Summary
Olfactory receptors (ORs), responsible for the sense of smell, play an essential role in various physiological processes outside the nasal epithelium, including cancer. In breast cancer, however, the expression and function of ORs remain understudied. We examined the significance of OR transcript abundance in primary and metastatic breast cancer to the brain, bone, and lung. Although 20 OR transcripts were differentially expressed in distant metastases, OR5B21 displayed an increased transcript abundance in all three metastatic sites compared with the primary tumor. Knockdown of OR5B21 significantly decreased the invasion and migration of breast cancer cells as well as metastasis to different organs especially the brain, whereas increasing of OR5B21 transcript abundance had the opposite effect. Mechanistically, OR5B21 expression was associated with epithelial to mesenchymal transition through the STAT3/NF-κB/CEBPβ signaling axis. We propose OR5B21 (and potentially other ORs) as a novel oncogene contributing to breast cancer metastasis and a potential target for adjuvant therapy.
Subject areas: Classification Description: Molecular biology, Cell biology, Cancer
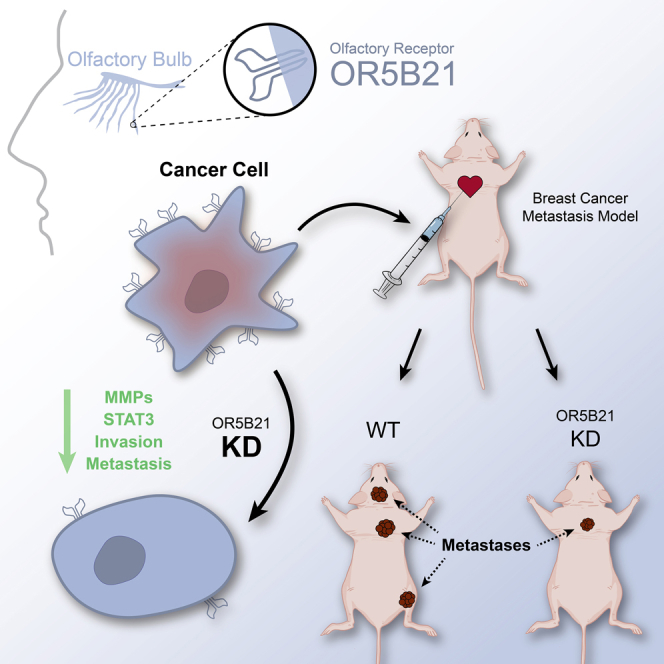
Graphical abstract
Highlights
-
•
OR5B21 overexpress in human breast cancer MDA-MB-231 metastatic cells
-
•
OR5B21 regulates invasion and migration ability of breast cancer cells
-
•
Knockdown of OR5B21 decreases breast cancer metastasis
-
•
OR5B21 induces EMT via the STAT3/NFκB/CEBPβ signaling axis
Classification Description: Molecular biology; Cell biology; Cancer
Introduction
Breast cancer is the second most frequently diagnosed malignancy, just behind lung cancer, and the leading cause of cancer death in women with more than 2 million new cases annually (Siegel et al., 2021). Although standard therapy, consisting of surgical removal of the tumor followed by adjuvant radiation and chemotherapy, has decreased breast cancer mortality, around 12% of patients are still at high risk of relapse (DeSantis et al., 2016). Breast cancer metastasis is one of the major causes of eventual mortality; thus, novel therapeutic targets to halt or delay metastasis are of critical need.
Generally, it is recognized that cancer cells responsible for developing distant organ metastases have changed their phenotype to adjust and facilitate colonization (Lambert et al. 2017). G protein-coupled receptors (GPCRs) are overexpressed and aberrantly activated during different metastatic phases including dissemination from the primary site and colonization at other sites (Dorsam and Gutkind 2007; O'Hayre et al., 2013). GPCRs are activated by a diverse array of ligands including inorganic ions, proteins, small molecules, and GPCR sensory stimuli such as light, taste, and odorants (Neves et al. 2002). This activation transduces a wide range of extracellular signals into intracellular messages resulting in cell proliferation, invasion, migration, survival, angiogenesis, and metastasis (Dorsam and Gutkind 2007; Hutchings et al., 2017; Wu et al., 2019).
The human olfactory receptor (OR) gene family is a crucial member of GPCRs typically expressed in the sensory neurons and plays an essential role in various physiological processes outside the olfactory epithelium, including cancer (Lee et al. 2019). For instance, OR51E2 (also called prostate-specific G-protein coupled receptor) is overexpressed in prostate cancer tissues with a crucial role in accelerating tumor cell invasiveness through loss of PTEN (Neuhaus et al., 2009). OR2AT4 regulates cell proliferation, apoptosis, and differentiation of human myelogenous leukemia (Manteniotis et al., 2016). OR2B6 and OR2W3 are abundant in invasive breast tumors, whereas OR2T6 is overexpressed in breast cancer tissues, contributing to metastasis (Li et al., 2019; Masjedi et al., 2019a, 2019b; Weber et al., 2018a). Thus, ORs might play a vital role in cancer progression and metastasis. Despite these observations, the function of the OR family in breast cancer metastasis and its underlying molecular mechanisms are still largely unknown.
In this study, we evaluated the expression profile of ORs in primary and different breast cancer metastatic sites including brain, lung, and bone. Among the 20 differentially expressed ORs, OR5B21 was upregulated in all organ metastases, with the highest being the brain. We show that OR5B21 plays a vital role in breast cancer cell invasion/migration and metastasis to different sites, especially the brain, by activating epithelial to mesenchymal transition through the STAT3/NF-κB/CEBPβ signaling axis.
Results
Olfactory receptors are associated with breast cancer metastasis
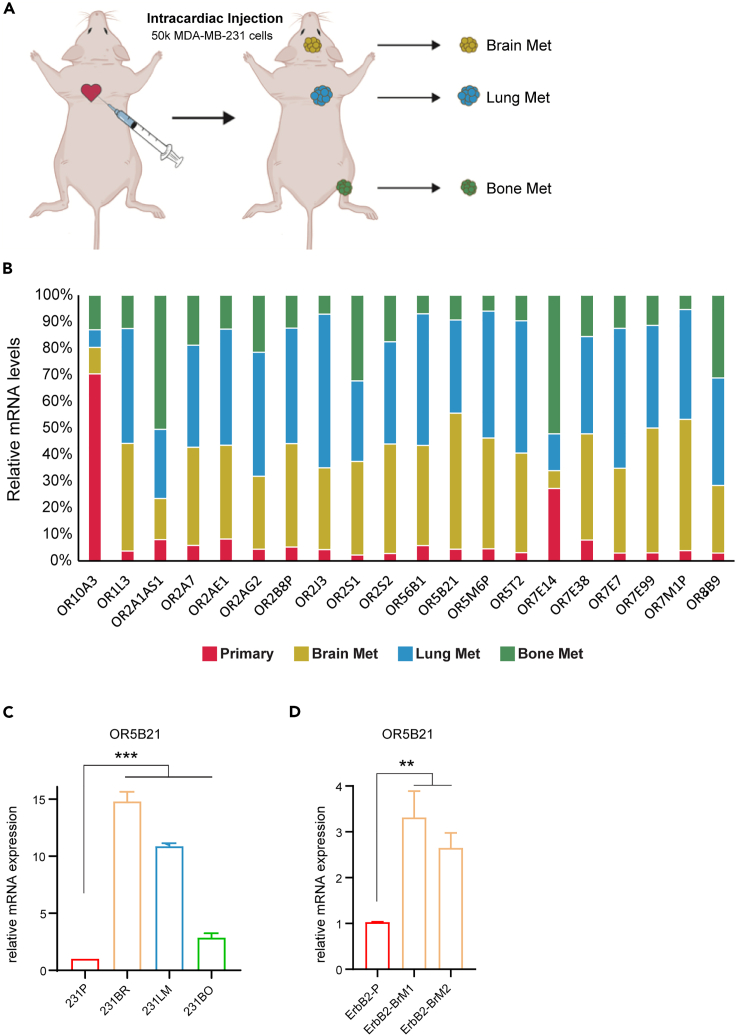
We established a breast cancer metastasis model by intracardiac injection of MDA-MB-231 (231P) cells in immunocompromised mice. We then produced a series of derivative cell lines from developed metastatic sites including the brain-seeking clone (231BR), the bone-seeking clone (231BO), and the lung-seeking clone (231LM) (Figure 1A). We screened these different cell lines for 20 common OR transcripts by quantitative real-time polymerase chain reaction (qRT-PCR) and found them to be differentially expressed in 231BR, 231BO, and/or 231LM, compared with the parental line (Figure 1B). OR5B21 transcript, in particular, was significantly increased in all three organ metastases, being 15-fold higher in 231BR (p < 0.0001), 10-fold higher in 231LM (P < 0.0001), and 3-fold higher in 231BO (p = 0.0082), compared with primary 231P (P < 0.0001; Figure 1C). These results were confirmed in another model where OR5B21 was enriched in two different clones of the brain-seeking mouse metastatic line, ErbB2-BrM1 and ErbB2-BrM2, compared with the primary ErbB2-P line (P < 0.01; Figure 1D). In addition, we analyzed differential expression of OR5B21 in healthy and tumor tissue using the Database: TCGA-BRCA and found OR5B21 to be upregulated in breast cancer compared with normal breast tissue (P < 0.001, Figure S1A). We therefore chose to focus on OR5B21 for further analyses in breast cancer metastasis.
