We read with great interest the recent article published in Gut in which Yeoh et al demonstrated that gut microbiota composition of recovered patients with COVID-19 remained significantly distinct from uninfected controls.1 Persisting symptoms, also known as ‘long COVID-19’, have been reported in a significant proportion of patients following hospital discharge.2 3 Gut dysbiosis might link to long COVID-19 risks.1 Few studies have focused on the recovery process of gut microbiota following SARS-CoV-2 infection.
Here, we conducted a prospective study to longitudinally monitor alterations of gut microbiota in patients with COVID-19 using 16S rDNA sequencing (detailed methods in online supplementary materials). Faecal microbiota was monitored at three timepoints, acute phase (from illness onset to viral clearance), convalescence (from viral clearance to 2 weeks after hospital discharge), postconvalescence (6 months after hospital discharge).
gutjnl-2021-324090supp001.pdf (106.9KB, pdf)
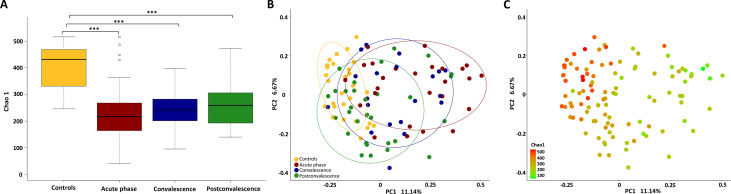
The gut microbiota richness, measured by Chao 1 index, was obviously lower (p<0.01, Wilcoxon rank-sum test; figure 1A) in the acute phase of COVID-19 (median 217, IQR 164–266) as compared with uninfected controls (median 432, IQR 332–468). There was a non-significant increase of the Chao 1 index from the acute phase (median 217, IQR 164–266) to the convalescence (median 241, IQR 202–279) and postconvalescence (median 259, IQR 193–302). A Bray-Curtis based principal coordinated analysis revealed that the overall microbial composition of patients with COVID-19 deviated from the uninfected controls (analysis of similarities, R = – 0.20, p=0.001, figure 1B). There was a tendency of the gut microbiota composition moving toward the controls from the acute phase to recovery phase along the first principal coordinate. Notably, the species richness as estimated by Chao 1 index, can explain the differences along the first principal coordinate (figure 1C).
Figure 1.
Changes of faecal microbial communities in different stages (acute, convalescence, postconvalescence) of patients with COVID-19 (n=30), compared with uninfected controls (n=30). (A) α-Diversity, illustrated by microbiota richness (Chao 1 index), was reduced in COVID-19 (p<0.01, Wilcoxon rank-sum test). Boxes represent the 25th–75th percentile of the distribution; the median is shown as a thick line in the middle of the box; whiskers extend to values with 1.5 times the difference between the 25th and 75th percentiles. ***P<0.001. (B) Principal coordinate analysis (PCoA) of Bray-Curtis distance analysis demonstrated that the overall microbial composition of patients with COVID-19 deviated from the uninfected controls (analysis of similarities, R = – 0.201, p=0.001). (C) The same PCoA plot as (B), coloured by α-diversity measured by Chao 1 index.
The median Chao 1 index in postconvalescence was 259. Patients were further divided into two subgroups according to their Chao 1 index in postconvalescence: low (≤259, n=15) and high (>259, n=15) (table 1). Patients with reduced postconvalescence richness had higher level of CRP (p=0.01), as well as higher occurrence of intensive care unit admission (p=0.03) and high flow nasal catheter oxygen therapy therapy (p=0.03) during the acute phase. In postconvalescence, low richness was associated with reduced pulmonary function of forced vital capacity (p=0.03), forced expiratory volume in the first 1 s of expiration (p=0.02), inspiratory vital capacity (p=0.05) and total lung capacity (p=0.05).
Table 1.
