Abstract
The rapid, global spread of the SARS-CoV-2 virus during the current pandemic has triggered numerous efforts in clinical and research settings to better understand the host genetics’ interactions and the severity of COVID-19. Due to the established major role played by MHC/HLA polymorphism in infectious disease course and susceptibility, immunologists and geneticists have teamed up to investigate its contribution to the SARS-CoV-2 infection and COVID-19 progression. A major goal of the Covid-19|HLA & Immunogenetics Consortium is to support and unify these efforts. Here, we present a review of HLA immunogenomics studies in the SARS-CoV-2 pandemic and reflect on the role of various HLA data, their limitation and future perspectives.
Keywords: MHC, HLA, association analysis, SARS- CoV-2, COVID-19, immunogenetics
The Role of Immunogenetics in Infectious Diseases
The SARS-CoV-2 Pandemic
In late 2019, hospitals in Wuhan, China, received patients with pneumonia symptoms of unknown origin (Zhu et al., 2020). Researchers quickly identified the cause of this disease, a novel member of the coronavirus family, a single-strand RNA virus further named SARS-CoV-2 by the WHO on February 11th, 2020. This infection lead to COVID-19 disease. It can progress towards the development of an acute respiratory distress syndrome (ARDS) which can be lethal, especially but not exclusively, in older patients and patients with comorbidities (Ruan et al., 2020; Zhou et al., 2020). Previous coronavirus outbreaks, in 2003 with SARS-CoV and 2012 with MERS, had already demonstrated the danger of these known zoonotic viruses for humans (Shi and Hu, 2008). Contrary to SARS-CoV and MERS, which were successfully contained, but caused almost a thousand deaths each, SARS-CoV-2 is still active and endangering human health. In the span of almost 2 years, the virus spread to at least 240 million individuals, leading to more than 4.8 million deaths across the globe (O (2021).oronavir, 2021). The greater scale of this pandemic may be explained by the higher rates of transmission observed, the common asymptomatic carriers and the various severity of infected people (Syangtan et al., 2021).
Researchers determined that SARS-CoV-2 shares 50–79.5% of global sequence similarity with MERS and SARS-CoV, respectively, and that the mechanism of SARS-CoV-2 infection is similar to SARS-CoV, such as highlighted by Guo et al. (Guo et al., 2020). Their viral spike protein, found on the envelope, binds to the ACE2 receptor to enter human cells. While the virus spread globally and on a large scale, multiple SARS-CoV-2 strains have now emerged as the virus mutates, particularly presenting variations in the spike protein, such as the Gamma variant (P.1) in Brazil and the Delta variant (B.1.617.2) in India. These new strains provide a great incentive to assess the possible effects on immunity of such modifications (Burki, 2021), mainly because vaccines were designed to target the original spike protein.
Understanding the host response and the effect of host genomics is key for understanding variation in disease course subsequent to SARS-CoV-2 infection. Initial reports about COVID-19 suggested a pathogenic role of the immune system in the disease, damaging the lungs in a cytokine-storm provoked by CD4+ T lymphocytes and monocytes (Zhou et al., 2020). This excessive reaction in the wake of SARS-CoV-2 infection seems to be confirmed in non-human primates with less severe illness in animals with anti-inflammatory responses (Fahlberg et al., 2020). The COVID Human Genetic Effort has investigated these cellular responses at the genetic and genomic levels, describing rare variants in the IFN and TLR genes in patients with severe symptoms (Zhang et al., 2020a; Bastard et al., 2020; Zhang et al., 2020b; Casanova et al., 2020). Additionally, association studies have identified polymorphisms in the chemokine receptors and IFN, validating their role (Pairo-Castineira et al., 2020; The Severe Covid-19, 2020; D-19 Host Genetics In, 2021). On the genomic level, multiple studies have identified potentially important genes for COVID-19 severity and susceptibility, and researchers organized in different consortia, such as the COVID-19 Host Genetics Initiative, have collected association studies for meta-analyses (Pairo-Castineira et al., 2020; The Severe Covid-19, 2020; Mayoral et al., 2020; Ganna et al., 2020; Castro de Moura et al., 2021). In the same collective spirit, the COVID-19|HLA & Immunogenetics Consortium was created to investigate the role of the most polymorphic region of the human genome, the Major Histocompatibility Complex (MHC), in particular the Human Leukocyte Antigen (HLA) genes which are known to be highly associated with infectious diseases (Chen et al., 2011; Garcia et al., 2013; Spínola, 2016; Sawai et al., 2018; Thoens et al., 2018; Sanchez-Mazas, 2020a). In this review, we acknowledge recent advances linking HLA variation with COVID-19 and advocate for further progress in these efforts.
Linking HLA and Infectious Diseases: From SNP to HLA Allele
In the past decade, genome-wide association studies (GWAS) have become an essential tool for exploring the link between genetic background and complex phenotypes (Visscher et al., 2017). Rather than focusing efforts on candidate genes, DNA genotyping chips recover Single Nucleotide Polymorphisms (SNP) genotypes along the entire genome. Significant genotype-phenotype associations can be identified by comparing the SNP frequency in one population with a continuous trait (e.g., height, viral load) or between two populations differing by a binary trait or disease (e.g., HIV-1 infected patients vs general population). Contrary to Mendelian genetics, GWAS results are characterized by common genetic variants (allelic frequency ≥0.5–1%) associated with a low to moderate effect size on the outcome of interest, illustrating the “common variant-common disease” hypothesis. Identification of individual SNP contributions allows an overall burden evaluation of the disease genetic risk (Khera et al., 2018) (or protection) and a better understanding of molecular pathophysiological pathways. The GWAS catalog (EMBL-EBI, 2021) was created in 2008 to compile all GWAS results (Welter et al., 2014; MacArthur et al., 2017) and now contain 300,000 associations from 5,000 independent studies (October 6th, 2021).
