Abstract
The cap-binding translation initiation factor eukaryotic initiation factor 4E (eIF4E) is phosphorylated in vivo at Ser209 in response to a variety of stimuli. In this paper, we show that the mitogen-activated protein kinase (MAPK) signal-integrating kinase Mnk2 phosphorylates eIF4E at this residue. Mnk2 binds to the scaffolding protein eIF4G, and overexpression of Mnk2 results in increased phosphorylation of endogenous eIF4E, showing that it can act as an eIF4E kinase in vivo. We have identified eight phosphorylation sites in Mnk2, of which at least three potential MAPK sites are likely to be essential for Mnk2 activity. In contrast to that of Mnk1, the activity of overexpressed Mnk2 is high under control conditions and could only be reduced substantially by a combination of PD98059 and SB203580, while the activity of endogenous Mnk2 in Swiss 3T3 cells was hardly affected upon treatment with these inhibitors. These compounds did not abolish phosphorylation of eIF4E, implying that Mnk2 may mediate phosphorylation of eIF4E in Swiss 3T3 cells. In vitro phosphorylation studies show that Mnk2 is a significantly better substrate than Mnk1 for extracellular signal-regulated kinase 2 (ERK2), p38MAPKα, and p38MAPKβ. Therefore, the high levels of activity of Mnk2 under several conditions may be explained by efficient activation of Mnk2 by low levels of activity of the upstream kinases. Interestingly, we found that the association of both Mnk1 and Mnk2 with eIF4G increased upon inhibition of the MAPK pathways while activation of ERK resulted in decreased binding to eIF4G. This might reflect a mechanism to ensure rapid, but transient, phosphorylation of eIF4E upon stimulation of the MAPK pathways.
All eukaryotic cellular cytoplasmic mRNAs contain an m7GpppG cap structure at the 5′ end that is specifically bound by eukaryotic initiation factor 4E (eIF4E). Subsequent binding of eIF4G and eIF4A to this factor leads to the formation of the eIF4F complex. The N terminus of eIF4G harbors the binding sites for eIF4E and the poly(A)-binding protein (12, 19), while the C terminus binds to eIF3, eIF4A, and Mnk1 (discussed below) (19, 33). Association of all of these factors and the small ribosomal subunit brings together the components of the 48S preinitiation complex.
The importance of tight regulation of the activity of factors involved in cap binding has been shown by experiments with cells overexpressing such factors (4, 7, 21, 22). Over expression of eIF4E led to malignant transformation of cells and conferred on them the ability to grow in soft agar. The exact mechanism by which this occurs remains unclear but is thought to involve the increased expression of various growth-stimulating factors. Many such factors are encoded by mRNAs that have highly structured 5′ untranslated regions (18), and increased levels of eIF4E activity are thought to enhance especially the translation of such mRNAs (17, 24). It is particularly significant that a rapidly growing amount of literature indicates a role for increased levels of eIF4E in naturally occurring human cancers (3, 15, 23, 30, 31).
eIF4E undergoes phosphorylation at Ser209, but the exact role of this phosphorylation is still unclear. In 48S preinitiation complexes, most of the eIF4E is phosphorylated, implying that the phosphorylation step is required for ongoing initiation (14, 20). Based on the crystal structure, it is thought that phosphorylation at Ser209 may lead to an interaction between the negatively charged phosphate group and the positive side chain of Lys159, on opposite sides of the mRNA-binding cleft in eIF4E (25). This could stabilize the interaction between the cap structure and eIF4E, but direct evidence for this is lacking. One report has, indeed, claimed that phosphorylated eIF4E has a higher affinity for cap structures than does its unphosphorylated form (26).
The recently discovered enzyme mitogen-activated protein kinase (MAPK) signal-integrating kinase 1 (Mnk1) is now considered to be one of the main kinases that phosphorylate eIF4E in vivo (recently reviewed in reference 32). Mnk1 phosphorylates eIF4E specifically and only at Ser209 (38), the site that is phosphorylated in vivo (6, 13). Mnk1 is activated by both the classical extracellular signal-regulated kinase (ERK) pathway and the stress- and cytokine-activated p38MAPK pathway (8, 38, 40), and the involvement of the ERK and p38MAPK pathways in the phosphorylation of eIF4E has been demonstrated in vivo (5, 28, 37). Furthermore, a requirement for eIF4E-eIF4G binding for eIF4E phosphorylation in vivo has been reported, again strongly supporting the idea that Mnk1 is a physiological eIF4E kinase (33).
Mnk2 was cloned simultaneously with Mnk1 (38) but has not previously been studied in detail. We show here that Mnk2 phosphorylates eIF4E in vitro and in vivo, that it also binds to eIF4G, and that it phosphorylates eIF4E at Ser209. Mnk2 is phosphorylated and activated by ERK and p38MAPKα and -β in vitro but not by two other forms of p38MAPK or by JNK. Several in vitro and in vivo phosphorylation sites in Mnk2 have been identified, and their importance for the function of Mnk2 was studied by expressing phosphorylation site mutant proteins in transfected 293 cells. We show that Mnk2 has a high basal activity in vivo that is unaffected by stimulation of the ERK pathway with phorbol ester. However, treatment of cells with inhibitors of the ERK and p38MAPK signaling pathways reduced the activity of over expressed Mnk2, showing that these pathways can affect this kinase in vivo. Mnk2 appears to be an eIF4E kinase with a high basal level of activity that requires only low levels of ERK or p38MAPK activity to attain this activity. Binding of eIF4G to Mnk1 and Mnk2 was reduced upon stimulation of cells with phorbol ester. This suggests that not only the activity of the Mnks but also their interaction with eIF4G may control the level of phosphorylation of eIF4E.
MATERIALS AND METHODS
Plasmids.
pEBG-Mnk1 has been previously described (38). pEBG-Mnk2 (a gift from A. Waskiewicz, Seattle, Wash.), pCS3MT-Mnk2, and pGEX3X-Mnk2 were created by cloning the Mnk2 coding sequence with adjacent Bc1I or EcoRI sites, created by PCR, respectively, into pEBG, pCS3MT, and pGEX3X. The various mutants were made using the Stratagene Quickchange kit with pCS3MT-Mnk2 as template DNA. pGEX3X-Mnk2T197A and pGEX3X-Mnk2T202A were made by replacing the wild-type Mnk2 sequence in pGEX3X-Mnk2 with the mutated sequence from pCS3MT-Mnk2T197A or T202A as an EcoRI fragment.
Cell culture and transfections.
Human embryonic kidney (HEK) 293 and Swiss 3T3 cells were grown in 10-cm-diameter plates in DMEM; Dulbecco modified Eagle medium (Gibco BRL) supplemented with 10% fetal bovine serum (Gibco BRL). Transient transfections were carried out by calcium phosphate precipitation of the DNA in HEPES-buffered saline (10). PD98059 and SB203580 (both from Calbiochem) were used at concentrations of 50 and 10 μM, respectively.
Cell harvesting.
After treatments, cells were washed once with phosphate-buffered saline and harvested in 400 μl of harvesting buffer (20 mM HEPES · KOH [pH 7.5], 50 mM β-glycerophosphate, 0.2 mM EDTA, 10% glycerol, 1% Triton X-100, 1 mM dithiothreitol, 0.5 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride: 1 mM benzamidine, 1 μg of leupeptin per ml, 1 μg of antipain per ml, 1 μg of pepstatin per ml). Cell debris and nuclei were spun for 1 min at 12,000 × g, and the supernatant was transferred to new tubes.
GST pulldowns, immunoprecipitation, and m7GTP-Sepharose chromatography.
For glutathione S-transferase (GST) pulldowns, glutathione-Sepharose beads (Pharmacia) were washed in wash buffer (harvesting buffer without Triton X-100). A 15-μl volume of packed beads and cell extracts was incubated at 4°C for at least 1 h and washed afterward three times with 500 μl of wash buffer. For anti-myc immunoprecipitations, the antibody (9E10 from Sigma) was bound to protein G beads (20 μl of packed beads per immunoprecipitation). After incubation for 1 h at 4°C in wash buffer, the beads were washed three times in 500 μl of wash buffer and subsequently added to the cell extracts. Binding of proteins to the antibodies was carried out for at least 2 h at 4°C. The beads were subsequently washed as described for the GST pulldowns. Mnk1 and Mnk2 antibodies were raised in sheep against the peptides NELAEEQEALAEGLC (Mnk1 residues 365 to 379) and DAGQDQPVVIRATSRC (Mnk2 residues 365 to 380). The antibodies, which do not cross-react, were purified and used for immunoprecipitation of Mnk1 and Mnk2 from Swiss 3T3 cells. Four milligrams of total protein was used per immunoprecipitation. Antibodies were bound to protein G-Sepharose and subsequently incubated with the cell extracts for 2 h at 4°C. The activity of the bound kinases was tested as described below.
Expression and purification of recombinant proteins.
GST fusion proteins of Mnk1 and Mnk2 were expressed from pGEX3X plasmids in Escherichia coli BL21 DE3 as previously described (38). Human eIF4E was expressed from a pET11d plasmid in E. coli BL21 DE3 and purified as described previously (36).
Kinase assays.