Comparison of clinical characteristics between patients with high or low microbial richness in the recovery phase
| All (n=30) | Low (n=15) | High (n=15) | P value | |
| Age, years | 53.5 (39.75, 59) | 53 (40, 57.5) | 53 (37, 59.75) | 0.72 |
| Male, n (%) | 19 (63.3%) | 10 (66.7%) | 9 (60%) | 0.70 |
| BMI of acute phase, kg/m2 | 24.2 (21.6, 25.2) | 24.9 (22.3, 25.8) | 23.8 (21.3, 25.0) | 0.36 |
| BMI of postconvalescence, kg/m2 | 24.1 (21.1, 26.6) | 24.7 (22.2, 27.2) | 23.1 (21.0, 25.3) | 0.41 |
| Severe illness during hospitalisation, n (%) | 10 (33.3%) | 7 (46.7%) | 3 (20.0%) | 0.12 |
| White cell count, ×109/L * | 5.7 (4.1, 8.9) | 5.7 (4.3, 9.9) | 6.5 (4.1, 8.0) | 0.79 |
| Haemoglobin, g/L * | 138 (128, 149) | 146 (128, 152) | 137 (129, 141) | 0.28 |
| Platelet count, ×109/L * | 187 (158, 231) | 188 (168, 238) | 182 (139, 225) | 0.53 |
| Neutrophil count, ×109/L * | 3.9 (2.6, 7.1) | 4.3 (2.9, 8.7) | 3.9 (2.5, 6.7) | 0.75 |
| Lymphocyte count, ×109/L * | 0.8 (0.6, 1.2) | 0.7 (0.5, 1.2) | 0.9 (0.6, 1.1) | 0.59 |
| D-dimer, mg/L * | 236 (170, 467) | 407 (175, 913) | 199 (170, 320) | 0.08 |
| CRP, mg/L * | 10.8 (5.9, 21.5) | 15.4 (10.5, 45.7) | 7.5 (2.2, 11.4)‡ | 0.01 |
| HFNC during hospitalisation | 7 (23.3%) | 6 (40.0%) | 1 (6.7%)‡ | 0.03 |
| ICU admission | 4 (13.3%) | 4 (26.7%) | 0 (0%)‡ | 0.03 |
| Duration from illness onset to hospital admission, d | 6 (4, 9.7) | 6 (4.5, 10.5) | 6 (1.75, 7.5) | 0.55 |
| Duration of viral shedding in respiratory tract, d | 17.5 (14, 23.7) | 18 (14, 22.5) | 17.5 (14.5, 26.25) | 0.98 |
| Days of hospitalisation, d | 17 (14.2, 23.7) | 17 (15, 22) | 19.5 (13.75, 25.75) | 0.65 |
| PFTs† | ||||
| FVC | 95.5 (89, 105) | 93 (84, 96) | 101.5 (94.7, 107.2)‡ | 0.03 |
| FEV1 | 95.5 (86.2, 107) | 91 (82.5, 97) | 103 (90.5, 112)‡ | 0.02 |
| PEF | 82 (71.2, 101) | 77 (71.5, 90) | 95.5 (75.2, 101) | 0.41 |
| FEV1/FVC ratio | 80.25 (74.4, 88.0) | 80.7 (76.4, 87.9) | 80.25 (73.8, 88.0) | 0.84 |
| FEF25%–75% | 87.5 (65, 120.2) | 87 (69.5, 101) | 87 (65, 135) | 0.63 |
| MEF 75% | 86 (72.7, 111) | 80 (74.5, 89.5) | 104 (77, 111.7) | 0.55 |
| MEF 50% | 80.5 (70, 102.7) | 81 (70, 91) | 79.5 (71.7, 118.7) | 0.88 |
| MEF 25% | 74.5 (57.2, 114) | 74 (50, 100) | 73 (58.7, 127.2) | 0.48 |
| MVV | 88 (67.2, 103.7) | 75 (64, 95.5) | 96 (78.7, 109.2) | 0.08 |
| DLCO | 88.5 (78.5, 95) | 86 (78, 92) | 94.5 (79.5, 99) | 0.07 |
| DLCO/VA | 73 (67, 79.7) | 73 (67, 83.5) | 73 (68.2, 79.2) | 0.82 |
| IVC | 83.5 (77.2, 92) | 82 (70.5, 86.5) | 88.5 (82.5, 93.5)‡ | 0.05 |
