Abstract
To determine the influence of posttranscriptional modifications on 3′ end processing and RNA stability in plant mitochondria, pea atp9 and Oenothera atp1 transcripts were investigated for the presence and function of 3′ nonencoded nucleotides. A 3′ rapid amplification of cDNA ends approach initiated at oligo(dT)-adapter primers finds the expected poly(A) tails predominantly attached within the second stem or downstream of the double stem-loop structures at sites of previously mapped 3′ ends. Functional studies in a pea mitochondrial in vitro processing system reveal a rapid removal of the poly(A) tails up to termini at the stem-loop structure but little if any influence on further degradation of the RNA. In contrast 3′ poly(A) tracts at RNAs without such stem-loop structures significantly promote total degradation in vitro. To determine the in vivo identity of 3′ nonencoded nucleotides more accurately, pea atp9 transcripts were analyzed by a direct anchor primer ligation-reverse transcriptase PCR approach. This analysis identified maximally 3-nucleotide-long nonencoded extensions most frequently of adenosines combined with cytidines. Processing assays with substrates containing homopolymer stretches of different lengths showed that 10 or more adenosines accelerate RNA processivity, while 3 adenosines have no impact on RNA life span. Thus polyadenylation can generally stimulate the decay of RNAs, but processivity of degradation is almost annihilated by the stabilizing effect of the stem-loop structures. These antagonistic actions thus result in the efficient formation of 3′ processed and stable transcripts.
Control of the amount of translatable mRNA is a common parameter to regulate gene expression in many organisms. The available quantity of an RNA depends largely on the rate of synthesis and/or posttranscriptional processes which control the stability of a transcript by preventing it from degradation. Such posttranscriptional processes have been studied in prokaryotes and different subcellular compartments of eukaryotic cells. In spite of the many variations, some common features behind these processes emerge in all organisms and systems. These include the involvement of mRNA secondary structures and 3′ polyadenylation of RNA. While organized RNA secondary structures generally support transcript stabilization, polyadenylation has opposing functions in eukaryotes and prokaryotes.
In the nucleus of eukaryotic cells, pre-mRNAs are polyadenylated at 3′ ends that have been generated by endonucleolytic cleavages. These poly(A) tails play important roles in translation initiation and mRNA export from the nucleus and also have a major function in stabilizing mRNAs (27). A completely different function has been attributed to polyadenylation in prokaryotes, where the degradation of mRNA is stimulated by the presence of 3′ poly(A) tracts (4). The degradation-promoting effect of polyadenylation was even found in transcripts, which were otherwise protected and stabilized by stem-loop structures (3). A similar effect has also been observed in chloroplasts, where polyadenylation also accelerates the decay of mRNA (15, 25).
Polyadenylation has also been found in mitochondria of various organisms. The addition of adenosines to mRNAs in mammalian mitochondria has been suggested to create functional translation stop codons, which are not encoded in the DNA (22). Short oligo(A) tails were also detected at the 3′ ends of RNA in yeast mitochondria. These are about 8 nucleotides (nt) long and predominantly found at the large rRNAs (29). Similarly, oligo(A) tracts were found at some RNA species of the fragmented rRNA in Plasmodium falciparum (12). While the function of the oligo(A) tails in yeast is still unclear, those in Plasmodium were suggested to protect these RNA fragments from exonucleolytic digestion. Recently, polyadenylation was also reported for RNAs in plant mitochondria. In sunflower a dicistronic mRNA associated with cytoplasmic male sterility was suggested to be specifically destabilized by polyadenylation in fertility-restored lines (10). In maize, polyadenylation was detected at multiple sites of the cox2 transcripts. As in sunflower, poly(A) tracts were indicated to decrease mRNA stability, although the effect observed in maize mitochondrial extracts was very weak (19). We report here nonencoded nucleotides at the 3′ ends of pea and Oenothera mitochondrial transcripts containing double inverted repeats in their 3′ untranslated regions (UTRs). These structures have been suggested to increase the stability of the RNA, and the stem-loop structures of pea atp9 transcripts as well as of orf138 mRNAs in Brassica were found to act as processing signals and/or stability elements at least in vitro (1, 5, 6, 16, 23). In this report special emphasis was dedicated to the interaction of the stabilizing effect of such double stem-loop structures and degradation-enhancing polyadenylation. In addition to adenosines, nonencoded cytidines and, to a lesser extent, uridines were detected in vivo. The combination of nonencoded cytidines with adenosines may indicate an involvement of the tRNA terminal nucleotidyltransferase in the addition of nonencoded nucleotides to 3′ ends of plant mitochondrial mRNAs.
MATERIALS AND METHODS
3′ RACE analysis of mtRNA.
The 3′ rapid amplification of cDNA ends (RACE) was carried out in principle as described by Frohmann et al. (9). First-strand synthesis was initiated on 5 μg of total pea mitochondrial RNA (mtRNA) with an oligo(dT)17-adapter primer (DTXSC) using Superscript II reverse transcriptase (RT) (GIBCO-BRL) under conditions given in the manufacturer's manual. About one-fourth of the total cDNA pool was used as the template in a PCR with adapter primer XSC and atp9-specific primer PA-1 (−349 to −329) with KlenTaq polymerase in a buffer supplied by the manufacturer (Clontech). After 35 cycles (30 s at 94°C, 1 min at 40°C, and 1 min at 68°C), PCR fragments were size fractionated on a 1.5% agarose gel and distinct products with sizes of 0.3, 0.4, 0.8, and 1.0 kb were observed above a smear ranging from about 0.2 to 2.0 kb. Since the PCR fragments generated from pea atp9 transcripts, which are polyadenylated downstream of the double stem-loops, are expected to be at least 665 bp long, cDNA fragments between 0.5 and 0.9 kb in length were eluted from the gel and used as templates in a PCR with primers XSC and PA-10, a nested atp9-specific primer (−98 to −84 with a 5′-attached EcoRI restriction site). After 35 cycles (30 s at 94°C, 1 min at 50°C, and 1 min at 68°C), reaction products were digested with SalI and EcoRI and run on an agarose gel. cDNA fragments between about 0.35 and 0.55 kb long including the expected fragment of about 0.43 kb, were eluted from the gel and cloned into pBluescript vectors (Stratagene) following standard protocols (24). In an analogous approach, oligo(dT)-adapter primer DTXSC and adapter primer XSC were replaced by DTNSX-1 and NSX-1 oligonucleotides, and atp9-specific primers PA-10 and PAX-1 (+259 to +274 with a 5′-attached XbaI restriction site) were used. cDNA clones generated with the NSX primer set carry NsiI restriction sites immediately downstream of the poly(A) tracts. Linearization at this site allows the in vitro transcription of mRNAs ending with poly(A) tracts.
