Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of coronavirus disease-2019 (COVID-19), a respiratory disease that varies in severity from mild to severe/fatal. Several risk factors for severe disease have been identified, notably age, male sex, and pre-existing conditions such as diabetes, obesity, and hypertension. Several advancements in clinical care have been achieved over the past year, including the use of corticosteroids (e.g., corticosteroids) and other immune-modulatory treatments that have now become standard of care for patients with acute severe COVID-19. While the understanding of the mechanisms that underlie increased disease severity with age has improved over the past few months, it remains incomplete. Furthermore, the molecular impact of corticosteroid treatment on host response to acute SARS-CoV-2 infection has not been investigated. In this study, a cross-sectional and longitudinal analysis of Ab, soluble immune mediators, and transcriptional responses in young (65 ≤ years) and aged (≥ 65 years) diabetic males with obesity hospitalized with acute severe COVID-19 was conducted. Additionally, the transcriptional profiles in samples obtained before and after corticosteroids became standard of care were compared. The analysis indicates that severe COVID-19 is characterized by robust Ab responses, heightened systemic inflammation, increased expression of genes related to inflammatory and pro-apoptotic processes, and reduced expression of those important for adaptive immunity regardless of age. In contrast, COVID-19 patients receiving steroids did not show high levels of systemic immune mediators and lacked transcriptional indicators of heightened inflammatory and apoptotic responses. Overall, these data suggest that inflammation and cell death are key drivers of severe COVID-19 pathogenesis in the absence of corticosteroid therapy.
Keywords: antibodies, COVID19, comorbidity, corticosteroid, inflammation, male, SARS-CoV-2, transcriptional
Graphical Abstract
Corticosteroids treatment significantly dampens the dysregulated inflammatory and apoptotic responses seen in acute severe COVID-19
Graphical Abstract.
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease-2019 (COVID-19), is responsible for millions of deaths globally.1 Most infections result in asymptomatic or mild disease (∼80%).2–4 However, advanced age (>65 years) and pre-existing comorbidities, such as obesity, diabetes, and pulmonary and cardio vascular disease, increase the probability of severe disease.5,6 Severe COVID-19 is characterized by pneumonia, lung fibrosis that often require ventilation, and a dysregulated inflammatory response that is followed by either recovery without complications, recovery with complications (“long COVID-19″), or fatality.7,8 Our understanding of COVID-19 disease progression is continuously evolving as more studies investigate longitudinal changes in immune responses during severe COVID-19.9–11
Infection with SARS-CoV-2 results in the sequential induction of the innate and adaptive immune responses.12,13 Innate immunity following infection by the 4 endemic human coronaviruses (hCoVs) and closely related SARS-CoV and Middle Eastern Respiratory Syndrome Coronavirus (MERS-CoV) is induced by detection of viral RNA by pattern recognition receptors (PRRs) RIG-1 and MDA-5 and TLRs 3, 7, 8, and 9.14–17 Recognition by these PRRs leads to production of type I IFN and transcription of IFN stimulated genes (ISGs) that are important for establishing an antiviral state. Importantly, SARS-CoV-2 is known to antagonize the type I IFN response, leading to dysregulated and altered immune responses.18,19 Furthermore, a feature of severe COVID-19 has been shown to be NLRP3 inflammasome activation leading to translation of IL-1B, IL-18, and pyroptosis, all of which enhance the pro-inflammatory cytokine storm.12,20 During the initiation of the adaptive immune response, antigen-specific T and B cells are activated and recruited to the site of infection.13,21,22 The spike and nucleocapsid protein are the dominant targets of the antibody (Ab) response, while the main target of neutralizing antibodies is believed to be the receptor binding domain (RBD) of the spike protein.23,24 In contrast, antigen-specific T cells detect a variety of SARS-CoV-2 structural and nonstructural proteins.25–27 Multiple studies have performed correlative analyses that showed that severe disease is associated with heightened innate and dysregulated adaptive immune responses, however, the molecular signatures of these responses are unclear.28
Several risk factors are associated with the development of severe COVID-19.29 One of the most significant factors is age, with the overwhelming majority of hospitalizations and deaths occurring in individuals older than 65 years of age.29–32 Additionally, obesity, diabetes, hypertension, and the male sex have also been associated with increased susceptibility to severe COVID-19.32–35 Earlier studies suggest that increased basal levels of systemic inflammation (“inflammaging”), reduced frequencies of naïve T and B cells, and dysregulated adaptive immune responses may facilitate disease progression.36–39 For instance, a recent study found impaired cytotoxic CD8 T cell responses in aged, but not young patients, while others reported a positive correlation between age and inflammatory cytokines during acute COVID-19.39,40 However, the mechanisms underlying increased susceptibility to severe disease and dysregulated host responses to infection with age remain poorly defined.
In this study, we undertook a multiplatform approach to interrogate the host response in young (≤65 years) and aged (>65 years) males with severe COVID-19 who also suffered from comorbidities known to increase susceptibility to severe disease (e.g., diabetes, obesity, cardiovascular diseases) during the acute phase of COVID-19. Furthermore, we interrogated the impact of corticosteroid therapy, which has proven to be highly effective in suppressing the host inflammatory response.41 Specifically, we characterized humoral, immune mediators, and transcriptional responses using whole blood samples collected 1–3 weeks post symptoms onset. Additionally, RNA sequencing data were leveraged to infer changes in circulating immune cell populations. Collectively, the data presented here show dampened inflammatory and apoptotic responses in young and aged corticosteroid-treated patients at the protein and transcriptional levels, suggesting that aberrant inflammation is a key driver of severe COVID-19.
RESULTS
Severe COVID-19 is associated with robust humoral responses and systemic inflammation that is modulated by corticosteroids
Fifty-two blood samples were obtained from 41 male subjects categorized as “young” (≤65 years, n = 19) or “aged” (>65 years, n = 22) hospitalized with several COVID-19. Diabetes, obesity, and cardiovascular disease were the most common comorbidities amongst our study participants (Supplementary Table S1). Patients whose samples were obtained between January and February 2021 had received corticosteroids, which had become standard of care by then (Graphical Abstract; Supplementary Table S1). To perform a longitudinal analysis, samples were grouped into “young” and “aged” groups and categorized into 2 different time (T) periods based on days post symptom onset (DPSO): T1 encompasses 6–11 DPSO, while T2 includes 15–21 DPSO. SARS-CoV-2-specific antibodies (Abs) and soluble mediators were quantified longitudinally in plasma using ELISA and Luminex assay respectively, while the focus reduction neutralization test (FRNT) was used to measure neutralizing Ab titers against SARS-CoV-2 (Graphical Abstract; Supplementary Fig. S1A). Anti-RBD IgG were readily detected at T1 in both age groups, with the modest increase at T2 in young patients only (Supplementary Fig. S1A). Neutralizing Ab titers showed no differences between age groups or timepoints (Supplementary Fig. S1B). Early in the pandemic, it has been reported that the bulk of the polyclonal neutralizing response targets the RBD region of SARS-CoV-2 spike protein and that RBD-specific Ab titers correlate with neutralization.21,42–45 In line with these findings, we observed a positive correlation between anti-RBD IgG endpoint titers and SARS-CoV-2 neutralization (r = 0.3902, P = 0.008) (Supplementary Fig. S1C). However, when we stratified patients by age, we observed a modest correlation between the RBD endpoint titers and live virus neutralization only in the young (r = 0.385, P = 0.070) compared to the aged cohort (r = 0.325, P = 0.140) suggesting that the polyclonal Ab response in the aged population differs from the adult population (Supplementary Fig. S1C).
We next measured levels of circulating immune mediators since severe COVID-19 is associated with a cytokine storm (Supplementary Fig. S1D and E). Additionally, patients were divided into two groups based on corticosteroid treatment (CORT) treatment or not (NC), and immune mediator levels were compared to those measured in age-matched male healthy donors (HDs). Samples obtained before corticosteroid became standard of care exhibited significantly elevated levels of numerous cytokines, chemokines, and growth factors compared to HDs (Supplementary Fig. S1D). Levels of both pro-inflammatory (e.g., IL-1α/b, IL-6, IL-17a, IL-33) and anti-inflammatory (e.g., IL-13) cytokines were significantly increased in both age groups (Supplementary Fig. S1D). Additionally, cytokines important for leukocyte proliferation and survival (e.g., IL-2, IL-3, IL-7, IL-12p70) were higher in both young and aged patients (Supplementary Fig. S1D). Markers of T cell-mediated immunity including CD40L, granzyme B, and PDL1 were elevated in young and aged patients, with CD40L produced to a significantly greater level in young compared to aged patients. Although there was no significant increase in levels of interferon-α (IFNα) relative to HD, we observed enhanced levels of IFN-β and IFN-γ in both age groups (Supplementary Fig. S1D). Chemokines primarily involved in the recruitment of monocytes (e.g., fractalkine, G-CSF, MIP-1α) were elevated regardless of age, while MIP1b was exclusively elevated in aged patients. Mitogenic growth factors, such as PDGF-AA and VEGF, were notably elevated in all patients (Supplementary Fig. S1D). In contrast, fewer immune mediators were increased in CORT-treated young and aged patients (Supplementary Fig. S1E). We detected significantly higher concentrations of IL10, TNFα, IP10, RANTES, and EGF in both young and aged CORT-treated patients compared to HD (Supplementary Fig. S1E). Two cytokines involved in anti-inflammatory and cellular cytotoxic responses, IL1RA and IL15 respectively, were significantly elevated in young CORT-treated patients only. On the other hand, levels of the chemokine, MIP1β were significantly higher in aged CORT-treated patients only (Supplementary Fig. S1E). Unlike NC patients, no significant changes in levels of IFN-γ and IFN-β were noted in CORT-treated young and aged patients.