Five-microliter aliquots of the beads with bound GST or myc fusion proteins, obtained as described above, were incubated in a total volume of 30 μl in 20 mM HEPES · KOH (pH 7.5)–50 mM KCl–2 mM MgCl2–200 μM ATP–1 μCi of [γ-32P]ATP for 1 h at 30°C. In eIF4E phosphorylation studies, 100 ng of recombinant eIF4E was added to the assay mixtures. The reaction was stopped by adding sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and heating the sample for 5 min at 95°C. Samples were analyzed by SDS-PAGE and autoradiography.
Isoelectric focusing.
Endogenous eIF4E was purified by m7GTP-Sepharose chromatography, and isoelectric focusing was performed as previously described (16). Phosphorylated and unphosphorylated forms of eIF4E were detected by Western blotting.
Phosphorylation site mapping in vitro.
GST-Mnk2 was expressed in E. coli as already described, and 3 μg of the purified protein was phosphorylated with activated ERK (30 U/ml against myelin basic protein [MBP]) in the presence of 10 μCi of [γ-32P]ATP in the same kinase buffer as described above. The reaction was stopped by adding 1% SDS–1% β-mercaptoethanol and heating for 5 min at 95°C. Cysteines were then alkylated with 4-vinylpyridine for 45 min at 30°C, and the proteins were separated by SDS-PAGE. Radioactively labeled GST-Mnk2 was excised from the gel, and in-gel trypsin (sequencing grade; Roche) digestion was performed overnight at 30°C in 20 mM ammonium bicarbonate–0.1% n-octyl glucopyranoside. Gel pieces were removed by using Spin-X columns (Costar), and the remaining SDS was precipitated with guanidine hydrochloride. The samples were acidified by adding 3 volumes of 0.1% trifluoroacetic acid (TFA), and peptides were purified by high-pressure liquid chromatography (HPLC; Gilson) on a Vydac C18 reversed-phase column (250 by 4.6 mm [inside diameter]) with 0.1% TFA in water as the starting buffer (buffer A) and 0.1% TFA in acetonitrile as buffer B. Fractions containing radioactivity were collected and analyzed by matrix-assisted laser desorption ionization–time of flight mass spectrometry and solid-phase sequencing.
Phosphorylation site mapping in vivo.
HEK 293 cells were transfected with pEBG-Mnk2. After 40 h, the cells were washed once with phosphate-free DMEM (Gibco BRL) and then grown in phosphate-free DMEM in the presence of 2 mCi of [32P]orthophosphate for 4 h. The cells were harvested, and GST-Mnk2 was purified on glutathione-Sepharose. After treatment with 4-vinylpyridine, radioactively labeled GST-Mnk2 was subsequently treated as described for the in vitro site mapping.
RESULTS
Mnk2 phosphorylates eIF4E in vitro at the physiological phosphorylation site, Ser209.
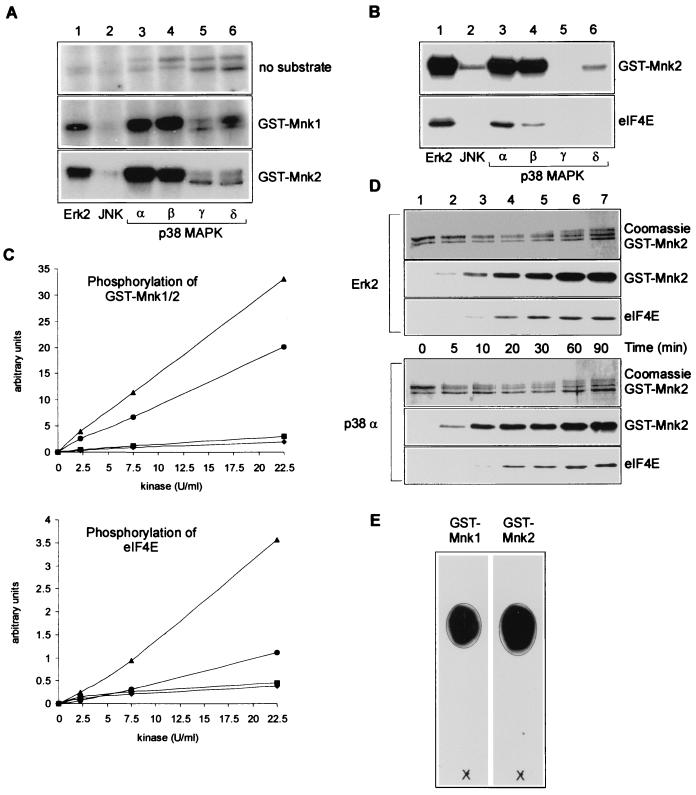
Mnk2 was identified and cloned simultaneously with Mnk1, but its function and regulation have not previously been characterized. In particular, it was not known whether Mnk2 could phosphorylate eIF4E and how its activity was controlled. First, to establish which upstream kinases phosphorylate Mnk1 and Mnk2, recombinant and activated ERK2, JNK, and the four isoforms of p38MAPK were each tested for the ability to phosphorylate Mnk1 and Mnk2 expressed in E. coli (Fig. 1A). The activated kinases were each used at the same activity (tested with myelin basic protein as the substrate). GST-Mnk1 and GST-Mnk2 were each phosphorylated by activated ERK2 and the α and β forms of p38MAPK (Fig. 1A, lanes 1, 3, and 4). Shorter exposure times of the autoradiograms for the phosphorylation of GST-Mnk2 sufficed to get the same intensity of the bands as seen for phosphorylation of GST-Mnk1, suggesting that Mnk2 is a better substrate for the upstream kinases (see below). This is probably not due to possible differences in the proper folding of the recombinant proteins, as the GST-tagged proteins were expressed and purified simultaneously. Depending on the experiment, a very low and variable efficiency of phosphorylation of Mnk1 or Mnk2 by the SB203580-insensitive forms of p38MAPK (γ and δ) was found (Fig. 1A, lanes 5 and 6, and B, lane 6). However, in comparison to ERK2 or the α and β forms of p38MAPK, neither these two kinases nor JNK significantly phosphorylated GST-Mnk1 or GST-Mnk2. The radiolabeled bands in the top section of Fig. 1A are due to autophosphorylation of the MAPKs being tested.
FIG. 1.
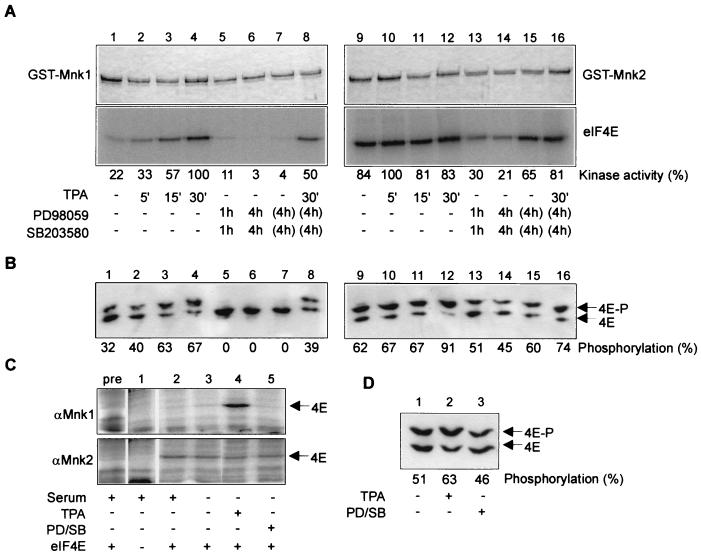
In vitro phosphorylation and activation of Mnk2. (A) Mnk1 and Mnk2 are phosphorylated by ERK and the α and β forms of p38MAPK. GST-MnK1 and GST-Mnk2 were expressed in E. coli, purified, and used in the in vitro kinase assays described in Materials and Methods. Bacterially expressed and in vitro-activated MAPKs were added as indicated below the lanes (all at a concentration of 0.4 U/ml against MBP). Phosphorylation of the GST fusion proteins was analyzed by SDS-PAGE and autoradiography. For the part showing phosphorylation of GST-Mnk2, a 6-h exposure of the autoradiogram is shown, while for the two other parts, a 24-h exposure is shown. (B) Phosphorylation of Mnk2 leads to its activation. Recombinant eIF4E was included in each incubation described for panel A to assess the activity of the kinase. Samples were analyzed by SDS-PAGE and autoradiography. (C) Mnk2 is a better substrate for ERK2 and p38MAPKβ than is Mnk1. Recombinant GST-Mnk1 or GST-Mnk2 was incubated with increasing amounts of recombinant activated ERK2 (Mnk1, diamonds; Mnk2, triangles) or p38MAPKβ (Mnk1, squares; Mnk2, circles). After 20 min, recombinant eIF4E was added, the mixture was incubated for an additional 10 min, and the samples were analyzed after SDS-PAGE using a Fuji Phosphor-Imager and Aida2000 software. The upper graph shows the results for phosphorylation of GST-Mnk1 or GST-Mnk2, and the lower graph represents phosphorylation of eIF4E. (D) Time course of phosphorylation of Mnk2 and eIF4E. GST-Mnk2 was incubated with activated ERK2 or p38MAPKα as described above. Aliquots were taken at the indicated time points. Phosphorylation of GST-Mnk2 and eIF4E was subsequently analyzed by SDS-PAGE and autoradiography. The top portion of each part shows the Coomassie staining of GST-Mnk2. The middle portion shows the autoradiogram of GST-Mnk2, and the lowest portion of each part shows the autoradiogram of phosphorylated eIF4E. (E) Mnk2 phosphorylates eIF4E at Ser209. GST-Mnk1 and GST-Mnk2 were purified from transfected HEK 293 cells as described in Materials and Methods (including two extra wash steps with lithium chloride to prevent copurification of ERK) and used to phosphorylate recombinant eIF4E. Radioactively labeled eIF4E was purified from SDS-polyacrylamide gels and digested with trypsin. The peptide mixture obtained was purified by HPLC, and the single peak containing radioactivity was analyzed by thin-layer electrophoresis as described in reference 1 using pH 1.9 buffer as the running buffer. A synthetic peptide containing the sequence SGS(P)TTK (with the latter P indicating the position of the phosphorylated serine) was applied to the thin-layer chromatography plates and visualized with ninhydrin (indicated by the circle) as described before (38). The symbol × indicates the origin, and the migration was toward the cathode.
