Abstract
Mass spectrometry (MS) is an essential technology in natural products research with MS fragmentation (MS/MS) approaches becoming a key tool. Recent advancements in MS yield dense metabolomics datasets which have been, conventionally, used by individual labs for individual projects; however, a shift is brewing. The movement towards open MS data (and other structural characterization data) and accessible data mining tools is emerging in natural products research. Over the past 5 years, this movement has rapidly expanded and evolved with no slowdown in sight; the capabilities of today vastly exceed those of 5 years ago. Herein, we address the analysis of individual datasets, a situation we are calling the ‘2021 status quo’, and the emergent framework to systematically capture sample information (metadata) and perform repository-scale analyses. We evaluate public data deposition, discuss the challenges of working in the repository scale, highlight the challenges of metadata capture and provide illustrative examples of the power of utilizing repository data and the tools that enable it. We conclude that the advancements in MS data collection must be met with advancements in how we utilize data; therefore, we argue that open data and data mining is the next evolution in obtaining the maximum potential in natural products research.
Graphical Abstract
This review covers the current and potential use of mass spectrometry data mining in natural products. Public data, metadata, databases, and data analysis tools are critical. The value and success of data mining rely on community participation.
1.0. Introduction
The fields of natural products (NPs) and mass spectrometry (MS) intertwine and as a natural consequence, new MS techniques and approaches are rapidly adopted. MS is a premier tool for dereplication and characterization of new molecules, measuring molecular formulae, isotopic patterns, spectral characteristics (MS/MS, MSn), etc.; moreover, untargeted MS approaches such as metabolomics strive to maximize the amount of structural information in a single analysis. A challenge with untargeted MS approaches is the immense amount and complexity of the data. Diverse chemicals are observed including natural product constituents, environmental chemicals (e.g. pesticides), contaminants, analysis artifacts, etc. Oftentimes, the ability to annotate or provide a putative identification of a chemical is limited. In this review, we aim to engage the natural products community and to encourage the dissemination of information including raw data and adopting the FAIR (Findability, Accessibility, Interoperability and Reusability) principles1, to further the field of natural products together.
The NP community has widely adopted dereplication practices to reduce the re-isolation of ‘knowns’. Metabolomics-based analysis of NP extracts is an emerging approach that should follow the successful implementation of dereplication practices. Metabolomics (i.e. untargeted MS) has proven to be an extremely viable strategy and has been applied to high throughput screening procedures, including the National Cancer Institute Program for Natural Products Discovery (NPNPD).2 The state of metabolomics and multi-’omics’ analysis in natural products has been covered extensively in recent years.3,4 Furthermore, recent reviews5,6 have highlighted the rapid development in tools and databases in 2020. Looking beyond dereplication and the current applications of metabolomics in NPs, there is room for growth and improvement especially in mining data (substructure- and network-based approaches)7 and repository-scale analysis. Global Natural Products Social molecular networking8 (GNPS), a tool mentioned throughout, has demonstrated how an open access and democratized platform can enhance research.9–11
While the focus of this review is on data analysis, we would be remiss not to mention the improvements to instrumentation hardware, data acquisition methods and chromatography. A comprehensive discussion of the seemingly innumerable MS techniques and approaches is not covered herein, but, we would like to highlight: the emerging potential of imaging mass spectrometry12 and ambient ionization13,14, the continued use of high-resolution mass spectrometry coupled to gas and liquid chromatography15 and the coupling of ion mobility measurement prior to mass analysis16 in the field of natural products.
2.0. The 2021 Status Quo: Data Analysis of Individual Datasets
The comparison between the use (and analysis) of individual datasets versus public data repositories is of vital importance to the arguments made in this review. We define an individual dataset as data that are generated from a single experiment or compilation of experiments under (nearly) the same conditions (e.g., model organism, experimental protocol and instrumentation). This type of data is by far the most commonly analyzed and reported in literature, whether it be the mass spectral files from liquid chromatography coupled to MS (LC-MS) or an FID file from an NMR experiment. In the following sections we discuss current approaches that would benefit from (or are extensible) processing individual datasets and seek to question if the NP community is leveraging data to its greatest potential.
2.1. Dereplication using in-house or public databases
Dereplication is one of the foundational processes in NPs research and due attention has been placed on trying to improve usability and availability.3,17 Commonly, dereplication occurs via commercial or in-house databases/extract libraries that focus on matching information such as retention time, UV/Vis spectra or mass.18–20 NPs relies heavily on databases like Dictionary of Natural Products (DNP), MarinLit, Antibase, Scifinder, Beilstein, etc., that are information repositories where searches are dictated by compound characteristics (e.g. exact mass, 𝛌max, C-H δ, organism, etc.) rather than data matching. Two reviews cover all NPs databases to date and are great resources to determine what is relevant to each researcher.3,21
Beyond choosing a database that fits, commercialization is a major factor, with private database licenses costing significant resources annually. Overall, the community is moving towards open access or public databases, like NP Atlas22 and COCONUT23, to facilitate wider accessibility and mitigating funding inequalities. Admittedly, open access resources are far from perfect but are becoming more useful. At the same time, they are only as good as the community participation and available support for them. Links between databases and related resources is vital, such as those found in NP Atlas22, GNPS8, SIRIUS24, Cytoscape25, Qiime226, Qiita27, MS2LDA28, MZmine229, MS-DIAL30 and MiBIG31. Data repositories such as Metabolights32 and Metabolomics Workbench33 are establishing connections as well. The growth of information and the links between resources should facilitate improved structural characterization and dereplication. The LOTUS34 database is a good model of what should be done via automated and manual curation of >500,000 organism-structure pairs of metabolites, mostly produced by the plant kingdom.
2.2. Mass-directed Fractionation
Mass-directed fractionation is used to speed up the discovery process. Knestrick et al. showed the advantage to layering multiple stages of mass spectrometry-based dereplication and chromatogram editing in the earliest stages of fractionation.35 MS-guided Solid-Phase Extraction of NPs yielded in-depth structural annotations and identifications through complementary structural information from NMR.36,37 Since this is a technique that requires an extensive setup, the literature is scarce, although a few studies show the power of this coupled technique.38–41 Mass-directed fractionation would benefit from reduced equipment costs and high performance large-scale chromatographic separations.39 A limitation of current MS-directed fractionation approaches is the reliance on in-house databases and mass-based queries which can lead to false identifications (via isobaric compounds). Nonetheless, MS-based separations remain a fast, robust technique for early-stage fractionation, making it highly attractive for drug discovery efforts and NP discovery.
2.3. Molecular Networking in GNPS
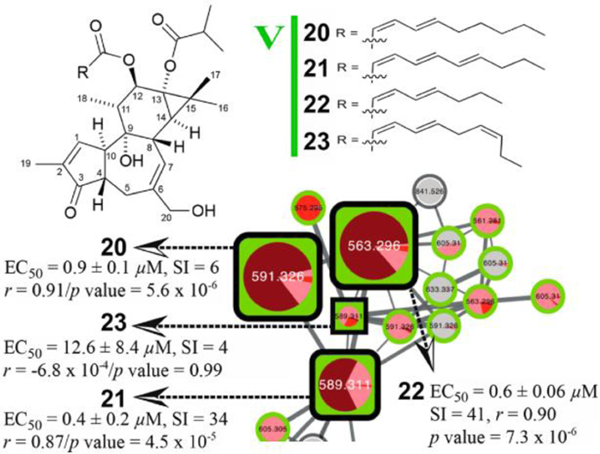
Molecular networking has become a prevalent NPs research tool. It was first introduced as a technique in 201242 and is one of the original cornerstone tools of the ~50 tools now available within GNPS molecular networking. To facilitate its usage, the GNPS interface arrived in 20168 and currently receives >300,000 accessions per month. A step-by-step protocol for various instrumental setups followed in 2020.43 Studies utilizing the GNPS workflows have ranged from drug discovery 37,44–53 (Fig 1) and strain prioritization54–58 to chemical ecology59–67 and a range of applications where molecular networking was used as part of the metabolomics analysis workflow. 68–80 In the past few years, additional workflows have been added to the molecular networking platform in GNPS such as MolNetEnhancer61 (MS/MS annotation) and MS2LDA28 (substructure mining). Feature Based Molecular Networking (FBMN)81 adds the ability to compare the relative abundance of features detected and clustered together using molecular networking. While it is possible to incorporate multiple datasets into a molecular network analysis, multi-dataset analyses are infrequently reported.
Fig 1.
GNPS molecular networking connected deoxyphorbol ester derivatives. Bioactivity scores for inhibition of chikungunya viral replication are plotted (larger nodes indicate a greater score). Node coloration indicates relative abundances in different fractions. Additional information is available in the manuscript. Reused with permission82 (https://pubs.acs.org/doi/10.1021/acs.jnatprod.7b00737); permissions related to the use of this material should be directed to the American Chemical Society.
2.4. Data Analysis using Uni- and Multivariate Statistics
Uni- and multi-variate statistics is a commonly utilized metabolomics technique employed in NPs. Such statistical strategies are typically implemented to differentiate extracts or metabolites and prioritize a subset for further analyses (viz. the composite score approach as demonstrated on Hydrastis Canadensis extracts).83 The review by Stuart et al. covers the application of metabolomics to marine NPs exclusively84 and serves as a good primer for statistical analysis of data from NP extracts. Worth highlighting is the recent example that appeared in Science by Zhang et al. which showed the power of using multivariate analysis to prioritize and leverage LC-MS data.50 Starting from 1482 actinobacterial isolates, 174 were deemed interesting based on hierarchical cluster analysis principal components analysis (HCAPCA). Micromonospora sp. WMMC-415 separated from the group, leading to the discovery of turbinmicin which is acutely bioactive against Candida auris, the “killer fungus”.53 Several LC-MS metabolomics tools exist to aid in the statistical analysis and visualization of data, like Qiime226, Qiita27, MZmine229, MS-DIAL85 and MetaboAnalyst86.
3. The Emergence of Public Data Deposition and Metadata Capture
Genomics, proteomics and other –omics fields have embraced the idea of public data deposition, but NPs research (and small molecule metabolomics) has been slow to adopt this mentality. The authors believe this is due to a myriad of challenges and entrenched misconceptions about ‘public data’. Previous reviews have highlighted the importance of publicly available LC-MS87 and NMR88 data and the limitations placed on scientific progress when there is a lack of transparency. We informally polled 79 participants via social media, from various career levels focusing on a variety of natural product sources. Overwhelmingly, the respondents indicated that they access and use public information (96.2%), either knowledgebases or databases, and free software is used such as XCMS89 and MZmine229(89.9%). Furthermore, a majority (78.5%) deem LC-MS repositories like GNPS/MassIVE a 5/5 on importance. However, there is a strong bias in this poll as 77.2% of respondents have contributed data to a LC-MS data repository; 50.6% have reused data from such repositories suggesting that mass spectrometry data is getting to the point that people find value in reuse. The top three responses when queried about not using public data were: “don’t know a tool to facilitate this” (27%), “no benefit to their research” (21.6%) and “data accessibility is poor” (18.9%). Certainly, an informal survey does not reflect the comprehensive views of the field. It does however indicate that many researchers are starting to think seriously about public data deposition and sample information (a.k.a. metadata) capture, something that should become the norm for the field. In the following section, we will highlight which data repositories exist for LC-MS and LC-MS/MSMS data and how systematic sample information would open up the use of such repositories.
