Abstract
Haemonchus contortus is arguably one of the most economically important and ubiquitous parasites of livestock globally and commonly involved in cases of anthelmintic resistance. Here, we performed reciprocal genetic crosses using susceptible (MHco3(ISE)) and multiple anthelmintic resistant (MHco18(UGA2004)) H. contortus isolates. Resultant admixed populations were designated MHco3/18 or MHco18/3, where the lead isolate reflects the origin of the females. Three independent filial generations were generated for each cross, which were subjected to bioassays, molecular approaches and population genetic analyses to investigate the phenotypic and genotypic inheritance of benzimidazole (BZ) resistance at each stage. A panel of microsatellite markers confirmed the success of the genetic cross as markers from both parents were seen in the F1 crosses. Egg hatch tests revealed a stark difference between the two F1 crosses with ED50 estimates for MHco18/3 being 9 times greater than those for MHco3/18. Resistance factors based on ED50 estimates ranged from 6 to 57 fold in the filial progeny compared to MHco3(ISE) parents. Molecular analysis of the F167Y and F200Y SNP markers associated with BZ resistance were analysed by pyrosequencing and MiSeq deep amplicon sequencing, which showed that MHco3/18.F1 and MHco18/3.F1 both had similar frequencies of the F200Y resistant allele (45.3% and 44.3%, respectively), whereas for F167Y, MHco18/3.F1 had a two-fold greater frequency of the resistant-allele compared to MHco3/18.F1 (18.2% and 8.8%, respectively). Comparison between pyrosequencing and MiSeq amplicon sequencing revealed that the allele frequencies derived from both methods were concordant at codon 200 (rc = 0.97), but were less comparable for codon 167 (rc = 0.55). The use of controlled reciprocal genetic crosses have revealed a potential difference in BZ resistance phenotype dependent on whether the resistant allele is paternally or maternally inherited. These findings provide new insight and prompt further investigation into the inheritance of BZ resistance in H. contortus.
Keywords: Benzimidazole resistance, Deep amplicon sequencing, Egg hatch test, Reciprocal genetic cross, Pyrosequencing, Single nucleotide polymorphisms
Graphical abstract
Highlights
-
•
Reciprocal cross used to investigate benzimidazole (BZ) resistance.
-
•
Phenotypic and genotypic tools combined for analysis.
-
•
Inheritance of BZ resistance influenced by maternal &/or cytoplasmic mechanisms.
-
•
Double homozygous resistant genotypes at F167Y and F200Y detected on β−tubulin gene.
1. Introduction
Resistance to each of the major broad-spectrum classes of anthelmintics has emerged rapidly in parasitic nematodes of small ruminants and is now widespread in several species, including Teladorsagia circumcincta and Haemonchus contortus (Kaplan, 2004). Resistance to the benzimidazole (BZ) class of anthelmintics was first reported in H. contortus in 1964 (Drudge et al., 1964) and is now prevalent worldwide (Fitzpatrick, 2013).
Understanding the evolution and inheritance of anthelmintic resistance has been a global research focus for many years. Developments in the field of BZ resistance are the most advanced for any of the anthelmintic classes, although questions relating to the inheritance of resistance genes still exist. A number of non-synonymous single nucleotide polymorphisms (SNPs) on the β-tubulin isotype 1 gene have been associated with BZ resistance in several helminth species, and include a phenylalanine to tyrosine substitution at codon 200 (F200Y) (Kwa et al., 1994), phenylalanine to tyrosine (F167Y) (Ghisi et al., 2007) or phenylalanine to histidine at codon 167 (F167H) (Prichard, 2001; Silvestre and Cabaret, 2002), and glutamic acid to alanine at codon 198 (E198A) (Silvestre and Cabaret, 2002). Most recently, a change at codon 198 was reported in H. contortus and T. circumincta where glutamic acid switched to leucine (E198L) and was found to be independent of F167Y and F200Y (Mohammedsalih et al. 2020; Martínez-Valladares et al., 2020). Similarly, other changes were observed in H. contortus where gulatmic acid changed to either valine, lysine or isoleucine (E198L/E198V/E198K/E198I) (Mohammedsalih et al., 2020). In H. contortus, the β-tubulin isotype 1 gene (HCON_00005260) is autosomal and is located on chromosome 1 at position 7027492-7031447; in globally distributed populations, this genomic locus remains highly differentiated as a result of longterm and widespread use of BZ drugs that have selected for resistance (Doyle et al., 2020). The presence of the β-tubulin SNPs appears to be well correlated with phenotypic expression of BZ resistance (von Samson-Himmelstjerna et al., 2007), however, the relative impact of the different SNPs towards the resistance phenotype and interactions between them remain unclear (Kotze et al., 2014).
Various worm mating protocols have been used to explore the genetics and inheritance of anthelmintic resistance, often with conflicting findings. BZ resistance in H. contortus was reported to be semi-dominant (Le Jambre et al., 1979), and that a matroclinous influence on the in vitro expression of BZ resistance was observed, putatively due to the maternal contribution to egg cytoplasm and shell formation. They also suggested extra-chromosomal inheritance of some traits. Martin et al. (1988) also identified a strong maternal effect in the inheritance of resistance to the BZ drug, thiabendazole (TBZ) in Trichostrongylus colubriformis. However, Sangster et al. (1998) found, using different isolates, that resistance was an incompletely recessive, autosomal trait suggesting that more than one gene was involved in resistance (Le Jambre et al., 1979; Herlich et al., 1981), and found little to no evidence for maternal effects on inheritance in F1 generations of H. contortus. However, these previous reciprocal F2 genetic crosses have not been pursued with molecular analysis of markers associated with drug resistance.
Here, we describe the phenotypic and genotypic analysis of BZ resistance in reciprocal genetic crosses of the susceptible (MHco3(ISE)) and multiple anthelmintic class resistant (MHco18(UGA2004)) H. contortus isolates. Using combined applied in vivo and in vitro techniques to follow phenotypic traits of the crosses together with genetic analyses using microsatellite makers, pyrosequencing and deep amplicon sequencing, we sought to investigate: (i) the influence of maternal versus paternal inheritance of resistant alleles on the phenotypic expression of BZ resistance; and (ii) how the two most common BZ resistance associated SNPs (F167Y and F200Y) are inherited following reciprocal genetic crosses between the resistant MHco18(UGA2004) and susceptible MHco3(ISE) isolate.
2. Materials and methods
2.1. H. contortus isolates
Two parental isolates were selected to undertake the initial cross. MHco3(ISE) is an anthelmintic drug susceptible H. contortus isolate that was inbred over 15 generations of half sibling matings and has been maintained in the lab at Moredun Research Institute, UK since 2004 (Roos et al., 1990, 2004). MHco18(UGA2004) is a BZ, levamisole and ivermectin resistant H. contortus isolate that was recovered from sheep in 2004 and maintained in the lab at University of Georgia, USA (Williamson et al., 2011).
2.2. Setup of reciprocal genetic crosses
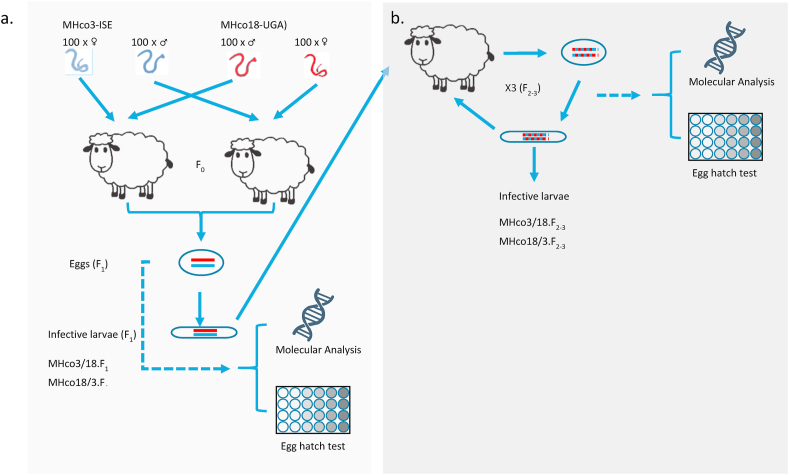
Reciprocal crosses are designed to examine the role that each parental sex plays in the inheritance of traits. Using the reciprocal F2 genetic cross approach outlined below meant that any sex-linked traits, either in phenotype or genotype, associated with BZ resistance could be identified. Reciprocal genetic crosses between MHco3(ISE) and MHco18(UGA2004) were carried out as previously described by Doyle et al., (2019). However, in this case, both isolates were used as the dam of a cross to allow for comparison and any sex-linked traits to be highlighted.
One hundred MHco3(ISE) females and 100 MHco18(UGA2004) males were surgically transferred to one parasite-naive male recipient lamb, and 100 MHco18(UGA2004) females and 100 MHco3(ISE) males were surgically transferred to another parasite-naive male lamb to achieve reciprocal genetic crosses (Fig. 1). The genetic crosses were designated MHco3/18 and MHco18/3, representing female/male parents. Faeces were collected from recipients seven days post-surgery to collect eggs and culture L3, designated F1. 5000 F1 L3 from the two different crosses were administered per os to parasite naïve male lambs to generate F2 populations. Faecal material was collected, from which parasites were cultured to infect two further worm-naive male lambs to generate a F3 population.
