Abstract
Adenovirus (Ad) is a non-enveloped linear double-stranded DNA virus with >50 serotypes in humans. Ad vectors have been used as gene delivery vehicles to express transgenes, small interfering RNAs (siRNAs) for gene silencing, or CRISPR/Cas and designer nucleases for genome editing. Although several methods are used to generate Ad vectors, the Ad-making process remains technically challenging and time consuming. Moreover, the Ad-making techniques have not been improved for the past two decades. Gibson DNA Assembly (GDA) technology allows one-step isothermal DNA assembly of multiple overlapping fragments. Here, we developed a one-step construction of Ad (OSCA) system using GDA technology. Specifically, we first engineered several adenoviral recipient vectors that contain the ccdB suicide gene flanked with two 20-bp unique sequences, which serve as universal sites for GDA reactions in the Ad genome ΔE1 region. In two proof-of-principle experiments, we demonstrated that the GDA reactions were highly efficient and that the resulting Ad plasmids could be effectively packaged into Ads. Ad-mediated expression of mouse BMP9 in mesenchymal stem cells was shown to effectively induce osteogenic differentiation both in vitro and in vivo. Collectively, our results demonstrate that the OSCA system drastically streamlines the Ad-making process and should facilitate Ad-based applications in basic, translational, and clinical research.
Keywords: recombinant adenovirus, adenoviral vectors, Gibson Assembly, gene delivery, gene therapy, viral vectors, oncolytic virus, vaccine, BMP9 osteogenic signaling, mesenchymal stem cells
Graphical Abstract
While adenovirus (Ad) vectors are one of the most popular gene delivery vehicles, the Ad-making process remains technically challenging and time consuming. Here, the authors developed a simplified and yet efficient one-step construction of adenovirus (OSCA) system using the Gibson DNA Assembly technology to drastically streamline the Ad-making process.
Introduction
Adenovirus (Ad) has received tremendous attention as a gene delivery vehicle for several decades due to its well-defined virology and biology, its non-integrating property and viral genetic stability, its high gene transduction efficiency, and its ease of large-scale production.1, 2, 3, 4 In fact, adenoviral vectors are not only used to deliver transgene expression but also employed to express small interfering RNAs (siRNAs) for gene silencing and/or CRISPR/Cas and designer nucleases systems for genome editing.1,4,5 Ad is a non-enveloped, linear double-stranded DNA virus, and differences in viral capsids delineate tropisms among serotypes. Adenoviral capsids, which are composed of capsid proteins, core proteins, and cement proteins, delineate tropisms among serotypes, and thus give rise to a vast range of therapeutic candidate viruses.1, 2, 3, 4 Human Ads type 2 and type 5 are the most commonly used serotypes.
Compared with other viral vectors used for gene delivery, adenoviral vectors offer several distinct advantages.1, 2, 3 First, Ad is one of the most efficient and non-integrating gene delivery systems since most mammalian cells express Ad primary receptor and secondary integrin receptors.1, 2, 3 Second, Ad vectors provide a versatile platform to modify viral capsids in order to optimize therapeutic features and targeting specificity of the Ad.1, 2, 3 Third, well-understood Ad virology and extensive experimentations with Ad vectors in preclinical and clinical studies make Ad vectors one of the most commonly used viral vectors in clinical trials worldwide.1, 2, 3 Lastly, even the inherent shortcomings, such as evoked host immunity, have been proved beneficial for anticancer immunotherapies, vaccination, and/or oncolytic therapies.1, 2, 3, 4
Even though the use of recombinant Ad vectors has been widespread, the initial construction and production of a given Ad vector (especially the first-generation Ad vectors) is still a challenging and time-consuming process.1, 2, 3 Recombinant Ad vectors are usually generated in four approaches. The classic method is to explore the homologous recombination occurring between an adenoviral shuttle vector carrying transgenes and an Ad backbone genome vector in E1-expressing packaging cells such as HEK-293 cells.1,6 An alternative technique is to use unique restriction enzymes for direct ligation of transgene-containing fragments to the linear E1/E3-deleted adenoviral genome DNA fragment.7 The third approach is to use site-specific recombinase and transposase systems such as the CRE/LOX and FLIP/FRT site-specific integration and Gateway transposon systems.8,9 The fourth approach is to take advantage of more efficient homologous recombination reactions in microorganisms such as bacteria and yeast to generate transgene-containing Ad vectors.10, 11, 12, 13 In fact, our previously developed AdEasy system has become one of the most commonly used techniques worldwide to generate Ad vectors.1,2,12,13 Nonetheless, making Ad vectors remains a technical challenge to many investigators, while the Ad-making technology has not been improved for the past two decades. Thus, it is highly desirable to make the Ad-making process as efficient and simple as possible.
Gibson DNA Assembly (GDA), named after its developer, Daniel G. Gibson,14 is a commonly used synthetic biology technique that allows the one-step isothermal DNA assembly of multiple overlapping fragments in a restriction enzyme-free, seamless, and sequence-independent fashion. A typical GDA in vitro recombination system contains three essential isothermal enzymes: 5′-exonuclease to remove nucleotides from the ends of double-stranded DNA molecules and expose complementary single-stranded DNA (ssDNA) overhangs for specific annealing; DNA polymerase to fill in the ssDNA gaps of the joined molecules; and DNA ligase to covalently seal the nicks.15 Thus, the GDA method is a useful molecular engineering tool to seamlessly assemble synthetic and natural genes, genetic pathways, and even entire genomes.14, 15, 16
In this study, we took the advantage of the GDA technology and developed a highly simplified one-step construction of Ad (OSCA) system. Interestingly, several recent studies explored the use of the GDA technology to construct Ad.17, 18, 19 However, those reports were mainly involved in the construction of either specific oncolytic Ad or rarely used Ad viruses without broad utilities for conventional Ad generation. For the OSCA system, we first engineered novel adenoviral recipient vectors that contain two 20-bp unique sequences, namely modified one-step site 1 (MOS1) and MOS2, which serve as the universal overlapping sites for Gibson Assembly reactions at the ΔE1 region of the Ad genome. To reduce cloning background, we inserted the bacterial suicide gene ccdB between MOS1 and MOS2. By carrying out two proof-of-principle experiments to express copGFP and mouse BMP9 (mBMP9), we demonstrated that the GDA reactions were highly efficient and yielded >95% positive colonies. The resultant recombinant Ad plasmids were effectively packaged into Ads within 7 days in 293pTP cells. Ad-mediated expression of mBMP9 in mesenchymal stem cells (MSCs) was shown to effectively induce osteogenic differentiation both in vitro and in vivo. Collectively, these results demonstrate that the reported OSCA system significantly simplifies the Ad-making process, which should further facilitate Ad-based applications in basic, translational, and clinical research.
