Abstract
Steroid hormone receptors act to regulate specific gene transcription primarily as steroid-specific dimers bound to palindromic DNA response elements. DNA-dependent dimerization contacts mediated between the receptor DNA binding domains stabilize DNA binding. Additionally, some steroid receptors dimerize prior to their arrival on DNA through interactions mediated through the receptor ligand binding domain. In this report, we describe the steroid-induced homomeric interaction of the rat glucocorticoid receptor (GR) in solution in vivo. Our results demonstrate that GR interacts in solution at least as a dimer, and we have delimited this interaction to a novel interface within the hinge region of GR that appears to be both necessary and sufficient for direct binding. Strikingly, we also demonstrate an interaction between GR and the mineralocorticoid receptor in solution in vivo that is dependent on the ligand binding domain of GR alone and is separable from homodimerization of the glucocorticoid receptor. These results indicate that functional interactions between the glucocorticoid and mineralocorticoid receptors in activating specific gene transcription are probably more complex than has been previously appreciated.
The effects of corticosteroids are determined through asymmetric distribution of the mineralocorticoid and glucocorticoid nuclear hormone receptors (MR and GR) and the protective effects of 11β-hydroxysteroid dehydrogenase, which selectively metabolizes glucocorticoids (2, 20, 31). MR is highly sensitive to both mineralocorticoids and glucocorticoids, while GR responds only to higher levels of glucocorticoids and is mostly insensitive to mineralocorticoids.
Coordinate signaling by GR and MR is specifically relevant to tissues such as the brain, where an abundance of MR and GR in areas such as the hippocampus is accompanied by an absence of 11β-hydroxysteroid dehydrogenase (14). Indeed, the effects of GR and MR are critical for homeostatic control of CAl pyramidal neurons, where the two receptors differentially mediate the control of ion regulation and transmitter responsiveness (27). Thus, MR and GR signaling influence memory, mood, and neuronal survival. Elevated cortisol levels correlate with depression and other stress-related psychopathologies and with a long-term attenuation of serotonin signaling (28, 29, 61).
GR and MR function predominantly to regulate specific gene expression patterns through palindromic response elements that accommodate receptor dimers (1). The DNA binding domains (DBDs) of the steroid hormone receptors are highly conserved. As a result, GR and MR, as well as progesterone receptors (PR) and androgen receptor (AR), bind in closely related ways to broadly overlapping response elements. Homodimerization contacts mediated through the receptor DBDs occur on DNA binding and are mediated through specific contacts involving residues in the second zinc finger of the receptor DBDs (38).
The potential for transcriptional regulation via heteromeric complexes of these steroid receptors has recently been substantiated by reports that GR and MR can function as DNA-bound heterodimers to modulate transcription in ways that are distinct from the GR and MR homodimers (37, 60). In vitro experiments have demonstrated the potential of GR and MR to form heterodimers on palindrominc response elements, while regulatory experiments have demonstrated that composite transcriptional responses are possible on costimulation of MR and GR in the cells. Another report has since demonstrated a similar potential for GR to regulate transcription as a DNA-bound heterodimer with AR (6).
For the majority of nuclear hormone receptors, however, the possibilities for DNA-dependent dimerization (39) may be restricted by additional dimerization contacts that form in solution between the receptor ligand binding domains (LBDs) (42). Thus, retinoid, thryoid, vitamin D, and orphan nuclear receptors act primarily as heterocomplexes with retinoic acid X receptors (RXRs) but can also form homodimers in solution under certain conditions (39). For example, the binding of 3,5,3′-l-triiodothyronine to thyroid hormone receptor (TR) destabilizes TR homodimers in favor of TR-RXR heterodimers, while the binding of 9-cis-retinoic acid to RXR decreases heterodimerization with TR in favor of RXR homodimers (35).
Dimerization of steroid hormone receptors in solution prior to DNA binding also has been described. However, the proclivity of these receptors for heterodimerization is considerably less and seems to encourage their functioning as steroid-specific dimers. The α and β estrogen receptors (ERs) form homo- and heterodimers in solution through a motif in the LBD and make subsequent DNA-dependent contacts within their zinc fingers on DNA binding. Similarly, the PR A and B isoforms homo-and heterodimerize in solution and on DNA binding.
For ERα, biochemical and crystallographic studies have demonstrated that solution dimerization occurs through a motif at the C-terminal region of the LBD anchored by α-helix 10 but also involving helices 7, 8 and 9 (5, 18). This interface aligns closely with that observed for solution dimerization of RXR (4). The dimerization interface for PR is also localized to the C terminus of the receptor LBD but is only about half the size of that for ER (56, 66). This decrease in surface area is reflected by a decreased stability of PR LBD dimers in biochemical experiments (15, 66). Additional contacts that have been reported to occur between the hinge region of PR (59) may act to stabilize PR dimers. In addition, individual domains within PR, AR, and ER α appear to be able to form intramolecular contacts (30, 32, 58).
The ability of GR to form homodimers in solution has been debated extensively without resolution, while the prospects for MR homodimerization and GR-MR heterodimerization prior to DNA contact have not been considered. Initial biochemical studies indicated that liganded GR migrated in sucrose gradients at 4S in a monomeric form (36). More careful preparations or the inclusion of cross-linking agents revealed the presence of a 6S form with enhanced DNA binding activity that was suggested to reflect the presence of GR homodimers in solution (67). However, the results of studies measuring the DNA binding of wild-type (WT) GR and GR peptides under a variety of experimental conditions have alternatively supported the cooperative binding of GR monomers or the coordinate binding of preformed GR dimers (10–12, 17, 43, 54, 62, 63).
Since the ability of GR and MR to form homo- and heterodimers in solution prior to their arrival on hormone response elements may be a determining factor for the coordination of corticosteroid signaling through GR and MR, we have undertaken a directed analysis of the ability of GR to form homomeric complexes in solution and to heterodimerize with MR in the cell. Our results demonstrate that steroid treatment induces the association of GR in solution into at least a receptor dimer, through an interface within a 35-amino-acid region of the receptor hinge that is not featured in this way in the dimerization of other nuclear receptors. The occurrence of this interaction in vivo is demonstrated in nuclear cotransport experiments in which the nuclear accumulation GR mutants deficient in nuclear localization is shown to be dependent on this short region of the GR hinge. Using the same assay, we have also determined that GR can enter into a heteromeric interaction in solution with MR through determinants in the GR LBD that are separable from the amino acids required for the homomeric interaction of GR in solution. These results suggest the potential for higher-order corticosteroid receptor complexes in the cell.
MATERIALS AND METHODS
Plasmids.
The compositions of the GR and MR peptides employed are summarized in the figures. Many of the plasmids used have been described previously (19, 22, 47, 51, 52). All other plasmids were constructed by standard restriction enzyme cloning or through PCR amplification of the inserts. Reading frame reconstruction and mutations were verified by DNA sequencing. All plasmids constructed for in vitro translation were either in a pGEM-7Z backbone (Promega) or a pTL2 backbone (44). Plasmids expressing glutathione S-transferase fusion proteins were constructed in a pGEX-3X or pGEX-2T (Pharmacia) backbone. Yeast expression plasmids were constructed in pGAD and pAS2 backgrounds (Clontech). The c-myc epitope tag employed in many experiments has been utilized previously (3, 46). MR with a BuGR2 (buGR) epitope tag (buMR) was constructed by insertion of the BuGR antibody epitope of amino acids 407 to 423 from rat GR at the N terminus of the rat MR expression plasmid pTL2MR, which was derived from p6RMR. The pKA epitope has been utilized previously (65).
Mammalian culture.
Sf7 cells stably transfected to express myGR (46) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% 10% FBS in the presence of 50 μg of G418 per ml. The parental cell line was grown in DMEM–10% FBS in the absence of G418. COS7 cells were maintained in DMEM–10% FBS. Transient transfections were performed using Lipofectamine (10 μl per 60-mm dish) (GIBCO BRL) and an 8-h incubation time as previously described (51). Transfected cells were maintained for 16 h in complete serum and were withdrawn from serum for 21 h prior to treatment. Hormone treatments with 10−6 M dexamethasone (Dex) or cortisol were carried out for 1 h.
In vitro protein binding assays.
The detailed protocol for the immunoprecipitation binding assay for GR-interacting proteins has been described in detail previously (46). Whole-cell extracts for immunoprecipitation binding assays were prepared from Sf7 cells stably expressing myGR, or the control parental cell line, by sonication in TEGD buffer (10 mM Tris-HCl [pH 7.4], 1 mM EDTA, 10% glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride) followed by centrifugation for 5 min at 16,000 × g. To dissociate the heat shock protein complex after steroid binding, extracts were incubated with 10−6 M Dex for 2 h at 4°C and then for 30 min at 25°C. Salt-induced hsp release was accomplished separately by incubation of the extracts in (0.4 M) NaCl for 2 h at 4°C. In experiments where the GR-hsp interaction was maintained through the binding assay, the GR-chaperone complex was stabilized by the addition of Na2MoO4 to 20 mM in all buffers. To prepare the immunoprecipitates, the molybdate-stabilized and the Dex-treated extracts were diluted threefold with binding buffer (25 mM HEPES [pH 7.9], 60 mM KCl, 0.5 mM EDTA, 12% glycerol, 0.1% NP-40, 0.2 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride) in the presence and absence of 20 mM Na2MoO4 respectively, while the salt-treated extracts were diluted with binding buffer without KCl to a final salt concentration of 60 mM. Immunoprecipitations for myGR and control Sf7 extracts were performed with the anti-myc antibody, 9E10, as described previously (46) and included at least three washes with binding buffer. Receptor concentrations used in subsequent binding assays were verified by quantitative Western blotting using a Bio-Rad GS525 molecular imager with a CH screen.
