Abstract
Manganese is an essential micronutrient for plant growth but can be toxic to plants when it reaches excessive levels. Although metal tolerance proteins (MTPs), which belong to the cation diffusion facilitator (CDF) family, have been demonstrated to play critical roles in manganese (Mn) tolerance in plants, the characteristics and functions of GmMTP members in the response of soybean (Glycine max) to Mn toxicity have not been documented. In this study, growth inhibition was observed in soybean plants that were exposed to a toxic level of Mn in hydroponics, as reflected by the generation of brown spots, and decreased leaf chlorophyll concentration and plant fresh weight. Subsequent genome-wide analysis resulted in the identification of a total of 14 GmMTP genes in the soybean genome. Among these GmMTPs, 9 and 12 were found to be regulated by excess Mn in leaves and roots, respectively. Furthermore, the function of GmMTP8.1, a Mn-CDF homologue of ShMTP8 identified in the legume Stylosanthes hamata that is involved in Mn detoxification, was characterized. Subcellular localization analysis showed that GmMTP8.1 was localized to the endoplasmic reticulum (ER). Heterologous expression of GmMTP8.1 led to the restoration of growth of the Mn-hypersensitive yeast (Saccharomyces cerevisiae) mutant Δpmr1, which is made defective in Mn transport into the Golgi apparatus by P-type Ca/Mn-ATPase. Furthermore, GmMTP8.1 overexpression conferred tolerance to the toxic level of Mn in Arabidopsis (Arabidopsis thaliana). Under excess Mn conditions, concentrations of Mn in shoots but not roots were decreased in transgenic Arabidopsis, overexpressing GmMTP8.1 compared to the wild type. The overexpression of GmMTP8.1 also led to the upregulation of several transporter genes responsible for Mn efflux and sequestration in Arabidopsis, such as AtMTP8/11. Taken together, these results suggest that GmMTP8.1 is an ER-localized Mn transporter contributing to confer Mn tolerance by stimulating the export of Mn out of leaf cells and increasing the sequestration of Mn into intracellular compartments.
Keywords: metal tolerance protein, cation diffusion facilitator, Mn transporter, Mn toxicity, Mn detoxification, Glycine max
Introduction
As one of the essential micronutrients for plant growth, manganese (Mn) acts as an activator of many enzymes, serving various functions in a set of physiological and biochemical processes (Millaleo et al., 2010; Marschner, 2012; Long et al., 2021). Being a trace element, Mn is required by plants at a low dose and can cause phytotoxicity when present in excess (Millaleo et al., 2010; Shao et al., 2017). Mn toxicity is an important factor limiting plant growth on acid, poorly drained soils, and sterilized soils. Previous studies have shown that excess Mn can trigger oxidative stress by the generation of reactive oxygen species (ROS) (e.g., OH and O2–) from Mn ions via the Fenton reaction, resulting in disrupted metabolic pathways and damaged thylakoid membrane structure and macromolecules in plant cells (Millaleo et al., 2010; Dragišić Maksimović et al., 2012). Therefore, Mn toxicity inhibits enzyme activity, declines chlorophyll biosynthesis and photosynthesis, and impairs the uptake and translocation of other mineral elements (Fernando and Lynch, 2015; Li et al., 2019). In general, visible symptoms of Mn toxicity for most plants include generation of brown spots, chlorosis, and necrosis in leaves, ultimately inhibiting plant growth (Millaleo et al., 2010; Li et al., 2019). Thus, better understanding of mechanisms underlying the response of plants to Mn toxicity is of great importance for breeding Mn-tolerant crop varieties.
To cope with Mn toxicity, plants have developed comprehensive mechanisms to regulate Mn homeostasis by mediating Mn uptake, efflux, and trafficking at both cellular and tissue levels (Shao et al., 2017; Alejandro et al., 2020). For example, the cation diffusion facilitator (CDF) family is an integral membrane divalent cation transporter that plays critical roles in metal tolerance by regulating metal homeostasis in plant cells (Migocka et al., 2014, 2015). Most CDF proteins include an N-terminal signature sequence, a conserved C-terminal cation efflux domain, and several transmembrane domains (TMDs), which are critical for metal transport activities (Montanini et al., 2007; Chen X. et al., 2016). CDF transporters are involved in the transport of divalent cations, such as zinc (Zn), iron (Fe), and Mn, from the cytoplasm to extracellular space or to subcellular compartments (Gustin et al., 2011). Thus, the CDF members identified from numerous organisms can be classified into three major substrate-specific clusters, namely, Zn-CDF, Fe/Zn-CDF, and Mn-CDF, according to their transported substrate specificities (Montanini et al., 2007). Plant CDF proteins are generally designated as metal tolerance proteins (MTPs), which can be further categorized into seven subgroups based on the initial phylogenetic analysis and annotation of the Arabidopsis (Arabidopsis thaliana) MTP family (Gustin et al., 2011). For example, of the 12 MTP members in Arabidopsis, six AtMTPs (AtMTP1, AtMTP2, AtMTP3, AtMTP4, AtMTP5, and AtMTP12) are assigned to groups 1, 5, and 12 within the Zn-CDF cluster, while AtMTP6 and AtMTP7 belong to groups 6 and 7 within the Fe/Zn-CDF cluster. The remaining four AtMTPs (AtMTP8, AtMTP9, AtMTP10, and AtMTP11) are categorized into groups 8 and 9 within the Mn-CDF cluster (Gustin et al., 2011).
Among the three substrate-specific CDF clusters, MTP members of groups 8 and 9 within the Mn-CDF cluster have been characterized to be involved in regulating Mn tolerance either by sequestration of Mn or export of Mn out of the cell. For example, group 8 includes ShMTP8 from Stylosanthes hamata, AtMTP8 from Arabidopsis, OsMTP8.1/8.2 from rice (Oryza sativa), and CsMTP8 from cucumber (Cucumis sativus). These MTP members exhibit substrate-specificity to Mn and are implicated in the transport of Mn into vacuoles (Delhaize et al., 2003; Chen et al., 2013; Migocka et al., 2014; Eroglu et al., 2016; Takemoto et al., 2017). In addition, a variety of MTP members of group 9 have also been involved in the regulation of Mn homeostasis, as observed in CsMTP9 from cucumber, OsMTP9/11 from rice, BmMTP10/11 from Beta vulgaris ssp. maritima, AtMTP11 from Arabidopsis, and PtMTP11.1/11.2 from poplar (Populus trichocarpa); these MTPs are localized to the plasma membrane, Golgi apparatus, or pre-vacuolar compartments and have also been documented to regulate Mn homeostasis and tolerance (Delhaize et al., 2007; Peiter et al., 2007; Erbasol et al., 2013; Migocka et al., 2015; Ueno et al., 2015; Tsunemitsu et al., 2018b). Although the conserved functions of MTPs within the Mn-CDF cluster indicate similar roles of MTPs in Mn detoxification, it is still unknown whether MTP homologues in this specific cluster from different plants fulfill the same function and contribute to Mn tolerance.
Soybean (Glycine max) is recognized as one of the economically important nitrogen fixing legume crops and provides more than 60% of high-quality protein and oil for humans and animals in the world (Herridge et al., 2008; Lu et al., 2017). Although several studies have been conducted to investigate the response of soybean to Mn toxicity (Lavres et al., 2009; Yang et al., 2009; Chen Z. et al., 2016; Santos et al., 2017; Liu et al., 2020), underlying molecular mechanisms, through the involvement of MTPs, have not been elucidated. Key participants within the specific Mn-CDF cluster involved in Mn detoxification in soybean remain undetermined. In this study, a total of 14 MTP members were identified in the soybean genome. The expression pattern of each GmMTP gene in response to various metal treatments was analyzed. Furthermore, GmMTP8.1 was identified as a novel MTP8-like protein and displayed high similarity with ShMTP8 from S. hamata within the Mn-CDF cluster. Thus, GmMTP8.1 was studied in more detail to characterize its potential functions in Mn detoxification.
Materials and Methods
Plant Materials and Growth Conditions
The soybean cultivar Zhonghuang 13 was used in this study. Seeds were surface-sterilized with 10% (v/v) H2O2 and germinated in paper rolls moistened with a half-strength Hoagland nutrient solution, and were cultivated in a growth chamber under conditions of 14-h light at 26°C/10-h dark at 23°C, 120 μmol m–2 s–1 photon flux density, and 70% relative humidity. After seed germination for 7 days, the soybean seedlings were transferred into a 15-L hydroponic box containing a 13-L full-strength Hoagland nutrient solution, as described by Chen Z. et al. (2016). The Hoagland nutrient solution contained 1500 μM KNO3, 1200 μM Ca(NO3)2, 400 μM NH4NO3, 500 μM KH2PO4, 1000 μM MgSO4, 25 μM MgCl2, 300 μM K2SO4, 300 μM (NH4)2SO4, 5 μM MnSO4, 0.38 μM ZnSO4, 1.57 μM CuSO4, 0.09 μM (NH4)6Mo7O24, 2.5 μM NaB4O7, and 80 μM Fe-EDTA. The pH value of the solution was adjusted to 5.8 using HCl or KOH every 2 days. The solution was aerated every 20 min per hour and refreshed weekly. Plants were cultivated in a greenhouse at temperatures ranging from 2 to 30°C under natural sunlight with a photoperiod of about 14 h and 50-70% relative humidity. Each hydroponic box containing four soybean seedlings served as one biological replicate, and the experiment included three biological replicates. After 14 days of growth, leaves, stems, and roots were separately cut from the plants. Samples from two plants of each biological replicate were pooled for gene expression analysis. All the samples were frozen with liquid nitrogen, and then stored at −80°C prior to RNA extraction.
To investigate the effects of excess Mn on soybean growth, after seed germination for 7 days, the seedlings were transferred to the full-strength Hoagland nutrient solution for 7 days as described above. Then, 14-day-old seedlings were transferred into the nutrient solution supplied with 5, 50, or 100 μM MnSO4, according to Chen Z. et al. (2016). Five μM MnSO4 was used as the control in this experiment. Each hydroponic box containing four soybean seedlings served as one biological replicate, and each treatment included three biological replicates. After 7 days of Mn treatments, shoots or roots from two plants of each biological replicate were pooled for determination of plant fresh weight and Mn concentration. A soil plant analysis development (SPAD)-502 chlorophyll meter (Konica-Minolta, Japan) was used to detect chlorophyll concentration according to the instructions of the manufacturer. Leaves or roots from the remaining two plants of each biological replicate were pooled for gene expression analysis. The samples were then stored at −80°C prior to RNA extraction.
