Abstract
Severe Acute Respiratory Coronavirus (SARS-CoV-2) has been emerging in the form of different variants since its first emergence in early December 2019. A new Variant of Concern (VOC) named the Omicron variant (B.1.1.529) was reported recently. This variant has a large number of mutations in the S protein. To date, there exists a limited information on the Omicron variant. Here we present the analyses of mutation distribution, the evolutionary relationship of Omicron with previous variants, and probable structural impact of mutations on antibody binding. Our analyses show the presence of 46 high prevalence mutations specific to Omicron. Twenty-three of these are localized within the spike (S) protein and the rest localized to the other 3 structural proteins of the virus, the envelope (E), membrane (M), and nucleocapsid (N). Phylogenetic analysis showed that the Omicron is closely related to the Gamma (P.1) variant. The structural analyses showed that several mutations are localized to the region of the S protein that is the major target of antibodies, suggesting that the mutations in the Omicron variant may affect the binding affinities of antibodies to the S protein.
Keywords: B.1.1.529, Omicron variant, SARS-CoV-2, COVID-19, Variants
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), the pathogen causing Coronavirus Disease 19 (COVID-19), first emerged in Wuhan city in China nearly two years ago [1]. SARS-CoV-2 is a positive-strand RNA virus of ∼30 kb nucleotides that encode 29 proteins from 15 open reading frames (ORFs) [2,3]. As with all RNA viruses, SARS-CoV-2 has been continuously evolving through mutations in different viral genes, presumably resulting in increased transmissibility and infectivity as observed among recent variants of concerns (VOCs). A striking example is the emergence of the Delta variant that caused a surge in infections in many parts of the world, leading to millions of causalities [4].
Recently, a new SARS-CoV-2 variant named Omicron has been reported [[5], [6], [7]]. Initially identified in Botswana (Nov. 11, 2021), the Omicron variant quickly spread to neighboring countries, and now it has been found in at least 26 countries around the world, including the first case reported in California, USA, as of December 1, 2021. An unprecedented number of mutations, particularly in the Spike (S) protein of the Omicron variant, has been related to its high transmissibility and infectivity. Therefore, we analyzed the available sequences of Omicron variant and the structural data on the Spike protein either in an apo form or in complex with neutralizing antibodies to gain insights into (i) the prevalence of mutations in SARS-CoV-2 genes, (ii) co-existence of mutations in Spike and other viral genes, (iii) evolutionary relationship with other VOCs, and (iv) possible impact of mutations on the binding of antibodies to the S protein.
We used the high-quality and complete sequences (n = 77) of the Omicron (B.1.1.529) variant downloaded from the GISAID repository (November 26, 2021) [8], and processed the sequences using the NextClade CLI [9] and/or an in-house Python script to calculate the prevalence of mutations. Considering mutations with >50% prevalence as signature mutations, a total of 30 mutations (A76V, T95I, Y145del, G339D, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F, L212I, S371L, S373P, S375F, K417N) were identified as signature mutations in the Omicron variant. Of these 30 signature mutations, 23 (bold-faced) are unique to the Omicron variant, i.e., these mutations were not identified in any of the previously reported variants. Further, we also identified nine additional mutations in other genes that were >85% prevalent in all Omicron (n = 70) sequences. These mutations are ORF1a:K856R, ORF1a:L2084I, ORF1a:A2710T, ORF1a:T3255I, ORF1a:P3395H, ORF1a:I3758V, ORF1b:P314L, ORF1b:I1566V, and ORF9b:P10S. Of these, only two mutations (ORF1a:T3255I or nsp4:T492I and ORF1b:P314L or nsp12:P323L) were observed in Delta and Delta Plus variants that were present with significant prevalence (>40%) [4]. Mutation P323L in nsp12 has co-evolved with D614G [10,11]. Altogether, our results showed 46 unique mutations in ORF1a, ORF1b, and S genes of the SARS-CoV-2 Omicron variant prevalent at more than 50% frequency. Additionally, we identified E: T9I, M:D3G, M:Q19E, M:A63T, N:P13L, N:R203K, and N:G204R mutations in nearly all sequences analyzed (i.e., ∼100% prevalent).
