Abstract
The ATPase ISWI can be considered the catalytic core of several multiprotein nucleosome remodeling machines. Alone or in the context of nucleosome remodeling factor, the chromatin accessibility complex (CHRAC), or ACF, ISWI catalyzes a number of ATP-dependent transitions of chromatin structure that are currently best explained by its ability to induce nucleosome sliding. In addition, ISWI can function as a nucleosome spacing factor during chromatin assembly, where it will trigger the ordering of newly assembled nucleosomes into regular arrays. Both nucleosome remodeling and nucleosome spacing reactions are mechanistically unexplained. As a step toward defining the interaction of ISWI with its substrate during nucleosome remodeling and chromatin assembly we generated a set of nucleosomes lacking individual histone N termini from recombinant histones. We found the conserved N termini (the N-terminal tails) of histone H4 essential to stimulate ISWI ATPase activity, in contrast to other histone tails. Remarkably, the H4 N terminus, but none of the other tails, was critical for CHRAC-induced nucleosome sliding and for the generation of regularity in nucleosomal arrays by ISWI. Direct nucleosome binding studies did not reflect a dependence on the H4 tail for ISWI-nucleosome interactions. We conclude that the H4 tail is critically required for nucleosome remodeling and spacing at a step subsequent to interaction with the substrate.
The assembly of eukaryotic genomes into chromatin is a highly complex and delicate task; the cell must efficiently package and condense the DNA into the eukaryotic nucleus while maintaining specific regions of accessible chromatin to enable important functions with chromatin substrates. While the chromatin structure must remain highly dynamic in order to accommodate changes in the expression of some genes, it also serves to stably maintain the functional states of other genes through epigenetic mechanisms (45, 51). Recently, genetic and biochemical analyses have identified a broad class of multisubunit chromatin remodeling complexes which are likely to play important roles both in the process of chromatin opening and in the maintenance of chromatin in a dynamic or flexible state (26, 48, 53). These complexes remodel or reorganize nucleosomes in a wide range of in vitro assays which test for altered accessibility of nucleosomal DNA.
Nucleosome remodeling complexes are modular entities. The nucleosome remodeling reaction is catalyzed by a dedicated ATPase in concert with only a subset of associated subunits (24, 37) or even by the ATPase subunit alone (6). This catalytic core, or engine, of the remodeling complexes is associated with other subunits, which are likely to contribute regulatory or targeting roles (6, 24, 37). Interestingly, some imitation switch (ISWI)-containing nucleosome remodeling complexes are also involved in the assembly of regular nucleosomal arrays (23, 47). This suggests that the processes of chromatin assembly and nucleosome remodeling may involve a common nucleosomal intermediate which is stabilized by ISWI-containing factors, thereby facilitating the interconversion of different chromatin configurations.
Chromatin remodeling complexes can be divided into several broad classes according to their core ATPase subunit, all of which belong to the superfamily of SNF2-type ATPases (10). The yeast SWI-SNF and RSC (for remodels the structure of chromatin) complexes and related machineries in Drosophila melanogaster and mammals are driven by SNF2-SWI2-type ATPases (36). The chromo-helicase–ATPase–DNA binding group (52) contains several related ATPases, most prominently the Mi-2 proteins that drive nucleosome remodeling reactions of the so-called NuRD or NRD complexes (for nucleosome remodeling and deacetylation) (41, 54, 55). Another family of complexes contains the ISWI ATPase, including the Drosophila nucleosome remodeling factor (NURF), the ATP-utilizing chromatin assembly and remodeling factor (ACF), and the chromatin accessibility complex (CHRAC) (23, 43, 47), and related chromatin remodeling complexes have been identified in organisms ranging from yeast to humans (28, 44).
Nucleosome remodeling ATPases share little sequence similarity beyond the ATPase domain that originally defined the family. Even the ATPase domains, the most related parts of these proteins, cannot substitute for each other (11). These differences also seem to be reflected in the general mechanism as well as the molecular details of chromatin remodeling by the three classes of complexes. While all types of complexes are able to induce nucleosome sliding on DNA (4, 16, 18, 27, 49), stable remodeled intermediates (30, 39) and nucleosome displacement in trans (nucleosome eviction) (31) have so far been described only for SWI2-SNF2-type complexes. Nucleosome interaction studies, ATPase assays, and quantitative nucleosome remodeling have been employed to identify similarities and differences between various nucleosome remodeling machines, and distinct requirements for free DNA and histone N termini for substrate recognition have been noted (3, 4, 16).
Ab initio, consideration of the process of nucleosome remodeling suggested that the histone N-terminal tails constitute obvious contact points on the surface of the nucleosome that could interact with chromatin remodeling complexes. These extensions protrude from the otherwise rather compact particle and reach out beyond the nucleosome to contact sites in the adjacent chromatin or nonhistone regulators. They are involved in a variety of important processes as diverse as gene activation and silencing, nucleosome positioning, and the folding of the nucleosomal fiber (for reviews, see references 12, 15, and 19). While Mi-2-containing complexes have little or no requirement for histone tails (3, 4), ISWI activity depends on the integrity of these structures (6, 14). Nucleosome remodeling by the SWI2-SNF2 complex does not absolutely require the tail domains (3, 17), although these are necessary for the catalytic remodeling activity of the SWI-SNF complex, possibly by playing a role in the release of the complex from remodeled nucleosomes (29).