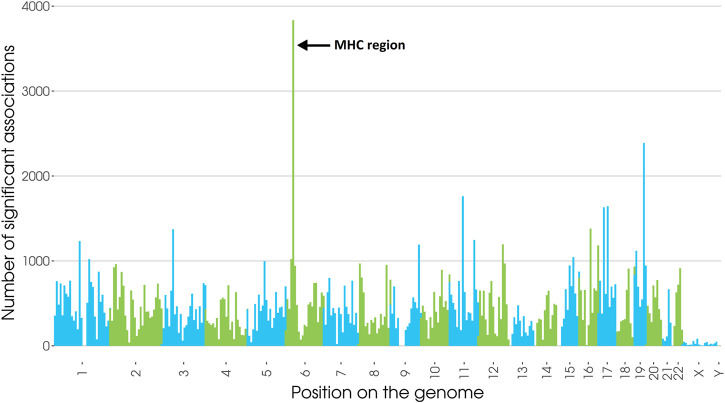
Numerous SNPs in the vicinity of HLA genes were confirmed to be associated with diseases (Price et al., 1999), and, the extended MHC accounts for 2.5% of all significant associations (Figure 1), and a third of significant chromosome 6 associations.
FIGURE 1.
Number of significant SNP associations to any trait or pathology from the GWAS catalog within the whole genome divided in 350 bins (inspired from Lenz TL et al., 2016 (Lenz et al., 2016) & Kennedy et al., 2017 (Kennedy et al., 2017)). 4,080 associations fall in the extended MHC region (25–34Mb, GRCh38), 2,784 of which are located between 30 and 34 Mb where most HLA genes are found. Updated on January 14th, 2021.155,456 associations with p-value<5 × 10−8.
Additionally, 21% of all traits in the catalog have at least one association in the extended MHC, illustrating the crucial role of MHC polymorphisms in human health. As expected, associations near the MHC region are immunity-related, from infectious diseases (Sanchez-Mazas, 2020a), to auto-immunity (Dendrou et al., 2018).
For example, one of the most significant HLA associations with an infectious disease is for HLA-B*57 tagging SNPs (SNPs not in an HLA gene but in linkage disequilibrium, LD, with specific HLA alleles) with HIV (OR = 3.47) (Fellay et al., 2007; Limou et al., 2008; Le Clerc et al., 2009; Limou et al., 2009). The rs2395029 SNP, which is almost in complete LD with the HLA-B*57:01 allele, was associated with HIV viral control in Europeans (Limou and Zagury, 2013), and symmetrically, the rs2523608 SNP, likely tagging the HLA-B*57:03 allele was discovered in African-American patients (Pelak et al., 2010). Other viral diseases showed associations with HLA SNPs include Hepatitis B virus (Hu et al., 2013; Jiang et al., 2015), Hepatitis C virus (Lee et al., 2018; Vergara et al., 2019), Epstein-Barr virus (Rubicz et al., 2013), and SARS-CoV (Sanchez-Mazas, 2020b). For a review, please see the extensive report by Sanchez and others (Sanchez-Mazas, 2020a).
However, the fact that GWAS identify a large genetic region associated with an outcome, without directly pinpointing functional, causal variants represents an important challenge for interpreting GWAS results. Such interpretation is made even more difficult by the complex LD patterns of the extended MHC region. Additional HLA typing and statistical inference of HLA alleles can refine the SNP association signals to specific HLA alleles, reflecting specific molecular functions and pathways. Such a strategy was successfully implemented for infectious diseases such as HIV, HPV, Dengue, and Ebola (Lin et al., 2003; Wang et al., 2011; Nishida et al., 2016; Adebamowo and Adeyemo, 2019; Chen et al., 2019; Ekenberg et al., 2019; Butler-Laporte et al., 2020; Chaisri et al., 2020; Huang et al., 2020; Ursu et al., 2020; Yengo et al., 2020).
Scope of the Review
Despite the central roles played by the MHC region and HLA molecules for the study of immune-related disease, understanding the underlying mechanisms of susceptibility and protection is far from complete (Trowsdale and Knight, 2013). The current pandemic raises questions regarding the role of HLA in recognition of or immune responses to a new virus. In this report, we review the first HLA-related investigations of SARS-CoV-2 and advocate for further efforts in HLA and COVID-19 analyses, using modern algorithms and resources, in order to enhance present and future research.
COVID-19 and HLA Association Studies
HLA polymorphisms have previously been closely associated with viral infections and disease outcomes, whether they are associated with protection or susceptibility. The intrinsic diversity of HLA molecules and the many possibilities to investigate their link to diseases sparked researchers’ interests during this novel pandemic. Researchers have investigated the interaction of host HLA diversity on both the infection by SARS-CoV-2 and the severity of the resulting COVID-19.
In Silico Peptide Binding and HLA Allele Frequencies
Studies using in silico peptide binding and HLA allele frequencies rely on available databases which do not require to generate data; thus, they are the first actionable steps to HLA analysis. Nguyen et al. proposed the first in silico HLA approaches in early 2020 by using the reference amino acid sequence of the SARS-CoV-2 (with NCBI accession number, NCBI:txid2697049) along with the netMHCpan software to predict the class I HLA alleles most susceptible to presenting SARS-CoV-2 peptides (Nguyen et al., 2020). They identified HLA-B*46:01 as the least presenting allele and HLA-B*15:03 as the most presenting one, possible risk and protective factors of infection, respectively. This publication was highlighted in the immunogenetics section of Nature, creating a starting point for HLA researchers (Zahn, 2020). Later, La Porta and others used Artificial Neural Networks to predict the binding capacity of each HLA class I allele, also demonstrating B*46:01 and others as a weak binder, and B*15:03 as a strong binder (La Porta and Zapperi, 2020). However, their results do not entirely overlap, demonstrating that functional studies should be performed. Barquera and others performed a similar analysis also considering HLA-DRB1 and HLA-DQA1/DQB1, indicating many HLA alleles (some highly frequent) among the best presenters, including B*15:03, and another list of worse presenters, including B*46:01 (Barquera et al., 2020).
Interestingly, B*15:03 frequency varies across the globe, with high frequencies in African populations and admixed ones (such as Brazilians), but low frequencies in Asia and Europe. Conversely, B*46:01 is highly frequent in Asia and rare in the rest of the world. The same dynamics can be observed for most of the alleles in the strong or weak presenter list.
