interacting with GLY248, THR341, HIS235, HIS250, SER294, GLN255 and also binding at the hydrophobic site with Van der Waals attraction to GLU340, LYS345, PRO344, CYS293, LYS290 and VAL292. 23-trans-p-Coumaroyloxytormentic acid has a significant role as an inhibitor and it is showing strong binding affinity with nsp15 catalytic sites. It showed interactions with LYS290, GLY248, HIS250 and THR341, as well as Pi-Alkyl and Pi-Pi interactions with TYR343, MET331 residues, with minimum binding energy and inhibitory concentrations of −9 kcal/mol and 2.72 μM, respectively. The 23-trans-p-coumaroyloxytormentic acid found in the citrus-like fruits of Eriobotrya japonica interacts with the nsp15 active sites, and it is the sweetest fruit generally used in Japanese traditional medicine.
The dynamics properties of the nsp15 and its protein-ligand complexes with limonin, 25-O-anhydrocimigenol 3-O-alpha-l-arabinopyranoside and aspergenin were provided by the Gromacs MD simulation for 100 ns. In total, the system for nsp15 found 475 donors and 970 acceptors, including the ligand hetero atoms. The RMSD plot of the molecular dynamics calculations are shown in Fig. 6a. The protein nsp15 and its complexes with limonin, 25-O-anhydrocimigenol-3-O-alpha-l-arabinopyranoside, and asperagenin have an average root mean square deviations of 0.44 nm, 0.621 nm, 0.669 nm and 0.626 nm, respectively. Nsp15 is stabilised after the binding of lead compounds and tightly associated at the protein catalytic site during the 100 ns simulation time. The equilibrated complex simulation trajectories revealed that the ligands bound to nsp15 protein were more stable. The residual fluctuations during the ligand bound to protein dynamics simulation were calculated as RMSF (Fig. 6b). The fluctuations were noticed in the ligand interactions with protein active site residues. The RMSF peaks showed that the nsp15 behavior concerning ligands (limonin, 25-O-anhydrocimigenol, 3-O-alpha-l-arabinopyranoside, and asperagenin) bound to residues, i.e., 1–45, 70–90, 170–180, 260–280, 330–345. The maximum peaks between 0.3 and 0.7 nm are of Mg2+ binding sites, NendoU binding sites, N-terminal sites, and catalytic sites of the endoribonuclease protein. The free energy calculation of limonin, 25-O-anhydrocimigenol 3-O-alpha-l-arabinopyranoside, and asperagenin with nsp15 is −1371.42, −1372.20 and −1372.44 kcal/mol, respectively, whereas the estimated inhibition constant (Ki) is 1.98 μM, 260.39 nM and 965.14 nM, respectively. The computed radius of gyration, solvent-accessible surface area, and average number of H-bonds in each frame over time of the nsp15 protein-ligand complexes with limonin, 25-O-anhydrocimigenol 3-O-alpha-l-arabinoside and asperagenin are shown in Fig. S15, Fig. S16 and Fig. S17, respectively.
Fig. 6.
MD simulation of enoribonuclease nsp15 protein and its protein-ligand complexes with limonin, 25-O-anhydrocimigenol 3-O-alpha-l-arabinopyranoside and asperagenin for 100 ns: (a) RMSD of solvated nsp15 protein (green colour) and its complexes with limonin, 25-O-anhydrocimigenol 3-O-alpha-l-arabinopyranoside and asperagenin during 100 ns MD simulation represented in pink, light green and blue respectively. (b) RMSF fluctuation peaks of nsp15 protein with limonin, 25-O-anhydrocimigenol 3-O-alpha-l-arabinopyranoside, and asperagenin complexes.