3.1. Where is the information?
For sake of argument, we separate repositories into two archetypes: information-based and spectral-based. The former, information-based repositories (a.k.a. knowledgebases), would include relevant, largely textual or graphical information such as structures, organisms, bioactivity, images of spectra, etc. in the specific case of NPs. Whereas the latter, spectral-based repositories (a.k.a. databases or data repositories), would include largely numerical information such as mass spectral data, NMR data, UV/Vis data, etc. which would be used in numeric comparisons. In practice, there is overlap and most resources are some combination of the two. Fortunately, there is a multitude of repositories, however, they vary in functionality and utility for the NPs community, and it is not as simple as ‘X repository contains MS/MS spectra and is searchable’. Ultimately, it is worth highlighting that although these are all useful resources that aid the everyday research laboratory, one consistent challenge is retrieving and utilizing information. The information is present (and growing), now we must focus on how to access, effectively combine and use such information.
The typical information repositories in NPs are DNP and others mentioned in section 2.1 and recently reviewed.3,21 Information repositories like DNP mainly contain compound characteristics which can either be pulled from publications or sometimes theoretically generated (as is the case for some with NMR data). Running a search is information-based, often textual and requires manual interpretation of data. For example, searching an exact mass from a mass spectrum or the 𝛌max of a UV/Vis trace. Search functions are immensely useful; however, it is tedious for even small individual datasets let alone re-analyzing public data. MS data repositories should emulate the comprehensiveness of information in NPs structure databases. Vice versa, NP knowledgebases should reflect the accessibility of MS databases and develop data-driven searches. Improved means of utilizing knowledgebases and linking textual and spectral information are needed to maximize our current resources.
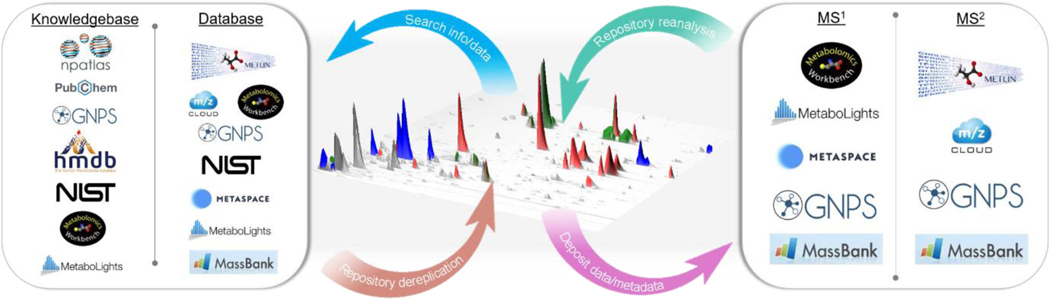
3.2. Mass Spectrometry Data Repositories for Natural Products
Data repositories range from general repositories like Zenodo90 or figshare91 (additional repositories - https://www.nature.com/sdata/policies/repositories) to specialized repositories like GNPS/MassIVE8, Metabolights32 and Metabolomics Workbench33. In the scope of this review, it is relevant to catalog these repositories and highlight their respective capabilities (Fig. 2). The METLIN92 database, started in the early 2000s, was one of the first to provide an online search function to the community. Since then, many information-based and spectral-based repositories arose (e.g. HMBD93, MetaSpace94, MassBank95, mzCloud96, NIST97, Metabolights32, GNPS/MassIVE98 and Metabolomics Workbench33). Some databases provide free analysis interfaces and the ability to download data for reuse, while others are available for purchase. The authors believe that open access to data has been a key and the critical step for recent computational advances in the structural characterization of metabolites and NPs.
Fig. 2.
(Left) Example knowledge and data repositories which can be searched using text or data as well as being used to annotate chemicals for dereplication. (Right) Illustrative MS data repositories categorized by their primary type of MS data stored. Data and metadata deposition is increasing; however, the reuse of repository data is less common.
The Human Metabolome Database (HMDB) is an open-access database containing information on metabolites and freely accessible MS/MS spectra of molecules found in the human body (exogenous and endogenous). HMDB links with additional databases such as PubChem and has informative links to drugs and drug metabolites (DrugBank), toxins and pollutants (TEDB), metabolic and disease pathways (MarkerDB), and food components (FooDB). LC-MS/MS data is sometimes available, as well as NMR data, that can serve as a valuable reference. Overall, HMDB serves as a knowledgebase, with the ability to search based on text, manual data input, structure, and sequence.
MetaboLights serves as an open-access repository for raw data and associated metadata. Multiple types of data are supported (LC-MS, NMR, imaging, etc.) and can be linked. A recent update to the online interface streamlined submission and curation of data.32 Metabolights hosts mainly biomedical metabolomics data with more than 50% of the data derived from human or mouse-based studies. Raw data from human (and mammalian) metabolomics make it unique compared to HMDB. MetaboLights serves an important role as a free and public knowledgebase for NPs99 with notable amounts of NMR and MS data.
The Metabolomics Workbench database, supported by the National Institutes of Health (NIH), is similar to MetaboLights while also containing entries pertaining to metabolites.33 Metabolomics Workbench entries are linked to many databases like the HMDB93, NP Atlas22, PubChem100 and KEGG101, amongst many others. Furthermore, it links with RefMet102 (A Reference list of Metabolite names) to provide standardized nomenclature and chemical information. A unique characteristic of Metabolomics Workbench is re-analysis of study results (often tables of peak areas) including statistics. While possible to access raw MS data, re-analysis of the raw data is not support to the same extent as re-analyzing study results.
METASPACE94 is a recently reported repository that focuses on spatial metabolomes via MS imaging experiments. It consists largely of MS imaging data of tissue cross-sections analyzed by matrix-assisted laser desorption ionization (MALDI) or desorption electrospray ionization (DESI). The majority of the data is collected from biomedical and pharmaceutical applications. As MS imaging becomes more common in NPs, METASPACE will become more pertinent for analyzing and re-analyzing spatial questions in NP studies.
MS spectral libraries (containing MS and MS/MS data) are currently a primary source of data reuse in natural products. While much attention is given to open source and free resources for analysis, it would be a disservice to not mention the extensive capabilities of databases for purchase such as METLIN92, mzCloud96 and NIST97. METLIN92 is one of the most extensive experimental mass spectral libraries curated with positive and negative mode data and MS/MS using different collision energy. Furthermore, neutral loss clustering enabling analog searches was recently reported.103 The NIST97 spectral library contains large amounts of EI spectra for GC-MS as well as MS/MS via collision-induced dissociation for over 30,000 compounds in the 2020 release. mzCloud96 curates high resolution spectra across multiple disciplines and their spectral tree interface allows for user-friendly analysis of MS/MS spectra. mzCloud libraries are not open and can only be used with commercial ThermoFisher software. Importantly, all three aforementioned repositories serve as dereplication tools using MS/MS spectra instead of information-based searches, an important distinction that generally provides more robust results. Many of the spectral libraries offer free online search tools with usage limitations. To the detriment to the community, these spectral libraries are not available in third party tools.
Open source MS spectral libraries are community-driven and growing. MassBank94 provides users access to a large repository of MS/MS data that is searchable in various query formats. MassBank of North America (MoNA - https://mona.fiehnlab.ucdavis.edu/) and its European counterpart (MassBank Europe - https://massbank.eu/MassBank/) provide users with the ability to search for spectra based on various mass spectrometry-based metadata (e.g. instrumentation, MS level, ionization mode). MoNA offers users the ability to query spectra based on raw data whereas MassBank Europe offers it in a more traditional peak list format. MassBank allows deposition of spectra as well as the ability to download the spectral libraries. For example, the MoNA spectral library can be downloaded and incorporated into dereplication workflows. The GNPS analysis ecosystem currently offers a comprehensive workflow for the NPs community; this has been covered extensively in the review by Fox Ramos et al.99 GNPS provides data and metadata deposition (publicly archived via MassIVE), free MS/MS spectral libraries and data analysis. Due to the extensive use of GNPS by the NPs community, it boasts the most MS/MS reference spectra of natural products. GNPS is known for molecular networking but new tools including, MASST9, ReDU10 and the GNPS dashboard11 aim at re-analysis and searches across the GNPS/MassIVE repository. For example, MS data in MetaboLights and Metabolomics Workbench can now be viewed and analyzed using the GNPS dashboard11 without the need for installation of software, further facilitating reuse and re-analysis of mass spectrometry datasets
3.3. Relevance of Metadata and Challenges in Recording Integral Information
Metadata provides context to data and are an important to align with FAIR concepts.1 One could argue that without metadata, data deposition and reuse is time prohibitive if not impossible. NP researchers search for interesting chemistry from the Mariana’s trench to the Atacama Desert or from the plants of Madagascar to the plants of your garden. Most often, the natural conditions (e.g., geography, climate) or controlled laboratory conditions yield unique chemistry that is in and of itself an insight into the biology of complex systems. Such conditions are reported in manuscripts but are frequently divorced from the data itself, rendering comparisons or analogies difficult when the context is lost. Thus, when operating at the repository-scale it is the union of the metadata and the data that is required for reanalysis. Therefore, we argue, it is the metadata that makes data in the repositories useful.
Metadata capture is an area in need of improvement. Previous attempts to address the problems with metadata capture and deposition were started with the Metabolomics Standards Initiative.104 Many others have also addressed this issue with metabolomics data in the past 15 years but few solutions have been systematically implemented.105–109 Ultimately, metadata is necessary for understanding and reuse of the data in most contexts. Regarding NPs, the challenge becomes more manageable when the advantages of metadata capture are recognized and adopted as a tenet. The NP field would benefit from MS data and knowledge (annotations, metadata, etc.) from all NP laboratories in the world being accessible as multiple, interoperable tools.
There are four main challenges to metadata capture. First, some information is not easily captured in simple descriptors. For example, conveying information about an unknown metabolite’s structure when only a substructure can be confidently determined based on the MS and MS/MS spectra. Certainly, adding this information to a dataset, as well as all the other confident chemical annotations, would be immensely beneficial in that other research could help piece complementary information and provide a more confident structure. Further, regio- and stereochemistry of substructure or incomplete structures are hard to convey when a common name, IUPAC name or other identifier cannot be assigned. Second, a lack of uniformity plagues metadata vocabulary. For example, a metadata label may be entered as Bacteria, Bacterium, bacteria, or bacterium. While these words are interpreted as the same by the human reader, they are not the same from a computer readability standpoint. Thus, when a search is performed all synonyms need to be searched and this quickly becomes intractable. Without standardization, searching for data let alone understanding the context within one repository can be tedious. Therefore, nearly all data repositories that require or suggest metadata have now started to use pragmatic (as well as controlled) vocabularies or ontologies, such as UBERON and DOID ontology (organ and biofluid ontology and disease ontology respectively). The field of NPs lacks a universally adopted and used ontology or vocabulary. Clear starting points for agreement include descriptors like depth, latitude and longitude, NCBI taxonomy and soil chemistry properties. Third, metadata are not stored in a consistent manner. Metadata exists in many types of files (e.g., .txt, .json, .xlsx) which makes its use complicated. Lastly, the generation and curation of metadata is currently a manual process, a seemingly negligible immediate (and apparent) benefit versus time cost. Standardization of the information desired by the NPs community coupled with automated capture of metadata from instrumentation or text-mining from written documents (e.g., electronic notebooks and manuscripts) would immensely benefit the metadata generation and curation effort.