Fig. 1.
Outline of the reciprocal crosses, filial crosses and in vivo passage. a. One hundred L4/immature female adults of a multi resistant population MHco18(UGA2004) (“resistant” haplotypes depicted as red lines) were crossed with 100 L4/immature male adults of a susceptible population MHco3(ISE) (“susceptible” haplotypes as blue lines) to generate heterozygous F1 progeny. A reciprocal cross was also initiated with 100 L4/immature female MHco3(ISE) which were crossed with 100 L4/immature male MHco18(UGA2004). Eggs were collected and cultured to generate infective larvae for subsequent infections from each cross b. Three more filial generations were derived by infecting parasite naïve lambs with 5000 infective larvae that were derived from the previous infection. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The cross was undertaken with a view to the future, as other markers may be developed for resistance to other drug families, and materials were archived accordingly for further investigation. At all stages in the process parasitic material was preserved. Adults and L3 from each generation were snap frozen in liquid nitrogen and stored at −70 °C for subsequent molecular analysis. Additionally, sufficient L3 from each generation were stored in liquid nitrogen so they could be resurrected for future experimental infections.
2.3. Faecal worm egg counts
A modification of the salt flotation faecal worm egg count (FWEC) method described by Jackson (1974) was used, with a sensitivity of up to one egg per gram. All FWECs presented are from single male lambs and, therefore, statistical analysis is not possible other than to calculate the basic range, arithmetic mean, and cumulative egg output for each animal. All filial generation FWECs were performed weekly from 18 to 21 days post infection.
2.4. Coproculture
Faecal material was collected and cultured from lambs throughout the study to generate infective larvae (L3) for subsequent infections using the protocols as outlined previously (Coop et al., 1982). The H. contortus larvae were stored at 8-10 °C, and used within 6 months of collection.
2.5. Benzimiadazole efficacy
To confirm the BZ drug sensitivity profile and to generate material for analysis, the MHco3/18.F3 and MHco18/3.F3 generations were treated at the manufacturer's recommended dose rate with fenbendazole (FBZ; Panacur, MSD Animal Health; 5 mg/kg bodyweight). The FBZ treated genetic cross populations were subsequently referred to as MHco3/18.F3.BZ and MHco18/3.F3.BZ. Lambs (n = 1 for each genetic cross). Efficacies of drug treatment were calculated using the following faecal egg count reduction test (FECRT) calculation (Kochapakdee et al., 1995):
| Percentage efficacy = (1 − [ FWECDay10 / FWECDay0 ]) x 100 |
where FWECDay0 and FWECDay10 are the FWECs of the lambs on day of FBZ administration and 10 days later, respectively.
2.6. Egg hatch test and determination of dominance
Egg hatch tests were performed for each filial generation and for the F3 population treated with a BZ drug on day 53 post infection.
The egg hatch test was conducted as previously described (von Samson-Himmelstjerna et al., 2009a). The final thiabendazole (TBZ) drug concentrations examined ranged between 0 and 5 μg/ml TBZ. Each drug concentration was made up independently from a stock solution of 1 mg/ml TBZ in dimethyl sulfoxide (DMSO) and set up in triplicate for each test. Each egg hatch test was set up three times during the course of each patent infection. An additional test was also carried out post-FBZ treatment of the F3 generation. Eggs/first stage larvae within each test well were fixed in ethanol (70% v/v) prior to analysis and to preserve material for future molecular studies. Egg death 50 (ED50) estimates were calculated for each generation of genetic cross. Probit analysis (Minitab 16 statistical software, Minitab LLC, USA) was used to calculate the ED50 estimates and standard error of the mean (SEM) for the egg hatch tests. Resistance factors were calculated by dividing the ED50 estimates with the susceptible parent MHco3(ISE) ED50.
Degrees of dominance (D) based on the log transformed egg hatch ED50 estimates were calculated using the previously published Falconer's equation (Stone, 1968).
In this equation, represent ED50 estimates of the F1 crosses, MHco18(UGA2004), and MHco3(ISE), respectively. D = 1 would be indicative of complete dominance; 0 < D < 1 of incomplete dominance; −1 < D < 0 of incomplete recessivity; and D = −1 of complete recessivity.
2.7. Single nucleotide polymorphism (SNP) and allele quantification
2.7.1. Pyrosequencing
Individual infective larvae (n = 88) from each filial generation of the reciprocal cross and the BZ treated F3 populations were picked into separate wells of a 96-well plate (Axygen, USA) containing 25 μl of worm lysis buffer (Kwa et al., 1995). Proteinase K (Promega, UK) was added to each well to create a final concentration of 100 μg/ml enzyme. Eight non-template control wells per plate were included. Plates were placed at −80 °C for 4 h before incubation at 56 °C overnight to allow for lysis of the parasite material. Lysates were heated to 90 °C for 30 min to deactivate the proteinase K. Lysates were stored at −20 °C prior to analysis.
The primers used to analyse the F167Y and F200Y SNPs of the β-tubulin isotype 1 gene have been previously described (von Samson-Himmelstjerna et al., 2009b). The codon 198 SNPs (E198A, E198L, E198V, E198K or E198I) were not examined as they are not present in the MHco18(UGA2004) and MHco3(ISE) isolates, confirmed by whole-genome sequencing (Doyle et al., 2021). For pyrosequencing PCR, 4 μl of gDNA crude lysate was included in each 50 μl reaction. NovaTaq™ Hot start master mix (Merck, UK) was used for the PCR step containing 0.2 μM Forward primer, 0.185 μM Reverse biotinylated primer, 1.5 mM MgCl2, 25 μl 2 × buffer and made up to 50 μl using DNA/RNA-free water.
Following a 15 min 95 °C polymerase activation step, amplification was performed using 45 cycles of 94 °C for 30 s, 53 °C for 30 s and 72 °C for 30 s, followed by a final extension step at 72 °C for 10 min. To confirm amplification, 10 μl of each PCR reaction was examined by gel electrophoresis on 2% agarose gels stained with gel red (Biotium, California, USA). The remaining 40 μl of the reaction was analysed by pyrosequencing on a PyroMark ID instrument (Qiagen, Germany) following the manufacturer's protocol.
2.7.2. Genotyping based on pyrosequencing results
SNP's at codons 167 and 200 were noted for each L3 (TT – homozygous susceptible - SS; AT heterozygote – SR; and AA – homozygous resistant - RR). Resistant (R) and susceptible (S) allele frequencies were counted and expressed as a percentage for each generation. Genotype combinations for both SNPs were counted and expressed as percentages. Pairwise comparison of genotypes was carried out using Shannon Diversity Indices where outputs are G test values and chi square probabilities. The populations were also checked for Hardy Weinberg Equilibrium and Fixation index (inbreeding coefficient) using the GenAlEx plug-in in Microsoft Excel (Microsoft corporation, USA) version 6.5 (Peakall and Smouse, 2012).
2.8. Deep amplicon sequencing
Pooled larvae from each filial generation derived from the reciprocal cross and the MHco3/18 BZ treated F3 population were used to create crude lysates for deep amplicon sequencing of a 328 bp fragment of the H. contortus β-tubulin isotype 1 gene that spanned the codons F200Y (TTC-TAC), E198A (GAA-GCA), F167Y (TTC-TAC) and the intervening intron. The modified primer sets, adapter/barcoded PCR amplification conditions and AMPure XP Magnetic Beads (1X) (Beckman Coulter, Inc.) purification were previously described by Ali et al. (2018). Ten μl of each barcoded PCR product was combined to make a pooled library. Pooled libraries were run on agarose gel electrophoresis to separate PCR products. The desired β-tubulin isotype 1 PCR amplicon was excised from the gel from which DNA was isolated by gel extraction purification (QIAquick Gel Extraction Kit, Qiagen, Germany). The eluted 20 μl DNA was then purified using AMPure XP Magnetic Beads (1X) (Beckman Coulter, Inc.) to produce a single purified DNA pool library. The library was first measured with a KAPA qPCR library quantification kit (KAPA Biosystems, USA) and then run on an Illumina MiSeq sequencer using a 600-cycle pair-end reagent kit (MiSeq Reagent Kits v2, MS-103-2003) at a concentration of 15 nM with the addition of 15% PhiX Control v3 (Illumina, FC-11-2003).
