Abstract
Background:
Phospholamban (PLN) is a critical regulator of calcium cycling and contractility in the heart. The loss of arginine at position 14 in PLN (R14del) is associated with dilated cardiomyopathy (DCM) with a high prevalence of ventricular arrhythmias. How the R14 deletion causes DCM is poorly understood and there are no disease-specific therapies.
Methods:
We used single-cell RNA sequencing to uncover PLN R14del disease-mechanisms in human induced pluripotent stem cells (hiPSC-CMs). We utilized both 2D and 3D functional contractility assays to evaluate the impact of modulating disease relevant pathways in PLN R14del hiPSC-CMs.
Results:
Modeling of the PLN R14del cardiomyopathy with isogenic pairs of hiPSC-CMs recapitulated the contractile deficit associated with the disease in vitro. Single-cell RNA sequencing revealed the induction of the unfolded protein response pathway (UPR) in PLN R14del compared to isogenic control hiPSC-CMs. The activation of UPR was also evident in the hearts from PLN R14del patients. Silencing of each of the three main UPR signaling branches (IRE1, ATF6, or PERK) by siRNA exacerbated the contractile dysfunction of PLN R14del hiPSC-CMs. We explored the therapeutic potential of activating the UPR with a small molecule activator, BiP protein Inducer X (BiX). PLN R14del hiPSC-CMs treated with BiX showed a dose-dependent amelioration of the contractility deficit of in both 2D cultures and 3D engineered heart tissues without affecting calcium homeostasis.
Conclusions:
Together, these findings suggest that the UPR exerts a protective effect in the setting of PLN R14del cardiomyopathy and that modulation of the UPR might be exploited therapeutically.
Keywords: induced pluripotent stem cells, phospholamban, dilated cardiomyopathy, single-cell RNA sequencing, UPR, disease modeling
Introduction
Phospholamban (PLN) encodes a critical regulatory protein of Ca2+ cycling and is a primary mediator of the beta-adrenergic effects, resulting in enhanced cardiac output1. The levels of PLN and its degree of phosphorylation profoundly influence the activation state of the sarcoplasmic reticulum calcium ATPase (SERCA2a). In the dephosphorylated state, PLN interacts with SERCA2a and shifts its Ca2+ activation toward lower apparent Ca2+ affinity. Upon PKA-mediated phosphorylation, the inhibitory interaction between PLN and SERCA2a is abolished, and the apparent Ca2+ affinity is raised. Thus, PLN is the rate-determining factor for Ca2+ re-uptake by SERCA2a and a key regulator of contractility in the heart.
Dilated cardiomyopathy (DCM) is the leading cause of heart failure, and familial DCM is responsible for up to a third of the reported cases2. Various pathogenic genetic variants have been linked to DCM, including mutations in the coding region of the PLN gene. DCM caused by the deletion of the arginine 14 codon in the PLN gene (R14del) is associated with prevalent ventricular arrhythmias, heart failure and sudden cardiac death3, 4. The limited mechanistic understanding of how the R14del contributes to the overall clinical presentation translates to the lack of disease-specific therapeutic strategies.
In this study, we investigated the molecular underpinnings of PLN R14del-induced cardiomyopathy by leveraging the power of human induced pluripotent stem cells (hiPSC), CRISPR/Cas9 genome editing and single-cell RNAseq (scRNA-seq) technologies. PLN R14del hiPSC-derived cardiomyocytes (hiPSC-CMs) faithfully recapitulated the contractile dysfunction observed of PLN R14del-induced cardiomyopathy. At the molecular level, we observed an elevated endoplasmic reticulum (ER) stress and unfolded protein response (UPR) response in the PLN R14del hiPSC-CMs compared to isogenic controls. Molecular and pharmacological modulation of the UPR pathway revealed a protective role of the UPR activation in PLN R14del hiPSC-CMs. Our findings suggest a mechanistic link between proteostasis and PLN R14del-induced pathophysiology that could be exploited to develop therapeutic strategies for PLN R14del cardiomyopathy.
Methods
Patients and healthy controls were enrolled in the study by informed consent approved by the Stanford Institutional Review Board and Stem Cell Research Oversight Committee. The histopathological part of the study met the criteria of the code of proper use of human tissue that is used in the Netherlands. The collection of human cardiac tissue was approved by the scientific advisory board of the biobank of the University Medical Center Utrecht, Utrecht, the Netherlands (protocol no. 12/387).
A detailed description of the Methods is available in the Supplemental Materials section. The data, analytic methods, and study materials are available to other researchers for purposes of reproducing the results or replicating the procedures on request. The iPSC lines have been deposited to the Stanford’s Cardiovascular Institute iPSC Biobank and are available upon request.
Statistical analysis.
Data were processed and analyzed using Microsoft Excel (Microsoft) and GraphPad Prism v.8.1.1 (GraphPad). For statistical analysis of two data sets, the Student’s t-test was used. One-way ANOVA was employed with Tukey’s multiple comparisons test to evaluate statistical differences among groups. Factorial designs were analyzed using two-way ANOVA for repeated measures in case of serial measurements, and was adjusted by Tukey’s post-hoc test in case of multiple comparisons. Data plots are displayed as mean ± SEM, unless specified otherwise, and a P-value <0.05 was set to determine statistical significance.
Data Availability.
Because of the sensitive nature of the data collected for this study, requests to access the dataset from qualified researchers may be sent to the corresponding author.
