Abstract
Platelets and platelet extracellular vesicles (pEV) are at the crossroads of coagulation and immunity. Extracellular vesicles are messengers that not only transmit signals between cells, but also provide information about the status of their cell of origin. Thus, pEVs have potential as both biomarkers of platelet activation and contributors to pathology. Coronavirus Disease‐19 (COVID‐19), caused by infection with severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), is a complex disease affecting multiple organs and is characterized by a high degree of inflammation and risk of thrombosis in some patients. In this review, we introduce pEVs as valuable biomarkers in disease with a special focus on their potential as predictors of and contributors to COVID‐19.
Keywords: calcium modulators, HDT, pCAMKII
Graphical Abstract
Review on platelet extracellular vesicles (pEVs); includes a special focus on pEV's potential as predictors of and contributors to COVID‐19.
Abbreviations
- ACE2
angiotensin‐converting enzyme 2
- CD147
cluster of Differentiation 147
- CLEC−2
C‐type lectin‐like receptor 2
- COVID‐19
Coronavirus Disease‐19
- DAMPs
damage‐associated molecular patterns
- EVs
extracellular vesicles
- HMGB1
high‐mobility Group Protein 1
- Ig
immunglobulin
- MDA
malondialdehyde
- OxLDL
oxidized low‐density lipoprotein
- pEVs
platelet extracellular vesicles
- PS
phosphatidylserine
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
- SLE
systemic lupus erythematosus
- TF
tissue‐factor
- TLR
toll‐like receptor.
1. INTRODUCTION
Extracellular vesicles (EV) are small membrane‐bound vesicles that contain molecules from their cell of origin. As EVs can be internalized by cellular recipients, they are suggested to mediate cellular signaling. 1 , 2 The two most described EV‐subtypes are microvesicles and exosomes. 3 Microvesicles are produced by plasma membrane budding and shedding, have a diameter ranging from approximately 100 to 1000 nm 2 , 4 , 5 , 6 and generally expose phosphatidylserine (PS) although there are exceptions. 7 , 8 , 9 , 10 They bud from cells activated by numerous inflammatory triggers. 11 Exosomes are typically smaller than microvesicles 9 and are released by cells from multivesicular bodies in an exocytosis‐dependent mechanism. 6 , 12 , 13
Platelets are anucleated cell fragments with a diameter of 1 to 3 μm and are produced by megakaryocytes. 14 , 15 , 16 They prevent bleeding and interact with pathogens and immune cells thereby assisting immune responses. 15 , 16 Platelets and megakaryocytes are the major sources of circulating EVs. 17 , 18 , 19 Similar to platelets, platelet EVs (pEVs) were first recognized as procoagulant entities. 20 , 21 , 22 However, their roles now appear to be more diverse and pEV subtypes may fine tune both coagulation and inflammation. 1 pEVs have also been identified in bone marrow, 23 lymph, 24 , 25 and synovial fluid. 26 , 27 This suggests that pEVs also enable platelets to transmit signals into tissues that are normally inaccessible to platelets.
Coronavirus Disease‐19 (COVID‐19) is caused by respiratory tract infection with coronavirus SARS‐CoV‐2. 28 COVID‐19 was declared a pandemic by the World Health Organization in March 2020 29 and is now recognized as a complex disease involving high levels of inflammation and thrombosis. 28 , 30 , 31 , 32 Platelet hyperactivation 33 , 34 , 35 and an increase in pEVs circulating 33 , 34 , 35 , 36 , 37 in blood of COVID‐19 patients is now documented. 33 , 34 , 35 , 36 , 37
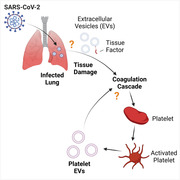
Herein, we review the current knowledge concerning pEVs and related blood‐borne EVs as biomarkers and contributors to pathologies. In particular, we will review (i) pEVs as a biomarker in COVID‐19, (ii) how they may be induced in COVID‐19, and (iii) how they may contribute to COVID‐19 pathology. Figure 1 recapitulates the concepts presented in the review.
FIGURE 1.
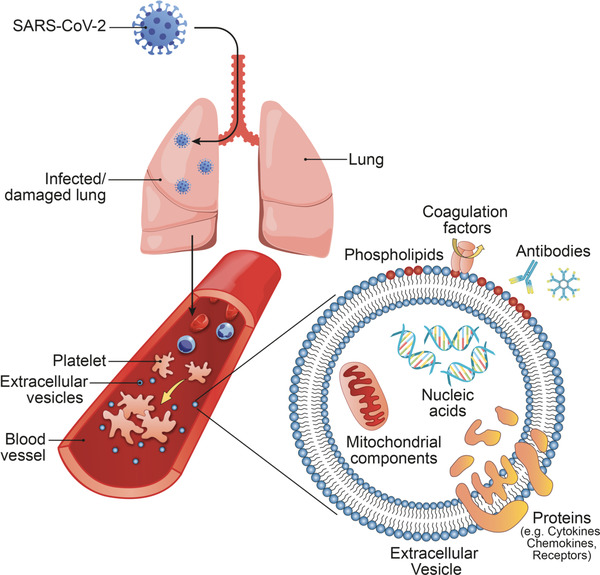
SARS‐CoV‐2 infection of lungs and subsequent damage may activate cells in the blood and induce platelet activation, aggregation, and extracellular vesicle release. Extracellular vesicles carry a diverse array of signaling molecules that can influence immune responses and coagulation
2. HISTORY OF EXTRACELLULAR‐VESICLE RESEARCH
Peter Wolf 22 provided the first description of EVs as small, procoagulant lipid particles that can be separated from platelets by differential centrifugation. Subsequent studies and electron microscopic analysis identified two main classes of vesicles released from cells: exosomes and microvesicles. 13 , 38 , 39 , 40 During the last 20 years, technological progress has transformed the study of EVs. Advances in flow cytometry—the most commonly used method to detect and quantify EVs—have enabled the analysis of EVs at a higher resolution than ever before, further revealing EV complexity. EVs have historically been categorized into major subclasses such as “microvesicle/microparticle” and “exosomes.” However, these narrow definitions have become problematic. Indeed, the vocabulary and methodology describing EVs have expanded at a rapid pace and may lead to confusion upon retrospective examination of EV studies. 3 , 41 , 42 Therefore, the International Society of Extracellular Vesicles (ISEV) recommends the use of the umbrella term “extracellular‐vesicle (EV)” unless specific investigations permit to determine whether EVs were liberated from the plasma membrane by budding or implicated exocytosis, and to include a detailed description of the isolation and detection methods used in the study of EVs. 3 , 41 , 42
Notably, interpretations of historical studies may have changed with advancements in the technologies and methods used to detect EVs. A common discrepancy is the concentration of EVs per microliter in healthy plasma, with a reported concentration of 200 up to 109 EVs/μL, which likely depends on the isolation and detection techniques used. 19 , 43 The pEV concentration in healthy plasma has been conservatively estimated at around 11,500/μL by cryo‐electron microscopy. 19 The most commonly used methods of EV isolation and detection are differential centrifugation and flow cytometry, respectively. However, common pitfalls associated with these techniques are the risk of co‐isolation and detection of EVs and lipoproteins, the potential of overlooking particularly small EVs due to insufficient resolution, and the risk of damaging EVs during isolation at high centrifugal forces. 43 , 44
3. pEV AS MARKERS AND MAKERS
pEVs, as a component of liquid biopsies, show potential as biomarkers in autoimmune diseases, cancer, cardiovascular diseases, and infectious diseases. 25 , 45 , 46 , 47 , 48 , 49 , 50 , 51 , 52 , 53 , 54 , 55 The presence of pEVs is documented in synovial fluid in rheumatoid arthritis, 26 and increased levels of circulating pEVs correlate with disease activity. 56 Moreover, an increase in pEV concentrations was found in lymph in murine models of atherosclerosis and autoimmune arthritis. 24 ‐ 27 The number of pEVs is increased in blood in systemic lupus erythematosus (SLE), and higher levels are suggested to associate with declining kidney function. 45 In addition to pEV concentration, pEV content can be used as a biomarker. The protein composition of EVs in disease can be distinguished from that of EVs in healthy controls. For instance, as activated platelets can translate and produce interleukin‐1 (IL‐1), this cytokine can be packaged into pEVs, which can augment inflammation. 26 , 57 Lipid mediators of inflammation, such as prostaglandins and leukotrienes, can also be transported or generated by pEVs given the latter's content of enzymatic machinery and fatty acids. 58 EVs can be released from activated and dying cells, and may therefore, carry self‐antigens and damage‐associated molecular patterns (DAMPs). Such EVs could have a role as potential biomarkers and may contribute to disease.
