Abstract
Formin homology (FH) proteins are implicated in cell polarization and cytokinesis through actin organization. There are two FH proteins in the yeast Saccharomyces cerevisiae, Bni1p and Bnr1p. Bni1p physically interacts with Rho family small G proteins (Rho1p and Cdc42p), actin, two actin-binding proteins (profilin and Bud6p), and a polarity protein (Spa2p). Here we analyzed the in vivo localization of Bni1p by using a time-lapse imaging system and investigated the regulatory mechanisms of Bni1p localization and function in relation to these interacting proteins. Bni1p fused with green fluorescent protein localized to the sites of cell growth throughout the cell cycle. In a small-budded cell, Bni1p moved along the bud cortex. This dynamic localization of Bni1p coincided with the apparent site of bud growth. A bni1-disrupted cell showed a defect in directed growth to the pre-bud site and to the bud tip (apical growth), causing its abnormally spherical cell shape and thick bud neck. Bni1p localization at the bud tips was absolutely dependent on Cdc42p, largely dependent on Spa2p and actin filaments, and partly dependent on Bud6p, but scarcely dependent on polarized cortical actin patches or Rho1p. These results indicate that Bni1p regulates polarized growth within the bud through its unique and dynamic pattern of localization, dependent on multiple factors, including Cdc42p, Spa2p, Bud6p, and the actin cytoskeleton.
In both unicellular and multicellular organisms, cell polarity is crucial for various cellular functions as diverse as differentiation, morphogenesis, motility, and signal transduction (11, 14). The budding yeast Saccharomyces cerevisiae undergoes polarized cell growth during budding in vegetative growth and during projection formation in mating response (36, 45). Polarized growth in a vegetative cell begins in late G1. The axis of polarization (i.e., the next bud site) is selected in either of two patterns, depending on the mating-type locus. A Mata or Matα haploid cell constructs the next bud site adjacent to the previous bud site (axial budding pattern), whereas a Mata/Matα diploid cell buds from sites that are either near the previous bud site or at the opposite end of the cell (bipolar budding pattern) (10). Cell growth occurs initially at the tip of the bud (apical growth) and then continues isotropically as the bud enlarges (37). Finally, just before cytokinesis, deposition of new cell wall and membrane occurs at the mother-bud neck.
Polarized cell growth in the yeast is a complex operation that involves (i) bud site selection and establishment, (ii) polarized organization of cytoskeletons, (iii) vectorial transport of secretory vesicles and organelles, (iv) local membrane growth, and (v) regulation of signal transduction cascades to communicate with other parts of the cell. In these processes, small GTP-binding proteins (small G proteins) and their interacting molecules play key signaling roles (7). A Ras family small G protein, Bud1p, is required for the bud site selection (9). A Rho family small G protein, Cdc42p, is essential for the organization of the bud site (1, 28). In the absence of Cdc42p, the actin cytoskeleton dose not become polarized and budding does not take place. Another Rho family small G protein, Rho1p, is required for activity of β(1 → 3) glucan synthase, the enzyme that catalyzes the synthesis of the major structural component of the yeast cell wall (13, 56). Rho3p and Rho4p are implicated in polarized growth, presumably regulating polarized secretion and reorganization of the actin cytoskeleton (46, 57). A Rab family small G protein, Sec4p, is essential for the late stage of a secretory pathway at the growth site (22, 52). The actin cytoskeleton also plays a prominent role in polarized cell growth. It appears as distinct structures during polarized cell growth (2, 31). Cortical actin patches are concentrated at the site of polarized growth, and actin cables run parallel to the mother-bud axis during budding. The actin cytoskeleton is thought to direct secretory vesicles containing growth components (e.g., new cell wall and membrane) to the growth site (5, 49, 51).
A newly discovered player in cell polarity is the formin homology (FH) protein family, which is conserved from yeast to mammal (63). This family includes Schizosaccharomyces pombe proteins fus1 and cdc12, Drosophila melanogaster proteins DIAPHANOUS and CAPPUCCINO, Aspergillus nidulans protein figA/speA, and vertebrate proteins Formin and mDia/hDia. In S. cerevisiae, there are two FH proteins, Bni1p and Bnr1p. BNI1 was first identified as a gene required for bipolar bud site selection (66). We have identified Bni1p as a protein interacting with the GTP-bound form of Rho1p (32). Bni1p has subsequently been shown to also interact with Cdc42p (16). Another FH protein, Bnr1p, interacts with the GTP-bound form of Rho4p (25). Bni1p and Bnr1p, at their proline-rich FH1 domains, bind an actin monomer-binding protein, profilin (16, 25), which is implicated in actin dynamics (24). At their C termini, Bni1p and Bnr1p bind Bud6p/Aip3p (16, 30), which has been identified as an actin-binding protein (3). Bni1p also binds Spa2p (19), a protein involved in proper morphogenesis of yeast cells (61). We have also shown that Bnr1p interacts with Hof1p (29), a protein involved in cytokinesis (41), and Smy1p (30), a kinesin-related protein which has been isolated as a dosage suppressor of the temperature sensitivity of myo2-66 (39). Bni1p and Bnr1p are implicated in reorganization of not only the actin cytoskeleton but also other cytoskeletal systems. Bni1p participates in microtubule function, since disruption of BNI1 causes defects in spindle orientation (34); localization of Kar9p (47), which has been reported to link the bud cortex to the plus ends of microtubules (33, 35); and a growth defect together with mutation either in DYN1, ASE1 (34), PAC1, or NIP100 (18), whose gene products are implicated in microtubule function (20). On the other hand, Bnr1p is functionally related to the septin system (30). These findings suggest that Bni1p and Bnr1p act as scaffold proteins juxtaposing Rho family G proteins and other important regulators with various cytoskeletal systems to achieve integrated cell polarity. However, the regulatory mechanisms of in vivo localization or function of Bni1p and Bnr1p have not thoroughly been explored.
In this study, we examined the in vivo behavior of Bni1p and investigated the regulatory mechanisms of its localization and function, especially in relation to Rho family small G proteins, Spa2p, Bud6p, and the actin cytoskeleton.
MATERIALS AND METHODS
Strains, media, and culture conditions.
Yeast strains used in this study are listed in Table 1. The Escherichia coli strain DH5α was used for construction and propagation of plasmids. Culture media were prepared as previously described (18). Yeast transformations were performed by the lithium acetate method (21). Transformants were selected on appropriate synthetic drop-out media. Standard yeast genetic manipulations were performed as previously described (23). Yeast cells were grown at 24°C unless otherwise specified.
TABLE 1.
Yeast strains used in this study
| Strainsa | Genotype (reference) |
|---|---|
| OHNY1 | MATaura3 leu2 trp1 his3 ade2 (50) |
| OHNY3 | MATa/MATα ura3/ura3 leu2/leu2 trp1/trp1 his3/his3 ade2/ade2 (53) |
| BTY3 | MATaura3 leu2 trp1 his3 ade2 bni1-Δ::HIS3 (19) |
| KY4 | MATa/MATα ura3/ura3 leu2/leu2 trp1/trp1 his3/his3 ade2/ade2 bni1-Δ::HIS3/bni1-Δ::HIS3 |
| KIBY1 | MATaura3 leu2 trp1 his3 ade2 bnr1-Δ::his5+ |
| DKBY80B | MATa/MATα ura3/ura3 leu2/leu2 trp1/trp1 his3/his3 ade2/ade2 bni1-Δ::URA3/BNI1 bnr1-Δ::his5+/BNR1 |
| KKC42-1 | MATaura3 leu2 trp1 his3 ade2 cdc42-1 |
| HNY21 | MATaura3 leu2 trp1 his3 ade2 rho1-104 (65) |
| TYSH1 | MATaura3 leu2 trp1 his3 ade2 spa2::HIS3 (19) |
| TFB6H1 | MATaura3 leu2 trp1 his3 ade2 bud6::HIS3 |
| TFB6H3 | MATa/MATα ura3/ura3 leu2/leu2 trp1/trp1 his3/his3 ade2/ade2 bud6::HIS3/bud6::HIS3 |
The strains used in this study are isogenic.
Yeast strain construction.
To construct a bnr1-Δ mutant strain, all but the first 4 amino acids and the last 20 amino acids of BNR1 were replaced with the S. pombe his5+ gene by using the one-step gene replacement method as previously described (44). The disruption fragment was generated by PCR using the following two oligonucleotides (BNR1 sequence is underlined): 5′-AGTGATGATGATCGTGACACAAAAGCAGATAAAAAAATAGCACAATCATCAGCGATGGACTCTTCCCGGATCCCCGGGTTAATTAA-3′ and 5′-CTATATATTTTGAATATCGTTCAGCATAGCATGCGTTCTCTCTAGTAAAACGTGATCTTCA TCCTTGAATTCGAGCTCGTTTAAAC-3′. Plasmid pFA6a-His3MX6 was used as the template for the his5+ portion of this construct. The PCR product was introduced into a diploid strain, OHNY3. To disrupt the BUD6 gene, pUC19-bud6::HIS3 was cut with PvuII and the digested DNA was introduced into OHNY3. The genomic DNA was isolated from each transformant, and the proper disruption of BNR1 or BUD6 was verified by PCR (data not shown). These strains were subjected to tetrad analysis to obtain KIBY1 or TFB6H1, respectively. The bnr1-Δ strain, KIBY1, grew normally at 14, 20, 24, 30, and 37°C and showed a normal axial budding pattern by the staining of bud scars (data not shown). We crossed the disruption mutant of BNI1, BTY12 (MATα bni1-Δ::URA3), with KIBY1. Resultant double-heterozygous strain DKBY80B (bni1-Δ/BNI1 bnr1-Δ/BNR1) was sporulated and subjected to tetrad analysis. In 28 tetrads dissected, no His+Leu+ segregant was recovered, indicating that the bni1-Δ bnr1-Δ double mutant is inviable. A segregant of the double mutant was germinated and arrested as a large round cell with one or more buds. Thus, we concluded that BNI1 and BNR1 are an essential gene pair for vegetative growth.