It is likely that a requirement for histone tails for nucleosome remodeling reflects the underlying mechanism. However, the four histone tails are not equivalent, and rather they participate in different global functions (19). Up to now the influence of histone tails could be demonstrated only by simultaneous tryptic removal of all eight tails, precluding an assessment of the importance of each individual structure. However, recently Luger et al. developed a system for the expression of histones in bacteria and the reconstitution of recombinant histone octamers (33, 34). Following these pioneering efforts, we generated a range of altered nucleosomes from recombinant histones which lack defined, individual histone N termini. We demonstrate that the histone H4 tails are essential to activate the ATPase activity of ISWI but that the other histone N-terminal tails are not required. Without the H4 tails ISWI is unable to induce nucleosome regularity, and CHRAC is unable to catalyze nucleosome sliding. Since ISWI interacts equally well with nucleosomes from which individual tails have been deleted, our data argue for the involvement of the H4 tail at a step subsequent to the interaction of ISWI with the nucleosomal substrate.
MATERIALS AND METHODS
Expression and refolding of Xenopus laevis histones into octamers.
The expression, purification, and refolding of histones were performed essentially as described in Luger et al. (33, 34), with a single modification to the purification scheme. Briefly, full-length and tailless Xenopus histones (gH4, Δ1-19; gH3, Δ1-26; gH2A, Δ1-12; gH2B, Δ1-26) were expressed in pET-3a vectors in BL21 pLysS cells, and inclusion bodies were prepared as described. These were subsequently dissolved in unfolding buffer (guanidine-HCl), and the debris was spun down, but the resultant supernatant was directly dialyzed three times into 1 liter of SAU-200 (7 M urea, 20 mM sodium acetate [pH 5.2], 200 mM sodium chloride, 5 mM β-mercaptoethanol, 1 mM EDTA). The histones were subsequently loaded onto an SP Sepharose FF column, eluted with SAU-600 (7 M urea, 20 mM sodium acetate [pH 5.2], 600 mM sodium chloride, 5 mM β-mercaptoethanol, 1 mM EDTA), pooled, and dialyzed into water as described previously (34). The refolding of histone octamers and their subsequent purification from misfolded histone aggregates and H3-H4 tetramers by gel filtration were also performed as described above. The histone preparations were not contaminated with bacterial protein to any significant extent.
ATPase assay.
ATPase assays were performed under ISWI remodeling conditions, essentially as previously described (6). Briefly, recombinant yNAP-1 containing an N-terminal His tag was expressed in Escherichia coli and purified using Ni-nitrilotriacetic acid agarose columns (Qiagen). Assays contained 300 ng of plasmid DNA, 450 ng of histones, as specified, and 35 ng of yNAP-1 in the presence of 150 mM ATP and 0.7 μCi of [γ-32P]ATP (3,000 Ci/mmol) and were incubated at 26°C. Unhydrolyzed ATP and free phosphate were separated after 1 h, within the linear range of ATP hydrolysis, by thin-layer chromatography using thin-layer chromatography cellulose Ready-Foils (Scheicher & Schüll). Spots were quantified by FluoroImager and Aida software.
Nucleosome assembly, mobility assay, and bandshift assay.
Mononucleosomes were reconstituted on a 248-bp DNA fragment representing sequences between −232 and +16 relative to the mouse ribosomal DNA transcription site (+1) (27). This DNA fragment was synthesized by PCR and body labeled by incorporating [α-32P]dATP during PCR. Nucleosome assembly by salt gradient dialysis was performed in the lid of siliconized Eppendorf tubes (38). A typical assembly reaction mixture (100 μl) contained 300 to 500 ng of histones, 500 ng of DNA, and 400 ng of bovine serum albumin in HI salt buffer (10 mM Tris-HCl [pH 7.6], 2 M NaCl, 1 mM EDTA, 1 mM β-mercaptoethanol, 0.05% Nonidet P-40). The salt concentration was continuously reduced to 50 mM NaCl during 16 to 24 h. The efficiency of nucleosome assembly was monitored by electrophoretic mobility shift assay (EMSA) in a 5% polyacrylamide gel in 0.5× Tris-borate-EDTA. Positioned nucleosomes were isolated for the mobility assay as described previously (4, 27). The mobility assay contained 60 fmol of positioned nucleosomes, which were incubated with 0.5 to 3 fmol of CHRAC or 3 to 6 fmol of ISWI for 90 min at room temperature.
For the EMSA, 5 to 75 fmol of ISWI was incubated with 50 fmol of nucleosomes for 5 min at room temperature, and the reaction was directly loaded onto a 5% polyacrylamide gel in 0.5× Tris-borate-EDTA without addition of competitor nucleosomes.
Chromatin assembly, MNase digestion, and supercoiling analysis.
Chromatin assembly with yNAP-1 was performed under the conditions of the ATPase assays but without labeled ATP. Reactions were incubated at 26°C for 4 h. For micrococcal nuclease (MNase) digestion, 300 ng of chromatinized DNA was digested for 20, 50, and 110 s, deproteinized with 50 μg of proteinase K at 55°C for 1 h, and precipitated prior to separation in a 1.3% Tris-glycine agarose gel. For supercoiling analysis, 300 ng of chromatinized DNA was purified in the same way as the MNase samples and was separated in a 1.3% Tris-glycine agarose gel containing 3.3 μM chloroquine, which was run in Tris-glycine buffer containing 2.8 μM chloroquine at 80 V for 10 h. For separation in the second dimension, the gel was then turned 90° and rerun in buffer containing 3.6 μM chloroquine at 80 V for 7 h. The bands were subsequently visualized with ethidium bromide.
RESULTS
The histone H4 tails are essential for CHRAC-induced nucleosome sliding.
