Abstract
Posttranscriptional regulation is important for tumor necrosis factor alpha (TNF-α) expression in monocytes and macrophages, and an AU-rich element (ARE) in the 3′ untranslated region (UTR) of TNF-α mRNA is implicated in control of its translation and mRNA stability. Regulation of mRNA turnover is thought to be mediated by trans-acting proteins, which bind the ARE and stabilize or destabilize the transcript. However, with the exception of the destabilizing factor tristetraprolin, the identity and function of the proteins binding the TNF-α mRNA ARE have not been established. To identify other proteins involved in the posttranscriptional control of TNF-α, the subcellular location of TNF-α mRNA was determined in the macrophage-like cell line RAW 264.7. TNF-α mRNA was located in the pellet following centrifugation of cytoplasm at 100,000 × g (P100 fraction). This fraction also contained proteins which formed two distinct ARE-specific complexes with the TNF-α mRNA 3′ UTR in electrophoretic mobility shift assays (EMSAs). A protein present in these two complexes was purified and identified by peptide mass mapping and tandem mass spectrometry as HuR. In EMSAs both complexes were supershifted by an anti-HuR antibody, while Western blotting also demonstrated the presence of HuR in the P100 extract. A HeLa cell tetracycline-regulated reporter system was used to determine the effect of HuR on mRNA stability. In this system, overexpression of HuR resulted in stabilization of an otherwise unstable reporter-mRNA containing the TNF-α ARE. These results demonstrate that the TNF-α ARE is a target of the mRNA-stabilizing factor HuR.
Tumor necrosis factor alpha (TNF-α) is an important inflammatory cytokine which orchestrates key features of the inflammatory response such as leukocyte migration, tissue resorption, the acute-phase response, and fever (5). Overproduction of TNF-α has been implicated in the pathogenesis of chronic inflammatory diseases such as rheumatoid arthritis and Crohn's disease (15).
The biosynthesis of TNF-α is tightly regulated at the transcriptional (45) and posttranscriptional (24, 31, 36, 37) levels. The TNF-α 3′ untranslated region (UTR) contains an AU-rich element (ARE), which was initially implicated in translational control (24). AREs are present in the 3′UTRs of many cytokine, inflammatory-gene, and oncogene mRNAs and confer instability (8, 11, 51; for a review, see reference 10). Mice with a targeted deletion of the TNF-α ARE show spontaneous production of TNF-α, resulting in chronic inflammatory arthritis and inflammatory bowel disease (31). Upon lipopolysaccharide (LPS) challenge, these mice produce elevated levels of TNF-α mRNA and protein (31), features which are consistent with an mRNA-destabilizing role for the ARE.
Certain inflammatory-gene mRNAs, including cyclooxygenase-2, interleukin-6 (IL-6), IL-8, and TNF-α mRNAs, are stabilized by activation of the p38 mitogen-activated protein kinase (MAPK) pathway by stimuli such as IL-1 and LPS (6, 12, 40, 49, 55). Studies with mRNA reporter constructs have shown that the p38 MAPK-mediated stabilization directly involves AREs (6, 33, 59). AREs thus confer instability on mRNAs; however, following activation of the p38 MAPK pathway, they allow mRNA stabilization and hence increased protein expression.
The stability of AU-rich mRNAs is thought to be controled by trans-acting proteins which bind to AREs. The proteins which bind the TNF-α ARE and regulate the stability of the mRNA in macrophages are not well understood. Early work (25) demonstrated the formation of a number of complexes between a 3′UTR mRNA probe and proteins in nuclear and cytoplasmic compartments. Subsequently it was shown that there was an LPS-inducible ARE-binding protein, as well as constitutive ones (21). Three proteins which bind to the TNF-α ARE have been identified. Tristetraprolin (TTP) is an LPS-inducible protein, which destabilizes the TNF-α mRNA (9, 32). TTP-null mice overexpress TNF-α protein and mRNA in the absence or presence of an LPS stimulus (9). TIAR and the related protein TIA-1 also bind the TNF-α ARE (20, 48). Macrophages from TIA-1-null mice produce more TNF-α protein than normal (48). TNF-α mRNA stability was found to be unchanged, but a greater proportion of the TNF-α mRNA was found to be polysome associated in these cells compared with wild-type macrophages. It was suggested that TIA-1 is a specific silencer of TNF-α translation (48).
A number of other proteins bind AREs from mRNAs other than TNF-α, such as AUF1 (or heterogeneous nuclear ribonucleoprotein [hnRNP] D) (61), HuR (39), hnRNP A0 (42), hnRNP A1 (22), hnRNP A2 (23), hnRNP C (22), hsp70 and hsp110 (26), enoyl coenzyme A hydratase (44), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (43). Of these, specific binding to the ARE and a physiological function has been demonstrated only for AUF1 and HuR, which destabilize (7, 38) and stabilize (14, 35, 46, 56, 57) AU-rich mRNAs, respectively.
To identify proteins which bind the TNF-α ARE and regulate TNF-α mRNA stability, we have investigated the proteins binding to it in an LPS-sensitive macrophage line. In a microsomal subcellular fraction containing the TNF-α mRNA, the major detectable protein that bound to the ARE with high affinity was HuR. When overexpressed, this protein stabilized an otherwise unstable reporter transcript containing the TNF-α ARE in its 3′UTR in manner similar to, but independent of, the p38 MAPK pathway.
MATERIALS AND METHODS
Plasmids.
Vectors used for synthesis of RNA probes for electrophoretic mobility shift assay (EMSA) and RNase protection assay (RPA) were constructed as follows. For the full-length murine TNF-α RPA probe, a 126-bp HincII-Eco47III fragment was excised from a murine TNF-α genomic construct (gift of A. Shakhov) and cloned into pBluescript (Stratagene) which had been linearized with EcoRV. For the β-globin RPA probe, a 269-bp SspI-BglII fragment was excised from pTetBBB (a gift from A.-B. Shyu) and cloned into pBluescript which had been cut with BamHI and EcoRV. The human 75-nucleotide (nt) TNF-α EMSA probe was prepared by PCR of human genomic DNA using the oligonucleotides CGACTAGTTCTATTTATGTTTGCACTTGTGA and GCTCTAGAAAATAAATACATTCATCTGTAAATAAAT. The PCR product was cut with SpeI and XbaI and cloned into pBluescript which had been linearized using SpeI. The murine full-length TNF-α 3′UTR EMSA probe vector was amplified by PCR from a plasmid containing the full murine TNF-α locus (a gift from A. Shakhov). The oligonucleotides used were 5′-GCGCCTCGAGGGAATGGGTGTTCATCCA-3′ and 5′-GCGCCTGCAGGCGATCTTTATTTCTCTCAATTGACTGATGGGC-3′. The product was cut with XhoI and PstI and cloned into pBluescript. The GAPDH 3′UTR EMSA probe was made by PCR of human genomic DNA using the oligonucleotides 5′-GGACTAGTGACCCCTGGACCACCAGC-3′ and 5′-GCTCTAGACACAGGGTACTTTATTGATGGTACATG-3′. The PCR product was restricted with SpeI and XbaI and cloned into pBluescript. The HuR expression vector used was CMV-FLAG HuR (a generous gift of J. Steitz). For mRNA stability measurements, an oligonucleotide spanning the TNF-α ARE (GATCCTTGTGATTATTTATTATTTATTTATTATTTATTTATTTACA GA) was cloned into the β-globin 3′UTR of pTetBBB. Taq polymerase, Vent polymerase, and restriction enzymes were from New England Biolabs and deoxynucleoside triphosphates were from Roche. The sequences of all novel plasmids were checked by automated DNA sequencing (ABC, London, United Kingdom).
Cell culture and transfection.