Indeed, pEVs are associated with autoantibodies in SLE, which suggests that they bear autoantigens and may facilitate cellular activation through activation of Fc receptors. 59 , 60 , 61 Moreover, platelets and their pEVs can contain DAMPs such as high‐mobility group protein 1 (HMGB1), 62 S100A8/9, 63 , 64 and mitochondrial DAMPs. 59 Another prominent DAMP that can be found on EVs are oxidation‐specific epitopes. 65 The latter result from oxidation of polyunsaturated fatty acids and are commonly found on oxidized low‐density lipids (OxLDL). 65 , 66 For instance, malondialdehyde (MDA)‐modifications of EVs may be the product of phosphatidylcholine peroxidation during EV‐biogenesis. These MDA epitopes are inflammatory/immunogenic and recognized by a subset of germline‐encoded (natural) IgM antibodies. 65 , 66 Low levels of natural IgM antibodies and high levels of MDA + EV and OxLDL are associated with an increased risk of cardiovascular disease. 65 EVs and MDA + EVs 65 are elevated in acute myocardial infarction (STEMI) 46 and in acute coronary syndrome, 48 and as such are likely indicative of tissue damage.
pEVs may be useful biomarkers in cancers associated with thrombotic risks and may enhance metastasis. 52 , 53 , 54 Of particular interest are prostate cancer cells, which are reported to release tissue‐factor (TF)‐associated EVs (TF + EV). 67 TF is the main initiator of coagulation, 68 and cancer cell TF + EVs may thereby induce platelet activation, 68 leading to the release of pEVs. In the case of coagulation initiated by cancer, TF + EVs are more likely the causative agents in this pathology as opposed to pEVs. However, detection of TF‐protein expressed on EVs (TF + EV) is notoriously difficult and generally only achieved through indirect determination of TF‐activity. 69 , 70 Considering these challenges, pEV‐quantification is a potential surrogate marker of TF‐activity and platelet activation in cancer and other diseases.
Table 1 provides an overview of the different techniques used to identify platelet EV and other blood‐borne EV in the literature cited in this section. As indicated in Table 1, pEVs are most commonly isolated from platelet‐poor or platelet‐free plasma obtained by differential centrifugation, subsequently labeled for platelet‐specific markers (primarily CD41) and detected by flow cytometry. Ideally, it is recommended to perform a two‐step centrifugation protocol on whole‐blood to obtain platelet‐free plasma. 44 Plasma prepared this way can be frozen and stored for long‐term. 44
TABLE 1.
Detection of platelet EV in different diseases
| Context | Refs. | Source | Isolation method | (p)EV‐identification | Detection |
|---|---|---|---|---|---|
| Arthritis | [ 25 ] | Lymph (Mouse) | 2 × 2500 × g, RT → EV in SUP |
CD41 (CellTracker: not‐platelet‐specific) |
Flow‐Cyt., Cryo‐EM |
| Arthritis | [ 26 ] | Synovial Fluid | 2 × 600 × g, 30 min, RT → EV in SUP | CD41 | Flow‐Cyt. |
| Arthritis | [ 27 ] | Blood, Synovial Fluid |
Blood: 2 × 2500 × g Synovial Fluid: 650 x g, 20 min →EV in SUP |
CD41 | Flow‐Cyt., EM, NTA, MassSpec |
| Arthritis | [ 47 ] | Blood, Platelets, Megakaryocytes | Blood → 2 × 2500 × g, 15 min, RT → EV in SUP | CD41, CLEC2, GPVI | Flow‐Cyt., Western‐Blot |
| Arthritis | [ 56 ] | Blood | 1550 × g, 20 min, RT → SUP 2 × 17,570 × g, 30 min, 20°C → EV in pellet | CD61 | Flow‐Cyt. |
| Atherosclerosis | [ 24 ] | Lymph (Mouse) | 1200 × g, 10 min, 4°C → analysis of EV in SUP | CD41 (AnV: not‐platelet‐specific) | Flow‐Cyt., Cryo‐EM |
| Cancer (Gastric) | [ 52 ] | Blood | 1550 × g, 15 min, RT → EV in SUP | CD41 | Flow‐Cyt. |
| Cancer (Prostate) | [ 53 ] | Blood | 2 × 28,00 × g, 15 min, RT → EV in SUP | CD41 | Flow‐Cyt. |
| Cancer (Breast) | [ 54 ] | Platelet Concentrates | 2000 × g, 15 min, 4°C → SUP 2400 × g, 15 min, RT → EV in pellet | CD41, CD62p, CXCR4 | Flow‐Cyt. |
| Myocardial Infarction | [ 46 ] | Blood | Plasma → 2500 × g, 10 min, RT → SUP 20,000 × g, 30 min, RT → EV in pellet | CD61, CD31, CD42b, (CD14, CD142, AnV: not‐platelet‐specific) | Flow‐Cyt. |
| Myocardial Infarction | [ 65 ] | Blood |
Blood: 2000 × g, 30 min → 21,000 × g, 30 min → EV in pellet Blood: 2000 ×g, 30 min → 13,000 × g, 2 min → 18,000 × g, 30 min → EV in pellet |
CD41 (CD235, CD14, CD31: not‐platelet‐specific) | Flow‐Cyt. |
| Multiple Sclerosis | [ 55 ] | Blood | 160 × g, 10 min, RT → SUP 2000 × g, 6 min → EV in SUP | CD41, CD62p | Flow‐Cyt. |
| SLE | [ 45 ] | Blood | Platelet‐poor plasma → 2000 ×g, 20 min, RT → SUP 13,000 × g, 2 min, RT → in SUP | CD41 (CD40L, VCAM, HMGB1, CD142, C4d, AnV: not‐platelet‐specific) | Flow‐Cyt. |
| SLE | [ 59 ] | Blood | 282 × g, 10 min → 2500 × g, 20 min → 3 × 3200 × g, 5 min, RT → EV in SUP | CD41 (mitotracker: not‐platelet‐specific) | Flow‐Cyt. |
| SLE | [ 60 ] | Blood | 1800 × g, 10 min, 21°C → 3000 × g, 10 min, 21°C → 18,890 × g, 30 min, RT → EV in pellet | Proteomics‐analysis | MassSpec |
| SLE | [ 61 ] | Blood | 1800 × g, 10 min, 21°C → 3000 × g, 10 min, 21°C → 19,000 × g, 30 min, 22°C → EV in pellet | CD41 (IgG1, IgM, C1q, AnV: not‐platelet‐specific) | Flow‐Cyt., MassSpec |
| Viral infection (H1N1) | [ 49 ] | Washed Platelets | In supernatant with platelets | CD41 | Flow‐Cyt. |
| Viral infection (HIV) | [ 50 ] | Washed Platelets | Platelet Suspension → 2 × 2000 × g, 15 min, 4°C → SUP 28,000 ×g, 1h, 4°C → EV in pellet | CD41, CXCR4 | Flow‐Cyt. |
| Viral infection (Dengue) | [ 51 ] | Washed Platelets | 1500 × g, 15 min, RT → 20,000 × g, 40 min, 4°C → EV in pellet; SUP 100,000 × g, 40 min, 4°C → EV in pellet | CD41, CD62p, CD63 | Flow‐Cyt., NTA |
Abbreviations. AnV, Annexin‐V; EM , electron microscopy; Flow‐Cyt., flow cytometry; MassSpec, mass spectrometry; NTA, nanoparticle tracking analysis; RT, room temperature; SUP, supernatant.
