Abstract
Prenatal testosterone (T)-treated female sheep manifest peripheral insulin resistance, ectopic lipid accumulation, and insulin signaling disruption in liver and muscle. This study investigated transcriptional changes and transcriptome signature of prenatal T excess–induced hepatic and muscle-specific metabolic disruptions. Genome-wide coding and noncoding (nc) RNA expression in liver and muscle from 21-month-old prenatal T-treated (T propionate 100 mg intramuscular twice weekly from days 30-90 of gestation; term: 147 days) and control females were compared. Prenatal T (1) induced differential expression of messenger RNAs (mRNAs) in liver (15 down, 17 up) and muscle (66 down, 176 up) (false discovery rate < 0.05, absolute log2 fold change > 0.5); (2) downregulated mitochondrial pathway genes in liver and muscle; (3) downregulated hepatic lipid catabolism and peroxisome proliferator-activated receptor (PPAR) signaling gene pathways; (4) modulated noncoding RNA (ncRNA) metabolic processes gene pathway in muscle; and (5) downregulated 5 uncharacterized long noncoding RNA (lncRNA) in the muscle but no ncRNA changes in the liver. Correlation analysis showed downregulation of lncRNAs LOC114112974 and LOC105607806 was associated with decreased TPK1, and LOC114113790 with increased ZNF470 expression. Orthogonal projections to latent structures discriminant analysis identified mRNAs HADHA and SLC25A45, and microRNAs MIR154A, MIR25, and MIR487B in the liver and ARIH1 and ITCH and miRNAs MIR369, MIR10A, and MIR10B in muscle as potential biomarkers of prenatal T excess. These findings suggest downregulation of mitochondria, lipid catabolism, and PPAR signaling genes in the liver and dysregulation of mitochondrial and ncRNA gene pathways in muscle are contributors of lipotoxic and insulin-resistant hepatic and muscle phenotype. Gestational T excess programming of metabolic dysfunctions involve tissue-specific ncRNA–modulated transcriptional changes.
Keywords: DOHAD, RNA sequencing, ovine, metabolic tissues
Epidemiological data from humans and experimental animal studies have established that exposure to insults (nutrition, stress, disease states, and environmental chemicals) during gestation alters the developmental trajectory and leads to persistent changes in physiology and the development of cardiometabolic disorders during adulthood (1-3). Inappropriate exposure to steroids during gestation can also reprogram the fetal developmental trajectory (4, 5). Because differentiation of cellular tissues and organs during development is tightly regulated by steroids during critical periods of gestation (6), inadvertent exposure to steroids can lead to organizational changes, culminating in long-term metabolic defects in the offspring. Steroid hormonal imbalances during gestation can occur as a result of maternal diseases, such as endocrine disorders (eg, hyperandrogenism) (7).
Gestational exposure to testosterone (T), an aromatizable androgen, programs metabolic disruptions in various animal models, including sheep (5, 8). Prenatal T treatment–induced changes in the female sheep offspring include peripheral insulin resistance leading to compensatory hyperinsulinemia, dyslipidemia, adipocyte defects (9, 10), and hypertension (11). The metabolic dysfunctions are also manifested at the tissue level with insulin resistance in the liver and muscle (12), and ectopic lipid accumulation and oxidative stress in the liver (13). Consistent with the tissue phenotype, prenatal T-treated animals also manifest transcriptional disruptions of insulin signaling pathway genes in the liver and muscle (14).
Prenatal T-treatment additionally causes differences in the gene expression of candidate genes in the insulin target tissues, liver and muscle. For example in the liver, prenatal T excess changed expression of inflammatory cytokines and antioxidants by upregulating tumor necrosis factor-α, the macrophage marker CD68, and superoxide dismutase (SOD) 1 and 2 (13). In muscle, prenatal T excess increased expression of the inflammatory genes chemokine ligand 2, tumor necrosis factor, and the antioxidant genes glutathione reductase and SOD1 (13) while downregulating SOD2, but not CD68, interleukin-6, or interleukin-1B (13). Expansion of these observations to genome-wide profiling of the transcriptome can identify pathways and mechanisms that produce the unique phenotypic outcomes induced by prenatal T.
Reprogramming of offspring trajectories may be linked to changes in epigenetic mechanisms (15-17). One type of epigenetic mechanism regulating gene expression is noncoding RNAs (ncRNAs). ncRNAs are a class of functional RNA molecules that regulate gene expression and based on their length can be classified into small ncRNAs (18-200 nucleotides [nt]) or long noncoding RNAs (> 200 nt). The small ncRNAs include microRNAs (miRNAs, 18-30 nt), small nuclear RNAs (snRNA, ~ 150 nt), and small nucleolar RNAs (snoRNA, 60-170 nt), among others whose dysregulation has been implicated in several diseases (18). The long noncoding RNAs (lncRNAs) are important for cellular differentiation and development (19) and are implicated in insulin resistance and type 2 diabetes (20). Further, ncRNAs like lncRNAs and miRNAs have been implicated in the pathogenesis of polycystic ovary syndrome (PCOS) (21-23), a major infertility disorder characterized by metabolic defects including peripheral insulin resistance (24-26). Because prenatal T-treated sheep emulate the metabolic characteristics of PCOS women (27), they offer a unique opportunity to probe the involvement of ncRNAs in establishing the metabolic phenotype. In support of this premise, aberrant miRNA expression has been shown to modulate metabolic defects in a dihydrotestosterone-induced PCOS phenotype in rats (28). As such, prenatal T-induced transcriptional changes in the liver and muscle may be regulated by changes in the epigenome, specifically differences in ncRNA expression, but this has not yet been tested.
Capitalizing on RNA transcriptomics in the liver and muscle from prenatal T-treated and control female sheep, the objectives of this study were to determine 1) changes in coding RNA and ncRNA; 2) gene pathways affected; 3) coding and ncRNA signatures of prenatal T exposure using predictive component analyses; and 4) putative ncRNA and coding RNA interactions through correlation of ncRNA and coding RNA expression. These findings may provide mechanistic insights into the developmental reprogramming of a hepatic and skeletal muscle insulin-resistant phenotype in this model of endocrine disruption.
Materials and Methods
Animals
All animal studies were conducted under approved protocols of the institutional animal care and use committee of the University of Michigan (approval No. PRO00000609). Experiments in this study used multiparous female Suffolk sheep, housed at the University of Michigan Sheep Research Facility (Ann Arbor, Michigan, USA) and met the National Research Council’s recommendations. Animals from both control and prenatal T-treatment groups coinhabited under similar conditions and were fed a similar maintenance diet to prevent obesity as described previously (29).
Prenatal Testosterone Treatment
Between gestational days 30 and 90, 100-mg T propionate (~ 1.2 mg/kg; Millipore Sigma) suspended in corn oil was administered intramuscularly twice a week to the prenatal T-treated group. Control animals did not receive any vehicle treatment, since our prior studies demonstrated no effects of corn oil in sheep (30). The effects of prenatal T-treatment on peripheral insulin sensitivity, adiposity, and tissue-specific changes in mediators of insulin sensitivity from this cohort have been published previously (10, 12, 13, 31, 32).
Tissue Collection
Metabolic tissues (liver and muscle) were collected during the second breeding season at approximately age 21 months following a 48-hour fast from prenatal T-treated animals and controls. Since prenatal T-treated animals were oligoanovulatory (33), to normalize their background steroid levels, both control and prenatal T-treated animals were ovariectomized, and tissue collection was carried out during the artificially induced follicular-phase steroid milieu, as described previously (34). Briefly, 2 weeks before euthanasia for tissue collection, animals were implanted with two controlled internal drug-releasing implants containing progesterone (CIDR-G; InterAg; implanted subcutaneously) for 14 days. On the fourteenth day, progesterone implants were removed and four 30-mm estradiol-containing Silastic (Dow Chemical Company, Midland, MI) implants were inserted subcutaneously to produce circulating estradiol concentrations in line with the follicular phase. Animals were euthanized 18 hours later during the presumptive late-follicular phase. From all animals, liver tissue was obtained from the tip of the left lobe and skeletal muscle was obtained from the vastus lateralis. Tissues were snap frozen and stored at –80 °C until processing for RNA isolation.
Total RNA Isolation, Library Construction, and Sequencing
Total RNA was extracted from the liver and muscle after homogenizing the tissues in Trizol reagent (Life Technologies) following the manufacturer’s recommendations. Residual contaminating DNA was removed using RNeasy binding column, treating the column with RNAse free DNAse (Qiagen) and eluting the RNA in nuclease-free water. The quality of purified RNA including the purity and RNA integrity were evaluated using an Agilent 2100 bioanalyzer (Agilent Technologies) at the University of Michigan Advanced Genomics Core. Liver from 5 randomly selected control and 4 prenatal T-treated sheep (RNA from 1 prenatal T-treated animal failed RNA integrity number quality standards for library preparation) and muscle from 5 control and 5 prenatal T-treated sheep were used for transcriptomic analysis. Library preparation was carried out using a TruSeq RNA Library Prep Kit (Illumina) and single-end sequenced at 50 cycles with single-read using a HiSeq2000 NextSeq platform.
Noncoding RNA Library Preparation and Sequencing
Libraries for ncRNA were prepared from total RNA using a NEBNext smallRNA kit (New England BioLabs) at the University of Michigan Advanced Genomics Core as per the manufacturer’s recommendations. The small RNA libraries were prepared from total RNA whose RNA integrity was determined using the Agilent 2100 bioanalyzer. Post–library preparation quality control metrics were determined and sequencing performed on an Illumina NextSeq platform. For ncRNA analysis of liver and muscle, 4 animals each from control and prenatal T-treated animals were used.
Total RNA Data Processing and Quality Control
For transcriptomic analysis, single-end reads were obtained from sequencing. An overview of the RNA sequencing (RNA-seq) analysis is summarized as a flowchart (Supplementary Fig. S1) (35). Raw fastq files from total RNA-seq data were trimmed using trimmomatic (36) with the following options: maximum mismatch count, palindrome clip threshold (not used in single-end reads, but must still be specified), and clip threshold score parameters for removing Illumina adapters: 2:30:10. Further, all leading and trailing bases with quality threshold below 3 and reads that dropped below a quality score of 15 while scanning bases with a 4–base pair window were also removed. Quality control metrics including mean read quality scores, duplicated reads, and GC content of sequences of both raw and trimmed files were evaluated using fastqc. The quality control metrics of samples were summarized using MultiQC (37). Trimmed reads from total RNA were further mapped to sheep reference genome (Oar_rambouillet_v1.0) using the Spliced Transcripts Alignment to a Reference (STAR) aligner (v2.6.0c). The quality control metrics of aligned reads were examined using the Quality of RNA-seq Tool-set (QoRTS) (v1.3.6). The reads that aligned to the exons of the reference genome were then counted using featureCounts (v1.6.1). While RNA sequencing of the total RNA led to sequencing of both coding RNA and ncRNA, during analysis emphasis was placed on the coding RNA from this data set as comprehensive sequencing for the ncRNA was carried out separately.
Noncoding RNA Data Processing and Quality Control
The raw fastq files from ncRNA sequencing were 5′ trimmed using cutadapt (v3.2) as per the manufacturer’s recommendation using the adapter sequence AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC. Using the built-in function for cutadapt, reads that were of low quality and less than 17 base pairs were removed from the sequencing data. Quality control metrics were assessed using fastqc and summarized using MultiQC. Similar to that used in total RNA, ncRNA was aligned to sheep reference genome (Oar_rambouillet_v1.0) using STAR. The counting of reads for each class of ncRNA evaluated in this study was achieved by first subsetting the whole genome annotation files for the reference genome into files that contained only references for a particular ncRNA. The aligned reads were then used in featureCounts (v1.6.1) with genes as the identifier, with all multimapping reads included in the analysis, along with annotation files specific for each ncRNA to obtain counts. The sequence of analytical approach is summarized as a flowchart (see Supplementary Fig. S1) (35).
Differential Expression Testing
To identify differentially expressed genes, DESeq2 (v1.24.0), which uses negative binomial distribution on counts, was used. Samples were normalized using default settings. For both total RNA and ncRNA, 2 levels of comparison were performed in this study: 1) comparison of tissues (liver vs muscle) in the control animals and 2) comparison of response to treatment (prenatal T vs control) both in liver and muscle. Differential expression analyses for each class of ncRNA were studied in a workflow similar to that of total RNA. Transcripts were prioritized based on statistical significance measured as false discovery rate–adjusted P values (FDR) and magnitude of association measured as log2fold change (log2FC). Coding RNA meeting FDR less than 0.05 (for differential gene expression analysis) and FDR less than 0.01 (for selecting genes to be included in the correlation analysis) and ncRNA meeting FDR less than 0.1 (for differential expression and correlation analysis), respectively, after multiple testing correction and absolute log2FC greater than 0.5 were considered significant. Finally, differentially expressed transcripts were visualized using volcano plots, and heat maps were generated using the heatmap.2 package. All differential expression testing and plots were processed using R statistical software (v3.5.1).
Data Reduction and Identification of Potential Biomarkers of Prenatal Testosterone Exposure
Data reduction and potential biomarker identification were performed using SIMCA P+ version 15 (Sartorius Stedim Data Analytics AB). Coding and ncRNA (lncRNA, miRNA, snoRNA, and snRNA)-normalized counts from liver and muscle tissues were imported to SIMCA P+ for the multivariate modeling and unit variance scaling (mean centered and divided by SD) was applied. Unsupervised principal component analysis (PCA) was performed to obtain an overview of the data and identify potential outliers and trends/groupings in the transcriptome data. The 2-dimensional (2D) and 3-dimensional (3D) PCA plots were explored to visualize patterns and groupings in the data. A tolerance ellipse based on Hotelling’s T2 (multivariate generalization of t distribution) was used to identify strong outliers outside the ellipses.
To further visualize differences in transcription between the control and prenatal T-treated animals, orthogonal projections to latent structures discriminant analysis or orthogonal partial least square discriminant analysis (OPLS-DA) was performed. OPLS-DA is a supervised technique by which the model is rotated so the variation related to treatment is the first predictive component, and the variation that is not related to treatment is explained in the orthogonal components. In the OPLS-DA models, prenatal T and control status was the outcome variable and the predictor variable matrix was the transcriptome data. All the OPLS-DA models used for potential biomarker selection had R2X values above 0.7 and Q2 values above 0.5. Each score/point on the score plot represents one observation, and clear separation and groupings indicate differences in transcriptome profiles between the groups. Score plots of the first predictive component (variation in the transcriptome data related to outcome) with the first orthogonal component (variation in the transcriptome data not related to outcome) were used. OPLS-DA loadings provide information about the variables responsible for these differences in profiles, with variables away from the origin with smaller CIs serving as better markers of the differences in the groups. Variable importance to projection (VIP) and S plots were investigated to identify potential biomarkers of prenatal T-treatment. Higher VIP value indicates higher importance to the model and responsible for the observed separation of the 2 groups. Additionally, VIP predictive plots were modified for the transcriptome data to add chromosome information for the coding genes. The S plots were plotted where the predictive component loading explaining model covariance is plotted against the model correlation to identify potential biomarkers with high significance, low variability, low FDR, and higher magnitude. Furthermore, OPLS DA model loadings and significance (based on differential expression FC) of these signatures were investigated to identify potential biomarkers of prenatal T-treatment in liver and muscle.