Results
Modeling DCM contractility defect with hiPSC-CMs
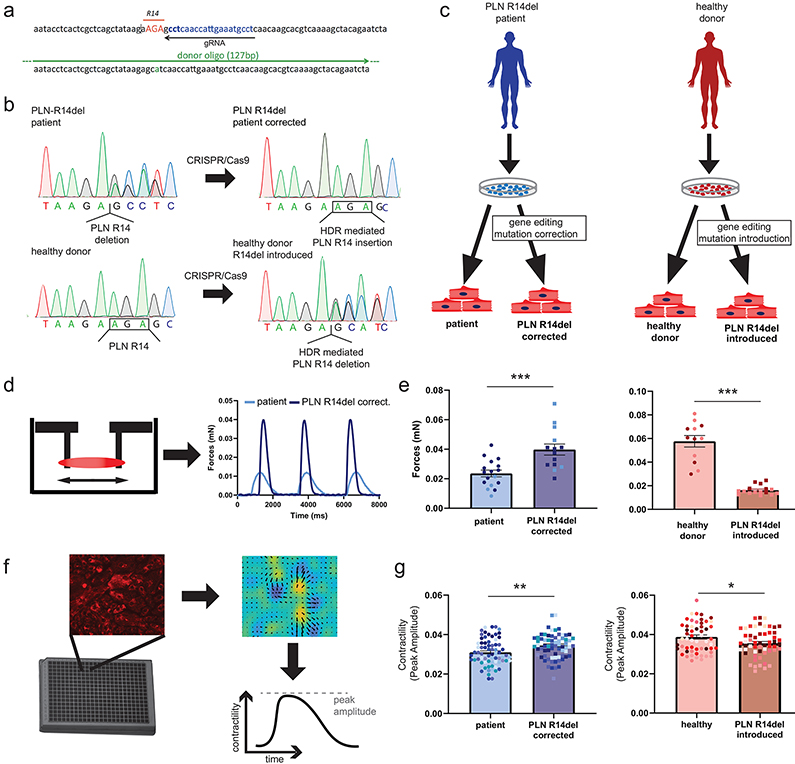
To model PLN R14del DCM, we recruited two unrelated families carrying the mutation (Figure I in the Supplement) to generate hiPSCs from carriers. Isogenic control lines in which the genetic background is identical are important for unequivocal assignment of phenotype to the underlying gene variant. Therefore, we corrected the R14del mutation in both patient hiPSC lines through CRISPR/Cas9-mediated genome editing, and similarly, introduced the R14del mutation in an hiPSC line derived from an individual with no history of heart disease (healthy donor, HD) (Figure 1a-b). We generated 3 pairs of isogenic models that differ only in the PLN R14del mutation (Figure 1c, Figure II in the Supplement). Impaired contractility is a pathological hallmark of DCM; therefore, we assessed the contractile function of the hiPSC-CMs. Consistent with previous studies5, hiPSC-CMs carrying the PLN R14del mutation (patient and HD R14del introduced) showed decreased contractility in three-dimensional engineered heart tissues (3D-EHTs) (Figure 1d-e) and two dimensional (2D) monolayer cultures (Figure 1f-g). Therefore, the isogenic paired hiPSCs lines recapitulated the contractile deficit, providing a quantifiable model for determining the molecular mechanism(s) that underlies the development of DCM.
Figure 1. PLN R14del hiPSC disease modeling and functional assessment of contractility.
a, Schematic overview of the strategy to precisely modify the PLN sequence using CRISPR/Cas9 and single-stranded donor oligonucleotides (ssODN) complementary to the guide RNA (gRNA).
b, Sanger sequencing analysis showing the correction and introduction of the R14del variant sequence in hiPSCs generated from DCM and healthy individual hiPSCs, respectively.
c, Schematic representation of hiPSC lines used in the study.
d-e, Assessment of force generation of hiPSC-CMs carrying the PLN R14del mutation and their corresponding isogenic controls in 3D-EHTs, (2 batches, n = 5-12 EHTs each). f-g, 2D monolayer contractility measurements of hiPSC-CMs carrying the PLN R14del mutation and their corresponding isogenic controls (12 batches, n = 5-10 wells each). Colors represent experimental batches.
Abbreviations: HDR, homology-directed repair; EHTs, engineered heart tissues. Data were presented as mean ± SEM. * p < 0.05, ** p < 0.005, *** p < 0.0005.
Single-cell RNAseq analysis reveals unfolded protein response (UPR) activation
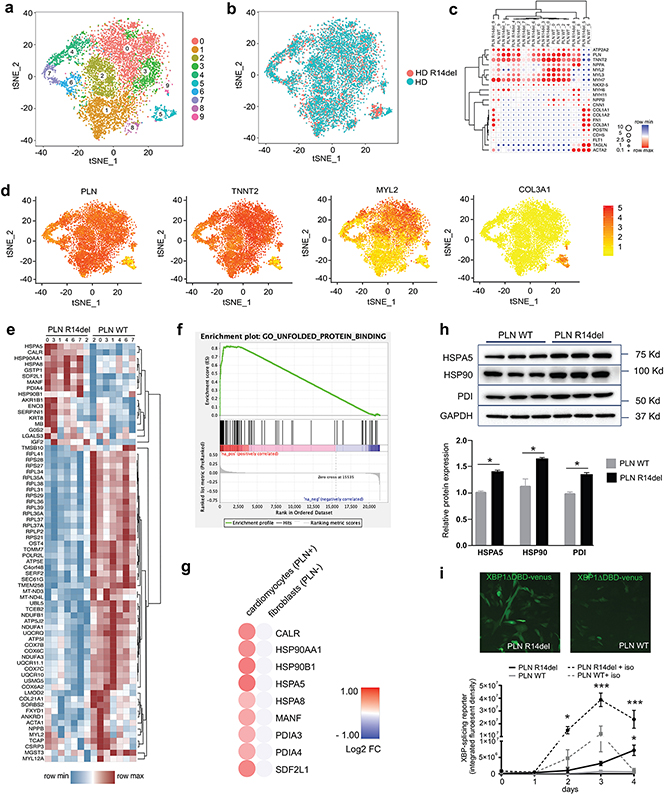
Single-cell transcriptomic analysis makes it possible to deconvolute the complex transcriptional responses that naturally occur across populations of cells into clusters of similarly responding cells. We used a high-throughput droplet-based scRNA-seq method (10X Genomics) to examine the transcriptional effects caused by introducing the PLN R14del mutation into a HD hiPSC line. Unbiased t-distributed stochastic neighbor embedding (tSNE) clustering parsed 9,244 single-cell transcriptomes from PLN WT hiPSC-CMs (5,279 cells) and PLN R14del hiPSC-CMs (3,965 cells) into ten distinct sub-populations (Figure 2a). The heterogeneity of the cardiomyocyte cultures was similar to prior scRNA-seq studies6. Despite this heterogeneity, t-SNE analysis revealed close clustering of the PLN mutant line with its isogenic counterpart, indicating that the transcriptomes and cell type composition are preserved across isogenic populations (Figure 2b). To assign cellular identity, subpopulations were classified on the basis of known marker genes. Most of the cell clusters appeared to be cardiomyocytes based on their specific patterns of gene expression (clusters 1-4 and 6-7 expressed marker genes TNNT2, MYL2, and MYH7). Two clusters, clusters 5 and 8, were identified as fibroblasts (expressing marker genes COL3A1, COL1A1, and FN1), while one cluster with very few cells, cluster 9, identified as smooth muscle cells (with marker genes ACTA2 and TAGLN) (Figure 2c-d).