To study whether phosphorylation by these kinases activated Mnk2 and whether Mnk2 could phosphorylate eIF4E, a similar experiment was performed in the presence of recombinant eIF4E (Fig. 1B). None of the six kinases tested was able to phosphorylate eIF4E directly (data not shown). When Mnk2 was incubated with ERK2 or p38MAPKα or -β, phosphorylation of elF4E was observed. This clearly shows both that phosphorylation of Mnk2 by these kinases leads to its activation and that Mnk2 can phosphorylate eIF4E in vitro. The low level of phosphorylation of Mnk2 by JNK and p38MAPKδ did not lead to activation. This is the first time that phosphorylation of eIF4E (or, indeed, any protein) by Mnk2 has been demonstrated. The findings in Fig. 1A and B are in agreement with the observed sensitivity of eIF4E phosphorylation in vivo to the inhibitors PD98059 and SB203580 (28, 29, 37). These compounds inhibit activation of ERK1 and ERK2 and the activities of p38MAPKα and -β, respectively, but do not affect JNK or the two other isoforms of p38MAPK.
To follow up on the apparent differences between the two Mnks as observed in Fig. 1A, more-detailed kinetic studies were performed on the phosphorylation of Mnk1 and Mnk2 by ERK2 and p38MAPKα and -β. We observed a higher level of phosphorylation of Mnk2 than of Mnk1 for any amount of upstream kinase used. This demonstrates that Mnk2 is a better substrate for these kinases than Mnk1 (Fig. 1C, top). Similarly, we also saw higher levels of phosphorylation of eIF4E, which are indicative of higher activity of Mnk2 than Mnk1 under each condition. As indicated by the data in Fig. 1B, p38MAPKβ did not seem to activate Mnk2 to the same extent as ERK2 (Fig. 1C, bottom). Similar conclusions could be drawn from experiments in which phosphorylation of Mnk1 and Mnk2 was monitored over time (data not shown). A time course experiment of phosphorylation of Mnk2 and its activation, as measured by phosphorylation of eIF4E, showed that there are no significant differences in the kinetics by which Mnk2 is phosphorylated and/or activated by ERK2 and p38MAPKα (Fig. 1D).
To assess whether Mnk2 phosphorylates eIF4E at the physiological site, Ser 209, recombinant eIF4E was phosphorylated with Mnk2 (and Mnk1 for comparison) and subsequently digested with trypsin. Thin-layer chromatographic analysis of the resulting phosphopeptides showed that Mnk1 and Mnk2 phosphorylate the same peptide and that this peptide comigrates with the synthetic peptide SGS(P)TTK, the expected tryptic peptide containing Ser209 (Fig. 1E). Solid-phase sequencing data for the purified peptides showed that the third residue in the peptide (corresponding to Ser209) is the only radiolabeled amino acid in the purified tryptic peptide (data not shown). These results prove that Mnk2, like Mnk1, phosphorylates recombinant eIF4E solely at the physiological site, Ser209.
Phosphorylation sites in Mnk2.
In order to understand how the activity of Mnk2 is regulated, it is important to know which residues in Mnk2 undergo phosphorylation. Mutation of such sites to either aspartate or alanine could then be used to create inactive or constitutively active forms, respectively, of the kinase, which should prove to be useful tools with which to study the cellular role of Mnk2, as shown before for Mnk1 (39). Two phosphorylation sites (Thr197 and Thr202) were identified in Mnk1 by a combination of two-dimensional (2D) phosphopeptide mapping and mutation of expected phosphorylation sites (39). Initially, we used a similar approach to study whether the two corresponding residues are phosphorylated in Mnk2. Numerous spots were seen on the autoradiograms of 2D peptide maps derived from tryptic digests of recombinant GST-Mnk2 labeled with ERK2, indicating the existence of a number of phosphorylation sites. Among many, one specific spot was present in the trypsin-GluC (which cuts between T197 and T202) digest from the T197A mutant but not in that from the T202A mutant. In a similar way, one specific phosphopeptide was found in the map for the T202A mutant that was not present in the T197A map (data not shown). These spots most likely correspond to the peptides containing Thr197 and Thr202. Although this suggests that T197 and T202 are phosphorylated in Mnk2, we were unable to prove this unambiguously.
Because of the complexity of the 2D maps and the apparent existence of several phosphorylation sites, we decided to identify phosphorylation sites in Mnk2 using techniques that involve their direct identification rather than by making mutations at possible sites of phosphorylation. To determine which sites are phosphorylated in Mnk2 in vivo, HEK 293 cells were transfected with a construct expressing GST-Mnk2 and labeled with 32P-orthophosphate.
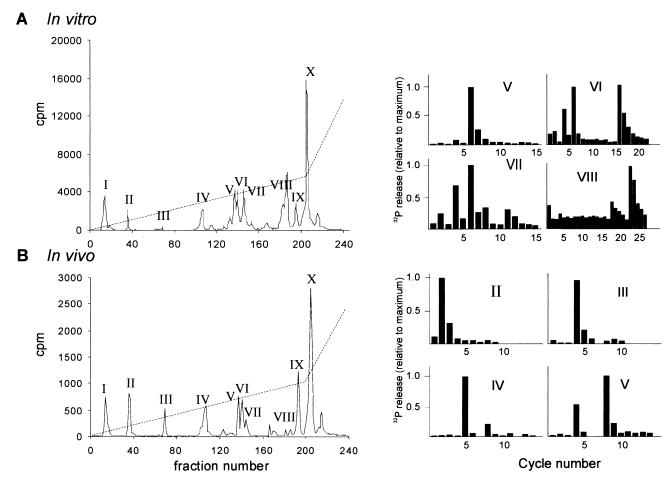
The HPLC profiles of the trypsin-digested proteins from either in vitro-phosphorylated GST-Mnk2 (Fig. 2A, left part) or in vivo-labeled GST-Mnk2 (Fig. 2B, left part) revealed 10 peaks of radioactivity. The phosphopeptides in the peak fractions of the HPLC were subsequently identified by mass spectrometry (data not shown), and the phosphorylated residues within these peptides were subsequently located by solid-phase sequencing (Fig. 2, right part). For example, analysis of the peptide in peak III from the in vivo analysis showed that it corresponds to residues 24 to 31 and the solid-phase sequencing data showed release of radioactive phosphate in the fourth cycle, corresponding to phosphorylation of Ser27. In the same way, we identified the other phosphorylated residues, as indicated in Fig. 3A. In this particular in vitro labeling experiment, only a very small amount of radioactive peptide was found in peak III, but results from other experiments showed that Ser27 is also phosphorylated in vitro (data not shown). In some in vivo labeling experiments, a very small amount of radioactivity was found in peak VIII, indicating that Thr332 is probably also phosphorylated in vivo. However, we have been unable to recover enough of this peptide to allow identification by mass spectrometry. The peptide in peak II was found in vivo and in vitro and showed a release of labeled phosphate at cycle 2 upon solid-phase sequencing. However, we have been unable to identify the phosphopeptide in this peak.
FIG. 2.
Identification of phosphorylated residues in Mnk2. (A) Identification of phosphorylation sites in vitro. Recombinant GST-Mnk2 was phosphorylated by ERK2 and purified and digested as described in Materials and Methods. Radioactive peptides were subsequently purified by HPLC (left part). A gradient of 0 to 30% acetonitrile over 90 min and 30 to 70% over the final 30 min was employed to elute peptides from the column, as indicated by the dotted line. Each fraction was analyzed by Cerenkov counting (solid line). The main peaks are indicated by Roman numerals. Purified peptides from the HPLC runs were analyzed by matrix-assisted laser desorption ionization–time of flight mass spectrometry and solid-phase sequencing as previously described (2). The bar graphs on the right show the release of radioactive phosphate at successive cycles of the Edman degradation for each peptide (as indicated by the peak number). Solid-phase sequencing results similar to those obtained for the corresponding peaks of the in vivo labeling were obtained for peaks II and IV (shown in panel B). (B) Identification of phosphorylation sites in vivo. GST-Mnk2 from radiolabeled transfected cells was purified on glutathione-Sepharose and SDS-PAGE and digested as described in Materials and Methods. Peptides were separated by HPLC using the same gradient as used for panel A (left part). Phosphorylated residues were identified as mentioned above (right part).