Gene Set Enrichment Analysis
We obtained human orthologs of sheep genes using BioMart (38) (Supplementary Table S1) (35). To test enrichment of pathways for top expressed genes in response to prenatal T-treatment in each tissue, RNA-enrich (39) was used. RNA-enrich uses a P value cutoff free enrichment methodology and genes were tested for enrichment by comparison against 6 databases including BioCarta pathway, EHMN metabolic pathways, Gene Ontology, KEGG pathway, Panther pathway, and transcription factor databases. Pathways with an FDR less than 0.01 were selected for comparison between tissues in response to prenatal T. Odds ratios (ORs) of enrichment are reported. Overlapping and unique findings were visualized using upset plots, built using the UpsetR (40) R package. Gene set enrichment analysis and UpsetR were processed using R statistical software (v3.5.1).
Correlation Between Noncoding RNA and Total RNA
To identify ncRNA–messenger RNA (mRNA) pairs in liver or muscle, ncRNA and genes differentially expressed in response to prenatal T-treatment were selected and matched within their respective samples from the same set of animals. The correlation matrices for control and prenatal T-treatment groups were generated by differential gene correlation analysis (41). The ncRNA-mRNA pairs with Pearson correlation coefficients with P values less than .05 across either control or treatment were considered significantly correlated within that group. To visualize the relationship between ncRNA-mRNA pairs, correlation plots and box plots were generated.
Data are available online at the Gene Expression Omnibus (GEO accession: GSE178777). Code for analysis is available online (https://github.com/bakulskilab).
Quantitative Polymerase Chain Reaction Analysis
Total RNA isolated as described above was reverse transcribed (RT) into complementary DNA using the SuperScrit Vilo cDNA synthesis kit (Invitrogen) for quantitative polymerase chain reaction (qPCR)-based gene expression analysis. A SYBRgreen-based real-time RT-PCR assay was used to determine the gene expression of TPK1 and ZNF470 in muscle and HADHA in liver on a BioRad myiQ iCycler real-time PCR instrument. Oligonucleotide primers for the genes under study were designed using Primer Express software (Life Technologies), and the primer sequences for the primers used are shown in Supplemental Table S2 (35). The relative amount of each transcript was calculated using the ΔΔ CT method and normalized to the endogenous reference gene ribosomal protein L19 (RPL19).
Assessment of Hepatic Ectopic Lipid Accumulation
Lipid accumulation in liver was assessed by Oil Red O staining and triglyceride content measured using a commercially available Wako L-Type TG M kit (Wako Diagnostics) as described previously (13).
Assessment of Hepatic Fibrosis
Collagen deposition as a marker of fibrosis was assessed through picrosirius red staining of hepatic sections and acid-soluble collagen content in tissue lysates using the Sirius Red dye binding (Sircol) assay following the protocol described previously (31).
Statistical Analyses
Data from qPCR, and lipid and collagen deposition between control and prenatal T-treated groups were analyzed by 2-tailed t test using Prism software (version 7.0, GraphPad software). A P value less than .05 was considered significant. Data were also analyzed by Cohen effect size analysis, and effect size with a Cohen d value of 0.8 and above were considered as large-magnitude changes, which reflect the effect size of difference between the control and prenatal T groups.
Results
Descriptive Statistics
Quality control plots generated by MultiQC demonstrated that samples across both mRNA and noncoding RNA after trimming had high mean read quality scores across all base pairs, per sequence GC content ranging from 30% to 70% and had sequence duplication levels ranging between 20% and 60% (Supplementary Figs. S2 and S3) (35).
Tissue-specific Coding RNA Gene Expression in Liver and Muscle
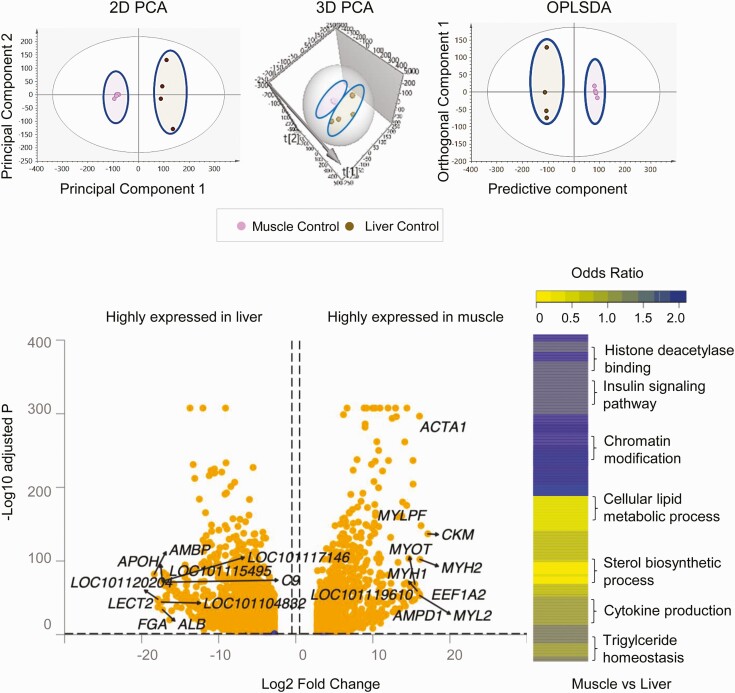
Using an unsupervised PCA model for the control animals, clear separation of liver and muscle transcriptome profiles (coding RNA) (Fig. 1) was observed in PCA 2D and 3D plots, supportive of differences in transcriptome profiles between the 2 tissues. Supervised OPLS DA performed to filter orthogonal to projection unrelated variance in the data improved the model only slightly as the 2 groups were already well defined and separated in the PCA model (see Fig. 1).
Figure 1.
Coding RNA sample clustering and tissue-specific enrichment in control animals. Principal component analysis (PCA) 2-dimensional (2D) and 3-dimensional (3D) score plots and orthogonal partial least square discriminant analysis (OPLS-DA) score plots for the liver and muscle from control animals are shown at the top. For the PCA the 2D and 3D plots are plotted with principal component 1 on the x axis and principal component 2 on the y axis, and for the OPLS-DA score plot with predictive component on the x axis and first orthogonal component on the y axis, showing separation between the liver (brown) and muscle (pink) from control animals. Each point represents one animal. The volcano plot showing differential gene expression comparing liver and muscle in control animals are shown at the bottom left. Genes are plotted by log2 fold change and –log10 adjusted P values. The orange points represent genes that have an absolute log2 fold change greater than 0.5 and false discovery rate (FDR) less than 0.05. Heat map of differentially regulated gene pathways across both liver and muscle at FDR less than 0.01 are shown at the bottom right.
Similarly, gene expression comparison between liver and muscle showed tissue-specific distinct gene expression profiles (see Fig. 1; Supplementary Table S3 (35)). The top 10 differentially expressed genes from the liver and muscle are shown in Table 1. The genes highly expressed in the liver that related to its plasma protein synthesis function include alpha-1-microglobulin/bikunin precursor (AMBP), fibrinogen alpha chain (FGA), apolipoprotein H (APOH), albumin (ALB), and complement 9 (C9). Other genes such as uncharacterized (LOC1011120204), leukocyte cell–derived chemotaxin 2 (LECT2), amine sulfotransferase-like (LOC101104832), serpin A3-7-like (LOC101117146), and C-reactive protein (LOC101115495) were also among the top 10 highly enriched genes in the liver.
Table 1.
The top 10 highly expressed genes in muscle and liver from control female sheep
| Liver | Muscle | ||||
|---|---|---|---|---|---|
| Gene | log2FC | Padj | Gene | log2FC | Padj |
| AMBP | –18.36 | .000 | CKM | 17.13 | .000 |
| LOC101120204 | –17.91 | .000 | MYLPF | 16.26 | .000 |
| FGA | –17.89 | .000 | MYH2 | 16.11 | .000 |
| LECT2 | –17.88 | .000 | MYL2 | 16.08 | .000 |
| LOC101104832 | –17.42 | .000 | ACTA1 | 16.05 | .000 |
| APOH | –17.33 | .000 | EEF1A2 | 16.04 | .000 |
| ALB | –17.29 | .000 | MYH1 | 15.74 | .000 |
| C9 | –17.24 | .000 | MYOT | 15.45 | .000 |
| LOC101117146 | –17.23 | .000 | LOC101119610 | 15.42 | .000 |
| LOC101115495 | –17.16 | .000 | AMPD1 | 15.24 | .000 |
Genes with adjusted P value(Padj) less than .05 comparing muscle vs liver gene expression in control tissues of the prenatal testosterone cohort. Positive values in fold change (FC) denote enriched in muscle and negative values represent downregulation in muscle compared to the liver (enriched in liver).
In muscle, the highly expressed genes were those conferring contractility to the muscle such as myosin light chain, phosphorylatable, fast skeletal muscle, (MYLPF), myosin heavy chain 1 (MYH1), MYH2, and myosin light chain 2 (MYL2) and commonly used markers of skeletal muscle function namely creatine kinase M-type (CKM) and actin alpha 1 (ACTA1). In addition, eukaryotic translation elongation factor 1 alpha 2 (EEF1A2), myotilin (MYOT), myozenin-1 (LOC101119610), and adenosine monophosphate deaminase 1 (AMPD1) were also highly enriched in the muscle tissue.
Analysis of those genes differentially expressed between liver and muscle in control animals revealed genes belonging to 578 gene pathways (FDR < 0.01) (see Fig. 1; Supplementary Table S4) (35). Of these, 293 gene pathways were specifically enriched in the liver and included those related to metabolism of amino acids, lipids, steroids, and organic acids that are in line with its functional role. In the muscle, 285 gene pathways were enriched including myofibrillar, contractile, and skeletal muscle development–related pathways that are key to the skeletal muscle function and development.
Tissue-specific Differences in Noncoding RNA Expression Between Liver and Muscle
Comparison of tissue-specific expression of ncRNAs (lncRNAs, miRNAs, snoRNAs, and snRNAs) between the liver and muscle in control animals are shown in Supplementary Table S5 (35), and the top 10 ncRNAs from each class that are enriched in the liver or muscle are shown in Table 2. Both in liver and muscle, several uncharacterized lncRNA and snoRNAs were enriched among the top 10 genes. Among snRNAs, LOC114110196 (adjusted P = .090) was found to be highly expressed in muscle but not in the liver. The top miRNAs enriched in the liver included MIR200A, MIR194, MIR148A, MIR411A, MIR655, and MIR369, while in the muscle they included MIR133, MIR10B, MIR181A-2, MIR362, MIR150, and MIR30C.
Table 2.
The top 10 genes noncoding RNA differentially expressed between liver and muscle from control female sheep
| Highly expressed in liver | Highly expressed in muscle | ||||
|---|---|---|---|---|---|
| Gene | log2FC | Padj | Gene | log2FC | Padj |
| Long ncRNA | |||||
| LOC114110111 | –6.23 | .000 | LOC105603743 | 11.09 | .000 |
| LOC105603426 | –5.5 | .000 | LOC105604459 | 7.92 | .000 |
| LOC114110424 | –4.61 | .004 | LOC114113790 | 6.18 | .000 |
| LOC114110280 | –4.15 | .02 | LOC114112617 | 5.25 | .01 |
| LOC105616344 | –3.79 | .000 | LOC105604726 | 5 | .006 |
| LOC105608433 | –3.72 | .05 | LOC114109081 | 4.97 | .001 |
| LOC114116879 | –3.7 | .05 | LOC105616014 | 4.85 | .001 |
| LOC114114068 | –3.59 | .000 | LOC114114816 | 4.83 | .003 |
| LOC114116820 | –3.56 | .06 | LOC105613565 | 4.53 | .02 |
| LOC101116687 | –3.42 | .000 | LOC114116861 | 4.48 | .05 |
| MicroRNA | |||||
| MIR200B | –7.77 | .000 | MIR133 | 5.3 | .001 |
| MIR200A | –6.57 | .000 | MIR10B | 5.3 | .000 |
| MIR194 | –4.95 | .000 | MIR181A-2 | 3.26 | .000 |
| MIR369 | –4.4 | .000 | MIR362 | 3.08 | .000 |
| MIR148A | –3.5 | .000 | MIR150 | 2.91 | .000 |
| MIR200C | –3.28 | .000 | MIR3955 | 1.98 | .09 |
| MIR152 | –2.57 | .000 | MIRLET7B | 1.78 | .000 |
| MIR411A | –2.5 | .000 | MIR30D | 1.68 | .000 |
| MIR655 | –2.44 | .003 | MIR30C | 1.65 | .000 |
| MIR410 | –2.28 | .10 | MIR30B | 1.58 | .000 |
| snoRNA | |||||
| LOC114116284 | –4.92 | .000 | LOC114111821 | 4.87 | .000 |
| LOC114117649 | –4.62 | .02 | LOC114115466 | 4.59 | .05 |
| LOC114116761 | –4.2 | .002 | LOC114114231 | 4.31 | .09 |
| LOC114110519 | –4.17 | .000 | LOC114111320 | 3.98 | .000 |
| LOC114110194 | –4.08 | .000 | LOC114116763 | 3.95 | .08 |
| LOC114117064 | –4.03 | .000 | LOC114115071 | 3.34 | .000 |
| LOC114115928 | –3.68 | .000 | LOC114114261 | 3.31 | .000 |
| LOC114111381 | –3.48 | .000 | LOC114114654 | 3.09 | .000 |
| LOC114113382 | –3.48 | .000 | LOC114111371 | 2.77 | .08 |
| LOC114117378 | –3.38 | .06 | LOC114110700 | 2.41 | .07 |
| snRNA | |||||
| LOC114110196 | 2.55 | 0.09 | |||
ncRNA genes with adjusted P value (Padj) less than .1 comparing muscle vs liver gene expression in control tissues of the prenatal testosterone cohort. Positive values in fold change (FC) denote enriched in muscle and negative values represent ncRNA that enriched in liver.
Abbreviations: snoRNA, small nucleolar RNA; snRNA, small nuclear RNA.
