Abstract
Ssy1p and Ptr3p are known components of a yeast plasma membrane system that functions to sense the presence of amino acids in the extracellular environment. In response to amino acids, this sensing system initiates metabolic signals that ultimately regulate the functional expression of several amino acid-metabolizing enzymes and transport proteins, including multiple, genetically distinct amino acid permeases. We have found that SSY5 encodes a third component of this amino acid sensing system. Mutations in SSY5 manifest phenotypes that are indistinguishable from those resulting from either single ssy1 and ptr3 mutations or ssy5 ssy1 and ssy5 ptr3 double mutations. Although Ssy5p is predicted to be a soluble protein, it exhibits properties indicating that it is a peripherally associated plasma membrane protein. Each of the three sensor components, Ssy1p, Ptr3p, and Ssy5p, adopts conformations and modifications that are dependent upon the availability of amino acids and on the presence of the other two components. These results suggest that these components function as part of a sensor complex localized to the plasma membrane. Consistent with a sensor complex, the overexpression of SSY1 or the unique N-terminal extension of this amino acid permease homologue inactivates the amino acid sensor in a dominant-negative manner. Each of the components of the Ssy1p-Ptr3p-Ssy5p (SPS) signaling system undergoes rapid physical changes, reflected in altered electrophoretic mobility, when leucine is added to cells grown in media lacking amino acids. Furthermore, the levels of each SPS sensor component present in whole-cell extracts diminish upon leucine addition. The rapid physical alterations and reduced levels of sensor components are consistent with their being downregulated in response to amino acid availability. These results reveal the dynamic nature of the amino acid-initiated signals transduced by the SPS sensor.
The ability of Saccharomyces cerevisiae cells to rapidly respond and adapt to changing environmental conditions is essential for viability. A prerequisite for the generation of a proper physiological response is the ability to sense and subsequently transduce information regarding the extra- and intracellular environments. Sensor-initiated signals are used to make dynamic adjustments in patterns of gene expression and protein turnover, processes that enable cells to express the necessary components appropriate for prevailing conditions. For example, in response to nutrient availability, yeasts regulate the expression and activity of proteins involved in nutrient uptake and utilization. Recently, several plasma membrane-localized nutritional sensors that monitor nutrient availability in the extracellular environment have been identified in yeast. These include two glucose sensors, SNF3 and RGT2 (41, 49); a G-protein-coupled receptor that is activated by the presence of fermentable sugars, GPR1 (35, 44, 67); a high-affinity ammonium transporter (MEP2) (45) that may function as an ammonium sensor (43); and an amino acid sensor, SSY1 (19, 30, 33, 34). Little is known regarding whether these primary sensors function alone or in complexes together with other proteins, and the mechanisms by which these sensors transduce nutritionally derived signals remain to be elucidated.
SNF3 and RGT2 encode unique members of the hexose transporter (HXT) family that possess unusually long C-terminal domains. These proteins are poorly expressed compared to the other known functional hexose transporters and pleiotropically affect the expression of multiple HXTs (41, 49). The expression of SNF3 is repressed by the presence of glucose (10), whereas RGT2 is constitutively expressed (49). Recent reports have shown that the cytoplasmically oriented C-terminal extensions of Snf3p and Rgt2p have important roles in signaling. The C terminus of Snf3p expressed as a soluble protein (62), or as a hybrid with the Hxt2p hexose transporter (48), can partly suppress phenotypes associated with an snf3 deletion. The C-terminal extensions of Snf3p and Rgt2p physically interact with two homologous proteins, Std1p and Mth1p (37, 56). Std1p and Mth1p are suggested to act as repressors of Snf3p and Rgt2p target genes in the absence of signaling. Green fluorescent protein-Std1p fusion proteins are localized to the cell periphery and the cell nucleus; thus the possibility exists that Std1p mediates information directly from the cell surface to the nucleus (56).
SSY1 encodes a unique member of the amino acid permease protein family that functions as a sensor of extracellular amino acids (19, 30, 33, 34). Ssy1p has a unique 200-amino-acid N-terminal extension that is required for SSY1 activity (34); however, its precise function remains obscure. Ssy1p is localized to the plasma membrane and is, as are the other members of the amino acid permease family, dependent upon the amino acid-specific packaging chaperone Shr3p (25, 42) to exit the endoplasmic reticulum (34). SSY1 is required for the transcriptional induction of multiple genes encoding amino acid permeases (AGP1, BAP2, BAP3, GNP1, VAP2, and TAT2), the peptide transporter (PTR2), and arginase (CAR1) in response to extracellular amino acids (19, 30, 34). SSY1-mediated signals are also required for full transcriptional repression of the general amino acid permease (GAP1) on ammonium-based media in the presence of amino acids (34). Ssy1p is dependent upon PTR3 to mediate amino acid-derived signals (34). Ptr3p is a peripherally associated plasma membrane protein and contains a domain that shares homology with amino acid permeases and Gcn4p (34).
Several factors required for transcription of SSY1-controlled genes have been identified, including STP1, STP2, and ABF1 (15, 16) and UGA35 (also known as DAL81) and GRR1 (30). STP1 and STP2 were originally identified as genes required for pre-tRNA maturation (66). Abf1p is a general transcription factor involved in global gene activation and silencing (17, 54, 57). UGA35/DAL81 encodes a nonspecific factor required for the full induction of several genes active in nitrogen utilization, including γ-aminobutyric acid and allophanate-inducible genes (8, 14, 64). Some of the SSY1-responsive genes, including BAP2, also carry promoter binding sites for Gcn4p and Leu3p (18). GCN4 encodes a transcription factor that is translationally regulated by the general amino acid control pathway that monitors intracellular levels of free amino acids. The general control pathway coordinately upregulates biosynthetic genes and transporters in response to amino acid deprivation (28). Leu3p, a transcription factor that is capable of acting as a repressor or an inducer, participates in regulating the transcription of several genes within the branched-chain amino acid biosynthetic pathways (9, 65). GRR1 was previously reported to be involved in glucose signaling and cell cycle control (5, 40). Grr1p is an F-box-containing component of discrete Skp1–Cullin–F-box (SCF) ubiquitin ligase complexes that mark proteins for degradation via the proteasome. Several F-box-containing proteins have been identified in yeast, and they are thought to provide specificity by recruiting distinct protein substrates to SCF complexes for ubiquitination (51). Presumably the inability of grr1 mutants to properly respond to nutritional signals is a consequence of aberrant patterns of regulated protein degradation (32, 50). The large number of factors that affect the transcription of target genes indicates a complex network of regulatory processes that most likely integrate signals derived from different primary nutritional sensors.
Although several of the downstream components required for the transmission of amino acid-induced signals have been identified, little is known regarding the primary component composition of the plasma membrane amino acid-sensing system. As previously indicated, this system not only depends upon Ssy1p, but also requires Ptr3p (34). Even less is known regarding the mechanisms associated with initial signaling events that occur within the sensing system in response to extracellular amino acids. We have focused our efforts on characterizing the proteins comprising the plasma membrane amino acid nutritional sensor. In this paper, we present results that identify Ssy5p as a third component of this signaling system. Mutations in SSY5 have earlier been described to interfere with amino acid uptake (33). We show that ssy5 mutants belong to the same phenotypic epistasis group as both ssy1 and ptr3 mutants. Ssy5p is a peripheral membrane protein that binds to the plasma membrane and is dependent upon SSY1 and PTR3 for wild-type expression. Additionally, Ssy1p and Ptr3p exhibit altered electrophoretic mobilities dependent on the presence of the other sensing components and on the availability of amino acids. Finally, we have documented the dynamic nature of amino acid-induced signaling and have found that each of the components of the Ssy1p1-Ptr3p-Ssy5p (SPS) signaling system is rapidly downregulated in response to amino acid availability.
MATERIALS AND METHODS
Strains and media.
The yeast strains used in this study are listed in Table 1. Strains carrying a null mutation of SSY5 were constructed as follows. PLY1 and PLY126 were transformed with a linear 9-kb XbaI fragment from pHK049 (see Table 2; plasmids are described in the following section) containing the ssy5Δ1::hisG-URA3-hisG deletion construct. Strain HKY75 was propagated on media containing 5-fluoroorotic acid (5-FOA) (26) to attain strain HKY77 carrying the unmarked ssy5Δ2 deletion. Strain HKY82 was obtained by transforming HKY77 with a linear SalI-SpeI fragment of pHK031 containing ssy1Δ12::hisG-URA3-hisG. Similarly, strain HKY83 was obtained by transforming HKY77 with a linear EcoRI fragment of pPL341 containing ptr3Δ14::hisG-URA3-hisG (34). Strains HKY82 and HKY83 were propagated on 5-FOA, resulting in strains HKY84 and HKY85 with unmarked ssy1Δ13 and ptr3Δ15 alleles, respectively. The correct integration of each gene replacement was confirmed by Southern analysis. The yeast strain cdc25H was obtained from Stratagene (La Jolla, Calif.).
TABLE 1.
Characteristics of the cerevisiae strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| cdc25H | MATaura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3, 112 cdc25-2 GAL+ | 53 |
| Isogenic derivatives of PLY1 | ||
| PLY1 | MATa ura3-52 his4Δ29 | 42 |
| PLY4 | MATα ura3-52 his4Δ29 ade2Δ1::URA3 | 42 |
| PLAS10-8C | MATaura3-52 his4Δ29 shr4-10 (ssy5-410) | This work |
| PLAS10-9A | MATα ura3-52 his4Δ29 shr4-10 (ssy5-410) | This work |
| PLAS7-4C | MATaura3-52 his4Δ29 shr10-7 (ssy1-107) | 34 |
| PLAS14-1A | MATaura3-52 his4Δ29 shr6-14 (ptr3-614) | 34 |
| HKY37 | MATaura3-52 his4Δ29 ssy1Δ13 | 34 |
| HKY71 | MATaura3-52 his4Δ29 ssy5Δ1::hisG-URA3-kanr-hisG | This work |
| Isogenic derivatives of PLY126 | ||
| PLY126 | MATaura3-52 lys2Δ201 | 34 |
| HKY20 | MATaura3-52 lys2Δ201 ssy1Δ13 | 34 |
| HKY31 | MATaura3-52 lys2Δ201 ptr3Δ15 | 34 |
| HKY33 | MATaura3-52 lys2Δ201 ssy1Δ13 ptr3Δ15 | 34 |
| HKY75 | MATaura3-52 lys2Δ201 ssy5Δ1::hisG-URA3-kanr-hisG | This work |
| HKY77 | MATaura3-52 lys2Δ201 ssy5Δ2 | This work |
| HKY84 | MATaura3-52 lys2Δ201 ssy1Δ13 ssy5Δ2 | This work |
| HKY85 | MATaura3-52 lys2Δ201 ptr3Δ15 ssy5Δ2 | This work |
TABLE 2.
