Abstract
Advances in biotechnology to treat and cure human disease have markedly improved human health and the development of modern societies. However, substantial challenges remain to overcome innate biological factors that thwart the activity and efficacy of pharmaceutical therapeutics. Until recently, the importance of extracellular DNA (eDNA) in biofilms was overlooked. New data reveal its extensive role in biofilm formation, adhesion, and structural integrity. Different approaches to target eDNA as anti-biofilm therapies have been proposed, but eDNA and the corresponding biofilm barriers are still difficult to disrupt. Therefore, more creative approaches to eradicate biofilms are needed. The production of eDNA often originates with the genetic material of bacterial cells through cell lysis. However, genomic DNA and eDNA are not necessarily structurally or compositionally identical. Variations are noteworthy because they dictate important interactions within the biofilm. Interactions between eDNA and biofilm components may as well be exploited as alternative anti-biofilm strategies. In this review, we discuss recent developments in eDNA research, emphasizing potential ways to disrupt biofilms. This review also highlights proteins, exopolysaccharides, and other molecules interacting with eDNA that can serve as anti-biofilm therapeutic targets. Overall, the array of diverse interactions with eDNA is important in biofilm structure, architecture, and stability.
Keywords: anti-biofilm therapies, biofilms, eDNA, eDNA therapy, eDNA-interactions
1 |. BACKGROUND
A global health crisis is growing due to antibiotic resistance. Antibiotics were first prescribed to treat severe infections after Alexander Fleming discovered penicillin in the 1940’s. Over time, various classes of antibiotics that target different bacterial machineries were introduced to the market. However, in recent years, the rate of discovery of new antibiotic classes has stagnated whereas the rate of antibiotic consumption has continued to increase (Silver, 2011). As a result, antibiotic resistance can emerge as bacteria respond to therapeutic treatments. The Centers for Disease Control and Prevention reports that at least 2.8 million people get infected and at least 35,000 die because of antibiotic resistant bacteria in the United States annually (CDC, 2019). In Europe, an estimated number of 25,000 deaths are associated to antibiotic resistant bacteria (“Annual report of the European Medicines Agency,” 2010). Greater than 30,000 deaths per year are reported in countries like Thailand as well as increasing variants of antibiotic resistant bacteria emerging in South America, the Middle East, and Asia (Howell, 2013). In addition to these statistics, neonatal sepsis attributed to antibiotic resistance are increasing in Tanzania and Mozambique, and approximately 58,000 neonatal sepsis deaths related to antibiotic resistance are reported in India (Hellen et al., 2015).
Bacteria have acquired different resistance mechanisms to overcome clinical treatments and one of these is the development of biofilms. Biofilms are dynamic communities of bacterial organisms encapsulated in a thick matrix of extracellular polymeric substance (EPS) that affixes the cells in close proximity to each other and facilitates the transfer of nutrients, waste, quorum sensing (QS), and genetic material (Donlan, 2002; Kostakioti et al., 2013). Bacteria in biofilms have stronger resilience compared to planktonic bacteria because the biofilm EPS serves as a physical shield against potential dangers in their environment (i.e., antibiotics, antibacterial agents, shear stress). Bacterial aggregation and eventual biofilm maturation occur in different stages and require species-specific conditions (Kostakioti et al., 2013). The first step of biofilm formation involves bacteria adhering on to a surface (either biotic or abiotic)—a step driven by a delicate balance of attractive or repelling forces. Bacterial adhesion on an abiotic surface is primarily driven by nonspecific (i.e., hydrophobic interactions) forces while adhesion to biotic surfaces is usually driven by molecular mechanisms (i.e., adhesins, lectins, etc.) (Dunne, 2002). The nonspecific initial attachment is reversible, which allows the bacteria to return to the planktonic population (Koo et al., 2017). In addition, electrostatic interactions can cause charge-charge attraction or repulsion between the cells and surface. Irreversible attachment can be achieved by species-specific adhesins on the cell surface that bind to the substrate and withstand detaching forces. After attachment, cell-to-cell adhesion occurs as adjacent cells are connected by various EPS components (Kostakioti et al., 2013). These physiochemical interactions (hydrogen bonding, Van der Waals, electrostatic, etc.) between cells and EPS components create a stable biofilm matrix. As cell-to-cell adhesion increases, biofilm proliferation enables a dynamic biofilm community to grow, and as the biofilms develop to maturity, cell dispersal becomes possible. Cell dispersal is achieved by shedding of daughter cells of actively growing cells, nutrient deficiency, QS, and/or shear stress (Donlan, 2002). Controlled cell-dispersal allows bacteria to scatter in a free-floating planktonic state and establish other biofilm populations elsewhere. Controlled dispersal benefits the pathogens and their ability to invade and overrun the host’s immune responses. Moreover, bacteria can also respond to their environment and gauge whether it is more beneficial to reside in the biofilm or join the planktonic population (Kostakioti et al., 2013). Through the development of these biofilms, resilience against antimicrobial agents increases, making pathogens difficult to eliminate. Therefore, advances in understanding the structure and molecular mechanisms of biofilm organization are critical in the fight against these deadly pathogens (Magana et al., 2018).
The EPS of biofilms is composed of proteins, exopolysaccharides, extracellular DNA (eDNA), RNA, water, secondary metabolites (e.g., pyocyanin [PYO], rhamnolipids), and, in some cases, iron-scavenging siderophores (Harrison & Buckling, 2009; Taylor et al., 2017; Wood et al., 2018). These components assemble into a rigid matrix that protects the encased bacterial cells (Figure 1). Biofilm matrix proteins help cells attach to surfaces, stabilize the biofilm, and modulate the structure of the matrix (Fong & Yildiz, 2015). Exopolysaccharides are natural polymers composed of biomacromolecules that can have positive or negative charges. Exopolysaccharides like poly N-acetylglucosamine and alginate are crucial to intercellular adhesion, structural support, and protection from the environment (Boyd & Chakrabarty, 1995; Jennings et al., 2015; M. H. Lin et al., 2015). Some exopolysaccharides are pathogen specific. Pseudomonas aeruginosa can encode exopolysaccharides, Pel and Psl, in their biofilms, which are important in cell–cell and cell–surface interactions (Jennings et al., 2015). eDNA are extracellular nucleic acid biopolymers critical to the integrity of the biofilm matrix by stabilizing charges, providing structural rigidity, and protecting the matrix from host defense responses. Lastly, the secondary metabolites and RNA found in the biofilm matrix are linked to nutrient maintenance, housekeeping, gene expression and biofilm fitness (Das et al., 2013; Taylor et al., 2017).
FIGURE 1.
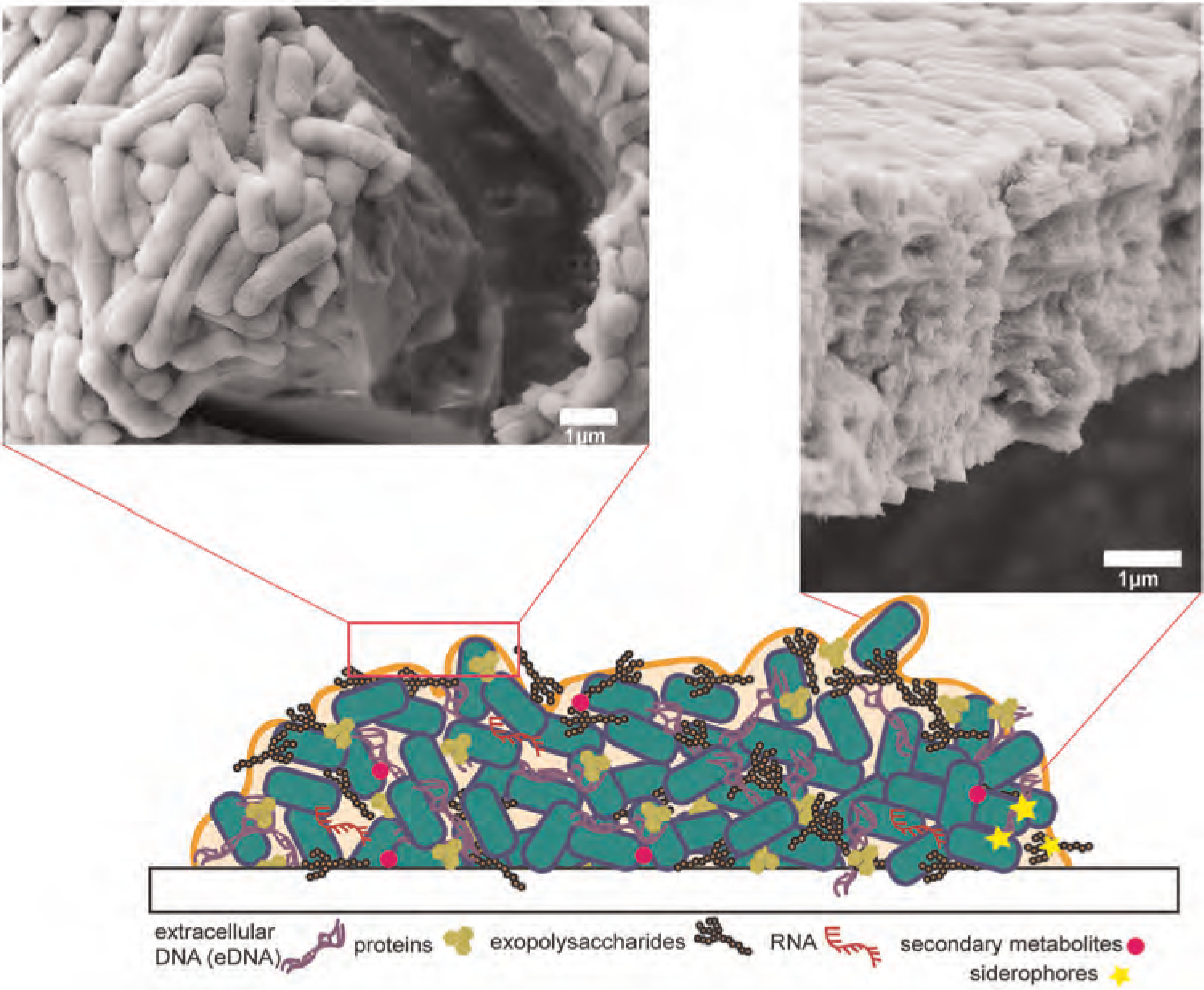
Illustration of a biofilm secreted by bacterial cells (green) showing proteins, exopolysaccharides, RNA, secondary metabolites, siderophores, and eDNA in the matrix. Scanning electron microscopy images of Escherichia coli biofilms show the high density of cells imbedded in EPS and the abundance of bacterial cell layers within the biofilm. eDNA, extracellular DNA; EPS, extracellular polymeric substance
Recently, the importance of eDNA in biofilm formation, cell-to-cell adhesion, cell signaling, and maintaining the structural stability of the biofilm matrix was better recognized. Its significance was overlooked until 2002, when Whitchurch et al. (2002) used DNAse I to disrupt biofilms. Disrupting biofilms allowed antibiotics to reach the cells, which resulted in enhanced bactericidal efficiency. Since then, eDNA has become the focus of many research efforts. In a review by Okshevsky et al. (2015), the importance of eDNA in biofilms was highlighted along with detailed descriptions of its production, adhesion, and role in microbial biofilms. In addition, the review suggested that researchers should examine strategies that destabilize the interactions between eDNA and other biomolecules in the biofilm matrix to disrupt biofilms. Over the last 5 years, additional work has been done to investigate and target eDNA interactions in biofilms. In this review, we will include recent developments, emphasizing potential routes that can be employed to disrupt biofilms. We will focus on works that used eDNA as a target and those that elucidated the interactions of eDNA with other biofilm components.