Prenatal Testosterone Treatment–induced Gene Expression Changes in Liver and Muscle
Liver
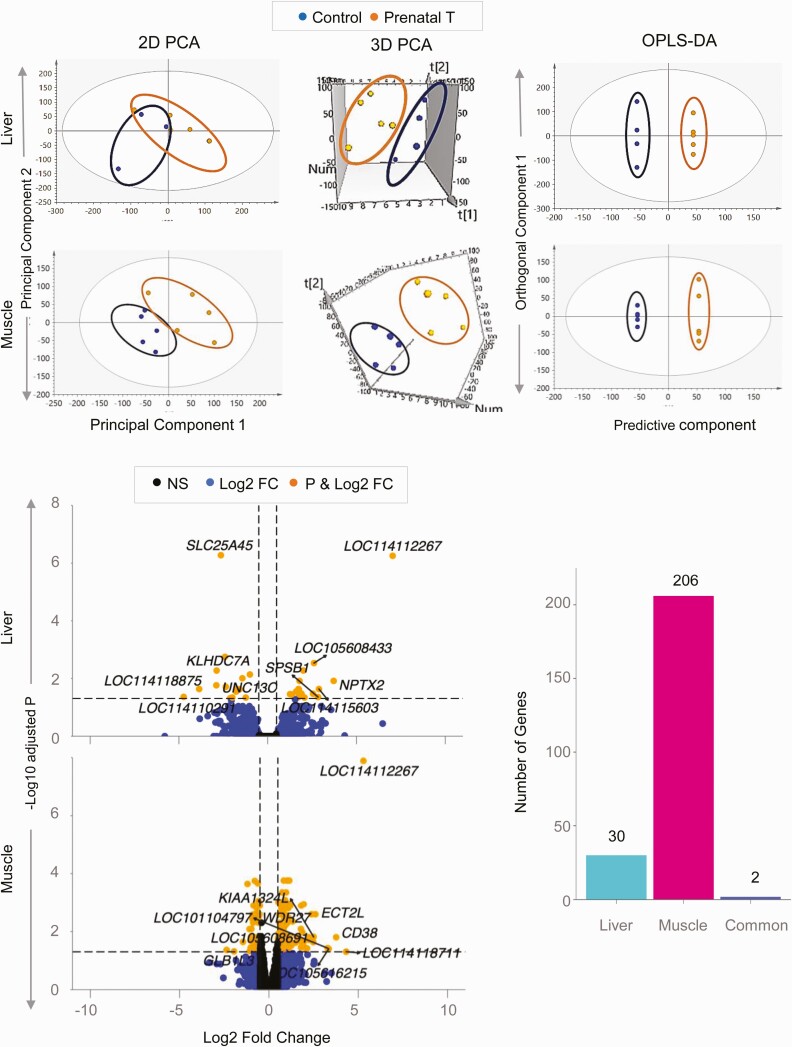
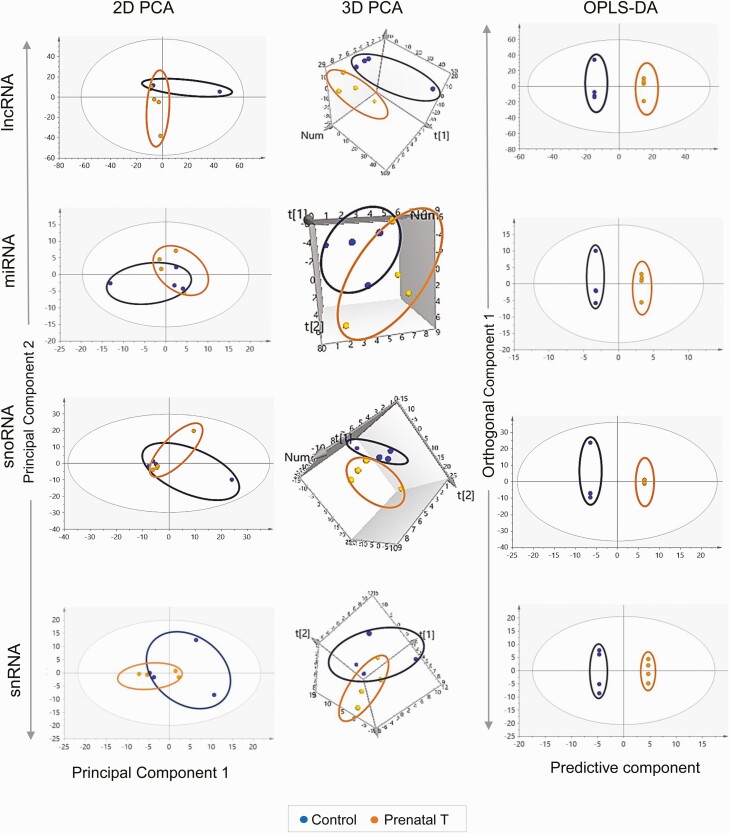
The 2D PCA score plot showed an overlap between the control and prenatal T-treated groups (Fig. 2). Further examination using the 3D PCA score plot showed clear separation between the 2 groups. The OPLS DA that accounted for the variance between the 2 groups also showed clear separation between the control and prenatal T-treated groups (see Fig. 2).
Figure 2.
Coding RNA sample clustering and differential expression in liver and muscle from prenatal testosterone (T)-treated animals. The 2-dimensional (2D) and 3-dimensional (3D) principal component analysis (PCA) and orthogonal partial least square discriminant analysis (OPLS-DA) score plots showing groupings and separation between control (blue) and prenatal T-treated (orange) groups in the coding RNA from liver and muscle tissue are shown at the top. For the PCA the 2D and 3D plots are plotted with principal component 1 on the x axis and principal component 2 on the y axis, and for the OPLS-DA score plot with predictive component on the x axis and first orthogonal component on the y axis with each point representing one animal. The volcano plot showing differential gene expression in the liver or muscle comparing prenatal T-treated animals against control animals are shown at the bottom left. Genes are plotted by log2 fold change on the x axis and –log10 P adjusted values on the y axis. Orange points denote the genes that have an absolute log2 fold change greater than 0.5 and P adjusted values less than .05. Black dots represent genes that did not meet P-adjusted cutoff of less than .05 and absolute log2 fold change greater than .5, and blue dots represent genes that met the absolute log2 fold change greater than 0.5 but did not meet the P-adjusted cutoff of less than .05. The bar plots at the bottom right represent the number of genes differentially modulated that are unique to either liver and muscle or common to both tissues.
Comparison of gene expression in the liver between control vs prenatal T-treated females revealed 32 differentially expressed genes at FDR less than 0.05 and absolute log2FC greater than 0.5, out of which 15 genes were downregulated and 17 genes were upregulated (see Fig. 2; Supplementary Table S6) (35). Among the top 10 dysregulated coding genes filtered by absolute magnitude (Table 3), genes with increased expression included sialomucin core protein 24 (LOC114112267; log2FC = 7.04, FDR = 5.6×10–7), neuronal pentraxin (NPTX2; log2FC = 3.72, FDR = 0.01), splA/ryanodine receptor domain and SOCS box containing 1 (SPSB1; log2FC = 2.84, FDR = 0.04), and uncharacterized genes (LOC114115603; log2FC = 2.89, FDR = 0.02) and (LOC105608433; log2FC = 2.61, FDR = 3.0×10–3). Genes with decreased expression included solute carrier family 25 member 45 (SLC25A45; log2FC = –2.64, FDR = 5.3×10–7), kelch domain containing 7A (KLHDC7A; log2FC = 2.88, FDR = 5.4×10-3), unc-13 homolog C (UNC13C; log2FC = –2.89, FDR = 0.01), immunoglobulin lambda variable 1-40-like (LOC114118875; log2FC = –3.86, FDR = 0.02), and deleted in malignant brain tumors 1 protein-like (LOC114110291; log2FC = –4.73, FDR = 0.04).
Table 3.
The top 10 genes differentially expressed in response to prenatal testosterone treatment in muscle and liver
| Liver | Muscle | ||||
|---|---|---|---|---|---|
| Gene | log2FC | Padj | Gene | log2FC | Padj |
| LOC114112267 | 7.04 | .000 | LOC114112267 | 5.29 | .000 |
| NPTX2 | 3.72 | .01 | LOC114118711 | 4.32 | .05 |
| LOC114115603 | 2.89 | .02 | CD38 | 3.76 | .02 |
| SPSB1 | 2.84 | .04 | LOC105616215 | 3.35 | .04 |
| LOC105608433 | 2.61 | .003 | LOC101104797 | 3.29 | .04 |
| SLC25A45 | –2.64 | .000 | ECT2L | 2.6 | .003 |
| KLHDC7A | –2.88 | .005 | KIAA1324L | 2.5 | .02 |
| UNC13C | –2.89 | .02 | LOC105608691 | 2.48 | .02 |
| LOC114118875 | –3.86 | .02 | WDR27 | 2.38 | .003 |
| LOC114110291 | –4.73 | .04 | GLB1L3 | –2.37 | .04 |
Genes with adjusted P value (Padj) less than .05 and absolute log2 fold change (FC) greater than 0.5. The top 10 differentially expressed genes based on absolute magnitude are listed in the table with their direction of effect.
Muscle
The 2D PCA plot showed clear differences in the coding RNA profiles between control and prenatal T-treated groups (see Fig. 2) with the separation becoming more distinct in the 3D PCA plot and OPLS DA model (see Fig. 2). Prenatal T-treatment altered expression of 208 genes with FDR less than 0.05 and absolute log2FC greater than 0.5 (143 upregulated and 65 downregulated) (see Fig. 2; Supplementary Table S7) (35). Of the top 10 genes sorted by absolute magnitude, 9 genes were upregulated and 1 gene was downregulated (see Table 3). The 9 upregulated genes included sialomucin core protein 24 (LOC114112267; log2FC = 5.29, FDR = 1.30×10–8), FUN14 domain-containing protein 2 (LOC114118711; log2FC = 4.32, FDR = 0.05), CD38 molecule (CD38; log2FC = 3.76, FDR = 0.01), multidrug resistance-associated protein 4-like (LOC105616215; log2FC = 3.35, FDR = 0.04), ubiquitin-like protein FUBI pseudogene (LOC101104797; log2FC = 3.29, FDR = 0.03), epithelial cell transforming 2 (ECT2L; log2FC = 2.60, FDR = 2.6×10–3), endosome-lysosome associated apoptosis and autophagy regulator family member 2 (KIAA1324L; log2FC = 2.00, FDR = 0.01), uncharacterized (LOC105608691; log2FC = 2.48, FDR = 0.02), and WD repeat domain 27 (WDR27; log2FC = 2.38, FDR = 2.6×10-3). The only downregulated gene in the top 10 was galactosidase beta 1 like 3 (GLB1L3; log2FC = –2.37, FDR = 0.04).
Prenatal T-induced dysregulation of genes in the liver and muscle were unique to tissue type barring 2 genes that were dysregulated both in liver and muscle (see Fig. 2). Both genes, uncharacterized gene (LOC114112267; log2FC = 5.28 in muscle and log2FC = 7.03 in liver) and G-protein coupled receptor-associated sorting protein1 (GPRASP1; log2FC = –1.08 in muscle and log2FC = –1.5 in liver) were concordant in magnitude and direction of change.
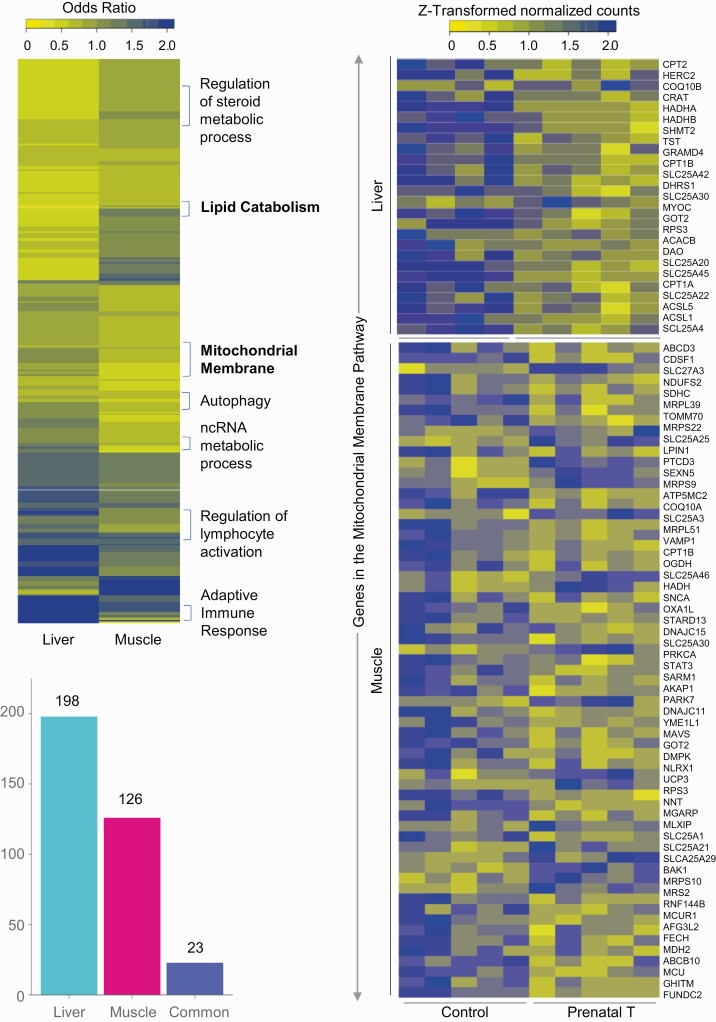
Functional Enrichment of Genes Differentially Affected by Prenatal Testosterone Treatment
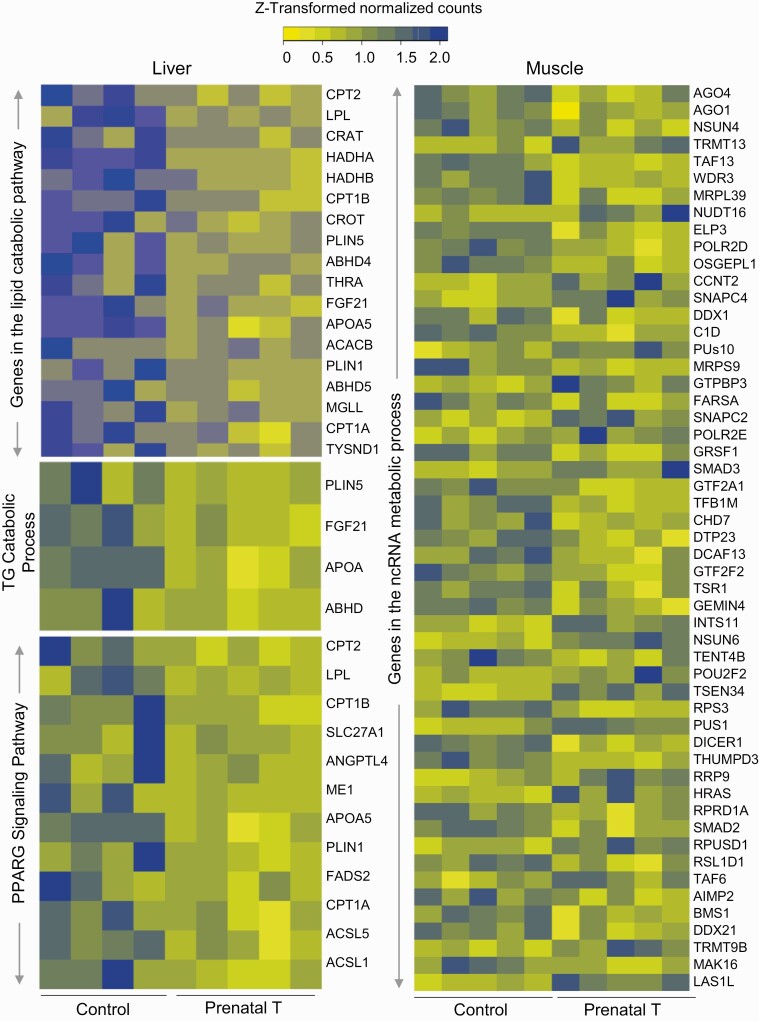
The number of pathways identified at FDR less than 0.01 influenced by prenatal T-treatment were lower in the muscle than in the liver (Fig. 3; Supplementary Table) S8 (35). Prenatal T-treatment induced enrichment of 23 common gene pathways among liver and muscle tissues as opposed to 198 gene pathways specifically enriched in the liver and 126 pathways in the muscle (see Fig. 3). Specifically, pathways related to mitochondrial membrane pathway (liver OR = 0.62, muscle OR = 0.79) were downregulated both in the liver and muscle (see Fig. 3). Among gene pathways that were uniquely enriched in a tissue-specific manner, dysregulation of gene pathways related to lipid catabolic pathway, triglyceride catabolic process, and peroxisome proliferator-activated receptor γ (PPARG) signaling were evident in the liver (Fig. 4), while pathways involved in the ncRNA metabolic process (see Fig. 4) and transcription elongation from RNA polymerase II promoter characterized the muscle (Supplementary Fig. S4) (35) from prenatal T-treated female sheep.