Figure 1.
ORs are differentially expressed in breast cancer metastasis
(A) Breast cancer metastasis model was developed by intracardiac injection of human breast cancer cell line MDA-MB-231 (231P). Brain-seeking clone (231BR), lung-seeking clone (231LM), and bone-seeking clone (231BO) were harvested and cultured.
(B) Transcript abundance of 20 olfactory receptor genes in primary and different metastatic clones.
(C) Relative transcript abundance of OR5B21 in various MDA-MB-231 metastatic sites with respect to the primary tumor.
(D) OR5B21 transcript abundance in two different brain-seeking clones of ErbB2 versus primary cells.
Data presented as mean ± SEM (n = 3); ∗∗p < 0.01, ∗∗∗p < 0.001 by one-way ANOVA.
OR5B21 induces breast cancer invasion/migration
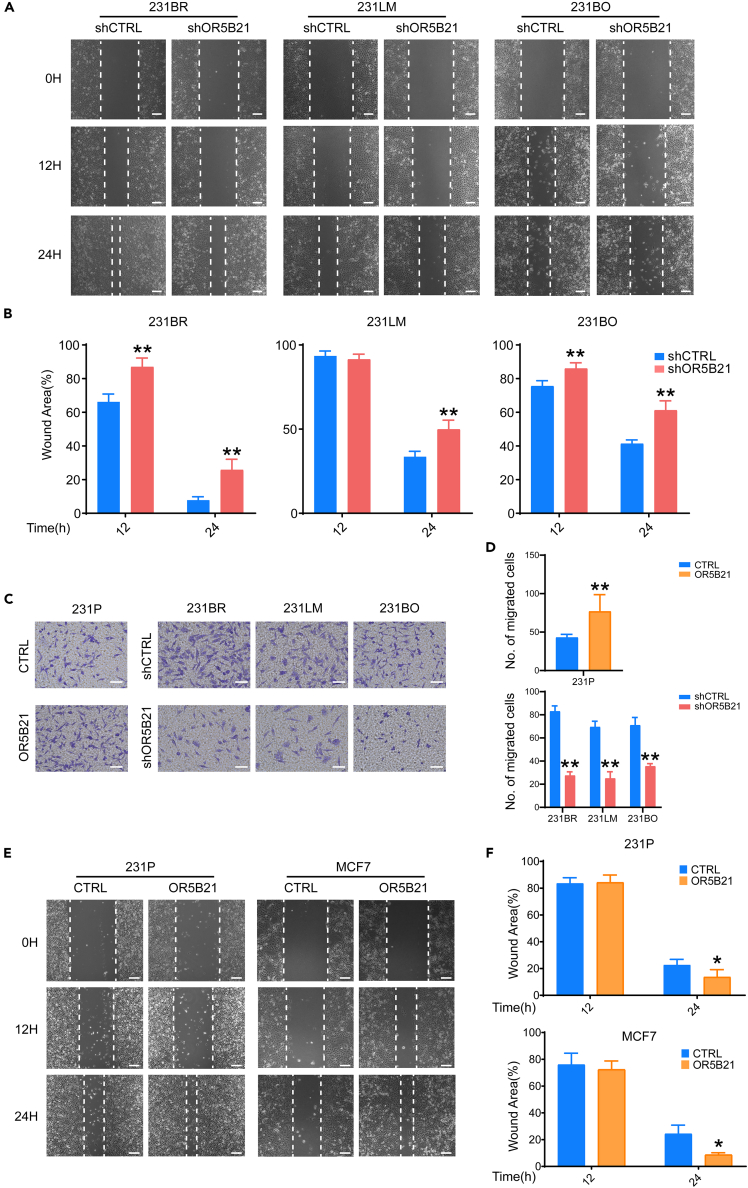
To evaluate the role of OR5B21 in breast cancer invasion/migration, we knocked down OR5B21 in different metastatic lines (231BR, 231LM, and 231BO; Figure S1B) and performed a scratch wound-healing assay. OR5B21 knockdown significantly reduced the invasion ability of all three metastatic cell lines (p < 0.01; Figures 2A and 2B). Similar results were obtained using Transwell migration assay (Figures 2C and 2D). To further validate our results, we increased OR5B21 transcription level in 231P as well as MCF7, a luminal non-metastatic human breast cancer cell line (Figure S1C), and analyzed cell invasion and migration. The increasing of OR5B21 transcript abundance led to a significant increase in cell migration and invasion in both cell lines (p < 0.05; Figures 2E and 2F). Interestingly, OR5B21 overexpression in 231P or knockdown in 231BR, 231LM, and 231BO did not affect cell proliferation as analyzed by colony formation assay and the secreted Vluc reporter assay (Figures S2A and S2B). These data demonstrate that OR5B21 participates in the invasion and migration of breast cancer cells.
Figure 2.
OR5B21 regulates breast cancer invasion/migration
(A and B) 231BR, 231LM, and 231BO expressing either shCTRL or shOR5B21 were plated as a monolayer, scratched, and analyzed by bright-field microscopy at different time points. Representative images (A) and relative quantification of migratory cells (B) are shown (n = 4).
(C and D) 231P cells overexpressing OR5B21 (or control cells) or 231BR, 231LM, 231BO cells transduced with shOR5B21 or scrambled shRNA (shCTRL) were plated on Matrigel-coated Transwell chambers. Forty-eight hours later, cells that migrated to the bottom well were stained with crystal violet and counted (n = 4).
(E and F) 231P and MCF7 control cells or overexpressing OR5B21 were plated, scratched, and analyzed at different time points (n = 3).
Data presented as mean ± SEM; ∗p < 0.05, ∗∗p < 0.01 by unpaired Student's t test. Scale bar, 150 μm (A and E), 100 μm (C).