4. pEVS AS BIOMARKERS IN COVID‐19
While pEVs have been most extensively studied as biomarkers in noninfectious diseases, pEVs are also found in association with viral infections. 49 , 50 , 51 , 71 , 72 Influenza virus H1N1 activates platelets and induces the release of pEVs by a mechanism that implicates thrombin and the activation of FcgRIIa by antibodies directed against this virus. 49 pEVs may contribute to the propagation of HIV, as they can transport C‐X‐C chemokine receptor type 4 (CXCR4), a coreceptor for HIV, to other cells. 50 In dengue virus infection, pEVs may be released in a C‐type lectin‐like receptor 2 (CLEC−2)‐dependent manner by platelets 51 and thereby show potential as biomarkers of disease severity 71 by distinguishing between patients who may or may not require platelet transfusion. 72 pEVs in infections and sepsis are discussed in more detail elsewhere. 73
Several studies point to EVs and pEVs as potential biomarkers in COVID‐19. 33 , 36 , 37 Increased levels of circulating pEVs have been observed in patients with SARS‐CoV‐2 infection. 33 , 37 One particular study reported higher numbers of circulating pEVs in patients with nonsevere and severe COVID‐19 in comparison with healthy individuals after determining pEV levels in platelet‐free plasma. 33 It is intriguing that the increase in pEVs was less pronounced in patients with severe disease relative to those with nonsevere disease. 33 Normalization of pEV numbers to platelet counts still revealed significantly increased levels of pEVs in severe COVID‐19, pointing to increased EV‐biogenesis during COVID‐19. 33 Cappellano et al. 37 evaluated pEVs as a biomarker for SARS‐CoV‐2 infection in hospitalized patients and found significantly elevated levels of pEVs in COVID‐19 patients compared with healthy controls. Moreover, they reported that increased levels of circulating pEVs could distinguish SARS‐CoV‐2‐infected patients from patients with suspected COVID‐19 who tested negative for SARS‐CoV‐2 infection at the time of hospitalization. Despite a significant difference in pEV concentration, detection of viral infection by pEV‐quantification is unlikely to replace PCR as the gold standard to detect SARS‐CoV‐2 infection. For these analyses, the investigators quantified pEVs in whole blood in the absence of additional purification steps to reduce sample manipulation. The reduction of preanalytical procedures may minimize the risk of damaging EVs, although the use of whole blood may lead to the generation of EVs after collection. Thus, although the approach used by Cappellano et al. 37 may be useful for laboratory analyses performed within hours of blood sampling, as intended by the authors, platelet‐free plasma by two‐step centrifugation protocols 44 is necessary for long‐term storage and sample biobanking.
It is interesting to note that we and others observed that absolute numbers of pEVs 33 , 36 and PS‐exposing (PS+) pEVs 33 were significantly lower in severe COVID‐19 when compared with nonsevere COVID‐19. 33 , 36 PS + EVs are considered procoagulatory since they provide a negatively charged surface for initiation and maintenance of coagulation. 74 Notably, a decrease in PS+ EVs has also been reported in multiple organ dysfunction syndrome and sepsis, which might point to common mechanisms in these diseases. 75 COVID‐19 is indeed a complex disease affecting many organs distant from the lungs and is characterized by a high degree of thrombosis. Thus, the observed decrease in PS+ pEVs may suggest increased consumption of such EVs in patients with more severe disease as PS is also an “eat‐me” signal for cellular removal. 33 , 36 , 76 In summary, PS+ pEVs may be a biomarker in COVID‐19 that distinguishes different stages of disease activity, especially with regard to coagulation and organ damage. 30 , 31 , 32
Table 2 provides an overview of the isolation and detection techniques used for the analysis of platelet EV in COVID‐19 in the literature cited in this section.
TABLE 2.
Detection of platelet EVs in COVID‐19
| Context | Ref | Source | Isolation Method | EV‐identification | Detection |
|---|---|---|---|---|---|
| EV‐quantification | [ 33 ] | Blood | 200 × g , 15 min → 1000 × g, 10 min → EV in SUP | CD41 (AnV: not‐platelet specific) | Flow‐Cyt. |
| EV‐quantification and EV‐TF‐activity | [ 36 ] | Blood |
FACS: 2 × 2500 × g, 15 min, RT → EV in SUP TF‐activity assay: 2 × 2500 × g, 15 min, RT → 3 × 70,000 × g, 90 min, 4°C → EV in pellet |
CD41 (CD31, CD146, CD15, IgG1: not‐platelet specific) | Flow‐Cyt., Other: TF‐activity assay |
| EV‐quantification | [ 37 ] | Blood | EV were analyzed directly in whole‐blood | CD41 (CD31, Phalloidin: not‐platelet specific) | Flow‐Cyt. |
Abbreviations. AnV, Annexin‐V; Flow‐Cyt., flow cytometry; RT, room temperature; SUP, supernatant; TF, tissue factor.
5. WHAT TRIGGERS THE RELEASE OF pEVS IN COVID‐19?
5.1. SARS‐CoV‐2‐mediated platelet activation via receptors of innate and adaptive immunity
Considering the high abundance of platelets in blood and their ability to detect pathogens, platelets have also become recognized as an important component of the immune response to microbial invasion. Platelets express various pattern‐recognition receptors, including functional expression of toll‐like receptors (TLR1, TLR2, TLR3, TLR4, TLR6, TLR7, TLR9) 77 , 78 and mRNA for all 10 TLRs. 79 TLR2 and TLR4 are not primarily known to recognize viruses, but a role for TLR4 and TLR2 signaling on platelets has been shown for dengue virus and cytomegalovirus infection. 80 , 81 The TLR4 ligand lipopolysaccharide induces EV‐release by monocytic cells, 10 but it is not known if TLR2 or TLR4 activation induces EV release by platelets. In fact, TLR2 and TLR4 engagement is not sufficient for platelet activation, but instead primes or sensitizes platelets to respond to other stimuli, or induces protein translation by platelets. 57 , 77 Since SARS‐CoV‐2 is a single‐stranded RNA (ssRNA) virus 82 and double‐stranded RNA (dsRNA) is considered to be an intermediate product of viral replication, 82 TLR3 (dsRNA) and TLR7 (ssRNA) are candidate receptors for SARS‐CoV‐2 recognition by platelets. Engagement of ssRNA by platelet TLR7 83 , 84 , 85 and ssRNA by platelet TLR3 86 is also known. TLR3 activation has been associated with EV release by different cell types, 87 , 88 , 89 but has not been described for platelets. Moreover, activation of TLR3 and TLR7 does not induce typical platelet activation, such as granule content release and exposure of activated GPIIbGPIIIa, as seen in response to thrombin stimulation. These interactions may be more similar to the priming or sensitization effect of TLR2 and TLR4. 77 , 90 Of note is that SARS‐CoV‐2 RNA has been found in association with platelets in some COVID‐19 patients. 33 , 34 Thus, TLR3 and TLR7 may be attractive targets for SARS‐CoV‐2 platelet interactions and priming of platelets in COVID‐19. In a recent study, 91 transmission‐electron‐microscopy revealed that SARS‐CoV‐2 may be taken up by platelets and locate in phagosome‐like structures. TLR3 and TLR7 are endosomal receptors and could thereby come in contact with SARS‐CoV‐2 RNA, but an actual interaction has not been shown in platelets. Damaged capillaries in the alveolar wall could be contact sites for SARS‐CoV‐2 and the blood. DAMPs, such as DNA liberated by activated or NETosing cells, 92 may also contribute to platelet activation and pEV release in COVID‐19. Indeed, while stimulation of TLR9 (recognizes unmethylated CpG DNA) induced oxidative stress in platelets, it also enhanced pEV release when platelets were activated by immune complexes recognized by FcγRIIa. 59 Moreover, antibody‐mediated recognition of SARS‐CoV‐2 via FcγRIIa may play a role in platelet activation and pEV‐release, as has been shown for H1N1. 49