In vitro-translated, 35S-labeled GR peptides were prepared in rabbit reticulocyte lysate (Promega) using [35S]methionine (1 mCi/mmol; Amersham/Pharmacia). Dex and salt treatments and Na2MoO4 stabilization were performed exactly as described for the whole-cell extracts. hsp dissociation following Dex and NaCl treatments was confirmed by sucrose gradient centrifugation analysis. In vitro-translated GRs were quantified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and molecular imager analysis
The 9E10 immunoprecipates were prepared for binding to in vitro-translated GRs by preincubation with unprogrammed reticulocyte lysate (10% solution in binding buffer with Na2MoO4 as required) at 4°C for 2 h followed by incubation with the 35S-labeled in vitro-translated proteins in binding buffer in the presence or absence of Na2MoO4 as required. Specific binding was revealed by three subsequent washes with 500 μl of ice-cold binding buffer and SDS-PAGE and molecular imager analysis of the bound proteins.
GST-GR fusion proteins were prepared as previously described (46, 51), with yields and purity determined by scans of Coomassie blue-stained SDS-PAGE gels. Binding reactions with 35S-labeled, in vitro-translated GRs were performed, and the products were analyzed by exactly the same protocol used for the immunoprecipitation binding reactions.
For direct binding studies, purified GST-GRX550 (GR amino acids 407 to 550) with a C-terminal extension (LARRASYP) containing a protein kinase A phosphorylation site was labeled with 32P using protein kinase A to a specific activity of 2.3 × 107 dpm/mg. The 32P-labeled GRX550 moiety was released from the GST purification tag by thrombin cleavage and recovered, and binding to GST GRs was performed by the same method as the immunoprecipition binding assays.
Cross-linking of GR and FTZ-F1 peptides was performed by the protocol described previously to study the multimerization of p53 (55). The GR505–550 (amino acids 505 to 550) and Drosophila FTZ-F1 peptides (amino acids 575 to 620, exactly analogous to GR amino acids 505 to 550) were expressed as GST fusion proteins with the C-terminal protein kinase A motif, labeled, cleaved from the GST, and purified as for GST-GRX550. Equal counts of each peptide were incubated in 200 mM Na3PO4 (pH 7.4) buffer at 4°C for 30 min in the presence of increasing concentrations of glutaraldehyde. The cross-linked peptides were resolved by Tris-Tricine SDS–16.5% PAGE (Bio-Rad) and visualized by autoradiography.
Yeast two-hybrid Assays.
The yeast strain Y190 was grown in yeast extract-peptone-dextrose (YEPD). Transformation was carried out using the lithium acetate method with plasmid DNA (53). Yeast colonies transformed with fusion constructs were grown in synthetic media lacking either leucine or tryptophan or both. Transformed yeast were selected and cultured overnight in the absence of hormone. The yeast cultures were then subcultured (1:10) in fresh selective media that contained either ethanol or 10−6 M desacetylcortivazol (DAC) (22) and grown for a further 16 h. The optical density at 600 nm (OD600) was determined, and the cultures were then assayed for β-galactosidase activity.
β-Galactosidase assays were performed as described elsewhere (52). β-Galactosidase units were calculated using the equation (1,000 OD420)/(t × v × OD600), where t is the reaction time at 30°C (in minutes) and v is the initial volume of culture used (in milliliters) (52).
Indirect and direct immunofluorescence.
GR and MR expression vectors were expressed from recombinant plasmids in COS7 cells by Lipofectamine-mediated transfection singly or in combination. Relative levels of expression were determined by Western blotting of whole-cell extracts using the BuGR2 antibody that recognized all of the constructs used in this study. For cotransport assays, a minimum 4:1 ratio of transporting receptor to passenger was confirmed prior to immunofluorescence. The amounts of plasmid transfected varied from 60 to 2,500 ng. To monitor the subcellular distribution by indirect immunofluorescence, transfected cells were plated onto poly-l-lysine-coated glass coverslips 24 h following transfection and incubated for a further 8 h in DMEM containing 10% charcoal-stripped fetal calf serum. The cells were synchronized to G0 by incubation in serum-free DMEM for a further 21 h prior to the initiation of treatment. Vehicle, Dex, or cortisol was added to a final concentration of 10−6 M in serum-free medium for 1 h prior to fixation of the cells. Indirect immunofluorescence was carried out exactly as described previously (48, 51, 68), with either primary anti-GR antibody BuGR2 (Affinity BioReagents, Inc.) for detection of WT GR and buMR or the anti-myc antibody, 9E10, for detection of myc epitope-tagged GR derivatives. In most experiments, fluorescein-conjugated anti-mouse sheep immunoglobulin (Boehringer Mannheim) was the secondary antibody used. However, to detect 9E10 signals in the presence of green fluorescent protein (GFP) we employed a rhodamine red-conjugated donkey anti-mouse secondary antibody (Jackson ImmunoResearch laboratories). Slides were examined for subcellular localization of GRs on a Zeiss Axioskop microscope, and the images were captured using Northern Eclipse 5 software (Empix Imaging Inc.). Cells were classified into one of five categories ranging from exclusively nuclear (N) to exclusively cytoplasmic (C) by visual observation, as we and others have previously established (48, 51, 68). Quantification was performed using double-blind encryption. All experiments were repeated in triplicate in at least three independent trials. Visualization of GFP-GRN524NL1− expressed alone and in combination with full-length GR and MR constructs was performed by direct fluorescence observation of live cells and quantified as for indirect immunofluorescence.
RESULTS
Oligomerization of GR in solution in vitro is dependent on the receptor hinge.
To begin to assess the potential for steroid-activated GRs to oligomerize in the absence of DNA, we tested the ability of in vitro-translated WT rat GR to bind to GR immunoprecipitated from whole-cell extracts of Dex-treated murine Sf7 fibroblasts (Fig. 1). An N-terminal c-myc epitope tag on GR stably expressed in Sf7 cells allowed for selective discrimination of the cellular receptor from the in vitro-translated GR peptides. Previously, we had shown that this assay accurately mapped a protein-protein interaction between GR and octamer transcription factors 1 and 2 that leads to the recruitment of the octamer factors to the mouse mammary tumor virus promoter in tissue culture cells (46).
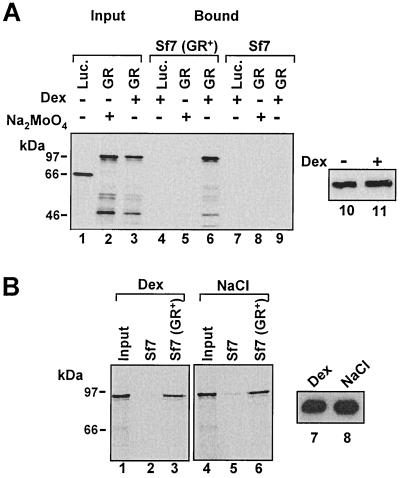
FIG. 1.
Binding of in vitro-translated, 35S-labeled GR to myGR immunoprecipitated from whole-cell extracts is dependent on hsp dissociation. (A) Immunoprecipitates with myc epitope antibody 9E10 from whole-cell extracts prepared from Sf7 cells expressing myGR (lanes 4 to 6) or control cells lacking GR (lanes 7 to 9) were tested for binding to in vitro-translated firefly luciferase (Luc.) or WT GR. Both the cells and in vitro-translated receptors were treated with 10−6 M Dex, as indicated in the figure and described in detail in Materials and Methods. For binding reactions performed in the absence of steroid, association of GR with the chaperone complex was stabilized in cell extracts prior to immunoprecipitation and in the binding assays through the inclusion of 20 mM Na2MoO4 in all buffers as indicated. Specific myGR binding (Bound, lanes 4 to 9) was revealed by SDS-PAGE and fluorography and is compared to 10% of the in vitro-translated proteins added to the binding-assay mixture (Input, lanes 1 to 3). Loading of the GR immunoprecipitates with or without hormone is revealed by Western blotting in lanes 10 and 11. (B) 9E10 immunoprecipitates from Sf7 and Sf7 (GR+) cells were tested for binding to in vitro-translated GR. In lanes 1 to 3, the cells and in vitro-translated receptors were treated with 10−6 M Dex, while in lanes 4 to 6, the immunoprecipitates and in vitro-translated GRs were treated with 0.4 M NaCl to strip the hsp-immunophilin complex from the receptor prior to binding. Binding of in vitro-translated GR is compared to 10% of the input from the in vitro translation. Loading of the GR immunoprecipitates in the binding assay is revealed by Western blot analysis in lanes 7 and 8.
In vitro-translated, Dex-treated GR was observed to bind efficiently to immunoprecipitated myGR (Fig. 1A, lane 6) but was not retained by immunoprecipitates prepared from the parental cell line lacking myGR (lane 9). Binding occurred independently of added DNA and was fully resistant to treatment of the binding-reaction mixture with DNase I (50). We also obtained very similar binding in experiments employing the antagonist RU486 as the GR ligand, indicating that GR-GR interaction was unlikely to be influenced significantly by agonist or antagonist-specific receptor conformations (50).
Prior to exposure to ligand, GR exists in the cytoplasm as a monomer in a chaperone complex featuring hsp90 and other heat shock proteins and immunophilins (45). The chaperone complex can be maintained in vitro through the stabilizing effects of sodium molybdate (33). Chaperone association appeared to effectively block GR dimerization in this assay, since no binding of the in vitro-translated GR to myGR was observed for molybdate-stablized receptors (Fig. 1A, lane 5).
To determine whether the GR-GR interaction we observed was strictly dependent on ligand or merely required the dissociation of GR from the chaperone complex, we assessed the ability of free GRs to associate in the absence of steroid (Fig. 1B). In this experiment, the immunopreciptitated myGR and the in vitro-translated receptor were dissociated from the hsp-immunophilin complex by treatment with 0.4 M NaCl at 30°C prior to binding (16). Again, no GR bound to immunoprecipitates from the parental Sf7 cells (lanes 2 and 5). However, unliganded GRs, stripped free of hsps, interacted with the same efficiency as we observed for the Dex-treated receptors (lanes 3 and 6).