Steroid treatment modulates host transcriptional responses during acute COVID-19
We next investigated the host transcriptional response to severe COVID-19 in both age groups. Transcriptional profiles of the patients were compared to that of sex- and age-matched healthy donors (HD, NCBI BioProject PRJNA511612). Principal component analysis (PCA) showed a distinct separation between HD and COVID-19 patients, as well as between CORT and NC patients regardless of age, with no clear distinction between T1 and T2 (Supplementary Fig. S2A).
We first analyzed the transcriptional response in NC patients compared to age-matched HD (Fig. 1; Supplementary Fig. S2). Acute SARS-CoV-2 infection results in substantial gene expression changes with ∼2000 differentiatlly expressed genes (DEGs) (1344 upregulated; 933 downregulated in young ND; and 1397 upregulated; 569 downregulated in aged ND), with a large overlap between the 2 age groups (Fig. 1A). Functional enrichment of DEGs expressed by both groups showed over-representation of Gene Ontology (GO) terms characteristic of antiviral host defense, including “viral gene expression” (e.g., RPL24, RPS7), “myeloid leukocyte activation” (e.g., CCR2, CD177, MMMP8/9, CXCL1/8, S100A8/11/12), “lymphocyte activation” (e.g., BCL6, CD28, CD3D, IGHA1/A2/G1/G2/G3/4, LYN), and “inflammatory response” (e.g., IL17RA, IL1B, RELB, RIPK2, TGFB1) (Fig. 1B; Supplementary Fig. S2A and B). Only DEGs from young patients enriched to GO term “antigen processing and presentation,” which included a number of genes important for MHC class I/II presentation (e.g., CD1D/74, HLA-G/DR/DOA/DMB/DQA1//DQB1) and peptide processing (e.g., TAP2, TAPBP/L, ERAP1/2, CTSD, PSMA5) (Fig. 1B and C). In contrast, only DEGs detected in the aged patients enriched to GO terms associated with adaptive immunity (e.g., “B cell mediated immunity,” “CD4 αβ T cell activation”). These DEGs encoded proteins with roles in T cell differentiation (e.g., IL4R, JAK3, RORA, ZBTB7B), activation (e.g., ARS, CD86), and regulation (e.g., FOXP1, TCIRG1), and were primarily upregulated relative to HD (Fig. 1D).
FIGURE 1.
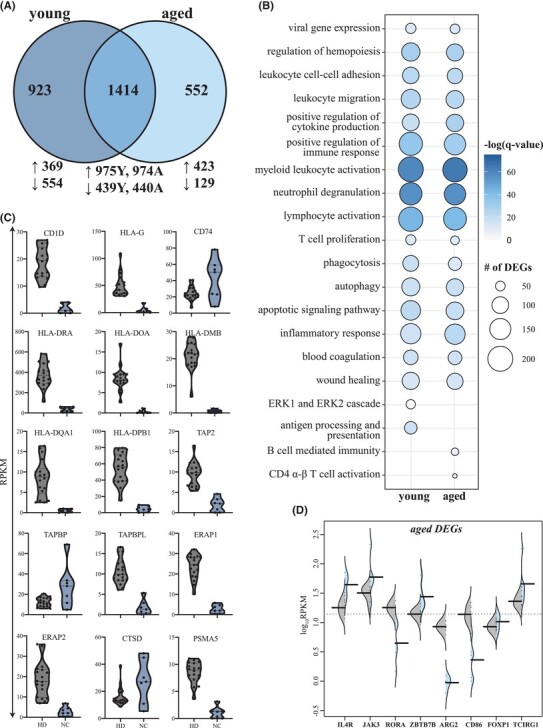
Transcriptional profiling of severe COVID-19 reflects innate and adaptive immune dysregulation. (A) Venn diagram of differentially expressed genes (DEGs) computed for young or aged patients not treated with corticosteroids (ND) relative to age- and sex-matched HD. (B) Bubbleplot representing the functional enrichment of DEGs detected in young and aged patients identified in panel (A). The size of each circle represents the number of DEGs belonging to the indicated gene ontology (GO) term while color represents the FDR-corrected P-value (q-value). (C) Violin plots depicting normalized transcript counts of DEGs belonging to GO term “antigen processing and presentation” (young patients); grey color represents healthy donors (HD) while the blue color represents the non-CORT treated group (NC) (D) Violin plots depicting normalized transcript counts of DEGs belonging to GO terms “CD4 alpha-beta T cell activation” and “B cell-mediated immunity” (aged patients); grey color represents healthy donors (HD) while the blue color represents the non-CORT treated group (NC)
Next, we carried out the same analysis for the larger cohort of CORT-treated patients to examine the effects of CORT on transcriptional changes (Supplementary Fig. 2; Supplementary Fig. S3). Comparisons of each CORT-treated age group to age-matched HD identified nearly 1500 DEGs in each group (Fig. 2A). The majority of DEGs were upregulated in both young (84%) and aged (89%) patients with a significant overlap but were not differentially expressed between the 2 age groups (Fig. 2A). DEGs expressed in young and aged patients enriched to GO terms primarily associated with antiviral innate immunity (e.g., “response to virus”; DDX58, EIF2AK2A, IFI16, IRF7, MX1, OAS2/3, STAT2); inflammation (“myeloid leukocyte activation”; C5AR1, CCR1, CD177, MMP8/9/25, MPO, S100A9/11); and cell death (“positive regulation of programmed cell death”; ADAM8, BCL2L1, GZMA) (Fig. 2B,D; Supplementary Fig. S3A). Additionally, DEGs detected in both age groups enriched to GO terms related to adaptive immunity, such as “B cell mediated immunity” (e.g., IGHA, IGHM, IGHG1/2/3/4, IGHV3-21, IGKC, IGLC3) and “lymphocyte activation” (e.g., ARG1, BCL6, CCR7, CD3D, HLA-B, LCK, JAK3; Fig. 2C; Supplementary Fig. S3B). DEGs detected only in young patients enriched to GO terms related to inflammation such as “interleukin-6 production” and “wound healing” (e.g., CCR2, CX3CR1, IL17RA, TGFB1, HBB, ITGA5, PLAUR, THBS1; Supplementary Fig. S3C). While DEGs detected only in aged patients also enriched to inflammation-related GO terms (e.g. “I-kappaB kinase/NF-kappaB signaling”; BCL3, NFKBIA, NFKBID, RELB) as well as adaptive immunity (“antimicrobial humoral response”, “CD4 αβ T cell differentiation”; CD83, SATSH3, ZBTB7B; Fig. 2C; Supplementary S3D).
FIGURE 2.