2 |. SCOPE
2.1 |. eDNA in biofilms
2.1.1 |. eDNA versus chromosomal DNA
DNA is a ubiquitous molecule that contains genetic material of living cells. The DNA of a bacterial cell is packaged as a single-looped double-stranded molecule called the genome. This chromosomal DNA, which is usually found in the cell, is also referred to as genomic DNA (gDNA). In addition to gDNA, bacteria also have small circular DNA called plasmids. Plasmids are extrachromosomal stretches of DNA that replicate distinctly from the gDNA. Furthermore, DNA can exist outside the cell; “eDNA” is a term generally used to denote DNA not enclosed in the cell. However, for the purposes of this review, we use the term “eDNA” to indicate genetic material within the biofilm matrix.
gDNA and eDNA may originate from the same cell, but they are not always structurally or compositionally identical. Upon release to the biofilm environment, DNA may or may not undergo structural fragmentations that can alter its function. For example, explosive lysis of P. aeruginosa causes the release of intact (nonfragmented) eDNA (Turnbull et al., 2016). Cell lysis refers to the breaking down of the cell membrane caused by environmental stress or other triggers. Lysis happens when the bacterial cell wall is weakened allowing osmotic pressure to rupture the cell membrane (Shehadul Islam et al., 2017). In a study conducted by Deng et al., it was observed that hyperbiofilm-forming Rugose small colony variants (RSCV) of P. aeruginosa cells release mostly fragmented eDNA. These RSCV isolated from chronic infections exhibit high resistance to antibiotics and are mostly unaffected by DNA enzymatic treatment (DNase I). In this study, it appears that biofilms form more abundantly around fragmented eDNA than around intact eDNA. In addition, when digested gDNA was added on the ATCC strain PAO1, biofilm formation was enhanced. Deng et al. attributed this enhancement to the increased interaction of fragmented eDNA with biofilm matrix proteins. Nevertheless, the specific fragmentation mechanism of eDNA—whether it occurs inside the cell or outside the cell within the biofilm matrix—still remains unclear and requires further investigation (Deng et al., 2020). Similarly, in Acinetobacter sp., eDNA originates from gDNA, but the two types of eDNA are not structurally identical (Wu & Xi, 2009). Using random amplification of polymorphic DNA (RAPD) analysis, gDNA and eDNA extracted enzymatically were compared. This analysis revealed different patterns of DNA bands between the two yet having identical genomic sequences. This further suggests that gDNA is released through cell lysis but may undergo physical/chemical changes. These eDNA structural differences may be caused by environmental stresses or other interactions in the biofilm that can degrade the nucleic acids. For compositional similarity, chemical analysis of P. aeruginosa biofilms revealed no distinction between eDNA and gDNA (Allesen-Holm et al., 2006) but when comparing eDNA and gDNA of multispecies biofilms, differences in composition were observed (Steinberger & Holden, 2005). Overall, these studies suggest that, although eDNA and gDNA show structural differences, their compositional similarities (primary sequences) indicate that eDNA likely originates from the bacteria themselves.
2.1.2 |. eDNA in Gram-positive bacteria
eDNA is important for Gram-positive Staphylococcus biofilms. Most, if not all, strains of Staphylococcus aureus have eDNA in their biofilms with varying levels depending on culture conditions (Sugimoto et al., 2018). Similarly, in a closely related species, Staphylococcus epidermidis, eDNA is also important in biofilm-formation. In a review by Montanaro et al. (2011), comparison of two clinical isolates of S. epidermidis from implant infections showed that a biofilm-forming strain (RP62A) produce more eDNA relative to the weak-biofilm-forming one (ATCC 12228) after 72 h. Comparing these two Staphylococcal species, it was reported that more eDNA was produced in S. epidermidis biofilms after 24 h in comparison to S. aureus (Zatorska et al., 2017). Further comparison of the two species shows that there is a time-based differential production of eDNA between them. The difference in eDNA production is due to the mechanism of eDNA release of each species—S. aureus bacterium releases eDNA through cell lysis, whereas S. epidermidis bacterium uses an autolysin protein (AtlE) for eDNA release (Dusane, 2017; Zatorska et al., 2017). This means that eDNA in S. epidermidis biofilms is released earlier compared to S. aureus biofilms. The early release of eDNA may cause the difference in observed eDNA concentrations.
eDNA is also critical for other Gram-positive bacteria. In addition to the Staphylococcus genus, the biofilms of another Gram-positive bacterium, Bacillus licheniformis, are composed mainly of polymers and eDNA (Randrianjatovo-Gbalou et al., 2017). Biofilms of Listeria monocytogenes also contained eDNA, with higher concentrations observed in diluted media compared to nutrient-rich media. Cell lysis and subsequent eDNA release are triggered by low-nutrient concentrations of media that create low ionic strength environments (Zetzmann et al., 2015). Pathogens likely release eDNA to help scavenge nutrients to survive. Similar importance of eDNA was also noted for Streptococcus intermedius (Petersen et al., 2004), Streptococcus mutans (Petersen et al., 2005), and Clostridioides difficile (Slater et al., 2019) biofilms.
2.1.3 |. eDNA in Gram-negative bacteria
Similar to Gram-positive bacteria, eDNA is also important for Gram-negatives. The importance of eDNA was noticed by Whitchurch et al. (2002), who discovered that eDNA is an essential component of the Gram-negative, P. aeruginosa biofilms. This finding was further confirmed after identifying eDNA as an abundant component of the flow-grown EPS of P. aeruginosa biofilms (Matsukawa & Greenberg, 2004). Additionally, the amount of eDNA was positively correlated with the biomass thickness of Burkholderia pseudomallei biofilms, regardless of the ratio of living and dead bacterial cells within the biofilms. This finding suggests that it is not only through cell death/lysis that eDNA is released (Pakkulnan et al., 2019). Uropathogenic Escherichia coli (UPEC) also incorporate eDNA in their biofilms, of which bacterial DNA-binding protein (DNABII) are also critical components (Devaraj et al., 2015).
2.1.4 |. eDNA production and release
Biofilm eDNA is secreted in various ways (Figure 2), and different microorganisms have different modes of releasing eDNA. Most eDNA production mechanisms in bacterial organisms is lysis-related (Ibáñez de Aldecoa et al., 2017). Lysis is the most obvious route of releasing DNA since the bacterial genome is often stored within the cytoplasm. Furthermore, a bacterial cell population can control this lytic process. Autolysis is a self-mediated destruction of the cell by its own enzymes (i.e., lysosomal enzymes like gelatinase, serine protease, autolysins), which can be beneficial to the overall population of surviving species (Lackie, 2013; V. C. Thomas et al., 2008). In addition to lysis, eDNA can also be released into the biofilm matrix via membrane vesicles (Sahu et al., 2012). This phenomenon is not surprising since bacteria usually use membrane vesicles to transport macromolecules (i.e., DNA, RNA, proteins, etc.) (Toyofuku et al., 2019). In addition, prophage-mediated eDNA release was reported where lysogenic phages caused lysis of planktonic Streptococcus pneumoniae releasing more eDNA into the biofilm matrix (Carrolo et al., 2010). In this study, about sixfold increase in eDNA was observed in strains carrying prophages and phage lysins. Spontaneous phage induction contribute greatly to the abundance and localization of eDNA in these biofilms (Carrolo et al., 2010). Similarly, autolysis does not seem to occur for Bacillus subtilis. Early competence genes are found to regulate eDNA release for undomesticated B. subtilis cells, and eDNA production is controlled by the ComX-ComP-ComA system (Zafra et al., 2012). In the same study, B. subtilis cells were transformed by eDNA suggesting the function of eDNA in horizontal gene transfer and social behavior. In addition to these mechanisms, many unknown pathways of eDNA production still need to be identified.
FIGURE 2.