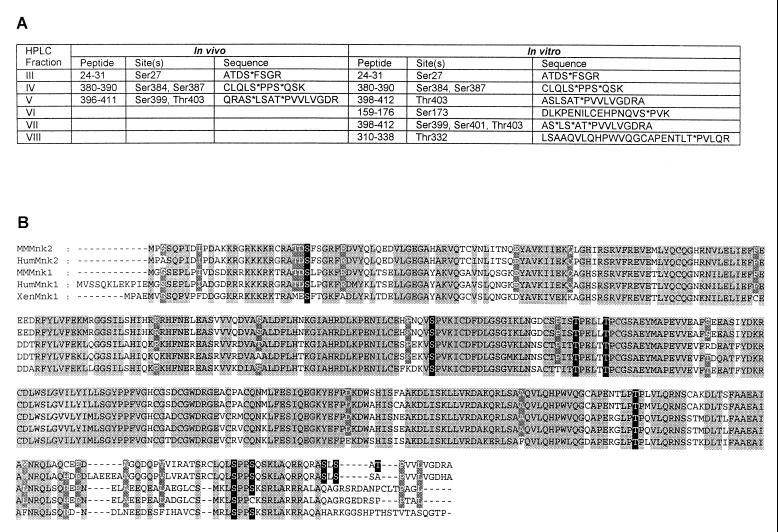
FIG. 3.
Phosphorylation sites in Mnk2. (A) Phosphorylated tryptic peptides and residues. The results shown in Fig. 2 are summarized. The HPLC peak numbers are given in the first column, followed by the position within Mnk2 of the peptides that were identified by mass spectrometry, the phosphorylation sites that were identified by solid-phase sequencing, and the sequences of the tryptic peptides (phosphorylation sites are indicated by asterisks). (B) Conservation of phosphorylation sites in Mnk1 and Mnk2. The mouse (MMMnk1 and MMMnk2), human (HumMnk1 and HumMnk2), and Xenopus (XenMnk1) Mnk1 and Mnk2 sequences were aligned using ClustalW software. Amino acids conserved in all five sequences are black letters on a light grey background, and residues conserved among four of the sequences are in white letters on a dark grey background. The phosphorylation sites identified in Fig. 2 are in white letters on a black background. The human Mnk2 amino acid sequence was taken from the EMBL database (accession no. AC007136). This sequence represents the putative human Mnk2 derived from translation of the genomic DNA sequence. For this alignment, a long N-terminal extension in the putative human Mnk2 sequence was omitted. The human Mnk2 sequence can also be fully assembled from several translated expressed sequence tags (not shown).
A peak of apparently undigested or partially digested material was routinely found in peak X. This material most likely also contains the tryptic peptide containing Thr197 and Thr202, as we could not detect this particular peptide in any of the other fractions. Furthermore, the expected size of this peptide is relatively large (reducing its mobility on the reversed-phase column). Analyses by mass spectrometry of smaller peptides generated by cleaving this undigested material with the protease GluC or AspN were unsuccessful.
The mass spectrometry data showed that several peptides were derived from incomplete tryptic digestion. Especially the C-terminal tryptic peptide appears in multiple forms. A singly phosphorylated form that was not cleaved at R397 was found in peak V, and a triply phosphorylated form that was not cleaved at R411 was present in peak VII. Finally a third form, apparently doubly phosphorylated, was found in peak VI together with the phosphopeptide containing Ser173. Another example is the phosphopeptide in peak IX. This peak contained the peptide corresponding to amino acids 24 to 49, and solid-phase sequencing yielded results similar to those obtained for peak III, again identifying Ser27 as a phosphorylation site.
In vitro phosphorylation of recombinant GST-Mnk2 with activated recombinant p38MAPKα and subsequent tryptic digestion and analysis by HPLC yielded a profile that was almost identical to that shown in Fig. 3A (data not shown). Thus, ERK and p38MAPKα appear to phosphorylate the same sites in Mnk2.
Alignment of human, mouse, and Xenopus Mnk sequences shows that, except for the most C-terminal phosphorylation sites (Ser399, Ser401, and Thr403), the identified phosphorylation sites are conserved between Mnk1 and Mnk2 (Fig. 3B). This is consistent with the idea that they are important for the activity or regulation of the kinases.
Residues at positions 197, 202, and 332 are directly involved in modulating Mnk2 activity.
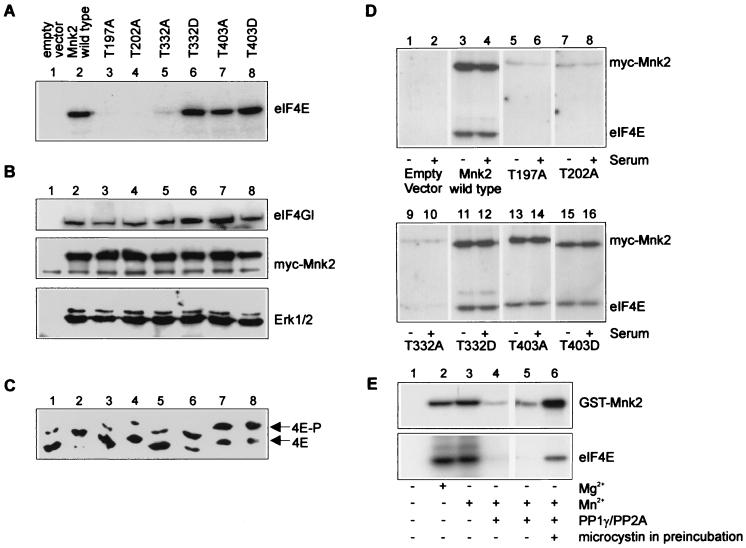
To test the importance of some of the possible MAPK sites for the activity and regulation of Mnk2, a number of mutants were created: Mnk2T197A, Mnk2T202A, Mnk2 T332A, Mnk2T332D, Mnk2T403A, and Mnk2T403D. The reasoning behind this choice is as follows. Although we could not directly identify Thr197 and Thr202 as phosphorylation sites, these residues are in the activation domain of the kinase and corresponding mutations in Mnk1 resulted in loss of activity. It has been reported that in Mnk1, although Thr332 does not appear to be a phosphorylation site, mutation of this residue to aspartate renders the kinase constitutively active while mutation to alanine did not inhibit its activity or even slightly stimulated it (39). Therefore, T332A and T332D mutants were also made for Mnk2 to study the importance of this residue in the regulation of Mnk2 activity. Thr-to-Ala or -Asp Mutants with a change at position 403 were also created, for three reasons. (i) This amino acid resides in the extreme C terminus of the kinase, a region that is often involved in regulating the activity of protein kinases. (ii) Thr403 is situated close to a region that has been proposed to allow the binding of ERK and p38MAPK (9, 35). (iii) This is the only canonical MAPK site that is not conserved between Mnk1 and Mnk2 and might therefore be important for the differences in the activities of these two kinases (as described in Fig. 4 to 6). Wild-type Mnk2 and the mutants were expressed as Myc-tagged fusion proteins in 293 cells, and the subsequently purified kinases were tested for the ability to phosphorylate eIF4E in vitro (Fig. 4A).
FIG. 4.
Analysis of phosphorylation site mutant forms of Mnk2. (A) eIF4E kinase activity of mutant Mnk2. 293 cells were transfected with plasmids encoding Myc-tagged Mnk2 proteins as indicated above each lane. After 48 h, the cells were lysed, Myc-tagged proteins were immunoprecipitated, and aliquots of the purified proteins were used in kinase assays as described in Materials and Methods. Phosphorylation of eIF4E was analyzed by SDS-PAGE and autoradiography. (B) Binding of eIF4G and ERK to Mnk2. The rest of the purified Myc-tagged proteins, as described above, were analyzed by SDS-PAGE and Western blotting with antibodies against ERK1 and -2 and eIF4GI. (C) eIF4E phosphorylation in transfected cells. Supernatants of the anti-Myc immunoprecipitations, as described for panel A, were used to purify eIF4E by m7GTP-Sepharose chromatography as described in Materials and Methods. eIF4E phosphorylation was assessed by one-dimensional isoelectric focusing and Western blotting with antibodies directed against human eIF4E. 4E and 4E-P indicate the positions of the unphosphorylated and phosphorylated forms of eIF4E. (D) The activity of wild-type Mnk2 or active mutant forms of Mnk2 is not decreased upon serum starvation. 293 cells were transfected with the constructs indicated below the lanes. Cells were grown for 42 h in serum-containing medium (+) or for 26 h in serum-containing medium, followed by 16 h in serum-free medium (−) as indicated below the lanes. Myc-tagged proteins were immunoprecipitated from cell extracts, and eIF4E kinase activity was determined as described in Materials and Methods. (E) Phosphorylation is required for Mnk2 activity. GST-Mnk2 from transfected 293 cells was purified as described in Materials and Methods. The beads were then washed three more times with 20 mM HEPES · KOH (pH 7.5)–100 mM KCl–1 mM dithiothreitol to remove β-glycerophosphate. After preincubation either in the absence (lanes 1 to 3) or in the presence (lanes 4 to 6) of the phosphatases PP1γ and PP2A for 30 min at 30°C, microcystin was added to block further phosphatase activity (final concentration, 20 μM). In lane 6, microcystin was included in the preincubation. Subsequently, recombinant eIF4E was added as the substrate together with unlabeled ATP and [γ-32P]ATP and the reaction mixtures were incubated for an additional 30 min at 30°C. Phosphorylation of Mnk2 and eIF4E was analyzed by SDS-PAGE and autoradiography. Lanes 1 to 4 were derived from the same experiment, but the upper part was exposed to film for 16 h and the lower part was exposed for 4 days. Lanes 5 and 6 were taken from a separate experiment, and both parts were exposed for 16 h.