A post MiSeq analysis separates all the sequence by sample via the recognised barcoded indices and generates the FASTQ files. The data analysis was performed with a bespoke pipeline using Mothur v1.39.5 software (Schloss et al., 2009) with modifications in the standard operating procedures of Illumina MiSeq in the previously described Command Prompt pipeline (Ali et al., 2018; Sargison et al., 2019b). Briefly, the ‘make.contigs’ command was run on raw paired-end reads from each sample to combine the two sets of reads. The command extracted sequence and quality scores from the FASTQ files, creating the complement of the reverse and forward reads, and then joined the read pairs into contigs. After removing long or ambiguous sequence reads (>328 bp) using the ‘screen.seqs’ command, the data was aligned with the H. contortus β-tubulin isotype 1 reference sequence library using the ‘align.seqs’ command. The sequences that did not match with the H. contortus β-tubulin isotype 1 reference sequence library were removed and the ‘summary.seqs’ command was used to summarise the 328 bp sequence reads of the H. contortus β-tubulin isotype 1 locus. The sequence reads were further run on the ‘screen.seqs’ command to generate the H. contortus β-tubulin isotype 1 FASTQ file. Once the sequence reads were classified as β-tubulin isotype 1, a count list of the consensus sequences of each sample was created using the ‘unique.seqs’ command. The count list was further used to create FASTQ files (Mendeley database at https://doi.org/10.17632/57n7p8gxh2.1) of the consensus sequences of each sample using the ‘split.abund’ command to sort data into groups of rare and abundant based on the cutoff value (1000 reads), followed by the ‘split.groups’ command. Consensus sequences for H. contortus β-tubulin isotype 1 were generated from the count list using Geneious Prime 2020.1 software (Kearse et al., 2012). These consensus sequences were used for the calculation of the relative allele frequencies of β-tubulin isotype 1 resistance-associated mutations. To achieve this, H. contortus β-tubulin isotype 1 were first assigned to susceptible and the relevant resistance mutations based on known SNPs at codons F200Y (TTC-TAC), E198A (GAA-GCA), F167Y (TTC-TAC), followed by dividing the number of sequences reads of each sample that contained the mutation by the total number of reads (R Core Team, 2014).
Genotyping results obtained from the analysis of individual larvae by pyrosequencing and pooled parasite material using deep amplicon sequencing were compared by Lin's Concordance Correlation Coefficient, calculated using the epiR program in R (version 3.6.3).
2.9. Microsatellite genotyping of the parental isolates and genetic crossing progeny
Thirty individual larvae of each population including the parental isolate, derived from the initial donor lamb infections described in section 2.2 [MHco3(ISE), MHco18(UGA2004)] and three of the genetic crossing progeny [MHco3/18.F1, MHco18/3.F1 and MHco3/18.F3(BZ)] were analysed. Individual larvae were added into a single 0.2 μl tube containing 20 μl of 10 mg/ml proteinase K (New England Biolabs) and Lysis Reagent (Viagen) (Redman et al., 2008; Chaudhry et al., 2015).
One μl of neat individual worm lysate was used as PCR template and identical dilutions of lysate buffer, made in parallel, were used as negative controls. A panel of six microsatellites (Hcms8a20, Hcms22c03 (Redman et al., 2008); Hcms25, Hcms33 (Otsen et al., 2000); Hcms22193 and Hcms53265 (Redman et al., 2015)) was selected to include potentially useful markers across the genome of H. contortus. The forward primer of each microsatellite primer pair was 5′-labelled with a fluorescent dye (IDT, UK) and the GeneScan ROX 400 internal size standard was used on the ABI Prism 3100 genetic analyser (Applied Biosystems, UK). Individual chromatograms were analysed using Gene Mapper software version 4.0 (Applied Biosystems, UK) to accurately size the amplicons and determine genotypes. Fixation index (pairwise FST) values were calculated from the multi-locus microsatellite genotype data by random permutation in Arlequin 3.11 (Excoffier et al., 2005). Principal coordinate analysis (PCA) was performed using GenAlEx version 6.5 preserving individual worm genotypes (Peakall and Smouse, 2012). A summary of primer sequences, allele ranges and PCR conditions for each marker as used in our hands is given in Supplementary Table S1.
3. Ethics statement
All experimental procedures described in this manuscript were examined and approved by the Moredun Research Institute Animal Welfare and Ethical Review Board (E30/14, E47/17 & E27/19) and were conducted under approved British Home Office licenses in accordance with the Animals (Scientific Procedures) Act of 1986.
4. Results
4.1. Faecal worm egg counts
FWECs from all stages of the crosses are illustrated in Fig. 4 and separated into three panels a, b and c.
Fig. 4.
Shows Faecal worm egg counts (eggs/g) of the individual donor animals used in this study split over 3 panels a–c. a. Shows an overview of all the donor animals used in this study (original recipients – F1; F1 passaged –F2; F2 passaged – F3). Where the blue line represents MHco3/18.F1; the yellow line represents MHco18/3.F1; the orange line represents MHco3/18.F2; the purple line represents MHco18/3.F2; the grey line represents MHco3/18.F3 and the green line represents MHco18/3.F3. The red circles highlight the area of the overview graph being shown in greater detail in panel b & c. The blue dashed arrow depicts when the F3 generation was treated with fenbendazole (FBZ). b. Faecal egg counts of the recipient donors used in the original reciprocal crosses. c. Faecal egg count of the individual donors infected with each genetic cross of the F2 generation after treatment with FBZ. The text box shows the efficacy of FBZ at 10 days post treatment. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The FWECs of both the MHco3/18.F1 and MHco18/3.F1 recipient lambs were zero eggs per gram (epg) on day 18 post infection (4 days post-surgery) and 306 epg (for both lambs) on day 28 (10 days post-surgery). The FWECs of the MHco3/18.F1 and MHco18/3.F1 recipient lambs are highlighted in Fig. 4b as this data is lost when looking at the overall picture in Fig. 4a due to their egg counts being lower compared to the other filial generations. The lambs producing the F2 populations showed an upward trend in FWEC with a peak in both crosses at day 47 post infection (week 6). Each of the MHco18/3 filial generations maintained a higher FWEC compared to the equivalent MHco3/18 generations (between 19 and 247 days post infection).
The F3 populations pre-treatment (day 21–46 post infection) had a similar trend where MHco18/3.F3 peaked at day 35 with 8028 epg, 1.5 times higher than MHco3/18.F3 FWEC at that timepoint (Fig. 4a). Post FBZ treatment, the FWECs of the lambs infected with both reciprocal cross F3 remained positive (Fig. 4c). FBZ treatment showed 77% efficacy against the MHco3/18.F3 at 10 days post treatment, whereas the equivalent efficacy against the MHco18/3.F3 was 87%.
The cumulative FWEC output was greater in MHco18/3 compared to the MHco3/18 filial populations. The difference between the parental isolates cumulative FWEC was 1.1 where MHco3(ISE) had 423,011 and MHco18(UGA2004) had 472,644. In the F1 population MHco18/3 cumulative FWEC was 1.4 times greater than MHco3/18.F1 (5232 and 3879, respectively). In the F2 population, the cumulative FWEC of MHco18/3.F2 was 3.3 times greater than MHco3/18.F2 (35,500 and 10,665, respectively). Lastly, in the F3 population, the cumulative FWEC of MHco18/3.F3 was 1.5 times greater compared to MHco3.18.F3 (156,287 and 106,627, respectively).
4.2. Egg hatch test
The parental isolate's ED50 estimates were 0.026 μg/ml TBZ for MHco3(ISE) and >5 μg/ml TBZ for MHco18(UGA2004). A precise ED50 estimate could not be calculated for MHco18(UGA2004) as more than 50% of the eggs hatched at all drug concentrations tested even at the highest concentration tested (Table 2). Generally, the MHco18/3 genetic cross filial generations had higher ED50 estimates (with the exception of the F3 untreated population) than their reciprocal counterparts, indicative of a higher level of the resistance phenotype. The greatest difference observed between the ED50 estimates was between the F1 populations where MHco18/3.F1 had a 9.8-fold greater ED50 estimate compared to the MHco3/18.F1. This was reduced to being a 1.2-fold difference in the F2 populations. Resistance based on ED50 estimates ranged from 6- to 57-fold higher compared to the susceptible MHco3(ISE) parental isolate (Table 2).
Table 2.
The ED50 estimates (μg/ml TBZ) along with standard error of the mean (SEM) for each of the reciprocal genetic crosses and filial generations as calculated using probit analysis. The resistance factors are calculated against the MHco3 (ISE) susceptible parent.
| Population | Designation | ED50 Estimate ± SEM (TBZ μg/ml) | Resistance Factor |
|---|---|---|---|
| Parent | MHco3 (ISE) | 0.026 ± 0.001 | 1 |
| Parent | MHco18 (UGA2004) | >5 | >192 |
| F1 | MHco3/18.F1 | 0.150 ± 0.004 | 6 |
| F2 | MHco3/18.F2 | 0.185 ± 0.002 | 7 |
| F3 | MHco3/18.F3 | 0.182 ± 0.007 | 7 |
| F3 BZ | MHco3/18.F3.BZ | 0.494 ± 0.020 | 19 |
| F1 | MHco18/3.F1 | 1.470 ± 0.160 | 57 |
| F2 | MHco18/3.F2 | 0.229 ± 0.003 | 9 |
| F3 | MHco18/3.F3 | 0.177 ± 0.009 | 7 |
| F3 BZ | MHco18/3.F3.BZ | 0.850 ± 0.030 | 33 |
4.2.1. Degrees of dominance
Degrees of dominance estimates from the two reciprocal crosses F1 were not in agreement, where MHco3/18 D = −0.389 suggested incomplete recessivity whereas MHco18/3 D = 0.416 suggested incomplete dominance.
