Abstract
Vaccination and the emergence of SARS-CoV-2 variants mark the second year of the pandemic. Variants have amino acid mutations at the spike region, a viral protein central in the understanding of COVID-19 pathogenesis and vaccine response. Variants may dominate local epidemics, as Gamma (P.1) in Brazil, emerging in 2020 and prevailing until mid-2021. Different obstacles hinder a wider use of Next-Generation Sequencing for genomic surveillance. We describe Sanger based sequencing protocols: i) Semi-nested RT-PCR covering up to 3.684 kb (>96 %) spike gene; ii) One-Step RT-PCR for key Receptor Binding Domain (RBD) mutations (codons 417–501); iii) One-Step RT-PCR of partial N region to improve genomic capability. Protocols use leftovers of RNA extracted from nasopharyngeal swabs for quantitative RT-PCR diagnosis; with retro-transcribed DNA sequenced at ABI 3500 using dye termination chemistry. Analyses of sequences from 95 individuals (late 2020/early 2021) identified extensive amino acid variation, 57 % with at least one key mutation at the Receptor Binding Domain, with B.1.1.28 lineage most prevalent, followed by Gamma and Zeta variants, with no Delta variant observed. The relatively low cost and simplicity may provide an accessible tool to improve surveillance of SARS-CoV-2 evolution, monitor new variants and vaccinated breakthroughs.
Keywords: SARS-CoV-2, COVID-19, Variants, Gamma, Brazil, Genomic surveillance
1. Introduction
As the COVID-19 pandemic entered its second year, optimism emerging from vaccination coverage and easing restrictions in the developed world occur amid an unsettled scenario of new infection waves in some areas, initially in resource-limited settings. New variants have been increasingly detected and some have been associated with these surges in incidence (Kirby, 2021; Mallapaty, 2021). Certain viral variations have been associated with enhanced infectivity (Korber et al., 2020; Khan et al., 2021), transmissibility (Kirby, 2021), and reinfection potential (Naveca et al., 2020; Harrington et al., 2021). Although the severity of disease and mortality may be influenced by an overstretched health care capacity, some studies suggested also an increase in the lethality of some variants (Public Health England, 2021). WHO has proposed a nomenclature for the variants of concern (VOC) and variants of interest (VOI), and provided some guidelines to improve surveillance to SARS-CoV-2 and to better prepare for future threats (WHO, 2021). With the growing complexity of variants new categories, as Variant Under Monitoring (VUM) are also being used for variants with less robust (suspected) evidence for increase transmissibility, impact on virulence, interference with diagnostic performance and/or therapeutic / vaccine efficacy. Specific surveillance of Mutations (Mutations of Concern or Mutations of Interest) are also targets of surveillance. Variants importance may be de-escalated by public health agencies based on at least one the following criteria: (1) the variant is no longer circulating, (2) the variant has been circulating for a long time without any impact on the overall epidemiological situation, (3) scientific evidence demonstrates that the variant is not associated with any concerning properties (Mullen et al., 2020).The swiftness that variations take over SARS-CoV-2 circulating in different areas is overwhelming. In few months, variants that represent minority infections may become almost unique, as happened with the variant of concern (VOC) Gamma (P.1) in Brazil in the first half of 2021,as had occurred for the S:D614G mutation worldwide (Mullen et al., 2020).
Information on viral variation is also needed to properly control during vaccine escalation and to monitor breakthroughs after adequate vaccination coverage, as the impact of current and future variants in vaccine response may be key to pandemic control. Additional concerns were elicited from in vitro neutralization studies showing a reduction in neutralization by some monoclonal antibody treatments, convalescent, and post-vaccination sera (Liu et al., 2021; Garcia-Beltran et al., 2021; Edara et al., 2021; CDC, 2021a), as well as evidence from vaccine trials (Madhi et al., 2021). Therefore, molecular epidemiology has a role not only in monitoring the evolution of the virus but also to inform on the potential impact of viral variation on monoclonal antibodies therapies as well as in vaccine strategies.
Sequencing of isolates is a key to inform on viral variation. Next-Generation Sequencing (NGS) is the standard technique to study SARS-CoV-2 genome variability. However, the worldwide capacity is limited, the technique demands specific requirements and it is not available in many areas. Real-time PCR protocols have been developed and may identify specific mutations, but unpredictable SARS-CoV-2 variability affects tests sensitivity (Peñarrubia, 2021). Sanger protocol, central to molecular epidemiology of HIV and other agents before the NGS area, may provide genomic sequencing information and complement NGS. Sanger can both help in screening samples for further NGS analysis as well as provide an alternative, simpler tool, to obtain key information from RNA leftovers of RNA test, allowing more extensive sequencing as in case-control and other studies at a local level. This information may be integrated into large-scale epidemiological surveillance based on a random sampling of SARS-CoV-2 positive cases.
We developed simple protocols using both semi-nested and One-Step RT-PCR to amplify DNA and robust Sanger sequencing protocols targeting relevant genetic regions of the SARS-CoV-2, as Spike and orf8-Nucleocapsid (N) protein.
2. Material and methods
2.1. Sample collection
Nasopharyngeal swab (NPS) samples, collected for diagnosis proposes, were processed at the Virology Center or Santos Regional Adolfo Lutz Center. All samples were tested by quantitative RT-PCR (qRT-PCR) and confirmed as SARS-CoV-2 infection.
2.2. Nucleic acid extraction
SARS-CoV-2 RNA was extracted from NPS samples with QIAmp® viral RNA mini kit (Qiagen, Hilden, Germany), BioGene kit (Bioclean Quibasa, MG, Brazil), or Loccus kit (Loccus, SP, Brazil) according to manufacturer’s protocol. Extraction followed the ongoing diagnosis routines at the laboratories and RNA leftovers from these routines were kept at -70 °C until use.
2.3. Diagnosis qRT-PCR method
Two assays, based on multiplex qRT-PCR that enables simultaneous amplification and detection of target nucleic acids of SARS-CoV-2, were used for diagnosis propose: i) Manual of Kit Molecular SARS-CoV-2 (E/RP) Bio-Manguinhos (2021); and ii) Manual of Allplex™ SARS-CoV-2 Assay, 2021 (Allplex™, Seegene, Korea).