The 3′ RACE analysis of Oenothera atp1 transcripts was essentially performed as described above for pea atp9 with the following modifications. About 10 μg of Oenothera mtRNA and DTNSX, NSX, and atp1-specific primers OeatpA-RI (+1362 to +1380 with a 5′-attached EcoRI restriction site) and OeAXI (+1721 to +1736 with a 5′-attached XbaI restriction site) were used for the amplification of the 3′ regions of atp1 transcripts. DNA fragments obtained in these PCRs were digested with XbaI and SalI and were cloned into pBluescript II vectors. Locations of all oligonucleotides are given relative to the translation start codon.
Direct ligation of anchor primers to 3′ ends of in vitro processing products and steady-state mtRNA.
In vitro processing assays were carried out under standard conditions, and reaction products were separated on a 6% polyacrylamide gel. Radioactive bands were visualized by autoradiography and were excised from the gel. The RNAs were eluted from the gel in a buffer containing 0.5 M ammonium acetate, 0.1 mM EDTA, and 0.1% sodium dodecyl sulfate. After phenol-chloroform extraction, the in vitro processing products were ethanol precipitated and ligated to 20 pmol of anchor primer (RiboNSX) in the presence of 20 U of RNA ligase (GIBCO-BRL), 1% (vol/vol) dimethyl sulfoxide, 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 10 mM dithiothreitol, 0.25 mM ATP, and 0.6 μg of bovine serum albumin per μl. The ligation reaction was performed for 24 h at 15°C in a 10-μl total reaction volume. Following two rounds of phenol-chloroform extraction, nonligated anchor primers were removed by centrifugation in Microcon 30 microfiltration units at 14,000 × g to a final volume of 10 μl. First-strand synthesis was primed with oligonucleotide NSX-1 using the complete ligation reaction as the template and Superscript II RT. Subsequent amplification with primers PAX-1 and NSX-1 was performed with KlenTaq polymerase (Clontech) in a buffer supplied by the manufacturer. After 35 cycles (1 min at 94°C, 1 min at 50°C, and 30 s at 68°C), PCR products were digested with XbaI and SalI and were cloned into pBluescript vectors. Respective clones were identified in a colony hybridization using Hybond-N membranes (Amersham Pharmacia Biotech) and oligonucleotide PA-17hyb (+266 to +284) as probe, following a protocol given by the manufacturer.
For the 3′ end analysis of steady-state transcripts, the procedure described above was used with the following modifications. One microgram of total mtRNA was added to the ligation reaction mixture, and primer PA-10 was used as the atp9-specific primer in the amplification reaction.
Lysate preparation and in vitro processing reactions.
Pea mitochondrial lysate preparation and the in vitro processing reactions were performed as described previously (6).
DNA templates and in vitro transcription.
Polyadenylated pea atp9 precursor molecules are transcribed from cDNA clone 9.17 generated in the 3′ RACE approach with primers PAX-1 and NSX-1 on in vitro-transcribed RNA obtained from clone no. 9. Plasmid 9.17 was linearized with NsiI and transcribed with T7 RNA polymerase. The resulting transcript (117 nt) contains the pea atp9 double stem-loop and 43-nt upstream and 23-nt downstream sequences. The upstream part corresponds to vector sequences from the T7 transcription start point to the XbaI restriction site. The downstream moieties comprise 21 adenosines at the 3′ end. Transcripts containing (A)3 or (A)10 poly(A) tracts downstream of the inverted repeat are directly transcribed from DNA fragments obtained by amplification with primer M13 universal and primers IR+dT3 [(A)3] and IR+dT10 [(A)10] on clone 9.17 as the DNA template.
The nonpolyadenylated standard substrate with original mitochondrial upstream sequences is generated by the transcription of PCR products as previously described by Dombrowski et al. (6).
The pea atp9 RNA substrate without inverted repeat (101 nt) is transcribed from a DNA template amplified by PCR with primers T7IR− (+159 to +179 with a T7 promoter attached at the 5′ end) and PIR− (+241 to +258). To generate analogous substrates with (A)21 (122 nt), (A)10 (111 nt), and (A)3 (104 nt) 3′ homopolymer sequences, respectively, primers IR−dT, IR−dT10, and IR−dT3 were used instead of PIR−.
All cloned DNA templates were inspected by complete sequence analysis. In vitro transcription reactions were performed with T7 RNA polymerase (MBI Fermentas) in a buffer supplied by the manufacturer. Purification of all in vitro transcripts by polyacrylamide gel electrophoresis was carried out as described elsewhere (6).
Nucleic acids.
Pea and Oenothera mtRNAs were isolated from respective organelles enriched by differential centrifugation and purified on Percoll (Amersham Pharmacia Biotech) gradients as described previously (2). Sequences of the primers used are available on request. All oligonucleotides were purchased from GIBCO-BRL.
Miscellaneous methods.
Sequencing reactions were carried out with the Thermo Sequenase fluorescent labeling kit (Amersham Pharmacia Biotech) according to instructions given by the manufacturer. All standard methods were used as described by Sambrook et al. (24).
RESULTS
Poly(A) tails at the 3′ ends of atp9 mRNAs in pea mitochondria.