Plasmid construction and other molecular biological techniques.
Standard molecular biological techniques were used for construction of plasmids, PCR, and DNA sequencing (58). Plasmids used in this study are listed in Table 2. PCRs were performed using a GeneAmp PCR System 2400 (Perkin-Elmer, Norwalk, Conn.) and DNA sequences were determined using an ALFred DNA sequencer (Amersham Pharmacia, Little Chalfont, United Kingdom). The relative steady-state protein levels for the truncated mutants of Bni1p were detected by Western blot analysis with an anti-green fluorescent protein (GFP) polyclonal antibody (MBL, Nagoya, Japan) and measured by densitometry using a ScanImager densitometer (Amersham Pharmacia).
TABLE 2.
Plasmids used in this study
| Plasmids | Characteristics (reference) |
|---|---|
| pRS314 | TRP1 CEN6 (60) |
| pRS316 | URA3 CEN6 (60) |
| pKO11 | PGAL1-HA∗2-TTDH3 URA3 CEN6 (29) |
| pGGX1 | PGAL1-EGFP-TTDH3 URA3 CEN6; made by replacing the 90-bp EcoRI-BamHI fragment containing two HA epitopes of pK011 with the 0.72-kb EcoRI-BamHI fragment containing the enhanced GFP coding region without the termination codon, which was amplified by PCR using pEGFP-N1 (Clontech) as a template |
| pAGX1 | PACT1-EGFP-TTDH3 URA3 CEN6; made by replacing the 720-bp NotI-EcoRI fragment containing PGAL1 of pGGX1 by the 0.47-kb NotI-EcoRI PCR fragment containing PACT1 |
| pAGX2 | PACT1-EGFP-(A4GA4G)-TTDH3 URA3 CEN6; made by inserting the BglII-BamHI synthetic double-stragonucleotide of 5′-AGATCTGCTGCCGCTGCTGGTACCGCTGCTGCCGCTGGATCC-3′ (each underlined region corresponds to the recognition site of BglII, KpnI, and BamHI, respectively), which encodes the linkerpolypeptide of RSAAAAGTAAAAGS at the BamHI site of pAGX1 |
| pTAGX2 | PACT1-EGFP-(A4GA4G)-TTDH3 TRP1 CEN6; made by replacing the NotI-XhoI fragment of the multicloning site of pRS314 by the NotI-XhoI fragment containing PACT1-EGFP-(A4GA4G)-TTDH3 of pAGX2 |
| pAGX2-BNI1 | PACT1-EGFP-(A4GA4G)-BNI1-TTDH3 URA3 CEN6; made by inserting the 5.9-kb BamHI-SmaI fragment from pRS314-PBNI1-myc-BNI1 containing the BNI1 open reading frame into the corresponding sites of pAGX2 |
| pTAGX2-BNI1 | PACT1-EGFP-(A4GA4G)-BNI1-TTDH3 TRP1 CEN6; made by inserting the 5.9-kb BamHI-SmaI fragment from pRS314-PBNI1-myc-BNI1 containing the BNI1 open reading frame into the corresponding sites of pTAGX2 |
| pGGX1-BNI1 | PGAL1-EGFP-BNI1-TTDH3 URA3 CEN6; made by inserting the 5.9-kb BamHI-SmaI fragment from pRS314-PBNI1-myc-BNI1 containing the BNI1 open reading frame into the corresponding sites of pGGX1 |
| pAGX1-ABP1 | PACT1-EGFP-ABP1-TTDH3 URA3 CEN6; made by inserting the 1.8-kb SmaI-SmaI PCR fragment containing the ABP1 open reading frame into the corresponding site of pAGX1 |
| pFA6a-His3MX6 | PTEF1-his5+-TTEF1 (44) |
| pRS314-PBNI1-myc | PBNI1-myc∗3-TTDH3 TRP1 CEN6 (18) |
| pRS314-PBNI1-myc-BNI1 | PBNI1-myc∗3-BNI1-TTDH3 TRP1 CEN6 (18) |
| pRS314-PBNI1-myc-bni1 (490-1954) | PBNI1-myc∗3-bni1 (490-1954)-TTDH3 TRP1 CEN6 (18) |
| pRS314-PBNI1-myc-bni1 (Δ133-245) | PBNI1-myc∗3-bni1 (Δ133-245)-TTDH3 TRP1 CEN6; made by deleting the 339-bp BsaHI-BsaHI fragment from pRS314-PBNI1-myc-BNI1 |
| pRS314-PBNI1-myc-bni1 (Δ826-987) | PBNI1-myc∗3-bni1 (Δ826-987)-TTDH3 TRP1 CEN6 (18) |
| pRS314-PBNI1-myc-bni1 (Δ1239-1328) | PBNI1-myc∗3-bni1 (Δ1239-1328)-TTDH3 TRP1 CEN6 (18) |
| pRS314-PBNI1-myc-bni1 (1-1750) | PBNI1-myc∗3-bni1 (1-1750)-TTDH3 TRP1 CEN6 (18) |
| pUC19-BUD6 | BUD6; made by inserting the 3.8-kb SalI-SmaI PCR fragment containing 0.74-kb BUD6 upstream and 0.66-kb downstream noncoding regions into the SalI-SacI (blunted) site of pUC19 |
| pUC19-bud6::HIS3 | a derivative of pUC19-BUD6; made by replacing the 2.2-kb NheI-SacI fragment of BUD6 with the 1.8-kb HIS3 fragment |
Time-lapse imaging of GFP fluorescence.
All experiments described here were performed with an enhanced GFP gene from the plasmid pEGFP-N1 (Clontech, Palo Alto, Calif.). To observe in vivo behavior of GFP-fusion proteins, cells expressing GFP-fusion proteins were grown aerobically and exponentially for 6 to 8 h in the appropriate drop-out media. Samples of 80 to 100 μl of the molten liquid medium containing 1% agarose were applied to a glass slide and quickly flattened by a coverslip. Where indicated, an appropriate amount of sodium azide (Wako, Kyoto, Japan), sodium chloride, latrunculin-A (LAT-A) (Wako), or dimethylsulfonyl oxide (DMSO) was added both in culture media and molten liquid media containing agarose. Cultured cells were immobilized onto another coverslip coated with 1 mg of concanavalin A (ConA) (Sigma-Aldrich, St. Louis, Mo.)/ml. Around the edges of the coverslip, a small amount of silicon grease was applied for sealing. The coverslip was put onto the flat pad of medium. Living cells were observed using an ECLIPSE TE300-2EF microscope (Nikon, Tokyo, Japan) equipped with a 100×/1.4-numerical-aperture Plan Apo oil-immersion objective lens (Nikon), a 100-W xenon arc lamp (Nikon), and an EGFP and pass filter set (excitation wavelength, 460 to 500 nm; dichroic wavelength, 505 nm; emission wavelength, 510 to 560 nm; Chroma Optics, Brattleboro, Vt.). Images were acquired at various time intervals (the interval was slightly varied for the occasional necessity of fixing the focus) by an ARGUS/DTRS four-dimensional time-lapse imaging system (Hamamatsu Photonics, Hamamatsu, Japan), including a C4742-95-12NR cooled charge-coupled device camera and a fluorescence illumination shutter. For each image, cells were illuminated for 0.5 to 1.5 s. Photobleaching and phototoxicity were minimized by closing the excitation shutter between image collections and by reducing the illumination intensity with a 1/4 or 1/8 neutral density filter. Still, phototoxicity retarded the growth speed of a few cells, and we omitted such cells from the observations. All these procedures were performed at room temperature unless otherwise specified. Images were analyzed using Aquacosmos v1.0 Software (Hamamatsu).
Fluorescent staining and cytological techniques.
Fluorescein isothiocyanate (FITC)-ConA (Sigma-Aldrich) staining of living cells was performed essentially as previously described (55). Chitin was stained with Calcofluor White M2R New (Sigma-Aldrich) as previously described (23). Assessment of budding pattern was performed as previously described (10). We classified the bud size with the relative length of the long axis of the bud to that of the mother (R) as follows: an R of <1/3 is “small,” an R such that 1/3 < R < 2/3 is “middle,” and an R of >2/3 is “large.” In addition, we termed a bud “tiny” when the length from the bud tip to the bud neck was shorter than the width of the neck. Geometric definitions of some terms used in this study are described in Fig. 1.
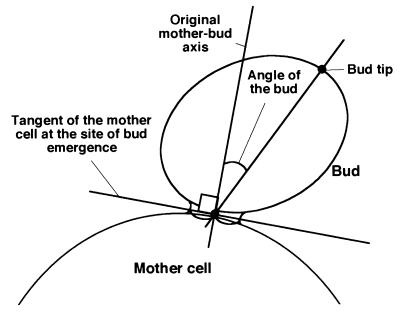
FIG. 1.
Geometric definition of terms used in this study. We define a “bud tip” as the farthest point in the bud from the midpoint of the mother-bud neck. The “original mother-bud axis” is the line perpendicular to the tangent of the mother cell at the site of bud emergence. The “angle of the bud” is the angle formed by (i) the line that intersects the bud tip and the midpoint of the mother-bud neck and (ii) the original mother-bud axis.