To analyze the effects of excess Fe, Zn, and copper (Cu) stress on gene expression, 14-day-old seedlings pre-cultured in the nutrient solution as described above were transferred into a fresh nutrient solution containing 800 μM Fe-EDTA, 20 μM ZnSO4, or 20 μM CuSO4, which were regarded as excess Fe, Zn, and Cu stress, respectively. Soybean seedlings grown in the full-strength nutrient solution were set as the control. Each hydroponic box containing four soybean seedlings served as one biological replicate, and the experiment included three biological replicates. After 7 days of metal treatments, samples of leaves or roots were separated from two plants of each biological replicate, and were then pooled for gene expression analysis. The samples were frozen with liquid nitrogen and stored at −80°C prior to RNA extraction.
Identification of Soybean GmMTP Genes
The sequences of 12 MTP genes in Arabidopsis obtained from the NCBI website1 were used separately as queries for BLAST search against the soybean genome in the Phytozome website.2 After removing redundant sequences, a total of 14 GmMTP members were identified in the soybean genome that contained the cation efflux domain (PF01545). Subsequently, these candidates were named from GmMTP4.1 to GmMTP11.2 based on the phylogenetic relationship and sequence identity between GmMTP and Arabidopsis AtMTPs. General information for each GmMTP gene was obtained from the Phytozome website. The protein molecular weight of each GmMTP member was calculated using the ExPASy software.3 Gene Structure Display Server4 was used for gene structure analysis. The Pfam and MEME programs were used to analyze GmMTP conserved domains and motifs, respectively, according to Liu et al. (2018). TMDs were predicted by the TMHMM Server v.2.0. Construction of the phylogenetic tree was based on entire protein sequence alignments through ClustalX using the method neighbor-joining with 1,000 bootstrap replicates with MEGA 4.1.5 The sequences of MTPs from other plant species were retrieved from corresponding databases, as described previously (Gustin et al., 2011; Migocka et al., 2014; Li et al., 2018; Liu J. et al., 2019).
Quantitative Real-Time Polymerase Chain Reaction Analysis of GmMTP Genes
Total RNA was extracted using TRIzol Reagent (Invitrogen, United States) according to the instructions of the manufacturer. First-strand cDNA was synthesized using a HiScript III cDNA (Vazyme, China) synthesis kit. SYBR Green Master Mix (Vazyme, China) and QuantStudio™ 6 Flex Real-Time System (Thermo Fisher Scientific, United States) were used for quantitative real-time polymerase chain reaction (qRT-PCR) analysis. qRT-PCR reaction was performed as follows: 95°C for 1 min, 40 cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 30 s. Fluorescence data were collected at 72°C. The qRT-PCR primers of genes are summarized in Supplementary Table 1. Relative gene expressions were calculated relative to the expression levels of the housekeeping gene, GmEF-1a or AtEF-1a. Gene expression analysis included three biological replicates.
Subcellular Localization of GmMTP8.1
The open reading frame (ORF) of GmMTP8.1 without stop codon was amplified by PCR from root cDNA using GmMTP8.1-GFP-F/R primers (Supplementary Table 1). The amplified product was digested with EcoRI and BamHI, and was then subcloned into the same sites after the cauliflower mosaic virus (CaMV) 35S promoter of the binary vector pBWA(V)HS-GLosgfp and fused with the N-terminal of green fluorescent protein (GFP). The GmMTP8.1-GFP construct and empty vector were introduced separately into Agrobacterium tumefaciens strain Gv3101 using the freeze-thaw method. The transformed Gv3101 cells were syringe-infiltrated into the abaxial side of near-fully expanded leaves of 5- to 6-week-old tobacco (Nicotiana benthamiana) plants, as described by Liu et al. (2016). The tonoplast marker (tandem-pore K+ channel, TPK1) and endoplasmic reticulum (ER) marker (auxin efflux carrier family protein, PIN5) (Voelker et al., 2006; Mravec et al., 2009), fused to the red fluorescent protein (mKATE), were used for co-localization with GmMTP8.1-GFP or empty vector. An empty vector with GFP alone was used as a control. After 3 days of cultivation, the GFP fluorescence in epidermal cells on the abaxial leaf side was imaged using a Zeiss LSM7 DUO (Zeiss, Germany) confocal microscope. Green fluorescence was stimulated at 488 nm and detected with filter sets at 500–530 nm. The red fluorescence was excited at 561 nm, and emission was captured at 580–630 nm.
Yeast Transformation and Metal Tolerance Analysis
Five Saccharomyces cerevisiae mutants, namely, the Mn-sensitive mutant Δpmr1 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YGL167c::kanMX4) defective in Mn transport into the Golgi apparatus; Zn-sensitive mutant Δzrc1 (MATa; ura3Δ0; leu2Δ0; his3Δ1; met15Δ0; YMR243c::kanMX4) defective in sequestering Zn into the vacuole; Cu-sensitive mutant Δcup2 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YGL166w::kanMX4) defective in a Cu-binding transcription factor for the activation of the metallothionein genes; cadmium (Cd)-sensitive mutant Δycf1 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YDR135c::kanMX4) defective in vacuolar Cd sequestration; and cobalt (Co)-sensitive mutant Δcot1 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YOR316c::kanMX4) defective in Co efflux into the vacuole, were purchased from Euroscarf.6 All strains are isogenic to wild-type BY4741 (MATa; his3D1; leu2D0; met15D0; ura3D0) and are able to grow under the same conditions.
To contract yeast expression plasmid, the ORF of GmMTP8.1 was amplified from root cDNA using GmMTP8.1-pYES2-F/R primers (Supplementary Table 1). The PCR product was digested with BamHI and EcoRI, and was then inserted in the same sites of the yeast expression vector pYES2 (Invitrogen, United States). After that, the pYES2-GmMTP8.1 construct and pYES2 empty vector were transformed separately into yeast cells using the LiOAc/PEG method (Gietz and Schiestl, 2007). For metal complementation analysis, yeast transformants were pre-cultured in a liquid synthetic complete medium containing a yeast nitrogen base, amino acids without uracil, and glucose (SC-U/Glu) at 30°C until the optical density (OD) at 600 nm reached a value of 0.6. Pre-cultured cells were diluted to an OD600 of 0.2, and 10-μl aliquots of dilutions (OD at 600 nm of 0.02, 0.002, and 0.0002) were spotted onto induction plates containing 2% (w) Gal, 1% (w/v) raffinose, 0.67% (w/v) yeast nitrogen base without amino acids, 0.1% (w/v) amino acids without uracil, and 2% (w/v) agar, according to Chen et al. (2015). The induction plates were added with 5 mM MnSO4, 15 mM ZnSO4, 100 μM CuSO4, 50 μM CdCl2, and 2 mM CoCl2 for metal complementation analyses of the yeast mutants Δpmr1, Δzrc1, Δcup2, Δycf1, and Δcot1, respectively. A control drop assay was performed on the same solid medium without the addition of metals. After incubation at 30°C for 2 days in the dark, the plates were photographed.
Functional Analysis of GmMTP8.1 in Arabidopsis
To construct the pYLRNAi-GmMTP8.1 plasmid, the ORF of GmMTP8.1 was amplified by PCR from root cDNA using GmMTP8.1-OE-F/R primers (Supplementary Table 1). The amplified product was digested with BamHI and MluI, and subcloned further into the same site of the pYLRNAi vector with the hygromycin phosphotransferase (HPT) gene as the selective marker, according to Liang et al. (2010). The pYLRNAi-GmMTP8.1 construct was transformed into A. tumefaciens strain Gv3101 using the freeze-thaw method. The transformed Gv3101 cells harboring the pYLRNAi-GmMTP8.1 construct were grown further in a yeast extract peptone (YEP) medium overnight at 28°C until OD600 reached 1 before transformation. Four-week-old Columbia Arabidopsis ecotype (wild type) was used for Agrobacterium-mediated transformation with the floral dip method, as described by Clough and Bent (1998). Seeds of the putative transformed Arabidopsis were selected in a half-strength Murashige and Skoog (MS) solid medium containing 20 mg/L hygromycin for two generations. More than 10 T2 transgenic Arabidopsis lines were resistant to hygromycin with a 3:1 ratio. Finally, two homozygous T3 transgenic lines overexpressing GmMTP8.1 were selected based on both qRT-PCR using GmMTP8.1-RT-F/R primers (Supplementary Table 1) and Western blot analysis using an anti-HPT antibody.
To assess the effects of excess Mn on the growth of Arabidopsis, seeds of the wild-type Arabidopsis and two GmMTP8.1-overexpression lines (OE1/2) were surface-sterilized and then germinated in an MS solid medium. Arabidopsis plants were grown in a growth chamber under conditions of 16-h light at 23°C/8-h dark at 20°C, 150 μmol m–2 s–1 photon flux density, and 70% relative humidity. After 7 days of normal growth, uniform seedlings with about 2-cm-long tap root were selected and transplanted into a fresh MS solid medium supplied with 0.1, 2 or 4 mM MnSO4, as described by Chen et al. (2015). Arabidopsis plants treated with 0.1 mM MnSO4 were set as the control. After 7 days of Mn treatments, the plants were thoroughly rinsed with 10 mM EDTA and dH2O. Shoots, roots, and whole plants were harvested separately to determine fresh weight and Mn concentration.
In addition, after 7 days of germination in the MS solid medium as described above, The Arabidopsis plants were transplanted into a hydroponic nutrient solution containing 0.25 mM Ca(NO3)2, 1 mM KH2PO4, 0.5 mM KNO3, 1 mM MgSO4, 50 μM Fe(III)-Na-EDTA, 50 μM H3BO3, 18 μM MnSO4, 1 μM CuSO4, 10 μM ZnSO4, and 0.02 μM MoO4Na2 for 14 days, according to Cailliatte et al. (2010). Subsequently, 21-day-old Arabidopsis plants were transplanted into a nutrient solution containing 18 and 400 μM MnSO4 as control and excess Mn treatments, respectively. After 7 days of Mn treatments, the plants were rinsed with 10 mM EDTA and dH2O. Shoots and roots were harvested separately to determine fresh weight and Mn concentration. All the treatments included three biological replicates.
Analysis of Manganese Concentration
The plant samples were oven dried at 105°C for 30 min and dried further at 75°C for 5 days. Dry samples were ground into powder before Mn assay. Approximately 0.07 g of the dry samples was thoroughly burned to ash at 600°C for 10 h in a muffle furnace, according to Chen Z. et al. (2016). After cooling to room temperature, the ash sample was mixed with 7 ml of 100 mM HCl. The mixture was fully dissolved by continuous shaking for 2 days. The solution was then used to analyze Mn concentration via atomic absorption spectroscopy. A certified reference material, GBW07603 (bush branches and leaves) approved by Institute of Geophysical and Geochemical Exploration, Chinese Academy of Geological Sciences, China, was used to validate the extraction and measurement. Mn concentration was calculated by comparison with a standard curve.