The RAW 264.7 murine macrophage-like cell line was cultured in Dulbecco's modified Eagle's medium (PAA Laboratories) supplemented with 10% fetal calf serum (Sigma-Aldrich). Cells were maintained at 37°C in the presence of 5% CO2. HeLa Tet-off cells (Clontech) were maintained as above, with G418 (100 ng ml−1; Life Technologies) included in the medium. HeLa Tet-off cells were seeded, transfected, and harvested as described previously (33).
Preparation of RNA transcripts.
Labeled probes were prepared by in vitro transcription. In general, reaction mixtures consisted of 1× T7 RNA polymerase buffer, 10 mM dithiothreitol (DTT), 2.5 mM each ATP, GTP, and CTP, 12 μM UTP (low-concentration probes) or 120 μM UTP (high-concentration probes), 50 μCi of [α-32P]UTP (800 Ci mmol−1) (Amersham-Pharmacia), 1 μg of linearized template DNA, 20 U of T7 RNA polymerase (Epicentre Technologies), and 20 U of recombinant RNasin RNase inhibitor (Promega). Reactions were stopped by addition of RNase-free DNase I (Promega). Unincorporated nucleotides were removed using S-200 spin columns (Amersham-Pharmacia Biotech) as specified by the manufacturer. “Cold” RNAs for competition assays were prepared as above using an Ampliscribe kit (Epicentre Technologies), except that only 0.25 μCi of [α-32P]UTP (800 Ci mmol−1) was used. Specific activites of probes were determined by scintillation counting.
RPA.
RPA was performed as described previously (33) using a kit from Ambion, except that for synthesis of the GAPDH probe the concentration of unlabeled UTP was 12 μM. Murine and human GAPDH RPA probes were from Ambion and Pharmingen, respectively. Samples were electrophoresed on 6% polyacrylamide gels and were visualized and quantified by phoshorimaging (Fuji BAS 2000).
EMSA, UV cross-linking, and antibody supershifts.
Typically between 5 and 10 μl of sample was incubated at room temperature (10 min) with bandshift buffer (10 mM HEPES [pH 7.6], 3 mM MgCl2, 20 mM KCl, 1 mM DTT, 5% glycerol [final concentrations]) and 20 or 200 fmol of labeled RNA probe as indicated in a final volume of 20 μl. For antibody supershift experiments, extract, bandshift buffer, and antibody (murine immunoglobulin G (Sigma-Aldrich) or 19F12 monoclonal antibody (raised against the first 13 N-terminal residues of HuR, a kind gift of H. Furneaux, Memorial Sloan-Kettering Cancer Center, New York), 3.25 μg each) were mixed, incubated for 10 min at room temperature (RT), and then incubated for 1 h on ice following probe addition. For UV cross-linking, extract was mixed with probe and bandshift buffer, incubated for 10 min at RT, and then exposed to 120 mJ of UV (on ice) using a Stratalinker (Stratagene). To EMSA, supershift, or UV-cross-linked samples, RNase T1 (Roche) and heparin sulfate were added to final concentrations of 50 U ml−1 and 5 mg ml−1, respectively, and the reaction mixture was incubated for 5 min at RT. Loading buffer (5 μl) containing 80% glycerol and 0.1% bromophenol blue was added to EMSA and supershift samples, which were then electrophoresed (150 V for 3 h at 4°C) on 4% acrylamide gels containing 0.5× Tris-borate-EDTA and 2.5% glycerol (which had been prerun for 1 h at 150 V). UV-cross-linked samples were mixed with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and electrophoresed on SDS–10% polyacrylamide gels. The gels were dried on 3 MM blotting paper (Whatman) and phosphorimaged. The EMSA figures shown were gamma corrected to improve the contrast in order to visualize complexes more clearly.
Cell fractionation.
Confluent RAW 264.7 cells from a 175-cm2 flask were rinsed with ice-cold phosphate-buffered saline and then harvested by scraping in ice-cold phosphate-buffered saline and centrifuging at 1,500 × g for 10 min at 4°C. The cells were resuspended in 1 ml of lysis buffer (10 mM HEPES [pH 7.6], 3 mM MgCl2, 40 mM KCl, 2 mM DTT, 5% glycerol, 0.5% NP-40, protease inhibitors [1 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin per ml, 10 μM E-64, and 1 μg of pepstatin per ml]) and lysed for 10 min on ice. The extract was centrifuged at 1500 × g for 10 min at 4°C, and pelleted nuclei were washed twice with 5 ml of nuclear wash buffer (10 mM HEPES [pH 7.6], 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT, protease inhibitors). This step was repeated, and the nuclei were lysed by being resuspended in 0.4 ml of 20 mM HEPES (pH 7.6)–0.42 M NaCl–25% glycerol–1.5 mM MgCl2–0.2 mM EDTA–1 mM DTT–inhibitors and agitated on a rotating wheel for 30 min at 4°C. The resulting extract was clarified by centrifugation at 12,000 × g for 20 min. The postnuclear supernatant or cytoplasmic fraction was fractionated by centrifugation at 100,000 × g (Beckman TL-100 ultracentrifuge; TLA 100.2 rotor) for 1 h to yield supernatant (S100) and pellet (P100) fractions. The pellet was sonicated three times for 10 s each at 200 W in 0.25 ml of lysis buffer, and the resulting extract was clarified by centrifugation at 10,000 × g for 10 min. All aliquots were snap-frozen immediately after preparation and stored at −70°C until use.
Mono-Q anion-exchange chromatography.
Nuclear extract (10 ml) was freshly prepared as above from approximately 2.5 × 109 cells. It was dialyzed against 1 liter of buffer A (20 mM Tris HCl [pH 8.3], 1 mM DTT, 1 mM EDTA, 0.05% Brij 35), filtered on a 0.2-μm-pore-site filter, and loaded onto a 1-ml Mono-Q column (Amersham-Pharmacia Biotech) equilibrated with buffer A on a fast protein liquid chromatography system (Amersham-Pharmacia Biotech). All chromatography was performed at room temperature. Protein was eluted with a linear 20-ml gradient (0 to 0.5 M NaCl) with buffer A which contained 1 M NaCl. Fractions (1 ml) were collected, 10-μl aliquots were removed for assaying, and the fractions were stored at −20°C.
Mono-S cation-exchange chromatography.
The Mono-S cation-exchange chromatography step was performed on a SMART chromatography system (Amersham-Pharmacia Biotech). Fractions 5 to 10 from the Mono-Q chromatography were pooled, dialyzed against buffer B (50 mM malonic acid [pH 6.0], 1 mM EDTA, 1 mM DTT, 0.05% Brij 35) for 2 h at room temperature, and loaded onto a 100-μl Mono-S column (Amersham-Pharmacia Biotech) equilibrated with buffer B. The column was developed with a linear gradient (0 to 1 M NaCl) of buffer (2 ml) containing 1 M NaCl, and 100-μl fractions were collected. A 5-μl volume of 1 M Tris HCl (pH 8.3) was added to each fraction to bring the pH close to neutral. A 10-μl volume of each fraction was removed for assaying, and the fractions were stored at −20°C.
Poly(U) RNA affinity chromatography.
Individual fractions from the Mono-S column were diluted by addition of 80 μl of buffer C (25 mM HEPES [pH 7.6], 1 mM DTT, 1 M KCl, 0.1% Brij 35). Poly(U)-Sepharose 4B beads (Amersham-Pharmacia Biotech) were rehydrated and washed in buffer C, and 20 μl of a 50% bead slurry was added to each fraction. The fractions were then agitated for 10 min at room temperature. The beads were pelleted by centrifugation at 1,500 × g for 3 min, and the supernatants were removed for assay. The pellets were washed once in buffer C containing only 0.5 M KCl and then once with buffer C with no KCl added. A 30-μl volume of SDS-PAGE sample buffer was added, and the samples were boiled for 5 min. The samples were electrophoresed on an SDS–10% polyacrylamide gel, which was then silver stained (52).
Preparation of samples for mass spectrometry.