4.3. Microsatellites
The reciprocal genetic crosses using the susceptible MHco3(ISE) and BZ resistant MHco18(UGA2004) H. contortus isolates were validated using a panel of six microsatellite markers. The presence or absence of the microsatellite marker alleles allowed the genetic crosses to be monitored, and provided confirmation that they were progressing as expected. Thirty individual L3 from each parental isolate, F1 progeny and drug selected F3 progeny were genotyped. A total of 16 different isolate specific alleles were identified from three out of the six microsatellite markers including five alleles that were present in MHco3(ISE), but absent in MHco18(UGA2004), and 11 alleles that were present in MHco18(UGA2004), but absent in MHco3(ISE) (Supplementary Fig. S1 a & b). The MHco18/3.F1 progeny carried all five alleles derived from the MHco3(ISE) parental isolate and eight from MHco18(UGA2004). Three alleles [252 (Hcms8a20), 211 (Hcms53265) and 258 (Hcms22c03)] from the MHco18(UGA2004) parental isolate were absent in the MHco18/3 F1 progeny (Fig. 2a). In contrast, MHco3/18.F1 progeny carried all 11 alleles derived from the MHco18(UGA2004) parent and two out of five alleles [196 (Hcms53265) and 211 (Hcms25)] from the MHco3(ISE) parental isolate (Fig. 2b). Five MHco3(ISE) specific alleles and two out of 11 alleles [220 (Hcms8a20) and 199 (Hcms53265)] of the MHco18(UGA2004) parental isolates were retained in the MHco3/18.F3(BZ) progeny (Fig. 2c).
Fig. 2.
Alleles present (different colour shades) in three crossing progeny [MHco18/3.F1, and MHco3/18.F1, MHco3/18.F3(BZ)] using six microsatellite markers. Panel a–c: Individual worm genotyping has been performed based on 30 individual L3 stage larvae of MHco18/3.F1, MHco3/18.F1 and MHco3/18.F3(BZ) respectively. In each panel, the dotted square icons represents the alleles unique to MHco18(UGA2004) and dotted circles represent those unique to MHco3(ISE) parental isolates, and the unmarked columns refer to alleles found in both parental isolates. X-axis represent the frequency of the alleles, Y-axis represents the bases pair of the alleles in each markers and name of the markers were shown on the top of the figure. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
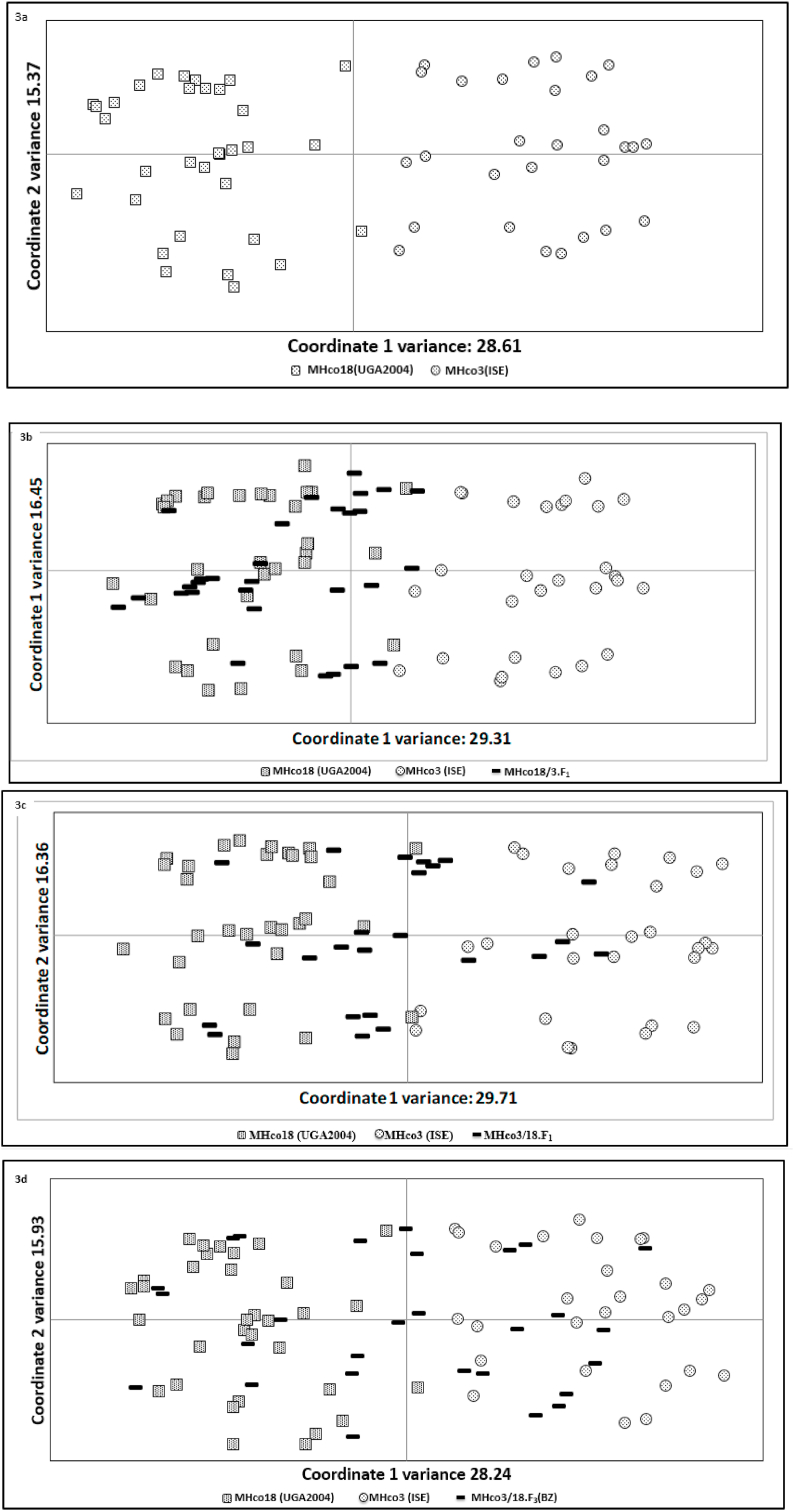
Genotyping of the MHco3(ISE) and MHco18(UGA2004) parental isolates using microsatellite markers confirmed a high level of genetic differentiation between MHco3(ISE) and MHco18(UGA2004). The principal coordinate analysis (PCA) of MHco3(ISE) and MHco18(UGA2004) microsatellite data revealed two separate clusters in the plot (Fig. 3a). This genetic differentiation was reflected by the high pairwise FST estimates calculated between both isolates (FST 0.2022) (Table 1).
Fig. 3.
Principle coordinate analysis of microsatellite markers for MHco3(ISE) & MHco18(UGA2004) parental isolates and progeny [MHco18/3.F1, MHco3/18.F1, MHco3/18.F3(BZ)] as shown in panel a–d respectively. Panel a shows the PCA plot comparing the microsatellite markers for the parental isolates. Panels b–d shows the PCA plot comparing the parental isolates with the respective genetic cross filial generation added shown in the black bar. In each panel the square represents MHco18(UGA2004) alleles, the circle represents MHco3(ISE) alleles and black bar represents the alleles which come from the genetic cross: MHco18/3.F1 (b), MHco3/18.F1 (c) and MHco3/18.F3(BZ) (d) generations.
Table 1.
Pairwise FST values based on genotyping 30 individual worms from parental isolates, F1 progeny [MHco18/3.F1, MHco3/18.F1] and drug selected F3 progeny [MHco3/18.F3(BZ)] with six microsatellite markers. Pairwise comparisons with statistically significant (p < 0.001).
| MHco3 (ISE) | MHco18 (UGA2004) | MHco3/18.F1 | |
|---|---|---|---|
| MHco18 (UGA2004) | 0.2022 | – | – |
| MHco18/3.F1 | 0.0731 | 0.0823 | – |
| MHco3/18.F1 | 0.0780 | 0.0770 | – |
| MHco3/18.F3.BZ | 0.0686 | 0.0542 | 0.0372 |
The F1 generation were, however, much more closely related to each of the MHco3(ISE) and MHco18(UGA2004) parental isolates; in the PCA MHco18/3.F1, MHco3/18.F1 and MHco3/18.F3(BZ) progeny showed clustering of alleles with MHco3(ISE) or MHco18(UGA2004) parental isolates (Fig. 3b, c and d), and were less distinct by measures of FST [ MHco18/3.F1 (FST = 0.0731 and FST = 0.0823) and MHco3/18.F1 (FST = 0.0780 and FST = 0.0770), respectively]. A similar pattern was seen in the comparison MHco3/18.F3(BZ) and MHco3(ISE) (FST = 0.0686) and MHco18(UGA2004) (FST = 0.0542) parental isolates. Genetic differentiation was lower between the reciprocal MHco3/18.F1 and MHco3/18.F3(BZ) progenies (FST = 0.0372) (Table 1).
