Abstract
SARS-CoV-2 infection produces neuroinflammation as well as neurological, cognitive (i.e., brain fog), and neuropsychiatric symptoms (e.g., depression, anxiety), which can persist for an extended period (6 months) after resolution of the infection. The neuroimmune mechanism(s) that produces SARS-CoV-2-induced neuroinflammation has not been characterized. Proposed mechanisms include peripheral cytokine signaling to the brain and/or direct viral infection of the CNS. Here, we explore the novel hypothesis that a structural protein (S1) derived from SARS-CoV-2 functions as a pathogen-associated molecular pattern (PAMP) to induce neuroinflammatory processes independent of viral infection. Prior evidence suggests that the S1 subunit of the SARS-CoV-2 spike protein is inflammatory in vitro and signals through the pattern recognition receptor TLR4. Therefore, we examined whether the S1 subunit is sufficient to drive 1) a behavioral sickness response, 2) a neuroinflammatory response, 3) direct activation of microglia in vitro, and 4) activation of transgenic human TLR2 and TLR4 HEK293 cells. Adult male Sprague-Dawley rats were injected intra-cisterna magna (ICM) with vehicle or S1. In-cage behavioral monitoring (8 h post-ICM) demonstrated that S1 reduced several behaviors, including total activity, self-grooming, and wall-rearing. S1 also increased social avoidance in the juvenile social exploration test (24 h post-ICM). S1 increased and/or modulated neuroimmune gene expression (Iba1, Cd11b, MhcIIα, Cd200r1, Gfap, Tlr2, Tlr4, Nlrp3, Il1b, Hmgb1) and protein levels (IFNγ, IL-1β, TNF, CXCL1, IL-2, IL-10), which varied across brain regions (hypothalamus, hippocampus, and frontal cortex) and time (24 h and 7d) post-S1 treatment. Direct exposure of microglia to S1 resulted in increased gene expression (Il1b, Il6, Tnf, Nlrp3) and protein levels (IL-1β, IL-6, TNF, CXCL1, IL-10). S1 also activated TLR2 and TLR4 receptor signaling in HEK293 transgenic cells. Taken together, these findings suggest that structural proteins derived from SARS-CoV-2 might function independently as PAMPs to induce neuroinflammatory processes via pattern recognition receptor engagement.
Keywords: SARS-CoV-2, Spike protein, S1 subunit, PAMP, TLR, Neuroinflammation, Microglia, Sickness behavior
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an RNA betacoronavirus (Wu et al., 2020) that is the causative agent of the global coronavirus disease 2019 (Covid-19) pandemic (Zhou et al., 2020). The spectrum of Covid-19 clinical symptoms ranges from asymptomatic infection to severe critical illness characterized by flu-like symptoms, dyspnea, hypoxemia, and a high risk of respiratory failure (Berlin et al., 2020; Gandhi et al., 2020). Notably, approximately one-third of Covid-19 patients develop neurological and neuropsychiatric symptoms, including anxiety, depression, PTSD, cognitive deficits, fatigue, and sleep disturbances (Schou et al., 2021). The mechanism(s) mediating these neurological and neuropsychiatric effects of SARS-CoV-2 infection has yet to be established. Several studies have demonstrated neuroinflammation in Covid-19 patients (Benameur et al., 2020, Bodro et al., 2020, Boroujeni et al., 2021, Eden et al., 2021, Farhadian et al., 2020, Nuovo et al., 2021, Pilotto et al., 2020, Song et al., 2020, Thakur et al., 2021, Yang et al., 2021), which might underpin these neurological and neuropsychiatric symptoms. Here, we explore the possibility that SARS-CoV-2 viral proteins, independent of viral CNS infection, function as pathogen-associated molecular patterns (PAMPs), which directly induce neuroinflammatory/behavioral effects via pattern recognition receptors (PRR) such as toll-like receptor (TLR)2 and TLR4 in the CNS.
Several mechanisms have been proposed that might mediate the neurological and neuropsychiatric sequelae of SARS-CoV-2 infection. These mechanisms include peripheral cytokine signaling to the CNS via immune-to-brain signaling pathways and/or direct SARS-CoV-2 infection of the CNS (Boldrini et al., 2021, Kempuraj et al., 2020), which might induce neuroinflammatory processes and the neuropsychiatric consequences of those processes. Indeed, a peripheral “cytokine storm” or pathological peripheral inflammation occurs in some Covid-19 patients (Merad et al., 2020; Del Valle et al., 2020). However, regarding direct viral infection of the CNS, there is little evidence in support of this mechanism (Lewis et al., 2021). Moreover, viral infection of the CNS appears to occur largely in patients with underlying chronic inflammatory diseases (Matschke et al., 2020). Alternatively, there is mounting evidence that viral-derived proteins, independent of SARS-CoV-2 viral infection itself, function as PAMPs, which then might drive inflammatory processes via PRR engagement (see below).
SARS-CoV-2 is an enveloped virus that contains positive, single-stranded RNA contained within a capsid comprised of the spike (S), envelope (E), membrane (M) and nucleocapsid (N) structural proteins (Mariano et al., 2020). CoV-2 infects cells after the S protein binds angiotensin-converting-enzyme 2 (ACE2) expressed on cells throughout the body with high expression in human respiratory epithelium (Donoghue et al., 2000). The S protein is comprised of S1 and S2 subunits. The S1 N-terminal domain of the S protein binds ACE2, and the C-terminal S2 domain is responsible for viral-cellular fusion (Hoffmann et al., 2020). Detachment of S1 from S2 facilitates the viral-cellular fusion machinery (Hu et al., 2021) and is critical for cell entry.
In addition, there is evidence that alternate receptors (e.g., neuropilin) mediate viral entry (Wenzel et al., 2021). Interestingly, the S1 subunit can undergo spontaneous dissociation from the mature virion in the absence of ACE2 binding (Cai et al., 2020) and that S1 is spontaneously shed from SARS-CoV-2 virions (Zhang et al., 2020). In support of these findings, the S1 subunit as well as the N protein have been detected at high levels in plasma of Covid-19 positive patients (Deng et al., 2021, Hingrat et al., 2020, Ogata et al., 2020, Perna et al., 2021). Notably, Perna et al. found that N protein levels were highly correlated with inflammatory status (C-reactive protein serum levels) and disease severity (Perna et al., 2021). These findings suggest that viral proteins are released from the SARS-CoV-2 virus and enter the circulation, which would allow access to the CNS. Indeed, Nuovo et al., using immunohistochemistry, detected the S protein in post-mortem human brain of patients who died of Covid-19. In brain, the S protein was largely co-localized with endothelial cells (Nuovo et al., 2021). They also found that proinflammatory mediators (caspase 3, IL-6, and TNF) co-localized with S staining. Intriguingly, viral RNA was largely not detected. Consistent with these findings, Matschke et al. detected the S and N protein in the absence of viral RNA in post-mortem brain in a subset of Covid-19 patients (Matschke et al., 2020). Moreover, Rhea et al. demonstrated that S1 injected IV crossed the BBB (Rhea et al., 2021). Likewise, Nuovo et al. injected the full-length S1 subunit intravenously (IV) into mice and 5 days after injection, found that it associated with endothelial cells in the brain along with activated caspase 3, IL-6, TNF, and the complement protein C5b-9. The S2 subunit was also injected IV but was not detected in brain nor did S2 induce proinflammatory mediators in brain endothelial cells (caspase 3, IL-6, TNF and C5b-9). Of note, S1 also induced brain endothelial cell damage suggesting that S1 might be capable of inducing damage-associated molecular patterns (DAMPs), for example high mobility group box 1 (HMGB1), which in turn are proinflammatory (Bianchi, 2007). These findings suggest that viral proteins, independent of SARS-CoV-2, might enter the brain, and function as PAMPs and/or induce DAMPs to elicit neuroinflammatory immune responses, and thus play a role in the pathogenesis of Covid-19.
Several studies have now examined the possibility that SARS-CoV-2 proteins might function as PAMPs to elicit inflammatory responses via signaling through TLRs including TLR2 and TLR4. TLRs are germ-line encoded receptors expressed by innate immune cells including macrophages and dendritic cells, as well as microglia, and function as PRRs to recognize molecular motifs common to bacterial and viral pathogens. For example, TLR4 recognizes lipopolysaccharide (Kumar et al., 2011). Many TLRs, including TLR2 and TLR4, signal via intracellular Toll-IL1 receptor domains. There then ensues a cascade that triggers degradation of IκB and activation of JNK, and the downstream transcription factors NF-κB and AP-1 that control numerous inflammation-related genes (Kawai and Akira, 2010). While TLR2 and TLR4 bind PAMPs lipoteichoic acid and lipopolysaccharides respectively, these PRRs also bind DAMPs including HMGB1 to elicit proinflammatory responses (Bianchi, 2007). Importantly, Olajide et al. reported that the S1 subunit of the S protein induced a proinflammatory response in human peripheral blood mononuclear cells, which was characterized by increased levels of cytokines, chemokines, NF-κβ, NLRP3 and caspase-1 (Olajide et al., 2021b). Utilizing the microglia cell line BV-2, Olajide also found that S1 induced a similar proinflammatory response via TLR4 signaling (Olajide et al., 2021a). Likewise, Shirato et al. demonstrated that the S1 subunit was sufficient to induce proinflammatory cytokines via TLR4 signaling in murine and human macrophages (Shirato and Kizaki, 2021). Consistent with this finding, the S protein binds to TLR4 with high affinity and induces a proinflammatory response in THP-1 and RAW 264.7 cells, which was blocked by a TLR4 inhibitor (Zhao et al., 2021). Alternatively, Zheng et al. reported that the E protein was sufficient to induce a proinflammatory response in bone marrow-derived macrophages, an effect blocked in TLR2 KO cells (Zheng et al., 2021). Zheng et al. also reported that the E protein, when delivered intratracheally, was sufficient to induce a proinflammatory response in lungs of mice, which was abrogated in TLR2 KO mice.
Taken together, these findings suggest that structural proteins derived from SARS-CoV-2 might be sufficient to function as PAMPs to elicit neuroinflammatory effects independent of SARS-CoV-2 cellular infection. To date, however, a systematic study of the neuroinflammatory/microglial and behavioral effects of SARS-CoV-2 structural proteins has not been conducted. Of the several structural proteins available for study, we chose to investigate the neuroinflammatory properties of the S1 subunit given that 1) the S protein has been detected in post-mortem brains of Covid-19 patients, which co-localized with inflammatory proteins (Nuovo et al., 2021), 2) the S1 but not the S2 subunit is sufficient to induce proinflammatory mediators in endothelial cells (Nuovo et al., 2021), 3) the S1 subunit is sufficient to induce proinflammatory responses in vitro (Olajide et al., 2021b) via TLR4 (Olajide et al., 2021a, Shirato and Kizaki, 2021) and 3) when injected IV, the S1 subunit crosses the BBB (Rhea et al., 2021), which would allow it to directly signal at PRRs on microglia and other cell types within the parenchyma of the brain. Therefore, we tested if the S1 subunit in vivo, is sufficient to produce behavioral sickness and neuroinflammatory responses. Using in vitro models, we also determined if S1 can directly activate adult primary microglia and transgenic human TLR2 and TLR4 HEK293 cells.
2. Methods
2.1. Animals
Male Sprague-Dawley rats (60 – 90 d of age; Envigo, Indianapolis, IN) were pair housed on a 12-h light–dark cycle (lights on at 0700 h). Food (standard laboratory chow) and water were available ad libitum. Rats were allowed to acclimate to colony conditions for at least one week prior to experimentation. All experiments were approved by the Institutional Animal Care and Use Committee of the University of Colorado Boulder in compliance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals as well as the ARRIVE guidelines for animal use.