Western Blot Analysis
Proteins from wild-type and GmMTP8.1-overexpression Arabidopsis (OE1/2) were extracted separately using an extraction buffer containing 100 mM Tris–HCl (pH 8), 0.5% (w/v) PVPP, 1 mM EDTA, and 1 mM PMSF. After centrifugation at 12,000 × g for 15 min at 4°C, supernatants were collected. Thirty μg of extracted protein was resolved by 12% SDS-PAGE and electrophoretically transferred further to a polyvinylidene difluoride (PVDF) membrane (Merck Millipore, United States) through a Trans-Blot system (Bio-Rad, United States), according to Liang et al. (2010). After that, the PVDF membrane was incubated in a 10-mM Tris–HCl buffer (pH 8) containing 5% (w/v) non-fat dry milk and 150 mM NaCl for 12 h. The PVDF membrane was then washed three times with a washing solution containing 10 mM Tris–HCl (pH 8), 150 mM NaCl, and 0.1% (v/v) Tween 20. Blotting of the PVDF membrane was performed by incubation with an anti-HPT primary antibody (1:1,000 dilution) in a blotting buffer containing 10 mM Tris–HCl (pH 8), 150 mM NaCl, 0.1% (v/v) Tween 20, and 3% (w/v) non-fat dry milk for 1 h, followed by three washes with a washing buffer. An alkaline phosphatase-tagged secondary antibody was then added into the fresh blotting buffer, where the PVDF membrane was incubated for 1 h. After three washes with the washing buffer, the band of target protein in the PVDF membrane was observed after alkaline phosphatase reaction.
Statistical Analyses
The SPSS program v13.0 (SPSS Institute, United States) was used to perform one-way ANOVA and Student’s t-test analyses.
Results
Soybean Growth Inhibited by Manganese Toxicity
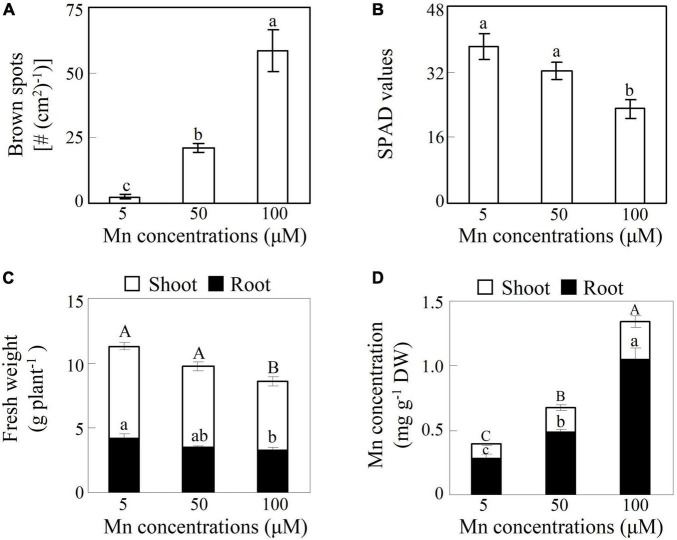
To investigate the toxic effects of Mn on soybean growth, 14-day-old soybean seedlings were exposed to different Mn treatments for 7 days. The results showed that excess Mn treatments (50 and 100 μM) resulted in the generation of brown spots on mature leaves compared to the control (5 μM Mn) (Figure 1A). Moreover, the number of brown spots with the 100 μM Mn treatment was 1.8-fold higher than that with the 50-μM Mn treatment (Figure 1A). In contrast, chlorophyll levels, indicated by SPAD values in leaves, were decreased by 15.7 and 39.9% with the 50- and 100-μM Mn treatments, respectively, compared to the control (Figure 1B). Although both shoot and root fresh weights were unaffected by the 50-μM Mn treatment, they were significantly decreased by more than 24% with100 μM Mn compared to the control (Figure 1C). In addition, increases in Mn concentrations were observed in shoots and roots of soybean under excess Mn treatments. Mn concentrations in shoots at 50 and 100 μM Mn were 79.8–175.3% higher than 5-μM Mn treatments, while Mn levels in roots at 50 and 100 μM Mn were 70.7–269.1% higher than the controls (Figure 1D). Furthermore, Mn concentrations in roots were more than 155% higher than in shoots under excess Mn stress (50 and 100 μM) (Figure 1D).
FIGURE 1.
Effects of excess manganese (Mn) treatment on soybean growth. (A) Number of brown spots. (B) Soil plant analysis development (SPAD) values. (C) Fresh weight. (D) Mn concentrations. Fourteen-day-old soybean seedlings were treated with 5, 50, and 100 μM MnSO4 for 7 days. Five μM MnSO4 was used as the control. Chlorophyll levels were detected with SPAD-502 meter. Each bar represents the means of four independent replicates with standard error. Different lower case letters indicate significant differences at P < 0.05 among Mn treatments in panels (A,B). Different upper case or lower case letters indicate significant differences at P < 0.05 among Mn treatments in shoot or root in panels (C,D). White and black bars represent shoot and root, respectively.
Identification of GmMTP Genes in Soybean
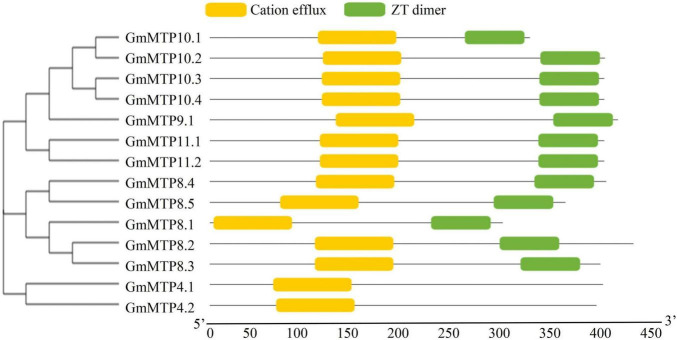
In this study, a total of 14 putative GmMTP genes were identified in the soybean genome. The 14 GmMTP genes were named from GmMTP4.1 to GmMTP11.2 based on the phylogenetic relationship and sequence identity between GmMTP and Arabidopsis AtMTPs (Supplementary Table 2). General information for the 14 GmMTP members is summarized in Supplementary Table 2. Most of the GmMTP members contained three to six conserved TMDs, except GmMTP10.1, which harbored only two TMDs (Supplementary Table 2). The 14 GmMTP genes can be classified into three distinct groups according to their sequence structure features (Supplementary Figure 1). Furthermore, most GmMTPs contained both the cation efflux (PF01545) and zinc transporter (ZT) dimer (PF16916) conserved motifs, except GmMTP4.1 and GmMTP4.2, which only contained the cation efflux motif (Figure 2 and Supplementary Table 3).
FIGURE 2.
Conserved motifs in GmMTPs. Yellow and green boxes indicate cation efflux and ZT dimer motifs, respectively. The neighbor-joining phylogenetic tree of GmMTPs was constructed using MEGA4.1.
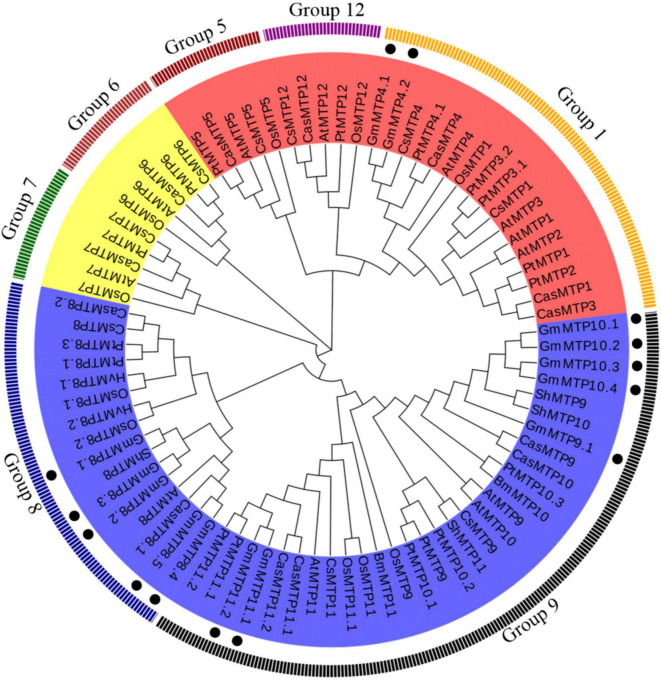
Phylogenetic analysis showed that plant MTPs can be classified into three distinct clusters, namely, Zn-CDF, Zn/Fe-CDF, and Mn-CDF, which were further divided into seven subgroups (Figure 3). Of the seven subgroups, group 9 had the largest number of plant MTPs, followed by groups 1 and 8. A total of 12 GmMTPs were classified into groups 8 and 9, both of which belonged to the Mn-CDF cluster (Figure 3). Among them, five GmMTPs (GmMTP8.1, GmMTP8.2, GmMTP8.3, GmMTP8.4, and GmMTP8.5) clustered with ShMTP8 from S. hamata, AtMTP8 from Arabidopsis, OsMTP8.1/8.2 from rice, CsMTP8 from cucumber, HvMTP8.1/8.2 from barley (Hordeum vulgare), and CasMTP8.1/8.2 from tea plant (Camellia sinensis) in group 8. Seven GmMTPs, namely, GmMTP9.1, GmMTP10.1, GmMTP10.2, GmMTP10.3, GmMTP10.4, GmMTP11.1, and GmMTP11.2, together with OsMTP9/11/11.1 from rice, CsMTP9 from cucumber, AtMTP11 from Arabidopsis, PtMTP11.1/11.2 from poplar, and BmMTP10/11 from B. vulgaris ssp. maritima, were divided into group 9 (Figure 3). In addition, GmMTP4.1 and GmMTP4.2 were classified into group 1 within the Zn-CDF cluster (Figure 3). Interestingly, none of the GmMTP members were classified into the Fe/Zn-CDF cluster (Figure 3).
FIGURE 3.
Phylogenetic analysis of metal tolerance proteins (MTPs) in soybean and other plants. The phylogenetic tree was constructed using the MEGA4.1 program. Except for CasMTPs derived from tea plant (Camellia sinensis), the first two letters of each protein represent the abbreviated species name. At: Arabidopsis thaliana, Os: Oryza sativa, Gm: Glycine max, Cs: Cucumis sativus, Hv: Hordeum vulgare, Pt: Populus trichocarpa, Sh: Stylosanthes hamata, and Bm: Beta vulgaris. ssp. maritima. MTPs from various plant species are divided into the Zn-CDF, Fe/Zn-CDF, and Mn-CDF clusters, which are highlighted in red, yellow, and blue, respectively. Plant MTPs can be further classified into seven subgroups. The solid circles indicate MTPs from soybean.