4.4. Pyrosequencing and deep amplicon sequencing
4.4.1. Genotype variability
We determined the frequency of all possible genotype combinations between the F167Y and F200Y loci using pyrosequecing of individual larvae from each stage of both crosses. In total, 9 different genotype combinations were possible (Table 3). MHco18(UGA2004) had a variety of β-tubulin codon 167 and 200 SNP genotype combinations present, with six out of the possible nine combinations detected; it contained no double susceptible genotypes, and a low level of double homozygous resistant genotypes (3.53%) at both SNPs (Table 3). The MHco3(ISE) parental population had 100% homozygous susceptible genotypes at both SNPs. Both reciprocal F1 genetic crosses had the same three codon 167-200 genotype combinations (SS-RS, RS-SS and RS-RS). MHco18/3.F1 appeared to have more heterozygotes at F167Y compared to MHco3/18.F1 (Table 3). In the F2 generations, the genotype combinations increased from three to six for MHco3/18.F2 and seven for MHco18/3.F2 where MHco3/18.F2 did not have any genotypes with homozygous resistant F167Y present. In the F3 generations pre treatment, both genetic crosses had seven genotype combinations. For MHco3/18.F3, the seven genotype combinations remained the same post FBZ treatment. However, there was a significant shift towards the F200Y homozygous resistant genotype after treatment (Shannon diversity indices chi square probability p < 0.001) with 46.0% of the larvae having SS-RR (codon167-200) genotypes, and a reduction in homozygous SS-SS genotypes (Table 3). Where homozygous resistant genotypes occurred at codon 167, this was observed in combination with SR genotypes at codon 200. MHco18/3.F3 had the same number of genotype combinations (n = 7) post FBZ treatment, but there was a shift from 34.6% to 45.7% of the genotypes occurring with the either heterozygous or homozygous resistant F167Y. The homozygous susceptible F200Y genotype was only seen in combination with the homozygous resistant F167Y genotype post FBZ treatment. For the homozygous resistant F167Y genotype, all possible F200Y genotypes were observed at a similar level to parental MHco18, including homozygous resistant genotypes, i.e. RR-RR, at both SNPs (3.7%).
Table 3.
Genotype frequency with percentages shown in parenthesis of the nine possible F167Y/F200Y genotype combinations observed for the parental isolates and genetic cross generations using pyrosequencing on individual larvae; where F3 generations have results for untreated and post fenbendazole (BZ) drug treatment.
| Genotype | MHco3 (ISE) | MHco18 (UGA2004) | MHco3/18.F1 | MHco3/18.F2 | MHco3/18.F3 | MHco3/18.F3.BZ | MHco18/3.F1 | MHco18/3.F2 | MHco18/3.F3 | MHco18/3.F3.BZ | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SS-SS | TT-TT | 84 (100.0) | 0 (0.0) | 0 (0.0) | 23 (33.3) | 25 (32.5) | 2 (2.6) | 0 (0.0) | 17 (21.3) | 17 (30.9) | 0 (0.0) |
| SS-SR | TT-AT | 0 (0.0) | 0 (0.0) | 66 (81.5) | 25 (36.2) | 26 (33.8) | 22 (29.0) | 56 (63.6) | 33 (41.3) | 12 (21.8) | 15 (18.5) |
| SS-RR | TT-AA | 0 (0.0) | 36 (42.4) | 0 (0.0) | 8 (11.6) | 9 (11.7) | 35 (46.1) | 0 (0.0) | 5 (6.3) | 7 (12.7) | 29 (35.8) |
| RS-SS | AT-TT | 0 (0.0) | 0 (0.0) | 3 (3.7) | 1 (1.5) | 4 (5.2) | 2 (2.7) | 10 (11.4) | 3 (3.8) | 6 (10.9) | 0 (0.0) |
| RS-RS | AT-AT | 0 (0.0) | 15 (17.7) | 12 (14.8) | 7 (10.1) | 11 (14.3) | 8 (10.5) | 22 (25.0) | 8 (10.0) | 10 (18.2) | 14 (17.3) |
| RS-RR | AT-AA | 0 (0.0) | 28 (33.0) | 0 (0.0) | 5 (7.3) | 1 (1.3) | 6 (7.9) | 0 (0.0) | 13 (16.3) | 2 (3.6) | 14 (17.3) |
| RR-SS | AA-TT | 0 (0.0) | 2 (2.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (1.8) | 2 (2.5) |
| RR-RS | AA-AT | 0 (0.0) | 1 (1.2) | 0 (0.0) | 0 (0.0) | 1 (1.3) | 1 (1.3) | 0 (0.0) | 1 (1.3) | 0 (0.0) | 4 (4.9) |
| RR-RR | AA-AA | 0 (0.0) | 3 (3.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (3.7) |
4.4.2. Hardy Weinberg Equilibrium and Fixation index
The individual genotype frequencies for both F167Y and F200Y obtained from each of the filial generations and BZ treated F3 populations by pyrosequencing were analysed using a chi square test for agreement with the Hardy Weinberg Equilibrium (HWE). The F1 generations of the genetic crosses had higher than expected frequencies of heterozygote genotypes at both SNPs. MHco3/18.F1 observed heterozygote frequency of 15 (13.6 expected according to HWE) at codon 167 was not significant but at codon 200 was highly significant (χ2 = 66.47, p < 0.001) with 77 observed heterozygotes (40 expected). MHco18/3.F1 had a significant number of heterozygotes at both SNPs with 32 observed at codon 167 (26.18 expected; χ2 = 4.34, p < 0.05) and 78 heterozygotes observed at codon 200 (43.43 expected; χ2 = 55.75, p < 0.001). These were the only genotypes from all that were analysed by pyrosequencing of individuals to show any significance in the chi square test that deviate from the HWE. The F1 results would corroborate with the Fixation Index ‘F’ value being close to −1 (where negative values indicate heterozygosity) for both F1 crosses (MHco3/18.F1 F = −0.906, MHco18/3.F1 F = −0.796) at the F200Y loci. It also supports the success of both genetic crosses, given the predominant genotypes are heterozygous.
4.4.3. Allele frequency
The pyrosequencing and MiSeq results showed MHco3(ISE) had almost 100% susceptible alleles for the F167Y and F200Y SNPs (1.0% F200Y R with MiSeq). MHco18(UGA2004) had the highest resistant allele frequency at 88.2% and 77.8% for F200Y resistant (R), whereas the F167Y R allele frequency was lower at 32.4% and 21.6% with pyrosequencing and MiSeq, respectively (Fig. 5a–d). MHco3/18 F167Y R allele showed a slight decrease from F1 but remained consistent across all the other filial generations. For the F200Y R allele, MHco3/18 has a similar pattern to F167Y with a slight decrease from F1, however, after FBZ treatment there was almost a two-fold increase seen in the resistance allele. The MHco18/3 F167Y R allele levels were inconsistent between the methods as pyrosequencing individuals showed a slight decrease between F1 and F2 generations (18.2 and 16.3% respectively) and MiSeq on pools showed a slight increase for F167Y R between F1 and F2 (9.2 and 11.3%, respectively). For the F200Y R allele, both methods were in agreement with a slight increase between F1 and F2 generations. MHco18/3.F3 was only analysed by pyrosequencing and showed a slight decline in the R allele at codon 200 between F2–F3 (48.7 and 38.4%, respectively), however, this increased over two-fold post FBZ treatment (Fig. 5b). An increase of 1.5-fold was also seen at the F167Y R allele post-FBZ treatment in MHco18/3.F3 (Fig. 5a). MiSeq data observed the F200Y R and F167Y R double mutants were present at 0.1% in MHco18(UGA2004). For MHco3/18.F1, MHco3/18.F2, MHco3/18.F3, and MHco3/18.F3 BZ crosses, the F200Y R and F167Y R double mutants were present at 2.2%, 0.7%, 1.6% and 0.03%, respectively. For MHco18/3.F1, and MHco18/3.F2 crosses, the F200Y R and F167Y R double mutants were present at 1.3% and 0.2%, respectively (data not shown). The MiSeq data also confirmed that no changes were observed at codon 198 in either of the parents or genetic cross populations (Doyle et al., 2021).
Fig. 5.
Mean Allele frequencies split over 4 panels a–d with ED50 estimates displayed in white dots with the exception of MHco18 where it is displayed above the column in text. a. Shows the mean allele frequencies of F167Y using pyrosequencing on individual larvae. b. Shows the mean allele frequencies of F200Y using pyrosequencing on individual larvae. c. Shows the mean allele frequencies of F167Y using Miseq on pools of larvae where ND = no data. d. Shows the mean allele frequencies of F200Y using Miseq on pools of larvae where ND = no data.
Overall, the allele frequency at both SNPs using deep amplicon sequencing on pools and pyrosequencing individual larvae from the parents and each filial generation of the crosses produced similar results (Fig. 5). To test the concordance of the two genotyping approaches, we compared the allele frequency from each using the Lin's Concordance Correlation Coefficient (CCC). The results from each method were comparable, with the CCC showing good correlation for F200Y (CCC = 0.97; with 95% CI = 0.89–0.99) and moderately correlated for F167Y (CCC = 0.55; with 95% CI = −0.17-0.89).