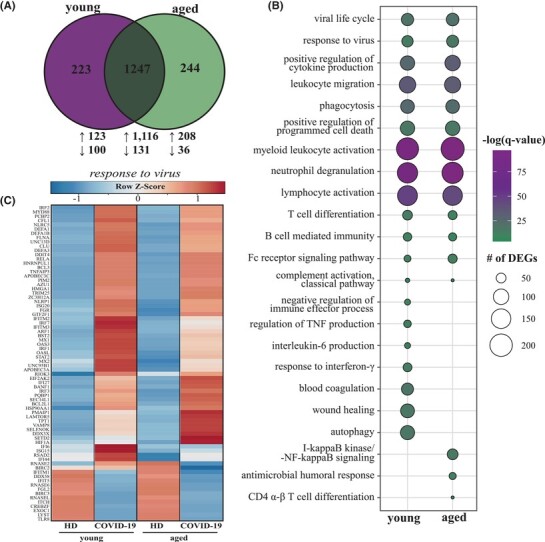
Young and aged corticosteroid-treated COVID-19 patients exhibit overlapping and distinct transcriptomes. (A) Venn diagram of differentially expressed genes (DEGs) computed for CORT-treated patients relative to age- and sex-matched HD in panel (A). (B) Bubbleplot representing the functional enrichment of DEGs detected in young and aged patients identified in panel (B). The size of each circle represents the number of DEGs belonging to the indicated gene ontology (GO) term while color represents the FDR-corrected P-value (q-value). (C) Heatmap representing DEGs enriching to all DEGs enriching to “response to virus” in young and aged patients. Each column represents the average rpkm for the given group. Range of colors is based on row-scaled and centered rpkm values of the represented DEGs. Red represents high expression while blue represents low expression
To further investigate the impact of CORT treatment on COVID-19 progression at the molecular level, we compared the transcriptional profiles between CORT-treated and NC patients (Fig. 3). Significant overlap was detected between the transcriptional response to SARS-CoV-2 infection generated by untreated and CORT-treated patients within each age group (Fig. 3A and B). DEGs unique to or shared between the treatment groups enriched to similar GO terms indicative of robust innate (e.g. “myeloid leukocyte activation/differentiation”, “response to bacterium”) and adaptive (e.g., “T cell differentiation”) immunity (Fig. 3C and D). For instance, in young patients (regardless of treatment) expression of genes important for T cell immunity such as CD28, and HLA-DMB were downregulated while that of B cell genes (e.g., IGHA1/2, IGHG1/2/3/4) and pro-apoptotic genes (e.g., BCL2A1, CD177) were upregulated (Fig. S4A). In untreated young patients, other genes important for antiviral defense were downregulated (e.g., CD27, CD3G, DDX58), while others were upregulated in CORT-treated patients (e.g., HLA-B/E/F, IL4R, RSAD2; Supplementary Fig. S4A). A subset of apoptotic genes, notably CASP8 and FADD, were highly induced only in untreated young patients. Aged NC and CORT-treated patients also shared a core of highly expressed lymphocyte activation genes such as FOS and IGHG1/2/3/4 (Supplementary Fig. S4A). A subset of T- and B-cell genes (e.g., HLA-A/B/E/F, IGHV3-21) as well as inflammatory genes such as IL18RA and S100A9/P associated with neutrophil-mediated immunity were uniquely upregulated in CORT-treated aged patients (Supplementary Fig. S4B). Genes encoding pro-inflammatory factors and ISGs, including BCL2L11, BIRC2, IFIH1, and IFNGR2, were exclusively expressed to high levels in only aged NC patients.
FIGURE 3.
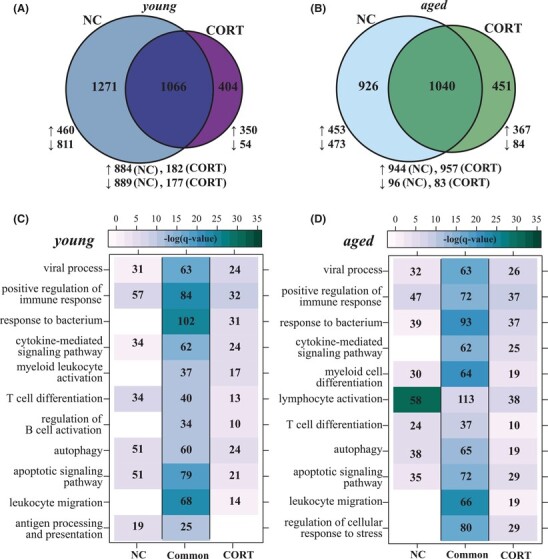
Corticosteroids treatment modulates transcriptional responses in acute COVID-19. Venn diagrams representing DEGs between corticosteroid (CORT)-treated and untreated (NC) for (A) young and (B) aged patients. Functional enrichment of CORT-unique, NC-unique and shared DEGs in panels (A) and (B) for (C) young and (D) aged cohorts. Color intensity represents the statistical significance (shown as the negative log of the FDR-adjust P-value, -log(q-value)) with a range of colors based on the GO terms with the lowest and highest –log(q-value) values for the entire set of genes per cluster. Numbers of genes enriching to each GO term per cluster are represented in each box; blank boxes indicate no statistical significance
Longitudinal regression analyses identify genes involved in antiviral and immune processes
In order to identify clusters of genes that are similarly regulated overtime from T1 to T2 in both young and aged CORT-treated patients, we employed a two-way forward stepwise regression analysis (Fig. 4). This analysis was only carried out using samples from CORT-treated patients since we had a large number of samples in this category. Genes considered significant in a minimum of 16 comparisons were retained and clustered by temporal expression patterns, which resulted in two clusters (Fig. 4A). The 768 genes in cluster 1 were downregulated at T1 and their expression remained reduced from T1 to T2, while genes in cluster 2 (n = 73) displayed the opposite pattern (Fig. 4A). Genes in cluster 1 enriched to several terms illustrating lymphocyte-mediated immunity (e.g., “lymphocyte differentiation,” “TCR signaling”) and response to signaling/stress (e.g., “response to peptide,” “macroautophagy”; Fig. 4B). Likewise, we detected several genes encoding subunits of the T cell receptor (e.g., TRAJ12, TRAC) as well as T cell coreceptors like CD28 and CD247 (Fig. 4C). Several apoptosis mediators (e.g., TNFRSF10B) and genes regulating B cell immunity (e.g., MEF2C) also belonged to cluster 2. The few genes in cluster 2 included growth factor TGFB1, protease FURIN, and NFKB transcription factor component RELA, all of which were expressed to similar levels by young and aged patients at T1 and T2 (Fig. 4D).
FIGURE 4.
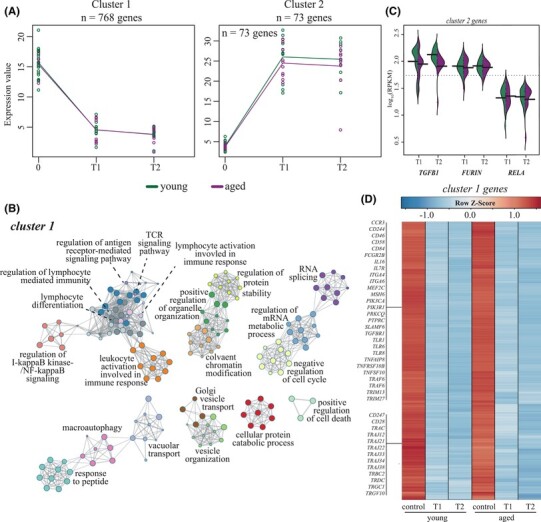
Regression analysis identifies longitudinal downregulation of key antiviral and T cell genes in both young and aged CORT-treated patients. (A) Gene expression (in rpkm) of 2 gene clusters identified by 2-ways forward regression analysis. (B) Gene ontology (GO) network depicting functional enrichment of the 500 genes contributing the most to variation seen in PCA using Metascape. Clustered nodes of identical color correspond to one GO term. Node size represents the number of DEGs associated with the GO term. Gray lines represent shared interactions between GO terms, with density and number indicating the strengths of connections between closely related GO terms. (C) Heatmap depicting gene expression of cluster 1 genes. Exemplar genes are labeled. Each column represents the average rpkm for the given group and timepoint. Range of colors is based on row-scaled and centered rpkm values of the all the DEGs in the heatmap. Red represents increased while blue represents decreased expression relative to the median. (D) Beanplot representing expression of exemplar genes identified in cluster 2 (panel A) in young and aged patients at T1 and T2
Next, we refined our longitudinal analysis by performing a regression against DPSO to identify longitudinally regulated genes (defined as protein-coding genes with FDR ≤ 0.05; Fig. 5). We first carried out this regression independently for each age group (Fig. 5A and B). This analysis uncovered 108 genes in the CORT-treated aged group only. These genes were mostly downregulated overtime and enriched to GO terms related to classical complement (e.g., IGHV1-18, IGHV2-70), TLR signaling pathways (e.g., BIRC2, IRF4), as well as epigenetic modification and cell division (e.g., DNAJC6, ESCO1, H2AC12) (Fig. 5A and B). We then carried out these analyses using age as a co-variate. We identified 102 genes, 3 of which, DDX6 (viral RNA sensing), RAB2A (intracellular trafficking), and UBE2S (modulation of viral replication), showed opposite expression trends in the 2 age groups (Fig. 5C). VAC14, which encodes a scaffold protein for intracellular vesicle transport, was downregulated in both young and aged patients over time, albeit to a greater magnitude in aged patients (Fig. 5C).
FIGURE 5.