Figure 3.
Functional enrichment of pathways enriched in liver and muscle and expression levels of genes in commonly dysregulated pathways. Heat map (top left) representing the differentially regulated gene pathways in liver and muscle from prenatal testosterone (T)-treated animals compared against control animals and enriched at a false discovery rate (FDR) of less than 0.01. The bar plots (bottom left) represent the number of gene pathways differentially modulated that are unique to either liver and muscle or commonly dysregulated in both tissues. Heat map (right) showing genes involved in the mitochondrial membrane pathway that is dysregulated both in liver (C = 4, T = 5) and muscle (C = 5, T = 5). The genes associated with the gene pathway in controls animals and prenatal T–treated animals are plotted along a gradient of colors, with blue representing the highest and yellow the lowest normalized counts.
Figure 4.
Expression levels of genes in dysregulated pathways enriched in either liver or muscle. Heat map showing genes involved in the lipid and triglyceride catabolism and peroxisome proliferator-activated receptor γ (PPARG) signaling in the liver (left; C = 4, T = 5) and noncoding RNA (ncRNA) metabolic process in the muscle (right; C = 5, T = 5) that are dysregulated in a tissue-specific manner by prenatal testosterone (T) treatment at a false discovery rate of less than 0.01. The genes associated with pathways in each tissue from control and prenatal T-treated animals are represented as a gradient of colors with blue representing the highest and yellow representing the lowest normalized counts.
Coding RNA Transcriptome Signatures of Prenatal Testosterone Treatment in Liver and Muscle
Liver
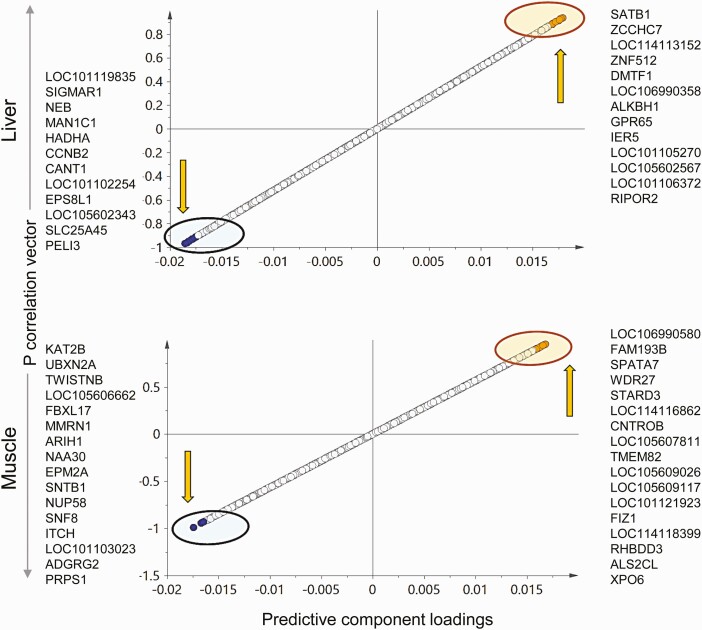
The OPLS DA S plot (Fig. 5) and VIP plot (Fig. 6) for the predictive component and model loadings were used to identify potential biomarkers of prenatal T-treatment in the liver. Twelve genes that were downregulated and 14 genes that were upregulated with the prenatal T-treatment had the potential to serve as biomarkers/transcriptome signatures from the S plot (see Fig. 5). The genes with high VIP values (see Fig. 6) and significance (FC without FDR correction) that showed decreased expression in the liver from prenatal T-treated female sheep included sigma nonopioid intracellular receptor 1 (SIGMAR1), nebulin (NEB), mannosidase alpha class 1C member 1 (MAN1C1), hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha (HADHA), cyclin B2 (CCNB2), calcium-activated nucleotidase 1 (CANT1), and solute carrier family 25 member 45 (SLC25A45I). The genes that showed increased expression include uncharacterized genes LOC114113152, LOC106990358, LOC101105270, and LOC101106372, alkB homolog 1, histone H2A dioxygenase (ALKBH1), G protein-coupled receptor 65 (GPR65), immediate early response 5 (IER5), and RHO family interacting cell polarization regulator 2 (RIPOR2). Some of these showed significant FCs based on the FDR cutoff of less than 0.05 (see Supplementary Table S6) (35): NEB, HADHA, CANT1, SLC25A45, LOC106990358, and LOC101106372. SIGMAR1 gene and other markers identified did not meet the FDR cutoff of less than 0.05 but were very important to the model based on loadings and VIP plots.
Figure 5.
Liver or muscle specific prenatal testosterone (T)-induced transcriptional signatures. The orthogonal partial least square discriminant analysis (OPLS-DA) plots for coding RNA from liver (top) and muscle (bottom) showing potential biomarkers of prenatal T-treatment. The up and down arrow represents the upregulated and downregulated genes with prenatal T-treatment, respectively. Each point represents one RNA with orange and blue points representing the potential biomarkers of prenatal T-treatment that are upregulated and downregulated, respectively.
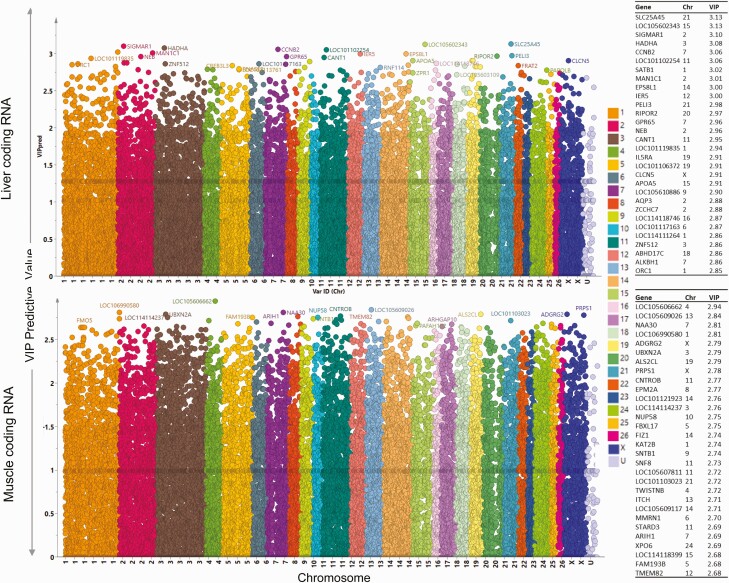
Figure 6.
Variable importance in the projection (VIP) for predictive component plots. Plots showing the VIP for total RNA in liver (top) and muscle (bottom). The plots are plotted with chromosome identification on the x axis (each color representing a chromosome) and VIP values on the y axis. The higher VIP value indicates high importance to the model and is responsible for the separation of prenatal T and control animals on the orthogonal partial least square discriminant analysis (OPLS-DA) score plot. The top 30 genes-based VIP values are listed on the right side of the plots.
Muscle
Both OPLS DA S plot (see Fig. 5) and VIP plot (see Fig. 6) together with model loading were used to identify potential biomarkers of prenatal T-treatment in the muscle. Sixteen genes with lower expression and 18 genes with higher expression had the potential to serve as signatures of prenatal T excess based on the S plot (see Fig. 5). Most of these genes showed significant FC compared to controls at the FDR cutoff of less than 0.05 (see Supplemental Table S7) (35). Among these, prenatal T-treatment downregulated the lysine acetyltransferase 2B (KAT2B), UBX domain protein 2A (UBXN2A), TWIST neighbor (TWISTNB), F-box and leucine rich repeat protein 17 (FBXL17), multimerin 1 (MMRN1), ariadne RBR E3 ubiquitin protein ligase 1 (ARIH1), N-alpha-acetyltransferase 30, NatC catalytic subunit (NAA30), EPM2A glucan phosphatase, laforin (EPM2A), syntrophin beta 1 (SNTB1), nucleoporin 58 (NUP58), SNF8 subunit of ESCRT-II (SNF8), itchy E3 ubiquitin protein ligase (ITCH), phosphoribosyl pyrophosphate synthetase 1 (PRPS1), while upregulating the family with sequence similarity 193 member B (FAM193B), spermatogenesis-associated 7 (SPATA7), WD repeat domain 27 (WDR27), StAR-related lipid transfer domain containing 3 (STARD3), centrobin, centriole duplication and spindle assembly protein (CNTROB), transmembrane protein 82 (TMEM82), FLT3 interacting zinc finger 1 (FIZ1), rhomboid domain containing 3 (RHBDD3), ALS2 C-terminal like (ALS2CL), and uncharacterized genes LOC105609026, LOC114116862, and LOC101121923.
Prenatal Testosterone Treatment Mediated Changes in Noncoding RNA Expression in Liver and Muscle
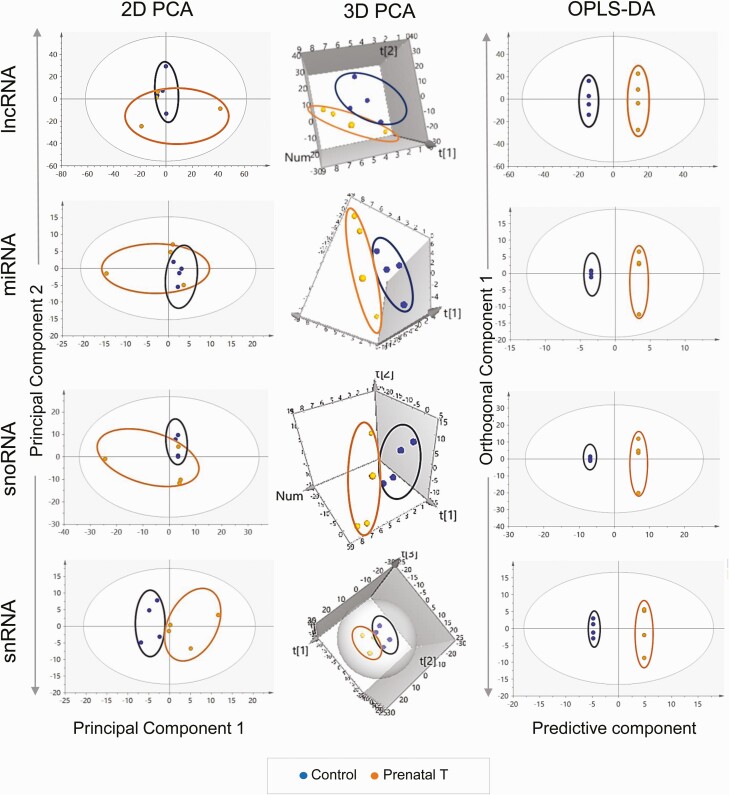
Liver
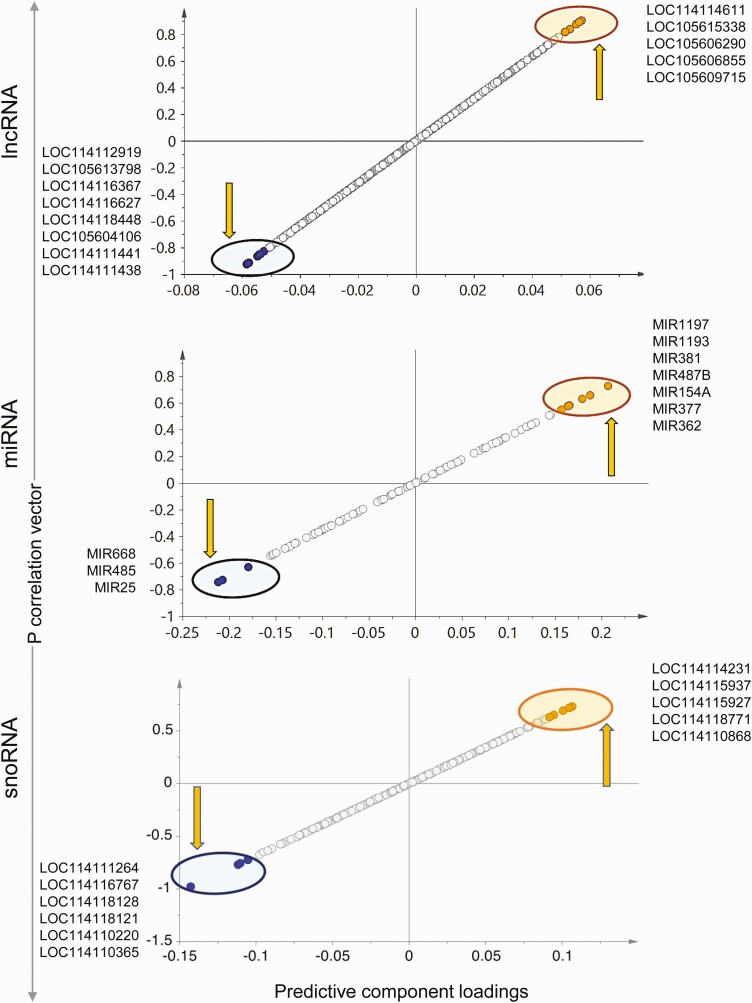
Long noncoding RNA.