FIG. 6.
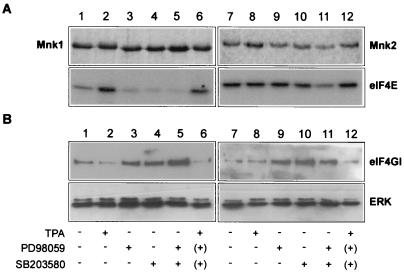
Decreased binding of eIF4G to Mnk1 and Mnk2 upon stimulation of the ERK pathway. (A) Stimulation or inhibition of specific MAPK pathways only modestly affects Mnk2 activity. 293 cells were transfected with either pEBG-Mnk1 (lanes 1 to 6) or pEBG-Mnk2 (lanes 7 to 12), expressing, respectively, wild-type Mnk1 or Mnk2 as GST fusion proteins. After 24 h, the medium was replaced with serum-free medium and cells were grown for an additional 16 h. The cells were harvested either without further treatment (lanes 1 and 7) or after treatment with TPA at 500 nM for 30 min (lanes 2 and 8), PD98059 (lanes 3 and 9), SB203580 (lanes 4 and 10), PD98059 and SB203580 (lanes 5 and 11), or PD98059 and SB203580 followed by TPA treatment (500 nM for 30 min) after washing of the cells to remove the inhibitors (as indicated by parentheses) (lanes 6 and 12). Treatment with PD98059 or SB203580 lasted 1 h. The different conditions are indicated below panel B. GST fusion proteins were purified, and their kinase activity against eIF4E was analyzed as described in Materials and Methods. At the top are the Coomassie-stained gels for GST-Mnk1 and GST-Mnk2 that were used in the kinase assays. At the bottom is the autoradiogram of the phosphorylated eIF4E. (B) Binding of eIF4G to Mnk1 and Mnk2 is reduced upon stimulation with phorbol ester. Aliquots of the samples obtained as shown in panel A were analyzed by SDS-PAGE and Western blotting with antibodies against ERK 1 and -2 (bottom) or eIF4GI (top).
The fusion protein containing wild-type Mnk2 isolated from 293 cells phosphorylated eIF4E in vitro (Fig. 4A, lane 2), as found in Fig. 1 for in vitro-activated recombinant GST-Mnk2. Mnk2 activity was completely abrogated by mutation of either Thr197 or Thr202 to Ala (Fig. 4A, lanes 3 and 4). The single mutations are each sufficient to abolish the kinase activity of Mnk2, demonstrating that both residues are required for the activation of Mnk2. Substitution of Ala for Thr197 in Mnk1 without mutation of Thr202 also inactivates the protein (data not shown). Thr332 also seems to play a major role in the activation of Mnk2, as its mutation to Ala almost completely abolished eIF4E kinase activity. However, in contrast to Mnk1 (39), mutation of Thr332 to Asp did not affect the activity of Mnk2 expressed in 293 cells (lane 6). Thus, the T332D mutation failed to enhance the activity of Mnk2 expressed in 293 cells, suggesting that this site is already phosphorylated in vivo. So, wild-type Mnk2 may already have maximal activity under these conditions, which is consistent with data discussed below. Phosphorylation of Thr403 does not seem to be a major determinant of Mnk2 activity, as mutation of this residue to either Ala or Asp did not affect Mnk2 activity significantly. To establish whether wild-type Mnk2 and its mutant forms could bind to eIF4G and ERK, the anti-Myc-immunoprecipitated fusion proteins and their binding partners were analyzed by SDS-PAGE and Western blotting (Fig. 4B). First, all of the Myc-tagged proteins were expressed at similar levels (Fig. 4B, middle), indicating that the decreased activity observed for some of the mutants (Fig. 4A) was not due to differences in their expression levels. Importantly, Mnk2 binds eIF4GI (Fig. 4B, top), strengthening the idea that Mnk2 is, indeed, a physiological eIF4E kinase. None of the mutations significantly affected the binding of eIF4GI (Fig. 4B, top) or ERK (Fig. 4B, bottom) to Mnk2. The site for interaction of Mnk1 and Mnk2 with eIF4G is thought to reside in a stretch of basic residues in the N terminus of the kinases (39), and therefore, the mutations used in this study were not expected to interfere with this interaction. The ERK binding data show that neither a threonyl residue at position 403 nor phosphorylation of this site affects ERK binding.
As we routinely obtain transfection efficiencies of 80% or higher (as determined by transfection of 293 cells with a control plasmid expressing the green fluorescent protein), we were able to study the effects of overexpression of Mnk2 and its mutant forms on the phosphorylation of endogenous eIF4E. eIF4E was purified from cell extracts by m7GTP-Sepharose chromatography, and its phosphorylation state was analyzed by one-dimensional isoelectric focusing (Fig. 4C).
Expression of wild-type Mnk2 in 293 cells caused a marked increase in phosphorylation of endogenous eIF4E, showing for the first time that Mnk2 acts as an eIF4E kinase in vivo (Fig. 4C, lane 2). Similar results were found for the three mutant proteins with unaffected kinase activity (lanes 6 to 8). Transfection of cells with Mnk2T197A, Mnk2T202A, and Mnk2T332A did not increase eIF4E phosphorylation in vivo (Fig. 4C, lanes 3 to 5), which is consistent with the lack of eIF4E kinase activity of these mutants in vitro (Fig. 4A). The level of eIF4E phosphorylation in the transfected cells thus closely matches the in vitro kinase activity of the purified Mnk2 proteins. We have obtained similar results for Mnk1, as presented below (Fig. 5 and 6).
FIG. 5.
Regulation of Mnk1 and Mnk2 activity. (A) eIF4E kinase activity of over expressed GST-Mnk1 and GST-Mnk2. 293 cells were transfected with either pEBG-Mnk1 or pEBG-Mnk2 for 26 h, followed by 16 h of serum starvation of the cells. The cells were either untreated (lanes 1 and 9); treated with TPA (500 nM) for 5, 15, or 30 min (lanes 2 to 4 and 10 to 12); or treated with PD98059 and SB203580 (lanes 5 to 8 and 13 to 16) for the indicated times. Cells treated for 4 h with the two inhibitors were either harvested directly (lanes 6 and 14), incubated with serum-free medium for 30 min after washing out of the inhibitors (as indicated by parentheses) (lanes 7 and 15), or incubated with serum-free medium in the presence of 500 nM TPA for 30 min after washing out of the inhibitors (lanes 8 and 16). The treatments are indicated below the lanes and apply also to panel B. The GST fusion proteins were purified from the extracts with glutathione-Sepharose as described in Materials and Methods, and one-fifth of the sample was used to study the kinase activity against eIF4E. The upper part shows the Coomassie staining of the fusion proteins, and the lower part shows the autoradiogram representing phosphorylated eIF4E. Radiolabeling was quantified using a Fuji Phosphor-Imager and Aida2000 software and expressed relative to the control (which was 1.0). (B) Phosphorylation of endogenous eIF4E in transfected cells. After the GST pulldown, the extracts were incubated with m7GTP-Sepharose to pull down eIF4E from the extracts as described in Materials and Methods. Unphosphorylated and phosphorylated forms of eIF4E (marked by arrows) were separated by isoelectric focusing and visualized by Western blotting with antibodies to eIF4E (16). The percentages of eIF4E in the phosphorylated form were determined using NIH Image software and are indicated below the lanes. (C) Activities of endogenous Mnk1 and Mnk2 from Swiss 3T3 cells. Swiss 3T3 cells were grown in the presence of serum or starved for serum for 16 h as indicated below the lanes. Prior to harvesting, cells were either untreated, incubated with 500 nM TPA for 30 min, or treated with 50 μM PD98059 and 10 μM SB203580 (PD/SB) for 4 h and harvested. Endogenous Mnk1 (top) and Mnk2 (bottom) were immunoprecipitated with specific antibodies, and kinase activity was determined as described in Materials and Methods using eIf4E as the substrate. In lane pre, the immunoprecipitation was performed with sheep preimmune serum. Lane 1 represents a control in which recombinant eIF4E, as a substrate for the kinase assay, was omitted. (D) Phosphorylation of endogenous eIF4E is hardly affected by MAPK signaling pathway inhibitors in Swiss 3T3 cells. Swiss 3T3 cells were either untreated (lane 1), treated with 500 nM TPA for 30 min (lane 2), or treated with 50 μM PD98059 and 10 μM SB203580 (lanes 3) and harvested. eIF4E was purified from the cell extracts by m7GTP-Sepharose chromatography and analyzed by isoelectric focusing as described in Materials and Methods. The percentages of eIF4E in the phosphorylated form were determined using NIH Image software and are indicated below the lanes.