Protein in the envelope of viruses are often heavily glycosylated, which may aid in the evasion of adaptive immune responses. 93 Indeed, the SARS‐CoV‐2 spike (S) protein has multiple glycosylation sites 94 that may be relevant to its function and interactions with target cells. In multiple variants of SARS‐CoV‐2, mutations to glycosylation sites affect infectiveness. 95 Of note, is that platelets express CLEC‐2 (C‐type lectin‐like type II) and DC‐SIGN, 96 which are relevant in HIV‐1 and dengue virus infections 51 , 96 and platelets may release EVs in a CLEC‐2‐dependent manner. 51
5.2. Direct SARS‐CoV‐2 platelet interaction via known and putative SARS‐CoV‐2 receptors
The primary receptor for SARS‐CoV‐2 in humans is ACE2 (Angiotensin‐converting enzyme 2), 97 , 98 which is ubiquitously expressed by type II epithelial cells in the upper and lower respiratory tract. 99 The typical cellular entry route for SARS‐CoV‐2 is engagement with ACE2 via the Spike protein, subsequent cleavage of the latter by the serine protease TMPRSS2 to enable fusion of the viral and cellular membrane resulting in infection of the target cell. 100 While expression of ACE2 has been shown for vascular endothelial cells and lung macrophages, 99 physiological expression by platelets or megakaryocytes is controversial. 101 Moreover, Koupenova et al. 91 recently reported that SARS‐CoV‐2 may be taken up by platelets through both ACE2‐dependent and independent mechanisms. Cluster of Differentiation 147 (CD147) has been proposed as an alternative receptor for SARS‐CoV‐2, 102 is commonly expressed in circulating cells, and is associated with risk factors of severe COVID‐19 such as obesity, asthma, and chronic obstructive pulmonary disease (COPD). 102 , 103 , CD147 is indeed functionally expressed on platelets, 104 , 105 but its relevance to SARS‐CoV‐2 infection has been called into question. 106 Other intriguing targets for direct SARS‐CoV‐2 platelet interaction are the integrins. Platelet integrin GPIIbGPIIIa binds the three amino acid motif Arg‐Gly‐Asp (RGD) present in physiological ligands (e.g., fibrinogen, von Willebrand Factor), which is crucial for platelet aggregation. 107 , 108 Platelet integrin GPIIbGPIIIa is important in platelet responses and is implicated in pEV‐release triggered by FcγRIIA or GPVI receptor activation. 25 , 26 , Furthermore, the Spike protein sequence of SARS‐CoV‐2 contains an RGD‐motif (403‐405: Arg‐Gly‐Asp) within the receptor‐binding domain. 109 , 110 Thus, platelet integrin GPIIbGPIIIa presents another alternative target receptor for SARS‐CoV‐2.
5.3. pEV‐release independent of direct SARS‐CoV‐2 platelet interaction
Platelets may not be stimulated to release pEVs solely upon direct interaction with SARS‐CoV‐2. Indeed, excessive inflammatory responses and tissue damage, particularly, in the lung and lung microvasculature, are observed in COVID‐19 patients. 111 , 112 Of interest is that autopsies of 21 patients 113 revealed inflammatory damage and microthrombi in multiple organs (lungs, heart, liver, kindeys, brain), while SARS‐CoV‐2 infected cells were absent from most of the affected tissues. Notably, SARS‐CoV‐2 viral RNA copies in a range of 63 to 6,310 copies per milliliter of blood have been detected in a quarter of hospitalized COVID‐19 patients. 114 Given that the concentration range of platelets in blood is 150 × 10^8 to 450^8 per mL of blood, platelets would outnumber virus by a factor of 23,771 to 7.14 × 10^6. This would make direct interactions of SARS‐CoV‐2 with platelets rare events, although the amount of SARS‐CoV‐2 in the blood may be underestimated viral as RNA may have been degraded. 91 Dissemination of SARS‐CoV‐2 throughout the circulation is not excluded, but may not be the primary path taken by the virus to affect platelet function and pEV release. As discussed by Chen et al., 115 infection of cells in the lungs (pneumocytes, epithelial cells) and nearby vasculature (endothelial cells) causes the production of inflammatory cytokines contributing to an immune response leading to tissue damage. 116 , 117 At the same time, damage to tissue may lead to the liberation of DAMPs, including, but not limited to, mitochondria and mitochondrial components and oxidized phospholipids, and TF associated with EVs, which would further amplify the inflammatory and coagulation cascade that stimulates platelet activation and subsequent pEV release. Moreover, the overwhelming inflammation may affect endothelial barrier integrity and thereby lead to increased expression of soluble thrombomodulin, soluble P‐selectin, and von Willebrand factor. 117 Indeed, elevated levels of TF activity associated with EVs have been reported in COVID‐19, which may directly contribute to excessive coagulation. 36 , 70 , 118 In addition, ACE2, the primary receptor for SARS‐CoV‐2 infection in humans, is important in the regulation of the renin–angiotensin–aldosterone system and a deficiency of ACE2 is linked to enhanced risk of inflammation and thrombosis. 98 As COVID‐19 is increasingly viewed as a thromboinflammatory disease, 32 it is conceivable that platelet activation in COVID‐19 may be a consequence of the inflammation, organ damage, and pathological activation of the coagulation cascade, rather than a consequence of direct virus–platelet interaction. Moreover, at later stages of the disease, secondary effects caused by the tissue damage and inflammatory response, as opposed to viral presence, might become decisive in determining disease outcome. Thus, changes to the presence and activity of pEVs may represent potential risk markers, as in other diseases. 25 , 45 , 46 , 47 , 48 , 49 , 50 , 51 , 119
6. THE CONTRIBUTION OF pEVS TO COVID‐19
The first descriptions of EVs referred to their procoagulant abilities, which are primarily attributed to exposure of the negatively charged phospholipid, PS. 120 , 121 , 122 PS exposure on platelets and pEVs supports propagation of coagulation. 120 , 121 , 122 In COVID‐19, studies report the presence of antiphospholipid antibodies, such as anticardiolipin antibodies, similar to those seen in antiphospholipid syndrome and SLE. 123 , 124 , 125 , 126 These antibodies may target PS‐exposing pEVs, thereby, forming immune complexes for cellular activation through Fc receptors. Negatively charged surfaces may also initiate the “intrinsic pathway” of coagulation. 121 However, the main cellular initiator of coagulation is TF. 121 While some procoagulant activity of circulating pEVs has been associated with TF in the past, 127 it has been suggested that platelets may acquire TF from other cells via TF‐expressing EVs in a P‐selectin glycoprotein ligand‐1‐dependent manner. 128 However, expression of TF by platelets and pEVs is controversial and direct detection of TF displayed by EVs is challenging. 69 The concentration of TF may be below the detection limit of flow cytometric approaches, but may be sufficiently high to initiate coagulation as concentrations as low as 20 fM suffice to initiate coagulation. 129 Furthermore, TF may be present in an inactive (“encrypted”) or active (“decrypted”) state. 121 Therefore, it is necessary to determine TF biological activity to confirm its role in a biological process.