To begin to localize the determinants required for solution oligomerization of GR, we examined the binding of a series of truncated in vitro-translated GR peptides to the myc-tagged GR expressed in fibroblasts (Fig. 2). Notably, GR-GR binding was unaffected by deletion of the receptor LBD as a peptide encoding amino acids 1 to 556 of GR was retained by myGR with the same efficiency as full-length, liganded receptor (lanes 5 and 6). Truncation of GR to amino acid 523 resulted in a small, but reproducible decrease in binding (lane 7), suggesting that 523 was immediately adjacent to or just within the beginning of the binding interface. Further truncation of the receptor through the hinge region to amino acid 494 completely abrogated binding (lane 8).
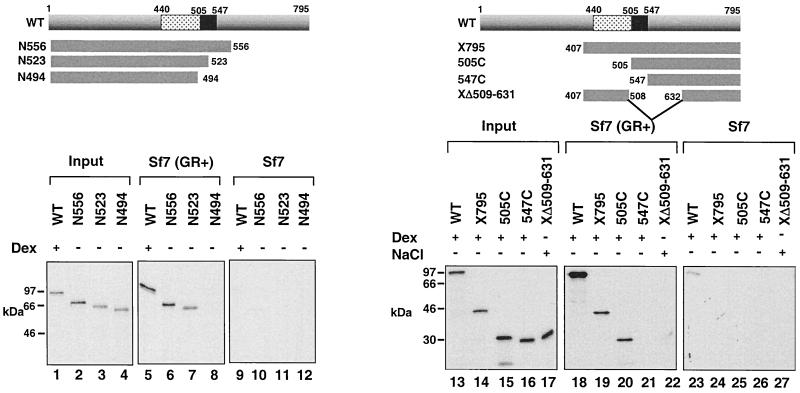
FIG. 2.
Binding of in vitro-translated, 35S-labeled GR to liganded myGR immunoprecipitated from whole-cell extracts is dependent on the receptor hinge region. Immunoprecipitates from whole-cell extracts prepared from Sf7 cells expressing myGR and treated with 10−6 M Dex for 1 h prior to harvesting [Sf7 (GR+), lanes 5 to 8 and 18 to 22] or control Dex-treated cells lacking myGR (Sf7, lanes 9 to 12 and 23 to 27) were tested for binding to in vitro-translated GR derivatives (lanes 5 to 12 and 18 to 27), whose composition is summarized schematically at the top of each panel. In vitro-translated WT GR and GR peptides X795, 505C, and 547C were treated with 10−6 M Dex, while GR peptide XΔ509–631 was treated with 0.4 M NaCl to strip away the chaperone complex prior to incubation with liganded, immunoprecipitated myGR. Dissociation of the in vitro-translated GRs from the hsp complex on Dex and NaCl treatment was confirmed by sedimentation analysis of the receptor over 15 to 30% sucrose gradients (Savory et al., unpublished). Lanes 1 to 4 and 13 to 17 show 10% of the input from the in vitro translations.
Large deletions from the N terminus of GR also had little effect on binding to myGR until they affected the hinge region. Truncation through the GR DBD to amino acid 505 left a GR peptide that still bound to myGR strongly (Fig. 2, lanes 18 to 20). This result conclusively excluded the possibility that binding might be stabilized by DNA. Further truncation through the hinge region of the receptor to amino acid 547 left an LBD peptide that was unable to bind to the myc-tagged receptor in the presence of Dex (lane 21). These results suggested that the primary determinants for GR-GR binding in solution reside within the receptor hinge region between amino acids 505 and 523. The inability of a GR peptide containing an internal truncation between amino acids 509 to 631 to bind myGR following NaCl-mediated stripping of the hsp complex from the GR peptide (lane 22) provided further evidence supporting the involvement of the hinge region in the GR-GR interaction.
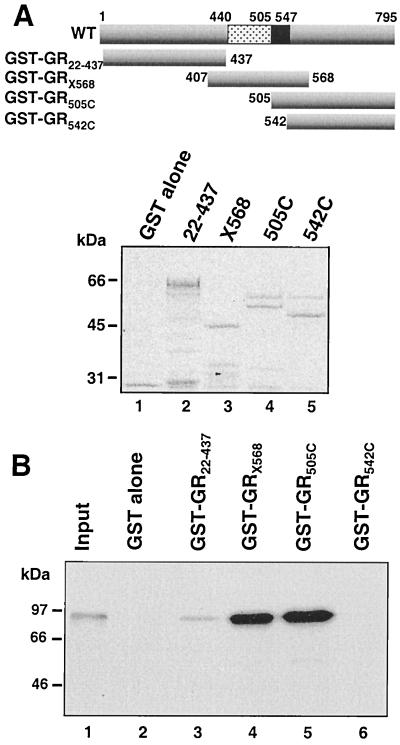
To ensure that our results were not biased by the nature of the assay employed, we reexamined GR-GR binding in solution in a GST pulldown experiment (Fig. 3). Four GR peptides were expressed as GST fusion proteins (Fig. 3A) and tested for their ability to bind in vitro-translated, liganded, WT GR. The results obtained (Fig. 3B) closely mirrored those obtained in immunoprecipitation binding experiments. In vitro-translated GR bound very strongly to the two GR peptides containing only the hinge region in common, X568 (amino acids 407 to 568) and 505C (amino acids 505 to 795) (lanes 4 and 5). By contrast, no binding was obtained to the LBD peptide (542C) (lane 6) or to GST alone (lane 2). Lastly, the increased sensitivity of this assay compared to that of the immunopreciptation experiments allowed for the visualization of a much weaker interaction between the in vitro translated GR and the receptor N terminus (lane 3). However, the significance of this interaction, which was not confirmed in other assays, remains to be established.
FIG. 3.
Amino acids 505 to 568 are required for GR-GR binding in a GST pulldown assay. Binding of in vitro-translated, 35S-labeled, Dex-treated, WT GR to GST-GR fusion proteins (B), whose composition is summarized schematically in the top panel A, is compared to 10% of the input 35S-labeled GR from the in vitro translation (lane 1). A Coomassie blue-stained SDS-PAGE gel of the loading of the GST fusion proteins on the Sepharose beads is shown in the middle panel.
The GR hinge is sufficient for direct GR-GR binding in vitro.
To examine whether the association of GR was direct and to determine whether the GR hinge region might be sufficient for GR-GR binding in solution, we performed two experiments. In the first, we examined the ability of a GR construct containing amino acids 407 to 550 (GRX550), expressed in and purified from bacteria, to bind to GST-GR fusion peptides bound to glutathione-Sepharose (Fig. 4A). To visualize binding, the purified peptide was 32P labeled using protein kinase A at an ectopic recognition motif included at its C terminus.
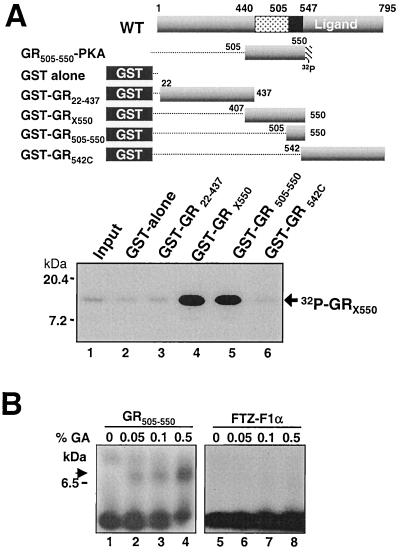
FIG. 4.
Amino acids 505 to 550 are sufficient for GR-GR binding in vitro. (A) GST-GRX550 containing a protein kinase A (PKA) recognition site purified free of the GST moiety and labeled with 32P by protein kinase A was tested for binding to GST-GR fusion proteins. The compositions of all of the proteins used in the assays are summarized at the top. GRX550 binding is compared to 10% of the input peptide shown in lane 1 and was resolved by autoradiography of an SDS-PAGE gel (18% polyacrylamide). (B). Tricine-Tris PAGE (16.5% polyacrylamide) of 32P-labeled GR505–550 (lanes 1 to 4) and the analogous peptide from the monomeric nuclear receptor FTZ-F1 from Drosophila (lanes 5 to 8) following binding reactions performed in increasing concentrations of the glutaraldehyde cross-linking agent as indicated. The arrow indicates the position of migration of the cross-linked GR peptides.
32P-labeled GRX550 bound efficiently to GST fusion proteins encoding the X550 and to a shorter GR peptide containing only amino acids 505 to 550 from the GR hinge region (GR505–550) (Fig. 4A, lanes 4 and 5). By contrast, no binding was detected to GST alone or to GST-GR22–437 or GST-GR542C (lanes 2, 3, and 6).
In a second assay (Fig. 4B), we assessed whether GR505–550 peptide dimers could be directly visualized through addition of a cross-linking agent. As a control for nonspecific cross-linking, the analogous peptide from the monomeric Drosophila FTZ-F1α nuclear receptor was tested in parallel with GR505–550. A band in high-percentage SDS-PAGE gels representative of at least GR peptide dimers increased in intensity in proportion to the concentration of the glutaraldehyde cross-linking agent included in the incubation of 32P-labeled GR505–550 (Fig. 4B, lanes 1 to 4). Indeed, the interaction observed for the GR peptide in this experiment is similar to the dimerization observed for a peptide from the tetramerization domain of p53 in a similar assay (55). By contrast, in the same experiment, the analogous FTZ-F1α peptide displayed no association, even at the highest level of cross-linking agent (lanes 5 to 8). Together, these data provide strong biochemical evidence implicating the hinge region of GR in a direct homomeric protein-protein interaction that is required and may be sufficient for the formation of GR dimers or higher-order oligomers in solution.
Yeast two-hybrid GR-GR interactions converge at the receptor hinge.