Romero-López et al. expanded this investigation to class II HLA alleles and identified multiple HLA-DP and HLA-DR HLA alleles as well as HLA-A*02:03 as the allele with the most binding affinity to a viral peptide (Romero-López et al., 2020). Further research by de Sousa et al. of the most frequent HLA alleles of people from Europe, Asia and Africa and their interaction with variants and seems to point towards a selective pressure of class II MHC only regarding the binding of the ORF8 protein in SARS-CoV-2 (de Sousa et al., 2020).
The first studies only displayed correlations between COVID-19 phenotypes (e.g., incidence, severity, mortality) and HLA allele frequencies obtained in the allelefrequencies.net database or from local bone marrow donor registries, notably in Italy, an important European cluster. Correale et al. investigated class I correlations at one-field resolution (Correale et al., 2020). Pisanti et al. took a closer look at HLA haplotypes with an Italian registry, and identified HLA-A*01:01 g ∼ B*08:01g ∼ C*07:01g ∼ DRB1*03:01g as positively correlated with incidence and HLA-A*02:01g ∼ B*18:01g ∼ C*07:01g ∼ DRB1*11:04g as negatively correlated with incidence (Pisanti et al., 2020). Some studies took a more global approach by comparing the COVID-19 statistics of every country to their known HLA allele frequencies, providing discordant and mostly non-significant results (Ishii, 2020; Sakuraba et al., 2020; Tomita et al., 2020; Toyoshima et al., 2020). Other studies focused on a cellular level and identified a preponderance of monocytes with low expression of HLA-DR in infection and severity of SARS-CoV-2 (Benlyamani et al., 2020; Zmijewski and Pittet, 2020; Kreutmair et al., 2021; Roussel et al., 2021).
HLA Association Studies
Later, HLA association studies of various sample sizes tried to evaluate the direct link between HLA and different COVID-19 phenotypes. Wang et al. inferred the HLA class I and class II genotypes of 332 Chinese individuals to compare severe and mild cases of COVID-19, using xHLA (Xie et al., 2017) and SOAP-HLA (Cao et al., 2013), two software which allow HLA genotyping from sequencing data. HLA-A*11:01 (p-value = 0.009, OR 2.3), HLA-B*51:01 (p-value = 0.007, OR 3.3), and HLA-C*14:02 (p-value = 0.003, OR 4.7) were identified as top signals in the HLA class I region (Wang et al., 2020a). Direct HLA typing cohorts were also investigated across the world, but with small sample sizes going as high as 190 individuals. No associations were found by Iturrieta-Zuazo et al. in 45 Spanish patients between COVID-19 severity and HLA supertypes (Iturrieta-Zuazo et al., 2020), and none was found between mortality at 30 days and HLA one-field genotypes from 72 individuals from Canary Islands by Lorente et al. (2021). Three different groups conducted association analyses against a healthy control group to identify susceptibility of infection to SARS-CoV-2: Wang et al. (2020b) compared 82 COVID-19 vs 3,548 controls from China and found HLA-B*15:27 as associated (p-value = 0.001, OR 3.6), Novelli et al. (2020) compared 99 COVID-19 vs 1,017 controls from Italy and found 3 significant association (HLA-B*27:07, p-value = 0.00001; HLA-DRB1*15:01, p-value = 0.002; HLA-DQB1*06:02, p-value = 0.0001), and Yung et al. (2020) compared 190 COVID-19 vs 3892. controls from Hong-Kong but did not identify any significant association. More recently, Khor et al. (2021) also identified HLA-A*11:01:01:01 as a risk factor for COVID-19 severity (p-value = 0.003, OR 3.4), in a study involving 190 patients and 423 controls, after controlling for comorbidities and other confounding factors. Shachar et al. (2021)showed no association between COVID-19 severity and HLA alleles in a large-scale study of HLA typed Israelis (n = 20,937), though it was limited to two-field information. Finally, Castro de Moura et al. investigated the relationship between the epigenome of patients and COVID-19 severity from 407 patients and showed differentially methylated probes in HLA-C associated with the response of interferon in the viral response (Castro de Moura et al., 2021).
In addition to these studies, the Severe COVID-19 Consortium conducted a genome-wide association study of 1,980 patients of European ancestry and notably investigated HLA with classical SNP association, and HLA association by NGS genotyping in a subset of individuals. This was the first high-scale genomics initiative. However, chromosome 3 (SLC6A20, LZTFL1, CCR9, FYC O 1, CXCR6, and XCR1) as well as in the ABO locus (with A as risk and O protective) were the only significantly associated loci (The Severe Covid-19, 2020). The absence of HLA association was also shown by the meta-analysis on COVID-19 severity performed by the COVID-19 Human Genetic Initiative (HGI), where a variant in HLA-G was found but not replicated (Pairo-Castineira et al., 2020). However, the HGI release 6 in June 2021 identified 5 variants reaching statistical significance within the CCHCR1 gene, situated 110 kb downstream of HLA-C (top SNP: rs111837807, p-value = 2.2 × 10−11, ORmeta 1.23) as well as a variant within HLA-DPB1 3′UTR (rs9501257, p-value = 4.1 × 10−8, ORmeta 1.19), when comparing the general population to patients with critical COVID-19 (ncases = 8,779, ncontrol = 1,001,875, from 25 studies of various ancestries). It is notable that multiple variants linked to HLA genes seemed consistent, but not significant, between studies (D-19 Host Genetics In, 2021), which suggests that increasing cohort sizes in the future or running in-depth HLA-centric explorations may reveal additional significant signals.
Conclusion
Classical large GWAS meta-analysis recently reported SNP associations in the MHC region, mostly with critical COVID-19 illness, however the impact of HLA molecules might not be as imagined for this novel infectious disease. Unlike HIV-1 infection where HLA is the driving signal of viral control and disease progression, impact of HLA in SARS-CoV-2 infection seems milder and mostly restricted to severity symptoms, and its role has yet to be fully understood.