Given the high prevalence of thrombosis in COVID‐19, 30 , 31 , 32 TF activity associated with EVs and its involvement in COVID‐19 pathology is of high interest. 36 , 70 , 118 , 130 Several studies 36 , 70 , 118 , 131 report a significant increase in TF activity associated with EVs from COVID‐19 patients when compared with healthy controls. Moreover, EV‐TF activity was significantly higher in COVID‐19 than in sepsis. 36 , 118 Guervilly et al. 36 found a higher EV‐TF activity in severe compared with nonsevere (moderate) COVID‐19 and a TF activity of more than 78.3 fM associated with thromboembolic events. Moreover, they observed fibrinolytic activity of EVs in COVID‐19, but found no significant change between nonsevere (moderate) and severe disease, indicating that the balance is shifted toward coagulation in severe disease. 36 Krishnamachary et al. 131 reported enrichment of TF‐protein and TF‐activity associated with EVs isolated from COVID‐19 patients with severe disease. When stratifying COVID‐19 patients with severe disease by mortality, no differences in EV‐TF‐activity was noted, as the level of EV‐TF‐activity was approximately 50 pM in both survivors and nonsurvivors. 131 These data appear to contrast with Guervilly et al., 36 who reported far lower levels of EV‐associated TF‐activity in COVID‐19. While the discrepancy might be explained by an overestimation of TF activity measured with commercial assays in absence of blocking antibodies to ensure of the specificity of the measurements, 132 , 133 , 134 these studies suggest that high EV‐TF activity may be associated with increased severity and mortality in COVID‐19. 36 , 70 , 131
Guervilly et al. 36 reported no differences in overall EV levels after comparing moderate and severe COVID‐19, with the exception of significantly lower levels of pEVs in severe COVID‐19. 36 This is consistent with observations made by Zaid et al. who also reported significantly lower levels of total and PS+ pEVs in severe COVID‐19 compared with nonsevere COVID‐19. 33 However, the procoagulant activity of EVs was increased in COVID‐19, and was thought to depend on the expression of active TF given that the fibrinolytic activity of EVs remained unchanged in these patients. 36 A recent interesting in vitro study by Wang et al. 130 found that infection of human monocyte‐derived macrophages with SARS‐CoV‐2 spike protein pseudovirus increased TF activity at the cell surface and stimulated the release of EVs with associated TF activity. 130 This study suggests that TF activity depended on “decryption” (activation) of TF by hydrolysis of sphingomyelin via acid sphingomyelinase (ASMase), and not on increased TF protein expression or externalization of PS. 130 This lends further support to a role for SARS‐CoV‐2 in the “decryption”’ of TF and the subsequent release of EVs with associated, active TF. 130 These observations are of clinical importance given the low femtomolar concentrations of EV‐associated TF activity reported in COVID‐19 patients 36 that are nevertheless sufficient to predict an increased thrombotic risk. The absolute quantification of TF + EVs, or identification of the cellular origin of TF‐exposing EVs is challenging and requires careful study. Thus, the distinction of “decrypted” from “encrypted” TF in association with EVs, by the quantification of TF activity, may be a more relevant risk marker in COVID‐19.
7. CONCLUDING REMARKS
EVs have attracted interest as biomarkers and players in COVID‐19 pathology. As COVID‐19 is a complex disease affecting multiple organs and is characterized by a high degree of thrombosis, the study of platelet activation and the involvement of procoagulant EVs has drawn interest. A growing number of independent studies found that quantification and characterization of pEVs and other types of blood‐borne EVs carry potential as biomarkers of COVID‐19 disease severity and as predictors of outcome. While high levels of circulating EVs appear to be found during all stages of the disease, the presence of TF activity associated with EVs may predict disease severity and initiate the “extrinsic” or “TF” pathway of coagulation and, thereby, directly contribute to the high thrombotic risk in COVID‐19. Conversely, the PS‐exposing pEVs may stimulate inflammation if targeted by antiphospholipid antibodies, or support the propagation of coagulation due to the exposure of negatively charged surfaces. However, what remains unclear, is the pathway of pEV biogenesis in COVID‐19 as the relevance of direct SARS‐CoV‐2‐platelet interaction has not been established. Excessive production of active TF and damaged surfaces, culminating in coagulation initiation and propagation, may be the most probable trigger for high levels of platelet activation and pEV biogenesis in COVID‐19. Thus, determination of plasma pEV levels provides an opportunity for a surrogate marker for excessive coagulation and inflammation. Important challenges to overcome are the clear identification of circulating EVs and their physical separation into distinct populations to assign specific functions to individual EV‐populations and to define their role in COVID‐19. In summary, these observations indicate directions for future studies of EV involvement in COVID‐19.
AUTHOR CONTRIBUTIONS
FP wrote the manuscript. EB supervised and reviewed the manuscript. LF reviewed the manuscript. All authors read and approved the final version of the manuscript for submitting to the Journal of Leukocyte Biology
CONFLICT OF INTEREST
The authors have no conflict of interest to disclose.
ACKNOWLEDGMENT
FP is recipient of a Postdoctoral fellowship from FRQS. EB is recipient of senior award from the Fonds de Recherche du Québec en Santé (FRQS).
Puhm F, Flamand L, Boilard E. Platelet extracellular vesicles in COVID‐19: Potential markers and makers.. J Leukoc Biol. 2022;111:63–74. 10.1002/JLB.3MIR0221-100R
REFERENCES
- 1. Puhm F, Boilard E, MacHlus KR. Platelet extracellular vesicles: beyond the blood. Arterioscler Thromb Vasc Biol. 2020;41:87‐96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jeppesen DK, Fenix AM, Franklin JL, et al. Reassessment of exosome composition. Cell. 2019;177:428‐445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Théry C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Todorova D, Simoncini S, Lacroix R, et al. Extracellular vesicles in angiogenesis. Circ Res. 2017;120:1658‐1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buzas EI, György B, Nagy G, et al. Emerging role of extracellular vesicles in inflammatory diseases. Nat Rev Rheumatol. 2014;10:356‐64. [DOI] [PubMed] [Google Scholar]
- 6. Van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213‐228. [DOI] [PubMed] [Google Scholar]
- 7. Dachary‐Prigent J, Freyssinet JM, Pasquet JM, et al. Annexin V as a probe of aminophospholipid exposure and platelet membrane vesiculation: a flow cytometry study showing a role for free sulfhydryl groups. Blood. 1993;81:2554‐2565. [PubMed] [Google Scholar]
- 8. Marcoux G, Duchez A‐C, Cloutier N, et al. Revealing the diversity of extracellular vesicles using high‐dimensional flow cytometry analyses. Sci Rep. 2016;6:35928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thery C, Boussac M, Veron P, et al. Proteomic analysis of dendritic cell‐derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166:7309‐7318. [DOI] [PubMed] [Google Scholar]
- 10. Puhm F, Afonyushkin T, Resch U, et al. Mitochondria are a subset of extracellular vesicles released by activated monocytes and induce type I IFN and TNF responses in endothelial cells. Circ Res. 2019;125:43‐52. [DOI] [PubMed] [Google Scholar]
- 11. Morel O, Morel N, Jesel L, et al. Microparticles: a critical component in the nexus between inflammation, immunity, and thrombosis. Semin Immunopathol. 2011;33:469‐86. [DOI] [PubMed] [Google Scholar]
- 12. Mathieu M, Martin‐Jaular L, Lavieu G, et al. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell‐to‐cell communication. Nat Cell Biol. 2019;21:9‐17. [DOI] [PubMed] [Google Scholar]
- 13. Heijnen HFG, Schiel AE, Fijnheer R, et al. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha‐granules. Blood. 1999;94:3791‐3799. [PubMed] [Google Scholar]
- 14. Jenne CN, Urrutia R, Kubes P. Platelets: bridging hemostasis, inflammation, and immunity. Int J Lab Hematol. 2013;35:254‐261. [DOI] [PubMed] [Google Scholar]
- 15. Machlus KR, Italiano JE. The incredible journey: from megakaryocyte development to platelet formation. J Cell Biol. 2013;201:785‐796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marcoux G, Laroche A, Espinoza Romero J, et al. Role of platelets and megakaryocytes in adaptive immunity. Platelets. 2020;32:340‐351. [DOI] [PubMed] [Google Scholar]
- 17. Flaumenhaft R, Dilks JR, Richardson J, et al. Megakaryocyte‐derived microparticles: direct visualization and distinction from platelet‐derived microparticles. Blood. 2009;113:1112‐11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aatonen MT, Ohman T, Nyman TA, et al. Isolation and characterization of platelet‐derived extracellular vesicles. J Extracell vesicles. 2014;3. 10.3402/jev.v3.24692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Arraud N, Linares R, Tan S, et al. Extracellular vesicles from blood plasma: determination of their morphology, size, phenotype and concentration. J Thromb Haemost. 2014;12:614‐627. [DOI] [PubMed] [Google Scholar]
- 20. Chargaff E, West R. The biological significance of the thromboplastic protein of blood. J Biol Chem. 1946;166:189‐197. [PubMed] [Google Scholar]
- 21. O'Brien JR. The platelet‐like activity of serum. Br J Haematol. 1955;1:223‐228. [PubMed] [Google Scholar]
- 22. Wolf P. The nature and significance of platelet products in human plasma. Br J Haematol. 1967;13:269‐288. [DOI] [PubMed] [Google Scholar]
- 23. French SL, Butov KR, Allaeys I, et al. Platelet‐derived extracellular vesicles infiltrate and modify the bone marrow during inflammation. Blood Adv. 2020;4:3011‐3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Milasan A, Tessandier N, Tan S, et al. Extracellular vesicles are present in mouse lymph and their level differs in atherosclerosis. J Extracell Vesicles. 2016;5:31427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tessandier N, Melki I, Cloutier N, et al. Platelets disseminate extracellular vesicles in lymph in rheumatoid arthritis. Arterioscler Thromb Vasc Biol. 2020;40:929‐942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boilard E, Nigrovic PA, Larabee K, et al. Platelets amplify inflammation in arthritis via collagen‐dependent microparticle production. Science (80‐). 2010;327:580‐583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. György B, Szabó TG, Turiák L, et al. Improved flow cytometric assessment reveals distinct microvesicle (cell‐derived microparticle) signatures in joint diseases. PLoS One. 2012;7:e49726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tay MZ, Poh CM, Rénia L, et al. The trinity of COVID‐19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20: 363‐374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. World Health Organization . World Health Organization. Dashboard. Feb20 2021. WHO Coronavirus Disease (COVID‐19) Dashboard. 2020 . Available from https://covid19.who.int. Accessed March 1, 2021.