These challenges require focused and concerted effort to address much like has been done recently with ReDU, the first-generation controlled metadata capture strategy within GNPS.10 MetaboLights uses a modified version of Sequence Read Archive submission to accomplish a related task. Metadata from MetaboLights can be converted into ReDU compatible formats and entries, providing an example of how metadata from disparate data repositories can be used. Once the ReDU metadata table has been added to the GNPS public data set it is possible to search using controlled vocabulary metadata terms or using MS/MS spectral searches using MASST to find metadata associations that link back to the public data sets as described in section 4.1.
4. The Emergence of the Need for Repository-scale Data Analysis
A natural outcome of having publicly deposited data with context-providing metadata is for researchers to mine this resource. While neither NPs nor MS has tapped into this resource extensively, genetics provides a roadmap for how it can be done effectively. The concept of Basic Local Alignment Search Tool110 (BLAST) has become ubiquitous and natural to the field of genetics. NPs (and MS) would benefit from analogous data processing tools.
In recent years, several examples have appeared in the literature that began to explore the idea that the inclusion of additional datasets aids in discovery. One of these explored data pertaining specifically to Pseudomonads in the attempt to shift away from the idea of ‘one-molecule-one-microbe’.111 By taking 260 ecological diverse strains of Pseudomonas and subjecting the data to molecular networking, the researchers were successful in the identification of four new lipopeptides with biosynthetic genes similar to each other yet divergent to all others. Furthermore, supplementing this new data with that of 370 additional wheat-associated Pseudomonas strains showed the dispersion of data across the original 260 strains, the 370 wheat strains, and common metabolites in both datasets. The mapping of data from one lab onto another to compare discoveries and metabolic overlap shows the potential for comparing and relating data, while also highlighting the paucity and limited accessibility of this type of data in repositories. Using a similar approach, Crüsemann et al. analyzed a large-scale molecular network containing 603 samples from 146 marine Actinobacteria. Through the evaluation of metabolomes originating from various conditions, the large-scale molecular networking study linked ‘taxonomy, culture conditions, and extraction methods, as well as informing the most valuable growth and extraction conditions’.54 We envision that upon investment of time, data and metadata, optimization of culture conditions through repository comparison would be a major functionality of repository scale re-analysis.
The work from Olivon et al. took a large data approach, analyzing 292 extracts from 107 New Caledonian Euphorbiaceae species.36 In addition to obtaining LC-MS/MS based information and conducting molecular networking, the layering of biological and taxonomic information led to the generation of prioritized natural product families and the subsequent identification of a new daphne diterpene orthoester. 38 Demonstrating the power of re-analysis, Olivon et al. returned to the same dataset to include additional preprocessing using MZmine2 to discover chloroaustralasines.112 Similar molecular network layering was published in the same year by Nothias et al., displaying the combination of bioassay and molecular networking and the use of MZmine2 as a pre-processing aid in data deconvolution.82
One of the first examples to highlight the power of repository re-analysis is a recent investigation into algal lipids in which public data were reanalyzed and a 40% increase in lipid annotations was obtained.113 A study in which soil samples were taken in 14 USA states, 188 soil samples collected from five distinct climate regions, evaluated the ‘city, state and regional process on backyard soil metabolite composition’.107 Localities dictated similarities and differences within metabolite composition and how certain processes shape soil composition. Additionally, it shed light on the plant, microbial, and human influences on the environment; sunscreen constituents, pesticides, herbicides, and medication were detectable from the soil samples.114 So how do we move further towards repository scale analysis and what advantages are gained?
Overall, there are currently very few studies that demonstrate the power of repository-scale re-analysis. Molecular networking, specifically cosine-based spectral similarity scoring, is likely to be used for data re-analysis as well. Complementary tools have emerged that take advantage of spectral data in repositories and improve networking of large datasets such as Spec2Vec115, MS2DeepScore116, and falcon117. The previously mentioned studies illustrate the possibilities, largely using molecular networking. MASST and ReDU, recently reported tools, aim to mitigate the challenges of re-analysis and are developed with the intent to facilitate re-analysis.
4.1. MASST
MASST9, inspired by NCBI’s BLAST tool, provides the ability to query a MS/MS spectrum against all public data files with MS/MS (and MS/MS spectral libraries) contained within GNPS/MassIVE (~1.2 billion MS/MS spectra). The tool operates via the creation of a searchable network generated from all MS/MS spectra which can be compared by spectral similar to a queried MS/MS spectrum. In the supplementary of the MASST publication, the authors provided multiple examples of the applicability to NPs. Example #5 examined the presence/absence of a Pseudomonas derived NP, orfamide, in non-laboratory settings. The subsequent MASST search revealed it was present in four datasets in the GNPS/MassIVE data repository. A matching MS/MS spectrum was observed in a Pseudomonas culture collection as would be expected. Unexpectedly, a match was observed in Trachymyrmex septentrionalis fungus gardens, suggesting a role for Pseudomonads and this natural product in the ecology of ant fungus gardens. Further examination of the fungus garden sample data from NCBI revealed the presence of Pseudomonas and subsequently, several Pseudomonads were isolated from the gardens. Example #8 evaluated the presence of staurosporine analogs in datasets, with 14 datasets matching its MS/MS. From marine and soil sediments, putative derivatives were suggested with additional CH2, NO, and CHN2O modifications. One of the first studies to show the functionality of MASST is work by Lybbert et al.118 They mined public data to aid in discovery of numerous derivatives from Pseudomonas spp. One of their most interesting results stemmed from a search on rhamnolipids, which surprisingly linked to datasets from ant-fungal mutualist dens, soil, plants, human teeth, feces, various lung mucus samples, and cultured laboratory isolates.
4.2. ReDU
Leveraging repository data in an effective and straightforward manner is a principal challenge. The Reanalysis of Data User (ReDU) Interface10 addresses the challenges of metadata via consistent formatting, the use of ontologies and a controlled vocabulary, as well as validation steps. Furthermore, ReDU is integrated with GNPS/MassIVE and other data analysis tools in GNPS. MetSummarizer119, a new method for systematic prediction of biological phenotypes as well as decomposition of complex extracts to their raw components, has utilized the power of ReDU. Training the MetSummarizer tool with repository data from ReDU improved annotations from 25 to 32.5%, with future periodic updates occurring to increase the accuracy of predictions. Preliminar y attempts at repository-like analyses were covered in the introductory section. ReDU’s repository-scale analysis capabilities are illustrated in subsequent examples using Group Comparator, Chemical Explorer and repository-scale molecular networking.
Group Comparator facilitates qualitative comparison of annotations (MS/MS matching of public MS/MS spectra via GNPS) between user-defined groups. The file selection interface allows one to select files based on metadata, such as SampleType_bacterial or SampleType_environmental, and then the Group Comparator tool can be launched. The resulting table displays a list of annotations, the number of files in which the annotation was observed and the proportion of files in which the annotation was observed with respect to the total number of files in the group. The annotations are performed periodically via GNPS on the data publicly deposited in MassIVE, which we termed ‘de novo annotation’. An important caveat is that the information is only as accurate as the data in ReDU and the means by which it was studied (e.g., extractions, instrumentation, and chromatography); additionally, the qualitative comparison accuracy should grow as the public MS/MS spectra grow in number and coverage. Like any tool, results should be interpreted with care and rigorously scrutinized. In spite of these caveats, there are meaningful insights to be gained by utilizing repository data to develop or enhance a hypothesis or observation.
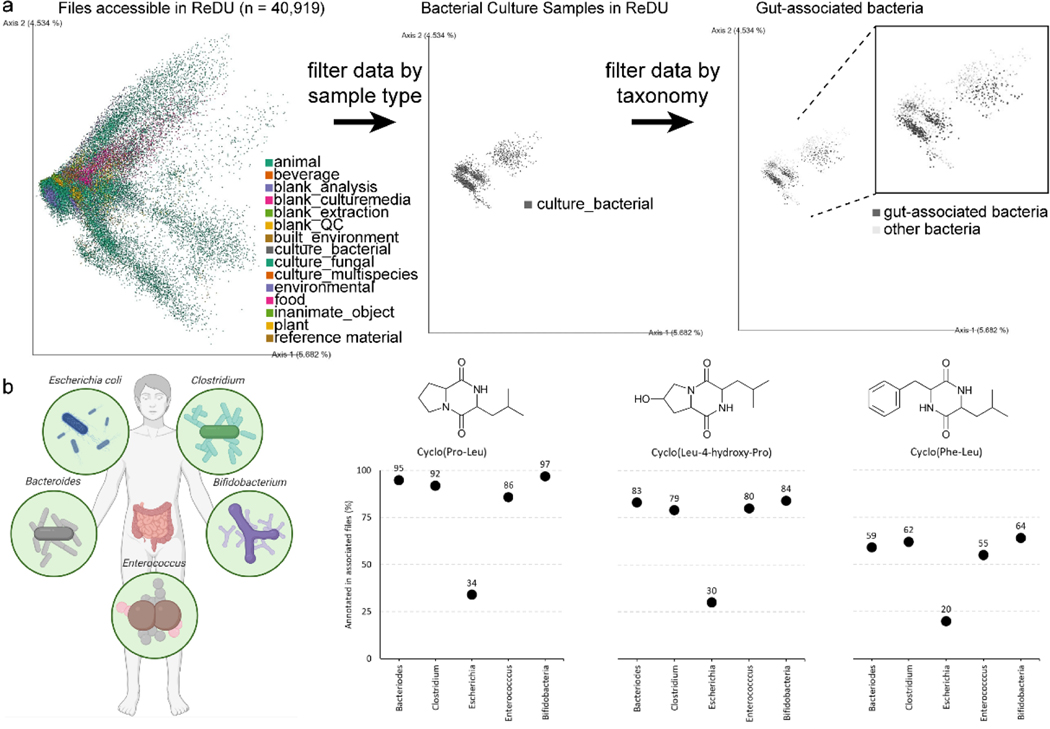
In an illustrative case study of Group Comparator (Fig. 3), bacterial files present in ReDU of gut-associated microbes (n= 465) were selected (Bacteroides spp., Escherichia coli, Enterococcus spp., Bifidobacteria spp. and Clostridium spp.). One of the most common secondary metabolites present in each group were various 2,5-diketopiperazines (DKPs). Gut microbes produce an array of small molecules yet the presence of DKPs has remained underexplored.120 DKPs occur in a variety of NPs ranging across bacteria, fungi, plants and mammals, whilst also having a broad biological purpose. One of the most intriguing prospects for these metabolites is for chemical communication where a few studies have shown DKPs serve as potential quorum sensing molecules.121,122 Here, we observe the presence of numerous DKPs across each group, with cyclo(Pro-Leu), cyclo(Leu-4-hydroxy-Pro) and cyclo(Phe-Leu) among the most abundant in the data (Fig. 3b), along with several additional DKPs present in varying percentages. Furthermore, the three main DKPs are less observed in Escherichia coli datasets, possibly pointing to a reduced role in this species. In summary, DKPs were observed, empirically, in many different datasets which supports that DKPs are widespread and lends further credence to the hypothesis that DKPs could play a role in gut flora communication.