2.2. SARS-CoV-2 S protein subunit proteins
Recombinant SARS-CoV-2 full-length S1 and S2 subunits were obtained from RayBiotech (cat#: 230–30161 and 230–30163, respectively). The genomic sequence of these subunits was derived from the original Wuhan-Hu-1 isolate (Wu et al., 2020). Of note, the S1 (Val16-Gln690) and S2 (Met697-Pro1213) proteins used here are also derived from HEK293 cells and are devoid of the immunogen LPS. The S1 protein is identical to the protein (Raybiotech) used by Rhea et al. who found that it crossed the BBB and entered the brain parenchyma after IV injection (Rhea et al., 2021).
2.3. Intra-cisterna magna (ICM) injection
We chose to inject the S1 subunit (1.0 μg) or vehicle (2.5 μl, 0.2 μm filtered, sterile 1x PBS, pH 7.4) ICM to directly administer the subunit into the CNS and thus examine direct effects in CNS. This route of administration obviates peripheral inflammatory effects that would be induced by IV injection, which would introduce a potential confound. Indeed, Nuovo et al. found that IV injection of the S1 subunit resulted in substantial levels of S1 in liver (Nuovo et al., 2021). The dose of the S1 subunit used here was based on this prior study in mice demonstrating the neuroinflammatory effects of the S1 subunit (Nuovo et al., 2021). Nuovo et al. injected mice IV with 10 μg S1 (Nuovo et al., 2021). Given that we chose to inject S1 ICM, we selected a dose 10-fold lower than the dose used by Nuovo et al. We have demonstrated that ICM injected substances reach forebrain regions in the CNS consistent with more typical ICV procedures, and this procedure produces no detectable inflammatory responses (Frank et al., 2012). Rats were anesthetized with 5% isoflurane in oxygen and then maintained on 3% isoflurane during the brief procedure (∼3 min). The dorsal aspect of the skull was shaved and swabbed with 70% EtOH. A sterile 27-gauge needle attached via sterile PE50 tubing to a 25 μl Hamilton syringe was inserted into the cisterna magna (verified by withdrawing 2 μl of clear CSF) and drug injected over a 30 s period. After injection, the needle was left in place for 30 s to allow for diffusion of drug.
2.4. Behavioral measures
After ICM injection, in-cage behavioral monitoring (8 h post-ICM) and juvenile social exploration (JSE; 24 h post-ICM) were conducted blind to treatment condition.
2.4.1. In-cage monitoring
In-cage behaviors were recorded for 2 h (1 h pre to 1 h post lights off) using a camera equipped with infrared video acquisition (Axis, M3104). Infrared cameras were placed in proximity to the rats' home cages, which allowed side-to-side 24 h video recordings of home cage behavior. These cameras introduce no light, provide excellent visual resolution of the animals during the light and dark cycles, and do not disrupt rat sleep or diurnal rhythmicity. All tails were marked, allowing the observer to assess individual rat behavior. A Synology Surveillance Station was used to acquire video recordings. Duration of behaviors (total activity, drinking, eating, self-grooming, allo-grooming, wall rearing and wrestling) were manually scored blind to treatment condition using the Behavioral Observation Research Interactive Software (Friard and Gamba, 2016). Home cage behaviors are based on the Stanford Ethogram definitions (https://mousebehavior.org ).
2.4.2. Juvenile social exploration (JSE)
JSE is a widely used measure of social avoidance and validated measure of anxiety (File and Seth, 2003) and is sensitive to the neuroinflammatory effects of stress and immunogenic agents such as LPS (Frank et al., 2012, Goshen and Yirmiya, 2009). Here, JSE was measured 1 day prior to (baseline) and 24 h post-S1/vehicle treatment. Each experimental rat was transferred to a novel cage with shaved wood bedding in a dimly lit room (40 lx). After a 15-min habituation period, a 28 to 32-day-old juvenile male rat was introduced to the adult rat’s cage for 5 min. The adult rat was then tested for time exploring the juvenile rat. Exploratory behaviors of the adult (sniffing, pinning, licking, and allo-grooming of the juvenile) were timed by an observer blind to treatment condition. After the test, the juvenile was removed, and the experimental adult rat was returned to its home cage. Although juvenile stimulus rats were reused for multiple tests, an adult rat was never re-tested with the same juvenile. For each adult rat, amount of time exploring the juvenile (JSE) was reported and quantified as a percent of baseline JSE.
2.5. Tissue dissection of brain regions
Immediately after the last behavioral test (JSE), animals were given a lethal dose of sodium pentobarbital. Animals were fully anesthetized and transcardially perfused with ice-cold saline (0.9%) for 3 min to remove peripheral immune leukocytes from the CNS vasculature. Brain was rapidly extracted, and hippocampus, hypothalamus and frontal cortex was dissected. These regions were chosen because they are involved in a broad array of process and will inform on regional differences. For hippocampus, one hemisphere was used for protein assays and the other hemisphere for RT-PCR of gene expression. RT-PCR was only conducted on hypothalamus and frontal cortex. Tissues were flash frozen in liquid nitrogen and stored at −80 °C.
2.6. Tissue processing for protein assays
Hippocampal samples were sonicated on ice using a tissue extraction reagent (Invitrogen, cat#: FNN0071) supplemented with a protease inhibitor cocktail (Sigma-Aldrich, cat#: P2714). Homogenates were centrifuged (14,000 × g for 10 min at 4 °C) and supernatants collected and stored at − 80 °C. Total protein was quantified using a Bradford assay.
2.7. Enzyme-linked immunosorbent assay (ELISA)
A multiplex ELISA was run to assay protein levels of rat IFNγ, IL-1β, IL-6, TNF, CXCL1, IL-2 and IL-10 using the Simoa Rat Cytokine Panel 1 7-Plex kit (Quanterix; cat#: 85–0015). An ELISA for rat IL-6 (R&D Systems, cat#: RLB00) was conducted as IL-6 protein was not detected using the multiplex ELISA. ELISAs were run according to the manufacturer's instructions and protein levels normalized to total protein.
2.8. Real time RT-PCR measurement of gene expression
Total RNA was isolated from hippocampus using TRI Reagent (MilliPore Sigma, cat#: 93289) and a standard method of phenol:chloroform extraction (Chomczynski and Sacchi, 1987). Total RNA was quantified using a NanoDrop 2000 spectrophotometer (ThermoFisher). cDNA synthesis was performed using the SuperScript II Reverse Transcriptase kit (ThermoFisher, cat#: 18064014). A detailed description of the PCR amplification protocol has been published previously (Frank et al., 2006). cDNA sequences were obtained from Genbank at the National Center for Biotechnology Information (NCBI; www.ncbi.nlm.nih.gov). Primer sequences were designed using the Operon Oligo Analysis Tool (http://www.operon.com/tools/oligo-analysis-tool.aspx) and tested for sequence specificity using the Basic Local Alignment Search Tool at NCBI (Altschul et al., 1997). Primers were obtained from ThermoFisher. Primer specificity was verified by melt curve analyses. All primers were designed to span exon/exon boundaries and thus exclude amplification of genomic DNA. Primer sequences are detailed in Supplemental data (Table 1 ). PCR amplification of cDNA was performed using the Quantitect SYBR Green PCR Kit (Qiagen, cat#: 204145). Formation of PCR product was monitored in real time using the CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Relative gene expression was determined using Actb as the housekeeping gene and the 2-ΔΔCT method (Livak and Schmittgen, 2001).
Table 1.
Primer sequences. Abbreviations: Actb, β-actin; Cd, Cluster of Differentiation; Cx3cr1, CX3C Chemokine Receptor 1; Gfap, Glial Fibrillary Acidic Protein; Hmgb1, High Mobility Group Box 1; Il, Interleukin; Iba1, Ionized calcium-binding adaptor molecule-1; MhcII, Major Histocompatibility Complex II; Nlrp3, NACHT Domain-, Leucine-Rich Repeat-, And PYD-Containing Protein 3; Tlr, Toll-Like Receptor; Tnf, Tumor Necrosis Factor.
| Gene | Primer Sequence5′ → 3′ | Function |
|---|---|---|
| Actb | F: TTCCTTCCTGGGTATGGAAT R: GAGGAGCAATGATCTTGATC |
Cytoskeletal protein (Housekeeping gene) |
| Cd11b | F: CTGGTACATCGAGACTTCTC R: TTGGTCTCTGTCTGAGCCTT |
Microglia/Macrophage antigen |
| Cd163 | F: GTAGTAGTCATTCAACCCTCAC R: CGGCTTACAGTTTCCTCAAG |
Hemoglobin receptor expressed by macrophages, but not microglia |
| Cd200r1 | F: TAGAGGGGGTGACCAATTAT R: TACATTTTCTGCAGCCACTG |
Cognate receptor for CD200 |
| Cx3cr1 | F: TCAGGACCTCACCATGCCTA R: CGAACGTGAAGACAAGGGAG |
Cognate receptor for CX3CL1 |
| Gfap | F: AGATCCGAGAAACCAGCCTG R: CCTTAATGACCTCGCCATCC |
Astrocyte antigen |
| Hmgb1 | F: GAGGTGGAAGACCATGTCTG R: AAGAAGAAGGCCGAAGGAGG |
DAMP |
| Il1b | F: CCTTGTGCAAGTGTCTGAAG R: GGGCTTGGAAGCAATCCTTA |
Pro-inflammatory cytokine |
| Il6 | F: AGAAAAGAGTTGTGCAATGGCA R: GGCAAATTTCCTGGTTATATCC |
Pro-inflammatory cytokine |
| Iba1 | F: GGCAATGGAGATATCGATAT R: AGAATCATTCTCAAGATGGC |
Microglia/Macrophage antigen |
| MhcIIα | F: AGCACTGGGAGTTTGAAGAG R: AAGCCATCACCTCCTGGTAT |
Microglia/Macrophage antigen |
| Nlrp3 | F: AGAAGCTGGGGTTGGTGAATT R: GTTGTCTAACTCCAGCATCTG |
Inflammasome component mediating caspase-1/IL-1β activation |
| Tlr2 | F: TGGAGGTCTCCAGGTCAAATC R: ACAGAGATGCCTGGGCAGAAT |
Receptor for lipotechoic acid and DAMPs |
| Tlr4 | F: TCCCTGCATAGAGGTACTTC R: CACACCTGGATAAATCCAGC |
Receptor for LPS and DAMPs |
| Tnf | F: CAAGGAGGAGAAGTTCCCA R: TTGGTGGTTTGCTACGACG |
Pro-inflammatory cytokine |
2.9. HEK293 cells transfected with human (h)TLR2 and hTLR4
HEK293 cells transfected with hTLR2 (HEK-Blue hTLR2, cat#: hkb-htlr2) and hTLR4 (HEK-Blue hTLR4, cat#: hkb-tlr4) genes were obtained from Invivogen. hTLR4 cells were co-transfected with the MD-2/CD14 co-receptor gene, while hTLR2 cells were co-transfected with the CD14 gene to enhance TLR2 responsiveness, respectively. These cell lines are also co-transfected with a secreted embryonic alkaline phosphatase (SEAP) reporter gene, which is placed under the control of an interferon-β minimal promoter fused to five NF-κβ (nuclear factor kappa-light-chain-enhancer of activated B cells) and AP-1 (activator protein-1) binding sites. Stimulation with a TLR2 or TLR4 ligand activates NF-κβ and AP-1, which induce the production and extracellular release of SEAP. Extracellular levels of SEAP are thus indicative of TLR signaling. Of note, HEK293 cells express endogenous levels of the PRRs TLR3, TLR5 and NOD1. Therefore, the parental cell line lacking hTLR2 and hTLR4 (null cells; HEK-Blue Null1, cat#: hkb-null1), but expressing the SEAP reporter gene, was assayed to control for effects via HEK293 endogenous PRRs.
hTLR2, hTLR4 and null cells were cultured in T-25 tissue culture flasks (Corning, cat#: 353108) in DMEM(1x) + GlutaMax-1 (Gibco, cat#: 10569–010) supplemented with 10% heat inactivated (HI) FBS (Gibco, cat#: 0082–139), 100U/ml Penicillin-Streptomycin (Gibco, 15140–148), 100 μg/ml Normocin (InvivoGen, cat#: ant-nr-1), and 1x HEK-Blue™ Selection (InvivoGen, cat#: hb-sel) at 37 °C, 5% CO2. Cells were cultured to 50–80% confluence prior to experiments. Cells were counted and cell viability determined by trypan blue exclusion. For all experiments, cell viability was greater than 95%. Cells were centrifuged (1000 × g, 10 min) and suspended in DMEM(1x) + GlutaMax-1 supplemented with 10% HI FBS to yield 1 × 10^5 cells/90 μl. Cells (90 μl) were then plated in a 96 well flat-bottom tissue culture treated plate with white walls (Corning, cat#: 3610). S1 and S2 were diluted in culture media (DMEM(1x), GlutaMax-1, 10% HI FBS) and added (10 μl) to cells to yield final concentrations of protein (0, 0.01, 0.1, and 1 μg/ml). Each concentration of S1 and S2 were assayed in duplicate. Cells were incubated for 4 h at 37 °C, 5% CO2.