Figure 2. Single-cell RNA-seq of isogenic hiPSC-CMs carrying the PLN R14del mutation.
a-b, Unbiased identification of cell clusters using t-SNE-based clustering of single-cell transcriptomes showing a two-dimensional visualization with distinctly isolated cell subpopulations (n = 5,279 cells, healthy donor (PLN WT); n = 3,965 cells, healthy donor PLN R14del introduced (PLN R14del))
c-d, Subpopulations were classified based on canonical marker gene expression.
e, Heatmap display of 77 differentially expressed genes in the cardiomyocyte subpopulations.
f, GSEA pathway enrichment analysis.
g, Comparison of UPR hallmark differential gene expression between cardiomyocyte and non-cardiomyocyte subpopulations in HD and R14del introduced hiPSC-CMs
h, Western blot expression analysis of UPR proteins from paired isogenic hiPSC-CMs lines (n = 3 batches).
i, Assessment of the UPR activity in living hiPSC-CMs (patient PLN14del (PLN R14del) and patient PLN R14del corrected (PLN WT)) transduced with AAV-F-XBP1ΔDBD (3 batches, n = 4-6 wells each). Statistical significance represented as differences between PLN WT vs PLN R14del in either control or isoproterenol (iso) conditions.
Data were presented as mean ± SEM. * p < 0.05, *** p < 0.0005.
Next, we explored the transcriptional changes associated with the R14del mutation. An advantage of single-cell over bulk RNAseq is that it affords greater statistical power to resolve differential expression (DE) because it focuses within cell-identity clusters rather than across the broader heterogeneous population in which cell-type-specific information remains obscure. Based on the tSNE clustering and canonical marker expression, we restricted the analysis to the cardiomyocyte subclusters 1-4 and 6-7. 77 genes (Wilcoxon rank-sum test) were differentially expressed in PLN R14del hiPSC-CMs when compared to the isogenic control hiPSC-CMs (Figure 2e). Gene enrichment analysis revealed genes associated with pathways related to ER stress and UPR signaling pathway (Figure 2f). Further bioinformatics analysis showed that several hallmark genes related to UPR pathway were activated in PLN R14del hiPSC-CMs, including gene members of the protein disulfide isomerase (PDIA3 and PDIA4) and heat shock protein 90 (HSP90A1 and HSP90B1), and HSPA5 (also known as Binding Immunoglobulin Protein (BiP)) when compared to isogenic PLN WT controls. Importantly, comparison of the gene expression profiles of the cell cluster identified as fibroblasts showed no significant differences in the expression of the UPR hallmark genes between the PLN R14del and PLN WT cells (Figure 2g). We further corroborated the activation of UPR markers in the hiPSC-CMs at the protein level using western blot analysis (Figure 2h). Finally, we monitored the activity of the UPR pathway in living hiPSC-CMs using an XBP1-splicing reporter construct7 (Figure 2i) and observed a significant increase in UPR activity in patient PLN R14del cells. Since adrenergic drive activates UPR response in cardiomyocytes8, 9, we also measured the UPR reporter activity upon isoproterenol stimulation. Under adrenergic stress conditions (72 hours treatment with 1 μM) we noticed an increase in UPR signaling in PLN WT cells, but an exaggerated (4-fold higher) response in hiPSC-CMs containing the mutation. Taken together, these data suggest that the PLN R14del mutation activates the UPR transcriptional program and sensitizes R14del hiPSC-CMs to adrenergic stress.
Histopathological characterization of the UPR in PLN-R14del patient myocardium
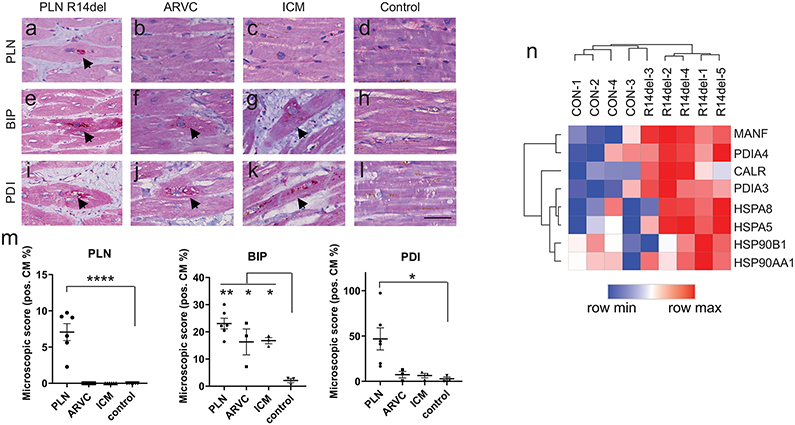
The accumulation of protein aggregates is often associated with dysfunctional UPR responses, for instance in neurodegenerative diseases10. Protein aggregation has been noted as a histopathological characteristic of end-stage PLN R14del DCM hearts but not in hearts representative of other forms of DCM11-13. Therefore, we examined human myocardial samples of PLN R14del hearts for evidence of UPR involvement. We also analyzed myocardial tissue from desmosomal arrhythmogenic right ventricular cardiomyopathy (ARVC) and ischemic cardiomyopathy (ICM) hearts (Figure III and Table I in the Supplement). Consistent with previous studies13, PLN immunolabeling revealed perinuclear aggregations in PLN R14del tissues (7±2%) but were absent in ARVC, ICM, and control hearts (Figure 3a-d,m). BiP is an essential regulator of the UPR that monitors ER stress and its expression is markedly increased in the presence of unfolded proteins in the ER14, 15. Accordingly, we observed highest BiP levels in PLN R14del (23±5%), but also detected comparable BiP expression in desmosomal ARVC (16±5%), and ICM (17±2%), relative to control hearts (2±1%) (Figure 3e-h,m), consistent with UPR activation in failing hearts16-18. Protein disulfide isomerase (PDI), a protein folding facilitator, is another chaperone that is induced by protein misfolding19. A striking PDI presence was detected in the myocardium of PLN R14del patient (46±29%), relative to desmosomal ARVC (7±6%), ICM (6±4%), and controls (3±3%) (Figure 3i-m). In addition, RNA sequencing showed that UPR genes are upregulated in the myocardium of R14del patient cardiac tissue relative to healthy individuals (Figure 3n). Taken together, the histological manifestations of PLN R14del shares hallmarks of neurodegenerative disease wherein the accumulation of protein aggregates is associated with abnormal levels of ER stress response20, 21.