Mnk2 has high basal activity that is only partially affected by PD98059 or SB203580.
As shown in Fig. 4A, the cells were grown in medium containing serum, and wild-type Mnk2 and some of the mutant proteins are apparently active under these conditions. The fact that none of the mutations further increased the activity of Mnk2, as shown for a T332D mutant form of Mnk1 (39), could reflect a high basal activity of wild-type Mnk2 that cannot be further enhanced. Therefore, we tested whether withdrawal of serum from the medium revealed any differences between wild-type Mnk2 and its mutant forms. Surprisingly, the activities of all active forms were not affected by serum starvation (Fig. 4D, compare lanes 3 and 4, 11 and 12, 13 and 14, and 15 and 16).
To study whether the high basal level of activity of GST-Mnk2 that is expressed in 293 cells is due to its already being phosphorylated, the purified fusion protein was treated with protein phosphatases. The resulting (dephosphorylated) Mnk2 was then tested for the ability to phosphorylate eIF4E (Fig. 4E). Mnk2 remained active in the presence of either magnesium or manganese, which was used to stimulate the activity of recombinant PP1γ (Fig. 4E, lanes 2 and 3). Pretreatment of the purified GST-Mnk2 with PP1γ and PP2A caused a shift in the mobility of GST-Mnk2, which is suggestive of dephosphorylation (data not shown), and almost completely abolished both autophosphorylation of Mnk2 and phosphorylation of eIF4E by Mnk2 (lanes 4 and 5). As a control to show that the activities of the phosphatases were indeed blocked by addition of the phosphatase inhibitor microcystin, this compound was added during the pretreatment and in this case Mnk2 activity was not inhibited (lane 6). Thus, Mnk2 activity is dependent on its phosphorylation and these results suggest that the basal level of phosphorylation of Mnk2 is high in serum-starved cells.
Next, we tested whether the activity of Mnk2 was altered upon treatment of cells with agents that affect the ERK and p38MAPK pathways, i.e., using tetradecanoyl phorbol acetate (TPA) as a stimulus to activate the ERK pathway and PD98059 and SB203580 as inhibitors of these pathways (Fig. 5 and 6). In these transfection studies, we also transfected cells with constructs expressing GST-Mnk1 to allow direct comparison of the regulation of the activities of the two kinases. GST-Mnk1 purified from serum-starved cells showed some eIF4E kinase activity (Fig. 5A, bottom, lane 1) but clearly less than GST-Mnk2 (lane 9). While TPA treatment increased Mnk1 activity (lanes 2 to 4), no significant change was seen in the activity of Mnk2 (lanes 10 to 12). Arsenite or hydrogen peroxide treatment of serum-starved cells to stimulate the p38MAPK pathway also failed to raise the activity of Mnk2 (data not shown), in contrast to data obtained earlier for Mnk1 (37). However, inhibition of the ERK and p38MAPK pathways using a combination of the inhibitors PD98059 and SB203580 reduced the activity of overexpressed Mnk2 approximately three- to fourfold (lanes 13 and 14). The effect of these two compounds on Mnk2 activity is somewhat variable, as we sometimes found a smaller reduction in kinase activity. Subsequently, after washing out these inhibitors, we incubated the cells in serum-free medium (lanes 7 and 15) or stimulated them with TPA (lanes 8 and 16). Mnk2 activity recovered substantially after removal of PD98059 and SB203580 without stimulation of the cells, while Mnk2 activity was fully restored in the presence of TPA. The data show that stimulation of the ERK pathway with TPA does lead to activation of Mnk2 and suggest a very low requirement by Mnk2 for upstream kinase activity to attain a high level of activity. The level of phosphorylation of endogenous eIF4E in transfected cells closely matched the activity of either Mnk1 or Mnk2 (Fig. 5B), and the increase in Mnk2 activity upon removal of PD98059 and SB203580 was reflected in an increase in eIF4E phosphorylation in these cells (lane 15). Preliminary data show that upon overexpression in 293 cells, the activity of the human homologue Mnk2 is regulated in a very similar manner (data not shown).
The complete inhibition of eIF4E phosphorylation in 293 cells that were untransfected (data not shown) or transfected with Mnk1 upon treatment with the MAPK signaling inhibitors implies that in 293 cells, Mnk2 activity must be virtually absent.
To assess whether the activities of endogenous Mnk1 and Mnk2 are regulated similarly to the expressed fusion proteins in transfected cells, endogenous Mnk1 and Mnk2 were immunoprecipitated from untransfected cells and tested for kinase activity (Fig. 5C). Swiss 3T3 cells were used for these immunoprecipitation assays, as the antibodies were raised using peptides based on sequences in the C termini of the murine kinases that are not conserved either between Mnk1 and Mnk2 or between humans and mice.
These results show that endogenous Mnk1 was regulated very similarly to the over expressed kinase in the transfected 293 cells. Hardly any Mnk1 activity was found in control cells (Fig. 5C, top: lanes 2 and 3), while TPA treatment resulted in rapid activation of endogenous Mnk1 (lane 4), as found for overexpressed GST-Mnk1 (Fig. 5A). Due to the lack of basal Mnk1 activity in the control cells, we could not analyze the effects of PD98059 and SB203580 on Mnk1 activity (lane 5). The activity of Mnk2 remained almost constant upon TPA treatment (Fig. 5C, bottom), implying that the endogenous kinase also has high basal activity. However, the threefold decrease upon treatment with the inhibitors, as seen in some experiments for the overexpressed kinase, was not found for endogenous Mnk2. Thus, endogenous Mnk2 seems to be less sensitive to inhibition by PD98059 and SB203580 than the overexpressed form.
A prediction that can be made based upon the unchanged activity of endogenous Mnk2 in Swiss 3T3 cells, even upon treatment with both PD98059 and SB203580, is that eIF4E should also be relatively insensitive to these inhibitors in these cells. Therefore, eIF4E was purified from the cell extracts using m7GTP-Sepharose and its phosphorylation state was analyzed by iso electric focusing (Fig. 5D). Indeed, incubation of Swiss 3T3 cells with a combination of PD98059 and SB203580 only slightly affected the level of phophorylation of eIF4E. These results strongly indicate, in contrast to the results obtained with 293 cells, where eIF4E phosphorylation is completely abolished by treatment with PD98059 and SB203580, that in Swiss 3T3 cells, Mnk2 can act as a physiological kinase and thus determine the level of eIF4E phosphorylation.
The differences between the activities of Mnk1 and Mnk2 under these various conditions might be explained by differences in their binding to eIF4G or ERK. For example, Mnk2 may be bound to eIF4G and members of the MAPK family constitutively while binding of these factors to Mnk1 might be subject to regulation and change, depending on the condition. Therefore, experiments similar to those described in Fig. 5 were performed and used to study the binding of ERK and eIF4G to either Mnk1 or Mnk2 under various conditions (Fig. 6). The kinase assay data again showed that Mnk2 activity was only reduced by a combination of PD98059 and SB203580 (Fig. 6A, compare lanes 7 and 9 to 11), while Mnk1 activity was reduced by each inhibitor on its own (lanes 1 and 3 to 5).
ERK1 or -2 binding to Mnk1 or Mnk2 was not affected either by stimulation of the ERK pathway (by TPA) or by inhibition of the ERK and p38MAPK pathways (by treatment with PD98059 and SB203580) (Fig. 6B, bottom). However, we consistently found that TPA treatment of cells resulted in decreased association of Mnk1 with eIF4G (Fig. 6B, top, lanes 2 and 6). In this particular experiment, the dissociation of Mnk2 from eIF4G in TPA-treated cells is not evident, but in similar experiments we have seen a small reduction in binding. Treatment with the two inhibitors increased the binding of eIF4G to either Mnk (lanes 3 to 5 and 9 to 11). There seem to be no major differences in the binding of Mnk1 and Mnk2 to either ERK1 or -2 or eIF4G. Other possible mechanisms underlying the differences between Mnk1 and Mnk2 in the regulation of their activities will be discussed below.
DISCUSSION
In this paper, we show for the first time that Mnk2 is an eIF4E kinase, based on three complementary lines of evidence: (i) Mnk2 phosphorylates eIF4E at Ser209 in vitro, (ii) overexpression of Mnk2 in HEK 293 cells leads to increased phosphorylation of eIF4E in vivo, and (iii) Mnk2 binds to eIF4G. It shares these characteristics with Mnk1, but the basal activities of the two enzymes are quite different in vivo.
