Abstract
This research presents the design and synthesis of a novel series of phthalazine derivatives as Topo II inhibitors, DNA intercalators, and cytotoxic agents. In vitro testing of the new compounds against HepG-2, MCF-7, and HCT-116 cell lines confirmed their potent cytotoxic activity with low IC50 values. Topo II inhibition and DNA intercalating activities were evaluated for the most cytotoxic members. IC50 values determination demonstrated Topo II inhibitory activities and DNA intercalating affinities of the tested compounds at a micromolar level. Amongst, compound 9d was the most potent member. It inhibited Topo II enzyme at IC50 value of 7.02 ± 0.54 µM with DNA intercalating IC50 of 26.19 ± 1.14 µM. Compound 9d was then subjected to an in vivo antitumor examination. It inhibited tumour proliferation reducing solid tumour volume and mass. Additionally, it restored liver enzymes, proteins, and CBC parameters near-normal, indicating a remarkable amelioration in their functions along with histopathological examinations.
Keywords: Topo II, DNA, antitumer, phthalazine, intercalators
1. Introduction
Cancer is characterised by uncontrolled cell growth and proliferation following genetic mutation. It represents one of the most important health issues worldwide and is the second leading cause of death1,2. Therefore, it represents one of the greatest challenges to medical researchers, especially with the continued failure of current therapies from one side and the development of drug resistance from the other side3–5.
The current search and discovery of new drug candidates with anticancer activities have become one of the most important issues for medicinal chemists nowadays6–11. Among the most important chemotherapeutic agents applied for cancer treatment are those that interact with DNA. Anticancer agents in the previously mentioned class belong to either alkylating agents, groove binders, or intercalating agents12. DNA intercalating agents got great attention from scientists due to their promising antitumoral activity13–18. They are classified into two major groups of compounds that intercalate between DNA base pairs (especially G and C, 70%) without covalent binding: 1) acridines and related compounds and 2) anthracyclines and related compounds19. These compounds produce local structural changes to the DNA molecule, including the lengthening of the DNA strand following the unwinding of its double helix. So, DNA intercalators are mutagenic due to their retardation or even inhibition of DNA transcription and replication20.
Doxorubicin is one of the two first isolated and introduced anthracyclines as antitumor agents. It works through two mechanisms of action; 1) intercalates into the DNA double helix without covalent binding, and 2) binds covalently to topoisomerase II (involved in DNA replication and transcription), poisons the cleavable complex of DNA and prevent its re-ligation, and finally results in an apoptotic action21,22.
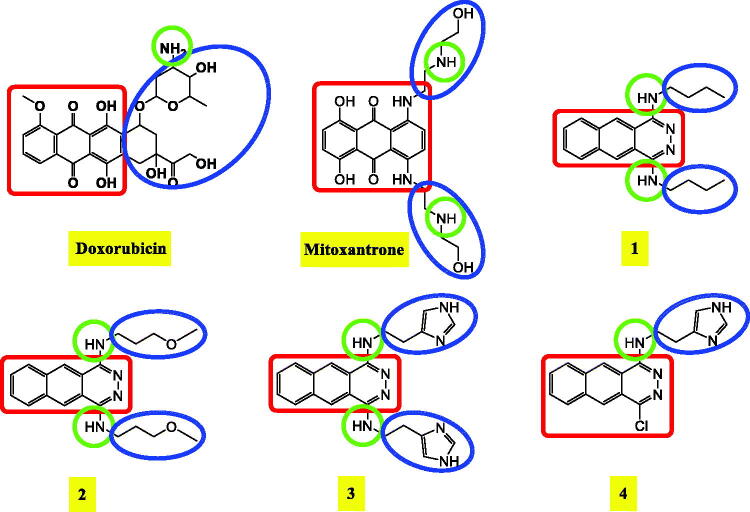
Phthalazine moiety was recommended in the area of medicinal chemistry to have promising antitumor activity and primarily to act as DNA intercalator and topoisomerase II inhibitors as well16,23,24. On the other hand, many other organic moieties like triazoles, hydrazine amides, hydrazine thioacetamides, benzylidene hydrazones, sulphonamides, benzoic acid, and thioacetamides derivatives were identified and introduced as potential antitumor agents25–29. Some reported DNA intercalators and topoisomerase II inhibitors showing their common pharmacophoric features were depicted in Figure 1.
Figure 1.
Some reported DNA intercalators and topoisomerase II inhibitors showing their common pharmacophoric features.
1.1. The rationale of molecular design
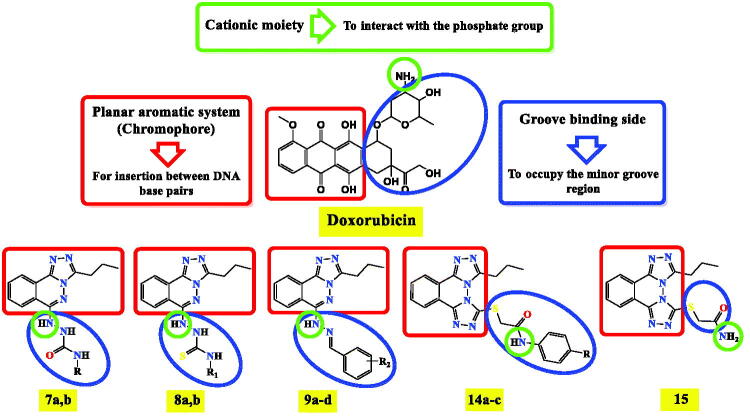
A ligand-based drug design approach30,31 was performed to design a new wave of promising DNA intercalators and topoisomerase II inhibitors taking into consideration the basic pharmacophoric features of doxorubicin. It is worth mentioning that there are three crucial pharmacophoric features present in doxorubicin which guided our rationale. The first one is the planar polyaromatic system (chromophore) inserted in between the DNA base pairs. The second one is the presence of a groove binding side to occupy the minor groove of DNA. The third part is the cationic moiety, or a species having the ability to be protonated in the physiological PH to interact with the negatively charged phosphate group of DNA sugar moiety32,33.
Molecular hybridisation of triazolo phthalazine moieties instead of the planar aromatic system of doxorubicin with different recommended anticancer moieties (hydrazine amides, hydrazine thioacetamides, benzylidene hydrazones, sulphonamides, benzoic acid, and thioacetamides derivatives) as the groove binding site with the presence of -NH- or -NH2 group to act as a cationic site were designed and synthesised as depicted in Figure 2.
Figure 2.
Molecular hybridisation of triazolo phthalazine moieties with different recommended anticancer moieties based on the basic pharmacophoric features of doxorubicin as DNA intercalator and topoisomerase II inhibitors.
2. Results and discussion:
2.1. Chemistry
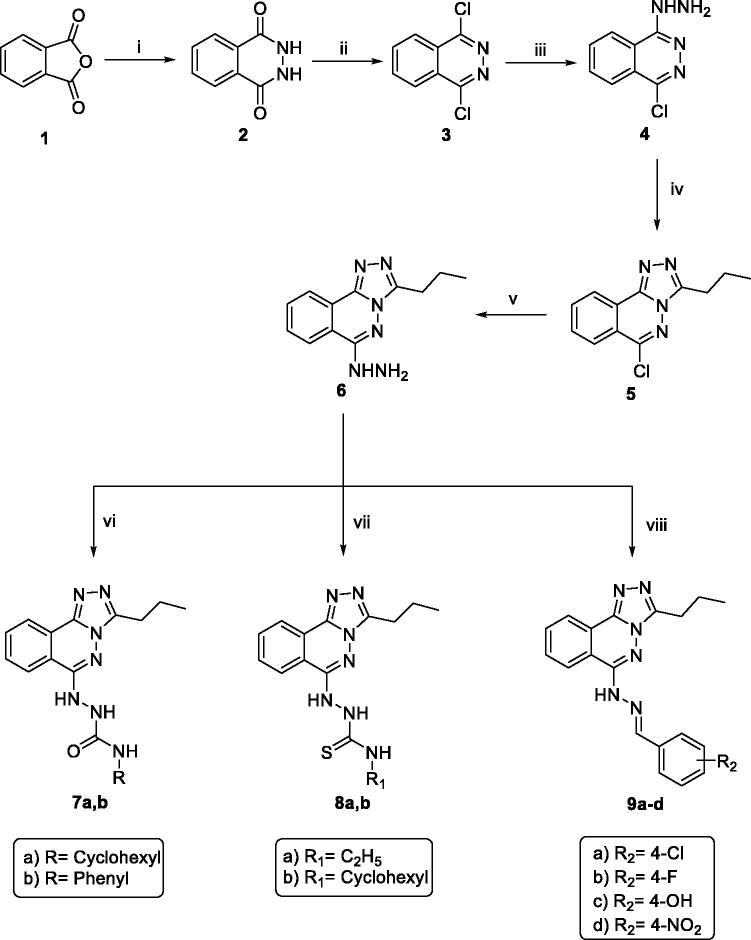
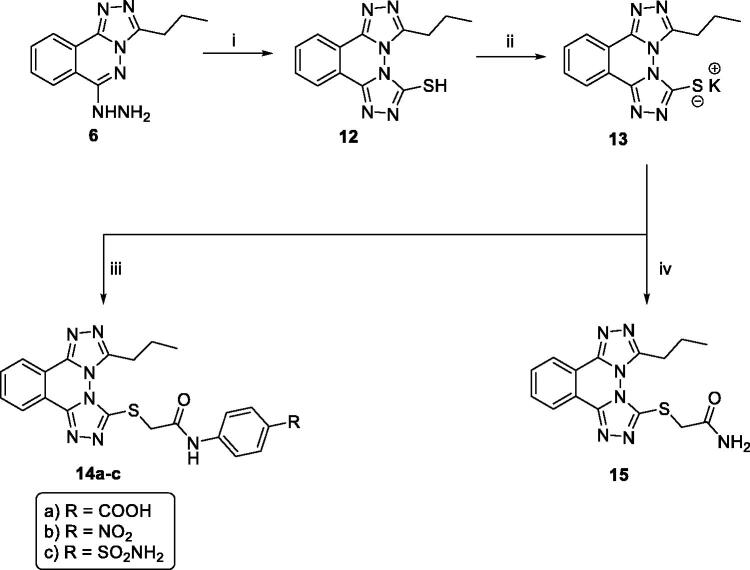
The new triazolo phthalazine members were synthesised following the reactions outlined in Schemes 1 and 2. 2,3-Dihydrophthalazine-1,4-dione 2 was prepared by reaction of phthalic anhydride 1 with hydrazine hydrate in absolute ethanol34. Compound 2 was then chlorinated with phosphorus oxychloride to afford 1,4-dichlorophthalazine 335, which was then heated with hydrazine hydrate in boiling ethanol36 to furnish 1-chloro-4-hydrazinylphthalazine 4. A solvent-free reaction was performed to cyclize compound 4; thus, compound 4 was heated with butyric anhydride to give the cyclized member 537. Reflux of compound 5 with hydrazine hydrate in boiling ethanol afforded the target hydrazinyl triazolo derivative 6. Compound 6, however, was allowed to react with different isocyanates and/or isothiocyanates to afford the corresponding semicarbazides 7a,b, and/or thiosemicarbazides 8a,b, respectively. Furthermore, treating the hydrazinyl compound 6 with appropriate substituted benzaldehyde derivatives with a catalytic amount of glacial acetic acid afforded the corresponding imines (Schiff's bases) 9a-d. IR charts of the later compounds revealed the loss of NH2 absorption band of compound 6 and the presence of NH absorption bands in the range of 3180 to 3242 cm−1. In contrast, 1H NMR spectra of members 9a-d displayed characteristic singlet signals in the range of δ 8.12 − 8.72 ppm representing the new benzylidene protons. The 13 C NMR spectra of compounds 9a-d, however, showed a characteristic downfield peak around δ 141 ppm corresponding to the new benzyledine carbon (Scheme 1).