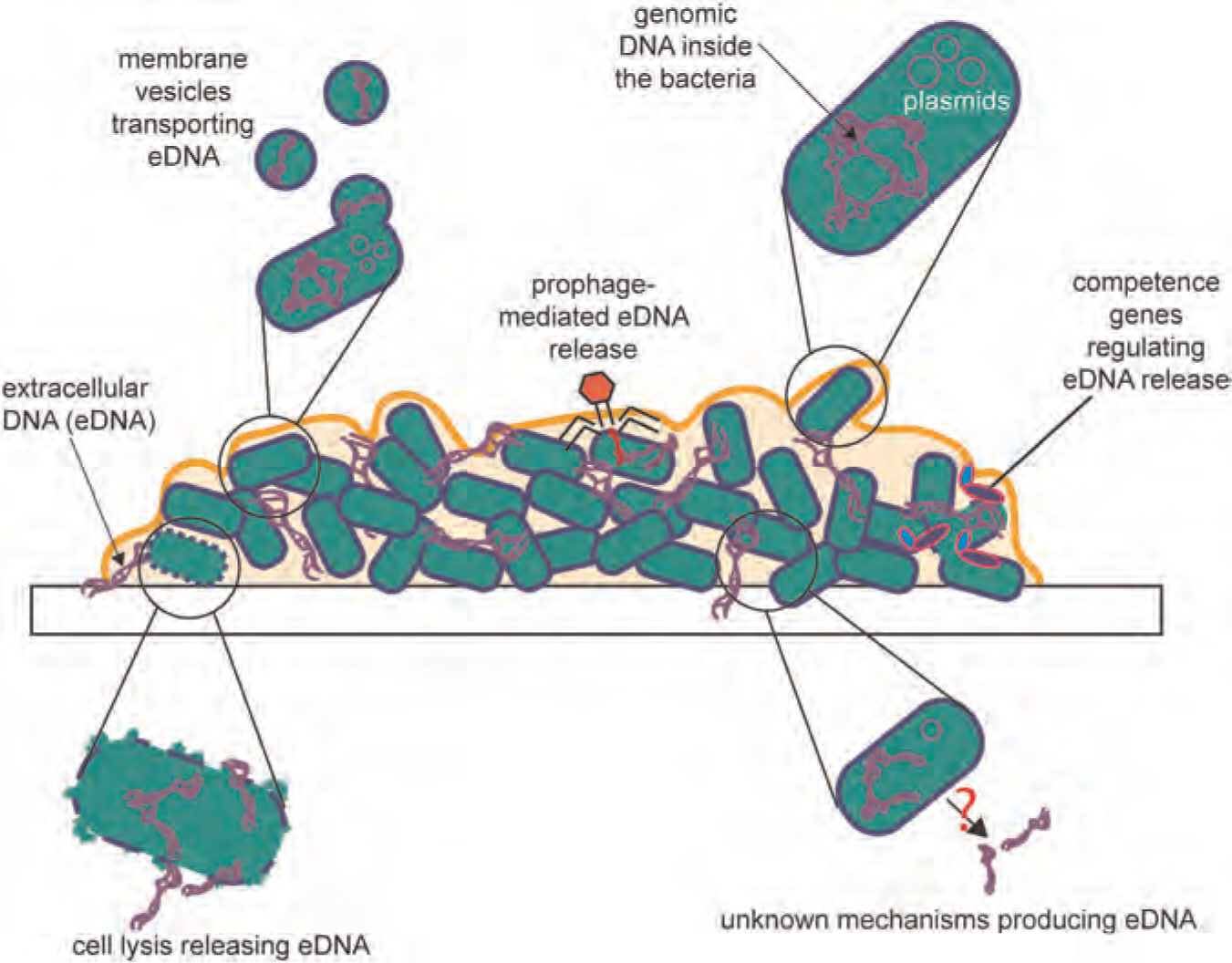
Illustration of eDNA in biofilms of bacterial cells (green) and how it is secreted into the biofilm matrix. Exopolysaccharides and other biofilm components not shown for simplicity. eDNA, extracellular DNA
For many bacterial species, autolytic eDNA release is reported to be facilitated by QS. QS regulates bacterial communication within the biofilm, which relies on signaling molecules that are detected by the surrounding bacterial population (Rutherford & Bassler, 2012). For S. epidermidis, QS-dependent eDNA secretion is mediated by autolysins (Qin et al., 2007) and for E. faecalis, by both gelatinase (GelE) and serine protease (SprE) (V. C. Thomas et al., 2008). Large amounts of eDNA were also observed to be released in P. aeruginosa biofilms through a QS-dependent mechanisms involving N-acyl-l-homoserine lactones and the Pseudomonas quinolone signal in planktonic cell cultures (Allesen-Holm et al., 2006). In a recent study, it was reported that 4-hydroxy-2-alkylquinolines (HAQs) molecules and PrrF small noncoding RNAs both play important roles in signaling eDNA release within P. aeruginosa biofilms (Tahrioui et al., 2019). According to their findings, sub-MIC (below the minimum inhibitory concentration) exposure to tobramycin, led to increased levels of HAQs in P. aeruginosa H103 strain, and the ΔprrF mutant released less eDNA compared to the wild type strain. One of the HAQ’s observed in this study is 2-n-heptyl-4-hydroxyquinoline-N-oxide (HQNO), a P. aeruginosa QS-regulated molecule, that disrupts the flow of electrons through the respiratory chain (Hazan et al., 2016). HQNO induces the production of reactive oxygen species (ROS), which results to cell membrane damage and consequent eDNA release (Tahrioui et al., 2019). QS also regulates the production of phenazines in P. aeruginosa species. Phenazines are nitrogen-containing heterocyclic compounds produced by bacteria with a wide array of biological functions (e.g., electron shuttles, cell signaling molecules, etc.) (Pierson & Pierson, 2010). One phenazine to take note of is PYO, which is activated by the LasR/LasI and RhlR/RhlI QS systems (Pierson & Pierson, 2010). PYO is reported to promote P. aeruginosa biofilm formation by also producing ROS such as H2O2, which results to cell lysis and subsequent eDNA release (T. Das & Manefield, 2012). Lastly, for Pseudomonas putida biofilms, recent findings suggest that eDNA release is induced under Cu2+ stress. The amount of eDNA produced in the biofilm is increased significantly, which is attributed to increase copper resistance (H. Lin et al., 2018). This response shows social behavior and the ability of the bacterial population to detect and react to its environment.
It is widely accepted eDNA originates from chromosomal DNA but it is also proposed that, for infectious biofilms that develop in vivo, eDNA also comes from the host (Chiang et al., 2013; Ikuma et al., 2013; Walker et al., 2005). For example, human neutrophils were shown to enhance the formation of P. aeruginosa biofilms by (a) decreasing the number of viable bacteria in the first 4 h of incubation, which increases nutrient availability for the surviving more-fit bacteria, and (b) providing actin and DNA to the biofilm matrix, which enhances polymerization (Walker et al., 2005). On another hand, it was recently observed that P. aeruginosa grown in murine implants formed biofilms with eDNA only localizing outside the matrix rather than inside the biofilm (Alhede et al., 2020). This observation greatly challenges many of the in vitro studies mentioned in this review that show localization of eDNA within the matrix. In this study, polymorphonuclear leukocytes (PMNs), a type of immune cells, showed cell membraned damage due to exposure to the infectious bacteria (Alhede et al., 2020). Based on their observations, the authors suggest that PMN-derived (or host-derived) DNA is not incorporated into the biofilms in vivo. Instead, the eDNA from necrotic PMN of the host aggregates outside the biofilms. Further investigations and comparisons still need to address this gap in our knowledge. It could be that DNA of PMNs inhibit eDNA release or simply that eDNA is less detectable than PMN DNA.
Identifying specific eDNA release mechanisms of pathogens is important in understanding its role in the biofilms. For example, the production of eDNA is different between the two Staphylococcal species, S. aureus and S. epidermidis. It was previously reported that eDNA is a major structural component of mature S. aureus biofilms but only a minor component for S. epidermidis. In the same study, DNAse I treatment inhibited biofilm formation of both Staphylococcal strains but detached only the 24-h old pre-formed S. aureus biofilms and not the 24-h old pre-formed S. epidermidis biofilms (Izano et al., 2008). The observed results resulted to an assumption that eDNA is not a substantial component of mature S. epidermidis biofilms. However, in another study, Qin et al. (2007) dispersed about half of the 6-h old pre-formed S. epidermidis biofilms while also using DNAse I. Qin et al. also gave further evidence that the eDNA release in S. epidermidis is mainly mediated by the autolysin protein, AtlE. As discussed previously, eDNA is released earlier in S. epidermidis; therefore, dispersal of its young biofilms with DNAse treatments is more effective. Moreover, this finding emphasizes the significance of understanding eDNA release mechanisms in applying anti-biofilm treatments.
Investigations were also conducted to understand if increased eDNA production meant abundant biofilm formation. Tang et al. (2013) found no correlation between the two, but they still proposed that eDNA is important to the biofilms even in low concentrations. Regardless of the quantity, the importance of eDNA in the initial adhesion and biofilm formation is observed throughout various bacterial genera.
2.1.5 |. Known functions of eDNA
eDNA aids in the structural integrity of the biofilm matrix, as wells as bacterial adhesion, metal chelation, metabolic vitality, antibiotic resistance, and horizontal gene transfer (Figure 3). It is critical in bacterial attachment and aggregation of biofilms on surfaces especially in the early stages of biofilm formation (Das et al., 2010; M. Okshevsky & Meyer, 2015). It is hypothesized that eDNA increases the hydrophobicity of the cell envelope, which allows the cells to adhere more easily to surfaces (M. Okshevsky & Meyer, 2015). Other than serving as a biofilm adhesive, eDNA was also observed to promote biofilm dispersion by preventing swarmer motile cells to settle in the existing biofilm (Berne et al., 2010). When eDNA interacts with the “sticky” surface-binding regions of the cells, dispersal of bacterial population become possible, and allows them to form bacterial population to form new biofilms elsewhere. Another function of eDNA is facilitating phenazine (i.e., PYO) retention and redox recycling for extracellular electron transfer (EET) in the biofilm (Saunders et al., 2020). This finding highlights the function of eDNA in the overall biofilm fitness. EET and redox cycling allow metabolic vitality to bacterial populations within the biofilm that lack access to electron acceptors and donors (Saunders et al., 2020). Additionally, eDNA can chelate cations activating resistance mechanisms in P. aeruginosa strains (Johnson et al., 2013). By chelating divalent metal ions that stabilize the LPS layer of Gram-negative bacteria, LPS modifications are triggered thereby “hiding” the bacteria from host defense systems and antibiotics (Mulcahy et al., 2008). Furthermore, eDNA contributes to antibiotic resistance by directly inactivating cationic antibiotics (Chiang et al., 2013; Saxena et al., 2019). Besides physiochemical interactions, eDNA also participate in horizontal gene transfer, which is the nonsexual transfer of genetic information between genomes (Keeling & Palmer, 2008; Zafra et al., 2012). This process is important in the virulence and the evolutionary fate of the bacterial population.