Protein bands were excised from the gel and chopped into 1-mm2 cubes. In-gel digestion with trypsin was performed by published methods (28, 58), with the incorporation of a destaining step (18) to remove silver. Cysteine residues were reduced with DTT and derivatized by treatment with iodoacetamide. The gel pieces were then dehydrated with acetonitrile and dried on a vacuum centrifuge prior to addition of modified trypsin (6.5 ng μl−1 in 25 mM ammonium hydrogen carbonate [Promega]) at 4°C. Excess trypsin was removed after 40 min, and the digestion was allowed to proceed overnight at 37°C). An aliquot (0.5 μl) was removed for direct matrix-assisted laser desorption ionization (MALDI) analysis, while the remainder of the digestion products were recovered by sequential extraction with 25 mM ammonium hydrogen carbonate, 5% formic acid, and acetonitrile.
MALDI mass spectrometry.
MALDI mass spectra were recorded with a Micromass (Manchester, United Kingdom) TofSpec 2E spectrometer, equipped with a 337-nm nitrogen laser. The instrument was operated in the positive-ion reflectron mode at an accelerating voltage of 20 kV with “time-lag focusing” enabled. The matrix was a mixture of α-cyano-4-hydroxycinnamic acid and nitrocellulose, applied to the target as a microcrystalline thin film by a modification of the procedure of Vorm et al. (54). Aliquots (0.5 μl) of the digest supernatant were injected into an equal volume of 10% formic acid previously deposited on the matrix film, allowed to dry, and briefly washed with a few microlitres of 0.1% trifluoroacetic acid. Spectra were internally calibrated using trypsin autolysis products, and the resulting peptide masses were searched against Swiss Prot/TREMBL release 35, using the Protein Probe program (Micromass), or against a nonredundant database maintained by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov), using the Mascot search engine (47). An initial mass tolerance of 100 ppm was used in all searches.
Electrospray mass spectrometry.
Samples were desalted on C18 zip tips (Millipore) or on home-packed Poros R2 microcolumns (PerSeptive Biosystems). The desalted samples were dissolved in 1 to 2 μl of 50% methanol–0.1% aqueous formic acid, loaded into palladium-coated borosilicate nanoelectrospray needles (Protana Inc., Odense, Denmark), and mounted in the source of a Micromass Q-Tof hybrid quadrupole/orthogonal-acceleration time-of-flight spectrometer. Stable flow was usually obtained with a capillary voltage between 900 and 1,200 V. The collision gas was argon, and collision energies and argon pressure were tuned to optimize the fragmentation pattern of individual precursor ions. Daughter ion spectra were charge-state deencrypted and deisotoped using a proprietary maximum-entropy algorithm (Micromass), and amino acid sequences were deduced with the assistance of the Micromass peptide-sequencing program. The resulting sequences were searched against the National Center for Biotechnology Information nonredundant database using the BLAST program (1).
Western blotting.
Whole-cell lysates were prepared using a buffer containing 50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 25 mM sodium β-glycerophosphate, 10 mM sodium tetrapyrophosphate, and 1- mM sodium orthovanadate. Cell fractions (20 μg protein) or whole-cell lysates (5 μg of protein) were Western blotted on polyvinylidene difluoride membrane (NEN). The blots were stained with anti-HuR mouse monoclonal antibody (2.5 μg μl−1), the murine M2 anti-FLAG antibody (Sigma-Aldrich [1:1,000 dilution]), a murine monoclonal anti-α-tubulin antibody DM-1A (Sigma-Aldrich [1:1,000 dilution]) and a horseradish peroxidase-conjugated rabbit anti-mouse antibody (1:2,000 dilution) using an enhanced chemiluminescence kit (Amersham-Pharmacia Biotech).
RESULTS
TNF-α mRNA is located in the P100 fraction following LPS treatment of RAW 264.7 cells.
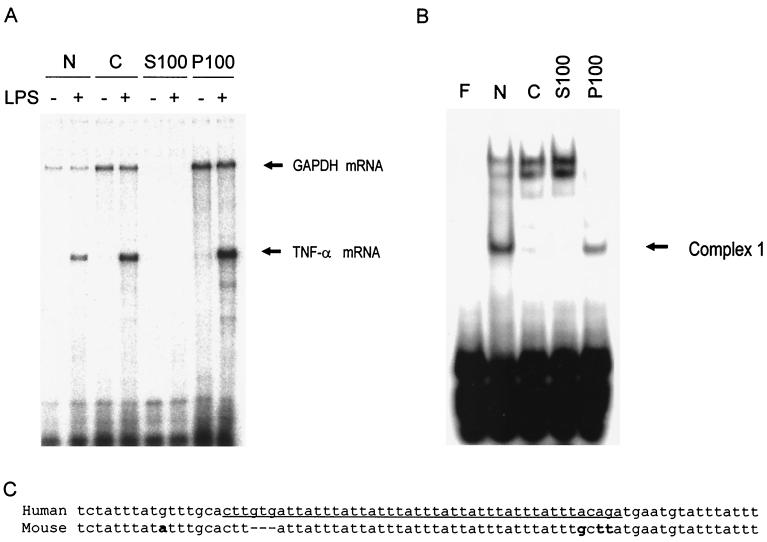
RAW 264.7 cells were either left untreated or treated for 2 h with LPS and then lysed. The subcellular location of TNF-α mRNA was determined as follows. The lysates were centrifuged to yield nuclear (N) and cytoplasmic (C) fractions. The latter was then centrifuged for 1 h at 100,000 × g to give supernatant (S100) and pellet (P100) fractions. The presence of TNF-α mRNA in the extracts was determined by RPA (Fig. 1A). A small amount of TNF-α mRNA was found in the nuclear fraction of LPS-treated cells, but the majority was in the cytoplasmic fraction. Following centrifugation at 100,000 × g, most of this TNF-α mRNA was found in the pellet (P100). None was found in untreated cells.
FIG. 1.
TNF-α mRNA is located in the P100 fraction of LPS-treated RAW 246.7 cells, and the P100 fraction also contains a protein(s) which binds to the TNF-α 3′UTR AU-rich region. Cells were either left untreated or treated for 2 h with LPS. The cells were lysed and fractionated into nuclei (N) and cytoplasm (C), which was centrifuged at 100,000 × g to yield supernatant (S100) and pellet (P100) fractions. (A) RNA levels were measured by an RPA using radiolabeled TNF-α and GAPDH probes. (B) EMSA using the 75-nt probe ([200 fmol] derived from the AU-rich region of the human TNF-α mRNA 3′UTR) on lysates of untreated cells, fractionated as for panel A. The results in both panels are representative of several experiments. (C) Alignment of the human TNF-α ARE sequence contained in the 75-nt probe with the murine TNF-α ARE. Differences are marked in bold, and the sequence inserted into the β-globin reporter used in overexpression experiments (see Fig. 7) is underlined.
EMSA indicates that the P100 fraction contains an AU-rich RNA binding protein.
The same fractions as described above were also analyzed by EMSA using approximately 200 fmol of an RNA probe containing 75 nt from the 3′UTR of human TNF-α mRNA (Fig. 1B). This probe contains the major AU-rich region of the TNF-α 3′UTR, which has eight copies of the AUUUA motif, some of them overlapping (Fig. 1C). The P100 extract contained protein which formed a complex of high electrophoretic mobility (complex 1) and which also appeared to be present in the nuclear extract. The nuclear extract also formed three lower-mobility minor complexes (Fig. 1B). EMSAs performed using the 75-nt probe (approximately 200 fmol) showed little difference in the pattern or intensity of complexes between fractions prepared from resting or LPS-treated cells. However, in some experiments, the intensity of complex 1 was increased by approximately twofold in the P100 fraction from cells treated with LPS for 2 h.
