Abstract
Mammals express two isoforms of arginase, designated types I and II. Arginase I is a component of the urea cycle, and inherited defects in arginase I have deleterious consequences in humans. In contrast, the physiologic role of arginase II has not been defined, and no deficiencies in arginase II have been identified in humans. Mice with a disruption in the arginase II gene were created to investigate the role of this enzyme. Homozygous arginase II-deficient mice were viable and apparently indistinguishable from wild-type mice, except for an elevated plasma arginine level which indicates that arginase II plays an important role in arginine homeostasis.
Recognition of the importance of arginine in mammalian physiology has risen dramatically with the demonstration that it is the substrate for synthesis of nitric oxide. Arginine is not generally considered to be an essential amino acid in healthy adults, but it is essential in several species of newborn animals (15, 16). Moreover, arginine depletion can occur in pathophysiologic conditions such as sepsis and trauma (8, 9) or following surgical resection of the small bowel (23). Regulation of arginine homeostasis is complex and involves dietary intake, synthesis in the kidney, and its utilization by arginases and other arginine catabolic enzymes (1, 17).
Arginase (l-arginine ureohydrolase, EC 3.4.3.1) catalyzes the hydrolysis of arginine to ornithine and urea. In mammals, there are two isoforms of arginase. Arginase I is found in the cytosol and is the isoform that participates in the urea cycle and is expressed at high levels in the liver of ureotelic animals. The arginase I cDNA and gene have been cloned (4, 5, 11), and extensive reviews are available (7, 10). A deficiency in arginase I in humans results in the clinical disease argininemia (2). In contrast to arginase I, arginase II is localized in the mitochondria. It has a wide tissue distribution, being most highly expressed in the prostate and kidney and poorly expressed in the liver (18, 22). The role of arginase II in mammalian physiology is not completely understood but is speculated to involve the synthesis of proline and glutamate and/or polyamines (24). No human disease has been associated with a deficiency in arginase II. The cDNA and gene for arginase II have been cloned (6, 18, 20, 22). In an effort to provide insight into the physiological role of arginase II, we constructed a murine model for the deficiency of this gene.
MATERIALS AND METHODS
DNA was prepared from tail clippings of 3-week-old pups. Genotyping was performed by Southern hybridization and PCR. Primers used for PCRs were the following: forward primer for both the wild-type and mutant genes, TCCTTTCTCCTGTCTAATTC, reverse primer for the wild-type gene, CTAGCATCTAATTGACTGCC; and reverse primer for the mutant gene, CCATGATGGATACTTTCTC. The PCR product for the wild-type gene was 510 bp in length, and for the mutant it was 900 bp in length.
Determination of arginase activity.
Mice were bred until 8 to 10 weeks old. Single kidneys were removed from the animals at autopsy, weighed, and placed in a fivefold volume (weight per volume) of lysis buffer (0.1 M NaCl, 10 mM Tris [pH 8.0]). Samples were homogenized thoroughly with a PRO Scientific, Inc., model PRO250 homogenizer in a glass tube. The enzyme assay protocol was adapted from that of Nuzum and Snodgrass (19).
Plasma and urine arginine analysis.
Plasma was taken from the animals by orbital sinus bleeding with heparinized capillary tubes (Fisher catalog no. 2501). Amino acid levels were determined on a Beckman model 6300 amino acid analyzer.
Statistical analysis.
Data were analyzed using analysis of variance tests. A P value of less than 0.01 was considered to be significant.
RESULTS
Generation of mice lacking a functional arginase II gene.
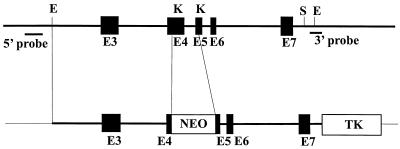
A 20-kb fragment of mouse genomic DNA containing the arginase II gene was isolated by screening a 129sv/Ev mouse genomic library in lambda FIXII vector (Stratagene catalog no. 946305) (20). This phage contained exons 3 to 7 of the arginase II gene as confirmed by sequencing. The targeting vector was constructed by removing an internal KpnI fragment and replacing it with a neomycin resistance cassette containing an RNA polymerase II promoter and bovine growth hormone polyadenylation sequence (12, 21) (Fig. 1). The construction also included the herpes simplex virus thymidine kinase gene for selection (3, 13). In the final construct, parts of exons 4 and 5 of the arginase gene were deleted, and the result was a frame shift in the final mRNA product. The embryonic stem cell line was electroporated with the linearized targeting vector DNA, and neomycin-resistant colonies were selected in G418-containing medium Successfully targeted cell lines were identified by Southern blot hybridization with both a 5′ flanking probe and a 3′ flanking probe.