The molecular SARS−COV-2 (E/RP) Bio-Manguinhos assay is intended for the diagnosis and epidemiological surveillance of Coronavirus. The primers and probes sequences of the SARS−COV-2 E gene and RNAse P (internal control) are from the Berlin Protocol (Charité/Berlin-WHO) (Corman et al., 2020), executed in a single qRT-PCR assay. The Manual of Allplex™ SARS-CoV-2 Assay, 2021 (Allplex™, Seegene, Korea) enables simultaneous amplification and detection of E, RdRP, and N gene of SARS-CoV-2 and internal control.
In both assays, the internal control was used to track the entire process from, nucleic acid extraction, and to verify any possible PCR inhibition.
Clinical samples were considered positive at Allplex™ kit and Bio-Manguinhos when at least one gene was detected with cycle threshold (Ct) under 40.
2.4. Primers design
In order to design primer suitable PCR sets to partial SARS-CoV-2 Spike (S1 and S2 region) protein, 3 sequences (NC_045512.2); SARS-CoV-2 Wuhan-Hu-1, and two sequences from the first case reported in Brazil, MT350282.1 and MT126808.1; (Araujo, 2020) were obtained from NCBI and imported into BioEdit sequence alignment editor (version 7.0.5.2) program. The N primer sequences were obtained from WHO.
The process of primer designing was conducted manually, without automated software packages. The primers were designed to conduct a nested reverse transcription-polymerase chain reaction (RT-PCR) protocol.
2.5. RT-PCR and semi-nested PCR protocols
To amplify the spike and nucleocapsid (N) regions, three protocols were used: i) long protocol suitable to amplify subunit S1 and partial S2 protein (Fig. 1 a); ii) short protocol, used to obtain part of the RBD region at S1 subunit (Fig. 1b) and iii) N gene protocol, that amplifies partial N gene (Fig. 1c). All reactions were performed in dedicated equipment at separate pre and post amplification areas.
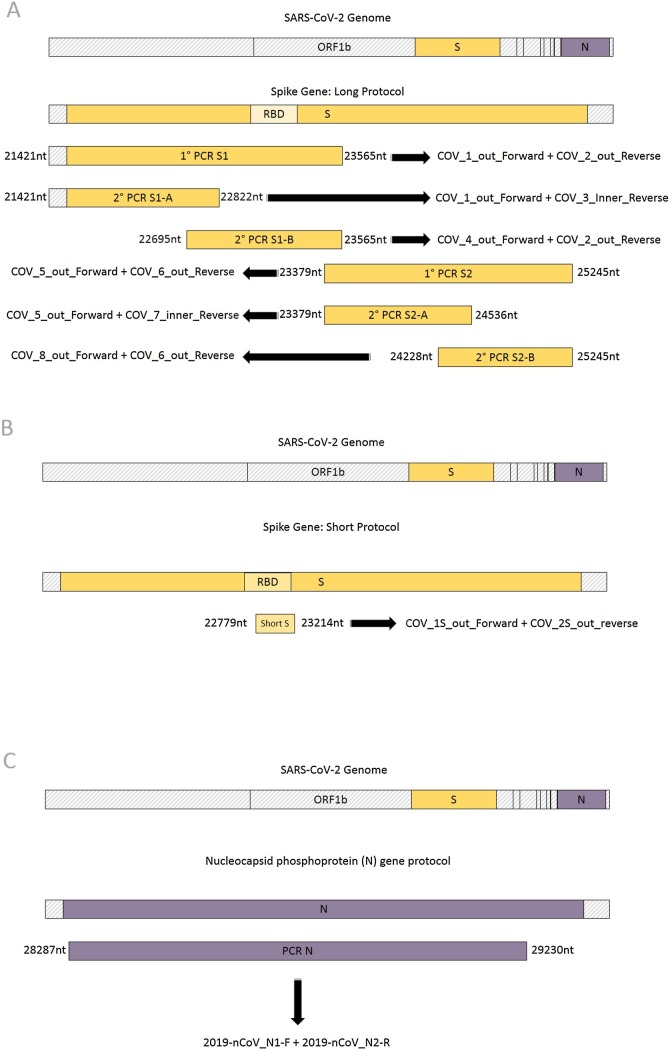
Fig. 1.
Schematic representation of PCR amplified fragments using three protocols described in the methodology. Illustrative sketch of the A) long protocol to obtain most of the spike region of SARS-CoV-2, with primers annealing region and the expected size of the amplified fragment; B) short protocol, to obtain part of the spike region of SARS-CoV-2, covering the Receptor Binding Domain and the; C) protocol to obtain the nucleocapsid (N) region of SARS-CoV-2.
2.5.1. Spike gene: long protocol
This protocol allows the amplification of up 96 % (up to 3684 bp of the 3821 bp of SARS-CoV-2 Wuhan-Hu-1 strain) comprising nucleotide position 21563–25245 nt, of spike gene using two One-Step RT-PCR. Tocover S1 and S2 subunits four semi-nested PCR: S1-A, S1-B, S2-A,and S2-B are used (Fig. 1a). In spike S1 protein gene amplification, the primers set used were: i) First round (One-Step RT-PCR); COV_1_out_Forward AGGGGTACTGCTGTTATGTC (21421−21440 nt) and COV_2_out_Reverse GCACCAATGGGTATGTCACA (23546−23565 nt), resulting in a 2144 bp product; and ii) Second round (semi-nested PCR) COV_1_out_Forward AGGGGTACTGCTGTTATGTC (21421−21440 nt) and COV_3_inner_Reverse ATCAGCAATCTTTCCAGTTTGC (22801−22822 nt), generating a fragment of 1401bp (S1-A). COV_4_out_Forward AGTGTTATGGAGTGTCTCCTACT (22695−22717 nt) COV_2_out_Reverse GCACCAATGGGTATGTCACA (23546−23565 nt) generating a fragment of 870 bp (S1-B). Primer pairs allowed the amplification of S1 protein, comprising nucleotide position 21599–23185 nt based on SARS-CoV-2 Wuhan-Hu-1 strain (accession number NC_045512.2).