Results
Development of the destination vectors for the one-step construction of adenovirus (OSCA) system using GDA technology
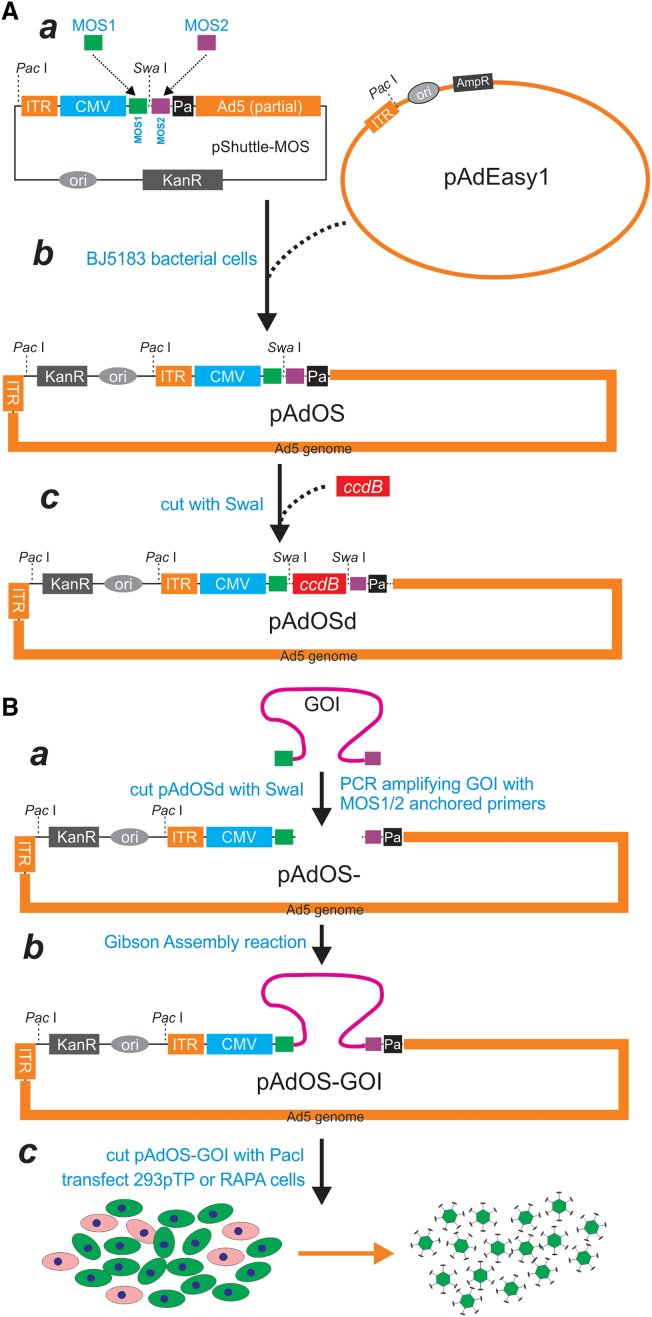
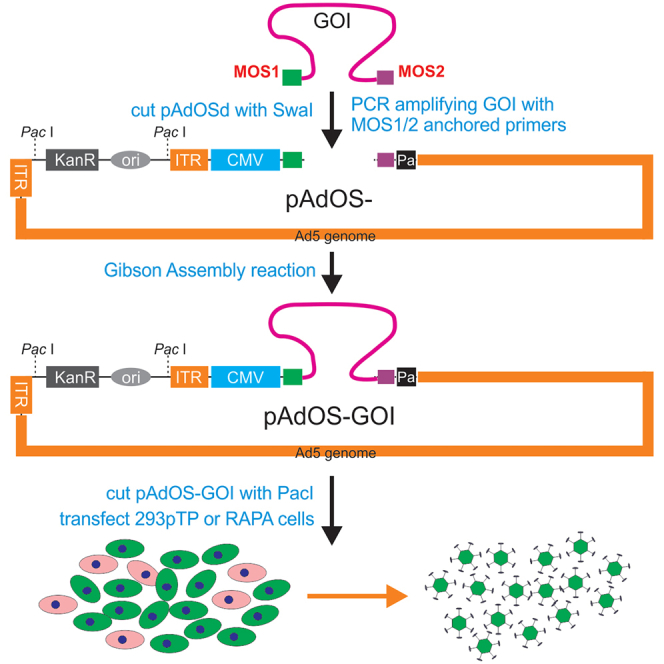
To develop a panel of adenoviral vectors that can serve as common recipients for GDA reactions, we modified three of the first-generation adenoviral shuttle vectors of the AdEasy system,12,13 pShuttle-CMV, pAdTrack-CMV, and pAdTrace-CMV, by subcloning an oligo cassette that contains a SwaI site flanked by two unique 20-bp sequences, namely MOS1 and MOS2 (Figure 1A, a; Table S1), to generate pShuttle-MOS, pAdTrack-MOS, and pAdTrace-MOS vectors. In order to increase the efficiency and accuracy of homologous recombination, these shuttle plasmids were linearized with PmeI and then transformed into BJ5183/pAdEasy1 bacterial cells, which contain the backbone of the adenoviral genome, for homologous recombination (Figure 1A, b). The kanamycin-resistant clones were picked up and subsequently confirmed by PCR and sequencing, resulting in pAdOS, pAdGOS, and pAdROS vectors.
Figure 1.
Schematic depiction of the OSCA system using the GDA technology
(A) Construction of the destination/recipient vectors for the OSCA system. Two unique sequences MOS1 and MOS2, flanked the unique SwaI site, were first engineered in a first-generation adenoviral shuttle vector, resulting in pShuttle-MOS (a). Linearized pShuttle-MOS was transformed into the pAdEasy-1-containing BJ5183 bacterial cells and selected for Kan-resistant pAdOS plasmid (b), which was subsequently confirmed by PCR and sequencing. The ccdB gene fragment flanked with SwaI sites was directly subcloned into the SwaI-cut pAdOS and grown in DB3.1 bacterial cells, resulting in the OSCA destination/recipient pAdOSd vector (c). Two alternative destination vectors pAdGOSd and pAdROSd, which co-express GFP and RFP, respectively, were constructed in a similar fashion (Figures S2 and S3). (B) Gibson Assembly-mediated one-step construction of recombinant Ads. The coding region of the GOI is first PCR amplified with MOS1- and MOS2-anchored primers (a), and the purified PCR fragment is assembled with the SwaI-digested destination vector, e.g., pAdOSd, pAdGOSd, or pAdROSd, through Gibson Assembly reactions (b). The resultant plasmids are verified, linearized by PacI digestion, and transfected into packaging cells such as 293pTP, leading to robust Ad generation in 5–7 days (c). The adenoviral lysate can be further amplified in HEK-293 cells to accomplish high titers.
To reduce potential background in GDA reactions, we further modified the above vectors by inserting the suicide gene ccdB expression cassette flanked with SwaI sites through GDA reactions, and grown in DB3.1 bacterial cells, resulting in the OSCA destination/recipient vectors, pAdOSd, pAdGOSd, and pAdROSd (Figure 1A, c). As shown in Figure S1 a versus b, the inclusion of ccdB in pAdGOS effectively eliminated any bacterial colony formation by pAdGOSd when transformed into competent DH10B cells. The vector maps and full-length sequences for pAdGOSd and pAdROSd are presented in Figures S2 and S3.
The practical use of the GDA-based OSCA system for transgene expression is illustrated in Figure 1B. Briefly, the MOS1 and MOS2-anchored primers are used to amplify the coding region of the gene of interest (GOI) (Figure 1B, a). The resulting PCR fragment is gel purified, and mixed with the SwaI-linearized destination vector, such as pAdOSd, pAdGOSd, or pAdROSd, for GDA reactions to generate the GOI-containing recombinant adenoviral plasmid, pAdOS-GOI (Figure 1B, b). This recombinant adenoviral plasmid is digested with PacI and then transfected into the optimized Ad packaging cells, such as 293pTP and RAPA as described,20,21 leading to robust Ad packaging and production in 5–7 days (Figure 1B, c). The initial adenoviral lysate can be further amplified in HEK-293, 293pTP, or RAPA cells to attain high titers for in vitro or in vivo use.