FIGURE 3.
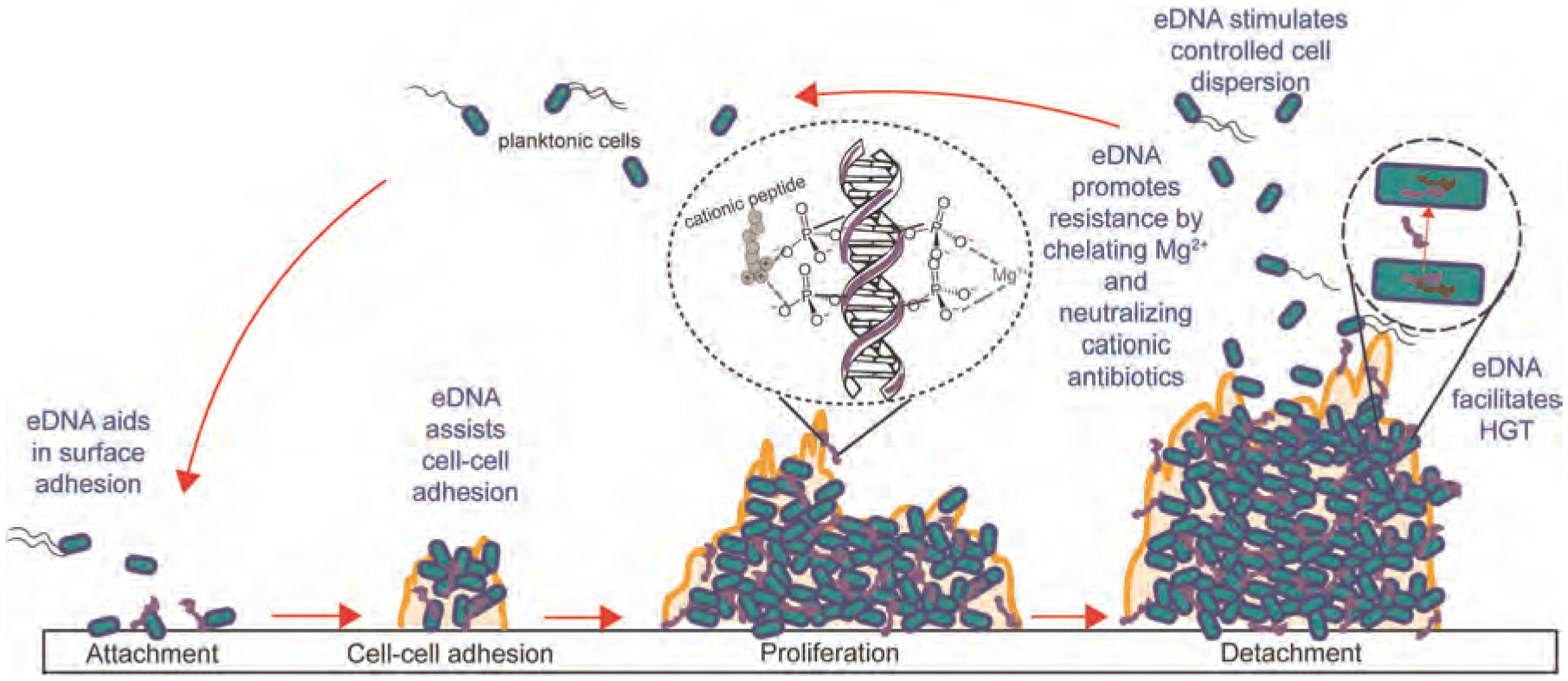
Life cycle of biofilms depicting functions of eDNA in the attachment, cell-cell adhesion, biofilm proliferation, detachment/dispersion, antibiotic resistance, and HGT. eDNA, extracellular DNA; HGT, horizontal gene transfer
2.1.6 |. Structural and chemical modifications of DNA
DNA, in general, can exist in different structural conformations: A-DNA, B-DNA, and Z-DNA forms (Ussery, 2001). The most common form in most living cells is B-DNA. To the best of our knowledge, B- and Z-forms are the only conformations directly found in functional living systems. Increasing evidence suggest that non-B conformations result to a number of diseases (Bacolla & Wells, 2004). The changes in conformation (e.g., from B to non-B) are induced by a variety of conditions (i.e., hydration level, chemical modifications of bases, presence of metal ions, and polyamine interactions) (Basu et al., 1988). Polyamines like spermine and spermidine can cause a B- to Z-DNA transition in the presence of low ionic strength buffers (T. J. Thomas & Messner, 1986). Moreover, eDNA within the biofilms may also undergo change in structural conformations (Rosenberg et al., 2019), suggesting that biofilms are dynamic worlds that continue to respond to their environments.
Changes in chemical and structural conformations are important to understand because they dictate the interactions between eDNA and biofilm constituents. As far as we know, conformational changes of eDNA within the biofilms have not been directly observed. However, it is very possible since the behaviors of eDNA are dynamic within the biofilm; for example, eDNA can chemically modulate its interactions to regulate its contribution to the overall viscoelastic relaxation of the biofilm (Peterson et al., 2013). It was also observed that the composition of eDNA was altered when Cu2+ stress was applied. Using RAPD analysis, Lin et al. observed smaller- and medium-sized fragments of eDNA compared to gDNA; the smaller sizes are possibly due to eDNA fragmentation or nucleotide deletion in the biofilm matrix. In addition, they also found some banding patterns specific to the eDNA samples subjected to Cu2+ stress (H. Lin et al., 2018). These new bands suggest possible nucleotide additions, deletions, or modifications when eDNA is exposed to Cu2+. This observation shows how eDNA aids in biofilm response and adaptation. These interactions are crucial when considering anti-biofilm therapies.
While DNase treatments are widely used for anti-biofilm therapies, this approach still faces limitations. For example, DNase will not likely work for DNA conformations other than B-DNA. DNase treatments also are not likely to work against DNA in the Z-form (Ramesh & Brahmachari, 1989). However, some groups still suggest that any type of DNase can disperse biofilms (Okshevsky et al., 2015). Although DNase may not likely work with Z-form DNA, it is possible that eDNA can flip between Z, B, and other forms. The eDNA digestion via DNase may also affect the conformation of the remaining Z-DNA rendering the Z-DNA susceptible to enzymatic degradation. Nevertheless, DNase is still currently expensive so translating this treatment into large-scale settings will be costly. Furthermore, rising cases of biofilm resistance against DNase are already being reported (Blesa & Berenguer, 2015).
2.2 |. eDNA interactions with biofilm components
The biofilm matrix is composed of various components that can interact with eDNA. It may be helpful to distinguish such interactions into specific and nonspecific. Specific interactions include molecules and macromolecules with high affinity to particular sites or sequences of DNA. Examples of these are various DNA-specific-binding proteins. In contrast, nonspecific DNA interactions involve lower affinity, for example, positively charged molecules that electrostatically interact with the negative phosphate-backbone of DNA.
2.2.1 |. eDNA interactions with proteins
Proteins are complex molecules that serve many cellular purposes; they may function as part of the immune system, catalyze reactions as enzymes, transmit signals as messengers, provide support as structural components, or help supply the cell with molecules required for survival as transporters and storage units (Alberts et al., 2002). In addition to these cellular purposes, proteins also have further functions within biofilms. Cellular proteins (e.g., virulence factors and ribosomal proteins) released into the extracellular environment seem to promote biofilm formation by also binding to eDNA (Graf et al., 2019). Many cellular proteins naturally interact with gDNA; for example condensins and cohesins are protein complexes that tether DNA to produce DNA loops for chromosome assembly (Skibbens, 2019). In the biofilms, eDNA–protein interaction can promote bacterial aggregation by stabilizing anionic eDNA through electrostatic forces and creating a favorable network for biofilm stability (Kavanaugh et al., 2019). When proteins bind eDNA via charge–charge interactions, eDNA can function as an adhesive between the bacteria (Arenas & Tommassen, 2017; Dengler et al., 2015).
Proteins are also important for intracellular bacterial nucleoid structure and function. The DNABII family of proteins bind and pack DNA; these proteins are usually located within the cell, but they also occur in the extracellular matrix of the biofilm. Within this protein family are integration host factor (IHF) and histone-like protein (HU), that bind DNA with high-affinity (Devaraj et al., 2018). Targeting DNABII proteins in biofilms using monoclonal antibodies (MAbs) disrupted diverse bacterial biofilms in vitro and in vivo (Novotny et al., 2016). The two in vivo models used for this finding are nontypeable Haemophilus influenza infection on chinchilla ear and P. aeruginosa infection of murine lung. DNABII proteins and eDNA are both observed to be universal biofilm components (i.e., found across multiple species), and are not only crucial to the biofilm structure but are also important in understanding the mechanisms of multispecies interactions in mixed microbial biofilm populations (Goodman et al., 2011). Consequently, DNABII proteins can serve as anti-biofilm targets. However, little is known about the extracellular localization mechanism of the DNA-binding proteins, thereby demanding more research in this area (Fong & Yildiz, 2015). Other than DNABII-type proteins, amyloid accumulation can also promote biofilm stability. Amyloids are aggregates of proteins usually associated with protein misfolding and many neurodegenerative disorders (Schwartz & Boles, 2013). Amyloids can provide additional support to the biofilm matrix through their inherent resistance to protease degradation thereby protecting the matrix from destruction. In S. aureus biofilms grown in peptone media, amyloids comprise of small peptides called phenol soluble modulins (PSMs) (Schwartz et al., 2012). These cationic PSMs interact with eDNA resulting in polymerization and increased amyloid aggregation. Furthermore, biofilms that have PSMs but lack eDNA, fail to assemble PSM amyloids (Schwartz et al., 2016). Collectively, these studies suggest that protein–eDNA interactions can serve as worthy therapeutic targets to disrupt biofilms.