| TLC | 96 (91.2, 105.7) | 92 (84.5, 98.5) | 98 (96, 106)‡ | 0.05 |
| RV | 120 (106.2, 130.7) | 123 (102.5, 130.5) | 120 (106.7, 129) | 0.90 |
| RV/TLC | 124.5 (111.2, 142.7) | 132 (113.5, 150) | 120.5 (110.7, 133.5) | 0.23 |
| Exercise capacity | ||||
| Pre-6WMT heart rate | 84 (75.7, 91) | 85 (81.5, 96) | 82 (74.5, 86.5) | 0.15 |
| Pre-6WMT systolic blood pressure | 130.5 (116.2, 142.7) | 133 (114, 156.5) | 123 (115.5, 136.2) | 0.40 |
| Pre-6WMT diastolic blood pressure | 77.5 (69, 90.7) | 71 (66, 99) | 77.5 (70.5, 86.2) | 0.85 |
| Pre-6WMT O2 saturation, % | 98 (97, 99) | 98 (98, 99) | 98 (97, 99) | 0.88 |
| 6WMT distance, m | 600 (540, 640) | 620 (575, 640) | 560 (515, 654) | 0.77 |
| Post-6WMT heart rate | 103.5 (98.2, 113.7) | 106 (100, 115) | 101.5 (97.5, 106.75) | 0.20 |
| Post-6WMT systolic blood pressure | 129 (122, 142.5) | 140 (124, 158.5) | 127.5 (121.2, 132.2) | 0.06 |
| Post-6WMT diastolic blood pressure | 80 (70.7, 86) | 81 (69, 92.5) | 78.5 (73.7, 84) | 0.71 |
| Post-6WMT O2 saturation, % | 98 (97, 98) | 98 (97, 98) | 98 (97, 98) | 0.71 |
The quantitative data are shown as median data and IQR data in brackets.
The occurrence data are shown as no. (%). Values indicate no. of positive results/total no. of patients with available assay results.
Between-group comparisons of continuous variable in patients with low and high richness were tested by Kruskal-Wallis test. For categorical variable, χ² test test was used for comparison between groups.
Statistically significance with a p value ≤0.05 was marked as bold.
*The results of laboratory test in the acute phase were compared, usually the first day after hospital admission.
†Pulmonary function tests were expressed as per cent of the predicted value.
‡ A p value ≤0.05 was denoted as statistically significant.
MEF 25%, mean expiratory flow at 25%; MEF 50%, mean expiratory flow at 50%; MEF 75%, mean expiratory flow at 75%; BMI, body mass index; CRP, C reactive protein; DLCO, diffusing capacity of the lung for carbon monoxide; DLCO/VA, diffusing capacity divided by the alveolar volume; FEF25%–75%, forced expiratory flow at 25%–75%; FEV1, forced expiratory volume in the first 1 s of expiration; FVC, forced vital capacity; HFNC, high flow nasal catheter oxygen therapy; ICU, intensive care unit; IVC, inspiratory vital capacity; MVV, maximal voluntary ventilation; PEF, peak expiratory flow; PFTs, pulmonary function tests; RV, residual volume; RV/TLC, residual volume divided by the total lung capacity; TLC, total lung capacity; 6WMT, 6 min walk tests.