Scheme 1.
General procedure for synthesis of target compounds 7a,b, 8a,b and 9a-d; Reagents and conditions: (i) NH2NH2.H2O / EtOH/reflux/5 h, (ii) POCl3/heating/1 h, (iii) NH2NH2.H2O / EtOH/reflux/0.5 h, (iv) Butyric anhydride/reflux/1 h, (v) NH2NH2.H2O / EtOH/reflux/0.5 h, (vi) The appropriate Isocyanates/EtOH/reflux/3 h, (vii) The appropriate Isothiocyanate/EtOH/reflux/3h, (viii) The appropriate Aromatic aldehydes / EtOH / gl. acetic acid / reflux/4h.
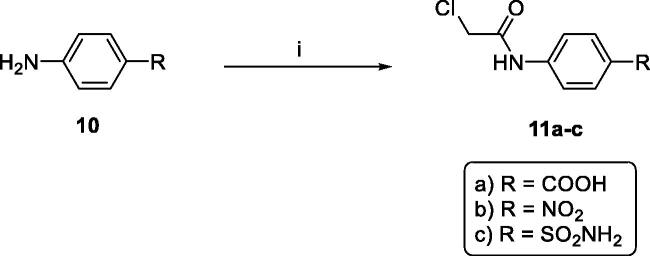
Scheme 2.
General procedure for synthesis of intermediates 11a-c, Reagents and conditions: (i) chloroacetyl chloride/DMF/stirring/1.5h.
Upon cyclisation of hydrazinyl triazolo derivative 6 with carbon disulphide in alcoholic potassium hydroxide, the corresponding mercaptotriazole derivative 12 was afforded.16 1H NMR spectrum of 12 displayed a singlet D2O exchangeable signal at δ 14.24 ppm corresponding to the SH proton. The potassium salt 13 was then obtained upon heating compound 12 with potassium hydroxide in absolute ethanol29. The potassium salt 13 was heated with the appropriate N-aryl-2-chloroacetamide derivatives 11a-c and/or 2-chloroacetamide in the presence of a catalytic amount of potassium iodide in DMF following the reported procedure to afford the corresponding thioacetamide derivatives, 14a-c and 15, respectively (Scheme 3).
Scheme 3.
General procedure for synthesis of target compounds 14a-d and 15; Reagents and conditions: (i) 1) CS2/KOH/EtOH/reflux/3 h, 2) HCl, (ii) KOH/absolute EtOH/reflux/0.5 h, (iii) N-Aryl-2-chloroacetamide derivatives 11a-c/DMF/heating/KI/heating/3 h. (iv) Chloroacetamide/DMF/heating over water bath/KI/heating/3 h.
2.2. Biological evaluation
2.2.1. In vitro anti-proliferative activities
Anti-proliferative activities of the target compounds were assessed via the standard MTT method38–40 against three cancer cell lines, namely, hepatocellular carcinoma (HepG-2), colorectal carcinoma (HCT-116), and human breast adenocarcinoma (MCF-7). Doxorubicin was used in this test as a positive control.
As illustrated in Table 1, the obtained results revealed that most synthesised compounds showed remarkable anti-proliferative activities against the tested cell lines.
Table 1.
Anti-proliferative activities towards HepG2, HCT-116, and MCF-7 cell lines.
| Comp. No. |
In vitro Cytotoxicity IC50 (µM)a |
||
|---|---|---|---|
| HepG-2 | HCT-116 | MCF-7 | |
| 7a | 24.92 ± 0.8 | 23.8 ± 0.81 | 19.45 ± 0.62 |
| 7b | 44.96 ± 1.1 | 48.58 ± 1.60 | 50.69 ± 1.55 |
| 8a | 28.86 ± 0.67 | 25.04 ± 0.70 | 16.48 ± 0.43 |
| 8b | 10.92 ± 0.34 | 13.79 ± 0.44 | 12.54 ± 0.32 |
| 9a | 25.16 ± 0.70 | 36.29 ± 1.10 | 45.52 ± 1.22 |
| 9b | 63.86 ± 1.02 | 61.71 ± 1.89 | 35.99 ± 0.98 |
| 9c | 56.32 ± 1.73 | 78.49 ± 2.04 | 57.76 ± 1.56 |
| 9d | 5.08 ± 0.17 | 4.74 ± 0.15 | 4.95 ± 0.10 |
| 14a | 5.65 ± 0.25 | 4.35 ± 0.19 | 4.36 ± 0.20 |
| 14b | 13.40 ± 0.40 | 12.64 ± 0.45 | 14.94 ± 0.55 |
| 14c | 24.75 ± 0.88 | 30.99 ± 0.11 | 20.64 ± 0.88 |
| 15 | 38.63 ± 1.00 | 53.54 ± 1.55 | 26.18 ± 0.77 |
| Doxorubicin | 8.28 ± 0.32 | 9.62 ± 0.50 | 7.67 ± 0.37 |
aIC50 values are the mean ± SD of three separate experiments.
In general, compounds 9d and 14a were found to be more active than the reference drug, doxorubicin, against the three tested cell lines. In particular, compound 9d was the most potent counterpart with IC50 values of 5.08, 4.74, and 4.95 µM as it was 1.63, 2.03, and 1.34 times more active than doxorubicin (IC50 = 8.28, 9.62, and 7.67 µM) against HepG2, HCT‐116, and MCF‐7 cell lines, respectively. While, compound 14a was about 1.46, 2.28, and 1.75 times as active as doxorubicin with IC50 values of 5.65, 4.35, and 4.36 µM. Moreover, compounds 8b and 14b were found to have satisfactory cytotoxicity against HepG2, HCT-116, and MCF-7 cell lines with IC50 values ranging from 10.92 to 14.94 µM. The rest of the compounds exhibited moderate anti-proliferative activities against the three tested cell lines.
2.2.2. Structure activity relationship (SAR)
The biological testing results provided us with a valuable SAR. Regarding the cyclohexyl bearing derivatives, it was noticed that compound 7a (incorporating N-cyclohexylsemicarbazide moiety) was more potent than compound 8b (incorporating N-cyclohexylthiosemicarbazide moiety) in both cytotoxic and Topo II inhibitory effects, as well. However, the later compounds were more active that their counterparts 7b (bearing a phenylsemicarbazide moiety) and 8a (bearing an ethylthiosemicarbazide moiety), respectively. For benzylidenehydrazine derivatives (compounds 9a-d), the effect of the substitution on the aromatic moieties in the order of 4-NO2 (9b) > 4-Cl (9a) > 4-OH (9c) > 4-F (9b). With regard to bis([1, 2, 4]triazolo)[3,4-a:4′,3′-c]phthalazine-3-thiol derivatives (compounds 14a-c), the activities decreased in the order of the substitution with 4-COOH (14a) > 4-NO2 (14b) > 4-SO2NH2 (14c).
2.2.3. Dna intercalation assay (DNA/methyl green colorimetric assay)
DNA/methyl green assay was carried out for the synthesised derivatives using doxorubicin as a positive control following the reported procedure described by Burre et al.41, to give extra quantitative data about the binding affinity of the target compounds towards the DNA molecules. DNA‐binding affinities of the target compounds were represented as IC50 values and are summarised in Table 2.
Table 2.
DNA intercalating affinity and IC50 values of the tested compounds against DNA and Topo II, respectively.
| Comp. No. | DNA/methyl green (IC50) (µM)a,b |
Topoisomerase II (IC50) (µM)a,c |
|---|---|---|
| 7a | 37.14 ± 2.0 | NTd |
| 7b | 36.57 ± 1.82 | NTd |
| 8a | 29.63 ± 1.41 | 22.28 ± 2.00 |
| 8b | 34.65 ± 1.10 | 8.91 ± 0.77 |
| 9a | 43.81 ± 2.22 | 27.66 ± 2-51 |
| 9b | 49.93 ± 2.53 | NTd |
| 9c | 62.18 ± 2 .20 | 21.39 ± 1.90 |
| 9d | 26.19 ± 1.14 | 7.02 ± 0.54 |
| 14a | 28.74 ± 1.71 | 7.64 ± 0.66 |
| 14b | 34.35 ± 2.80 | 13.66 ± 1.02 |
| 14c | 46.34 ± 2.30 | NTd |
| 15 | 71.15 ± 3.11 | NTd |
| Doxorubicin | 31.27 ± 1.8 | 9.65 ± 0.77 |
aThree independent experiments were performed for each concentration.
b50% Inhibition concentration values of DNA/methyl green assay.
c50% Inhibition of Topo II.
dNot tested.
Compounds 8a, 9d, and 14a exhibited excellent DNA binding affinities more than the reference drug with IC50 values of 29.63 ± 1.41, 26.19 ± 1.10, and 28.74 ± 1.71 µM, respectively. In addition, compounds 7a, 7b, 8b, and 14b showed remarkable activities but slightly less than the reference drug with IC50 values of 37.14 ± 2.0, 36.57 ± 1.8, 34.65 ± 1.1, and 34.35 ± 2.80 µM, respectively. Moreover, some compounds as 9a, 9b, and 14c showed moderate activities with IC50 values ranging from 43.81 ± 2.20 to 49.93 ± 2.53 µM. Finally, compounds 9c and 15 exhibited weak affinities towards DNA with IC50 values ranging from 62.18 ± 2.20 to 71.15 ± 3.11 µM, respectively.
2.2.3. Topoisomerase II inhibitory activity
Seven compounds that exhibited significant DNA binding affinities (8a, 8b, 9a, 9c, 9d, 14a, and 14b) were further estimated to determine their inhibitory activities towards topoisomerase II. The activity of topoisomerase II was determined according to the reported procedure described by Patra et al.42. Doxorubicin was utilised as a positive control in this test. The results were reported as IC50 values and summarised in Table 2. Compounds 8b, 9d, and 14a was found to be the most potent derivatives with IC50 values of 8.91 ± 0.77, 7.02 ± 0.54, and 7.64 ± 0.66 µM, which were more active than the reference drug, doxorubicin (IC50 = 9.65 ± 0.77 µM). The other tested compounds, 8a, 9a, 9c, and 14 b, exhibited moderate to weak activities with high IC50 values ranging from 13.66 ± 1.02 to 13.66 ± 1.02 µM.
2.2.4. In vivo antitumor activity
To examine the in vivo anticancer activity of compound 9d, adult female Swiss albino mice (30 mice) inoculated with I.P. injection of Solid Ehrlich Carcinoma (SEC) tumour cell lines in a volume of 0.2 ml physiological saline contains 1 × 106 viable cells for 24 h.