2.2.2 |. eDNA interactions with exopolysaccharides
Exopolysaccharides are high-molecular weight (10–1000 kDa) carbohydrate polymers secreted by microorganisms in the biofilms. Functions of exopolysaccharides in biofilms include cellular adhesion, stress protection, water retention, and absorption of excess energy (Nwodo et al., 2012). In a recent study, the interaction between the exopolysaccharide, Psl, and eDNA was observed in P. aeruginosa biofilms; the two components crosslink to form a skeleton that allows bacteria to adhere and grow (Wang et al., 2015). Using molecular modeling, the authors were able to mimic the physical interaction between Psl and DNA. Psl was able to fit into the minor groove of DNA double helix forming hydrogen bonds with an estimated −2.18 kcal/mol interaction energy. Additionally, in Caulobacter crescentus biofilms, eDNA can bind to polar polysaccharide “hold-fast” regions of cells, which are necessary for surface adhesion, thereby promoting biofilm dispersion (Berne et al., 2010). It is critical to note here that controlled biofilm dispersion is an essential stage in the biofilm life cycle since it contributes to bacterial dispersal and disease transmission (Kaplan, 2010). In summary, eDNA-interactions with biofilm exopolysaccharides appear essential to bacterial aggregation, adhesion, dispersion, and vary in detail from species to species.
2.2.3 |. eDNA interactions with other biofilm components
Within the biofilms, interacting with eDNA, are various metabolites and cations crucial for bacterial survival and biofilm development (Bellin et al., 2016). For example, PYO is a redox-active metabolite that provides P. aeruginosa a blue-green color. PYO is a phenazine that has a number of significant roles in cell-signaling, biofilm development, iron-acquisition, cell metabolism, and antibiotic tolerance (Schiessl et al., 2019; Zhu et al., 2019). In P. aeruginosa biofilms, PYO interacts with eDNA resulting to: enhanced electron transfer within the biofilm components, better eDNA binding to P. aeruginosa cell surface, and increased DNA and sputum viscosity (Das et al., 2013, 2015). Since PYO–eDNA interactions are vital to the biofilm development, the disruption of such interactions can present a favorable target for biofilm biomass reduction and decreased biofilm exudates in infections. Das et al. (2015) therefore suggested the potential of elevating antioxidant levels (e.g., glutathione and ascorbic acid) to disrupt PYO-eDNA intercalation in P. aeruginosa biofilms.
Besides metabolites, cations in the biofilms also interact with eDNA. eDNA can sequester divalent cations and trigger resistance mechanisms that increase the pathogen’s virulence (Johnson et al., 2013; Mulcahy et al., 2008). Cation limitation destabilizes the negative charges of the lipopolysaccharides on the outer membrane of Gram-negative bacteria, which can cause cell lysis. Consequently, this event triggers a virulence response. Therefore, subinhibitory concentrations of eDNA can induce increased antibiotic resistance. Additionally, in a study investigating heavy metal stress and eDNA formation, it was identified that extracellular nucleic acids in the EPS of unsaturated P. putida CZ1 biofilm also bind Cu2+. The phosphate groups of DNA have a cation-binding ability that allows biofilms to store cation and nutrient reservoirs for the cells. The increase of eDNA contents under Cu2+ stress may also be attributed to expression of resistance mechanisms (H. Lin et al., 2018). Using a different approach, external addition of metal chelators (e.g., EDTA) was used to disperse PAO1 biofilms (Banin et al., 2006). Exposing P. aeruginosa biofilms to EDTA induced bacterial cell detachment from biofilms likely caused by chelation of several divalent ions required to stabilize the biofilm matrix. It is, however, important to note that high concentrations of chelators are needed for treatments to be effective; subinhibitory concentrations can only trigger resistance mechanisms. Hence, cytotoxicity becomes a challenge for this approach.
2.3 |. eDNA as anti-biofilm target
eDNA may serve as an effective anti-biofilm target. Research has focused on DNase treatments ever since DNase was used to degrade eDNA and disrupt biofilms (Matsukawa & Greenberg, 2004; Okshevsky et al., 2015; Whitchurch et al., 2002). By applying nucleases directly into biofilms, eDNA is degraded and the biofilm matrix is weakened. Likewise, disturbing eDNA interactions can threaten the matrix. It is important to realize that even if the biofilm is not completely disrupted, access points for other treatments can still be created. Access points and disturbances in the biofilm matrix can ultimately increase antibiotic susceptibility of the pathogens living within.
2.3.1 |. Nuclease treatments
Nucleases are enzymes that degrade nucleic acids. The most common anti-biofilm treatment in vitro that uses nucleases is DNase I, a DNA specific endonuclease (Eun, 1996). DNase I cleaves single- and double-stranded DNA into oligonucleotides with 5′-phosphorylated and 3′-hydroxylated ends (Hartmann, 2017). Okshevsky et al. (2015) reported a comprehensive study of various bacterial species where DNase treatments were implemented for biofilm control. Overall, it is noted that DNase treatments may prevent biofilm formation, but they do not always disrupt pre-formed biofilms. The growth phase of the biofilm dictates the effectiveness of this therapy; only younger biofilms (6-h old or less) are disrupted. This therapeutic approach is also applied in clinical settings; cystic fibrosis (CF) patients are commonly treated with Dornase alfa (pulmozyme, recombinant human DNAse I, rhDNAse) (Wagener & Kupfer, 2012). CF patients accumulate sputum in their lungs, thereby reducing pulmonary function. The accumulated sputum for these patients is rich with actin and eDNA; hence, treatment with dornase alfa results to degradation of eDNA and reduced sputum viscosity (Wagener & Kupfer, 2012). This therapeutic strategy can also be applied in bacterial infections by cleaving eDNA in the biofilms and creating access points for coadministered antibiotic treatments.
Pathogens like S. aureus can also produce extracellular nucleases. They release nucleases to signal controlled biofilm dispersion and also to defend themselves from neutrophil extracellular traps (NETs). These NETs are composed of the host’s nuclear DNA associated with antimicrobial peptides, histones and proteases (Berends et al., 2010); they are released by the host’s neutrophils to kill bacteria. Furthermore, as pathogens fight for survival, surface colonization and nutrient availability become very important. Some bacteria, therefore, release nucleases to prevent biofilm formation of other pathogens. For example, nucleases released by B. licheniformis have anti-biofilm properties (Nijland et al., 2010). Hence, identifying more extracellular nucleases secreted by biofilm-forming pathogens may have future therapeutic use.
Alternatively, Okshevsky et al. suggested the upregulation of the nuclease production of a singular species in a multicolony biofilm for biofilm control. However, since nucleases serve a variety of purposes (e.g., transformation, bacterial dispersal, protection against NETs) that can increase biofilm production, this approach still needs to be investigated (Okshevsky et al., 2015). We still do not know if upregulating nucleases would be beneficial or destructive to the overall biofilm life cycle. Furthermore, few studies so far have used nuclease treatments to target biofilms in vivo (Koo et al., 2017).
2.3.2 |. eDNA and eDNA-interactions as targets for anti-biofilm therapy
We have detailed various eDNA interactions within the biofilm matrix in this review (e.g., proteins, exopolysaccharides, metabolites, etc.) but it is essential to find and investigate more. Targeting eDNA interactions can cause biofilm matrix disturbances that can lead to eradication of a bacterial colony. However, many of these interactions still need to be investigated as anti-biofilm targets. For example, although eDNA-protein complexes are found in B. licheniformis biofilms, only the DNase treatment increased biofilm permeability; the proteinase treatment (proteinase K) did not (Randrianjatovo-Gbalou et al., 2017). In this case, the proteinaceous components of the biofilm were digested by Proteinase K but the overall permeability of the biofilm matrix was largely unaffected. It could be that targeting eDNA alone increases permeability of the matrix and targeting the matrix proteins structurally weakens the framework. This explanation support findings that report DNABII targeting can collapse diverse bacterial biofilms both in vitro and in vivo (Novotny et al., 2016). Here, highly specific MAbs against protective epitopes of a DNABII protein disrupted pre-formed biofilms and released the encapsulated bacterial population, rendering them less resistant. It is also important to note that DNABII proteins are positioned at the vertices of strands of eDNA within the biofilm so targeting them induced a collapse of the whole matrix (Novotny et al., 2016). By applying this strategy, both proteins and eDNA (and their interactions) are compromised, disrupting the overall structural integrity of the biofilm. This approach can be very effective in disrupting and even preventing biofilm formation. With this strategy in mind, there may possibly be more lynchpin proteins like DNABII that contribute significantly to the biofilm matrix. Therefore, we need to identify more eDNA-interacting proteins that can be targeted.