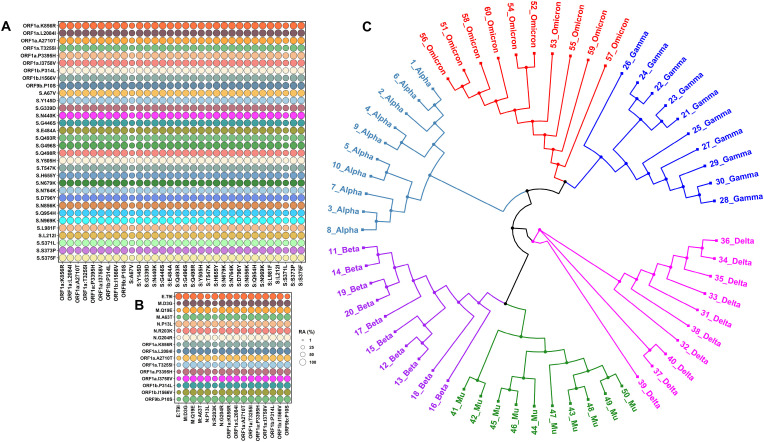
Thirty signature mutations in S protein were used to identify co-evolving mutations and their prevalence in all (n = 77) sequences analyzed. We calculated relative abundance using an in-house Python script to examine if the aforementioned mutations have co-evolved. As shown in Fig. 1 A, all Omicron-specific mutations in S protein appear to have co-evolved. Additionally, high prevalent mutations in genes other than S protein also appear to have co-evolved (Fig. 1B). Next, in order to determine evolutionary relationships of Omicron with other variants, we randomly selected 10 high-quality and high-coverage sequences from our dataset (n = 77) of Omicron and aligned with the latest, high-quality, high-coverage sequences of the Alpha (n = 10), Beta (n = 10), Gamma (n = 10), Delta (n = 10), and Mu (n = 10) variants using the MAFFT program [12]. Phylogenetic analyses suggest that the Omicron variant is closely related to the Gamma (P.1) variant (Fig. 1C) that emerged simultaneously in Brazil and Japan, as reported elsewhere [13]. It is important to note that some more recent studies indicate that the SARS-CoV-2 Spike protein may have insertion sequences that may have been derived from either other coronaviruses or host derived [14]. These findings have important implications for the use of different receptors for viral entry and for the potential failure of antibodies to neutralize this new variant. Clearly, additional studies are needed to confirm these new findings.
Fig. 1.