Plasmids and oligonucleotides used in this study
| Plasmid or oligonucleotide | Description or sequence | Reference |
|---|---|---|
| Plasmids | ||
| pPL193 | 5-kb EcoRI fragment containing PTR3 in pRS316 | 34 |
| pPL341 | ptr3Δ14::hisG-URA3-kanr-hisG in pUC118 (ΔB-ΔH) | 34 |
| pPL356 | 3.9-kb SpeI-ClaI fragment containing SSY1 in pRS316 | 34 |
| pPL496 | 7.8-kb Sau3A fragment containing SSY5 in pRS202 | This work |
| pHK003 | pPL356 with all XbaI sites removed | 34 |
| pHK010 | SSY1-HA1 in pRS316 | 34 |
| pHK012 | SSY1 in pRS202 | This work |
| pHK018 | PTR3-HA1 in pRS316 | 34 |
| pHK026 | PTR3 in pRS202 | This work |
| pHK031 | ssy1Δ12::hisG-URA3-kanr-hisG in pBluescript | This work |
| pHK035 | 4.9-kb BstXI-BamHI fragment containing SSY5 in pRS316 | This work |
| pHK036 | Same as pHK035, except SSY5 fragment inserted in opposite orientation | This work |
| pHK037 | SSY5 in pRS202 | This work |
| pHK038 | SSY1-NT(1–206) in pRS316 | This work |
| pHK039 | SSY1-NT(1–206) in pRS202 | This work |
| pHK040 | pPL193 with BamHI site in MCS removed | This work |
| pHK041 | pHK036 with BamHI site in MCS removed | This work |
| pHK042 | pHK040 with BamHI site (b) introduced after ATG in PTR3 | This work |
| pHK043 | pHK041 with BamHI site (b) introduced after ATG in SSY5 | This work |
| pHK044 | PTR3 in pSOS | This work |
| pHK045 | SSY5 in pSOS | This work |
| pHK047 | pHK041 with BamHI site (a) introduced after the ATG of SSY5 ORF | This work |
| pHK048 | SSY5-c-myc in pRS316 | This work |
| pHK049 | ssy5Δ1::hisG-URA3-kanr-hisG in pRS316 | This work |
| pHK050 | SSY5 in pAD40 | This work |
| Oligonucleotides | ||
| POL95-039 | 5′-GATTGAGTTGAATTCTTCTACCACTAC-3′ | |
| POL96-021 | 5′-CCTGGCTGATTTCTAGATAGGGTTATATG-3′ | |
| POL99-026 | 5′-GATATACCGATGGGATCCGTCAGATTTTTTGG-3′ | |
| POL00-004 | 5′-GATATACCGATGGGGATCCTCAGATTTTTTGG-3′ | |
| POL00-005 | 5′-TAGGCGTTCATGGGGATCCACTCTCATAG-3′ |
Standard yeast media were prepared as described by Guthrie and Fink (26). Nonstandard synthetic medium with proline as the nitrogen source (SPD) was prepared as follows. Proline (4 g/liter) and Difco yeast nitrogen base (26.8 g/liter) were added together to make a 4× stock solution that was filter sterilized. Other components were autoclaved as separate stock solutions (40% glucose and 4% Difco Bacto agar). Stock solutions and sterile water were mixed to make a 2× solution containing 4% glucose, and an equal volume of molten 4% agar was added. Where required, SPD was supplemented as indicated (e.g., 30 mM l-histidine or l-methionine). The concentration of yeast nitrogen base in these synthetic media is fourfold higher than the amount used in other standard synthetic media. Yeast transformations were performed as described by Ito et al. (31) with 50 μg of heat-denatured calf thymus DNA. Transformants were selected on solid complete synthetic dextrose medium (SC) lacking either uracil, leucine, or lysine as required.
Plasmids.
The plasmids and oligonucleotides used in this study are listed in Table 2. In separate reactions, a 4.9-kb BamHI-BstEII fragment from pPL496 containing SSY5 was ligated to BamHI-digested pRS316 (59) and pRS202 (13), resulting in pHK036 (CEN, ori1), pHK037 (CEN, ori2), and pHK037 (2μm, ori2). A blunt-ended 5-kb BamHI-BglII fragment isolated from pSE1076 (1) containing a hisG URA3 kanr hisG blaster cassette was inserted into BsrGI-digested pHK036 made blunt by using T4 DNA polymerase. This ssy5Δ construct (pHK049) removes nucleotides (nt) −28 to +1108 (nucleotide designations are in relation to the first nucleotide of the initiator ATG codon). Plasmid pHK031 was constructed by inserting a SalI-SpeI fragment from pHK030 carrying the ssy1Δ12::hisG URA3 kanr hisG deletion allele (34) into SalI-SpeI-digested pBluescript II KS(+) (Stratagene, La Jolla, Calif.). To remove the BamHI sites within their multicloning sequences, plasmids pPL193 (34) and pHK036 were digested with BamHI, made blunt with T4 DNA polymerase, and religated, creating pHK040 and pHK041, respectively. The SSY5–c-myc epitope-tagged allele in pHK048 was constructed in two steps. First, a BamHI site (reading frame a) was introduced immediately after the ATG initiation codon of SSY5 by using site-directed mutagenesis (36, 63) with single-stranded pHK041 as a template and oligonucleotide POL99-026 as mutagenic primer, resulting in pHK047. In step 2, a BamHI-flanked cloning cassette, encoding the c-myc epitope reiterated three times (c-myc3) was inserted into the unique BamHI site of pHK047, creating pHK048. The construction of plasmids encoding SOS hybrid proteins required multiple steps. Site-directed mutagenesis with single-stranded pHK040 and pHK041 as a template and mutagenic primers POL00-005 and POL00-004 was used to create unique BamHI sites (reading frame b) immediately following the ATG of PTR3 and the ATG of SSY5, resulting in plasmids pHK042 and pHK043, respectively. The vectors contained in the CytoTrap kit were obtained from Stratagene. The BamHI-SalI fragments from plasmids pHK042 and pHK043 were inserted into BamHI-SalI-digested pSOS (Stratagene), creating pHK044 and pHK045. Plasmid pHK050 was constructed by moving the SacI-SalI fragment from pHK035 containing SSY5 into SacI-SalI-digested pAD40 (obtained from M. Wigler, Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.). In a single reaction, two XbaI sites were introduced into the SSY1 coding sequence with single-stranded pHK003 as a template (34) and mutagenic primers POL95-039 and POL96-021. The resulting plasmid was digested with XbaI to release the internal fragment of SSY1 and religated, resulting in pHK038. pHK038 encodes only the first 206 N-terminal amino acids of Ssy1p. The 2μm plasmids pHK039 and pHK012 were constructed by inserting the SacII-KpnI fragments from pPL356 (34) or pHK038 into SacII-KpnI-digested pRS202, respectively. Plasmid pHK026 (2μm PTR3) was created by ligating a SalI-SacII fragment containing PTR3 obtained from pPL193 into SalI-SacII-digested pRS202.
Genetic analysis.
Strain PLY1 was used to isolate spontaneous mutants resistant to 30 mM histidine (42). The super-high-histidine-resistant (shr) mutants were backcrossed to PLY4 (MATα his4Δ29 ura3-52 ade2Δ1::URA3), an isogenic derivative of PLY1. Tetrad analysis indicated that the mutant phenotypes segregated 2:2. Strain PLAS10-8C (shr4-10) was obtained as a meiotic segregant from one of these crosses. SHR4 was cloned by complementation of the 30 mM histidine-resistant phenotype; strain PLAS10-8C (shr4-10) was transformed with a plasmid library (pRS202 library, obtained from Philip Hieter [13]), and Ura+ transformants unable to grow on selective medium containing 30 mM histidine were identified. Complementing plasmids were isolated and further analyzed. Plasmid pHK035, which contains a 4.9-kb BstEII-BamHI insert with a single open reading frame (ORF) (YJL156c), fully complemented shr4 mutant alleles.
Northern analysis.
Steady-state levels of AGP1 and PTR2 mRNA (see Fig. 2) were determined in strains PLY126 and HKY77 transformed with YCp405 and pRS316. Cells from overnight cultures grown in SD were harvested, washed once, and resuspended in 10× volume of fresh SD at an optical density at 600 nm (OD600) of 0.2. Cultures were grown to an OD600 of 0.8, one-half of the culture was induced by addition of leucine to a concentration of 0.15 mM, and an identical aliquot of water was added to the remaining half of the culture (uninduced control). After 45 min, 30 ml of cells was harvested, RNA was prepared, and Northern analysis was performed with aliquots (10 μg) of RNA as described previously (34). SSY5 mRNA levels were analyzed in strains HKY77, HKY84, and HKY85 transformed with pHK048 and YCp405 (Fig. 4B). Cells were grown overnight in SD or SD supplemented with 1.3 mM leucine, washed once, and resuspended in a 10× volume of fresh medium at an OD600 of 0.2. RNA was isolated when cultures reached a cell density of an OD600 of 0.8. Radioactive probes were prepared with the following template DNA fragments: a 1.2-kb EcoRV internal fragment of SSY5 and a 339-bp PCR fragment from AGP1 amplified with oligonucleotide primer pairs POL00-006 and POL00-007. Additionally, the previously described 569-bp PCR fragment of CAR1, 530-bp PCR-fragment from PTR2, and a 1.65-kb BamHI-HindIII fragment containing ACT1 were used (34). The DNA fragments were labeled with [α-32P]dCTP (3,000 Ci/mmol, Amersham, United Kingdom) by using a random-primed DNA labeling kit (MBI Fermentas Molecular Biology) and purified by using Bio-Rad Bio Spin columns. After hybridization, blots were rinsed once with 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS), washed two times with 5× SSC–0.1% SDS, one time with 1× SSC–0.1% SDS, and one time with 0.5× SSC–0.1% SDS if required. Washings were performed at 55°C for 20 min. After washings, blots were visualized and quantified with a Fujix Bio-Image Analyzer BAS1500 (Fuji Photo Film Co., Ltd., Tokyo, Japan).
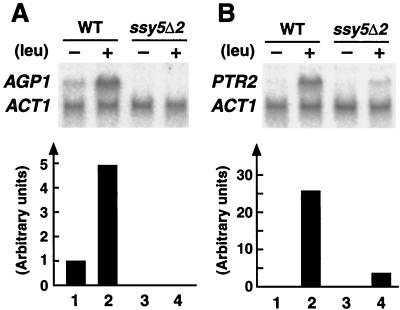
FIG. 2.