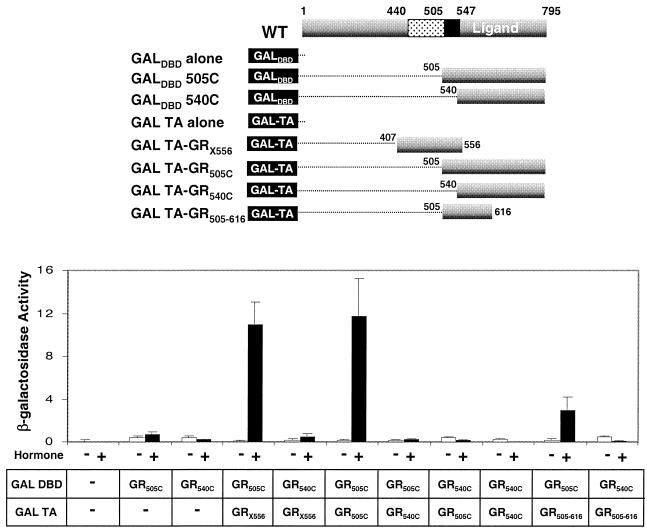
As a first step toward assessing the involvement of the GR hinge region in DNA-independent oligomerization of GR within the cell, we assessed the interaction between GR peptides in a two-hybrid analysis in yeast (Fig. 5). The GR LBD in the presence or absence of the hinge region (GR505C/GR540C) and the GR DBD including the hinge region (X556) were expressed as a series of five fusion proteins with the Gal4 activation domain or the Gal4 DBD. A sixth fusion protein, with GRX556 fused to the Gal4 DBD was toxic to the cells and thus could not be tested (34).
FIG. 5.
Amino acids 505 to 556 are required for GR-GR binding in a yeast two-hybrid assay. Relative activation of a β-galactosidase reporter gene from Gal4 response elements on coexpression of the indicated Gal-GR fusion proteins, whose composition is summarized at the top, following treatment of liquid cultures for 16 h with 10−6 M DAC or vehicle. The error bars indicate the standard errors of the means from three independent experiments performed in duplicate. All Gal-GR fusion proteins were expressed to similar levels as determined by Western blot analysis of yeast extracts (Savory et al., unpublished).
The GR LBD constructs (GR505C/GR540C) expressed at low levels in yeast fused to the Gal4 DBD activated transcription poorly in response to the synthetic steroid DAC. Coexpression of a second GR540C linked to the Gal activation domain had no further effect on β-galactosidase activity, confirming that the GR LBD is not sufficient for dimerization. The lack of interaction between the GR540C peptides was not due to a lack of responsiveness to DAC, since the Gal DBD-GR540C construct interacted strongly with the p160 transcriptional coactivator TIF2 in a DAC-dependent manner in the same assay (M. Liao, Y. A. Lefebvre, and R. J. G. Haché, unpublished data).
By contrast, when the hinge region of GR was included with the LBD in both GAL4-GR constructs (GR505C), a strong ligand-dependent activation of lacZ transcription reflecting the association of the two GR peptides was observed. Similarly, the lacZ gene was also strongly activated when the Gal activation domain–GR DBD-hinge fusion protein (GalTA-GRX556) was coexpressed with the Gal DBD-GR505C construct. Moreover, a short GALTA-GR construct containing only amino acids 505 to 616 of GR also interacted strongly with GALDBD-GR505C. Thus, these results exactly mirrored the results of our in vitro assays in implicating the hinge region of GR in receptor oligomerization of the GR in solution in vivo
The hinge region of GR is required for GR-GR binding in solution in mammalian cells.
To assess whether the hinge region of GR functioned to promote receptor oligomerization in mammalian cells prior to its arrival on DNA, we examined GR-GR interactions in a nuclear cotransport assay.
GR exchanges between a steroid-free, hsp-complexed cytoplasmic form and a liganded hsp-free nuclear form. Transport of GR into the nucleus is mediated by two nuclear localization sequences (NLSs), NL1 and NL2. NL2 occurs within the receptor LBD and mediates the partial transfer of GR to the nucleus in many cell types including simian COS7 cells, while NL1 is a short basic motif in the hinge region of GR that is sufficient for complete nuclear transfer of the receptor in the same cell lines (51). Mutations in NL1 dramatically impede the translocation of the receptor to the nucleus upon exposure to ligand (51). We hypothesized that if GR-GR binding could occur in the cytoplasm prior to nuclear uptake, then coexpression of WT GR could be expected to promote an increase in the transfer of NL1− GRs to the nucleus. This hypothesis was elegantly validated for PR several years ago (23).
Therefore, we examined whether coexpression of WT GR could increase the nuclear localization of full-length GR with an inactivating substitution of 3 amino acids in NL1 (GRNL1−) (51) and a second GR construct with a deletion in the hinge region of GR from amino acids 511 to 539 (GRΔ511–539) including NL1, but that also would be expected to disrupt the oligomerization of GR in solution.
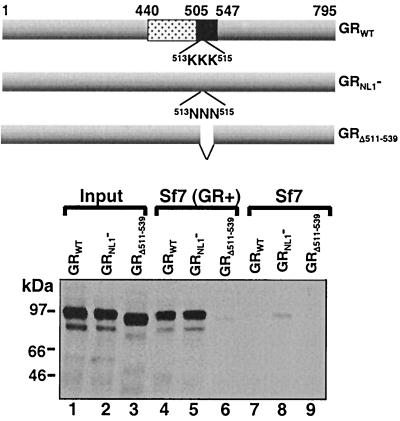
Because NL1 occurs within the GR hinge, it was necessary to first ensure that site-directed mutagenesis of NL1 did not also affect GR-GR binding. Therefore we compared the binding of two mutated forms of GR, GRNL1− and GRΔ511–539, to myGR in our immunoprecipitation binding assay (Fig. 6). Steroid-treated GRNL1− was retained on the myGR beads with the same efficiency as the WT receptor was (lanes 4 and 5). By contrast, GRΔ511–539 in dex-treated cells was unable to bind myGR (lane 6). Thus, NL1 did not appear to overlap significantly with determinants within the receptor hinge required for solution dimerization of GR in vitro.
FIG. 6.
Binding of in vitro-translated 35S-labeled GR to myGR immunoprecipitated from whole-cell extracts requires amino acids 511 to 539. Immunoprecipitates from whole-cell extracts prepared from Sf7 cells expressing myGR [Sf7 (GR+); lanes 4 to 6] or control cells (Sf7, lanes 7 to 9) as indicated were tested for binding to the in vitro-translated GRs, whose composition is illustrated schematically at the top. All samples were treated with 10−6 M Dex. GR binding is compared to 10% of the input from the in vitro translations shown in lanes 1 to 3.
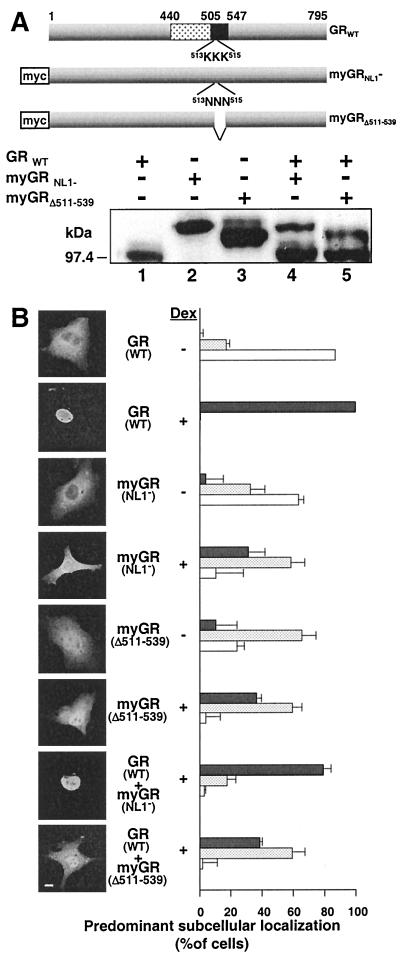
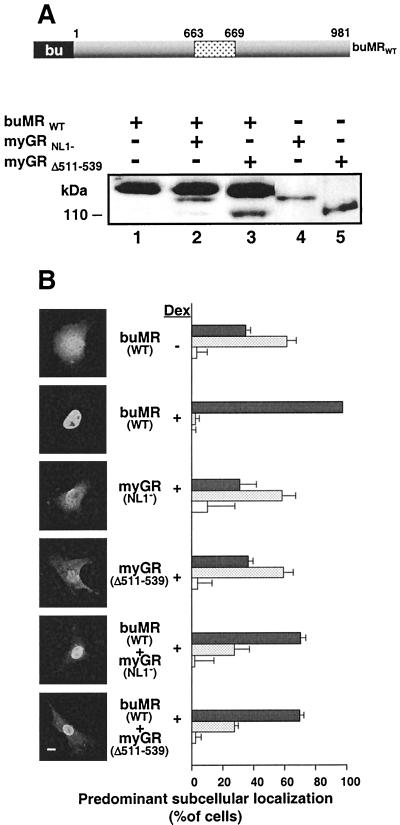
Cotransport of GRNL1− and GRΔ511–539 into the nucleus by WT GR was examined in the experiment in Fig. 7. GR expression was accomplished by transient transfection, and localization was monitored by indirect immunofluorescence. For this experiment, a c-myc epitope tag was introduced onto the N terminus of the two mutant GRs (myGRNL1− and myGRΔ511–539) but tag was absent from the WT receptor. In this configuration, the myc tag antibody 9E10 could be used to show the subcellular localization of the mutant GRs in the presence of the WT receptor. Second, Western blotting was used to ensure that the ratio of WT GR to mutated receptor was at least 4:1 in all experiments (Fig. 7A). This ratio enhanced the opportunity for dimerization of the mutated receptors with WT GR while ensuring that differences in subcellular localization between different constructs did not arise due to differences in the relative amounts of the GRs expressed. The WT/mutated GR ratio was obtained by decreasing the amount of mutated receptor plasmids expressed rather than overexpressing the WT GR.