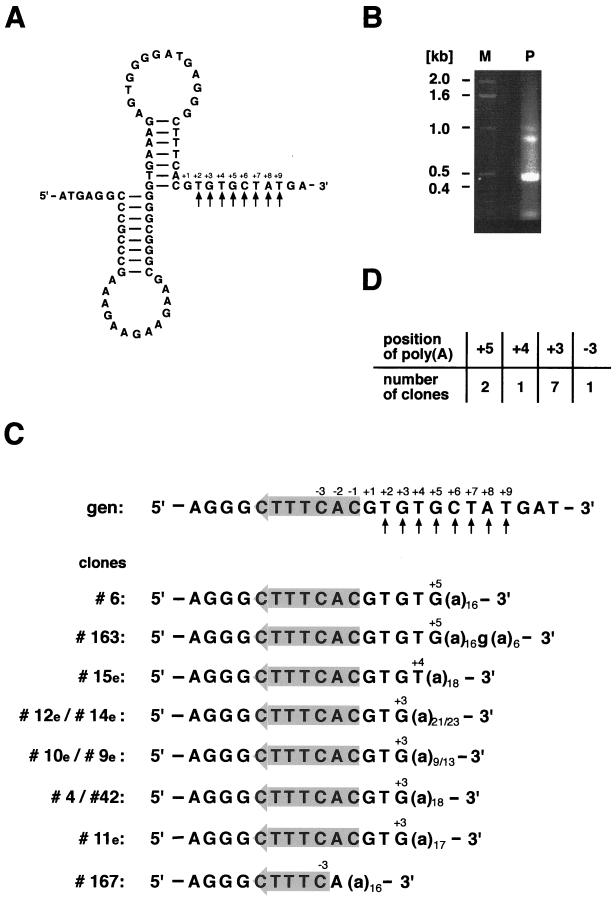
The pea atp9 gene is transcribed into three different mRNA species with differing 5′ termini but identical 3′ ends. These 3′ termini were mapped in two independent S1 protection analyses within the 3′ part of the second of two consecutive inverted repeats and immediately downstream thereof (6, 21). Recently it was found that this double inverted repeat with the potential to fold into a double stem-loop structure does not terminate transcription in vitro but rather functions as a processing signal and stabilizing element during a controlled 3′ processing event (5, 6). To investigate whether nonencoded poly(A) tails are present at these transcript termini, a 3′ RACE analysis, including two rounds of PCR with an oligo(dT)-adapter primer and two nested atp9-specific primers, was performed with total pea mtRNA (for details see Materials and Methods). The sequence analysis of 18 clones indicated the presence of nonencoded poly(A) tails at the 3′ ends of these mRNAs. In four clones, polyadenylation sites are located at positions +6 and +2 relative to the first nonpaired nucleotide (+1) downstream of the second stem (Fig. 1). The majority of the poly(A) tracts are found at positions −2 to −5 within the downstream part of the second stem. These polyadenylation sites coincide with previously mapped 3′ ends. Three clones contained poly(A) sequences attached within the second loop, while two others are added at the beginning and end of the upstream part of the second stem, respectively. The lengths of the poly(A) stretches vary between 14 and 25 adenosines, with 10 of the 18 clones containing more than the 17 adenosines derived from the oligo(dT)-adapter primer. In four clones, nonencoded cytidine, guanosine, and thymidine (corresponding to uridine in the RNA) nucleotides are found predominantly in the 5′ part of the attached sequences. Almost all clones investigated are edited at position +20 (relative to the translation start codon [+1]), while a cytidine-to-uridine alteration at position +50 is found less frequently. Both editing events change the codon identity from serine to leucine (data not shown).
FIG. 1.
3′ RACE analysis of pea mitochondrial atp9 transcripts. (A) Previously mapped 3′ ends are indicated within the pea atp9 potential double stem-loop structure (arrows). (B) Nucleotide sequences of 3′ RACE clones containing poly(A) tails. Nonencoded nucleotides found in 18 analyzed cDNA clones (designations are shown on the left) are given in lowercase letters. The positions of the polyadenylation sites (indicated above the sequences) correspond to the 3′-most nucleotide. Numbering refers to the first nucleotide (+1) downstream of the inverted repeats (indicated by oppositely oriented grey shadowing arrows) in the genomic sequence (gen). Vertical arrows beneath the genomic sequence indicate previously mapped 3′ ends. (C) The numbers of clones identified are given for each polyadenylation site.
Potential poly(A) tracts in Oenothera atp1 transcripts.
The analysis of Oenothera atp1 transcripts extends this study to other double-stem-loop-containing mRNAs from mitochondria of another plant species (Fig. 2). Amplification with an atp1-specific primer and an adapter primer on a total cDNA pool primed with an oligo(dT)-adapter primer generated a dominant product of about 0.45 kb expected of nonencoded poly(A) sequences attached downstream of the stem-loop structure (Fig. 2B). This fragment was eluted from the gel and was either used as a template in a second PCR with a nested, atp1-specific primer and an adapter primer or directly cloned. Sequencing of 10 cDNA clones obtained from both amplification reactions revealed poly(A) tracts attached at positions +5, +4, and +3 immediately downstream of the double stem-loop structure. These polyadenylation sites are consistent with previously mapped 3′ ends (26). Only a single clone indicated a polyadenylation site at position −3 within the downstream part of the second stem (Fig. 2C). These results show that the distribution of the polyadenylation sites in Oenothera atp1 mRNAs is less variable than in pea atp9 transcripts. The lengths of the poly(A) tails observed in the Oenothera atp1 cDNA clones range from 16 to 23 adenosines with only a single guanosine insertion within a poly(A) sequence.
FIG. 2.
Identification of potential polyadenylation sites in Oenothera atp1 transcripts. (A) Potential double stem-loop structure found in the 3′ UTR of Oenothera atp1 mRNAs. Previously mapped 3′ termini are indicated by arrows. (B) DNA fragments generated in a PCR with adapter primer (NSX) and an atp1-specific oligonucleotide (OeatpA-RI) on oligo(dT)-adapter-primed cDNA are size fractionated on a 1.5% agarose gel (lane P). Besides minor fragments of about 1.0, 0.85, and 0.2 kb, a strong cDNA fragment (0.45 kb) corresponding to RNAs polyadenylated downstream of the stem-loop is detected. Lengths of coelectrophoresed DNA marker fragments (lane M) are given in kilobase pairs on the left. (C) Partial sequences of Oenothera atp1 cDNA clones containing nonencoded poly(A) tails (indicated in lowercase letters). Designations and numbering are analogous to those for Fig. 1. (D) Correlation of the number of clones with poly(A) addition sites.
Rapid and efficient processing of polyadenylated pea atp9 transcripts in vitro.