Expression Analysis of GmMTP Genes
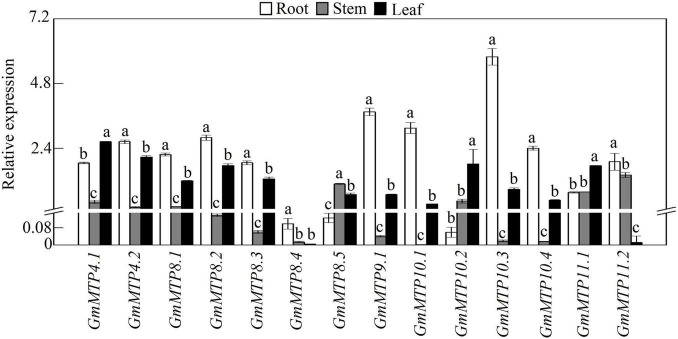
We subsequently analyzed the expression patterns of GmMTP genes in leaves, stems, and roots of 21-day-old soybean seedlings under normal growth conditions. The results showed that GmMTPs exhibited differential expressions in various tissues of soybean (Figure 4). Among them, three GmMTPs (GmMTP4.1, GmMTP10.2, and GmMTP11.1) showed highest expression in leaves, while 10 GmMTPs (GmMTP4.2, GmMTP8.1, GmMTP8.2, GmMTP8.3, GmMTP8.4, GmMTP9.1, GmMTP10.1, GmMTP10.3, GmMTP10.4, and GmMTP11.2) were mainly expressed in roots (Figure 4). Interestingly, most of the GmMTP genes exhibited low expressions in stems, except GmMTP8.5 with the highest expression in stems compared to leaves or roots (Figure 4). Furthermore, the transcript of GmMTP4.1 in leaves was higher than that of the other GmMTPs, while GmMTP10.3 exhibited the highest expression in roots compared to the other GmMTPs (Figure 4).
FIGURE 4.
Expressions of GmMTPs in leaf, stem, and root of soybean. After seeds were germinated for 7 days, the seedlings were transplanted into a full-strength Hoagland solution. Leaf, stem, and root were harvested separately after 14 days of growth for gene expression analysis. Each bar represents the mean value of three independent replicates with standard error. White, gray, and black bars represent root, stem, and leaf, respectively. Different lower case letters indicate significant differences at P < 0.05 in the same tissue.
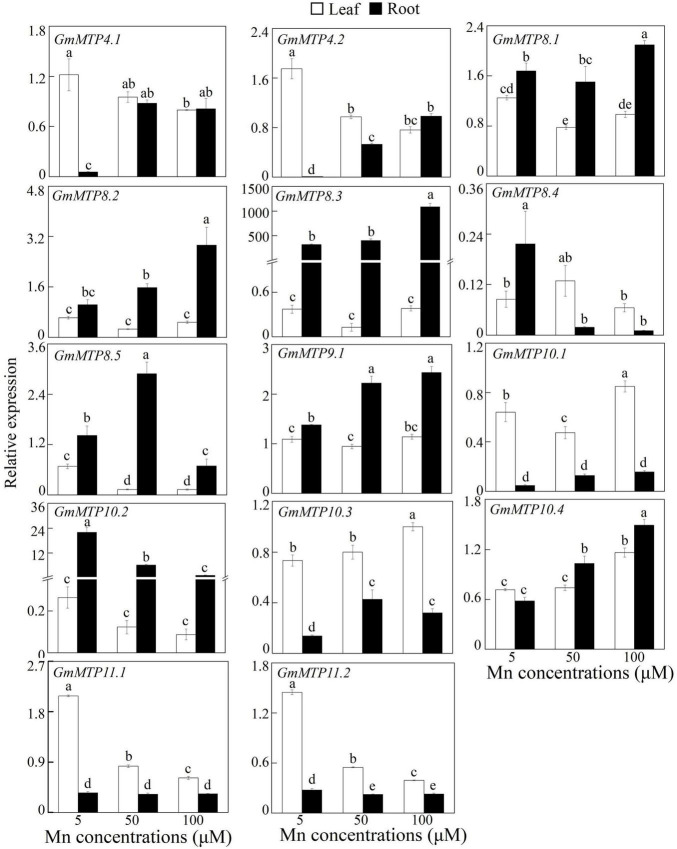
Expressions of the GmMTPs in leaves and roots in response to Mn toxicity were investigated. As shown in Figure 5, all GmMTPs responded to excess Mn in leaves and/or roots, and the number of GmMTPs regulated by Mn stress in roots (12 GmMTPs) was higher than those in leaves (9 GmMTPs). For example, two GmMTPs (GmMTP10.3 and 10.4) were enhanced and six GmMTPs (GmMTP4.1, GmMTP4.2, GmMTP8.1, GmMTP8.5, GmMTP11.1, and GmMTP11.2) were suppressed in leaves by the 50- and/or 100-μM Mn treatments. In contrast, most GmMTPs (GmMTP4.1, GmMTP4.2, GmMTP8.1, GmMTP8.2, GmMTP8.3, GmMTP8.5, GmMTP9.1, GmMTP10.3, and GmMTP10.4) were enhanced and only three GmMTPs (GmMTP8.4, GmMTP10.2, and GmMTP11.2) were suppressed in roots by the 50- and/or 100-μM Mn treatments (Figure 5). Interestingly, the transcripts of GmMTP10.3, GmMTP10.4, and GmMTP11.2 regulated by Mn in roots were similar to those in leaves (Figure 5).
FIGURE 5.
Expressions of GmMTPs in leaf and root of soybean under excess Mn treatments. Fourteen-day-old soybean seedlings were transferred into a nutrient solution containing 5, 50, or 100 μM MnSO4. Five μM MnSO4 was used as the control. After 7 days of Mn treatment, leaves and roots were harvested separately for gene expression analysis. Each bar represents the mean value of three independent replicates with standard error. White and black bars represent leaf and root, respectively. Different lower case letters indicate significant differences at P < 0.05.
Similar to the regulation by excess Mn stress, the number of GmMTPs regulated by excess Fe, Zn, and Cu stresses in roots was higher than those in leaves (Supplementary Figure 2). For example, in roots, a total of 13, 12, and 11 GmMTPs were regulated by Fe, Zn, and Cu stresses, respectively. In leaves, 7, 9, and 9 GmMTPs were found to be regulated by Fe, Zn, and Cu stresses, respectively. Furthermore, most of the GmMTP genes were regulated by at least one metal stress in both leaves and roots, except GmMTP9.1, GmMTP10.4, and GmMTP11.2 that were not responsive in leaves to any of the three tested metals (Supplementary Figure 2). Interestingly, five GmMTPs (GmMTP4.1, GmMTP4.2, GmMTP8.3, GmMTP8.4, and GmMTP10.2) were simultaneously suppressed, and three GmMTPs (GmMTP4.1, GmMT8.5, and GmMT10.1) were concurrently enhanced in leaves and roots by all the three metals, respectively (Supplementary Figure 2).
Subcellular Localization of GmMTP8.1
In this study, GmMTP8.1, a member of group 8 within the Mn-CDF cluster (Figure 3), displayed highest similarity with ShMTP8 in S. hamata that has been well-characterized for its involvement in Mn detoxification by the sequestration of Mn into vacuoles, was further selected to dissect its function in Mn detoxification. The GmMTP8.1 protein includes 294 amino acid residues (33.1 kDa) and possesses common features of Mn-CDF transporters, such as four putative TMDs, cation efflux, and ZT dimer domains (Figure 2 and Supplementary Table 2). The consensus sequence DxxxD (x = any amino acid) localizes to the first TMD and the cytosolic loop of GmMTP8.1 (Supplementary Figure 3). In addition, based on amino acid sequence comparisons, GmMTP8.1 shares a high degree of homology identity (90.8%) with ShMTP8 in S. hamata, and shares 83.3% identity with HvMTP8.1 in barley, 83% identity with OsMTP8.1 in rice, 83% identity with CsMTP8 in cucumber, and 81% identity with AtMTP8 in Arabidopsis, which were the representative MTP8-like proteins included in group 8 (Supplementary Figures 3–5). This suggests that the role of GmMTP8.1 is similar to that of the MTP homologs.
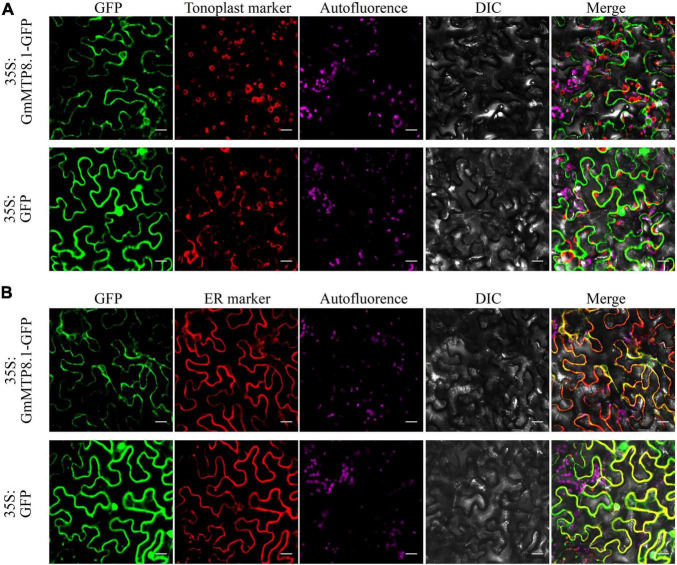
We further analyzed the subcellular localization of GmMTP8.1 in tobacco epidermis cells. The results showed that the GFP signal of GmMTP8.1 was found to be co-localized with that of the ER marker but not localized in the tonoplast of tobacco leaf epidermal cells, whereas the fluorescence of cells transformed with GFP alone was observed in whole cells, such as plasma membrane, cytoplasm, and nucleus (Figures 6A,B). Thus, these results suggest that GmMTP8.1 is localized to the ER.
FIGURE 6.