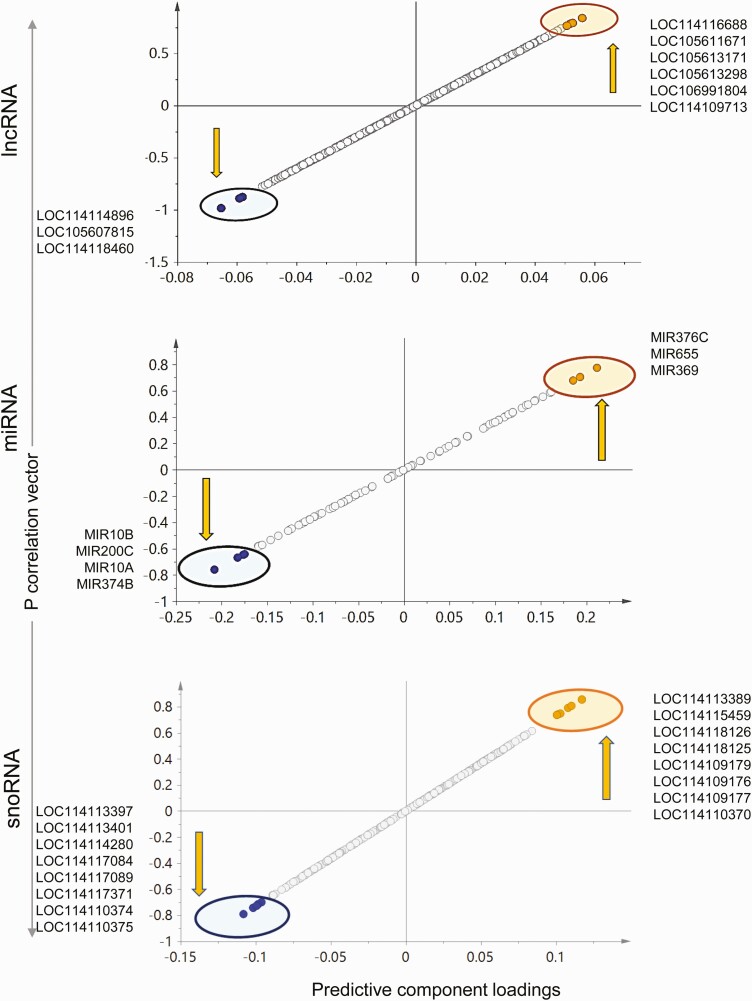
The 2D PCA score plot (Fig. 7) shows an overlap between the control and prenatal T-treated group with separation of the 2 groups improving in the 3D score plot (see Fig. 7). The OPLS DA performed to identify lncRNA potential biomarkers of prenatal T exposure showed 8 lncRNAs that were expressed in lower levels and 5 lncRNAs that were expressed at higher levels in response to prenatal T-treatment, all of which have the potential to serve as signatures of prenatal T exposure based on S plot (Fig. 8), model loadings, and VIP predictive values. These potential markers did not meet the criteria of FDR cutoff of less than 0.1 because of within-group variations (see Supplementary Fig. S5 and Table S9) (35) but LOC114116367 had an unadjusted FC P value of less than .05 (P = .0197), and 3 lncRNAs—LOC105604106, LOC114111441 and LOC114111438—having a P value of less than 0.1 were expressed at lower levels in prenatal T-treated liver.
Figure 7.
Clustering and orthogonal partial least square discriminant analysis (OPLS-DA) plots for noncoding RNA in the liver. The 2-dimensional (2D) and 3-dimensional (3D) principal component analysis (PCA) and OPLS-DA score plots for long noncoding RNA (lncRNA), microRNA (miRNA), small nucleolar RNA (snoRNA), and small nuclear RNA (snRNA) in liver from control (blue) and prenatal testosterone (T)-treated (orange) animals. For the PCA the 2D and 3D plots are plotted with principal component 1 on the x axis and principal component 2 on the y axis, and for the OPLS-DA score plot with predictive component on the x axis and first orthogonal component on the y axis with each point representing one animal.
Figure 8.
Prenatal testosterone (T) treatment–altered liver–specific noncoding RNA (ncRNA) potential biomarkers. The orthogonal partial least square discriminant analysis (OPLS-DA) plots for ncRNA (long noncoding RNA [lncRNA], microRNA [miRNA], and small nucleolar RNA [snoRNA]) from liver showing potential biomarkers of prenatal T-treatment. The up and down arrows represent the upregulated and downregulated genes with prenatal T treatment, respectively. Each point represents a noncoding RNA with orange and blue points representing the potential biomarkers of prenatal T treatment that are upregulated and downregulated, respectively.
Micro RNA.
The 2D PCA score plot showed partial overlap of the 2 groups, and this grouping was not improved in the 3D model. OPLS DA showed 3 miRNAs that were downregulated and 7 miRNAs that were upregulated that had the potential to be used as signatures of prenatal T exposure (Fig. 8). No miRNA qualified for the FC FDR cutoff criteria of less than 0.1 (see Supplementary Fig. S5 and Table S9) (35) and only MIR485 had an FC unadjusted P value of less than .1. Although MIR381 was identified as very important to the model based on loadings and VIP values, the FC P value for this miRNA was less than 0.2 (see Supplementary Table S9) (35).
Small nucleolar RNA.
Overlap of the control animals and prenatal T-treated animals were observed for this class of ncRNA in the 2D PCA score plot (Fig. 7) with separation improving in the 3D PCA score plot where the 2 groups were well separated (see Fig. 7). Supervised OPLS DA score plot also showed clear separation between the control and prenatal T-treated groups (see Fig. 7). Six snoRNAs that were downregulated and 5 snoRNAs that were upregulated with prenatal T-treatment had the potential to serve as signatures of prenatal T-treatment based on S plot (Fig. 8), loadings, and VIP values. No snoRNAs met the criteria of FDR less than 0.1 cutoff (see Supplementary Fig. S5 and Table S9) (35). However, 2 snoRNAs—LOC114115927 and LOC114118771—showed higher expression with prenatal T-treatment with an unadjusted FC P value of less than .1 (see Supplemental Table S9) (35).
Small nuclear RNA.
The PCA 2D score plot showed tighter grouping for prenatal T-treated animals and a large within group variation for control animals (see Fig. 7). Additionally, there was also no clear separation of the 2 groups in the 3D score plot for this PCA model (see Fig. 7). The variation not related to prenatal T-treatment was filtered in the OPLS DA model leading to considerable improvement in the separation of the 2 groups in this model (see Fig. 7). Six snRNAs showing higher expression and 11 snRNAs showing lower expression had the potential to serve as snRNA signatures of prenatal T exposure based on the S plot (see Supplementary Fig. S7) (35); however, no snRNA showing lower expression was confirmed by models loading CIs. Among the ones that were selected and upregulated by prenatal T-treatment, 2 snRNAs (LOC114109175 and LOC114109186) had an FC-adjusted P value of less than .1, but none of the snRNAs met the FDR cutoff of less than 0.1 to serve as potential biomarker (see Supplementary Table S9) (35).
Muscle
Long noncoding RNA.
There were no clear groupings of the control and prenatal T-treated animal profiles in the 2D score plot (Fig. 9), with samples from the prenatal T group showing more within-group variation; however, the two groups showed a clear separation on the 3D PCA plot. The OPLS DA model also showed a clear separation of the 2 groups (see Fig. 9). Three lncRNAs that were downregulated and 6 that were upregulated by prenatal T-treatment had the potential to serve as specific signatures of prenatal T-treatment based on S plot, loadings, and VIP values (Fig. 10). Only one lncRNA—LOC114118460, downregulated with prenatal T-treatment—met the FDR cutoff criteria of less than 0.1 (Supplementary Table S10) (35). The lncRNA LOC105607815 was found to be downregulated but did not reach FC significance but was very influential to the model based on S plot, loadings, and VIP values.
Figure 9.
Clustering and orthogonal partial least square discriminant analysis (OPLS-DA) plots for noncoding RNA (ncRNA) in muscle. The 2-dimensional (2D) and 3-dimensional (3D) principal component analysis (PCA) and OPLS-DA score plots for long noncoding RNA (lncRNA), microRNA (miRNA), small nucleolar RNA (snoRNA), and small nuclear RNA (snRNA) in muscle from control (blue) and prenatal testosterone (T)-treated (orange) animals. For the PCA the 2D and 3D plots are plotted with principal component 1 on the x axis and principal component 2 on the y axis, and for the OPLS-DA score plot with predictive component on the x axis and first orthogonal component on the y axis with each point representing one animal.
Figure 10.
Prenatal testosterone (T) treatment–altered muscle-specific noncoding RNA (ncRNA) potential biomarkers. The orthogonal partial least square discriminant analysis (OPLS-DA) S plots for ncRNA (long noncoding RNA [lncRNA], microRNA [miRNA], and small nucleolar RNA [snoRNA]) from muscle showing potential biomarkers of prenatal T treatment. The up and down arrows represent the upregulated and downregulated genes with prenatal T treatment, respectively. Each point represents an ncRNA with orange and blue points representing the potential biomarkers of prenatal T treatment that are upregulated and downregulated, respectively.
Only 5 lncRNAs showed differential expression and met the P-adjusted criteria of less than .1 in the muscle (see Supplemental Figure S6 and Table S10) (35). All of the 5 differentially expressed lncRNAs were downregulated by prenatal T-treatment (Table 4) and included uncharacterized noncoding RNA genes (LOC105607806; log2FC = –2.63, Padj = 2.14E-02, LOC114112617; log2FC = –2.22, Padj = 8.13E-04, LOC114112974; log2FC = –1.72, Padj = 3.65E-02, LOC114113790; log2FC = –2.21, Padj = 8.98E-04, and LOC114118460; log2FC = –2.18, Padj = 8.78E-04).
Table 4.
Long noncoding RNA modulated (with adjusted P value [Padj] < .1) in response to prenatal testosterone treatment in muscle
| lncRNA | log2FC | Padj |
|---|---|---|
| LOC114112974 | –1.72 | .04 |
| LOC114118460 | –2.18 | .001 |
| LOC114112617 | –2.2 | .001 |
| LOC114113790 | –2.21 | .001 |
| LOC105607806 | –2.63 | .02 |
Abbreviations: FC, fold change; lncRNA, long noncoding RNA.
Micro RNA.
Whereas there was no separation of the 2 groups in the 2D PCA plot and the prenatal T animals showed within-group variance, clear separation was evident in the 3D PCA plot (see Fig. 9). OPLS DA score plot also showed clear separation of the 2 groups. Four miRNAs that were downregulated and 3 miRNAs that showed higher expression with prenatal T-treatment had the potential to serve as miRNA signatures of prenatal T exposure based on S plot (see Fig. 10), but MIR376C and MIR655, which were upregulated, were not confirmed by loading CIs. No miRNA selected met the FC FDR cutoff of less than 0.1 (see Supplementary Table S10) (35) although 1 miRNA (MIR10B) that was downregulated had an unadjusted FC P value of less than .05. Two downregulated miRNAs (MIR10A and MIR374B) and 1 upregulated miRNA (MIR369) had an unadjusted FC P value of less than .1.
Small nucleolar RNA.
There was no separation between the control and prenatal T-treated groups on the 2D PCA score plot, with more within group variation in the prenatal T-treated group (see Fig. 9). However, 3D PCA score plot showed clear groupings and good separation between the 2 groups (see Fig. 9). The OPLS DA score plot showed improvement and a clear separation of prenatal T and control animals (see Fig. 9). Eight snoRNAs with lower expression and 8 snoRNAs with higher expression had potential to serve as signatures of prenatal T exposure based on S plot (see Fig. 10). No snoRNA met the FDR criteria of less than 0.1 (see Supplementary Table S10) (35), with only one downregulated snoRNA (LOC114113401) with an unadjusted P value of less than .1 (P = .05).
Small nuclear RNA.
The control and prenatal T-treated groups were clearly separated in 2D and 3D PCA score plots (see Fig. 9). OPLS DA score plot also showed clear separation and groupings of control and prenatal T-exposed animals (see Fig. 9). Six snRNAs with lower expression with prenatal T exposure and 10 snRNAs showing higher expression had the potential to serve as signatures of prenatal T-treatment based on S plot (see Supplemental Fig. S7) (35). Four snRNAs that were downregulated (LOC114111412, LOC114111395, LOC114115462, and LOC114116759) and 4 that were upregulated (LOC114117108, LOC114117109, LOC114117110, and LOC114108902) were confirmed by loadings. No snRNA met the FC FDR criteria (see Supplementary Fig S6 and Table S10) (35); however, 4 downregulated identified markers had unadjusted P values of less than .05 (LOC114111412, LOC114111395, LOC114117093, and LOC114117060).
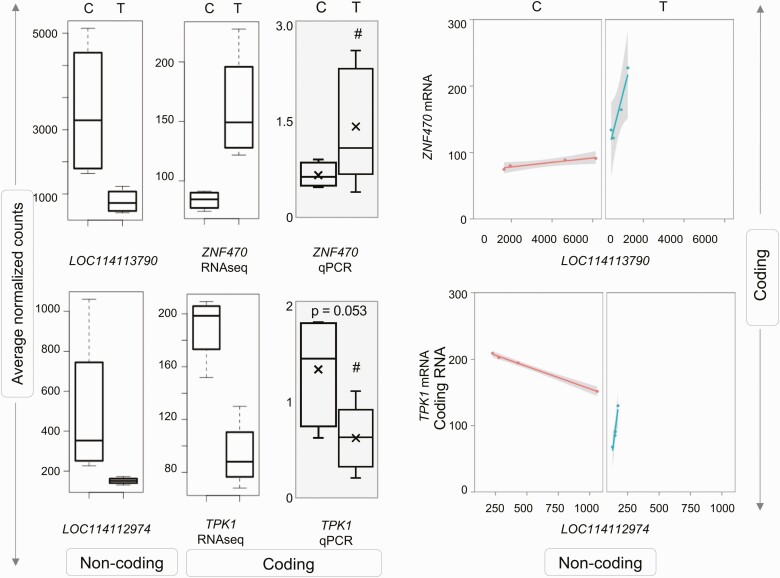
Correlation of Noncoding RNA With Messenger RNA (mRNA)/Correlation of Potential Biomarkers With mRNA Expression
Since none of the ncRNA were differentially expressed by prenatal T-treatment in the liver (Supplementary Fig. S5 and Table S9) (35), no correlation between ncRNA-mRNA was performed for this tissue. In the muscle, analysis of ncRNA interactions with mRNA using differential gene correlation analysis revealed significant correlation in 3 ncRNA-mRNA pairs in prenatal T-treated female sheep (Table 5). Of these, the downregulated uncharacterized lncRNA LOC114113790 correlated with increased expression for transcription factor zinc finger protein 470 (ZNF470), while the downregulation of 2 lncRNAs—LOC114112974 (Fig. 11) and LOC105607806 (see Supplementary Fig. S8)—correlated with the reduced expression of thiamin pyrophosphokinase 1 (TPK1). The mRNA expression of ZNF470 and TPK1 were validated by qPCR analysis. The gene expression changes for both genes were in line with RNA-seq analysis with a large magnitude increase in expression for ZNF470 and a trend (P = .05) toward decreased expression of TPK1 with prenatal T-treatment (see Fig. 11).