Expression of AGP1 and PTR2 in wild-type (WT) and ssy5 null mutant strains. Strains PLY126 (wild type) and HKY77 (ssy5Δ2) were grown in SD to an OD600 of 0.8, and total RNA was isolated 45 min after the addition of water (lanes 1 and 3) or 0.15 mM leucine (lanes 2 and 4). Expression levels of AGP1 (A) and PTR2 (B) were determined by Northern analysis and quantitated by phosphorimaging. The levels of actin (ACT1) transcript were used to standardize quantitations (lower panels). The relative expression levels were normalized to the expression levels in the uninduced wild-type strain.
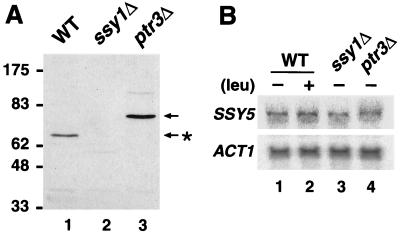
FIG. 4.
Functional expression of Ssy5p requires SSY1 and PTR3. (A) Strains HKY77 (wild type [WT]; lane 1), HKY84 (ssy1Δ13; lane 2), and HKY85 (ptr3Δ15; lane3) expressing SSY5-c-myc were grown in SD to an OD600 of 0.8, and the levels of Ssy5p-c-myc were analyzed in whole-cell lysates by SDS-PAGE and immunoblotting. Prior to electrophoresis, samples were denatured for 10 min at 37°C. (B) Total RNA was prepared from strains HKY77 (wild-type; lanes 1 and 2), HKY84 (ssy1Δ13; lane 3), and HKY85 (ptr3Δ15; lane 4) expressing SSY5-c-myc grown to an OD600 of 0.8 in SD (lanes 1, 3, and 4) or SD supplemented with 1.3 mM leucine (lane 2). The levels of SSY5 mRNA were analyzed by Northern blotting, and the levels of actin (ACT1) transcripts were used to control loading.
Protein manipulations.
Protein was determined by the method of Markwell et al. (46). Whole-cell protein was examined with lysates prepared from 100 ml of cells by the glass-bead method described by Chang and Slayman (11). The membrane association of Ssy5p in strain HKY77 transformed with plasmids pHK048 and YCp405 was examined as follows. An aliquot of a lysate (100 μg of protein) in low-salt BB buffer (0.3 M sorbitol, 5 mM MgCl2, 5 mM Tris [pH 7.5]) was diluted 1:1 with either H2O, 1.6 M urea, or 2 mM EDTA; mixed; and incubated on ice for 30 min. Samples were centrifuged at 100,000 × g for 45 min at 4°C, and protein pellets were resuspended in 2× sample buffer containing low-salt BB buffer. After sonication and denaturation and then incubation for 10 min at 37°C, aliquots (10 μg of protein) were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and analyzed by immunoblotting.
The levels of expression and electrophoretic properties of Ssy5p, Ssy1p, and Ptr3p were examined in whole-cell lysates prepared from strains HKY77(pHK048, YCp405), HKY84(pHK048, YCp405), HKY85(pHK048, YCp405), HKY20(pHK010, YCp405), HKY33(pHK010, YCp405), HKY84(pHK010, YCp405), HKY31(pHK018, YCp405), HKY33(pHK018, YCp405), and HKY85(pHK018, YCp405). In each case, lysates were prepared from 100 ml of cells grown to an OD600 of 0.8 in SD or SC lacking uracil and lysine, as indicated. After denaturation of protein preparations (10 min at 37°C for Ssy5p and Ssy1p, 3 min at 95°C for Ptr3p) in sample buffer, aliquots (10 to 20 μg of protein) were resolved by SDS-PAGE and analyzed by immunoblotting.
Immunoblots were probed with 1:1,000 dilutions of monoclonal antibodies recognizing the hemagglutinin (HA) (12CA5) or c-myc (9E10) epitopes; or rabbit anti-Pma1p diluted 1:3,000 as described by Klasson et al. (34). Chemiluminescent signals were visualized by enhanced chemiluminescence (ECL-PLUS system; Amersham) and quantitated by using the LAS1000 system (Fuji Photo Film Co., Ltd.).
Amino acid pool size determination.
Whole-cell and vacuolar amino acid pool concentrations were determined with cells grown in YPD to an OD600 of ≈1 essentially as described by Ohsumi et al. (47). Appropriate quantities of cultures (3 × 108 cells) were harvested by centrifugation, and cell pellets were washed twice with 1.5 ml of water and resuspended in 1.5 ml of AA buffer (0.6 M sorbitol, 2.5 mM potassium phosphate buffer [pH 6]) containing 10 mM glucose. For the determination of vacuolar amino acid pools, the cells were resuspended in the same buffer containing 0.8 mM CuCl2 and incubated for 10 min at 30°C. One-milliliter aliquots of cell suspensions were filtered (Whatman GF/F filters), and filters were washed four times with AA buffer. The washed filters were boiled in 3 ml of water for 15 min, and 1-ml aliquots were centrifuged to remove particles of filter. The concentrations of amino acids in 30-μl aliquots were determined.
Time course experiments.
Cells from overnight cultures of strains HKY31(pHK018, YCp405), HKY77(pHK048, YCp405), and HKY20(pHK010, YCp405) grown in SD were washed once and resuspended in a 10× volume of fresh SD to an OD600 of 0.1 to 0.2. After cultures reached a cell density of OD600 of 0.5, the cultures were split into two equal volumes. One-half of the cultures received an addition of l-leucine to a concentration of 1.3 mM (induced); the other half received an aliquot of water (uninduced control). Subsamples (130 ml) were withdrawn immediately prior to the addition of l-leucine (t = 0) and at 10, 30, 60, 120, and 180 min after l-leucine addition. Subsamples were rapidly chilled on ice, total cell protein was prepared from 100 ml of culture, and RNA was isolated from 30 ml of culture.
RESULTS
ssy5 mutations result in histidine resistance and increased vacuolar pools of arginine and histidine.
Yeast strains carrying mutations in SHR4 were isolated in a genetic selection for shr mutants resistant to 30 mM histidine (42). SHR4 was cloned by complementation of the recessive 30 mM histidine-resistant phenotype of strain PLAS10-8C (shr4-10) (Materials and Methods). Subsequent sequence analysis indicated that SHR4 is identical to ORF YJL156c, previously identified as SSY5 (33). In addition to exhibiting resistance to 30 mM histidine, ssy5 mutant strains have increased vacuolar pools of arginine and histidine (Table 3). Compared to the wild-type strain PLY1, the vacuolar levels of histidine and arginine in mutant strain PLAS10-8C (ssy5-410) were increased by two- and threefold, respectively. In contrast, the levels of lysine remained unchanged. The observed increases in vacuolar amino acid pools resulting from the ssy5 mutation are similar to those observed in strains carrying mutations in SSY1 and PTR3 (Table 3) (34).
TABLE 3.
Concentrations of basic amino acids in whole cells and vacuoles of the wild-type and ssy5, ssy1, and ptr3 mutant strainsa
| Amino acid | Concn of wild type (PLY1)
|
Concn of ssy5-410 (PLAS10-8C)
|
Fold | Concn of ssy1-107 (PLAS7-4C)
|
Fold | Concn of ptr3-614 (PLAS14-1A)
|
Fold | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| WC | Vac | WC | Vac | WC | Vac | WC | Vac | ||||
| Histidine | 27 | 24 | 58 | 52 | 2.2 | 72 | 62 | 2.6 | 102 | 78 | 3.5 |
| Arginine | 87 | 81 | 268 | 256 | 3.2 | 294 | 266 | 3.3 | 429 | 371 | 4.6 |
| Lysine | 82 | 80 | 89 | 87 | 1.1 | 89 | 85 | 1.1 | 100 | 95 | 1.2 |
Concentrations are nanomoles per 108 cells. Fold, fold wild-type vacuolar amino acid concentration; WC, whole cells; Vac, vacuoles.
A deletion allele of SSY5 was created by replacing the major portion of the coding region with the selectable URA3 marker. This construct, ssy5Δ1::hisG-URA3 kanr hisG, was introduced into haploid strains PLY1 (ura3-52 his4Δ29) and PLY126 (ura3-52 lys2Δ201) by transformation. Viable transformants were readily obtained, indicating that SSY5 is not an essential gene. The resulting ssy5Δ1 null mutant strains, HKY71 and HKY75, were resistant to 30 mM histidine. A diploid resulting from crossing HKY71 and the wild-type strain PLY4 did not grow in the presence of toxic levels of histidine, indicating that the ssy5 null mutation is recessive. In contrast, the diploid strain obtained by crossing strains HKY71 (MATa ura3-52 his4Δ29 ssy5Δ1) and PLAS10-9A [MATα ura3-52 his4Δ29 shr4-10 (ssy5-410)] grew well on medium containing 30 mM histidine. This diploid was sporulated, and meiotic segregants were analyzed by tetrad analysis. In all cases, the mutant phenotype segregated 4:0; all spore-derived colonies from 15 tetrads exhibited resistance to toxic levels of histidine. These results indicate that SSY5 and SHR4 are identical and that the originally isolated shr4 alleles are likely to be loss-of-function mutations.
ssy5 null mutant strains exhibit levels of resistance to toxic amino acids and azetidine carboxylate similar to those of ssy1 and ptr3 mutants.
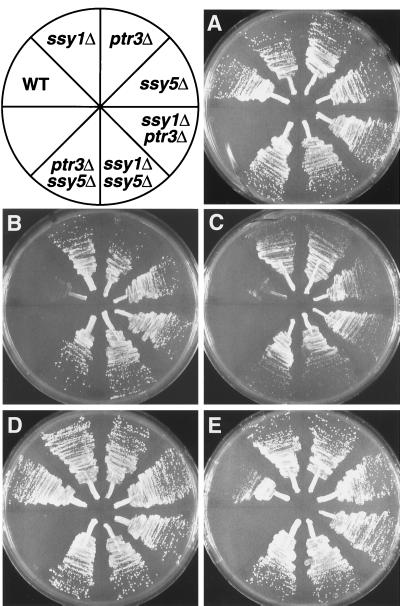
The growth characteristics of isogenic wild-type, ssy1Δ13, ptr3Δ15, ssy5Δ2, ssy1Δ13 ptr3Δ15, ssy5Δ2 ssy1Δ13, and ssy5Δ2 ptr3Δ15 strains were examined. All strains grew well on SPD and SD (Fig. 1A and D, respectively). Only the growth of the wild-type strain was inhibited on medium containing toxic levels of histidine (Fig. 1B), methionine (Fig. 1C), or l-azetidine-2-carboxylate (Fig. 1E). l-Azetidine-2-carboxylate is a proline analogue (38) that pleiotropically inhibits multiple amino acid permeases, including the general amino acid permease (GAP1) (29). In contrast to the wild-type strain, the ssy5Δ2, ssy1Δ13, and ptr3Δ15 mutant strains grew equally well on each of the selective media and formed colonies of similar size. Thus, the ssy5 null mutation manifests identical levels of resistance to either ssy1 or ptr3 null mutations. Additionally, strains carrying the possible double mutant combinations ssy1Δ13 ptr3Δ15, ssy5Δ2 ssy1Δ13, and ssy5Δ2 ptr3Δ15 did not exhibit any additive affects.