FIG. 1.
Generation of a targeted disruption of the murine arginase II locus. The structure of the arginase II gene used for construction is illustrated on the upper line, and the final targeting construct is shown below. The final construct spans a total of 6.7 kb from the 5′ EcoRI site to the 3′ SacI site. The KpnI-to-KpnI fragment that was targeted contains portions of exons 4 and 5. The positions of the 5′ and 3′ probes used for delineation of the targeted event are illustrated. Restriction enzymes are designated as follows: E, EcoRI; K, KpnI; S, SacI. The selectable markers are neomycin acetyltransferase (NEO) and herpes simplex virus thymidylate kinase (TK). Exons are represented by shaded boxes and are designated E3 through E7.
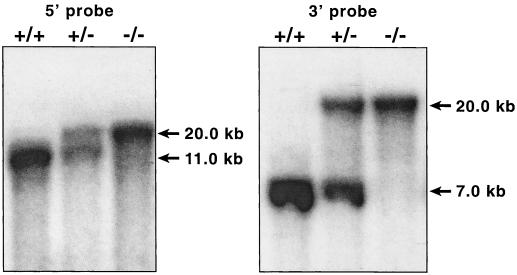
Two positive colonies were expanded. Homologous recombination was confirmed by Southern blotting. Cells from two of the clones were injected into C57BL/6J blastocysts and gave rise to germ line chimeras. Male chimeras with germ line transmission were bred to C57BL/6J females to establish hybrid F1 progeny. Confirmation that the appropriate genomic rearrangement had occurred was provided by Southern analysis of the DNA from all genotypes (Fig. 2).
FIG. 2.
Proof of transmittal of the disrupted allele. A Southern blot of DNA, isolated as described, digested with KpnI, and hybridized with radioactively labeled 5′ probe or 3′ probe is shown. The genotypes of the samples are designated above the lanes, and the sizes of the fragments are shown on the right of each panel.
Heterozygous (argII/argIImut) mice were interbred and produced the expected 1:2:1 ratio of wild-type, heterozygous, and homozygous offspring (327 animals analyzed). Functional inactivation of the arginase II gene was confirmed by assaying the enzyme in the kidney. The data shown in Table 1 demonstrate that enzymatic activity for arginase is completely deficient in the kidney of homozygous arginase II-deficient animals. Interestingly, the level of activity in heterozygous animals is less than the normal amount but not the expected half-normal level. The significance of this observation, if any, is difficult to assess due to the large variation encountered in this assay. Northern blotting of mRNA from the mutant animals revealed the presence of a band similar in both intensity and size to the normal arginase II mRNA, as expected for a transcript of the modified gene.
TABLE 1.
Arginase activity in adult kidney
| Genotype | No. of animals | Activitya |
|---|---|---|
| argII/argII | 20 | 295.6 ± 212 |
| argII/argIImut | 20 | 238.8 ± 186 |
| argIImut/argIImut | 20 | 3.9 ± 4.1 |
Activity is expressed as micromoles of urea formed per hour per gram (wet weight) of tissue ± 1 standard deviation.
Homozygous arginase II-deficient animals are completely fertile and do not have a readily distinguishable phenotype. The results of all anatomical and histochemical studies on mutant animals were within normal limits.
Polyamine and amino acid analysis.
Given the potential role of arginase in amino acid and polyamine biosynthesis, we evaluated these pathways with metabolite assays. Polyamines were measured in several tissues including brain, liver, kidney, and testes. No significant differences were detected between mutant and normal animals. We also analyzed polyamines in cultured skin fibroblasts from both mutant and normal animals and found no significant differences. Plasma amino acid levels in adult mice were measured at 8 to 10 weeks of age. These data are provided in Table 2. There was a significant difference in plasma arginine levels between wild-type and homozygous arginase II-deficient animals (P = 0.0017) and between wild-type and heterozygous animals (P = 0.0008). There was a trend toward elevated ornithine levels in the homozygous arginase II-deficient mice, but the differences were not statistically significant. The other amino acids showed no significant differences among the genotypes. Analysis of urinary amino acids did not reveal any significant differences between wild-type and mutant animals.