SEAP was directly detected in microplate wells according to the manufacturer’s instructions (NovaBright™ SEAP Enzyme Reporter Gene Chemiluminescent Detection System 2.0, Invitrogen, cat#: N10577). Chemiluminescence was measured using an Infinite M200Pro multimode plate reader (Tecan). The plate reader was preheated to 37 °C prior to chemiluminescent detection. Immediately upon addition of the chemiluminescent substrate, the plate was loaded into the plate reader and chemiluminescence measured once every 60 s over a 10 min interval.
2.10. Isolation of adult primary rat microglia
Whole brain microglia were isolated using a Percoll density gradient as previously described (Frank et al., 2006). We have previously shown (Frank et al., 2006) that this microglia isolation procedure yields highly pure microglia (Iba-1+/MHCII+/CD163-/GFAP-). In the present experiments, immunophenotype and purity of microglia was assessed using real time RT-PCR. Microglia were suspended in DMEM + GlutaMax-1 + 10% HI FBS and microglia concentration determined by trypan blue exclusion. Microglia concentration was adjusted to a density of 4–6 × 104 cells/100 μl and 100 μl added to individual wells of a 96-well v-bottom plate. Cells were incubated with the S1 subunit (0, 0.01, 0.1, and 1 μg/ml) for 24 h at 37 °C, 5% CO2. Supernatants were collected for protein assays and cells lysed for RT-PCR. The plate was centrifuged at 1000 × g for 10 min at 4 °C to pellet cells. Supernatant was collected and stored at −80 °C and cells washed 1x in ice cold PBS and centrifuged at 1000 × g for 10 min at 4 °C. Cell lysis was conducted using the CellsDirect procedure (Invitrogen; cat#: 117390) and cDNA synthesis was performed according to the manufacturer’s protocol using SuperScript IV (Invitrogen, cat#: 18091050).
2.11. Statistical analysis
All data are presented as mean + SEM. Statistical analyses consisted of the following: t-tests for behavior, a 2 (drug; vehicle vs S1) × 3 (brain region; hypothalamus vs hippocampus vs frontal cortex) mixed ANOVA for neuroinflammatory gene expression, t-tests for hippocampal protein levels and a one-way ANOVA for in vitro experiments. The Bonferroni-Holm post-hoc test was used to correct for multiple testing. Analyses were conducted using Prism 8 (Graphpad Software, LLC). Threshold for statistical significance was set at α = 0.05. Sample sizes are provided in figure captions.
3. Results
3.1. Behavioral effects of S1
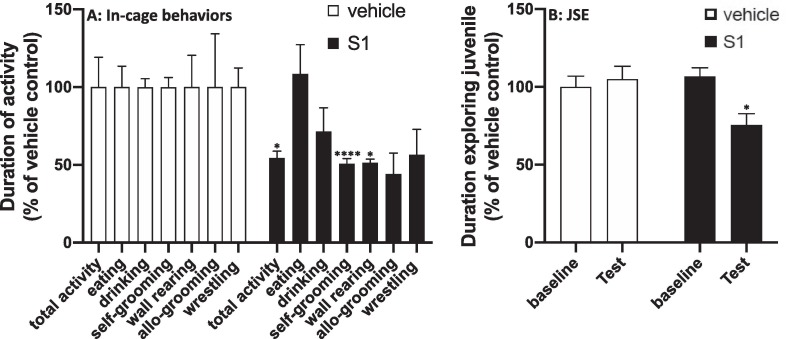
S1 treatment significantly reduced the duration of several in-cage behaviors (Fig. 1 A) compared to vehicle treatment including total activity (df = 10, t = 2.31, p = 0.04), self-grooming (df = 10, t = 6.98, p < 0.0001) and wall rearing (df = 10, t = 2.37, p = 0.04), however eating (df = 10, t = 0.36, p = 0.72.), drinking (df = 10, t = 1.76, p = 0.11) and allo-grooming (df = 10, t = 2.37, p = 0.16) were not significantly affected. The effect of S1 on wrestling behavior was marginally significant (df = 10, t = 2.13, p = 0.06).
Fig. 1.
Effect of S1 on behavior. Rats were injected ICM with vehicle (1x PBS) or S1 (1 μg). (A) 8 h after ICM injection, home cage infrared video recordings were made of in-cage behaviors from 1 h pre- to 1 h post-lights off and duration of behaviors scored based on Stanford Ethogram definitions. (B) JSE was scored 24 h prior to ICM injection (baseline) and 24 h after ICM injection (test). Data are presented as the mean + SEM. N = 6/group. S1 vs vehicle, *p < 0.05, ****p < 0.0001.
JSE was then conducted the next morning as a measure of social avoidance (Fig. 1 B). Baseline JSE was not significantly different between treatment groups prior to ICM injection (df = 10, t = 0.76, p = 0.46), however S1 treatment significantly reduced the amount of time the adult rat engaged or socially explored the juvenile compared to vehicle control (df = 10, t = 2.69, p = 0.02). Engagement in most of these behaviors (i.e., motor activity, social behavior, and self-grooming) is reduced during the sickness response to infection, which is thought to reflect a shift in the motivational state of the organism (Dantzer et al., 2008). Furthermore, proinflammatory cytokines are both necessary and sufficient to elicit this sickness response (Dantzer, 2009). Thus, these behavioral effects of S1 suggest that S1 might induce a neuroinflammatory response. Therefore, we determined whether S1 administration induces neuroinflammatory effects in several brain regions.
3.2. Neuroinflammatory effects of S1 (mRNA)
Towards examining the neuroinflammatory effects of S1, we measured expression of an array of genes including microglia/brain macrophage activation markers (Iba1, Cd11b, MhcIIα (RT1-Da)), microglia/brain macrophage checkpoint receptors (Cd200r1 and Cx3cr1), an astrocyte activation marker (Gfap), pattern recognition receptors (Tlr2, Tlr4), inflammasomes (Nlrp3), proinflammatory cytokines (Il1b, Il6, Tnf) and damage-associated molecular patterns (Hmgb1).
3.2.1. 24 h after ICM injection
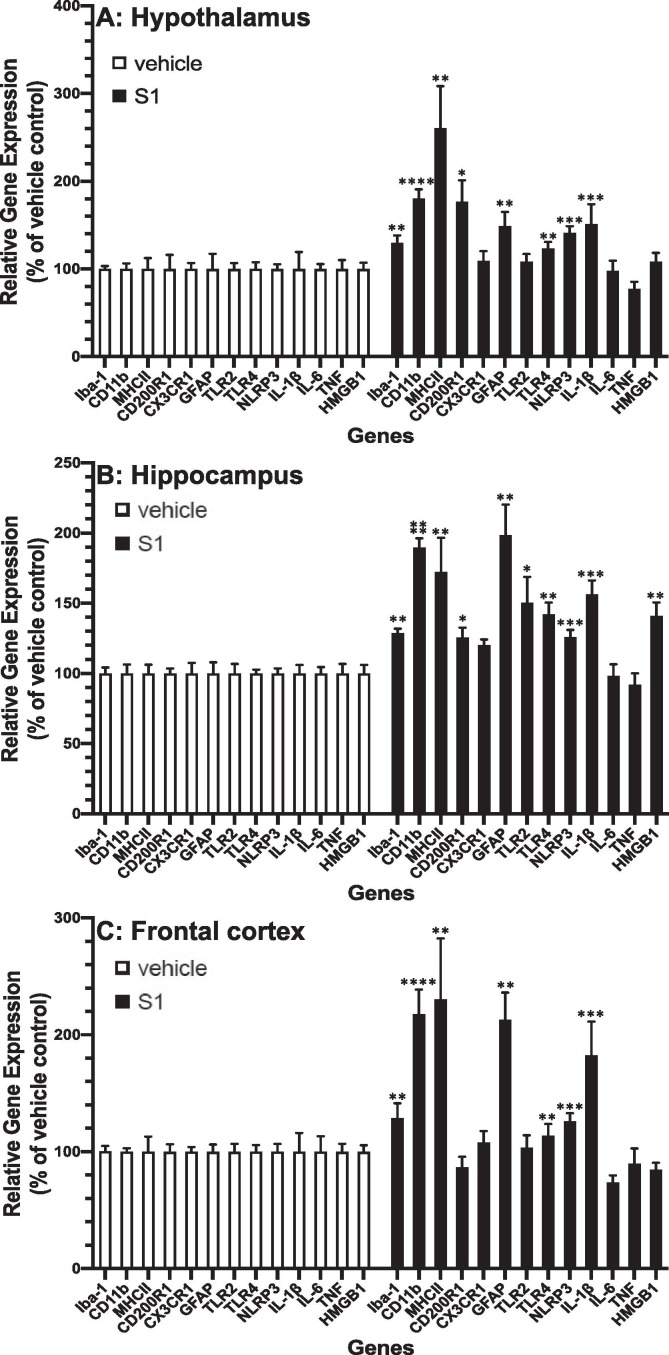
In hypothalamus (Fig. 2 A), hippocampus (Fig. 2 B) and frontal cortex (Fig. 2 C), we found that across all brain regions S1 increased gene expression of Iba1 (S1 main effect; df = 1, 10, F = 16.19, p = 0.002), Cd11b (S1 main effect; df = 1, 10, F = 70.4, p < 0.0001), MhcIIα (S1 main effect; df = 1, 10, F = 17.2, p = 0.002), Tlr4 (S1 main effect; df = 1, 10, F = 19.77, p = 0.001), Nlrp3 (S1 main effect; df = 1, 10, F = 25.47, p = 0.0005), Gfap (S1 main effect; df = 1, 10, F = 18.17, p = 0.0017) and Il1b (S1 main effect; df = 1, 10, F = 29.62, p = 0.0003). Several genes were differentially expressed in brain regions. S1 increased Cd200r1 (brain region × S1 interaction; df = 2, 20, F = 7.34, p = 0.004) in hippocampus (p < 0.05) and hypothalamus (p < 0.05). S1 also increased the DAMP Hmgb1 (brain region × S1 interaction; df = 2, 20, F = 9.33, p = 0.0014) and Tlr2 (brain region × S1 interaction; df = 2, 20, F = 6.07, p = 0.009) in hippocampus (Tlr2, p < 0.05; Hmgb1, p < 0.05).