FIG. 1.
Growth characteristics of strains carrying single and double mutant combinations of ssy5, ssy1, and ptr3 null alleles. Strains PLY126 (wild type [WT]), HKY20 (ssy1Δ13), HKY31 (ptr3Δ15), HKY77 (ssy5Δ2), HKY33 (ssy1Δ13 ptr3Δ15), HKY84 (ssy5Δ2 ssy1Δ13), and HKY85 (ssy5Δ2 ptr3Δ15) were streaked onto the following media: SPD (plus uracil and lysine) (A), SPD (plus uracil and lysine) containing 30 mM l-histidine (B), SPD (plus uracil and lysine) containing 30 mM l-methionine (C), SD (plus uracil and lysine) (D), and SD (plus uracil and lysine) containing 100 μg of l-azetidine-2-carboxylate ml−1 (E). Plates were incubated for 3 days at room temperature and photographed.
SSY5 is required for amino acid-induced expression of permeases.
Wild-type (PLY126) and ssy5Δ2 (HKY77) strains carrying plasmids pRS316 and YCp405, which complement the ura3-52 and lys2Δ201 auxotrophies, respectively, were grown on SD medium without nutritional supplements to an OD600 of 0.8. Total RNA was isolated 45 min after the addition of 0.15 mM leucine or an equal volume of water, and the transcript levels of the broad-range amino acid permease (AGP1) (30) and the peptide transporter (PTR2) (52) were determined by Northern analysis (Fig. 2). The levels of expression were quantitated by phosphorimaging, and ACT1 transcript levels were used to standardize quantitations. AGP1 transcripts were detected in wild-type cells (Fig. 2A, lanes 1 and 2), but not in ssy5 null mutant cells (Fig. 2A, lanes 3 and 4). The lack of detectable AGP1 transcripts in ssy5 mutant cells indicates that SSY5 is required to maintain the basal AGP1 expression observed in wild-type cells grown in the absence of exogenously added amino acids (Fig. 2, compare lanes 1 and 3). When leucine was added to wild-type cells, AGP1 mRNA levels increased by fivefold (Fig. 2A, compare lanes 1 and 2). Similarly, leucine-induced wild-type cells have approximately 20-fold more PTR2 transcripts than uninduced cells (Fig. 2B, compare lanes 1 and 2). In contrast, when leucine was added to ssy5Δ2 cells, the levels of AGP1 transcripts did not increase (Fig. 2A, lane 4), and PTR2 (Fig. 2B, lane 4) did not accumulate to wild-type levels. These results indicate that ssy5 mutations mimic the observed transcriptional defects exhibited by ssy1 and ptr3 mutations (4, 19, 30, 34).
Ssy5p is a peripheral membrane protein that associates with the plasma membrane.
SSY5 encodes a 76-kDa protein comprised of 687 amino acids that does not share significant sequence homology with other known proteins. A functional epitope-tagged version of Ssy5p was created by introducing the c-myc epitope at the extreme N terminus (see Materials and Methods). The SSY5-c-myc allele was judged to be functional based on its ability to complement the 30 mM histidine-resistant phenotype of ssy5 mutants. The level of Ssy5p-c-myc present in whole-cell extracts was examined by SDS-PAGE and immunoblotting. Ssy5p-c-myc migrated as a 67-kDa protein, significantly faster than its predicted molecular weight, and the signal strength of the immunoreactive Ssy5p-c-myc band was rather weak. The low levels of Ssy5p within extracts are consistent with the low codon index bias (0.017) of the SSY5 ORF.
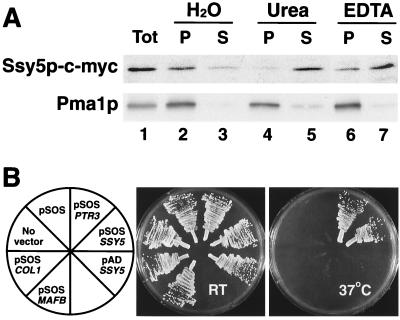
Ssy5p is predicted to be a hydrophilic protein that lacks identifiable transmembrane domains and N-terminal ER targeting signal sequences. However, Ssy5p-c-myc was found to be enriched in the membrane fraction of whole-cell lysates together with the integral plasma membrane ATPase (Pma1p) (Fig. 3A, lanes 2 and 3), Ssy5p-c-myc could be displaced from membranes by treatment with urea (Fig. 3A, lanes 4 and 5) and the chelating agent EDTA (Fig. 3A, lanes 6 and 7). Pma1p was not extracted from the membrane under these conditions. These results suggest that Ssy5p is a peripherally associated membrane protein.
FIG. 3.
SSY5 encodes a peripherally associated PM protein. (A) The membrane association of Ssy5p was examined by using whole-cell lysates prepared from strain HKY77 expressing SSY5-c-myc. Aliquots of total protein lysate (Tot) were diluted 1:1 with H2O, 1.6 M urea, or 2 mM EDTA; mixed; and incubated on ice for 30 min. Membrane pellet (P) and soluble (S) fractions, obtained after centrifugation at 100,000 × g for 45 min at 4°C, were resolved by SDS-PAGE and analyzed by immunoblotting. As a control, the membrane association of the PM ATPase (Pma1p) was monitored. (B) The ability of Ssy5p to associate with the PM was assessed by using the SOS membrane recruitment system. Strains cdc25H (No vector) and cdc25H transformed with plasmids pSOS, pSOS-PTR3 (pHK044), pSOS-SSY5 (pHK045), pAD-SSY5 (pHK050), pSOS-Col1, and pSOS-MAFB were grown on YPD. Culture plates were incubated at room temperature (RT [permissive]) and 37°C (nonpermissive) as indicated, and after 4 days, the plates were photographed.
We examined whether Ssy5p was able to associate with the plasma membrane by using the Sos recruitment system (3). The Sos recruitment system exploits the ability of the human Cdc25p homologue, h-SOSp, to suppress the temperature-sensitive cdc25-2 mutation in strain cdc25H (53). Fusion proteins that direct h-SOSp to the cytosolic face of the plasma membrane enable cdc25H cells (cdc25-2) to grow at 37°C (2). Strain cdc25H was transformed with pSOS, pSOS-PTR3 (pHK044), pSOS-SSY5 (pHK045), and pAD40-SSY5 (pHK050) and negative control plasmids pSOS-Coll (murine type IV collagenase, amino acids 148 to 357) (12) and pSOS-MAFB (full-length MafB) (3). Transformants were selected at room temperature on SC (no Leu). Leu+ transformants were streaked on to two YPD plates. One plate was incubated at room temperature, and the other was incubated at 37°C. After 4 days, the plates were photographed (Fig. 3B). All transformants grew at similar rates on the YPD plate incubated at room temperature. Only transformants carrying plasmids expressing h-SOSp as a fusion protein with either Ssy5p (pSOS-SSY5) or Ptr3p (pSOS-PTR3) grew at 37°C. Transformants carrying the other plasmids were unable to form colonies at the nonpermissive temperature.
These results indicate that h-SOSp expressed alone (pSOS), or fused to control proteins that do not interact with the plasma membrane (PM), such as transcription factor MafB (pSOS-MAFB) and collagenase 1 (pSOS-Coll), is unable to suppress the cdc25-2 mutation. Additionally, transformants carrying pAD-SSY5 did not grow at 37°C, indicating that by itself, Ssy5p is not able to activate the essential Ras signaling pathway. These findings demonstrate that h-SOSp fusion proteins containing either Ssy5p or Ptr3p associate with the PM, thereby enabling h-SOSp to carry out its function. The ability of Ssy5p and Ptr3p to recruit h-SOSp to the PM is consistent with the finding that Ssy5p fractionates as a peripheral membrane protein (Fig. 3A) and previous localization studies regarding Ptr3p (34).
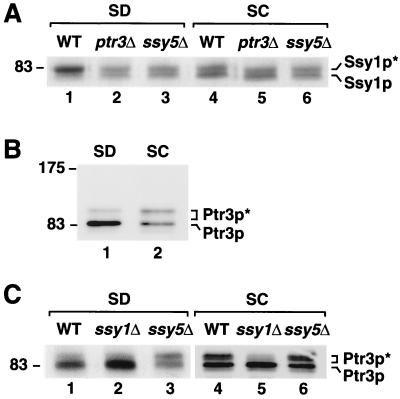
SSY1 and PTR3 are required for the proper expression of Ssy5p.
We compared the levels and electrophoretic properties of Ssy5p-c-myc in whole-cell extracts isolated from wild-type (HKY77), ssy1Δ13 (HKY84), and ptr3Δ15 (HKY85) null mutant strains (Fig. 4A). As previously indicated, Ssy5p-c-myc migrates as a 67-kDa protein (Ssy5p∗) in extracts isolated from wild-type cells (Fig. 4A, lane 1). We were unable to detect Ssy5p-c-myc in extracts prepared from ssy1 null mutant cells (Fig. 4A, lane 2). In extracts derived from ptr3 null mutants, Ssy5p-c-myc migrated as a 76-kDa protein, a mobility that corresponds to the predicted molecular mass of Ssy5p (Fig. 4A, lane 3). Although the data presented in Fig. 4 were obtained by using extracts isolated from strains grown in SD medium without amino acids, similar observations regarding the behavior of Ssy5p-c-myc in ssy1 and ptr3 null mutants were made when strains were grown in SC medium. Faint immunoreactive protein bands, corresponding to twice the molecular weight of the expressed Ssy5p, were observed in extracts prepared from wild-type and ptr3Δ cells (most easily seen in Fig. 4A, lane 3). Consistent with Ssy5p being a membrane protein, the intensity of the slower-migrating bands varied, dependent upon the denaturing conditions used, and increased when samples were subjected to higher denaturing temperatures.