Since in LPS-treated cells the cytoplasmic TNF-α mRNA was in the P100 fraction, it seemed likely that proteins regulating its stability and translation would also be present in this fraction. Thus, the protein(s) forming complex 1 would be strong candidates for being involved in these processes. Since a complex of identical mobility (but greater apparent abundance) was formed by nuclear extracts and since many mRNA binding proteins are known to shuttle between the nucleus and the cytoplasm, complex 1 formed by nuclear extracts was characterized further with a view to purifying and identifying the protein(s) responsible.
The pattern of complex formation in EMSA is dependent on the RNA probe and protein concentrations.
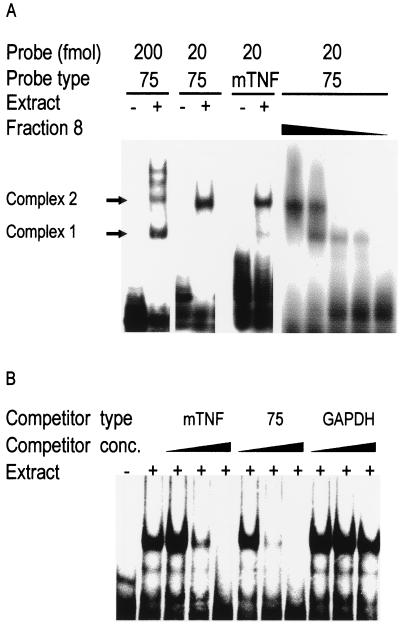
EMSAs were performed using a nuclear extract of resting cells with the 75-nt probe either at high concentration (approximately 200 fmol in the assay) or at low concentration (approximately 20 fmol in the assay) (Fig. 2A). With a high concentration of probe, complex 1 predominated, with complex 2 and a pair of lower-mobility bands also visible. With a low probe concentration, complex 2 alone was observed. A similar probe concentration-dependent conversion between complexes 1 and 2 was observed when the P100 extract was used (data not shown). The predominance of complex 1 or 2 was also dependent on the protein concentration (Fig. 2A). Starting with a fraction in which complex 2 alone was detected (see Fig. 3A), progressive dilution resulted in the appearance of complex 1 followed by the disappearance of complex 2. These observations suggest that complexes 1 and 2 might be oligomers containing the same protein (but in different stoichiometries) and exhibiting different affinities for RNA.
FIG. 2.
A lower RNA probe concentration produces a slower-mobility complex, complex formation is specific for the TNF-α ARE, and dilution causes complex 2 to shift to the complex 1 position. (A) (Left panel) EMSA on nuclear extract using 20 fmol (low probe concentration) or 200 fmol (high probe concentration) of the 75-nt probe or full-length murine TNF-α 3′UTR probe as indicated. (Right panel) Dilutions of fraction 8 (Fig. 3A) from the Mono-S column (1:2, 1:4, 1:20, 1:40, and no addition) were analyzed by EMSA using 20 fmol of the 75-nt probe. (B) Competition study in EMSA between radiolabeled full-length murine TNF-α probe and increasing concentrations (1-fold, 10-fold, and 100-fold excesses) of unlabeled murine TNF-α, 75-nt, or GAPDH 3′UTR probes. Similar results were obtained for each panel in two separate experiments.
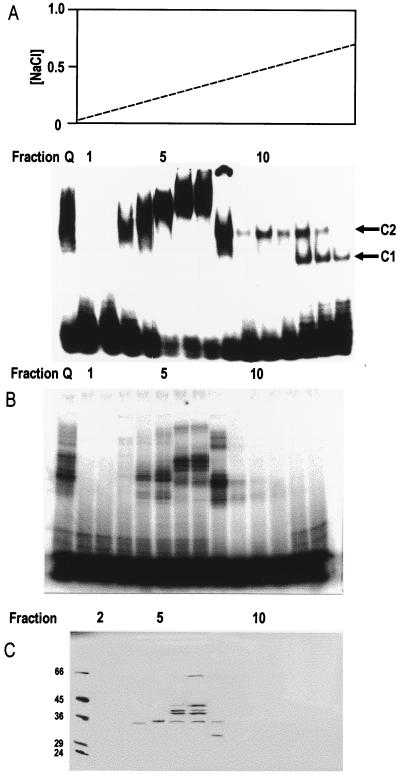
FIG. 3.
Purification by chromatography on a Mono-S column of the protein(s) responsible for complex 2. (A) The upper panel shows a salt gradient eluting the Mono-S column. The lower panel shows EMSA using 20 fmol of the 75-nt probe per assay. Pooled fractions from Mono-Q chromatography after dialysis and Mono-S fractions 1 to 14 are shown. Complex 1 (C1) and complex 2 (C2) are indicated. (B) Phosphorimage of SDS-PAGE of proteins UV cross-linked to the radiolabeled 75-nt probe (following digestion of unprotected probe with RNase T1) in the pooled Mono-Q fractions after dialysis and in Mono-S fractions 1 to 13. (C) SDS-PAGE (silver stained) of proteins of Mono-S fractions 2 to 13 absorbed with poly (U)-Sepharose 4B beads. The beads were washed, and proteins were eluted in SDS sample buffer for electrophoresis. Similar results were seen in two independent experiments.
Complex formation is specific and involves the TNF-α mRNA 3′UTR AU-rich region.
To assess the specificity of complex formation, EMSAs were performed on nuclear extracts using labeled murine full-length 3′UTR probe in the presence of unlabeled competitor RNAs (Fig. 2B). Strong self-competition by the full-length murine 3′UTR probe was observed with a 10- and 100-fold excess over labeled probe. The 75-nt AU-rich RNA derived from the human TNF-α 3′UTR was similarly effective as a competitor, while the GAPDH 3′UTR did not compete significantly even at a 100-fold excess. Although the murine and human TNF-α AREs are not identical (Fig. 1C), the full-length murine 3′UTR and the 75-nt human AU-rich probe display almost identical patterns of complex formation (Fig. 2A) and cross-compete for RNA binding proteins (Fig. 2B). The shorter, human-derived probe was used to assay for RNA binding proteins in the subsequent purification steps.
Purification of a protein forming complexes 1 and 2.
The identical mobilities of complexes 1 and 2 in nuclear and P100 fractions and their similar dependence on the concentration of probe suggested that the same proteins may be involved in both fractions. To identify the proteins responsible for forming complexes 1 and 2, they were purified from the nuclear extract, in which they were relatively abundant. Their presence in chromatography fractions was monitored by EMSA with the 75-nt probe at the lower concentration (20 fmol in the assay [favoring the formation of complex 2]). A nuclear extract of RAW 264.7 cells was dialyzed and chromatographed on a Mono-Q anion-exchange column, which was developed with a salt gradient. Only protein forming complex 2 was detected, and this eluted between 0.125 and 0.25 M NaCl. This material was pooled and is shown in the left lane in Fig. 3A. It was dialyzed and loaded on to a Mono-S cation-exchange column and eluted with a salt gradient. The column fractions were assayed for complex formation with the 75-nt probe (Fig. 3A). Protein forming complex 2 eluted very sharply in fraction 8, with fractions 10 to 12 containing smaller amounts; fractions 12 to 13 gave rise to complex 1. The early fractions (fractions 3 to 7) formed slower-migrating complexes, which were not generated by the original extract or the Mono-Q fractions. Why these proteins are unmasked by the chromatography is not known. Possibly they are concentrated by the Mono-S chromatography and so become visible.
The Mono-S fractions were further analyzed by SDS-PAGE following UV cross-linking to the radiolabeled RNA and digestion with RNase T1 (Fig. 3B). A number of protein bands were seen in fractions 4 to 7, and fraction 8 contained a major band with an apparent molecular mass of 45 kDa. Subtracting the molecular mass of the probe fragment produced following digestion with RNase T1 (∼12 kDa) gave an estimate of 33 kDa for this protein. Since the assays used in this purification were designed to monitor complex 2 formation and therefore used a low, subsaturating concentration of probe (20 fmol), it was not possible to quantitate RNA binding activity for the early steps of the purification.