In spike S2 protein gene amplification, the primers set used were: i) First round (One-Step RT-PCR); COV_5_out_Forward ACCAGGTTGCTGTTCTTTATCAG (23379−23401 nt) and COV_6_out_Reverse ACTATGGCAATCAAGCCAGCT (25225−25245 nt), producing an 1866 bp fragment; and ii) second round (semi-nested PCR) COV_5_out_Forward ACCAGGTTGCTGTTCTTTATCAG (23379−23401 nt) and COV_7_inner_Reverse GCACTTCAGCCTCAACTTTGT (24516−24536 nt), generating a fragment of 1157 bp (S2-A) and; ii) COV_8_out_forward GTGCAGGTGCTGCATTACA (24228–24246 nt) and COV_6_out_reverse ACTATGGCAATCAAGCCAGCT (25225−25245 nt) generating a fragment of 1017 bp each (S2-B). These pairs of primers permitted the partial amplification of the S2 protein, corresponding to 1866 bp (nucleotide position 23379–25245 comprising to 622 amino acid) compared to complete S2 region 2190 bp (nucleotide position 23194–25384 comprising 730 amino acid) based on SARS-CoV-2 Wuhan-Hu-1 strain (accession number NC_045512).
For One-Step RT-PCR reactions (S1 and S2) the extracted RNA was reverse-transcribed and amplified using SuperScript® III One-Step RT-PCR System with Platinum Taq High Fidelity® (Life Technologies, USA). The total reaction mixture volume of 50 μL contained the following: 2x reaction mix (25 μL), 10 μM primers (1 μL each), enzyme mix (reverse transcriptase and Taq polymerase, 1 μL), extracted viral RNA template (10 μL), and RNase-free water (12 μL). RT-PCR conditions for amplification were as follows: reverse transcription at 50 °C for 30 min, initial PCR activation at 94 °C for 2 min, 35 amplification cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, extension at 68 °C for 2 min 45 s, and a final extension at 68 °C for 10 min.
The semi-nested PCR reactions (S1-A, S1-B, S2-A, S2-B) the RT-PCR product (2.5 μL), 10 μM primers (1 μL each), and RNase-free water (8 μL) were added to a Go Taq® Green Master Mix 2X (12.5 μL) (Promega Biosciences, USA). PCR conditions were as follows: initial denaturation at 94 °C for 3 min, 35 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s extension at 72 °C for 1 min 30 s, and a final extension at 72 °C for 10 min.
2.5.2. Spike gene: short protocol
To allow the sequence of a minimally informative region of the spike gene, including already described mutations of current variants at codons as 417, 452, 477, 478, 484, and 501. To monitor these mutations we designed a even simpler, low-cost, One-Step RT-PCR, followed by two cycle sequencing reactions that covers the region of these mutations.
This protocol allows the amplification of up to 436 bp of the spike protein, comprising nucleotide position 22779–23214 nt (based on Wuhan-CoV-2 strain). For the amplification, the primers set used were: COV_1S_out_AAGTCAGACAAATCGCTCCAG (22779−22799 nt) and COV_2S_out_reverse ACACCTGTGCCTGTTAAACC (23195−23214 nt). (Fig. 1b).
For the One-Step reaction, the extracted RNA was reverse-transcribed and amplified using SuperScript® III One-Step RT-PCR system with Platinum Taq High Fidelity® (Life Technologies, USA). The total reaction mixture volume of 50 μL contained the following: 2x reaction mix (25 μL), 10 μM primers (1 μL each), enzyme mix (reverse transcriptase and Taq polymerase, 1 μL), extracted viral RNA template (10 μL), and RNase-free water (12 μL). RT-PCR conditions for amplification were as follows: reverse transcription at 50 °C for 30 min, 94 °C for 2 min, 35 amplification cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, extension at 68 °C for 2 min, and a final extension at 68 °C for 10 min.
2.5.3. Nucleocapsid (N) gene protocol
This protocol allows the amplification of up 943 bp, up to 75 %, of partial N gene, comprising nucleotide position 28287–29230 (position of N gene on Wuhan-CoV-2 strain 28274–29533) (Fig. 1c). The protocol may be used as a supportive tool for cases in which the SARS-CoV-2 variant could not be classified with genomic information from spike region and to improve signal for phylogeny. For the amplification, the primers set used were: 2019-nCoV_N1-F_GACCCCAAAATCAGCGAAAT (28287−28306 nt) and 2019-nCoV_N2-R_GCGCGACATTCCGAAGAA (29213−29230 nt) (WHO primer suggestion based on Wuhan-CoV-2 strain).
For One-Step RT-PCR reaction, it was used the same spike gene long protocol products, volume, and PCR conditions: the extracted RNA was reverse-transcribed and amplified using SuperScript® III One-Step RT-PCR system with Platinum Taq High Fidelity® (Life Technologies, USA). The total reaction mixture volume of 50 μL contained the following: 2x reaction mix (25 μL), 10 μM primers (1 μL each), enzyme mix (reverse transcriptase and Taq polymerase, 1 μL), extracted viral RNA template (10 μL), and RNase-free water (12 μL). RT-PCR conditions for amplification were as follows: reverse transcription at 50 °C for 30 min, 94 °C for 2 min, 35 amplification cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, extension at 68 °C for 2 min 45 s, and a final extension at 68 °C for 10 min.
2.6. Sequencing
The 3684 bp and 436 bp PCR products of respectively, long and short protocols of spike gene and the 943 bp PCR product of partial N gene were sequenced using two or four primers for each region (Table 1 ). Each sequencing reaction was performed using 2.5−20 ng of PCR product (lower input for the small fragments and higher input for the longer fragments), 0.5 μL of BigDye Terminator v3.1 cycle sequencing kit® (Applied Biosystems, USA), 4 μL of 2.5x Sequencing Buffer (Applied Biosystems, USA), and 1.6 μL for each 1 μM primer and water to a final volume 10 μL per reaction. Dye-labeled products were sequenced using a Genetic Analyzer ABI 3500 (Applied Biosystems, USA). Sequencing chromatograms were edited manually using Sequencher 4.7 software (Gene Codes, USA).