Efficient generation of the copGFP-expressing adenoviral vector using the one-step construction of adenovirus system
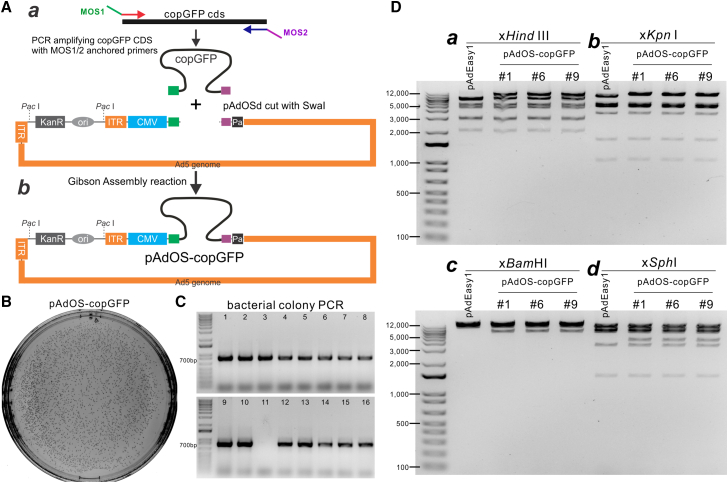
To carry out a proof-of-principle experiment, we sought to use the OSCA system to make an adenoviral vector expressing the marker gene copGFP. We first amplified the coding sequence of copGFP with MOS1- and MOS2-anchored primers, and purified the fragment for GDA reactions (Figure 2A,s a and b), yielding adenoviral plasmid pAdOS-copGFP. Using the NEBuilder HiFi DNA Assembly kit, we found that the GDA reactions were generally very efficient as ∼10% of the assembly products yielded nearly thousands of colonies after direct plating (Figure 2B). PCR screening of randomly picked up clones indicated that 15 of 16 were positive for the presence of the copGFP transgene (Figure 2C). To further verify the structural integrity of adenoviral genome of the pAdOS-copGFP plasmids generated from the Gibson Assembly reactions, we digested the representative clones, in comparison with the adenoviral backbone vector pAdEasy1, with four restriction enzymes, Hind III (Figure 2D, a), Kpn I (Figure 2D, b), Bam HI (Figure 2D, c), and Sph I (Figure 2D, d). The restriction digestion results indicated that all three selected clones yielded the same expected digestion patterns (Figure 2D). The assembled junctions were further verified by DNA sequencing (Figure S4A). Collectively, these results demonstrate that the OSCA system is highly efficient for GDA-based rapid construction of recombinant Ad plasmids.
Figure 2.
Construction and characterization of copGFP-expressing adenoviral vector using the OSCA system
(A) Construction of AdOS-copGFP using the OSCA system. The copGFP coding sequence was PCR amplified with MOS1- and MOS2-anchored primers (a), followed by Gibson Assembly (b). (B) Bacterial colonies post the Gibson Assembly reaction. (C) Identification of pAdOS-copGFP using PCR screening of bacterial colonies. Randomly picked up 16 colonies were PCR amplified with copGFP specific primers, and all but one (#11) were positive for copGFP. (D) Validation of adenoviral recombinant pAdOS-copGFP clones. The representative three clones, along with the control adenoviral backbone vector pAdEasy1, were digested with Hind III (a), Kpn I (b), Bam HI (c), and Sph I (d). The digested plasmid DNA was resolved in 1% agarose gels.
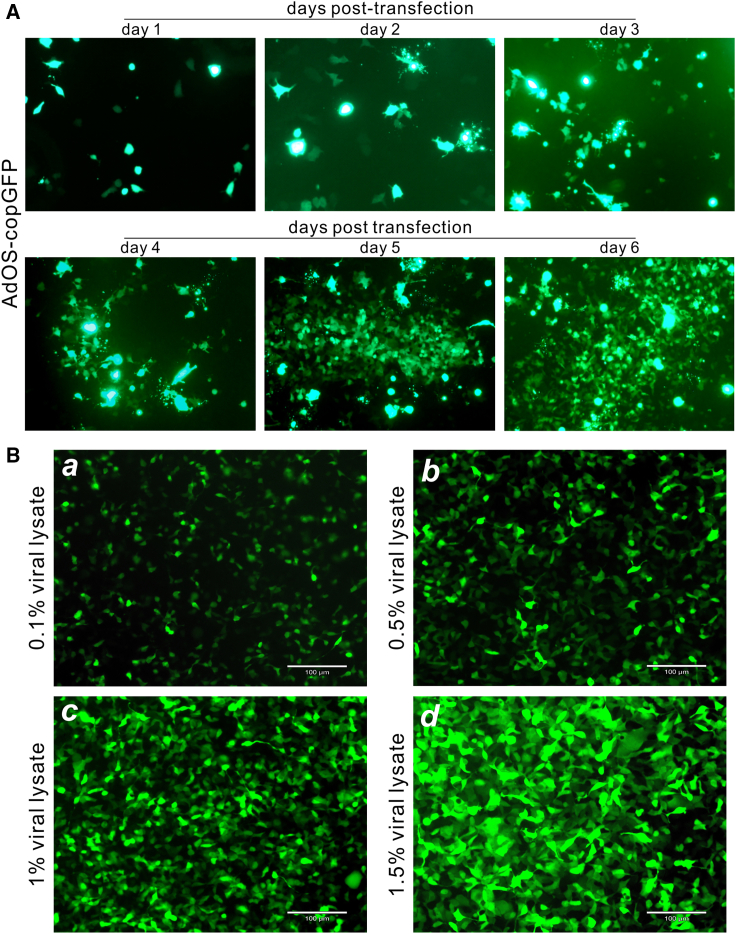
We next tested whether the pAdOS-copGFP plasmid could be effectively packaged into Ad. The pAdOS-copGFP plasmid was first linearized with Pac I restriction enzyme, and then transfected into 293pTP cells (or RAPA cells, data not shown). While the transfection efficiency was modest, the GFP signal became increasingly intensified, and formed comet-like foci (indicating active focal amplification and production of Ad) at 4 days after transfection, becoming apparent at day 7, which was also the endpoint of the Ad packaging (Figures 3A and S5A). When the viral lysate was prepared at 7 days after transfection, we infected HEK-293 cells with different titers (as measured by percentage of the collected viral lysate) and demonstrated that significant copGFP expression was observed in a dose-dependent manner and was detected at as low as 0.1% of viral lysate (Figure 3B,s a–d). High-titer AdOS-copGFP (e.g., >1012 pfu/mL) was obtained through two to four rounds of repeated infections of HEK-293, 293pTP or RAPA cells. These results indicate that the OSCA system should be highly efficient for construction of recombinant Ads.
Figure 3.
Packaging and production of recombinant Ads generated from the Gibson Assembly technology
(A) The initial production of AdOS-copGFP virus in 293pTP cells. At the indicated time points, GFP signal was also recorded. Comet-like Ad-producing foci were apparent at 5 days after transfection. Representative images are shown. Both GFP and bright field images were also recorded at a lower magnification (4×) (Figure S5A). (B) Transduction efficiency of adenoviral lysate. The collected adenoviral lysate was used to infect subconfluent HEK-293 cells at the indicated viral titers (percentage of viral lysate volume). GFP signals were recorded at 24 h post infection. Representative images are shown.