RESULTS
Localization of GFP-Bni1p in living cells.
To investigate the dynamics of Bni1p localization, we constructed a fusion of GFP to the N terminus of Bni1p. When this fusion protein was expressed under its own promoter, the GFP signal was too weak to detect. Then, we constructed pAGX2-BNI1 (Table 2) to express GFP-Bni1p under the ACT1 promoter. The cells harboring this plasmid displayed a clear GFP signal at bud tips and cytokinesis sites. This pattern of Bni1p localization was consistent with the results from indirect immunofluorescence studies of hemagglutinin (HA)-tagged Bni1p described previously (16, 26). Moreover, pAGX2-BNI1 was found to complement various phenotypes of the bni1-Δ mutant strain: a defect in cell shape, bipolar-specific random budding (66), and the lethal phenotype in combination with bnr1-Δ mutation (see Materials and Methods). These observations suggest that the addition of the GFP tag or the replacement of its promoter does not impair the function of Bni1p. Thus, we concluded that pAGX2-BNI1 can be used as a good marker for the native behavior of Bni1p with the caveat of overexpression.
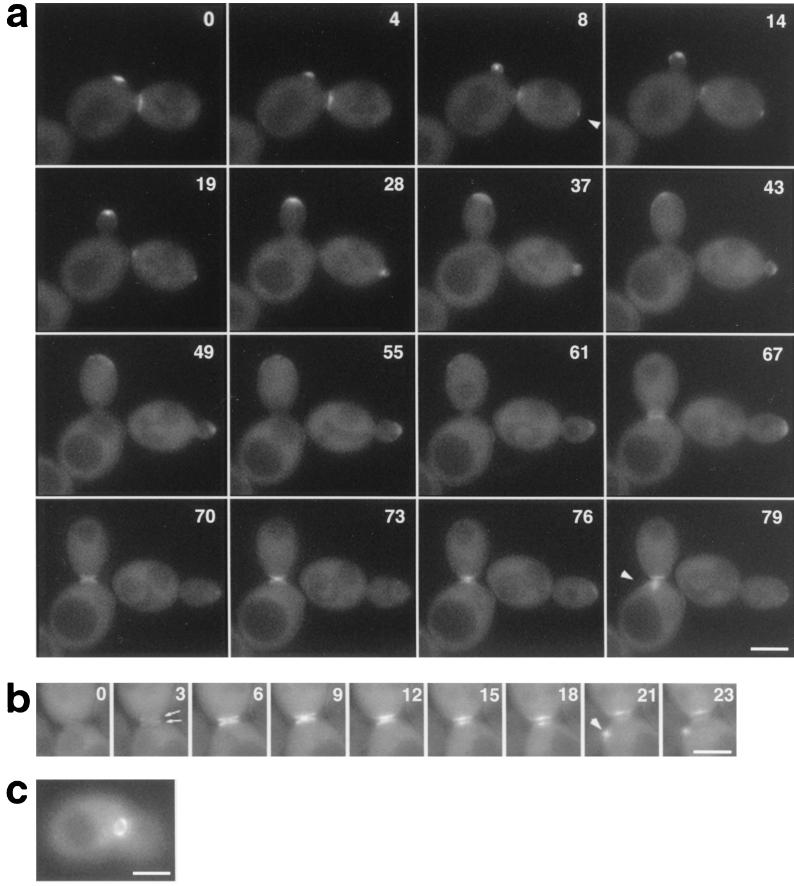
To examine the relationship between Bni1p localization and the cell cycle, we collected 14 sets of time-lapse images taken every 3 min over 1 to 2 h. The ability to follow a living cell throughout the cell cycle provided novel findings of Bni1p localization. Figure 2a shows an example in which GFP-Bni1p within two cells is observed. GFP-Bni1p appeared as a single spot or a patch at the pre-bud site 10 to 15 min before visible bud emergence (with a mean of 11.6 min for 11 clear records) (arrowhead), while the remnant of old localization at the previous cytokinesis site gradually disappeared. In a small-budded cell, the GFP-Bni1p spot often moved in the bud (Fig. 2a, right cell, 28 and 37 min; Fig. 3a, 20 to 56 min) (see below). As the bud enlarged, GFP-Bni1p localization became more diffuse and faint and formed a “cap” at the distal cortex of the bud (Fig. 2a, left cell, 19 to 55 min). Timing of the disappearance of GFP-Bni1p from the bud tip varied in each cell, but in some cells, the GFP-Bni1p signal could be detected until 10 min before its reappearance at the cytokinesis site, suggesting that Bni1p remains at the bud tip until very close to the time of cytokinesis, like Spa2p and Rho1p (38). Before cytokinesis, the GFP signal first appeared at the mother-bud neck as a couple of rings, one in the bud side and the other in the mother side (Fig. 2b, 3 min, arrows). The rings became brighter and were filled up to the two dish-like structures contacting each other at their center (Fig. 2b, 9 min). Then, the distance between the two patches became wider (15 min), indicative of septum formation. The diameter of this GFP-Bni1p structure was about 1.5 to 1.9 μm (mean, 1.7 μm; n = 12) and was not obviously changed during cytokinesis. Thus, this pattern of localization at the cytokinesis sites was distinct from that of Myo1p, which contracts during cytokinesis (6, 42), or from that of septins, which remain a ring during cytokinesis (43). The patch of the mother side disappeared earlier than that of the bud side and reappeared at the new pre-bud site (Fig. 2a, 79 min; Fig. 2b, 21 min). Cells expressing GFP-Bni1p also displayed general cytoplasmic fluorescence except for nuclei and vacuoles. We tested whether GFP-Bni1p spots were restricted to the cell surface or located in the cytoplasm by Z-section imaging of 17 cells by confocal laser microscopy. We found no obvious incidence of GFP-Bni1p spots located in the cytoplasm (data not shown). By conventional fluorescent microscopy, we did not find any distinct GFP spot in the cytoplasm of healthy cells. Thus, solid structures of Bni1p seemed to be restricted to the cell cortex.
FIG. 2.
GFP-Bni1p localization. (a) Time-lapse analysis of Bni1p localization throughout the cell cycle. Diploid strain KY4 (bni1-Δ) harboring the GFP-BNI1 plasmid, pAGX2-BNI1, was examined by time-lapse video microscopy as described in Materials and Methods. Images were taken every ∼3 min for 2 h. Numbers indicate the time (in minutes) since the beginning of observation. Visible bud emergence occurred at around 4 min (left cell) and 19 min (right cell). (b) Bni1p localization at cytokinesis; (c) GFP-Bni1p ring at the mother-bud neck. Arrowhead, GFP-Bni1p signal at pre-bud sites; arrows, a double ring formed at the mother-bud neck. Bar, 4 μm.
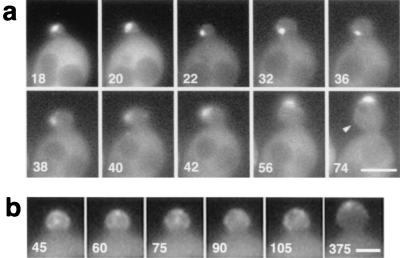
FIG. 3.
Movement of GFP-Bni1p in small buds. (a) Movement of a Bni1p spot from the bud tip to the bud neck and then to the bud tip again. Strain KY4 (bni1-Δ) harboring the GFP-BNI1 plasmid pAGX2-BNI1 was subjected to time-lapse imaging at 2-min intervals for 90 min. Numbers indicate the time (in minutes) since the beginning of observation. Note that the bud growth oriented first to the left side (22 to 42 min) and then reoriented to the top (56 to 74 min). Arrowhead, a protrusion of the bud cortex as a result of the bud growth at 22 to 42 min. Bar, 4 μm. (b) Movement of fuzzy dots of GFP-Bni1p in a small bud. The same strain as that in panel a was subjected to time-lapse imaging at 15-s intervals for 15 min. Numbers indicate the time (in seconds) since the beginning of observation. At 375 s, the GFP-Bni1p signal coalesced into a cap at the bud tip. Bar, 2 μm.
Movement of Bni1p spots in tiny- to small-budded cells.
Time-lapse analysis revealed the dynamic nature of Bni1p localization. Especially notable was that the GFP-Bni1p signal moved in tiny- to small-budded cells instead of staying still at the bud tips. To examine this movement of Bni1p, we analyzed 26 sets of time-lapse images at 1- or 2-min intervals over the period from the visible bud emergence to the formation of the middle-sized bud. Among them, seven cases showed clear movement of the spot from the bud tip to the bud neck, and then the spot returned to the bud tip (Fig. 3a). In eight cases, GFP-Bni1p first localized at the very tiny bud in which the precise location could not be determined, then stayed at the bud neck, and moved slowly to the tip as the bud grew (Fig. 2a, right cell). In three cases, GFP-Bni1p first localized at the tiny-bud tip, dispersed in the small bud diffusely or as a few fuzzy dots, and then accumulated at the bud tip as a single dot or a crescent. In three cases out of eight time-lapse images at 15-s intervals over 10 to 15 min, such fuzzy signals of Bni1p moved in the bud (Fig. 3b). The rest of the 8 cases displayed no or only slight movement of Bni1p around the bud tip. Thus, in at least 65% of the cases (n = 26), GFP-Bni1p somehow moved in the bud. Duration of the Bni1p movement varied from 3 to 30 min (mean, 14.0 min; n = 18). Drastic movement was not seen once after the spot returned to the bud tip and changed into a crescent.