Fig. 2.
Effects of S1 24 h post-ICM injection on neuroinflammatory genes. Rats were injected ICM with vehicle (1x PBS) or S1 (1 μg). 24 h after ICM injection, gene expression of glial activation markers and neuroinflammatory-related genes was measured in (A) hypothalamus, (B) hippocampus and (C) frontal cortex. Data are presented as the mean + SEM. N = 6/group. S1 vs vehicle, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
3.2.2. 7d after ICM injection
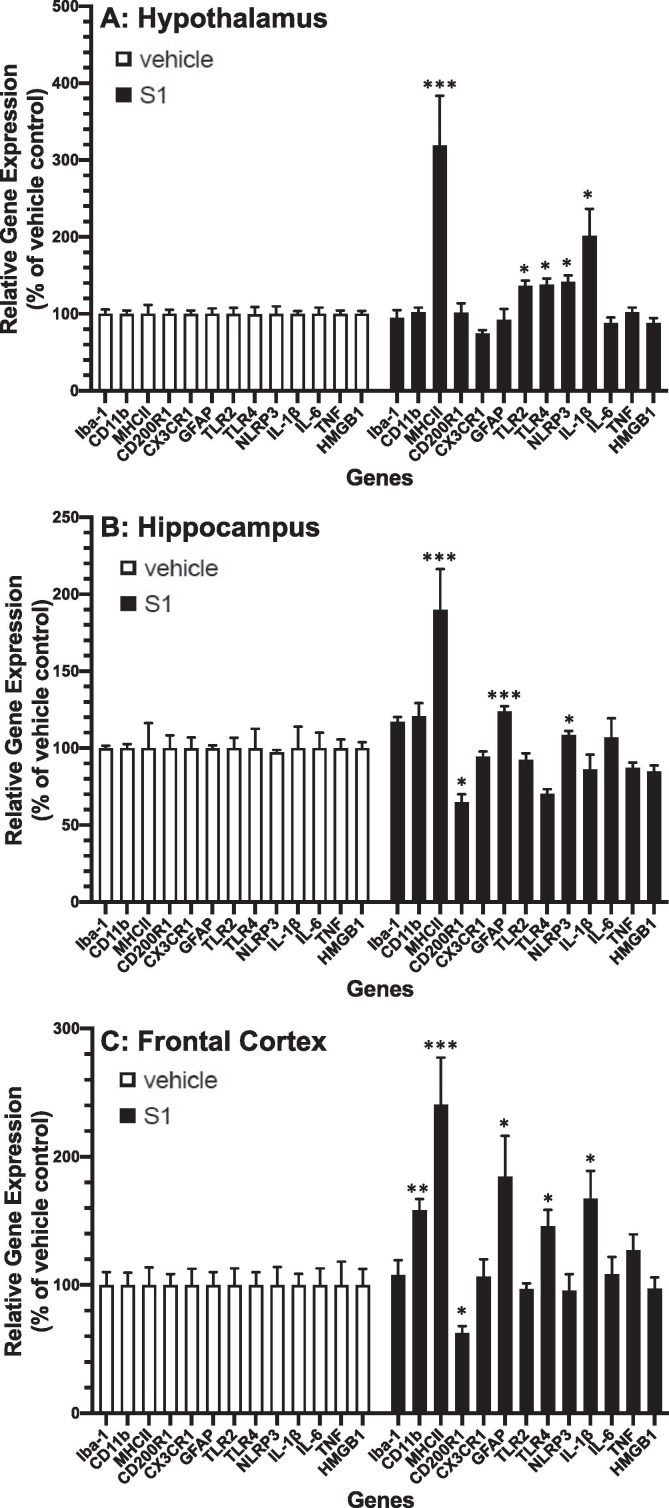
Given the effects of S1 observed 24 h post-ICM injection, we examined the durability of these neuroinflammatory effects in hypothalamus (Fig. 3 A), hippocampus (Fig. 3 B) and frontal cortex (Fig. 3 C). We found that at 7d after ICM injection, S1 increased gene expression across all brain regions for MhcIIα (S1 main effect; df = 1, 10, F = 25.03, p = 0.0005). Several genes were differentially expressed in brain regions. S1 increased Cd11b (brain region × S1 interaction; df = 2, 20, F = 10.54, p = 0.0007) in frontal cortex (p < 0.01), Gfap (brain region × S1 interaction; df = 2, 20, F = 5.61, p = 0.012) in hippocampus (p < 0.001), Tlr2 (brain region × S1 interaction; df = 2, 20, F = 6.02, p = 0.009) in hypothalamus (p < 0.05), Tlr4 (brain region × S1 interaction; df = 2, 20, F = 9.6, p = 0.0012) in hypothalamus (p < 0.05) and frontal cortex (p < 0.05), Nlrp3 (brain region × S1 interaction; df = 2, 20, F = 4.01, p = 0.034) in hypothalamus (p < 0.05) and hippocampus (p < 0.05) and Il1b (brain region × S1 interaction; df = 2, 20, F = 9.05, p = 0.0016) in hypothalamus (p < 0.05) and frontal cortex (p < 0.05). S1 decreased Cd200r1 (brain region × S1 interaction; df = 2, 20, F = 3.53, p = 0.049) in hippocampus (p < 0.05) and frontal cortex (p < 0.05).
Fig. 3.
Effects of S1 7d post-ICM injection on neuroinflammatory genes. Rats were injected ICM with vehicle (1x PBS) or S1 (1 μg). 7d after ICM injection, gene expression of glial activation markers and neuroinflammatory-related genes was measured in (A) hypothalamus, (B) hippocampus and (C) frontal cortex. Data are presented as the mean + SEM. N = 6/group. S1 vs vehicle, *p < 0.05, **p < 0.01, ***p < 0.001.
3.3. Neuroinflammatory effects of S1 (protein)
To examine whether the neuroinflammatory effects of S1 extended to the protein level, we measured several proteins in hippocampal tissue lysate including IL-1β, IL-6, TNF, CXCL1, IL-10, IFNγ and IL-2.
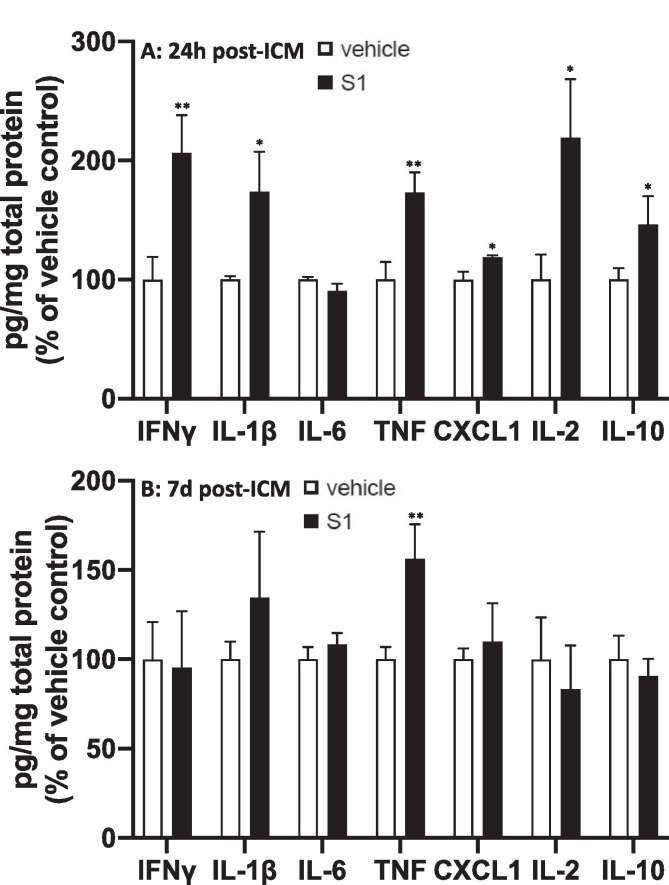
3.3.1. 24 h after ICM injection (Fig. 4A)
Fig. 4.
Effects of S1 24 h and 7d post-ICM injection on neuroinflammatory proteins. Rats were injected ICM with vehicle (1x PBS) or S1 (1 μg). Protein levels of neuroinflammatory-related proteins were measured in hippocampus (A) 24 h and (B) 7d after ICM injection. Data are presented as the mean + SEM. N = 5–6/group. S1 vs vehicle, *p < 0.05, **p < 0.01.
S1 treatment increased protein levels of IL-1β (df = 10, t = 2.96, p = 0.007), TNF (df = 10, t = 3.25, p = 0.005), CXCL1 (df = 10, t = 2.47, p = 0.015), IFNγ (df = 10, t = 2.98, p = 0.005), IL-2 (df = 10, t = 2.37, p = 0.02) and IL-10 (df = 10, t = 2.98, p = 0.04) compared to vehicle treatment. IL-6 was not significantly altered by S1.
3.3.2. 7d after ICM injection (Fig. 4B)
S1 treatment only increased protein levels of TNF (df = 10, t = 2.97, p = 0.007), whereas all other proteins were not significantly changed.
3.4. Proinflammatory effects of S1 on microglia in vitro
Given that microglia play a pivotal role in the neuroinflammatory response (Ransohoff and Perry, 2009) and that S1 induced a proinflammatory response in human peripheral blood mononuclear cells (Olajide et al., 2021b), BV2 cells (Olajide et al., 2021a) as well as human macrophages (Shirato and Kizaki, 2021), we examined the possibility that S1 might directly induce a proinflammatory response in primary microglia isolated from adult rat brain.
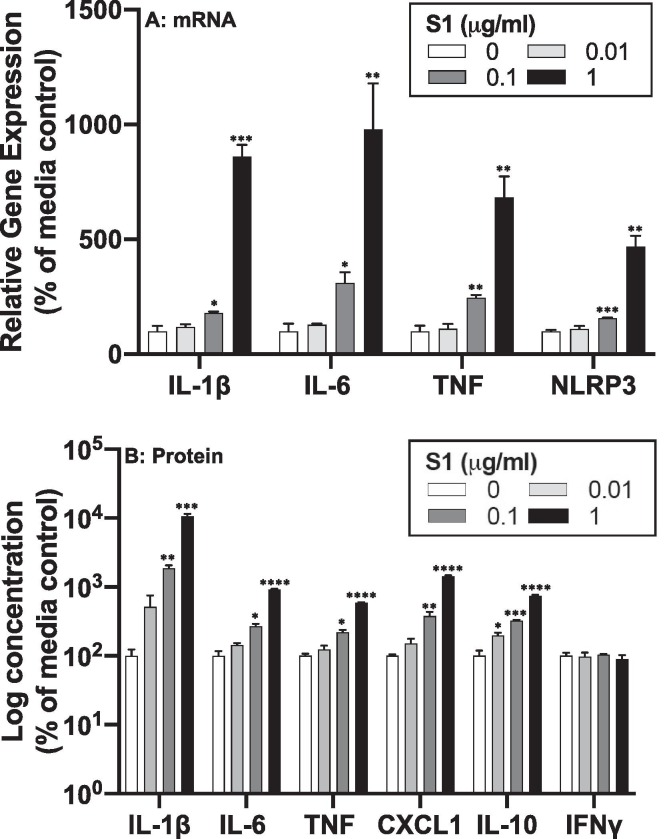
Microglia were directly exposed to S1 for 24 h, and cells analyzed for gene expression changes (Fig. 5 A) and cell culture supernatants assayed for protein levels (Fig. 5 B). S1 increased gene expression for Il1b (df = 3, 8, F = 159.0, p < 0.0001), Il6 (df = 3, 8, F = 15.5, p = 0.0011), Tnf (df = 3, 8, F = 31.71, p < 0.0001) and Nlrp3 (df = 3, 8, F = 50.81, p < 0.0001). S1 also increased protein levels for IL-1β (df = 3, 8, F = 130.5, p < 0.0001), IL-6 (df = 3, 8, F = 358.4, p < 0.0001), TNF (df = 3, 8, F = 313.2, p < 0.0001), CXCL1 (df = 3, 8, F = 187.8, p < 0.0001), and IL-10 (df = 3, 8, F = 181.0, p < 0.0001) in a concentration dependent manner. Post-hoc comparisons are depicted in Fig. 5.