FIG. 7.
Amino acids 511 to 539 are required for oligomerization of GR in mammalian cells in vivo. Physical interaction between GRs in transiently transfected COS7 cell was assessed by the ability of WT GR to promote the nuclear localization of myGR derivatives lacking the major GR NLS, NL1. (A) Western analysis of whole-cell extracts prepared from COS7 cells expressing WT GR and myGR derivatives singly (lanes 1 to 3) and in combination (lanes 4 and 5) with the common buGR antibody illustrating the minimum 4:1 ratio of expression for WT GR to myGR derivatives used in the immunofluorescence assays in panel B. A summary of the composition of the constructs employed in this experiment is given at the top. (B) In situ immunofluorescence analysis of the localization of WT GR using antibody buGR (GR WT) and of the myGR derivatives using anti-myc antibody 9E10 (remaining panels) before Dex treatment (− Dex) and following a 1-h treatment with 10−6 M Dex (+ Dex). Photomicrographs of the immunofluorescence pattern of representative cells are shown to the left, while quantification of observations of a minimum of 150 cells for each sample in each of a minimum of three independent experiments performed in triplicate are shown to the right. As described previously (48), GR localization in each cell was categorized as completely or mostly nuclear (solid grey bars), equally distributed throughout the cell (stippled bars), or localized predominantly or exclusively to the cytoplasm (white bars). The error bars indicate the standard errors of the means. Bar, 10 μm.
Prior to steroid treatment, WT GR and the mutated receptors were localized almost completely to the cytoplasm (Fig. 7B). Dex treatment of COS7 cells results in the rapid and complete transfer of WT GR to the nucleus. By contrast, both myGRNL1− and myGRΔ511–539 become only partially nuclear. Previously, we demonstrated that the localization and subcellular trafficking of GR in transiently transfected cells can be accurately monitored by manual scoring of GR localization in hundreds of transfected cells (24, 48, 51). The results accurately reflect the average behavior of GR in the cells. In these experiments, the GR in the transfected cells has been classified according to three categories, mostly or completely nuclear, equally distributed within the cell, and predominantly or exclusively cytoplasmic. Thus, following hormone treatment, WT GR was concentrated in the nucleus of virtually all cells while myGRNL1− and myGRΔ511–539 were equally distributed between the nucleus and cytoplasm in over 60% of the cells scored. Further, biochemical fractionation experiments indicated that myGRNL1− and myGRΔ511–539 associated similarly with chromatin (Savory et al., unpublished).
Coexpression of WT GR with myGRNL1− (GRWT + myGRNL1−) resulted in a sizable increase in the transfer of myGRNL1− to the nucleus following Dex treatment, such that myGRNL1− became concentrated in the nucleus of 80% of the cells scored. By contrast, the localization of myGRΔ511–539 was completely unaffected by coexpression of the WT GR (compare myGRΔ511–539 with GR + myGRΔ511–539). These results are completely consistent with the results in vitro and in yeast and provide strong evidence that liganded GR is able to oligomerize in the cytoplasm of the mammalian cell in a manner that requires the receptor hinge region.
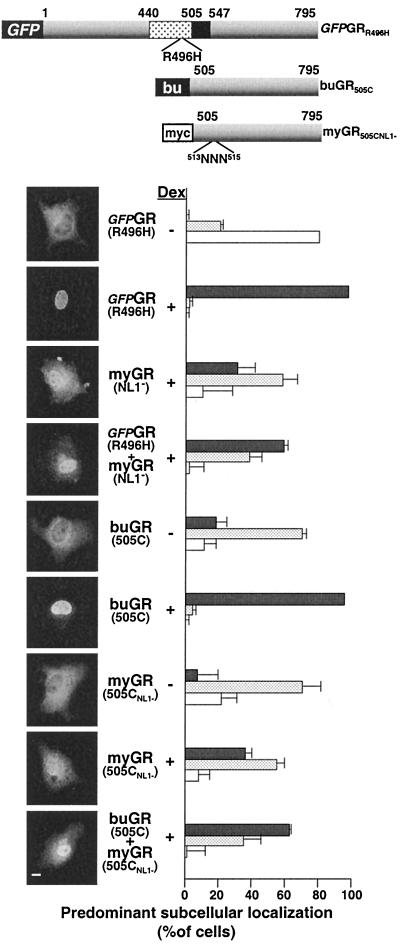
Since GR within the nucleus is targeted to DNA, it was possible that the increased nuclear localization of myGRNL1− reflected an increase in the DNA occupancy of this receptor in the nucleus that might serve to anchor the protein and decrease its rate of nuclear export, rather than directly through increased nuclear import through piggy-backing with the WT GR. To distinguish between these possibilities, we directly examined the effect of GR DNA binding in the cotransport assay (Fig. 8). In the first instance, a full-length GR containing a point mutation in the DBD that abrogates DNA binding by the receptor (GFP-GRR496H) remained competent to promote the nuclear accumulation of the full-length myGRNL1− clone. In the second instance, GR truncated from the N terminus through the DBD to amino acid 505 (buGR505C) was similarly able to promote an increase in the nuclear localization of the NL1− version of the same peptide (myGR505CNL1−). Thus, we conclude that the increased transfer of NL1− GRs to the nucleus that was dependent on amino acids 511 to 539 of GR occurs independently from the binding of GR to DNA and reflects the cotransport of GR oligomers into the nucleus.
FIG. 8.
Solution oligomerization of GR in mammalian cells is independent of the targeting of GR to DNA. (A) The new constructs (GFP-GRR496H, buGR505C, and myGR505CNl1−) used in this experiment are summarized schematically. A schema of the myGRNL1− construct is shown in Fig 7. (B) In situ immunofluorescence analysis of the localization of GFP-GRR496H, myGRNL1−, buGR505C, and myGR505CNL1− expressed singly and in the combinations shown. Dex treatment at 10−6 M for 1 h was performed as indicated. Specific localization of the GR constructs was visualized as follows: GFP-GRR496H using direct fluorescence; myGRNL1− by indirect immunofluorescence analysis using antibody 9E10 followed by a rhodamine-red-conjugated anti-mouse secondary antibody (allowing the detection of myGRNL1− in the presence of GFP-GRR496H); buGR505C by indirect immunofluorescence using antibody BuGR2 and a fluorescein-conju- gated anti-mouse secondary antibody; myGR505CNL1− by indirect immunofluorescence using antibody 9E10 followed by a fluorescein-conjugated anti-mouse secondary antibody. Photomicrographs of the immunofluorescence pattern of representative cells are shown to the left, while quantification of observations of a minimum of 150 cells for each sample in each of a minimum of three independent experiments performed in triplicate is shown to the right. The localization of myGRNL1− prior to steroid treatment is shown in Fig. 7 and is not repeated here. GR localization in each cell was categorized as completely or mostly nuclear (solid grey bars), equally distributed throughout the cell (stippled bars), or localized predominantly or exclusively to the cytoplasm (white bars). The error bars indicate the standard errors of the means. Bar, 10 μm.
Cotransport of GR into the nucleus by MR demonstrates a heteromeric interaction between GR and MR that is mediated through the GR LBD.
A central issue in corticosteroid hormone action is the potential for interaction between GR and MR following corticosteroid treatment. Recent work has suggested that GR and MR can converge on palindromic DNA response elements to form receptor heterodimers (37, 60). However, such effects could be precluded by homodimerization of GR in solution unless GR and MR also had a similar ability to form heterodimers. Therefore, to begin to investigate whether GR might also interact in solution with MR, we examined whether MR could substitute for GR in promoting the nuclear transfer of the NL1-deficient myGR constructs (Fig. 9).
FIG. 9.
MR promotes the nuclear uptake of GR irrespective of GR amino acids 511 to 539. (A). Western blot of COS7 cell extracts comparing the levels of the MR and GR constructs in panel B, using the antibody BuGR. A schematic of the buMR construct is shown at the top of the panel. The GR constructs used are summarized in Fig. 7. (B) In situ immunofluorescence analysis of GR and MR peptide localization in transfected COS7 cells prior to Dex treatment (− Dex) or following 1 h treatment with 10−6 M Dex (− Dex). The localization of the GR constructs prior to hormone treatment is shown in Fig. 7 and is not repeated here. Antibody buGR was used to identify the localization of buMR, while myc epitope antibody 9E10 was used to localize the myGR derivatives in both the absence and presence of buMR. Photomicrographs of the immunofluorescence pattern of representative cells are shown to the left, while quantification of our observations, performed as described in the legend to Fig. 7 is displayed to the right. MR-GR localization in each cell was categorized as completely or mostly nuclear (solid grey bars), equally distributed throughout the cell (stippled bars), or localized predominantly or exclusively to the cytoplasm (white bars). Bar, 10 μm.
First, to monitor the ratio of MR to GR expressed in cotransfections, we introduced an epitope tag for the GR antibody buGR into MR. We then titrated the relative levels of MR upon coexpression with myGRNL1− and myGRΔ511–539 to the minimum 4:1 ratio used with WT GR (Fig. 9A).
Prior to steroid treatment, MR expressed by transient transfection was distributed mostly equally in the cell. Dex, which binds MR and has agonist activity at 10−6 M (25), induced the rapid and complete transfer of MR to the nucleus, while the myGRNL1− and myGRΔ511–539 constructs were distributed mostly equally throughout the cells, as before (Fig. 9B).
MR substituted very efficiently for WT GR in the cotransport experiment with myGRNL1−, promoting the predominant nuclear occupancy of myGRNL1− in close to 80% of the cells scored (buMR + myGRNL1−). Thus, GR also appears to oligomerize with MR in the cytoplasm. However, by contrast to GR, buMR was equally efficient in promoting the nuclear accumulation of myGRΔ511–539 (buMR + myGRΔ511–539). Exactly the same result was obtained when the cells were treated with the natural corticosteroid cortisol (Savory et al., unpublished).