TABLE 2.
Concentrations of related plasma amino acids
| Genotype | No. of animals | Mean concn in plasma (μmol/liter) ± SD
|
|||
|---|---|---|---|---|---|
| Arginine | Ornithine | Citrulline | Proline | ||
| argII/argII | 15 | 65.5 ± 30.3 | 92.7 ± 48.7 | 60.6 ± 31.0 | 119.3 ± 49.5 |
| argII/argIImut | 15 | 115.9 ± 45.2 | 113.1 ± 54.1 | 74.7 ± 22.3 | 145.0 ± 50.4 |
| argIImut/argIImut | 18 | 136.3 ± 74.6 | 133.0 ± 114.7 | 54.6 ± 18.9 | 126.4 ± 54.5 |
DISCUSSION
In the present study, we demonstrate that we have created mice deficient in arginase II. The mice had hyperargininemia but were normal with respect to related amino acids (Table 2).
Results of metabolic-labeling studies have suggested that arginase degradation and/or dietary intake are the primary regulators of arginine homeostasis. In support of this notion, our results indicate that arginine catabolism via arginase II is an important factor in arginine homeostasis. Furthermore, the absence of any significant alteration in plasma citrulline levels (Table 2) suggests that there have been no compensatory changes in rates of arginine synthesis via the intestinal-renal axis in the homozygous arginase II-deficient mice.
The activity of arginase II in the mammary gland is greatly increased during lactogenesis, and it has been postulated that this occurs to enhance the capacity for synthesis of proline or glutamate for milk (14, 25). However, there was no apparent deficiency in nutrients in the milk of homozygous arginase II-deficient mice, because the growth rates of their pups prior to weaning were indistinguishable from those of wild-type pups.
Unlike the cases of arginase I deficiency, our results suggest that a deficiency in arginase II in humans is possibly a benign trait, at least in otherwise healthy individuals. However, it is possible that a deficiency in arginase II would be deleterious under conditions of disease or injury, and experiments are under way to test this possibility.
ACKNOWLEDGMENT
This work was supported in part by NIH grant GM57384 to W.E.O. and S.M.M.
REFERENCES
- 1.Beaumier L, Castillo L, Ajami A M, Young V R. Urea cycle intermediate kinetics and nitrate excretion at normal and “therapeutic” intakes of arginine in humans. Am J Physiol. 1995;269:E884–E896. doi: 10.1152/ajpendo.1995.269.5.E884. [DOI] [PubMed] [Google Scholar]
- 2.Brusilow S W, Horwich A L. Urea cycle enzymes. In: Scriver C R, Beaudet A L, Sly W L, editors. The metabolic and molecular bases of inherited disease. Vol. 1. New York, N.Y: McGraw-Hill, Inc.; 1995. pp. 1187–1232. [Google Scholar]
- 3.Castillo L, Chapman T E, Sanchez M, Yu Y M, Burke J F, Ajami A M, Vogt J, Young V R. Plasma arginine and citrulline kinetics in adults given adequate and arginine-free diets. Proc Natl Acad Sci USA. 1993;90:7749–7753. doi: 10.1073/pnas.90.16.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dizikes G J, Grody W W, Kern R M, Cederbaum S D. Isolation of human liver arginase cDNA and demonstration of nonhomology between the two human arginase genes. Biochem Biophys Res Commun. 1986;141:53–59. doi: 10.1016/s0006-291x(86)80333-3. [DOI] [PubMed] [Google Scholar]
- 5.Dizikes G J, Spector E B, Cederbaum S D. Cloning of rat liver arginase cDNA and elucidation of regulation of arginase gene expression in H4 rat hepatoma cells. Somat Cell Mol Genet. 1986;12:375–384. doi: 10.1007/BF01570732. [DOI] [PubMed] [Google Scholar]
- 6.Gotoh T, Sonoki T, Nagasaki A, Terada K, Takiguchi M, Mori M. Molecular cloning of cDNA for nonhepatic mitochondrial arginase (arginase II) and comparison of its induction with nitric oxide synthase in a murine macrophage-like cell line. FEBS Lett. 1996;395:119–122. doi: 10.1016/0014-5793(96)01015-0. [DOI] [PubMed] [Google Scholar]