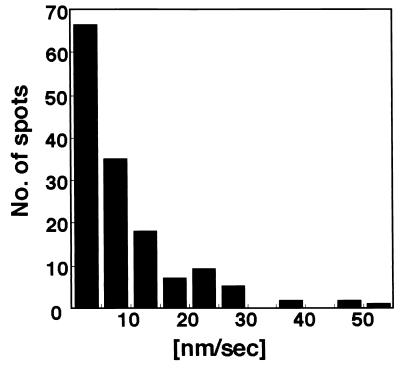
The rates of the movement of GFP-Bni1p spots were determined using six time-lapse images taken every 15 s over 8 to 15 min (Fig. 4). For simplicity, we limited our investigation to tiny- and small-budded cells with a single distinctive Bni1p spot. The mean of the speed of Bni1p movement was 9.22 nm/s (standard deviation, 9.65 nm/s), the median was 5.92 nm/s, the range was 0 to 55 nm/s, and n was 145. The average maximal velocity of six spots was 33 nm/s. GFP-Bni1p spots did not vibrate rapidly in a small area in the time-lapse images taken every 5 s (data not shown). These observations indicate that the motility of GFP-Bni1p spots is distinct from the rapid movement of cortical actin patches (maximal velocity of more than 1 μm/s) (12, 62). Rather, it is comparable to the slower movement of cdc12p (an average maximal velocity of 71 nm/s), an FH protein in S. pombe (8).
FIG. 4.
Rate of Bni1p movement. Total velocities reported are derived from a compilation of 145 displacements each over a 15-s intervals observed for six representative spots as assayed by time-lapse microscopy.
The Bni1p movement could be due to the delivery of newly synthesized Bni1p to the cell cortex and following rapid degradation. To address this issue, we constructed a single-copy plasmid, pGGX1-BNI1, to express GFP-Bni1p under the GAL1 promoter. Cells of wild-type strain OHNY3 were transformed with pGGX1-BNI1, cultured in a galactose-containing medium for 6 h, and then transferred to a glucose-containing medium to stop the expression of GFP-Bni1p. Three hours after the transfer, Bni1p still moved in these cells (data not shown). This observation shows that Bni1p movement cannot be due to the delivery of the newly synthesized Bni1p but to the true motility of Bni1p molecules.
The motility of the GFP-Bni1p spots might be due to the random diffusion or to energy-requiring active process. To address this issue, we performed the GFP-Bni1p time-lapse experiment in the presence of sodium azide, a metabolic inhibitor, as described previously (12). As a control experiment, wild-type cells expressing GFP-Abp1p were treated with 20 mM sodium azide. In our strain background, GFP-labeled cortical actin patches stopped moving completely in about 50% of cells (n = 26) 30 to 40 min after the addition of sodium azide, but their punctate appearance was not changed as previously reported (12) (data not shown). Sodium chloride had no effect on the motility of actin patches. Cells expressing GFP-Bni1p were treated with 20 mM sodium azide in the same way. Ten minutes after the addition of sodium azide, however, proper localization of Bni1p was observed at neither bud tips nor cytokinesis sites. GFP-Bni1p spots became diffuse, and the longer incubation (to 30 min) after the addition of sodium azide caused multiple aggregates of GFP signal to be distributed randomly in 94% (n = 100) of the cells (data not shown). These aggregates were not motile. The addition of sodium chloride did not change the Bni1p localization and movement. These observations suggest an energy-requiring active process that localizes the Bni1p molecules properly. However, the mechanism by which macroscopic spots of Bni1p move remains to be clarified.
Coincidence of Bni1p localization with the site of bud growth.
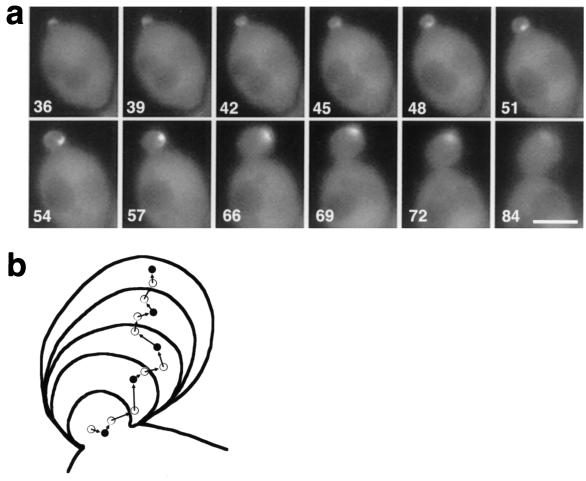
The most unique feature of Bni1p localization was its coincidence with the site of apparent bud growth. When a Bni1p spot stayed at one side of the bud or moved slowly back to the bud tip along one side of the bud, this side of the bud cortex swelled faster than the other side (Fig. 3a, 22 to 42 min and 74 min [arrowhead]; Fig. 5a, 54 to 69 min). Among the 26 sets of time-lapse images taken every 1 or 2 min, we examined three cases in which Bni1p localization inclined toward one side of the bud for 15 min or more. In all cases, the direction of bud growth during this period was inclined toward that side. The mean angle of the bud was 23.3°. On the other hand, the mean angle of the bud was 1.5° for five cells in which Bni1p did not lean to one side. Figure 5b shows a trace of the bud shape of Fig. 5a, taken every 9 min. The dots and arrows show the localization and movement of the GFP-Bni1p spot taken every 3 min. This trace clearly showed the parallel change of the direction of the Bni1p movement and the apparent bud growth at around 66 min. All of these observations indicate the tight coincidence of Bni1p localization and the apparent growth site of the bud.
FIG. 5.
Movement of a GFP-Bni1p spot and its colocalization with the apparent site of bud growth. (a) Time-lapse analysis of GFP-Bni1p movement. The visible bud emergence occurred at 27 min, and until 36 min the GFP-Bni1p spot was at the very tip of the bud (data not shown). The spot started to move to the bud neck at 39 min and stayed at the bud neck until 51 min. Then, the spot moved back to the bud tip along the right side of the bud cortex (54 to 72 min). During this period, the apparent bud growth occurred mainly on its right side, resulting in the angled bud. (b) Trace of the movement of Bni1p and outline of the bud in panel a. Each circle corresponds to the brightest point of the GFP-Bni1p signal at 3-min intervals from a 45-min time point. Each closed circle corresponds to the brightest point of GFP-Bni1p at 9-min intervals from a 48-min time point. Each curve corresponds to the outline of the bud at the same time points as the closed circles. The Bni1p localization was first strongly deviated to the right side of the bud, and after a 66-min time point, it was reoriented toward the center. Bar, 3 μm.
A defect of restricting the growth site in bni1 mutant cells.
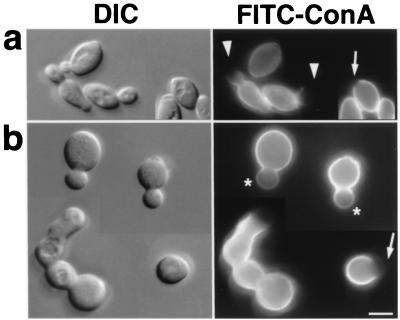
Since the localization of Bni1p and the apparent site of bud growth were tightly coupled, we assumed that Bni1p plays a role in directing and/or restricting the growth site of the bud. This assumption was supported by the observation of an abnormally spherical shape of bni1 mutant cells compared to ellipsoidal wild-type cells (Fig. 6b) (66). To analyze the morphological defect, we applied a pulse-chase technique of cell wall by using FITC-labeled ConA. It has been shown that newly synthesized mannoproteins, which are major components of yeast cell wall, appear at the bud tip during the early stage of the cell cycle (17). First, the entire cell surface was labeled with FITC-ConA, which binds mannose polymers. Then, the cells were allowed to grow further in the absence of FITC-ConA for 60 min. After that, 75% of the middle-sized buds (i.e., buds that were one-third to two-thirds of the length of the mother) of the wild-type diploid cells showed a staining pattern that faded out towards the bud tips (with an unstained region at the bud tips), indicative of focused growth at the bud tips (apical growth) (Fig. 6a, arrowheads). In contrast, only 22% of the bni1-Δ buds had unstained regions at the bud tips. The rest of the bni1-Δ middle-sized buds stained rather uniformly (Fig. 6b, asterisks). The staining of buds was weaker than the staining of their mother, indicating that these buds were indeed grown during the experiment. We also examined the unlabeled region of unbudded cells, which marked the pre-bud site (Fig. 6a, arrow). At the beginning of the cell cycle, polarized growth in wild-type cells is tightly restricted to the pre-bud site, which was marked by the ring of cortical actin patches. Such unlabeled regions were larger in bni1-Δ cells than in wild-type cells (Fig. 6b, arrows). Thus, tightly polarized growth to the pre-bud site was also defective, presumably resulting in imperfect bud-neck formation of bni1-Δ cells (Fig. 6b, left). It has been reported that either a spa2 or a bud6 mutation causes a similar defect in apical growth and bud neck formation. These mutants were subjected to the same experiments shown in Fig. 6. We found that these mutants also displayed a defect that was similar to, but milder than, that of a bni1-Δ mutant (data not shown).
FIG. 6.
A defect of bni1-Δ cells in cell morphology and apical growth. (a) OHNY3 (BNI1); (b) KY4 (bni1-Δ). Cells were cultured exponentially in YPGalactose medium, which generated a thinner cell shape in wild-type cells and helped to assess apical growth. Then, cells were subjected to living stain with FITC-ConA and returned to growth for 60 min. Fixed cells were visualized by differential interference contrast (DIC) and fluorescence microscopy (FITC-ConA). Arrowheads, staining that faded out towards one end; asterisks, staining without the unlabeled region (the gradient of brightness observed in the buds of Δbni1 cells is mostly a photographic artifact because of the halo of their brighter mother cells); arrows, unlabeled regions in unbudded cells, marking pre-bud sites. Bar, 5 μm.