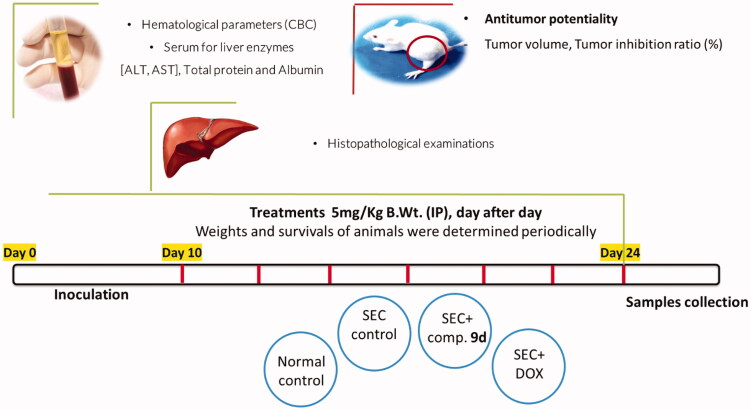
These mice were randomly divided into four groups (7 mice/group). The 1st group (normal saline-control group) was used as a negative control, the 2nd group (the SEC-control group) was injected with the SEC, the 3rd group (compound-treated group) was injected with SEC then with compound 9d, and the 4th group was injected with the SEC then with a standard anticancer drug, doxorubicin (DOX), as described in the experimental section. Bodyweight and survival were recorded daily until the 24th day in both treated and control groups. At the end of the experiment, the blood of each group was collected under light anaesthesia for the estimation of hematological and biochemical assays. The anaesthetised animals were then sacrificed to evaluate of the antitumor activity and to conduct hematological, biochemical, and histopathological assays, Figure 3.
Figure 3.
Methodology and Experimental design of the in vivo study.
2.2.4.1. Antitumor potentiality
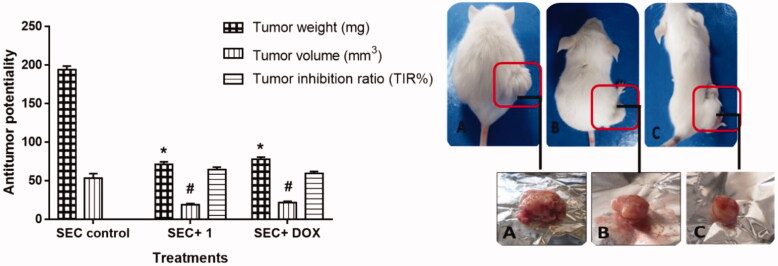
In vivo anticancer activity of the compound, 9d was estimated against SEC development. At first, the tumour development caused a 194 mg increase in solid tumour weight during the experimental period. During this study, treatment with compound 9d and doxorubicin significantly reduced the increase in the solid tumour mass by 63.4 (71 mg) and 59.8% (78 mg), compared to control as represented in Figure 3. Treatment with Compound 9d significantly inhibited tumour inhibition ratio (TIR) % by 64.5 in tumour volume (19 mm3) compared to doxorubicin (DOX) treatment with TIR% of 59 (22 mm3), compared to control. This indicated that compound 9d and doxorubicin had a significant antitumor effect, Figure 4.
Figure 4.
Left panel: Bar chart representation of the effect of compound 9d treatment on the proliferation of solid tumour mass in the SEC-bearing mice. Right panel: Morphological representation for the tumour mass volume of A: SEC-group, B: SEC+ 9d, and C: SEC + DOX. Values are expressed as Mean ± SEM values of mice in each group (n = 7). Signs of * and # are values with significant differences in tumour weight and tumour volume, respectively compared to SEC control using an unpaired t-test (P ≤ 0.05) using GraphPad prism.
2.2.4.2. Hematological and biochemical assays (Blood parameters assay)
At the end of the experiment, animals from different groups were sacrificed, and blood samples were collected for hematological parameters, including Hb, RBC’s, and WBC’s levels, and serum for determination of liver enzymes ALT, AST levels, and proteins.
Liver enzymes ALT and AST were significantly increased to 63.4, 64.67 (U/L), respectively, following tumour inoculation as shown in Table 3, compared with normal mice at 45.14 and 53.67 (U/L) because of hepatocellular damage. While liver protein and albumin were decreased to 6.13 and 2.97 (g/dL). Treatment with compound 9d substantially reduced liver enzymes to 42.9, 55.6 U/L, respectively, and increased liver protein and albumin to 8.04 and 6.25 (g/dL), indicating a remarkable amelioration in the hepatocellular functions.
Table 3.
Biochemical and hematological parameters in the tested groups.
| Parameter/ Treatment |
Biochemical parameters |
Hematological parameters |
|||||
|---|---|---|---|---|---|---|---|
| ALT (U/L) |
AST (U/L) |
Total Protein (g/dL) |
Albumin (g/dL) |
Hb (g/dL) |
RBCs count (×106/µL) |
WBCs count (×103/µL) |
|
| Normal control | 45.14 ± 2.69 | 53 ± 2.7 | 9.88 ± 0.35 | 5.95 ± 0.44 | 9.09 ± 0.61 | 6.08 ± 0.77 | 4.28 ± 0.44 |
| SEC control | 63.4 ± 4.53 | 64.67 ± 3.6 | 6.13 ± 0.24 | 2.97 ± 0.17 | 5.36 ± 0.41 | 3.33 ± 0.57 | 6.21 ± 0.57 |
| SEC + 9d (5 mg/kg BW) | 42.9#±1.01 | 55.6#±3.1 | 8.04#±0.41 | 6.25#±0.53 | 8.2#±0.31 | 5.37 ± 0.37 | 3.72#±0.46 |
| SEC + DOX (5 mg/kg BW) | 38.67#±1.6 | 48.4#±3.3 | 6.73 ± 0.26 | 6.01#±0.22 | 7.82 ± 0.27 | 5.26 ± 0.38 | 4.11 ± 0.57 |
Values are expressed as Mean ± SEM (n = 7).
#Significant difference between treated groups and SEC control using unpaired t-test (P ≤ 0.05) using the GraphPad prism7.
In terms of hematological parameters in SEC-bearing mice, all CBC parameters were changed in the SEC control, with Hb content and RBCs significantly decreased to 5.36 (g/dL) and 3.33 (106/µL), respectively. When compared to normal control levels, the WBC count was significantly increased to 6.21 (103/µL). Tumour propagation is routinely associated with decreased haemoglobin, RBC, and WBC counts43,44. After treatment with compound 9d, CBC levels were nearly restored to normal, where it elevated the Hb (8.2 g/dL), RBC’s (5.37 106/µL) and reduced the WBC’s (3.72 103/µl) levels.
Interestingly our results following previous studies45,46, illustrated the anticancer activity by improving hematological and biochemical parameters after treatment with the tested compound. Taken together, treatment of SEC mice with compound 9d improved hematological and biochemical parameters, as well as tumour weight and volume.
2.2.4.3. Histopathological examinations
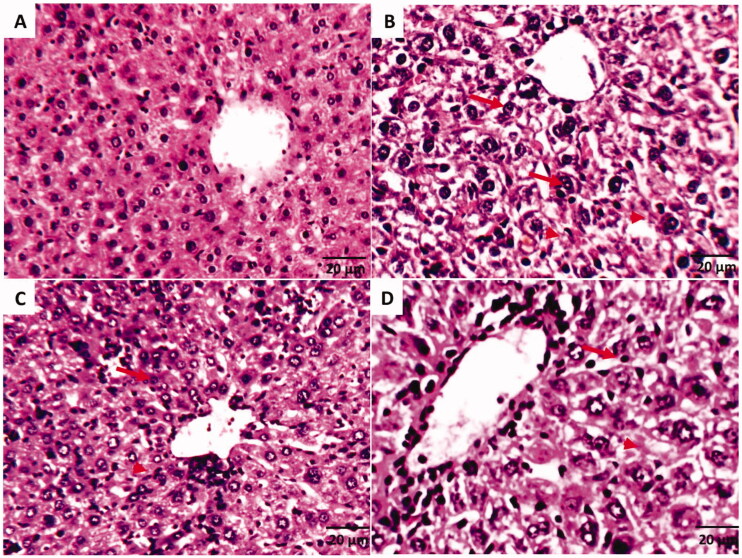
Histopathological examinations of liver tissues of the SEC-bearing mice in different treatments were illustrated in Figure 5. According to compound 9d ability to improve liver enzymes and proteins, its treatment was able to keep liver structure close to normal.
Figure 5.
Histopathological examinations of liver tissues of SEC-bearing mice in different treatments (A) Normal control group that shows the normal structure of central vein surrounded with hepatocytes. (B) SEC control group shows pyknosis (arrows) & karyolysis (arrowhead), hydropic degeneration of hepatocytes, and loss of cell boundaries. (C) SEC group treated with 9d (5 mg/Kg BW) that shows hepatic cells are near normal and show tissue improvement as compared with a little hydropic degeneration. (D) SEC group treated with DOX shows tissue enhancement like normal group, but still, some hydropic degeneration, pyknosis (arrows) and karyolysis (arrowhead) were shown. (H&E stain, magnification ×200).
2.3. In silico studies
2.3.1. Docking studies
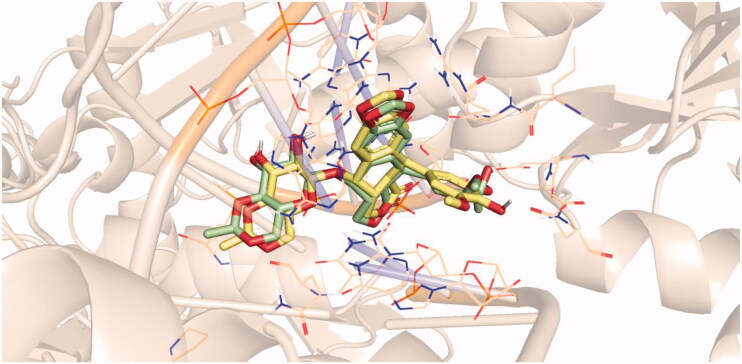
Molecular docking studies were performed to shed light on the binding modes of the newly synthesised compounds inside the DNA binding site of Topo II (PDB ID: 3qx3). Docking investigation was carried out using Discovery Studio 2.5 software. An X-ray crystallographic structure of Topo II with its co-crystallised ligand, etoposide, was downloaded from the Protein Data Bank (PDB). Re-docking of the co-crystalized ligand was initially performed aiming to validate the used docking protocol. The simulation of the re-docked ligand successfully regenerated the same binding mode of the co-crystalized one inside the DNA binding site of Topo II with RMSD of 0.81 Å, which indicates the validity of the docking process, Figure 6.
Figure 6.
Superimposition of the co-crystallised ligand (light green) and the docking pose (light yellow) of the same molecule.
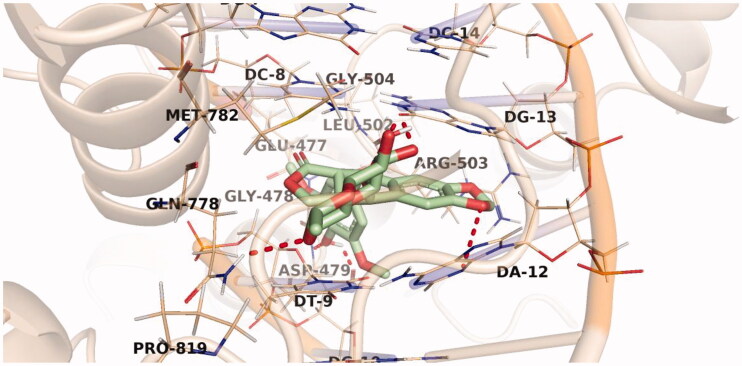
The predicted binding pattern of the co-crystallised ligand, etoposide, revealed an affinity value of −30.13 kcal/mol with the formation of six H-bonds. The planar aromatic system occupied the hydrophobic pocket formed by Glu477, Gly478, Asp479, Leu502, Arg503, Gln778, Met782, and Pro819. It was also stacked between different DNA nucleotides, namely, Cytosine (DC-8 and DC-14), Guanine (DG-7, DG-10, and DG-13), Adenine (DA-12), and Thymine (DT-9). The sugar moiety of etoposide was directed towards the DNA minor groove and stabilised by the formation of two H-bond interactions with Gln778 and DG-13. Similarly, its phenolic OH group formed two H-bond interactions with Asp479. Two H-bonds were also formed between the etoposide oxygen atoms and the DNA nucleotides DG-13 and DA-12 Figure 7.