4. Conclusions
The ion-dependent enzymes, i.e., helicases (nsp13), exonuclease (nsp14), and endoribonuclease (nsp15) of SARS-CoV-2 are targeted to search potential phytochemicals from the family of limonoids and triterpenoids. The present study screened 369 limonoids and triterpenoids by performing in silico molecular docking, predicting drug-likeness and molecular dynamics simulations to propose the potential lead compounds against the protein nsp13, nsp14 and nsp15. Most of the limonoids and triterpenoids bind effectively at the active site of the proteins, which can inhibit their catalytic activities. The phytochemicals 26-deoxyactein and 25-O-anhydrocimigenol 3-O-beta-d-xylopyranoside potentially bind at the catalytic sites LYS290 and SER291 of helicases, and their docked structures are stable within 100 ns of simulation time. Similarly, for nsp14, the lead compounds promoted the inhibitory characteristics via blocking of ASP90, GLU92, GLU191, HIS268 and ASP273 residues. The pharmacological features of the potential leads, i.e., tomentosolic acid, timosaponin A-I, and shizukaol A proved to be within the future scope of experimental studies. On the other hand, the limonoids and triterpenoids, i.e. limonin, 25-O-anhydrocimigenol, 3-O-alpha-l-arabinopyranoside, and asperagenin extracted from the plant parts pose potential actions against the endoribonuclease nsp15 catalytic activity. These compounds played a critical role in suppressing the enzyme activity by blocking HIS235, HIS250, LYS290, SER294 and TYR343 catalytic site residues. The computational studies provide a better platform to identify the lead compounds to suppress the expression of viral RNA in the host system and promote the activation of immune responses. More importantly, our computational studies with the various protein targets of SARS-CoV-2 revealed that the phytochemical limonin is binding effectively with multiple protein targets of SARS-CoV-2. Therefore, limonin and related phytochemicals mainly found in citrus fruits can be explored further to fight against COVID-19.
Declaration of competing interest
There is no conflict of interest.
Footnotes
Peer review under responsibility of The Center for Food and Biomolecules, National Taiwan University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jtcme.2021.12.002.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Abhilash K. Second wave of COVID-19: unrelenting rampage of the SARS CoV-2 variants. Curr Med Issues. 2021;19(3):129. doi: 10.4103/cmi.cmi_44_21. [DOI] [Google Scholar]
- 2.WHO Coronavirus (COVID-19) Dashboard Covid19.who.int. 2021. https://covid19.who.int/
- 3.Pal M., Berhanu G., Desalegn C., Kandi V. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): an update. Cureus. 2020;12(3) doi: 10.7759/cureus.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beeraka N., Sadhu S., Madhunapantula S., et al. Strategies for targeting SARS CoV-2: small molecule inhibitors-the current status. Front Immunol. 2020;11:552925. doi: 10.3389/fimmu.2020.552925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eroshenko N., Gill T., Keaveney M., Church G., Trevejo J., Rajaniemi H. Implications of antibody-dependent enhancement of infection for SARS-CoV-2 countermeasures. Nat Biotechnol. 2020;38(7):789–791. doi: 10.1038/s41587-020-0577-1. [DOI] [PubMed] [Google Scholar]
- 6.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586(7830):516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 7.Suraphan P., Ho C.-T., Sheen L.-Y. Dietary therapy and herbal medicine for COVID-19 prevention: a review and perspective. J Tradit Complement Med. 2020;10:420–427. doi: 10.1016/j.jtcme.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robson F., Khan K., Le T., et al. Coronavirus RNA proofreading: molecular basis and therapeutic targeting. Mol Cell. 2020;79(5):710–727. doi: 10.1016/j.molcel.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jang K., Jeong S., Kang D., Sp N., Yang Y., Kim D. A high ATP concentration enhances the cooperative translocation of the SARS coronavirus helicases nsP13 in the unwinding of duplex RNA. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-61432-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shu T., Huang M., Wu D., et al. SARS-Coronavirus-2 Nsp13 possesses NTPase and RNA helicases activities that can Be inhibited by bismuth salts. Virol Sin. 2020;35(3):321–329. doi: 10.1007/s12250-020-00242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo G., Gao M., Gao X., et al. SARS-CoV-2 non-structural protein 13 (nsp13) hijacks host deubiquitinase USP13 and counteracts host antiviral immune response. Signal Transduct Target Ther. 2021;6(1) doi: 10.1038/s41392-021-00509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White M., Lin W., Cheng X. Discovery of COVID-19 inhibitors targeting the SARS-CoV-2 Nsp13 helicases. J Phys Chem Lett. 2020;11(21):9144–9151. doi: 10.1021/acs.jpclett.0c02421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim Y., Wower J., Maltseva N., et al. Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2. Commun Biol. 2021;4(1):193. doi: 10.1038/s42003-021-01735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhardwaj K., Guarino L., Kao C. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J Virol. 2004;78(22):12218–12224. doi: 10.1128/jvi.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2020;19(3):155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tahir M. Coronavirus genomic nsp14-ExoN, structure, role, mechanism, and potential application as a drug target. J Med Virol. 2021;93(7):4258–4264. doi: 10.1002/jmv.27009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saramago M., Bárria C., Costa V., et al. New targets for drug design: importance of nsp14/nsp10 complex formation for the 3’-5’ exoribonucleolytic activity on SARS-CoV-2. FEBS J. 2021 doi: 10.1111/febs.15815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jockusch S., Tao C., Li X., et al. Sofosbuvir terminated RNA is more resistant to SARS-CoV-2 proofreader than RNA terminated by Remdesivir. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-73641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuzimoto A., Isidoro C. The antiviral and coronavirus-host protein pathways inhibiting properties of herbs and natural compounds - additional weapons in the fight against the COVID-19 pandemic? J Tradit Complement Med. 2020;10(4):405–419. doi: 10.1016/j.jtcme.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vardhan S., Sahoo S. In silico ADMET and molecular docking study on searching potential inhibitors from limonoids and triterpenoids for COVID-19. Comput Biol Med. 2020;124:103936. doi: 10.1016/j.compbiomed.2020.103936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mehany T., Khalifa I., Barakat H., Althwab S., Alharbi Y., El-Sohaimy S. Polyphenols as promising biologically active substances for preventing SARS-CoV-2: a review with research evidence and underlying mechanisms. Food Biosci. 2021;40:100891. doi: 10.1016/j.fbio.2021.100891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vidoni C., Fuzimoto A., Ferraresi A., Isidoro C. Targeting autophagy with natural products to prevent SARS-CoV-2 infection. J Tradit Complement Med. 2021 doi: 10.1016/j.jtcme.2021.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kousar K., Majeed A., Yasmin F., Hussain W., Rasool N. Phytochemicals from selective plants have promising potential against SARS-CoV-2: investigation and corroboration through molecular docking, MD simulations, and quantum computations. BioMed Res Int. 2020:1–15. doi: 10.1155/2020/6237160. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selvaraj C., Dinesh D., Panwar U., Abhirami R., Boura E., Singh S. Structure-based virtual screening and molecular dynamics simulation of SARS-CoV-2 Guanine-N7 methyltransferase (nsp14) for identifying antiviral inhibitors against COVID-19. J Biomol Struct Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1778535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fakhri S., Piri S., Majnooni M., Farzaei M., Echeverría J. Targeting neurological manifestations of coronaviruses by candidate phytochemicals: a mechanistic approach. Front Pharmacol. 2021;11 doi: 10.3389/fphar.2020.621099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Z., Xiao Y., Xu L., et al. Glycyrrhizic acid nanoparticles as antiviral and anti-inflammatory agents for COVID-19 treatment. ACS Appl Mater Interfaces. 2021;13(18):20995–21006. doi: 10.1021/acsami.1c02755. [DOI] [PubMed] [Google Scholar]
- 27.Stevaert A., Krasniqi B., Van Loy B., et al. Betulonic acid derivatives interfering with human coronavirus 229E replication via the nsp15 endoribonuclease. J Med Chem. 2021;64(9):5632–5644. doi: 10.1021/acs.jmedchem.0c02124. [DOI] [PubMed] [Google Scholar]