Since the protein(s) in crude nuclear extracts forming complexes 1 and 2 was strongly competed by poly(U) RNA but not by poly(A), poly(G), or poly(C) RNA (data not shown), the complexing proteins in the Mono-S fractions were absorbed with poly(U)-Sepharose 4B in the presence of 0.5 M NaCl. After absorption, the supernatants no longer formed complexes in the EMSA, indicating that the active protein(s) had bound to the beads (data not shown). The beads were washed and then boiled in SDS-PAGE sample buffer, and the eluted proteins were separated by SDS-PAGE and stained with silver (Fig. 3C). A number of protein bands were seen in fractions 4 to 7. Fraction 8 contained a protein of 32 kDa which was not visible in other fractions. The prominent 32-kDa band was a strong candidate for the protein forming complex 2, which eluted sharply in fraction 8 (Fig. 3A). Since the protein absorbed on poly(U)-Sepharose 4B was eluted under denaturing conditions, it was not possible to measure the RNA binding activity after this step, preventing calculation of the overall recovery and fold purification for the process.
Mass spectrometric identification of complex 2 as HuR.
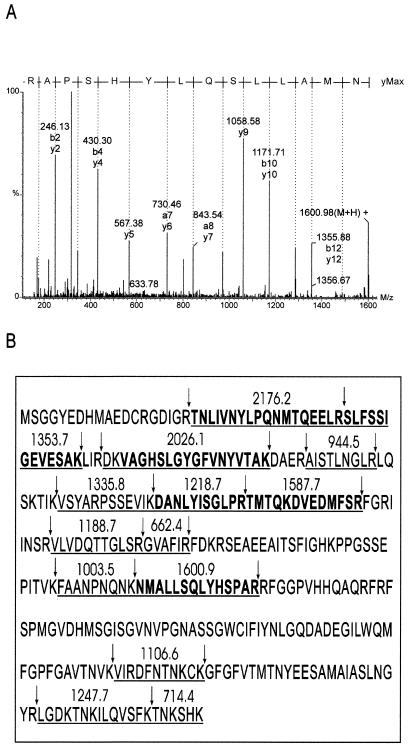
The silver-stained band from fraction 8 was excised, and the protein was subjected to in-gel digestion. An aliquot of the digest supernatant was analyzed by MALDI mass spectrometry, and the masses of the resulting protonated peptides were used to search a nonredundant database. The three top matches were, in order of statistical significance, the murine, human, and chicken homologues of Hu antigen R (HuR). In the case of the mouse protein, 19 digest peptides matched to better than 50 ppm accuracy, corresponding to coverage of 48% of the amino acid sequence.
This identification was confirmed by electrospray ionization tandem mass spectrometry of the tryptic digest. The deconvoluted tandem mass spectrum of a doubly charged precursor peptide of m/z 801 and its deduced amino acid sequence is shown in Fig. 4A. Six peptides were analyzed, from which a total of 75 amino acid residues matching the sequence of HuR were obtained. These MALDI and electrospray ionization mass spectrometry data are summarized in Fig. 4B.
FIG. 4.
Identification of HuR by mass spectrometry (A) Daughter ion spectrum of the doubly charged peptide of m/z 801 from an in-gel tryptic digest of the 32-kDa band of fraction 8 (Fig. 3C). The spectrum was transformed onto a singly charged mass axis as described in Materials and Methods. (B) HuR amino acid sequence showing protonated masses of peptides identified by MALDI–time-of-flight mass spectrometry (underlined). Boldface type represents peptides that were sequenced by tandem electrospray mass spectrometry, and arrows indicate tryptic cleavage sites.
Complexes 1 and 2 are supershifted by an anti-HuR antibody.
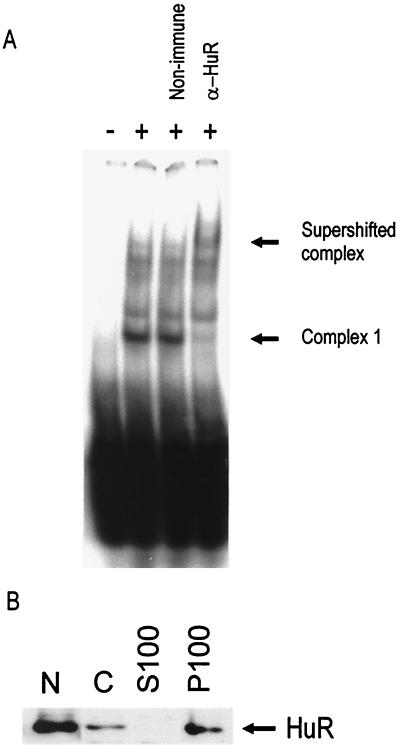
To confirm that complex 2 contained HuR, a monoclonal antibody to HuR was added to a sample of fraction 8 from the Mono-S column, which was then analyzed by EMSA (Fig. 5). The antibody produced a strong supershift, depleting all of the activity in the complex 2 position. No supershift was observed with a nonimmune murine antibody. Protein concentration-dependent formation of complexes 1 and 2 was apparent as before (Fig. 2B), and both complexes were clearly depleted in the presence of the anti-HuR antibody, confirming that HuR is present in both complexes. Thus, either an excess of probe or a lower concentration of HuR favored the formation of complex 1, while a high concentration of HuR or a low concentration of probe favored the formation of complex 2. These observations are consistent with the oligomerization of HuR on the ARE. However, since HuR was not purified to homogeneity, it is not clear whether other proteins are also present in these complexes.
FIG. 5.
The purified protein which forms complexes 1 and 2 is supershifted by an anti-HuR antibody. A sample of fraction 8 from the Mono-S column (as in Fig. 3) was mixed with 3.5 μg of anti-HuR antibody or murine immunoglobulin G and incubated for 10 min at RT. A 20-fmol portion of the 75-nt probe was added, and the sample was incubated for 1 h on ice. RNase T1 and heparin sulfate were added, and the sample was incubated for 5 min at RT. Samples were then electrophoresed as for EMSA. A high concentration (1:2 dilution [left panel]) and a lower concentration (1:4 dilution [right panel]) of fraction 8 were used.
Complex 1 of P100 extracts contains HuR.
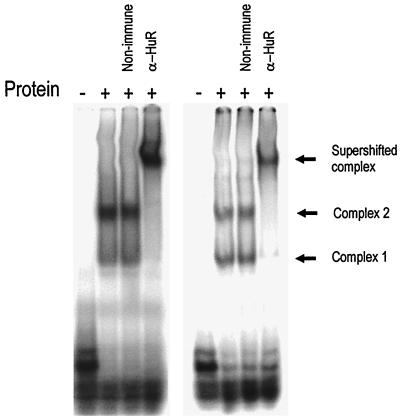
To test whether complex 1 of P100 extracts (Fig. 1B) also contains HuR, an antibody supershift experiment was performed (Fig. 6A). Incubation of a P100 extract with anti-HuR totally depleted the major complex (complex 1) and resulted in a weak supershifted band.
FIG. 6.
Complex 1 in the P100 fraction contains HuR. (A) An EMSA supershift experiment was performed on a sample of P100 extract which was incubated with purified murine IgG or anti-HuR antibody as described in the legend to Fig. 5 but using 200 fmol of the 75-nt probe. (B) Western blot using anti-HuR antibody on RAW 264.7 cells, fractionated as described in the legend to Fig. 1. A 20-μg sample of protein was loaded per lane. Both panels are representative of two independent experiments.
The subcellular distribution of HuR in RAW cells was determined by Western blotting (Fig. 6B). HuR was found to be most abundant in the nuclear extract but was also detectable in the cytoplasm, in which it was localized in the P100 fraction. There was no change in HuR levels in either the P100 or nuclear fractions following a 2-h LPS treatment of the cells (data not shown).
Overexpression of HuR in HeLa Tet-off cells stabilizes a β-globin reporter containing the TNF-αAU-rich region.