Multiple HLA-focused analyses performed during the last 2 years have had greatly varying results with inconsistent associations even in large studies [n = 20,937 in (Shachar et al., 2021)]. Further direct HLA allele association studies could provide the necessary power to carefully assess the role of HLA in immune response against SARS-CoV-2, but unfortunately, typing has not been conducted on large samples to date, leading to underpowered studies (most studies with less than 190 individuals). Indeed, HLA exploration requires large sample size; the HLA system has an important diversity, with thousands of alleles on multiple different genes. In a given population, a few numbers of these alleles are usually sufficient to represent the majority of individuals. However, to understand the role of the HLA system in diseases, it is important to also study alleles with a smaller frequency, which may be absent of cohorts with limited sample size.
HLA allele inference from sequencing (WGS and WES) and SNP genotyping data already generated for genome-wide analyses with the support of large biobanks and international consortia should therefore be given a high priority in the near future to provide a definitive answer on the impact of HLA molecules on COVID-19 phenotypes. Indeed, promising results from large association meta-analyses showed associations of both class I and class II HLA SNPs with severity, in the latest data release of the COVID-19 Host Genetic Initiative. Furthermore, the study of HLA 5-gene haplotype organization, and other immunogenetic parameters such as cell surface expression levels and interaction with KIR ligands may paint a bigger picture on the underlying immunogenetic mechanisms involved in the infection course.
HLA studies reported in this review rely on correlations and moderate size cohorts as stated. However, the COVID-19 crisis created an international collaboration to share data in order to explore host genetics risk factors for different COVID-19 outcomes (D-19 Host Genetics In, 2021). A vast amount of NGS and GWAS data have been generated: 49,562 COVID-19 positive cases vs >2M population controls with GWAS data in the COVID-19 Host Genetics Initiative (D-19 Host Genetics In, 2021); 20,952 cases vs 565,205 controls with WES data in the Regeneron study (Kosmicki et al., 2021). Thinking beyond COVID-19, the large national and international human genomics efforts represent a unique opportunity to promote large-scale HLA-centric analyses and to better describe HLA allele diversity across the globe by leveraging novel inference algorithms. These algorithms allow HLA typing from NGS and GWAS data (i.e., xHLA (Xie et al., 2017) and HIBAG (Zheng et al., 2014), respectively). Concerning other immunogenetics parameters, such as 5-gene HLA haplotypes or KIR ligands, it is now possible to infer them with HLA data (Geffard et al., 2020), with a detailed review of these tools in Douillard et al. (2021). Using these tools at a large scale on existing cohorts with GWAS and NGS data will clarify the role of HLA in COVID-19 outcomes and help understanding the mechanisms of the pathology.
The SARS-CoV-2 pandemic has had a huge global health toll, and has sparked a collective effort in the scientific community to identify candidate targets accounting for the diversity in response to the infection. HLA was quickly investigated for links with the SARS-CoV-2 infection and the resulting COVID-19 disease. The first studies, often underpowered, showed discordant results, and more robust association studies recently suggested a much milder effect of HLA SNPs and alleles on COVID-19 phenotypes as foreseen. The choice of the phenotype of interest was also proven to be crucial in association studies, as COVID-19 severity seems to be more closely linked to HLA. In this report, the COVID-19|HLA & Immunogenetics Consortium aimed to provide a critical view of current HLA analyses and their intrinsic power and limitations. We also hope this report will incite geneticists to run HLA-centric studies by expanding the pool of data available for HLA genotyping and genotypes imputation, in order to untangle the precise role of the Major Histocompatibility Complex in COVID-19 outcomes and other immune-related diseases.
Author Contributions
VD contributed in writing the review and produced figures. EC, SM, JH, P-A G, NV and SL contributed in writing and editing various sections of the review.
Funding
NV has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No. 846520. This work is supported by the ATIP-Avenir Inserm program, the Region Pays de Loire ConnectTalent. This work was also supported by National Institutes of Health (NIH) National Institute of Allergy and Infectious Disease (NIAID) grants R01AI128775 (JAH, SJM), and R01AI159260 (JAH).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Adebamowo S. N., Adeyemo A. A. (2019). Classical HLA Alleles Are Associated with Prevalent and Persistent Cervical High-Risk HPV Infection in African Women. Hum. Immunol. 80 (9), 723–730. 10.1016/j.humimm.2019.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barquera R., Collen E., Di D., Buhler S., Teixeira J., Llamas B., et al. (2020). Binding Affinities of 438 <scp>HLA</scp> Proteins to Complete Proteomes of Seven Pandemic Viruses and Distributions of Strongest and Weakest <scp>HLA</scp> Peptide Binders in Populations Worldwide. HLA 96 (3), 277–298. 10.1111/tan.13956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard P., Rosen L. B., Zhang Q., Michailidis E., Hoffmann H-H., Zhang Y., et al. (2020). Autoantibodies against Type I IFNs in Patients with Life-Threatening COVID-19. Science 370 (6515), eabd4585. 10.1126/science.abd4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlyamani I., Venet F., Coudereau R., Gossez M., Monneret G. (2020). Monocyte HLA‐DR Measurement by Flow Cytometry in COVID‐19 Patients: An Interim Review. Cytom A 97 (12), 1217–1221. 10.1002/cyto.a.24249 [DOI] [PubMed] [Google Scholar]