High osteogenic activity of mouse BMP9 expressed by the adenoviral vector generated using the one-step construction of adenovirus system
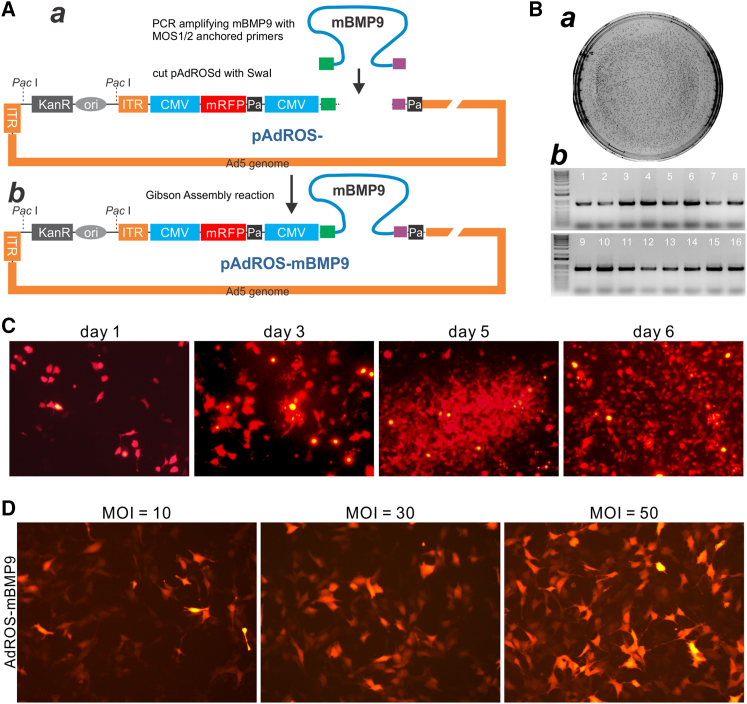
We further demonstrated the biological functionality of OSCA-produced Ad by constructing the adenoviral vector AdROS-mBMP9 to express mBMP9. We and others have demonstrated that, through a comprehensive analysis of the 14 types of human BMPs, human BMP9 is one of the most potent osteogenic factors in promoting bone formation from MSCs both in vitro and in vivo.22, 23, 24, 25 However, no studies were carried out to investigate the osteogenic activity of mBMP9. Here, we amplified the coding region of mBMP9 with MOS1- and MOS2-anchored primers, and generated pAdROS-mBMP9 using the OSCA system (Figure 4A, a and b). Consistent with the results shown in Figures 2B–2D, the GDA reactions were efficient and generated a high percentage of positive clones (Figure 4B, a and b; Figure S4B). Furthermore, the Pac I-linearized pAdROS-mBMP9 was shown to generate Ad with high efficiency in packaging cells (Figures 4C and S5B). High-titer AdROS-mBMP9 (e.g., >1012 pfu/mL) was obtained through two to four rounds of repeated infections of HEK-293, 293pTP, or RAPA cells. The generated AdROS-mBMP9 virus was shown to effectively transduce imBMSC MSCs in a titer-dependent fashion (Figure 4D).
Figure 4.
Construction and production of mBMP9-expressing Ad vector using the OSCA system
(A) Packaging and production of recombinant Ads generated from the Gibson Assembly technology. The coding region of mBMP9 was PCR amplified with gene-specific primers containing MOS1 and MOS2 sequences (a), and Gibson assembled with SwaI-digested pAdROSd vector to generate pAdROS-mBMP9 (b). (B) Bacterial colony verification. The Gibson Assembly product was transformed into DH10B (a) and subjected to colony PCR with BMP9-specific primers (b). (C) Packaging of AdROS-mBMP9 in 293pTP cells. At the indicated time points, RFP signal was also recorded. Comet-like Ad-producing foci were apparent at 5 days after transfection. Representative images are shown. Both RFP and bright field images were also recorded at a lower magnification (4×) (Figure S5B). (D) Transduction efficiency of AdROS-mBMP9 in MSCs. Subconfluent imBMSC cells were infected with the indicated titers of AdROS-mBMP9, and RFP signal was recorded at 36 h post infection. Representative images are shown. MOI, multiplicity of infection, indicating number of infectious Ads per cell.
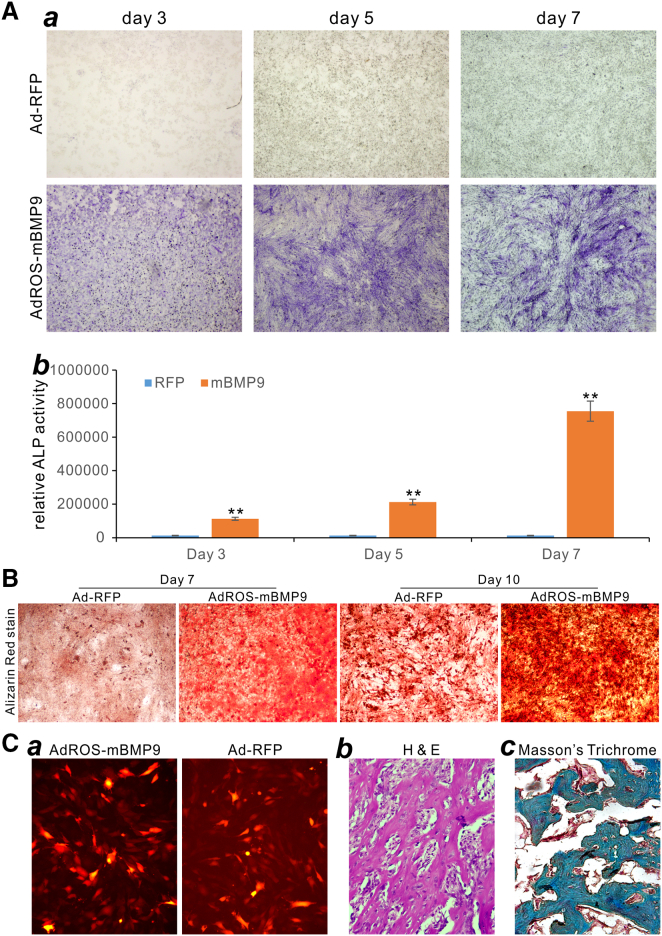
To test the biological function of mBMP9, we infected the mouse bone marrow-derived MSCs imBMSCs with AdROS-mBMP9 and Ad-RFP control viruses, and found that mBMP9 effectively induced alkaline phosphatase (ALP) activities in a time-course-dependent fashion, compared with that of the control Ad-RFP group (Figure 5A,s a and b), indicating that Ad-mediated expression of mBMP9 was able to induce early osteogenic marker ALP in MSCs. Furthermore, the Ad-mediated mBMP9 expression was shown to induce matrix mineralization (Figure 5B). When the MSCs were infected with AdROS-mBMP9 or Ad-RFP (Figure 5C, a), collected and subcutaneously injected into the athymic nude mice, robust bone formation was detected in the mBMP9 group, but not in the RFP control group, at 4 weeks after implantation as assessed by hematoxylin-eosin (H&E) staining (Figure 5C, b). Trichrome staining further revealed that mBMP9 induced robust trabecular bone formation with highly mineralized bone matrix (Figure 5C, c). Collectively, these results demonstrate that mBMP9 can be easily included into adenoviral vector, and the resultant Ad can effectively transduce MSCs to induce osteogenic differentiation.
Figure 5.