- 30. Bilaloglu S, Aphinyanaphongs Y, Jones S, et al. Thrombosis in hospitalized patients with COVID‐19 in a New York city health system. JAMA J Am Med Assoc. 2020;324:799‐801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Middleton EA, He X‐Y, Denorme F, et al. Neutrophil extracellular traps contribute to immunothrombosis in COVID‐19 acute respiratory distress syndrome. Blood. 2020;136:1169‐1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bonaventura A, Vecchié A, Dagna L, et al. Endothelial dysfunction and immunothrombosis as key pathogenic mechanisms in COVID‐19. Nat Rev Immunol. 2021;21: 319‐329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zaid Y, Puhm F, Allaeys I, et al. Platelets can associate with SARS‐Cov‐2 RNA and are hyperactivated in COVID‐19. Circ Res. 2020. 127:1404‐1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Manne BK, Denorme F, Middleton EA, et al. Platelet gene expression and function in patients with COVID‐19. Blood. 2020;136:1317‐1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hottz ED, Azevedo‐Quintanilha IG, Palhinha L, et al. Platelet activation and platelet‐monocyte aggregate formation trigger tissue factor expression in patients with severe COVID‐19. Blood. 2020;136:1330‐1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guervilly C, Bonifay A, Burtey S, et al. Dissemination of extreme levels of extracellular vesicles: tissue factor activity in patients with severe COVID‐19. Blood Adv. 2021;5:628‐634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cappellano G, Raineri D, Rolla R, et al. Circulating platelet‐derived extracellular vesicles are a hallmark of SARS‐Cov‐2 infection. Cells. 2021;10:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Webber AJ, Johnson SA. Platelet participation in blood coagulation aspects of hemostasis. Am J Pathol. 1970;60:19‐42. [PMC free article] [PubMed] [Google Scholar]
- 39. Crawford N. The presence of contractile proteins in platelet microparticles isolated from human and animal platelet‐free plasma. Br J Haematol. 1971;21:53‐69. [DOI] [PubMed] [Google Scholar]
- 40. Sandberg H, Bode AP, Dombrose FA, et al. Expression of coagulant activity in human platelets: release of membranous vesicles providing platelet factor 1 and platelet factor 3. Thromb Res. 1985;39:63‐79. [DOI] [PubMed] [Google Scholar]
- 41. Sedgwick AE, D'Souza‐Schorey C. The biology of extracellular microvesicles. Traffic. 2018;19:319‐327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lötvall J, Hill AF, Hochberg F, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for extracellular vesicles. J Extracell Vesicles. 2014;3:26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Johnsen KB, Gudbergsson JM, Andresen TL, et al. What is the blood concentration of extracellular vesicles? Implications for the use of extracellular vesicles as blood‐borne biomarkers of cancer. Biochim Biophys Acta—Rev Cancer. 2019;1871:109‐116. [DOI] [PubMed] [Google Scholar]
- 44. Berckmans RJ, Lacroix R, Hau CM, et al. Extracellular vesicles and coagulation in blood from healthy humans revisited. J Extracell Vesicles. 2019;8:1688936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mobarrez F, Vikerfors A, Gustafsson JT, et al. Microparticles in the blood of patients with systemic lupus erythematosus (SLE): phenotypic characterization and clinical associations. Sci Rep. 2016;6:36025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chiva‐Blanch G, Laake K, Myhre P, et al. Platelet‐, monocyte‐derived & tissue factorcarrying circulating microparticles are related to acute myocardial infarction severity. PLoS One. 2017;12:1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gitz E, Pollitt AY, Gitz‐Francois JJ, et al. CLEC‐2 expression is maintained on activated platelets and on platelet microparticles. Blood. 2014;124:2262‐2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nomura S, Uehata S, Saito S, et al. Enzyme immunoassay detection of platelet‐derived microparticles and RANTES in acute coronary syndrome. Thromb Haemost. 2003;89:506‐512. [PubMed] [Google Scholar]
- 49. Boilard E, Paré G, Rousseau M, et al. Influenza virus H1N1 activates platelets through FcγRIIA signaling and thrombin generation. Blood. 2014;123:2854‐2863. [DOI] [PubMed] [Google Scholar]
- 50. Rozmyslowicz T, Majka M, Kijowski J, et al. Platelet‐ and megakaryocyte‐derived microparticles transfer CXCR4 receptor to CXCR4‐null cells and make them susceptible to infection by X4‐HIV. Aids. 2003;17:33‐42. [DOI] [PubMed] [Google Scholar]
- 51. Sung P‐SS, Huang T‐FF, Hsieh S‐LL. Extracellular vesicles from CLEC2‐activated platelets enhance dengue virus‐induced lethality via CLEC5A/TLR2. Nat Commun. 2019;10:2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim HK, Song KS, Park YS, et al. Elevated levels of circulating platelet microparticles, VEGF, IL‐6 and RANTES in patients with gastric cancer: possible role of a metastasis predictor. Eur J Cancer. 2003;39:184‐191. [DOI] [PubMed] [Google Scholar]
- 53. Helley D, Banu E, Bouziane A, et al. Platelet microparticles: a potential predictive factor of survival in hormone‐refractory prostate cancer patients treated with docetaxel‐based chemotherapy. Eur Urol. 2009;56:479‐485. [DOI] [PubMed] [Google Scholar]
- 54. Janowska‐Wieczorek A, Marquez‐Curtis LA, Wysoczynski M, et al. Enhancing effect of platelet‐derived microvesicles on the invasive potential of breast cancer cells. Transfusion. 2006;46:1199‐1209. [DOI] [PubMed] [Google Scholar]
- 55. Sheremata WA, Jy W, Horstman LL, et al. Evidence of platelet activation in multiple sclerosis. J Neuroinflammation. 2008;5:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Knijff‐Dutmer EAJJ, Koerts J, Nieuwland R, et al. Elevated levels of platelet microparticles are associated with disease activity in rheumatoid arthritis. Arthritis Rheum. 2002;46:1498‐1503. [DOI] [PubMed] [Google Scholar]
- 57. Brown GT, McIntyre TM. Lipopolysaccharide signaling without a nucleus: kinase cascades stimulate platelet shedding of proinflammatory IL‐1β–rich microparticles. J Immunol. 2011;186:5489‐5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Boilard E. Thematic review series: exosomes and microvesicles: lipids as key components of their biogenesis and functions extracellular vesicles and their content in bioactive lipid mediators: more than a sack of microRNA. J Lipid Res. 2018;59:2037‐2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Melki I, Allaeys I, Tessandier N, et al. Platelets release mitochondrial antigens in systemic lupus erythematosus. Sci Transl Med. 2021;13:eaav5928. [DOI] [PubMed] [Google Scholar]
- 60. Østergaard O, Nielsen CT, Iversen LV, et al. Unique protein signature of circulating microparticles in systemic lupus erythematosus. Arthritis Rheum. 2013;65:2680‐2690. [DOI] [PubMed] [Google Scholar]
- 61. Nielsen CT, Østergaard O, Stener L, et al. Increased IgG on cell‐derived plasma microparticles in systemic lupus erythematosus is associated with autoantibodies and complement activation. Arthritis Rheum. 2012;64:1227‐1236. [DOI] [PubMed] [Google Scholar]
- 62. Harris HE, Andersson U, Pisetsky DS. HMGB1: a multifunctional alarmin driving autoimmune and inflammatory disease. Nat Rev Rheumatol. 2012;8:195‐202. [DOI] [PubMed] [Google Scholar]