Figure 7.
Binding of etoposide with DNA-Topo II, the hydrogen bonds are represented in red dashed lines.
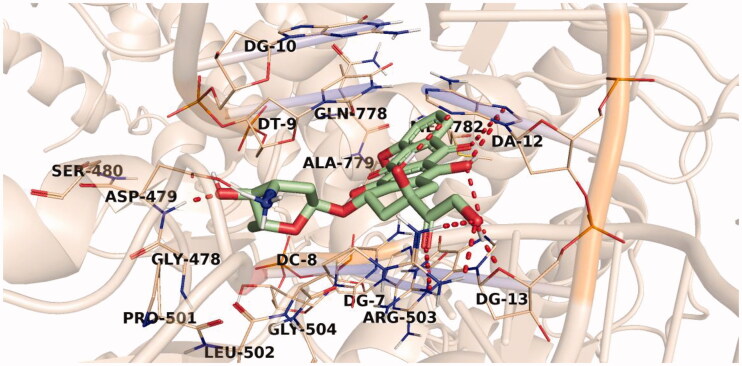
The proposed binding mode of doxorubicin, with an affinity value of −33.50 kcal/mol, revealed that the doxorubicin planar aromatic chromophore formed aromatic stacking interactions with the different key residues Glu477, Gly478, Asp479, Leu502, Arg503, Gln778, Met782 in addition to the DNA nucleotides DT-9, DC-8, DC-11, DG-13, and DA-12. The sugar moiety of doxorubicin was oriented into the minor groove of DNA and stabilised by two H-bonds with Asp479. The rest of the compound was involved in several H-bond interactions with Arg503, DG-13, and DA-12, Figure 8.
Figure 8.
Binding of doxorubicin with DNA-Topo II, the hydrogen bonds are represented in red dashed lines.
A general investigation of docking results revealed that the designed compounds displayed a binding pattern comparable to that of the native ligand with predicted binding energy scores ranging from −18.49 to −29.91 kcal/mol.
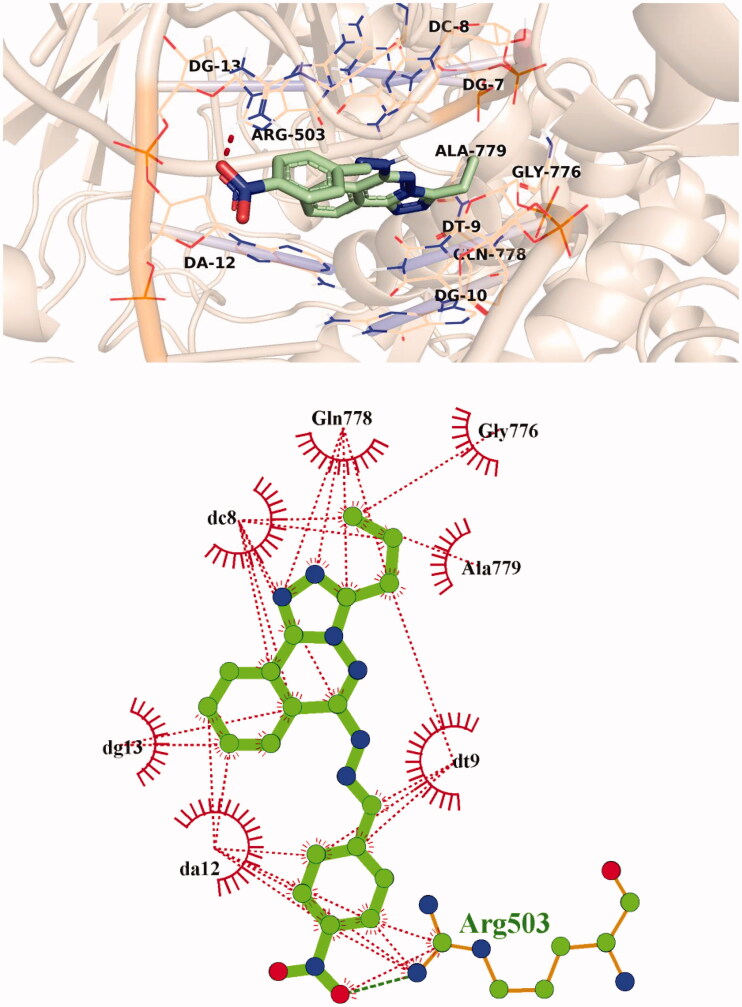
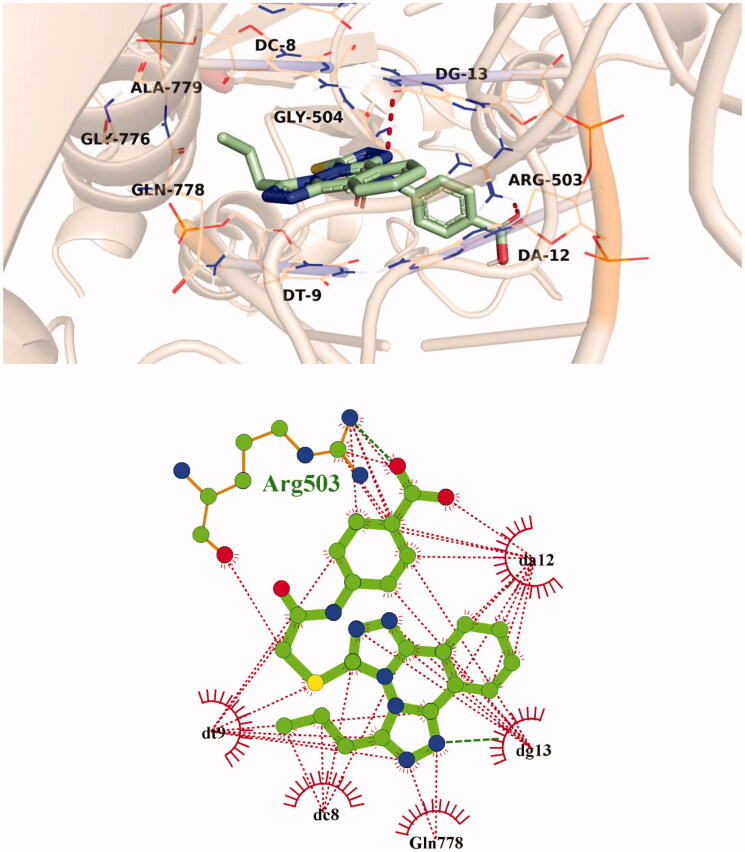
The predicted binding mode of compound 9d as illustrated in Figure 9. Its triazolo phthalazine planner moiety was inserted between the DNA nucleotides with the formation of many hydrophobic interactions with DT-9, DC-8, DG-13, and DA-12 as well as Gly776, Gln778, and Ala779 amino acids. In addition, the 4-nitrophenyl part was oriented in the minor groove of DNA, forming hydrophobic interactions with DA-12 and Arg503. The nitro group of 9d, however, interacted with Arg503 via an H-bond interaction.
Figure 9.
3D and 2D illustration of compound 9d in the Topo II active site.
Compound 14a showed an affinity value of −27.36 kcal/mol. The planar aromatic system occupied the hydrophobic pocket formed by DT-9, DC-8, DG-13, and DA-12 nucleotides in addition to Arg503, Gly504, Gly776, Gln778, and Ala779 residues forming several pi-pi interactions. The benzoic acid moiety was directed towards the DNA minor groove with the formation of H-bond interaction with Arg503 residue Figure 10.
Figure 10.
3D and 2D illustration of compound 14a in the Topo II active site.
2.3.2. In silico ADMET analysis
ADMET studies were carried out for the synthesised compounds using doxorubicin as a reference compound. The predicted ADMET parameters were listed in Table 4.
Table 4.
Predicted ADMET profile for the synthesised compounds
| Comp. | BBB levela | Solubility levelb | Absorption levelc | CYP2D6 predictiond | PPB predictione |
|---|---|---|---|---|---|
| 7a | 4 | 2 | 0 | false | true |
| 7b | 3 | 2 | 0 | false | true |
| 8a | 2 | 2 | 0 | false | true |
| 8b | 1 | 1 | 0 | false | true |
| 9a | 1 | 1 | 0 | true | true |
| 9b | 1 | 1 | 0 | true | true |
| 9c | 2 | 2 | 0 | false | false |
| 9d | 4 | 1 | 1 | false | true |
| 14a | 4 | 2 | 2 | false | false |
| 14b | 4 | 1 | 2 | false | false |
| 14c | 4 | 1 | 2 | false | true |
| 15 | 3 | 2 | 0 | false | false |
| Doxorubicin | 4 | 2 | 3 | false | false |
aBBB level, blood brain barrier level, 0 = very high, 1 = high, 2 = medium, 3 = low, 4 = very low.
bSolubility level, 1 = very low, 2 = low, 3 = good, 4 = optimal.
cAbsorption level, 0 = good, 1 = moderate, 2 = poor, 3 = very poor.
dCYP2D6, cytochrome P2D6, TRUE = inhibitor, FALSE = non inhibitor.
ePBB, plasma protein binding, FALSE means less than 90%, TRUE means more than 90%.
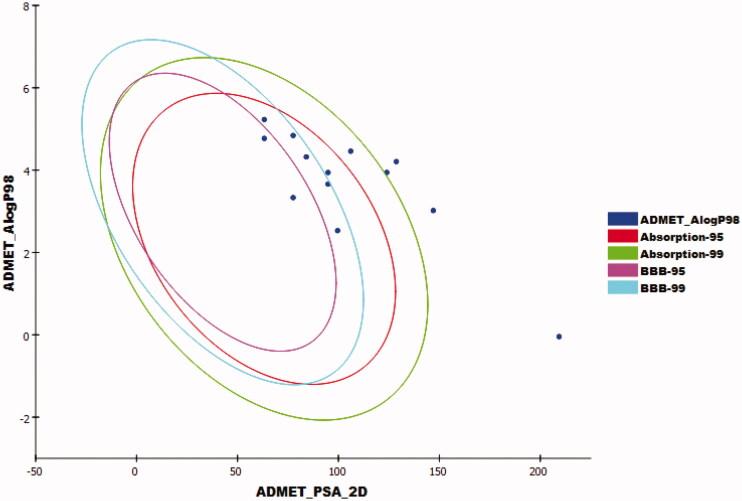
The results revealed that compounds 7a, 9d, 14a, 14 b, and 14c had very low Blood Brain Barrier penetration power. Accordingly, such compounds were expected to be safe to CNS. Aqueous solubility of the synthesised compounds ranged from low to very low. Compounds 7a, 7 b, 8a, 8 b, 9a, 9 b, 9c, and 15 showed good absorption level. Except compounds 9a and 9b, all members were predicted as non-inhibitors of CYP2D6. Except compounds 9c, 14a, 14 b, and 15, all compounds were expected to bind plasma protein more than 90% (Figure 11).