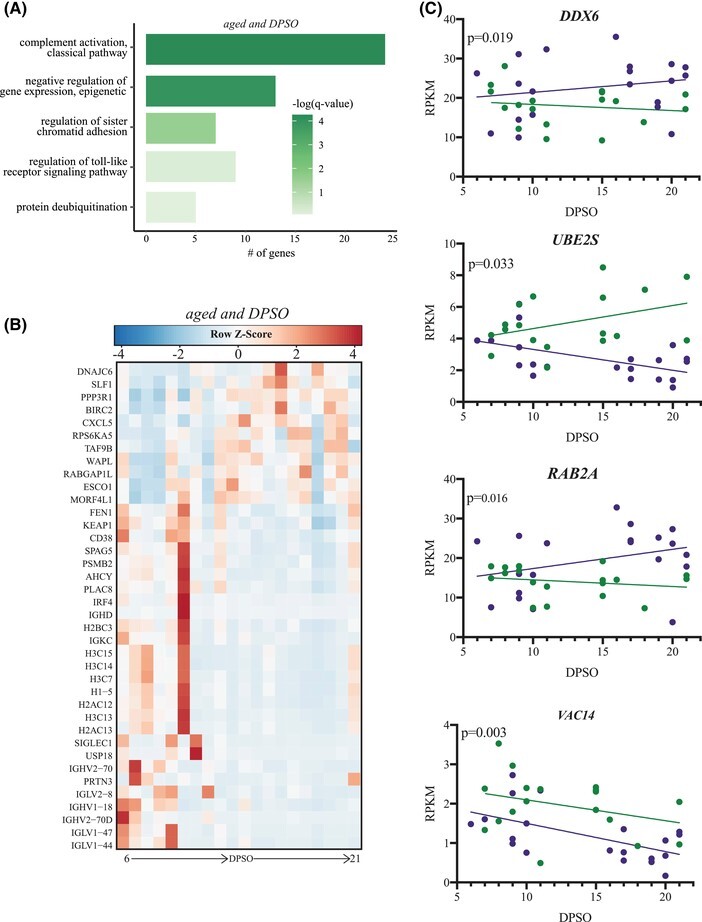
Genes related to humoral immunity and DNA metabolism are longitudinally downregulated in aged CORT-treated patients. (A) Functional enrichment of LRGs identified for aged CORT-treated patients only. Color of each bar color represents the FDR-corrected P-value (q-value). (B) Expression of LRGs from panel (B). Each column represents the average (or singular value) rpkm for the given timepoint. Range of colors is based on row-scaled and centered rpkm values of the represented DEGs. Red represents upregulation; blue represents down-regulation. (C) Expression of longitudinal-regulated genes (LRGs) (DD6, RAB2A, UBE2S, VAC14) longitudinally regulated in aged and young corticosteroids (CORT)-treated patients
Severe COVID-19 is associated with profound immune cell dysregulation
Since these blood samples were obtained from a biobank premixed with an RNA stabilizer, immune phenotyping of circulating immune cells could not be performed in this study. Therefore, we applied deconvolutional analysis to our bulk RNA sequencing (RNA-seq) data to infer changes in cell frequencies (Supplementary Fig. S5). To validate this approach, we obtained bulk RNA-seq and single-cell (sc)RNA-seq from a previously published study.46 We used scRNA-Seq cell type-specific marker genes to deconvolute the bulk RNAseq data from the same cohort. The deconvoluted cell type proportions matched the proportions in the single-cell data set remarkably well, giving credence to the validity of the deconvolution procedure (Supplementary Fig. S5A).46–49 We first compared cell frequencies among HD and CORT-treated patients. Although no significant differences were observed, there was a trend towards a decrease in CD4 memory cells and an increase in CD14 monocytes in severe COVID-19 patients irrespective of their age (Supplementary Fig. S5B and Table S3). No significant differences in the proportions of major immune cell types were detected between young and aged patients other than a modest decrease in CD8 naïve lymphocytes and CD16+ monocytes in aged patients (Supplementary Fig. S5C and Table S4).
To further delineate differences in cell subsets between HD and COVID-19 patients at a higher resolution, we next performed digital cell quantification (DCQ) using in silico flow cytometry. We used the bulk RNA-Seq data to infer cell proportions using ImmQuant with the IRIS database (Supplementary Fig. S5D and E).50,51 Our analysis revealed a dysregulation of both innate and adaptive immune cell subsets in young and aged patients compared to HD. Specifically, significant increases in bone marrow-derived plasma cells were predicted for all patients regardless of treatment (Supplementary Fig. S5D and E). We also observed a significant decrease in IL15-stimulated NK cells, activated day 7 monocytes, and CD4 Th1 cells, whereas there were significant increases in CD4 Th2 and plasma cells, young and aged NC patients only. Increases in monocytes and neutrophils were anticipated at later timepoints in CORT-treated patients. A unique decrease in IL2-stimulated NK cells was predicted for young CORT-treated patients at T1-T2 (Supplementary Fig. S5D and E). No significant differences were observed for cell frequencies between T1 and T2.
DISCUSSION
Our current understanding of COVID-19 is rapidly evolving. However, immune mechanisms governing severe disease remain elusive but critical for treating short- and long-term consequences of COVID-19 in vulnerable populations. In this study, we performed a longitudinal analysis of the functional and transcriptional host responses to severe COVID-19 in the peripheral blood of young (≤65 years) and aged (≥65 years) male patients who suffered from co-morbidities known to exacerbate COVID-19 severity. Furthermore, to our knowledge, this is the first study to investigate the effects of corticosteroid therapy (i.e., corticosteroids; CORT) on host responses during severe COVID-19.
Virus-specific Abs are critical determinants of disease progression and overall immunity. Multiple studies reported a strong induction of IgG against RBD of SARS-CoV-2 spike protein within 2 weeks post symptom onset, where anti-RBD and anti-spike IgG titer magnitudes are directly correlated with disease severity.28,42,52–54 Indeed, we observed an induction of RBD-specific IgG Abs within the first 6–11 days post symptom onset (DPSO) in both young and aged NC and CORT-treated patients that correlated with neutralizing Abs (nAbs) titers, primarily in young patients. These data support studies suggesting that many neutralizing Abs are directed toward the RBD of the spike to prevent viral entry.21,42–45 However, in the aged cohort there is no clear correlation between the level of RBD binding Abs and neutralization. This could be occurring due to an increase in non-neutralizing RBD-specific Abs or an increase in neutralizing Abs outside of RBD within the aged cohort. Interestingly, the development of antigen-specific IgG and neutralizing Abs occurred simultaneously with predicted increases in the frequencies of plasma B cells in untreated patients while upregulation of B cell-related genes was most notable in CORT-treated patients. The robust transcriptional inflammatory response accompanied by high levels of systemic immune mediators, and a significant downregulation of immunoglobulin genes by NC patients, suggest a potential for poor B cell responses and decreased Ab durability at later time points.21,55 Poor Ab durability in severe disease may be due to dysregulated germinal center responses driven by a milieu of inflammatory cytokines and extra-follicular B cell activation that generates short-lived plasma cells.56,57 We observed significantly elevated levels of TNF-α, a cytokine known to drive germinal center dysregulation, in all patients regardless of age or treatment56 and preliminary studies suggested beneficial effects of anti-TNF-α therapy.58–60 Protection offered by peripheral Abs is still unclear, but a correlation between plasma and mucosal Abs suggests that protective Ab responses also be detected at the site of infection.61
Consistent with previous studies, we detected a robust cytokine storm in severe COVID-19 patients that was significantly attenuated by corticosteroid treatment.27,62–74 In untreated patients, elevated levels of cytokines previously associated with severe COVID-19 were noted, including IL-1b, IL-6, IL-10, IL-12, IFN-γ, and TNF-α as previously reported.40,63–67,69,71,75 Moreover, levels of TNF-α and MIP3β were higher in aged patients relative to young patients, suggesting that some cytokines may serve as early, age-specific prognostic markers of severe disease.76 Indeed G-CSF and GM-CSF, which were significantly increased in NC patients, were previously identified as hallmarks of severe COVID-19 in other studies.66,67,77–79 In contrast, a limited number of pro- and anti-inflammatory cytokines, chemokines, and growth factors were detected in CORT-treated patients. Corticosteroids became standard of care at the end of June 2020 given the significant reduction in 28-day mortality and the need for mechanical ventilation they provide.82 In CORT-treated patients, levels of several inflamamtory cytokines and chemokines were elevated (TNF-α, eotaxin, IP-10, RANTES, and EGF). The anti-inflammatory IL-1RA, NK cell/memory T cell survival factor IL-15, and monocyte attractant MCP-1 were elevated in young patients only while MIP-1β, which is involved in the recruitment of NK and T cells, was elevated in CORT-treated aged patients. These observations indicate that despite comparable disease severity scores, age impacts the nature and magnitude of the cytokine response to infection. It remains unclear whether peripheral cytokines and chemokines originate from lung-resident versus peripheral blood cells.81
Our bivariate and regression transcriptional analyses of whole blood samples from CORT-treated patients revealed robust transcriptional changes indicative of dysregulated inflammatory responses despite reduced levels of systemic inflammation. Moreover, in contrast to the large transcriptional differences observed in blood samples obtained earlier in the pandemic before CORT treatment became standard of care, the gene expression profiles of young and aged samples were more uniform post CORT treatment. Genes encoding proteins that play a role in coagulation and wounding were also upregulated in CORT-treated patients, suggesting that long-term complications including thrombotic events may not be reduced with corticoid treatment. Genes encoding members of the NFKB signaling pathway were largely induced in the aged cohort (e.g., NFKBIA), thrombotic pathways in the young (e.g., THBS1), and complement pathway in both groups (e.g., C5AR1).83–87 These gene expression changes are predicted to parallel increased frequency of neutrophils and monocytes, which are a major sources of inflammatory mediators in COVID-19.68,88 Indeed, genes associated with neutrophil defense and extracellular traps (NETs) (e.g., MPO, S100A12) were highly upregulated in both CORT-treated age groups. Severe COVID-19, as well as other severe viral respiratory infections, have been associated with NET formation, which may lead to enhanced inflammation, pulmonary dysfunction and changes in vasculature.89–94 Interestingly, we did not detect significant increases in systemic levels of IL-8, a neutrophil-specific chemoattractant and inducer of NET formation.