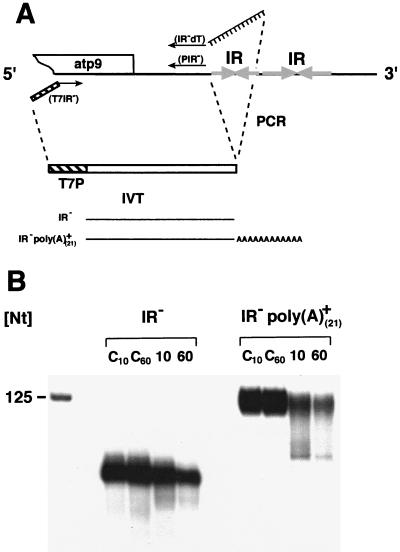
The results of the 3′ RACE analyses identified nonencoded poly(A) tracts at the 3′ ends of transcripts containing double stem-loop structures in their 3′ UTRs. Such poly(A) tails were recently suggested to decrease mRNA stability in plant mitochondria (10, 19). This destabilizing effect would counteract the function of the double stem-loop, which was found to act as a processing signal and stability element at least in vitro (5, 6). To solve this question, the consequence of poly(A) tails on double-stem-loop-containing mRNA was tested in vitro. Nonpolyadenylated synthetic transcripts (96 nt) used in these assays were transcribed from a T7 promoter-containing PCR product (Fig. 3A) and corresponded to the standard substrate shown to be correctly processed in a pea mitochondrial in vitro system (6). It contains the pea atp9 double stem-loop structure and 21 and 28 original mitochondrial nt upstream and downstream, respectively. In the polyadenylated mRNA (113 nt), the 3′ trailer downstream of the stem-loop structure consists of a 21-mer adenosine homopolymer downstream of two mitochondrially encoded nucleotides while the upstream leader (42 nt) represents pBluescript vector sequences. Details of the construction of the DNA templates for transcription of these substrates (Fig. 3B) are given in Materials and Methods. To test the influence of the regions upstream of the inverted repeat, an analogous RNA substrate, which contains original mitochondrial downstream sequences (28 nt) and the alien sequences deriving from the vector (42 nt), was also tested in vitro. After incubation of the nonpolyadenylated standard transcript for 60 min, a single processing product of about 70 nt was observed (Fig. 3C, left). This product corresponds to an RNA species where the downstream part is removed, as seen in previous processing experiments (6). An accelerated generation of processing products is observed upon incubation of polyadenylated substrates. After 10 min, two processing products of about 70 and 80 nt are generated. While the smaller product has a size consistent with a 3′ end similar to that of the product obtained in the reaction with the nonpolyadenylated standard substrate, an additional RNA, about 5 to 10 nt larger, is generated. Longer incubation (for 60 min) resulted in the generation of two products that seem to be slightly smaller than those observed after 10 min (Fig. 3C, right). A product very similar to the one generated from the nonpolyadenylated standard substrate and to the smaller one (70 nt) deriving from the polyadenylated substrate was observed in in vitro tests with the nonpolyadenylated substrate, with vector sequences upstream of the stem-loop structure. The temporal dynamic of the product formation is identical to that of the processing reaction of the nonpolyadenylated standard substrate, which indicates that sequences upstream of the stem-loop structure have no degradation-promoting function (data not shown).
FIG. 3.
In vitro processing of polyadenylated and nonpolyadenylated pea atp9 precursor molecules. (A) Generation of the pea atp9 standard transcript containing a double inverted repeat (grey horizontal arrows). The substrate is transcribed in vitro (IVT) from a PCR product obtained with primers PA-12 and T7IVR+, which contains a promoter for T7 RNA polymerase (T7P, hatched box). (B) The polyadenylated substrate is transcribed from the linearized clone 9.17, obtained by RT-PCR cloning of a polyadenylated pea atp9 mRNA with primers NSX-dT (adapter primer for cDNA synthesis), PAX-1, and NSX-1 (PCR). (C) Identical amounts of substrates (about 12,000 cpm) with [poly(A)+] and without [poly(A)−] poly(A) tracts are incubated for 10 and 60 min in a pea mitochondrial lysate. Resulting reaction products are separated on a 6% polyacrylamide gel (lanes 10 and 60). Control reactions (lanes C10 and C60) are performed under identical conditions without protein. Lengths of coelectrophoresed DNA marker fragments are given in nucleotides (Nt) in the right margin. Discrepancies between the sizes of RNA molecules and the DNA marker fragments are due to different migration velocities of RNA and DNA molecules.
A similar processing pattern was observed with heterologous synthetic polyadenylated and nonpolyadenylated Oenothera atp1 transcripts. Again, accelerated processing is observed with poly(A)+ substrates, while final degradation of this RNA seems to be unaffected (data not shown).
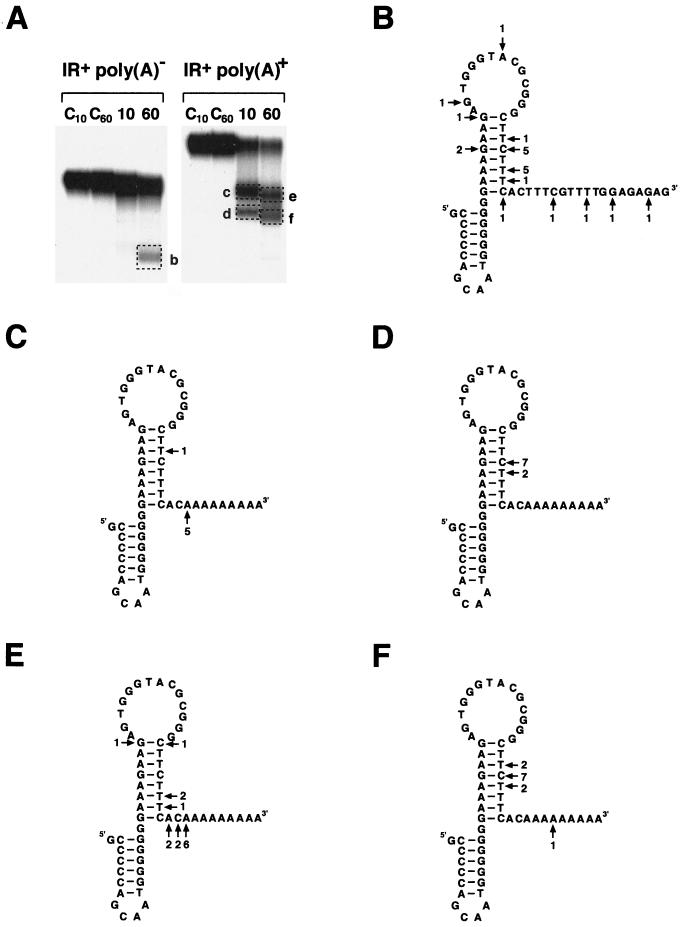
The precise 3′ ends of the detected products were determined by a direct ligation of anchor primers (RiboNSX) to eluted RNA species, followed by an RT-PCR analysis initiated with primer NSX complementary to the anchor primer (Fig. 4). Sequencing of 22 cDNA clones obtained from the processing products generated from the nonpolyadenylated standard substrate revealed 3′ ends which scatter over a broad range (from nucleotide positions −26 to +17) with reference to the first nucleotide (+1) downstream of the second stem-loop (Fig. 4B). The 3′ ends detected in 14 clones, however, are located within the downstream part of the second stem or immediately downstream of this structure and coincide with previously mapped transcript termini (6) (Fig. 1 and 4B). In the larger gel fragment excised, other termini are identified at low frequencies, which are attributed to the background of random termini in the RNA population of such an in vitro processing reaction (Fig. 4A). Transcript termini from the smaller processing products generated after 10 and 60 min from polyadenylated substrates are detected in 9 and 12 clones, respectively, and are almost exclusively located within the second stem, which is consistent with the majority of termini generated from the nonpolyadenylated precursor RNA (Fig. 4D and F). In contrast the bulk of the 3′ ends of the larger processing products are found immediately downstream of the second stem with slightly shorter products observed after 60 min (Fig. 4C and E). Interestingly, half of the 22 clones obtained from these products end with a nonencoded adenosine, indicating that the poly(A) tail is not completely removed. In summary the results of the in vitro processing assays indicate that polyadenylated transcripts are processed much faster than nonpolyadenylated substrates. Total degradation of the double-stem-loop-containing substrates, however, is not significantly accelerated.