Pairwise calculations of Shannon's Diversity Indices using pyrosequencing data and subsequent G-tests on the two loci using GenAlEx showed that all genetic cross populations, with the exception of one, were significantly different from the parents at both loci (p < 0.01) (see Supplementary Table S2). MHco18/3.F3.BZ showed no significant difference at F167Y compared to the MHco18(UGA2004) parent. The F200Y allele frequencies observed in the three filial generations of both genetic crosses were significantly different from both the F3.BZ post-treatment populations (p < 0.001). The F200Y allele frequency was also significantly different between the MHco18/3.F2 and both F3 untreated genetic cross generations (p < 0.05). For F167Y, the most significant differences were observed between the MHco3/18.F1, F2 and F3 (including F3.BZ) against MHco18/3.F3.BZ (p < 0.001). Significant differences (p < 0.001) were seen between MHco18/3.F2 and MHco18/3.F3.BZ. There were also significant differences between the F1 genetic crosses, including MHco18.3.F1 compared to MHco3/18.F2 and MHco18/3.F3.BZ and MHco18/3.F3 compared to MHco18/3.F3.BZ at codon 167 (p < 0.05) (See Supplementary Table S2).
5. Discussion
The study used classical reciprocal genetic crossing techniques combined with in-depth phenotypic and genotypic analyses to explore the inheritance of BZ resistance and two of the commonly associated resistance SNPs in the β-tubulin isotype 1 gene. A key finding of this work was that, unexpectedly, phenotypic differences in the levels of BZ resistance between the reciprocal crosses were observed. Both the overall total egg output and ED50 estimates of the MHco18/3 genetic cross filial progeny were 2.0- and 2.6-fold higher, respectively, than those of the corresponding MHco3/18 genetic cross filial progeny. With the exception of the BZ selected F3 populations, these phenotypic differences were not associated with genotypic variation at the β-tubulin isotype 1 locus. This implies differences in the phenotypic outcome was dependent on whether the resistant allele was maternally or paternally inherited.
The reciprocal genetic crosses using the susceptible MHco3(ISE) and BZ-resistant MHco18(UGA2004) H. contortus isolates were validated using microsatellite markers. Microsatellite analysis of the parent isolates was shown to cluster into two distinct populations with an FST value indicating high genetic differentiation (Wright, 1978; Hendrick, 2000; Peakall and Smouse, 2012). Analysis of the F1 genetic crosses demonstrates that the alleles have been successfully admixed from MHco3(ISE) and MHco18(UGA2004) H. contortus isolates. Both F1 genetic crosses show clustering with alleles observed in the parental isolates with FST reduced to around 0.08 or less. Importantly, when individual worms of F1 genetic crosses were genotyped with the panel of six microsatellite markers, their genetic background was intermediate of the MHco3(ISE) and MHco18(UGA2004) H. contortus isolates.
Analysis of the egg hatch data showed that both F1 generations had ED50 estimates higher than the susceptible isolate but lower than the resistant isolate which was in agreement with previous studies looking into the inheritance of BZ resistance using reciprocal crosses with H. contortus and T. colubriformis (Le Jambre et al., 1979; Herlich et al., 1981; Martin et al., 1988; Sangster et al., 1998; Hunt et al., 2010). In Haemonchus, different genetic mechanisms of BZ resistance have been reported, with some suggesting that BZ resistance is semi-dominant (Le Jambre et al., 1979), whilst others have suggested that it is a fully recessive trait (Herlich et al., 1981). Degrees of dominance estimates using Falconer's equation from the two reciprocal crosses suggested incomplete recessivity in one cross and incomplete dominance in the other. The findings illustrate the complexity involved in investigating anthelmintic resistance and that inherent inter- or intra-isolate differences may both play a role in the phenotypic expression of BZ resistance. Work on Caenorhabditis elegans has reported that genetically identical individuals can have differing phenotypic responses potentially due to heterogeneity in gene expression (Viney and Diaz, 2012). Additionally the process by which resistance is selected (e.g under or suboptimal dosing compared to frequent dosing) may influence the phenotypic outcome (Sargison, 2016). Studies on H. contortus from farms in USA found that the L198V variant of isotype 2 correlated to higher EC50 estimates of benzimidazole resistance than that conferred by the F200Y variant alone (Doyle et al., 2021).
The individual and cummaltive FWECs/outputs were significantly higher in the MHco18/3 isolate compared to MHco3/18 at all stages of selection, albeit with reduced impact at each subsequent generation. The findings highlight that phenotype and factors such as parasite fitness and plasticity may be interlinked. A similar finding with increased faecal worm egg outputs and reduced time to patency was observed in T. circumcincta isolates that were selected for monepantel resistance (Bartley et al., 2015). In general, the MHco18/3 genetic cross filial generations had higher ED50 estimates compared to the MHco3/18 filial generations, indicating greater phenotypic resistance in vitro. The nine-fold difference observed in ED50 estimates between the F1 generations with BZ resistant (MHco18/3) or susceptible (MHco3/18) female parents suggested that there was some positive influence on the resistance phenotype coming from the dam of the cross. The topic of matroclinous influence on in vitro expression of BZ resistance has been previously investigated using genetic cross studies, with equivocal findings. Sangster et al. (1998) reported little to no influence; the larval development test used in their study showed that the LC50 estimates of both of the F1 generations of the reciprocal genetic crosses were around three times greater than those of the susceptible parental isolate and it was noted that this difference was lost in the F2 generations. On the other hand, Le Jambre et al. (1979) reported that the progeny of resistant females crossed with non-resistant males had 2.2x higher EHT ED50 estimates than the progeny of the reciprocal cross, and that this was maintained through subsequent generations. These results led Le Jambre and colleagues to suggest in 1979 that there was an element of cytoplasmic/extra nuclear factor inheritance, also known as cytoplasmic inheritance, involved in BZ resistance rather than solely the traditional nuclear inheritance. A maternal or cytoplasmic effect has been proposed as a mechanism for inheritance of resistance to the macrocyclic lactone anthelmintic, abamectin, in the carmine spider mite Tetranychus cinnabarinus (He et al., 2009), but studies looking at macrocyclic lactone resistance in nematodes found no non-chromosomal influence (Le Jambre et al., 1999). Differences in phenotypic responses have also been observed at different developmental stages (Le Jambre et al., 1979; Kotze 1997), suggesting that the importance of different mechanisms may be at play throughout the life of the nematode, the crosses were undertaken through the transfer of juvenile worms whereas the egg hatch test is looking at egg to L1 development. The involvement of other non-specific mechanisms in the expression of BZ resistance has been proposed, including ABC transporters (Kerboeuf et al., 1999), cytochrome P450 enzymes (reviewed by Matouskova et al., 2016), and microRNAs (Devaney et al., 2010).
Assessment of the β-tubulin associated SNPs of each of the filial generations provided interesting findings. The F1 progeny of both reciprocal crosses show the expected genotypes associated with successful crossing, but these do not account for the greatly different EHT ED50 phenotype results. Genotyping individuals from each population offered an advantage over sequencing of pooled populations of being able to report actual genotypes and resultant combinations for each SNP. Consequently this allows us to present the first case of a double homozygous resistant genotype at both codons 167 and 200. This genotype combination was found in both the MHco18(UGA2004) parent L3 and in the MHco18/3.F3.BZ L3 population. The MiSeq assay results also intimated that this combination was also observed in pools of larvae. This has not been found previously despite the large number of genotyping studies conducted on Trichostrongylid nematodes (Mottier and Prichard, 2008; Hodgkinson et al., 2008; Barrere et al., 2012; Kotze et al., 2012; Redman et al., 2015; Atanásio-Nhacumbe et al., 2019); it has been considered that having a combination of two homozygous resistant genotypes in the β-tubulin isotype 1 gene would be lethal (Mottier and Prichard, 2008) and that even in the heterozyote, the resistant alleles are never on the same strand, i.e. the variants are always in trans, not in cis. In this study, these double homozygous resistant genotypes have only been reported in the L3 stage of the parasite and it has never been looked at in adults to see if these individuals can undergo normal development in the host and be sexually reproductive. The fact they have hatched from eggs and developed to L3 shows that the mutation is not lethal to this stage of development.
It has been reported by Barrere et al. (2012) that a heterozygous genotype at both F167Y and F200Y confers a resistant phenotype, capable of surviving three times the recommended dose rate of albendazole. In this study, the main difference between the two crosses in the F2 and F3 generations was that the MHco18/3 had higher resistance allele frequencies at F167Y, which was most noticeable after BZ treatment of the F3 population. In the resistant parent isolate 57.6% of the genotypes were either heterozygous or homozygous resistant at codon 167. Perhaps the F167Y in the MHco18(UGA2004) parental isolate is important in conferring a resistant phenotype. MHco18/3.F1 had 25.0% SR-SR genotype, whereas MHco3/18.F1 had 14.8%. The other genotype combinations for the F1 populations were heterozygous at either SNP position, potentially conferring a more susceptible phenotype. The other filial generations had similar levels of double heterozygotes to each other.
Previous work carried out by Sargison et al. (2019a) investigating mating barriers between different H. contortus isolates suggested that MHco3(ISE) females were more likely to produce progeny from matings with their own isolate when co-infected with two other genetically/geographically different isolates. However, it was noted that females from the two other H. contortus isolates did not show this attribute towards MHco3(ISE), and freely mated between the two isolates with the co-infections being tested. This study design did not allow for isolate choices in mating, but it is possible that sub-populations with particular morphological features influenced the mating behaviour, resulting in an apparent sex-linked difference that manifested in the resistance phenotype. It was not possible to investigate this within the present study. As mentioned above, the possibility of other non-specific or extra-nuclear mechanisms of resistance being involved can not be precluded and requires further investigation.