Figure 11.
The expected ADMET study of the target compounds.
2.3.3. Toxicity studies
Toxicity prediction was carried out based on the validated and constructed models in Discovery studio software47,48. As shown in Table 5, most compounds showed in silico low adverse effects and toxicity against the tested models. Regarding developmental toxicity potential (DTP), all the tested compounds were predicted to be non-toxic. For Carcinogenic Potency TD50 (Rat), all compounds showed higher values (from 0.873 to 34.570 mg/kg body weight/day) than that of doxorubicin (0.861 mg/kg body weight/day) except compound 7a (0.651 mg/kg body weight/day). For rat maximum tolerated dose model, compounds 7 b, 8a, 8 b, 9a, 9 b, and 9c showed higher levels than doxorubicin. The tested compounds showed high oral LD50 values ranging from 0. 0.229to 12.326 mg/kg body weight/day which were higher than that of doxorubicin (0.227 mg/kg body weight/day) Moreover, except compounds 9a and 9d, all compounds were predicted to be mild and non-irritant against ocular irritancy and skin irritancy models, respectively.
Table 5.
Toxicity properties of the synthesised compounds
| Comp. | DTP | Carcinogenic Potency TD50 (Rat)a |
Rat Maximum Tolerated Dose (Feed)b |
Rat Oral LD50b | Ocular Irritancy | Skin Irritancy |
|---|---|---|---|---|---|---|
| 7a | Non-Toxic | 0.651 | 0.222 | 0.861 | Mild | Non-Irritant |
| 7b | Non-Toxic | 19.886 | 0.369 | 2.513 | Mild | Non-Irritant |
| 8a | Non-Toxic | 34.570 | 0.366 | 0.742 | Mild | Non-Irritant |
| 8b | Non-Toxic | 0.873 | 0.260 | 0.297 | Mild | Non-Irritant |
| 9a | Non-Toxic | 2.505 | 0.290 | 0.447 | Mild | Irritant |
| 9b | Non-Toxic | 2.668 | 0.311 | 0.229 | Mild | Non-Irritant |
| 9c | Non-Toxic | 20.483 | 0.779 | 0.501 | Mild | Non-Irritant |
| 9d | Non-Toxic | 1.991 | 0.177 | 0.573 | Mild | Irritant |
| 14a | Non-Toxic | 25.171 | 0.429 | 2.371 | Mild | Non-Irritant |
| 14b | Non-Toxic | 5.431 | 0.090 | 1.863 | Mild | Non-Irritant |
| 14c | Non-Toxic | 22.020 | 0.095 | 12.326 | Mild | Non-Irritant |
| 15 | Non-Toxic | 23.275 | 0.118 | 0.542 | Mild | Non-Irritant |
| Doxorubicin | Toxic | 0.861 | 0.277 | 0.227 | Mild | Non-Irritant |
aUnit: mg/kg body weight/day.
bUnit: g/kg body weight.
3. Conclusion
A new series of phthalazine derivatives was designed hoping to discover novel Topo II inhibitor and DNA intercalator agents as well. Twelve compounds were synthesised and tested in vitro for their anti-proliferative activities against three human cancer cell lines, HepG-2, MCF-7, and HCT-116. The tested members exhibited a promising cytotoxic effect with IC50 values ranging from 4.35 ± 0.19 to 78.49 ± 2.04 µM. All compounds were further estimated for their in vitro DNA intercalating effects. Amongst, seven compounds were further examined for their in vitro inhibitory activity against Topo II enzyme. Three compounds, 8b, 9d, and 14a, out of the seven exhibited potent Topo II inhibitory activities with IC50 values of 8.91 ± 0.77, 7.02 ± 0.54, and 7.64 ± 0.66 µM, respectively. Finally, in vivo antitumor studies were carried out for compound 9d. In vivo study exhibited that treatment with compound 9d substantially inhibited tumour proliferation reducing solid tumour volume and mass. Additionally, it restored liver enzymes, proteins, and CBC parameters near-normal, indicating a remarkable amelioration in their functions along with histopathological examinations. Hence, compound 9d was investigated as a novel anti-cancer agent through Topo II inhibition and DNA-binding affinity. To conclude, compounds presented in the current study were proved to be potent Topo II inhibitors with DNA intercalating efficacy that can be further adopted for hit optimisation and/or lead discovery.
4. Experimental
4.1. Chemistry
Starting materials and reagents were purchased from Sigma-Aldrich and used without purification. Melting points measurement was carried out by a Gallen lamp melting point apparatus and are uncorrected. Reactions progress was monitored by TLC (Merck, Germany), the spots were detected by exposure to UV lamp at λ 254 nm. IR spectra were recorded by pye Unicam SP 1000 IR spectrophotometer using KBr discs and expressed in wavenumber (cm−1). 1H and 13 C NMR spectra were recorded with Bruker Advance 400 spectrophotometer operating at 400 MHz and 100 MHz, respectively and the chemical shifts were given in δ as parts per million (ppm) downfield from tetramethylsilane (TMS) as internal standard. The mass spectra were recorded on Varian MAT 311-A (70 e.v.).
The previously reported compounds 2,3-dihydrophthalazine-1,4-dione 234, 1,4-dichlorophthalazine 335 and 1-chloro-4-hydrazineylphthalazine 435, 6-chloro-3-propyl-[1, 2, 4]triazolo[3,4-a]phthalazine 549, and N-aryl-2-chloroacetamide 11a-c50 were synthesised following the described procedures.
4.1.1. 6-Hydrazineyl-3-propyl-[1, 2, 4]triazolo[3,4-a]phthalazine 6
To a boiling solution of hydrazine hydrate 70% (3.73 ml, 0.074 mol) in ethanol (50 ml), 6-chloro-3-propyl-[1, 2, 4]triazolo[3,4-a]phthalazine 5 (2.46 g, 0.01 mol) was added. The reaction mixture was refluxed for 0.5 h then cooled. The obtained precipitate was filtered, washed with petroleum ether (3 × 20 ml), dried, and recrystallized from ethanol to obtain compound 6.
White crystals (yield 83%); m.p. 259–261 °C; IR (KBr) ν cm-1: 3329, 3143, 3136; 1H NMR (DMSO-d6) δ ppm: 0.98 (t, J = 7.2 Hz, 3H, CH3), 1.36 (m, 2H, CH2), 3.01 (t, J = 7.2 Hz, 2H, CH2), 3.75 (br s, 2H, exchangeable with D2O, NH2), 7.77 (dd, J = 8.4, 7.2 Hz, 1H, Ar-H,), 7.89 (dd, J = 7.2, 8.0 Hz, 1H, Ar-H), 8.24 (d, J = 8.4 Hz, 1H, Ar-H), 8.36 (d, J = 8.0 Hz, 1H, Ar-H), 8.95 (s, 1H, exchangeable with D2O, NH); Mass (m/z): 242.51 (M+, 16%), 143.05 (100%).
4.1.2. General procedure for the synthesis of target compounds 7a,b
A mixture of the hydrazinyl compound 6 (0.242 g, 0.001 mol) and the appropriate isocyanate namely, cyclohexyl isocyanate and phenyl isocyanate (0.001 mol) were refluxed in absolute ethanol (25 ml) for 3 h. The solution was cooled. Then, the obtained solid was filtered and recrystallized from ethanol to produce compounds 7a,b, respectively.
4.1.2.1. N-Cyclohexyl-2–(3-propyl-[1, 2, 4]triazolo[3,4-a]phthalazin-6-yl)hydrazine-1-carboxamide 7a
White crystals (yield 89%); m.p. 269–271 °C; IR (KBr) ν cm−1: 3290, 3271, 3147, 1674; 1H NMR (DMSO-d6) δ ppm: 0.95 (t, 3H, CH3), 1.06 (m, 2H, CH2), 1.14 (m, 2H, CH2), 1.19 (m, 3H, CH3), 1.53 (m, 2H, CH2), 1.65 (m, 2H, CH2), 1.75 (m, 2H, CH2), 2.94 (t, J = 7.6 Hz, 2H, -CH2), 3.44 (m, 1H, CH), 6.48 (s, 1H, exchangeable with D2O, NH), 7.68 (s, 1H, exchangeable with D2O, -NH), 7.84 (dd, J = 8.0, 7.6 Hz, 1H, Ar-H), 7.91 (dd, J = 7.6, 7.6 Hz, 1H, Ar-H,), 8.35 (d, J = 8.0 Hz, 1H, Ar-H), 8.41 (d, J = 7.6 Hz, 1H, Ar-H), 9.41 (s, 1H, exchangeable with D2O, NH); MS (m/z): 367.19 (M+, 14.7%), 332 (100%, base peak).
4.1.2.2. N-Phenyl-2–(3-propyl-[1, 2, 4]triazolo[3,4-a]phthalazin-6-yl)hydrazine-1-carboxamide 7 b
White crystals (yield 82%); mp: 258–260 °C; IR (KBr) ν cm−1: 3271, 3217, 3147, 1662; 1H NMR (DMSO-d6) δ ppm: 0.97 (t, J = 7.6 Hz, 3H, CH3), 1.72 (m, 2H, CH2), 2.95 (t, J = 7.6 Hz, 2H, CH2), 6.95 (t, 1H, Ar-H), 7.25 (t, 2H, Ar-H), 7.4 (d, 2H, Ar-H), 7.88 (dd, J = 7.2, 8.0 Hz, 1H, Ar-H), 7.99 (dd, J = 8.0, 7.2 Hz, 1H, Ar-H), 8.40 (d, J = 8.0 Hz, 1H, Ar-H), 8.43 (d, J = 8.0 Hz, 1H, Ar-H), 8.18 (s, 1H, exchangeable with D2O, NH), 8.76 (s, 1H, exchangeable with D2O, NH), 9.64 (s, 1H, exchangeable with D2O, NH); 13 C NMR (DMSO-d6) δ ppm: 11.40, 13.61, 17.82, 117.82, 123.06, 124.17, 124.99, 127.20, 129.30 (2 C), 129.33, 130.03, 130.64, 133.60, 135.16, 141.93, 145.59, 148.75, 151.17.
4.1.3. General procedure for the synthesis of target compounds 8a,b
A mixture of compound 6 (0.242 g, 0.001 mol) and appropriate isothiocyanate namely, ethyl isothiocyanate, and cyclohexyl isothiocyanate (0.001 mol) in absolute ethanol (20 ml) were heated under reflux for 3 h. After cooling, the precipitate was collected, dried, and recrystallized from ethanol to afford compounds 8a,b, respectively.