Similarly, exopolysaccharides and metabolites may serve as anti-biofilm targets. The Psl polysaccharide interacts with eDNA to benefit the biofilm in many ways: to form a backbone skeleton support for the matrix, to encourage bacteria to produce more EPS, and to inhibit over-cation chelation that can lyse the cells within the biofilm (Wang et al., 2015). Like the strategy for eDNA-DNABII proteins, targeting eDNA–exopolysaccharide interactions may also collapse the biofilm structure. However, since polysaccharides are also ubiquitous in the biofilms of various species, targeting them directly has been the goal of different studies (Itoh et al., 2005; Izano et al., 2008). Most research would rather focus on direct polysaccharide-degradation than identifying interactions, which work as indirect anti-biofilm targets. In addition, metabolites and cations in the biofilm can dynamically affect the biofilm matrix. For example, eDNA can increase antibiotic resistance by sequestering Mg2+ ions and decreasing the pH of the biofilm. For P. aeruginosa, biofilms pH values range from pH 5.5–6.6 (Wilton et al., 2016). Such acidic conditions can upregulate two-component systems, which are related to antibiotic resistance. Identifying more metabolites and cations may introduce more anti-biofilm targets.
Biofilms create a barrier that is very difficult for antibiotics to pass through and access the bacterial cells. Antibiotic-conjugated molecules that inhibit the eDNA–biofilm component interactions may be utilized for anti-biofilm therapies. By attaching inhibitors to antibiotics, the biofilm can be disrupted, and antibiotic delivery becomes more efficient. Conjugating antibiotics to polymers, proteins, antibodies, and other molecules with high affinity to eDNA can directly interrupt and disturb the biofilm matrix. Hence, understanding the structural differences that occur with gDNA once secreted and becomes eDNA is vital. Once the matrix is dissolved, antibiotics delivered nearby can attack the pathogens.
The concept of antibiotic-conjugation has already been previously reported. For example, antibody-antibiotic conjugates are being used against S. aureus. Human MAbs designed to bind on the surface of S. aureus bacteria are conjugated with an antibiotic (dmDNA31), thereby directing the drug closer to the target pathogen (Zhou et al., 2016). In another study, chitosan, a derivative of the polysaccharide chitin, was linked to an aminoglycoside antibiotic (streptomycin) (Zhang et al., 2013). This chitosan-antibiotic conjugate reduced biofilm mass and suppressed biofilm formation of Gram-positive bacteria but not Gram-negative bacteria. Chitosan allowed streptomycin access into the biofilms secreted by Listeria monocytogenes (Zhang et al., 2013).
3 |. MAJOR CONCLUSIONS
Biofilms create barriers that challenge our current therapeutic approaches. eDNA may function as a worthwhile anti-biofilm target. In this review, we highlighted recent developments defining the importance, production mechanisms, and possible conformations of eDNA in biofilms. We described components in the matrix that interact with eDNA noting their potential as anti-biofilm targets. Overall, these various interactions, whether known or unknown, are very important in biofilm regulation. Therefore, we need to continue pursuing creative approaches in eradicating biofilms.
ACKNOWLEDGMENTS
We wish to express our warmest gratitude to Dr. Rikke Louise Meyer from the Interdisciplinary Nanoscience Center and Department of Bioscience, Aarhus University, Denmark for her insight and thoughtful suggestions regarding the content and organization of this manuscript. We also acknowledge the helpful contributions of Dr. Anh K. Lam (University of Oklahoma) during manuscript preparation. Finally, we acknowledge the contributions of Laura Gatch, Miranda Vesy, Mehrnaz Afkhami, Adwaita Parab, and Prof. Mariëlle H. Hoefnagels throughout the writing process. Funding was provided by the National Institutes of Health (C.V.R., R03AI142420-01 and The University of Oklahoma.
Funding information
National Institute of Allergy and Infectious Diseases, Grant/Award Number: R03AI142420-01
REFERENCES
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, & Walter P (2002). Protein function, Molecular biology of the cell (4th ed.). Garland Science. [Google Scholar]
- Alhede M, Alhede M, Qvortrup K, Kragh KN, Jensen PØ, Stewart PS, & Bjarnsholt T (2020). The origin of extracellular DNA in bacterial biofilm infections in vivo. Pathogens and Disease, 78(2), ftaa018. 10.1093/femspd/ftaa018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, & Tolker-Nielsen T (2006). A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Molecular Microbiology, 59(4), 1114–1128. 10.1111/j.1365-2958.2005.05008.x [DOI] [PubMed] [Google Scholar]
- Annual report of the European Medicines Agency. (2010). https://www.ema.europa.eu/en/documents/annual-report/annual-report-european-medicines-agency-2009_en.pdf
- Arenas J, & Tommassen J (2017). Meningococcal biofilm formation: Let’s stick together. Trends in Microbiology, 25(2), 113–124. 10.1016/j.tim.2016.09.005 [DOI] [PubMed] [Google Scholar]
- Bacolla A, & Wells RD (2004). Non-B DNA conformations, genomic rearrangements, and human disease. Journal of Biological Chemistry, 279(46), 47411–47414. [DOI] [PubMed] [Google Scholar]
- Banin E, Brady KM, & Greenberg EP (2006). Chelator-induced dispersal and killing of Pseudomonas aeruginosa cells in a biofilm. Applied and Environmental Microbiology, 72(3), 2064–2069. 10.1128/AEM.72.3.2064-2069.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu HS, Feuerstein BG, Zarling DA, Shaffer RH, & Marton LJ (1988). Recognition of Z-RNA and Z-DNA determinants by polyamines in solution: Experimental and theoretical studies. [DOI] [PubMed]
- Journal of Biomolecular Structure and Dynamics, 6(2), 299–309. 10.1080/07391102.1988.10507714 [DOI] [PubMed] [Google Scholar]
- Bellin DL, Sakhtah H, Zhang Y, Price-Whelan A, Dietrich LEP, & Shepard KL (2016). Electrochemical camera chip for simultaneous imaging of multiple metabolites in biofilms. Nature Communications, 7(1), 10535. 10.1038/ncomms10535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berends ETM, Horswill AR, Haste NM, Monestier M, Nizet V, & von Köckritz-Blickwede M (2010). Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. Journal of Innate Immunity, 2(6), 576–586. 10.1159/000319909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berne C, Kysela DT, & Brun YV (2010). A bacterial extracellular DNA inhibits settling of motile progeny cells within a biofilm. Molecular Microbiology, 77(4), 815–829. 10.1111/j.1365-2958.2010.07267.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blesa A, & Berenguer J (2015). Contribution of vesicle-protected extracellular DNA to horizontal gene transfer in Thermus spp. International Microbiology, 18(3), 177–187. [DOI] [PubMed] [Google Scholar]
- Boyd A, & Chakrabarty A (1995). Pseudomonas aeruginosa biofilms: Role of the alginate exopolysaccharide. Journal of Industrial Microbiology, 15(3), 162–168. [DOI] [PubMed] [Google Scholar]
- Carrolo M, Frias MJ, Pinto FR, Melo-Cristino J, & Ramirez M (2010). Prophage spontaneous activation promotes DNA release enhancing biofilm formation in Streptococcus pneumoniae. PLOS One, 5(12):e15678. 10.1371/journal.pone.0015678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. (2019). Biggest threats and data. https://www.cdc.gov/drugresistance/biggest-threats.html
- Chiang W-C, Nilsson M, Jensen PØ, Høiby N, Nielsen TE, Givskov M, & Tolker-Nielsen T (2013). Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrobial Agents and Chemotherapy, 57(5), 2352–2361. 10.1128/AAC.00001-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T, Kutty SK, Kumar N, & Manefield M (2013). Pyocyanin facilitates extracellular DNA binding to Pseudomonas aeruginosa influencing cell surface properties and aggregation. PLOS One, 8(3): e58299. 10.1371/journal.pone.0058299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T, Kutty SK, Tavallaie R, Ibugo AI, Panchompoo J, Sehar S, Aldous L, Yeung AWS, Thomas SR, Kumar N, Gooding JJ, & Manefield M (2015). Phenazine virulence factor binding to extracellular DNA is important for Pseudomonas aeruginosa biofilm formation. Scientific Reports, 5, 8398. 10.1038/srep08398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T, & Manefield M (2012). Pyocyanin promotes extracellular DNA release in Pseudomonas aeruginosa. PLOS One, 7(10), e46718. 10.1371/journal.pone.0046718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T, Sharma PK, Busscher HJ, van der Mei HC, & Krom BP (2010). Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Applied and Environmental Microbiology, 76(10), 3405–3408. 10.1128/AEM.03119-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng B, Ghatak S, Sarkar S, Singh K, Das Ghatak P, Mathew-Steiner SS, Roy S, Khanna S, Wozniak DJ, McComb DW, & Sen CK (2020). Novel bacterial diversity and fragmented eDNA identified in hyperbiofilm-forming Pseudomonas aeruginosa rugose small colony variant. iScience, 23(2), 100827. 10.1016/j.isci.2020.100827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengler V, Foulston L, DeFrancesco AS, & Losick R (2015). An electrostatic net model for the role of extracellular dna in biofilm formation by Staphylococcus aureus. Journal of Bacteriology, 197(24), 3779–3787. 10.1128/JB.00726-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj A, Buzzo J, Rocco CJ, Bakaletz LO, & Goodman SD (2018). The DNABII family of proteins is comprised of the only nucleoid associated proteins required for nontypeable Haemophilus influenzae biofilm structure. MicrobiologyOpen, 7(3), e00563. 10.1002/mbo3.563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj A, Justice SS, Bakaletz LO, & Goodman SD (2015). DNABII proteins play a central role in UPEC biofilm structure. Molecular Microbiology, 96(6), 1119–1135. 10.1111/mmi.12994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan RM (2002). Biofilms: Microbial life on surfaces. Emerging Infectious Diseases, 8(9), 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunne WM Jr. (2002). Bacterial adhesion: Seen any good biofilms lately? Clinical Microbiology Reviews, 15(2), 155–166. 10.1128/cmr.15.2.155-166.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusane DH (2017). CORR Insights®: Does extracellular DNA production vary in staphylococcal biofilms isolated from infected implants versus controls? Clinical Orthopaedics and Related Research, 475(8), 2114–2116. 10.1007/s11999-017-5360-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eun H-M (1996). 3—Nucleases. In Eun H-M (Ed.), Enzymology primer for recombinant DNA technology (pp. 145–232). Academic Press. [Google Scholar]