OR5B21 promotes breast cancer metastasis
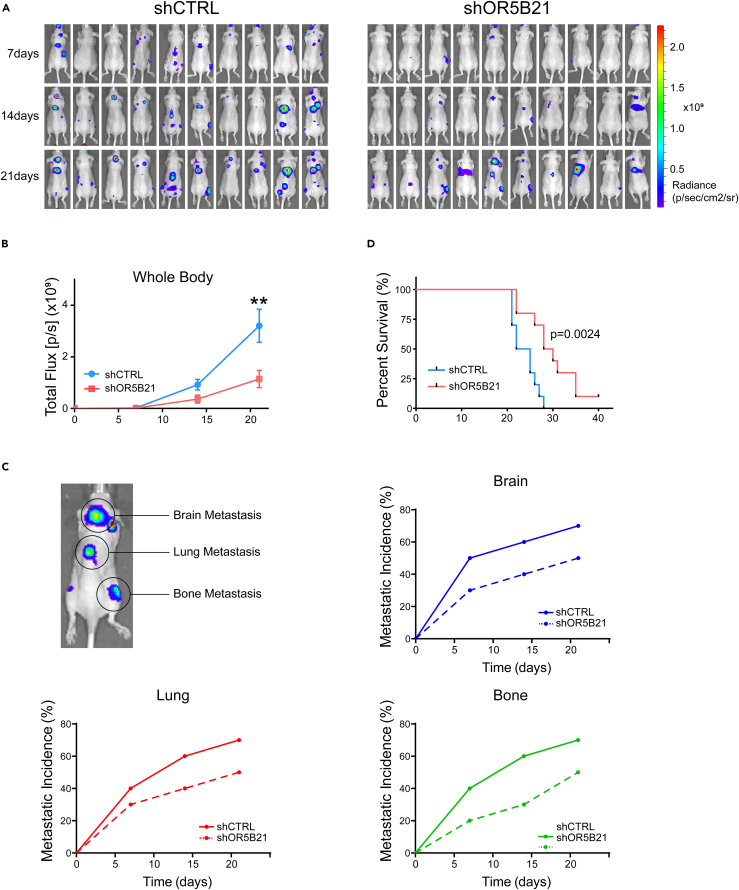
Our in vitro data revealed that OR5B21 was highly associated with pro-metastatic traits. To confirm the role of OR5B21 in breast cancer metastasis, we knocked down OR5B21 in the brain-seeking highly metastatic breast cancer line 231BR expressing firefly luciferase (Fluc) and injected them into the left ventricle of athymic mice. Tumor metastasis to different sites was monitored by bioluminescence imaging weekly. Although control mice formed multiple extensive metastases, OR5B21 knockdown led to a significant decrease in overall metastasis (Figures 3A and 3B). On day 21 post injection, the overall Fluc mean signal was 3.2 × 109 for the control group versus 1.14 × 109 for the OR5B21 knockdown group (p < 0.01, n = 10/group; Figure 3B). In addition, OR5B21 knockdown decreased the incidence rate of the brain, lung, and bone metastases (Figure 3C). For instance, 5/10, 6/10, and 7/10 mice formed brain metastasis in the control group compared with 3/10, 4/10, and 5/10 mice in the OR5B21 knockdown group, at days 7, 14, and 21 post tumor cells injection, respectively (Figure 3C). Importantly, OR5B21 knockdown resulted in a significant increase in the overall mice survival where the median survival for the shOR5B21 group was 29 days versus 23.5 days for the shCTRL group (n = 10/group; p = 0.0024; Figure 3D). Collectively, these data suggest that OR5B21 plays a vital role in breast cancer metastasis to different tissues, especially the brain.
Figure 3.
OR5B21 knockdown inhibits breast cancer metastasis
231BR cells expressing Fluc and either shCTRL or shOR5B21 were injected intracardially into athymic mice, and metastasis was monitored over time by bioluminescence imaging.
(A) Mouse images at days 7, 14, and 21 post cell injections.
(B) Overall whole-body bioluminescence signals at different time points; data presented as mean ± SEM; ∗∗p < 0.01 (n = 10 mice/group), unpaired Student’s t test.
(C) Brain, lung, and bone metastasis incidence rate for each group over time.
(D) Kaplan-Meir survival curves for each group, p = 0.0024 Log rank (Mantel-cox) test (n = 10 mice/group).
OR5B21 induces breast cancer metastasis by activating STAT3/NF-κB/CEBPβ signaling
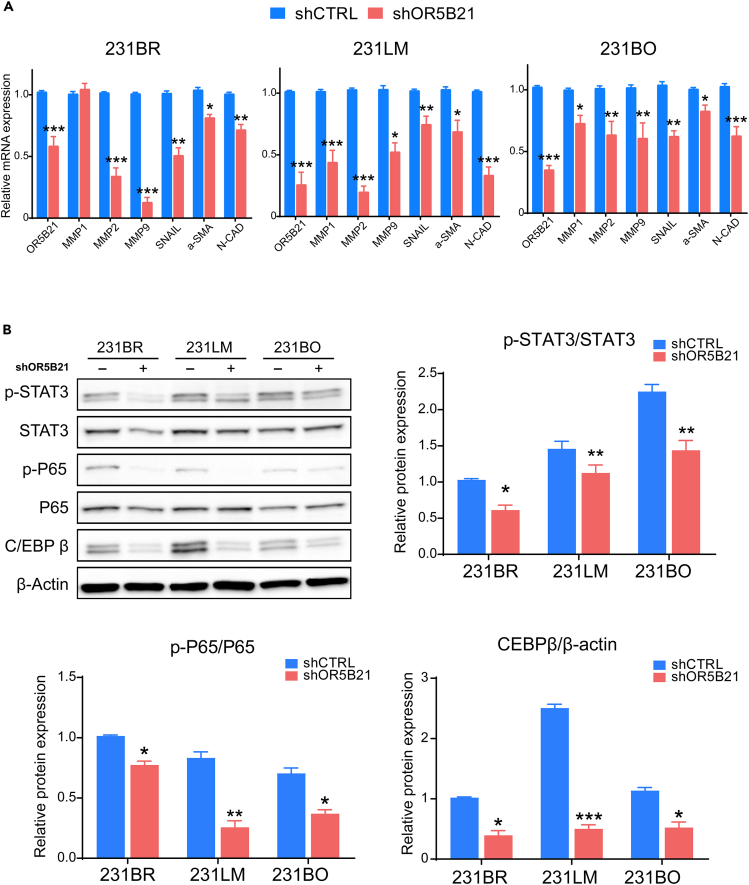
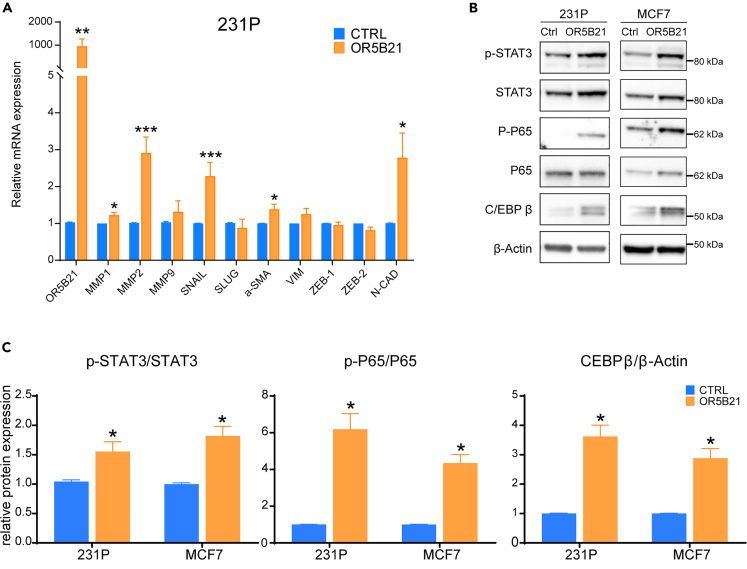
To identify the potential underlying mechanism responsible for the observed effect on breast cancer metastasis, we evaluated the outcome of OR5B21 genetic manipulation on markers associated with invasion/migration and epithelial to mesenchymal transition (EMT), typically involved in metastasis. As expected, MMP1, MMP2, MMP9, Snail, α-SMA, and/or N-cadherin were highly expressed in metastatic breast cancer lines compared with primary 231P (Figure S3). Knockdown of OR5B21 led to a significant decrease in MMP1, MMP2, and MMP9 (markers associated with invasion/migration) as well as SNAIL, α-SMA, and N-cadherin (associated with EMT) in 231BR, 231LM, and 231BO metastatic lines (Figure 4A). Since NF-κB activation is an important characteristic of EMT and cancer metastasis (Azam et al. 2020; Huber et al., 2004; Shostak and Chariot 2011; Smart et al. 2020), we evaluated the effect of OR5B21 on this pathway as well as two transcription factors known to co-operate with NF-κB as synergistic initiators and master regulators of EMT, the signal transducer and activator of transcription 3 (STAT3) and the CCAAT/enhancer binding protein b (C/EBPβ) (Bagnato and Rosanò 2019; Carro et al., 2010; Grivennikov and Karin 2010; Shackleford et al., 2011; Zahnow 2002, 2009). C/EBPβ promotes both STAT3 expression and NF-κB signaling (Canino et al., 2015; Cappello et al., 2009; Shackleford et al., 2011). Of interest, phosphorylation of STAT3 and upregulation of C/EBPβ in different metastatic breast cancer lines (231BR, 231LM, 231BO) coincided with phosphorylation of NF-κB p65 (Figure 4B). Knockdown of OR5B21 reversed STAT3 and p65 phosphorylation and decreased C/EBPβ expression in all three metastatic lines (Figure 4B). On the other hand, OR5B21 overexpression in 231P and MCF7 showed the opposite effect (Figures 5A–5C). Altogether, these data suggest that OR5B21 promotes breast cancer metastasis to different tissues by activating EMT through the STAT3/NF-κB/C/EBPβ signaling axis, revealing OR5B21 as a potential target for breast cancer therapy.