FIG. 4.
Precise determination of 3′ termini of in vitro processing products. (A) The in vitro processing products (b to f, indicated by dotted boxes) obtained from nonpolyadenylated [poly(A)−] and polyadenylated [poly(A)+] substrates with inverted repeats (IR+) were excised, and the RNA molecules were eluted from the gel. The 3′ ends of the different products were analyzed by direct anchor primer ligation followed by RT-PCR analysis. Lanes 10 and 60 represent results at 10 and 60 min; lanes C10 and C60 represent control reactions at 10 and 60 min. (B to F) Secondary structures of the pea atp9 double stem-loop with nonpolyadenylated (B) and polyadenylated (C to F) downstream moieties. Arrows indicate the 3′ terminal nucleotide found in individual clones. The numbers of cDNA clones containing certain 3′ ends are given at the arrows. Designations of the individual stem-loops (B to F) correspond to the designations of the analyzed processing products (b to f).
Temporal dynamics of the in vitro processing of polyadenylated pea atp9 transcripts.
The generation of an additional distinct product from polyadenylated transcripts raises the possibility that this could be an intermediate processing product. To investigate the temporal course of the in vitro processing of polyadenylated pea atp9 mRNAs (Fig. 1B), the generation of processing products was followed over a period of 90 min (Fig. 5). Already after 1 min, diffuse smaller RNAs appear, indicating the initiation of the processing reaction, which results in the detectable formation of the approximately 90- to 95-nt-long RNA fragment after 2.5 min. The smeared RNA between the substrate and this product indicates exonucleolytic processivity. This impression is substantiated by the continuously decreasing size of this 90- to 95-nt-long RNA, with several nucleotides being consecutively removed upon prolonged incubation. A similar effect is also seen with the 80- to 85-nt-long product, which first appears after 5 min. The substrate and the two processing products show distinct dynamics in their respective abundances. As expected, the substrate decreases continuously over 90 min. The larger product (90 to 95 nt) has its highest abundance after 10 min, while the smaller product (80 to 85 nt) reaches maximum abundance after 60 min. These results suggest that the large processing product, whose 3′ ends map immediately downstream of the double stem-loop, is a stabilized intermediate in the exonucleolytic generation of the small product, with its 3′ ends located within the downstream part of the second stem. Thus, an exoribonuclease seems to remove the poly(A) tails very rapidly but stalls or falls away from the RNA either at the 3′ end of the stem-loop structure or simply at the transition from the poly(A) stretch to the “normal” sequence. The smaller product most likely results from a second steric hindrance within the downstream part of the stem.
FIG. 5.
Time course of the in vitro processing of a polyadenylated pea atp9 precursor. A polyadenylated pea atp9 substrate is incubated over a period of 90 min, and probes are taken at 0 to 90 min, as indicated above the lines. Control reactions are performed under identical conditions in the absence of protein for 0 (C0) and 90 (C90) min. Sizes of coelectrophoresed DNA marker fragments are given in nucleotides (Nt).
Accelerated total degradation of RNA substrates with no inverted repeat.
The results presented above indicate that polyadenylation accelerates RNA processing of stem-loop-containing RNAs, while the total degradation of the transcripts remains almost unaffected. To investigate the stabilizing function of this stem-loop structure during the formation of the different processing products, polyadenylated and nonpolyadenylated RNAs without a stem-loop were tested in the in vitro processing system. The substrate without a stem-loop structure and poly(A) tract, representing 101-nt-long RNA molecules with 3′ ends just upstream of the first inverted repeat (Fig. 6A), is consecutively degraded without formation of any stable intermediate, indicating that the stem-loop is indeed responsible for the generation of stabilized products (Fig. 6B, left). A different processivity is seen with polyadenylated transcripts. Here the presence of a poly(A) tract in the substrate significantly accelerates the total turnover of the precursor RNA (Fig. 6B, center). Only a weak signal indicates the presence of an intermediate, which corresponds in size to the RNA without a poly(A) tail. The low intensity of this signal indicates only weak stability of this product.
FIG. 6.
Accelerated degradation of RNAs without stem-loops containing 3′ poly(A) tails. (A) Substrates with no inverted repeat containing no 3′ homopolymer tract (IR−) and a 3′ poly(A)21 [IR− poly(A)+] are transcribed in vitro (IVT) from PCR products with primer T7IR− containing a T7 promoter (T7P, hatched box) and primers PIR− and IR−dT (A)21, respectively. (B) Incubation of the respective substrates with pea mitochondrial protein extracts for 10 and 60 min. Designation of the lanes (10 and 60) corresponds to the length of incubation. Control reactions are performed in the absence of protein under otherwise identical conditions for 10 (C10) and 60 (C60) min. The sizes of DNA marker fragments are indicated in nucleotides (Nt).