Subcellular localization of GmMTP8.1 in tobacco leaf lower epidermal cells. The images are labeled to show the 35S:GFP and 35S:GFP-GmMTP8.1 constructs. 35S:GFP-GmMTP8.1 was co-expressed with the (A) tonoplast marker TPK1 and (B) endoplasmic reticulum marker PIN5 fused with red fluorescence protein (mKATE) in tobacco leaf epidermal cells. green fluorescent protein (GFP) fluorescence, red fluorescent protein (RFP) fluorescence, chloroplast autofluorescence, bright field images, and merged images are displayed from left to right. Fluorescence was observed by confocal microscopy. Scale bar is 20 μm.
Metal Transport Activity of GmMTP8.1 in Yeast Cells
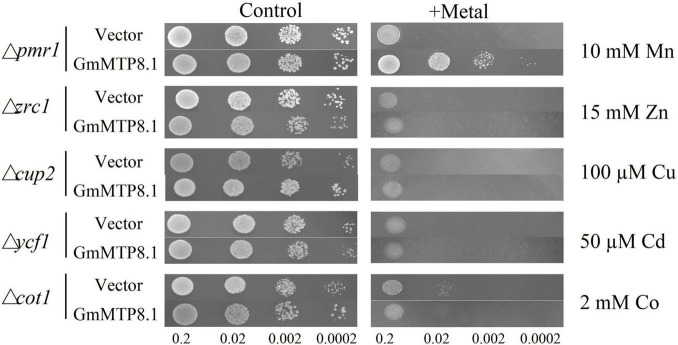
A complementation assay of yeast mutants was further conducted to investigate the metal transport activity of GmMTP8.1, which was heterologously expressed in five yeast mutants that are defective in transport activity of various metals, namely, Mn (Δpmr1), Zn (Δzrc1), Cu (Δcup2), Cd (Δycf1), and Co (Δcot1). The results showed that the yeast mutants carrying either the pYES2 empty vector or the pYES2-GmMTP8.1 construct grew similarly in the control medium (Figure 7). However, the growth of the five yeast mutants transformed with the pYES2 empty vector was inhibited in the medium containing high or toxic metal levels (Figure 7). Furthermore, although the growth of the yeast mutants Δzrc1, Δcup2, Δycf1, and Δcot1, transformed with the pYES2-GmMTP8.1 construct, was suppressed in the corresponding metal treatments, GmMTP8.1 expression rescued the sensitivities to excess Mn in the Δpmr1 mutant (Figure 7), suggesting that GmMTP8.1 is a specific Mn transporter.
FIGURE 7.
Heterologous expression of GmMTP8.1 in different yeast mutants. The yeast mutants Δpmr1, Δzrc1, Δcup2, Δycf1, and Δcot1 harboring the pYES2 empty vector or the pYES2-GmMTP8.1 construct were used. Yeast cells (10 μl) with OD600 of 0.2 and four serial dilutions (10-fold) were spotted in SC-U/Gal medium added with or without 5 mM MnSO4, 15 mM ZnSO4, 100 μM CuSO4, 50 μM CdCl2, and 2 mM CoCl2 for 2 day at 30°C.
Overexpression of GmMTP8.1 Enhanced Manganese Tolerance and Reduced Manganese Accumulation in Arabidopsis
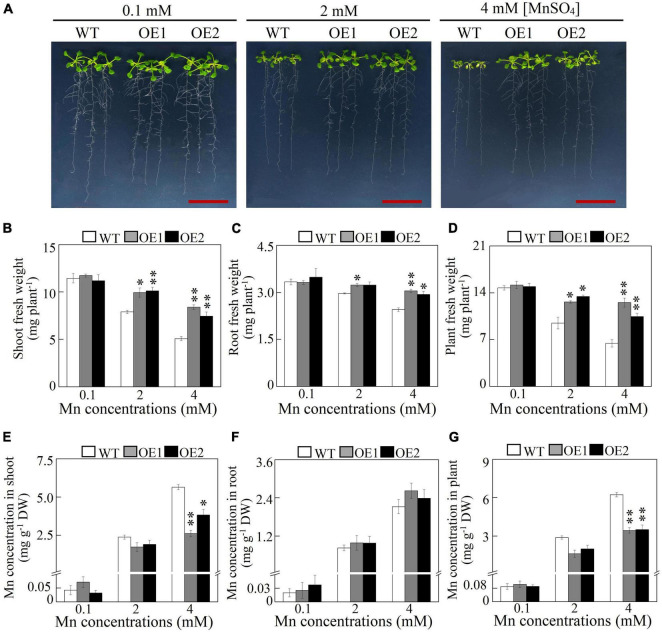
Subsequently, transgenic Arabidopsis overexpressing GmMTP8.1 was generated to investigate the roles of GmMTP8.1 in Mn detoxification in the plants. Increased transcripts of GmMTP8.1 in the overexpression lines (OE1 and OE2) compared to the wild-type (WT) were confirmed by RT-PCR analysis (Supplementary Figure 6A). Western-blot analysis indicated further that the GmMTP8.1 protein was expressed in transgenic Arabidopsis (Supplementary Figure 6B). WT and transgenic Arabidopsis were exposed to different levels of Mn, from 0.1 to 4 mM, in an MS solid medium for 7 days. As shown in Figure 8A, no difference in Arabidopsis growth was observed between the GmMTP8.1 overexpression lines (OE1 and OE2) and WT under control conditions containing 0.1 mM Mn. Although excess Mn (2 mM and 4 mM Mn) applications inhibited the growth of both overexpression lines and WT, the overexpression lines exhibited a higher level of tolerance to excess Mn than WT, especially with the 4-mM Mn treatment (Figure 8A). Shoot, root, and whole plant fresh weight of the overexpression lines were 46.6-65.2, 19.7-24.5, and 62.2-94.8%, respectively, higher than those of WT with the 4-mM Mn treatment (Figures 8B-D). In contrast, the overexpression of GmMTP8.1 resulted in significantly decreased Mn accumulation in shoots and whole plants of transgenic Arabidopsis exposed to the 4-mM Mn treatment, although no differences in root Mn accumulation were observed between WT and the transgenic lines (Figures 8E-G). Mn concentrations in shoots of the overexpression lines were 32.2–53.5% lower than those found in WT grown at 4 mM Mn (Figure 8E). Furthermore, total Mn concentrations in the transgenic Arabidopsis plants were 44–45% less than those in WT exposed to 4 mM Mn treatment (Figure 8G).
FIGURE 8.
Growth and Mn concentration of Arabidopsis overexpressing GmMTP8.1. (A) Phenotype of Arabidopsis grown with different Mn treatments. (B) Shoot fresh weight. (C) Root fresh weight. (D) Plant fresh weight. (E) Mn concentration in shoot. (F) Mn concentration in root. (G) Mn concentration in plant. Seven-day-old Arabidopsis seedlings were transferred to a solid Murashige and Skoog (MS) medium containing 0.1, 2, or 4 mM MnSO4. Arabidopsis plants treated with 0.1 mM MnSO4 were set as the control. After 7 days of Mn treatment, fresh weight and Mn concentration were determined. WT represents the wild type Arabidopsis. OE1 and OE2 are two transgenic lines overexpressing GmMTP8.1. Each bar represents the mean value of three independent replicates with standard error. White, gray, and black bars represent WT, OE1, and OE2, respectively. Asterisks indicate significant differences between the wild type and the overexpression lines with the same Mn treatment. *, 0.01 < P < 0.05. **, 0.001 < P < 0.01. DW, dry weight. Scale bar is 2 cm.
In addition, we also assessed the effects of excess Mn on the growth of transgenic Arabidopsis in hydroponics. After growing in a nutrient solution containing 18 or 400 μM MnSO4 for 7 days, the transgenic lines exhibited improved Mn tolerance compared to the WT (Supplementary Figure 7). Under the 400-μM Mn treatment, shoot fresh weight of the overexpression lines was more than 53% higher than that of the WT, although root fresh weight was similar between the transgenic lines and WT (Supplementary Figure 7). Consistent with Mn concentration in Arabidopsis grown in the MS solid medium, a low level of Mn concentration in shoots but not roots was observed in transgenic Arabidopsis compared to the WT, exposed to the 400-μM Mn treatment (Supplementary Figure 7). These results, together, suggest that the overexpression of GmMTP8.1 confers Mn tolerance in Arabidopsis mainly by decreasing accumulation of Mn.
Manganese Transporters Are Upregulated by Overexpression of GmMTP8.1 in Arabidopsis
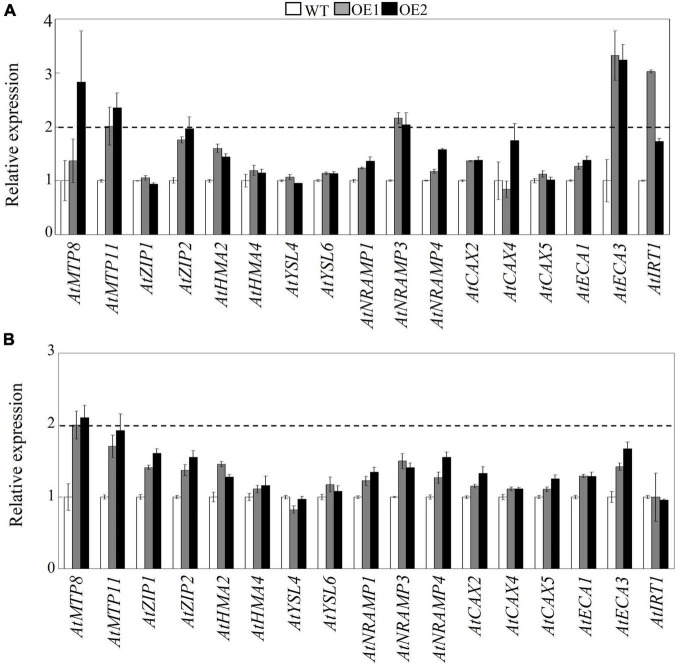
To investigate the mechanisms involved in decreased Mn in the shoots but unchanged level of Mn in the roots of transgenic Arabidopsis, the transcript levels of 17 Mn transporter genes were determined in transgenic Arabidopsis overexpressing GmMTP8.1. These Mn transporters included genes encoding metal tolerance proteins (AtMTP8/11), calcium exchangers (AtCAX2/4/5), ER-type calcium (Ca2+)-ATPases (AtECA1/3), iron-regulated transporter (AtIRT1), natural resistance-associated macrophage proteins (AtNRAMP1/3/4), zinc transporters (AtZIP1/2), yellow stripe-like proteins (AtYSL4/6), and heavy metal ATPases (AtHMA2/4). The qRT-PCR analysis showed that the overexpression of GmMTP8.1 led to a significant increase (>2-fold) in the transcripts of AtMTP11, AtNRAMP3, and AtECA3 in leaves of the two transgenic lines (Figure 9). However, among the tested genes, only AtMTP8 exhibited about 2.1-fold induction in roots of the transgenic Arabidopsis (Figure 9). The upregulation of these Mn transporters might contribute to the regulation of Mn homeostasis and tolerance in transgenic Arabidopsis.