A deficient antiviral IFN response has been proposed as the mechanism behind severe COVID-19.68,83,95,96 Levels of type I (IFN-β) and II (IFN-γ) IFN, but not IFN-α, were highly elevated in NC patients and accompanied by increases in cytokines that drive IFN production (e.g., IL-2, IL-12, and IL-13) and activation of IFN-secreting immune cells (e.g., IL-3, IL-7, IL-33). In contrast, levels of IFN-β and IFN-γ, were not significantly elevated compared to healthy donors in CORT-treated patients. However, the expression of IFN-stimulated genes (ISGs) was comparable between CORT-treated and untreated patients with only a few ISGs downregulated in untreated patients, such as STAT2 and STING1 recently implicated in RNA virus detection.97 These data suggest that corticosteroid treatment can modulate type I IFN production, but not enough to disrupt ISG expression, which is likely initiated early in the infection (before CORT treatment). IFN production in NC patients could be mediated by peripheral plasmacytoid dendritic cells (pDCs; major producers of types I IFNs) and T cells (major producers of type II IFNs) as has been reported by other studies.5,27,66,74,83,96,98–100
On the other hand, we detected robust changes in the expression of T cell activation genes in CORT-treated patients, suggesting dysregulation in cellular immunity and in line with our predicted T cell lymphopenia as reported by other studies.5,27,66,74,83,96,98–100 We also detected lower expression of genes critical for T cell-mediated immunity in NC patients including a number of MHC class I molecules (e.g., HLA-A) and T cell receptor signaling components (e.g., ZAP70). Additionally, we detected significantly elevated levels of soluble PDL1 in the plasma of NC patients, suggesting a dysregulation of adaptive immunity and potential exhaustion in these patients. The role of the antiviral response in the periphery is still unclear given that SARS-CoV-2 replication is restricted to the respiratory tract and that actively replicating virus has not been detected in the blood.
There are several limitations to our study. First, our study strictly examined severe COVID-19 in male individuals ages 46 to 76 with obesity and diabetes. Given the sex-specific differences in the immune responses to SARS-CoV-2 infection, we would therefore, caution against generalization of the data reported in this study.101 Second, given our focus on only severe COVID-19 patients, we are unable to identify statistically significant biomarkers that differentiate mild, moderate, and severe COVID-19 cases from each other. Furthermore, we lack a comparison to COVID-19 patients without co-morbidities to uncover mechanisms driven by pre-existing medical conditions, although such a cohort is unlikely given that obesity, diabetes and hypertension are significant risk factors for severe COVID-19.29,102 Third, our findings specifically examine the peripheral blood response in severe COVID-19 rather than the site of infection: the respiratory tract. Also, our study does not profile post-acute phase of COVID-19 to understand short- and long-term outcomes (e.g., “long COVID19”). Future studies should aim to compare longitudinal respiratory and peripheral samples collected from larger cohorts of individuals with varying disease severity to identify and characterize tissue-specific transcriptional and functional changes. This will enable the identification of biomarkers of different disease severities. Our study also provide data to inform future clinical trials of COVID-19 therapeutics and advocates for the use of corticosteroids, such as dexamethasone. We observed that levels of IL6, a prognostic marker for the development of severe COVID-19 and fatality, were unchanged in CORT-treated, but not NC patients.103 The long-term impact of CORT treatment on long-term immunity and vaccine responsiveness needs further investigation.
In summary, we performed a multi-platformed study incorporating both functional and transcriptional data to provide crucial insight into the molecular pathogenesis of severe COVID-19 in the peripheral blood of patients receiving and not receiving corticosteroids treatment. Our data support the hypothesis that severe COVID-19 is associated with pronounced dysregulation of all major immune cell subsets in blood, which confirms immunological misfiring in severe COVID-19. However, profound changes are mediated by corticosteroid therapy. The synergy of dysregulated innate and adaptive immunity fueled by an aberrant cytokine response may be key to driving systemic pathology in severe COVID-19.
MATERIALS AND METHODS
Study design
Remnant blood samples (n = 36) were collected from hospitalized male patients with severe COVID-19 from 2020 to 2021 through the COVID biobank of the university of California Irvine. COVID-19 diagnosis was confirmed positive with a nasopharyngeal positive qRT-PCR test. Samples were classified into 2 time points based on days post symptom onset (DPSO): T1 (6-11 DPSO) and T2 (15-21 DPSO) (Fig. 1A). Samples were further classified into young (≤65 years) and aged (≥65 years) cohorts based on age. Complete metadata for COVD19 patients and healthy donors can be found in Supplementary Table S1. This study was approved by University of California Irvine Institutional Review Board (HS# 2012–8716).
ELISA endpoint titer
SARS-CoV-2 specific IgG titers against the receptor-binding domain (RBD) of the spike protein was determined by ELISA. For this, MaxiSorp (ThermoFisher) plates were coated with 50 μL of a 1 μg/mL mixture of protein in carbonate buffer (0.1 M Na2CO3 0.1 M NaHCO3 pH 9.3) overnight at 4°C. Blocking buffer (PBS + 5%BSA + 0.5% Tween) was then added to pates for 1 h at 37°C followed by washing 3× with wash buffer. Serially diluted polyclonal sera were then added at a volume of 50uL/well. Polyclonal sera were incubated for 1 h at room temperature in the ELISA plate, washed 3x with wash buffer, followed by addition of goat-anti-human IgG HRP (Sigma) conjugated secondary (1:5000) for 1 h at room temperature. The plate was washed 3× with wash buffer and the ELISA was developed with 100 μL of TMB enhanced substrate (Neogen Diagnostics) and placed in a dark space for 15 min. The reaction was quenched with 1 N HCl and the plate was read for an optical density (OD) of 450 nm on a BioTek Epoch plate reader. IgG endpoint titer were calculated using GrapPad Prism 8.
Focus reduction neutralization test
Human sera were diluted 4-fold in a serial manner and mixed with ∼100 focus-forming units (FFU) of virus followed by incubation at 37°C for 1 h to allow for the formation of immune complexes. The sera and virus mixture were then added onto confluent Vero-WHO monolayers in 96-well plates for 1 h at 37°C to allow virus adsorption. Cells were then overlaid with 2% methylcellulose mixed with DMEM containing 5% FBS and incubated for 24 h at 37°C. After 24 h, the media was removed, and the monolayers were fixed with 5% paraformaldehyde in PBS for 15 min at room temperature, rinsed with PBS, and permeabilized in Perm Wash (PBS, 0.05% Triton-X). Infected cell foci were stained by incubating with polyclonal anti-SARS Guinea Pig sera (BEIresources) for 1 h at room temperature and then washed 3 times with Perm Wash. Detection of foci was completed by incubation with a 1:5000 dilution of horseradish peroxidase-conjugated goat anti-guinea pig IgG (Sigma) for 1 h. After 3 washes with Perm Wash, staining was visualized by addition of TrueBlue detection reagent (KPL). Infected foci were enumerated by CTL Elispot and FRNT curves were generated by log-transformation of the x-axis followed by non-linear curve fit regression analysis using Graphpad Prism 8.