4.1.3.1. N-Ethyl-2–(3-propyl-[1, 2, 4]triazolo[3,4-a]phthalazin-6-yl)hydrazine-1-carbothioamide 8a
White crystals (yield 69%); mp: 242–244 °C; IR (KBr) ν cm−1: 3365, 3273, 3228; 1H NMR (DMSO-d6) δ ppm: 0.95 (t, J = 6.4 Hz, 3H, CH3), 1.02 (t, J = 6.0 Hz, 3H, CH3), 1.67 (t, J = 7.6 Hz, 2H, CH2), 2.92 (q, J = 7.6 Hz, 2H, -CH2), 3.48 (q, J = 6.0 Hz, 2H, CH2), 7.80 (dd, J = 7.6, 7.2 Hz, 1H, Ar-H), 7.91 (dd, J = 7.2, 7.2 Hz, 1H, Ar-H), 8.25 (d, J = 7.6 Hz, 1H, Ar-H), 8.31 (s, 1H, exchangeable with D2O, NH), 8.34 (d, J = 7.2 Hz, 1H, Ar-H), 9.27 (s, 1H, exchangeable with D2O, NH), 9.68 (s, 1H, exchangeable with D2O, NH); 13 C NMR (DMSO-d6) δ ppm: 11.37, 14.94, 17.78, 19.56, 38.86, 117.66, 122.72, 123.62, 124.85, 130.45, 133.52, 142.02, 150.97, 151.44, 182.20.
4.1.3.2. N-Cyclohexyl-2–(3-propyl-[1, 2, 4]triazolo[3,4-a]phthalazin-6-yl)hydrazine-1-carbothioamide 8 b
White crystals (yield 80%); mp: 221–223 °C; IR (KBr) ν cm−1: 3367, 3242, 2924; 1H NMR (DMSO-d6) δ ppm: 0.94 (m, 1H, CH), 0.97 (t, 3H, CH3), 1.23 (m, 4H, 2CH2), 1.49 (m, 1H, CH), 1.62 (m, 2H, CH2), 1.71 (m, 2H, -CH2), 1.79 (m, 2H, CH2), 2.93 (t, J = 7.6 Hz, 2H, CH2), 4.23 (m, 1H, CH), 7.69 (s, 1H, exchangeable with D2O, NH), 7.82 (dd, J = 7.7, 7.5 Hz, 1H, Ar-H), 8.02 (dd, J = 7.8, 7.7 Hz, 1H, Ar-H), 8.14 (d, J = 7.5 Hz, 1H, Ar-H), 8.25 (d, J = 7.8 Hz, 1H, Ar-H), 9.22 (s, 1H, exchangeable with D2O, NH), 9.63 (s, 1H, exchangeable with D2O, NH); MS (m/z): 383.21 (M+, 14.7%), 301.33 (100%, base peak).
4.1.4. General procedure for the synthesis of target compounds 9a-d
Equimolar amounts of compound 6 (0.242 g, 0.001 mol) and the appropriate aldehyde namely 4-chlorobenzaldehyde, 4-flourobenzaldehyde, 4-hydroxybenzaldehyde, 4-nitrobenzaldehyde, (0.001 mol) were refluxed in absolute ethanol (25 ml) with a catalytic amount of glacial acetic acid for 4 h. The reaction was followed up by TLC. After the completion of the reaction, the mixture was cooled. The formed precipitate was filtered, dried, and recrystallized from ethanol to afford compounds 9a-d, respectively.
4.1.4.1. 6-[2–(4-Chlorobenzylidene)hydrazineyl]-3-propyl-[1, 2, 4]triazolo[3,4-a]phthalazine 9a
White crystals (yield 79%); mp: 251–253 °C; IR (KBr) ν cm−1: 3242, 3072; 1H NMR (DMSO-d6) δ ppm: 0.96 (t, J = 7.0 Hz, 3H, CH3), 1.45 (m, 2H, CH2), 3.09 (t, J = 7.6 Hz, 2H, CH2), 7.56 (2d, J = 8.4 Hz, 2H, Ar-H), 7.81 (2d, J = 8.8 Hz, 2H, Ar-H), 7.97 (dd, J = 8.0, 7.2 Hz, 1H, Ar-H), 8.02 (dd, J = 8.0, 8.8 Hz, 1H, Ar-H), 8.51 (d, J = 7.2 Hz, 1H, Ar-H), 8.54 (s, 1H, CH), 8.60 (d, J = 8.8 Hz, 1H, Ar-H), 11.56 (s, 1H, exchangeable with D2O, NH); 13 C NMR (DMSO-d6) δ ppm: 11.40, 17.83, 21.61, 117.85, 123.08, 124.16, 124.96, 127.20, 128.75, 129.91, 130.68, 132.42, 133.61, 139.82, 141.94, 145.80, 148.76, 151.19, 161.68.
4.1.4.2. 6-[2–(4-Fluorobenzylidene)hydrazineyl]-3-propyl-[1, 2, 4]triazolo[3,4-a]phthalazine 9 b
Yellowish white crystals (yield 71%); mp: 267–269 °C; IR (KBr) ν cm−1: 3180, 3064, 3031; 1H NMR (DMSO-d6) δ ppm: 0.99 (t, 3H, CH3), 1.59 (m, 2H, CH2), 3.10 (t, J = 7.6 Hz, 2H, CH2), 7.32 (2d, J = 8.8 Hz, 2H, Ar-H),7.83 (dd, J = 7.2, 7.6 Hz, 1H, Ar-H), 7.93 (2d, 2H, J = 8.8 Hz, Ar-H), 8.01 (dd, J = 7.6, 7.6 Hz, 1H, Ar-H), 8.52 (d, J = 7.2 Hz, 1H, Ar-H), 8.54 (d, J = 7.6 Hz, 1H, Ar-H), 8.72 (s, 1H, CH), 11.41 (s, 1H, exchangeable with D2O, NH).
4.1.4.3. 4-[(2–(3-Propyl-[1, 2, 4]triazolo[3,4-a]phthalazin-6-yl)hydrazineylidene)methyl]-phenol 9c
Reddish white crystals (yield 81%); mp: 249–251 °C; IR (KBr) ν cm−1: 3421, 3213, 3066; 1H NMR (DMSO-d6) δ ppm: 0.96 (t, 3H, CH3), 1.59 (m, 2H, CH2), 3.04 (t, J = 7.6 Hz, 2H, CH2), 6.86 (2d, J = 8.0 Hz, 2H, Ar-H), 7.61 (2d, J = 8.0 Hz, 2H, Ar-H), 7.86 (dd, J = 7.2, 7.6 Hz, 1H, Ar-H), 7.99 (dd, J = 7.2, 7.6 Hz, 1H, Ar-H), 8.39 (s, 1H, CH), 8.44 (d, J = 7.6 Hz, 1H, Ar-H), 8.48 (d, J = 7.6 Hz, 1H, Ar-H), 9.90 (s, 1H, exchangeable with D2O, OH), 11.03 (s, 1H, exchangeable with D2O, NH); 13 C NMR (DMSO-d6) δ ppm: 11.43, 17.84, 21.52, 117.90, 123.13, 124.24, 125.04, 127.23 (2 C), 129.96 (2 C), 130.73, 132.46, 133.68, 139.87, 141.98, 145.83, 148.82, 151.22.
4.1.4.4. 6-[2–(4-Nitrobenzylidene)hydrazineyl]-3-propyl-[1, 2, 4]triazolo[3,4-a]phthalazine 9d
Yellow crystals (yield 83%); mp: 251–253 °C; IR (KBr) ν cm−1: 3217, 3078, 3047, 2931; 1H NMR (DMSO-d6) δ ppm: 0.98 (t, 3H, CH3), 1.59 (m, 2H, CH2), 2.62 (t, J = 6.8 Hz, 2H, CH2), 7.46 (dd, J = 8.0, 7.6 Hz, 1H, Ar-H), 7.56 (2d, 2H, J = 8.8 Hz, Ar-H), 7.58 (dd, J = 8.0, 8.4 Hz, 1H, Ar-H), 7.87 (2d, J = 8.8 Hz, 2H, Ar-H), 8.03 (d, J = 7.6 Hz, 1H, Ar-H), 8.08 (d, J = 8.4 Hz, 1H, Ar-H), 8.12 (s, 1H, CH), 11.90 (s, 1H, exchangeable with D2O, NH); 13 C NMR (DMSO-d6) δ ppm: 11.40, 17.53, 21.55, 117.82, 123.06, 124.17, 124.99, 127.20 (2 C), 129.30 (2 C), 130.03, 130.64, 133.60, 135.16, 141.93, 145.59, 148.75, 151.17.
4.1.5. 6-Propylbis([1, 2, 4]triazolo)[3,4-a:4',3'-c]phthalazine-3-thiol 12
A mixture of compound 6 (2.42 g, 0.01 mol), carbon disulphide (0.71 ml, 0.01 mol) and potassium hydroxide (0.56 g, 0.01 mol) was refluxed in absolute ethanol (20 ml) for 3 h. The mixture was then cooled to room temperature and poured onto 1 N HCl (l20 ml). The yellow precipitated product was filtered, washed with distilled water, dried, and crystallised from ethanol to give compound 12.
Yellowish white crystal (yield 72%); mp > 300 °C; IR (KBr) ν cm−1: 3067, 2919, 2563, 1599; 1H NMR (DMSO-d6) δ ppm: 0.96 (t, 3H, CH3), 1.60 (m, 2H, CH2), 3.40 (t, J = 6.8 Hz, 2H, CH2), 7.38 (dd, J = 6.4, 7.6 Hz, 1H, Ar-H), 7.43 (dd, J = 7.6, 6.4 Hz, 1H, Ar-H), 7.77 (d, J = 7.6 Hz, 1H, Ar-H), 7.94 (d, J = 7.6 Hz, 1H, Ar-H), 14.24 (s, 1H, exchangeable with D2O, SH); MS (m/z): 284.11 (M+, 16.90%), 173.33 (100%, base peak).
4.1.6. Potassium 6-propylbis([1, 2, 4]triazolo)[3,4-a:4',3'-c]phthalazine-3-thiolate 13
A mixture of 20 (2.84 g, 0.01 mol) and potassium hydroxide (0.56 g, 0.01 mol) in absolute ethanol (20 ml) was heated with continuous stirring for 0.5 h. After cooling, a precipitate was produced. The precipitate was collected and washed with diethyl ether to afford the corresponding potassium salt 13.
4.1.7. General procedure for the synthesis of target compounds 14a-c and 15
A mixture of the potassium salt 13 (0.322 g, 0.001 mol) and the appropriate chloroacetanilides namely, 4–(2-chloroacetamido)benzoic acid 11a, 2-chloro-N-(4-nitrophenyl)acetamide 11b, 2-chloro-N-(4-sulfamoylphenyl) acetamide 11c, or 2-chloroacetamide (0.001 mol) in dry DMF (20 ml) with a catalytic amount of potassium iodide was heated over a water bath for 3 h. The reaction mixture was then cooled, poured into ice water (50 ml) and stirred well for 1 h. The separated solid was filtered, washed with water, dried, and crystallised from ethanol to afford the corresponding derivatives 14a-c and 15, respectively.
4.1.7.1. 4-[2-((6-Propylbis([1, 2, 4]triazolo)[3,4-a:4',3'-c]phthalazin-3-yl)thio)acetamido]benzoic acid 14a
White (yield 81%); mp: 244–246 °C; IR (KBr) ν cm−1: 3425, 3248, 3178, 1685, 1673; 1H NMR (DMSO-d6) δ ppm: 0.99 (t, 3H, CH3), 1.67 (m, 2H, CH2), 3.47 (t, J = 6.8 Hz, 2H, CH2), 4.51 (s, 2H, SCH2), 7.79 (2d, J = 6.0 Hz, 2H, Ar-H), 8.03 (m, 4H, Ar-H), 8.56 (2d, J = 6.0 Hz, 2H, Ar-H), 10.64 (s, 1H, exchangeable with D2O, NH).