Figure 3. Determination of the UPR status in PLN R14del disease and other forms of cardiomyopathies.
Histological analysis of human myocardium from patients suffering from PLN R14del, desmosomal arrhythmogenic cardiomyopathy (ARVC), and ischemic cardiomyopathy (ICM) versus control (healthy) hearts.
a–d, Abnormal accumulation of PLN in perinuclear aggregates (arrows) in severely affected cardiomyocytes in PLN R14del and absent in ARVC, ICM, control.
e-h, Diffuse moderate immunolabeling (arrows) for BIP in PLN 14del, and present to a lesser extent in ARVC and ICM.
i-l, High immunolabeling for dotted cytoplasmic PDI (arrows) in PLN 14del, low PDI presence in ARVC, and ICM (scale bar = 25 μm).
m, Quantification of immunostaining in PLN R14del (n = 6 patients), ARVC (n = 3 patients), ICM (n = 3 patients), and control (n = 3 patients).
n, RNA sequence analysis of UPR gene expression from healthy and PLN R14del myocardium from patient samples.
Data were presented as mean ± SEM. * p < 0.05, ** p < 0.005, **** p < 0.00005.
UPR activity protects contractile function in hiPSC-CMs harboring PLN-R14del
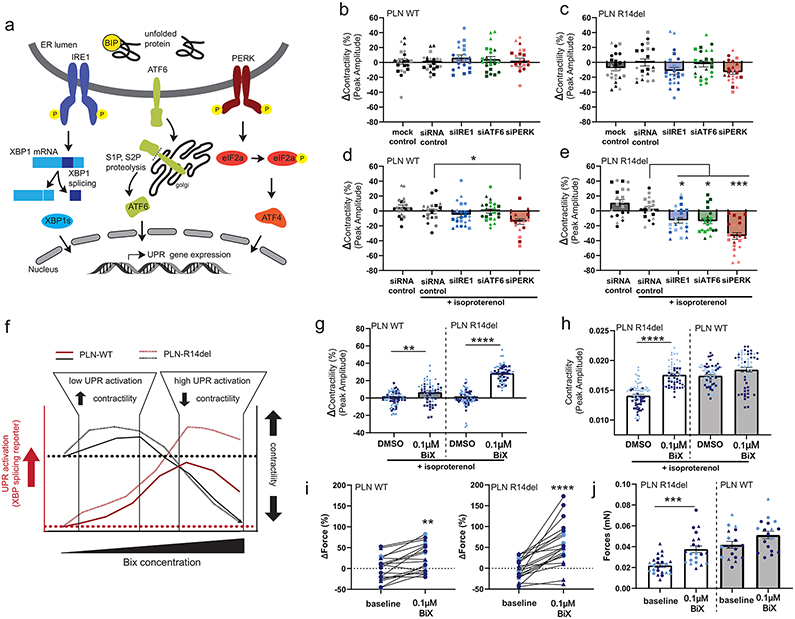
In principle, UPR could have a pathological or beneficial effect on the PLN R14del cardiomyocytes. To establish the role of the UPR, we carried out a loss-of-function experiment in which we attenuated the main transducers of the UPR (IRE1, ATF6, and PERK) (Figure 4a). These interconnected signaling branches of the UPR provide the cells with an adaptive response to ER stress in order to restore proteostasis22. Under baseline conditions, targeting each of the three branches by siRNA did not significantly affect the contractility in either PLN WT (HD and patient R14del corrected) or PLN R14del (patient and HD R14del introduced) hiPSC-CMs, although a trend towards decreased function (~10%) was observed with siRNA against IRE1 and PERK (siIRE1 and siPERK) in PLN R14del (Figure 4b-c) hiPSC-CMs. Given that chronic adrenergic stimulation dramatically increased UPR fluxes in hiPSC-CMs harboring PLN R14del (Figure 2i), we next evaluated the impact of the loss-of-function in the presence of isoproterenol. Under adrenergic stress, the contractility of PLN WT hiPSC-CMs remained unchanged after the siRNA-mediated knockdown of IRE1 and ATF6, but we observed significantly decreased contractile function after PERK knockdown (14.3±3.7%) (Figure 4d). By contrast, we observed a significant contractility deficit in PLN R14del hiPSC-CMs upon knockdown of each of the three UPR arms compared to control siRNA treated (siIRE1, −15.5 ± 4.2%; siATF6, -,16.7 ± 4.1%; siPERK, −36.6 ± 5%), indicating that the mutant hiPSC-CMs were sensitized to a loss of UPR signaling in a genotype-specific manner (Figure 4e). Taken together, these data suggest that activation of the UPR in PLN R14del hiPSC-CMs preserves cell function, and therefore plays a protective role in alleviating ER stress and potentially blunts disease pathogenesis.