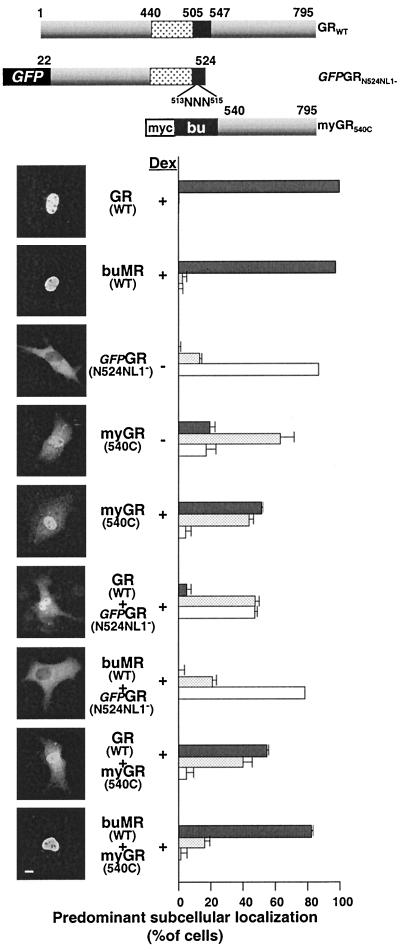
Since this result suggested that heteromeric interaction between GR and MR occurred through a surface on GR distinct from that required for GR-GR binding, we began to delimit this difference by comparing the ability of WT GR and buMR to promote the nuclear occupancy of two additional GR constructs (Fig. 10A). The first construct (GFP-GRN524 NL1−) contained the N terminus of GR to amino acid 524, included the site-directed elimination of NL1, and contained an N-terminal GFP tag to allow direct visualization of the protein in the cell. The second construct contained the GR LBD from amino acids 540 to 795 with sequential N-terminal myc and buGR tags (myGR540C). Titrations were again performed to ensure the minimum 4:1 ratio between WT GR, buMR, and the mutated GR constructs (52). As predicted from the absence of both GR NLSs, GFP-GRN524 NL1− was exclusively localized to the cytoplasm in almost all cells while the NL2-containing myGR540C was partially nuclear (Fig. 10B). Addition of WT GR to cells expressing GFP-GRN524 NL1− promoted a strong shift in the distribution of GFP-GRN524 NL1− toward the nucleus (GR + GFP-GRN524 NL1−) but had no effect on the distribution of the GR LBD (GR+myGR540C). By contrast, the effect of coexpression of MR was exactly reversed. Coexpression of buMR strongly enhanced the nuclear import of myGR540C (buMR+NL1− myGR540C), but had no effect on the localization of GFPGRN524 NL− (buMR + GFP-GRN524 NL1−).
FIG. 10.
MR-dependent nuclear uptake of GR is mediated through the GR LBD. In situ immunofluorescence analysis of the ability of full-length GR and MR (buMR) to influence the subcellular localization of GFP-GRN524NL1− and myGR540C (summarized schematically at the top) in COS7 cells is shown. Cells examined in the absence of Dex treatment are indicated (− Dex), while all other samples were examined following a 1-h treatment with 10−6 M Dex (− Dex). The receptor combinations expressed are as indicated in the middle of each data set. The localization of GRWT and buMRWT prior to hormone treatment was shown previously and is not repeated here. Localization of GFPGRN524 NL1− was determined by observation of direct fluorescence from fixed cells, while localization of GR and buMR was done by indirect immunofluorescence using antibody buGR2 and localization of myGR540C was done by indirect immunofluorescence using antibody 9E10. Quantification of our observations, performed as described in the legend to Fig. 7, is displayed to the right, while representative photomicrographs are shown to the left. MR-GR localization in each cell was categorized as completely or mostly nuclear (solid gray bars), equally distributed throughout the cell (stippled bars), or localized predominantly or exclusively to the cytoplasm (white bars). Bar, 10 μm.
This striking contrast in the interaction of these GR peptides with WT GR and WT MR substantiates the likelihood that GR oligomerization and heteromeric interactions between GR and MR originate in the cytoplasm of the cell through distinct interfaces in the hinge region and LBD of GR, respectively.
DISCUSSION
How, where, and with what nuclear hormone receptors partner in the cell can predetermine their ability to regulate gene expression prior to their arrival on DNA (39, 40). In this work, we have demonstrated the potential for corticosteroid receptor multimers to form in solution in the cytoplasm and to be maintained through the transport of these receptors into the nucleus. Oligomerization of GR in solution required determinants within a short region of the receptor hinge distinct from the DNA-dependent dimerization interface in the receptor DBD. By contrast, a separate interaction with MR appeared to be dependent solely on the GR LBD. These results suggest that corticosteroid signaling may involve an interplay between GR and MR that is more complex than has previously been appreciated.
Evidence has been presented supporting the DNA-independent oligomerization of GR in mammalian tissue culture cells and in yeast through an interface within the receptor hinge region. Immunoprecipitation binding experiments, GST pulldown assays, and direct-binding experiments have shown that a short region of the GR hinge is necessary and sufficient for GR-GR binding and is likely to be involved in direct protein-protein contacts between receptor monomers. It seems most likely that the GR-GR interaction reflects simple dimerization of the receptor. In particular, this appears to be supported by the results of the peptide cross-linking experiments. However, at present we cannot exclude the potential formation of higher-order complexes of GR. Indeed, the RXR nuclear receptor has been shown previously to form a tetrameric complex prior to ligand binding (7, 21).
Our results provide the first indication of a requirement for amino acids within the hinge region of a nuclear receptor for oligomerization and sets the determinants for solution oligomerization of GR apart from those required for the dimerization of the other steroid and nuclear hormone receptors. Only PR exhibits limited similarity to GR in solution dimerization, in that its hinge region has been proposed to stabilize dimerization of PR mediated through the receptor LBD (59, 66).
Although our results demonstrate a requirement for the GR hinge in solution dimerization of GR, they do not exclude the potential for additional protein-protein contacts to occur between monomers that could further stabilize the interaction and be important to signaling downstream from GR. Indeed, the minimum region of GR that appears to be required for GR-GR binding in solution, amino acids 505 to 524, may reflect a central core requirement rather than a complete oligomerization domain. While a direct interaction was demonstrated in vitro with high concentrations of the peptide from amino acids 505 to 550, all of the interactions detected in vivo were with GR peptides that included at least portions of the receptor LBD or DBD in addition to the minimal domain. A more complete description of the nature of the complete surface mediating GR oliogmerization in solution awaits a mutagenic survey or direct structural analysis.
The possibility of additional intramolecular contacts outside of the minimal core domain of GR would be consistent with observations that have been made for the PR, ER, and AR receptors (30, 32, 58, 59). For GR, the potential for additional communication between receptor domains has been demonstrated in several studies. For example, Lefstin et al. have established that the receptor LBD communicates directly with the DBD (34), while the synergistic nature of the AF-1 and AF-2 transcriptional activation functions within GR suggests communication between the receptor N terminus and LBD (26).
Like other nuclear hormone receptors, GR is a shuttling protein that traffics continuously between the nucleus and cytoplasm (13). Further, NL1− GRs redistribute rapidly to the cytoplasm (50). Thus, our cotransport experiments present a strong argument that GR oligomers form in the cytoplasm and are transported together into the nucleus and that the NL1− derivatives of GR accumulate further through a continuous facilitation of their transport into the nucleus. Indeed, cotransport efficiency was maintained between GR peptides containing only the receptor hinge and LBDs. The ability to form receptor dimers prior to the arrival on DNA could be expected to have several advantages for GR, including an increased ability to recognize and bind cognate DNA sequences within chromatin and the potential for more efficient interaction with transcriptional coregulatory molecules.
We find it intriguing and potentially highly significant that the solution dimerization domain of GR overlaps closely with its basic NL1 nuclear import signal. These results suggest that a close juxatposition of the basic NL1 NLSs within the GR dimer might play a significant role in promoting the transport of GR into the nucleus. The GR NL1 contains a core basic motif typical of NLSs that are targeted to the nucleus by importin α proteins (50, 57). This central core motif is required for the binding of GR to importin α in vitro and in two-hybrid experiments and is required for NL1 function (50).
Importin α proteins bind to basic nuclear import sequences through a series of armadilio (arm) repeats within a large central domain of the proteins (9). Specificity for binding is found within individual arm repeats, and each importin α has the potential to bind at least two basic motifs (8). Thus, dimerization of GR in a manner that would closely juxtapose the basic NL1 motifs could increase the attraction of the receptor for importin α. Alternatively, a closely spaced interaction with multiple importin α proteins could facilitate transport by increasing interactions with importin β.
In additional experiments that are being prepared separately for publication (T. Antakly et al., unpublished data), immunogold electron microscopy has been used to detect the intracellular localization of GR in rat liver cells. The results of these experiments confirm the presence of GR dimers in the cytoplasm and nucleus. Intriguingly, however, GRs associated with the nuclear pore were predominantly detected as higher-order multimers, mostly containing four molecules of GR. These results suggest that the juxaposition of basic NLSs within the GR dimer, as well as the potential convergence of importin α proteins at importin β, may reflect the predominant mechanism for the transport of GR into the nucleus.
However, oligomerization of GR would not seem to be required for NL1-mediated nuclear transport, since hsp-associated GR monomers are transferred to the nucleus (41, 49) in an NL1-dependent manner (50). Nonetheless, in our experiments it remains possible that at least some of the decreased nuclear occupancy observed for the cotransported NL1− GRs (80% nuclear) compared to the WT receptor (98% nuclear) reflects transport efficiency of receptor dimer, in addition to being a reflection of the efficiency or stability of dimerization.