4.1.7.2. N-(4-Nitrophenyl)-2-[(6-propylbis([1, 2, 4]triazolo)[3,4-a:4',3'-c]phthalazin-3-yl)thio]acetamide 14 b
Yellowish white crystals (yield 74%); mp: 277–279 °C; IR (KBr) ν cm−1: 3278, 3082, 1701; 1H NMR (DMSO-d6) δ ppm: 0.97 (t, 3H, CH3), 1.55 (m, 2H, CH2), 3.65 (t, J = 6.8 Hz, 2H, CH2), 4.38 (s, 1H, SCH2), 7.78 (dd, J = 8.4 Hz, 2H), 7.93 (m, 2H, Ar-H), 8.24 (dd, J = 8.4, 2H, Ar-H), 8.45 (m, 2H, Ar-H), 10.95 (s, 1H, exchangeable with D2O, -NH); 13 C NMR (DMSO-d6) δ ppm: 11.46, 17.84, 23.20, 40.76, 119.20 (2 C), 119.91, 120.39, 123.72, 123.66, 127.22 (2 C), 131.98, 132.29, 139.14, 141.91, 143.67, 145.52, 147.07, 149.78, 166.40.
4.1.7.3. 2-[(6-Propylbis([1, 2, 4]triazolo)[3,4-a:4',3'-c]phthalazin-3-yl)thio]-N-(4-sulfamoyl-phenyl)acetamide 14c
Yellowish white crystals (yield 76%); mp: 245–247 °C; IR (KBr) ν cm−1: 3297, 3243, 3194, 1676; 1H NMR (DMSO-d6) δ ppm: 0.97 (t, 3H, CH3), 1.57 (m, 2H, CH2), 3.67 (t, J = 6.0 Hz, 2H, CH2), 4.40 (s, 1H, SCH2), 7.39 (s, 2H, exchangeable with D2O, NH2), 7.73 (dd, J = 8.0 Hz, 2H, Ar-H), 7.84 (dd, J = 8.0, 2H, Ar-H), 7.91 (m, 2H, Ar-H), 8.39 (m, 2H, Ar-H), 10.74 (s, 1H, exchangeable with D2O, NH); MS (m/z): 496 (M+, 12.91%), 320 (100% base peak).
4.1.7.4. 2-[(6-Propylbis([1, 2, 4]triazolo)[3,4-a:4',3'-c]phthalazin-3-yl)thio]acetamide 15
Yellowish white crystals (yield 73%); mp: 238–240 °C; IR (KBr) ν cm−1: 3194, 3084, 1655; 1H NMR (DMSO-d6) δ ppm: 0.95 (t, 3H, CH3), 1.60 (m, 2H, CH2), 3.68 (t, J = 7.0 Hz, 2H, CH2), 4.18 (s, 2H, SCH2), 7.41 (s, 1H, exchangeable with D2O, H-N-H), 7.81 (s, 1H, exchangeable with D2O, H-N-H), 7.94 (m, 2H, Ar-H), 8.45 (d, 2H, Ar-H).
4.2. Biological evaluation
4.2.1. In vitro anti-proliferative activity
Anti-proliferative activity of the synthesised compounds was estimated using the MTT assay protocol8,38–40 as shown in Supplementary data.
4.2.2. Dna intercalation assay (DNA/methyl green colorimetric assay)
The DNA/methyl green assay was estimated in vitro for all the target derivatives using doxorubicin as a reference drug, adopting the protocol described by Burres et al.41 as shown in the Supplementary data.
4.2.3. Measurement of topoisomerase II activity
Compounds (8a, 8b, 9a, 9c, 9d, 12a, and 12b) that showed the better results in anti-proliferative and DNA/methyl green assay were further evaluated for their in vitro inhibitory activities against Topoisomerase II using doxorubicin as a reference drug following to reported procedure described by Patra et al.42 as shown in the Supplementary data.
4.2.4. In vivo antitumor activity
4.2.4.1. Animals and tumour cell line
Adult female Swiss albino mice purchased from Theodor Bilharzia Research Institute, Giza, Egypt, with an average bodyweight of (18–23) g were used. Mice were housed under constant conditions of 12 h light/dark cycle in a temperature under conditions of controlled humidity (22 ± 2 °C), with free access to standard laboratory mice food and water. All procedures related to care and maintenance of the animals were performed according to the international guiding principles for animal research and approved by the Faculty of Science, Suez Canal University bioethics and animal ethics committee (Approval number REC-07–2021).
Solid Ehrlich carcinoma (SEC) was purchased from the National Cancer Institute (Cairo University, Egypt). The tumour cell line was proliferated in mice through serial intraperitoneal (I.P.) transplantation of a volume of 0.2 ml physiological saline contains 1 × 106 viable cells for 24 h. SEC cells were collected 7 days after I.P. implantation. The harvested cells were diluted with saline to obtain a concentration of 5 × 106 viable SEC cells/mL. A volume of 0.2 ml saline contains 1 × 106 SEC cells that were I.P. implanted into each normal mouse. SEC cells (1 × 106 tumour cells/mouse) were implanted subcutaneously into the right thigh of the hind limb.
The experimental animals were randomly divided into four groups. Group 1 served as the normal saline control (5 ml/kg B.Wt., I.P.). Group 2 served as the SEC control (1 × 106 cells/mouse). Group 3 served as the compound-treated group (5 mg/kg B.Wt., I.P.). Group 4 received the standard anticancer drug doxorubicin (5 mg/kg BW, I.P.) and is considered as a reference control. Bodyweight and survival were recorded daily until the 24th day in both treated and control groups. At the end of the experiment, the blood of each group was collected under light anaesthesia to the estimate of hematological and biochemical assays. The anaesthetised animals were then sacrificed for evaluation of the antitumor activity and histopathological examination.
4.2.4.2. Antitumor potentiality
It includes tumour volume, weight, and tumour inhibition ratio (TIR%). Time interval measurements of tumour volume using digital Vernier calliper (Tricle Brand, Shanghai, China). Measure tumour length and width using a clipper and then calculate tumour volume using formulations V = (L × W × W)/2, where V is tumour volume, W is tumour width, L is tumour length. While TIR% was calculated according to the following equation
4.2.4.3. Blood assays
At the end of the experiment, animals from different groups were sacrificed, and blood samples were collected for hematological parameters including, Hb, RBC’s, and WBC’s levels, and serum for determination of liver enzymes ALT, AST levels, and proteins. Complete blood count (CBC) was investigated using the Abbott CELL-DYN®1800 automated haematology analyser (USA) using ready-made kits (Abbott Laboratories, Abbott Park, IL, USA). Activities of aspartate aminotransferase (AST) and alanine aminotransferase (ALT) were evaluated using commercial kits (ELITech clinical systems, France). Serum albumin level was determined by kit purchased from STANBIO Company (USA). Protein content was determined by colorimetric method using ready-made kits produced by Instrumentation Laboratory SpA, Inova diagnostics, Milano, Italy.
4.2.4.4. Histopathological study
Specimens of liver-sacrificed mice were fixed in 10% saline formalin. The fixed liver specimens were dehydrated in ascending series of ethyl alcohol and embedded in paraffin. Sections at 5 mm thicknesses were stained with haematoxylin and eosin and examined under the light microscope.
4.3. In silico studies
4.3.1. Docking study
Discovery Studio 2.5 software was used to perform docking and visualisation according to the described protocol.16
4.3.2. In silico ADMET analysis
ADMET studies were performed according to the reported procedure as adescribed in Supplementary data51–53.
4.3.3. Toxicity studies
Toxicity studies were performed according to the reported procedure as adescribed in Supplementary data54–56
Supplementary Material
Funding Statement
The authors extend their appreciation to the Research center at AlMaarefa University for funding this work under TUMA project number “TUMA-2021-4”.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Siegel RL, Miller KD, Jemal A.. Cancer statistics, 2019. A Cancer J Clinic 2019;69:7–34. [DOI] [PubMed] [Google Scholar]
- 2.Eliaa SG, Al-Karmalawy AA, Saleh RM, Elshal MF.. Empagliflozin and doxorubicin synergistically inhibit the survival of triple-negative breast cancer cells via interfering with the mtor pathway and inhibition of calmodulin: in vitro and molecular docking studies. ACS Pharmacol Transl Sci 2020;3:1330–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khattab M, Al‐Karmalawy AA.. Revisiting activity of some nocodazole analogues as a potential anticancer drugs using molecular docking and DFT calculations. Front Chem 2021;9:628398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Karmalawy AA, Khattab MJNJOC.. Molecular modelling of mebendazole polymorphs as a potential colchicine binding site inhibitor. N J Chem 2020;44:13990–6. [Google Scholar]
- 5.Eldehna WM, Abo-Ashour MF, Nocentini A, et al. . Novel 4/3-((4-oxo-5-(2-oxoindolin-3-ylidene)thiazolidin-2-ylidene)amino) benzenesulfonamides: synthesis, carbonic anhydrase inhibitory activity, anticancer activity and molecular modelling studies. Eur J Med Chem. 2017;139:250–62. [DOI] [PubMed] [Google Scholar]
- 6.Eissa IH, El-Helby A-GA, Mahdy HA, et al. . Discovery of new quinazolin-4(3H)-ones as VEGFR-2 inhibitors: design, synthesis, and anti-proliferative evaluation. Bioorg Chem. 2020;105:104380. [DOI] [PubMed] [Google Scholar]
- 7.Eissa IH, Ibrahim MK, Metwaly AM, et al. . Design, molecular docking, in vitro, and in vivo studies of new quinazolin-4(3H)-ones as VEGFR-2 inhibitors with potential activity against hepatocellular carcinoma. Bioorg Chem 2021;107:104532. [DOI] [PubMed] [Google Scholar]
- 8.Ran F, Li W, Qin Y, et al. . Inhibition of vascular smooth muscle and cancer cell proliferation by new VEGFR inhibitors and their immunomodulator effect: design, synthesis, and biological evaluation. Oxidat Med Cell Long 2021;2021:8321400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eldehna WM, Al-Wabli RI, Almutairi MS, et al. . Synthesis and biological evaluation of certain hydrazonoindolin-2-one derivatives as new potent anti-proliferative agents. J Enzyme Inhib Med Chem 2018;33:867–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdelsalam EA, Zaghary WA, Amin KM, et al. . Synthesis and in vitro anticancer evaluation of some fused indazoles, quinazolines and quinolines as potential EGFR inhibitors. Bioorg Chem 2019;89:102985. [DOI] [PubMed] [Google Scholar]
- 11.Saleh NM, Abdel‐Rahman AAH, Omar AM, et al. . Pyridine‐derived VEGFR‐2 inhibitors: rational design, synthesis, anticancer evaluations, in silico ADMET profile, and molecular docking. Arch Pharm 2021;354:e2100085. [DOI] [PubMed] [Google Scholar]