Fig. 5.
Proinflammatory effects of S1 in isolated microglia. Whole brain microglia were isolated from adult rats and exposed to several concentrations of S1 (0, 0.01, 0.1, and 1 μg/ml) for 24 h. (A) RNA was isolated from cells and proinflammatory gene expression measured and (B) protein levels were measured in cell culture supernatants. Data are presented as the mean + SEM. N = 3 replications. S1 concentration vs media control, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
As noted in the Introduction, S1 contains the receptor binding domain for the ACE2 receptor, which SARS-CoV-2 predominantly utilizes to enter ACE2 + cells. Hernandez et al. found that ACE2 protein was found throughout the brain and colocalized largely with endothelial cells, neurons, and astrocytes (Hernández et al., 2021). An analysis of transcriptome databases also found widespread ACE2 expression in brain endothelial cells, neurons, astrocytes, and oligodendrocytes (Chen et al., 2020), however expression in microglia and other CNS macrophages was not found. Rhea et al. found that S1 bound ACE2 to a much higher degree in lung compared to brain, however there was no evidence of S1 binding ACE2 in liver, kidney, or spleen (Rhea et al., 2021). We examined ACE2 gene expression in whole brain primary microglia and failed to detect expression up to 40 cycles of PCR (data not shown) suggesting that microglia do not express the ACE2 receptor. This finding suggests that S1 signals through an alternate receptor on microglia. Given the findings of Shirato and Kizaki (2021) and Olajide et al. (2021) that demonstrated that S1 signals through the PRR TLR4 (Olajide et al., 2021a, Shirato and Kizaki, 2021), we explored the possibility that S1 might utilize alternate PRRs (i.e., TLR2) in addition to TLR4 to induce an inflammatory response.
3.5. Effects of S1 on hTLR2 and hTLR4 cells
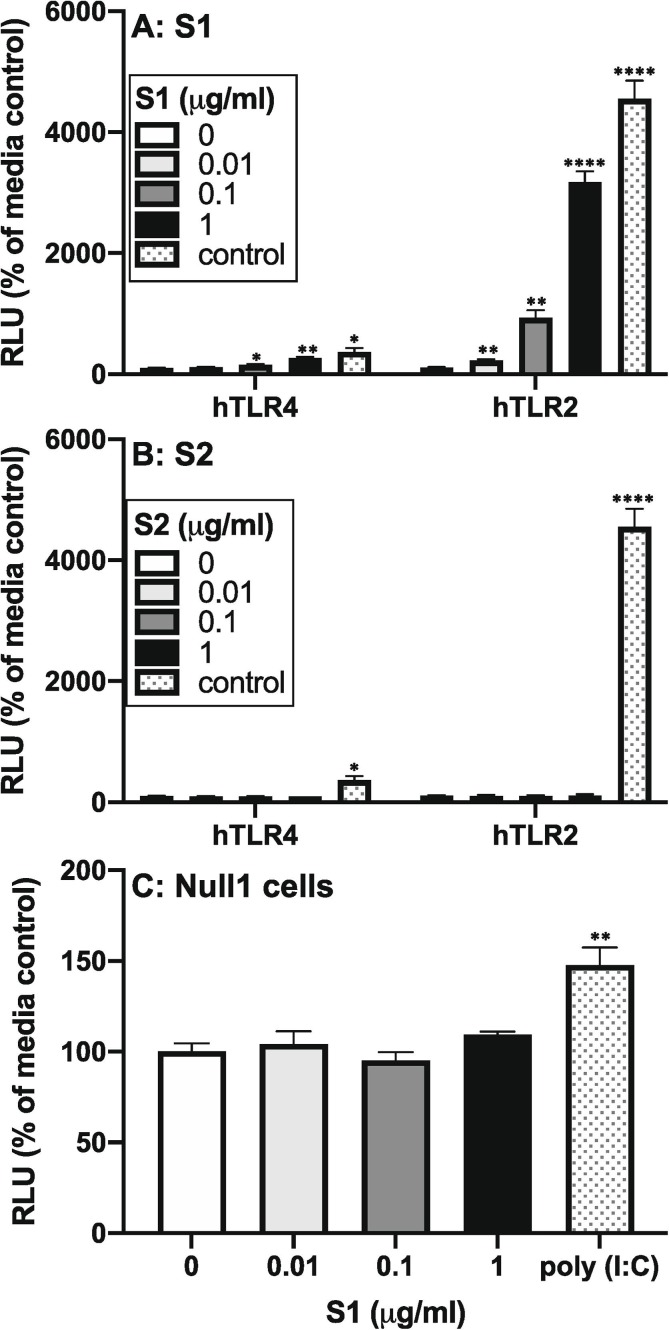
HEK293 cells expressing the hTLR2 or hTLR4 receptor were exposed to S1 (Fig. 6 A) for 4 h. Here, we also assessed the effects of the S2 subunit (Fig. 6 B) to examine specificity of S1 effects. As noted, secreted embryonic alkaline phosphatase (SEAP) is measured in supernatant of cultured HEK293 cells and reflects TLR-induced NF-κB and AP-1 signaling. Null1 cells, which lack hTLR2 and hTLR4, but express the SEAP reporter gene, were also exposed to S1 (Fig. 6 C) and assayed to control for effects via HEK293 endogenous PRRs. S1 significantly increased SEAP expression in hTLR2 (df = 3, 8, F = 170.0, p < 0.0001) and hTLR4 (df = 3, 8, F = 24.98, p < 0.0002) cells, but failed to alter SEAP expression in Null1 cells. S2 failed to affect SEAP expression at all concentrations in hTLR2 and hTLR4 cells. Positive controls for hTLR2 (PAM3csk4) and hTLR4 (LPS) also increased SEAP expression indicating that the cell lines were functional. The positive control for Null1 cells (poly I:C) also increased SEAP expression indicating that Null1 cells were functional. Post-hoc comparisons are depicted in Fig. 6.
Fig. 6.
Effect of S1 and S2 on hTLR2 and hTLR4 signaling. HEK293 cells expressing hTLR2 or hTLR4 were exposed to several concentrations of (A) S1 (0, 0.01, 0.1, and 1 μg/ml) or (B) S2 (0, 0.01, 0.1, and 1 μg/ml) for 4 h and SEAP expression measured in supernatants. (C) Null1 cells were exposed to S1 (0, 0.01, 0.1, and 1 μg/ml) for 4 h and SEAP expression measured in supernatants. hTLR2 control = 100 ng/ml PAM3csk4; hTLR4 control = 100 ng/ml LPS; Null1 control (TLR3) = 100 ng/ml poly I:C. Data are presented as the mean + SEM. N = 3 replications. S1 concentration vs media control, *p < 0.05, **p < 0.01, ****p < 0.0001.
4. Discussion
The present set of findings provide converging evidence in support of the notion that the S1 subunit of the SARS-CoV-2 spike protein might function as a PAMP, independent of viral infection, to induce a neuroinflammatory response and the behavioral sequelae of that response. Prior studies demonstrated that S1 has proinflammatory properties (Nuovo et al., 2021, Olajide et al., 2021a, Olajide et al., 2021b, Shirato and Kizaki, 2021), which were largely restricted to examining in vitro effects. Given these proinflammatory properties of S1 and that neuroinflammatory processes produce sickness behaviors (Dantzer et al., 2008), we examined whether S1 treatment was sufficient to induce a behavioral sickness response. Indeed, S1 treatment reduced time of engagement in several behaviors (activity, self-grooming, wall rearing, JSE) suggesting that S1 induced a sickness response. However, S1 failed to affect eating and drinking behavior, which is not consistent with a classic sickness response (Hart, 1988). Dantzer has proposed that cytokine-induced shifts in motivational state mediate the behavioral sickness response to inflammatory insults (Dantzer et al., 2008). Therefore, we explored the neuroinflammatory/neuroimmune effects of S1, which have largely not been characterized aside from the study of Nuovo et al. (Nuovo et al., 2021) who examined a small number of inflammatory proteins using immunohistochemistry in brain endothelial cells.
Consistent with its behavioral effects, S1 treatment resulted in an array of neuroinflammatory/neuroimmune effects in hypothalamus, hippocampus, and frontal cortex. 24 h after ICM injection, S1 treatment increased gene expression of microglia/brain macrophage activation markers (Iba1, Cd11b, MhcIIα), astrocyte activation markers (Gfap), PRRs (Tlr4), inflammasomes (Nlrp3), and proinflammatory cytokines (Il1b). These effects of S1 occurred in all brain regions. Several genes were differentially affected in these brain regions by S1 treatment. S1 increased expression of the microglial/macrophage checkpoint receptor Cd200r1 in hypothalamus and hippocampus, while S1 increased expression of the PRR Tlr2 and the DAMP Hmgb1 in hippocampus. To determine the persistence of these effects, we examined the neuroinflammatory/neuroimmune effects of S1 at 7 days post-injection. We found that S1 treatment resulted in a robust increase in MhcIIα expression in all brain regions. Notably, the magnitude of the effect of S1 on MhcIIα at 7 days post-injection was comparable to the magnitude observed at 24 h post-injection. For the other genes tested, several effects of S1 persisted, but varied more across brain regions compared to the 24 h timepoint. S1 increased Cd11b (frontal cortex), Gfap (hippocampus), Tlr4 (hypothalamus, frontal cortex), Nlrp3 (hypothalamus, hippocampus), and Il1b (hypothalamus, frontal cortex). Interestingly, Tlr2 was increased in hypothalamus at the 7d timepoint, but was not increased at the 24 h timepoint. In addition, Cd200r1 was decreased (hippocampus, frontal cortex) at the 7d timepoint, whereas it was increased at the 24 h timepoint (hypothalamus, hippocampus) indicating that the effects of S1 are not monotonic. In hippocampus, we tested whether S1 induced proteins involved in neuroinflammatory processes. Indeed, S1 induced several proteins including IL-1β, TNF, CXCL1, IFNγ, IL-10 and IL-2 at 24 h post-treatment. IL-1β and TNF are necessary and sufficient to elicit a sickness response (Dantzer et al., 2008) and thus might have mediated the effects of S1 on sickness behavior here. TNF protein remained elevated 7d after S1 treatment. These findings corroborate, in part, and extend the findings of Nuovo et al. who demonstrated that S1 induced a neuroinflammatory response (caspase 3, IL-6, TNF and C5b-9 protein) in brain endothelial cells 5 days after S1 exposure (Nuovo et al., 2021). Consistent with Nuovo et al., we found that S1 increased TNF protein at 24 h and 7d after ICM injection, however we failed to find effects on IL-6 protein at these time points.