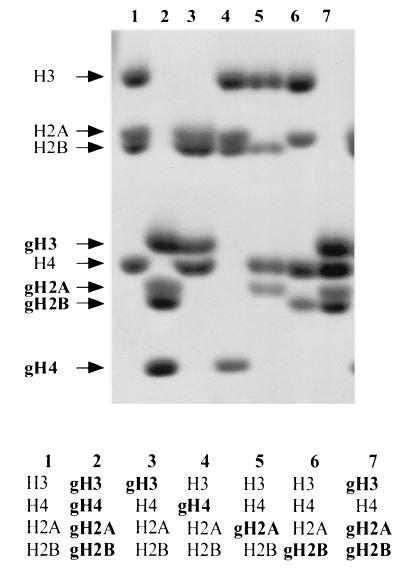
It was recently reported that CHRAC is able to catalyze an energy-dependent sliding of intact histone octamers on a small DNA fragment (27). In order to establish whether any specific histone N terminus was required for induced nucleosome sliding, we adapted the techniques of Luger et al. (33, 34) to express and refold full-length Xenopus histones or histones lacking the trypsin-sensitive N termini into a variety of histone octamers in which one or several of the histone tails were deleted. In short, full-length or tailless histones were individually expressed in bacteria, purified under denaturing conditions, mixed in the appropriate stoichiometry, and refolded by slow removal of the denaturant. Properly folded histone octamers were separated from aggregates and subnucleosomal assemblies by gel filtration. Figure 1 shows the protein composition of various hybrid nucleosomes. In this and in the following figures, truncated histones lacking N termini are indicated with the prefix “g” (for globular, in accordance with the nomenclature by Luger et al. [33]), although the trypsin-sensitive C-terminal tails of H2A and H2B are still present (e.g., gH4 indicates histone H4 lacking the N terminus).
FIG. 1.
Generation of histone octamers. Histone octamers were reconstituted from appropriate combinations of full-length and tailless Xenopus histones (as indicated below the figure), purified by gel filtration, and separated in a 15% sodium dodecyl sulfate gel which was stained with Coomassie blue. The mobility of the histones is indicated on the left of the figure. Tailless histones, of which only the globular part contributes to the nucleosome, are indicated with the prefix “g” (e.g., gH4 indicates a tailless H4).
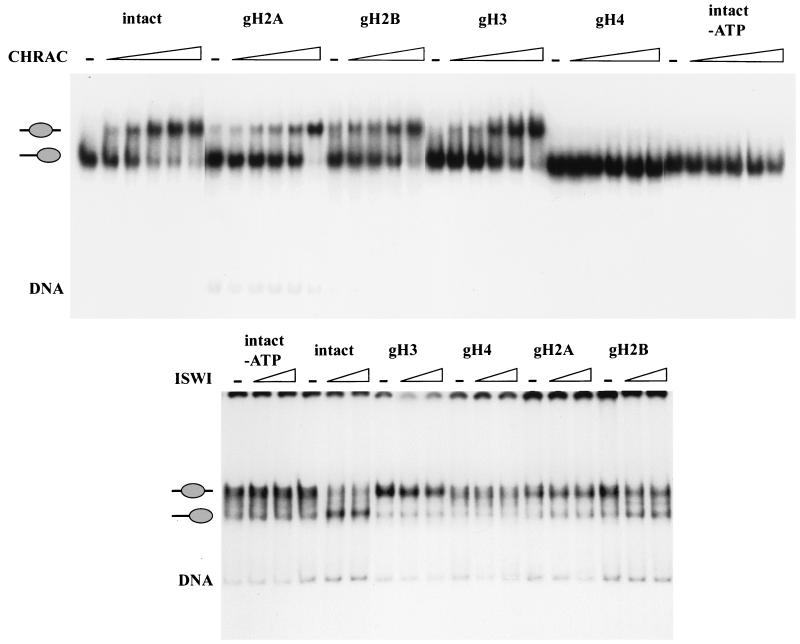
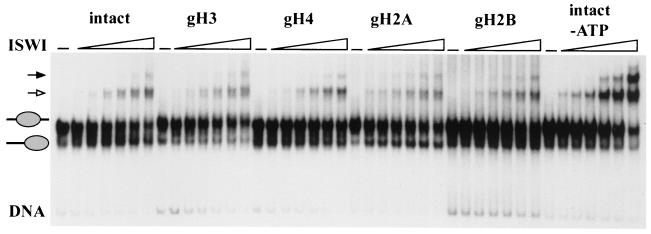
Intact nucleosomes or nucleosomes lacking individual histone tails were reconstituted by salt gradient dialysis from appropriate histone mixtures on a 248-bp DNA fragment. When the products of the reconstitutions were analyzed on a native gel, the two protein-DNA complexes diagnostic for the two previously characterized translational positions (27) were obtained, although the ratio between the two positions varied depending on the tail complement of the histone octamer (data not shown). Under these conditions, centrally located nucleosomes migrate more slowly, and nucleosomes which abut the two opposite ends comigrate as a single faster mobility band in the gel. Previous studies showed that CHRAC could mobilize purified translationally positioned nucleosomes from fast-migrating end positions to slow-migrating central positions (27). Nucleosomes abutting the fragment ends were purified and analyzed for CHRAC-dependent mobilization (Fig. 2, top). Nucleosomes composed of all four intact histones were induced to slide from the peripheral to the central position as a function of CHRAC concentration (Fig. 2 top, intact). In the absence of ATP, no nucleosome sliding occurred (Fig. 2 top, intact, −ATP). This is consistent with previous results (27) and confirms that the refolded octamers could effectively substitute for native octamers. Subsequent experiments examined the effect of CHRAC on nucleosomes in which the histone tails had been individually removed. CHRAC was able to induce nucleosome sliding if the nucleosomal substrate lacked either the histone H3, H2A, or H2B tails, although less efficiently since higher concentrations of CHRAC were required (Fig. 2, top). However, deletion of the H4 tails completely abolished nucleosome sliding (Fig. 2, top, gH4), highlighting the importance of this domain for nucleosome mobilization.
FIG. 2.