- Burki T. (20211027). Understanding Variants of SARS-CoV-2. Lancet 397, 462. 10.1016/s0140-6736(21)00298-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler-Laporte G., Kreuzer D., Nakanishi T., Harroud A., Forgetta V., Richards J. B. (2020). Genetic Determinants of Antibody-Mediated Immune Responses to Infectious Diseases Agents: A Genome-wide and HLA Association Study. Open Forum Infect. Dis. 7 (11), 1–14. 10.1093/ofid/ofaa450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Wu J., Wang Y., Jiang H., Zhang T., Liu X., et al. (2013). An Integrated Tool to Study MHC Region: Accurate SNV Detection and HLA Genes Typing in Human MHC Region Using Targeted High-Throughput Sequencing. PLoS One 8 (7). e69388. 10.1371/journal.pone.0069388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J-L., Su H. C., Abel L., Aiuti A., Almuhsen S., Arias A. A., et al. (2020). A Global Effort to Define the Human Genetics of Protective Immunity to SARS-CoV-2 Infection. Cell 181 (6), 1194–1199. 10.1016/j.cell.2020.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro de Moura M., Davalos V., Planas-Serra L., Alvarez-Errico D., Arribas C., Ruiz M., et al. (2021). Epigenome-wide Association Study of COVID-19 Severity with Respiratory Failure. EBioMedicine 66, 103339. 10.1016/j.ebiom.2021.103339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisri S., Jumnainsong A., Romphruk A., Leelayuwat C. (2020). The Effect of KIR and HLA Polymorphisms on Dengue Infection and Disease Severity in Northeastern Thais. Med. Microbiol. Immunol. 209 (5), 613–620. 10.1007/s00430-020-00685-z [DOI] [PubMed] [Google Scholar]
- Chen D., McKay J. D., Clifford G., Gaborieau V., Chabrier A., Waterboer T., et al. (2011). Genome-wide Association Study of HPV Seropositivity. Hum. Mol. Genet. [Internet 20 (23), 4714–4723. 10.1093/hmg/ddr383 [DOI] [PubMed] [Google Scholar]
- Chen Y., Liao Y., Yuan K., Wu A., Liu L. (2019). HLA-A, -B, -DRB1 Alleles as Genetic Predictive Factors for Dengue Disease: A Systematic Review and Meta-Analysis. Viral Immunol. 32 (3), 121–130. 10.1089/vim.2018.0151 [DOI] [PubMed] [Google Scholar]
- Correale P., Mutti L., Pentimalli F., Baglio G., Saladino R. E., Sileri P., et al. (2020). HLA-B*44 and C*01 Prevalence Correlates with Covid19 Spreading across Italy. Int. J. Mol. Sci. [Internet 21 (15), 5205. 10.3390/ijms21155205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- COVID-19 Host Genetics Initiative (2021). Mapping the Human Genetic Architecture of COVID-19. Nature. 10.1038/s41586-021-03767-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa E., Ligeiro D., Lérias J. R., Zhang C., Agrati C., Osman M., et al. (2020). Mortality in COVID-19 Disease Patients: Correlating the Association of Major Histocompatibility Complex (MHC) with Severe Acute Respiratory Syndrome 2 (SARS-CoV-2) Variants. Int. J. Infect. Dis. 98, 454–459. 10.1016/j.ijid.2020.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dendrou C. A., Petersen J., Rossjohn J., Fugger L. (2018). HLA Variation and Disease. Nat. Rev. Immunol 18 (5), 325–339. 10.1038/nri.2017.143 [DOI] [PubMed] [Google Scholar]
- Douillard V., Castelli E., Mack S. J., Hollenbach J., Gourraud P-A., Vince N., et al. (2021). Approaching Genetics through the MHC Lens: Tools and Methods for HLA Research. Front. Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekenberg C., Tang M-H., Zucco A. G., Murray D. D., MacPherson C. R., Hu X., et al. (2019). Association between Single-Nucleotide Polymorphisms in HLA Alleles and Human Immunodeficiency Virus Type 1 Viral Load in Demographically Diverse, Antiretroviral Therapy–Naive Participants from the Strategic Timing of AntiRetroviral Treatment Trial. J. Infect. Dis. 220 (8), 1325–1334. 10.1093/infdis/jiz294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embl-Ebi N. I. H. (2021). The GWAS Catalog. Available from:https://www.ebi.ac.uk/gwas .
- Fahlberg M., Blair R., Doyle-Meyers L., Midkiff C., Zenere G., Russell-Lodrigue K., et al. (2020). Cellular Events of Acute, Resolving or Progressive COVID-19 in SARS-CoV-2 Infected Non-human Primates. Nat. Commun. 11 (1), 6078. 10.1038/s41467-020-19967-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellay J., Shianna K. V., Ge D., Colombo S., Ledergerber B., Weale M., et al. (2007). Study of Major Determinants for Host Control of HIV-1. Science 317 (August), 944–947. 10.1126/science.1143767) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganna A., Unit T. G., General M. (2020). The COVID-19 Host Genetics Initiative, a Global Initiative to Elucidate the Role of Host Genetic Factors in Susceptibility and Severity of the SARS-CoV-2 Virus Pandemic. Eur. J. Hum. Genet. [Internet 28 (6), 715–718. 10.1038/s41431-020-0636-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia A., Milet J., Courtin D., Sabbagh A., Massaro J. D., Castelli E. C., et al. (2013). Association of HLA-G 3′UTR Polymorphisms with Response to Malaria Infection: A First Insight. Infect. Genet. Evol. 16, 263–269. 10.1016/j.meegid.2013.02.021 [DOI] [PubMed] [Google Scholar]