Figure 4. Defining the role of the UPR pathway in R14del hiPSC-CMs.
a, Schematics of UPR signaling including stress sensors IRE1, ATF6, and PERK at the ER membrane and the three corresponding downstream transcriptional effectors.
b-e, Functional analysis of UPR pathway perturbation in isogenic hiPSC-CMs. Cells were transfected with the indicated siRNAs and 2D monolayer contractility was measured after four days either under baseline conditions (b,c) or with 0.1 μM isoproterenol (d,e). Contractility represented as percent change from control siRNA (6 batches, n = 2-3 wells each). Circles(●), squares(■), triangles(▲), diamonds (♦) represent experimental batches; dark-colored symbols represent hiPSC-CMs from healthy donor background, light-colored symbols from the patient background. All data are presented as mean ± SEM. * p < 0.05, *** p < 0.0005 vs siRNA controls (d,e).
f-h, Assessment of contractility in hiPSC-CMs after pharmacological activation of the UPR with the small molecule BiX. Overview of BiX dose-response kinetics in hiPSC-CMs showing effects on UPR activation and contractility (f, see also Figure IV in the Supplement). Assessment of 2D contractility measurements after 72 hours BiX (or DMSO vehicle control) treatment with 0.1 μM isoproterenol in isogenic hiPSC-CMs (g,h; 4 batches, n = 12-16 wells each). Contractility is represented as percent change from the DMSO control per batch (g) or actual peak contraction amplitude (h). Circles(●), squares(■), triangles(▲), diamonds (♦) represent experimental batches; dark-colored symbols represent hiPSC-CMs from healthy donor background, light-colored symbols from the patient background. All data are presented as mean ± SEM. ** p < 0.005, **** p < 0.00005 vs DMSO vehicle controls (g,h).
i-j, Force generation of 3D-EHTs derived from isogenic hiPSC-CM at baseline and following treatment with BiX for 72h. Force generation is represented as percent change from baseline (i) or as total generated force (j) (4 batches, n=2-8 EHT each). ** p < 0.005, *** p < 0.0005, **** p < 0.00005 by paired student t-test.
Pharmacological targeting the UPR in PLN-R14del ameliorates contractility
Next, we evaluated whether pharmacologically stimulating the UPR pathway in PLN R14del hiPSC-CMs beyond basal levels would have a positive effect on cardiomyocyte function. BiP protein Inducer X (BiX) is a small molecule proteostasis regulator that increases expression of the BiP protein to consequently induce UPR and protect neurons from ER stress23, 24. In both PLN R14del (patient and HD R14del introduced) and PLN WT (HD and patient R14del corrected) hiPSC-CMs, treatment with BiX induced a dose-dependent increase in a reporter for XBP1-splicing (indicative of activated UPR). The maximal effect was higher in PLN R14del than in corresponding isogenic control hiPSC-CMs (Emax = 33.1 ± 7% vs. 56.8 ± 7% XBP-1 splice-positive cells in normal vs. R14del hiPSC-CMs, respectively) (Figure IVb,c in the Supplement). BiX (0.1 μM) significantly enhanced contractility in PLN R14del and PLN WT hiPSC-CMs as detected in 2D cell sheets (Figure 4g,h). In the EHTs, this translated to an increase in force from about 20 μN in untreated EHTs to 40 μN in treated EHTs, corresponding to a level statistically indistinguishable from isogenic control EHTs (Figure 4i,j). Remarkably, the increase in peak contractility was more pronounced in the PLN R14del hiPSC-CMs, and BiX treatment (0.1 μM) led to a nearly complete restoration of the contractile deficit without increasing the expression levels of myofilament proteins (Figure V in the Supplement). At high concentrations, BiX decreased contractility in both the PLN R14del and isogenic controls (Emax = −25% at 10 μM for both R14del and isogenic controls) (Figure IVa in the Supplement). We considered whether the beneficial effect of BiX is mediated through the modulation of the calcium handling properties of PLN R14del hiPSC-CMs. Compared to DMSO control, there was no significant difference in the intracellular calcium handling kinetics in PLN R14del hiPSC-CMs treated with BiX (Figure VI in the Supplement). In agreement, the expression levels of PLN and other key calcium handling proteins, including SERCA2a, NCX1, RYR2 and CASQ2, were similar between BiX and DMSO treated PLN R14del hiPSC-CMs (Figure VII in the Supplement). Collectively, these data suggest that stimulating the UPR pathway in PLN R14del hiPSC-CMs restored contractility to levels of isogenic controls without affecting calcium homeostasis.
Discussion
Human iPSCs provide an opportunity for modeling DCM in vitro to understand the molecular consequences of pathogenic mutation such as PLN R14del. However, the inherited heterogeneity caused by a mixture of cell types in hiPSC-CM differentiation cultures and the donor’s genetic background present confounding factors for defining disease-specific phenotypes. To overcome these challenges, we generated multiple isogenic hiPSC lines carrying the R14del mutation and performed scRNA-seq on these genotype-defined hiPSC lines. This experimental approach can be widely employed to define the specific contributions of pathogenic mutation in DCM and cardiomyopathies more broadly.
Using this approach, we revealed that the PLN R14del mutation activates the UPR, an integrative intracellular signalling pathway that plays a critical role in the maintenance of proteostasis in the ER25. Proteostasis, the balance between protein synthesis, folding, re-folding and degradation, is essential for the long-term preservation of cell and tissue function. With age, the ability of many cells and organs to maintain proteostasis is gradually compromised26. Accumulating evidence suggests that UPR is activated in response to a loss of proteostasis in the ER and the corresponding accumulation of protein aggregates, which characterizes age-related diseases and protein folding disorders, like Alzheimer’s Parkinson’s and Amyotrophic Lateral Sclerosis10. Reminiscent of these diseases, aggregation of PLN protein is a hallmark of PLN R14del DCM12, suggesting that the disruption of proteostasis plays a role in disease presentation. It is tempting to speculate that the altered stability and folding kinetics of the PLN R14del mutant directly activates the UPR in mutant hiPSC-CMs and human heart tissues. It will be of interest to explore this hypothesis further in the future.
Our isogenic models allowed us to perform high throughput genetic and pharmacological assays to both understand the underlying pathological mechanisms and to identify therapeutic targets to prevent or treat such diseases. We found that activation of UPR in PLN R14del hiPSC-CMs is protective as molecular inhibition of each of the three UPR sensors, IRE1, PERK, and ATF6, exacerbated the contractile dysfunction. Conversely, pharmacological enhancement of UPR by the small molecule agonist BiX, ameliorated the contractility defect in PLN R14del hiPSC-CMs. BiX activates the chaperone BiP thereby increasing the activity of the UPR pathway. These observations are consistent with previous studies showing that activation of UPR suppressed disease onset and progression in a cellular and animal models of neurodegenerative diseases27-31, and prevent damage in the heart caused by ischemia/reperfusion32-34. These findings suggest that enhancing the expression or activity of individual proteostasis network components could be therapeutically beneficial in PLN R14del DCM.