The signaling pathways involved in the regulation of Mnk1 and Mnk2 seem to be very similar. Like Mnk1, Mnk2 is phosphorylated and activated by both ERK2 and p38MAPKα and -β in vitro (Fig. 1), but Mnk2 was a better substrate for the various upstream kinases than Mnk1. Waskiewiez et al. (37) reported that Mnk2 is phosphorylated efficiently by p38MAPK, even though it did not show stable binding to Mnk2. Mnk2 activity was decreased upon treatment of cells with both PD98059 and SB203580 (Fig. 5), but over expressed Mnk2, especially endogenous Mnk2, seems to be less sensitive to agents that alter signaling through the MAPK pathways than Mnk1. Taken together with the fact that washing out the inhibitors PD98059 and SB203580 without further adding any stimulus results in almost complete reactivation of Mnk2 but not of Mnk1 (Fig. 5), these data indicate that the high basal activity of Mnk2 can be maintained by low levels of activity of the upstream kinases. Consistent with these findings, the HPLC profiles of radiolabeled peptides from trypsin-digested Mnk2 purified from [32P]orthophosphate-labeled transfected cells that had been treated with TPA, PD98059, SB203580, or a combination of these two inhibitors (data not shown) were almost indistinguishable from the one shown for untreated cells (Fig. 2).
An alternative explanation for the high basal activity of Mnk2 could be that an additional kinase or pathway regulates Mnk2 activity in vivo, which would explain why Mnk2 retains significant activity even in the presence of PD98059 and SB203580 (Fig. 5) and is active in serum-starved cells. However, inhibitors of phosphatidylinositol 3-kinase, mTOR, and protein kinase C did not affect Mnk2 activity in vivo (data not shown), implying that none of these major signaling components is involved in the regulation of Mnk2 activity.
Despite the complexity of the phosphorylation site mapping, we have been able to identify the phosphopeptides in 7 of the 10 major HPLC peaks that were found upon digestion with trypsin (Fig. 2). By solid-phase sequencing, eight in vitro phosphorylation sites in Mnk2 were identified. The similarity between the HPLC profiles of the tryptic digests of in vitro- and in vivo-labeled GST-Mnk2 (Fig. 2) suggests that all of these sites are also phosphorylated in vivo and five of these in vivo phosphorylation sites were confirmed directly using mass spectrometry and solid-phase sequencing.
Several of the phosphorylation sites identified in vivo (i.e., Ser384 and Thr403) and in vitro (i.e., Ser173, Thr332, Ser384, and Thr403) are within TP or SP motifs, and these residues are thus most likely phosphorylated by ERK and p38MAPK. All of the major spots that contain an SP or TP site are still present in the 2D maps of the tryptic digests of the inactive T197A and T202A mutant proteins that were phosphorylated in vitro (data not shown), supporting the idea that these sites are indeed MAPK phosphorylation sites, rather than autophosphorylation sites. The other residues, which lack the adjacent prolyl residue, are probably autophosphorylation sites, as the MAPKs are solely proline directed.
Mutation of Thr332 to Ala almost completely abolishes the activity of Mnk2 in vivo, while mutation to Asp does not affect its activity. In contrast, in Mnk1, mutation of Thr332 to Asp caused it to be constitutively active while a change to Ala did not affect kinase activity (38). This shows that in both these kinases, this residue, and most likely its phosphorylation, plays a key role in regulating their activities. The data from the T332 mutants can be interpreted to suggest that this site is constitutively phosphorylated in Mnk2, rendering it basally active. The role of the three possible phosphorylation sites in the C-terminal region of Mnk2 (Ser399, Ser401, and Thr403) is unclear. Although these sites are not found in Mnk1 and were therefore candidates for mediating the differences between Mnk1 and Mnk2, mutation of Thr403 to either Ala or Asp did not significantly affect Mnk2 activity. Whether phosphorylation of Ser399, Ser401, or any of the other sites identified plays a role in the high basal activity of Mnk2 awaits further investigation. Preliminary data indicate that phosphorylation at Ser27 is not a prerequisite for Mnk2 activity (B. van Kollenburg and G. C. Scheper, data not shown).
Two lines of evidence do indicate that Thr197 and Thr202 are phosphorylation sites in Mnk2: (i) mutation of either residue to alanine abolishes Mnk2 activity (Fig. 4), and (ii) specific, and different, peptides are missing on trypsin-GluC 2D maps of the T197A and T202A mutant proteins (data not shown). In many kinases, phosphorylation of serine or threonine residues in the positions corresponding to T197 and T202 (in the so-called T loop) plays an important role in activating these enzymes (11). In particular, in Mnk1 (38), phosphorylation of residues corresponding to Thr197 and Thr202 has been shown to be essential for activity. A mutant in which both Thr residues were changed to Asp (which might mimic phosphothreonine) showed no activity upon expression in 293 cells (data not shown), perhaps arguing against the notion that these sites are phosphorylation sites. Despite substantial effort using HPLC and mass spectrometry techniques, we were unable to prove unambiguously that these sites are phosphorylated in Mnk2.
It is unclear why mammalian cells should possess two kinases that each phosphorylate eIF4E but show different basal activities. Our data suggest that changes in the phosphorylation state of eIF4E in response to growth stimuli and stressful agents are likely caused by changes in the activity of Mnk1. Mnk2 might be required to maintain the low level of eIF4E phosphorylation under conditions where Mnk1 is (almost) inactive, such as serum starvation. One could speculate that the synthesis of certain proteins that are essential for cell survival must occur under most conditions, and Mnk2 activity would ensure phosphorylation of eIF4E and thereby the translation of the mRNAs encoding such proteins. Northern blot analysis has shown that the RNAs encoding Mnk1 and Mnk2 are expressed in a variety of tissues (38), but nothing is known about the amounts of the (active) proteins in cells. On the one hand, the very low level of eIF4E phosphorylation in serum-starved 293 cells suggests that these cells possess very little, if any, Mnk2, while on the other hand, in Swiss 3T3 cells, Mnk2 appears to be involved in maintaining a high level of phosphorylated eIF4E.
Mnk1 and Mnk2 each bind to eIF4G (Fig. 6). The extreme C terminus of eIF4G and the N-terminal 23 amino acids of Mnk1 are required for the interaction of Mnk1 and eIF4G (27, 39). Within this region of Mnk1, a stretch of basic residues in the N terminus of Mnk1 is probably important for binding to eIF4G and this is conserved in Mnk2 (Fig. 3B). Therefore, Mnk2 most likely binds to the same region on eIF4G as Mnk1 and binding of Mnk1 and Mnk2 to eIF4G would then be mutually exclusive, potentially giving an extra level of control to regulate the level of eIF4E phosphorylation in cells.
The differences between the activities of Mnk1 and Mnk2 do not seem to be due to changes in their binding to ERK: the amounts of total ERK1 and ERK2 bound to these kinases were very similar under a wide variety of conditions that affect the MAPK signaling pathways. Also, phosphorylated ERK bound to both kinases (as detected by using phosphospecific antibodies), and again, no obvious differences were found between the two Mnks (not shown). We have found that the amount of eIF4G bound to either Mnk1 or Mnk2 did vary under differing conditions. Stimulation of the classical ERK pathway with TPA led to decreased eIF4G-Mnk association, while treatment with PD98059 and SB20380 had the opposite effect. The increased binding of eIF4G to Mnk1 when its activity is very low or completely inhibited could be a way to ensure rapid phosphorylation of eIF4E upon Mnk1 activation. Subsequent dissociation may serve to limit—in extent or duration—the consequent phosphorylation of eIF4E. This seems to apply only partly to Mnk2: increased binding was found upon treatment with MAPK pathway inhibitors, but the effects of TPA on the Mnk2-eIF4G association was smaller than that seen for Mnk1. This may play a role in the observed high basal activity of Mnk2. It has been reported that phorbol ester treatment of cells leads to reduced phosphorylation of the residues within eIF4G that can be phosphorylated by Mnk1 (34). The reduction in phosphorylation at these particular sites might be due to the decreased binding of Mnk1 or Mnk2 to eIF4G, even though Mnk1 is activated under these conditions (as shown in Fig. 6). It remains to be established whether the phosphorylation at these sites in eIF4G underlies the reduced association with Mnk1 (and possibly Mnk2).
ACKNOWLEDGMENTS
This was work supported by an EU Marie Curie fellowship awarded to G.C.S.
We thank Thomas Schulz (University of Utrecht) for the GST monoclonal antibodies; Linda Campbell for technical assistance; Pat Eyers, Anudharan Balendran, Jane Leitch, Grahame Hardie, and Nick Helps (University of Dundee) for helpful advice and their kind gifts of recombinant MAPKs and protein phosphatases; and Graham Pavitt for critical reading of the manuscript.