Fig. 3.
(a) Left: Two-dimensional emperor plots displaying the prinicipal component analysis (based on MS/MS data) of files in ReDU (n = 40,919) colored by SampleType. Middle: Highlighting via filtering of bacterial files in ReDU, n = 2,246 files. Right: Highlighting specific bacterial files based on taxonomy in ReDU with gut-associated bacterial in dark grey and all other bacterial files in light grey. (b) Left: Illustration displaying the gut-associated bacterial genera selected for Group Comparator analysis (created with BioRender.com). Right: Dot plots displaying the Group Comparator results (percentage of files in which an annotated spectrum was observed) of three of the most abundant diketopiperazines: cyclo(Pro-Leu), cyclo(Leu-4-hydroxy-Pro) and cyclo(Phe-Leu).
Another tool in ReDU is Chemical Explorer which tabulates de novo annotation search results on GNPS/MassIVE repository data while tracking the metadata attached to the files in which the annotations are observed. The result allows one to query specific chemicals and determine how many times a chemical annotation has been observed in the repository data, the files in which it was observed and the metadata associated with the annotation. Haffner et al. have very recently showed the utility of Chemical Explorer by evaluating their core metabolites identified in the study of 6 diverse populations against 10 datasets present (n= 1,286 samples) in ReDU. The repository-mined results further substantiate their results showing industrialized populations share commonalities in their fecal metabolomes, despite ‘geographic, dietary, or behavioral’ differences.123
To further show the utility of Chemical Explorer, we queried the small molecule talaromycin A in another case study. The talaromycins were originally isolated from Talaromyces stipitatus124, known endophytes of multiple plants.125 When Chemical Explorer was run, the metabolite was found in data files associated to Gossypium hirsutum, a known host for Talaromyces spp.126, as well as their herbivorous predators, Helicoverpa virescens. Moreover, Arabidopsis thaliana was suggested in the Chemical Explorer analysis; interestingly, a relationship between A. thaliana and talaromycins has not been reported in the literature. Talaromyces spp. are excellent plant colonizers and have been described in a number of endophytic relationships. Similar to the orfamide example in MASST, Chemical Explorer was used to mine the entire collection of data in ReDU in order to find meaningful relationships between various datasets, and in this case revealed a potential expanded ecological role of Talaromyces spp. as an Arabidopsis thaliana endophyte.
Repository-scale molecular networking facilitated via ReDU is a derivation of molecular networking as previously discussed in section 2.3; however, the inclusion of repository-scale data is immensely facilitated via ReDU’s file selection interface. The illustrative example represented in Fig. 2 of the ReDU publication showcased the power of molecular networking with the enhancement of repository-scale analysis.10 Through mining relevant files from blood, urine and fecal samples, various clindamycin analogues (itself derived from the natural product, lincomycin) were easily identifiable. Discovery remains the most commonplace aim of molecular networking in the NPs community and therefore, inspiring translation of this capability to ReDU.
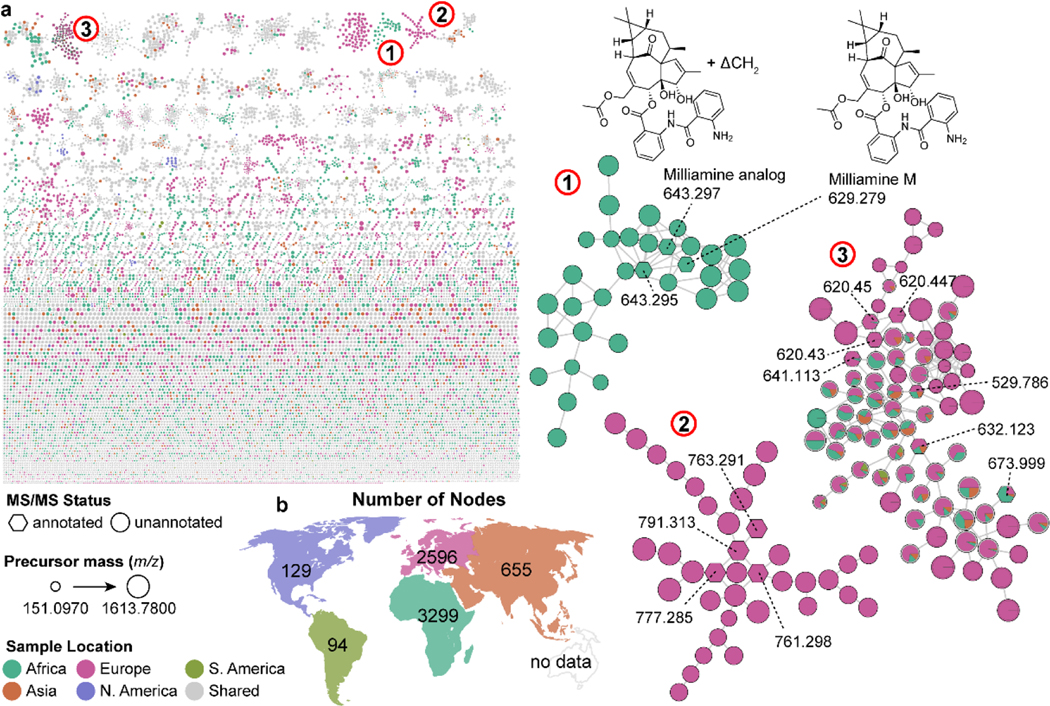
Euphorbia spp. were selected as a case study to explore the relationship between the geographical location of these species and the chemicals detected (specially the MS/MS) in the repository, similar to what was done by Ernst et al.126 These data files were previously used to exhibit the competence of MolNetEnhancer61, FBMN81 and CANOPUS128; therefore, they have been well characterized. ReDU’s straightforward file selection interface aided in the selection of 236 files (5 complete datasets) originating from 8 Euphorbia spp. Unfortunately, the metadata for which continent the species are native to was not available, especially since many species were cultivated in botanical gardens throughout Europe. Therefore, we manually curated the geographic information provided in ReDU (latitude and longitude) such that the correct native continent was assigned to each Euphorbia spp. Molecular networking was performed on the selected Euphorbia data and the distribution of files pertaining to each continent are indicated in Fig 4.
Fig. 4.
(a) Repository-scale molecular networking via ReDU with Euphorbia spp. with highlighted molecular families 1–3 containing milliamines, terracinolides and diterpenes, respecitvely. An illustrative connection between Milliamine M and a putative Milliamine analog differing in mass by ΔCH2 via molecular networking is displayed. (b) Number of nodes in repository-scale molecular network observed as occuring from samples native to the continent.
The resulting molecular network contained 18,898 nodes (clusters of similar MS/MS spectra observed between datasets) of which 5.6% of nodes and 11.7% of spectra were annotated. Milliamines (Fig. 4, group 1), previously observed by Ernst et al.61, appear endemic to Africa (Madagascar, specifically) when overlaid with geographic information. Previous studies used material originating in Brazil129 and Japan130 for discovery, but these locations reflect the ornamental value of the flower rather than the origin of the species. Jatrophane diterpenes (Fig. 4, group 2), such as the terracinolides, cluster similarly in the network as in the study by Nothias et al.82 When applying the geographic origin parsed into G1-G5 onto the molecular network nodes as pie charts, we see that these metabolites in the molecular family are supposedly endemic to Europe (Euphorbia dendriodes originating from Corsica). Further investigation into the literature shows that terracinolides were originally isolated from Californian Euphorbia terracina131, but the lack of deposited samples from this location and others does not allow for the multi-continental connection to be observed.
The comprehensiveness of interpretation is limited by the extent to which data is deposited for analysis; therefore, we hope these examples serve to demonstrate what could be done rather than what is currently possible. However, the limitations of the data present today for data-repository scale analysis does not limit interesting insights, such as Euphorbia milli and the milliamines, which was only observed in the African continent. This observation supports its known origin of Madagascar and our analysis resulted in numerous molecular families that are geographically-enriched. While NPs research typically hunts for unique chemistry, the proportion of chemicals or abundance is equally of value, such as the diterpene molecular family is shared amongst species from multiple continents (Fig. 4, group 3); however, not all nodes were found equally observed. These types of relationships are vital for comparing potentially biodiversity-rich ecological niches, and possible geographically-specific biotransformations. One could imagine larger scale geographic distribution studies being conducted on many sources, similar to the recent work by Gericke et al.132
5. Conclusions and Perspective
Repository-scale analysis and data mining are the next steps in the field of natural products. Such approaches efficiently utilize the cost, time and resources of research and enable new methods of discovery and understanding. In this review, we have highlighted the challenges and demonstrated how repository-scale analysis opens up new questions that can be asked using MS-based metabolomics. The potential benefits are wide-ranging: from validating drug metabolite occurrence in independent studies, to probing chemical diversity under different conditions, toward identifying strains based on chemical similarity. While techniques like bioassay-guided fractionation and the ‘grind and find’ mentality still play important roles in NPs, they disregard the resource consumptive nature that many researchers simply cannot entertain and thus demand new, innovative, and openly accessible approaches. However, these end goals require a community effort to not only curate data and metadata but to also establish and adhere to FAIR standards, which encouragingly, has begun.
So, we can ask the question: what would an ideal scenario look like for repository-scale analysis? The principal challenges for use of metadata that were highlighted in section 3.3 offer a starting point for achieving the overarching goal of efficient repository-scale analysis. Addressing controlled metadata vocabularies has been started with tools like ReDU with future changes possible to adapt to community needs. Furthermore, as the community continues to move towards using openly accessible MS analysis tools, universal file formatting has started to be addressed. One of the challenges not yet mentioned in our review is the composition of the data present in repositories and its lack of reflecting the ongoing research in the community.
Strikingly, only 0.3% of all datasets, still representing ~15,000 LC-MS/MS files, present in GNPS/MassIVE pertain to Actinomycetes, the widely studied and prolific producers of natural product-derived antibiotics. In an ideal world, the data files represented in repositories should reflect the research conducted on such organisms, resulting in less rediscovery and accelerated discovery. Unfortunately, the secretive nature of drug discovery is partially to blame for the lack of transparency for depositing NP data and we hope platforms like GNPS are helping to combat this inclination. Now that natural molecules are no longer patentable and only the natural product in connection to disease treatment or biological phenotype is, there is no longer an excuse for not making natural products discovery data publicly available. The reward for participation in public data deposition is of immediate benefit to your own research, providing one with new insights and leading to new discoveries. Concurrently, providing information to the public furthers everyone’s insight and discovery, which will be required to combat some of the major health problems we will face in the decades to come.
Improving structure annotations and moving towards automation are more difficult challenges to address but are by no means impossible. Repository-scale analysis and dataset context (leading to a reduction of false positives) could lead to improved ‘tag’ information via consistent metadata capture. Importantly, complementary information about NPs can be collected from various sources. For example, NMR offers some advantages over MS-based approaches, especially as it typically allows for non-destructive de novo structural elucidation of a natural product - something that is almost impossible based on MS alone. Therefore, the two techniques must complement each other: combining the structural information can boost annotation and identification efforts. Therefore, we look forward to joint analyses by MS and NMR of natural product mixtures taking the benefits of the growing MS and NMR repositories.