Here, we showed that the small molecule BiX improved contractile performance without affecting Ca2+ transients in PLN R14del hiPSC-CMs. BiX preferentially activates ER stress response elements upstream of the BiP gene, a molecular chaperone, thereby increasing the activity of the UPR pathway22. BiX showed a detrimental effect at high doses in hiPSC-CMs, suggesting a narrow therapeutic window between the effective doses and those at which it causes adverse toxic effects. The underlying mechanism for the biphasic response is unclear. It is possible that BiX is a ubiquitous ER stressor, limiting its application as a therapeutic strategy because it may activate several pathways of the UPR, including the ER stress-induced apoptotic pathways. Although the inotropic mechanisms of BiX are unprecedented, our findings suggest that selective modulation of UPR components could improve cardiac function in PLN R14del DCM and potentially avoid the deleterious effects that can occur with drugs that target calcium signaling or other upstream regulators of contraction.
In conclusion, by combining scRNA-seq, human cardiac tissue samples, and disease modeling in vitro using isogenic hiPSC-CMs, our findings implicates the UPR pathway in PLN R14del DCM pathogenesis. Furthermore, stimulating the UPR with the small molecule BiX lead to functional rescue of the contractility deficit in PLN R14del hiPSC-CMs in vitro in a genotype-specific manner, suggesting UPR as a potential new therapeutic target. Finally, as disease penetrance in PLN R14del DCM is age-related35, we theorize that the decline in the proteostasis network capacity associated with ageing36 may enhance the propensity of R14del PLN proteins to form aggregates, increasing disease susceptibility. Our study provides important proof-of-concept that activation of proteostasis mechanisms has a protective effect on PLN R14del-associated DCM in vitro and could be harnessed therapeutically to delay the onset or slow the progression of disease.
Study limitations.
Although patient-specific hiPSC-CMs offer an attractive experimental model, it has certain limitations. Despite the significant progress in the field over the past few years, the hiPSC-CMs are developmentally immature and their phenotype is more similar to human fetal cardiomyocyte. Further studies in relevant animal models of PLN R14del are needed to translate the in vitro findings and evaluate the therapeutic potential of targeting UPR signaling network in PLN R14del DCM. Nevertheless, our studies demonstrated the potential of using complex physiological models of patient-derived hiPSC-CMs to better understand the mechanisms of PLN R14del DCM pathogenesis towards the development of mechanism-based therapies.
Supplementary Material
CLINICAL PERSPECTIVE.
What is new?
Employing human induced pluripotent stem cells (hiPSCs) and CRISPR-Cas9 gene editing technologies, we created an in vitro model of DCM associated with the phospholamban (PLN) R14del mutation.
Single-cell RNAseq revealed the activation of the unfolded protein response (UPR) pathway in PLN R14del hiPSC-derived cardiomyocytes (hiPSC-CMs).
UPR activation was also evident by significant upregulation of marker gene in the hearts of PLN R14del patients.
Pharmacological and molecular modulation of the UPR suggest a compensatory role in PLN R14del DCM.
Augmentation of UPR by the small molecule BiX ameliorated contractile dysfunction.
What are the clinical implications?
The findings suggest a mechanistic link between proteostasis and PLN R14del-induced pathophysiology that could be exploited to develop therapeutic strategies for PLN R14del cardiomyopathy.
Targeting of the UPR might be a viable therapeutic strategy for the treatment of PLN R14del DCM.
The study highlights hiPSC-CM modeling combined with small molecule testing as a paradigm for studying genotype-phenotype associations in heart disease with the potential for the development of mechanism-based therapeutic strategies.
Acknowledgements
We are very thankful to the patients and their families for providing valuable material for research purposes. We thank the Stanford CVI iPSC Biobank for providing the patient hiPSC lines, and Dr. Masayuki Miura for providing the XBP1-reporter vector. We gratefully acknowledge the help of Christian Snijders Blok and Dr. Jan Willem Buikema.
Sources of Funding
This research was supported by grants from the NIH R01 HL139679, HL15041401 and R00 HL104002 (to IK); R01HL113006, R01HL130840, R01HL128072, R01HL132225 and P01HL141084 (to MM); the Foundation Leducq (CurePLaN) to MM, IK, MH, and FWA; The American Heart Association (AHA 17IRG33410532, to IK), and the Joan and Sanford I. Weill Scholars Endowment (to MM and IK). FWA is supported by UCL Hospitals NIHR Biomedical Research Centre. DF was funded by the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 708459 and the PLN Foundation. JS was supported by EVICARE (No. 725229) of the European Research Council (ERC) and ZonMw program No. 116006102. IPG was supported by the American Heart Association postdoctoral fellowship AHA 19POST34380920. WR is supported by the Dutch Heart Foundation (CVON PREDICT Young Talent Program) and PLN Foundation. MH is funded by the Dutch Research Council (NOW) VENI grant no. 016.176.136.