Since the diffuse RNAs between the substrate and this intermediate are suggestive of an exonucleolytic digestion of the substrates, a potential involvement of a 3′-to-5′ exoribonuclease activity was monitored with 5′- and 3′-labeled transcripts containing the inverted repeat (Fig. 3A). While a stable product of the expected size is seen with the 5′-labeled substrates, the 3′-labeled transcripts disappear without formation of a detectable processing product or intermediate (data not shown). This suggests that indeed a 3′-to-5′ exoribonuclease takes part in the 3′ processing and/or degradation event investigated here.
Detection of nonencoded nucleotides at 3′ transcript termini of steady-state RNA.
The 3′ RACE analysis using an oligo(dT)-adapter primer does indicate the presence of poly(A) sequences at the 3′ ends of mtRNAs. Several questions are, however, left unanswered in this approach. First, the lengths of the poly(A) tails cannot be determined, and second, the majority of other nonencoded nucleotides (i.e., C, G, or U) might remain undetected. This may be a major drawback, since, for example, considerable uridylyltransferase activity has been detected in plant mitochondrial protein extracts (S. Binder, unpublished results).
For an alternative approach, anchor primers were ligated directly to 3′ ends of total steady-state mtRNA from pea. The products of an RT-PCR initiated at an oligonucleotide complementary to the anchor primer were cloned and analyzed (Fig. 7A and B).
FIG. 7.
A direct anchor primer ligation–RT-PCR analysis identifies nonencoded nucleotides at pea atp9 steady-state transcripts. (A) Scheme of the direct anchor primer ligation–RT-PCR approach as outlined in Materials and Methods. (B) Ethidium bromide-stained agarose gel with DNA fragments obtained from a PCR carried out with primers PA-10 and NSX-1. Sizes of DNA marker fragments are given in kilobase pairs on the right. (C) Sequences of the downstream inverted repeat of the pea atp9 3′ UTR (indicated by grey shading) obtained from a direct anchor primer ligation–RT-PCR approach. Nonencoded nucleotides are given in lowercase letters. Previously mapped 3′ transcript termini are indicated by vertical arrows beneath the genomic sequence (gen). The numbering of the sequence is given with respect to the first nucleotide (+1) downstream of the inverted repeat. (D) Number of clones containing encoded 3′ terminal nucleotides and clones with nonencoded nucleotides found at certain positions relative to the first nucleotide downstream of the inverted repeat (+1).
From 39 clones sequenced, 13 contain 3′ nonencoded nucleotides at sites previously mapped as 3′ termini (Fig. 7C and D). Adenosines were found in 10 clones, but in addition, cytidines are detected in 5 clones and thymidines (corresponding to uridines) are detected in 2 clones. The cytidines are almost exclusively combined with adenosines. Sole adenosines are found in seven clones, albeit five of these clones contain only single nonencoded adenosines. This experiment confirms that cytidines and, less frequently, uridines are added in vivo to the 3′ ends of plant mitochondrial mRNAs. RNA fragments deriving from the 18S rRNA and the 5′ UTR of the atp9 transcript are most likely coincidentally ligated to the 3′ ends of atp9 mRNA, although identical atp9 5′ UTR fragments are found in two clones derived from independent ligation–RT-PCR approaches.
Poly(A) tracts with 10 or more adenosines stimulate RNA processing and degradation.
The anchor primer ligation–RT-PCR analysis detects only short nonencoded extensions up to 3 nt. Two explanations are possible for such relatively short tracts. Either the extensions per se are indeed only a few nucleotides long, or they are the result of the processing reaction. To test the latter assumption, substrates with no inverted repeat containing 0, 3, 10, or 21 additional adenosines were incubated with the pea mitochondrial lysate. While both 10 or 21 oligo(A) tails significantly accelerate degradation, no difference is observed in the processing of substrates with 3 or 0 additional adenosines (Fig. 8A). An analogous effect is seen with substrates containing inverted repeats. An extension of 3 adenosines has no impact on processing, whereas a processing product is rapidly formed from a precursor with 10 adenosines (Fig. 8B).
FIG. 8.
Oligo(A) tails with 10 or more adenosines promote RNA degradation or processing. (A) Incubation of synthetic transcripts with no inverted repeat with either no extension (IR−), a 3-nt extension [IR−(A)3], a 10-nt tract [IR−(A)10], or a 21-nt tail [IR−(A)21] are incubated with a pea mitochondrial lysate for 10 and 60 min. Clear differences are observed after 60 min with strong degradation-stimulating effects for the tails with 10 and 21 adenosines. In comparison, no significant differences in the decay are observed in reactions with substrates without extensions or with three additional terminal adenosines. Nt, nucleotides. (B) Substrates containing the pea atp9 inverted repeat with 3 [IR+poly(A)3] or 10 [IR+poly(A)10] adenosines attached 2 nt downstream of the second stem were incubated with a pea mitochondrial lysate. The product generated from the latter substrate where sequences downstream of the stem-loops have been removed clearly indicates a processing-promoting effect of the 10-adenosine extension. In comparison, only a minor shortening of the substrate with three terminal adenosines is observed after 10 and 60 min (lanes 10 and 60). No degradation or processing is observed after incubation of the substrates for 10 and 60 min in the absence of protein (lanes C10 and C60). Nt, nucleotides.
These results and the fairly fragment in vitro processing products containing one or more residual nonencoded adenosines (Fig. 4C to F) strongly suggest that the short extensions detected in vivo are the product of RNA processing and/or degradation.
DISCUSSION
3′ nonencoded nucleotides in plant mitochondrial transcripts.
Two independent approaches were used to identify nonencoded nucleotides at the 3′ ends of plant mitochondrial transcripts. While the 3′ RACE analysis initiated with an oligo(dT)-adapter primer indirectly shows the presence of nonencoded adenosines at the 3′ transcript termini, the direct anchor primer ligation–RT-PCR approach provides straightforward evidence for the existence of such nonencoded 3′ extensions without any inherent experimental bias for certain nucleotides.