FIGURE 9.
Effects of GmMTP8.1 overexpression on the transcript levels of Mn transporter genes in Arabidopsis. (A) Gene expression in leaves. (B) Gene expression in roots. Seven-day-old seedlings were transferred to a solid MS medium containing 4 mM MnSO4 for 7 days. Leaves and roots were sampled for RNA extraction and real-time (RT)-polymerase chain reaction (PCR) analysis. WT represents the wild type Arabidopsis. OE1 and OE2 are two transgenic lines overexpressing GmMTP8.1. Each bar represents the mean value of three independent replicates with standard error. White, gray, and black bars represent WT, OE1, and OE2, respectively. The relative expression level of gene of more than 2-fold was regarded as significant difference between the wild type and the overexpression lines.
Discussion
Manganese is a trace element that can cause phytotoxicity when present in excess (Millaleo et al., 2010; Li et al., 2019). Excess Mn disrupts various physiological processes in plants. For example, Mn toxicity causes oxidative stress and lipid peroxidation in cowpea (Vigna unguiculata) and cucumber (Fecht-Christoffers et al., 2006; Dragišić Maksimović et al., 2012), inhibits chlorophyll biosynthesis and photosynthesis in Stylosanthes guianensis (Liu P. et al., 2019; Jia et al., 2020), and disturbs the homeostasis of other nutrients in polish wheat (Triticum polonicum) (Sheng et al., 2015). In this study, excess Mn treatments led to increase in Mn levels in soybean, which was accompanied by increased number of brown spots and decreased chlorophyll concentrations, ultimately reducing plant fresh weight of the 14-day-old soybean seedlings (Figure 1). Similar results have been found in previous studies where soybean growth was inhibited by Mn toxicity (Chen Z. et al., 2016; Santos et al., 2017; Liu et al., 2020), although different growth stages of soybean were applied. Furthermore, important roles of antioxidant systems, such as antioxidant enzymes and non-enzymatic components, have been suggested in the response of soybean to Mn toxicity (Chen Z. et al., 2016; Santos et al., 2017; Liu et al., 2020). Although some advances have been made in investigating the response of soybean to Mn toxicity, its potential molecular mechanisms involved in Mn tolerance have not been elucidated.
Plant MTPs have been documented to play critical roles in Mn detoxification by Mn sequestration or efflux from plant cells (Li et al., 2019, 2021; Alejandro et al., 2020). In order to investigate the involvement of GmMTPs in the tolerance of soybean to Mn toxicity, we first identified 14 GmMTP genes in the soybean genome. Most of them were found to contain the conserved cation efflux and ZT dimer motifs (Figure 2). These motifs are reported to be essential for metal transport activities of MTPs (Montanini et al., 2007; Kolaj-Robin et al., 2015; Chen Z. et al., 2016). Cumulative studies demonstrated conserved functions of the MTP homologues in Mn detoxification in various plant species, suggesting similar roles for GmMTPs in Mn tolerance of soybean. According to the phylogenetic analysis, five GmMTPs (GmMTP8.1, GmMTP8.2, GmMTP8.3, GmMTP8.4, and GmMTP8.5) were classified into group 8, and seven GmMTPs (GmMTP9.1, GmMTP10.1, GmMTP10.2, GmMTP10.3, GmMTP10.4, GmMTP11.1, and GmMTP11.2) were classified into group 9 (Figure 3). Group 8 contained representative MTPs from other plant species implicated in Mn detoxification mainly through sequestration of Mn by transport into vacuoles, such as ShMTP8 from S. hamata, OsMTP8.1/8.2 from rice, CsMTP8 from cucumber (Delhaize et al., 2003; Chen et al., 2013; Migocka et al., 2014; Takemoto et al., 2017). In addition, two MTP8-like proteins from the tea plant have recently been reported to participate in Mn detoxification via efflux of Mn from plant cells (Li et al., 2017; Zhang et al., 2020). On the other hand, group 9 also included MTP9/10/11 homologues from Arabidopsis, rice, poplar, and B. vulgaris with similar functions in sequestration of Mn into endomembrane compartments or export of Mn out of cells (Peiter et al., 2007; Erbasol et al., 2013; Tsunemitsu et al., 2018a).
Subsequent expression analysis showed that nine GmMTPs (GmMTP4.1, GmMTP4.2, GmMTP8.1, GmMTP8.2, GmMTP8.3, GmMTP8.5, GmMTP9.1, GmMTP10.3, and GmMTP10.4) and two GmMTPs (GmMTP10.3 and GmMTP10.4) were enhanced by excess Mn in roots and leaves, respectively (Figure 5). Similarly, a set of MTP homologues engaged in Mn detoxification are also upregulated by excess Mn in leaves or roots, such as AtMTP8 in Arabidopsis (Eroglu et al., 2016), CsMTP8/9 in cucumber (Migocka et al., 2014, 2015), HvMTP8.1 in barley (Pedas et al., 2014), CasMTP8.1/8.2 in tea plant (Li et al., 2017; Zhang et al., 2020), and BmMTP10 in B. vulgaris (Erbasol et al., 2013). However, the transcripts of AtMTP11 in Arabidopsis and OsMTP8.1/8.2 in rice, which are also involved in Mn detoxification (Delhaize et al., 2007; Peiter et al., 2007; Takemoto et al., 2017), are unaffected by excess Mn levels. It is estimated that functions of some of MTP members might not depend on external Mn at transcriptional levels but at post-transcriptional levels. For example, although the transcript of OsMTP8.1 in rice is not affected by excess Mn, the accumulation of OsMTP8.1 protein is significantly increased under excess Mn conditions, contributing to Mn tolerance (Chen et al., 2013).
In this study, five MTP8-like members (GmMTP8.1 to GmMTP8.5) in soybean that clustered closely showed similar structures and homology identities (Figure 2 and Supplementary Figures 1, 5), which is consistent with the results of phylogenetic analysis (Figure 3). However, GmMTP8.1 to GmMTP8.5 possessed different numbers of exons and introns, and GmMTP8.1 had the lowest molecular size (Supplementary Table 2). Furthermore, variations in the expression levels of these five GmMTP8s were observed in leaves or roots of soybean response to different Mn levels (Figure 5). Sequence and gene expression differences among the GmMTP8s suggest that these genes may have distinct or redundant biological functions in Mn detoxification. Similarly, although OsMTP8.1 and OsMTP8.2 are two MTP8-like homologues in rice, the abundance of OsMTP8.1 in shoots is regulated by external Mn levels, and OsMTP8.2 does not respond to the Mn treatments (Chen et al., 2013; Takemoto et al., 2017). Furthermore, although the disruption of MTP8.2 does not affect Mn tolerance in rice, the knockdown of MTP8.2 in the mtp8.1 mutant results in severe growth inhibition under Mn toxicity conditions (Takemoto et al., 2017), suggesting that OsMTP8.1 coordinates with OsMTP8.2 to increase Mn tolerance in rice. Thus, the five GmMTP8s might possess various functions in soybean response to Mn toxicity, which merits further verification.
A detailed functional analysis of GmMTP8.1, which shares a high degree of homology identity with ShMTP8 (90.8%), OsMTP8.1 (83%), CsMTP8 (83%), and AtMTP8 (81%) (Supplementary Figures 3–5), was performed further. GmMTP8.1 contained four TMDs, a signature sequence specific to the CDF family, and two short motifs (DSLLD and DHYFD) specific to the Mn-CDF cluster (Supplementary Figure 3). These common features are also present in the reported Mn transporters, such as ShMTP8, CsMTP8, and OsMTP8.1/8.2 (Delhaize et al., 2003; Chen et al., 2013; Migocka et al., 2014; Takemoto et al., 2017), suggesting similar roles of GmMTP8.1 in conferring Mn tolerance. It has been demonstrated that ShMTP8 in S. hamata can complement the phenotype of yeast mutant Δpmr1 defective in Mn transport under excess Mn conditions; the overexpression of ShMTP8 increases Mn tolerance in Arabidopsis by sequestration of Mn into vacuoles (Delhaize et al., 2003). Furthermore, in Arabidopsis, AtMTP8 is found to localize in the tonoplast, and the T-DNA insertion Arabidopsis mtp8 mutant displays greater sensitivity to excess Mn than the wild type (Eroglu et al., 2016). Similar functions of MTP8-like proteins have also been observed in OsMTP8.1/8.2 in rice and CsMTP8 in cucumber (Chen et al., 2013; Migocka et al., 2014; Takemoto et al., 2017; Zhang et al., 2021). In this study, heterologous expression of GmMTP8.1 led to restore the growth of the yeast mutant pmr1 to excess Mn (Figure 7), suggesting that GmMTP8.1 is responsible for Mn transport. Furthermore, the overexpression of GmMTP8.1 conferred Mn tolerance in transgenic Arabidopsis grown in both MS solid medium and hydroponic system (Figure 8 and Supplementary Figure 7). Therefore, GmMTP8.1 contributes to increase tolerance to Mn in soybean through regulation of Mn transport.
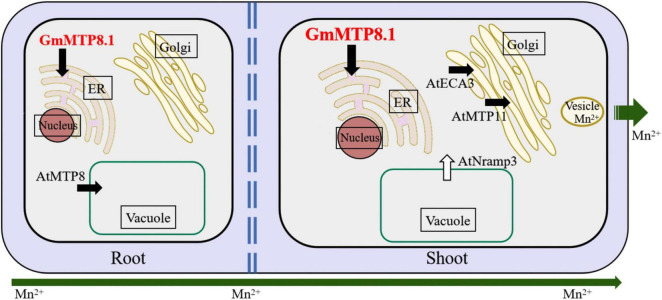
Although GmMTP8.1 exhibited high similarity with the MTP8-like proteins, some functional variations are observed among these homologues. For example, the representative MTP8 members, namely, ShMTP8, CsMTP8, OsMTP8.1/8.2, and AtMTP8, are found to be localized to the tonoplast, detoxifying Mn by sequestration of Mn into the vacuoles (Delhaize et al., 2003; Chen et al., 2013; Migocka et al., 2014; Eroglu et al., 2016; Takemoto et al., 2017). However, HvMTP8.1 and HvMTP8.2 from barley are suggested to be involved in delivering Mn to the Golgi apparatus, regulating Mn homeostasis (Pedas et al., 2014). Furthermore, two MTP8 homologues from the tea plant are localized to the plasma membrane and participated in Mn efflux from plant cells (Li et al., 2017; Zhang et al., 2020). Unexpectedly, different from those identified MTP8-like proteins, GmMTP8.1 was found to be localized to the ER in tobacco leaf epidermis cells (Figure 6), suggesting that GmMTP8.1 is involved in Mn transport into the ER, which has probably contributed to reduction in cytosolic Mn and maintenance of Mn homeostasis in transgenic Arabidopsis overexpressing GmMTP8.1 (Figure 10). Similarly, the ER-localized AtECA1 has been found to promote Arabidopsis growth under Mn toxicity conditions by pumping Mn into the ER, reducing cytosolic Mn to levels that do not disrupt other elements homeostasis (Wu et al., 2002).