Non-standard Abbreviations and Acronyms
- 3D-EHT
three dimensional-engineered heart tissue
- ARVC
arrhythmogenic right ventricular cardiomyopathy
- BiX
BiP protein Inducer X
- DCM
dilated cardiomyopathy
- DE
differential expression
- ER
endoplasmic reticulum
- HD
healthy donor
- HDR
homologous directed repair
- hiPSC-CM
human induced pluripotent stem cell-derived cardiomyocyte
- ICM
ischemic cardiomyopathy
- PDI
protein disulfide isomerase
- scRNA-seq
single-cell RNA sequencing
- tSNE
t-distributed stochastic neighbor embedding
- UPR
unfolded protein response
Footnotes
Disclosures
None
References
- 1.MacLennan DH and Kranias EG. Phospholamban: a crucial regulator of cardiac contractility. Nat Rev Mol Cell Biol. 2003;4:566–577. [DOI] [PubMed] [Google Scholar]
- 2.Rosenbaum AN, Agre KE and Pereira NL. Genetics of dilated cardiomyopathy: practical implications for heart failure management. Nat Rev Cardiol. 2020;17:286–297. [DOI] [PubMed] [Google Scholar]
- 3.Haghighi K, Kolokathis F, Gramolini AO, Waggoner JR, Pater L, Lynch RA, Fan GC, Tsiapras D, Parekh RR, Dorn GW 2nd, et al. A mutation in the human phospholamban gene, deleting arginine 14, results in lethal, hereditary cardiomyopathy. Proc Natl Acad Sci U S A. 2006;103:1388–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Zwaag PA, van Rijsingen IA, Asimaki A, Jongbloed JD, van Veldhuisen DJ, Wiesfeld AC, Cox MG, van Lochem LT, de Boer RA, Hofstra RM, et al. Phospholamban R14del mutation in patients diagnosed with dilated cardiomyopathy or arrhythmogenic right ventricular cardiomyopathy: evidence supporting the concept of arrhythmogenic cardiomyopathy. Eur J Heart Fail. 2012;14:1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stillitano F, Turnbull IC, Karakikes I, Nonnenmacher M, Backeris P, Hulot JS, Kranias EG, Hajjar RJ and Costa KD. Genomic correction of familial cardiomyopathy in human engineered cardiac tissues. Eur Heart J. 2016;37:3282–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman CE, Nguyen Q, Lukowski SW, Helfer A, Chiu HS, Miklas J, Levy S, Suo S, Han JJ, Osteil P, et al. Single-Cell Transcriptomic Analysis of Cardiac Differentiation from Human PSCs Reveals HOPX-Dependent Cardiomyocyte Maturation. Cell Stem Cell. 2018;23:586–598 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwawaki T, Akai R, Kohno K and Miura M. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat Med. 2004;10:98–102. [DOI] [PubMed] [Google Scholar]
- 8.Ayala P, Montenegro J, Vivar R, Letelier A, Urroz PA, Copaja M, Pivet D, Humeres C, Troncoso R, Vicencio JM, et al. Attenuation of endoplasmic reticulum stress using the chemical chaperone 4-phenylbutyric acid prevents cardiac fibrosis induced by isoproterenol. Exp Mol Pathol. 2012;92:97–104. [DOI] [PubMed] [Google Scholar]
- 9.Zhuo XZ, Wu Y, Ni YJ, Liu JH, Gong M, Wang XH, Wei F, Wang TZ, Yuan Z, Ma AQ, et al. Isoproterenol instigates cardiomyocyte apoptosis and heart failure via AMPK inactivation-mediated endoplasmic reticulum stress. Apoptosis. 2013;18:800–810. [DOI] [PubMed] [Google Scholar]
- 10.Hetz C and Saxena S. ER stress and the unfolded protein response in neurodegeneration. Nat Rev Neurol. 2017;13:477–491. [DOI] [PubMed] [Google Scholar]
- 11.Karakikes I, Stillitano F, Nonnenmacher M, Tzimas C, Sanoudou D, Termglinchan V, Kong CW, Rushing S, Hansen J, Ceholski D, et al. Correction of human phospholamban R14del mutation associated with cardiomyopathy using targeted nucleases and combination therapy. Nat Commun. 2015;6:6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Te Rijdt WP, van der Klooster ZJ, Hoorntje ET, Jongbloed JDH, van der Zwaag PA, Asselbergs FW, Dooijes D, de Boer RA, van Tintelen JP, van den Berg MP, et al. Phospholamban immunostaining is a highly sensitive and specific method for diagnosing phospholamban p.Arg14del cardiomyopathy. Cardiovasc Pathol. 2017;30:23–26. [DOI] [PubMed] [Google Scholar]
- 13.Te Rijdt WP, van Tintelen JP, Vink A, van der Wal AC, de Boer RA, van den Berg MP and Suurmeijer AJ. Phospholamban p.Arg14del cardiomyopathy is characterized by phospholamban aggregates, aggresomes, and autophagic degradation. Histopathology. 2016;69:542–550. [DOI] [PubMed] [Google Scholar]
- 14.Bertolotti A, Zhang Y, Hendershot LM, Harding HP and Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. [DOI] [PubMed] [Google Scholar]
- 15.Patil C and Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr Opin Cell Biol. 2001;13:349–355. [DOI] [PubMed] [Google Scholar]
- 16.Gao G, Xie A, Zhang J, Herman AM, Jeong EM, Gu L, Liu M, Yang KC, Kamp TJ and Dudley SC. Unfolded protein response regulates cardiac sodium current in systolic human heart failure. Circ Arrhythm Electrophysiol. 2013;6:1018–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okada K, Minamino T, Tsukamoto Y, Liao Y, Tsukamoto O, Takashima S, Hirata A, Fujita M, Nagamachi Y, Nakatani T, et al. Prolonged endoplasmic reticulum stress in hypertrophic and failing heart after aortic constriction: possible contribution of endoplasmic reticulum stress to cardiac myocyte apoptosis. Circulation. 2004;110:705–712. [DOI] [PubMed] [Google Scholar]
- 18.Azfer A, Niu J, Rogers LM, Adamski FM and Kolattukudy PE. Activation of endoplasmic reticulum stress response during the development of ischemic heart disease. Am J Physiol Heart Circ Physiol. 2006;291:H1411–H1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perri ER, Thomas CJ, Parakh S, Spencer DM and Atkin JD. The Unfolded Protein Response and the Role of Protein Disulfide Isomerase in Neurodegeneration. Front Cell Dev Biol. 2015;3:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoozemans JJ, van Haastert ES, Nijholt DA, Rozemuller AJ, Eikelenboom P and Scheper W. The unfolded protein response is activated in pretangle neurons in Alzheimer's disease hippocampus. Am J Pathol. 2009;174:1241–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoozemans JJ, van Haastert ES, Eikelenboom P, de Vos RA, Rozemuller JM and Scheper W. Activation of the unfolded protein response in Parkinson's disease. Biochem Biophys Res Commun. 2007;354:707–711. [DOI] [PubMed] [Google Scholar]