- Geffard E., Limou S., Walencik A., Daya M., Watson H., Torgerson D., et al. (2020). Easy-HLA: a Validated Web Application Suite to Reveal the Full Details of HLA Typing. Bioinformatics 36 (7), 2157–2164. 10.1093/bioinformatics/btz875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y-R., Cao Q-D., Hong Z-S., Tan Y-Y., Chen S-D., Jin H-J., et al. (2020). The Origin, Transmission and Clinical Therapies on Coronavirus Disease 2019 (COVID-19) Outbreak – an Update on the Status. Mil. Med. Res. 7 (1), 11. 10.1186/s40779-020-00240-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Liu Y., Zhai X., Dai J., Jin G., Wang L., et al. (2013). New Loci Associated with Chronic Hepatitis B Virus Infection in Han Chinese. Nat. Genet. 45 (12), 1499–1503. 10.1038/ng.2809 [DOI] [PubMed] [Google Scholar]
- Huang Y-H., Liao S-F., Khor S-S., Lin Y-J., Chen H-Y., Chang Y-H., et al. (2020). Large-scale Genome-wide Association Study Identifies HLA Class II Variants Associated with Chronic HBV Infection: a Study from Taiwan Biobank. Aliment. Pharmacol. Ther. 52 (4), 682–691. 10.1111/apt.15887 [DOI] [PubMed] [Google Scholar]
- Ishii T. (2020). Human Leukocyte Antigen (HLA) Class I Susceptible Alleles against COVID-19 Increase Both Infection and Severity Rate. Cureus 12 (12), e12239. 10.7759/cureus.12239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturrieta-Zuazo I., Rita C. G., García-Soidán A., de Malet Pintos-Fonseca A., Alonso-Alarcón N., Pariente-Rodríguez R., et al. (2020). Possible Role of HLA Class-I Genotype in SARS-CoV-2 Infection and Progression: A Pilot Study in a Cohort of Covid-19 Spanish Patients. Clin. Immunol. 219, 108572. 10.1016/j.clim.2020.108572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Ma X., Yu H., Cao G., Ding D., Chen H., et al. (2015). Genetic Variants in Five Novel Loci Including CFB and CD40 Predispose to Chronic Hepatitis B. Hepatology 62 (1), 118–128. 10.1002/hep.27794 [DOI] [PubMed] [Google Scholar]
- Kennedy A. E., Ozbek U., Dorak M. T. (2017). What Has GWAS Done for HLA and Disease Associations? Int. J. Immunogenet. 44 (5), 195–211. 10.1111/iji.12332 [DOI] [PubMed] [Google Scholar]
- Khera A. V., Chaffin M., Aragam K. G., Haas M. E., Roselli C., Choi S. H., et al. (2018). Genome-wide Polygenic Scores for Common Diseases Identify Individuals with Risk Equivalent to Monogenic Mutations. Nat. Genet. 50 (9), 1219–1224. 10.1038/s41588-018-0183-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor S., Omae Y., Nishida N., Sugiyama M., Kinoshita N., Suzuki T., et al. (2021). HLA-A*11:01:01:01, HLA-C*12:02:02:01-HLA-B*52:01:02:02, Age and Sex Are Associated with Severity of Japanese COVID-19 with Respiratory Failure. Front. Immunol. 12. 10.1101/2021.01.26.21250349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmicki J. A., Horowitz J. E., Banerjee N., Lanche R., Marcketta A., Maxwell E., et al. (2021). Pan-ancestry Exome-wide Association Analyses of COVID-19 Outcomes in 586,157 Individuals. Am. J. Hum. Genet. 108 (7), 1350–1355. 10.1016/j.ajhg.2021.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreutmair S., Unger S., Núñez N. G., Ingelfinger F., Alberti C., De Feo D., et al. (2021). Distinct Immunological Signatures Discriminate Severe COVID-19 from Non-SARS-CoV-2-driven Critical Pneumonia. Immunity, 1–16. 10.1016/j.immuni.2021.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Porta C. A. M., Zapperi S. (2020). Estimating the Binding of Sars-CoV-2 Peptides to HLA Class I in Human Subpopulations Using Artificial Neural Networks. Cell Syst 11 (4), 412–417. 10.1016/j.cels.2020.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Clerc S., Limou S., Coulonges C., Carpentier W., Dina C., Taing L., et al. (2009). Genomewide Association Study of a Rapid Progression Cohort Identifies New Susceptibility Alleles for AIDS (ANRS Genomewide Association Study 03). J. Infect. Dis. 200 (8), 1194–1201. 10.1086/605892 [DOI] [PubMed] [Google Scholar]
- Lee M., Huang Y., Chen H., Khor S., Chang Y., Lin Y., et al. (2018). Human Leukocyte Antigen Variants and Risk of Hepatocellular Carcinoma Modified by Hepatitis C Virus Genotypes: A Genome‐wide Association Study. Hepatology 67 (2), 651–661. 10.1002/hep.29531 [DOI] [PubMed] [Google Scholar]
- Lenz T. L., Spirin V., Jordan D. M., Sunyaev S. R. (2016). Excess of Deleterious Mutations Around HLA Genes Reveals Evolutionary Cost of Balancing Selection. Mol. Biol. Evol. 33 (10), 2555–2564. 10.1093/molbev/msw127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limou S., Coulonges C., Foglio M., Heath S., Diop G., Leclerc S., et al. (2008). Exploration of Associations between Phospholipase A2 Gene Family Polymorphisms and AIDS Progression Using the SNPlexTM Method. Biomed. Pharmacother. 62 (1), 31–40. 10.1016/j.biopha.2007.11.001 [DOI] [PubMed] [Google Scholar]
- Limou S., Le Clerc S., Coulonges C., Carpentier W., Dina C., Delaneau O., et al. (2009). Genomewide Association Study of an AIDS‐Nonprogression Cohort Emphasizes the Role Played by HLA Genes (ANRS Genomewide Association Study 02). J. Infect. Dis. 199 (3), 419–426. 10.1086/596067 [DOI] [PubMed] [Google Scholar]