In considering the neuroinflammatory effects of S1, there are several effects that are particularly noteworthy. First, the effect of S1 on MhcIIα persisted across time and brain regions. In one of the most comprehensive neuropathological studies in deceased Covid-19 patients, Matschke et al. found upregulated MHCII in post-mortem brain of patients (Matschke et al., 2020). Increased MHCII is considered a marker of microglial priming in conditions such aging, neurodegenerative disease, traumatic brain injury and stress exposure (Perry and Holmes, 2014). Interestingly, the T helper type 1 cell (Th1) cytokine IFNγ upregulates microglial MHCII (Grau et al., 1997, Neumann et al., 1996, Neumann et al., 1998, Ta et al., 2019, Wong et al., 1984) and primes microglia (Chao et al., 1995, Hu et al., 1995, Spencer et al., 2016, Ta et al., 2019) to subsequent proinflammatory stimuli. Interestingly, S1 increased hippocampal IFNγ protein, which might have played a mediating role in S1 effects on MhcIIα. Th1 cells and NK cells are considered the predominant source of IFNγ in the CNS (e.g., meninges and choroid plexus), which increases peripheral leukocyte trafficking into the brain parenchyma (Deczkowska et al., 2016). Also, it is important to consider that the effects of S1 on MhcIIα were measured in whole tissue, thus it is unclear if the effects were microglia/macrophage specific. Nevertheless, as MHCII is largely expressed by microglia and other brain macrophages, the effect of S1 on MhcIIα likely reflects a considerable shift away from a surveillant or homeostatic state in CNS macrophages. Also, S1 increased expression of the microglia/macrophage markers Cd11b and Iba1 in all brain regions at the 24 h timepoint, additional evidence that S1 induced a shift in the activation state of CNS macrophages. Furthermore, S1 increased expression of the chemokine CXCL1 in hippocampus, which is a potent chemoattractant for neutrophils via the CXCR2 receptor (Ravindran et al., 2013). This raises the possibility that S1 might recruit peripheral leukocytes into the brain considering that IFNγ facilitates leukocyte trafficking.
Second, S1 increased the Th1 cytokine IL-2 in hippocampus suggesting that T cells might be involved in the effects of S1. Also, cytotoxic CD8 + T cells have been found in post-mortem brain of Covid-19 patients (Matschke et al., 2020). This finding is particularly relevant here given that activated CD8 + T cells are a source of both IFNγ and TNF (Behr et al., 2018). Thus, the S1-induced increase in these proinflammatory cytokines might reflect activation of this subset of T cells. In addition, the neuroinflammatory effects of S1 were not restricted to CNS macrophages as we found upregulation of the astrocyte marker Gfap. Interestingly, evidence of astrogliosis was found in post-mortem brain of all patients who died of Covid-19 (Matschke et al., 2020). However, GFAP protein levels were not assessed here, thus it is unclear whether S1 effects on GFAP mRNA reflects astrogliosis.
Third, S1 increased Cd200r1 at the 24 h timepoint, and decreased it 7d post-treatment in hippocampus and frontal cortex. CD200R1 is a macrophage/microglia checkpoint receptor expressed almost exclusively on microglia as well as other CNS macrophages (Koning et al., 2009, Wright et al., 2000) and CD200 is a membrane glycoprotein expressed by neurons and endothelial cells. CD200 binding CD200R1 constitutively inhibits microglia/macrophage cell function and proinflammatory cytokine responses (Gorczynski et al., 2008, Jenmalm et al., 2006, Zhang et al., 2004). Disruption of CD200:CD200R1 signaling potentiates the proinflammatory response of microglia (Costello et al., 2011, Denieffe et al., 2013, Frank et al., 2018). The decrease in Cd200r1 7d post-treatment in hippocampus and frontal cortex suggests that S1 might induce a protracted primed state in CNS macrophages.
Fourth, in further support of this possible priming effect of S1, Nlrp3 (hypothalamus, hippocampus) and Il1b (hypothalamus, frontal cortex) gene expression were also increased 7d post-S1. Priming of the NLRP3 inflammasome involves upregulation of Nlrp3 and Il1b expression (Lamkanfi and Kanneganti, 2010). Taken together, these effects of S1, though not uniform across brain regions and time, on MhcIIα, Cd200r1, Nlrp3, Il1b and IFNγ suggest that S1 might prime CNS macrophages for a prolonged period after S1 exposure. If so, a primed neuroinflammatory response might play a pivotal role in the pathogenesis of post-acute sequelae of SARS-CoV-2 (PASC; long-Covid), which is characterized by fatigue, cognitive deficits, and neuropsychiatric symptoms, which persist up to 6 months after SARS-CoV-2 infection (Nalbandian et al., 2021).
A key question arising from the present results is what receptor(s) mediates the neuroinflammatory effects of S1. S1 binds largely ACE2 (Hoffmann et al., 2020) and ACE2 protein is found throughout the brain and colocalizes with endothelial cells, neurons, and astrocytes (Hernández et al., 2021). An analysis of transcriptome databases also found widespread ACE2 expression in brain endothelial cells, neurons, astrocytes, and oligodendrocytes (Chen et al., 2020), however expression in microglia and other CNS macrophages was not found. It is possible that ACE2 mediates, in part, some of the neuroinflammatory effects of S1, for example via astrocyte activation. However, to our knowledge, there is no evidence that S1 signaling via ACE2 induces inflammatory mediators. In addition, we failed to detect ACE2 gene expression in adult rat primary microglia and S1 directly induced a proinflammatory response in primary microglia, which do not express ACE2. This direct effect of S1 on microglia suggests that S1 signals through a receptor apart from ACE2. Microglia express several TLRs including TLR2 and TLR4 (Kumar, 2019), thus S1 might signal through these PRRs to produce a neuroinflammatory response in microglia.
S1 does induce endothelial cell damage (Nuovo et al., 2021). It is feasible, therefore, that S1 indirectly induces a neuroinflammatory response via the release of DAMPs from damaged cells. Matschke et al. found evidence of tissue damage in post-mortem brain of Covid-19 patients (Matschke et al., 2020). In addition, Wenzel et al. found evidence of endothelial cell death in post-mortem brain of patients (Wenzel et al., 2021). DAMPs, for example HMGB1, induce an inflammatory response via TLRs in myeloid cells (Yang et al., 2013), and S1 signals through TLR4 in THP-1 and RAW264.7 cells (Shirato and Kizaki, 2021) and BV2 cells (Olajide et al., 2021a) to induce a proinflammatory response. Our work corroborates and extends these previous findings. We report that S1 increased expression of the DAMP Hmgb1, which was restricted to the hippocampus 24 h after treatment; and S1 activated transgenic HEK293 cells expressing human TLR2 and TLR4. Interestingly, S1 induced a greater response in hTLR2 cells than in hTLR4 cells suggesting that S1 might utilize both TLR2 and TLR4 to drive proinflammatory processes. We also found that the S2 subunit failed to induce signaling via hTLR2 and hTLR4 suggesting that the effects are specific to S1. This finding is consistent with the study of Nuovo et al. who found that the S1 subunit, but not the S2 subunit generates an inflammatory response in brain endothelial cells (Nuovo et al., 2021).
We found that S1 upregulated expression of TLR2 (hippocampus) and TLR4 (hypothalamus, hippocampus, frontal cortex) at the 24 h time point and TLR4 (hypothalamus, frontal cortex) at the 7d time point. Consistent with this effect of S1 in vivo, S1 induced a profound upregulation of TLR4 protein in BV2 cells (Olajide et al., 2021a). This S1-induced upregulation of TLR2 and TLR4 might play a role in the protracted neuroimmune response to S1. The neuroinflammatory effects of S1 on TNF protein (hippocampus) and Il1b mRNA (hypothalamus, frontal cortex) persisted 7d after S1 treatment, which are products of TLR2/4 signaling. Thus, the implication is that an S1-induced molecule is likely still signaling through innate immune receptors such as TLR2/4 to produce these protracted increases in proinflammatory cytokines. It is also possible that, in addition to neuroinflammation via direct TLR signaling, S1-induced cellular damage in the CNS might chronically elevate DAMPs, which then produce this protracted proinflammatory response via TLR2/4 signaling. We surmise that S1 likely signals directly through multiple receptors (e.g., TLR2, TLR4) to elicit a neuroinflammatory response and might indirectly elicit a neuroinflammatory response through the release of DAMPs from damaged cells via ACE2.
The present set of findings add to a growing body of evidence (Nuovo et al., 2021, Olajide et al., 2021a, Olajide et al., 2021b, Shirato and Kizaki, 2021, Zheng et al., 2021) that structural proteins derived from SARS-CoV-2 might function independently as PAMPs to induce inflammatory processes via PRR engagement. Here, we provide evidence that the S1 subunit might function as a PAMP in the CNS to drive neuroinflammatory processes and the behavioral consequences of those processes, and thus play a role in the pathogenesis of SARS-CoV-2 infection. However, S1 signaling at TLRs was not demonstrated in vivo, thus the present findings are suggestive even though we found that S1 signals through TLR2/4 in vitro. Further, a control protein such as S2 was not assessed in vivo, thus specificity of S1 effects in vivo are unclear. In addition, behavioral endpoints did not include measures of depressive-like behavior, therefore it is unclear if the effects of S1 recapitulate this neuropsychiatric phenotype. It is important to consider that the neuroinflammatory effects of S1 likely occur in concert with effects of SARS-CoV-2 on peripheral proinflammatory processes, which might also drive neuroinflammation via immune-to-brain signaling pathways (McCusker and Kelley, 2013). Moreover, there is some evidence of SARS-CoV-2 infection of the brain (Matschke et al., 2020), which would drive neuroinflammatory processes concomitant with the direct CNS effects of S1 and/or peripheral cytokine effects. These facets of SARS-CoV-2 induced neuroinflammation likely synergize to drive the neurological, cognitive, and neuropsychiatric symptoms during infection as well as the protracted CNS effects that are observed many months after resolution of SARS-CoV-2 infection (Groff et al., 2021, Nalbandian et al., 2021, Taquet et al., 2021).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The present work was supported by an NIH grant (R01MH108523) to MGF and SFM.