- Fong JN, & Yildiz FH (2015). Biofilm matrix proteins. In Ghannoum M, Parsek M, Whiteley M, & Mukherjee P (Eds.), Microbial biofilms (pp. 201–222). ASM Press. [Google Scholar]
- Goodman S, Obergfell K, Jurcisek J, Novotny LA, Downey JS, Ayala EA, Tjokro N, Li B, Justice SS, & Bakaletz LO (2011). Biofilms can be dispersed by focusing the immune system on a common family of bacterial nucleoid-associated proteins. Mucosal Immunol, 4, 625–637. 10.1038/mi.2011.27 [DOI] [PubMed] [Google Scholar]
- Graf AC, Leonard A, Schäuble M, Rieckmann LM, Hoyer J, Maass S, Lalk M, Becher D, Pané-Farré J, & Riedel K (2019). Virulence factors produced by Staphylococcus aureus biofilms have a moonlighting function contributing to biofilm integrity. Molecular and Cellular Proteomics, 18(6), 1036–1053. 10.1074/mcp.RA118.001120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison F, & Buckling A (2009). Siderophore production and biofilm formation as linked social traits. The ISME Journal, 3(5), 632–634. 10.1038/ismej.2009.9 [DOI] [PubMed] [Google Scholar]
- Hartmann G (2017). Chapter four—Nucleic acid immunity. In Alt FW (Ed.), Advances in immunology (Vol. 133, pp. 121–169). Academic Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazan R, Que YA, Maura D, Strobel B, Majcherczyk PA, Hopper LR, Wilbur DJ, Hreha TN, Barquera B, & Rahme LG (2016). Auto poisoning of the respiratory chain by a quorum-sensing-regulated molecule favors biofilm formation and antibiotic tolerance. Current Biology, 26(2), 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen G, Molly M-P, Suraj P, Sumanth G, Jordan L, Devra B, & Ramanan L (2015). State of the world’s antibiotics. Center for Disease Dynamics, Economics & Policy: Washington, DC. [Google Scholar]
- Howell L (2013). Global risks 2013—Eight edition. Cologny/Geneva, Switzerland: Paper presented at the World Economic Forum. [Google Scholar]
- Ibáñez de Aldecoa AL, Zafra O, & González-Pastor JE (2017). Mechanisms and regulation of extracellular DNA release and its biological roles in microbial communities. Frontiers in Microbiology, 8, 1390. 10.3389/fmicb.2017.01390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikuma K, Decho AW, & Lau BL (2013). The extracellular bastions of bacteria—A biofilm way of life. Nature Education Knowledge, 4(2), 2–19.10. [Google Scholar]
- Itoh Y, Wang X, Hinnebusch BJ, Preston JF 3rd, & Romeo T (2005). Depolymerization of beta-1,6-N-acetyl-D-glucosamine disrupts the integrity of diverse bacterial biofilms. Journal of Bacteriology, 187(1), 382–387. 10.1128/JB.187.1.382-387.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izano EA, Amarante MA, Kher WB, & Kaplan JB (2008). Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Applied and Environmental Microbiology, 74(2), 470–476. 10.1128/aem.02073-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings LK, Storek KM, Ledvina HE, Coulon C, Marmont LS, Sadovskaya I, Secor PR, Tseng BS, Scian M, Filloux A, Wozniak DJ, Howell PL, & Parsek MR (2015). Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proceedings of the National Academy of Sciences of the United States of America, 112(36), 11353–11358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L, Horsman SR, Charron-Mazenod L, Turnbull AL, Mulcahy H, Surette MG, & Lewenza S (2013). Extracellular DNA-induced antimicrobial peptide resistance in Salmonella enterica serovar Typhimurium. BMC Microbiology, 13, 115. 10.1186/1471-2180-13-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan JB (2010). Biofilm dispersal: Mechanisms, clinical implications, and potential therapeutic uses. Journal of Dental Research, 89(3), 205–218. 10.1177/0022034509359403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanaugh JS, Flack CE, Lister J, Ricker EB, Ibberson CB, Jenul C, Moormeier DE, Delmain EA, Bayles KW, & Horswill AR (2019). Identification of extracellular DNA-binding proteins in the biofilm matrix. mBio, 10(3), e01137–19. 10.1128/mBio.01137-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, & Palmer JD (2008). Horizontal gene transfer in eukaryotic evolution. Nature Reviews Genetics, 9(8), 605–618. 10.1038/nrg2386 [DOI] [PubMed] [Google Scholar]
- Koo H, Allan RN, Howlin RP, Stoodley P, & Hall-Stoodley L (2017). Targeting microbial biofilms: Current and prospective therapeutic strategies. Nature Reviews Microbiology, 15(12), 740–755. 10.1038/nrmicro.2017.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostakioti M, Hadjifrangiskou M, & Hultgren SJ (2013). Bacterial biofilms: Development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harbor Perspectives in Medicine, 3(4), a010306. 10.1101/cshperspect.a010306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackie JM (2013). Numerical entries and A. In Lackie JM (Ed.), The dictionary of cell & molecular biology (5th ed., pp. 1–61). Academic Press. [Google Scholar]
- Lin H, Wang C, Zhao H, Chen G, & Chen X (2018). Interaction between copper and extracellular nucleic acids in the EPS of unsaturated Pseudomonas putida CZ1 biofilm. Environmental Science and Pollution Research, 25(24), 24172–24180. [DOI] [PubMed] [Google Scholar]
- Lin MH, Shu JC, Lin LP, Chong KY, Cheng YW, Du JF, & Liu S-T (2015). Elucidating the crucial role of poly N-acetylglucosamine from Staphylococcus aureus in cellular adhesion and pathogenesis. PLOS One, 10(4), e0124216. 10.1371/journal.pone.0124216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magana M, Sereti C, Ioannidis A, Mitchell CA, Ball AR, Magiorkinis E, Chatzipanagiotou S, Hamblin MR, Hadjifrangiskou M, & Tegos GP (2018). Options and limitations in clinical investigation of bacterial biofilms. Clinical Microbiology Reviews, 31(3), e00084–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukawa M, & Greenberg EP (2004). Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. Journal of Bacteriology, 186(14), 4449–4456. 10.1128/jb.186.14.4449-4456.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanaro L, Poggi A, Visai L, Ravaioli S, Campoccia D, Speziale P, & Arciola CR (2011). Extracellular DNA in biofilms. The International Journal of Artificial Organs, 34(9), 824–831. 10.5301/ijao.5000051 [DOI] [PubMed] [Google Scholar]
- Mulcahy H, Charron-Mazenod L, & Lewenza S (2008). Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLOS Pathogens, 4(11), e1000213. 10.1371/journal.ppat.1000213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijland R, Hall MJ, & Burgess JG (2010). Dispersal of biofilms by secreted, matrix degrading, bacterial DNase. PLOS One, 5(12), e15668. 10.1371/journal.pone.0015668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny LA, Jurcisek JA, Goodman SD, & Bakaletz LO (2016). Monoclonal antibodies against DNA-binding tips of DNABII proteins disrupt biofilms in vitro and induce bacterial clearance in vivo. EBioMedicine, 10, 33–44. 10.1016/j.ebiom.2016.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwodo UU, Green E, & Okoh AI (2012). Bacterial exopolysaccharides: Functionality and prospects. International Journal of Molecular Sciences, 13(11), 14002–14015. 10.3390/ijms131114002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okshevsky M, & Meyer RL (2015). The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Critical Reviews in Microbiology, 41(3), 341–352. 10.3109/1040841X.2013.841639 [DOI] [PubMed] [Google Scholar]
- Okshevsky M, Regina VR, & Meyer RL (2015). Extracellular DNA as a target for biofilm control. Current Opinion in Biotechnology, 33, 73–80. 10.1016/j.copbio.2014.12.002 [DOI] [PubMed] [Google Scholar]
- Pakkulnan R, Anutrakunchai C, Kanthawong S, Taweechaisupapong S, Chareonsudjai P, & Chareonsudjai S (2019). Extracellular DNA facilitates bacterial adhesion during Burkholderia pseudomallei biofilm formation. PLOS One, 14(3):e0213288. 10.1371/journal.pone.0213288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen FC, Pecharki D, & Scheie AA (2004). Biofilm mode of growth of Streptococcus intermedius favored by a competence-stimulating signaling peptide. Journal of Bacteriology, 186(18), 6327–6331. 10.1128/JB.186.18.6327-6331.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen FC, Tao L, & Scheie AA (2005). DNA binding-uptake system: A link between cell-to-cell communication and biofilm formation. Journal of Bacteriology, 187(13), 4392–4400. 10.1128/JB.187.13.4392-4400.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson BW, van der Mei HC, Sjollema J, Busscher HJ, & Sharma PK (2013). A distinguishable role of eDNA in the viscoelastic relaxation of biofilms. mBio, 4(5), e00497–13. 10.1128/mBio.00497-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson LS 3rd, & Pierson EA (2010). Metabolism and function of phenazines in bacteria: Impacts on the behavior of bacteria in the environment and biotechnological processes. Applied Microbiology and Biotechnology, 86(6), 1659–1670. 10.1007/s00253-010-2509-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z, Ou Y, Yang L, Zhu Y, Tolker-Nielsen T, Molin S, & Qu D (2007). Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology, 153(7), 2083–2092. 10.1099/mic.0.2007/006031-0 [DOI] [PubMed] [Google Scholar]