Ad-mediated BMP9 transgene expression induces osteogenic differentiation of MSCs
(A) Ad-mediated expression of BMP9 effectively induces osteogenic marker ALP in MSCs. Mouse imBMSCs were infected with AdROS-mBMP9 or Ad-RFP. At the indicated time points, the ALP activities of the infected cells were assessed histochemically (a) and quantitatively (b). ∗∗p < 0.01, compared with respective RFP group. (B) Ad-mediated expression of BMP9 effectively induces matrix mineralization in MSCs. AdROS-mBMP9- or Ad-RFP-infected imBMSCs were cultured in mineralization medium and subjected to Alizarin Red S staining at the indicated time points. Representative results are shown. (C) Ad-mediated expression of BMP9 effectively induces ectopic bone formation in MSCs. Subconfluent imBMSCs were infected with AdROS-mBMP9 or Ad-RFP (a). The infected cells were collected and injected subcutaneously into athymic nude mice. While no masses were formed in the Ad-RFP group, bony masses were retrieved from the AdROS-mBMP9 group at 4 weeks after implantation and subjected to H&E staining (b) and Masson’s trichrome staining (c). Representative results are shown.
Discussion
In order to simplify the Ad-making process, we developed the OSCA system by taking advantage of the GDA technology. Using the engineered Ad recipient vectors that contain the bacterial suicide gene ccdB flanked with MOS1 and MOS2 sites at the ΔE1 region, we demonstrated that the GDA reactions were highly efficient and yielded >95% positive colonies. The resultant recombinant Ad plasmids were effectively packaged into Ads within 7 days in 293pTP cells. Ad-mediated expression of mBMP9 in MSCs was shown to effectively induce osteogenic differentiation. These results demonstrate that, like its human counterpart, mBMP9 exhibits high osteogenic activity in MSCs. Therefore, our results demonstrate that the OSCA system can significantly streamline the Ad-making process and should further facilitate Ad-based applications in basic, translational, and clinical research.
Even though the second and third generations of Ad vectors have been developed, the first-generation Ad vectors remain among the most commonly used Ad vectors for basic and translational research. Many efforts have been devoted to developing techniques for rapid and efficient production of the first-generation Ad vectors.1,2 Interestingly, several recent studies explored the use of the GDA technology for the construction of special Ad vectors.17, 18, 19 Freedman et al. used the GDA technology to generate a modified oncolytic group B Ad EnAdenotucirev (EnAd) to express a bispecific single-chain antibody, which was controlled by the virus major late promoter.17 Pan et al. exploited the GDA technology and constructed a replication-competent infectious clone of human Ad type 14.18 Furthermore, Zou et al. utilized GDA technique to generate an infectious clone of fowl Ad 4 (FAdV-4).19 However, those reported systems were mainly designed to construct specific oncolytic Ad, or rarely used Ad type 14 and fowl Ad 4, which, unlike the OSCA system, are not suitable for conventional Ad generation. Two decades ago, we and others took advantage of highly efficient homologous recombination machinery in certain microorganisms, notably bacteria and yeast, and developed several systems to generate transgene-containing Ad vectors with high efficiency.10, 11, 12, 13 In fact, our previously developed AdEasy system remains as one of the most commonly used techniques worldwide to generate Ad vectors.1,2,12,13 An essential component of the AdEasy system is the RecA+ Escherichia coli strain BJ5183 cells, which exhibit a high rate of homologous recombination, yet still allow the generation of stable large recombinants.1,12,13 However, the BJ5183 cells exhibit a relatively low transformation efficiency, compared with conventional strains used for molecular cloning, which poses technical challenges to many researchers with average cloning experience. It is thus highly desirable to make the Ad-making process as efficient and simple as possible. Therefore, the OSCA system reported here provides a timely technical upgrade of the Ad-making process.
It is noteworthy that, while our work mainly focused on Ad type 5, the OSCA system can be easily further modified and adapted for other Ad serotypes, such as type 2. While the reported OSCA system is mainly designed to construct Ads that overexpress transgenes, it is conceivable that the OSCA system can be further modified to accommodate the expression of siRNAs for gene silencing. In this case, the siRNA expression cassette may need to be constructed in an MOS1/2-containing shuttle vector, followed by GDA assembly to complete the construction of siRNA-expressing Ad vector. Lastly, it is worth pointing out that, while it is critical to develop techniques for efficient Ad making, it is equally important to enhance Ad packaging and infection efficiency. We have found that the cationic polymer polybrene can drastically enhance Ad infection efficiency in mammalian cells.26 We also demonstrated that overexpression of Ad5 precursor terminal protein (pTP), or both pTP and E1A in HEK-293 cells, namely 293pTP and RAPA cell lines, respectively, dramatically accelerates Ad packaging and amplification processes.20,21 Thus, a combined use of the OSCA system and 293pTP or RAPA cells should significantly expedite Ad production.
Conclusions
In order to simplify the Ad-making process, we developed the OSCA system by taking advantage of the GDA technology. The essential component of this OSCA system is the Ad recipient vectors that contain the bacterial suicide gene ccdB flanked with MOS1 and MOS2 sites in the ΔE1 region. In two proof-of-principle experiments, we demonstrated that the GDA reactions were highly efficient, and that the resultant recombinant Ad plasmids were effectively packaged into Ads. Ad-mediated expression of mBMP9 in MSCs was shown to effectively induce osteogenic differentiation both in vitro and in vivo. Collectively, our results demonstrate that the OSCA system drastically simplifies the Ad-making process and should facilitate Ad-based applications in basic, translational, and clinical research.
Materials and methods
Cell culture, enzymes, and chemicals
Human HEK-293 derivative lines 293pTP and RAPA cells were used for Ad packaging and amplification as previously described.20,21 Mouse bone marrow-derived MSCs imBMSCs were previously characterized.27 All cells were maintained in DMEM (Dulbecco's Modified Eagle Medium) containing 10% fetal bovine serum (FBS; Gemini Bio-Products), 100 U/mL penicillin, and 100 μg/mL streptomycin at 37°C in 5% CO2 as described previously.28, 29, 30 All restriction endonucleases, and the Gibson Assembly Master Mix or the NEBuilder HiFi DNA Assembly kit, were purchased from New England Biolabs (NEB; Ipswich, MA). Unless indicated otherwise, other chemicals were purchased from Thermo Fisher Scientific (Waltham, MA) or Millipore Sigma (St Louis, MO).
Construction of the adenoviral backbone-containing Gibson DNA Assembly recipient vectors pAdOSd, pAdROSd, and pAdGOSd
The CMV-PA expression cassette of the pShuttle-CMV, pAdTrack-CMV, or pAdTrace-CMV shuttle vectors from the AdEasy system1,12,13,31 were first modified by inserting an oligo cassette containing a SwaI restriction site flanked with two unique 20-bp sequences, namely MOS1 and MOS2, at the BamHI and XbaI sites of the parental shuttle vectors, resulting in the pShuttle-MOS vector (e.g., from pShuttle-CMV) (Table S1). This vector was linearized with PmeI and subjected to homologous recombination reactions in pAdEasy1-containing BJ5183 bacterial cells. The kanamycin-resistant colonies were grown up and verified by PCR and restriction digestion to generate the pAdOS vector.