The present study found that microbiota richness was not restored to normal levels after 6-month recovery. Patients with lower postconvalescence richness showed higher level of CRP and illness severity during the acute phase, suggesting close correlations between inflammatory response and gut dysbiosis in COVID-19, as illustrated in previous studies.1 4 Microbial diversity is a critical determinant of microbial ecosystem stability.5 Stable ecosystems provide colonisation resistance to opportunistic pathogens.6 Therefore, the persistent reduction of gut microbiota richness may have long-term biological influence during the COVID-19 pandemic.7 Follow-up studies of 3 months and 6 months have shown pulmonary function impairment along with cardiac abnormalities in patients with COVID-19.2 8 The results here indicated that postconvalescence patients with lower microbial richness had worse pulmonary functions. Gut microbiota is implicated in the pathogenesis of acute lung injury via several potential mechanisms, including direct translocation of bacteria from gut to the lung and immune modulation effects of microbes related metabolites.9 10 Our study corroborates the growing evidence that gut dysbiosis is associated with the recovery process of COVID-19. Due to the relatively small sample size, our results need to be confirmed in further studies with larger sample size and more techniques. Targeted manipulation to promote the microbial diversity could be an important strategy to treat long COVID-19 and speed up recovery.
Acknowledgments
We thank Ling Yu, Cheng Ren, and Ting Xu, who are staff in the infectious department of the First Affiliated Hospital, School of Medicine, Zhejiang University for their efforts in organising patients’ follow-up visits.
Footnotes
Twitter: @Baohong Wang
YC and SG contributed equally.
Contributors: Concept and design: LL. Acquisition and interpretation of data: all authors. Drafting of the manuscript: YC and SG. Critical revision of the manuscript: LL. Final approval: all authors.
Funding: This study was funded by National Natural Science Foundation of China (U20A20343); Zhejiang Province key research and development plan emergency project (No. 2020C03123); National Science and Technology Major Project (No. 2017Zx10204401).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
The State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, National Clinical Research Centre for Infectious Diseases, Collaborative Innovation Centre for Diagnosis and Treatment of Infectious Diseases, Department of Infectious Diseases, The First Affiliated Hospital, College of Medicine, Zhejiang University. Ethics approval was obtained from the Institutional Review Board of the First Affiliated Hospital, Zhejiang University School of Medicine (IIT20200069A-R1).
References
- 1. Yeoh YK, Zuo T, GC-Y L. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 2021;70:698–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang C, Huang L, Wang Y, et al. 6-Month consequences of COVID-19 in patients discharged from Hospital: a cohort study. Lancet 2021;397:220–32. 10.1016/S0140-6736(20)32656-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nabavi N. Long covid: how to define it and how to manage it. BMJ 2020;370:m3489. 10.1136/bmj.m3489 [DOI] [PubMed] [Google Scholar]
- 4. Gu S, Chen Y, Wu Z. Alterations of the gut microbiota in patients with COVID-19 or H1N1 influenza. Clin Infect Dis 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lahti L, Salojärvi J, Salonen A, et al. Tipping elements in the human intestinal ecosystem. Nat Commun 2014;5:4344. 10.1038/ncomms5344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol 2013;13:790–801. 10.1038/nri3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Finlay BB, Amato KR, Azad M, et al. The hygiene hypothesis, the COVID pandemic, and consequences for the human microbiome. Proc Natl Acad Sci U S A 2021;118. 10.1073/pnas.2010217118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sonnweber T, Sahanic S, Pizzini A, et al. Cardiopulmonary recovery after COVID-19 - an observational prospective multi-center trial. Eur Respir J 2020. 10.1183/13993003.03481-2020. [Epub ahead of print: 10 Dec 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dickson RP, Singer BH, Newstead MW, et al. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol 2016;1:16113. 10.1038/nmicrobiol.2016.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang Q, Ran X, He Y, et al. Acetate downregulates the activation of NLRP3 inflammasomes and attenuates lung injury in neonatal mice with bronchopulmonary dysplasia. Front Pediatr 2020;8:595157. 10.3389/fped.2020.595157 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2021-324090supp001.pdf (106.9KB, pdf)