Luminex assay
Soluble immune mediators in plasma were quantified with Human XL Cytokine Luminex Performance Panel Kit (Thermo Scientific) per manufacturer's instructions. This panel quantified levels of the following soluble mediators: IL1ra, IL1b, IL1a, IL2, IL3, IL33, IL4, IL5, IL6, IL7, IL8, IL10, IL12p70, IL13, IL15, IL17A, IL17E, RANTES (i.e., CCL5), TNF-α, IFN-α, IFN-β, IFN-γ, eotaxin, fractalkine, G-CSF, GM-CSF, IFN-γ-induced protein 10 (IP10), Mϕ chemoattractant protein-1 (MCP1), Mϕ inflammatory protein 1α (MIP1α), Mϕ inflammatory protein 1β (MIP1β), Mϕ inflammatory protein 3α (MIP3α), growth-regulated oncogene α (GROα), growth-regulated oncogene β (GROβ), platelet-derived growth factor A (PDGF-A), platelet-derived growth factor AB/BB (PDGF-AB/BB), transforming growth factor α (TGF-α), tumor necrosis family-related apoptosis-inducing ligand (TRAIL), vascular endothelial growth factor (VEGF), endothelial growth factor (EGF), basic fibroblast growth factor (FGF-basic), Fms-related tyrosine kinase 3 ligand (Flt-3 L), CD40L, and granzyme B. Analytes with undetectable levels in the majority of patients were omitted from analysis. COVID-19 patients were compared to age-matched healthy male donors.
Library construction and sequencing
RNA was extracted from whole blood using the RiboPure™-Blood Kit (Life Technologies) per the manufacturer's instructions. Quality and quantity of RNA from whole blood was determined using an Agilent 2100 Bioanalyzer. cDNA libraries were constructed using the NEB Next Ultra II Direction RNA Library Prep Kit (Thermo Fischer). RNA was treated with RNase H and DNase I after depletion of ribosomal RNA (rRNA). Adapters were ligated to cDNA products and the subsequent ∼300 base pair (bp) amplicon was PCR-amplified and selected by size exclusion. cDNA libraries were assessed for quality and quantity on an Agilent 2100 Bioanalyzer prior to 150 bp single-end sequencing using the Illumina NovaSeq platform.
Bioinformatic analysis
Preliminary data analysis was performed with RNA-Seq workflow module of systemPipeR.104 RNA-Seq reads were demultiplexed, quality-filtered and trimmed using Trim Galore (average Phred score cut-off of 30, minimum length of 50 bp). FastQC was used to generate quality reports. Hisat2 was used to align reads to the reference genome Homo sapiens (Homo_sapiens.GRCh38.dna.primary_assembly.fa) and the Homo_sapiens.GRCh38.85.gtf file was used for annotation. Raw expression values (gene-level read counts) were generated using the summarizeOverlaps function and normalized (read per kilobase of transcript per million mapped reads, rpkm) using the edgeR (v.3.30.3) and systemPipeR (v1.22.0) packages. Differentially expressed genes (DEGs) were defined as protein-coding genes with an average rpkm ≥ 5, FDR-corrected P-value ≤ 0.05 and log2(fold-change) ≥ 1 in either direction. To identify significant patterns of longitudinal gene expression, we applied a two-ways forward regression analysis using MaSigPro.105 Only clusters containing genes that were considered statistically significant in at least 16 comparisons were retained. as the total number of normalized reads mapping to all viral genes.
Longitudinal analysis for CORT-treated young and age patients over T1 and T2 was performed with short time-series expression miner (STEM).106 STEM specifically integrates a clustering algorithm based on the sole assumption that experiments can be naturally and sequentially ordered. This software further differentiates between random and chronological patterns. Identification of longitudinally regulated genes (LRGs) with age as a factor or by age cohort in CORT-treated patients was performed with DESeq2.
Functional enrichment of genes was performed with Metascape to identify significant Gene Ontology (GO) biological processes.107 The Cytoscape network data integration and visualization tool was used to generate GO (biological process) term network.108 Bar graphs, beanplots, bubbleplots, heatmaps, Venn diagrams and volcano plots were created in R. Graphs were generated with GraphPad Prism software (version 8).
Digital cell quantification (DCQ), (i.e., in silico flow cytometry) using gene expression data was carried out with ImmQuant using the IRIS database. Transcriptional data for age- and sex-matched healthy donors were retrieved from NBCI BioProject PRJNA511612.
Deconvolution of RNA-Seq data
In order to estimate the proportion of major cell subsets in blood samples, cell type-specific marker genes were identified using a reference scRNA-Seq dataset and used to deconvolute bulk RNAseq expression data. The scRNAseq data and cluster annotations were obtained from a recent study; bulk RNAseq data were generated in the present study.46 Both the bulk and single cell count data were normalized using the vst function from the sctransform R package.48 Cell type-specific markers for each scRNAseq cluster that were used for deconvolutional analysis were found using the NS-Forest machine learning method.47 Marker gene by sample bulk and marker gene by cell single-cell matrices were used for deconvolution using the damped weighted least squares method.49
Statistical analysis
Statistical significance between groups in panels 1A, 1B, 1D and 1E were computed with Mann-Whitney U-test panels S5A and S5B with one-way ANOVA with Tukey's HSD. Spearman correlation was used to calculate correlations in panel 1C. *P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, **** P ≤ 0.0001.
Supplementary Material
Figure S1. Corticosteroids attenuates the cytokine storm in severe COVID-19.
Figure S2. Transcriptional analysis of acute COVID-19 in young and aged patients.
Figure S3. CORT-treated young COVID-19 patients feature unique upregulation of genes modulating coagulation, thrombosis and inflammatory responses.
Figure S4. ND COVID-19 patients are associated with unique upregulation of inflammatory and pro-apoptotic genes.
Figure S5. Digital cell quantification predicts severe lymphopenia.
Table S1. Participant demographics and clinical data.
Table S2. Concentrations (pg/mL) of soluble immune mediators in young and aged COVID-19 patients and healthy donors. NA, not detected or quantifiable.
Table S3. Statistical analysis of deconvolution results between HD and CORT-treated COVID19 patients.
Table S4. Statistical analysis of deconvolution results between young and aged CORT-treated COVID19 patients.
ACKNOWLEDGMENTS
The authors are grateful to all the study participants. The authors wish to acknowledge the support of the Chao Family Comprehensive Tissue Shared Resource, supported by the National Cancer Institutes of Health under the award number P30CA062203. This work was supported by in part by UL1TR001414 – 06 (D.C.), 1U01CA260541-01 (A.K.P. and J.D.B.), COVID-19 Basic, Translational and Clinical Research Fund from UC Irvine Office of the Vice Chancellor for Research as well as a Saint Louis University COVID-19 Research Seed Funding.
Abbreviations used in this paper
- Ab, antibody; COVID-19
Coronavirus Disease 2019
- CORT
corticosteroids
- CoV
coronavirus
- DPSO
days post symptom onset
- MERS
Middle eastern respiratory syndrome
- ND
no corticosteroids treatment
- RBD
receptor binding domain
- SARS
severe acute respiratory syndrome
Contributor Information
Amanda N Pinski, Department of Molecular Biology and Biochemistry, University of California, Irvine, Irvine, California, USA; Center for Virus Research, University of California, Irvine, Irvine, California, USA; Institute for Immunology, University of California, Irvine, California, USA.
Tara L Steffen, Department of Molecular Microbiology and Immunology, Saint Louis University, St Louis, Missouri, USA.
Michael Z Zulu, Department of Molecular Biology and Biochemistry, University of California, Irvine, Irvine, California, USA; Center for Virus Research, University of California, Irvine, Irvine, California, USA; Institute for Immunology, University of California, Irvine, California, USA.
Sarah L George, Department of Medicine, Division of Infectious Diseases, Saint Louis University, St Louis, Missouri, USA.
Alexandria Dickson, Department of Molecular Microbiology and Immunology, Saint Louis University, St Louis, Missouri, USA.
Delia Tifrea, Department of Pathology and Laboratory Medicine, University of California, Irvine, California, USA.
Kevin J Maroney, Department of Molecular Biology and Biochemistry, University of California, Irvine, Irvine, California, USA.
Neil Tedeschi, J. Craig Venter Institute, La Jolla, California, USA.
Yun Zhang, J. Craig Venter Institute, La Jolla, California, USA.
Richard H Scheuermann, J. Craig Venter Institute, La Jolla, California, USA.
Amelia K Pinto, Department of Molecular Microbiology and Immunology, Saint Louis University, St Louis, Missouri, USA.
James D Brien, Department of Molecular Microbiology and Immunology, Saint Louis University, St Louis, Missouri, USA.
Ilhem Messaoudi, Department of Molecular Biology and Biochemistry, University of California, Irvine, Irvine, California, USA; Center for Virus Research, University of California, Irvine, Irvine, California, USA; Institute for Immunology, University of California, Irvine, California, USA.