Table 5.
Long noncoding RNAs that correlated with messenger RNA in muscle from prenatal testosterone-treated animals
| lncRNA | mRNA | Control correlation | Control P | Prenatal T correlation | Prenatal T P | Z score diff | P diff | Classes |
|---|---|---|---|---|---|---|---|---|
| LOC114113790 | ZNF470 | 0.959 | .041 | 0.952 | .048 | –0.064 | .949 | +/+ |
| LOC114112974 | TPK1 | –0.998 | .002 | 0.972 | .028 | 4.048 | .000 | –/+ |
| LOC105607806 | TPK1 | –0.988 | .012 | 0.988 | .012 | 3.604 | .000 | –/+ |
Abbreviations: diff, difference; lncRNA, long noncoding RNA; mRNA, messenger RNA; T, testosterone.
Figure 11.
Correlation of coding and noncoding RNA (ncRNA) expression in muscle from prenatal testosterone (T)-treated animals. Box plots representing the average normalized counts for long noncoding RNA (lncRNA) and messenger RNA (mRNA) pairs that showed significant correlation are shown on the left. The treatment groups—control (C) or prenatal T (T)—are denoted on the x axis and the average normalized counts and fold change vs C on the y axis are denoted by RNA sequencing (RNA-seq) and quantitative polymerase chain reaction (qPCR; gray-shaded box) analyses, respectively. The line plots representing the correlation of expression between the lncRNA-mRNA in treatment group are shown at right. Correlation between lncRNA-mRNA for each pair is represented in orange line for C animals and as a green line for prenatal T-treated animals.
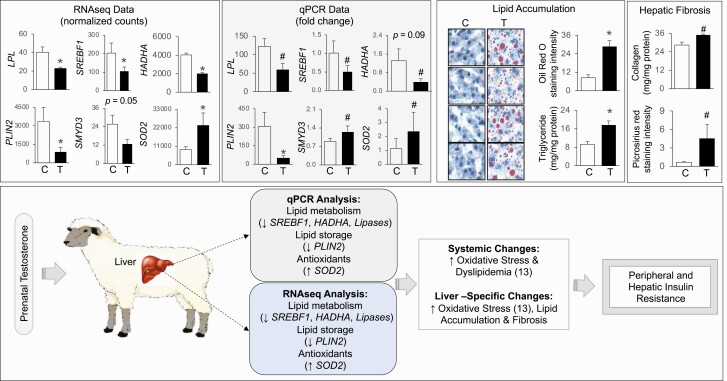
Validation of Differentially Expressed Gene Expression and Gene Pathways
To validate the RNA-seq determined differentially expressed genes involved in lipid metabolism—LPL, SREBF1, HADHA, and PLIN2, histone methylating enzyme SMYD3, and antioxidants SOD2—were compared with results from qPCR analysis. Measures of SREBF1, LPL, PLIN2, SMYD3, and SOD2 represent results for the animals used in this study, which were part of a previously published study involving a larger cohort of animals (13, 32). In line with the observations of the RNA-seq analysis, results from qPCR analysis also showed significant decrease in PLIN2 expression, large magnitude decrease in LPL, SREBF1, and HADHA and large magnitude increase in SOD2 mRNA expression (Fig. 12). However, SMYD3 changes noted by RNA-seq analysis did not parallel results from PCR analysis (see Fig. 12).
Figure 12.
Validation of RNA sequencing (RNA-seq)-determined differential gene expression in liver from prenatal testosterone (T)-treated sheep. Histograms representing the average fold change for messenger RNA (mRNA) from RNA-seq and quantitative polymerase chain reaction (qPCR) analysis. The treatment groups—control (C) or prenatal T (T)—are denoted on the x axis and the average normalized counts and fold change vs C on the y axis are denoted by RNA-seq and qPCR (gray-shaded box) analyses, respectively, at the top left. Lipid accumulation from oil red O staining and triglyceride assay and collagen deposition by collagen assay and picrosirius red staining from C and T animals are shown at the top right. The data from the larger cohort for some of these genes, lipid accumulation, and collagen deposition were previously published (13, 31, 32). The gene expression changes in prenatal T-treated animals from qPCR and RNA-seq analysis and its link with systemic and hepatic specific changes are summarized (bottom).
Since RNA sequence analyses revealed downregulation of pathways involved in lipid catabolism, the effect on hepatic lipid accumulation was examined. Consistent with what was reported previously (13), prenatal T-treatment–induced increase in lipid accumulation by both Oil Red O staining and triglyceride content were evident in the subset of animals used for RNA-seq analysis (see Fig. 12). Likewise, in line with the increased expression of collagen type XVIII alpha 1 chain (COL18A1; see Supplementary Table S6) (35), collagen content in the liver from prenatal T-treated animals was also increased as evident by picrosirius red staining and acid soluble collagen measurement (see Fig. 12).
Discussion
Using high-throughput next-generation sequencing, the findings in this study point to similar as well as distinct patterns of mRNA and ncRNA expression in the liver and muscle. The tissue-specific expression of coding RNA is in line with the physiological functions of each tissue. Prenatal T-treatment culminated in similar, as well as tissue-specific, changes in gene networks in liver and muscle. Both metabolic tissues manifested dysregulation of genes related to mitochondrial function that powers the cell’s biochemical reactions. Prenatal T excess downregulated genes related to lipid and triglyceride catabolism and PPARG signaling, in keeping with hepatic lipid accumulation and insulin resistant state of the liver (12, 13). In the muscle, 5 discrete lncRNAs were downregulated by prenatal T-treatment, with 2 lncRNAs correlated with an antioxidant gene and a third lncRNA correlated with a zinc finger transcription factor involved in the regulation of many cellular processes and differentiation of several tissues. Leveraging the predictive ability of OPLS-DA, VIP plots identified tissue-specific coding and ncRNAs (HADHA and SLC25A45, and miRNAs 154A, 25, and 487B in the liver and ARIH1 and ITCH and miRNAs 369, 10A, and 10B in the muscle, respectively) that have the potential to serve as signatures of prenatal T treatment. The relevance of these findings and their relationships with the metabolic changes induced by prenatal T excess are discussed next.
Liver- and Muscle-specific Gene Expression in Control Female Sheep
Although both liver and skeletal muscle tissues play a role in the maintenance of energy homeostasis and insulin sensitivity of the organism (42), considerable divergences in gene expression patterns are evident between liver and skeletal muscle, likely a reflection of their functional role and embryonic origin. These findings in sheep, a precocial, large-animal model, agree with earlier reports from mice (43), pigs (44), and humans (45). Liver is a vital organ involved in the efficient uptake and metabolism of amino acids, carbohydrates, bile acids, cholesterol, proteins, lipids and vitamins, biotransformation of endogenous and exogenous chemicals, and synthesis of plasma proteins (46), and genes involved in these processes are highly expressed. Similarly, consistent with the presence of actin and myosin proteins in muscle fibers (47), myosin family genes and myofibrillar protein genes are highly enriched in skeletal muscle.
As with coding RNA, tissue-specific changes in ncRNA were evident in all classes. Although evidence exists regarding the importance of many ncRNAs in the development and progression of hepatic and muscular defects (48-50), the understanding of the physiological role played by these ncRNAs is limited. Other studies have shown a role for MIR200 in the maintenance of hepatic function through regulation of fatty acid metabolism and control of hepatic stellate cells proliferation (51, 52).
Effect of Prenatal Testosterone Treatment on Liver Gene Expression
Prenatal T-treatment programs hepatic insulin resistance (12, 53), accompanied by increases in negative mediators of insulin signaling, namely increases in oxidative stress, lipotoxicity, and collagen accumulation (13, 31) in female sheep. These changes reflect metabolic injury–associated increases in hepatic fibrosis (54). In agreement with this functional phenotype, prenatal T-treated livers had increased expression of COL18A1. However, prenatal T-treated animals also manifested increased expression of SPSB1 (see Table 3), a negative regulator of profibrotic transforming growth factor β (TGFB) 1 signaling pathway (55), and NPTX2, a modulator of immunological responses (56) that is associated with amelioration of obesity-associated liver steatosis in rats (57). These changes suggest a remedial response to overcome the fibrotic and inflammatory phenotype typically associated with hepatic steatosis.
Pathway analysis indicated clear dysregulation of gene pathways associated with mitochondrial function, PPAR signaling, and lipid catabolism, which are related with hepatic insulin resistance and steatosis phenotype. Considering the majority of fatty acid oxidation in the liver occurs in mitochondria (58), downregulation of mitochondrial membrane proteins including carnitine palmitoyltransferase (CPT), an enzyme required for the transport of long-chain fatty acyl-CoAs from the cytoplasm into the mitochondria (59), along with triglyceride- and lipid catabolism–related genes are consistent with prevention of lipid metabolism. In the liver, PPARG, in addition to regulating glucose sensitivity, fatty acid uptake and mobilization, and triglyceride synthesis, appears to have an anti-inflammatory and antifibrotic role (60, 61). Loss of hepatic PPARG in mice is associated with increased blood triglycerides, ectopic lipid accumulation in other tissues, and insulin resistance in muscle (62, 63). The downregulation of the PPARG gene pathway in the liver from prenatal T-treated sheep parallels findings from the mice study supporting the contributory role for PPARG in the development of the pathological phenotype in the liver and muscle. Taken together these observations indicate dysregulation at multiple levels ranging from transcriptional control to lipid metabolism and transport as contributors to the development and maintenance of hepatic steatosis in the liver of prenatal T-treated sheep.
Effect of Prenatal Testosterone Treatment on Muscle Transcriptome
As is the case with the liver, skeletal muscle from prenatal T-treated sheep is characterized by insulin resistance and lipid accumulation (12, 13, 53). Furthermore, skeletal muscle of prenatal T-treated sheep manifested mitochondrial dysfunction, reflected as reduced mitochondrial copy number (31). Consistent with prenatal T excess–induced mitochondrial dysfunction, RNA-seq analyses (this study) found dysregulation of mitochondrial pathway genes that included downregulation of CPT1B, a muscle-specific mitochondrial enzyme involved in lipid transport into mitochondria (59). RNA-seq analysis also revealed downregulation of genes involved in cellular bioenergetics (synthesis of nicotinamide adenine dinucleotides NAD+ and NADP and their reduced forms NADH and NADPH) (64, 65) and mitochondrial biogenesis (66), such as nicotinamide phosphoribosyltransferase (NAMPT; see Supplementary Table S7) (35). Downregulation of CPT1B and NAMPT, coupled with reduced mitochondrial copy number (59), may underlie the increased triglyceride accumulation in the skeletal muscle of the prenatal T-treated sheep. Dysregulation of multiple gene pathways involved in RNA and ncRNA metabolism suggests involvement of epigenetic or transcription factor mechanisms in programming the metabolic defect in this model.
Potential Transcriptional Signatures of Prenatal Testosterone Exposure
Analytic Considerations
A strength of our study is the use of 2 complementary analytic approaches, traditional single transcript differential expression analysis (DEseq2), and dimension reduction multivariate modeling (PCA and OPLS-DA). Although RNA-seq is a very sensitive technique that can detect transcripts as low as only a few copies (67), measurement of thousands of coding and nc transcripts with few observations presents analytic challenges. Traditional single-transcript differential expression analysis with DEseq2 is an unbiased discovery that can identify many correlated associations, while dimension reduction multivariate modeling techniques (PCA and OPLS-DA) can identify informative transcriptional signatures of prenatal T-treatment. By reducing the data into fewer weighted variables (components) by combining related information, this approach helps reduce model complexity and retain prediction power (68). Such approaches that have been successfully used in the analyses of metabolomics data (69) to uncover patterns in big data sets were used in this study to help identify specific mRNA signatures.
The dimension reduction multivariate modeling technique also can be applied to ncRNA, important modulators of metabolic programming, that pose additional challenges due to lower expression levels (70). When the normalized expression values in the sample are very low (sometimes below one) with many zero values in the data, significance testing using the traditional negative binomial distribution can lack power (71, 72). By using unit variance scaling to give importance to these low-expressing transcripts, we identified potential ncRNA signatures of prenatal T-treatment. Many of the ncRNAs identified are yet to be characterized, and thus their functional significances are unknown. Advancement in functional genomics and the bioinformatics fields will aid in investigating the roles played by these nc transcriptome signatures on molecular mechanisms.
Functional Considerations
Consistent with differential gene expression, predictive component plots both in the liver and muscle showed gene expression patterns reflective of the functional outcomes programmed by prenatal T-treatment (Fig. 12). In the liver, predictive plots identified prenatal T-treatment downregulation of HADHA and SLC25A45, genes involved in mitochondrial function. The HADHA gene encodes a mitochondrial protein that catalyzes the last 3 steps of mitochondrial β-oxidation of long chain fatty acids (73). The SLC25A45 gene belongs to the SLC25 mitochondrial solute carrier family and is a transmembrane protein located in the mitochondrial outer membrane involved in lipid transfer from the endoplasmic reticulum to mitochondria (74, 75). Similar downregulation of HADHA (76) and SLC25 member SLC25A47 (77) expression have been shown to be associated with lipid accumulation in the high-fat diet–fed rodent liver. In muscle, predictive component plots identified downregulation of ARIH1 and ITCH, members of E3 ubiquitin ligases that catalyze the ubiquitination of proteins for degradation by the 26S proteasome (78). These ligases have a role in the development of insulin resistance either by directly degrading insulin signaling molecules, or indirectly by regulating proinflammatory cytokines (79). Downregulation of Pellino3, an E3 ubiquitin ligase family member, is associated with insulin resistance in high-fat diet–fed mice (80).
Changes in ncRNA with prenatal T-treatment were also relevant to hepatic insulin resistance, steatosis, and fibrosis. In the liver, increased MIR487B expression may contribute to the hepatic insulin resistance observed in the model, as this miRNA can ablate insulin signaling by silencing insulin receptor substrate (81). Likewise, increased MIR154A and lower MIR25 may be linked to hepatic fibrosis. Increased MIR154A was observed with myocardial fibrosis (82), while the reduction in MIR25 levels promote TGFB signaling, an inducer of collagen expression in liver (83). In muscle, the upregulation of MIR369 and downregulation of MIR10A and MIR10B parallel observations in skeletal muscle from an insulin resistant high fat diet-fed mouse model (84). These findings, along with downregulation of genes involved in lipid catabolism (this study), the presence of hepatic steatosis in the liver (13), dysregulation of mitochondrial gene pathways (this study), and presence of insulin resistance in liver and muscle (12, 13, 53) support these genes as potential biomarkers associated with prenatal T excess induced metabolic dysfunctions.