The H4 tail is required for CHRAC-mediated nucleosome sliding. (Top) End-positioned nucleosomes reconstituted from histone octamers either containing all four full-length histones (intact) or with one tail missing (e.g., the H3 tail in the gH3 sample) were incubated with increasing concentrations of CHRAC, in which the CHRAC-to-nucleosome ratio was from 1:120 to 1:20. All reactions contained ATP, except for the one presented in the last panel. The reaction mixture was separated by native polyacrylamide gel electrophoresis. The position of traces of free DNA is indicated (DNA). (Bottom) Centrally positioned nucleosomes were mobilized with ISWI, and the ISWI-to-nucleosome ratio was from 1:20 to 1:10. Nucleosome sliding was analyzed as described in the legend for the top panel.
The molecular engine that drives the ability of CHRAC to induce nucleosome sliding is the ATPase ISWI. We previously observed that recombinant ISWI is able to mobilize nucleosomes, however, with altered directionality compared to that of CHRAC. ISWI can induce nucleosome sliding only from a central to a peripheral position (6). In order to assess the tail dependence of nucleosome sliding induced by recombinant ISWI, the centrally positioned nucleosome was purified and used as the starting material for the sliding assay. Nucleosome sliding by ISWI was abolished if the H4 N terminus was deleted (Fig. 2, bottom), as was seen for CHRAC. However, in contrast to the results obtained with purified CHRAC, recombinant ISWI was more sensitive to the deletion of the H3 and H2A N termini. Possible reasons for the observed difference in the tail requirement between recombinant ISWI and ISWI in the context of CHRAC are considered in the Discussion.
The histone H4 tail is necessary and sufficient to induce ISWI ATPase activity.
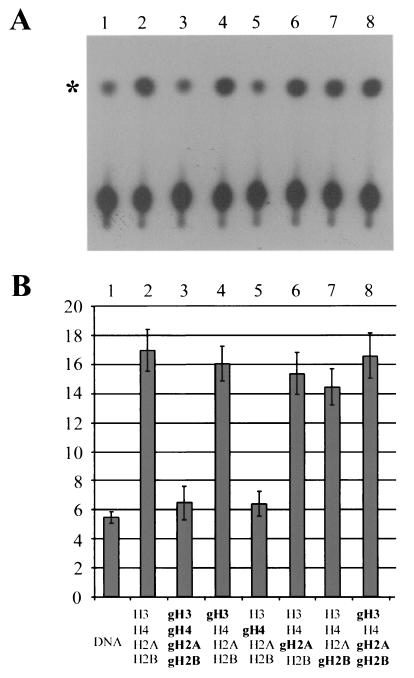
The low level of basal ATPase activity of ISWI is stimulated by the presence of properly folded nucleosomes but not by free histones (6). Measuring ATPase activity therefore allows us to assess nucleosomal features that affect the substrate recognition by the enzyme. In order to gain further insight into the role of individual histone tails for ISWI activity, we compared the ATPase activity of ISWI under conditions of nucleosome assembly (see below) with that of reactions containing only DNA but no histones, thus establishing the degree of nucleosome-dependent stimulation. Whereas the degree of stimulation by DNA alone was variable, considerable stimulation by intact nucleosomes was consistently observed (Fig. 3, panels 1 and 2). In contrast, nucleosomes in which all of the tails had been removed failed to stimulate the ATPase above the basal level seen with DNA alone (Fig. 3, panel 3). We subsequently assessed the contribution of individual histone tails on ATP hydrolysis by programming the assembly with histone octamers from which specific tails were absent (Fig. 3, panels 4 to 7). This showed that the removal of the histone H4 tails (Fig. 3, panel 5) reduced ATPase activity to the level seen with deletion of all the tails, whereas individual removal of the H2A, H2B, or H3 tails had no apparent effect on the amount of ATP hydrolyzed. This result clearly established a requirement for substrate recognition by ISWI. In order to examine whether the other histone N termini contributed to ISWI ATPase activity, we reconstituted a histone octamer in which only the H4 tail was present, the remaining three histone tails being removed. Remarkably, this octamer induced a level of ATP hydrolysis equivalent to that of fully intact octamers (Fig. 3, compare panels 2 and 8), establishing that the ISWI ATPase responds solely to the histone H4 tail. This result highlights the critical role of the H4 N terminus for ATPase function.
FIG. 3.
ISWI ATPase is activated by the histone H4 tail. (A) ATPase assays were performed under conditions where ISWI generates regular nucleosome ladders and contained DNA, the recombinant histones indicated, and purified yNAP-1 and ISWI. The asterisk indicates the signal derived from free phosphate during the 1-h incubation. (B) Quantitation of ISWI ATPase as described for panel A in the presence of either DNA alone (lane 1) or the indicated histone octamers. The ATPase activity is displayed as the percentage of ATP hydrolyzed during the assay. The bars represent the average of three independent experiments, and the variability is indicated by the error bars.
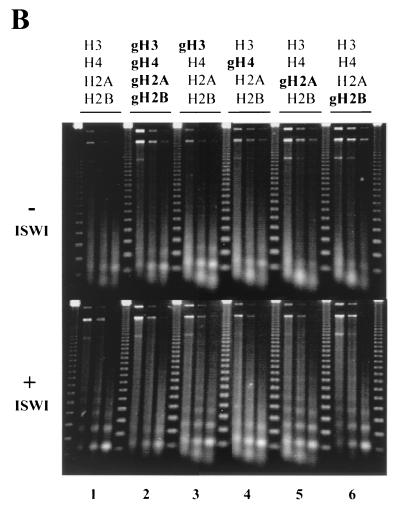
A requirement for the H4 tails for the generation of regular nucleosomal arrays.