- 63. Lood C, Tydén H, Gullstrand B, et al. Platelet‐derived S100A8/A9 and cardiovascular disease in systemic lupus erythematosus. Arthritis Rheumatol. 2016;68:1970‐1980. [DOI] [PubMed] [Google Scholar]
- 64. Wang Y, Fang C, Gao H, et al. Platelet‐derived S100 family member myeloid‐related protein‐14 regulates thrombosis. J Clin Invest. 2014;124:2160‐2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tsiantoulas D, Perkmann T, Afonyushkin T, et al. Circulating microparticles carry oxidation‐specific epitopes and are recognized by natural IgM antibodies. J Lipid Res. 2015;56:440‐448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Obermayer G, Afonyushkin T, Binder CJ. Oxidized low‐density lipoprotein in inflammation‐driven thrombosis. J Thromb Haemost. 2018;16:418‐428. [DOI] [PubMed] [Google Scholar]
- 67. Hell L, Däullary T, Burghart V, et al. Extracellular vesicle‐associated tissue factor activity in prostate cancer patients with disseminated intravascular coagulation. Cancers (Basel). 2021;13:1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Smith SA, Travers RJ, Morrissey JH. How it all starts: initiation of the clotting cascade. Crit Rev Biochem Mol Biol. 2015;50:326‐336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Østerud B, Bouchard BA. Detection of tissue factor in platelets: why is it so troublesome? Platelets. 2019;30:957‐961. [DOI] [PubMed] [Google Scholar]
- 70. Rosell A, Havervall S, von Meijenfeldt F, et al. Patients with COVID‐19 have elevated levels of circulating extracellular vesicle tissue factor activity that is associated with severity and mortality. Arterioscler Thromb Vasc Biol. 2020;41:ATVBAHA120315547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Punyadee N, Mairiang D, Thiemmeca S, et al. Microparticles provide a novel biomarker to predict severe clinical outcomes of dengue virus infection. J Virol. 2015;89:1587‐1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Patil R, Bajpai S, Ghosh K, et al. Microparticles as prognostic biomarkers in dengue virus infection. Acta Trop. 2018;181:21‐24. [DOI] [PubMed] [Google Scholar]
- 73. Kerris EWJJ, Hoptay C, Calderon T, et al. Platelets and platelet extracellular vesicles in hemostasis and sepsis. J Investig Med. 2020;68:813‐820. [DOI] [PubMed] [Google Scholar]
- 74. Morel O, Toti F, Hugel B, et al. Procoagulant microparticles: disrupting the vascular homeostasis equation?. Arterioscler Thromb Vasc Biol. 2006;26:2594‐2604. [DOI] [PubMed] [Google Scholar]
- 75. Joop K, Berckmans RJ, Nieuwland R, et al. Microparticles from patients with multiple organ dysfunction syndrome and sepsis support coagulation through multiple mechanisms. Thromb Haemost. 2001;85:810‐820. [PubMed] [Google Scholar]
- 76. Ravichandran KS. Find‐me and eat‐me signals in apoptotic cell clearance: progress and conundrums. J Exp Med. 2010;207:1807‐1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cognasse F, Laradi S, Berthelot P, et al. Platelet inflammatory response to stress. Front Immunol. 2019;10:1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Portier I, Campbell RA. Role of platelets in detection and regulation of infection. Arterioscler Thromb Vasc Biol. 2020;41:70‐78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Koupenova M, Mick E, Mikhalev E, et al. Sex differences in platelet toll‐like receptors and their association with cardiovascular risk factors. Arterioscler Thromb Vasc Biol. 2015;35:1030‐1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Quirino‐Teixeira AC, Rozini SV, Barbosa‐Lima G, et al. Inflammatory signaling in dengue‐infected platelets requires translation and secretion of nonstructural protein 1. Blood Adv. 2020;4:2018‐2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Assinger A, Kral JB, Yaiw KC, et al. Human cytomegalovirus‐platelet interaction triggers toll‐like receptor 2‐dependent proinflammatory and proangiogenic responses. Arterioscler Thromb Vasc Biol. 2014;34:801‐809. [DOI] [PubMed] [Google Scholar]
- 82. V'kovski P, Kratzel A, Steiner S, et al. Coronavirus biology and replication: implications for SARS‐CoV‐2. Nat Rev Microbiol. 2021;19:155‐170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Koupenova M, Corkrey HA, Vitseva O, et al. The role of platelets in mediating a response to human influenza infection. Nat Commun;10:1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Youssefian T, Drouin A, Massé JM, et al. Host defense role of platelets: engulfment of HIV and Staphylococcus aureus occurs in a specific subcellular compartment and is enhanced by platelet activation. Blood. 2002;99:4021‐4029. [DOI] [PubMed] [Google Scholar]
- 85. Simon AY, Sutherland MR, Pryzdial ELG. Dengue virus binding and replication by platelets. Blood. 2015;126:378‐385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Anabel AS, Eduardo PC, Pedro Antonio HC, et al. Human platelets express Toll‐like receptor 3 and respond to poly I:c. Hum Immunol. 2014;75:1244‐1251. [DOI] [PubMed] [Google Scholar]
- 87. Sun L, Wang X, Zhou Y, et al. Exosomes contribute to the transmission of anti‐HIV activity from TLR3‐activated brain microvascular endothelial cells to macrophages. Antiviral Res. 2016;134:167‐171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Fukushima Y, Okamoto M, Ishikawa K, et al. Activation of TLR3 and its adaptor TICAM‐1 increases miR‐21 levels in extracellular vesicles released from human cells. Biochem Biophys Res Commun. 2018;500:744‐750. [DOI] [PubMed] [Google Scholar]
- 89. Mills JT, Schwenzer A, Marsh EK, et al. Airway epithelial cells generate pro‐inflammatory tenascin‐C and small extracellular vesicles in response to TLR3 stimuli and rhinovirus infection. Front Immunol. 2019;10:1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Koupenova M, Clancy L, Corkrey HA, et al. Circulating platelets as mediators of immunity, inflammation, and thrombosis. Circ Res. 2018;122:337‐351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Koupenova M, Corkrey HA, Vitseva O, et al. SARS‐COV‐2 initiates programmed cell death in platelets. Circ Res. 2021;129:631‐646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zuo Y, Zuo M, Yalavarthi S, et al. Neutrophil extracellular traps and thrombosis in COVID‐19. J Thromb Thrombolysis. 2021;51:446‐453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Watanabe Y, Bowden TA, Wilson IA, et al. Exploitation of glycosylation in enveloped virus pathobiology. Biochim Biophys Acta–Gen Subj. 2019;1863:1480‐1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Watanabe Y, Allen JD, Wrapp D, et al. Site‐specific glycan analysis of the SARS‐CoV‐2 spike. Science (80‐). 2020;369:330‐333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Li Q, Wu J, Nie J, et al. The impact of mutations in SARS‐CoV‐2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284‐1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Chaipan C, Soilleux EJ, Simpson P, et al. DC‐SIGN and CLEC‐2 mediate human immunodeficiency virus type 1 capture by platelets. J Virol. 2006;80:8951‐8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS‐CoV‐2. Nature. 2020;581:221‐224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Verdecchia P, Cavallini C, Spanevello A, et al. The pivotal link between ACE2 deficiency and SARS‐CoV‐2 infection. Eur J Intern Med. 2020;76:14‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Xu H, Zhong L, Deng J, et al. High expression of ACE2 receptor of 2019‐nCoV on the epithelial cells of oral mucosa. Int J Oral Sci. 2020;12:1‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271‐280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Campbell RA, Boilard E, Rondina MT. Is there a role for the ACE2 receptor in SARS‐CoV‐2 interactions with platelets? J Thromb Haemost. 2021;19:46‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wang K, Chen W, Zhou Y‐S, et al. SARS‐CoV‐2 invades host cells via a novel route: cD147‐spike protein. bioRxiv. 2020. 10.2020/03/14/988345 [DOI] [Google Scholar]