- 28.Vardhan S., Sahoo S. Virtual screening by targeting proteolytic sites of furin and TMPRSS2 to propose potential compounds obstructing the entry of SARS-CoV-2 virus into human host cells. J Tradit Complement Med. 2021 doi: 10.1016/j.jtcme.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eswar N., Webb B., Marti-Renom M., et al. Comparative protein structure modeling using modeller. Curr Protoc Bioinformatics. 2006;15(1) doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khor B., Tye G., Lim T., Noordin R., Choong Y. The structure and dynamics of BmR1 protein from Brugia malayi: in silico approaches. Int J Mol Sci. 2014;15(6):11082–11099. doi: 10.3390/ijms150611082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7(1) doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris G., Huey R., Lindstrom W., et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh S., Bajpai U., Michael Lynn A. Structure based virtual screening to identify inhibitors against MurE Enzyme of Mycobacterium tuberculosis using AutoDock Vina. Bioinformation. 2014;10(11):697–702. doi: 10.6026/97320630010697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris G., Huey R., Lindstrom W., et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huey R., Morris G., Olson A., Goodsell D. A semiempirical free energy force field with charge-based desolvation. J Comput Chem. 2007;28(6):1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- 36.Hess B., Kutzner C., van der Spoel D., Lindahl E. GROMACS 4:algorithms for highly efficient ,load -Balanced,and scalable molecular simulation .J. Chem Theory Comput. 2008;4(3):435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 37.Jöhrer Stuppner, Greil Çiçek. Structure-guided identification of black cohosh (actaea racemosa) triterpenoids with in vitro activity against multiple myeloma. Molecules. 2020;25(4):766. doi: 10.3390/molecules25040766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu C., Yen G. Ganoderic acid and lucidenic acid (triterpenoid) Enzymes. 2014:33–56. doi: 10.1016/b978-0-12-802215-3.00003-3. [DOI] [PubMed] [Google Scholar]
- 39.Tian Z., Zhou L., Huang F., et al. Anti-cancer activity and mechanisms of 25-anhydrocimigenol-3-O-D-xylopyranoside isolated from Souliea vaginata on hepatomas. Anti Cancer Drugs. 2006;17(5):545–551. doi: 10.1097/00001813-200606000-00008. [DOI] [PubMed] [Google Scholar]
- 40.Cicek S., Khom S., Taferner B., Hering S., Stuppner H. Bioactivity-guided isolation of GABAAReceptor modulating constituents from the rhizomes ofActaea racemosa. J Nat Prod. 2010;73(12):2024–2028. doi: 10.1021/np100479w. [DOI] [PubMed] [Google Scholar]
- 41.Screening and identification of DPP-4 inhibitors from Xiaokean formula by a fluorescent probe. China J Chin Mater Med. 2016 doi: 10.4268/cjcmm20160714. [DOI] [PubMed] [Google Scholar]
- 42.Talmon M., Bosso L., Quaregna M., et al. Anti-inflammatory activity of absinthin and derivatives in human bronchoepithelial cells. J Nat Prod. 2020;83(6):1740–1750. doi: 10.1021/acs.jnatprod.9b00685. [DOI] [PubMed] [Google Scholar]
- 43.Korinek M., Hsieh P., Chen Y., et al. Randialic acid B and tomentosolic acid block formyl peptide receptor 1 in human neutrophils and attenuate psoriasis-like inflammation in vivo. Biochem Pharmacol. 2021;190:114596. doi: 10.1016/j.bcp.2021.114596. [DOI] [PubMed] [Google Scholar]
- 44.Tang P., Li Q., Liao S., et al. Shizukaol A exerts anti-inflammatory effect by regulating HMGB1/Nrf2/HO-1 pathway. Phytomedicine. 2021;82:153472. doi: 10.1016/j.phymed.2021.153472. [DOI] [PubMed] [Google Scholar]
- 45.Fang P., Cao Y., Yan H., et al. Lindenane disesquiterpenoids with anti-HIV-1 activity fromChloranthus japonicus. J Nat Prod. 2011;74(6):1408–1413. doi: 10.1021/np200087d. [DOI] [PubMed] [Google Scholar]
- 46.Effect of limonin and nomilin on HIV-1 replication on infected human mononuclear cells. Planta Med. 2003;69(10):910–913. doi: 10.1055/s-2003-45099. [DOI] [PubMed] [Google Scholar]
- 47.Fan Zhang, Luo, et al. Limonin: a review of its pharmacology, toxicity, and pharmacokinetics. Molecules. 2019;24(20):3679. doi: 10.3390/molecules24203679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frick D., Lam A. Understanding helicase as a means of virus control. Curr Pharmaceut Des. 2006;12(11):1315–1338. doi: 10.2174/138161206776361147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Byrd A. Superfamily 2 helicases. Front Biosci. 2012;17(7):2070. doi: 10.2741/4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jia Z., Yan L., Ren Z., et al. Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019;47(12):6538–6550. doi: 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jankowsky E., Fairman M. RNA helicase — one fold for many functions. Curr Opin Struct Biol. 2007;17(3):316–324. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 52.Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells. 2020;9(5):1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar K., Lupoli T. Exploiting existing molecular scaffolds for long-term COVID treatment. ACS Med Chem Lett. 2020;11(7):1357–1360. doi: 10.1021/acsmedchemlett.0c00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adedeji A., Singh K., Kassim A., et al. Evaluation of SSYA10-001 as a replication inhibitor of severe acute respiratory syndrome, mouse hepatitis, and Middle East respiratory syndrome coronaviruses. Antimicrob Agents Chemother. 2014;58(8):4894–4898. doi: 10.1128/aac.02994-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weng C., Chau C., Chen K., Chen D., Yen G. The anti-invasive effect of lucidenic acids isolated from a newGanoderma lucidum strain. Mol Nutr Food Res. 2007;51(12):1472–1477. doi: 10.1002/mnfr.200700155. [DOI] [PubMed] [Google Scholar]
- 56.Sahoo A., Dash U., Kanhar S., Mahapatra A. In vitro biological assessment of Homalium zeylanicum and isolation of lucidenic acid A triterpenoid. Toxicol Rep. 2017;4:274–281. doi: 10.1016/j.toxrep.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu D., Yao Q., Chen Y., Hu X., Qing C., Qiu M. The in vitro and in vivo antitumor activities of tetracyclic triterpenoids compounds actein and 26-deoxyactein isolated from rhizome of cimicifuga foetida L. Molecules. 2016;21(8):1001. doi: 10.3390/molecules21081001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ogando N., Zevenhoven-Dobbe J., van der Meer Y., Bredenbeek P., Posthuma C., Snijder E. The enzymatic activity of the nsp14 exoribonuclease is critical for replication of MERS-CoV and SARS-CoV-2. J Virol. 2020;94(23) doi: 10.1128/jvi.01246-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Narayanan N., Nair D. Ritonavir may inhibit exoribonuclease activity of nsp14 from the SARS-CoV-2 virus and potentiate the activity of chain terminating drugs. Int J Biol Macromol. 2021;168:272–278. doi: 10.1016/j.ijbiomac.2020.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Squeglia F., Romano M., Ruggiero A., Maga G., Berisio R. Host DDX helicase as possible SARS-CoV-2 proviral factors: a structural overview of their hijacking through multiple viral proteins. Front Chem. 2020;8 doi: 10.3389/fchem.2020.602162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pillon M., Frazier M., Dillard L., et al. Cryo-EM structures of the SARS-CoV-2 endoribonuclease Nsp15 reveal insight into nuclease specificity and dynamics. Nat Commun. 2021;12(1) doi: 10.1038/s41467-020-20608-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim Y., Jedrzejczak R., Maltseva N., et al. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV -2. Protein Sci. 2020;29(7):1596–1605. doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bhardwaj K., Palaninathan S., Alcantara J., et al. Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15. J Biol Chem. 2008;283(6):3655–3664. doi: 10.1074/jbc.m708375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frazier M., Dillard L., Krahn J., et al. 2021. Characterization of SARS2 Nsp15 Nuclease Activity Reveals It's Mad about U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mariano G., Farthing R., Lale-Farjat S., Bergeron J. Structural characterization of SARS-CoV-2: where we are, and where we need to Be. Front Mol Biosci. 2020;7 doi: 10.3389/fmolb.2020.605236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van de Sand L., Bormann M., Alt M., et al. Glycyrrhizin effectively inhibits SARS-CoV-2 replication by inhibiting the viral main protease. Viruses. 2021;13(4):609. doi: 10.3390/v13040609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361(9374):2045–2046. doi: 10.1016/s0140-6736(03)13615-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wen C., Kuo Y., Jan J., et al. Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. J Med Chem. 2007;50(17):4087–4095. doi: 10.1021/jm070295s. [DOI] [PubMed] [Google Scholar]
- 69.Kumar S., Kashyap P., Chowdhury S., Kumar S., Panwar A., Kumar A. Identification of phytochemicals as potential therapeutic agents that binds to Nsp15 protein target of coronavirus (SARS-CoV-2) that are capable of inhibiting virus replication. Phytomedicine. 2021;85:153317. doi: 10.1016/j.phymed.2020.153317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Balestrieri E., Pizzimenti F., Ferlazzo A., et al. Antiviral activity of seed extract from Citrus bergamia towards human retroviruses. Bioorg Med Chem. 2011;19(6):2084–2089. doi: 10.1016/j.bmc.2011.01.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.