ISWI can also function as a chromatin assembly factor in the context of both CHRAC and ACF (6, 24, 44). The establishment of regular nucleosomal arrays by ISWI can be analyzed in a minimal nucleosome assembly reaction consisting of histones, DNA, topoisomerase I, and a histone chaperone, NAP-1. Briefly, NAP-1-mediated transfer of histone octamers onto DNA generates a polynucleosomal fiber that lacks obvious regularity when analyzed by MNase cleavage (see below). In the presence of ATP, ISWI induces regular spacing of nucleosomes in this array, which can be deduced from the appearance of a ladder of fragments above the general smear upon MNase digestion (6, 23). The regularity of the array could be brought about by the repositioning of nucleosomes as a result of ISWI-induced nucleosome sliding (6), in which case we would expect to see an effect from deleting the H4, H3, and H2A tails. However, the ATPase activity of ISWI under assembly conditions is solely affected by deletion of the H4 tail (Fig. 3). We therefore examined the tail dependence of the spacing activity of ISWI. If refolded histones (H2A-H2B dimers and H3-H4 tetramers at this ionic strength) are introduced into the minimal chromatin assembly reaction, long, regular nucleosomal arrays are created in the presence of ATP (Fig. 4B) (6).
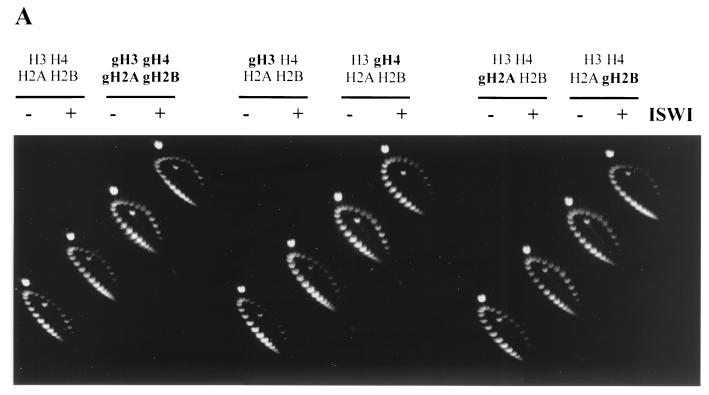
FIG. 4.
The H4 tail is required for ISWI to generate regular chromatin. (A) Two-dimensional supercoiling assay. Nucleosomes were reconstituted from the indicated mixtures of recombinant histones on circular plasmids using NAP-1 as a chaperone in the absence (−) or presence (+) of ISWI, and the resulting superhelicity was relaxed with topoisomerase I. The topoisomer distribution in the purified DNA was visualized by two-dimensional gel electrophoresis. (B) MNase digestion. Histone octamers of the indicated type were assembled into chromatin in a NAP-1 chromatin assembly system and subjected to MNase digestion. The resulting DNA fragments were purified and visualized by agarose gel electrophoresis and ethidium bromide staining. ISWI-generated regularity of nucleosomal arrays can be evaluated from a comparison of the patterns without (− ISWI) and with (+ ISWI) ISWI. The marker is a 123-bp DNA ladder.
A critical parameter in this comparative analysis is the extent of nucleosome deposition, which can be conveniently assessed by monitoring the topology of the resulting minichromosomes. The assembly of a nucleosome by the winding of DNA around a histone octamer introduces one negative superhelical turn into the plasmid in the presence of topoisomerase. Parallel reactions were programmed with histone octamers, where either all histones were intact, all histones had their N-terminal tails deleted, or mixtures of histones had individual tails deleted. Measuring the superhelical density of the deproteinized DNA by two-dimensional gel electrophoresis allowed us to verify that chromatin assembly with the various histone mixtures led to equivalent nucleosomal densities (Fig. 4A). This showed that the deletion of individual histone tails, or indeed all tails, did not affect the efficiency of NAP-1-mediated nucleosome deposition, whether or not ISWI was present. Addition of ISWI correlated with a small increase (2 to 3) in the number of supercoils constrained. ISWI-dependent generation of regularity was monitored under conditions of equivalent nucleosome density by MNase digestion (6). In the absence of ISWI (Fig. 4B, upper panel) MNase digestion generated a continuum (smear) of fragments, indicating the random arrangement of histone octamers on the plasmid independent of the presence of histone tails. Adding ISWI to a reaction containing intact histones generated a regular fragment pattern upon MNase digestion, indicating that the deposited nucleosomes had formed an ordered array (Fig. 4B, bottom panel 1). This result is consistent with earlier observations using native Drosophila histones, indicating that recombinant histones can substitute for native histones in this assay. In contrast, assembly of histone octamers in which all the tails had been removed did not yield a regular nucleosome ladder in the presence of ISWI (Fig. 4B, panel 2). Deletion of individual tails affected regularity to a different extent. Deletion of the H4 tails prevented the establishment of regularity (Fig. 4B, panel 4), while deletion of each of the other tails had a much less severe impact (compare the upper and lower panels of Fig. 4B for each case). Nucleosomes containing just the H4 tails gave rise to better regularity than nucleosomes from which all the tails had been deleted, although the regularity of chromatin reconstituted from intact histone was not reached (data not shown). These results further support our notion that the N terminus of histone H4 is particularly critical for ISWI function. The distinct requirements of the other tails for nucleosome sliding and nucleosome spacing may reflect true mechanistic differences or the particularities of the assays.
The H4 tail is required at a step subsequent to substrate binding.
H4 tails may be required for the interaction of ISWI with its nucleosomal substrate. We recently established an EMSA to analyze the binding of ISWI to nucleosomes. ISWI-nucleosome interactions can be visualized if small DNA segments protrude beyond the realm of the histone octamer but not with nucleosomes reconstituted onto 147-bp fragments where all DNA is likely to be in contact with histone (4). ISWI retards nucleosomes with protruding linker DNA upon gel electrophoresis to give rise to two bands of lower mobility, presumably representing defined species containing one and two molecules of ISWI (Fig. 5). Individual deletion of each of the histone N termini, including, significantly, the histone H4 tails, did not affect the ability of ISWI to interact with the nucleosome qualitatively or quantitatively in this assay, whether or not ATP was included in the reaction (Fig. 5 and data not shown). While this experiment does not rule out a more transient interaction of the H4 tail with ISWI, it argues against a critical role for the H4 tail in the stable interaction of ISWI with the nucleosomal substrate.