Relationship Between Coding and Noncoding RNA
Coding RNA that regulates biological processes can be regulated by ncRNA. In skeletal muscle from prenatal T-treated animals, lncRNAs correlated with coding RNA ZNF470 and TPK1 expression. There is limited information regarding the role of ZNF470, a member of the zinc finger family of transcription factors; however, a closely related member, ZNF479, is involved in transcriptional control of metallothioneins (85), which, owing to their high affinity, serve as scavengers of heavy metals and reactive oxygen species and hence help prevent cellular damage (86). In contrast, TPK1, an antioxidant that exerts its action by generating thiamine pyrophosphate, a suppressor of superoxide, hydroperoxide, and hydroxyl radical (87), had lower expression with prenatal T-treatment. Since lipid accumulation and mitochondrial dysfunction lead to the production of reactive oxygen species, and the overall oxidative state is dictated by the balance between oxidants and antioxidants, how the changes in ZNF470 and TPK1 expression contribute to maintenance of skeletal muscle oxidative status requires further study.
Translational Significance
Prenatal T-treated sheep manifest metabolic dysfunction characteristic of nonalcoholic fatty liver disease (NAFLD) (88), PCOS (89, 90), and metabolic syndrome (91). Peripheral insulin resistance, dyslipidemia, oxidative stress, and hepatic steatosis and fibrosis in the prenatal T-treated female sheep (see Fig. 12) (13, 31) are typically present in NAFLD (92). Therefore, the findings from this study of changes in expression of coding RNA and ncRNA with functional linkage to insulin resistance, lipid metabolism, mitochondrial dysfunction, and fibrosis are of translational significance. Among NAFLD patients, the progression to liver fibrosis occurs in one-third of patients within 4 and 5 years of diagnosis, along with increased expression of collagen and profibrinogenic TGFB signaling (54, 92), a feature also observed in prenatal T-treated sheep. Similarly, mitochondrial dysfunction has an important role in the pathogenesis of PCOS, NAFLD, and other metabolic dysfunctions (93-98). In agreement with this, a decrease in mitochondrial oxidative metabolism and β-oxidation gene expression were observed in PCOS skeletal muscle (99) and in liver and skeletal muscle from NAFLD patients (100, 101). These changes may be regulated by ncRNA, as emerging studies have linked multiple miRNAs with progression of NAFLD and PCOS (21, 102). The dysregulation of multiple miRNAs targeting β-oxidation, glucose metabolism, and oxidative and inflammatory status in obesity-associated NAFLD (103, 104) provide support for the premise. Since the phenotype of prenatal T-treated sheep mimics the NAFLD and PCOS phenotype, these transcriptional signatures of coding and ncRNA linked with the lipid metabolism and fibrosis may provide potential therapeutic targets that may aid in ameliorating these conditions.
Strengths and Limitations
Animal models cannot fully replicate the physiological environment of heterogeneous human patient populations; however, these models are designed to reduce heterogeneity by controlling potential confounders arising from genetic, nutritional, and environmental backgrounds. As such, the use of animal models with similar physiology will likely be relevant if the mechanisms understood are fully translatable. In this regard, the major strengths of this study are the use of an animal model of translational significance to human PCOS and NAFLD (105-108), using 2 metabolically relevant organs, comprehensive RNA-seq of coding RNA and ncRNA coupled with bioinformatic approaches and validation of phenotypically relevant differentially expressed genes in liver and muscle by qPCR. The findings from this study should be interpreted while considering a few limitations. One limitation is that these studies were performed on whole liver and muscle, which in addition to hepatocytes and myocytes, respectively, are composed of a diverse array of immune and vascular cells. Therefore, future studies need to capitalize on these findings to understand how these identified mRNA and ncRNA have mechanistic roles at the cellular level. In addition, although there is evidence supporting a strong correlation between differentially expressed mRNAs and their protein products (109), while functional evidence with increase in lipid and collagen accumulation is evident, further studies encompassing protein and activity of the identified targets are necessary. These studies were carried out in female sheep from ovariectomized animals, and tissues were collected during the artificial follicular phase with higher estrogenic milieu. Future studies need to expand into how these changes occur in male offspring and under different steroidal backgrounds.
Conclusions
The findings from this study expand our previous phenotypic and functional characterization of the effects of prenatal T-treatment in the liver and muscle, and provide mechanistic understandings into the development of hepatic and muscle metabolic dysfunctions. Identification of several coding RNA and ncRNA signatures of prenatal T excess offer possible diagnostic and therapeutic targets. Future studies are needed to further understand and ameliorate the metabolic dysfunctions in the offspring of hyperandrogenic pregnancies.
Acknowledgments
We thank Mr. Douglas Doop for his valuable assistance in breeding, lambing, and animal care; and Dr Almudena Veiga-Lopez, Dr Bachir Abi Salloum, Mr Evan Beckett, Mrs Carol Herkimer, and students supported through the University of Michigan Undergraduate Research Opportunity Program for help with administration of treatments and tissue harvest.
Financial Support: This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (award Nos. R01 HD099096 and P01 HD44232 to V.P.). M.P. was supported by a Ruth L. Kirschstein Institutional Training Grant (No. T32 ES007062). N.S. was supported by a National Institute of Diabetes and Digestive and Kidney Diseases Institutional Training Grant (No. T32 DK071212).
Glossary
Abbreviations
- 2D
2-dimensional
- 3D
3-dimensional
- CPT
carnitine palmitoyltransferase
- FC
fold change
- FDR
false discovery rate
- lncRNA
long noncoding RNA
- mRNA
messenger RNA
- miRNA
microRNA
- NAFLD
nonalcoholic fatty liver disease
- ncRNA
noncoding RNA
- nt
nucleotide
- OPLS-DA
orthogonal partial least square discriminant analysis/orthogonal projections to latent structures discriminant analysis
- OR
odds ratio
- PCA
principal component analysis
- PCOS
polycystic ovary syndrome
- PPARG
peroxisome proliferator-activated receptor γ
- qPCR
quantitative polymerase chain reaction
- RNA-seq
RNA sequencing
- snoRNA
small nucleolar RNA
- snRNA
small nuclear RNA
- SOD
superoxide dismutase
- STAR
Spliced Transcripts Alignment to a Reference
- T
testosterone
- TGFB
transforming growth factor β
- VIP
variable importance to projection
Additional Information
Disclosures: The authors have nothing to disclose.
Data Availability
Some or all data generated or analyzed during this study are included in this published article or in the data repositories listed in “References.”
References
- 1. Barker DJ. Fetal origins of coronary heart disease. BMJ. 1995;311(6998):171-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Heindel JJ, Newbold R, Schug TT. Endocrine disruptors and obesity. Nat Rev Endocrinol. 2015;11(11):653-661. [DOI] [PubMed] [Google Scholar]
- 3. Padmanabhan V, Cardoso RC, Puttabyatappa M. Developmental programming, a pathway to disease. Endocrinology. 2016;157(4):1328-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moisiadis VG, Matthews SG. Glucocorticoids and fetal programming part 1: outcomes. Nat Rev Endocrinol. 2014;10(7):391-402. [DOI] [PubMed] [Google Scholar]
- 5. Stener-Victorin E, Padmanabhan V, Walters KA, et al. . Animal models to understand the etiology and pathophysiology of polycystic ovary syndrome. Endocr Rev. 2020;41(4):538-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Markey CM, Coombs MA, Sonnenschein C, Soto AM. Mammalian development in a changing environment: exposure to endocrine disruptors reveals the developmental plasticity of steroid-hormone target organs. Evol Dev. 2003;5(1):67-75. [DOI] [PubMed] [Google Scholar]
- 7. Hakim C, Padmanabhan V, Vyas AK. Gestational hyperandrogenism in developmental programming. Endocrinology. 2017;158(2):199-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Puttabyatappa M, Sargis RM, Padmanabhan V. Developmental programming of insulin resistance: are androgens the culprits? J Endocrinol. 2020;245(3):R23-R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Veiga-Lopez A, Moeller J, Patel D, et al. . Developmental programming: impact of prenatal testosterone excess on insulin sensitivity, adiposity, and free fatty acid profile in postpubertal female sheep. Endocrinology. 2013;154(5):1731-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cardoso RC, Veiga-Lopez A, Moeller J, et al. . Developmental programming: impact of gestational steroid and metabolic milieus on adiposity and insulin sensitivity in prenatal testosterone-treated female sheep. Endocrinology. 2016;157(2):522-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. King AJ, Olivier NB, Mohankumar PS, Lee JS, Padmanabhan V, Fink GD. Hypertension caused by prenatal testosterone excess in female sheep. Am J Physiol Endocrinol Metab. 2007;292(6): E1837-E1841. [DOI] [PubMed] [Google Scholar]
- 12. Lu C, Cardoso RC, Puttabyatappa M, Padmanabhan V. Developmental programming: prenatal testosterone excess and insulin signaling disruptions in female sheep. Biol Reprod. 2016;94(5):113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Puttabyatappa M, Andriessen V, Mesquitta M, Zeng L, Pennathur S, Padmanabhan V. Developmental programming: impact of gestational steroid and metabolic milieus on mediators of insulin sensitivity in prenatal testosterone-treated female sheep. Endocrinology. 2017;158(9):2783-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nada SE, Thompson RC, Padmanabhan V. Developmental programming: differential effects of prenatal testosterone excess on insulin target tissues. Endocrinology. 2010;151(11):5165-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lecoutre S, Maqdasy S, Breton C. Maternal obesity as a risk factor for developing diabetes in offspring: an epigenetic point of view. World J Diabetes. 2021;12(4):366-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gabory A, Attig L, Junien C. Developmental programming and epigenetics. Am J Clin Nutr. 2011;94(6 Suppl):1943S-1952S. [DOI] [PubMed] [Google Scholar]
- 17. Bansal A, Simmons RA. Epigenetics and developmental origins of diabetes: correlation or causation? Am J Physiol Endocrinol Metab. 2018;315(1):E15-E28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96(4):1297-1325. [DOI] [PubMed] [Google Scholar]
- 19. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7-21. [DOI] [PubMed] [Google Scholar]
- 20. He X, Ou C, Xiao Y, Han Q, Li H, Zhou S. LncRNAs: key players and novel insights into diabetes mellitus. Oncotarget. 2017;8(41):71325-71341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen B, Xu P, Wang J, Zhang C. The role of MiRNA in polycystic ovary syndrome (PCOS). Gene. 2019;706:91-96. [DOI] [PubMed] [Google Scholar]
- 22. Ilie IR, Georgescu CE. Polycystic ovary syndrome—epigenetic mechanisms and aberrant microRNA. Adv Clin Chem. 2015;71:25-45. [DOI] [PubMed] [Google Scholar]
- 23. Zhao J, Huang J, Geng X, et al. . Polycystic ovary syndrome: novel and hub lncRNAs in the insulin resistance-associated lncRNA-mRNA network. Front Genet. 2019;10:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chang S, Dunaif A. Diagnosis of polycystic ovary syndrome: which criteria to use and when? Endocrinol Metab Clin North Am. 2021;50(1):11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barber TM, Dimitriadis GK, Andreou A, Franks S. Polycystic ovary syndrome: insight into pathogenesis and a common association with insulin resistance. Clin Med (Lond). 2016;16(3):262-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Escobar-Morreale HF. Polycystic ovary syndrome: definition, aetiology, diagnosis and treatment. Nat Rev Endocrinol. 2018;14(5):270-284. [DOI] [PubMed] [Google Scholar]
- 27. Cardoso RC, Padmanabhan V. Developmental programming of PCOS traits: insights from the sheep. Med Sci (Basel). 2019;7(7):79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hossain MM, Cao M, Wang Q, et al. . Altered expression of miRNAs in a dihydrotestosterone-induced rat PCOS model. J Ovarian Res. 2013;6(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Manikkam M, Crespi EJ, Doop DD, et al. . Fetal programming: prenatal testosterone excess leads to fetal growth retardation and postnatal catch-up growth in sheep. Endocrinology. 2004;145(2):790-798. [DOI] [PubMed] [Google Scholar]
- 30. Veiga-Lopez A, Ye W, Phillips DJ, Herkimer C, Knight PG, Padmanabhan V. Developmental programming: deficits in reproductive hormone dynamics and ovulatory outcomes in prenatal, testosterone-treated sheep. Biol Reprod. 2008;78(4):636-647. [DOI] [PubMed] [Google Scholar]
- 31. Puttabyatappa M, Ciarelli JN, Chatoff AG, Padmanabhan V. Developmental programming: metabolic tissue-specific changes in endoplasmic reticulum stress, mitochondrial oxidative and telomere length status induced by prenatal testosterone excess in the female sheep. Mol Cell Endocrinol. 2021;526:111207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guo X, Puttabyatappa M, Domino SE, Padmanabhan V. Developmental programming: prenatal testosterone-induced changes in epigenetic modulators and gene expression in metabolic tissues of female sheep. Mol Cell Endocrinol. 2020;514:110913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Manikkam M, Steckler TL, Welch KB, Inskeep EK, Padmanabhan V. Fetal programming: prenatal testosterone treatment leads to follicular persistence/luteal defects; partial restoration of ovarian function by cyclic progesterone treatment. Endocrinology. 2006;147(4):1997-2007. [DOI] [PubMed] [Google Scholar]
- 34. Goodman RL, Legan SJ, Ryan KD, Foster DL, Karsch FJ. Importance of variations in behavioural and feedback actions of oestradiol to the control of seasonal breeding in the ewe. J Endocrinol. 1981;89(2):229-240. [DOI] [PubMed] [Google Scholar]
- 35. Saadat N, Puttabyatappa M, Elangovan VR, et al. . Supplemental data for Developmental programming: impact of prenatal testosterone excess on liver and muscle transcriptome and the relationship between noncoding and coding RNA networks in female sheep. Posted October 14, 2021. 10.6084/m9.figshare.14923092 [DOI]