- 103. Radzikowska U, Ding M, Tan G, et al. Distribution of ACE2, CD147, CD26, and other SARS‐CoV‐2 associated molecules in tissues and immune cells in health and in asthma, COPD, obesity, hypertension, and COVID‐19 risk factors. Allergy. 2020;75:2829‐2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Seizer P, Borst O, Langer HF, et al. EMMPRIN (CD147) is a novel receptor for platelet GPVI and mediates platelet rolling via GPVI‐EMMPRIN interaction. Thromb Haemost. 2009;101:682‐686. [DOI] [PubMed] [Google Scholar]
- 105. Schmidt R, Bültmann A, Fischel S, et al. Extracellular matrix metalloproteinase inducer (CD147) is a novel receptor on platelets, activates platelets, and augments nuclear factor kappaB‐dependent inflammation in monocytes. Circ Res. 2008;102:302‐309. [DOI] [PubMed] [Google Scholar]
- 106. Shilts J, Crozier TWM, Greenwood EJD, et al. No evidence for basigin/CD147 as a direct SARS‐CoV‐2 spike binding receptor. Sci Rep. 2021;11:1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Flaujac C, Boukour S, Cramer‐Bordé E. Platelets and viruses: an ambivalent relationship. Cell Mol Life Sci. 2010;67:545‐556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Ali RA, Wuescher LM, Worth RG. Platelets: essential components of the immune system. Curr Trends Immunol. 2015;16:65‐78. [PMC free article] [PubMed] [Google Scholar]
- 109. Sigrist CJ, Bridge A, Le Mercier P. A potential role for integrins in host cell entry by SARS‐CoV‐2. Antiviral Res. 2020;177:104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Makowski L, Olson‐Sidford W, W‐Weisel J. Biological and clinical consequences of integrin binding via a rogue RGD motif in the SARS CoV‐2 spike protein. Viruses. 2021;13:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. McGonagle D, O'Donnell JS, Sharif K, et al. Immune mechanisms of pulmonary intravascular coagulopathy in COVID‐19 pneumonia. Lancet Rheumatol. 2020;2:e437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Huertas A, Montani D, Savale L, et al. Endothelial cell dysfunction: a major player in SARS‐CoV‐2 infection (COVID‐19)?. Eur Respir J;56:2001634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Schurink B, Roos E, Radonic T, et al. Viral presence and immunopathology in patients with lethal COVID‐19: a prospective autopsy cohort study. Lancet Microbe. 2020;1:e290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Fajnzylber J, Regan J, Coxen K, et al. SARS‐CoV‐2 viral load is associated with increased disease severity and mortality. Nat Commun. 2020;11:5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Chen LYC, Hoiland RL, Stukas S, et al. Confronting the controversy: interleukin‐6 and the COVID‐19 cytokine storm syndrome. Eur Respir J. 2020;56:2003006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Colling ME, Kanthi Y. COVID–19‐associated coagulopathy: an exploration of mechanisms. Vasc Med (United Kingdom). 2020;25:471‐478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Goshua G, Pine AB, Meizlish ML, et al. Endotheliopathy in COVID‐19‐associated coagulopathy: evidence from a single‐centre, cross‐sectional study. Lancet Haematol. 2020;7:e575‐e582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Campbell RA, Hisada Y, Denorme F, et al. Comparison of the coagulopathies associated with COVID‐19 and sepsis. Res Pract Thromb Haemost. 2021;5:e12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Boulanger CM, Loyer X, Rautou P‐EE, et al. Extracellular vesicles in coronary artery disease. Nat Rev Cardiol. 2017;14:259‐272. [DOI] [PubMed] [Google Scholar]
- 120. Lentz BR. Exposure of platelet membrane phosphatidylserine regulates blood coagulation. Prog Lipid Res. 2003;42:423‐438. [DOI] [PubMed] [Google Scholar]
- 121. Smith SA, Travers RJ, Morrissey JH. Initiation of clotting cascade. Crit Rev Biochem Mol Biol. 2016;50:326‐336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Zelaya H, Rothmeier AS, Ruf W. Tissue factor at the crossroad of coagulation and cell signaling. J Thromb Haemost. 2018;16:1941‐1952. [DOI] [PubMed] [Google Scholar]
- 123. Zuo Y, Estes SK, Ali RA, et al. Prothrombotic autoantibodies in serum from patients hospitalized with COVID‐19. Sci Transl Med. 2020;12:eabd3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Zhang Y, Cao W, Jiang W, et al. Profile of natural anticoagulant, coagulant factor and anti‐phospholipid antibody in critically ill COVID‐19 patients. J Thromb Thrombolysis. 2020;50:580‐586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Escher R, Breakey N, Lämmle B. Severe COVID‐19 infection associated with endothelial activation. Thromb Res. 2020;190:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Hossri S, Shadi M, Hamarsha Z, et al. Clinically significant anticardiolipin antibodies associated with COVID‐19. J Crit Care. 2020;59:32‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Mallat Z, Hugel BB, Ohan J, et al. Shed membrane microparticles with procoagulant potential in human atherosclerotic plaques. Circulation. 1999;99:348‐353. [DOI] [PubMed] [Google Scholar]
- 128. del Conde I, Shrimpton CN, Thiagarajan P, et al. Tissue‐factor–bearing microvesicles arise from lipid rafts and fuse with activated platelets to initiate coagulation. Blood. 2005;106:1604‐1611. [DOI] [PubMed] [Google Scholar]
- 129. Butenas S, Bouchard BA, Brummel‐Ziedins KE, et al. Tissue factor activity in whole blood. Blood. 2005;105:2764‐2770. [DOI] [PubMed] [Google Scholar]
- 130. Wang J, Pendurthi UR, Yi G, et al. SARS‐CoV‐2 infection induces the activation of tissue factor‐mediated coagulation by activation of acid sphingomyelinase. Blood. 138: 344‐349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Krishnamachary B, Cook C, Kumar A, et al. Extracellular vesicle‐mediated endothelial apoptosis and EV‐associated proteins correlate with COVID‐19 disease severity. J Extracell vesicles. 2021;10:e12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Mackman N, Hisada Y, Grover SP, et al. Response by Mackman et al to letter regarding article, “Patients with covid‐19 have elevated levels of circulating extracellular vesicle tissue factor activity that is associated with severity and mortality‐brief report.” Arterioscler Thromb Vasc Biol. 2021;41:e381‐e382. [DOI] [PubMed] [Google Scholar]
- 133. Rosell A, Moser B, Hisada Y, et al. Evaluation of different commercial antibodies for their ability to detect human and mouse tissue factor by western blotting. Res Pract Thromb Haemost. 2020;4:1013‐1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Hisada Y, Mackman N. Measurement of tissue factor activity in extracellular vesicles from human plasma samples. Res Pract Thromb Haemost. 2019;3:44‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]