Polarized actin patch-independent, filamentous actin (F-actin)-dependent localization of Bni1p.
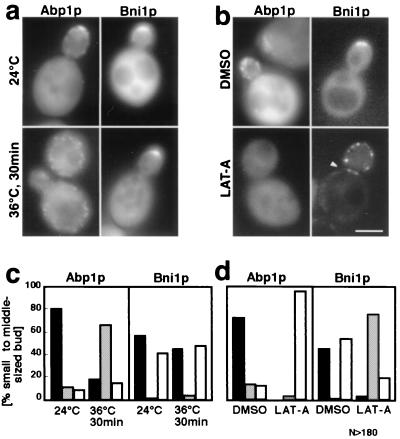
The actin cytoskeleton has been shown to be concentrated in regions of active cell growth (2). Bni1p has been reported to interact with the actin cytoskeleton directly and via the actin-binding proteins, including Pfy1p (16). To assess the possible role of the actin cytoskeleton in regulating Bni1p localization, the cells expressing GFP-Bni1p were exposed to mild heat shock, which caused temporal depolarization of cortical actin patches (40). In our strain background, the temperature shift from 24 to 36°C caused depolarization of cortical actin patches within 30 min and repolarization within 120 min (data not shown). Wild-type cells expressing GFP-Bni1p were subjected to the mild heat shock along with the control cells expressing GFP-Abp1p (12), which colocalized with cortical actin patches (Fig. 7a and c). Thirty minutes after the shift to 36°C, the actin patches marked by GFP-Abp1p were depolarized and distributed throughout the cells. However, the GFP-Bni1p signal was apparently unperturbed, indicating that the localization of Bni1p does not depend on polarized actin patches.
FIG. 7.
Polarized actin patch-independent but F-actin-dependent Bni1p localization. (a and c) Depolarization of cortical actin patches but not of Bni1p by mild heat shock. Cells of strain OHNY3 expressing GFP-Abp1p or GFP-Bni1p were cultured exponentially at 24°C and then shifted to 36°C for 30 min. Then, the cells were directly examined by fluorescent microscopy. (c) Each cell was classified either as a polarized GFP signal (solid bar), a depolarized GFP signal (hatched bar), or no concentration of GFP signal (open bar). (b and d) Alteration of the GFP-Bni1p localization by LAT-A-induced complete disruption of the actin cytoskeleton. Cells of strain OHNY3 expressing GFP-Abp1p or GFP-Bni1p were cultured exponentially at 24°C, and LAT-A was added from a 10 mM DMSO stock to a final concentration of 100 μM. An equal volume of DMSO alone was added to the control population of cells. After a 30-min incubation, the cells were directly observed by conventional fluorescent microscopy as was done in panels a and c. Bar, 5 μm.
Complete disruption of F-actin structures can be accomplished by treating cells with LAT-A, which sequesters actin monomers and hence extinguishes F-actin rapidly (5). Wild-type cells expressing GFP-Bni1p or GFP-Abp1p were treated with 100 μM LAT-A for 30 min (Fig. 7b and d). The GFP-Abplp localization was changed from patches at the cell cortex to general cytosolic fluorescence essentially in all cells. The GFP-Bni1p localization was dramatically changed in an opposite way: while the cytoplasmic signal was reduced, the signal at the bud cortex changed into highly concentrated bright dots. These dots were not motile (data not shown). Among these cells showing abnormal localization, 30% (n = 51) of the cells displayed simultaneous localization of GFP-Bni1p at the bud tip and at the bud neck as a ring (or a double ring) early in the cell cycle (arrowhead). Such a ring was never seen in untreated middle-sized-budded cells. Thus, we concluded that F-actin structures, but not polarized cortical actin patches, play an important role in the proper localization of Bni1p.
Role of Cdc42p and Rho1p in GFP-Bni1p localization.
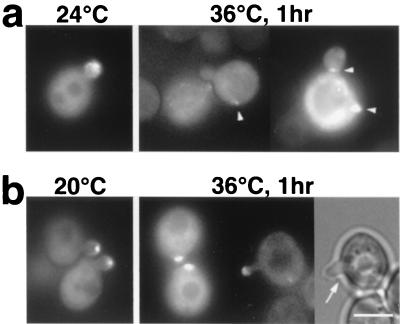
Rho family small G proteins have been implicated in regulation of the actin cytoskeleton. In S. cerevisiae, Rho1p and Cdc42p have been shown to interact with Bni1p (16, 32). To investigate their role in Bni1p localization, we examined the GFP-Bni1p localization in cdc42 or rho1 temperature-sensitive mutant cells (Fig. 8a or b). At 24°C, 53% (n = 102) of the small-budded cdc42-1 cells displayed GFP-Bni1p concentrated at the tips. One hour after the shift to 36°C, only 4% (n = 115) showed a GFP-Bni1p signal at bud tips, 61% displayed no obvious concentration of the GFP signal, and the rest (35%) showed the GFP signal as a few bright aggregates randomly distributed throughout the cells (Fig. 8a, arrowheads). Note that this abnormal pattern of Bni1p localization was distinct from that in LAT-A-treated cells: in cdc42-1 cells, Bni1p spots were distributed in the mother cell as well as in the bud, indicative of complete polarity loss. As for rho1-104 cells, 39% (n = 101) of the small buds displayed GFP-Bni1p concentrated at the tips at 24°C. One hour after the shift to 36°C, 33% (n = 98) of the small buds still displayed GFP-Bni1p localization at tips. During further observation until 3 h after the shift to 36°C, dead cells accumulated in the population as well as small-budded cells as previously reported (64). GFP-Bni1p still localized at the bud tips of small-budded live cells in a similar proportion. Thus, Rho1p was not necessary for the localization of Bni1p at bud tips. These Bni1p spots, however, did not display obvious movement in any of the five sets of time-lapse images taken every 1 min (data not shown). It might be a secondary effect because these cells were dying. Alternatively, Rho1p might be required for the proper movement of Bni1p. This possibility was attractive because rho1 mutant cells often had a thin basal part of the bud (Fig. 8b, arrow). Loss of the movement of tightly concentrated Bni1p spots could cause insufficient development of the bud curvature.
FIG. 8.
Bni1p localization in cdc42 or rho1 mutant cells. (a) cdc42-1 temperature-sensitive mutant cells. Cells of strain KKC42-1 (cdc42-1) harboring pTAGX2-BNI1 were cultured exponentially in synthetic dextrose supplemented with 5% Casamino Acids liquid media at 24°C, shifted to 36°C for 1 h, and examined by fluorescent microscopy. Arrowheads, abnormal GFP dots in mother cells or at the bud neck. (b) rho1-104 temperature-sensitive mutant cells. Cells of strain HNY21 (rho1-104) harboring pTAGX2-BNI1 were cultured exponentially in SDA-AU liquid media at 20°C and shifted to 36°C for 1 h. Then, the cells were directly examined by fluorescent microscopy. Shown is a cell examined by light microscopy (right) and fluorescent microscopy (middle). Arrow, an abnormally thin basal part of the bud. Bar, 4 μm.
Role of Spa2p and Bud6p in GFP-Bni1p localization.
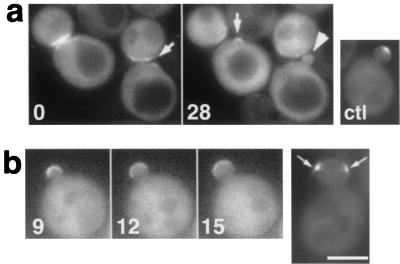
Spa2p and Bud6p have been shown to localize at the bud tips and cytokinesis sites and to interact with Bni1p (16, 19, 61). Our previous study has shown that the bud tip localization of myc-tagged Bni1p is dependent on Spa2p (19). We examined the GFP-Bni1p localization in spa2 mutant cells and found that the GFP signal at small bud tips was significantly reduced (Fig. 9a, arrowhead). Time-lapse images of seven cells around visible bud emergence at 2- or 3-min intervals demonstrated that Bni1p localization at the pre-bud site was visible (arrows) but soon became dispersed; i.e., although spa2 cells were not grossly defective in the initial polarization of Bni1p at bud emergence, they appeared to lose it quickly as the bud enlarged. This was not simply due to instability of Bni1p in spa2 cells, because the rapid and intense relocalization of Bni1p at cytokinesis sites was intact. We next examined the GFP-Bni1p localization in bud6 mutant cells. Roughly, GFP-Bni1p localized at bud tips and cytokinesis sites throughout the cell cycle. However, the GFP-Bni1p localization in tiny to small buds was more diffuse (Fig. 9b; compare Fig. 3a). Moreover, in three cases of six clear records of small- to middle-sized buds at 2- or 3-min intervals, the GFP-Bni1p signal separated into two discrete patches at the bud cortex for the period of 6 min or more (Fig. 9b, arrows), and the buds became rectangular. These observations suggest that Bni1p localization in the bud tips is dependent largely on Spa2p and partly on Bud6p.
FIG. 9.