Relative abundance (RA) of signature Omicron variant mutations.Panel A shows the RA of Spike signature mutations and high prevalent mutations in OFR1a and ORF1b. Panel B shows the RA of Omicron variant mutations in ORF1a and ORF1b and signature mutations in structural proteins E, M, and N. Please note ORF1b:P314L corresponds to nsp12 mutation nsp12:P323L. Panel C shows the phylogenetic relationship among different SARS-CoV-2 variants. The GISAID identification numbers for the sequences used in Panel B are as below. 1_Alpha to 10_Alpha: EPI_ISL_5803029, EPI_ISL_6000214, EPI_ISL_6026865, EPI_ISL_6027306, EPI_ISL_6141708, EPI_ISL_6227805, EPI_ISL_6229383, EPI_ISL_6251101, EPI_ISL_6383583, EPI_ISL_675143; 1_Beta1 to 10_Beta: EPI_ISL_5053750, EPI_ISL_5274500, EPI_ISL_5430264, EPI_ISL_5515861, EPI_ISL_5524663, EPI_ISL_6422293, EPI_ISL_6699711, EPI_ISL_6751445, EPI_ISL_6774033, EPI_ISL_6774035; 1_Gamma to 10_Gamma: EPI_ISL_6121588, EPI_ISL_6121598, EPI_ISL_6121603, EPI_ISL_6569634, EPI_ISL_6689781, EPI_ISL_6689782, EPI_ISL_6689786, EPI_ISL_6689787, EPI_ISL_6689788, EPI_ISL_6689789; 1_Delta to 10_Delta: EPI_ISL_6739692, EPI_ISL_6739693, EPI_ISL_6761790, EPI_ISL_6763188, EPI_ISL_6769723, EPI_ISL_6772657, EPI_ISL_6775864, EPI_ISL_6775870, EPI_ISL_6795204, EPI_ISL_6809412; 1_ Mu to 10_Mu: EPI_ISL_6526278, EPI_ISL_6526285, EPI_ISL_6569586, EPI_ISL_6569593, EPI_ISL_6569599, EPI_ISL_6569609, EPI_ISL_6569625, EPI_ISL_6569673, EPI_ISL_6675615, EPI_ISL_6675624, and 1_Omicron to 10_ Omicron: EPI_ISL_6752026, EPI_ISL_6774086, EPI_ISL_6699747, EPI_ISL_6699744, EPI_ISL_6699751, EPI_ISL_6752027, EPI_ISL_6699728, EPI_ISL_6699764, EPI_ISL_6699734, EPI_ISL_6698790.
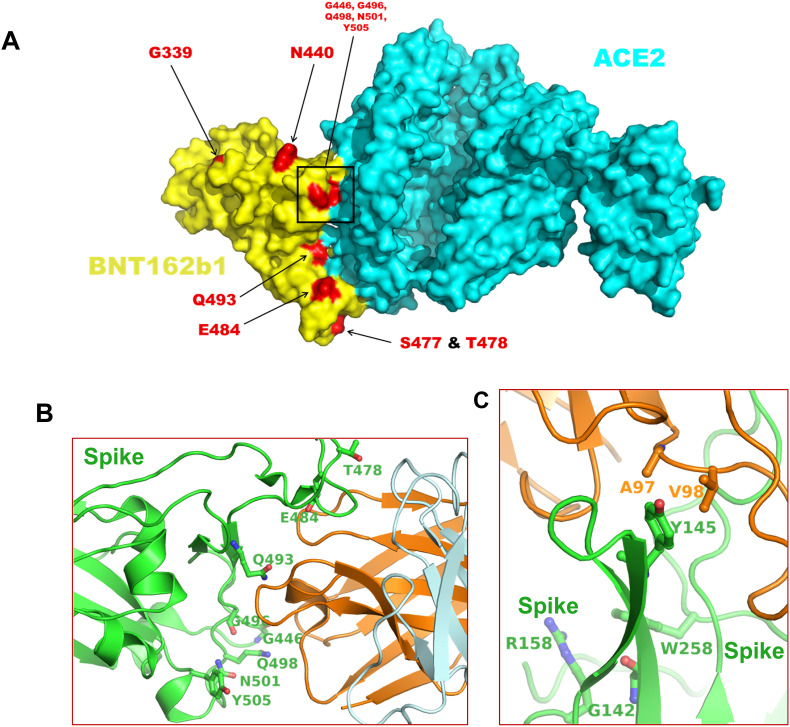
We next examined whether the mutations in the Omicron variant would affect the binding of antibodies generated by previous infection or immunizations. We used an in-house R-script to search the Protein Data Bank (PDB) (www.rcsb.org) for S protein structures that would affect antibody binding. As of December 1, a total of 194 structures of the S protein or receptor-binding domain of S protein (S-RBD) in complex with antibodies/nanobodies have been deposited within the PDB. Of these, 81 structures have been determined by the X-ray crystallography and the remaining by the cryo-Electron Microscopy (cryo-EM). We selected the crystal structure of the IGHV3-53 antibody in complex with the SARS-CoV-2 S-RBD since this structure was solved at a high resolution (1.71 Å) (PDB entry 7JMP) [15] to assess the implications of Omicron S-RBD mutations on the binding of antibodies. The rationale behind selecting this structure is based on the fact that the Cα atoms of the S-RBD from this structure when superimposed onto the Cα atoms of S-RBD encoded by Pfizer vaccine candidate BNT162b1 (abandoned for use after clinical trials) in complex with ACE-2 (PDB file 7L7F) [16] resulted in a root-mean-square-deviation (RMSD) of 1.26 Å. Our analysis showed that the positions of 8 residues corresponding to the Omicron variant signature mutations (T478, E484, Q493, G496, G446, Q498, N501, and Y505) are at S-RBD (encoded by BNT162b1)/ACE2 interface (Fig. 2 A). The identical 8 residues (T478, E484, Q493, G496, G446, Q498, N501, and Y505) are also present at the interface of the S-RBD/IGHV3-53 complex (Fig. 