Abstract
Darevskia rock lizards include 29 sexual and seven parthenogenetic species of hybrid origin distributed in the Caucasus. All seven parthenogenetic species of the genus Darevskia were formed as a result of interspecific hybridization of only four sexual species. It remains unknown what are the main advantages of interspecific hybridization along with switching on parthenogenetic reproduction in evolution of reptiles. Data on whole transcriptome sequencing of parthenogens and their parental ancestors can provide value impact in solving this problem. Here we have sequenced ovary tissue transcriptomes from unisexual parthenogenetic lizard D. unisexualis and its parental bisexual ancestors to facilitate the subsequent annotation and to obtain the collinear characteristics for comparison with other lizard species. Here we report generated RNAseq data from total mRNA of ovary tissues of D. unisexualis, D. valentini and D. raddei with 58932755, 51634041 and 62788216 reads. Obtained RNA reads were assembled by Trinity assembler and 95141, 62123, 61836 contigs were identified with N50 values of 2409, 2801 and 2827 respectively. For further analysis top Gene Ontology terms were annotated for all species and transcript number was calculated. The raw data were deposited in the NCBI SRA database (BioProject PRJNA773939). The assemblies are available in Mendeley Data and can be accessed via doi:10.17632/rtd8cx7zc3.1.
Keywords: Darevskia lizards, Parthenogenesis, Transcriptome analysis, Ovaries, AMP
Specifications Table
| Subject | Biology |
| Specific subject area | Transcriptomics |
| Type of data | Transcriptome assemblies, raw sequences |
| How data were acquired | Ovary RNA from three lizard species were isolated and used for sequencing by the Macrogen Inc. (Korea) |
| Data format | Analyzed, Raw |
| Parameters for data collection | Data collection contains raw transcriptome data for ovary tissues of three lizard species: unisexual (parthenogenetic) D. unisexualis and parental bisexual D. valentini and D. raddei nairensis |
| Description of data collection | Data collection includes total Illumina HiSeq2500 generated transcriptome reads, transcripts, TRINITY contigs, predicted proteins, and ORFs. |
| Data source location | All lizards were collected from Armenia populations. D. unisexualis from the Hrazdan population (40.503493 N 44.748097 E) D. r. nairensis from Vahramaberd population (40.844394 N, 43.755720 E) D. valentini from Sepasar population (41.027492 N, 43.816634 E) |
| Data accessibility | Raw data - BioProject PRJNA773939 in NCBI SRA database. Trinity assemblies - doi:https://doi.org/10.17632/rtd8cx7zc3.1 in Mendeley Data |
Value of the Data
-
•
Data provides information about the first assembled ovary transcriptomes of three genetically related Darevskia lizards species and information about their genes and proteins.
-
•
This data may benefit evolutionary biologists because it shows genetic differences between unisexual (parthenogenetic) and bisexual parental lizards.
-
•
The data may provide insight into the genetic underpinning of parthenogenetic reproduction and can be used in further study of these genes.
1. Data Description