Defective Bni1p localization in spa2 or bud6 mutant cells. The strains used were TYSH1 (spa2) (a) and TFB6H3 (bud6) (b). Mutant cells harboring GFP-BNI1 plasmid pAGX2-BNI1 were examined by time-lapse video microscopy. Numbers indicate the time (in minutes) since the beginning of observation. With spa2 diploid strains, we obtained results similar to those in panel a. (a) ctl, cell of wild-type strain OHNY1 harboring pAGX2-BNI1 as a control; arrows, GFP-Bni1p signal at incipient bud sites; arrowhead, the diffuse GFP-Bni1p signal in the small bud. (b) Arrows, GFP-Bni1p signal separated into two distinct patches. Bar, 5 μm.
Regions of Bni1p required for its localization.
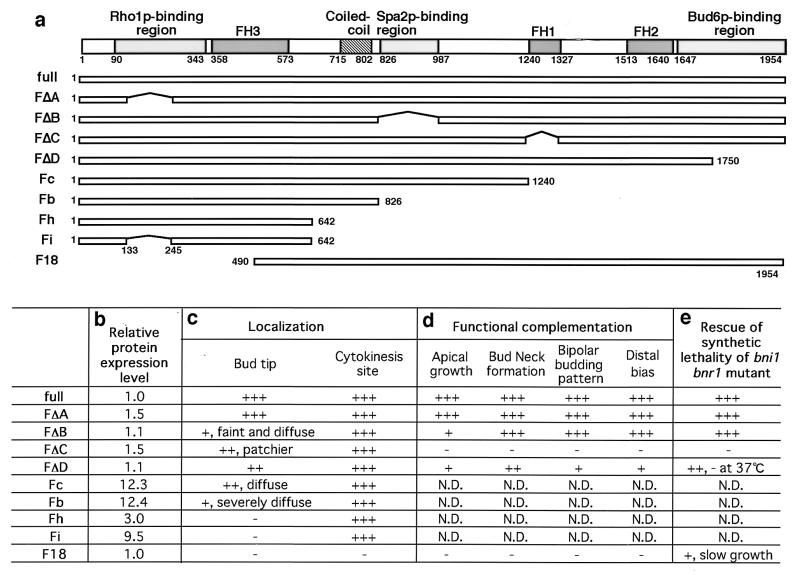
As a member of the FH protein family, Bni1p has three conserved FH domains: FH1, FH2, and FH3 domains (Fig. 10a). The FH1 domain is rich in proline and binds profilin (16). The FH1 domain of Bni1p has also been reported to interact with Act1p (16) and Myo3p (27), suggesting that this is the core region for the Bni1p function regulating the actin cytoskeleton. The FH2 domain is defined by a consensus sequence among FH proteins (15), but its function is still unclear. The FH3 domain is conserved weakly among FH proteins and that of fus1p has been shown to be necessary and sufficient for its localization in S. pombe (54). The regions of Bni1p required for interaction with Rho1p, Spa2p, and Bud6p have been mapped by the yeast two-hybrid method (16, 19, 32). Cdc42p has been reported to bind at the 1-to-1,214 amino acid (aa) region of Bni1p, as estimated by the yeast two-hybrid method (16). However, high-background β-galactosidase activity caused by DBD-CdC42p (G12V) and weak activity of AD-Cdc42p (G12V) precluded closer investigation of the Cdc42p-binding region of Bni1p (data not shown).
FIG. 10.
Mapping of the regions of Bni1p required for its proper localization and function. (a) Structures of the truncated constructs of Bni1p. Each number under the domain structure of Bni1p indicates either the first or the last amino acid residue of the region. Each horizontal bar represents a segment of Bni1p: full (aa 1 to 1954), FΔA (aa Δ133 to 245), FΔB (aa Δ826 to 987), FΔC (aa Δ1239 to 1328), FΔD (aa 1 to 1750), Fc (aa 1 to 1240), Fb (aa1 to 826), Fh (aa 1 to 642), Fi (aa 1 to 642, Δ133 to 245), and F18 (aa 490 to 1954). (b) Relative expression levels of truncated Bni1p. Various DNA fragments encoding truncated Bni1p's were cloned into pAGX2, and the resultant plasmids were used to transform cells of wild-type strain OHNY3. Transformants were subjected to Western blotting using the anti-GFP polyclonal antibody. Expression levels of mutant Bni1p relative to those of full-length Bni1p were determined by densitometry. (c) Localization of GFP-fused truncated Bni1p at the bud tip. The cells expressing truncated Bni1p were examined for GFP localization by conventional fluorescent microscopy. (d) Functional complementation of the bni1-Δ mutant phenotypes by the truncated proteins. Various DNA fragments encoding truncated Bni1p's were cloned into pRS314-PBNI1-myc (18), and the resultant plasmids were used to transform KY4 (bni1-Δ). Transformants were subjected to morphological examinations of apical growth by the same FITC-ConA living stain used in Fig. 6, of bud neck formation by phase-contrast microscopy, and of budding pattern by chitin staining. (e) Rescue of the synthetic lethality of the bni1-Δ bnr1-Δ cells by truncated Bni1p. The same plasmids used in panel d were used to transform DKBY80B (bni1-Δ/BNI1, bnr1-Δ/BNR1). Transformants were subjected to tetrad analysis. +++, indistinguishable from the wild type; ++, mild defect; +, severe defect; −, no signal or function-like negative control. N.D., not determined.
Asking which interaction is required for the proper localization of Bni1p, we examined the localization of truncated proteins fused to GFP (Fig. 10c). FΔA, FΔB, FΔC, and FΔD lacked interaction with Rho1p, Spa2p, Pfy1p, and Bud6p, respectively, as estimated by the yeast two-hybrid method (data not shown). Among these truncated proteins, FΔA showed normal localization and movement, suggesting that the direct interaction of Bni1p with Rho1p is not required for the proper localization or movement of Bni1p. Roughly, FΔB, FΔC, and FΔD were also localized to bud tips and cytokinesis sites. However, neither of them displayed the clear movement of Bni1p. Deletion of the Spa2p-binding region had a strong effect on making Bni1p localization diffuse at bud tips. Fc and Fb still localized to bud tips and cytokinesis sites. The Fb localization at bud tips was not concentrated as a spot but always as a broad crescent. Fh or Fi did not locate to bud tips at all, indicating that the coiled-coil region is required for Bni1p localization at bud tips. On the other hand, the rapid and intense relocalization at mother-bud necks was not affected in all of these truncated proteins. Only F18, which lacked the N-terminal region including half of the FH3 domain, did not display the localization at cytokinesis sites.
Regions of Bni1p required for its functions.
To investigate the region required for Bni1p functions, we used the truncated constructs of FΔA, FΔB, FΔC, FΔD, and F18. These DNA fragments were subcloned into the plasmid pRS314-PBNI1-myc, resulting in expression of myc-tagged truncated proteins under the BNI1 promoter. The bni1-Δ diploid cells were transformed with these plasmids and examined for their phenotypes. The bni1-Δ diploid cells had morphological defects of (i) apical growth, (ii) bud neck formation, (iii) bipolar bud site selection, and (iv) distal bias at first bud site selection (34, 66) (Fig. 10d). FΔA rescued the defects of bni1-Δ cells as well as full-length Bni1p. FΔB and FΔD significantly rescued the imperfect bud neck formation of bni1-Δ cells, but scarcely rescued the defect in apical growth. FΔB also rescued the defect of bipolar bud-site selection, while FΔD did so only poorly. Neither F18 nor FΔC rescued the morphological defects of bni1-Δ cells at all.
In the course of this study, we found that the bnr1 null mutation showed the lethal phenotype in combination with the bni1-Δ mutation (see Materials and Methods). It indicates that BNI1 and BNR1 are an essential gene pair for vegetative growth. The five truncated constructs of Bni1p were also tested for their ability to complement the synthetic lethality of the bni1-Δ bnr1-Δ double-mutant cells (Fig. 10e). FΔA and FΔB complemented the synthetic lethality, FΔD did mostly, F18 did poorly, and FΔC did not at all.
We concluded that (i) the FH1 domain is essential for any function of Bni1p as far as it was tested, (ii) F18, which does not localize to growth sites, loses most of the functions of Bni1p, (iii) the interaction of Bni1p with Spa2p or Bud6p is not absolutely essential for the viability but is important for the regulation of the localization and/or the function of Bni1p, especially in relation to the polarity in the bud, and (iv) the Rho1p-binding region of Bni1p is apparently not required for all the aspects of the Bni1p localization and function as far as it was tested.
DISCUSSION
In this study, we have investigated in vivo localization of Bni1p and analyzed some of the complex regulatory mechanisms underlying the Bni1p localization and functions. There are a number of proteins that localize to presumptive bud sites, to small-bud tips, and subsequently to mother-bud necks during cytokinesis in S. cerevisiae (38). Some of these proteins are involved in the establishment of polarity (Bem1p, Cdc42p), others are involved in growth of the cell surface (Rho1p, Myo2p, Sec4p), and yet others control actin assembly (Cap2p, Abp1p, Sac6p, Cof1p). The functions of others are still elusive (Spa2p, Smy1p). Recent progress of techniques to visualize in vivo behavior of proteins has enabled us to perform detailed observations and to discriminate their localization patterns from each other.