- Limou S., Zagury J-F. (2013). Immunogenetics: Genome-wide Association of Non-progressive HIV and Viral Load Control: HLA Genes and beyond. Front. Immunol. 4 (MAY), 1–13. 10.3389/fimmu.2013.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M., Tseng H-K., Trejaut J. A., Lee H-L., Loo J-H., Chu C-C., et al. (2003). Association of HLA Class I with Severe Acute Respiratory Syndrome Coronavirus Infection. BMC Med. Genet. 4 (1), 9. 10.1186/1471-2350-4-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorente L., Martín M. M., Franco A., Barrios Y., Cáceres J. J., Solé-Violán J., et al. HLA Genetic Polymorphisms and Prognosis of Patients with COVID-19. Med Intensiva. 2021;45(2):96–103. 10.1016/j.medin.2020.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., et al. (2017). The New NHGRI-EBI Catalog of Published Genome-wide Association Studies (GWAS Catalog). Nucleic Acids Res. 45 (D1), D896–D901. 10.1093/nar/gkw1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayoral E. P. C., Hernández-Huerta M. T., Pérez-Campos Mayoral L., Matias-Cervantes C. A., Mayoral-Andrade G., Barrios L. Á. L., et al. (2020). Factors Related to Asymptomatic or Severe COVID-19 Infection. Med. Hypotheses [Internet] 144 (August), 110296. 10.1016/j.mehy.2020.110296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A., David J. K., Maden S. K., Wood M. A., Weeder B. R., Nellore A., et al. (2020). Human Leukocyte Antigen Susceptibility Map for SARS-CoV-2. J. Virol. 94 (13), 1–12. 10.1128/jvi.00510-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida N., Ohashi J., Khor S. S., Sugiyama M., Tsuchiura T., Sawai H., et al. (2016). Understanding of HLA-Conferred Susceptibility to Chronic Hepatitis B Infection Requires HLA Genotyping-Based Association Analysis. Sci. Rep. 6 (April), 1–7. 10.1038/srep24767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novelli A., Andreani M., Biancolella M., Liberatoscioli L., Passarelli C., Colona V. L., et al. (2020). HLA Allele Frequencies and Susceptibility to COVID‐19 in a Group of 99 Italian Patients. HLA 96 (5), 610–614. 10.1111/tan.14047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A. D., Rawlik K., Pasko D., et al. (2020). Genetic Mechanisms of Critical Illness in Covid-19. Nature 591(7848), 92-98. 10.1038/s41586-020-03065-y [DOI] [PubMed] [Google Scholar]
- Pelak K., Goldstein D. B., Walley N. M., Fellay J., Ge D., Shianna K. V., et al. (2010). Host Determinants of HIV‐1 Control in African Americans. J. Infect. Dis. 201 (8), 1141–1149. 10.1086/651382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisanti S., Deelen J., Gallina A. M., Caputo M., Citro M., Abate M., et al. (2020). Correlation of the Two Most Frequent HLA Haplotypes in the Italian Population to the Differential Regional Incidence of Covid-19. J. Transl Med. 18 (1), 1–16. 10.1186/s12967-020-02515-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price P., Witt C., Allock R., Sayer D., Garlepp M., Kok C. C., et al. (1999). The Genetic Basis for the Association of the 8.1 Ancestral Haplotype (A1, B8, DR3) with Multiple Immunopathological Diseases. Immunol. Rev. 167 (1), 257–274. 10.1111/j.1600-065x.1999.tb01398.x [DOI] [PubMed] [Google Scholar]
- Romero-López J. P., Carnalla-Cortés M., Pacheco-Olvera D. L., Ocampo-Godínez J. M., Oliva-Ramírez J., Moreno-Manjón J., et al. (2020). A Bioinformatic Prediction of Antigen Presentation from SARS-CoV-2 Spike Protein Revealed a Theoretical Correlation of HLA-Drb1*01 with COVID-19 Fatality in Mexican Population: An Ecological Approach. J. Med. Virol. 93 (4), 2029–2038. 10.1002/jmv.26561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel M., Ferrant J., Reizine F., Le Gallou S., Dulong J., Carl S., et al. (2021). Comparative Immune Profiling of Acute Respiratory Distress Syndrome Patients with or without SARS-CoV-2 Infection. Cell Rep. Med 2 (6), 100291. 10.1016/j.xcrm.2021.100291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical Predictors of Mortality Due to COVID-19 Based on an Analysis of Data of 150 Patients from Wuhan, China. Intensive Care Med. [Internet]. 2020;46 (5):846–848. 10.1007/s00134-020-05991-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubicz R., Yolken R., Drigalenko E., Carless M. A., Dyer T. D., Bauman L., et al. (2013). A Genome-wide Integrative Genomic Study Localizes Genetic Factors Influencing Antibodies against Epstein-Barr Virus Nuclear Antigen 1 (EBNA-1). Plos Genet. 9 (1), e1003147. 10.1371/journal.pgen.1003147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba A., Haider H., Sato T. (2020). Population Difference in Allele Frequency of HLA-C*05 and its Correlation with COVID-19 Mortality. Viruses 12 (11), 1333. 10.3390/v12111333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Mazas A. (2020). A Review of HLA Allele and SNP Associations with Highly Prevalent Infectious Diseases in Human Populations. Swiss Med. Wkly [Internet 150, w20214. 10.4414/smw.2020.20214 [DOI] [PubMed] [Google Scholar]
- Sanchez-Mazas A. (2020). HLA Studies in the Context of Coronavirus Outbreaks. Swiss Med. Wkly 150, w20248. 10.4414/smw.2020.20248 [DOI] [PubMed] [Google Scholar]
- Sawai H., Nishida N., Khor S-S., Honda M., Sugiyama M., Baba N., et al. (2018). Genome-wide Association Study Identified New Susceptible Genetic Variants in HLA Class I Region for Hepatitis B Virus-Related Hepatocellular Carcinoma. Sci. Rep. 8 (1), 7958. 10.1038/s41598-018-26217-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shachar S. Ben., Barda N., Manor S., Israeli S., Dagan N., Carmi S., et al. (2021). MHC Haplotyping of SARS-CoV-2 Patients: HLA Subtypes Are Not Associated with the Presence and Severity of COVID-19 in the Israeli Population. J. Clin. Immunol. 10.1007/s10875-021-01071-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z., Hu Z. (2008). A Review of Studies on Animal Reservoirs of the SARS Coronavirus. Virus. Res. 133 (1), 74–87. 10.1016/j.virusres.2007.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spínola H. (2016). HLA Loci and Respiratory Infectious Diseases. J. Respir. Res. 2 (3), 56–66. 10.17554/j.issn.2412-2424.2016.02.15 [DOI] [Google Scholar]