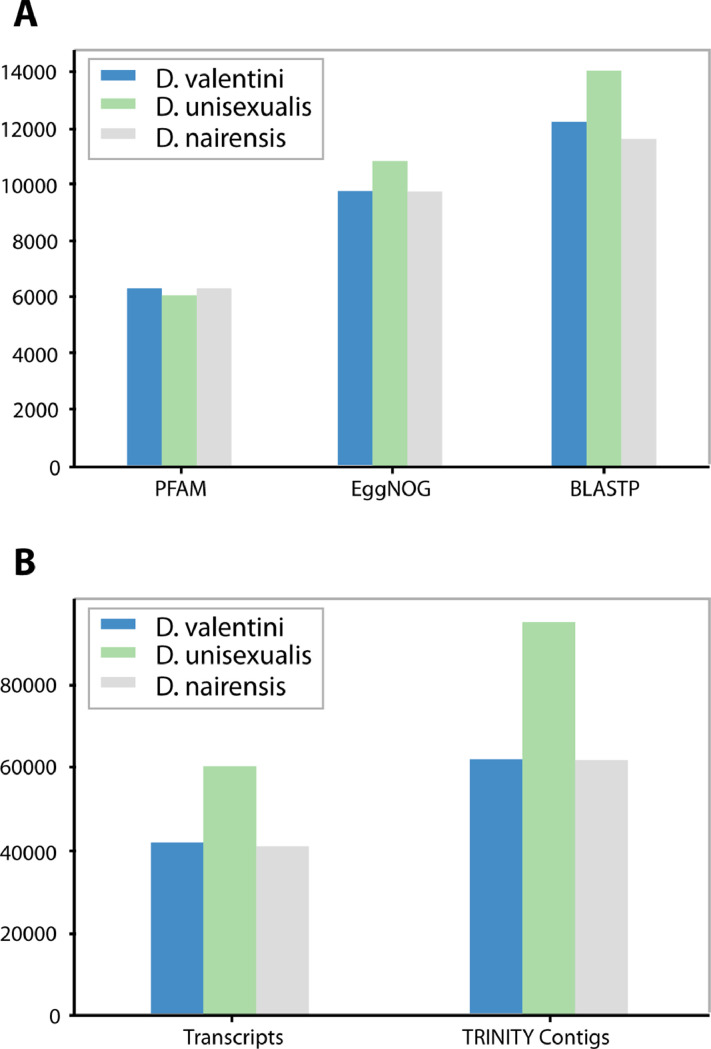
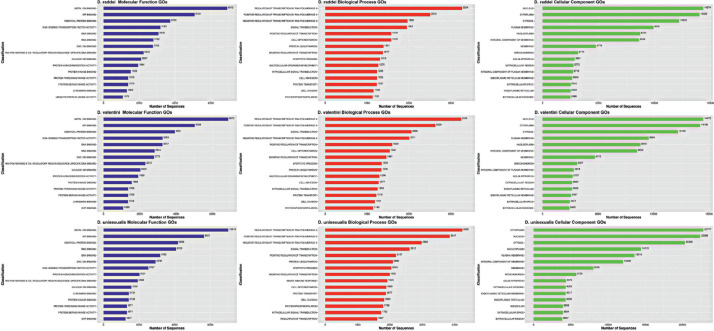
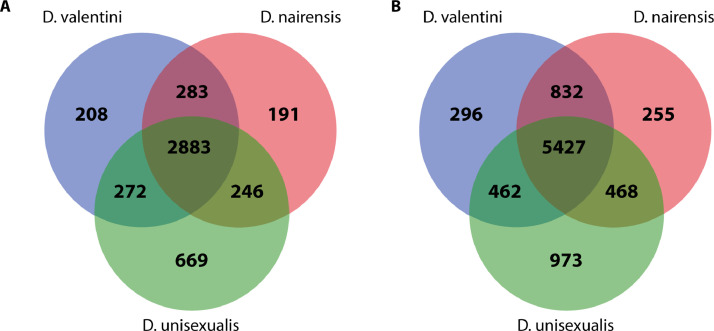
Ovary RNA from three individuals of each species was pooled together and used to prepare the three cDNA libraries: D. unisexualis, D. raddei nairensis, D. valentini. Table 1 shows the total number of bases, reads, GC (%), Q20 (%), and Q30 (%) that were calculated for the three samples. The characteristics of assembled transcriptome sequences are presented in Table 2. Structural characteristics of three transcriptomes are shown in Fig. 1. Obtained Trinity assemblies contain 60132 transcripts for D. unisexualis, 41680 for D. valentini, and 413664 for D. r. nairensis. TransDecoder peptide output was used for BLASTP, Pfam, and EggNOG search (Fig. 1A, Supplementary 1). BLASTP v. 2.9.0+ revealed 14049, 12331, and 11865 proteins for D. unisexualis, D. valentini, and D. nairensis respectively (Fig. 1A). Parthenogenetic species D. unisexualis showed greater TRINITY contigs (> 81.4% and > 87.2%) and transcripts (> 44.3% and > 45.4%) numbers than D. valentini and D. r. raddei respectively (Fig. 1B). The D. unisexualis showed more hits for each of the searching instruments. Top 10 GO terms taken from all GO terms datasets and distribution graphs are presented in Fig. 2 (Supplementary 2). The biggest number of annotated genes and the most annotated category was a cellular component, biological processes were less annotated. In the molecular functions category, most genes were related to binding. The most highly enriched genes in biological processes were related to the regulation of transcription of RNA polymerase II. It was found that in cellular components over-represented molecules were the nucleus and cytoplasm origin. In total, 38844, 38756, 63219 transcripts with GO terms were annotated in Table 3 for D. valentini, D.raddei, and D. unisexualis respectively. The summary of Trinotate shows a more prevalent number of annotated transcripts with GO in D. unisexualis than in D. valentini and D. raddei nairensis (> 62.8% and > 63.1%). The total number of GO in the parthenogenetic sample exceeds D. valentini and D. raddei nairensis on 58.9% and 60% respectively (Table 3). The final TransDecoder stats are presented in Table 4. The overall number of ORFs in D. unisexualis was 45.7% more than in bisexual parental samples, according to the TransDecoder results.. The analysis of common and unique genes on Venn diagrams (Fig. 3A, B) displays that D. unisexualis has more unique genes in BLASTP (> 221.6% and > 250%) and EggNOG (> 228.7% and > 281.6%) than D. valentini and D. nairensis respectively (Supplementary 4). The antimicrobial peptides have been searched in D. valentini, D. nairensis, and D. unisexualis with 59, 81, and 70 possible matches respectively. These sequences were found in 29, 34, and 36 transcripts. The antimicrobial peptides detected have antibacterial activity against Gram+, Gram- as well as against fungi (Supplementary 5). The raw RNA sequence reads for each lizard are available in the NCBI SRA database (PRJNA773939). The assembled transcripts are available in the Mendeley data (doi:https://doi.org/10.17632/rtd8cx7zc3.1).