In order to reduce the background of GDA reactions, the bacterial suicide gene ccdB expression cassette was PCR amplified with both primers anchored with SwaI sites, ligated into the SwaI-digested pAdOS vector, and transformed into competent DB3.1 bacterial cells. Bacterial colonies were PCR screened, and positive candidate clones were grown up and further verified by PCR amplification, restriction digestions, and DNA sequencing. The resultant GDA recipient vector was designated as pAdOSd. Similar recipient vectors were also constructed from pAdTrack-CMV and pAdTrace-CMV shuttle vectors, and designated as pAdGOSd and pAdROSd, respectively. All oligo sequences are listed in Table S1. The vector maps and sequences for pAdGOSd and pAdROSd are shown in Figures S2 and S3. All cloning and assembly junctions were verified by DNA sequencing.
Gibson DNA Assembly reactions
The GDA reactions were conducted by using the Gibson Assembly Master Mix or NEBuilder HiFi DNA Assembly kit from NEB as described.32 The coding region for the GOI (see below) was PCR amplified using the Phusion High-Fidelity PCR kit. Each assembly reaction (usually in a reaction volume of 10–15 μL) contained approximately 100 ng of insert DNA and 50 ng of the SwaI-linearized pAdOSd, pAdGOSd, or pAdROSd vector, and was incubated at 50°C for 40–60 min. After the GDA reaction was completed, the reaction mix was processed by 7.5 M ammonium acetate/ethanol precipitation. Alternatively, the GDA reaction mix was digested with SwaI in a 100-μL reaction at 25°C for 10 min, followed by 7.5 M ammonium acetate/ethanol precipitation. The pellet was resuspended in 30 μL of double-distilled H2O (ddH2O), and 15μL were used to transform electro-competent DH10B cells. Then 500 μL of LB (Lysogeny Broth) were added to the transformation mix, and 100 μL were plated onto LB/Kan plates, followed by 37°C incubation overnight. Colony PCR screening was carried out using primers specific for the GOI. Positive clones were further verified by DNA sequencing.
Generation and amplification of recombinant Ads expressing copGFP and mouse BMP9 using one-step construction of adenovirus
The coding sequences for copGFP (from pCDF1-MCS1-EF1-copGFP) and mBMP9 (EST DNA clone from TransOmic Technologies, Huntsville, AL) were PCR amplified with forward primers anchored with the MOS1 and kozak sequences and reverse primers anchored with the MOS2 sequence (Table S1). The PCR fragments were gel purified and used for GDA reactions. Positive candidate clones were screened by colony PCR and validated by restriction digestions and DNA sequencing. The resultant recombinant Ad plasmids were designated as pAdOS-copGFP and pAdROS-mBMP9, respectively.
For making recombinant Ads, these adenoviral plasmids were first linearized with PacI to liberate adenoviral inverted terminal repeat (ITR) sequences at both ends, and then transfected into 293pTP or RAPA cells as described.20,21 Apparent Ad packaging and production was obtained 5–7 days after transfection. Adenoviral lysates were prepared by multiple cycles of freeze-thaw as described.13,31 High titer Ads (e.g., >1012 pfu/mL) were usually obtained through two to four rounds of repeated infections of HEK-293, 293pTP, or RAPA cells, and the resulting Ads were designated as AdOS-copGFP and AdROS-mBMP9, respectively. Analogous Ad expressing only RFP (Ad-RFP) was used as a control.25,33, 34, 35, 36, 37 For the adenoviral infections, polybrene (4–8 μg/mL) was added to enhance infection efficiency as previously reported.26
Qualitative and quantitative assays of alkaline phosphatase activity
ALP activity was assessed quantitatively with a modified assay using the Great Escape SEAP Chemiluminescence assay kit (BD Clontech, Mountain View, CA) and qualitatively with histochemical staining assay (using a mixture of 0.1 mg/mL napthol AS-MX phosphate and 0.6 mg/mL Fast Blue BB salt), as previously described.38, 39, 40, 41 Each assay condition was performed in triplicate and the results were repeated in at least three independent experiments.
Ectopic bone formation assay
The animal studies were conducted by following the guidelines approved by the Institutional Animal Care and Use Committee (IACUC) of The University of Chicago. Stem cell-mediated ectopic bone formation via MSC implantation was performed as described.36,42,43 Specifically, subconfluent imBMSC cells were infected with AdROS-mBMP9 or Ad-RFP for 16 h, harvested, and resuspended in PBS for subcutaneous injection (5 × 106/injection) into the flanks of athymic nude mice (five per group, 4–6 weeks old, female; ENVIGO, Indianapolis, IN). At 4 weeks after implantation, animals were sacrificed, and the implantation sites were retrieved for histologic evaluation and trichrome staining as described below.
Hematoxylin-eosin (H&E) analysis and trichrome staining
Retrieved bony tissues were fixed, decalcified in 10% buffered formalin, and embedded in paraffin. Serial sections of the embedded specimens were stained with H&E. Trichrome staining was carried out as previously described.44, 45, 46, 47, 48
Statistical analysis
Quantitative ALP assays were performed in triplicate. Statistical significance was determined by one-way analysis of variance and the student's t test. A value of p < 0.05 was defined statistically significant.
Data availability
The data that support this study are available from the corresponding authors upon reasonable request. Source data are provided with this paper.
Acknowledgments
A small amount of T.C.H.'s and R.R.R.'s efforts in the reported work was funded by research grants from the National Institutes of Health (CA226303 to T.C.H. and DE030480s to R.R.R.). X.H. was funded by a research grant from Chongqing Science and Technology Commission, China (cstc2018jcyjAX0561). W.W. was supported by the Medical Scientist Training Program of the National Institutes of Health (T32 GM007281). This project was also supported in part by The University of Chicago Cancer Center Support Grant (P30CA014599) and the National Center for Advancing Translational Sciences of the National Institutes of Health through grant number UL1 TR000430. T.C.H. was primarily supported by the Mabel Green Myers Research Endowment Fund and The University of Chicago Orthopaedics Alumni Fund. Funding sources were not involved in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the paper for publication.
Author contributions
N.N., F.H., J.F., F.D., and T.-C.H. conceived and designed the study. N.N., F.H., F.D., H.W., D.S., J.L., Y.Z., and Hongwei Wang performed the experiments and collected data. X.H., P.Z., C.C., D.A.H., M.S., K.H.Q., W.W., D.Q., and B.H.-S. participated in experiments and/or provided essential experimental materials. T.-C.H., N.N., F.H., R.C.H., H.H.L., R.R.R. and L.S. drafted and revised the manuscript. All authors reviewed, edited, and approved the manuscript.
Declarartion of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omto.2021.11.011.
Contributor Information
Tong-Chuan He, Email: tche@uchicago.edu.
Jiaming Fan, Email: fanjiaming1988@cqmu.edu.cn.