We examined the possibility that the inability to detect Ssy5p-c-myc in extracts of ssy1Δ null mutants was due to the lack of transcription of SSY5 in these mutants. Northern blot analysis showed that the amounts of SSY5-c-myc mRNA were similar in both wild-type and ssy1Δ cells (Fig. 4B, compare lanes 1 and 3). This finding indicates that SSY5 transcription is independent of sensor function, a conclusion that is supported by the fact that SSY5 is transcribed normally in the absence of PTR3 (Fig. 4B, lane 4). Furthermore, wild-type cells grown in the presence of leucine, at concentrations known to affect the transcription of amino acid permease genes (e.g., AGP1 [see Fig. 2]), did not exhibit altered levels of SSY5 expression (Fig. 4B, lanes 1 and 2). Together the data presented in both panels of Fig. 4 indicate that Ssy5p is unstable in the absence of Ssy1p and that Ssy5p is likely to be posttranscriptionally modified in a Ptr3p-dependent manner.
Ssy1p and Ptr3p exhibit altered electrophoretic properties dependent upon amino acid availability and sensor component interactions.
Our finding that the functional expression of Ssy5p requires both SSY1 and PTR3 (Fig. 4) prompted us to examine the levels and electrophoretic properties of Ssy1p and Ptr3p. Whole-cell extracts containing functional epitope-tagged Ssy1p (Ssy1p-HA1) (34) were isolated from wild-type strains grown in media without (SD) and with (SC) added amino acids. In addition, extracts were prepared from various mutant strains lacking sensor function. In wild-type (HKY20) cells grown in SD (Fig. 5A, lane 1), Ssy1p migrates as a single major band (Ssy1p∗). In cells grown in SC, a second lower band (Ssy1p) becomes evident, and the intensity of the upper Ssy1p∗ band is clearly diminished (Fig. 5A, lane 4). The pattern of Ssy1p staining observed in ptr3 and ssy5 null mutant strains is similar to that observed in wild-type strains grown in SC (Fig. 5A, compare lanes 5 and 6 with lane 4). This pattern is also seen in mutant strains grown in the absence of amino acids (Fig. 5A, lanes 2 and 3).
FIG. 5.
SPS sensor component interactions. (A) The pattern of Ssy1p migration during SDS-PAGE is dependent upon Ptr3p and Ssy5p and is altered in response to amino acids. Whole-cell lysates were prepared from strains HKY20 (wild-type [WT]; lanes 1 and 4), HKY33 (ptr3Δ15; lanes 2 and 5), and HKY84 (ssy5Δ2; lanes 3 and 6) transformed with pHK010 (SSY1-HA1) grown in SD (lanes 1 to 3) and SC (lanes 4 to 6) to an OD600 of 0.8. The levels of Ssy1p-HA1 in extracts were analyzed by SDS-PAGE and immunoblotting. (B) The electrophoretic mobility of Ptr3p is altered in response to amino acids. Whole-cell lysates were prepared from wild-type strain HKY31 transformed with pHK018 (PTR3-HA1) grown in SD (lane 1) and SC (lane 2) to an OD600 of 0.8. The levels of Ptr3p-HA1 in extracts were analyzed by SDS-PAGE and immunoblotting. (C) The pattern of Ptr3p migration during SDS-PAGE is dependent upon Ssy1p and Ssy5p. Whole-cell lysates were prepared from strains HKY31 (wild type; lanes 1 and 4), HKY33 (ssy1Δ; lanes 2 and 5), and HKY85 (ssy5Δ; lanes 3 and 6) transformed with pHK018 (PTR3-HA1) grown in SD (lanes 1 to 3) and SC (lanes 4 to 6) to an OD600 of 0.8. The levels of Ptr3p-HA1 in extracts were analyzed by SDS-PAGE and immunoblotting. Note that lane 2 in panel B and lane 4 in panel C are derived from the same sample. A longer exposure time was used for a more detailed analysis of the protein bands in lanes 4 to 6.
The expression of functional epitope-tagged Ptr3p (Ptr3p-HA1) (34) was similarly examined. Multiple forms of Ptr3p were observed in extracts obtained from all strains, regardless of the amino acid content of the growth medium. In SD-grown wild-type cells (HKY31), the majority of Ptr3p is present in a faster-migrating band (Ptr3p); however, a faint and slower-migrating band (Ptr3p∗) is also evident (Fig. 5B, lane 1). The difference in apparent molecular mass between the slower- and faster-migrating forms of Ptr3p is approximately 15 to 20 kDa. In cells grown on SC, the relative intensity of the upper Ptr3p∗ band increased, and that of the lower band diminished (Fig. 5B, lane 2). When immunoblots were exposed for longer times (Fig. 5C, lane 4), the upper band was shown to be comprised of at least two bands. In SC-grown ssy1 null mutant cells, Ptr3p exhibited the same pattern of expression observed in both SD-grown wild-type cells and ssy1 null mutant cells (Fig. 5C, compare lane 5 with lanes 1 and 2). The slower-migrating bands (Ptr3p∗) were barely detectable in SSY1-deleted cells (Fig. 5C, lanes 2 and 5). In contrast, the electrophoretic properties of Ptr3p in SD-grown ssy5 null mutant cells appear similar to those observed in SC-grown wild-type cells (Fig. 5C, compare lane 3 with lane 4).
The data presented in Fig. 5 indicate that Ssy1p and Ptr3p are differentially modified and that multiple forms of Ssy1p and Ptr3p exist in cells. The electrophoretic properties of the slower-migrating forms of Ssy1p and Ptr3p, Ssy1p∗ and Ptr3p∗, respectively, are likely to result from posttranslational modifications. We examined the possibility that Ssy1p∗ and Ptr3p∗ were phosphorylated by incubating whole-cell extracts in the presence of various amounts of alkaline phosphatase (25 to 250 U per mg of protein). Under the conditions used, phosphatase treatment did not diminish the intensity of the Ssy1p∗ or Ptr3p∗ bands. We also examined the possibility that the large difference in the apparent molecular weight of Ptr3p and Ptr3p∗ was due to ubiquitination by overexpressing c-myc-tagged ubiquitin (22). We did not observe an upward shift of either Ptr3p or Ptr3p∗ in strains overexpressing the tagged ubiquitin construct. Although these results appear to rule out the possibility of phosphate and ubiquitin modification, they are negative in nature and thus need to be confirmed by further experimentation. Finally, we have observed that the total levels of Ssy1p and Ptr3p are consistently reduced in extracts from cells grown in the presence of amino acids. The quantitative analysis of the immunoreactive bands in Fig. 5 indicates that the levels of Ssy1p and Ptr3p are reduced by 15 and 50%, respectively, in SC-grown cells (Fig. 5A, lanes 1 and 4; B, lanes 1 and 2).
Overexpression of Ssy1p or the N terminus of Ssy1p disrupts amino acid sensor function.
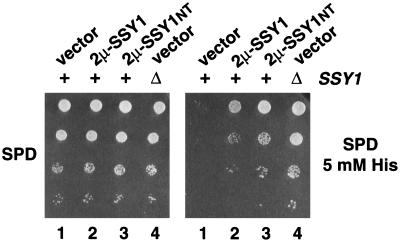
We examined the effects of individually overproducing SSY1, PTR3, and SSY5. The wild-type strain PLY1 was separately transformed with 2μm-based plasmids containing these genes as inserts. Ura+ transformants were selected on SC − Ura, cells derived from single colonies were resuspended in water, and dilution series were analyzed on minimal SPD medium supplemented with 0.16 mM histidine (SPD) or toxic levels of histidine (SPD plus 5 mM His). Transformants carrying plasmids PTR3 (pHK026) or SSY5 (pHK037) grew well on SPD containing 0.16 mM histidine, indicating that the overexpression of Ptr3p and Ssy5p did not have any deleterious effects on growth; however, these strains were unable to grow on SPD supplemented with toxic levels of histidine.
In contrast, strains carrying SSY1 (pHK012) grew well on SPD and on SPD containing 5 mM histidine (Fig. 6, dilution series 2). Similarly, transformants overexpressing only the first 206 N-terminal amino acids of Ssy1p (pHK039, 2μm-SSY1NT) grew well on both SPD and SPD (plus 5 mM His) (Fig. 6, dilution series 3). Transformants overexpressing the N-terminal domain grew nearly as well as the ssy1 null mutant strain (Fig. 6, dilution series 4). The ability of these transformants to grow in the presence of toxic levels of histidine indicates that the overexpression of Ssy1p or the extended hydrophilic N-terminal portion of Ssy1p disrupts sensor function. The fact that plasmids pHK012 and pHK039 enabled the growth of wild-type cells, which are otherwise unable to grow on SPD containing 5 mM histidine (Fig. 6, dilution series 1), demonstrates that overexpression of Ssy1p or the N-terminal region of Ssy1p exerts dominant-negative effects. The expression of only the N terminus of Ssy1p was not able to suppress ssy1 mutant alleles. These results are consistent with out previous findings that the N terminus of Ssy1p, the portion of Ssy1p absent from the other members of the amino acid permease gene family, has an important role in sensor function (34).
FIG. 6.
Overexpression of Ssy1p or the N terminus of Ssy1p exerts a dominant-negative effect on amino acid sensor function. Dilution series of strain PLY1 transformed with pRS202 (vector; lane 1), pHK012 (2μm-SSY1; lane 2), pHK039 (2μm-SSY1NT; lane 3), and strain HKY37 (ssy1Δ13) transformed with pRS202 (vector; lane 4) were spotted onto SPD and SPD supplemented with 5 mM histidine as indicated. Culture plates were incubated for 4 days at room temperature and photographed.
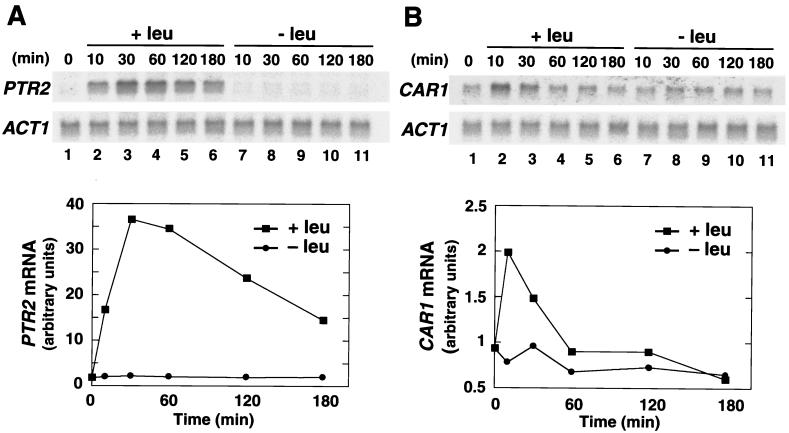
Time course of amino acid sensor-dependent transcriptional induction.