Figure 4.
OR5B21 regulates invasion/migration through STAT3/NF-κB/CEBPβ signaling pathway
(A) Relative mRNA levels of MMP1, MMP2, MMP9, SNAIL, α-SMA, N-CAD (normalized to β-Actin) in 231BR, 231LM, and 231BO expressing shCTRL or shOR5B21 (n = 7).
(B) 231BR, 231LM, and 231BO cells expressing shCTRL or shOR5B21 were analyzed by western blotting for p-STAT3, STAT3, p-P65, P65, C/EBP-β, and β-Actin loading control. Representative blots from three independent experiments are shown, while bar graphs represent relative quantification of each target, normalized to β-Actin.
Data presented as mean ± SEM (n = 3); ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 by unpaired Student's t test.
Figure 5.
OR5B21 overexpression increases invasion/migration of BC cells
(A) Relative mRNA levels of MMP1, MMP2, MMP9, SNAIL, α-SMA, N-CAD (normalized to β-Actin) in 231P control cells (CTRL) or overexpressing OR5B21.
(B and C) 231P and MCF7 control cells or overexpressing OR5B21 were analyzed by western blotting for p-STAT3, STAT3, p-P65, P65, C/EBP-β, and β-Actin loading control. Representative blots from three independent experiments are shown (B), while bar graphs represent relative quantification of each target, normalized to β-Actin (C).
Data presented as mean ± SEM (n = 3); ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 by unpaired Student's t test.
Discussion
The human OR repertoire contains 851 loci and mediates olfactory recognition by activating a G-protein-mediated transduction cascade in olfactory sensory neurons (Firestein 2001; Kato and Touhara 2009). In recent years, it has become clear that ORs play an essential role in physiological processes outside the nasal epithelium, some of which are ectopically overexpressed in different cancers (Flegel et al., 2013; Gelis et al., 2016; Kalra et al., 2020; Neuhaus et al., 2009; Rodriguez et al., 2014). Thus, ORs have emerged as markers for cancer diagnostics and potential therapeutic targets (Lee et al., 2019). For instance, 34 ORs were observed in breast cancer with the CHEK2 1100delC mutation (Muranen et al., 2011), whereas OR2T6 showed a key role in breast cancer progression (Li et al., 2019). OR transcript abundance revealed that OR2W3 is associated with invasion and basal-like subtype, whereas OR2B6 correlates with proliferation and luminal A subtype (Masjedi et al., 2019a, 2019b). Activation of different ORs can regulate various signaling pathways leading to either enhanced migration and proliferation or apoptosis. Expression of OR51E1 and closely related OR51E2 is upregulated in prostate cancer (Lee et al., 2019; Maβberg et al., 2016; Neuhaus et al., 2009; Sanz et al., 2014; Xia et al., 2001; Xu et al., 2000). Overexpression or stimulation of OR51E2 promotes prostate cancer invasiveness and progression (Neuhaus et al., 2009; Rodriguez et al., 2016; Sanz et al., 2014). OR51E2 was identified as a potential tumor antigen for CD8+ cytotoxic T cells with implications in cancer immunotherapy (Matsueda et al., 2012). OR51E1 activation negatively regulates prostate cancer cell growth (Maβberg et al., 2016), whereas OR51E2 inhibits melanoma cell proliferation/migration and induces apoptosis (Gelis et al., 2017). Similarly, OR2J3 is overexpressed in lung cancer, and its activation results in apoptosis through ERK1/ERK2/PI3K signaling (Kalbe et al., 2017). Activation of OR1A2 inhibits cell proliferation of liver cancer cells via p38 MAPK signaling (Maβberg et al., 2015). OR51B4 activation in colorectal cancer and OR2AT4 in acute and chronic myelogenous leukemia inhibits cell migration and induces apoptosis (Manteniotis et al., 2016; Weber et al., 2017). In addition to these, several other olfactory receptors serve as potential biomarkers or therapeutic targets in different cancers, such as OR2C3 for melanoma and OR10H1 for bladder cancer (Lee et al., 2019; Weber et al., 2018b). Here, we provide evidence that OR5B21 is ectopically expressed in different breast cancer metastatic sites, with the most upregulation observed in the brain. Interfering with OR5B21 expression leads to inhibition of breast cancer invasion, migration, and metastasis through the STAT3/NF-κB/C/EBPβ signaling axis, serving as a potential therapeutic target for breast cancer.
ORs are a member of GPCR family known to directly or indirectly stimulate STAT transcription factor activity (Ram et al. 2000; Wu et al. 2003; Yu et al., 2014). STAT3, one of the seven members of the STAT family, is constitutively activated in all breast cancer subtypes and is most often associated with triple-negative breast cancer (Furth 2014; Song et al., 2008; Yu et al., 2014). Aberrant STAT3 signaling promotes breast tumor metastasis by interacting with the MMP9 promoter through Fra-1 and c-Jun (Song et al., 2008). On the other hand, STAT3 activation is associated with Snail expression, an interlined event (Saitoh et al., 2016; Xie et al., 2020). Furthermore, several studies showed that STAT3 and C/EBPβ transcription factors act synergistically and co-operate with NF-κB to induce mesenchymal transformation, stimulating cancer metastasis (Cappello et al., 2009; Carro et al., 2010; Schweiger et al., 2020; Shackleford et al., 2011; Xia et al., 1997). In our breast cancer model, aberrant STAT3 signaling correlated with overexpression of MMP9, Snail, and activation of NF-κB in breast cancer metastasis to different tissues (brain, bone, and lung). Of importance, OR5B21 knockdown inhibited phosphorylation of STAT3 and reversed the expression of MMP2, MMP9, and Snail, which coincided with a decrease in both C/EBPβ levels and activation of NF-κB, whereas overexpression of OR5B21 had the opposite effect, endorsing the role of C/EBPβ/STAT3 in OR5B21-induced metastasis. Certainly, additional mechanisms could also be involved. The ligands to which OR5B21 responds to are unknown, and how exactly these ligands affect breast cancer metastasis through OR5B21 remains unclear.