Supplemental information
References
- 1.Lee C.S., Bishop E.S., Zhang R., Yu X., Farina E.M., Yan S., Zhao C., Zheng Z., Shu Y., Wu X., et al. Adenovirus-mediated gene delivery: potential applications for gene and cell-based therapies in the New era of personalized medicine. Genes Dis. 2017;4:43–63. doi: 10.1016/j.gendis.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breyer B., Jiang W., Cheng H., Zhou L., Paul R., Feng T., He T.C. Adenoviral vector-mediated gene transfer for human gene therapy. Curr. Gene Ther. 2001;1:149–162. doi: 10.2174/1566523013348689. [DOI] [PubMed] [Google Scholar]
- 3.Crystal R.G. Adenovirus: the first effective in vivo gene delivery vector. Hum. Gene Ther. 2014;25:3–11. doi: 10.1089/hum.2013.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ehrke-Schulz E., Zhang W., Gao J., Ehrhardt A. Recent advances in preclinical developments using adenovirus hybrid vectors. Hum. Gene Ther. 2017;28:833–841. doi: 10.1089/hum.2017.140. [DOI] [PubMed] [Google Scholar]
- 5.Maggio I., Stefanucci L., Janssen J.M., Liu J., Chen X., Mouly V., Goncalves M.A. Selection-free gene repair after adenoviral vector transduction of designer nucleases: rescue of dystrophin synthesis in DMD muscle cell populations. Nucleic Acids Res. 2016;44:1449–1470. doi: 10.1093/nar/gkv1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham F.L., Prevec L. Methods for construction of adenovirus vectors. Mol. Biotechnol. 1995;3:207–220. doi: 10.1007/BF02789331. [DOI] [PubMed] [Google Scholar]
- 7.Mizuguchi H., Kay M.A. Efficient construction of a recombinant adenovirus vector by an improved in vitro ligation method. Hum. Gene Ther. 1998;9:2577–2583. doi: 10.1089/hum.1998.9.17-2577. [DOI] [PubMed] [Google Scholar]
- 8.Hardy S., Kitamura M., Harris-Stansil T., Dai Y., Phipps M.L. Construction of adenovirus vectors through Cre-lox recombination. J. Virol. 1997;71:1842–1849. doi: 10.1128/jvi.71.3.1842-1849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng P., Parks R.J., Cummings D.T., Evelegh C.M., Sankar U., Graham F.L. A high-efficiency Cre/loxP-based system for construction of adenoviral vectors. Hum. Gene Ther. 1999;10:2667–2672. doi: 10.1089/10430349950016708. [DOI] [PubMed] [Google Scholar]
- 10.Ketner G., Spencer F., Tugendreich S., Connelly C., Hieter P. Efficient manipulation of the human adenovirus genome as an infectious yeast artificial chromosome clone. Proc. Natl. Acad. Sci. U S A. 1994;91:6186–6190. doi: 10.1073/pnas.91.13.6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chartier C., Degryse E., Gantzer M., Dieterle A., Pavirani A., Mehtali M. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 1996;70:4805–4810. doi: 10.1128/jvi.70.7.4805-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He T.C., Zhou S., da Costa L.T., Yu J., Kinzler K.W., Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. U S A. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo J., Deng Z.L., Luo X., Tang N., Song W.X., Chen J., Sharff K.A., Luu H.H., Haydon R.C., Kinzler K.W., et al. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat. Protoc. 2007;2:1236–1247. doi: 10.1038/nprot.2007.135. [DOI] [PubMed] [Google Scholar]
- 14.Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., 3rd, Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 15.Gibson D.G. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 2011;498:349–361. doi: 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lienert F., Lohmueller J.J., Garg A., Silver P.A. Synthetic biology in mammalian cells: next generation research tools and therapeutics. Nat. Rev. Mol. Cell Biol. 2014;15:95–107. doi: 10.1038/nrm3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freedman J.D., Hagel J., Scott E.M., Psallidas I., Gupta A., Spiers L., Miller P., Kanellakis N., Ashfield R., Fisher K.D., et al. Oncolytic adenovirus expressing bispecific antibody targets T-cell cytotoxicity in cancer biopsies. EMBO Mol. Med. 2017;9:1067–1087. doi: 10.15252/emmm.201707567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan H., Yan Y., Zhang J., Zhao S., Feng L., Ou J., Cao N., Li M., Zhao W., Wan C., et al. Rapid construction of a replication-competent infectious clone of human adenovirus type 14 by Gibson assembly. Viruses. 2018;10:568. doi: 10.3390/v10100568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou X.H., Bi Z.X., Guo X.J., Zhang Z., Zhao Y., Wang M., Zhu Y.L., Jie H.Y., Yu Y., Hung T., et al. DNA assembly technique simplifies the construction of infectious clone of fowl adenovirus. J. Virol. Methods. 2018;257:85–92. doi: 10.1016/j.jviromet.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 20.Wu N., Zhang H., Deng F., Li R., Zhang W., Chen X., Wen S., Wang N., Zhang J., Yin L., et al. Overexpression of Ad5 precursor terminal protein accelerates recombinant adenovirus packaging and amplification in HEK-293 packaging cells. Gene Ther. 2014;21:629–637. doi: 10.1038/gt.2014.40. [DOI] [PubMed] [Google Scholar]
- 21.Wei Q., Fan J., Liao J., Zou Y., Song D., Liu J., Cui J., Liu F., Ma C., Hu X., et al. Engineering the rapid adenovirus production and amplification (RAPA) cell line to expedite the generation of recombinant adenoviruses. Cell Physiol. Biochem. 2017;41:2383–2398. doi: 10.1159/000475909. [DOI] [PubMed] [Google Scholar]
- 22.Cheng H., Jiang W., Phillips F.M., Haydon R.C., Peng Y., Zhou L., Luu H.H., An N., Breyer B., Vanichakarn P., et al. Osteogenic activity of the fourteen types of human bone morphogenetic proteins (BMPs) J. Bone Joint Surg. Am. 2003;85:1544–1552. doi: 10.2106/00004623-200308000-00017. [DOI] [PubMed] [Google Scholar]
- 23.Mostafa S., Pakvasa M., Coalson E., Zhu A., Alverdy A., Castillo H., Fan J., Li A., Feng Y., Wu D., et al. The wonders of BMP9: from mesenchymal stem cell differentiation, angiogenesis, neurogenesis, tumorigenesis, and metabolism to regenerative medicine. Genes Dis. 2019;6:201–223. doi: 10.1016/j.gendis.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu W., Deng Z., Zeng Z., Fan J., Feng Y., Wang X., Cao D., Zhang B., Yang L., Liu B., et al. Highly expressed BMP9/GDF2 in postnatal mouse liver and lungs may account for its pleiotropic effects on stem cell differentiation, angiogenesis, tumor growth and metabolism. Genes Dis. 2020;7:235–244. doi: 10.1016/j.gendis.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L., Luo Q., Shu Y., Zeng Z., Huang B., Feng Y., Zhang B., Wang X., Lei Y., Ye Z., et al. Transcriptomic landscape regulated by the 14 types of bone morphogenetic proteins (BMPs) in lineage commitment and differentiation of mesenchymal stem cells (MSCs) Genes Dis. 2019;6:258–275. doi: 10.1016/j.gendis.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao C., Wu N., Deng F., Zhang H., Wang N., Zhang W., Chen X., Wen S., Zhang J., Yin L., et al. Adenovirus-mediated gene transfer in mesenchymal stem cells can be significantly enhanced by the cationic polymer polybrene. PLoS One. 2014;9:e92908. doi: 10.1371/journal.pone.0092908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu X., Li L., Yu X., Zhang R., Yan S., Zeng Z., Shu Y., Zhao C., Wu X., Lei J., et al. CRISPR/Cas9-mediated reversibly immortalized mouse bone marrow stromal stem cells (BMSCs) retain multipotent features of mesenchymal stem cells (MSCs) Oncotarget. 