- 36. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Durinck S, Moreau Y, Kasprzyk A, et al. . BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21(16):3439-3440. [DOI] [PubMed] [Google Scholar]
- 39. Lee C, Patil S, Sartor MA. RNA-Enrich: a cut-off free functional enrichment testing method for RNA-seq with improved detection power. Bioinformatics. 2016;32(7):1100-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Conway JR, Lex A, Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33(18):2938-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McKenzie AT, Katsyv I, Song WM, Wang M, Zhang B. DGCA: a comprehensive R package for differential gene correlation analysis. BMC Syst Biol. 2016;10(1):106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Saltiel AR. Insulin signaling in the control of glucose and lipid homeostasis. Handb Exp Pharmacol. 2016;233:51-71. [DOI] [PubMed] [Google Scholar]
- 43. Li B, Qing T, Zhu J, et al. . A comprehensive mouse transcriptomic BodyMap across 17 tissues by RNA-seq. Sci Rep. 2017;7(1):4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. González-Prendes R, Mármol-Sánchez E, Quintanilla R, et al. . About the existence of common determinants of gene expression in the porcine liver and skeletal muscle. BMC Genomics. 2019;20(1):518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Malarkey DE, Johnson K, Ryan L, Boorman G, Maronpot RR. New insights into functional aspects of liver morphology. Toxicol Pathol. 2005;33(1):27-34. [DOI] [PubMed] [Google Scholar]
- 47. Squire J. Special issue: the actin-myosin interaction in muscle: background and overview. Int J Mol Sci. 2019;20(22):5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Goljanek-Whysall K, Sweetman D, Münsterberg AE. microRNAs in skeletal muscle differentiation and disease. Clin Sci (Lond). 2012;123(11):611-625. [DOI] [PubMed] [Google Scholar]
- 49. Szabo G, Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol. 2013;10(9):542-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Takahashi K, Yan I, Haga H, Patel T. Long noncoding RNA in liver diseases. Hepatology. 2014;60(2):744-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen M, Wang S, Li X, Storey KB, Zhang X. The potential contribution of miRNA-200-3p to the fatty acid metabolism by regulating AjEHHADH during aestivation in sea cucumber. PeerJ. 2018;6:e5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang JJ, Tao H, Hu W, et al. . MicroRNA-200a controls Nrf2 activation by target Keap1 in hepatic stellate cell proliferation and fibrosis. Cell Signal. 2014;26(11):2381-2389. [DOI] [PubMed] [Google Scholar]
- 53. Puttabyatappa M, Padmanabhan V. Prenatal testosterone programming of insulin resistance in the female sheep. Adv Exp Med Biol. 2017;1043:575-596. [DOI] [PubMed] [Google Scholar]
- 54. Roehlen N, Crouchet E, Baumert TF. Liver fibrosis: mechanistic concepts and therapeutic perspectives. Cells. 2020;9(4):875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu S, Nheu T, Luwor R, Nicholson SE, Zhu HJ. SPSB1, a novel negative regulator of the transforming growth factor-β signaling pathway targeting the type II receptor. J Biol Chem. 2015;290(29):17894-17908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gewurz H, Zhang XH, Lint TF. Structure and function of the pentraxins. Curr Opin Immunol. 1995;7(1):54-64. [DOI] [PubMed] [Google Scholar]
- 57. Kozaczek M, Bottje W, Greene E, et al. . Comparison of liver gene expression by RNAseq and PCR analysis after 8 weeks of feeding soy protein isolate- or casein-based diets in an obese liver steatosis rat model. Food Funct. 2019;10(12):8218-8229. [DOI] [PubMed] [Google Scholar]
- 58. Sozio MS, Liangpunsakul S, Crabb D. The role of lipid metabolism in the pathogenesis of alcoholic and nonalcoholic hepatic steatosis. Semin Liver Dis. 2010;30(4):378-390. [DOI] [PubMed] [Google Scholar]
- 59. Schreurs M, Kuipers F, van der Leij FR. Regulatory enzymes of mitochondrial beta-oxidation as targets for treatment of the metabolic syndrome. Obes Rev. 2010;11(5):380-388. [DOI] [PubMed] [Google Scholar]
- 60. Dixon ED, Nardo AD, Claudel T, Trauner M. The role of lipid sensing nuclear receptors (PPARs and LXR) and metabolic lipases in obesity, diabetes and NAFLD. Genes (Basel). 2021;12(5):645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mello T, Materozzi M, Galli A. PPARs and mitochondrial metabolism: from NAFLD to HCC. PPAR Res. 2016;2016:7403230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gavrilova O, Haluzik M, Matsusue K, et al. . Liver peroxisome proliferator-activated receptor gamma contributes to hepatic steatosis, triglyceride clearance, and regulation of body fat mass. J Biol Chem. 2003;278(36):34268-34276. [DOI] [PubMed] [Google Scholar]
- 63. Matsusue K, Haluzik M, Lambert G, et al. . Liver-specific disruption of PPARgamma in leptin-deficient mice improves fatty liver but aggravates diabetic phenotypes. J Clin Invest. 2003;111(5):737-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yang Y, Sauve AA. NAD(+) metabolism: bioenergetics, signaling and manipulation for therapy. Biochim Biophys Acta. 2016;1864(12):1787-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ying W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid Redox Signal. 2008;10(2):179-206. [DOI] [PubMed] [Google Scholar]
- 66. Kim JS, Yoon CS, Park DR. NAMPT regulates mitochondria biogenesis via NAD metabolism and calcium binding proteins during skeletal muscle contraction. J Exerc Nutrition Biochem. 2014;18(3):259-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rao MS, Van Vleet TR, Ciurlionis R, et al. . Comparison of RNA-Seq and microarray gene expression platforms for the toxicogenomic evaluation of liver from short-term rat toxicity studies. Front Genet. 2018;9:636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Trygg J, Wold S. Orthogonal projections to latent structures (O-PLS). J Chemometr. 2002;16(3):119-128. [Google Scholar]
- 69. Lamichhane S, Sen P, Dickens AM, Hyötyläinen T, Orešič M. An overview of metabolomics data analysis: current tools and future perspectives. Comprehensive analytical chemistry. 2018; 82:387-413. [Google Scholar]
- 70. Weikard R, Demasius W, Kuehn C. Mining long noncoding RNA in livestock. Anim Genet. 2017;48(1):3-18. [DOI] [PubMed] [Google Scholar]
- 71. Ran D, Daye ZJ. Gene expression variability and the analysis of large-scale RNA-seq studies with the MDSeq. Nucleic Acids Res. 2017;45(13):e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Li Q, Yu X, Chaudhary R, Slebos RJC, Chung CH, Wang X. lncDIFF: a novel quasi-likelihood method for differential expression analysis of non-coding RNA. BMC Genomics. 2019;20(1):539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fletcher AL, Pennesi ME, Harding CO, Weleber RG, Gillingham MB. Observations regarding retinopathy in mitochondrial trifunctional protein deficiencies. Mol Genet Metab. 2012;106(1):18-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Clémençon B, Babot M, Trézéguet V. The mitochondrial ADP/ATP carrier (SLC25 family): pathological implications of its dysfunction. Mol Aspects Med. 2013;34(2-3):485-493. [DOI] [PubMed] [Google Scholar]
- 75. Janer A, Prudent J, Paupe V, et al. . SLC25A46 is required for mitochondrial lipid homeostasis and cristae maintenance and is responsible for Leigh syndrome. EMBO Mol Med. 2016;8(9):1019-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Li L, Lu DZ, Li YM, Zhang XQ, Zhou XX, Jin X. Proteomic analysis of liver mitochondria from rats with nonalcoholic steatohepatitis. World J Gastroenterol. 2014;20(16):4778-4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Da Silva VML. LRH-1 signaling and control of liver intermediary metabolism: impact on nonalcoholic fatty liver disease. Institute of Bioengineering: École polytechnique fédérale de Lausanne. Accessed October 14, 2021. https://core.ac.uk/reader/159366548
- 78. Zheng N, Shabek N. Ubiquitin ligases: structure, function, and regulation. Annu Rev Biochem. 2017;86:129-157. [DOI] [PubMed] [Google Scholar]
- 79. Yang XD, Xiang DX, Yang YY. Role of E3 ubiquitin ligases in insulin resistance. Diabetes Obes Metab. 2016;18(8):747-754. [DOI] [PubMed] [Google Scholar]
- 80. Yang S, Wang B, Humphries F, Hogan AE, O’Shea D, Moynagh PN. The E3 ubiquitin ligase Pellino3 protects against obesity-induced inflammation and insulin resistance. Immunity. 2014;41(6):973-987. [DOI] [PubMed] [Google Scholar]
- 81. Wang J, Tan J, Qi Q, et al. . miR-487b-3p suppresses the proliferation and differentiation of myoblasts by targeting IRS1 in skeletal muscle myogenesis. Int J Biol Sci. 2018;14(7):760-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Dong P, Liu WJ, Wang ZH. MiR-154 promotes myocardial fibrosis through β-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22(7):2052-2060. [DOI] [PubMed] [Google Scholar]
- 83.Genz B, Coleman MA, Irvine KM, et al. Overexpression of miRNA-25-3p inhibits Notch1 signaling and TGF-β-induced collagen expression in hepatic stellate cells. Sci Rep. 2019; 9:8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Massart J, Katayama M, Krook A. microManaging glucose and lipid metabolism in skeletal muscle: Role of microRNAs. Biochim Biophys Acta. 2016;1861(12 Pt B):2130-2138. [DOI] [PubMed] [Google Scholar]
- 85. Wu YJ, Ko BS, Liang SM, et al. . ZNF479 downregulates metallothionein-1 expression by regulating ASH2L and DNMT1 in hepatocellular carcinoma. Cell Death Dis. 2019;10(6):408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Coyle P, Philcox JC, Carey LC, Rofe AM. Metallothionein: the multipurpose protein. Cell Mol Life Sci. 2002;59(4):627-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Okai Y, Higashi-Okai K, F Sato E, Konaka R, Inoue M. Potent radical-scavenging activities of thiamin and thiamin diphosphate. J Clin Biochem Nutr. 2007;40(1):42-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Robinson KE, Shah VH. Pathogenesis and pathways: nonalcoholic fatty liver disease & alcoholic liver disease. Transl Gastroenterol Hepatol. 2020;5:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Padmanabhan V, Veiga-Lopez A. Sheep models of polycystic ovary syndrome phenotype. Mol Cell Endocrinol. 2013;373(1-2):8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Anagnostis P, Tarlatzis BC, Kauffman RP. Polycystic ovarian syndrome (PCOS): long-term metabolic consequences. Metabolism. 2018;86:33-43. [DOI] [PubMed] [Google Scholar]
- 91. Rochlani Y, Pothineni NV, Mehta JL. Metabolic syndrome: does it differ between women and men? Cardiovasc Drugs Ther. 2015;29(4):329-338. [DOI] [PubMed] [Google Scholar]
- 92. Karkucinska-Wieckowska A, Simoes ICM, Kalinowski P, et al. . Mitochondria, oxidative stress and nonalcoholic fatty liver disease: a complex relationship. Eur J Clin Invest. Published online May 29, 2021. doi: 10.1111/eci.13622 [DOI] [PubMed] [Google Scholar]
- 93. Corbould A. Insulin resistance in skeletal muscle and adipose tissue in polycystic ovary syndrome: are the molecular mechanisms distinct from type 2 diabetes? Panminerva Med. 2008;50(4):279-294. [PubMed] [Google Scholar]
- 94. Lee J, Park JS, Roh YS. Molecular insights into the role of mitochondria in non-alcoholic fatty liver disease. Arch Pharm Res. 2019;42(11):935-946. [DOI] [PubMed] [Google Scholar]
- 95. Zeng X, Huang Q, Long SL, Zhong Q, Mo Z. Mitochondrial dysfunction in polycystic ovary syndrome. DNA Cell Biol. 2020;39(8):1401-1409. [DOI] [PubMed] [Google Scholar]
- 96. Merhi Z, Kandaraki EA, Diamanti-Kandarakis E. Implications and future perspectives of AGEs in PCOS pathophysiology. Trends Endocrinol Metab. 2019;30(3):150-162. [DOI] [PubMed] [Google Scholar]
- 97. Schröder M, Sutcliffe L. Consequences of stress in the secretory pathway: the ER stress response and its role in the metabolic syndrome. Methods Mol Biol. 2010;648:43-62. [DOI] [PubMed] [Google Scholar]
- 98. Xia SW, Wang ZM, Sun SM, et al. . Endoplasmic reticulum stress and protein degradation in chronic liver disease. Pharmacol Res. 2020;161:105218. [DOI] [PubMed] [Google Scholar]
- 99. Skov V, Glintborg D, Knudsen S, et al. . Reduced expression of nuclear-encoded genes involved in mitochondrial oxidative metabolism in skeletal muscle of insulin-resistant women with polycystic ovary syndrome. Diabetes. 2007;56(9):2349-2355. [DOI] [PubMed] [Google Scholar]
- 100. Nakamuta M, Kohjima M, Morizono S, et al. . Evaluation of fatty acid metabolism-related gene expression in nonalcoholic fatty liver disease. Int J Mol Med. 2005;16(4):631-635. [PubMed] [Google Scholar]
- 101. García-Ruiz C, Baulies A, Mari M, García-Rovés PM, Fernandez-Checa JC. Mitochondrial dysfunction in non-alcoholic fatty liver disease and insulin resistance: cause or consequence? Free Radic Res. 2013;47(11):854-868. [DOI] [PubMed] [Google Scholar]
- 102. Su Q, Kumar V, Sud N, Mahato RI. MicroRNAs in the pathogenesis and treatment of progressive liver injury in NAFLD and liver fibrosis. Adv Drug Deliv Rev. 2018;129:54-63. [DOI] [PubMed] [Google Scholar]
- 103. Latorre J, Moreno-Navarrete JM, Mercader JM, et al. . Decreased lipid metabolism but increased FA biosynthesis are coupled with changes in liver microRNAs in obese subjects with NAFLD. Int J Obes (Lond). 2017;41(4):620-630. [DOI] [PubMed] [Google Scholar]
- 104. Soronen J, Yki-Jarvinen H, Zhou Y, et al. . Novel hepatic microRNAs upregulated in human nonalcoholic fatty liver disease. Physiol Rep. 2016;4(1):e12661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Padmanabhan V, Veiga-Lopez A. Reproduction symposium: developmental programming of reproductive and metabolic health. J Anim Sci. 2014;92(8):3199-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Gonzalez-Bulnes A, Chavatte-Palmer P. Contribution of large animals to translational research on prenatal programming of obesity and associated diseases. Curr Pharm Biotechnol. 2017;18(7):541-551. [DOI] [PubMed] [Google Scholar]
- 107. Fuller-Jackson JP, Henry BA. Adipose and skeletal muscle thermogenesis: studies from large animals. J Endocrinol. 2018;237(3):R99-R115. [DOI] [PubMed] [Google Scholar]
- 108. Symonds ME, Pope M, Budge H. The ontogeny of brown adipose tissue. Annu Rev Nutr. 2015;35:295-320. [DOI] [PubMed] [Google Scholar]
- 109. Koussounadis A, Langdon SP, Um IH, Harrison DJ, Smith VA. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci Rep. 2015;5:10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Some or all data generated or analyzed during this study are included in this published article or in the data repositories listed in “References.”