Table 1.
Sars-CoV-2 virus partial Spike (S1 and S2) region and partial N region primer sets designed for Sanger.
| Primers | Sequence (5′ - 3′) | Location (nt) | Region | Protocol |
|---|---|---|---|---|
| COV_1_out_Forward | AGGGGTACTGCTGTTATGTC | 21421−21440 | S1-A | Amplification and sequencing |
| COV_11_out_Forward | AGAGGCTGGATTTTTGGTACTACT | 21866−21889 | S1-A | Sequencing |
| COV_12_inner_Reverse | GCTGTCCAACCTGAAGAAGAATC | 22319−22341 | S1-A | Sequencing |
| COV_3_inner_Reverse | ATCAGCAATCTTTCCAGTTTGC | 22801−22822 | S1-A | Amplification and sequencing |
| COV_4_out_Forward | AGTGTTATGGAGTGTCTCCTACT | 22695−22717 | S1-B | Amplification and sequencing |
| COV_13_out_Forward | GGCTGCGTTATAGCTTGGAAT | 22853−22873 | S1-B | Sequencing |
| COV_14_inner_Reverse | ACACCTGTGCCTGTTAAACC | 23195−23214 | S1-B | Sequencing |
| COV_2_out_Reverse | GCACCAATGGGTATGTCACA | 23546−23565 | S1-B | Amplification and sequencing |
| COV_5_out_Forward | ACCAGGTTGCTGTTCTTTATCAG | 23379−23401 | S2-A | Amplification and sequencing |
| COV_9_out_Forward | CCGTGCTTTAACTGGAATAGCT | 23854−23875 | S2-A | Sequencing |
| COV_10_out_Reverse | TGACCTCTTGCTTGGTTTTGA | 23990−24010 | S2-A | Sequencing |
| COV_7_inner_Reverse | GCACTTCAGCCTCAACTTTGT | 24516−24536 | S2-A | Amplification and sequencing |
| COV_8_out_Forward | GTGCAGGTGCTGCATTACA | 24,228–24,246 | S2-B | Amplification and sequencing |
| COV_15_inner_Reverse | CAAACCAGTGTGTGCCATTTG | 24852−24872 | S2-B | Sequencing |
| COV_16_out_Forward | AGCTTCTGCTAATCTTGCTGC | 24619−24639 | S2-B | Sequencing |
| COV_6_out_Reverse | ACTATGGCAATCAAGCCAGCT | 25225−25245 | S2-B | Amplification and sequencing |
| COV_1S_out_Forward | AAGTCAGACAAATCGCTCCAG | 22779−22799 | S1 | Amplification and sequencing |
| COV_2S_out_Reverse | ACACCTGTGCCTGTTAAACC | 23195−23214 | S1 | Amplification and sequencing |
| 2019-nCoV_N1-F* | GACCCCAAAATCAGCGAAAT | 28287−28306 | N | Amplification and sequencing |
| 2019-nCoV_N2-R* | GCGCGACATTCCGAAGAA | 29213−29230 | N | Amplification and sequencing |
| 2019-nCoV_N3-F* | GGGAGCCTTGAATACACCAAAA | 28681−28702 | N | Sequencing |
| 2019-nCoV_N3-R* | TGTAGCACGATTGCAGCATTG | 28732−28752 | N | Sequencing |
nt: Nucleotide.
Primer obtained form WHO.
2.7. Variant definition
Sequences were analyzed using GISAID (https://www.gisaid.org/epiflu-applications/covsurver-mutations-app/; Elbe and Buckland-Merrett, 2017), and at OUTBREAK platform (https://outbreak.info/; Mullen et al., 2020).
2.8. Statistical analysis
Continuous variables were described as median and 25th-75th interquartile range (IQR) unless noted, and differences among groups evaluated with Mann Whitney or Kruskal Wallis rank tests. Fischer exact test, two-tailed, was used for dichotomized or categorical variables. The proportion of detected mutations was calculated using as denominator only sequences that included the region of the respective amino acid. The lower Ct values obtained from the original real-time PCR reaction performed for diagnostic purposes. The lower value of one to two or three (N, RdRp, E) targets at each reaction was used to evaluate sequencing performance. A phylogenetic tree was build using Bayesian phylogenetic tree inference implemented with BEAST v.1.7.4 under GTR (G + I) with concatenated spike and nucleocapsid regions of SARS-CoV-2 partial sequences joined through BioEdit, along with references sequences obtained at the GISAID platform. Sequences were aligned with ClustalW. A coronavirus HKU1 sequence was used as outgroup.
2.9. Ethical approval
This study was carried out following the Declaration of Helsinki as revised in 2000 and approved by the Ethics Committee of the Adolfo Lutz Institute, Sao Paulo, Brazil. The study was registered at the Institute, CTC 18 M/2020 and CTC 39 M/2020 and at the institutional ethical committee - CAAE: 31924420.8.0000.0059 and CAAE: 43250620.4.1001.0059.
All study participants were tested for SARS-CoV-2 at a public laboratory and has results made available to patients through the federal health laboratory data management system. Samples from those that were not reached to provide informed consent had data anonymized before analysis and information used only for surveillance purposes.
3. Results
We evaluated 113 RNA extracted NPS samples, obtaining 95 sequences from individuals living in the coastal cities of Sao Paulo (63 % - 60/95) and other areas of Sao Paulo (37 % - 35/95) (Table 2 ). Collection of most samples was in January or February 2021, with 6 samples from 2020. The median age of patients was 41 (24–50) years old and 59 % female. At the time of the study, a symptomatic clinical setting was required for testing. Symptoms occurred 4 days (3–5) before the collection date.
Table 2.
City of collection of samples.
| All sequences (n = 95) | |
|---|---|
| Coastal cities | 60 (63 %) |
| Guaruja | 38 (40 %) |
| Praia Grande | 8 (8%) |
| Santos | 8 (8%) |
| Other coastal cities | 6 (7%) |
| Sao Paulo Metropolitan area | 35 (37 %) |
| Sao Paulo | 8 (8%) |
| Francisco Morato | 5 (6%) |
| Caieras | 3 (3%) |
| Taboao da Serra | 3 (3%) |
| Other Cities | 16 (17 %) |
Distribution of samples analyzed according to geographic area in the State of São Paulo as coastal cities (Baixada Santista), cities at the metropolitan area of São Paulo and others within the State.