Additionally, genomics is beginning to play a larger role in aiding metabolite annotations. The MIBiG database31 collects validated links between biosynthetic gene clusters and molecular structures. This information is invaluable in linking metabolomics data with biological context. Structural information such as stereochemistry cannot easily be determined from metabolomics data; however, specific genes may provide an inroad into how certain bonds are positioned in space. Furthermore, linking the biosynthetic machinery to the structures, and indeed the spectral data, also facilitates confident labels and “tags” as to who is the producer (and source) of molecules in complex mixtures. Recently, a community effort was launched to record publicly available paired genome and metabolome data (i.e., from the same biological source) as well as validated links between gene clusters and metabolite spectra and structures therein (https://pairedomicsdata.bioinformatics.nl). Such paired data provides a complementary dimension needed to answer questions by applying linking strategies that are currently available and will be developed based on such initiatives.133–135 The first software framework that capitalizes on these developments has also appeared; NPLinker136 utilizes genomics and metabolomics data from public repositories, runs genome and metabolome mining and network analyses, and ranks links between biosynthetic gene clusters and metabolite spectra.
Finally, knowing the structure is one thing - knowing what they do is yet something else. Here, repository-scale analysis can start to help paint the picture and add functional labels to metabolites found in complex metabolite extracts. Connecting paired genomics and metabolomics datasets to paired proteomics and transcriptomics datasets will allow for a more complete picture to come alive as complementary information illuminates the active genetic machinery under specific conditions. Furthermore, with the increasing availability of well-curated structure-organism knowledgebases, taxonomic considerations will also become more valuable to aid in structural and functional annotations.137
Compared to five years ago, the mass spectrometry tools and methods employed by natural product researchers are currently undergoing a mini revolution akin to the revolutions seen when sequence repositories became the norm. Another generation of science and scientists is now flourishing and open data and data mining tools will continue to enhance NP science. There are technical impediments which need to be addressed; however, more importantly, and the takeaway message of this review, is that there is clear benefit to participate as individual natural product researchers and engage the community in making natural products datasets publicly available and curating sample information in order to perform metadata-guided repository-scale analyses.
6. Methods
Group Comparator
Files were initially filtered based on SampleType_culture_bacterial to reduce the overall number of possible files. NCBITaxonomy was then used as the selection category, with known gut-associated obligate bacteria in mind for selection. The most well represented genera in ReDU were, in descending order, Bacteroides spp. (185 files), Enterococcus spp. (94 files), Clostridium spp. (82 files), Bifidobacterium spp. (79 files) and Escherichia coli (70 files). These genera were separated into respective Groups and run through the Group Comparator workflow. 2,5-Diketopiperazines were among the highest abundance metabolites annotated in each group and therefore were selected for comparison.
Molecular Networking
Files were initially filtered based on SampleType_plant to reduce the overall number of files. Euphorbia spp. files were then targeted using NCBITaxonomy for molecular networking. Evaluation of geography metadata pointed to many of the files originating from botanical gardens in Europe, therefore, native geographic niche was manually identified and appropriate files were divided into Groups pertaining to the 5 continents represented by the deposited Euphorbia spp. (G1-North America, G2-South America, G3-Europe, G4-Africa and G5-Asia). The species represented in ReDU include: Euphorbia dendroides, Euphorbia pithyusa, Euphorbia lathyris, Euphorbia horrida, Euphorbia kansai, Euphorbia pekinensis, Euphorbia hirta and Euphorbia milli.
A molecular network was created using the online workflow (https://ccms-ucsd.github.io/GNPSDocumentation/) on the GNPS website (http://gnps.ucsd.edu) using default settings. The data was filtered by removing all MS/MS fragment ions within +/− 17 Da of the precursor m/z. MS/MS spectra were window filtered by choosing only the top 6 fragment ions in the +/− 50Da window throughout the spectrum. The precursor ion mass tolerance was set to 2.0 Da and a MS/MS fragment ion tolerance of 0.5 Da. A network was then created where edges were filtered to have a cosine score above 0.7 and more than 6 matched peaks. Further, edges between two nodes were kept in the network if and only if each of the nodes appeared in each other’s respective top 10 most similar nodes. Finally, the maximum size of a molecular family was set to 100, and the lowest scoring edges were removed from molecular families until the molecular family size was below this threshold. The spectra in the network were then searched against GNPS’ spectral libraries. The library spectra were filtered in the same manner as the input data. All matches kept between network spectra and library spectra were required to have a score above 0.7 and at least 6 matched peaks.
7. Acknowledgements
This work was supported by the Danish National Research Foundation (DNRF137) and NIH GM107550. J.J.J.v.d.H. was funded by an ASDI eScience grant, ASDI.2017.030, from the Netherlands eScience Center. This research was supported in part by the Intramural Research Program of National Institute of Environmental Health Sciences of the NIH (ES103363–01).
Conflicts of Interest
PCD is on the scientific advisory board of Sirenas, Cybele Microbiome, Galileo and founder and scientific advisor of Ometa Labs LLC and Enveda (with approval by UC San Diego).
References
- 1.Wilkinson MD, Scientific Data, 2016, 3, 160018. [Google Scholar]
- 2.Grkovic T, Akee RK, Thornburg CC, Trinh SK, Britt JR, Harris MJ, Evans JR, Kang U, Ensel S, Henrich CJ, Gustafson KR, Schneider JP and O’Keefe BR, ACS Chemical Biology, 2020, 15, 1104–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Santen JA, Kautsar SA, Medema MH and Linington RG, Natural Product Reports, 2020, 38, 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Demarque DP, Dusi RG, de Sousa FDM, Grossi SM, Silvério MRS, Lopes NP and Espindola LS, Scientific Reports, 2020, 10, 1051-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medema MH, Natural Product Reports, 2021, 301–306. [DOI] [PubMed] [Google Scholar]
- 6.Misra BB, Metabolomics, 2021, 17, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beniddir MA, bin Kang K, Genta-Jouve G, Huber F, Rogers S. and van der Hooft JJJ, Natural Product Reports DOI: 10.1039/d1np00023c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang M, Carver JJ, Phelan V. v., Sanchez LM, Garg N, Peng Y, Nguyen DTDD, Watrous J, Kapono CA, Luzzatto-Knaan T, Porto C, Bouslimani A, Melnik A. v., Meehan MJ, Liu WT, Crüsemann M, Boudreau PD, Esquenazi E, Sandoval-Calderón M, Kersten RD, Pace LA, Quinn RA, Duncan KR, Hsu CC, Floros DJ, Gavilan RG, Kleigrewe K, Northen T, Dutton RJ, Parrot D, Carlson EE, Aigle B, Michelsen CF, Jelsbak L, Sohlenkamp C, Pevzner P, Edlund A, McLean J, Piel J, Murphy BT, Gerwick L, Liaw CC, Yang YL, Humpf HU, Maansson M, Keyzers RA, Sims AC, Johnson AR, Sidebottom AM, Sedio BE, Klitgaard A, Larson CB, Boya CAPP, Torres-Mendoza D, Gonzalez DJ, Silva DB, Marques LM, Demarque DP, Pociute E, O’Neill EC, Briand E, Helfrich EJNN, Granatosky EA, Glukhov E, Ryffel F, Houson H, Mohimani H, Kharbush JJ, Zeng Y, Vorholt JA, Kurita KL, Charusanti P, McPhail KL, Nielsen KF, Vuong L, Elfeki M, Traxler MF, Engene N, Koyama N, Vining OB, Baric R, Silva RR, Mascuch SJ, Tomasi S, Jenkins S, Macherla V, Hoffman T, Agarwal V, Williams PG, Dai J, Neupane R, Gurr J, Rodríguez AMCC, Lamsa A, Zhang C, Dorrestein K, Duggan BM, Almaliti J, Allard PM, Phapale P, Nothias LF, Alexandrov T, Litaudon M, Wolfender JL, Kyle JE, Metz TO, Peryea T, Nguyen DTDD, VanLeer D, Shinn P, Jadhav A, Müller R, Waters KM, Shi W, Liu X, Zhang L, Knight R, Jensen PR, Palsson B, Pogliano K, Linington RG, Gutiérrez M, Lopes NP, Gerwick WH, Moore BS, Dorrestein PC and Bandeira N, Nature Biotechnology, 2016, 34, 828–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang M, Jarmusch AK, Vargas F, Aksenov AA, Gauglitz JM, Weldon K, Petras D, da Silva R, Quinn R, Melnik A. v., van der Hooft JJJJ, Caraballo-Rodríguez AM, Nothias LF, Aceves CM, Panitchpakdi M, Brown E, di Ottavio F, Sikora N, Elijah EO, Labarta-Bajo L, Gentry EC, Shalapour S, Kyle KE, Puckett SP, Watrous JD, Carpenter CS, Bouslimani A, Ernst M, Swafford AD, Zúñiga EI, Balunas MJ, Klassen JL, Loomba R, Knight R, Bandeira N. and Dorrestein PC, Nature Biotechnology, 2020, 38, 23–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarmusch AK, Wang M, Aceves CM, Advani RS, Aguirre S, Aksenov AA, Aleti G, Aron AT, Bauermeister A, Bolleddu S, Bouslimani A, Caraballo Rodriguez AM, Chaar R, Coras R, Elijah EO, Ernst M, Gauglitz JM, Gentry EC, Husband M, Jarmusch SA, Jones KL, Kamenik Z, le Gouellec A, Lu A, McCall LI, McPhail KL, Meehan MJ, Melnik A. v., Menezes RC, Montoya Giraldo YA, Nguyen NH, Nothias LF, Nothias-Esposito M, Panitchpakdi M, Petras D, Quinn RA, Sikora N, van der Hooft JJJ, Vargas F, Vrbanac A, Weldon KC, Knight R, Bandeira N. and Dorrestein PC, Nature Methods, 2020, 17, 901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petras D, v Phelan V, Acharya D, Allen AE, Aron AT, Bandeira N, Bowen BP, Belle-Oudry D, Boecker S, Cummings DA, Deutsch JM, Fahy E, Garg N, Gregor R, Handelsman J, Navarro-Hoyos M, Jarmusch AK, Jarmusch SA, Louie K, Maloney KN, Marty MT, Meijler MM, Mizrahi I, Neve RL, Northen TR, Molina-Santiago C, Panitchpakdi M, Pullman B, Puri AW, Schmid R, Subramaniam S, Thukral M, Vasquez-Castro F, Dorrestein PC and Wang M, bioRxiv DOI: 10.1101/2021.04.05.438475. [DOI] [Google Scholar]