- 7.Iyer R K, Bando J M, Jenkinson C P, Vockley J G, Kim P S, Kern R M, Cederbaum S D, Grody W W. Cloning and characterization of the mouse and rat type II arginase genes. Mol Gen Metab. 1998;63:168–175. doi: 10.1006/mgme.1997.2669. [DOI] [PubMed] [Google Scholar]
- 8.Jeevanandam M, Ramias L, Schiller W R. Altered plasma free amino acid levels in obese traumatized man. Metabolism. 1991;40:385–390. doi: 10.1016/0026-0495(91)90149-q. [DOI] [PubMed] [Google Scholar]
- 9.Jeevanandam M, Young D H, Ramias L, Schiller W R. Effect of major trauma on plasma free amino acid concentrations in geriatric patients. Am J Clin Nutr. 1990;51:1040–1045. doi: 10.1093/ajcn/51.6.1040. [DOI] [PubMed] [Google Scholar]
- 10.Jenkinson C P, Grody W W, Cederbaum S D. Comparative properties of arginases. Comp Biochem Physiol B. 1996;114B:107–132. doi: 10.1016/0305-0491(95)02138-8. [DOI] [PubMed] [Google Scholar]
- 11.Kawamoto S, Amaya Y, Murakami K, Tokunaga F, Iwanaga S, Kobayashi K, Saheki T, Kimura S, Mori M. Complete nucleotide sequence of cDNA and deduced amino acid sequence of rat liver arginase. J Biol Chem. 1987;262:6280–6283. [PubMed] [Google Scholar]
- 12.Mansour S L, Thomas K R, Capecchi M R. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- 13.McMahon A P, Bradley A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990;62:1073–1085. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- 14.Mezl V A, Knox W E. Metabolism of arginine in lactating rat mammary gland. Biochem J. 1977;166:105–113. doi: 10.1042/bj1660105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris J G, Rogers Q R. Ammonia intoxication in the near-adult cat as a result of a dietary deficiency of arginine. Science. 1978;199:431–432. doi: 10.1126/science.619464. [DOI] [PubMed] [Google Scholar]
- 16.Morris J G, Rogers Q R, Winterrowd D L, Kamikawa E M. The utilization of ornithine and citrulline by the growing kitten. J Nutr. 1979;109:724–729. doi: 10.1093/jn/109.4.724. [DOI] [PubMed] [Google Scholar]
- 17.Morris S M., Jr . Regulation of arginine availability and its impact on NO synthesis. In: Ignarro L J, editor. Nitric oxide: biology and pathobiology. San Diego, Calif: Academic Press; 2000. pp. 187–197. [Google Scholar]
- 18.Morris S M, Jr, Bhamidipati D, Kepka-Lenhart D. Human type II arginase: sequence analysis and tissue-specific expression. Gene. 1997;193:157–161. doi: 10.1016/s0378-1119(97)00099-1. [DOI] [PubMed] [Google Scholar]
- 19.Nuzum C T, Snodgrass P J. Multiple assays of the five urea cycle enzymes in human liver homogenates. In: Grisolia S, Baguena R, Mayor F, editors. The urea cycle. New York, N.Y: John Wiley & Sons; 1976. pp. 325–349. [Google Scholar]
- 20.Shi O, Kepka-Lenhart D, Morris S M J, O'Brien W E. Structure of the murine arginase II gene. Mamm Genome. 1998;9:822–824. doi: 10.1007/s003359900874. [DOI] [PubMed] [Google Scholar]
- 21.Soriano P, Montgomery C, Geske R, Bradley A. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- 22.Vockley J G, Jenkinson C P, Shukla H, Kern R M, Grody W W, Cederbaum S D. Cloning and characterization of the human type II arginase gene. Genomics. 1996;38:118–123. doi: 10.1006/geno.1996.0606. [DOI] [PubMed] [Google Scholar]
- 23.Wakabayashi Y, Yamada E, Yoshida T, Takahashi H. Arginine becomes an essential amino acid after massive resection of rat small intestine. J Biol Chem. 1994;269:32667–32671. [PubMed] [Google Scholar]
- 24.Wu G, Morris S M., Jr Arginine metabolism: nitric oxide and beyond. Biochem J. 1998;336(Pt. 1):1–17. doi: 10.1042/bj3360001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yip M C, Knox W E. Function of arginase in lactating mammary gland. Biochem J. 1972;127:893–899. doi: 10.1042/bj1270893. [DOI] [PMC free article] [PubMed] [Google Scholar]