Evidence obtained for the heterodimerization of MR and GR provides an additional new level of support for the coordinate action of these receptors in regulating transcription. Previous transient transfection assays have shown that full length MR and GR could cooperate to activate transcription as DNA-bound heterodimers (37, 60). However, solution dimerization of GR in the absence of heteromeric interactions with MR, would have been expected to significantly limit the potential of DNA-bound GR-MR heterodimers to form.
By contrast, the separation of the surfaces required for homodimerization of GR and heterodimerization of GR with MR suggests exciting new possibilities for functional cooperation between corticorticosteroid receptors. In particular, our results suggest the potential for the formation of higher-order GR-MR regulatory complexes consisting of two molecules of GR for each molecule or more of MR. This possibility is supported by the observation that MR promoted the cotransport of NL1− GR, which formed homodimers, as efficiently as it promoted the nuclear transfer of GR540C, which did not. Establishing whether MR is also able to homodimerize in solution and how this interaction influences its interactions with GR becomes an obvious objective. How these DNA-independent interactions are affected by binding to hormone response elements also remains to be determined.
Finally, experiments examining the overlap in localization between GR and MR in the nucleus of hippocampal neurons (64) introduces the possibility that association of GR and MR in solution may be subject to regulation. If GR and MR form heterodimers in solution and GR-MR heterodimers target DNA indistinguishably from homodimers, it would be expected that stimulation of hippocampal neurons with corticosteroid concentrations high enough to activate both receptors would result in a complete overlap in the localization of the two receptors in the nucleus. However, one careful study of the localization of GR and MR in hippocampal neurons revealed only a partial overlap in the localization of GR and MR to discrete clusters within the nucleus (64). These results suggest that heterodimerization of GR with MR is not completely permissive but is subject to additional constraints in the cell whose nature remains to be revealed.
ACKNOWLEDGMENTS
We are grateful to K. Yamamoto and M. Petkovitch for providing plasmids used in this study. We also thank our colleagues in the Haché and Lefebvre laboratories for their helpful comments and assistance.
This work was supported from an operating grant from the Medical Research Council of Canada to Y. A. Lefebvre. R. J. G. Haché is a Scientist of the Medical Research Council of Canada, while G. G. Préfontaine holds an MRC Studentship.
REFERENCES
- 1.Beato M, Herrlich P, Schütz G. Steroid hormone receptors: many actors in search of a plot. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 2.Benediktsson R, Edwards C R. 11-beta-hydroxysteroid dehydrogenases: tissue-specific dictators of glucocorticoid action. Essays Biochem. 1996;31:23–36. [PubMed] [Google Scholar]
- 3.Boruk M, Savory J G, Haché R J. AF-2-dependent potentiation of CCAAT enhancer binding protein beta-mediated transcriptional activation by glucocorticoid receptor. Mol Endocrinol. 1998;12:1749–1763. doi: 10.1210/mend.12.11.0191. [DOI] [PubMed] [Google Scholar]
- 4.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature. 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 5.Brzozowski A M, Pike A C, Dauter Z, Hubbard R E, Bonn T, Engstrom O, Ohman L, Greene G L, Gustafsson J, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 6.Chen S, Wang J, Yu G, Liu W, Pearce D. Androgen and glucocorticoid receptor heterodimer formation. A possible mechanism for mutual inhibition of transcriptional activity. J Biol Chem. 1997;272:14087–14092. doi: 10.1074/jbc.272.22.14087. [DOI] [PubMed] [Google Scholar]
- 7.Chen Z, Iyer J, Bourguet W, Held P, Mioskowski C, Lebeau L, Noy N, Chambon P, Gronemeyer H. Ligand- and DNA-induced dissociation of RXR tetramers. J Mol Biol. 1998;275:55–65. doi: 10.1006/jmbi.1997.1413. [DOI] [PubMed] [Google Scholar]
- 8.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 9.Cortes P, Ye Z S, Baltimore D. RAG-1 interacts with the repeated amino acid motif of the human homologue of the yeast protein SRP1. Proc Natl Acad Sci USA. 1994;91:7633–7637. doi: 10.1073/pnas.91.16.7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dahlman-Wright K, Siltala-Roos H, Carlstedt-Duke J, Gustafsson J. Protein-protein interactions facilitate DNA binding by the glucocorticoid receptor DNA-binding domain. J Biol Chem. 1990;265:14030–14035. [PubMed] [Google Scholar]
- 11.Dahlman-Wright K, Wright A, Gustafsson J, Carlstedt-Duke J. Interaction of the glucocorticoid receptor DNA-binding domain with DNA as a dimer is mediated by a short segment of five amino acids. J Biol Chem. 1991;266:3107–3112. [PubMed] [Google Scholar]
- 12.Dahlman-Wright K, Wright A P, Gustafsson J. Determinants of high-affinity DNA binding by the glucocorticoid receptor: evaluation of receptor domains outside the DNA-binding domain. Biochemistry. 1992;31:9040–9044. doi: 10.1021/bi00152a047. [DOI] [PubMed] [Google Scholar]
- 13.Defranco D B, Madan A P, Tang Y, Chandran U R, Xiao N, Yang J. Nucleocytoplasmic shuttling of steroid receptors. Vitam Horm. 1995;51:315–338. doi: 10.1016/s0083-6729(08)61043-2. [DOI] [PubMed] [Google Scholar]
- 14.De Kloet E R, Vreugdenhil E, Oitzl M S, Joels M. Brain corticosteroid receptor balance in health and disease. Endocr Rev. 1998;19:269–301. doi: 10.1210/edrv.19.3.0331. [DOI] [PubMed] [Google Scholar]
- 15.DeMarzo A M, Beck C A, Onate S A, Edwards D P. Dimerization of mammalian progesterone receptors occurs in the absence of DNA and is related to the release of the 90-kDa heat shock protein. Proc Natl Acad Sci USA. 1991;88:72–76. doi: 10.1073/pnas.88.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denis M, Gustafsson J. Translation of glucocorticoid receptor mRNA in vitro yields a nonactivated protein. J Biol Chem. 1989;264:6005–6008. [PubMed] [Google Scholar]
- 17.Drouin J, Sun Y L, Tremblay S, Lavender P, Schmidt T J, de Lean A, Nemer M. Homodimer formation is rate-limiting for high affinity DNA binding by glucocorticoid receptor. Mol Endocrinol. 1992;6:1299–1309. doi: 10.1210/mend.6.8.1406707. [DOI] [PubMed] [Google Scholar]
- 18.Fawell S E, Lees J A, White R, Parker M G. Characterization and colocalization of steroid binding and dimerization activities in the mouse estrogen receptor. Cell. 1990;60:953–962. doi: 10.1016/0092-8674(90)90343-d. [DOI] [PubMed] [Google Scholar]
- 19.Freedman L P, Yoshinaga S K, Vanderbilt J N, Yamamoto K R. In vitro transcription enhancement by purified derivatives of the glucocorticoid receptor. Science. 1989;245:298–301. doi: 10.1126/science.2473529. [DOI] [PubMed] [Google Scholar]
- 20.Funder J W. Glucocorticoid and mineralocorticoid receptors: biology and clinical relevance. Annu Rev Med. 1997;48:231–240. doi: 10.1146/annurev.med.48.1.231. [DOI] [PubMed] [Google Scholar]
- 21.Gampe R T, Jr, Montana V G, Lambert M H, Wisely G B, Milburn M V, Xu H E. Structural basis for autorepression of retinoid X receptor by tetramer formation and the AF-2 helix. Genes Dev. 2000;14:2229–2241. doi: 10.1101/gad.802300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garabedian M J, Yamamoto K R. Genetic dissection of the signaling domain of a mammalian steroid receptor in yeast. Mol Biol Cell. 1992;3:1245–1257. doi: 10.1091/mbc.3.11.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guiochon-Mantel A, Loosfelt H, Lescop P, Sar S, Atger M, Perrot-Applanat M, Milgrom E. Mechanisms of nuclear localization of the progesterone receptor: evidence for interaction between monomers. Cell. 1989;57:1147–1154. doi: 10.1016/0092-8674(89)90052-4. [DOI] [PubMed] [Google Scholar]
- 24.Haché R J, Tse R, Reich T, Savory J G, Lefebvre Y A. Nucleocytoplasmic trafficking of steroid-free glucocorticoid receptor. J Biol Chem. 1999;274:1432–1439. doi: 10.1074/jbc.274.3.1432. [DOI] [PubMed] [Google Scholar]
- 25.Hellal-Levy C, Couette B, Fagart J, Souque A, Gomez-Sanchez C, Rafestin-Oblin M. Specific hydroxylations determine selective corticosteroid recognition by human glucocorticoid and mineralocorticoid receptors. FEBS Lett. 1999;464:9–13. doi: 10.1016/s0014-5793(99)01667-1. [DOI] [PubMed] [Google Scholar]
- 26.Hittelman A B, Burakov D, Iñiguez-Lluhí J A, Freedman L P, Garabedian M J. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J. 1999;18:5380–5388. doi: 10.1093/emboj/18.19.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joels M. Steroid hormones and excitability in the mammalian brain. Front Neuroendocrinol. 1997;18:2–48. doi: 10.1006/frne.1996.0144. [DOI] [PubMed] [Google Scholar]
- 28.Karten Y J, Nair S M, van Essen L, Sibug R, Joels M. Long-term exposure to high corticosterone levels attenuates serotonin responses in rat hippocampal CA1 neurons. Proc Natl Acad Sci USA. 1999;96:13456–134561. doi: 10.1073/pnas.96.23.13456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karten Y J, Slagter E, Joels M. Effect of long-term elevated corticosteroid levels on field responses to synaptic stimulation, in the rat CA1 hippocampal area. Neurosci Lett. 1999;265:41–44. doi: 10.1016/s0304-3940(99)00211-6. [DOI] [PubMed] [Google Scholar]