- 12.Spraker JE, Luu GT and Sanchez LM, Natural Product Reports, 2020, 37, 150–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarmusch AK and Cooks RG, Natural Product Reports, 2014, 31, 730–738. [DOI] [PubMed] [Google Scholar]
- 14.Oberlies NH, Knowles SL, Amrine CSM, Kao D, Kertesz V. and Raja HA, Natural Product Reports, 2019, 36, 944–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petras D, Koester I, da Silva R, Stephens BM, Haas AF, Nelson CE, Kelly LW, Aluwihare LI and Dorrestein PC, Frontiers in Marine Science, 2017, 4, 405. [Google Scholar]
- 16.Schrimpe-Rutledge AC, Sherrod SD and McLean JA, Current Opinion in Chemical Biology, 2018, 42, 160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hubert J, Nuzillard JM and Renault JH, Phytochemistry Reviews, 2017, 16, 55–95. [Google Scholar]
- 18.Chervin J, Stierhof M, Tong MH, Peace D, Hansen KØ, Urgast DS, Andersen JH, Yu Y, Ebel R, Kyeremeh K, Paget V, Cimpan G, van Wyk A, Deng H, Jaspars M. and Tabudravu JN, Journal of Natural Products, 2017, 80, 1370–1377. [DOI] [PubMed] [Google Scholar]
- 19.Zani CL and Carroll AR, Journal of Natural Products, 2017, 80, 1758–1766. [DOI] [PubMed] [Google Scholar]
- 20.Pérez-Victoria I, Martín J. and Reyes F, Planta Medica, 2016, 82, 857–871. [DOI] [PubMed] [Google Scholar]
- 21.Sorokina M. and Steinbeck C, Journal of Cheminformatics DOI: 10.1186/s13321-020-00424-9. [DOI] [Google Scholar]
- 22.van Santen JA, Jacob G, Singh AL, Aniebok V, Balunas MJ, Bunsko D, Neto FC, Castaño-Espriu L, Chang C, Clark TN, Cleary Little JL, Delgadillo DA, Dorrestein PC, Duncan KR, Egan JM, Galey MM, Haeckl FPJ, Hua A, Hughes AH, Iskakova D, Khadilkar A, Lee JH, Lee S, Legrow N, Liu DY, Macho JM, McCaughey CS, Medema MH, Neupane RP, O’Donnell TJ, Paula JS, Sanchez LM, Shaikh AF, Soldatou S, Terlouw BR, Tran TA, Valentine M, van der Hooft JJJ, Vo DA, Wang M, Wilson D, Zink KE and Linington RG, ACS Central Science, 2019, 5, 1824–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sorokina M, Merseburger P, Rajan K, Yirik MA and Steinbeck C, Journal of Cheminformatics, 2021, 13, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dührkop K, Fleischauer M, Ludwig M, Aksenov AA, Melnik A. v., Meusel M, Dorrestein PC, Rousu J. and Böcker S, Nature Methods, 2019, 16, 299–302. [DOI] [PubMed] [Google Scholar]
- 25.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B. and Ideker T, Genome Research, 2003, 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Estaki M, Jiang L, Bokulich NA, McDonald D, González A, Kosciolek T, Martino C, Zhu Q, Birmingham A, Vázquez-Baeza Y, Dillon MR, Bolyen E, Caporaso JG and Knight R, Current Protocols in Bioinformatics, 2020, 70, e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzalez A, Navas-Molina JA, Kosciolek T, McDonald D, Vázquez-Baeza Y, Ackermann G, DeReus J, Janssen S, Swafford AD, Orchanian SB, Sanders JG, Shorenstein J, Holste H, Petrus S, Robbins-Pianka A, Brislawn CJ, Wang M, Rideout JR, Bolyen E, Dillon M, Caporaso JG, Dorrestein PC and Knight R, Nature Methods, 2018, 15, 796–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Hooft JJJ, Wandy J, Barrett MP, Burgess KEV and Rogers S, Proceedings of the National Academy of Sciences of the United States of America, 2016, 113, 13738–13743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pluskal T, Castillo S, Villar-Briones A. and Orešič M, BMC Bioinformatics, 2010, 11, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsugawa H, Ikeda K, Takahashi M, Satoh A, Mori Y, Uchino H, Okahashi N, Yamada Y, Tada I, Bonini P, Higashi Y, Okazaki Y, Zhou Z, Zhu ZJ, Koelmel J, Cajka T, Fiehn O, Saito K, Arita M. and Arita M, Nature Biotechnology, 2020, 38, 1159–1163. [DOI] [PubMed] [Google Scholar]
- 31.Kautsar SA, Blin K, Shaw S, Navarro-Muñoz JC, Terlouw BR, van der Hooft JJJ, van Santen JA, Tracanna V, Suarez Duran HG, Pascal Andreu V, Selem-Mojica N, Alanjary M, Robinson SL, Lund G, Epstein SC, Sisto AC, Charkoudian LK, Collemare J, Linington RG, Weber T. and Medema MH, Nucleic Acids Research, 2020, 48, D454–D458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haug K, Cochrane K, Nainala VC, Williams M, Chang J, Jayaseelan KV and O’Donovan C, Nucleic Acids Research, 2020, 48, D440–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sud M, Fahy E, Cotter D, Azam K, Vadivelu I, Burant C, Edison A, Fiehn O, Higashi R, Nair KS, Sumner S. and Subramaniam S, Nucleic Acids Research, 2016, 44, D463–D470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rutz A, Sorokina M, Galgonek J, Mietchen D, Willighagen E, Graham J, Stephan R, Page R, Vondrášek J, Steinbeck C, Pauli GF, Wolfender J-L, Bisson J. and Allard P-M, bioRxiv DOI: 10.1101/2021.02.28.433265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knestrick MA, Tawfik R, Shaw LN and Baker BJ, Journal of Pharmaceutical and Biomedical Analysis, 2019, 176, 112831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Hooft JJJ, Mihaleva V, de Vos RCH, Bino RJ and Vervoort J, Magnetic Resonance in Chemistry, 2011, 49, S55–S60. [DOI] [PubMed] [Google Scholar]
- 37.Wolfender JL, Nuzillard JM, van der Hooft JJJ, Renault JH and Bertrand S, Analytical Chemistry, 2019, 91, 704–742. [DOI] [PubMed] [Google Scholar]
- 38.Olivon F, Allard PM, Koval A, Righi D, Genta-Jouve G, Neyts J, Apel C, Pannecouque C, Nothias LF, Cachet X, Marcourt L, Roussi F, Katanaev VL, Touboul D, Wolfender JL and Litaudon M, ACS Chemical Biology, 2017, 12, 2644–2651. [DOI] [PubMed] [Google Scholar]
- 39.Bu X, Regalado EL, Hamilton SE and Welch CJ, TrAC - Trends in Analytical Chemistry, 2016, 82, 22–34. [Google Scholar]
- 40.Barbieri M. and Heard CM, Journal of Pharmaceutical and Biomedical Analysis, 2019, 166, 90–94. [DOI] [PubMed] [Google Scholar]
- 41.Lima R. de C. L., Gramsbergen SM, van Staden J, Jäger AK, Kongstad KT and Staerk D, Journal of Natural Products, 2017, 80, 1020–1027. [DOI] [PubMed] [Google Scholar]
- 42.Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, van der Voort M, Pogliano K, Gross H, Raaijmakers JM, Moore BS, Laskin J, Bandeira N. and Dorrestein PC, Proceedings of the National Academy of Sciences of the United States of America, 2012, 109, E1743–E1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aron AT, Gentry EC, McPhail KL, Nothias LF, Nothias-Esposito M, Bouslimani A, Petras D, Gauglitz JM, Sikora N, Vargas F, van der Hooft JJJJ, Ernst M, bin Kang K, Aceves CM, Caraballo-Rodríguez AM, Koester I, Weldon KC, Bertrand S, Roullier C, Sun K, Tehan RM, Boya P CA, Christian MH, Gutiérrez M, Ulloa AM, Tejeda Mora JA, Mojica-Flores R, Lakey-Beitia J, Vásquez-Chaves V, Zhang Y, Calderón AI, Tayler N, Keyzers RA, Tugizimana F, Ndlovu N, Aksenov AA, Jarmusch AK, Schmid R, Truman AW, Bandeira N, Wang M. and Dorrestein PC, Nature Protocols, 2020, 15, 1954–1991. [DOI] [PubMed] [Google Scholar]
- 44.Quinn RA, Nothias LF, Vining O, Meehan M, Esquenazi E. and Dorrestein PC, Trends in Pharmacological Sciences, 2017, 38, 143–154. [DOI] [PubMed] [Google Scholar]
- 45.bin Kang K, Park EJ, da Silva RR, Kim HW, Dorrestein PC and Sung SH, Journal of Natural Products, 2018, 81, 1819–1828. [DOI] [PubMed] [Google Scholar]
- 46.Paz WHP, de Oliveira RN, Heerdt G, Angolini CFF, De Medeiros LS, Silva VR, Santos LS, Soares MBP, Bezerra DP, Morgon NH, Almeida JRGS, da Silva FMA, Costa E. v. and Koolen HHF, Journal of Natural Products, 2019, 82, 2220–2228. [DOI] [PubMed] [Google Scholar]
- 47.Reher R, Kuschak M, Heycke N, Annala S, Kehraus S, Dai HF, Müller CE, Kostenis E, König GM and Crüsemann M, Journal of Natural Products, 2018, 81, 1628–1635. [DOI] [PubMed] [Google Scholar]
- 48.Mudalungu CM, von Törne WJ, Voigt K, Rückert C, Schmitz S, Sekurova ON, Zotchev SB and Süssmuth RD, Journal of Natural Products, 2019, 82, 1478–1486. [DOI] [PubMed] [Google Scholar]
- 49.Woo S, bin Kang K, Kim J. and Sung SH, Journal of Natural Products, 2019, 82, 1820–1830. [DOI] [PubMed] [Google Scholar]
- 50.Zhu G, Hou C, Yuan W, Wang Z, Zhang J, Jiang L, Karthik L, Li B, Ren B, Lv K, Lu W, Cong Z, Dai H, Hsiang T, Zhang L. and Liu X, Chemical Communications, 2020, 56, 10171–10174. [DOI] [PubMed] [Google Scholar]
- 51.Alcover CF, Bernadat G, Kabran FA, le Pogam P, Leblanc K, Fox Ramos AE, Gallard JF, Mouray E, Grellier P, Poupon E. and Beniddir MA, Journal of Natural Products, 2020, 83, 1207–1216. [DOI] [PubMed] [Google Scholar]
- 52.Li Y, Yu HB, Zhang Y, Leao T, Glukhov E, Pierce ML, Zhang C, Kim H, Mao HH, Fang F, Cottrell GW, Murray TF, Gerwick L, Guan H. and Gerwick WH, Journal of Natural Products, 2020, 83, 617–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang F, Zhao M, Braun DR, Ericksen SS, Piotrowski JS, Nelson J, Peng J, Ananiev GE, Chanana S, Barns K, Fossen J, Sanchez H, Chevrette MG, Guzei IA, Zhao C, Guo L, Tang W, Currie CR, Rajski SR, Audhya A, Andes DR and Bugni TS, Science, 2020, 370, 974–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crüsemann M, O’Neill EC, Larson CB, Melnik A. v., Floros DJ, da Silva RR, Jensen PR, Dorrestein PC and Moore BS, Journal of Natural Products, 2017, 80, 588–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tangerina MMP, Furtado LC, Leite VMB, Bauermeister A, Velasco-Alzate K, Jimenez PC, Garrido LM, Padilla G, Lopes NP, Costa-Lotufo L. v. and Pena Ferreira MJ, PLoS ONE, 2021, 15, e0244385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zdouc MM, Iorio M, Maffioli SI, Crüsemann M, Donadio S. and Sosio M, Journal of Natural Products, 2021, 84, 204–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pham HT, Lee KH, Jeong E, Woo S, Yu J, Kim W-Y, Lim YW, Kim KH and bin Kang K, Journal of Natural Products, 2021, 84, 298–309. [DOI] [PubMed] [Google Scholar]