In summary, we identified that OR5B21 plays a vital role in breast cancer metastasis by inducing EMT through the STAT3/NF-κB/C/EBPβ signaling pathway. Downregulation of OR5B21 significantly inhibited breast cancer metastasis to different tissues, especially the brain. Thus, we propose OR5B21 as a potential target for adjuvant breast cancer therapy.
Limitations of the study
There are three major limitations in this study that could be addressed in future research. First, an absence of adequate antibodies against OR5B21 limits the evaluation of OR5B21 protein expression levels in various tissues. In this study, we analyzed OR5B21 expression in normal and primary breast cancer tissue using the TCGA-BRCA dataset, but the expression and clinicopathological characteristics of OR5B21 in breast cancer metastasis versus the primary tumor are still unknown. The role of olfactory receptors in cancer biology is gaining more attraction, and future studies may overcome this challenge with commercially available antibodies. Second, to establish a model that mimics the human pathophysiology, the widely established human breast cancer cell line MDA-MB-231 in immunocompromised mice was used for in vivo studies, but since olfactory receptors have been reported to be involved in cancer immune regulation (Matsueda et al., 2012), future work should investigate the role of OR5B21 in an immunocompetent background to further elucidate its role in anti-cancer immunity. Third, the vast majority of ORs are not examined in this work and screening the olfactory receptor family expressions in primary and metastatic breast cancer from patient tissue by proteomics or genomic sequencing would unravel information on the role of additional olfactory receptors in breast cancer metastasis.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Phospho-Stat3 (Tyr705) (D3A7) XP® Rabbit mAb | Cell Signaling Technology | Cat#9145; RRID: AB_2491009 |
| Stat3 (79D7) Rabbit mAb | Cell Signaling Technology | Cat#4904; RRID: AB_331269 |
| Phospho-NF-κB p65 (Ser536) (93H1) Rabbit mAb | Cell Signaling Technology | Cat#3033; RRID: AB_331284 |
| NF-κB p65 (C22B4) Rabbit mAb | Cell Signaling Technology | Cat#4764; RRID: AB_823578 |
| β-Actin (8H10D10) Mouse mAb | Cell Signaling Technology | Cat#3700; RRID: AB_2242334 |
| C/EBPβ (LAP) Antibody | Cell Signaling Technology | Cat#3087; RRID: AB_2078052 |
| Anti-rabbit IgG, HRP-linked Antibody | Cell Signaling Technology | Cat#7074; RRID: AB_2099233 |
| Anti-mouse IgG, HRP-linked Antibody | Cell Signaling Technology | Cat#7076; RRID: AB_330924 |
| Chemicals, peptides, and recombinant proteins | ||
| D-Luciferin, Potassium Salt | Gold Biotechnology | LUCK-100 |
| Cypridina Luciferin TFA salt | Nanolight | 305-1 |
| SuperSignal™ West Pico PLUS Chemiluminescent Substrate | ThermoFisher | 34577 |
| SuperSignal™ West Femto PLUS Chemiluminescent Substrate | ThermoFisher | 34095 |
| Matrigel matrix | Corning | 356234 |
| Crystal violet solution | Sigma | V5265 |
| Critical commercial assays | ||
| RNeasy Mini Kit | QIAGEN | 217004 |
| 5×All-In-One RT MasterMix Kit | Abm | G485 |
| PowerUp™ SYBR™ Green Master Mix | ThermoFisher | A25742 |
| Novex™ WedgeWell™ 10%, Tris-Glycine gel | ThermoFisher | XP00100PK2 |
| Nitrocellulose Membrane, 0.45 μm | Bio-Rad | 1620115 |
| Pierce™ BCA Protein Assay Kit | ThermoFisher | 23225 |
| RIPA Buffer | Boston Bioproducts | #BP-115 |
| Halt™ Protease and Phosphatase Inhibitor Cocktail | ThermoFisher | 78441 |
| 24 mm Transwell® with 8.0 μm Pore Polycarbonate Membrane Insert | Corning | 3428 |
| Deposited data | ||
| TCGA breast cancer dataset | TCGA-BRCA | https://portal.gdc.cancer.gov/projects/TCGA-BRCA |
| Experimental models: Cell lines | ||
| MDA-MB-231 | ATCC | HTB-26 |
| ErbB2 parental and brain metastatic cell lines | Joan Massague laboratory | Muller et al. 1988; Valiente et al. 2014 |
| MDA-MB-231parental and metastatic cell lines | Ichiro Nakano laboratory | Yamashita et al. 2020 |
| MCF7 | ATCC | HTB-22 |
| Experimental models: Organisms/strains | ||
| Mouse: Athymic Nude-Foxn1nu | Envigo | 6904F |
| Oligonucleotides | ||
| Primers for qPCR, see Table S1 | MGH primerbank | https://pga.mgh.harvard.edu/primerbank/ |
| Recombinant DNA | ||
| pLKO.1-puro Mammalian shOR5B21 Transduction Particles | Sigma | TRCN0000203498 |
| pLKO.1-puro Mammalian shRNA Control Transduction Particles | Sigma | SHC016 |
| pLenti-GIII-CMV-GFP-2A-Puro-OR5B21 | Abm | LV247930 |
| pLenti-CMV-GFP-2A-Puro-Blank Vector | Abm | LV590 |
| Software and algorithms | ||
| imageJ | National Institutes of Health | https://imagej.nih.gov/ij/ |
| Graphpad prime 7.0a | Graphpad | https://www.graphpad.com/ |
| Adobe Illustrator CS6 | Adobe | https://www.adobe.com/products/illustrator/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Bakhos A. Tannous (btannous@hms.harvard.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Cell culture
MDA-MB-231 metastatic cell lines were established and previously reported (Yamashita et al., 2020). In brief, the metastatic murine model was established via cardiac injection of MDA-MB-231 in immunocompromised mice and which then produced a series of derivative cell lines from the resultant tumors within the brain (231-BR), lung (231-LM), and bone (231-BO). The human breast cancer cell line MDA-MB-231 (231P, ATCC, HTB-26), its metastatic sublines (231BR, 231LM, 231BO), and MCF7(ATCC, HTB-22) cell line were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. All experiments were performed with cells passaged <10 times. ErbB2-P activated mouse breast cancer line [generated from FVB-Tg(MMTV-Erbb2)NK1Mul/J transgenic mice and its brain-seeking clones ErbB2-BrM1 and ErbB2-BrM2 were a kind gift from Dr. Massague (MSKCC) (Muller et al., 1988; Valiente et al., 2014).
In vivo breast cancer metastasis model
All animal studies were approved by the Massachusetts General Hospital Subcommittee on Research Animal Care following guidelines set forth by the National Institutes of Health Guide for the Care and Use of Laboratory Animals. 500,000 231BR cells expressing Fluc were engineered to express shCTRL or shOR5B21 and injected into the left ventricles of 6-weeks old female athymic nude mice (Chung et al., 2009). The growth/dissemination of tumors was monitored weekly by bioluminescence imaging after injecting the mice intraperitoneally with D-luciferin (150mg/kg body weight; Gold Biotechnology) using a Xenogen IVIS 200 Imaging System (PerkinElmer), and quantifying image intensity using Living Image software 4.3.1 (PerkinElmer). We defined a metastatic event as any detectable luciferase signal above the background.