2017;8:111847–111865. doi: 10.18632/oncotarget.22915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang X., Chen Q., Luo W., Pakvasa M., Zhang Y., Zheng L., Li S., Yang Z., Zeng H., Liang F., et al. SATB2: a versatile transcriptional regulator of craniofacial and skeleton development, neurogenesis and tumorigenesis, and its applications in regenerative medicine. Genes Dis. 2020 doi: 10.1016/j.gendis.2020.10.003. Available Online 17 October 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu X., Li Z., Zhang H., He F., Qiao M., Luo H., Zhang J., Zhang M., Mao Y., Wagstaff W., et al. Modeling colorectal tumorigenesis using the organoids derived from conditionally immortalized mouse intestinal crypt cells (ciMICs) Genes Dis. 2021;8:814–826. doi: 10.1016/j.gendis.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X., Zhao L., Wu X., Luo H., Wu D., Zhang M., Zhang J., Pakvasa M., Wagstaff W., He F., et al. Development of a simplified and inexpensive RNA depletion method for plasmid DNA purification using size selection magnetic beads (SSMBs) Genes Dis. 2021;8:298–306. doi: 10.1016/j.gendis.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He T.-C. John Wiley & Sons, Inc.; 2004. Adenoviral Vectors in Current Protocols in Human Genetics. Vol. Unit 12.4 Vectors for Gene Therapy 12.14.11-12.14.25. [Google Scholar]
- 32.Deng F., Chen X., Liao Z., Yan Z., Wang Z., Deng Y., Zhang Q., Zhang Z., Ye J., Qiao M., et al. A simplified and versatile system for the simultaneous expression of multiple siRNAs in mammalian cells using Gibson DNA Assembly. PLoS One. 2014;9:e113064. doi: 10.1371/journal.pone.0113064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mao Y., Ni N., Huang L., Fan J., Wang H., He F., Liu Q., Shi D., Fu K., Pakvasa M., et al. Argonaute (AGO) proteins play an essential role in mediating BMP9-induced osteogenic signaling in mesenchymal stem cells (MSCs) Genes Dis. 2021;8:918–930. doi: 10.1016/j.gendis.2021.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan J., Feng Y., Zhang R., Zhang W., Shu Y., Zeng Z., Huang S., Zhang L., Huang B., Wu D., et al. A simplified system for the effective expression and delivery of functional mature microRNAs in mammalian cells. Cancer Gene Ther. 2020;27:424–437. doi: 10.1038/s41417-019-0113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan J., Wei Q., Liao J., Zou Y., Song D., Xiong D., Ma C., Hu X., Qu X., Chen L., et al. Noncanonical Wnt signaling plays an important role in modulating canonical Wnt-regulated stemness, proliferation and terminal differentiation of hepatic progenitors. Oncotarget. 2017;8:27105–27119. doi: 10.18632/oncotarget.15637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li R., Zhang W., Cui J., Shui W., Yin L., Wang Y., Zhang H., Wang N., Wu N., Nan G., et al. Targeting BMP9-promoted human osteosarcoma growth by inactivation of notch signaling. Curr. Cancer Drug Targets. 2014;14:274–285. doi: 10.2174/1568009614666140305105805. [DOI] [PubMed] [Google Scholar]
- 37.Cao D., Lei Y., Ye Z., Zhao L., Wang H., Zhang J., He F., Huang L., Shi D., Liu Q., et al. Blockade of IGF/IGF-1R signaling axis with soluble IGF-1R mutants suppresses the cell proliferation and tumor growth of human osteosarcoma. Am. J. Cancer Res. 2020;10:3248–3266. [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang B., Yang L., Zeng Z., Feng Y., Wang X., Wu X., Luo H., Zhang J., Zhang M., Pakvasa M., et al. Leptin potentiates BMP9-induced osteogenic differentiation of mesenchymal stem cells through the activation of JAK/STAT signaling. Stem Cells Dev. 2020;29:498–510. doi: 10.1089/scd.2019.0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo W., Zhang L., Huang B., Zhang H., Zhang Y., Zhang F., Liang P., Chen Q., Cheng Q., Tan D., et al. BMP9-initiated osteogenic/odontogenic differentiation of mouse tooth germ mesenchymal cells (TGMCS) requires Wnt/beta-catenin signalling activity. J. Cell Mol. Med. 2021;25:2666–2678. doi: 10.1111/jcmm.16293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He F., Ni N., Zeng Z., Wu D., Feng Y., Li A.J., Luu B., Li A.F., Qin K., Wang E., et al. FAMSi: a synthetic biology approach to the fast assembly of multiplex siRNAs for silencing gene expression in mammalian cells. Mol. Ther. Nucleic Acids. 2020;22:885–899. doi: 10.1016/j.omtn.2020.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z., Liu J., Zeng Z., Fan J., Huang S., Zhang L., Zhang B., Wang X., Feng Y., Ye Z., et al. lncRNA Rmst acts as an important mediator of BMP9-induced osteogenic differentiation of mesenchymal stem cells (MSCs) by antagonizing Notch-targeting microRNAs. Aging (Albany NY) 2019;11:12476–12496. doi: 10.18632/aging.102583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang Q., Sun M.H., Cheng H., Peng Y., Montag A.G., Deyrup A.T., Jiang W., Luu H.H., Luo J., Szatkowski J.P., et al. Characterization of the distinct orthotopic bone-forming activity of 14 BMPs using recombinant adenovirus-mediated gene delivery. Gene Ther. 2004;11:1312–1320. doi: 10.1038/sj.gt.3302298. [DOI] [PubMed] [Google Scholar]
- 43.Li R., Zhang W., Yan Z., Liu W., Fan J., Feng Y., Zeng Z., Cao D., Haydon R.C., Luu H.H., et al. Long non-coding RNA (LncRNA) HOTAIR regulates BMP9-induced osteogenic differentiation by targeting the proliferation of mesenchymal stem cells (MSCs) Aging. 2021;13:4199–4214. doi: 10.18632/aging.202384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen L., Jiang W., Huang J., He B.C., Zuo G.W., Zhang W., Luo Q., Shi Q., Zhang B.Q., Wagner E.R., et al. Insulin-like growth factor 2 (IGF-2) potentiates BMP-9-induced osteogenic differentiation and bone formation. J. Bone Miner Res. 2010;25:2447–2459. doi: 10.1002/jbmr.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan S., Zhang R., Wu K., Cui J., Huang S., Ji X., An L., Yuan C., Gong C., Zhang L., et al. Characterization of the essential role of bone morphogenetic protein 9 (BMP9) in osteogenic differentiation of mesenchymal stem cells (MSCs) through RNA interference. Genes Dis. 2018;5:172–184. doi: 10.1016/j.gendis.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song D., Zhang F., Reid R.R., Ye J., Wei Q., Liao J., Zou Y., Fan J., Ma C., Hu X., et al. BMP9 induces osteogenesis and adipogenesis in the immortalized human cranial suture progenitors from the patent sutures of craniosynostosis patients. J. Cell Mol. Med. 2017;21:2782–2795. doi: 10.1111/jcmm.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye J., Wang J., Zhu Y., Wei Q., Wang X., Yang J., Tang S., Liu H., Fan J., Zhang F., et al. A thermoresponsive polydiolcitrate-gelatin scaffold and delivery system mediates effective bone formation from BMP9-transduced mesenchymal stem cells. Biomed. Mater. 2016;11:025021. doi: 10.1088/1748-6041/11/2/025021. [DOI] [PubMed] [Google Scholar]
- 48.Zhang H., Wang J., Deng F., Huang E., Yan Z., Wang Z., Deng Y., Zhang Q., Zhang Z., Ye J., et al. Canonical Wnt signaling acts synergistically on BMP9-induced osteo/odontoblastic differentiation of stem cells of dental apical papilla (SCAPs) Biomaterials. 2015;39:145–154. doi: 10.1016/j.biomaterials.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support this study are available from the corresponding authors upon reasonable request. Source data are provided with this paper.