- Syangtan G., Bista S., Dawadi P., Rayamajhee B., Shrestha L. B., Tuladhar R., et al. (2021). Asymptomatic SARS-CoV-2 Carriers: A Systematic Review and Meta-Analysis. Front. Public Heal 8, 1–10. 10.3389/fpubh.2020.587374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Severe Covid-19 GWAS Group (2020). Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N. Engl. J. Med. 383 (16), 1522–1534. 10.1056/NEJMoa2020283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoens C., Heinold A., Lindemann M., Horn P. A., Chang D-I., Scherbaum N., et al. (2018). A Single-Nucleotide Polymorphism Upstream of the HLA-C Locus Is Associated with an Anti–hepatitis C Virus–Seronegative State in a High-Risk Exposed Cohort. J. Infect. Dis. 218 (12), 2016–2019. 10.1093/infdis/jiy492 [DOI] [PubMed] [Google Scholar]
- Tomita Y., Ikeda T., Sato R., Sakagami T. (2020). Association between HLA Gene Polymorphisms and Mortality of COVID‐19: An In Silico Analysis. Immunity, Inflamm. Dis. 8 (4), 684–694. 10.1002/iid3.358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima Y., Nemoto K., Matsumoto S., Nakamura Y., Kiyotani K. (2020). SARS-CoV-2 Genomic Variations Associated with Mortality Rate of COVID-19. J. Hum. Genet. 65 (12), 1075–1082. 10.1038/s10038-020-0808-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowsdale J., Knight J. C. (2013). Major Histocompatibility Complex Genomics and Human Disease. Annu. Rev. Genomics Hum. Genet. [Internet] 14 (1), 301–323. 10.1146/annurev-genom-091212-153455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursu L., Calenic B., Diculescu M., Dima A., Constantinescu I. (2020). HLA Alleles and KIR Genes in Romanian Patients with Chronic Hepatitis C. J. Gastrointest. Liver Dis. 29 (4), 595–601. 10.15403/jgld-2546 [DOI] [PubMed] [Google Scholar]
- Vergara C., Thio C. L., Johnson E., Kral A. H., O’Brien T. R., Goedert J. J., et al. (2019). Multi-Ancestry Genome-wide Association Study of Spontaneous Clearance of Hepatitis C Virus. Gastroenterology 156 (5), 1496–1507. 10.1053/j.gastro.2018.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher P. M., Wray N. R., Zhang Q., Sklar P., McCarthy M. I., Brown M. A., et al. (2017). 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet. [Internet 101 (1), 5–22. 10.1016/j.ajhg.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Huang S., Gao R., Zhou Y., Lai C., Li Z., et al. (2020). Initial Whole-Genome Sequencing and Analysis of the Host Genetic Contribution to COVID-19 Severity and Susceptibility. Cell Discov 6(1), 83. 10.1038/s41421-020-00231-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S-F., Chen K-H., Chen M., Li W-Y., Chen Y-J., Tsao C-H., et al. (2011). Human-Leukocyte Antigen Class I Cw 1502 and Class II DR 0301 Genotypes Are Associated with Resistance to Severe Acute Respiratory Syndrome (SARS) Infection. Viral Immunol. 24 (5), 421–426. 10.1089/vim.2011.0024 [DOI] [PubMed] [Google Scholar]
- Wang W., Zhang W., Zhang J., He J., Zhu F. (2020). Distribution of <scp>HLA</scp> Allele Frequencies in 82 Chinese Individuals with Coronavirus Disease‐2019 (COVID‐19). HLA 96 (2), 194–196. 10.1111/tan.13941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., et al. (2014). The NHGRI GWAS Catalog, a Curated Resource of SNP-Trait Associations. Nucleic Acids Res. 42 (D1), D1001–D1006. 10.1093/nar/gkt1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2021). WHO Coronavirus (COVID-19) Dashboard. Available from:https://covid19.who.int/.
- Xie C., Yeo Z. X., Wong M., Piper J., Long T., Kirkness E. F., et al. (2017). Fast and Accurate HLA Typing from Short-Read Next-Generation Sequence Data with xHLA. Proc. Natl. Acad. Sci. [Internet] 114 (30), 8059–8064. 10.1073/pnas.1707945114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yengo C. K., Torimiro J., Kowo M., Lebon P. A., Tiedeu B. A., Luma H., et al. (2020). Variation of HLA Class I (-A and -C) Genes in Individuals Infected with Hepatitis B or Hepatitis C Virus in Cameroon. Heliyon 6 (10), e05232. 10.1016/j.heliyon.2020.e05232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung Y., Cheng C., Chan H., Xia J. T., Lau K., Wong R. S. M., et al. (2020). Association of HLA‐B22 Serotype with SARS‐CoV ‐2 Susceptibility in Hong Kong Chinese Patients. Hla, tan.14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn L. M. (2020). HLA Genetics and COVID-19. Science 368 (6493), 841–2841. 10.1126/science.368.6493.841-b [DOI] [Google Scholar]
- Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., et al. (2020). Inborn Errors of Type I IFN Immunity in Patients with Life-Threatening COVID-19. Science 370 (6515), eabd4570. 10.1126/science.abd4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S-Y., Zhang Q., Casanova J-L., Su H. C. (2020). Severe COVID-19 in the Young and Healthy: Monogenic Inborn Errors of Immunity? Nat. Rev. Immunol. 20 (8), 455–456. 10.1038/s41577-020-0373-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Shen J., Cox C., Wakefield J. C., Ehm M. G., Nelson M. R., et al. (2014). HIBAG - HLA Genotype Imputation with Attribute Bagging. Pharmacogenomics J. 14 (2), 192–200. 10.1038/tpj.2013.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Fu B., Zheng X., Wang D., Zhao C., Qi Y., et al. (2020). Pathogenic T-Cells and Inflammatory Monocytes Incite Inflammatory Storms in Severe COVID-19 Patients. Natl. Sci. Rev. 7 (6), 998–1002. 10.1093/nsr/nwaa041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. (2020). A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 382 (8), 727–733. 10.1056/nejmoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmijewski J. W., Pittet J-F. (2020). Human Leukocyte Antigen-DR Deficiency and Immunosuppression-Related End-Organ Failure in SARS-CoV2 Infection. Anesth. Analg 131 (4), 989–992. 10.1213/ane.0000000000005140 [DOI] [PMC free article] [PubMed] [Google Scholar]