Table 1.
Statistics of the RNA-seq generated from three lizards.
| Species | Total reads | Total bases | Q20 basesa | Q30 basesb | GC content |
|---|---|---|---|---|---|
| D. unisexualis | 58.932755 M | 17,313222 G | 97.98% | 94.34% | 48.05% |
| D. valentini | 51.634041 M | 15,593480 G | 97.65% | 93.35% | 46.59% |
| D. r. nairensis | 62.788216 M | 18,962041 G | 97.13% | 92.33% | 46.77% |
Q20 - ratio of bases with probability of containing no more than one error in 100 bases.
Q30 - ratio of bases with probability of containing no more than one error in 1,000 bases.
Table 2.
Summary characteristics of transcriptome sequence assembly of all three samples data.
| D. raddei nairensis | D. valentini | D. unisexualis | |
|---|---|---|---|
| # contigs (>= 0 bp) | 122746 | 126141 | 228862 |
| # contigs (>= 1000 bp) | 39398 | 39145 | 57984 |
| # contigs (>= 5000 bp) | 3517 | 3533 | 2447 |
| # contigs (>= 10000 bp) | 134 | 118 | 8 |
| # contigs (>= 25000 bp) | 0 | 0 | 0 |
| # contigs (>= 50000 bp) | 0 | 0 | 0 |
| Total length (>= 0 bp) | 139903614 | 140424203 | 204321108 |
| Total length (>= 1000 bp) | 105256962 | 104453618 | 138481341 |
| Total length (>= 5000 bp) | 22937215 | 22851989 | 14738047 |
| Total length (>= 10000 bp) | 1524044 | 1315073 | 90998 |
| Total length (>= 25000 bp) | 0 | 0 | 0 |
| Total length (>= 50000 bp) | 0 | 0 | 0 |
| # contigs | 61836 | 62123 | 95141 |
| Largest contig | 16187 | 14170 | 12181 |
| Total length | 121008445 | 120525010 | 164367910 |
| GC (%) | 46,23 | 46,19 | 46,06 |
| N50 | 2827 | 2801 | 2409 |
| N75 | 1567 | 1558 | 1378 |
| L50 | 13677 | 13716 | 22778 |
| L75 | 27887 | 27951 | 45070 |
| # N's per 100 kbp | 0.0 | 0.0 | 0.0 |
Fig. 1.
Structural characteristics of three transcriptomes. A number of annotated proteins (A), and TRINITY contigs, and transcripts (B).
Fig. 2.
Distribution of the top 10 GO terms for each of the three species.
Table 3.
Summary of Trinotate/Gene Ontologies.
| Species | Total transcripts with GO | Total transcripts with only one GO | Total transcripts with multiple GO | Total GO in the file | Total unique GO in the file |
|---|---|---|---|---|---|
| D. valentini | 38844 | 1241 | 37603 | 553827 | 16934 |
| D. nairensis | 38756 | 1195 | 37561 | 550189 | 16885 |
| D. unisexualis | 63219 | 1796 | 61423 | 880200 | 17771 |
Table 4.