3.1. High Ct values were related to a lower performance of the short protocol
Overall, the protocols described in this study were able to amplify the spike protein gene in 84 % (95/113). We tested samples with a Ct lower than 30 to obtain the longer, semi-nested spike sequences, with 93 % (67/72) of success. Aiming to test the sensitivity of a single One-Step PCR protocol, we included samples with higher Ct, up to 32, and performed the shorter sequences. Lower performance was observed with 63 % (32/51) of positivity. The impact of Ct was more pronounced for this single One-Step protocol, with no sequence obtained in cases with Ct above 27, whereas the semi-nested protocols up to 30. For the nested PCR, only one sample with high Ct (Ct>36) was tested and not amplified. Although we did not evaluate the actual sample Ct from the extracted material used in the reaction, the Ct value initially obtained for diagnosis purposes was associated with the success of amplification and sequencing, with more positive reactions for samples with lower Ct (19, IQR 17–22) vs. higher Ct (31, IQR 23–32) (p < 0.001). Using the median Ct of the samples (20) to dichotomize cases, a lower Ct was associated with sequencing success in 94 % of attempts vs. 75 % (p = 0.001).
3.2. Spike gene: long protocol
The Spike gene long protocol had 93 % (67/72) positivity for some S1 or S2 fragments, and a fragment covering at least codons 417–1176 was obtained in 84 % (56/67) samples. From all sequences, 34 % (23/67) are B.1.1.28 variants, Gamma in 27 % (18/67), Zeta in 19 % (13/67), and no clear variant could be defined in 13 (19 %) of these spike sequences. Eleven sequences did not have enough fragment coverage for proper analysis.
3.3. Nucleocapsid (N) gene protocol
Partial N protocol was used as a supportive tool to provide or confirm variant classification. This protocol was used in eight cases of which 4 were not classified with spike gene long protocol, one sequence with an atypical Gamma pattern of spike variant mutation (see below), and 3 already classified as Gamma that had failed at the N gene reaction of qRT-PCR. Four P.1 were reconfirmed, including the atypical Gamma pattern of spike variant mutation, two sequences as Zeta variant and another two remaining unclassified.
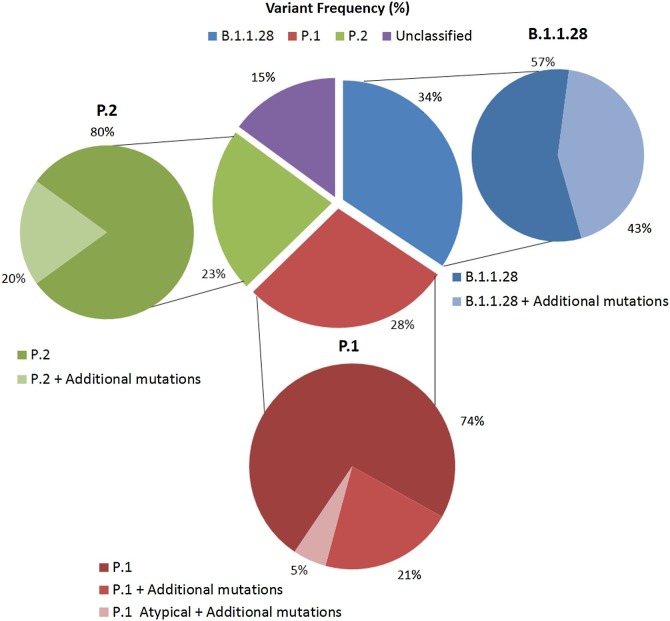
Considering all spike sequences (n = 67) with the addition of N sequences, as observed in Fig. 2 , 34 % (23/67) were classified as B.1.1.28 variant, followed by P.1 (Gamma variant) 28 % (19/67), P.2 (Zeta variant) 23 % (15/67) and 15 % (10/67) remained unclassified.
Fig. 2.
Variant classification of SARS-CoV-2 sequences obtained in our study, most collected in January and February 2021. Sequences were further classified according to the absence or presence of additional mutations, not associated with the variant definition.
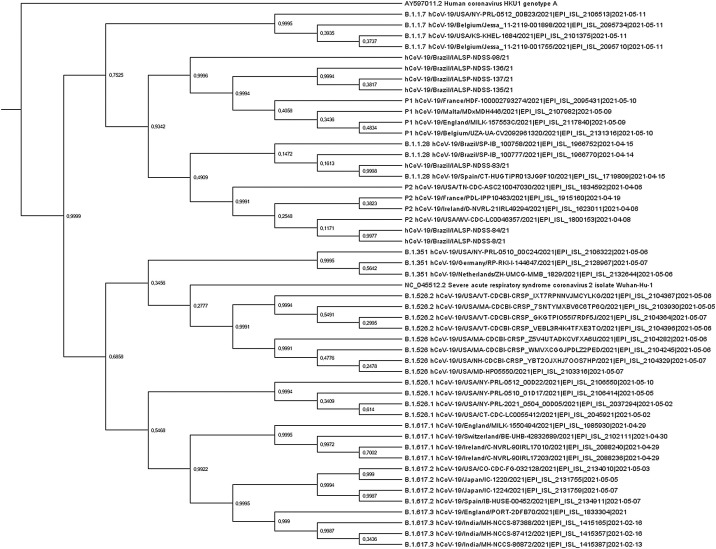
Concatenated spike and N regions evaluated in phylogenetic trees along with different variants obtained from GISAID. As shown in Fig. 3 , the combined spike and N regions provide reasonable phylogenetic signal and allow the discrimination of most variants with high posterior probability.
Fig. 3.
Bayesian phylogenetic tree inference implemented with BEAST v.1.7.4 under GTR (G + I) with concatenated Spike and Nucleocapsid regions of SARS-CoV-2 partial sequences joined through BioEdit, along with references sequences obtained at the GISAID platform. The sequences were aligned with ClustalW. A coronavirus HKU1 sequence was used as outgroup.