- 30.Kraus W L, McInerney E M, Katzenellenbogen B S. Ligand-dependent, transcriptionally productive association of the amino- and carboxyl-terminal regions of a steroid hormone nuclear receptor. Proc Natl Acad Sci USA. 1995;92:12314–12318. doi: 10.1073/pnas.92.26.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krozowski Z, Li K X, Koyama K, Smith R E, Obeyesekere V R, Stein-Oakley A, Sasano H, Coulter C, Cole T, Sheppard K E. The type I and type II 11beta-hydroxysteroid dehydrogenase enzymes. J Steroid Biochem Mol Biol. 1999;69:391–401. doi: 10.1016/s0960-0760(99)00074-6. [DOI] [PubMed] [Google Scholar]
- 32.Langley E, Zhou Z X, Wilson E M. Evidence for an anti-parallel orientation of the ligand-activated human androgen receptor dimer. J Biol Chem. 1995;270:29983–29990. doi: 10.1074/jbc.270.50.29983. [DOI] [PubMed] [Google Scholar]
- 33.Leach K L, Dahmer M K, Hammond N D, Sando J J, Pratt W B. Molybdate inhibition of glucocorticoid receptor inactivation and transformation. J Biol Chem. 1979;254:11884–11890. [PubMed] [Google Scholar]
- 34.Lefstin J A, Thomas J R, Yamamoto K R. Influence of a steroid receptor DNA-binding domain on transcriptional regulatory functions. Genes Dev. 1994;8:2842–2856. doi: 10.1101/gad.8.23.2842. [DOI] [PubMed] [Google Scholar]
- 35.Lehmann J M, Zhang X K, Graupner G, Lee M O, Hermann T, Hoffmann B, Pfahl M. Formation of retinoid X receptor homodimers leads to repression of T3 response: hormonal cross talk by ligand-induced squelching. Mol Cell Biol. 1993;13:7698–7707. doi: 10.1128/mcb.13.12.7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Litwack G, Cake M H, Filler R, Taylor K. Physical measurements of the liver glucocorticoid receptor. Biochem J. 1978;169:445–448. doi: 10.1042/bj1690445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu W, Wang J, Sauter N K, Pearce D. Steroid receptor heterodimerization demonstrated in vitro and in vivo. Proc Natl Acad Sci USA. 1995;92:12480–12484. doi: 10.1073/pnas.92.26.12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luisi B F, Xu W X, Otwinowski Z, Freedman L P, Yamamoto K R, Sigler P B. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- 39.Mangelsdorf D J, Evans R M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 40.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martins V R, Pratt W B, Terracio L, Hirst M A, Ringold G M, Housley P R. Demonstration by confocal microscopy that unliganded overexpressed glucocorticoid receptors are distributed in a nonrandom manner throughout all planes of the nucleus. Mol Endocrinol. 1991;5:217–225. doi: 10.1210/mend-5-2-217. [DOI] [PubMed] [Google Scholar]
- 42.Moras D, Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- 43.Oakley R H, Jewell C M, Yudt M R, Bofetiado D M, Cidlowski J A. The dominant negative activity of the human glucocorticoid receptor beta isoform. Specificity and mechanisms of action. J Biol Chem. 1999;274:27857–27866. doi: 10.1074/jbc.274.39.27857. [DOI] [PubMed] [Google Scholar]
- 44.Ohno C K, Petkovich M. FTZ-F1 beta, a novel member of the Drosophila nuclear receptor family. Mech Dev. 1993;40:13–24. doi: 10.1016/0925-4773(93)90084-b. [DOI] [PubMed] [Google Scholar]
- 45.Pratt W B, Toft D O. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 46.Préfontaine G G, Lemieux M E, Giffin W, Schild-Poulter C, Pope L, LaCasse E, Walker P, Haché R J G. Recruitment of octamer transcription factors to DNA by glucocorticoid receptor. Mol Cell Biol. 1998;18:3416–3430. doi: 10.1128/mcb.18.6.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rusconi S, Yamamoto K R. Functional dissection of the hormone and DNA binding activities of the glucocorticoid receptor. EMBO J. 1987;6:1309–1315. doi: 10.1002/j.1460-2075.1987.tb02369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sackey F N, Haché R J G, Reich T, Kwast-Welfeld J, Lefebvre Y A. Determinants of subcellular distribution of the glucocorticoid receptor. Mol Endocrinol. 1996;10:1191–1205. doi: 10.1210/mend.10.10.9121487. [DOI] [PubMed] [Google Scholar]
- 49.Sanchez E R, Hirst M, Scherrer L C, Tang H Y, Welsh M J, Harmon J M, Simons S S, Jr, Ringold G M, Pratt W B. Hormone-free mouse glucocorticoid receptors overexpressed in Chinese hamster ovary cells are localized to the nucleus and are associated with both hsp70 and hsp90. J Biol Chem. 1990;265:20123–20130. [PubMed] [Google Scholar]
- 50.Savory J G. Ph.D. thesis. Ottawa, Ontario, Canada: University of Ottawa; 1999. [Google Scholar]
- 51.Savory J G, Hsu B, Laquian I R, Giffin W, Reich T, Haché R J G, Lefebvre Y A. Discrimination between NL1- and NL2-mediated nuclear localization of the glucocorticoid receptor. Mol Cell Biol. 1999;19:1025–1037. doi: 10.1128/mcb.19.2.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schena M, Yamamoto K R. Mammalian glucocorticoid receptor derivatives enhance transcription in yeast. Science. 1988;241:965–967. doi: 10.1126/science.3043665. [DOI] [PubMed] [Google Scholar]
- 53.Schiestl R H, Gietz R D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 54.Segard-Maurel I, Rajkowski K, Jibard N, Schweizer-Groyer G, Baulieu E E, Cadepond F. Glucocorticosteroid receptor dimerization investigated by analysis of receptor binding to glucocorticosteroid responsive elements using a monomer-dimer equilibrium model. Biochemistry. 1996;35:1634–1642. doi: 10.1021/bi951369h. [DOI] [PubMed] [Google Scholar]
- 55.Stommel J M, Marchenko N D, Jimenez G S, Moll U M, Hope T J, Wahl G M. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanenbaum D M, Wang Y, Williams S P, Sigler P B. Crystallographic comparison of the estrogen and progesterone receptor's ligand binding domains. Proc Natl Acad Sci USA. 1998;95:5998–6003. doi: 10.1073/pnas.95.11.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang Y, Ramakrishnan C, Thomas J, DeFranco D B. A role for HDJ-2/HSDJ in correcting subnuclear trafficking, transactivation, and transrepression defects of a glucocorticoid receptor zinc finger mutant. Mol Biol Cell. 1997;8:795–809. doi: 10.1091/mbc.8.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tetel M J, Giangrande P H, Leonhardt S A, McDonnell D P, Edwards D P. Hormone-dependent interaction between the amino- and carboxyl-terminal domains of progesterone receptor in vitro and in vivo. Mol Endocrinol. 1999;13:910–924. doi: 10.1210/mend.13.6.0300. [DOI] [PubMed] [Google Scholar]
- 59.Tetel M J, Jung S, Carbajo P, Ladtkow T, Skafar D F, Edwards D P. Hinge and amino-terminal sequences contribute to solution dimerization of human progesterone receptor. Mol Endocrinol. 1997;11:1114–1128. doi: 10.1210/mend.11.8.9963. [DOI] [PubMed] [Google Scholar]
- 60.Trapp T, Rupprecht R, Castren M, Reul J M, Holsboer F. Heterodimerization between mineralocorticoid and glucocorticoid receptor: a new principle of glucocorticoid action in the CNS. Neuron. 1994;13:1457–1462. doi: 10.1016/0896-6273(94)90431-6. [DOI] [PubMed] [Google Scholar]
- 61.Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban P C, Bock R, Klein R, Schutz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- 62.Truss M, Chalepakis G, Slater E P, Mader S, Beato M. Functional interaction of hybrid response elements with wild-type and mutant steroid hormone receptors. Mol Cell Biol. 1991;11:3247–3258. doi: 10.1128/mcb.11.6.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsai S Y, Carlstedt-Duke J, Weigel N L, Dahlman K, Gustafsson J, Tsai M J, O'Malley B W. Molecular interactions of steroid hormone receptor with its enhancer element: evidence for receptor dimer formation. Cell. 1988;55:361–369. doi: 10.1016/0092-8674(88)90059-1. [DOI] [PubMed] [Google Scholar]
- 64.van Steensel B, van Binnendijk E P, Hornsby C D, van der Voort H T, Krozowski Z S, de Kloet E R, van Driel R. Partial colocalization of glucocorticoid and mineralocorticoid receptors in discrete compartments in nuclei of rat hippocampus neurons. J Cell Sci. 1996;109:787–792. doi: 10.1242/jcs.109.4.787. [DOI] [PubMed] [Google Scholar]
- 65.Wang J M, Préfontaine G G, Lemieux M E, Pope L, Akimenko M A, Haché R J. Developmental effects of ectopic expression of the glucocorticoid receptor DNA binding domain are alleviated by an amino acid substitution that interferes with homeodomain binding. Mol Cell Biol. 1999;19:7106–7122. doi: 10.1128/mcb.19.10.7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams S P, Sigler P B. Atomic structure of progesterone complexed with its receptor. Nature. 1998;393:392–396. doi: 10.1038/30775. [DOI] [PubMed] [Google Scholar]
- 67.Wrange Ö, Eriksson P, Perlmann T. The purified activated glucocorticoid receptor is a homodimer. J Biol Chem. 1989;264:5253–5259. [PubMed] [Google Scholar]
- 68.Ylikomi T, Bocquel M T, Berry M, Gronemeyer H, Chambon P. Cooperation of proto-signals for nuclear accumulation of estrogen and progesterone receptors. EMBO J. 1992;11:3681–3694. doi: 10.1002/j.1460-2075.1992.tb05453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]