Open Reading Frames (ORFs) prediction numbers using TransDecoder.
| Species | Total | Complete | 5-prime partial | 3-prime partial | Internal |
|---|---|---|---|---|---|
| D. valentini | 55816 | 34333 | 12663 | 3133 | 22136 |
| D. nairensis | 55808 | 34499 | 12893 | 3026 | 5327 |
| D. unisexualis | 81344 | 43408 | 22136 | 5390 | 10473 |
Fig. 3.
Venn diagrams showing overlapping hits for three species (A) and overlapping of EggNOG genes (B).
2. Experimental Design, Materials and Methods
2.1. Species sampling and tissues collection
Samples of D. valentini, D. r. nairensis, and D. unisexualis for transcriptome analysis were collected in Armenia in 2019, outside of the protected areas. Several adult lizards of female D. unisexualis from the Hrazdan population (40.503493 N 44.748097 E), females D. r. nairensis from the Vahramaberd population (40.844394 N, 43.755720 E), and females D. valentini from the Sepasar population (41.027492 N, 43.816634 E) were used to surgically extract ovary. Before dissecting out the organs, the animals were subjected to chloroform euthanasia. All tissue samples were stored in RNAlater® reagent at −20°C according to the manufacturer's recommended protocol (Qiagen Inc.) until they were shipped to Macrogen Inc. (Korea) for RNA extraction and further transcriptome preparation.
2.2. RNA sequencing and raw data quality control
Total RNA was isolated from an organ/tissue using standard Trizol Tissue RNA Extraction protocol (Standard protocol for QIAzol Lysis Reagent, Qiagen). RNA RIN scores ranged from 6.4 to 6.7. Ovary RNA from three individuals of each species was pooled together and used to prepare the three cDNA libraries: D. unisexualis, D. r. nairensis, D. valentini. Inside the procedure was a cleanup on a carrier with polyT and random primers from TruSeq Stranded mRNA kit were used for preparation cDNA. The paired-end sequencing libraries were prepared by random fragmentation of the cDNA samples into 350-500 bp fragments, followed by 5′ and 3′ adapter ligation using TruSeq RNA Sample Prep Kit v2 (Illumina Inc.) according to TruSeq RNA Sample Preparation Guide (Version 2, Part #15026495 Rev.F).
Sequencing of transcriptome libraries was performed on Illumina HiSeq2500 with a mean read length of 101 bp. The Illumina Hiseq generated raw sequencing data utilizing HiSeq Control Software v2.2 for system control and base calling through an integrated primary analysis software. The BCL (base calls) binaries were converted into FASTQ by the Illumina package bcl2fastq (v1.8.4) [1] (RRID:SCR_015058). Raw transcriptome data were trimmed by Trimmomatic v0.39 [2] to remove adapters. Optical duplicates from reads were removed by the rmdup tool [3]. Raw transcriptomes contained 58932755, 51634041, and 62788216 reads for D. unisexualis, D. valentini and D. r. nairensis with GC content of 48.05%, 46.59%, and 46.77% respectively. Filtered reads quality was estimated by FastQC v0.11.9 [4] and became prepared for assembling.
2.3. Transcriptome annotation and assembly
Reads obtained after trimming and quality estimating by FastQC and Seq2fun pipeline [5] were assembled using Trinity v2.1.1 [6]. Transcriptome assembly with Trinity can be divided into several parts: searching and calculating k-mers, assembling contigs from k-mers, clustering contigs into components. For Trinity assembler, the default parameters were taken, where the minimum contig length value was 200, k-mer size was 25. TransDecoder v5.5.0 [7] program was used to predict translated proteins and ORFs (open reading frames) from assembled transcripts with at least 100 amino acids length. NCBI-blast-2.9.0+ [8] was used for homology search and protein domain identification on TransDecoder predicted proteins with such parameters as e-value < 1e-5 and percentage of similarity > 95%.
The total number of bases, reads, GC (%), Q20 (%), and Q30 (%) were calculated for the three samples (Table 1). Obtained protein sequences from TransDecoder were cross-referenced with the Gene Ontology (GO) [9,10] database using the EggNog v2.0.1 [11] tool. This tool provides functional information in the context of structure, molecular functions, the biological process of query sequences, search matches, and performs them as GO terms. Top GO terms were determined and visualized using the Trinotate package in the TransPi [12] pipeline. PFAM [13] and BLASTP searches were also performed by the TransPi pipeline with OnlyAnn (only annotation) option. This mode used such databases as Swissprot, Uniprot custom database (available under request), and Pfam.