REFERENCES
- 1.Boyle W J, van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 2.Casamayor A, Morrice N A, Alessi D R. Phosphorylation of Ser-241 is essential for the activity of 3-phosphoinositide-dependent protein kinase-1: identification of five sites of phosphorylation in vivo. Biochem J. 1999;342:287–292. [PMC free article] [PubMed] [Google Scholar]
- 3.De Benedetti A, Harris A L. eIF4E expression in tumors: its possible role in progression of malignancies. Int J Biochem Cell Biol. 1999;31:59–72. doi: 10.1016/s1357-2725(98)00132-0. [DOI] [PubMed] [Google Scholar]
- 4.De Benedetti A, Rhoads R E. Overexpression of eukaryotic protein synthesis initiation factor 4E in HeLa cells results in aberrant growth and morphology. Proc Natl Acad Sci USA. 1990;87:8212–8216. doi: 10.1073/pnas.87.21.8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flynn A, Proud C G. Insulin and phorbol ester stimulate initiation factor eIF-4E phosphorylation by distinct pathways in Chinese hamster ovary cells overexpressing the insulin receptor. Eur J Biochem. 1996;236:40–47. doi: 10.1111/j.1432-1033.1996.00040.x. [DOI] [PubMed] [Google Scholar]
- 6.Flynn A, Proud C G. Serine 209, not serine 53, is the major site of phosphorylation in initiation factor eIF-4E in serum-treated Chinese hamster ovary cells. J Biol Chem. 1995;270:21684–21688. doi: 10.1074/jbc.270.37.21684. [DOI] [PubMed] [Google Scholar]
- 7.Fukuchi-Shimogori T, Ishii I, Kashiwagi K, Mashiba H, Ekimoto H, Igarashi K. Malignant transformation by overproduction of translation initiation factor eIF4G. Cancer Res. 1997;57:5041–5044. [PubMed] [Google Scholar]
- 8.Fukunaga R, Hunter T. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 1997;16:1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gavin A C, Nebreda A R. A MAP kinase docking site is required for phosphorylation and activation of p90(rsk)/MAPKAP kinase-1. Curr Biol. 1999;9:281–284. doi: 10.1016/s0960-9822(99)80120-1. [DOI] [PubMed] [Google Scholar]
- 10.Hall-Jackson C A, Cross D A, Morrice N, Smythe C. ATR is a caffeine-sensitive, DNA-activated protein kinase with a substrate specificity distinct from DNA-PK. Oncogene. 1999;18:6707–6713. doi: 10.1038/sj.onc.1203077. [DOI] [PubMed] [Google Scholar]
- 11.Hardie G, Hanks T. The protein kinase facts book. London, United Kingdom: Academic Press; 1995. [Google Scholar]
- 12.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joshi B, Cai A L, Keiper B D, Minich W B, Mendez R, Beach C M, Stepinski J, Stolarski R, Darzynkiewicz E, Rhoads R E. Phosphorylation of eukaryotic protein synthesis initiation factor 4E at Ser-209. J Biol Chem. 1995;270:14597–14603. doi: 10.1074/jbc.270.24.14597. [DOI] [PubMed] [Google Scholar]
- 14.Joshi-Barve S, Rychlik W, Rhoads R E. Alteration of the major phosphorylation site of eukaryotic protein synthesis initiation factor 4E prevents its association with the 48S initiation complex. J Biol Chem. 1990;265:2979–2983. [PubMed] [Google Scholar]
- 15.Kerekatte V, Smiley K, Hu B, Smith A, Gelder F, De Benedetti A. The proto-oncogene/translation factor eIF4E: a survey of its expression in breast carcinomas. Int J Cancer. 1995;64:27–31. doi: 10.1002/ijc.2910640107. [DOI] [PubMed] [Google Scholar]
- 16.Kleijn M, Voorma H O, Thomas A A M. Phosphorylation of eIF-4E and initiation of protein synthesis in P19 embryonal carcinoma cells. J Cell Biochem. 1995;59:443–452. doi: 10.1002/jcb.240590405. [DOI] [PubMed] [Google Scholar]
- 17.Koromilas A E, Lazaris-Karatzas A, Sonenberg N. mRNAs containing extensive secondary structure in their 5′ non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J. 1992;11:4153–4158. doi: 10.1002/j.1460-2075.1992.tb05508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamphear B J, Kirchweger R, Skern T, Rhoads R E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 20.Lamphear B J, Panniers R. Cap binding protein complex that restores protein synthesis in heat-shocked Ehrlich cell lysates contains highly phosphorylated eIF-4E. J Biol Chem. 1990;265:5333–5336. [PubMed] [Google Scholar]
- 21.Lazaris-Karatzas A, Montine K S, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 22.Lazaris-Karatzas A, Smith M R, Frederickson R M, Jaramillo M L, Liu Y L, Kung H F, Sonenberg N. Ras mediates translation initiation factor 4E-induced malignant transformation. Genes Dev. 1992;6:1631–1642. doi: 10.1101/gad.6.9.1631. [DOI] [PubMed] [Google Scholar]
- 23.Li B D, Liu L, Dawson M, De Benedetti A. Overexpression of eukaryotic initiation factor 4E (eIF4E) in breast carcinoma. Cancer. 1997;79:2385–2390. [PubMed] [Google Scholar]
- 24.Manzella J M, Rychlik W, Rhoads R E, Hershey J W, Blackshear P J. Insulin induction of ornithine decarboxylase. Importance of mRNA secondary structure and phosphorylation of eucaryotic initiation factors eIF-4B and eIF-4E. J Biol Chem. 1991;266:2383–2389. [PubMed] [Google Scholar]
- 25.Marcotrigiano J, Gingras A C, Sonenberg N, Burley S K. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89:951–961. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 26.Minich W B, Balasta M L, Goss D J, Rhoads R E. Chromatographic resolution of in vivo phosphorylated and nonphosphorylated eukaryotic translation initiation factor eIF-4E: increased cap affinity of the phosphorylated form. Proc Natl Acad Sci USA. 1994;91:7668–7672. doi: 10.1073/pnas.91.16.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morino S, Imataka H, Svitkin Y V, Pestova T V, Sonenberg N. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol Cell Biol. 2000;20:468–477. doi: 10.1128/mcb.20.2.468-477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morley S J. Signalling through either the p38 or ERK mitogen-activated protein (MAP) kinase pathway is obligatory for phorbol ester and T cell receptor complex (TCR-CD3)-stimulated phosphorylation of initiation factor (eIF) 4E in Jurkat T cells. FEBS Lett. 1997;418:327–332. doi: 10.1016/s0014-5793(97)01405-1. [DOI] [PubMed] [Google Scholar]
- 29.Morley S J, McKendrick L. Involvement of stress-activated protein kinase and p38/RK mitogen-activated protein kinase signaling pathways in the enhanced phosphorylation of initiation factor 4E in NIH 3T3 cells. J Biol Chem. 1997;272:17887–17893. doi: 10.1074/jbc.272.28.17887. [DOI] [PubMed] [Google Scholar]
- 30.Nathan C A, Carter P, Liu L, Li B D, Abreo F, Tudor A, Zimmer S G, De Benedetti A. Elevated expression of eIF4E and FGF-2 isoforms during vascularization of breast carcinomas. Oncogene. 1997;15:1087–1094. doi: 10.1038/sj.onc.1201272. [DOI] [PubMed] [Google Scholar]
- 31.Nathan C A, Liu L, Li B D, Abreo F W, Nandy I, De Benedetti A. Detection of the proto-oncogene eIF4E in surgical margins may predict recurrence in head and neck cancer. Oncogene. 1997;15:579–584. doi: 10.1038/sj.onc.1201216. [DOI] [PubMed] [Google Scholar]
- 32.Pyronnet S. Phosphorylation of the cap-binding protein eIF4E by the MAPK-activated protein kinase Mnk1. Biochem Pharmacol. 2000;60:1237–1243. doi: 10.1016/s0006-2952(00)00429-9. [DOI] [PubMed] [Google Scholar]
- 33.Pyronnet S, Imataka H, Gingras A C, Fukunaga R, Hunter T, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits Mnk1 to phosphorylate eIF4E. EMBO J. 1999;18:270–279. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raught B, Gingras A C, Gygi S P, Imataka H, Morino S, Gradi A, Aebersold R, Sonenberg N. Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. EMBO J. 2000;19:434–444. doi: 10.1093/emboj/19.3.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith J A, Poteet-Smith C E, Malarkey K, Sturgill T W. Identification of an extracellular signal-regulated kinase (ERK) docking site in ribosomal S6 kinase, a sequence critical for activation by ERK in vivo. J Biol Chem. 1999;274:2893–2898. doi: 10.1074/jbc.274.5.2893. [DOI] [PubMed] [Google Scholar]
- 36.Stern B D, Wilson M, Jagus R. Use of nonreducing SDS-PAGE for monitoring renaturation of recombinant protein synthesis initiation factor, eIF-4 alpha. Protein Expr Purif. 1993;4:320–327. doi: 10.1006/prep.1993.1041. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, Flynn A, Waskiewicz A J, Webb B L, Vries R G, Baines I A, Cooper J A, Proud C G. The phosphorylation of eukaryotic initiation factor eIF4E in response to phorbol esters, cell stresses, and cytokines is mediated by distinct MAP kinase pathways. J Biol Chem. 1998;273:9373–9377. doi: 10.1074/jbc.273.16.9373. [DOI] [PubMed] [Google Scholar]
- 38.Waskiewicz A J, Flynn A, Proud C G, Cooper J A. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 1997;16:1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waskiewicz A J, Johnson J C, Penn B, Mahalingam M, Kimball S R, Cooper J A. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol Cell Biol. 1999;19:1871–1880. doi: 10.1128/mcb.19.3.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wysk M, Yang D D, Lu H T, Flavell R A, Davis R J. Requirement of mitogen-activated protein kinase kinase 3 (MKK3) for tumor necrosis factor-induced cytokine expression. Proc Natl Acad Sci USA. 1999;96:3763–3768. doi: 10.1073/pnas.96.7.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]