Method details
Plasmid transfection and lentiviral transduction
Lentiviral vectors expressing shRNA for OR5B21 and shCTRL were purchased from Sigma (SIGMA TRCN0000203498; sequence: CCGGCTATGGCACAATCATCTTCATCTCGA GATGAAGATGATTGTGCCATAGTTTTTTG). Overexpression of OR5B21 was achieved by lentivirus vector expressing this receptor under the control of a constitutively active promoter (Abm, LV247930, NM_001005218). Lentivirus vector stocks were produced by triple transfection of 293T cells with the lentivirus vector plasmid, the packaging genome plasmid (pCMVΔR8.91) and plasmid encoding the vesicular stomatitis virus envelope glycoprotein (pVSV-G); viral particles were collected from conditioned medium, concentrated by ultracentrifugation, and tittered as transducing units per ml (Maguire et al., 2013). Cells were transduced with each of these lentivirus vector at a multiplicity of infection of 10 transducing units per cell in the presence of 10 ug/ml of polybrene and OR5B21 transcript abundance was analyzed 48 hrs post-transduction by qPCR.
Cell invasion/migration assay
For scratchy invasion assay, cells engineered with shOR5B21 or shCTRL or to overexpress OR5B21 were starved for 18 hrs in DMEM containing 1% FBS and seeded at 5 × 106 cells/well in a six-well plate until forming a monolayer. A 200ul pipette tip was used to create a scratch wound, and the medium was immediately refreshed. Cells migrating from the leading edge were photographed at different time points using phase-contrast microscopy. The wound healing area was measured in a 10× field using ImageJ (National Institutes of Health). For transwell migration assay, Matrigel matrix (Corning, 356234) was thawed at 4°C overnight, and diluted in DMEM (1:1). Next, each transwell insert (8μm pore size, Corning) was coated with 300ul of diluted Matrigel matrix and incubated at 37°C for 1 hour. 20%FBS DMEM was added to the lower chambers as chemoattractant. MDA-MB-231 cells were trypsinized, resuspended in 1%FBS DMEM and seeded in Matrigel matrix coated transwell inserts with a final density of 1 × 104/well. 48 hrs later, transwell inserts were washed with PBS and upper side of inserts were gently swabbed by cotton swabs, cells that migrated to the bottom well were fixed in 20% methanol stained with 1% crystal violet (Sigma) and imaged under a microscope.
Cell viability assay
For colony formation assay, MDA-MB-231 cell lines engineered with shOR5B21 or shCTRL or to overexpress OR5B21 were seeded with density of 1,000/well in 6-well plate. In 14 days, the cells were fixed in 20% methanol and stained with 1% crystal violet (Sigma). Colony images were captured under microscope and the colony was defined to consist of at least 50 cells. Using secreted Vluc reporter assay to monitor cell viability were established and previously reported. In brief, OR5B21 was knocked down in 231BR cells expressing Vargula hilgendorfii luciferase (Vluc) under a constitutively active SV40 minimal promoter, and cell proliferation was measured over time by evaluating the Vluc levels in 50 μL conditioned medium using 50 μL vargulin substrate (5 ng mL−1; Nanolight).
Western blotting
Total protein was extracted from cells using RIPA buffer (Boston Bioproducts) containing 1× protease/phosphatase inhibitors cocktails (ThermoFisher) and quantified using the BCA reagent kit (ThermoFisher). Thirty μg of protein were electrophoresed in 10% Novex™ Tris-Glycine Gels (ThermoFisher), transferred to nitrocellulose membranes (Bio-Rad) followed by blocking in 5% non-fat milk in TBS/0.1% TWEEN for 1 hour. Membranes were then probed overnight with the primary antibody in TBS/0.1% TWEEN. The primary antibodies used were: Phospho-p65 and total p65, phospho-STAT3 and total STAT3, C/EBP-β, and β-Actin as a loading control (Cell Signaling). Corresponding secondary antibody-HRP conjugate were used (Cell Signaling). Proteins were visualized using SuperSignal™ West Pico PLUS Chemiluminescent Substrate or SuperSignal™ West Femto PLUS Chemiluminescent Substrate (ThermoFisher).
RNA isolation and RT-qPCR
Cell pellets were collected, and RNA was isolated using RNeasy Mini Kit (QIAGEN). 2 μg total RNA from individual samples were reverse transcribed into cDNA using 5×All-In-One RT MasterMix Kit (Abm). Expression of different genes was analyzed by quantitative real-time PCR using specific primers, with β-actin mRNA as an internal control using a QuantStudio 3 PCR system (Applied Biosystems) with SYBR Green Master Mix (ThermoFisher). Primer sequences are listed in Table S1.
Quantification and statistical analysis
GraphPad Prism v7.0a software (LaJolla, CA) was used for statistical analysis of all data. Graphs and figures were generated using GraphPad Prism v.7.0a and Adobe Illustrator CS6. All cell culture experiments consisted of a minimum of three independent replicates and were repeated at least three times. The results are presented as the mean ± SEM. A p value less than 0.05 was considered to be statistically significant (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001). Normality of distribution was verified by applying a Shapiro–Wilks's test for normality. For analysis between 2 groups, a two-tailed Student's t-test (unpaired, two tails) was used. For analysis between multiple groups, one-way ANOVA was used. Survival was analyzed using Kaplan-Meier curves and log rank (Mantel-Cox) tests.
Acknowledgments
The authors thank Dr. Joan Massague (MSKCC) for providing the primary ErbB2-P breast cancer lines and corresponding brain-seeking clones ErbB2-BrM1 and ErbB2-BrM2, Elie E. Tabet for producing the viral vector in the MGH vector core (supported by NIH/NINDS P30NS04776), Katherine Xiang for help with figure illustrations, and 1S10RR015504 Shared Instrumentation grant for the IVIS imaging system. This work was supported by grants from the National Institutes of Health, the National Institute of Neurological Disorders and Stroke P30NS04776 (B.A.T.).
Author contributions
M.L., L.A.C., I.N., and B.A.T. designed the research and experiments and wrote and edited the manuscript; M.L., D.J.R., M.W.S., and L.A.C. performed experiments and collected data; M.L., M.W.S., L.A.C., and B.A.T. interpreted and analyzed the data.
Declaration of interests
The authors declare no competing interests.
Published: December 17, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103519.
Contributor Information
Litia A. Carvalho, Email: lcarvalho@mgh.harvard.edu.
Bakhos A. Tannous, Email: btannous@hms.harvard.edu.