2.4. AMPs identification
To identify antimicrobial peptides (AMP) in the transcriptome, we blasted the assembled transcripts against the known AMPs from the DRAMP 3.0 database (Data Repository of Antimicrobial Peptides) [14] using BLAST-2.2.26+ [15] with the similarity cutoff of 70%.
Ethics Statement
All individuals were hand-caught; alive-animal handling procedures were approved by Yerevan State University according to the ethical guidelines, capture permit Code 5/22.1/51043 was issued by the Ministry of Nature Protection of the Republic of Armenia for scientific studies. The study was approved by the Ethics Committee of the Moscow State University (Permit Number: 24–01) and conducted strictly according to ethical principles and scientific standards.
CRediT authorship contribution statement
Sergei S. Ryakhovsky: Formal analysis, Writing – original draft, Writing – review & editing. Victoria A. Dikaya: Formal analysis. Vitaly I. Korchagin: Data curation. Andrey A. Vergun: Writing – original draft, Writing – review & editing, Data curation, Methodology. Lavrentii G. Danilov: Formal analysis, Writing – original draft, Writing – review & editing. Sofia D. Ochkalova: . Anastasiya E. Girnyk: Data curation. Daria V. Zhernakova: Conceptualization, Visualization. Marine S. Arakelyan: Methodology, Supervision. Vladimir B. Brukhin: Writing – original draft, Writing – review & editing. Aleksey S. Komissarov: Project administration, Conceptualization, Visualization, Data curation, Writing – original draft, Writing – review & editing, Supervision. Alexey P. Ryskov: Project administration, Conceptualization, Writing – original draft, Writing – review & editing, Supervision, Funding acquisition.
Declaration of Competing Interest
All authors have read and approved the final manuscript. Consent for publication: Not applicable. The authors declare that they have no competing interests.
Acknowledgments
This research was funded by the Russian Science Foundation (RSF) Research Project № 19-14-00083. RNA characterization experiments were performed using the Center for Precision Genome Editing and Genetic Technologies for Biomedicine, IGB RAS.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.dib.2021.107685.
Contributor Information
Sergei S. Ryakhovsky, Email: ryakhovsky@scamt-itmo.ru.
Aleksey S. Komissarov, Email: komissarov@scamt-itmo.ru.
Alexey P. Ryskov, Email: ryskov@mail.ru.
Appendix. Supplementary materials
References
- 1.bcl2fastq Conversion Software, (n.d.). https://support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html. Accessed October 23, 2021.
- 2.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.aglabx/rmdup: Removes optical duplicates from raw Illumina sequence reads, GitHub. (n.d.). https://github.com/aglabx/rmdup (accessed October 23, 2021).
- 4.Babraham bioinformatics - FastQC a quality control tool for high throughput sequence data, (n.d.). https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed May 14, 2021).
- 5.Liu P., Ewald J., Galvez J.H., Head J., Crump D., Bourque G., Basu N., Xia J. Ultrafast functional profiling of RNA-seq data for nonmodel organisms. Genome Res. 2021;31:713–720. doi: 10.1101/gr.269894.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., Chen Z., Mauceli E., Hacohen N., Gnirke A., Rhind N., di Palma F., Birren B.W., Nusbaum C., Lindblad-Toh K., Friedman N., Regev A. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Home • TransDecoder/TransDecoder Wiki, GitHub. (n.d.). https://github.com/TransDecoder/TransDecoder (accessed October 23, 2021).
- 8.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 9.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., Sherlock G. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gene ontology consortium, the gene ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49:D325–D334. doi: 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huerta-Cepas J., Szklarczyk D., Heller D., Hernández-Plaza A., Forslund S.K., Cook H., Mende D.R., Letunic I., Rattei T., Jensen L.J., von Mering C., Bork P. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47:D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.R.E. Rivera-Vicéns, C.G. Escudero, N. Conci, M. Eitel, G. Wörheide, TransPi – a comprehensive TRanscriptome ANalysiS PIpeline for de novo transcriptome assembly, 2021. doi: 10.1101/2021.02.18.431773. [DOI] [PubMed]
- 13.Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J., Finn R.D., Bateman A. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021;49:D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi G., Kang X., Dong F., Liu Y., Zhu N., Hu Y., Xu H., Lao X., Zheng H. DRAMP 3.0: an enhanced comprehensive data repository of antimicrobial peptides. Nucleic Acids Res. 2021 doi: 10.1093/nar/gkab651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.