5.1. Conclusions
This is the first trichostrongyle gastrointestinal nematode genetic crossing study where individual genotypes at the β-tubulin isotype 1 gene were investigated alongside phenotypic indices. The apparent less than perfect correlation between phenotype and genotype demonstrated that their relationship is complex and that multiple genes/mechanisms may be involved in BZ resistance and that β-tubulin only explains part of the phenotypic variance, and/or that the phenotypic tools used for assessing ED50 are too insensitive to correlate with what is observed genotypically. This study has confirmed previous studies’ findings in that the inheritance of BZ resistance is influenced by maternal and/or cytoplasmic mechanisms. The work has for the first time demonstrated that, albeit extremely rare, double homozygous resistant genotypes at positions 167 and 200 on the β-tubulin isotype 1 gene are viable and do not preclude the development from egg to infective larvae stage and further work investigating the potential for the further development of these individual L3 to progress to fertile adults is required to assess whether it is a unique characteristic for the MHco18(UGA2004) isolate.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
We gratefully acknowledge funding from The Scottish Government's Rural and Environment Science and Analytical Services Division (RESAS) Strategic Research Programme 2016-2021 and BBSRC (BB/M003949/1 -The BUG consortium Building Upon the Genome). We are grateful to the Bioservices Division, Moredun Research Institute, for expert care and assistance with animals. For the purpose of Open Access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpddr.2021.11.001.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Ali Q., Rashid I., Shabbir M.Z., Shahzad K., Ashraf K., Sargison N.D., Chaudhry U. Population genetics of benzimidazole-resistant Haemonchus contortus and Haemonchus placei from buffalo and cattle: implications for the emergence and spread of resistance mutations. Parasitol. Res. 2018;117:3575–3583. doi: 10.1007/s00436-018-6055-8. [DOI] [PubMed] [Google Scholar]
- Atanásio-Nhacumbe A., Mota Lambert S., Maria Paraná da Silva Souza B., Consuêlo Carybé Ayres M. Molecular detection of benzimidazole resistance levels associated with F167Y and F200Y polymorphisms in Haemonchus contortus of goats from Mozambique. Parasitol. Res. 2019;118:245–253. doi: 10.1007/s00436-018-6162-6. [DOI] [PubMed] [Google Scholar]
- Barrere V., Alvarez L., Suarez G., Ceballos L., Moreno L., Lanusse C., Prichard R. Relationship between increased albendazole systemic exposure and changes in single nucleotide polymorphisms on the B-tubulin isotype 1 encoding gene in Haemonchus contortus. Vet. Parasitol. 2012;186:344–349. doi: 10.1016/j.vetpar.2011.11.068. [DOI] [PubMed] [Google Scholar]
- Bartley D.J., Devin L., Nath M., Morrison A.A. Selection and characterisation of monepantel resistance in Teladorsagia circumcincta isolates. Internet J. Parasit. Dis. 2015;5(2):69–76. doi: 10.1016/j.ijpddr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry U., Redman E.M., Raman M., Gilleard J.S. Genetic evidence for the spread of a benzimidazole resistance mutation across southern India from a single origin in the parasitic nematode Haemonchus contortus. Int. J. Parasitol. 2015;45(11):721–728. doi: 10.1016/j.ijpara.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Coop R.L., Sykes A.R., Angus K.W. The effect of three levels of intake of Ostertagia circumcincta larvae on growth rate, food intake and body composition of growing lambs. J. Agri Sci. Camb. 1982;98:247–255. [Google Scholar]
- Devaney E., Winter A.D., Britton C. MicroRNAs: a role in drug resistance in parasitic nematodes? Trends Parasitol. 2010;26(9):428–433. doi: 10.1016/j.pt.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drudge J.H., Szanto J., Wyant Z.N., Elam G. Field studies on parasite control in sheep: comparison of thiabendazole, ruelene, and phenothiazine. Am. J. Vet. Res. 1964;25:1512–1518. [PubMed] [Google Scholar]
- Doyle S.R., Illingworth C.J.R., Laing R., Bartley D.J., Redman E., Martinelli A., Holroyd N., Morrison A.A., Rezansoff A., Tracey A., Devaney E., Berriman M., Sargison N., Cotton J.A., Gilleard J.S. Population genomic and evolutionary modelling analyses reveal a single major QTL for ivermectin drug resistance in the pathogenic nematode, Haemonchus contortus. BMC Genom. 2019;20:218. doi: 10.1186/s12864-019-5592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle S.R., Tracey A., Laing R., Holroyd N., Bartley D., Bazant W., Beasley H., Beech R., Britton C., Brooks K., Chaudhry U. Genomic and transcriptomic variation defines the chromosome-scale assembly of Haemonchus contortus, a model gastrointestinal worm. Comm. Biol. 2020;3(1):1–16. doi: 10.1038/s42003-020-01377-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle S.R., Laing R., Bartley D., Morrison A., Holroyd N., Maitland K., Antonopoulos A., Chaudhry U., Kaplan R., Sargison N., Britton C. Genomic landscape of drug response reveals novel mediators of anthelmintic resistance. Preprint from bioRxiv. 2021 doi: 10.1101/2021.11.12.465712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L., Laval G., Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick J.L. Global food security: the impact of veterinary parasites and parasitologists. Vet. Parasitol. 2013;195(3–4):233–248. doi: 10.1016/j.vetpar.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Ghisi M., Kaminsky R., Mäser P. Phenotyping and genotyping of Haemonchus contortus isolates reveals a new putative candidate mutation for benzimidazole resistance in nematodes. Vet. Parasitol. 2007;144(3–4):313–320. doi: 10.1016/j.vetpar.2006.10.003. [DOI] [PubMed] [Google Scholar]
- He L., Gao X., Wang J., Zhao Z., Liu N. Genetic analysis of abamectin resistance in Tetranychus cinnabarinus. Pestic. Biochem. Physiol. 2009;95(3):147–151. [Google Scholar]
- Hendrick P.W. second ed. Jones and Bartlett; Boston: 2000. Genetics of Populations. [Google Scholar]
- Herlich H., Rew R.S., Colglazier M.L. Inheritance of cambendazole resistance in Haemonchus contortus. Am. J. Vet. Res. 1981;42:1342–1344. [PubMed] [Google Scholar]
- Hodgkinson J.E., Clark H.J., Kaplan R.M., Lake S.L., Matthews J.B. The role of polymorphisms at β tubulin isotype 1 codons 167 and 200 in benzimidazole resistance in cyathostomins. Int. J. Parasitol. 2008;38:1149–1160. doi: 10.1016/j.ijpara.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Hunt Kotze AC., Knox M.R., Anderson L.J., McNally J., Lf L.J. The use of DNA markers to map anthelmintic resistance loci in an intraspecific cross of Haemonchus contortus. Parasitology. 2010;137:705–717. doi: 10.1017/S0031182009991521. [DOI] [PubMed] [Google Scholar]
- Jackson F. New technique for obtaining nematode ova from sheep faeces. Lab. Pract. 1974;23(2):65–66. [PubMed] [Google Scholar]
- Kaplan R.M. Drug resistance in nematodes of veterinary importance: a status report. Trends Parasitol. 2004;20:477–481. doi: 10.1016/j.pt.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerboeuf D., Chambrier P., Le Vern Y., Aycardi J. Flow cytometry analysis of drug transport mechanisms in Haemonchus contortus susceptible or resistant to anthelmintics. Parasitol. Res. 1999;85(2):118–123. doi: 10.1007/s004360050519. [DOI] [PubMed] [Google Scholar]
- Kochapakdee S., Pandey V.S., Pralomkarm W., Choldumrongkul S., Ngampongsai W., Lawpetchara A. Anthelmintic resistance in goats in southern Thailand. Vet. Rec. 1995:124–125. doi: 10.1136/vr.137.5.124. [DOI] [PubMed] [Google Scholar]
- Kotze A.C. Cytochrome P450 monooxygenase activity in Haemonchus contortus (Nematoda) Int. J. Parasitol. 1997;27:33–40. doi: 10.1016/s0020-7519(96)00161-0. [DOI] [PubMed] [Google Scholar]