AUTHORSHIP
I.M., A.N.P., A.K.P., and J.D.B. conceptualized the work; T.L.S., J.D.B., A.N.P., K.J.M., and M.Z.Z. carried out the experiments; T.L.S., J.D.B., A.N.P., M.Z.Z., A.K.P., Y.Z., N.T., R.H.S., and I.M. analyzed the data; T.L.S., A.K.P., J.D.B., A.N.P., M.Z.Z., and I.M. wrote and edited the manuscript. A.D., S.L.G., and D.T. were responsible for the collection of patient samples. S.L.G. is the PI for the ACTTI trial. All authors reviewed and approved the final version of the manuscript.
DISCLOSURE
The authors declare no competing interest.
AVAILABILITY
The accession numbers for the RNA-Sequencing data reported in this paper is available in the NCBI BioProject PRJNA751085.
REFERENCES
- 1. Woolf SH, Chapman DA, Lee JH. COVID-19 as the leading cause of death in the United States. JAMA. 2021;325:123–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Johansson MA, Quandelacy TM, Kada S et al. SARS-CoV-2 transmission from people without COVID-19 symptoms. JAMA Network Open. 2021;4:e2035057–e2035057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sami S, Akinbami LJ, Petersen LR et al. Prevalence of SARS-CoV-2 antibodies in first responders and public safety personnel, New York City, New York, USA, May-July 2020. Emerg Infect Dis. 2021;27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lumley SF, Wei J, O'donnell D et al. The duration, dynamics and determinants of SARS-CoV-2 antibody responses in individual healthcare workers. Clin Infect Dis. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu C, Chen X, Cai Y et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020;180:934–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deng G, Yin M, Chen X, Zeng F. Clinical determinants for fatality of 44,672 patients with COVID-19. Critical Care. 2020;24:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mudd PA, Crawford JC, Turner JS et al. Distinct inflammatory profiles distinguish COVID-19 from influenza with limited contributions from cytokine storm. Sci Adv. 2020;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carfì A, Bernabei R, Landi F. Persistent symptoms in patients after acute COVID-19. JAMA. 2020;324:603–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lucas C, Wong P, Klein J et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Arunachalam PS, Wimmers F, Mok CKaP et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Blanco-Melo D, Nilsson-Payant BE, Liu W-C et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rodrigues TS, De Sá KSG, Ishimoto AY et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J Exp Med. 2021;218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang F, Gan R, Zhen Z et al. Adaptive immune responses to SARS-CoV-2 infection in severe versus mild individuals. Signal Transduction and Targeted Therapy. 2020;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lim Y, Ng Y, Tam J, Liu D. Human coronaviruses: a review of virus-host interactions. Diseases. 2016;4:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fitzgerald KA, Kagan JC. Toll-like receptors and the control of immunity. Cell. 2020;180:1044–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yin X, Riva L, Pu Y et al. MDA5 governs the innate immune response to SARS-CoV-2 in lung epithelial cells. Cell Reports. 2021;34:108628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frieman M, Heise M, Baric R. SARS coronavirus and innate immunity. Virus Research. 2008;133:101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lei X, Dong X, Ma R et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nature Communications. 2020;11:3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zheng Yi, Zhuang M-W, Han L et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling. Signal Transduction and Targeted Therapy. 2020;5:299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Freeman TL, Swartz TH. Targeting the NLRP3 inflammasome in severe COVID-19. Front Immunol. 2020;11:1518–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rodda LB, Netland J, Shehata L et al. Functional SARS-CoV-2-specific immune memory persists after mild COVID-19. Cell. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Weiskopf D, Schmitz KS, Raadsen MP et al. Phenotype and kinetics of SARS-CoV-2–specific T cells in COVID-19 patients with acute respiratory distress syndrome. Science Immunology. 2020;5:eabd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Piccoli L, Park Y-J, Tortorici MA et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Steffen TL et al. The receptor binding domain of SARS-CoV-2 spike is the key target of neutralizing antibody in human polyclonal sera. bioRxiv. 2020:2020.2008.2021.261727. bioRxiv. bioRxiv. bioRxiv. [Google Scholar]
- 25. Grifoni A, Weiskopf D, Ramirez SI et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501 e1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Le Bert N, Tan AT, Kunasegaran K et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020. [DOI] [PubMed] [Google Scholar]
- 27. Weiskopf D, Schmitz KS, Raadsen MP et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol. 2020;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang P, Liu L, Nair MS et al. SARS-CoV-2 neutralizing antibody responses are more robust in patients with severe disease. Emerg Microbes Infect. 2020;9:2091–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Callender LA, Curran M, Bates SM et al. The impact of pre-existing comorbidities and therapeutic interventions on COVID-19. Front Immunol. 2020;11:1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Levin AT, Hanage WP, Owusu-Boaitey N et al. Assessing the age specificity of infection fatality rates for COVID-19: systematic review, meta-analysis, and public policy implications. Eur J Epidemiol. 2020;35:1123–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Romero Starke K, Petereit-Haack G, Schubert M et al. The age-related risk of severe outcomes due to COVID-19 infection: a rapid review, meta-analysis, and meta-regression. Int J Environ Res Public Health. 2020;17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. O'brien J, Du KY, Peng C. Correction to: incidence, clinical features, and outcomes of COVID-19 in Canada: impact of sex and age. J Ovarian Res. 2021;14:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hamer M, Gale CR, Kivimäki M, Batty GD. Overweight, obesity, and risk of hospitalization for COVID-19: a community-based cohort study of adults in the United Kingdom. Proc Natl Acad Sci U S A. 2020;117:21011–21013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang Yi, Lu Y, Huang Y-M et al. Obesity in patients with COVID-19: a systematic review and meta-analysis. Metabolism. 2020;113:154378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jin J-M, Bai P, He W et al. Gender differences in patients with COVID-19: focus on severity and mortality. Front Public Health. 2020;8:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bajaj V, Gadi N, Spihlman AP, Wu SC, Choi CH, Moulton VR. Aging, immunity, and COVID-19: how age influences the host immune response to coronavirus infections? Front Physiol. 2020;11:571416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goplen NP, Cheon InSu, Sun J. Age-related dynamics of lung-resident memory CD8(+) T cells in the age of COVID-19. Front Immunol. 2021;12:636118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mueller AL, Mcnamara MS, Sinclair DA. Why does COVID-19 disproportionately affect older people? Aging (Albany NY). 2020;12:9959–9981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Westmeier J, Paniskaki K, Karaköse Z et al. Impaired Cytotoxic CD8(+) T cell response in elderly COVID-19 patients. mBio. 2020;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Angioni R, Sánchez-Rodríguez R, Munari F et al. Age-severity matched cytokine profiling reveals specific signatures in Covid-19 patients. Cell Death Dis. 2020;11:957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Raju R, V P, Biatris PS, J S. Therapeutic role of corticosteroids in COVID-19: a systematic review of registered clinical trials. Futur J Pharm Sci. 2021;7:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Iyer AS, Jones FK, Nodoushani A et al. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci Immunol. 2020;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ju B, Zhang Qi, Ge J et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. [DOI] [PubMed] [Google Scholar]
- 44. Shi R, Shan C, Duan X et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. [DOI] [PubMed] [Google Scholar]
- 45. Wu Y, Wang F, Shen C et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lewis SA et al. Longitudinal analyses reveal age-specific immune correlates of COVID-19 severity. medRxiv. 2021:2021.2001.2025.21250189. medRxiv. medRxiv. medRxiv. [Google Scholar]
- 47. Aevermann B, Zhang Y, Novotny M et al. A machine learning method for the discovery of minimum marker gene combinations for cell-type identification from single-cell RNA sequencing. Genome Res. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tsoucas D, Dong R, Chen H et al. Accurate estimation of cell-type composition from gene expression data. Nat Commun. 2019;10:2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Frishberg A, Brodt A, Steuerman Y, Gat-Viks I. ImmQuant: a user-friendly tool for inferring immune cell-type composition from gene-expression data. Bioinformatics. 2016;32:3842–3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Klein S et al. Sex, age, and hospitalization drive antibody responses in a COVID-19 convalescent plasma donor population. medRxiv, 10.1101/2020.06.26.20139063 (2020). medRxiv medRxiv medRxiv [DOI] [PMC free article] [PubMed]