References
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behr F.M., Chuwonpad A., Stark R., van Gisbergen K. Armed and Ready: Transcriptional Regulation of Tissue-Resident Memory CD8 T Cells. Front. Immunol. 2018;9:1770. doi: 10.3389/fimmu.2018.01770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benameur K., Agarwal A., Auld S.C., Butters M.P., Webster A.S., Ozturk T., Howell J.C., Bassit L.C., Velasquez A., Schinazi R.F., Mullins M.E., Hu W.T. Encephalopathy and Encephalitis Associated with Cerebrospinal Fluid Cytokine Alterations and Coronavirus Disease, Atlanta, Georgia, USA, 2020. Emerg. Infect. Dis. 2020;26(9):2016–2021. doi: 10.3201/eid2609.202122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin D., Gulick R., Martinez F., et al. Severe Covid-19. New Eng. J. Med. 2020;383(25):2451–2460. doi: 10.1056/NEJMcp2009575. In this issue. [DOI] [PubMed] [Google Scholar]
- Bianchi M.E. DAMPs, PAMPs and alarmins: all we need to know about danger. J. Leukoc. Biol. 2007;81(1):1–5. doi: 10.1189/jlb.0306164. [DOI] [PubMed] [Google Scholar]
- Bodro, M., Compta, Y., Llanso, L., Esteller, D., Doncel-Moriano, A., Mesa, A., Rodriguez, A., Sarto, J., Martinez-Hernandez, E., Vlagea, A., Egri, N., Filella, X., Morales-Ruiz, M., Yague, J., Soriano, A., Graus, F., Garcia, F., Hospital Clinic Infecto, C., Hospital Clinic Neuro, C.-g., 2020. Increased CSF levels of IL-1beta, IL-6, and ACE in SARS-CoV-2-associated encephalitis. Neurol Neuroimmunol Neuroinflamm 7. [DOI] [PMC free article] [PubMed]
- Boldrini M., Canoll P.D., Klein R.S. How COVID-19 Affects the Brain. JAMA psychiatry. 2021;78(6):682. doi: 10.1001/jamapsychiatry.2021.0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroujeni M.E., Simani L., Bluyssen H.A.R., Samadikhah H.R., Zamanlui Benisi S., Hassani S., Akbari Dilmaghani N., Fathi M., Vakili K., Mahmoudiasl G.-R., Abbaszadeh H.A., Hassani Moghaddam M., Abdollahifar M.-A., Aliaghaei A. Inflammatory Response Leads to Neuronal Death in Human Post-Mortem Cerebral Cortex in Patients with COVID-19. ACS Chem. Neurosci. 2021;12(12):2143–2150. doi: 10.1021/acschemneuro.1c00111. [DOI] [PubMed] [Google Scholar]
- Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369(6511):1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao C.C., Hu S., Peterson P.K. Modulation of human microglial cell superoxide production by cytokines. J. Leukoc. Biol. 1995;58(1):65–70. doi: 10.1002/jlb.58.1.65. [DOI] [PubMed] [Google Scholar]
- Chen R., Wang K., Yu J., Howard D., French L., Chen Z., Wen C., Xu Z. The Spatial and Cell-Type Distribution of SARS-CoV-2 Receptor ACE2 in the Human and Mouse Brains. Front. Neurol. 2020;11:573095. doi: 10.3389/fneur.2020.573095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Costello D.A., Lyons A., Denieffe S., Browne T.C., Cox F.F., Lynch M.A. Long term potentiation is impaired in membrane glycoprotein CD200-deficient mice: a role for Toll-like receptor activation. J. Biol. Chem. 2011;286(40):34722–34732. doi: 10.1074/jbc.M111.280826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzer R. Cytokine, sickness behavior, and depression. Immunol. Allergy Clin. North Am. 2009;29(2):247–264. doi: 10.1016/j.iac.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzer R., O'Connor J.C., Freund G.G., Johnson R.W., Kelley K.W. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat. Rev. Neurosci. 2008;9(1):46–56. doi: 10.1038/nrn2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deczkowska A., Baruch K., Schwartz M. Type I/II Interferon Balance in the Regulation of Brain Physiology and Pathology. Trends Immunol. 2016;37(3):181–192. doi: 10.1016/j.it.2016.01.006. [DOI] [PubMed] [Google Scholar]
- Deng Q., Ye G., Pan Y., Xie W., Yang G., Li Z., Li Y. High Performance of SARS-Cov-2N Protein Antigen Chemiluminescence Immunoassay as Frontline Testing for Acute Phase COVID-19 Diagnosis: A Retrospective Cohort Study. Front. Med. (Lausanne) 2021;8:676560. doi: 10.3389/fmed.2021.676560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denieffe S., Kelly R.J., McDonald C., Lyons A., Lynch M.A. Classical activation of microglia in CD200-deficient mice is a consequence of blood brain barrier permeability and infiltration of peripheral cells. Brain Behav. Immun. 2013;34:86–97. doi: 10.1016/j.bbi.2013.07.174. [DOI] [PubMed] [Google Scholar]
- Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R., Breitbart R.E., Acton S. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ. Res. 2000;87:E1–E9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- Eden A., Kanberg N., Gostner J., Fuchs D., Hagberg L., Andersson L.M., Lindh M., Price R.W., Zetterberg H., Gisslen M. CSF Biomarkers in Patients With COVID-19 and Neurologic Symptoms: A Case Series. Neurology. 2021;96:e294–e300. doi: 10.1212/WNL.0000000000010977. [DOI] [PubMed] [Google Scholar]
- Farhadian S., Glick L.R., Vogels C.B.F., Thomas J., Chiarella J., Casanovas-Massana A., Zhou J., Odio C., Vijayakumar P., Geng B., Fournier J., Bermejo S., Fauver J.R., Alpert T., Wyllie A.L., Turcotte C., Steinle M., Paczkowski P., Dela Cruz C., Wilen C., Ko A.I., MacKay S., Grubaugh N.D., Spudich S., Barakat L.A. Acute encephalopathy with elevated CSF inflammatory markers as the initial presentation of COVID-19. BMC Neurol. 2020;20:248. doi: 10.1186/s12883-020-01812-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- File S.E., Seth P. A review of 25 years of the social interaction test. Eur. J. Pharmacol. 2003;463(1-3):35–53. doi: 10.1016/s0014-2999(03)01273-1. [DOI] [PubMed] [Google Scholar]
- Frank M.G., Barrientos R.M., Thompson B.M., Weber M.D., Watkins L.R., Maier S.F. IL-1RA injected intra-cisterna magna confers extended prophylaxis against lipopolysaccharide-induced neuroinflammatory and sickness responses. J. Neuroimmunol. 2012;252(1-2):33–39. doi: 10.1016/j.jneuroim.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M.G., Fonken L.K., Annis J.L., Watkins L.R., Maier S.F. Stress disinhibits microglia via down-regulation of CD200R: A mechanism of neuroinflammatory priming. Brain Behav. Immun. 2018;69:62–73. doi: 10.1016/j.bbi.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M.G., Wieseler-Frank J.L., Watkins L.R., Maier S.F. Rapid isolation of highly enriched and quiescent microglia from adult rat hippocampus: immunophenotypic and functional characteristics. J. Neurosci. Methods. 2006;151(2):121–130. doi: 10.1016/j.jneumeth.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Friard O., Gamba M. BORIS: a free, versatile open-source event-logging software for video/audio coding and live observations. Methods Ecol. Evol. 2016;7:1325–1330. [Google Scholar]
- Gandhi R., Lynch J., Del Rio C., et al. Mild or Moderate Covid-19. New Eng. J. Med. 2020;383(18):1757–1766. doi: 10.1056/NEJMcp2009249. In this issue. [DOI] [PubMed] [Google Scholar]
- Gorczynski R., Boudakov I., Khatri I. Peptides of CD200 modulate LPS-induced TNF-alpha induction and mortality in vivo. J. Surg. Res. 2008;145:87–96. doi: 10.1016/j.jss.2007.04.043. [DOI] [PubMed] [Google Scholar]
- Goshen I., Yirmiya R. Interleukin-1 (IL-1): a central regulator of stress responses. Front. Neuroendocrinol. 2009;30(1):30–45. doi: 10.1016/j.yfrne.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Grau V., Herbst B., Van der Meide P.H., Steiniger B. Activation of microglial and endothelial cells in the rat brain after treatment with interferon-gamma in vivo. Glia. 1997;19(3):181–189. doi: 10.1002/(sici)1098-1136(199703)19:3<181::aid-glia1>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Groff D., Sun A., Ssentongo A.E., Ba D.M., Parsons N., Poudel G.R., Lekoubou A., Oh J.S., Ericson J.E., Ssentongo P., Chinchilli V.M. Short-term and Long-term Rates of Postacute Sequelae of SARS-CoV-2 Infection: A Systematic Review. JAMA Netw Open. 2021;4(10):e2128568. doi: 10.1001/jamanetworkopen.2021.28568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart B.L. Biological basis of the behavior of sick animals. Neurosci. Biobehav. Rev. 1988;12(2):123–137. doi: 10.1016/s0149-7634(88)80004-6. [DOI] [PubMed] [Google Scholar]
- Hernández V.S., Zetter M.A., Guerra E.C., Hernández-Araiza I., Karuzin N., Hernández-Pérez O.R., Eiden L.E., Zhang L. ACE2 expression in rat brain: Implications for COVID-19 associated neurological manifestations. Exp. Neurol. 2021;345:113837. doi: 10.1016/j.expneurol.2021.113837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingrat, Q.L., Visseaux, B., Laouenan, C., Tubiana, S., Bouadma, L., Yazdanpanah, Y., Duval, X., Burdet, C., Ichou, H., Damond, F., Bertine, M., Benmalek, N., Choquet, C., Timsit, J.F., Ghosn, J., Charpentier, C., Descamps, D., Houhou-Fidouh, N., French Covid cohort management committee, C.-C.s.g., members of the French, C.c.s.g., member of the Co, V.C.s.g.P.i., Steering, C., Co, V.C.C.C., Coordination, statistical, a., Virological, L., Biological, C., Partners, Sponsor, Genetic, 2020. Detection of SARS-CoV-2 N-antigen in blood during acute COVID-19 provides a sensitive new marker and new testing alternatives. Clin Microbiol Infect. [DOI] [PMC free article] [PubMed]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Sheng W.S., Peterson P.K., Chao C.C. Cytokine modulation of murine microglial cell superoxide production. Glia. 1995;13(1):45–50. doi: 10.1002/glia.440130106. [DOI] [PubMed] [Google Scholar]
- Hu W., Zhang Y., Fei P., Zhang T., Yao D., Gao Y., Liu J., Chen H., Lu Q., Mudianto T., Zhang X., Xiao C., Ye Y., Sun Q., Zhang J., Xie Q.i., Wang P.-H., Wang J., Li Z., Lou J., Chen W. Mechanical activation of spike fosters SARS-CoV-2 viral infection. Cell Res. 2021;31(10):1047–1060. doi: 10.1038/s41422-021-00558-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenmalm M.C., Cherwinski H., Bowman E.P., Phillips J.H., Sedgwick J.D. Regulation of myeloid cell function through the CD200 receptor. J. Immunol. 2006;176(1):191–199. doi: 10.4049/jimmunol.176.1.191. [DOI] [PubMed] [Google Scholar]
- Kawai T., Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 2010;11(5):373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Kempuraj D., Selvakumar G.P., Ahmed M.E., Raikwar S.P., Thangavel R., Khan A., Zaheer S.A., Iyer S.S., Burton C., James D., Zaheer A. COVID-19, Mast Cells, Cytokine Storm, Psychological Stress, and Neuroinflammation. Neuroscientist. 2020;26(5-6):402–414. doi: 10.1177/1073858420941476. [DOI] [PubMed] [Google Scholar]
- Koning N., Swaab D.F., Hoek R.M., Huitinga I. Distribution of the immune inhibitory molecules CD200 and CD200R in the normal central nervous system and multiple sclerosis lesions suggests neuron-glia and glia-glia interactions. J. Neuropathol. Exp. Neurol. 2009;68(2):159–167. doi: 10.1097/NEN.0b013e3181964113. [DOI] [PubMed] [Google Scholar]