3.4. Spike gene: short protocol
Sequences covering amino acids 407–550 using the short protocol for spike gene region were obtained in 32/51 (63 %) of cases.
Considering these sequences (n = 32) along with those from the spike long protocol that covered this same region (n = 57), 89 sequences had this segment covered. Most sequences (57 %) shown some mutation at this segment, 56 % (50/89) was carrying the E484K mutation, common to Gamma and Zeta variants, and 24 % (21/89) shown all three Gamma related mutations (K417T, E484K, and N501Y). However, none with mutations at amino acid 452 (Variant Epsilon, de-escalated), 478 (VOC Delta), and or with the mutation to asparagine at amino acid 417, characteristic of the Beta variant.
3.5. A diversity of additional mutations was observed among variants
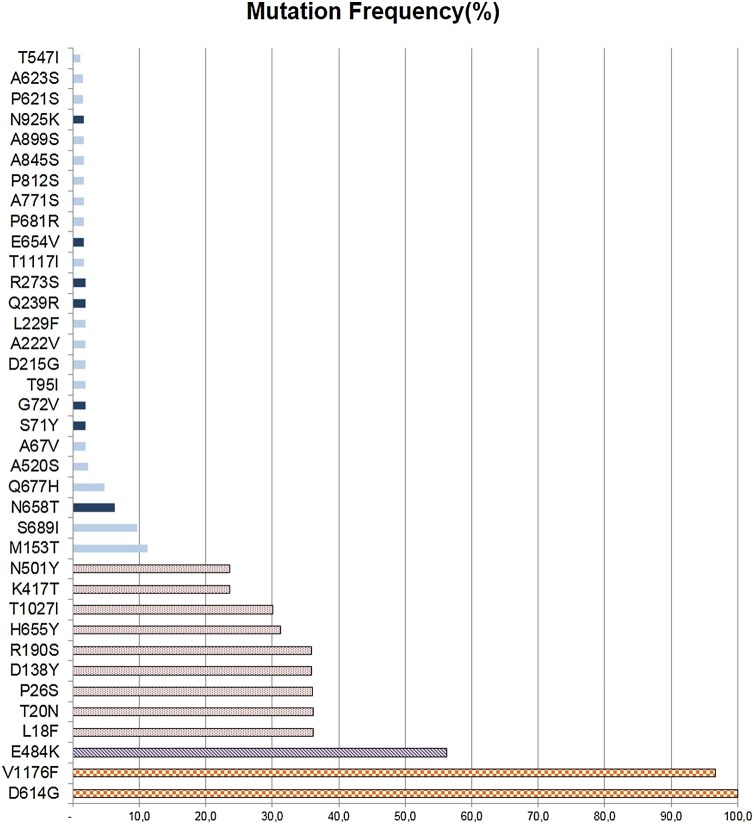
The presence of one or more additional mutations not related to lineage or variant definition was common. Most 43 % (10/23) samples of B.1.1.28 lineage had at least one additional mutation, most at codons 153 and 689. The proportion of additional mutations at Gamma 26 % (5/19) and Zeta 20 % (3/15) was also important. All sequences had the D614G mutation. To test if the proportion of additional variation on Gamma of this study (26 %) was distinct from that observed in other Gamma sequences, we compared our dataset to that of GISAID submission in January 2021 (n = 290 Gamma sequences). After excluding 29 with more than 5% unresolved nucleotides (N), 16 % (41/261) had one or more additional mutations, not significantly different from the 26 % (5/19) observed in our data set (p = 0.3). Fig. 4 lists the observed mutations from our dataset.
Fig. 4.
Frequency of SARS-CoV-2 mutations detected in spike protein according to the variant classification of sequences. Solid Light Blue: Unique mutation; Solid Dark Blue: Existing mutations; Dotted Red: P.1 variant mutations; Dashed Purple: Common mutations to P.1 and P.2 variants; Grid Orange: common mutations to P.1, P.2, and B.1.1.28. Mutations A845S, N925K, G72V, and L229F were present at P.1 variant; T547I, A623S, A899S, and P812S at P.2 variant; P621S, E654V, A771S, T1117I, A67V, T95I, D215G, A222V, Q239R, R273S, Q677H, N658T, S689I and M153T at B.1.1.28 variant.
Sequences GISAID accession numbers are: EPI_ISL_1182095 to EPI_ISL_1182104, EPI_ISL_1191781, EPI_ISL_2458064 to EPI_ISL_2458109, EPI_ISL_2467837 to EPI_ISL_2467864 and EPI_ISL_2474178 to EPI_ISL_2474186, EPI_ISL_2614385 and EPI_ISL_2614386.
4. Discussion
In this study, we describe simple Sanger protocols that allow sequencing partial spike and N regions of the SARS-CoV-2 genome and may help to study viral diversity. Three protocols are described: i) a spike short protocol covering key amino acid mutations at RBD; ii) a spike long protocol covering up to 96 % of spike gene; iii) a partial N protocol to improve variants discrimination and to investigate cases with real-time PCR failure in N gene reactions.
The short protocol represents a rapid and lower-cost alternative (about 25 US dollars) to identify key mutations associated with high transmissibility, infection, and mortality. It includes most of the RBD and covers major mutations associated with the most relevant variants of concern (VOC),variants of interest (VOI) and variants under monitoring (VUM), all listed at the outbreak tool (https://outbreak.info/situation-reports) and at international agencies (WHO, 2021; CDC, 2021a,b). It can be a tool for screening variants as P.1 (Gamma), which has three mutations, at this fragment (K417T, E484K, and N501Y). It can also monitor mutations associated with newer variants of concern, as the L452R, and E484Q (VOC Delta, B.1.617.2) initially described in India and mutations found in the US isolates as the S477 N (CDC, 2021a,b). It also allows the discrimination of South African B.1.351 (Beta) amino acid substitution, Asparagine at position 417 of Spike protein, whereas the Gamma, the Brazilian P.1 variant, has a mutation to amino acid Threonine.