The Bni1p localization described here was marked by its highly focused distribution and its movement in the bud at the early stage of the cell cycle (Fig. 2, 3, and 5). Although Bni1p has been shown to interact physically and functionally with Spa2p and Bud6p (16, 19), these proteins have their own localization pattern. GFP-Spa2p has been reported to localize more diffusely as a cap or a crescent of the bud cortex (4). GFP-Bud6p has been reported to localize at the bud tip and also at the bud neck early in the cell cycle (3). We confirmed these observations in our strain background and found no drastic movement of the peak point of GFP-Spa2p or GFP-Bud6p (data not shown). The same applies to patterns of localization at cytokinesis sites. For example, it has been reported that Myo1p and Igg1p/Cyk1p colocalize with actin contractile rings, which constrict during cytokinesis (6, 42). Spa2p has also been reported to be narrowed during cytokinesis (4), although it is not clear whether it colocalizes with contractile rings. We observed that the GFP-Bni1p signal was wider than the GFP-Spa2p signal at mother-bud necks (data not shown) and that the GFP-Bni1p signal was not constricted during cytokinesis (Fig. 2b). GFP-Bud6p localized to cytokinesis sites as a double ring and remained a ring until the daughter cells were separated from their mother cells (3) (our unpublished observation). Thus, although the Bni1p localization overlaps with localizations of Spa2p and Bud6p, they are not identical to each other. These findings suggest an unexpected variety of specialized regulatory mechanisms required for proper morphogenesis of the yeast cells.
Motile structures at the bud cortex also have their own rate and pattern of movement. Rapid movement of cortical actin patches has been well characterized (12, 62). Another example is GFP-Kar9p or the plus ends of cytoplasmic microtubules (48, 59). They move much faster (maximal velocity of more than 1 μm) than Bni1p does (D. Pellman, personal communication). Moreover, in large-budded cells, the small spot of Kar9p is kept moving; it is not restricted to the bud tip. This pattern of localization is in contrast to that of Bni1p, bound to the distal pole of the bud in large-budded cells. It should be emphasized, however, that such differences of localization patterns do not preclude the significance of the functional correlation or physical interaction of these proteins, which have been established by a number of previous studies (16, 19, 34).
The movement of Cdc12p, the FH protein of S. pombe, has been studied in detail (8). During interphase, a GFP-Cdc12p spot moves in the cytoplasm along with both actin cables and microtubules toward cytokinesis sites. At cytokinesis, it changes into a contractile ring. Although Bni1p did not seem to move in the cytoplasm, the pattern and the slow rate of the Bni1p movement are comparable to those of the Cdc12p movement. These results suggest a possible pattern of dynamic localization as a feature of FH proteins.
One of the notable features of the Bni1p localization demonstrated here is its coincidence with the focus of apical growth inside the bud (Fig. 5). It suggests the role of Bni1p in local membrane growth. Consistent with this assumption, bni1-Δ diploid mutant cells show a severe defect of directed growth (Fig. 6). Rings of cortical actin patches at the pre-bud sites were loosened in bni1-Δ cells (our unpublished observation). Thus, it seems logical to assume that Bni1p plays a specific role in restriction of the growth site by organizing the actin cytoskeleton.
Our analysis on local growth of the bud cortex indicates that the bud cortex does not grow uniformly. Instead, the focus of the apical growth moves inside the bud, especially in the early stage of the cell cycle. We found that Bni1p localization coincided with the apparent site of the bud growth (Fig. 5). Provided that Bni1p directs the site of bud growth and Bni1p does not move from the bud tip when it is tightly polarized as a dot, the basal part of the bud might become like a thin cylinder. This hypothetical function of Bni1p may explain the abnormal morphogenesis of thin buds in rho1 mutant cells (Fig. 8b).
We found that there are several mechanisms regulating Bni1p localization and function. First, we have found that F-actin structures other than cortical actin patches are responsible for the proper localization of Bni1p (Fig. 7). Conversely, Bni1p is thought to regulate the actin cytoskeleton by the interaction with Pfy1p, Act1p (16), and Myo3p (27) at the FH1 domain. The FH1 domain was essential for all of the Bni1p functions as far as was tested. This result is consistent with our present understanding that the core function of Bni1p is to regulate these actin-regulated proteins.
Second, we have found that polarity of the Bni1p localization is dependent on Cdc42p but not on Rho1p (Fig. 8). The polarity of cortical actin patches has also been reported to be lost in cdc42 mutant cells but not in rho1 mutant cells (1, 65). Cdc42p may regulate Bni1p localization by their direct binding and/or may regulate indirectly by depolarizing other proteins. It is interesting that Cdc42p and Rho1p may bind different regions. Unexpected results showed that the direct interaction of Bni1p with Rho1p is scarcely important for either Bni1p localization or function. As previously reported (19), the truncated mutant F18 (aa 490 to 1954), which lacks the N-terminal region including the Rho1p-binding region and half of the FH3 domain, localizes at neither bud tips nor cytokinesis sites. At that time, it was thought that the interaction of Rho1p and Bni1p was required for Bni1p localization since the existence of the FH3 domain was not known. Our present study suggests that the interaction with Cdc42p and/or the unknown interaction at the FH3 domain is more important for Bni1p localization than the interaction with Rho1p.
Third, we have found that Bni1p localization at bud tips is markedly dependent on Spa2p and modestly dependent on Bud6p (Fig. 10). The interaction of Spa2p with Bni1p did not participate in complementing the defect of bipolar bud site selection of the bni1-Δ mutant or the lethal phenotype of the bni1-Δ bnr1-Δ double mutation but was required for the regulation of apical growth and for the proper localization of Bni1p. Thus, it is suggested that the interaction of Bni1p with Spa2p is required mainly for the spatial regulation of Bni1p. The significance of the interaction of Bni1p with Bud6p is still more elusive. bni1 and bud6 mutations shared various defective phenotypes, including bipolar bud site selection, apical growth (66), spindle orientation (34), Kar9p localization (47), and the synthetic lethality with bck1 (19) (our unpublished observation). They seem to have partially overlapping, and partially cooperative, functions related to the actin cytoskeleton which remain to be clarified.
In conclusion, the present observations strongly suggest that the proper localization and function of Bni1p are not determined by a single factor but are achieved by concerted participation of Cdc42p, Spa2p, Bud6p, and the actin cytoskeleton.
ACKNOWLEDGMENTS
We thank Yoshinori Kobayashi for technical assistance. We are also grateful to David Pellman, Kunihiro Matsumoto, Yoshikazu Ohya, Yasushi Matsui, and Kazuma Tanaka for comments on the manuscript.
This investigation was supported by grants-in-aid for Scientific Research and for Cancer Research from the Ministry of Education, Science, Sports, and Culture, Japan (1999, 2000).
REFERENCES
- 1.Adams A E, Johnson D I, Longnecker R M, Sloat B F, Pringle J R. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J Cell Biol. 1990;111:131–142. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams A E, Pringle J R. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amberg D C, Zahner J E, Mulholland J W, Pringle J R, Botstein D. Aip3/Bud6, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol Biol Cell. 1997;8:729–753. doi: 10.1091/mbc.8.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arkowitz R A, Lowe N. A small conserved domain in the yeast Spa2p is necessary and sufficient for its polarized localization. J Cell Biol. 1997;138:17–36. doi: 10.1083/jcb.138.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayscough K R, Stryker J, Pokala N, Sanders M, Crews P, Drubin D G. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bi E, Maddox P, Lew D J, Salmon E D, McMillan J N, Yeh E, Pringle J R. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J Cell Biol. 1998;142:1301–1312. doi: 10.1083/jcb.142.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabib E, Drgonova J, Drgon T. Role of small G proteins in yeast cell polarization and wall biosynthesis. Annu Rev Biochem. 1998;67:307–333. doi: 10.1146/annurev.biochem.67.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang F. Movement of a cytokinesis factor cdc12p to the site of cell division. Curr Biol. 1999;9:849–852. doi: 10.1016/s0960-9822(99)80372-8. [DOI] [PubMed] [Google Scholar]
- 9.Chant J, Pringle J R. Budding and cell polarity in Saccharomyces cerevisiae. Curr Opin Genet Dev. 1991;1:342–350. doi: 10.1016/s0959-437x(05)80298-9. [DOI] [PubMed] [Google Scholar]
- 10.Chant J, Pringle J R. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J Cell Biol. 1995;129:751–765. doi: 10.1083/jcb.129.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cid V J, Durán A, del Rey F, Snyder M P, Nombela C, Sánchez M. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doyle T, Botstein D. Movement of yeast cortical actin cytoskeleton visualized in vivo. Proc Natl Acad Sci USA. 1996;93:3886–3891. doi: 10.1073/pnas.93.9.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drgonova J, Drgon T, Tanaka K, Kollar R, Chen G C, Ford R A, Chan C S, Takai Y, Cabib E. Rho1p, a yeast protein at the interface between cell polarization and morphogenesis. Science. 1996;272:277–279. doi: 10.1126/science.272.5259.277. [DOI] [PubMed] [Google Scholar]
- 14.Drubin D G, Nelson W J. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- 15.Emmons S, Phan H, Calley J, Chen W, James B, Manseau L. Cappuccino, a Drosophila maternal effect gene required for polarity of the egg and embryo, is related to the vertebrate limb deformity locus. Genes Dev. 1995;9:2482–2494. doi: 10.1101/gad.9.20.2482. [DOI] [PubMed] [Google Scholar]