- Kumar H., Kawai T., Akira S. Pathogen recognition by the innate immune system. Int. Rev. Immunol. 2011;30(1):16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- Kumar V. Toll-like receptors in the pathogenesis of neuroinflammation. J. Neuroimmunol. 2019;332:16–30. doi: 10.1016/j.jneuroim.2019.03.012. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M., Kanneganti T.-D. Nlrp3: an immune sensor of cellular stress and infection. Int. J. Biochem. Cell Biol. 2010;42(6):792–795. doi: 10.1016/j.biocel.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A., Frontera J., Placantonakis D.G., Lighter J., Galetta S., Balcer L., Melmed K.R. Cerebrospinal fluid in COVID-19: A systematic review of the literature. J. Neurol. Sci. 2021;421:117316. doi: 10.1016/j.jns.2021.117316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mariano G., Farthing R., Lale-Farjat S., Bergeron J., et al. Structural Characterization of SARS-CoV-2: Where We Are, and Where We Need to Be. Front. Mol. Biosci. 2020;7:1–28. doi: 10.3389/fmolb.2020.605236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matschke J., Lütgehetmann M., Hagel C., Sperhake J.P., Schröder A.S., Edler C., Mushumba H., Fitzek A., Allweiss L., Dandri M., Dottermusch M., Heinemann A., Pfefferle S., Schwabenland M., Sumner Magruder D., Bonn S., Prinz M., Gerloff C., Püschel K., Krasemann S., Aepfelbacher M., Glatzel M. Neuropathology of patients with COVID-19 in Germany: a post-mortem case series. Lancet Neurol. 2020;19(11):919–929. doi: 10.1016/S1474-4422(20)30308-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker R.H., Kelley K.W. Immune-neural connections: how the immune system's response to infectious agents influences behavior. J. Exp. Biol. 2013;216:84–98. doi: 10.1242/jeb.073411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbandian A., Sehgal K., Gupta A., Madhavan M.V., McGroder C., Stevens J.S., Cook J.R., Nordvig A.S., Shalev D., Sehrawat T.S., Ahluwalia N., Bikdeli B., Dietz D., Der-Nigoghossian C., Liyanage-Don N., Rosner G.F., Bernstein E.J., Mohan S., Beckley A.A., Seres D.S., Choueiri T.K., Uriel N., Ausiello J.C., Accili D., Freedberg D.E., Baldwin M., Schwartz A., Brodie D., Garcia C.K., Elkind M.S.V., Connors J.M., Bilezikian J.P., Landry D.W., Wan E.Y. Post-acute COVID-19 syndrome. Nat. Med. 2021;27(4):601–615. doi: 10.1038/s41591-021-01283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H., Boucraut J., Hahnel C., Misgeld T., Wekerle H. Neuronal control of MHC class II inducibility in rat astrocytes and microglia. Eur. J. Neurosci. 1996;8(12):2582–2590. doi: 10.1111/j.1460-9568.1996.tb01552.x. [DOI] [PubMed] [Google Scholar]
- Neumann H., Misgeld T., Matsumuro K., Wekerle H. Neurotrophins inhibit major histocompatibility class II inducibility of microglia: involvement of the p75 neurotrophin receptor. Proc. Natl. Acad. Sci. U.S.A. 1998;95(10):5779–5784. doi: 10.1073/pnas.95.10.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuovo G.J., Magro C., Shaffer T., Awad H., Suster D., Mikhail S., He B., Michaille J.-J., Liechty B., Tili E. Endothelial cell damage is the central part of COVID-19 and a mouse model induced by injection of the S1 subunit of the spike protein. Ann Diagn Pathol. 2021;51:151682. doi: 10.1016/j.anndiagpath.2020.151682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata A.F., Maley A.M., Wu C., Gilboa T., Norman M., Lazarovits R., Mao C.-P., Newton G., Chang M., Nguyen K., Kamkaew M., Zhu Q., Gibson T.E., Ryan E.T., Charles R.C., Marasco W.A., Walt D.R. Ultra-Sensitive Serial Profiling of SARS-CoV-2 Antigens and Antibodies in Plasma to Understand Disease Progression in COVID-19 Patients with Severe Disease. Clin. Chem. 2020;66(12):1562–1572. doi: 10.1093/clinchem/hvaa213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olajide O.A., Iwuanyanwu V.U., Adegbola O.D., Al-Hindawi A.A. SARS-CoV-2 Spike Glycoprotein S1 Induces Neuroinflammation in BV-2 Microglia. Mol. Neuro. 2021 doi: 10.1007/s12035-021-02593-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olajide O.A., Iwuanyanwu V.U., Lepiarz-Raba I., Al-Hindawi A.A. Induction of Exaggerated Cytokine Production in Human Peripheral Blood Mononuclear Cells by a Recombinant SARS-CoV-2 Spike Glycoprotein S1 and Its Inhibition by Dexamethasone. Inflammation. 2021 doi: 10.1007/s10753-021-01464-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna F., Bruzzaniti S., Piemonte E., Maddaloni V., Atripaldi L., Sale S., Sanduzzi A., Nicastro C., Pepe N., Bifulco M., Matarese G., Galgani M., Atripaldi L. Serum levels of SARS-CoV-2 nucleocapsid antigen associate with inflammatory status and disease severity in COVID-19 patients. Clin. Immunol. 2021;226:108720. doi: 10.1016/j.clim.2021.108720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry V.H., Holmes C. Microglial priming in neurodegenerative disease. Nat. Rev. Neurol. 2014;10(4):217–224. doi: 10.1038/nrneurol.2014.38. [DOI] [PubMed] [Google Scholar]
- Pilotto A., Odolini S., Masciocchi S., Comelli A., Volonghi I., Gazzina S., Nocivelli S., Pezzini A., Focà E., Caruso A., Leonardi M., Pasolini M.P., Gasparotti R., Castelli F., Ashton N.J., Blennow K., Zetterberg H., Padovani A. Steroid-Responsive Encephalitis in Coronavirus Disease 2019. Ann. Neurol. 2020;88(2):423–427. doi: 10.1002/ana.25783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff R.M., Perry V.H. Microglial physiology: unique stimuli, specialized responses. Annu. Rev. Immunol. 2009;27(1):119–145. doi: 10.1146/annurev.immunol.021908.132528. [DOI] [PubMed] [Google Scholar]
- Ravindran A., Sawant K.V., Sarmiento J., Navarro J., Rajarathnam K. Chemokine CXCL1 dimer is a potent agonist for the CXCR2 receptor. J. Biol. Chem. 2013;288(17):12244–12252. doi: 10.1074/jbc.M112.443762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhea E.M., Logsdon A.F., Hansen K.M., Williams L.M., Reed M.J., Baumann K.K., Holden S.J., Raber J., Banks W.A., Erickson M.A. The S1 protein of SARS-CoV-2 crosses the blood-brain barrier in mice. Nat. Neurosci. 2021;24:368–378. doi: 10.1038/s41593-020-00771-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schou T.M., Joca S., Wegener G., Bay-Richter C. Psychiatric and neuropsychiatric sequelae of COVID-19 - A systematic review. Brain Behav. Immun. 2021 doi: 10.1016/j.bbi.2021.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Kizaki T. SARS-CoV-2 spike protein S1 subunit induces pro-inflammatory responses via toll-like receptor 4 signaling in murine and human macrophages. Heliyon. 2021;7 doi: 10.1016/j.heliyon.2021.e06187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, E., Bartley, C.M., Chow, R.D., Ngo, T.T., Jiang, R., Zamecnik, C.R., Dandekar, R., Loudermilk, R.P., Dai, Y., Liu, F., Hawes, I.A., Alvarenga, B.D., Huynh, T., McAlpine, L., Rahman, N.T., Geng, B., Chiarella, J., Goldman-Israelow, B., Vogels, C.B.F., Grubaugh, N.D., Casanovas-Massana, A., Phinney, B.S., Salemi, M., Alexander, J., Gallego, J.A., Lencz, T., Walsh, H., Lucas, C., Klein, J., Mao, T., Oh, J., Ring, A., Spudich, S., Ko, A.I., Kleinstein, S.H., DeRisi, J.L., Iwasaki, A., Pleasure, S.J., Wilson, M.R., Farhadian, S.F., 2020. Exploratory neuroimmune profiling identifies CNS-specific alterations in COVID-19 patients with neurological involvement. bioRxiv.
- Spencer N.G., Schilling T., Miralles F., Eder C. Mechanisms Underlying Interferon-gamma-Induced Priming of Microglial Reactive Oxygen Species Production. PLoS ONE. 2016;11 doi: 10.1371/journal.pone.0162497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta T.T., Dikmen H.O., Schilling S., Chausse B., Lewen A., Hollnagel J.O., Kann O. Priming of microglia with IFN-gamma slows neuronal gamma oscillations in situ. Proc. Natl. Acad. Sci. U.S.A. 2019;116:4637–4642. doi: 10.1073/pnas.1813562116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taquet M., Dercon Q., Luciano S., Geddes J.R., Husain M., Harrison P.J. Incidence, co-occurrence, and evolution of long-COVID features: A 6-month retrospective cohort study of 273,618 survivors of COVID-19. PLoS Med. 2021;18 doi: 10.1371/journal.pmed.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur K.T., Miller E.H., Glendinning M.D., Al-Dalahmah O., Banu M.A., Boehme A.K., Boubour A.L., Bruce S.S., Chong A.M., Claassen J., Faust P.L., Hargus G., Hickman R.A., Jambawalikar S., Khandji A.G., Kim C.Y., Klein R.S., Lignelli-Dipple A., Lin C.C., Liu Y., Miller M.L., Moonis G., Nordvig A.S., Overdevest J.B., Prust M.L., Przedborski S., Roth W.H., Soung A., Tanji K., Teich A.F., Agalliu D., Uhlemann A.C., Goldman J.E., Canoll P. COVID-19 neuropathology at Columbia University Irving Medical Center/New York Presbyterian Hospital. Brain. 2021 doi: 10.1093/brain/awab148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel J., Lampe J., Muller-Fielitz H., Schuster R., Zille M., Muller K., Krohn M., Korbelin J., Zhang L., Ozorhan U., Neve V., Wagner J.U.G., Bojkova D., Shumliakivska M., Jiang Y., Fahnrich A., Ott F., Sencio V., Robil C., Pfefferle S., Sauve F., Coelho C.F.F., Franz J., Spiecker F., Lembrich B., Binder S., Feller N., Konig P., Busch H., Collin L., Villasenor R., Johren O., Altmeppen H.C., Pasparakis M., Dimmeler S., Cinatl J., Puschel K., Zelic M., Ofengeim D., Stadelmann C., Trottein F., Nogueiras R., Hilgenfeld R., Glatzel M., Prevot V., Schwaninger M. The SARS-CoV-2 main protease M(pro) causes microvascular brain pathology by cleaving NEMO in brain endothelial cells. Nat. Neurosci. 2021 doi: 10.1038/s41593-021-00926-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong G.H., Bartlett P.F., Clark-Lewis I., Battye F., Schrader J.W. Inducible expression of H-2 and Ia antigens on brain cells. Nature. 1984;310:688–691. doi: 10.1038/310688a0. [DOI] [PubMed] [Google Scholar]
- Wright G.J., Puklavec M.J., Willis A.C., Hoek R.M., Sedgwick J.D., Brown M.H., Barclay A.N. Lymphoid/neuronal cell surface OX2 glycoprotein recognizes a novel receptor on macrophages implicated in the control of their function. Immunity. 2000;13:233–242. doi: 10.1016/s1074-7613(00)00023-6. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A.C., Kern F., Losada P.M., Agam M.R., Maat C.A., Schmartz G.P., Fehlmann T., Stein J.A., Schaum N., Lee D.P., Calcuttawala K., Vest R.T., Berdnik D., Lu N., Hahn O., Gate D., McNerney M.W., Channappa D., Cobos I., Ludwig N., Schulz-Schaeffer W.J., Keller A., Wyss-Coray T. Dysregulation of brain and choroid plexus cell types in severe COVID-19. Nature. 2021;595:565–571. doi: 10.1038/s41586-021-03710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Antoine D.J., Andersson U., Tracey K.J. The many faces of HMGB1: molecular structure-functional activity in inflammation, apoptosis, and chemotaxis. J. Leukoc. Biol. 2013;93:865–873. doi: 10.1189/jlb.1212662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jackson C.B., Mou H., Ojha A., Peng H., Quinlan B.D., Rangarajan E.S., Pan A., Vanderheiden A., Suthar M.S., Li W., Izard T., Rader C., Farzan M., Choe H. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020;11:6013. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Cherwinski H., Sedgwick J.D., Phillips J.H. Molecular mechanisms of CD200 inhibition of mast cell activation. J. Immunol. 2004;173:6786–6793. doi: 10.4049/jimmunol.173.11.6786. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Kuang M., Li J., Zhu L., Jia Z., Guo X., Hu Y., Kong J., Yin H., Wang X., You F. SARS-CoV-2 spike protein interacts with and activates TLR41. Cell Res. 2021;31:818–820. doi: 10.1038/s41422-021-00495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Karki R., Williams E.P., Yang D., Fitzpatrick E., Vogel P., Jonsson C.B., Kanneganti T.D. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat. Immunol. 2021;22:829–838. doi: 10.1038/s41590-021-00937-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]