Misincorporation of base pairs during PCR, more commonly A to G and T to C transitions (Potapov and Ong, 2017), may lead to the identification of artificial amino acid signatures that do not reflect the actual viral genome. The use of high fidelity in all these protocols and at first round of the longer spike protocol minimizes this risk. Misidentification of variants that are defined by different amino acid signatures are very unlikely to occur due to PCR misincorporation, and therefore the full mutational profile of a VOC has a high probability of correctness. However, individual, isolated amino acid mutations, especially due to single nucleotide substitution, may be confirmed by a new, independent PCR reaction (using the extracted RNA) or by new viral isolation and sequencing from the patient or potential secondary infections that may carry a similar variant.
The protocol developed to obtain a longer segment of Spike covers almost the full Spike (up to 96 %) region and allows not only identify amino acid substitutions associated with new and emerging variants, but also can be useful to track the evolution of spike gene in a vaccinated population.
The fact that the P.1 (Gamma) variant was unnoticed in Brazil until identified in Tokyo airport, Japan, from passengers coming from Manaus, the major city at the Amazon Basin, Brazil, and only subsequently documented as widespread in this region illustrates the fragility of current surveillance capability in some resource-limited settings. When selecting samples to test our protocols, Gamma was described in some cities of the State of Sao Paulo, but not yet at coastal cities. After we documented this variant in the coastal cities of Sao Paulo (Cabral et al., 2021), Gamma has come to dominate the pandemic in Sao Paulo and other areas of Brazil (WHO, 2021). This scenario changed at the second half of 2021, with the Delta variant becoming the most prevalence. At our small dataset, Gamma related cases increased from zero in 2020 to 19 % in January and 36 % in February (p = 0.06), illustrating the speed that a variant may substitute older SARS-CoV-2 circulating variants.
In one case the sequenced spike gene harbored all mutations previously described for the Gamma variant but lacked the three (K417 T, E484 K, N501Y) mutations that have been associated with different biological advantages to the virus. Variations like this couldincrease with time, and local level monitor capability may provide additional layers of control before widespread dissemination occurs.
Overall, we found a high proportion of mutations at spike protein, with most of these sequences harboring at least one mutation. We also noticed that the B.1.1.28 lineage, present for a longer time in the region, showed more additional mutations (43 % - 10/23) than Gamma (26 % - 5/19) and Zeta (20 % - 3/15) variants (Fig. 4). This also suggests that these new variants are evolving fast in this region due to the high current levels of transmission in Brazil and elsewhere. This scenario favors the selection of variants that may show immune escape potential and/or longer binding to cell receptors. Variants prevalence and profile of additional mutations observed changes very fast and today these lineages are no longer predominant in the region. Without proper monitoring and non-pharmacological measures, newer variants may emerge and affect vaccine strategies.
Spike sequencing may be sufficient to classify variants, but in other instances, it may be necessary to add more sequencing information. Moreover, the spike region has an extensive length (3.8 kb) and in some cases, entire sequences cannot be fully obtained. The P.1 (Gamma variant), for example, has a complex mutational pattern of 12 amino acids changes occurring all along the spike region, and even without complementary information from other regions as the N region, it may alone support variant assignment. Sequences from these two viral genes may complement each other in an effort to improve viral surveillance, as the nucleoprotein genomic regions also concentrate the bulk of mutations identified in many variants. The partial N protocol help in providing genetic information in cases in which variant could not be classified using the spike long protocol. In a single One-Step methodology was possible to obtain a PCR product and posterior sequences that correspond to up to 75 % of the N region (amino acid 05–319). Partial N sequencing also contributed to a better resolution of phylogenetic analysis, providing additional signal to tree building.
Another utility of the N region protocol could be to inspect sequences for variation associated with failure at diagnostic tests. Real-time PCR protocols have been developed and are able to identify different variants (Peñarrubia et al., 2020), but PCR performance can be compromised if an additional or alternative variation is present in samples. N region is generally an important target for this test, and variations at this region may interfere with primer or probes annealing. Sequencing of nucleocapsidic (N) region is also useful for assessing the reliability of antigenic tests, which detects the virus by means of antibodies binding the N protein, and may be relevant for routine serological diagnosis.
We opt to use leftovers from regular, clinical diagnosis settings to more closely mimic real-world environments where this or a similar procedure could be implemented to improve surveillance capability. However, conditions of preservation of these small extracted RNA volumes and temperature may affect performance in some samples. In addition, the amount remaining of some samples compromised the repetition of reactions. The performance of PCR and sequencing assays tended to improve when extraction for our assays was performed directly from clinical isolates (data not shown).
Albeit marked by simplicity, these methods have a potential informative power for limited settings that may benefit from a simpler approach to genomic surveillance. If in one hand the methodology of a short protocol does not allow proper lineage evaluation, on the other hand, it can provide swift access to information on the presence of key mutations among the samples of a region or a subgroup of the population. If linked to proper contact tracing and preventive measures, it may provide a powerful tool to block variant expansion to new areas and populations.
The spike region seems to be a reasonable target to monitor the variants already identified and new emerging mutations that may give rise to new variants. Therefore, the RBD is the active immunogen of most vaccines products and harbors many of the mutations described in emerging and established variants as seen in the Alpha UK variant (B.1.1.7), the Beta, South African (B.1.351), the Gamma, Brazilian (P.1) and the VOC Delta (B.1.617.2) variants emerging along with the recent surge of cases in India; (CDC, 2021a,b, WHO, 2021). Surveillance cases of infections after immunization will be instrumental to monitor vaccine effectiveness as the evolution of new variants of the virus.
In conclusion, these Sanger protocols provide important information from key regions of the SARS-CoV-2 genome, may identify all major variants and can be a complementary tool in surveillance strategies to help monitor viral evolution and vaccine effectiveness.
Author statement
Gabriela Bastos Cabral1: Methodology, validation, writing original draft, review and editing.
Cintia Mayumi Ahagon2: Methodology, Validation, investigation, formal analysis, writing – review & editing.
Paula Morena de Souza Guimarães2: Validation, investigation, writing – review & editing.
Giselle Ibette Silva Lopez-Lopes2: Data curation, investigation – review & editing.