- 58.de Felício R, Ballone P, Bazzano CF, Alves LFG, Sigrist R, Infante GP, Niero H, Rodrigues-Costa F, Fernandes AZN, Tonon LAC, Paradela LS, Costa RKE, Dias SMG, Dessen A, Telles GP, da Silva MAC, Lima A. O. de S. and Trivella DBB, Metabolites, 2021, 11, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heine D, Holmes NA, Worsley SF, Santos ACA, Innocent TM, Scherlach K, Patrick EH, Yu DW, Murrell JC, Vieria PC, Boomsma JJ, Hertweck C, Hutchings MI and Wilkinson B, Nature Communications, 2018, 9, 2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peters K, Treutler H, Döll S, Kindt ASD, Hankemeier T. and Neumann S, Metabolites, 2019, 9, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ernst M, bin Kang K, Caraballo-Rodríguez AM, Nothias LF, Wandy J, Chen C, Wang M, Rogers S, Medema MH, Dorrestein PC and van der Hooft JJJJ, Metabolites, 2019, 9, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jarmusch SA, Lagos-Susaeta D, Diab E, Salazar O, Asenjo JA, Ebel R. and Jaspars M, Molecular Omics, 2021, 17, 95–107. [DOI] [PubMed] [Google Scholar]
- 63.Christian N, Sedio BE, Florez-Buitrago X, Ramírez-Camejo LA, Rojas EI, Mejía LC, Palmedo S, Rose A, Schroeder JW and Herre EA, American Journal of Botany, 2020, 107, 219–228. [DOI] [PubMed] [Google Scholar]
- 64.Gdaniec BG, Allard PM, Queiroz EF, Wolfender JL, van Delden C. and Köhler T, Environmental Microbiology, 2020, 22, 3572–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCall LI, Callewaert C, Zhu Q, Song SJ, Bouslimani A, Minich JJ, Ernst M, Ruiz-Calderon JF, Cavallin H, Pereira HS, Novoselac A, Hernandez J, Rios R, Branch OLH, Blaser MJ, Paulino LC, Dorrestein PC, Knight R. and Dominguez-Bello MG, Nature Microbiology, 2020, 5, 108–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sedio BE, Devaney JL, Pullen J, Parker GG, Wright SJ and Parker JD, Ecology and Evolution, 2020, 10, 8770–8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Defossez E, Pitteloud C, Descombes P, Glauser G, Allard PM, Walker TWN, Fernandez-Conradi P, Wolfender JL, Pellissier L. and Rasmann S, Proceedings of the National Academy of Sciences of the United States of America, 2021, 118, e2013344118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Caesar L, Kellogg JJ, v Kvalheim O, Cech R. and Cech NB, Planta Medica, 2018, 84, 721–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Silva E, da Graça JP, Porto C, Martin do Prado R, Hoffmann-Campo CB, Meyer MC, de Oliveira Nunes E. and Pilau EJ, Scientific Reports, 2020, 10, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hautbergue T, Jamin EL, Costantino R, Tadrist S, Meneghetti L, Tabet JC, Debrauwer L, Oswald IP and Puel O, Analytical Chemistry, 2019, 91, 12191–12202. [DOI] [PubMed] [Google Scholar]
- 71.Xu S, Wang JJ, Wei Y, Deng WW, Wan X, Bao GH, Xie Z, Ling TJ and Ning J, Journal of Agricultural and Food Chemistry, 2019, 67, 12084–12093. [DOI] [PubMed] [Google Scholar]
- 72.Raheem DJ, Tawfike AF, Abdelmohsen UR, Edrada-Ebel RA and Fitzsimmons-Thoss V, Scientific Reports, 2019, 9, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gao YL, Wang YJ, Chung HH, Chen KC, Shen TL and Hsu CC, Rapid Communications in Mass Spectrometry, 2020, 34, ee8549. [DOI] [PubMed] [Google Scholar]
- 74.Houriet J, Allard PM, Queiroz EF, Marcourt L, Gaudry A, Vallin L, Li S, Lin Y, Wang R, Kuchta K. and Wolfender JL, Frontiers in Pharmacology, 2020, 11, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Buedenbender L, Astone FA and Tasdemir D, Marine Drugs, 2020, 18, 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kazandjian TD, Petras D, Robinson SD, van Thiel J, Greene HW, Arbuckle K, Barlow A, Carter DA, Wouters RM, Whiteley G, Wagstaff SC, Arias AS, Albulescu L-O, Plettenberg Laing A, Hall C, Heap A, Penrhyn-Lowe S, v McCabe C, Ainsworth S, da Silva RR, Dorrestein PC, Richardson MK, Gutiérrez JM, Calvete JJ, Harrison RA, Vetter I, Undheim EAB, Wüster W. and Casewell NR, Science, 2021, 371, 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Petras D, Minich JJ, Cancelada LB, Torres RR, Kunselman E, Wang M, White ME, Allen EE, Prather KA, Aluwihare LI and Dorrestein PC, Chemosphere, 2021, 271, 129450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Soldatou S, Eldjárn GH, Ramsay A, van der Hooft JJJ, Hughes AH, Rogers S. and Duncan KR, Marine drugs, 2021, 19, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hughes AH, Magot F, Tawfike AF, Rad-Menéndez C, Thomas N, Young LC, Stucchi L, Carettoni D, Stanley MS, Edrada-Ebel R. and Duncan KR, Microorganisms, 2021, 9, 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Leão T, Wang M, Moss N, da Silva R, Sanders J, Nurk S, Gurevich A, Humphrey G, Reher R, Zhu Q, Belda-Ferre P, Glukhov E, Whitner S, Alexander KL, Rex R, Pevzner P, Dorrestein PC, Knight R, Bandeira N, Gerwick WH and Gerwick L, Marine Drugs, 2021, 19, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nothias LF, Petras D, Schmid R, Dührkop K, Rainer J, Sarvepalli A, Protsyuk I, Ernst M, Tsugawa H, Fleischauer M, Aicheler F, Aksenov AA, Alka O, Allard PM, Barsch A, Cachet X, Caraballo-Rodriguez AM, da Silva RR, Dang T, Garg N, Gauglitz JM, Gurevich A, Isaac G, Jarmusch AK, Kameník Z, bin Kang K, Kessler N, Koester I, Korf A, le Gouellec A, Ludwig M, Martin H C, McCall LI, McSayles J, Meyer SW, Mohimani H, Morsy M, Moyne O, Neumann S, Neuweger H, Nguyen NH, Nothias-Esposito M, Paolini J, Phelan V. v., Pluskal T, Quinn RA, Rogers S, Shrestha B, Tripathi A, van der Hooft JJJ, Vargas F, Weldon KC, Witting M, Yang H, Zhang Z, Zubeil F, Kohlbacher O, Böcker S, Alexandrov T, Bandeira N, Wang M. and Dorrestein PC, Nature Methods, 2020, 17, 905–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nothias LF, Nothias-Esposito M, da Silva R, Wang M, Protsyuk I, Zhang Z, Sarvepalli A, Leyssen P, Touboul D, Costa J, Paolini J, Alexandrov T, Litaudon M. and Dorrestein PC, Journal of Natural Products, 2018, 81, 758–767. [DOI] [PubMed] [Google Scholar]
- 83.Kellogg JJ, Kvalheim OM and Cech NB, Analytica Chimica Acta, 2020, 1095, 38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stuart KA, Welsh K, Walker MC and Edrada-Ebel RA, Expert Opinion on Drug Discovery, 2020, 15, 499–522. [DOI] [PubMed] [Google Scholar]
- 85.Tsugawa H, Cajka T, Kind T, Ma Y, Higgins B, Ikeda K, Kanazawa M, Vandergheynst J, Fiehn O. and Arita M, Nature Methods, 2015, 12, 523–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, Wishart DS and Xia J, Nucleic Acids Research, 2018, 46, W486–W494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Aksenov AA, da Silva R, Knight R, Lopes NP and Dorrestein PC, Nature Reviews Chemistry, 2017, 1, 0054. [Google Scholar]
- 88.McAlpine JB, Chen SN, Kutateladze A, Macmillan JB, Appendino G, Barison A, Beniddir MA, Biavatti MW, Bluml S, Boufridi A, Butler MS, Capon RJ, Choi YH, Coppage D, Crews P, Crimmins MT, Csete M, Dewapriya P, Egan JM, Garson MJ, Genta-Jouve G, Gerwick WH, Gross H, Harper MK, Hermanto P, Hook JM, Hunter L, Jeannerat D, Ji NY, Johnson TA, Kingston DGI, Koshino H, Lee HW, Lewin G, Li J, Linington RG, Liu M, McPhail KL, Molinski TF, Moore BS, Nam JW, Neupane RP, Niemitz M, Nuzillard JM, Oberlies NH, Ocampos FMM, Pan G, Quinn RJ, Reddy DS, Renault JH, Rivera-Chávez J, Robien W, Saunders CM, Schmidt TJ, Seger C, Shen B, Steinbeck C, Stuppner H, Sturm S, Taglialatela-Scafati O, Tantillo DJ, Verpoorte R, Wang BG, Williams CM, Williams PG, Wist J, Yue JM, Zhang C, Xu Z, Simmler C, Lankin DC, Bisson J. and Pauli GF, Natural Product Reports, 2019, 36, 35–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tautenhahn R, Patti GJ, Rinehart D. and Siuzdak G, Analytical Chemistry, 2012, 84, 5035–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. https://zenodo.org/.
- 91. https://figshare.com/.
- 92.Montenegro-Burke JR, Guijas C. and Siuzdak G, in Methods in Molecular Biology, 2020, vol. 2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Azquez-Fresno R. v, Sajed T, Johnson D, Li C, Karu N, Sayeeda Z, Lo E, Assempour N, Berjanskii M, Singhal S, Arndt D, Liang Y, Badran H, Grant J, Serra-Cayuela A, Liu Y, Mandal R, Neveu V, Pon A, Knox C, Wilson M, Manach C. and Scalbert A, Nucleic Acids Research, 2018, 46, D608–D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alexandrov T, Ovchinnikova K, Palmer A, Kovalev V, Tarasov A, Stuart L, Nigmetzianov R, Fay D, Gaudin M, Lopez CG, Vetter M, Swales J, Bokhart M, Kompauer M, McKenzie J, Rappez L, Velickovic D, Lavigne R, Zhang G, Thinagaran D, Ruhland E, Sans M, Triana S, Sammour DA, Aboulmagd S, Bagger C, Strittmatter N, Rigopoulos A, Gemperline E, Joensen AM, Geier B, Quiason C, Weaver E, Prasad M, Balluff B, Nagornov K, Li L, Linscheid M, Hopf C, Heintz D, Liebeke M, Spengler B, Boughton B, Janfelt C, Sharma K, Pineau C, Anderton C, Ellis S, Becker M, Pánczél J, da Violante G, Muddiman D, Goodwin R, Eberlin L, Takats Z. and Shahidi-Latham S, bioRxiv DOI: 10.1101/539478. [DOI] [Google Scholar]
- 95.Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K, Oda Y, Kakazu Y, Kusano M, Tohge T, Matsuda F, Sawada Y, Hirai MY, Nakanishi H, Ikeda K, Akimoto N, Maoka T, Takahashi H, Ara T, Sakurai N, Suzuki H, Shibata D, Neumann S, Iida T, Tanaka K, Funatsu K, Matsuura F, Soga T, Taguchi R, Saito K. and Nishioka T, Journal of Mass Spectrometry, 2010, 45, 703–714. [DOI] [PubMed] [Google Scholar]
- 96. HighChem LLC, mzCloud.