- 53. Ma H, Zeng W, He H et al. Serum IgA, IgM, and IgG responses in COVID-19. Cell Mol Immunol. 2020;17:773–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Prévost J, Gasser R, Beaudoin-Bussières G et al. Cross-sectional evaluation of humoral responses against SARS-CoV-2 spike. Cell Rep Med. 2020;1:100126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen Y, Zuiani A, Fischinger S et al. Quick COVID-19 healers sustain anti-SARS-CoV-2 antibody production. Cell. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kaneko N, Kuo H-H, Boucau J et al. Loss of Bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cell. 2020;183:143–157 e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Woodruff MC, Ramonell RP, Nguyen DC et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat Immunol. 2020;21:1506–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Feldmann M, Maini RN, Woody JN et al. Trials of anti-tumour necrosis factor therapy for COVID-19 are urgently needed. Lancet. 2020;395:1407–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Robinson PC, Liew DFL, Liew JW et al. The potential for repurposing anti-TNF as a therapy for the treatment of COVID-19. Med (N Y). 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kridin K, Schonmann Y, Damiani G et al. Tumor necrosis factor inhibitors are associated with a decreased risk of COVID-19-associated hospitalization in patients with psoriasis-a population-based cohort study. Dermatol Ther. 2021:e15003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Isho B, Abe KT, Zuo M et al. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci Immunol. 2020;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Blanco-Melo D, Nilsson-Payant BE, Liu W-C et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045 e1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chen G, Wu Di, Guo W et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130:2620–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Henry BM, De Oliveira MHS, Benoit S, Plebani M, Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chem Lab Med. 2020;58:1021–1028. [DOI] [PubMed] [Google Scholar]
- 65. Herold T, Jurinovic V, Arnreich C et al. Elevated levels of IL-6 and CRP predict the need for mechanical ventilation in COVID-19. J Allergy Clin Immunol. 2020;146:128–136 e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Huang C, Wang Y, Li X et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kwon Ji-S, Kim JiY, Kim M-C et al. Factors of severity in patients with COVID-19: cytokine/chemokine concentrations, viral load, and antibody responses. Am J Trop Med Hyg. 2020;103:2412–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lee JS, Park S, Jeong HW et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci Immunol. 2020;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Liu J, Li S, Liu J et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55:102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Petrey AC, Qeadan F, Middleton EA, Pinchuk IV, Campbell RA, Beswick EJ. Cytokine release syndrome in COVID-19: innate immune, vascular, and platelet pathogenic factors differ in severity of disease and sex. J Leukoc Biol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Qin C, Zhou L, Hu Z et al. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis. 2020;71:762–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tang Y, Liu J, Zhang D, Xu Z, Ji J, Wen C. Cytokine storm in COVID-19: the current evidence and treatment strategies. Front Immunol. 2020;11:1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang J, Jiang M, Chen X, Montaner LJ. Cytokine storm and leukocyte changes in mild versus severe SARS-CoV-2 infection: review of 3939 COVID-19 patients in China and emerging pathogenesis and therapy concepts. J Leukoc Biol. 2020;108:17–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wu Y, Huang X, Sun J et al. Clinical characteristics and immune injury mechanisms in 71 patients with COVID-19. mSphere. 2020;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kong M, Zhang H, Cao X, Mao X, Lu Z. Higher level of neutrophil-to-lymphocyte is associated with severe COVID-19. Epidemiol Infect. 2020;148:e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zheng H-Yi, Zhang Mi, Yang C-X et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol. 2020;17:541–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chi Y, Ge Y, Wu B et al. Serum cytokine and chemokine profile in relation to the severity of coronavirus disease 2019 in China. J Infect Dis. 2020;222:746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Xu Z-S, Shu T, Kang L et al. Temporal profiling of plasma cytokines, chemokines and growth factors from mild, severe and fatal COVID-19 patients. Signal Transduct Target Ther. 2020;5:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhao Y, Qin L, Zhang P et al. Longitudinal COVID-19 profiling associates IL-1RA and IL-10 with disease severity and RANTES with mild disease. JCI Insight. 2020;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Thwaites RS, Sanchez Sevilla Uruchurtu A, Siggins MK et al. Inflammatory profiles across the spectrum of disease reveal a distinct role for GM-CSF in severe COVID-19. Sci Immunol. 2021;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wu M, Chen Y, Xia H et al. Transcriptional and proteomic insights into the host response in fatal COVID-19 cases. Proc Natl Acad Sci U S A. 2020;117:28336–28343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Group RC et al. Corticosteroids in hospitalized patients with COVID-19. N Engl J Med. 2021;384:693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hadjadj J, Yatim N, Barnabei L et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Jobe F, Simpson J, Hawes P, Guzman E, Bailey D. Respiratory syncytial virus sequesters NF-kappa B Subunit p65 to Cytoplasmic inclusion bodies to inhibit innate immune signaling. Journal of Virology. 2020;94. doi:ARTN e01380-2010.1128/JVI.01380-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kumar N, Xin Z-T, Liang Y, Ly H, Liang Y. NF-kappaB signaling differentially regulates influenza virus RNA synthesis. J Virol. 2008;82:9880–9889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tian B, Zhang Y, Luxon BA et al. Identification of NF-kappaB-dependent gene networks in respiratory syncytial virus-infected cells. J Virol. 2002;76:6800–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. [DOI] [PubMed] [Google Scholar]
- 88. Giamarellos-Bourboulis EJ, Netea MG, Rovina N et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27:992–1000 e1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Arcanjo A, Logullo J, Menezes CCB et al. The emerging role of neutrophil extracellular traps in severe acute respiratory syndrome coronavirus 2 (COVID-19). Sci Rep. 2020;10:19630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Cortjens B, De Boer OJ, De Jong R et al. Neutrophil extracellular traps cause airway obstruction during respiratory syncytial virus disease. J Pathol. 2016;238:401–411. [DOI] [PubMed] [Google Scholar]
- 91. Papayannopoulos V. Neutrophil extracellular traps in immunity and disease. Nat Rev Immunol. 2018;18:134–147. [DOI] [PubMed] [Google Scholar]
- 92. Wang J, Li Q, Yin Y et al. Excessive neutrophils and neutrophil extracellular traps in COVID-19. Front Immunol. 2020;11:2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Zhu L, Liu Lu, Zhang Y et al. High level of neutrophil extracellular traps correlates with poor prognosis of severe influenza a infection. J Infect Dis. 2018;217:428–437. [DOI] [PubMed] [Google Scholar]
- 94. Zuo Yu, Yalavarthi S, Shi H et al. Neutrophil extracellular traps in COVID-19. JCI Insight. 2020;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Galani I-E, Rovina N, Lampropoulou V et al. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat Immunol. 2020. [DOI] [PubMed] [Google Scholar]
- 96. Arunachalam PS, Wimmers F, Mok CKaP et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Li Y, Renner DM, Comar CE et al. SARS-CoV-2 induces double-stranded RNA-mediated innate immune responses in respiratory epithelial-derived cells and cardiomyocytes. Proc Natl Acad Sci U S A. 2021;118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Kuri-Cervantes L, Pampena MB, Meng W et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci Immunol. 2020;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Tan Li, Wang Qi, Zhang D et al. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct Target Ther. 2020;5:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Bernardes JP, Mishra N, Tran F et al. Longitudinal multi-omics analyses identify responses of megakaryocytes, erythroid cells, and plasmablasts as hallmarks of severe COVID-19. Immunity. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Chakravarty D, Nair SS, Hammouda N et al. Sex differences in SARS-CoV-2 infection rates and the potential link to prostate cancer. Commun Biol. 2020;3:374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Zheng Z et al. Risk factors of critical & mortal COVID-19 cases: a systematic literature review and meta-analysis. J Infect. 2020;81:e16–e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Santa Cruz A, Mendes-Frias A, Oliveira AI et al. Interleukin-6 is a biomarker for the development of fatal severe acute respiratory syndrome coronavirus 2 pneumonia. Front Immunol. 2021;12:613422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Backman TWH, Girke T. systemPipeR: nGS workflow and report generation environment. BMC Bioinformatics. 2016;17:388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Conesa A, Nueda MJ, Ferrer A, Talon M. maSigPro: a method to identify significantly differential expression profiles in time-course microarray experiments. Bioinformatics. 2006;22:1096–1102. [DOI] [PubMed] [Google Scholar]
- 106. Ernst J, Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zhou Y, Zhou B, Pache L et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Shannon P. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Corticosteroids attenuates the cytokine storm in severe COVID-19.
Figure S2. Transcriptional analysis of acute COVID-19 in young and aged patients.
Figure S3. CORT-treated young COVID-19 patients feature unique upregulation of genes modulating coagulation, thrombosis and inflammatory responses.
Figure S4. ND COVID-19 patients are associated with unique upregulation of inflammatory and pro-apoptotic genes.
Figure S5. Digital cell quantification predicts severe lymphopenia.
Table S1. Participant demographics and clinical data.
Table S2. Concentrations (pg/mL) of soluble immune mediators in young and aged COVID-19 patients and healthy donors. NA, not detected or quantifiable.
Table S3. Statistical analysis of deconvolution results between HD and CORT-treated COVID19 patients.
Table S4. Statistical analysis of deconvolution results between young and aged CORT-treated COVID19 patients.