Igor Mohamed Hussein2: Methodology, Validation, investigation – review & editing.
Audrey Cilli2: Investigation, writing – review & editing.
Ivy de Jesus Alves1: Investigation, Resources.
Andréa Gobetti Coelho Bombonatte1: Investigation, Resources.
Maria do Carmo Sampaio Tavares Timenetsky2: Funding acquisition, Resources.
Jaqueline Helena da Silva Santos2: Investigation.
Katia Corrêa de Oliveira Santos2: Investigation.
Fabiana Cristina Pereira dos Santos2: Investigation.
Luís Fernando de Macedo Brígido2: Conceptualization, project administration, funding acquisition, supervision, formal analysis, visualization, writing – review & editing.
Ethical statement
This study was carried out following the Declaration of Helsinki as revised in 2000 and approved by the Ethics Committee of the Adolfo Lutz Institute, Sao Paulo, Brazil. The study was registered at the Institute, CTC 18 M/2020 and CTC 39 M/2020 and at the institutional ethical committee - CAAE: 31924420.8.0000.0059 and CAAE: 43250620.4.1001.0059.
All study participants were tested for SARS-CoV-2 at a public laboratory and has results made available to patients through the federal health laboratory data management system. Samples from those that were not reached to provide informed consent had data anonymized before analysis and information used only for surveillance purposes.
Funding
Partially funded by FAPESP 2016/25212-9; FAPESP 2018/14384-9; FESIMA CAF: 059/2021.
Declaration of Competing Interest
The authors have not identified potential conflicts of interest.
Data availability
Additional data can be made available upon request.
References
- Cabral G.B., Ahagon C.M., Lopez-Lopes G.I.S., et al. P1 variants and key amino acid mutations at the Spike gene identified using Sanger protocols. medRxiv. 2021 03.21.21253158; Published: 2021 March 24. [Google Scholar]
- Center for Disease Control and Prevention . 2021. Information for Laboratories About Coronavirus (COVID-19) Available at: < https://www.cdc.gov/coronavirus/2019-ncov/lab/virus-requests.html> (Accessed oct 09, ) [Google Scholar]
- Center for Disease Control and Prevention . 2021. Variants and Genomic Surveillance for SARS-CoV-2. Available on: < https://www.cdc.gov/coronavirus/2019-ncov/variants/index.html> (Accessed June 6, ) [Google Scholar]
- Corman V.M., Landt O., Kaiser M., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. 25.3.2000045. (Accessed oct 08, 2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edara V.V., Norwood C., Floyd K., et al. Infection- and vaccine-induced antibody binding and neutralization of the B.1.351 SARS-CoV-2 variant. Cell Host Microbe. 2021;29(April (4)):516–521.e3. doi: 10.1016/j.chom.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., St Denis K., et al. Circulating SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Preprint. medRxiv. 2021 2021.02.14.21251704. Published 2021 Feb 18. [Google Scholar]
- Harrington D., Kele B., Pereira S., et al. Confirmed reinfection with SARS-CoV-2 variant VOC-202012/01. Clin. Infect. Dis. 2021;(January 9) doi: 10.1093/cid/ciab014. ciab014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Zia T., Suleman M., et al. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: an insight from structural data [published online ahead of print, 2021 Mar 23] J. Cell. Physiol. 2021 doi: 10.1002/jcp.30367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby T. New variant of SARS-CoV-2 in UK causes surge of COVID-19. Lancet Respir. Med. 2021;9(2):e20–e21. doi: 10.1016/S2213-2600(21)00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu J., Xia H., et al. Neutralizing activity of BNT162b2-Elicited serum. N. Engl. J. Med. 2021;384(15):1466–1468. doi: 10.1056/NEJMc2102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhi S.A., Baillie V., Cutland C.L., et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B.1.351 variant. N. Engl. J. Med. 2021;384(20):1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty S. India’s massive COVID surge puzzles scientists. Nature. 2021;592(7856):667–668. doi: 10.1038/d41586-021-01059-y. [DOI] [PubMed] [Google Scholar]
- Manual of Allplex™ SARS-CoV-2 Assay . 2021. Manual of Allplex™ SARS-CoV-2 Assay. is available at: < https://seegenebrazil.com.br/wp-content/uploads/2021/01/RV10247Y_V1-15_PT-Manual.pdf>. (Accessed oct 08, ) [Google Scholar]
- Manual of Kit Molecular SARS-CoV-2 (E/RP) Bio-Manguinhos . 2021. Manual of Kit Molecular SARS-CoV-2 (E/RP) Bio-Manguinhos. is available at: < https://www.bio.fiocruz.br/images/molec-sars-cov2-e-rp-rox-2x48r-11-05-2020-lotes-01ao05.pdf>.(Accessed oct 08, ) [Google Scholar]
- Mullen J.L., Tsueng G., Latif A.A., et al. 2020. outbreak.info.https://outbreak.info/ Available online: [Google Scholar]
- Naveca, F., da Costa C., Nascimento V., et al., 2021. Three SARS-CoV-2 reinfection cases by the new Variant of Concern (VOC) P.1/501Y.V3. Preprint at Res. Sq. 10.21203/rs.3.rs-318392/v1. [DOI]
- Peñarrubia L., Ruiz M., Porco R., et al. Multiple assays in a real-time RT-PCR SARS-CoV-2 panel can mitigate the risk of loss of sensitivity by new genomic variants during the COVID-19 outbreak. Int. J. Infect. Dis. 2020;97:225–229. doi: 10.1016/j.ijid.2020.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapov V., Ong J.L. Examining sources of error in PCR by single-molecule sequencing. PLoS One. 2017;12(1) doi: 10.1371/journal.pone.0169774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Public Health England . 2021. SARS-CoV-2 Variants of Concern and Variants Under Investigation in England. Technical briefing 7, March 21available at: < https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/972247/Variants_of_Concern_VOC_Technical_Briefing_7_England.pdf>. [Google Scholar]
- World Health Organization . 2021. Tracking SARS-CoV-2 Variants. Available on: < https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/> (Accessed June 6, ) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Additional data can be made available upon request.