- 16.Evangelista M, Blundell K, Longtine M S, Chow C J, Adames N, Pringle J R, Peter M, Boone C. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- 17.Farkaš V, Kovařík J, Koŝinová A, Bauer Š. Autoradiographic study of mannan incorporation into the growing cell walls of Saccharomyces cerevisiae. J Bacteriol. 1974;117:265–269. doi: 10.1128/jb.117.1.265-269.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujiwara T, Tanaka K, Inoue E, Kikyo M, Takai Y. Bni1p regulates nicrotubule-dependent nuclear migration through the actin cytoskeleton in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:8016–8027. doi: 10.1128/mcb.19.12.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujiwara T, Tanaka K, Mino A, Kikyo M, Takahashi K, Shimizu K, Takai Y. Rho1p-Bni1p-Spa2p interactions: implication in localization of Bni1p at the bud site and regulation of the actin cytoskeleton in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1221–1233. doi: 10.1091/mbc.9.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geiser J R, Schott E J, Kingsbury T J, Cole N B, Totis L J, Bhattacharyya G, He L, Hoyt M A. Saccharomyces cerevisiae genes required in the absence of the CIN8-encoded spindle motor act in functionally diverse mitotic pathways. Mol Biol Cell. 1997;8:1035–1050. doi: 10.1091/mbc.8.6.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gietz D, Jean A S, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goud B, Salminen A, Walworth N C, Novick P J. A GTP-binding protein required for secretion rapidly associates with secretory vesicles and the plasma membrane in yeast. Cell. 1988;53:753–768. doi: 10.1016/0092-8674(88)90093-1. [DOI] [PubMed] [Google Scholar]
- 23.Guthrie C, Fink G R. Guide to yeast genetics and molecular biology. London, United Kingdom: Academic Press Limited; 1991. [Google Scholar]
- 24.Haarer B K, Lillie S H, Adams A E, Magdolen V, Bandlow W, Brown S S. Purification of profilin from Saccharomyces cerevisiae and analysis of profilin-deficient cells. J Cell Biol. 1990;110:105–114. doi: 10.1083/jcb.110.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, Takahashi K, Sasaki T, Takai Y. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 1997;16:2745–2755. doi: 10.1093/emboj/16.10.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jansen R-P, Dowzer C, Michaelis C, Galova M, Nasmyth K. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin myo4p and other cytoplasmic proteins. Cell. 1996;84:687–697. doi: 10.1016/s0092-8674(00)81047-8. [DOI] [PubMed] [Google Scholar]
- 27.Johnson D I. Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol Mol Biol Rev. 1999;63:54–105. doi: 10.1128/mmbr.63.1.54-105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson D I, Pringle J R. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J Cell Biol. 1990;111:143–152. doi: 10.1083/jcb.111.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamei T, Tanaka K, Hihara T, Umikawa M, Imamura H, Kikyo M, Ozaki K, Takai Y. Interaction of Bnr1p with a novel Src homology 3 domain-containing Hof1p. Implication in cytokinesis in Saccharomyces cerevisiae. J Biol Chem. 1998;273:28341–28345. doi: 10.1074/jbc.273.43.28341. [DOI] [PubMed] [Google Scholar]
- 30.Kikyo M, Tanaka K, Kamei T, Ozaki K, Fujiwara T, Inoue E, Takita Y, Ohya Y, Takai Y. An FH domain-containing Bnr1p is a multifunctional protein interacting with a variety of cytoskeletal proteins in Saccharomyces cerevisiae. Oncogene. 1999;18:7046–7054. doi: 10.1038/sj.onc.1203184. [DOI] [PubMed] [Google Scholar]
- 31.Kilmartin J V, Adams A E. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohno H, Tanaka K, Mino A, Umikawa M, Imamura H, Fujiwara T, Fujita Y, Hotta K, Qadota H, Watanabe T, Ohya Y, Takai Y. Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP-binding protein in Saccharomyces cerevisiae. EMBO J. 1996;15:6060–6068. [PMC free article] [PubMed] [Google Scholar]
- 33.Korinek W S, Copeland M J, Chaudhuri A, Chant J. Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science. 2000;287:2257–2259. doi: 10.1126/science.287.5461.2257. [DOI] [PubMed] [Google Scholar]
- 34.Lee L, Klee S K, Evangelista M, Boone C, Pellman D. Control of mitotic spindle position by the Saccharomyces cerevisiae formin Bni1p. J Cell Biol. 1999;144:947–961. doi: 10.1083/jcb.144.5.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee L, Tirnauer J S, Li J, Schuyler S C, Liu J Y, Pellman D. Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science. 2000;287:2260–2262. doi: 10.1126/science.287.5461.2260. [DOI] [PubMed] [Google Scholar]
- 36.Lew D, Weinert T, Pringle J. Cell cycle control in Saccharomyces cerevisiae. In: Pringle J, Broach J, Jones E, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 607–695. [Google Scholar]
- 37.Lew D J, Reed S I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lew D J, Reed S I. Cell cycle control of morphogenesis in budding yeast. Curr Opin Genet Dev. 1995;5:17–23. doi: 10.1016/s0959-437x(95)90048-9. [DOI] [PubMed] [Google Scholar]
- 39.Lillie S H, Brown S S. Suppression of a myosin defect by a kinesin-related gene. Nature. 1992;356:358–361. doi: 10.1038/356358a0. [DOI] [PubMed] [Google Scholar]
- 40.Lillie S H, Brown S S. Immunofluorescence localization of the unconventional myosin, Myo2p, and the putative kinesin-related protein, Smy1p, to the same regions of polarized growth in Saccharomyces cerevisiae. J Cell Biol. 1994;125:825–842. doi: 10.1083/jcb.125.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lippincott J, Li R. Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J Cell Biol. 1998;143:1947–1960. doi: 10.1083/jcb.143.7.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lippincott J, Li R. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J Cell Biol. 1998;140:355–366. doi: 10.1083/jcb.140.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Longtine M S, DeMarini D J, Valencik M L, Al-Awar O S, Fares H, De Virgilio C, Pringle J R. The septins: roles in cytokinesis and other processes. Curr Opin Cell Biol. 1996;8:106–119. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- 44.Longtine M S, McKenzie III A, Demarini D J, Shah N G, Wach A, Brachat A, Philippsen P, Pringle J R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 45.Madden K, Snyder M. Cell polarity and morphogenesis in budding yeast. Annu Rev Microbiol. 1998;52:687–744. doi: 10.1146/annurev.micro.52.1.687. [DOI] [PubMed] [Google Scholar]
- 46.Matsui Y, Toh-e A. Yeast RHO3 and RHO4 ras superfamily genes are necessary for bud growth, and their defect is suppressed by a high dose of bud formation genes CDC42 and BEM1. Mol Cell Biol. 1992;12:5690–5699. doi: 10.1128/mcb.12.12.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller R K, Matheos D, Rose M D. The cortical localization of the microtubule orientation protein, Kar9p, is dependent upon actin and proteins required for polarization. J Cell Biol. 1999;144:963–975. doi: 10.1083/jcb.144.5.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller R K, Rose M D. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J Cell Biol. 1998;140:377–390. doi: 10.1083/jcb.140.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mulholland J, Preuss D, Moon A, Wong A, Drubin D, Botstein D. Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J Cell Biol. 1994;125:381–391. doi: 10.1083/jcb.125.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nonaka H, Tanaka K, Hirano H, Fujiwara T, Kohno H, Umikawa M, Mino A, Takai Y. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J. 1995;14:5931–5938. doi: 10.1002/j.1460-2075.1995.tb00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Novick P, Botstein D. Phenotypic analysis of temperature-sensitive yeast actin mutants. Cell. 1985;40:405–416. doi: 10.1016/0092-8674(85)90154-0. [DOI] [PubMed] [Google Scholar]
- 52.Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- 53.Ozaki K, Tanaka K, Imamura H, Hihara T, Kameyama T, Nonaka H, Hirano H, Matsuura Y, Takai Y. Rom1p and Rom2p are GDP/GTP exchange proteins (GEPs) for the Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 1996;15:2196–2207. [PMC free article] [PubMed] [Google Scholar]
- 54.Petersen J, Nielsen O, Egel R, Hagan I M. FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J Cell Biol. 1998;141:1217–1228. doi: 10.1083/jcb.141.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pringle J R, Preston R A, Adams A E, Stearns T, Drubin D G, Haarer B K, Jones E W. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- 56.Qadota H, Python C P, Inoue S B, Arisawa M, Anraku Y, Zheng Y, Watanabe T, Levin D E, Ohya Y. Identification of yeast Rho1p GTPase as a regulatory subunit of 1, 3-beta-glucan synthase. Science. 1996;272:279–281. doi: 10.1126/science.272.5259.279. [DOI] [PubMed] [Google Scholar]
- 57.Robinson N G G, Guo L, Imai J, Toh-e A, Matsui Y, Tamanoi F. Rho3 of Saccharomyces cerevisiae, which regulates the actin cytoskeleton and exocytosis, is a GTPase which interacts with Myo2 and Exo70. Mol Cell Biol. 1999;19:3580–3587. doi: 10.1128/mcb.19.5.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 59.Shaw S L, Yeh E, Maddox P, Salmon E D, Bloom K. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J Cell Biol. 1997;139:985–994. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Snyder M. The SPA2 protein of yeast localizes to sites of cell growth. J Cell Biol. 1989;108:1419–1429. doi: 10.1083/jcb.108.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waddle J A, Karpova T S, Waterston R H, Cooper J A. Movement of cortical actin patches in yeast. J Cell Biol. 1996;132:861–870. doi: 10.1083/jcb.132.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wasserman S. FH proteins as cytoskeletal organizers. Trends Cell Biol. 1998;8:111–115. doi: 10.1016/s0962-8924(97)01217-8. [DOI] [PubMed] [Google Scholar]
- 64.Yamochi W, Tanaka K, Nonaka H, Maeda A, Musha T, Takai Y. Growth site localization of Rho1 small GTP-binding protein and its involvement in bud formation in Saccharomyces cerevisiae. J Cell Biol. 1994;125:1077–1093. doi: 10.1083/jcb.125.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yeh E, Skibbens R V, Cheng J W, Salmon E D, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zahner J E, Harkins H A, Pringle J R. Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1857–1870. doi: 10.1128/mcb.16.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]