Expression of TNF-α is strongest in cells of the monocyte/macrophage lineage, and the RAW 264.7 macrophage-like cell line from which HuR was purified in this work expresses high levels of TNF-α in response to LPS challenge. However, it was not possible to examine the effect of HuR overexpression on the production of endogenous TNF-α in RAW 264.7 cells because it is not possible to transfect them efficiently in a transient manner or to derive stably transfected lines. Therefore, a HeLa cell tetracycline-regulated β-globin reporter system designed and constructed by Xu et al. (60) was used to assess the regulation of TNF-α mRNA stability by HuR. This system also avoided the use of the toxic transcriptional inhibitor actinomycin D, which is known to alter the subcellular location of HuR (46).
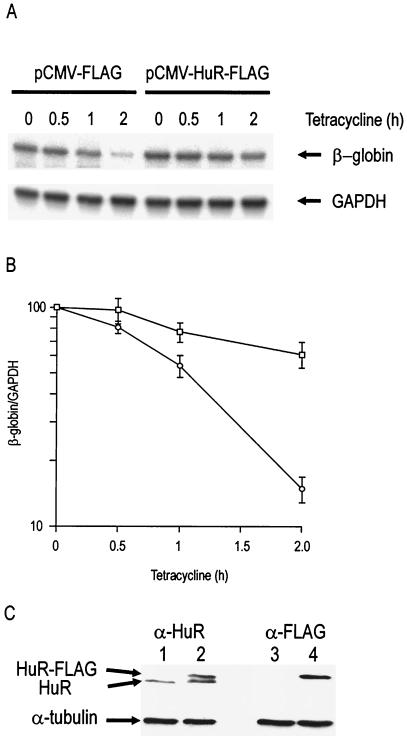
In HeLa Tet-off cells, the stable β-globin mRNA is expressed under the control of a tetracycline-responsive promoter, which can be rapidly switched off by the addition of 100 ng of tetracycline per ml to the medium. Decay of the β-globin mRNA is scarcely detectable (t1/2≃ 20 h) following the addition of tetracycline (33). The insertion of AU-rich elements into the 3′UTR of the β-globin reporter modulates the stability of the reporter mRNA in a specific manner (33, 59, 60). The insertion of the TNF-α ARE destabilized the reporter mRNA, reducing its half-life to about 1 h (Fig. 7). Transient cotransfection of 200 ng of an HuR construct (pCMV-HuR-FLAG) stabilized the chimeric β-globin/TNF-α reporter mRNA (Fig. 7A and B). The expression of FLAG-tagged HuR was checked by Western blotting with anti-HuR and anti-FLAG antibodies. Figure 7C shows that expression of recombinant HuR appeared approximately equal to that of endogenous HuR in lysates of cells transfected with 200 ng of expression vector. Equal protein loading was verified by immunoblotting the same membranes for α-tubulin, and the identity of the recombinant HuR was confirmed by immunoblotting with the anti-FLAG antibody (Fig. 7C). Since the transfection efficiency is 40% under the conditions used (data not shown), there is approximately a 3.5-fold increase in total HuR (including HuR-FLAG) protein levels in cells containing the pCMV-HuR-FLAG vector. Inhibition of p38 MAPK by addition of SB 203580 had no effect on the HuR-mediated stabilization (data not shown). Activation of p38 MAPK by transient cotransfection of MKK6EE (an active mutant which on its own stabilizes the TNF-α ARE reporter) DNA did not result in any increase in stability conferred by the transfected HuR overexpression vector (data not shown).
FIG. 7.
Transient overexpression of HuR in HeLa Tet-off cells stabilizes the mRNA of a reporter construct carrying the TNF-α 3′UTR AU-rich region. (A) HeLa Tet-off cells were transiently transfected with a construct consisting of the rabbit β-globin gene into which 44 nt of the TNF-α 3′UTR AU-rich region (Fig. 1C) had been inserted. These cells were cotransfected with either 200 ng of empty expression vector (pCMV-FLAG) or 200 ng of HuR expression vector (pCMV-HuR-FLAG). Tetracycline was added after 20 h, and the cells were harvested at various time points as indicated: β-globin and GAPDH mRNA levels were measured by RPA. (B) Graph of log β-globin mRNA/GAPDH mRNA levels plotted against time after addition of tetracycline in the presence (squares) or absence (circles) of HuR overexpression. Error bars indicate the standard error of the mean of three independent experiments. (C) Western blots of whole-cell lysates from cells transiently transfected with either 200 ng of empty expression vector pCMV-FLAG (lanes 1 and 3) or 200 ng of HuR expression vector pCMV-HuR-FLAG (lanes 2 and 4). The blots were stained with anti-HuR (lanes 1 and 2) or anti-FLAG (lanes 3 and 4) antibodies as indicated and with an anti-tubulin antibody to control for gel loading (lanes 1 to 4).
DISCUSSION
We set out to identify the major ARE binding protein(s) present in a P100 subcellular fraction which contained the TNF-α mRNA. We used a strategy of protein purification, monitored by native EMSA, with identification by mass spectrometry and immunological characterization. The use of native EMSAs as opposed to UV cross-linking (in which the specificity of RNA binding may be altered owing to preferential covalent linkage of particular amino acids and nucleotides) alone enabled specific RNA-protein interactions to be detected in the assay. This approach also allowed unequivocal identification of a protein by amino acid sequencing and MALDI mass mapping, which was confirmed by antibody supershift analysis.
The P100 fraction contained a prominent AU-rich RNA binding protein complex as judged by EMSA. A protein forming a complex of identical mobility was also present in the nuclear fraction. The more abundant nuclear protein was purified to near homogeneity and identified as HuR by MALDI mass mapping and was partially sequenced by tandem mass spectrometry. These procedures covered 48 and 23% of the molecule, respectively. The identification was confirmed by supershift analysis of purified material using an anti-HuR monoclonal antibody raised to the N-terminal 13 amino acids. In crude extracts or chromatographic fractions, two complexes, both of which contained HuR, could be detected under different assay conditions. In the presence of high concentrations of HuR relative to probe, a low-mobility complex was formed (complex 2), whereas under the reverse conditions a high-mobility complex was formed (complex 1). Whether HuR is the sole protein in either complex is not certain since it was not purified to homogeneity. However, the results are consistent with oligomerization of HuR either on its own or with other proteins on AU-rich RNA.
HuR is a member of the embryonic lethal abnormal vision (ELAV) family of RNA binding proteins, and an unidentified ELAV protein has been previously suggested to bind TNF-α mRNA (50). Unlike other family members such as Hel-N1 (or HuB), HuC, and HuD, which are developmentally regulated and tissue specific (2, 19, 30, 53), HuR is ubiquitous (39) and predominantly nuclear (14). In the present work, HuR was also found to be most abundant in nuclei in RAW 264.7 macrophages.
Hel-N1 was the first ELAV family member to be assigned a specific RNA binding sequence (34). A random RNA selection procedure revealed that Hel-N1 binds preferentially to AU-rich RNA (34). HuR was later independently cloned (39) and purified on the basis of its ability to bind AU-rich RNA (41), which is consistent with the present observation of its binding the TNF-α ARE.
HuR was initially thought to be an mRNA-destabilizing factor because of its affinity for various unstable AU-rich mRNAs (41). However, this idea was inconsistent with the finding that overexpression of Hel-N1 stabilized the AU-rich GLUT1 mRNA (27). It was later shown that vascular endothelial growth factor mRNA, which contains an ARE in its 3′UTR, was destabilized in cells stably expressing antisense HuR and stabilized in cells overexpressing HuR (35). HuR overexpression stabilizes reporter mRNAs containing sequences from granulocyte-macrophage colony-stimulating factor (14) and c-fos (14, 46), while expression of antisense HuR has also recently been shown to destabilize p21 mRNA (57) and cyclin A and B1 mRNAs (56). In the present study, we show that overexpression of HuR in a HeLa Tet-off cell line stabilizes an unstable reporter construct containing the AU-rich region from the TNF-α 3′UTR. This is consistent with in vitro experiments in which addition of a recombinant ELAV protein, Hel-N1, to an mRNA degradation assay mixture stabilized an in vitro-transcribed RNA containing the TNF-α ARE (16). This is the first in vivo evidence of an RNA binding protein stabilizing TNF-α mRNA. The fact that HuR represents the most abundant or highest-affinity protein binding the TNF-α ARE in a macrophage cell line, together with the fact that macrophages are the major TNF-α-producing cells, strongly suggests a role for HuR in stabilizing TNF-α mRNA.