FIGURE 10.
Hypothetical view of GmMTP8.1 affecting Mn tolerance in transgenic Arabidopsis. Endoplasmic reticulum (ER)-localized GmMTP8.1 is involved in Mn transport into the ER, which has probably contributed in reduction of cytosolic Mn and maintenance of Mn homeostasis in transgenic Arabidopsis overexpressing GmMTP8.1. Furthermore, cytosolic Mn in leaf can be transported into the Golgi apparatus by the involvement of Golgi-localized AtMTP11 and ECA3, and then sequestered into vesicles and removed from the cell via exocytosis, thereby decreasing cellular Mn levels in shoot. The upregulation of AtNRAMP3 might also help to maintain Mn homeostasis in leaves of transgenic Arabidopsis. In addition, GmMTP8.1 has likely mediated Mn tolerance, along with AtMTP8, by increasing Mn sequestration into root vacuoles, conferring Mn tolerance. White and black arrows represent import into and export out of the cytosol, respectively.
In this study, less Mn accumulation was observed in shoots but not roots of transgenic Arabidopsis overexpressing GmMTP8.1 than in those of WT under excess Mn conditions (Figure 8 and Supplementary Figure 7), suggesting that GmMTP8.1 participates in the extrusion of Mn from leaf cells. The involvement of MTP homologues in Mn efflux has been observed in several plants, such as CasMTP8.1/8.2 in tea plant, HvMTP8.1/8.2 in barley, CsMTP9 in cucumber, AtMTP11 in Arabidopsis, and OsMTP11 in rice (Peiter et al., 2007; Pedas et al., 2014; Migocka et al., 2015; Li et al., 2017; Ma et al., 2018; Zhang et al., 2020). These MTPs are generally localized to the Golgi apparatus or the plasma membrane (Pedas et al., 2014; Li et al., 2017; Zhang et al., 2020). However, ER-localized MTP8-like proteins involved in cellular Mn extrusion have not been reported, although ShMTP8 also seems to target the ER membrane in yeast cells (Delhaize et al., 2003). This study indicates the role of ER-localized GmMTP8.1 participating in the export of Mn out of the leaf cells (Figure 10). Such a cellular Mn extrusion pathway has previously been suggested for the participation of the Golgi-localized AtMTP11 in Arabidopsis and OsMTP11 in rice, in that Mn can be loaded into the trans-Golgi network (TGN), sequestered into vesicles, trafficked to the plasma membrane, and ultimately released to the extracellular space (Peiter et al., 2007; Ma et al., 2018). As GmMTP8.1 was localized to the ER and displayed less similarity with AtMTP11 (56.8%) and OsMTP11 (58.5%), it might indirectly participate in stimulation of the cellular extrusion of Mn. Thus, we speculated that GmMTP8.1 might influence Mn distribution and status, subsequently leading to transcriptional changes in transporter genes that are responsible for Mn efflux.
To test this hypothesis, we analyzed the expressions of Mn transporter genes in transgenic Arabidopsis. The results showed that the overexpression of GmMTP8.1 led to significant induction of AtMTP11, AtNRAMP3, and AtECA3 in leaves of transgenic Arabidopsis under excess Mn conditions (Figure 9). In Arabidopsis, the Golgi-localized AtMTP11 and ECA3 are found to play crucial roles in Mn homeostasis by transporting Mn into the Golgi apparatus (Peiter et al., 2007; Mills et al., 2008), which might be further released to the extracellular space via the exocytosis of secretory vesicles, maintaining Mn homeostasis (Figure 10). Moreover, AtNRAMP3 is involved in the transportation of Mn from the vacuole into chloroplasts of mesophyll cells (Lanquar et al., 2010). Thus, the upregulation of these transporter genes has possibly contributed to stimulation of the export of Mn out of leaf cells in the GmMTP8.1 overexpression lines. On the other hand, we found that Mn levels in roots of transgenic lines were similar to those in the WT under excess Mn conditions (Figure 8 and Supplementary Figure 7), and that the overexpression of GmMTP8.1 led to increase in the transcript of AtMTP8 in roots of transgenic Arabidopsis (Figure 9). The enhancement of AtMTP8 may contribute to increase the sequestration of Mn into root vacuoles (Figure 10), although a direct measurement of intracellular Mn distribution would help to support this hypothesis.
In conclusion, a total of 14 GmMTP genes were identified in the soybean genome. These GmMTPs exhibited different responses to Mn toxicity. The heterologous expression of ER-localized GmMTP8.1 facilitated the growth of yeast-sensitive mutant Δpmr1 and Arabidopsis under Mn toxicity conditions, which could be through the mechanisms of stimulating export of Mn out of cells and increasing sequestration of Mn in intracellular compartments.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
ZC designed the research and revised the manuscript. JL, RD, YJ, JH, XZ, NA, and JS performed the experiments and analyzed the data. JL and RD wrote the manuscript. All the authors approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors appreciate Rainer Schultze-Kraft and Idupulapati M. Rao for the critical comments and revisions. The authors also thank Guihua Wang, Linjie Wang, and Qin Liu for the technical help.
Footnotes
Funding
This research was funded by the Key Research and Development Program of Hainan (ZDYF2019039), the Modern Agro-Industry Technology Research System (CARS-34), and the Integrated Demonstration of Key Techniques for the Industrial Development of Featured Crops in Rocky Desertification Areas of Yunnan-Guangxi-Guizhou Provinces (SMH2019-2021).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.683813/full#supplementary-material
References
- Alejandro S., Höller S., Meier B., Peiter E. (2020). Manganese in plants: from acquisition to subcellular allocation. Front. Plant Sci. 11:300. 10.3389/fpls.2020.00300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cailliatte R., Schikora A., Briat J. F., Mari S., Curie C. (2010). High-affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions. Plant Cell 22 904–917. 10.1105/tpc.109.073023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Li J., Wang L., Ma G., Zhang W. (2016). A mutagenic study identifying critical residues for the structure and function of rice manganese transporter OsMTP8.1. Sci. Rep. 6:32073. 10.1038/srep32073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Fujii Y., Yamaji N., Masuda S., Takemoto Y., Kamiya T., et al. (2013). Mn tolerance in rice is mediated by MTP8.1, a member of the cation diffusion facilitator family. J. Exp. Bot. 64 4375–4387. 10.1093/jxb/ert243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Sun L., Liu P., Liu G., Tian J., Liao H. (2015). Malate synthesis and secretion mediated by a manganese-enhanced malate dehydrogenase confers superior manganese tolerance in Stylosanthes guianensis. Plant Physiol. 167 176–188. 10.1104/pp.114.251017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Yan W., Sun L., Tian J., Liao H. (2016). Proteomic analysis reveals growth inhibition of soybean roots by manganese toxicity is associated with alteration of cell wall structure and lignification. J. Proteomics 30 151–160. 10.1016/j.jprot.2016.03.037 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Delhaize E., Gruber B. D., Pittman J. K., White R. G., Leung H., Miao Y., et al. (2007). A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. Plant J. 51 198–210. 10.1111/j.1365-313x.2007.03138.x [DOI] [PubMed] [Google Scholar]
- Delhaize E., Kataoka T., Hebb D. M., White R. G., Ryan P. R. (2003). Genes encoding proteins of the cation diffusion facilitator family that confer manganese tolerance. Plant Cell 15 1131–1142. 10.1105/tpc.009134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragišić Maksimović J., Mojović M., Maksimović V., Römheld V., Nikolic M. (2012). Silicon ameliorates manganese toxicity in cucumber by decreasing hydroxyl radical accumulation in the leaf apoplast. J. Exp. Bot. 63 2411–2420. 10.1093/jxb/err359 [DOI] [PubMed] [Google Scholar]
- Erbasol I., Bozdag G. O., Koc A., Pedas P., Karakaya H. C. (2013). Characterization of two genes encoding metal tolerance proteins from Beta vulgaris subspecies maritima that confers manganese tolerance in yeast. Biometals 26 795–804. 10.1007/s10534-013-9658-7 [DOI] [PubMed] [Google Scholar]
- Eroglu S., Meier B., Wirén N. V., Peiter E. (2016). The vacuolar manganese transporter MTP8 determines tolerance to iron deficiency-induced chlorosis in Arabidopsis. Plant Physiol. 170 1030–1045. 10.1104/pp.15.01194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fecht-Christoffers M. M., Führs H., Braun H. P., Horst W. J. (2006). The role of hydrogen peroxide-producing and hydrogen peroxide-consuming peroxidases in the leaf apoplast of cowpea in manganese tolerance. Plant Physiol. 140 1451–1463. 10.1104/pp.105.070474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando D. R., Lynch J. P. (2015). Manganese phytotoxicity: new light on an old problem. Ann. Bot. 116 313–319. 10.1093/aob/mcv111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Schiestl R. H. (2007). High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2 31–34. 10.1038/nprot.2007.15 [DOI] [PubMed] [Google Scholar]
- Gustin J. L., Zanis M. J., Salt D. E. (2011). Structure and evolution of the plant location diffusion facilitator family of ion transporters. BMC Evol. Biol. 11:76. 10.1186/1471-2148-11-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herridge D. F., Peoples M. B., Boddey R. M. (2008). Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 311 1–18. 10.1007/s11104-008-9668-3 [DOI] [Google Scholar]
- Jia Y., Li X., Liu Q., Hu X., Li J., Dong R., et al. (2020). Physiological and transcriptomic analyses reveal the roles of secondary metabolism in the adaptive responses of Stylosanthes to manganese toxicity. BMC Genomics 21:861. 10.1186/s12864-020-07279-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaj-Robin O., Russell D., Hayes K. A., Pembroke J. T., Soulimane T. (2015). Cation diffusion facilitator family: structure and function. FEBS Lett. 589 1283–1295. 10.1016/j.febslet.2015.04.007 [DOI] [PubMed] [Google Scholar]