Previous work has demonstrated that leucine is a potent inducer of PTR2 (4) and CAR1 (21) transcription. We have shown that the leucine-induced transcription of these genes is dependent on all three sensor components, SSY1, PTR3, and SSY5 (Fig. 2) (34). The time course of leucine-induced transcription of PTR2 and CAR1 was examined. The level of PTR2 transcripts increased almost 40-fold within 30 min after cells received an aliquot of 1.3 mM leucine (Fig. 7A, lanes 2 to 6). After 30 min, the level of PTR2 transcripts gradually decreased, and after 180 min, the level of PTR2 transcripts was down to half of the maximum level. The time course of leucine-induced CAR1 transcription (Fig. 7B, lanes 2 to 6) exhibited a faster response; however, in general, the pattern of induction was similar. Ten minutes after the addition of leucine, the level of CAR1 transcripts increased twofold, and after 60 min, CAR1 transcripts were restored to basal levels. No increase in PTR2 or CAR1 transcription was observed in parallel uninduced cultures (Fig. 7, lanes 7 to 11). Thus, in the presence of inducing amino acids, cells transiently upregulate the expression of PTR2 and CAR1. Similar results regarding the transcription of the branched-chained amino acid transporter genes BAP2 and BAP3 have been reported (15).
FIG. 7.
Time course of l-leucine-induced transcription of PTR2 and CAR1. A liquid culture of strain HKY77 carrying plasmid pHK048 (SSY5-c-myc) was grown in SD to an OD600 of 0.5, and the culture was split into two parts (t = 0). One-half of the cell culture received an aliquot of l-leucine (+leu; lanes 2 to 6) to a final concentration of 1.3 mM, and the other received an equal volume of water (−leu; lanes 7 to 11). Both cultures were incubated at 30°C for an additional 180 min, and at the times indicated, subsamples were withdrawn and total RNA was isolated. The levels of PTR2 (A) and CAR1 (B) expression were analyzed by Northern blotting (upper panels), and after background correction, signal strengths (arbitrary units) relative to the levels of actin mRNA (ACT1) were quantitated (lower panels).
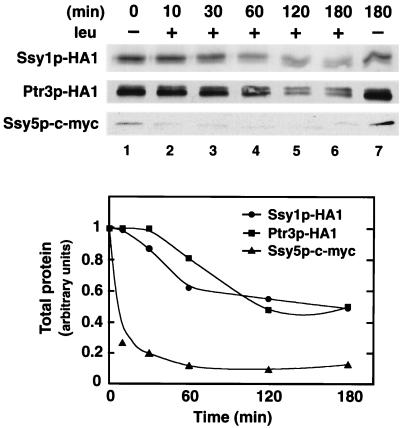
Time course of amino acid-induced sensor component modifications and sensor downregulation.
We examined whether the observed amino acid-induced changes in the electrophoretic mobility of Ssy1p and Ptr3p (Fig. 5) and Ssy5p occur in a similar time frame as sensor-dependent transcriptional induction (Fig. 7). Strains expressing SSY1-HA1, PTR3-HA1, and SSY5-c-myc were grown in liquid cultures of SD to an OD600 of 0.5. Each culture was divided into two culture flasks: one received an aliquot of leucine to a final concentration of 1.3 mM, and the other received an equal volume of water. At various times, subsamples were removed, whole-cell lysates were prepared, and the levels and electrophoretic characteristics of Ssy1p-HA1, Ptr3p-HA1, and Ssy5p-c-myc were analyzed by immunoblotting (Fig. 8).
FIG. 8.
Time course analysis of the physical alterations to Ssy1p, Ptr3p, and Ssy5p after induction of sensor function by l-leucine. Strains HKY20, HKY33, and HKY77 expressing SSY1-HA1, PTR3-HA1, and SSY5-c-myc, respectively, were grown in liquid cultures of SD to an OD600 of 0.5 (t = 0; lane 1). At t = 0, each culture received an aliquot of l-leucine to a final concentration of 1.3 mM (lanes 2 to 6). At the times indicated, subsamples were removed, whole-cell lysates were prepared, and proteins were analyzed by immunoblotting (upper panel). Lane 7 shows protein levels present in an uninduced control culture similarly incubated for 180 min. The corresponding chemiluminescent signals were quantitated (lower panel).
At time zero (Fig. 8, lane 1) and in a control culture that did not receive an aliquot of leucine (Fig. 8, lane 7), Ssy1p is predominantly expressed in its slower-migrating form (Ssy1p∗), Ptr3p is principally present in its faster-migrating from, and Ssy5p is readily detected in its Ptr3p-dependent processed form (Ssy5p∗). After the addition of leucine, the levels of immunologically detectable Ssy5p rapidly decreased; after 5 min, 25% of Ssy5p remained; and after 60 min, Ssy5p levels were 10-fold lower than in the starting culture. The levels of Ssy1p and Ptr3p also decreased—however, at a markedly slower rate and to a lesser extent than Ssy5p. After 120 min, the total levels of Ssy1p and Ptr3p were twofold lower than in the starting culture. It should be noted that the quantitations graphically presented in Fig. 8B were calculated based on the combined signal strength of both Ssy1p and Ssyp1∗ and Ptr3p and Ptr3p∗, respectively. Two hours after leucine was added, the sensor components exhibited similar characteristics to those isolated from SC-grown cells (Fig. 5). The level of each component was reduced, Ssy1p∗ was not the predominant species of Ssy1p, and the levels of Ptr3p∗ were similar to those of Ptr3p. Thus the addition of leucine induced rapid and long-term changes that apparently result in the down regulation of all three sensor components.
DISCUSSION
We have found that SSY5 encodes a third component of the yeast plasma membrane sensor of extracellular amino acids. Ssy5p functions together with the two previously characterized sensor components, Ssy1p1 and Ptr3p, to regulate diverse metabolic processes important for proper amino acid uptake and compartmentalization. The conclusion that Ssy5p is a component of the plasma membrane amino acid sensor is based on several observations. First, mutations in SSY5 belong to the same epistasis group as ssy1 and ptr3 mutations. Ssy5 mutants exhibit similar increases in vacuolar pools of histidine and arginine (Table 3). ssy5 null mutants display identical levels of resistance to toxic amino acids and azetidine carboxylate, and the ssy5Δ ssy1Δ, ssy5Δ ptr3Δ, and ssy1Δ ptr3Δ double mutant strains exhibit levels of resistance identical to those of each of the single mutant strains. Additionally, SSY5 is required for amino acid-induced transcription of two genes, AGP1 and PTR2, known to be controlled by SSY1 and PTR3 (Fig. 2). The resistance to toxic amino acids and amino acid analogues is likely to be a consequence of the altered uptake and increased capacity to compartmentalize amino acids (Table 3) (34). Second, functional epitope-tagged Ssy5p-c-myc fractionates as a peripherally bound membrane protein, and h-SOS-Ssy5p fusion proteins are recruited to the cytosolic face of the plasma membrane in an Ssy5p-dependent manner (Fig. 3). The observation that Ssy5p, which is predicted to be a soluble protein, is able to associate with the plasma membrane, directly implicates this protein as a constituent of this sensing system. Third, the proper expression of Ssy5p, but not the transcription of SSY5, requires both SSY1 and PTR3 (Fig. 4). Finally, the electrophoretic properties of both Ssy1p and Ptr3p are altered in ssy5 null mutant strains (Fig. 5A and C).
The demonstrated ability of Ssy1p, Ptr3p, and Ssy5p to localize to the plasma membrane is consistent with the possibility that these components associate within a sensor complex, although there is as yet no direct evidence for a physical association between them. Of the three identified sensor components, Ssy1p is the only integral membrane-spanning component; thus, Ssy1p is likely to be the component that transmits signals across the PM. The in vivo membrane topology of the general amino acid permease (Gap1p) has recently been determined; both the N- and C-terminal domains are oriented towards the cytoplasm (24). Based upon the sequence and structural homology that exists between Ssy1p and the other members of the amino acid permease gene family members, it is likely that the N terminus of Ssy1p is also cytoplasmically oriented. The large N-terminal extension of Ssy1p may serve to organize the assembly of the other peripheral membrane components. Ssy5p is a strong candidate for directly interacting with Ssy1p. In the absence of Ssy1p, Ssy5p is unstable, as evidenced by our inability to detect Ssy5p in protein lysates derived from ssy1 null mutants (Fig. 4A). The fact that Ssy5p is degraded in the absence of Ssy1p prevented us from directly assessing whether its localization to the PM is dependent upon Ssy1p. We have previously found that the Ptr3p localizes to the plasma membrane independently of Ssy1p (34).
We further investigated the functional relationships between the SPS sensor components and made several observations that indicate that these three components do indeed intimately interact with one another. We have found that each of the components displays physical properties that are dependent upon the availability of amino acids and on the presence of the other two components. In wild-type cells grown in SD, Ssy1p∗ is the predominating form of Ssy1p, whereas in ptr3Δ or ssy5Δ null mutants it is not (Fig. 5A). Thus, in mutant cells lacking either PTR3 and SSY5, Ssy1p exhibits characteristics that mimic those of Ssy1p isolated from wild-type cells grown in the presence of amino acids. Additionally, we have consistently observed that cells lacking either Ptr3p and Ssy5p express less Ssy1p (Fig. 5A). Ptr3p migrates as several bands that exhibit significant differences in mobility on SDS gels (Fig. 5B). The presence of the slower-migrating Ptr3p∗ species in amino acid-containing medium is dependent on SSY1, but not SSY5 (Fig. 5C); however, in SD-grown cells lacking SSY5, Ptr3p exhibits characteristics identical to those of SC-grown wild-type cells (Fig. 5C). In wild-type cells, Ssy5p is apparently proteolytically modified in a PTR3-dependent manner (Fig. 4A). Because the c-myc epitope used to visualize Ssy5p is located within the N terminus, the Ptr3p-dependent processing event is likely to occur within the C-terminal portion of Ssy5p. The inability to detect Ssy5p in whole-cell extracts prepared from strains carrying ssy1 mutations (Fig. 4A) is presumably the consequence of a proteolytic cleavage event that minimally removes its N terminus.
The observation that overexpression of the first 206 amino acids comprising the N-terminal extension of Ssy1p disrupts sensor function in a dominant-negative manner (Fig. 6) is also consistent with the existence of a multicomponent sensor complex. This finding clearly shows that the N-terminal domain of Ssy1p has an important role in sensor function, a conclusion supported by our previous observation that small in-frame mutations within the N-terminal domain abolish signaling (34). These results suggest that functional SPS sensor complexes assemble with a precise stoichiometry. Consequently, the overproduction of the N-terminus Ssy1p would interfere with SPS sensor function by forming nonproductive complexes with proteins normally interacting with Ssy1p. These nonproductive interactions would effectively decrease the availability of the limiting components to form functional sensor complexes. Similarly, dominant-negative phenotypes associated with the mutations in the cytoplasmically oriented C terminus of the G-protein-coupled alpha-factor receptor (STE2) have been observed to exert their effects by sequestering G-proteins (20, 39). The overexpression of full-length Ssy1p also exhibited dominant-negative effects; this unexpected observation may be the result of a fraction of Ssy1p being mislocalized. Accordingly, limiting sensor components would be sequestered at inappropriate intracellular membranes.