FIG. 5.
Recombinant ISWI binds to nucleosomes. ISWI (5 to 75 fmol) was incubated with mononucleosomes reconstituted with either four wild-type recombinant histones (intact) or three wild-type histones and one lacking the N-terminal tail (e.g., gH3 for globular H3) on a 248-bp radioactively labeled DNA fragment. Resulting complexes were separated by native polyacrylamide gel electrophoresis and visualized by autoradiography. Nucleosome-ISWI complexes are marked by arrows.
DISCUSSION
A critical role for the H4 tail for ISWI functions.
The nucleosome remodeling ATPase ISWI has been implicated in two prominent reactions in chromatin, the establishment of an ordered nucleosomal array during the process of chromatin assembly and the generation of access to specific DNA sequences once a nucleosomal array has been established by facilitating nucleosome relocation. Our study highlights the importance of the first 20 N-terminal amino acids on H4 for both ISWI functions. Perhaps surprisingly, none of the other histone tails contributes to the activation of the ISWI ATPase, although their deletion has additional effects depending on the type of reaction. Previously, the general importance of the histone N termini for the ATPase activity of the ISWI-containing NURF had been established (14), but this study did not address the role of individual tails. The use of recombinant histone mutants, pioneered by Richmond, Luger, and colleagues (33, 34), allowed us to define the functional interactions of a remodeling machine with its nucleosomal substrate in further detail. Our results point to critical roles of the histone H4 tail for several ISWI functions which have important implications in the context of current knowledge of chromatin structure and regulation (see below).
Whereas nucleosome sliding induced by CHRAC solely required the N termini of H4, ISWI-induced sliding was also sensitive to removal of the H3 and H2A tails (Fig. 2). While this discrepancy could in theory be explained by inherent differences in the experiments (e.g., the inevitable requirement to use either centrally or peripherally positioned nucleosomes as starting material), we recently obtained support for the hypothesis that the association of CHRAC subunits modulates the properties of ISWI. The interaction of recombinant ISWI with recombinant ACF1, a factor associated with ISWI in ACF (24) and CHRAC (Ferrari, Eberharter, Längst, and Becker, unpublished data), alters the tail requirements for nucleosome sliding such that it is no longer sensitive to the deletion of the H3 and H2A N termini. However, like in purified CHRAC, deletion of the H4 tail still abolishes remodeling activity under those circumstances (Ferrari et al., unpublished data). Although the different tail requirements for ISWI-induced nucleosome sliding and spacing might simply reflect differences in experimental setup (e.g., the presence of NAP-1 during nucleosome assembly), the possibility that they reflect mechanistic distinctions remains to be explored.
Multiple functions are integrated at the N terminus of histone H4.
The histone H4 N terminus reaches well beyond the globular core of the nucleosome that is represented in the crystal structure (32) but presumably adopts, at least under certain conditions, an α-helical structure (reviewed in reference 19). The importance of the tail presumably lies in its ability to reach out to contact other proteins, either histones in adjacent nucleosomes or nonhistone proteins that determine the functional state of the nucleosomal fiber (19). The histone tails contribute to the stabilization of the higher order folding of chromatin structure, which is associated with transcriptional repression (13, 20, 42). This is likely to reflect nucleosome-nucleosome contacts via the tails, possibly as visualized in the crystal structure of the nucleosome, where a portion of the H4 tail contacts an exposed surface of the H2A-H2B dimer on an adjacent nucleosome (32). Genetic experiments in yeast also revealed a contribution of the H4 tail to both transcriptional activation and repression (8, 25). These phenomena are presumably mediated by dedicated factors interacting with the H4 N termini. The bromodomain motif, which is found in a number of chromatin-associated proteins, including components of histone acetyltransferases and nucleosome remodeling ATPases, interacts with specific sites on the H3 and H4 N termini in vitro (7, 35, 50). Tail interactions of the heterochromatin proteins SIR3 and SIR4 (21) and TUP1-SSn6 (9, 22) are involved in the silencing of chromosomal domains, but the structural basis for this remains to be elucidated. Interactions of this kind may well affect the ISWI-H4 tail interactions, with consequences for the degree of chromatin fluidity at these sites.
The interaction between ISWI and its substrate, the nucleosome, may be further modulated by posttranslational modification of H4. Recently, the acetylation of conserved lysine residues in the H4 tail has received wider attention (40, 53). In Drosophila, H4 isoforms acetylated at individual lysines are enriched in chromatin with different functional states (46). While acetylation of lysine 12 characterizes the transcriptionally inactive, epicentric heterochromatin, acetylation of lysine 16 is crucial for the global activation of transcription from the male X chromosome (2). We recently showed that acetylation of H4 at lysine 16 by the acetyltransferase MOF causes derepression of transcription from chromatin templates in vitro and in vivo (1). It will be interesting to see whether the modification status of the H4 tail influences the substrate recognition and catalysis of nucleosome sliding by ISWI. Obviously, the H4 N terminus integrates a number of distinct functions. By analyzing more subtle mutations in the H4 N terminus in vitro and in vivo it should be possible to separate the requirements for remodeling by ISWI from other functions.
Implications for possible mechanisms of nucleosome remodeling.