- 22.Hetz C, Chevet E and Oakes SA. Proteostasis control by the unfolded protein response. Nat Cell Biol. 2015;17:829–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kudo T, Kanemoto S, Hara H, Morimoto N, Morihara T, Kimura R, Tabira T, Imaizumi K and Takeda M. A molecular chaperone inducer protects neurons from ER stress. Cell Death Differ. 2008;15:364–375. [DOI] [PubMed] [Google Scholar]
- 24.Oida Y, Izuta H, Oyagi A, Shimazawa M, Kudo T, Imaizumi K and Hara H. Induction of BiP, an ER-resident protein, prevents the neuronal death induced by transient forebrain ischemia in gerbil. Brain Res. 2008;1208:217–224. [DOI] [PubMed] [Google Scholar]
- 25.Walter P and Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. [DOI] [PubMed] [Google Scholar]
- 26.Kaushik S and Cuervo AM. Proteostasis and aging. Nat Med. 2015;21:1406–1415. [DOI] [PubMed] [Google Scholar]
- 27.Valenzuela V, Jackson KL, Sardi SP and Hetz C. Gene Therapy Strategies to Restore ER Proteostasis in Disease. Mol Ther. 2018;26:1404–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Y, Park KK, Yang L, Wei X, Yang Q, Cho KS, Thielen P, Lee AH, Cartoni R, Glimcher LH, et al. Differential effects of unfolded protein response pathways on axon injury-induced death of retinal ganglion cells. Neuron. 2012;73:445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuleta A, Vidal RL, Armentano D, Parsons G and Hetz C. AAV-mediated delivery of the transcription factor XBP1s into the striatum reduces mutant Huntingtin aggregation in a mouse model of Huntington's disease. Biochem Biophys Res Commun. 2012;420:558–563. [DOI] [PubMed] [Google Scholar]
- 30.Gorbatyuk MS, Shabashvili A, Chen W, Meyers C, Sullivan LF, Salganik M, Lin JH, Lewin AS, Muzyczka N and Gorbatyuk OS. Glucose regulated protein 78 diminishes alpha-synuclein neurotoxicity in a rat model of Parkinson disease. Mol Ther. 2012;20:1327–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sado M, Yamasaki Y, Iwanaga T, Onaka Y, Ibuki T, Nishihara S, Mizuguchi H, Momota H, Kishibuchi R, Hashimoto T, et al. Protective effect against Parkinson's disease-related insults through the activation of XBP1. Brain Res. 2009;1257:16–24. [DOI] [PubMed] [Google Scholar]
- 32.Jin JK, Blackwood EA, Azizi K, Thuerauf DJ, Fahem AG, Hofmann C, Kaufman RJ, Doroudgar S and Glembotski CC. ATF6 Decreases Myocardial Ischemia/Reperfusion Damage and Links ER Stress and Oxidative Stress Signaling Pathways in the Heart. Circ Res. 2017;120:862–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martindale JJ, Fernandez R, Thuerauf D, Whittaker R, Gude N, Sussman MA and Glembotski CC. Endoplasmic reticulum stress gene induction and protection from ischemia/reperfusion injury in the hearts of transgenic mice with a tamoxifen-regulated form of ATF6. Circ Res. 2006;98:1186–1193. [DOI] [PubMed] [Google Scholar]
- 34.Blackwood EA, Azizi K, Thuerauf DJ, Paxman RJ, Plate L, Kelly JW, Wiseman RL and Glembotski CC. Pharmacologic ATF6 activation confers global protection in widespread disease models by reprograming cellular proteostasis. Nat Commun. 2019;10:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Rijsingen IA, van der Zwaag PA, Groeneweg JA, Nannenberg EA, Jongbloed JD, Zwinderman AH, Pinto YM, Dit Deprez RH, Post JG, Tan HL, et al. Outcome in phospholamban R14del carriers: results of a large multicentre cohort study. Circ Cardiovasc Genet. 2014;7:455–465. [DOI] [PubMed] [Google Scholar]
- 36.Hipp MS, Kasturi P and Hartl FU. The proteostasis network and its decline in ageing. Nat Rev Mol Cell Biol. 2019;20:421–435. [DOI] [PubMed] [Google Scholar]
- 37.Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ and Palecek SP. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A. 2012;109:E1848–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levitas A, Muhammad E, Zhang Y, Perea Gil I, Serrano R, Diaz N, Arafat M, Gavidia AA, Kapiloff MS, Mercola M, et al. A Novel Recessive Mutation in SPEG Causes Early Onset Dilated Cardiomyopathy. PLoS Genet. 2020;16:e1009000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feyen DAM, McKeithan WL, Bruyneel AAN, Spiering S, Hormann L, Ulmer B, Zhang H, Briganti F, Schweizer M, Hegyi B, et al. Metabolic Maturation Media Improve Physiological Function of Human iPSC-Derived Cardiomyocytes. Cell Rep. 2020;32:107925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cradick TJ, Qiu P, Lee CM, Fine EJ and Bao G. COSMID: A Web-based Tool for Identifying and Validating CRISPR/Cas Off-target Sites. Mol Ther Nucleic Acids. 2014;3:e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang M, Wang J, Torre E, Dueck H, Shaffer S, Bonasio R, Murray JI, Raj A, Li M and Zhang NR. SAVER: gene expression recovery for single-cell RNA sequencing. Nat Methods. 2018;15:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Satija R, Farrell JA, Gennert D, Schier AF and Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaaf S, Eder A, Vollert I, Stohr A, Hansen A and Eschenhagen T. Generation of strip-format fibrin-based engineered heart tissue (EHT). Methods Mol Biol. 2014;1181:121–129. [DOI] [PubMed] [Google Scholar]
- 44.Sala L, van Meer BJ, Tertoolen LGJ, Bakkers J, Bellin M, Davis RP, Denning C, Dieben MAE, Eschenhagen T, Giacomelli E, et al. MUSCLEMOTION: A Versatile Open Software Tool to Quantify Cardiomyocyte and Cardiac Muscle Contraction In Vitro and In Vivo. Circ Res. 2018;122:e5–e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma A, Burridge PW, McKeithan WL, Serrano R, Shukla P, Sayed N, Churko JM, Kitani T, Wu H, Holmstrom A, et al. High-throughput screening of tyrosine kinase inhibitor cardiotoxicity with human induced pluripotent stem cells. Sci Transl Med. 2017;9: eaaf2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M and Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tarasov A, Vilella AJ, Cuppen E, Nijman IJ and Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31:2032–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anders S, Pyl PT and Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson MD, McCarthy DJ and Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Love MI, Huber W and Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Because of the sensitive nature of the data collected for this study, requests to access the dataset from qualified researchers may be sent to the corresponding author.