Supplemental information
Data and code availability
This paper analyzes existing, publicly available data. The accession number and link are listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Azam Z., To S.-S.T., Tannous B.A. Mesenchymal transformation: the rosetta stone of glioblastoma pathogenesis and therapy resistance. Adv. Sci. 2020;7:2002015. doi: 10.1002/advs.202002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnato A., Rosanò L. New routes in GPCR/β-arrestin-driven signaling in cancer progression and metastasis. Front. Pharmacol. 2019;10:114. doi: 10.3389/fphar.2019.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canino C., Luo Y.Y., Marcato P., Blandino G., Pass H.I., Cioce M. A STAT3-NFkB/DDIT3/CEBPβ axis modulates ALDH1A3 expression in chemoresistant cell subpopulations. Oncotarget. 2015;6:12637–12653. doi: 10.18632/oncotarget.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappello C., Zwergal A., Kanclerski S., Haas S.C., Kandemir J.D., Huber R., Page S., Brand K. C/EBPbeta enhances NF-KappaB-associated signalling by reducing the level of IkappaB-alpha. Cell Signal. 2009;21:1918–1924. doi: 10.1016/j.cellsig.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Carro M.S., Lim W.K., Alvarez M.J., Bollo R.J., Zhao X., Snyder E.Y., Sulman E.P., Anne S.L., Doetsch F., Colman H., et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E., Yamashita H., Au P., Tannous B.A., Fukumura D., Jain R.K. Secreted Gaussia luciferase as a biomarker for monitoring tumor progression and treatment response of systemic metastases. PLoS One. 2009;4:e8316. doi: 10.1371/journal.pone.0008316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis C.E., Fedewa S.A., Sauer A.G., Kramer J.L., Smith R.A., Jemal A. Breast Cancer Statistics, 2015: convergence of incidence rates between black and white women. CA: A Cancer J. Clinicians. 2016;66:31–42. doi: 10.3322/caac.21320. [DOI] [PubMed] [Google Scholar]
- Dorsam R.T., Gutkind J.S. G-protein-coupled receptors and cancer. Nat. Rev. Cancer. 2007;7:79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- Firestein S. How the olfactory system makes sense of scents. Nature. 2001;413:211–218. doi: 10.1038/35093026. [DOI] [PubMed] [Google Scholar]
- Flegel C., Manteniotis S., Osthold S., Hatt H., Gisselmann G. Expression profile of ectopic olfactory receptors determined by deep sequencing. PLoS One. 2013;8:e55368. doi: 10.1371/journal.pone.0055368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth P.A. STAT Signaling in different breast cancer sub-types. Mol. Cell Endocrinol. 2014;382:612–615. doi: 10.1016/j.mce.2013.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelis L., Jovancevic N., Bechara F.G., Neuhaus E.M., Hatt H. Functional expression of olfactory receptors in human primary melanoma and melanoma metastasis. Exp. Dermatol. 2017;26:569–576. doi: 10.1111/exd.13316. [DOI] [PubMed] [Google Scholar]
- Gelis L., Jovancevic N., Veitinger S., Mandal B., Arndt H.D., Neuhaus E.M., Hatt H. Functional characterization of the odorant receptor 51E2 in human melanocytes. J. Biol. Chem. 2016;291:17772–17786. doi: 10.1074/jbc.M116.734517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S.I., Karin M. Dangerous liaisons: STAT3 and NF-KappaB collaboration and crosstalk in cancer. Cytokine Growth Factor Rev. 2010;21:11–19. doi: 10.1016/j.cytogfr.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber M.A., Azoitei N., Baumann B., Grünert S., Sommer A., Pehamberger H., Kraut N., Beug H., Wirth T. NF-KappaB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J. Clin. Invest. 2004;114:569–581. doi: 10.1172/JCI21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchings C.J., Koglin M., Olson W.C., Marshall F.H. Opportunities for Therapeutic antibodies directed at g-protein-coupled receptors. Nat. Rev. Drug Discov. 2017;16:787–810. doi: 10.1038/nrd.2017.91. [DOI] [PubMed] [Google Scholar]
- Kalbe B., Schulz V.M., Schlimm M., Philippou S., Jovancevic N., Jansen F., Scholz P., Lübbert H., Jarocki M., Faissner A., et al. Helional-induced activation of human olfactory receptor 2J3 promotes apoptosis and inhibits proliferation in a non-small-cell lung cancer cell line. Eur. J. Cell Biol. 2017;96:34–46. doi: 10.1016/j.ejcb.2016.11.004. [DOI] [PubMed] [Google Scholar]
- Kalra S., Mittal A., Gupta K., Singhal V., Gupta A., Mishra T., Naidu S., Sengupta D., Ahuja G. Analysis of single-cell transcriptomes links enrichment of olfactory receptors with cancer cell differentiation status and prognosis. Commun. Biol. 2020;3:506. doi: 10.1038/s42003-020-01232-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A., Touhara K. Mammalian olfactory receptors: pharmacology, G protein coupling and desensitization. Cell Mol. Life Sci. 2009;66:3743–3753. doi: 10.1007/s00018-009-0111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert A.W., Pattabiraman D.R., Weinberg R.A. Emerging biological principles of metastasis. Cell. 2017;168:670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.-J., Depoortere I., Hatt H. Therapeutic potential of ectopic olfactory and taste receptors. Nat. Rev. Drug Discov. 2019;18:116–138. doi: 10.1038/s41573-018-0002-3. [DOI] [PubMed] [Google Scholar]
- Li M., Wang X., Ma R.-R., Shi D.-B., Wang Y.-W., Li X.-M., He J.-Y., Wang J., Gao P. The olfactory receptor family 2, subfamily t, member 6 (OR2T6) is involved in breast cancer progression via initiating epithelial-mesenchymal transition and MAPK/ERK pathway. Front. Oncol. 2019;9:1210. doi: 10.3389/fonc.2019.01210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire C.A., Bovenberg M.S., Crommentuijn M.H.W., Niers J.M., Kerami M., Teng J., Sena-Esteves M., Badr C.E., Tannous B.A. Triple bioluminescence imaging for in vivo monitoring of cellular processes. Mol. Ther. Nucleic Acids. 2013;2:e99. doi: 10.1038/mtna.2013.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manteniotis S., Wojcik S., Brauhoff P., Möllmann M., Petersen L., Göthert J.R., Schmiegel W., Dührsen U., Gisselmann G., Hatt H. Functional characterization of the ectopically expressed olfactory receptor 2AT4 in human myelogenous leukemia. Cell Death Discov. 2016;2:15070. doi: 10.1038/cddiscovery.2015.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masjedi S., Zwiebel L.J., Giorgio T.D. Olfactory receptor gene abundance in invasive breast carcinoma. Sci. Rep. 2019;9:13736. doi: 10.1038/s41598-019-50085-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masjedi S., Zwiebel L.J., Giorgio T.D. Olfactory receptor gene abundance in invasive breast carcinoma. Sci. Rep. 2019;9:1–12. doi: 10.1038/s41598-019-50085-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maßberg D., Jovancevic N., Offermann A., Simon A., Baniahmad A., Perner S., Pungsrinont T., Luko K., Philippou S., Ubrig B., et al. The activation of OR51E1 causes growth suppression of human prostate cancer cells. Oncotarget. 2016;7:48231–48249. doi: 10.18632/oncotarget.10197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maßberg D., Simon A., Häussinger D., Keitel V., Gisselmann G., Conrad H., Hatt H. Monoterpene (-)-citronellal affects hepatocarcinoma cell signaling via an olfactory receptor. Arch. Biochem. Biophys. 2015;566:100–109. doi: 10.1016/j.abb.2014.12.004. [DOI] [PubMed] [Google Scholar]
- Matsueda S., Wang M., Weng J., Li Y., Yin B., Zou J., Li Q., Zhao W., Peng W., Legras X., et al. Identification of prostate-specific G-protein coupled receptor as a tumor antigen recognized by CD8+ T cells for cancer immunotherapy. PLoS One. 2012;7:1–9. doi: 10.1371/journal.pone.0045756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller W.J., Sinn E., Pattengale P.K., Wallace R., Leder P. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell. 1988;54:105–115. doi: 10.1016/0092-8674(88)90184-5. [DOI] [PubMed] [Google Scholar]