The occurrence of HuR in both nuclei and cytoplasm is consistent with the previously reported nucleocytoplasmic shuttling properties of this protein (4, 13, 14). The presence of HuR in the P100 fraction could be due to its association with ribosome-bound mRNA, which would sediment at 100,000 × g. An alternative possibility is that HuR may sediment if it is present in stress granules which contain other RNA recognition motif proteins such as TIA-1/TIAR and poly(A) binding protein (29). HuR has been found to be present in cytoplasmic foci in heat-shocked HeLa cells (17) and in untreated cultured neurons (3) and recently has been shown to colocalize in the stress granules described by Kedersha et al. (29) (P. Anderson, personal communication).
The p38 MAPK pathway stabilizes mRNAs of several inflammatory proteins, including TNF-α. We therefore examined whether the effect of HuR was altered by activating or inhibiting this pathway. Inhibition of p38 MAPK had no effect on the stabilization of the TNF-α 3′UTR reporter by HuR. Activation of p38 MAPK by expression of MKK6EE (an active mutant) on its own stabilized the reporter, but coexpression with HuR did not result in any increase in reporter mRNA stability. Thus, the HuR-mediated stabilization does not appear to depend on p38 MAPK activity.
In summary, we have identified HuR as the major TNF-α ARE binding protein in macrophages and shown that it stabilizes an mRNA containing the TNF-α ARE. The only other protein which is known to regulate TNF-α mRNA stability is TTP, which destabilizes it (9, 32). Indeed, it is possible that stabilizing and destabilizing factors compete for the ARE and determine the stability and fate of mRNAs containing AREs. The p38 MAPK cascade stabilizes mRNAs through AREs by an unknown mechanism. Although p38 MAPK activation is not required for stabilization of TNF-α mRNA by HuR, this does not rule out the possibility that HuR is in fact involved in the p38 MAPK-mediated stabilization. The fact that HuR is the major TNF-α ARE binding protein in macrophages strongly suggests that HuR is involved in this process. To understand the mechanism of TNF-α mRNA stabilization in more detail, it will be necessary to identify proteins which interact with HuR and to identify relevant targets of the p38 MAPK cascade.
ACKNOWLEDGMENTS
We gratefully acknowledge support from the Medical Research Council, Arthritis Research Campaign, and Wellcome Trust.
We are extremely grateful to A. Shakov, A.-B. Shyu, J. Steitz, and H. Furneaux for providing reagents and to P. Anderson for sharing unpublished data with us.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antic D, Keene J D. Embryonic lethal abnormal visual RNA-binding proteins involved in growth, differentiation, and posttranscriptional gene expression. Am J Hum Genet. 1997;61:273–278. doi: 10.1086/514866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antic D, Keene J D. Messenger ribonucleoprotein complexes containing human ELAV proteins: interactions with cytoskeleton and translational apparatus. J Cell Sci. 1998;111:183–197. doi: 10.1242/jcs.111.2.183. [DOI] [PubMed] [Google Scholar]
- 4.Atasoy U, Watson J, Patel D, Keene J D. ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J Cell Sci. 1998;111:3145–3156. doi: 10.1242/jcs.111.21.3145. [DOI] [PubMed] [Google Scholar]
- 5.Beutler B, Cerami A. The biology of cachectin/TNF—a primary mediator of the host response. Annu Rev Immunol. 1989;7:625–655. doi: 10.1146/annurev.iy.07.040189.003205. [DOI] [PubMed] [Google Scholar]
- 6.Brook M, Sully G, Clark A R, Saklatvala J. Regulation of tumour necrosis factor alpha mRNA stability by the mitogen-activated protein kinase p38 signalling cascade. FEBS Lett. 2000;483:57–61. doi: 10.1016/s0014-5793(00)02084-6. [DOI] [PubMed] [Google Scholar]
- 7.Buzby J S, Brewer G, Nugent D J. Developmental regulation of RNA transcript destabilization by A + U-rich elements is AUF1-dependent. J Biol Chem. 1999;274:33973–33978. doi: 10.1074/jbc.274.48.33973. [DOI] [PubMed] [Google Scholar]
- 8.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carballo E, Lai W S, Blackshear P J. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 10.Chen C Y, Shyu A B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 11.Chen C Y, Shyu A B. Selective degradation of early-response-gene mRNAs: functional analyses of sequence features of the AU-rich elements. Mol Cell Biol. 1994;14:8471–8482. doi: 10.1128/mcb.14.12.8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dean J L, Brook M, Clark A R, Saklatvala J. p38 mitogen-activated protein kinase regulates cyclooxygenase-2 mRNA stability and transcription in lipopolysaccharide-treated human monocytes. J Biol Chem. 1999;274:264–269. doi: 10.1074/jbc.274.1.264. [DOI] [PubMed] [Google Scholar]
- 13.Fan X C, Steitz J A. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc Natl Acad Sci USA. 1998;95:15293–15298. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan X C, Steitz J A. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feldmann M, Brennan F M, Maini R N. Role of cytokines in rheumatoid arthritis. Annu Rev Immunol. 1996;14:397–440. doi: 10.1146/annurev.immunol.14.1.397. [DOI] [PubMed] [Google Scholar]
- 16.Ford L P, Watson J, Keene J D, Wilusz J. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev. 1999;13:188–201. doi: 10.1101/gad.13.2.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallouzi I E, Brennan C M, Stenberg M G, Swanson M S, Eversole A, Maizels N, Steitz J A. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc Natl Acad Sci USA. 2000;97:3073–3078. doi: 10.1073/pnas.97.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gharahdaghi F, Weinberg C R, Meagher D A, Imai B S, Mische S M. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis. 1999;20:601–605. doi: 10.1002/(SICI)1522-2683(19990301)20:3<601::AID-ELPS601>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 19.Good P J. A conserved family of elav-like genes in vertebrates. Proc Natl Acad Sci USA. 1995;92:4557–4561. doi: 10.1073/pnas.92.10.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gueydan C, Droogmans L, Chalon P, Huez G, Caput D, Kruys V. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor alpha mRNA. J Biol Chem. 1999;274:2322–2326. doi: 10.1074/jbc.274.4.2322. [DOI] [PubMed] [Google Scholar]
- 21.Gueydan C, Houzet L, Marchant A, Sels A, Huez G, Kruys V. Engagement of tumor necrosis factor mRNA by an endotoxin-inducible cytoplasmic protein. Mol Med. 1996;2:479–488. . (Erratum, 2:786.) [PMC free article] [PubMed] [Google Scholar]
- 22.Hamilton B J, Nagy E, Malter J S, Arrick B A, Rigby W F. Association of heterogeneous nuclear ribonucleoprotein A1 and C proteins with reiterated AUUUA sequences. J Biol Chem. 1993;268:8881–8887. [PubMed] [Google Scholar]
- 23.Hamilton B J, Nichols R C, Tsukamoto H, Boado R J, Pardridge W M, Rigby W F. hnRNP A2 and hnRNP L bind the 3′UTR of glucose transporter 1 mRNA and exist as a complex in vivo. Biochem Biophys Res Commun. 1999;261:646–651. doi: 10.1006/bbrc.1999.1040. [DOI] [PubMed] [Google Scholar]