2B). Mutations such as G446S, Q493R, and G496S can create steric interference for the binding of antibodies to the S-RBD, whereas mutations such as E484A and Y505H may result in the loss of interactions with the antibody. It appears that the net result of the mutations is the reduced affinity between the S-RBD and antibodies that bind to this domain, which suggests that the pre-existing immunization may not impart the necessary and optimal protection to prevent infection by the Omicron variant.
Fig. 2.
Distribution of amino acid residue position at Spike/ACE or Spike/antibody interface.Panel A shows the distribution of amino acid residues mutated in the Omicron variant in the complex formed between S-RBD encoded by BNT162b1 and ACE2. This figure has been generated by PDB entry 7L7F. Panel B shows the position of residues at the interface of S-RBD and antibody IGHV3-53, where the mutations in Omicron variant have been identified. The S protein is rendered in green ribbons, whereas the antibody is rendered in orange and light-blue ribbons. The amino acid residues representing the mutation sites in the Omicron variant are rendered as balls and sticks. Panel C shows the interaction Y145 of S protein with antibody (1–87) residues A97 and V98. A deletion mutation (as seen in Omicron variant) will abrogate these interactions. This figure also shows some residue positions in S protein where the mutations in Delta and Delta Plus variants are present. Figures for Panel D and E were generated from PDB entries 7JMP and 7L2D, respectively. In both panels, the atoms of S protein residues are colored as green – carbons, blue–nitrogens, and red – oxygens. Atoms of antibodies are colored as orange – carbons, blue – nitrogens, and red – oxygens.
We also analyzed the cryo-EM structure of an NTD-directed neutralizing antibody in complex with prefusion SARS-CoV-2 spike glycoprotein (PDB entry 7L2D) [15]. We selected this structure because the RMSD between Cα atoms of the antibody binding domain of this structure and corresponding Cα atoms of the S protein encoded by currently used Pfizer vaccine BNT162b2 (PDB entry 7L7K) [16] was 1.8 Å. It is important to note that the BNT162b2-encoded S protein structure does not include the bound antibody. As shown in Fig. 2C, Y145 forms hydrophobic interactions with antibody residues A97 and V98. A deletion mutation (Y145del), as seen in the Omicron variant, will result in the loss of these interactions and thereby reduce the binding affinity of the appropriate antibodies. Notably, Y145del is in close vicinity of positions where some signature mutations in Delta and Delta Plus have been reported [4].
In summary, in this report, we show that the Omicron variant has many mutations in the S protein. These mutations co-evolved with the mutations throughout the viral genome at a very high prevalence and the Omicron variant is closely related to the Gamma variant. The structural analyses suggest that the uniquely positioned mutations in the Omicron variant may reduce the binding of antibodies present in an individual induced by either prior infection or following vaccination against the SARS-CoV-2 virus. However, our structural analyses were restricted to monoclonal antibodies and their impact on polyclonal antibodies remains to be analyzed.