- 97. https://chemdata.nist.gov/.
- 98. https://massive.ucsd.edu/.
- 99.Fox Ramos AE, Evanno L, Poupon E, Champy P. and Beniddir MA, Natural Product Reports, 2019, 36, 960–980. [DOI] [PubMed] [Google Scholar]
- 100.Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B, Zaslavsky L, Zhang J. and Bolton EE, Nucleic Acids Research, 2021, 49, D1388–D1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kanehisa M, Furumichi M, Tanabe M, Sato Y. and Morishima K, Nucleic Acids Research, 2017, 45, D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fahy E. and Subramaniam S, Nature Methods, 2020, 17, 1173–1174. [DOI] [PubMed] [Google Scholar]
- 103.Aisporna A, Benton HP, Galano JM, Giera M. and Siuzdak G, bioRxiv DOI: 10.1101/2021.04.02.438066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fiehn O, Robertson D, Griffin J, vab der Werf M, Nikolau B, Morrison N, Sumner LW, Goodacre R, Hardy NW, Taylor C, Fostel J, Kristal B, Kaddurah-Daouk R, Mendes P, van Ommen B, Lindon JC and Sansone SA, Metabolomics, 2007, 3, 175–178. [Google Scholar]
- 105.Goodacre R, Broadhurst D, Smilde AK, Kristal BS, Baker JD, Beger R, Bessant C, Connor S, Capuani G, Craig A, Ebbels T, Kell DB, Manetti C, Newton J, Paternostro G, Somorjai R, Sjöström M, Trygg J. and Wulfert F, Metabolomics, 2007, 3, 231–241. [Google Scholar]
- 106.Hur M, Campbell AA, Almeida-De-Macedo M, Li L, Ransom N, Jose A, Crispin M, Nikolau BJ and Wurtele ES, Natural Product Reports DOI: 10.1039/c3np20111b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Salek RM, Steinbeck C, Viant MR, Goodacre R. and Dunn WB, GigaScience, 2013, 2, 2047–217X–2–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rocca-Serra P, Salek RM, Arita M, Correa E, Dayalan S, Gonzalez-Beltran A, Ebbels T, Goodacre R, Hastings J, Haug K, Koulman A, Nikolski M, Oresic M, Sansone SA, Schober D, Smith J, Steinbeck C, Viant MR and Neumann S, Metabolomics DOI: 10.1007/s11306-015-0879-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Alseekh S, Aharoni A, Brotman Y, Contrepois K, D’Auria J, Ewald J, Ewald JC, Fraser PD, Giavalisco P, Hall RD, Heinemann M, Link H, Luo J, Neumann S, Nielsen J, Perez de Souza L, Saito K, Sauer U, Schroeder FC, Schuster S, Siuzdak G, Skirycz A, Sumner LW, Snyder MP, Tang H, Tohge T, Wang Y, Wen W, Wu S, Xu G, Zamboni N. and Fernie AR, Nature Methods, 2021, 18, 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Altschul SF, Gish W, Miller W, Myers EW and Lipman DJ, Journal of Molecular Biology, 1990, 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 111.Nguyen DD, Melnik A. v., Koyama N, Lu X, Schorn M, Fang J, Aguinaldo K, Lincecum TL, Ghequire MGKK, Carrion VJ, Cheng TL, Duggan BM, Malone JG, Mauchline TH, Sanchez LM, Kilpatrick AM, Raaijmakers JM, de Mot R, Moore BS, Medema MH and Dorrestein PC, Nature Microbiology, 2016, 2, 16197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Olivon F, Apel C, Retailleau P, Allard PM, Wolfender JL, Touboul D, Roussi F, Litaudon M. and Desrat S, Organic Chemistry Frontiers, 2018, 5, 2171–2178. [Google Scholar]
- 113.Tsugawa H, Satoh A, Uchino H, Cajka T, Arita M. and Arita M, Metabolites, 2019, 9, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nguyen TD, Lesani M, Forrest I, Lan Y, Dean DA, Gibaut QMRR, Guo Y, Hossain E, Olvera M, Panlilio H, Parab AR, Wu C, Bernatchez JA, Cichewicz RH and McCall LI, Metabolites, 2020, 10, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Huber F, Ridder L, Verhoeven S, Spaaks JH, Diblen F, Rogers S. and van der Hooft JJJ, PLOS Computational Biology, 2021, 17, e1008724-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huber F, van der Burg S, van der Hooft JJJ and Ridder L, bioRxiv DOI: 10.1101/2021.04.18.440324. [DOI] [Google Scholar]
- 117.Bittremieux W, Laukens K, Noble WS and Dorrestein PC, bioRxiv DOI: 10.1101/2021.02.05.429957. [DOI] [Google Scholar]
- 118.Lybbert AC, Williams JL, Raghuvanshi R, Jones AD and Quinn RA, Metabolites, 2020, 10, 445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mongia M. and Mohimani H, Scientific Reports, 2021, 11, 8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Donia MS and Fischbach MA, Science, 2015, 349, 1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Park DK, Lee KE, Baek CH, Kim IH, Kwon JH, Lee WK, Lee KH, Kim BS, Choi SH and Kim KS, Journal of Bacteriology, 2006, 188, 2214–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bofinger MR, de Sousa LS, Fontes JEN and Marsaioli AJ, ACS Omega, 2017, 2, 1003–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Haffner JJ, Katemauswa M, Kagone TS, Hossain E, Jacobson D, Flores K, Parab AR, Obregon-Tito AJ, Tito RY, Reyes LM, Troncoso-Corzo L, Guija-Poma E, Meda N, Carabin H, Honap TP, Sankaranarayanan K, Lewis CM and McCall L-I, bioRxiv DOI: 10.1101/2021.05.08.442269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lynn DG, Phillips NJ, Hutton WC, Shabanowitz J, Fennell DI and Cole RJ, Journal of the American Chemical Society, 1982, 104, 7319–7322. [Google Scholar]
- 125.Bara R, Aly AH, Wray V, Lin W, Proksch P. and Debbab A, Tetrahedron Letters, 2013, 54, 1686–1689. [Google Scholar]
- 126.Marois JJ, Fravel DR and Papavizas GC, Soil Biology and Biochemistry, 1984, 16, 387–390. [Google Scholar]
- 127.Ernst M, Nothias LF, van der Hooft JJJ, Silva RR, Saslis-Lagoudakis CH, Grace OM, Martinez-Swatson K, Hassemer G, Funez LA, Simonsen HT, Medema MH, Staerk D, Nilsson N, Lovato P, Dorrestein PC and Rønsted N, Frontiers in Plant Science, 2019, 10, 846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dührkop K, Nothias LF, Fleischauer M, Reher R, Ludwig M, Hoffmann MA, Petras D, Gerwick WH, Rousu J, Dorrestein PC and Böcker S, Nature Biotechnology, 2020, 39, 462–471. [DOI] [PubMed] [Google Scholar]
- 129.Zani CL, Marston A, Hamburger M. and Hostettmann K, Phytochemistry, 1993, 34, 89–95. [Google Scholar]
- 130.Uemura D. and Hirata Y, Bulletin of the Chemical Society of Japan, 1977, 50, 2005–2009. [Google Scholar]
- 131.Marco JA, Sanz-Cervera JF, Yuste A, Jakupovic J. and Lex J, Journal of Organic Chemistry, 1996, 61, 1707–1709. [DOI] [PubMed] [Google Scholar]
- 132.Gericke O, Fowler RM, Heskes AM, Bayly MJ, Semple SJ, Ndi CP, Stærk D, Løland CJ, Murphy DJ, Buirchell BJ and Møller BL, bioRxiv, 2020, 2020.11.02.364471. [Google Scholar]
- 133.Schorn MA, Verhoeven S, Ridder L, Huber F, Acharya DD, Aksenov AA, Aleti G, Moghaddam JA, Aron AT, Aziz S, Bauermeister A, Bauman KD, Baunach M, Beemelmanns C, Beman JM, Berlanga-Clavero MV, Blacutt AA, Bode HB, Boullie A, Brejnrod A, Bugni TS, Calteau A, Cao L, Carrión VJ, Castelo-Branco R, Chanana S, Chase AB, Chevrette MG, v Costa-Lotufo L, Crawford JM, Currie CR, Cuypers B, Dang T, de Rond T, Demko AM, Dittmann E, Du C, Drozd C, Dujardin J-C, Dutton RJ, Edlund A, Fewer DP, Garg N, Gauglitz JM, Gentry EC, Gerwick L, Glukhov E, Gross H, Gugger M, Guillén Matus DG, Helfrich EJN, Hempel B-F, Hur J-S, Iorio M, Jensen PR, bin Kang K, Kaysser L, Kelleher NL, Kim CS, Kim KH, Koester I, König GM, Leao T, Lee SR, Lee Y-Y, Li X, Little JC, Maloney KN, Männle D, Martin H C, McAvoy AC, Metcalf WW, Mohimani H, Molina-Santiago C, Moore BS, Mullowney MW, Muskat M, Nothias L-F, O’Neill EC, Parkinson EI, Petras D, Piel J, Pierce EC, Pires K, Reher R, Romero D, Roper MC, Rust M, Saad H, Saenz C, Sanchez LM, Sørensen SJ, Sosio M, Süssmuth RD, Sweeney D, Tahlan K, Thomson RJ, Tobias NJ, Trindade-Silva AE, van Wezel GP, Wang M, Weldon KC, Zhang F, Ziemert N, Duncan KR, Crüsemann M, Rogers S, Dorrestein PC, Medema MH and van der Hooft JJJ, Nature Chemical Biology, 2021, 17, 363–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.van der Hooft JJJ, Mohimani H, Bauermeister A, Dorrestein PC, Duncan KR and Medema MH, Chemical Society Reviews, 2020, 49, 3297–3314. [DOI] [PubMed] [Google Scholar]
- 135.Louwen JJR and van der Hooft JJJ, mSystems, 2021, 6, e00726–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hjörleifsson Eldjárn G, Ramsay A, van der Hooft JJJ, Duncan KR, Soldatou S, Rousu J, Daly R, Wandy J. and Rogers S, PLOS Computational Biology, 2021, 17, e1008920-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rutz A, Dounoue-Kubo M, Ollivier S, Bisson J, Bagheri M, Saesong T, Ebrahimi SN, Ingkaninan K, Wolfender JL and Allard PM, Frontiers in Plant Science, 2019, 10, 1329. [DOI] [PMC free article] [PubMed] [Google Scholar]