The importance of the histone H4 tail for all aspects of ISWI function in vitro points to significant mechanistic differences to nucleosome remodeling by the Mi-2 ATPase (3, 4, 16), which does not require any of the histone N termini. Since both enzymes are capable of inducing nucleosome sliding, this process may be brought about by different mechanisms (4). The mechanistic principle underlying the H4 tail dependence of ISWI function is unclear at present. A role for H4 tails in substrate recognition by ISWI seems unlikely, since we observed stable complexes of ISWI with nucleosomes lacking H4 tails. Clearly, ISWI does not recognize the histone H4 tails in isolation but requires the context of a nucleosome, since neither refolded histones nor peptides corresponding to the H4 N terminus stimulate ATPase activity (6, 14). We also failed to detect an interaction between ISWI and glutathione S-transferase–H4 tail fusion proteins in standard pull-down assays (5, 21; data not shown).
However, more transient interactions between the H4 N terminus and ISWI may influence a rate-limiting step of the nucleosome remodeling and ATP hydrolysis reactions allosterically or may otherwise be involved in the mechanics of nucleosome sliding. Analyzing the remodeling reaction catalyzed by the SWI-SNF complex, Logie et al. (29) observed that while histone tails were not important for the remodeling step itself, they stimulated the remodeling rate by promoting the turnover of the enzyme-substrate complex. A more transient interaction of histone tails may have been observed in ATPase assays where the addition of histone N-terminal peptides as glutathione S-transferase fusions diminished the ATPase activities of NURF and recombinant ISWI some two- to fourfold (6, 14). However, these analyses did not reveal a specific effect of the H4 tail.
The ability to create variant nucleosome substrates from recombinant subunits should greatly facilitate the elucidation of the sequence of events that leads to nucleosome remodeling and to uncover the relationship between chromatin assembly and nucleosome remodeling.
ACKNOWLEDGMENTS
We thank K. Luger and T. Richmond (ETH, Zurich, Switzerland) for generously providing the Xenopus histone expression plasmids and advice on the procedures involved in expressing and purifying histones.
K.P.N. was supported by EMBO and the Wellcome Trust. We thank M. Mann for making funds available to support C.R.C. and the EMBL International Ph.D. Programme, as well as the Deutsche Forschungsgemeinschaft, for continuing support.
C.R.C. and G. L. contributed equally and should be considered joint first authors.
REFERENCES
- 1.Akhtar A, Becker P B. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential to dosage compensation in Drosophila. Mol Cell. 2000;5:367–375. doi: 10.1016/s1097-2765(00)80431-1. [DOI] [PubMed] [Google Scholar]
- 2.Bone J R, Lavender J, Richman R, Palmer M J, Turner B M, Kuroda M I. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes Dev. 1994;8:96–104. doi: 10.1101/gad.8.1.96. [DOI] [PubMed] [Google Scholar]
- 3.Boyer L A, Logie C, Bonte E, Becker P B, Wade P A, Wolffe A P, Wu C, Imbalzano A N, Peterson C L. Functional delineation of three groups of the ATP-dependent family of chromatin remodeling enzymes. J Biol Chem. 2000;275:18864–18870. doi: 10.1074/jbc.M002810200. [DOI] [PubMed] [Google Scholar]
- 4.Brehm A, Längst G, Kehle J, Clapier C R, Imhof A, Eberharter A, Müller J, Becker P B. dMi2 and ISWI represent two classes of chromatin remodeling factors characterized by distinct nucleosome binding and mobilization properties. EMBO J. 2000;19:4332–4341. doi: 10.1093/emboj/19.16.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breiling A, Bonte E, Ferrari S, Becker P B, Paro R. The Drosophila polycomb protein interacts with nucleosomal core particles in vitro via its repression domain. Mol Cell Biol. 1999;19:8451–8460. doi: 10.1128/mcb.19.12.8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corona D F V, Längst G, Clapier C R, Bonte E J, Ferrari S, Tamkun J W, Becker P B. ISWI is an ATP-dependent nucleosome remodeling factor. Mol Cell. 1999;3:239–245. doi: 10.1016/s1097-2765(00)80314-7. [DOI] [PubMed] [Google Scholar]
- 7.Dhalluin C, Carlson J E, Zeng L, He C, Aggarwal A K, Zhou M M. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 8.Durrin L K, Mann R K, Kayne P S, Grunstein M. Yeast histone H4 N-terminal sequence is required for promoter activation in vivo. Cell. 1991;65:1023–1031. doi: 10.1016/0092-8674(91)90554-c. [DOI] [PubMed] [Google Scholar]
- 9.Edmondson D G, Smith M M, Roth S Y. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- 10.Eisen J A, Sweder K S, Hanawalt P C. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;14:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elfring L K, Deuring R, McCallum C M, Peterson C L, Tamkun J W. Identification and characterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol Cell Biol. 1994;14:2225–2234. doi: 10.1128/mcb.14.4.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fletcher T M, Hansen J C. Core histone tail domains mediate oligonucleosome folding and nucleosomal DNA organization through distinct molecular mechanisms. J Biol Chem. 1995;270:25359–25362. doi: 10.1074/jbc.270.43.25359. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Ramirez M, Dong F, Ausio J. Role of the histone “tails” in the folding of oligonucleosomes depleted of histone H1. J Biol Chem. 1992;267:19587–19595. [PubMed] [Google Scholar]
- 14.Georgel P T, Tsukiyama T, Wu C. Role of histone tails in nucleosome remodeling by Drosophila NURF. EMBO J. 1997;16:4717–4726. doi: 10.1093/emboj/16.15.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 16.Guschin D, Wade P A, Kikyo N, Wolffe A P. ATP-dependent histone octamer mobilization and histone deacetylation mediated by the Mi-2 chromatin remodeling complex. Biochemistry. 2000;39:5238–5245. doi: 10.1021/bi000421t. [DOI] [PubMed] [Google Scholar]