When leucine is added to wild-type cells grown in medium without supplementary amino acids, the transcription of PTR2 and CAR1 is transiently induced (Fig. 7). The response is quite rapid; within 10 min, there is a 15-fold induction of PTR2 and a 2-fold induction of CAR1. After reaching maximum levels (PTR2, 30 min; CAR1, 10 min), their transcript levels slowly adjust back to basal levels. Similar patterns of induction have been reported for the branched-chain amino acid permeases (BAP2 and BAP3) (15). Concurrent with its effect on transcription, leucine stimulates the components of the SPS sensor to become physically modified and causes the levels of each of the SPS sensor components to diminish (Fig. 8). The rapid physical alterations and reduced levels of sensor components are consistent with their being downregulated in response to amino acid availability. It is important to note that in cells grown in medium supplemented with amino acids, the downregulated sensor components are necessary to maintain the steady-state transcript levels of AGP1 and PTR2 (Fig. 2) and the glutamine permease (GNP1) (34). Additionally, the downregulated SPS sensor is required to fully repress the functional expression of Gap1p (34).
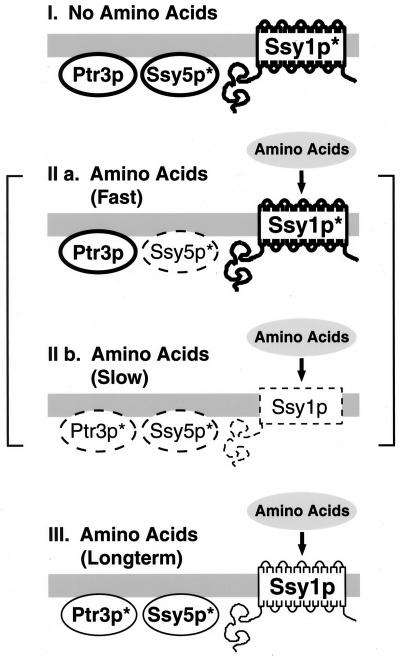
Regarding the leucine-induced mobility changes and downregulation of SPS sensor components (Fig. 8), we have defined three sensor states, each associated with defined patterns of component expression (see Fig. 9 for a schematic presentation). In the absence of extracellular amino acids, the state I, or preactivation, conformation, Ssy1p migrates predominantly in its slower-migrating form (Ssy1p∗), Ptr3p is primarily present in its faster-migrating form, and Ssy5p is readily detected in its Ptr3p-dependent low-molecular-mass processed form (Ssy5p∗). Within minutes after the addition of leucine, rapid changes are observed, resulting in the transient state IIa sensor complex (Fig. 9). The characteristics of each component within the state IIa sensor correspond to those observed after 10 min of leucine addition (Fig. 8). The state IIa sensor is defined by the low levels of Ssy5p; the electrophoretic characteristics of the other two components appear unchanged. With increasing time (30- and 60-min time points) (Fig. 8), shifts in the migration of Ssy1p and Ptr3p become increasingly obvious (Fig. 9, state IIb conformation). Within the transitional state IIb complex, the levels of Ssy1p∗ decrease and the relative proportion of the slower-migrating form of Ptr3p (Ptr3p∗) increases. The state III configuration, evident 120 min after leucine addition, is indistinguishable from the downregulated sensor conformation that exists in cells grown in SC media. It is possible that the downregulated sensor conformation is actually comprised of a mixed sensor population; the patterns of bands associated with the state I conformation are still evident in the state III sensor.
FIG. 9.
Schematic diagram summarizing the dynamic characteristics of the SPS sensor component interactions. The state I sensor is the complex present in cells grown in the absence of amino acids. The SPS components are present in high levels, represented by the heavy outlines. In analogy to the G-protein-coupled α-factor receptor complex in MATa cells (39), the state I conformation may represent a preactivation complex. States IIa and IIb are transient complexes that rapidly form when cells grown in the absence of amino acids are induced by amino acids. The components in the transient state IIa and IIb sensor undergoing dynamic changes in expression levels are represented by the dashed outlines. The state III conformation is the downregulated complex, and the diminished levels of the components are represented by light outlines. For additional details, see text.
In our analysis to date, we have defined three components of a plasma membrane sensor of extracellular amino acids. Further biochemical and genetic analysis is necessary to ascertain if these components represent the entire primary sensing complex or if other components exist. The isolation of dominant constitutively activated alleles in any of the genes encoding SPS sensor components would enable epistasis relationships to be better defined. Because the SPS sensor components are localized to the plasma membrane, it is possible that the SPS sensor components, particularly Ssy1p, are subject to regulation by posttranslational mechanisms known to regulate amino acid permeases and other metabolite transporters. In response to ammonium, Gap1p is dephosphorylated and polyubiquitinated (27, 60). Similar mechanisms, as well as substrate-induced feedback inhibition, operate to regulate the uracil permease (23, 58). Finally, other proteins associated with the plasma membrane regulate amino acid uptake. For example, the target of rapamycin pathway, in a manner resembling the SPS sensor, inversely regulates the activity of general and specific amino acid permeases (6, 7, 55). ScRheb (YCR027c) is a member of a new class of farnesylated small G-proteins of the Ras superfamily, that negatively regulates the uptake of arginine by Can1p (61). This regulation is thought to occur at the plasma membrane directly with or through other proteins interacting with the Can1p permease. The fact that yeast plasma membrane nutrient sensors have only recently been discovered reveals how little is understood regarding the molecular signals that enable yeast to adapt to constantly changing environments. Many more novel discoveries can be expected.
ACKNOWLEDGMENTS
We thank Annalena Moliner for technical assistance during the cloning of SHR4 and Carolyn Slayman for the generous gift of Pma1p antibodies. We thank members of the Ljungdahl laboratory and members of Morten Kielland-Brandt's group at the Carlsberg Laboratory (Copenhagen) for fruitful discussions.
This work was supported by the Ludwig Institute for Cancer Research. The cooperative research agreement between LICR-Stockholm Branch and Fuji Photo Film (Europe) is gratefully acknowledged.
REFERENCES
- 1.Allen J B, Elledge S J. A family of vectors that facilitate transposon and insertional mutagenesis of cloned genes in yeast. Yeast. 1994;10:1267–1272. doi: 10.1002/yea.320101003. [DOI] [PubMed] [Google Scholar]
- 2.Aronheim A, Engelberg D, Li N, al-Alawi N, Schlessinger J, Karin M. Membrane targeting of the nucleotide exchange factor Sos is sufficient for activating the Ras signaling pathway. Cell. 1994;78:949–961. doi: 10.1016/0092-8674(94)90271-2. [DOI] [PubMed] [Google Scholar]
- 3.Aronheim A, Zandi E, Hennemann H, Elledge S J, Karin M. Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol Cell Biol. 1997;17:3094–3102. doi: 10.1128/mcb.17.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes D, Lai W, Breslav M, Naider F, Becker J M. PTR3, a novel gene mediating amino acid-inducible regulation of peptide transport in Saccharomyces cerevisiae. Mol Microbiol. 1998;29:297–310. doi: 10.1046/j.1365-2958.1998.00931.x. [DOI] [PubMed] [Google Scholar]
- 5.Barral Y, Mann C. G1 cyclin degradation and cell differentiation in Saccharomyces cerevisiae. C R Acad Sci Sec III Life Sci. 1995;318:43–50. [PubMed] [Google Scholar]
- 6.Beck T, Hall M N. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 7.Beck T, Schmidt A, Hall M N. Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J Cell Biol. 1999;146:1227–1238. doi: 10.1083/jcb.146.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bricmont P A, Daugherty J R, Cooper T G. The DAL81 gene product is required for induced expression of two differently regulated nitrogen catabolic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:1161–1166. doi: 10.1128/mcb.11.2.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brisco P R, Cunningham T S, Kohlhaw G B. Cloning, disruption and chromosomal mapping of yeast LEU3, a putative regulatory gene. Genetics. 1987;115:91–99. doi: 10.1093/genetics/115.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Celenza J L, Marshall C L, Carlson M. The yeast SNF3 gene encodes a glucose transporter homologous to the mammalian protein. Proc Natl Acad Sci USA. 1988;85:2130–2134. doi: 10.1073/pnas.85.7.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang A, Slayman C W. Maturation of the yeast plasma membrane [H+]ATPase involves phosphorylation during intracellular transport. J Cell Biol. 1991;115:289–295. doi: 10.1083/jcb.115.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen S, Knoll B J, Little J W, Mount D W. Preferential cleavage of phage lambda repressor monomers by recA protease. Nature. 1981;294:182–184. doi: 10.1038/294182a0. [DOI] [PubMed] [Google Scholar]
- 13.Connelly C, Hieter P. Budding yeast SKP1 encodes an evolutionarily conserved kinetichore protein required for cell cycle progression. Cell. 1996;86:275–285. doi: 10.1016/S0092-8674(00)80099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coornaert D, Vissers S, André B. The pleiotropic UGA35(DURL) regulatory gene of Saccharomyces cerevisiae: cloning, sequence and identity with the DAL81 gene. Gene. 1991;97:163–171. doi: 10.1016/0378-1119(91)90048-g. [DOI] [PubMed] [Google Scholar]
- 15.de Boer M, Bebelman J P, Goncalves P M, Maat J, Van Heerikhuizen H, Planta R J. Regulation of expression of the amino acid transporter gene BAP3 in Saccharomyces cerevisiae. Mol Microbiol. 1998;30:603–613. doi: 10.1046/j.1365-2958.1998.01094.x. [DOI] [PubMed] [Google Scholar]
- 16.de Boer M, Nielsen P S, Bebelman J P, Heerikhuizen H, Andersen H A, Planta R J. Stp1p, Stp2p and Abf1p are involved in regulation of expression of the amino acid transporter gene BAP3 of Saccharomyces cerevisiae. Nucleic Acids Res. 2000;28:974–981. doi: 10.1093/nar/28.4.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Della Seta F, Treich I, Buhler J M, Sentenac A. ABF1 binding sites in yeast RNA polymerase genes. J Biol Chem. 1990;265:15168–15175. [PubMed] [Google Scholar]
- 18.Didion T, Grauslund M, Kielland-Brandt M C, Andersen H A. Amino acids induce expression of BAP2, a branched-chain amino acid permease in Saccharomyces cerevisiae. J Bacteriol. 1996;178:2025–2029. doi: 10.1128/jb.178.7.2025-2029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Didion T, Regenberg B, Jørgensen M U, Kielland-Brandt M C, Andersen H A. The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol Microbiol. 1998;27:643–650. doi: 10.1046/j.1365-2958.1998.00714.x. [DOI] [PubMed] [Google Scholar]