The 3′ RACE analysis predominantly detects poly(A) tracts within the 3′ part of the second stem or immediately downstream thereof. Since no adenosines are encoded in these regions downstream of both pea atp9 and Oenothera atp1 genes, all these clones should derive from annealing of the oligo(dT)-adapter primer at nonencoded, posttranscriptionally added adenosines (Fig. 1 and 2). Only the 3′ ends observed in pea atp9 clones no. 7 and j27 are located in regions which would allow an annealing of the oligo(dT) stretch to encoded sequences (Fig. 1). Besides the adenosines expected from the use of an oligo(dT)-adapter primer, cytidines, thymidines (corresponding to uridines in the RNA), and guanosines are also found in the 3′ RACE products of pea atp9 and Oenothera atp1 mRNAs. While the location of the guanosine within the 17-mer adenosine (corresponding to thymidines) stretch in the adapter primer rather suggests an artifact, several cytidines and a single thymidine are either found directly at or near the 3′-most nucleotides. The direct anchor primer ligation–RT-PCR approach confirms the presence of cytidines and uridines in more than half of the clones with nonencoded nucleotides. Interestingly, no guanosine is found in the clones obtained in this analysis. The detection of other RNA fragments at the 3′ ends of atp9 mRNAs is due to the high content of usually 18S rRNA fragments in mtRNA preparations. Such fragments are also often observed in chimeric clones of mitochondrial cDNA libraries (11). The validity of the method is indirectly supported by the analogous investigation of the in vitro processing products (Fig. 4B), where in 64 clones no nucleotides were detected that could be derived from artifacts generated in the ligation or amplification reactions.
The real in vivo length of the nonencoded extensions remains unclear. A diagnostic 3′ RACE analysis initiated by an oligo(dT)-adapter primer with 17 thymidines on an RNA with a 24-mer adenosine poly(A) tail results in clones containing between 15 and 24 adenosines, corroborating the length variation occurring under such experimental conditions (data not shown). Thus, such a 3′ RACE analysis does not allow any conclusion on the a priori lengths of the poly(A) tracts. In the alternative approach, the number of nonencoded nucleotides found is rather low. The majority of these clones contain a single nonencoded nucleotide, and the maximum of three posttranscriptionally added nucleotides is found in only three clones. No longer poly(A) tails are observed in this approach, suggesting a very low frequency of such longer extensions.
The in vitro assays of substrates with poly(A) tracts of different sizes indicate that tails with 10 nucleotides already promote a similar acceleration of degradation, as do longer poly(A) tails (Fig. 8A). In contrast, short tails with only three adenosines have no influence on transcript stability, suggesting that the termini seen in the direct anchor primer ligation–RT-PCR analysis derive from relatively stable end products rather than from substrates promoting and initiating degradation. This assumption is substantiated by the detection of one or more residual nonencoded adenosines at the processing products generated from polyadenylated substrates.
The observation that mitochondria can discriminate between transcripts with homopolymer tracts of different sizes and specifically degrade those with 10 or more nucleotides supports the functional and biological significance of these nonencoded nucleotides.
Function of polyadenylation in plant mitochondria.
Recent reports of polyadenylation of plant mitochondrial transcripts suggested that the presence of 3′ poly(A) tails accelerates the degradation of mRNAs (10, 19). Our results obtained for transcripts without a stem-loop structure are consistent with these results. A significant acceleration of RNA decay is caused by the presence of poly(A) tails in these transcripts. Surprisingly, an even stronger destabilizing effect is observed with poly(G)21 tails, which contradicts previous reports on chloroplasts and cannot be rationally explained at present (7, 8; data not shown). No effects of poly(G) stretches were detected in transcripts of nuclear reporter genes in plants, indicating that indeed the effects of poly(G) stretches might differ for different compartments of the plant cell (13).
In plant mitochondria we furthermore observe a differentiated response to polyadenylation of mRNAs with double stem-loop structures. With these substrates a significant acceleration of 3′ end processing is observed, while total degradation is only marginally increased (Fig. 3). In this reaction RNA moieties downstream of the stem-loop are rapidly removed, but the progress of degradation is stopped or stalled at the stem-loop structure (Fig. 4). The stabilizing effect of these structures then seems to be stronger than the stimulation of mRNA decay by polyadenylation. The identification of poly(A) addition sites within the stem-loop indicates that these are polyadenylated again and suggests repeated cycles of polyadenylation and removal of poly(A) tails together with several adjacent nucleotides. This may be an effective mechanism for eventually overcoming the stem-loop structure and initiating the total degradation of the RNA. The life span of such a transcript is thus balanced between the degradation-promoting polyadenylation and the stabilizing effect of the stem-loop. In plant mitochondria the former effect seems to be minor in comparison to that in bacteria and chloroplasts, where poly(A) tails have a much stronger destabilizing effect on stem-loop-containing transcripts (3, 17, 18).
How are the 3′ nonencoded nucleotides added to mRNAs?
The presence of nonencoded nucleotides at the 3′ ends of mRNAs raises the question of how these nucleotides are added. In other compartments of plant cells and in other organisms, poly(A) tails are generally synthesized by poly(A) polymerase, which has so far not been reported in plant mitochondria. It has recently been shown that this protein from Escherichia coli incorporates all 4 nt into the 3′ extensions, at least in vitro, so one can speculate that an analogous enzyme activity may add the nonencoded nucleotides to mRNA 3′ ends in plant mitochondria (28).
The presence of a significant number of cytidines among the nonencoded nucleotides also raises the possibility that a tRNA terminal nucleotidyltransferase is involved in the addition of the nonencoded nucleotides. This theory is strengthened by the observation that the cytidines are almost exclusively combined with adenosines and that complete CCA additions were identified in two clones (Fig. 1 and 7). This enzyme activity has been detected in plant mitochondrial extracts from potato and wheat and should now be tested for the potential to add short or even longer oligo(A) tracts to mRNA substrates (14, 20).
ACKNOWLEDGMENTS
This work was supported by grant Bi 590/3-3 from the Deutsche Forschungsgemeinschaft and a fellowship from the Studienstiftung des Deutschen Volkes to J.K.
We thank Axel Brennicke for fruitful discussions and his constructive comments on the manuscript.