- Muranen T.A., Greco D., Fagerholm R., Kilpivaara O., Kämpjärvi K., Aittomäki K., Blomqvist C., Heikkilä P., Borg Å., Nevanlinna H. Breast tumors from CHEK2 1100delC-mutation carriers: genomic landscape and clinical implications. Breast Cancer Res. 2011;13:R90. doi: 10.1186/bcr3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus E.M., Zhang W., Gelis L., Deng Y., Noldus J., Hatt H. Activation of an olfactory receptor inhibits proliferation of prostate cancer cells. J. Biol. Chem. 2009;284:16218–16225. doi: 10.1074/jbc.M109.012096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves S.R., Ram P.T., Iyengar R. G protein pathways. Science. 2002;296:1636–1639. doi: 10.1126/science.1071550. [DOI] [PubMed] [Google Scholar]
- O’Hayre M., Vázquez-Prado J., Kufareva I., Stawiski E.W., Handel T.M., Seshagiri S., Gutkind J.S. The emerging mutational landscape of G proteins and G-protein-coupled receptors in cancer. Nat. Rev. Cancer. 2013;13:412–424. doi: 10.1038/nrc3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram P.T., Horvath C.M., Iyengar R. Stat3-mediated transformation of NIH-3T3 cells by the constitutively active Q205L Galphao protein. Science. 2000;287:142–144. doi: 10.1126/science.287.5450.142. [DOI] [PubMed] [Google Scholar]
- Rodriguez M., Luo W., Weng J., Zeng L., Yi Z., Siwko S., Liu M. PSGR promotes prostatic intraepithelial neoplasia and prostate cancer xenograft growth through NF-ΚB. Oncogenesis. 2014;3:e114. doi: 10.1038/oncsis.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M., Siwko S., Zeng L., Li J., Yi Z., Liu M. Prostate-specific G-protein-coupled receptor collaborates with loss of PTEN to promote prostate cancer progression. Oncogene. 2016;35:1153–1162. doi: 10.1038/onc.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh M., Endo K., Furuya S., Minami M., Fukasawa A., Imamura T., Miyazawa K. STAT3 integrates cooperative ras and TGF-β signals that induce snail expression. Oncogene. 2016;35:1049–1057. doi: 10.1038/onc.2015.161. [DOI] [PubMed] [Google Scholar]
- Sanz G., Leray I., Dewaele A., Sobilo J., Lerondel S., Bouet S., Grébert D., Monnerie R., Pajot-Augy E., Mir L.M. Promotion of cancer cell invasiveness and metastasis emergence caused by olfactory receptor stimulation. PLoS One. 2014;9:e85110. doi: 10.1371/journal.pone.0085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger M.W., Li M., Giovanazzi A., Fleming R.L., Tabet E.I., Nakano I., Würdinger T., Chiocca E.A., Tian T., Tannous B.A. Extracellular vesicles induce mesenchymal transition and therapeutic resistance in glioblastomas through NF-ΚB/STAT3 signaling. Adv. Biosyst. 2020;4:1900312. doi: 10.1002/adbi.201900312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleford T.J., Zhang Q., Tian L., Vu T.T., Korapati A.L., Baumgartner A.M., Le X.-F., Liao W.S., Claret F.X. Stat3 and CCAAT/enhancer binding protein beta (C/EBP-beta) regulate Jab1/CSN5 expression in mammary carcinoma cells. Breast Cancer Res. 2011;13:R65. doi: 10.1186/bcr2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shostak K., Chariot A. NF-ΚB, stem cells and breast cancer: the links get stronger. Breast Cancer Res. 2011;13:214. doi: 10.1186/bcr2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2021. CA: A Cancer J. Clinicians. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- Smart E., Semina S.E., Frasor J. Update on the role of NFκB in promoting aggressive phenotypes of estrogen receptor-positive breast cancer. Endocrinology. 2020;161:bqaa152. doi: 10.1210/endocr/bqaa152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Qian L., Song S., Chen L., Zhang Y., Yuan G., Zhang H., Xia Q., Hu M., Yu M., et al. Fra-1 and Stat3 synergistically regulate activation of human MMP-9 gene. Mol. Immunol. 2008;45:137–143. doi: 10.1016/j.molimm.2007.04.031. [DOI] [PubMed] [Google Scholar]
- Valiente M., Obenauf A.C., Jin X., Chen Q., Zhang X.H.F., Lee D.J., Chaft J.E., Kris M.G., Huse J.T., Brogi E., Massagué J. Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell. 2014;156:1002–1016. doi: 10.1016/j.cell.2014.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber L., Al-Refae K., Ebbert J., Jägers P., Altmüller J., Becker C., Hahn S., Gisselmann G., Hatt H. Activation of odorant receptor in colorectal cancer cells leads to inhibition of cell proliferation and apoptosis edited by H. Matsunami. PLoS One. 2017;12:e0172491. doi: 10.1371/journal.pone.0172491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber L., Maßberg D., Becker C., Altmüller J., Ubrig B., Bonatz G., Wölk G., Philippou S., Tannapfel A., Hatt H., Gisselmann G. Olfactory receptors as biomarkers in human breast carcinoma tissues. Front. Oncol. 2018;8:33. doi: 10.3389/fonc.2018.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber L., Schulz W.A., Philippou S., Eckardt J., Ubrig B., Hoffmann M.J., Tannapfel A., Kalbe B., Gisselmann G., Hatt H. Characterization of the olfactory receptor OR10H1 in human urinary bladder cancer. Front. Physiol. 2018;9:456. doi: 10.3389/fphys.2018.00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu E.H.T., Lo R.K.H., Wong Y.H. Regulation of STAT3 activity by G16-coupled receptors. Biochem. Biophys.Res. Commun. 2003;303:920–925. doi: 10.1016/s0006-291x(03)00451-0. [DOI] [PubMed] [Google Scholar]
- Wu V., Yeerna H., Nohata N., Chiou J., Harismendy O., Raimondi F., Inoue A., Russell R.B., Tamayo P., Gutkind J.S. Illuminating the Onco-GPCRome: novel G protein-coupled receptor-driven oncocrine networks and targets for cancer immunotherapy. J. Biol. Chem. 2019;294:11062–11086. doi: 10.1074/jbc.REV119.005601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia C., Cheshire J.K., Patel H., Woo P. Cross-talk between transcription factors NF-kappa B and C/EBP in the transcriptional regulation of genes. Int. J. Biochem. Cell Biol. 1997;29:1525–1539. doi: 10.1016/s1357-2725(97)00083-6. [DOI] [PubMed] [Google Scholar]
- Xia C., Ma W., Wang F., Sb H., Liu M. Identification of a prostate-specific g-protein coupled receptor in prostate cancer. Oncogene. 2001;20:5903–5907. doi: 10.1038/sj.onc.1204803. [DOI] [PubMed] [Google Scholar]
- Xie Q., Zhu Z., He Y., Zhang Z., Zhang Y., Wang Y., Luo J., Peng T., Cheng F., Gao J., et al. A lactate-induced snail/STAT3 pathway drives GPR81 expression in lung cancer cells. Biochim. Biophys. Acta Mol. Basis Dis. 2020;1866:165576. doi: 10.1016/j.bbadis.2019.165576. [DOI] [PubMed] [Google Scholar]
- Xu L.L., Stackhouse B.G., Florence K., Zhang W., Shanmugam N., Sesterhenn I.A., Zou Z., Srikantan V., Augustus M., Roschke V., et al. PSGR, a novel prostate-specific gene with homology to a g protein-coupled receptor, is overexpressed in prostate cancer. Cancer Res. 2000;60:6568–6572. [PubMed] [Google Scholar]
- Yamashita D., Minata M., Ibrahim A.N., Yamaguchi S., Coviello V., Bernstock J.D., Harada S., Cerione R.A., Tannous B.A., La Motta C., Nakano I. Identification of ALDH1A3 as a viable therapeutic target in breast cancer metastasis-initiating cells. Mol. Cancer Ther. 2020;19:1134–1147. doi: 10.1158/1535-7163.MCT-19-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Lee H., Herrmann A., Buettner R., Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat. Rev. Cancer. 2014;14:736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- Zahnow C.A. CCAAT/enhancer binding proteins in normal mammary development and breast cancer. Breast Cancer Res. 2002;4:113–121. doi: 10.1186/bcr428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahnow C.A. CCAAT/enhancer-binding protein β: its role in breast cancer and associations with receptor tyrosine kinases. Expert Rev. Mol. Med. 2009;11:1–34. doi: 10.1017/S1462399409001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper analyzes existing, publicly available data. The accession number and link are listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.