- Lanquar V., Schnell Ramos M., Lelièvre F., Barbier-Brygoo H., Krieger-Liszkay A., Krämer U., et al. (2010). Export of vacuolar manganese by AtNRAMP3 and AtNRAMP4 is required for optimal photosynthesis and growth under manganese deficiency. Plant Physiol. 152 1986–1999. 10.1104/pp.109.150946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavres J., Malavolta E., Nogueira N. D., Moraes M. F., Reis A. R., Rossi M. L., et al. (2009). Changes in anatomy and root cell ultrastructure of soybean genotypes under manganese stress. Rev. Bras. Cienc. Solo 33 395–403. 10.1590/S0100-06832009000200017 [DOI] [Google Scholar]
- Li J. F., Jia Y. D., Dong R. S., Huang R., Liu P. D., Li X. Y., et al. (2019). Advances in the mechanisms of plant tolerance to manganese toxicity. Int. J. Mol. Sci. 20:5096. 10.3390/ijms20205096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Y., Zheng L., Fan Y., Wang Y. R., Ma Y. R., Gu D. F., et al. (2021). Pear metal transport protein PbMTP8.1 confers manganese tolerance when expressed in yeast and Arabidopsis thaliana. Ecotox. Environ. Safe. 208:111687. 10.1016/j.ecoenv.2020.111687 [DOI] [PubMed] [Google Scholar]
- Li Q., Li Y., Wu X., Zhou L., Zhu X., Fang W. (2017). Metal transport protein 8 in Camellia sinensis confers superior manganese tolerance when expressed in yeast and Arabidopsis thaliana. Sci. Rep. 7:39915. 10.1038/srep39915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wu Y., Li B., He W., Yang Y., Yang Y. (2018). Genome-wide identification and expression analysis of the cation diffusion facilitator gene family in turnip under diverse metal ion stresses. Front. Genet. 9:103. 10.3389/fgene.2018.00103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Tian J., Lam H., Lim B., Yan X., Liao H. (2010). Biochemical and molecular characterization of PvPAP3, a novel purple acid phosphatase isolated from common bean enhancing extracellular ATP utilization. Plant Physiol. 152 854–865. 10.1104/pp.109.147918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Gao Y., Tang Y., Wang D., Chen X., Yao Y., et al. (2019). Genome-wide identification, comprehensive gene feature, evolution, and expression analysis of plant metal tolerance proteins in tobacco under heavy metal toxicity. Front. Genet. 10:345. 10.3389/fgene.2019.00345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Huang R., Hu X., Jia Y., Li J., Luo J., et al. (2019). Physiological responses and proteomic changes reveal insights into Stylosanthes response to manganese toxicity. BMC Plant Biol. 19:212. 10.1186/s12870-019-1822-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Xue Y., Chen Z., Liu G., Tian J. (2016). Characterization of purple acid phosphatases involved in extracellular dNTP utilization in Stylosanthes. J. Exp. Bot. 67 4141–4154. 10.1093/jxb/erw190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Dang H., Chen Z., Wu J., Chen Y., Chen S., et al. (2018). Genome-wide identification, expression, and functional analysis of the sugar transporter gene family in Cassava (Manihot esculenta). Int. J. Mol. Sci. 19:987. 10.3390/ijms19040987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Xue Y., Xie B., Zhu S., Lu X., Liang C., et al. (2020). Complex gene regulation between young and old soybean leaves in responses to manganese toxicity. Plant Physiol. Bioch. 155 231–242. 10.1016/j.plaphy.2020.07.002 [DOI] [PubMed] [Google Scholar]
- Long L., Kristensen R. K., Guo J., Chen F., Pedas P. R., Zhang G., et al. (2021). Assessing the variation in traits for manganese deficiency tolerance among maize genotypes. Environ. Exp. Bot. 183:104344. 10.1016/j.envexpbot.2020.104344 [DOI] [Google Scholar]
- Lu S. J., Zhao X. H., Hu Y. L., Liu S. L., Nan H. Y., Li X. M., et al. (2017). Natural variation at the soybean j locus improves adaptation to the tropics and enhances yield. Nat. Genet. 49:773. 10.1038/ng.3819 [DOI] [PubMed] [Google Scholar]
- Ma G., Li J. Y., Li J. J., Li Y., Zhang W. (2018). OsMTP11, a trans-Golgi network localized transporter, is involved in manganese tolerance in rice. Plant Sci. 274 59–69. 10.1016/j.plantsci.2018.05.011 [DOI] [PubMed] [Google Scholar]
- Marschner H. (2012). Marschner’s Mineral Nutrition of Higher Plants. 3rd Edn. London: Academic Press. [Google Scholar]
- Migocka M., Papierniak A., Dziubińska E., Poździk P., Posyniak E., Garbiec A., et al. (2014). Cucumber metal transport protein MTP8 confers increased tolerance to manganese when expressed in yeast and Arabidopsis thaliana. J. Exp. Bot. 65 5367–5384. 10.1093/jxb/eru295 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Migocka M., Papierniak A., Kosieradzka A., Posyniak E., Maciaszczyk-Dziubinska E., Biskup R., et al. (2015). Cucumber metal tolerance protein CsMTP9 is a plasma membrane H+-coupled antiporter involved in the Mn2+ and Cd2+ efflux from root cells. Plant J. 84 1045–1058. 10.1111/tpj.13056 [DOI] [PubMed] [Google Scholar]
- Millaleo R., Reyes-Díaz M., Ivanov A. G., Mora M. L., Alberdi M. (2010). Manganese as essential and toxic element for plants: transport, accumulation and resistance mechanism. J. Soil Sci. Plant Nutr. 10 470–481. 10.4067/S0718-95162010000200008 27315006 [DOI] [Google Scholar]
- Mills R. F., Doherty M. L., López-Marqués R. L., Weimar T., Dupree P., Palmgren M. G., et al. (2008). ECA3, a Golgi-localized P2A-type ATPase, plays a crucial role in manganese nutrition in Arabidopsis. Plant Physiol. 146 116–128. 10.1104/pp.107.110817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanini B., Blaudez D., Jeandroz S., Sanders D., Chalot M. (2007). Phylogenetic and functional analysis of the cation diffusion facilitator (CDF) family: improved signature and prediction of substrate specificity. BMC Genomics 8:107. 10.1186/1471-2164-8-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravec J., Skùpa P., Bailly A., Hoyerová K., Krecek P., Bielach A., et al. (2009). Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459 1136–1140. 10.1038/nature08066 [DOI] [PubMed] [Google Scholar]
- Pedas P., Stokholm M. S., Hegelund J. N., Ladegård A. H., Schjoerring J. K., Husted S. (2014). Golgi localized barley MTP8 proteins facilitate Mn transport. PLoS One 9:e113759. 10.1371/journal.pone.0113759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiter E., Montanini B., Gobert A., Pedas P., Husted S., Maathuis F. J. M., et al. (2007). Secretory pathway-localized cation diffusion facilitator confers plant manganese tolerance. Proc. Natl. Acad. Sci. U.S.A. 104 8532–8537. 10.1073/pnas.0609507104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos E. F., Kondo Santini J. M., Paixão A. P., Júnior E. F., Lavres J., Campos M., et al. (2017). Physiological highlights of manganese toxicity symptoms in soybean plants: Mn toxicity responses. Plant Physiol. Biochem. 113 6–19. 10.1016/j.plaphy.2017.01.022 [DOI] [PubMed] [Google Scholar]
- Shao J. F., Yamaji N., Shen R. F., Ma J. F. (2017). The key to Mn homeostasis in plant: regulation of Mn transporters. Trends Plant Sci. 22 215–224. 10.1016/j.tplants.2016.12.005 [DOI] [PubMed] [Google Scholar]
- Sheng H., Zeng J., Yan F., Wang X., Wang Y., Kang H., et al. (2015). Effect of exogenous salicylic acid on manganese toxicity, mineral nutrients translocation and antioxidative system in polish wheat. Acta Physiol. Plant. 37:32. 10.1007/s11738-015-1783-1 [DOI] [Google Scholar]
- Takemoto Y., Tsunemitsu Y., Fujii K. M., Mitani U. N., Yamaji N., Ma J. F., et al. (2017). The tonoplast-localized transporter MTP8.2 contributes to manganese detoxification in the shoots and roots of Oryza sativa L. Plant Cell Physiol. 58 1573–1582. 10.1093/pcp/pcx082 [DOI] [PubMed] [Google Scholar]
- Tsunemitsu Y., Yamaji N., Ma J. F., Kato S. I., Iwasaki K., Ueno D. (2018b). Rice reduces Mn uptake in response to Mn stress. Plant Signal. Behav. 13:e1422466. 10.1080/15592324.2017.1422466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemitsu Y., Genga M., Okada T., Yamaji N., Ma J. F., Miyazaki A., et al. (2018a). A member of cation diffusion facilitator family, MTP11, is required for manganese tolerance and high fertility in rice. Planta 248 231–241. 10.1007/s00425-018-2890-1 [DOI] [PubMed] [Google Scholar]
- Ueno D., Sasaki A., Yamaji N., Fujii Y., Takemoto Y., Moriyama S., et al. (2015). A polarly localized transporter for efficient manganese uptake in rice. Nat. Plants 1:15170. 10.1038/nplants.2015.170 [DOI] [PubMed] [Google Scholar]
- Voelker C., Schmidt D., Mueller-Roeber B., Czempinski K. (2006). Members of the Arabidopsis AtTPK/KCO family form homomeric vacuolar channels in planta. Plant J. 48 296–306. 10.1111/j.1365-313X.2006.02868.x [DOI] [PubMed] [Google Scholar]
- Wu Z., Liang F., Hong B., Young J. C., Sussman M. R., Harper J. F., et al. (2002). An endoplasmic reticulum-bound Ca2+/Mn2+ pump, ECA1, supports plant growth and confers tolerance to Mn2+ stress. Plant Physiol. 130 128–137. 10.1104/pp.004440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., You J., Xu Y., Yang Z. (2009). Interaction between aluminum toxicity and manganese toxicity in soybean (Glycine max). Plant Soil 319 277–289. 10.1016/S0145-305X(01)00005-2 [DOI] [Google Scholar]
- Zhang X. Y., Li Q. H., Xu W. L., Zhao H., Guo F., Wang P., et al. (2020). Identification of MTP gene family in tea plant (Camellia sinensis L.) and characterization of CsMTP8.2 in manganese toxicity. Ecotox. Environ. Safe. 202:110904. 10.1016/j.ecoenv.2020.110904 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Fu D., Sun Z., Ju C., Miao C., Wang Z., et al. (2021). A tonoplast-associated calcium signaling regulates manganese homeostasis in Arabidopsis. Mol. Plant 21 78–72. 10.1016/j.molp.2021.03.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.