REFERENCES
- 1.Bellaoui M, Pelletier G, Budar F. The steady-state level of mRNA from the Ogura cytoplasmic male sterility locus in Brassica cybrids is determined post-transcriptionally by its 3′ region. EMBO J. 1997;16:5057–5068. doi: 10.1093/emboj/16.16.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Binder S, Brennicke A. Transcription initiation sites in mitochondria of Oenothera berteriana. J Biol Chem. 1993;268:7849–7855. [PubMed] [Google Scholar]
- 3.Blum E, Carpousis A J, Higgins C F. Polyadenylation promotes degradation of 3′-structured RNA by the Escherichia coli mRNA degradosome in vitro. J Biol Chem. 1999;274:4009–4016. doi: 10.1074/jbc.274.7.4009. [DOI] [PubMed] [Google Scholar]
- 4.Carpousis A J, Vanzo N F, Raynal L C. mRNA degradation. A tale of poly(A) and multiprotein machines. Trends Genet. 1999;15:24–28. doi: 10.1016/s0168-9525(98)01627-8. [DOI] [PubMed] [Google Scholar]
- 5.Dombrowski S, Binder S. 3′-inverted repeats in plant mitochondrial mRNAs act as processing and stabilizing elements but do not terminate transcription. In: Moller I M, Gardeström P, Glimelius K, Glaser E, editors. Plant mitochondria: from gene to function. Leiden, The Netherlands: Backhuys Publishers; 1998. pp. 9–12. [Google Scholar]
- 6.Dombrowski S, Brennicke A, Binder S. 3′-inverted repeats in plant mitochondrial mRNAs are processing signals rather than transcription terminators. EMBO J. 1997;16:5069–5076. doi: 10.1093/emboj/16.16.5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drager R G, Zeidler M, Simpson C L, Stern D B. A chloroplast transcript lacking the 3′ inverted repeat is degraded by 3′ to 5′ exoribonuclease activity. RNA. 1996;2:652–663. [PMC free article] [PubMed] [Google Scholar]
- 8.Drager R G, Girard-Bascou J, Choquet Y, Kindle K L, Stern D B. In vivo evidence for 5′ to 3′ exoribonuclease degradation of unstable chloroplast mRNA. Plant J. 1998;13:85–96. doi: 10.1046/j.1365-313x.1998.00016.x. [DOI] [PubMed] [Google Scholar]
- 9.Frohman M A, Dush M K, Martin G R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gagliardi D, Leaver C J. Polyadenylation accelerates the degradation of the mitochondrial mRNA associated with cytoplasmic male sterility in sunflower. EMBO J. 1999;18:3757–3766. doi: 10.1093/emboj/18.13.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giegé P, Brennicke A. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc Natl Acad Sci USA. 1999;96:15324–15329. doi: 10.1073/pnas.96.26.15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gillespie D E, Salazar N A, Rehkopf D H, Feagin J E. The fragmented mitochondrial ribosomal RNAs of Plasmodium falciparum have short A tails. Nucleic Acids Res. 1999;27:2416–2422. doi: 10.1093/nar/27.11.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutierrez R A, MacIntosh G C, Green P J. Current perspectives on mRNA stability in plants: multiple levels and mechanisms of control. Trends Plant Sci. 1999;4:429–438. doi: 10.1016/s1360-1385(99)01484-3. [DOI] [PubMed] [Google Scholar]
- 14.Hanic-Joyce P J, Gray M W. Processing of transfer RNA precursors in a wheat mitochondrial extract. J Biol Chem. 1990;265:13782–13791. [PubMed] [Google Scholar]
- 15.Hayes R, Kudla J, Gruissem W. Degrading chloroplast mRNA: the role of polyadenylation. Trends Biochem Sci. 1999;24:199–202. doi: 10.1016/s0968-0004(99)01388-2. [DOI] [PubMed] [Google Scholar]
- 16.Kaleikau E K, Andre C P, Walbot V. Structure and expression of rice mitochondrial apocytochrome b gene (cob-1) and pseudo-gene (cob-2) Curr Genet. 1992;22:463–470. doi: 10.1007/BF00326411. [DOI] [PubMed] [Google Scholar]
- 17.Kudla J, Hayes R, Gruissem W. Polyadenylation accelerates degradation of chloroplast mRNA. EMBO J. 1996;15:7137–7146. [PMC free article] [PubMed] [Google Scholar]
- 18.Lisitsky I, Klaff P, Schuster G. Addition of destabilizing poly(A)-rich sequences to endonuclease cleavage sites during the degradation of chloroplast mRNA. Proc Natl Acad Sci USA. 1996;93:13398–13403. doi: 10.1073/pnas.93.23.13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lupold D S, Caoile A G F S, Stern D B. Polyadenylation occurs at multiple sites in maize mitochondrial cox2 mRNA and is independent of editing status. Plant Cell. 1999;11:1565–1577. doi: 10.1105/tpc.11.8.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchfelder A, Brennicke A. Characterization and partial purification of tRNA processing activities from potato mitochondria. Plant Physiol (Rockville) 1994;105:1247–1254. doi: 10.1104/pp.105.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morikami A, Nakamura K. Transcript map of oppositely oriented pea mitochondrial genes encoding the α-subunit and the subunit 9 of F0F1-ATPase complex. Biosci Biotechnol Biochem. 1993;57:1530–1535. doi: 10.1271/bbb.57.1530. [DOI] [PubMed] [Google Scholar]
- 22.Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 23.Saalaoui E, Litvak S, Araya A. The apocytochrome b from an alloplasmic line of wheat (T. aestivum, cytoplasm-T. timopheevi) exists in two differently expressed forms. Plant Sci. 1990;66:237–246. [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Schuster D, Lisitsky I, Klaff P. Polyadenylation and degradation of mRNA in the chloroplast. Plant Physiol (Rockville) 1999;120:937–944. doi: 10.1104/pp.120.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schuster W, Hiesel R, Isaac P G, Leaver C J, Brennicke A. Transcript termini in messenger RNAs in higher plant mitochondria. Nucleic Acids Res. 1986;14:5943–5954. doi: 10.1093/nar/14.15.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wahle E, Rüegsegger U. 3′-end mRNA processing of pre-mRNA in eukaryotes. FEMS Microbiol Rev. 1999;23:277–295. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 28.Yehudai-Resheff S, Schuster G. Characterization of the E. coli poly(A) polymerase: nucleotide specificity, RNA binding affinities and RNA structure dependence. Nucleic Acids Res. 2000;28:1139–1144. doi: 10.1093/nar/28.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuckenberg P D, Phillips S L. Oligoadenylate is present in the mitochondrial RNA of Saccharomyces cerevisiae. Mol Cell Biol. 1982;2:450–456. doi: 10.1128/mcb.2.4.450. [DOI] [PMC free article] [PubMed] [Google Scholar]