- 20.Dosil M, Giot L, Davis C, Konopka J B. Dominant-negative mutations in the G-protein-coupled α-factor receptor map to the extracellular ends of the transmembrane segments. Mol Cell Biol. 1998;18:5981–5991. doi: 10.1128/mcb.18.10.5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubois E L, Wiame J-M. Non specific induction of arginase in Saccharomyces cerevisiae. Biochimie. 1976;58:207–211. doi: 10.1016/s0300-9084(76)80371-9. [DOI] [PubMed] [Google Scholar]
- 22.Ellison M J, Hochstrasser M. Epitope-tagged ubiquitin. A new probe for analyzing ubiquitin function. J Biol Chem. 1991;266:21150–21157. [PubMed] [Google Scholar]
- 23.Galan J M, Moreau V, André B, Volland C, Haguenauer-Tsapis R. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J Biol Chem. 1996;271:10946–10952. doi: 10.1074/jbc.271.18.10946. [DOI] [PubMed] [Google Scholar]
- 24.Gilstring C F, Ljungdahl P O. A method for determining the in vivo topology of yeast polytopic membrane proteins demonstrates that Gap1p fully integrates into the membrane independently of Shr3p. J Biol Chem. 2000;275:31488–31495. doi: 10.1074/jbc.M005047200. [DOI] [PubMed] [Google Scholar]
- 25.Gilstring C F, Melin-Larsson M, Ljungdahl P O. Shr3p mediates specific COPII coatomer-cargo interactions required for the packaging of amino acid permeases into ER-derived transport vesicles. Mol Biol Cell. 1999;10:3549–3565. doi: 10.1091/mbc.10.11.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guthrie C, Fink G R, editors. Guide to yeast genetics and molecular biology. Vol. 194. San Diego, Calif: Academic Press, Inc.; 1991. [Google Scholar]
- 27.Hein C, Springael J Y, Volland C, Haguenauer-Tsapis R, André B. NP11, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol Microbiol. 1995;18:77–87. doi: 10.1111/j.1365-2958.1995.mmi_18010077.x. [DOI] [PubMed] [Google Scholar]
- 28.Hinnebusch A G. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev. 1988;52:248–273. doi: 10.1128/mr.52.2.248-273.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iglesias R, Ferreras J M, Arias F J, Munoz R, Rojo M A, Girbes T. Effect of L-azetidine 2-carboxylic acid on the activity of the general amino-acid permease from Saccharomyces cerevisiae var. ellipsoideus. Arch Microbiol. 1991;155:320–324. doi: 10.1007/BF00243449. [DOI] [PubMed] [Google Scholar]
- 30.Iraqui I, Vissers S, Bernard F, de Craene J-O, Boles E, Urrestarazu A, André B. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol Cell Biol. 1999;19:989–1001. doi: 10.1128/mcb.19.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnston M. Feasting, fasting and fermenting. Trends Genet. 1999;15:29–33. doi: 10.1016/s0168-9525(98)01637-0. [DOI] [PubMed] [Google Scholar]
- 33.Jorgensen M U, Bruun M B, Didion T, Kielland-Brandt M C. Mutations in five loci affecting GAP1-independent uptake of neutral amino acids in yeast. Yeast. 1998;14:103–114. doi: 10.1002/(SICI)1097-0061(19980130)14:2<103::AID-YEA203>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 34.Klasson H, Fink G R, Ljungdahl P O. Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol Cell Biol. 1999;19:5405–5416. doi: 10.1128/mcb.19.8.5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraakman L, Lemaire K, Ma P, Teunissen A W, Donaton M C, Van Dijck P, Winderickx J, de Winde J H, Thevelein J M. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol Microbiol. 1999;32:1002–1012. doi: 10.1046/j.1365-2958.1999.01413.x. [DOI] [PubMed] [Google Scholar]
- 36.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 37.Lafuente M J, Gancedo C, Jauniaux J C, Gancedo J M. Mth1 receives the signal given by the glucose sensors Snf3 and Rgt2 in Saccharomyces cerevisiae. Mol Microbiol. 2000;35:161–172. doi: 10.1046/j.1365-2958.2000.01688.x. [DOI] [PubMed] [Google Scholar]
- 38.Lasko P F, Brandriss M C. Proline transport in Saccharomyces cerevisiae. J Bacteriol. 1981;148:241–247. doi: 10.1128/jb.148.1.241-247.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leavitt L M, Macaluso C R, Kim K S, Martin N P, Dumont M E. Dominant negative mutations in the alpha-factor receptor, a G protein-coupled receptor encoded by the STE2 gene of the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1999;261:917–932. doi: 10.1007/s004380051039. [DOI] [PubMed] [Google Scholar]
- 40.Li F N, Johnston M. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: coupling glucose sensing to gene expression and the cell cycle. EMBO J. 1997;16:5629–5638. doi: 10.1093/emboj/16.18.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang H, Gaber R F. A novel signal transduction pathway in Saccharomyces cerevisiae defined by Snf3-regulated expression of HXT6. Mol Biol Cell. 1996;7:1953–1966. doi: 10.1091/mbc.7.12.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ljungdahl P O, Gimeno C J, Styles C A, Fink G R. SHR3: a novel component of the secretory pathway specifically required for the localization of amino acid permeases in yeast. Cell. 1992;71:463–478. doi: 10.1016/0092-8674(92)90515-e. [DOI] [PubMed] [Google Scholar]
- 43.Lorenz M C, Heitman J. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 1998;17:1236–1247. doi: 10.1093/emboj/17.5.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lorenz M C, Pan X, Harashima T, Cardenas M E, Xue Y, Hirsch J P, Heitman J. The G protein-coupled receptor Gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics. 2000;154:609–622. doi: 10.1093/genetics/154.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marini A-M, Soussi-Boudekou S, Vissers S, Andre B. A family of ammonium transporters in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:4282–4293. doi: 10.1128/mcb.17.8.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Markwell M K, Haas S M, Bieber L L, Tolbert N E. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 47.Ohsumi Y, Kitamoto K, Anraku Y. Changes induced in the permeability barrier of the yeast plasma membrane by cupric ion. J Bacteriol. 1988;170:2676–2682. doi: 10.1128/jb.170.6.2676-2682.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Özcan S, Dover J, Johnston M. Glucose sensing and signaling by two glucose receptors in the yeast. EMBO J. 1998;17:2566–2573. doi: 10.1093/emboj/17.9.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Özcan S, Dover J, Rosenwald A G, Wölfl S, Johnston M. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc Natl Acad Sci USA. 1996;93:12428–12432. doi: 10.1073/pnas.93.22.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Özcan S, Johnston M. Function and regulation of yeast hexose transporters. Microbiol Mol Biol Rev. 1999;63:554–569. doi: 10.1128/mmbr.63.3.554-569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patton E E, Willems A R, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet. 1998;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- 52.Perry J R, Basral M A, Steiner H-Y, Naider F, Becker J M. Isolation and characterization of a Saccharomyces cerevisiae peptide transport gene. Mol Cell Biol. 1994;14:104–115. doi: 10.1128/mcb.14.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petitjean A, Hilger F, Tatchell K. Comparison of thermosensitive alleles of the CDC25 gene involved in the cAMP metabolism of Saccharomyces cerevisiae. Genetics. 1990;124:797–806. doi: 10.1093/genetics/124.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rivier D H, Ekena J L, Rine J. HMR-I is an origin of replication and a silencer in Saccharomyces cerevisiae. Genetics. 1999;151:521–529. doi: 10.1093/genetics/151.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmidt A, Beck T, Koller A, Kunz J, Hall M N. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmidt M C, McCartney R R, Zhang X, Tillman T S, Solimeo H, Wölfl S, Almonte C, Watkins S C. Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4561–4571. doi: 10.1128/mcb.19.7.4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schroeder S C, Weil P A. Genetic tests of the role of Abf1p in driving transcription of the yeast TATA box binding protein-encoding gene, SPT15. J Biol Chem. 1998;273:19884–19891. doi: 10.1074/jbc.273.31.19884. [DOI] [PubMed] [Google Scholar]
- 58.Séron K, Blondel M-O, Haguenauer-Tsapis R, Volland C. Uracil-induced down-regulation of the yeast uracil permease. J Bacteriol. 1999;181:1793–1800. doi: 10.1128/jb.181.6.1793-1800.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Springael J-Y, André B. Nitrogen-regulated ubiquitination of the Gap1 permease of Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1253–1263. doi: 10.1091/mbc.9.6.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Urano J, Tabancay A P, Yang W, Tamanoi F. The Saccharomyces cerevisiae Rheb G-protein is involved in regulating canavanine resistance and arginine uptake. J Biol Chem. 2000;275:11198–11206. doi: 10.1074/jbc.275.15.11198. [DOI] [PubMed] [Google Scholar]
- 62.Vagnoli P, Coons̀ D M, Bisson L F. The C-terminal domain of Snf3p mediates glucose-responsive signal transduction in Saccharomyces cerevisiae. FEMS Microbiol Lett. 1998;160:31–36. doi: 10.1111/j.1574-6968.1998.tb12886.x. [DOI] [PubMed] [Google Scholar]
- 63.Vieira J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 64.Vissers S, André B, Muyldermans F, Grenson M. Induction of the 4-aminobutyrate and urea-catabolic pathways in Saccharomyces cerevisiae. Specific and common transcriptional regulators. Eur J Biochem. 1990;187:611–616. doi: 10.1111/j.1432-1033.1990.tb15344.x. [DOI] [PubMed] [Google Scholar]
- 65.Wang D, Zheng F, Holmberg S, Kohlhaw G B. Yeast transcriptional regulator Leu3p. Self-masking, specificity of masking, and evidence for regulation by the intracellular level of Leu3p. J Biol Chem. 1999;274:19017–19024. doi: 10.1074/jbc.274.27.19017. [DOI] [PubMed] [Google Scholar]
- 66.Wang S S, Hopper A K. Isolation of a yeast gene involved in species-specific pre-tRNA processing. Mol Cell Biol. 1988;8:5140–5149. doi: 10.1128/mcb.8.12.5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xue Y, Batlle M, Hirsch J P. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Galpha subunit and functions in a Ras-independent pathway. EMBO J. 1998;17:1996–2007. doi: 10.1093/emboj/17.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]