- 24.Han J, Brown T, Beutler B. Endotoxin-responsive sequences control cachectin/tumor necrosis factor biosynthesis at the translational level. J Exp Med. 1990;171:465–475. doi: 10.1084/jem.171.2.465. . (Erratum, 171:971–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hel Z, Skamene E, Radzioch D. Two distinct regions in the 3′ untranslated region of tumor necrosis factor alpha mRNA form complexes with macrophage proteins. Mol Cell Biol. 1996;16:5579–5590. doi: 10.1128/mcb.16.10.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henics T, Nagy E, Oh H J, Csermely P, von Gabain A, Subjeck J R. Mammalian Hsp70 and Hsp110 proteins bind to RNA motifs involved in mRNA stability. J Biol Chem. 1999;274:17318–17324. doi: 10.1074/jbc.274.24.17318. [DOI] [PubMed] [Google Scholar]
- 27.Jain R G, Andrews L G, McGowan K M, Pekala P H, Keene J D. Ectopic expression of Hel-N1, an RNA-binding protein, increases glucose transporter (GLUT1) expression in 3T3–L1 adipocytes. Mol Cell Biol. 1997;17:954–962. doi: 10.1128/mcb.17.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeno P, Mini T, Moes S, Hintermann E, Horst M. Internal sequences from proteins digested in polyacrylamide gels. Anal Biochem. 1995;224:75–82. doi: 10.1006/abio.1995.1010. [DOI] [PubMed] [Google Scholar]
- 29.Kedersha N L, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King P H, Levine T D, Fremeau R T, Jr, Keene J D. Mammalian homologs of Drosophila ELAV localized to a neuronal subset can bind in vitro to the 3′UTR of mRNA encoding the Id transcriptional repressor. J Neurosci. 1994;14:1943–1952. doi: 10.1523/JNEUROSCI.14-04-01943.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kontoyiannis D, Pasparakis M, Pizarro T T, Cominelli F, Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity. 1999;10:387–398. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- 32.Lai W S, Carballo E, Strum J R, Kennington E A, Phillips R S, Blackshear P J. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lasa M, Mahtani K R, Finch A, Brewer G, Saklatvala J, Clark A R. Regulation of cyclooxygenase 2 mRNA stability by the mitogen-activated protein kinase p38 signaling cascade. Mol Cell Biol. 2000;20:4265–4274. doi: 10.1128/mcb.20.12.4265-4274.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levine T D, Gao F, King P H, Andrews L G, Keene J D. Hel-N1: an autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol Cell Biol. 1993;13:3494–3504. doi: 10.1128/mcb.13.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levy N S, Chung S, Furneaux H, Levy A P. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J Biol Chem. 1998;273:6417–6423. doi: 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- 36.Lieberman A P, Pitha P M, Shin M L. Poly(A) removal is the kinase-regulated step in tumor necrosis factor mRNA decay. J Biol Chem. 1992;267:2123–2126. [PubMed] [Google Scholar]
- 37.Lieberman A P, Pitha P M, Shin M L. Protein kinase regulates tumor necrosis factor mRNA stability in virus-stimulated astrocytes. J Exp Med. 1990;172:989–992. doi: 10.1084/jem.172.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loflin P, Chen C Y, Shyu A B. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 1999;13:1884–1897. doi: 10.1101/gad.13.14.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma W J, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 40.Miyazawa K, Mori A, Miyata H, Akahane M, Ajisawa Y, Okudaira H. Regulation of interleukin-1beta-induced interleukin-6 gene expression in human fibroblast-like synoviocytes by p38 mitogen-activated protein kinase. J Biol Chem. 1998;273:24832–24838. doi: 10.1074/jbc.273.38.24832. [DOI] [PubMed] [Google Scholar]
- 41.Myer V E, Fan X C, Steitz J A. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Myer V E, Steitz J A. Isolation and characterization of a novel, low abundance hnRNP protein: A0. RNA. 1995;1:171–182. [PMC free article] [PubMed] [Google Scholar]
- 43.Nagy E, Rigby W F. Glyceraldehyde-3-phosphate dehydrogenase selectively binds AU-rich RNA in the NAD(+)-binding region (Rossmann fold) J Biol Chem. 1995;270:2755–2763. doi: 10.1074/jbc.270.6.2755. [DOI] [PubMed] [Google Scholar]
- 44.Nakagawa J, Waldner H, Meyer-Monard S, Hofsteenge J, Jeno P, Moroni C. AUH, a gene encoding an AU-specific RNA binding protein with intrinsic enoyl-CoA hydratase activity. Proc Natl Acad Sci USA. 1995;92:2051–2055. doi: 10.1073/pnas.92.6.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pauli U. Control of tumor necrosis factor gene expression. Crit Rev Eukaryotic Gene Expression. 1994;4:323–344. doi: 10.1615/critreveukargeneexpr.v4.i2-3.20. [DOI] [PubMed] [Google Scholar]
- 46.Peng S S, Chen C Y, Xu N, Shyu A B. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perkins D N, Pappin D J, Creasy D M, Cottrell J S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 48.Piecyk M, Wax S, Beck A R, Kedersha N, Gupta M, Maritim B, Chen S, Gueydan C, Kruys V, Streuli M, Anderson P. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 2000;19:4154–4163. doi: 10.1093/emboj/19.15.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ridley S H, Dean J L, Sarsfield S J, Brook M, Clark A R, Saklatvala J. A p38 MAP kinase inhibitor regulates stability of interleukin-1-induced cyclooxygenase-2 mRNA. FEBS Lett. 1998;439:75–80. doi: 10.1016/s0014-5793(98)01342-8. [DOI] [PubMed] [Google Scholar]
- 50.Sakai K, Kitagawa Y, Hirose G. Binding of neuronal ELAV-like proteins to the uridine-rich sequence in the 3′-untranslated region of tumor necrosis factor-alpha messenger RNA. FEBS Lett. 1999;446:157–162. doi: 10.1016/s0014-5793(99)00206-9. [DOI] [PubMed] [Google Scholar]
- 51.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 52.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 53.Szabo A, Dalmau J, Manley G, Rosenfeld M, Wong E, Henson J, Posner J B, Furneaux H M. HuD, a paraneoplastic encephalomyelitis antigen, contains RNA-binding domains and is homologous to Elav and Sex-lethal. Cell. 1991;67:325–333. doi: 10.1016/0092-8674(91)90184-z. [DOI] [PubMed] [Google Scholar]
- 54.Vorm O, Mann M. Improved mass accuracy in matrix-assisted laser desorption/ionization time-of flight mass spectrometry of peptides. J Am Soc Mass Spectrom. 1994;5:955–958. doi: 10.1016/1044-0305(94)80013-8. [DOI] [PubMed] [Google Scholar]
- 55.Wang S W, Pawlowski J, Wathen S T, Kinney S D, Lichenstein H S, Manthey C L. Cytokine mRNA decay is accelerated by an inhibitor of p38-mitogen-activated protein kinase. Inflamm Res. 1999;48:533–538. doi: 10.1007/s000110050499. [DOI] [PubMed] [Google Scholar]
- 56.Wang W, Caldwell M C, Lin S, Furneaux H, Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 2000;19:2340–2350. doi: 10.1093/emboj/19.10.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang W, Furneaux H, Cheng H, Caldwell M C, Hutter D, Liu Y, Holbrook N, Gorospe M. HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol. 2000;20:760–769. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- 59.Winzen R, Kracht M, Ritter B, Wilhelm A, Chen C Y, Shyu A B, Muller M, Gaestel M, Resch K, Holtmann H. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 1999;18:4969–4980. doi: 10.1093/emboj/18.18.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu N, Loflin P, Chen C Y, Shyu A B. A broader role for AU-rich element-mediated mRNA turnover revealed by a new transcriptional pulse strategy. Nucleic Acids Res. 1998;26:558–565. doi: 10.1093/nar/26.2.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang W, Wagner B J, Ehrenman K, Schaefer A W, DeMaria C T, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]