- Ramesh N, & Brahmachari SK (1989). Structural alteration from non-B to B-form could reflect DNase I hypersensitivity. Journal of Biomolecular Structure and Dynamics, 6(5), 899–906. [DOI] [PubMed] [Google Scholar]
- Randrianjatovo-Gbalou I, Rouquette P, Lefebvre D, Girbal-Neuhauser E, & Marcato-Romain C-E (2017). In situ analysis of Bacillus licheniformis biofilms: Amyloid-like polymers and eDNA are involved in the adherence and aggregation of the extracellular matrix. Journal of Applied Microbiology, 122(5), 1262–1274. 10.1111/jam.13423 [DOI] [PubMed] [Google Scholar]
- Rosenberg M, Azevedo NF, & Ivask A (2019). Propidium iodide staining underestimates viability of adherent bacterial cells. Scientific Reports, 9(1), 6483. 10.1038/s41598-019-42906-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, & Bassler BL (2012). Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harbor Perspectives in Medicine, 2(11), a012427. 10.1101/cshperspect.a012427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu PK, Iyer PS, Oak AM, Pardesi KR, & Chopade BA (2012). Characterization of eDNA from the clinical strain Acinetobacter baumannii AIIMS 7 and its role in biofilm formation. The Scientific World Journal, 2012, 973436. 10.1100/2012/973436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders SH, Tse ECM, Yates MD, Otero FJ, Trammell SA, Stemp EDA, Barton JK, Tender LM, & Newman DK (2020). Extracellular DNA promotes efficient extracellular electron transfer by pyocyanin in Pseudomonas aeruginosa biofilms. Cell, 182(4), 919–932. 10.1016/j.cell.2020.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena P, Joshi Y, Rawat K, & Bisht R (2019). Biofilms: Architecture, resistance, quorum sensing and control mechanisms. Indian Journal of Microbiology, 59(1), 3–12. 10.1007/s12088-018-0757-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiessl KT, Hu F, Jo J, Nazia SZ, Wang B, Price-Whelan A, Min W, & Dietrich LEP (2019). Phenazine production promotes antibiotic tolerance and metabolic heterogeneity in Pseudomonas aeruginosa biofilms. Nature Communications, 10(1), 762. 10.1038/s41467-019-08733-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz K, & Boles BR (2013). Microbial amyloids—Functions and interactions within the host. Current Opinion in Microbiology, 16(1), 93–99. 10.1016/j.mib.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz K, Ganesan M, Payne DE, Solomon MJ, & Boles BR (2016). Extracellular DNA facilitates the formation of functional amyloids in Staphylococcus aureus biofilms. Molecular Microbiology, 99(1), 123–134. 10.1111/mmi.13219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz K, Syed AK, Stephenson RE, Rickard AH, & Boles BR (2012). Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLOS Pathogens, 8(6), e1002744. 10.1371/journal.ppat.1002744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehadul Islam M, Aryasomayajula A, & Selvaganapathy PR (2017). A review on macroscale and microscale cell lysis methods. Micromachines, 8(3), 83. 10.3390/mi8030083 [DOI] [Google Scholar]
- Silver LL (2011). Challenges of antibacterial discovery. Clinical Microbiology Reviews, 24(1), 71–109. 10.1128/CMR.00030-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens RV (2019). Condensins and cohesins—One of these things is not like the other! Journal of Cell Science, 132(3), jcs220491. 10.1242/jcs.220491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater RT, Frost LR, Jossi SE, Millard AD, & Unnikrishnan M (2019). Clostridioides difficile LuxS mediates inter-bacterial interactions within biofilms. Scientific Reports, 9(1), 9903. 10.1038/s41598-019-46143-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberger R, & Holden P (2005). Extracellular DNA in single-and multiple-species unsaturated biofilms. Applied and Environmental Microbiology, 71(9), 5404–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto S, Sato F, Miyakawa R, Chiba A, Onodera S, Hori S, & Mizunoe Y (2018). Broad impact of extracellular DNA on biofilm formation by clinically isolated methicillin-resistant and -sensitive strains of Staphylococcus aureus. Scientific Reports, 8(1), 2254. 10.1038/s41598-018-20485-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahrioui A, Duchesne R, Bouffartigues E, Rodrigues S, Maillot O, Tortuel D, Hardouin J, Taupin L, Groleau MC, Dufour A, Déziel E, Brenner-Weiss G, Feuilloley M, Orange N, Lesouhaitier O, Cornelis P, & Chevalier S (2019). Extracellular DNA release, quorum sensing, and PrrF1/F2 small RNAs are key players in Pseudomonas aeruginosa tobramycin-enhanced biofilm formation. NPJ Biofilms and Microbiomes, 5(1), 15. 10.1038/s41522-019-0088-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Schramm A, Neu TR, Revsbech NP, & Meyer RL (2013). Extracellular DNA in adhesion and biofilm formation of four environmental isolates: A quantitative study. FEMS Microbiology Ecology, 86(3), 394–403. 10.1111/1574-6941.12168 [DOI] [PubMed] [Google Scholar]
- Taylor PK, Van Kessel AT, Colavita A, Hancock RE, & Mah T-F (2017). A novel small RNA is important for biofilm formation and pathogenicity in Pseudomonas aeruginosa. PLOS One, 12(8), e0182582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas TJ, & Messner RP (1986). A left-handed (Z) conformation of poly(dA-dC).poly(dG-dT) induced by polyamines. Nucleic Acids Research, 14(16), 6721–6733. 10.1093/nar/14.16.6721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas VC, Thurlow LR, Boyle D, & Hancock LE (2008). Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. Journal of Bacteriology, 190(16), 5690–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku M, Nomura N, & Eberl L (2019). Types and origins of bacterial membrane vesicles. Nature Reviews Microbiology, 17(1), 13–24. 10.1038/s41579-018-0112-2 [DOI] [PubMed] [Google Scholar]
- Turnbull L, Toyofuku M, Hynen AL, Kurosawa M, Pessi G, Petty NK, Osvath SR, Cárcamo-Oyarce G, Gloag ES, Shimoni R, Omasits U, Ito S, Yap X, Monahan LG, Cavaliere R, Ahrens CH, Charles IG, Nomura N, Eberl L, & Whitchurch CB (2016). Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nature Communications, 7(1), 11220. 10.1038/ncomms11220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ussery DW (2001). DNA structure: A-, B-and Z-DNA helix families, eLS. John Wiley & Sons. [Google Scholar]
- Wagener JS, & Kupfer O (2012). Dornase alfa (pulmozyme). Current Opinion in Pulmonary Medicine, 18(6), 609–614. [DOI] [PubMed] [Google Scholar]
- Walker TS, Tomlin KL, Worthen GS, Poch KR, Lieber JG, Saavedra MT, Fessler MB, Malcolm KC, Vasil ML, & Nick JA (2005). Enhanced Pseudomonas aeruginosa biofilm development mediated by human neutrophils. Infection and Immunity, 73(6), 3693–3701. 10.1128/IAI.73.6.3693-3701.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Liu X, Liu H, Zhang L, Guo Y, Yu S, Wozniak DJ, & Ma LZ (2015). The exopolysaccharide Psl-eDNA interaction enables the formation of a biofilm skeleton in Pseudomonas aeruginosa. Environmental Microbiology Reports, 7(2), 330–340. 10.1111/1758-2229.12252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, & Mattick JS (2002). Extracellular dna required for bacterial biofilm formation. Science, 295(5559), 1487. 10.1126/science.295.5559.1487 [DOI] [PubMed] [Google Scholar]
- Wilton M, Charron-Mazenod L, Moore R, & Lewenza S (2016). Extracellular DNA acidifies biofilms and induces aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrobial Agents and Chemotherapy, 60(1), 544–553. 10.1128/aac.01650-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TL, Gong T, Zhu L, Miller J, Miller DS, Yin B, & Wood TK (2018). Rhamnolipids from Pseudomonas aeruginosa disperse the biofilms of sulfate-reducing bacteria. NPJ Biofilms and Microbiomes, 4(1), 22. 10.1038/s41522-018-0066-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, & Xi C (2009). Evaluation of different methods for extracting extracellular DNA from the biofilm matrix. Applied and Environmental Microbiology, 75(16), 5390–5395. 10.1128/AEM.00400-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafra O, Lamprecht-Grandío M, de Figueras CG, & González-Pastor JE (2012). Extracellular DNA release by undomesticated Bacillus subtilis is regulated by early competence. PLOS One, 7(11), e48716. 10.1371/journal.pone.0048716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatorska B, Groger M, Moser D, Diab-Elschahawi M, Lusignani LS, & Presterl E (2017). Does extracellular DNA production vary in staphylococcal biofilms isolated from infected implants versus controls? Clinical Orthopaedics and Related Research, 475(8), 2105–2113. 10.1007/s11999-017-5266-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetzmann M, Okshevsky M, Endres J, Sedlag A, Caccia N, Auchter M, Waidmann MS, Desvaux M, Meyer RL, & Riedel CU (2015). DNase-sensitive and -resistant modes of biofilm formation by Listeria monocytogenes. Frontiers in Microbiology, 6, 1428. 10.3389/fmicb.2015.01428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Mu H, Zhang W, Cui G, Zhu J, & Duan J (2013). Chitosan coupling makes microbial biofilms susceptible to antibiotics. Scientific Reports, 3(1), 3364. 10.1038/srep03364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Lehar S, Gutierrez J, Rosenberger CM, Ljumanovic N, Dinoso J, Koppada N, Hong K, Baruch A, Carrasco-Triguero M, Saad O, Mariathasan S, & Kamath AV (2016). Pharmacokinetics and pharmacodynamics of DSTA4637A: A novel THIOMAB™ antibody antibiotic conjugate against Staphylococcus aureus in mice. mAbs, 8(8), 1612–1619. 10.1080/19420862.2016.1229722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K, Chen S, Sysoeva TA, & You L (2019). Universal antibiotic tolerance arising from antibiotic-triggered accumulation of pyocyanin in Pseudomonas aeruginosa. PLOS Biology, 17(12), e3000573. 10.1371/journal.pbio.3000573 [DOI] [PMC free article] [PubMed] [Google Scholar]