- 17.Guyon J R, Narlikar G J, Sif S, Kingston R E. Stable remodeling of tailless nucleosomes by the human SWI-SNF complex. Mol Cell Biol. 1999;19:2088–2097. doi: 10.1128/mcb.19.3.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamiche A, Sandaltzopoulos R, Gdula D A, Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97:833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- 19.Hansen J C, Tse C, Wolffe A P. Structure and function of the core histone N-termini: more than meets the eye. Biochemistry. 1998;37:17637–17641. doi: 10.1021/bi982409v. [DOI] [PubMed] [Google Scholar]
- 20.Hansen J C, Wolffe A P. Influence of chromatin folding on transcription initiation and elongation by RNA polymerase III. Biochemistry. 1992;31:7977–7988. doi: 10.1021/bi00149a032. [DOI] [PubMed] [Google Scholar]
- 21.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 22.Huang L, Zhang W, Roth S Y. Amino termini of histones H3 and H4 are required for a1-α2 repression in yeast. Mol Cell Biol. 1997;17:6555–6562. doi: 10.1128/mcb.17.11.6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito T, Bulger M, Pazin M J, Kobayashi R, Kadonaga J T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 24.Ito T, Levenstein M E, Fyodorov D V, Kutach A K, Kobayashi R, Kadonaga J T. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 1999;13:1529–1539. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kayne P S, Kim U J, Han M, Mullen J R, Yoshizaki F, Grunstein M. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell. 1988;55:27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- 26.Kingston R E, Narlikar G J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 27.Längst G, Bonte E J, Corona D F V, Becker P B. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- 28.LeRoy G, Orphanides G, Lane W S, Reinberg D. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science. 1998;282:1900–1904. doi: 10.1126/science.282.5395.1900. [DOI] [PubMed] [Google Scholar]
- 29.Logie C, Tse C, Hansen J C, Peterson C L. The core histone N-terminal domains are required for multiple rounds of catalytic chromatin remodeling by the SWI/SNF and RSC complexes. Biochemistry. 1999;38:2514–2522. doi: 10.1021/bi982109d. [DOI] [PubMed] [Google Scholar]
- 30.Lorch Y, Cairns B R, Zhang M, Kornberg R D. Activated RSC-nucleosome complex and persistently altered form of the nucleosome. Cell. 1998;94:29–34. doi: 10.1016/s0092-8674(00)81218-0. [DOI] [PubMed] [Google Scholar]
- 31.Lorch Y, Zhang M, Kornberg R D. Histone octamer transfer by a chromatin-remodeling complex. Cell. 1999;96:389–392. doi: 10.1016/s0092-8674(00)80551-6. [DOI] [PubMed] [Google Scholar]
- 32.Luger K, Mäder A W, Richmond R K, Sargent D F, Richmond T J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 33.Luger K, Rechsteiner T J, Flaus A J, Waye M M, Richmond T J. Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 34.Luger K, Rechsteiner T J, Richmond T J. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 35.Ornaghi P, Ballario P, Lena A M, Gonzalez A, Filetici P. The bromodomain of Gcn5p interacts in vitro with specific residues in the N terminus of histone H4. J Mol Biol. 1999;287:1–7. doi: 10.1006/jmbi.1999.2577. [DOI] [PubMed] [Google Scholar]
- 36.Peterson C L. Multiple SWItches to turn on chromatin? Curr Opin Genet Dev. 1996;6:171–175. doi: 10.1016/s0959-437x(96)80047-5. [DOI] [PubMed] [Google Scholar]
- 37.Phelan M L, Sif S, Narlikar G J, Kingston R E. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 38.Sandaltzopoulos R, Becker P B. A solid phase approach for the analysis of reconstituted chromatin. In: Becker P B, editor. Chromatin protocols. Totowa, N.J: Humana Press; 1999. pp. 195–206. [DOI] [PubMed] [Google Scholar]
- 39.Schnitzler G, Sif S, Kingston R E. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell. 1998;94:17–27. doi: 10.1016/s0092-8674(00)81217-9. [DOI] [PubMed] [Google Scholar]
- 40.Strahl B D, Allis C D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 41.Tong J K, Hassig C A, Schnitzler G R, Kingston R E, Schreiber S L. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 42.Tse C, Sera T, Wolffe A P, Hansen J C. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol Cell Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsukiyama T, Daniel C, Tamkun J, Wu C. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kD subunit of the nucleosome remodeling factor. Cell. 1995;83:1021–1026. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- 44.Tsukiyama T, Palmer J, Landel C C, Shiloach J, Wu C. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turner B M. Histone acetylation as an epigenetic determinant of long-term transcriptional competence. Cell Mol Life Sci. 1998;54:21–31. doi: 10.1007/s000180050122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turner B M, Birley A J, Lavender J. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell. 1992;69:375–384. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
- 47.Varga-Weisz P D, Wilm M, Bonte E, Dumas K, Mann M, Becker P B. Chromatin remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 48.Vignali M, Hassan A H, Neely K E, Workman J L. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whitehouse I, Flaus A, Cairns B R, White M F, Workman J L, Owen-Hughes T. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400:784–787. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- 50.Winston F, Allis C D. The bromodomain: a chromatin-targeting module? Nat Struct Biol. 1999;6:601–604. doi: 10.1038/10640. [DOI] [PubMed] [Google Scholar]
- 51.Wolffe A P, Matzke M A. Epigenetics: regulation through repression. Science. 1999;286:481–486. doi: 10.1126/science.286.5439.481. [DOI] [PubMed] [Google Scholar]
- 52.Woodage T, Basrai M A, Baxevanis A D, Hieter P, Collins F S. Characterization of the CHD family of proteins. Proc Natl Acad Sci USA. 1997;94:11472–11477. doi: 10.1073/pnas.94.21.11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 54.Xue Y, Wong J, Moreno G T, Young M K, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y, LeRoy G, Seelig H, Lane W S, Reinberg D. The dermatosis-specific autoantigen Mi2 is a component of a protein complex containing deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]