Author statement
SNB and KS conceptualized the study; KS wrote the first draft, and KS and SNB edited the final manuscript. ANS, SRK, and KS conducted genetic analyses, wrote required programs either in R or in Python; KS conducted structural analysis of Spike/antibody complex; SNB, and HSC edited the manuscript and contributed to understanding the pathogenicity of CoVs. All authors approved the final manuscript.
Acknowledgments
This work was partially supported by National Institute of Allergy and Infectious Diseases grants R01 AI129745, R01 AI113883, and DA052845 to S.N.B. Further, S.N.B. acknowledges independent research and development (IRAD) funding from the National Strategic Research Institute (NSRI) at the University of Nebraska. K. Singh was partially funded by the Bond Life Sciences Center (Early Concept grant) and by the National Institute of Allergy and Infectious Diseases (R37 AI076119). HSC and SNB acknowledge the support by the National Institute of Allergy and Infectious Diseases (3R21AI144374-02S1). KS acknowledges the computation facilities of the Molecular Interactions Core at the University of Missouri, Columbia, MO 65212. We thank the laboratories that have generously deposited sequences into the GISAID database.
Data availability
Data will be made available on request.
References
- 1.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020;109 doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921 e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mariano G., Farthing R.J., Lale-Farjat S.L.M., Bergeron J.R.C. Structural characterization of SARS-CoV-2: where we are, and where we need to Be. Front Mol Biosci. 2020;7 doi: 10.3389/fmolb.2020.605236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kannan S.R., Spratt A.N., Cohen A.R., Naqvi S.H., Chand H.S., Quinn T.P., et al. Evolutionary analysis of the Delta and Delta Plus variants of the SARS-CoV-2 viruses. J. Autoimmun. 2021;124 doi: 10.1016/j.jaut.2021.102715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callaway E. Heavily mutated Omicron variant puts scientists on alert. Nature. 2021;600:21. doi: 10.1038/d41586-021-03552-w. [DOI] [PubMed] [Google Scholar]
- 6.Torjesen I. Covid-19: Omicron may be more transmissible than other variants and partly resistant to existing vaccines, scientists fear. BMJ. 2021;375:n2943. doi: 10.1136/bmj.n2943. [DOI] [PubMed] [Google Scholar]
- 7.Gao S.J., Guo H., Luo G. Omicron variant (B.1.1.529) of SARS-CoV-2, a global urgent public health alert. J. Med. Virol. 2021 doi: 10.1002/jmv.27491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kannan S.R., Spratt A.N., Quinn T.P., Heng X., Lorson C.L., Sonnerborg A., et al. Infectivity of SARS-CoV-2: there is something more than D614G? J. Neuroimmune Pharmacol. 2020;15:574–577. doi: 10.1007/s11481-020-09954-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827 e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spratt A.N., Kannan S.R., Woods L.T., Weisman G.A., Quinn T.P., Lorson C.L., et al. Evolution, correlation, structural impact and dynamics of emerging SARS-CoV-2 variants. Comput. Struct. Biotechnol. J. 2021;19:3799–3809. doi: 10.1016/j.csbj.2021.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venkatakrishnan A., Anand P., Lenehan P., Suratekar R., Raghunathan B., Niesen M.J., Soundararajan V. APA; MLA; Chicago: 2021. Omicron Variant of SARS-CoV-2 Harbors a Unique Insertion Mutation of Putative Viral or Human Genomic Origin. December 3. [Google Scholar]
- 15.Wu N.C., Yuan M., Liu H., Lee C.D., Zhu X., Bangaru S., et al. bioRxiv; 2020. An Alternative Binding Mode of IGHV3-53 Antibodies to the SARS-CoV-2 Receptor Binding Domain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogel A.B., Kanevsky I., Che Y., Swanson K.A., Muik A., Vormehr M., et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021;592:283–289. doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.