- Kotze A.C., Cowling K., Baghall N.H., Hines B.M., Ruffell A.P., Hunt P.W., Coleman G.T. Relative level of thaibendazole resistance associated with the E198A and F200Y SNPs in larvae of a multidrug resistant isolate of Haemonchus contortus. Internet J. Parasit. Dis. 2012;2:92–97. doi: 10.1016/j.ijpddr.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotze A.C., Hunt P.W., Skuce P., von Samson-Himmelstjerna G., Martin R.J., Sager H., Krücken J., Hodgkinson J., Lespine A., Jex A.R., Gilleard J.S., Beech R.N., Wolstenholme A.J., Demeler J., Robertson A.P., Charvet C.L., Neveum C., Kaminsky R., Prichard R.K. Recent advances in candidate-gene and whole-genome approaches to the discovery of anthelmintic resistance markers and the description of drug/receptor interactions. Internet J. Parasit. Dis. 2014;4(3):164–184. doi: 10.1016/j.ijpddr.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwa M.S., Veenstra J.G., Roos M.H. Benzimidazole resistance in Haemonchus contortus is correlated with a conserved mutation at amino acid 200 in beta-tubulin isotype 1. Mol. Biochem. Parasitol. 1994;63:299–303. doi: 10.1016/0166-6851(94)90066-3. [DOI] [PubMed] [Google Scholar]
- Kwa M.S., Veenstra J.G., Van Dijk M., Roos M.H. Beta-tubulin genes from the parasitic nematode Haemonchus contortus modulate drug resistance in Caenorhabditis elegans. J. Mol. Biol. 1995;246:500–510. doi: 10.1006/jmbi.1994.0102. [DOI] [PubMed] [Google Scholar]
- Le Jambre L.F., Royal W.M., Martin P.J. The inheritance of thiabendazole resistance in Haemonchus contortus. Parasitology. 1979;78:107–119. doi: 10.1017/s0031182000049179. [DOI] [PubMed] [Google Scholar]
- Le Jambre L.F., Ian J Lenane I.J., Wardrop A.J. A hybridisation technique to identify anthelmintic resistance genes in Haemonchus. Int. J. Parasitol. 1999;29(12):1979–1985. doi: 10.1016/s0020-7519(99)00157-5. [DOI] [PubMed] [Google Scholar]
- Martin P.J., McKenzie J.A., Stone R.A. The inheritance of thiabendazole resistance in Trichostrongylus colubrformis. Int. J. Parasitol. 1988;18:703–709. doi: 10.1016/0020-7519(88)90109-9. [DOI] [PubMed] [Google Scholar]
- Martínez-Valladares M., Valderas-García E., Gandasegui J., Skuce P., Morrison A., de Agüero V.C.G., Cambra-Pellejà M., Balaña-Fouce R., Rojo-Vázquez F.A. Teladorsagia circumcincta beta tubulin: the presence of the E198L polymorphism on its own is associated with benzimidazole resistance. Parasites Vectors. 2020;13(1):1–12. doi: 10.1186/s13071-020-04320-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouskova P., Vokral I., Lamka J., Skalova L. The role of xenobiotic-metabolizing enzymes in anthelmintic deactivation and resistance in helminths. Trends Parasitol. 2016;32:481–491. doi: 10.1016/j.pt.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Mohammedsalih K.M., Krücken J., Khalafalla A., Bashar A., Juma F.R., Abakar A., Abdalmalaik A.A.H., Coles G., von Samson-Himmelstjerna G. New codon 198 β-tubulin polymorphisms in highly benzimidazole resistant Haemonchus contortus from goats in three different states in Sudan. Parasites Vectors. 2020;13:114. doi: 10.1186/s13071-020-3978-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottier M.L., Roger K., Prichard R.K. Genetic analysis of a relationship between macrocyclic lactone and benzimidazole anthelmintic selection on Haemonchus contortus. Pharmacogenetics Genom. 2008;18:129–140. doi: 10.1097/FPC.0b013e3282f4711d. [DOI] [PubMed] [Google Scholar]
- Otsen M., Plas M.E., Groeneveld J., Roos M.H., Lenstra J.A., Hoekstra R. Genetic markers for the parasitic nematode Haemonchus contortus based on intron sequences. Exp. Parasitol. 2000;95:226–229. doi: 10.1006/expr.2000.4532. [DOI] [PubMed] [Google Scholar]
- Peakall R., Smouse P.E. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research--an update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichard R.K. Genetic variability following selection of Haemonchus contortus with anthelmintics. Trends Parasitol. 2001;17(9):445–453. doi: 10.1016/s1471-4922(01)01983-3. [DOI] [PubMed] [Google Scholar]
- Redman E., Packard E., Grillo V., Smith J., Jackson F., Gilleard J.S. Microsatellite analysis reveals marked genetic differentiation between Haemonchus contortus laboratory isolates and provides a rapid system of genetic fingerprinting. Int. J. Parasitol. 2008;38:111–122. doi: 10.1016/j.ijpara.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Redman E., Whitelaw F., Tait A., Burgess C., Bartley Y., Skuce P., Jackson F., Gilleard J. The emergence of resistance to the benzimidazole anthlemintics in parasitic nematodes of livestock is characterised by multiple independent hard and soft selective sweeps. PLoS Neglected Trop. Dis. 2015;6(9) doi: 10.1371/journal.pntd.0003494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos M.H., Boersema J.H., Borgsteede F.H., Cornelissen J., Taylor M., Ruitenberg E.J. Molecular analysis of selection for benzimidazole resistance in the sheep parasite Haemonchus contortus. Mol. Biochem. Parasitol. 1990;43:77–88. doi: 10.1016/0166-6851(90)90132-6. [DOI] [PubMed] [Google Scholar]
- Roos M.H., Otsenb M., Hoekstraa R., Veenstrab J.G., Lenstra J.A. Genetic analysis of inbreeding of two strains of the parasitic nematode Haemonchus contortus. Int. J. Parasitol. 2004;34:109–115. doi: 10.1016/j.ijpara.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Sangster N.C., Redwin J.M., Bjorn H. Inheritance of levamisole and benzimidazole resistance in an isolate of Haemonchus contortus. Int. J. Parasitol. 1998;28:503–510. doi: 10.1016/s0020-7519(97)00194-x. [DOI] [PubMed] [Google Scholar]
- Sargison N.D. Keys to solving health problems in small ruminants: anthelmintic resistance as a threat to sustainable nematode control. Small Rumin. Res. 2016;142:11–15. [Google Scholar]
- Sargison N.D., Redman E., Morrison A.A., Bartley D.J., Jackson F., Hoberg E., Gilleard J.S. Mating barriers between genetically divergent strains of the parasitic nematode Haemonchus contortus suggest incipient speciation. Int. J. Parasitol. 2019;49:531–540. doi: 10.1016/j.ijpara.2019.02.008. [DOI] [PubMed] [Google Scholar]
- Sargison N.D., MacLeay M., Morrison A.A., Bartley D.J., Evans M., Chaudhry U. Development of amplicon sequencing for the analysis of benzimidazole resistance allele frequencies in field populations of gastrointestinal nematodes. Internet J. Parasit. Dis. 2019;10:92–100. doi: 10.1016/j.ijpddr.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J., Sahl J.W., Stres B., Thallinger G.G., Van Horn D.J., Weber C.F. Introducing mothur: opensource, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestre A., Cabaret J. Mutation in position 167 of isotype 1 b-tubulin gene of Trichostrongylid nematodes: role in benzimidazole resistance? Mol. Biochem. Parasitol. 2002;120:297–300. doi: 10.1016/s0166-6851(01)00455-8. [DOI] [PubMed] [Google Scholar]
- Stone B.F. A formula for determining degree of dominance in cases of monofactorial inheritance of resistance to chemicals. Bull. World Health Organ. 1968;38(2):325–326. [PMC free article] [PubMed] [Google Scholar]
- Viney M., Diaz A. Phenotypic plasticity in nematodes: evolutionary and ecological significance. Worm. 2012;1(2):98–106. doi: 10.4161/worm.21086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Samson-Himmelstjerna G., Blackhall W.J., McCarthy J.S., Skuce P.J. Single nucleotide polymorphism (SNP) markers for benzimidazole resistance in veterinary nematodes. Parasitology. 2007;134:1077–1086. doi: 10.1017/S0031182007000054. [DOI] [PubMed] [Google Scholar]
- von Samson-Himmelstjerna G., Coles G.C., Jackson F., Bauer C., Borgsteede F., Cirak V.Y., Demeler J., Donnan A., Dorny P., Epe C., Harder A., Hoglund J., Kaminsky R., Kerboeuf D., Kuttler U., Papadopoulos E., Posedi J., Small J., Varady M., Vercruysse J., Wirtherle N. Standardization of the egg hatch test for the detection of benzimidazole resistance in parasitic nematodes. Parasitol. Res. 2009;105:825–834. doi: 10.1007/s00436-009-1466-1. [DOI] [PubMed] [Google Scholar]
- von Samson-Himmelstjerna G., Walsh T.K., Donnan A.A., Carriere S., Jackson F., Skuce P.J., Rohn K., Wolstenholme A.J. Molecular detection of benzimidazole resistance in Haemonchus contortus using real-time PCR and pyrosequencing. Parasitology. 2009;136:349–358. doi: 10.1017/S003118200800543X. [DOI] [PubMed] [Google Scholar]
- Williamson S.M., Storey B., Howell S., Harper K.M., Kaplan R.M., Wolstenholme A.J. Candidate anthelmintic resistance-associated gene expression and sequence polymorphisms in a triple-resistant field isolate of Haemonchus contortus. Mol. Biochem. Parasitol. 2011;180:99–105. doi: 10.1016/j.molbiopara.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Wright S. vol. 4. The University of Chiacago Press; 1978. (Evolution and the Genetics of Populations. Variabilty within and Among Natural Populations). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.