- 12.Martinez R, Chacon-Garcia L.. The search of DNA-intercalators as antitumoral drugs: what it worked and what did not work. Curr Med Chem 2005;12:127–51. [DOI] [PubMed] [Google Scholar]
- 13.Ibrahim M, Taghour M, Metwaly A, et al. . Design, synthesis, molecular modeling and anti-proliferative evaluation of novel quinoxaline derivatives as potential DNA intercalators and topoisomerase II inhibitors. Eur J Med Chem 2018;155:117–34. [DOI] [PubMed] [Google Scholar]
- 14.Eissa IH, El-Naggar AM, Abd El-Sattar NE, et al. . Design and discovery of novel quinoxaline derivatives as dual DNA intercalators and topoisomerase II inhibitors. Anticancer Agents Med Chem 2018;18:195–209. [DOI] [PubMed] [Google Scholar]
- 15.Eissa IH, Metwaly AM, Belal A, et al. . Discovery and antiproliferative evaluation of new quinoxalines as potential DNA intercalators and topoisomerase II inhibitors. Arch Pharm 2019;352:1900123. [DOI] [PubMed] [Google Scholar]
- 16.El-Helby A-GA, Sakr H, Ayyad RR, et al. . Design, synthesis, molecular modeling, in vivo studies and anticancer activity evaluation of new phthalazine derivatives as potential DNA intercalators and topoisomerase II inhibitors. Bioorg Chem 2020;103:104233. [DOI] [PubMed] [Google Scholar]
- 17.Abbass EM, Khalil AK, Mohamed MM, et al. . Design, efficient synthesis, docking studies, and anticancer evaluation of new quinoxalines as potential intercalative Topo II inhibitors and apoptosis inducers. Bioorg Chem 2020;104:104255. [DOI] [PubMed] [Google Scholar]
- 18.El-Adl K, Ibrahim M-K, Alesawy MS, et al. . [1, 2, 4] Triazolo [4, 3-c] quinazoline and bis ([1, 2, 4] triazolo)[4, 3-a: 4′, 3′-c] quinazoline derived DNA intercalators: design, synthesis, in silico ADMET profile, molecular docking and anti-proliferative evaluation studies. Bioorg Medic Chem 2021;30:115958. [DOI] [PubMed] [Google Scholar]
- 19.Alesawy MS, Al‐Karmalawy AA, Elkaeed EB, et al. . Design and discovery of new 1, 2, 4‐triazolo [4, 3‐c] quinazolines as potential DNA intercalators and topoisomerase II inhibitors. Archiv der Pharmazie 2020;354:e2000237. [DOI] [PubMed] [Google Scholar]
- 20.Ferguson LR, Denny WA.. Genotoxicity of non-covalent interactions: DNA intercalators. Mutat Res Fundamental Mol Mech Mutagen 2007;623:14–23. [DOI] [PubMed] [Google Scholar]
- 21.Carvalho C, Santos RX, Cardoso S, et al. . Doxorubicin: the good, the bad and the ugly effect. Curr Med Chem 2009;16:3267–85. [DOI] [PubMed] [Google Scholar]
- 22.Ghanem A, Emara HA, Muawia S, et al. . Tanshinone IIA synergistically enhances the antitumor activity of doxorubicin by interfering with the PI3K/AKT/mTOR pathway and inhibition of topoisomerase II: in vitro and molecular docking studies. N J Chem 2020;44:17374–81. [Google Scholar]
- 23.Kim JS, Rhee H-K, Park HJ, et al. . Synthesis of 1-/2-substituted-[1,2,3]triazolo[4,5-g]phthalazine-4,9-diones and evaluation of their cytotoxicity and topoisomerase II inhibition. Bioorg Med Chem 2008;16:4545–50. [DOI] [PubMed] [Google Scholar]
- 24.El-Shershaby MH, Ghiaty A, Bayoumi AH, et al. . From triazolophthalazines to triazoloquinazolines: a bioisosterism-guided approach toward the identification of novel PCAF inhibitors with potential anticancer activity. Bioorg Med Chem 2021;42:116266. [DOI] [PubMed] [Google Scholar]
- 25.Gaber M, El-Wakiel NA, El-Ghamry H, Fathalla SK.. Synthesis, spectroscopic characterization, DNA interaction and biological activities of Mn (II), Co (II), Ni (II) and Cu (II) complexes with [(1H-1, 2, 4-triazole-3-ylimino) methyl] naphthalene-2-ol. J Mol Struc 2014;1076:251–61. [Google Scholar]
- 26.Chen C-Y, Lee P-H, Lin Y-Y, et al. . Synthesis, DNA-binding abilities and anticancer activities of triazole-pyrrolo[2,1-c][1,4]benzodiazepines hybrid scaffolds. Bioorg Med Chem Lett 2013;23:6854–9. [DOI] [PubMed] [Google Scholar]
- 27.El‐Helby AGA, Sakr H, Eissa IH, et al. . Benzoxazole/benzothiazole‐derived VEGFR‐2 inhibitors: design, synthesis, molecular docking, and anticancer evaluations. Archiv Der Pharmazie 2019;352:1900178. [DOI] [PubMed] [Google Scholar]
- 28.El‐Helby AGA, Sakr H, Eissa IH, et al. . Design, synthesis, molecular docking, and anticancer activity of benzoxazole derivatives as VEGFR‐2 inhibitors. Arch Pharm 2019;352:1900113. [DOI] [PubMed] [Google Scholar]
- 29.Sakr H, Ayyad RR, El‐Helby AA, et al. . Discovery of novel triazolophthalazine derivatives as DNA intercalators and topoisomerase II inhibitors. Arch Pharm 2021;354:e2000456. [DOI] [PubMed] [Google Scholar]
- 30.Said MA, Eldehna WM, Nocentini A, et al. . Sulfonamide-based ring-fused analogues for CAN508 as novel carbonic anhydrase inhibitors endowed with antitumor activity: design, synthesis, and in vitro biological evaluation. Eur J Med Chem 2020;189:112019. [DOI] [PubMed] [Google Scholar]
- 31.Al-Sanea MM, Elkamhawy A, Paik S, et al. . Synthesis and biological evaluation of novel 3-(quinolin-4-ylamino)benzenesulfonamidesAQ3 as carbonic anhydrase isoforms I and II inhibitors. J Enzyme Inhib Med Chem 2019;34:1457–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Avendaño C, Menendez JC, Medicinal chemistry of anticancer drugs. Elsevier, UK; 2015. [Google Scholar]
- 33.Minotti G, Menna P, Salvatorelli E, et al. . Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol Rev 2004;56:185–229. [DOI] [PubMed] [Google Scholar]
- 34.El-Helby AGA, Ayyad RR, Sakr HM, et al. . Design, synthesis, molecular modeling and biological evaluation of novel 2, 3-dihydrophthalazine-1, 4-dione derivatives as potential anticonvulsant agents. J Mol Struct 2017;1130:333–51. [Google Scholar]
- 35.Xue D-Q, Zhang X-Y, Wang C-J, et al. . Synthesis and anticancer activities of novel 1,2,4-triazolo[3,4-a]phthalazine derivatives . Eur J Medic Chem 2014;85:235–44. [DOI] [PubMed] [Google Scholar]
- 36.El‐Helby AGA, Ayyad RR, Sakr H, et al. . Design, synthesis, molecular docking, and anticancer activity of phthalazine derivatives as VEGFR‐2 inhibitors. Archiv Der Pharmazie 2017;350:1700240. [DOI] [PubMed] [Google Scholar]
- 37.Badr M, El‐Sherief H, El‐Naggar G, Mahgoub S.. Substitution and ring closure reactions of phthalazine derivatives. J. Heterocycl. Chem 1984;21:471–5. [Google Scholar]
- 38.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 1983;65:55–63. [DOI] [PubMed] [Google Scholar]
- 39.Denizot F, Lang R.. Rapid colorimetric assay for cell growth and survival. Modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J Immunol Methods 1986;89:271–7. [DOI] [PubMed] [Google Scholar]
- 40.Thabrew M, Hughes RD, Mcfarlane IG.. Screening of hepatoprotective plant components using a HepG2 cell cytotoxicity assay. J Pharm Pharmacol 2011;49:1132–5. [DOI] [PubMed] [Google Scholar]
- 41.Burres NS, Frigo A, Rasmussen RR, McAlpine JB.. A colorimetric microassay for the detection of agents that interact with DNA. J Nat Prod 1992;55:1582–7. [DOI] [PubMed] [Google Scholar]
- 42.Patra N, De U, Kang J-A, et al. . A novel epoxypropoxy flavonoid derivative and topoisomerase II inhibitor, MHY336, induces apoptosis in prostate cancer cells. Euro J Pharmacol 2011;658:98–107. [DOI] [PubMed] [Google Scholar]
- 43.Nafie MS, Arafa K, Sedky NK, et al. . Triaryl dicationic DNA minor-groove binders with antioxidant activity display cytotoxicity and induce apoptosis in breast cancer. Chem Biol Interact 2020;324:109087. [DOI] [PubMed] [Google Scholar]
- 44.Dicato M, Plawny L, Diederich M.. Anemia in cancer. Ann. Oncol 2010;21:vii167–vii172. [DOI] [PubMed] [Google Scholar]
- 45.ElZahabi HS, Nafie MS, Osman D, et al. . Design, synthesis and evaluation of new quinazolin-4-one derivatives as apoptotic enhancers and autophagy inhibitors with potent antitumor activity. Euro J Med Chem 2021;222:113609. [DOI] [PubMed] [Google Scholar]
- 46.Boraei AT, Eltamany EH, Ali IA, et al. . Synthesis of new substituted pyridine derivatives as potent anti-liver cancer agents through apoptosis induction: in vitro, in vivo, and in silico integrated approaches. Bioorg Chem 2021;111:104877. [DOI] [PubMed] [Google Scholar]
- 47.Xia X, Maliski EG, Gallant P, Rogers D.. Classification of kinase inhibitors using a Bayesian model. J Med Chem 2004;47:4463–4470. [DOI] [PubMed] [Google Scholar]
- 48.BIOVIA QSAR, ADMET and predictive toxicology . 2020. https://www.3dsbiovia.com/products/collaborative-science/biovia-discovery-studio/qsar-admet-and-predictive-toxicology.html
- 49.Carling RW, Moore KW, Street LJ, et al. . 3-Phenyl-6-(2-pyridyl) methyloxy-1, 2, 4-triazolo [3, 4-a] phthalazines and analogues: high-affinity γ-aminobutyric acid-A benzodiazepine receptor ligands with α2, α3, and α5-subtype binding selectivity over α1. J Med Chem 2004;47:1807–1822. [DOI] [PubMed] [Google Scholar]
- 50.Alswah M, Ghiaty A, El-Morsy A, El-Gamal K.. Synthesis and biological evaluation of some [1, 2, 4] triazolo [4, 3-a] quinoxaline derivatives as novel anticonvulsant agents. Inter Scholarly Res Notices 2013;2013:587054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.El-Zahabi MA, Elbendary ER, Bamanie FH, et al. . Design, synthesis, molecular modeling and anti-hyperglycemic evaluation of phthalimide-sulfonylurea hybrids as PPARγ and SUR agonists. Bioorganic Chemistry 2019;91:103115. [DOI] [PubMed] [Google Scholar]
- 52.Ibrahim MK, Eissa IH, Alesawy MS, et al. . Design, synthesis, molecular modeling and anti-hyperglycemic evaluation of quinazolin-4(3H)-one derivatives as potential PPARγ and SUR agonists. Bioorg Med Chem 2017;25:4723–4744. [DOI] [PubMed] [Google Scholar]
- 53.Eissa IH, Dahab MA, Ibrahim MK, et al. . Design and discovery of new antiproliferative 1,2,4-triazin-3(2H)-ones as tubulin polymerization inhibitors targeting colchicine binding site. Bioorg Chem. 2021;112:104965. [DOI] [PubMed] [Google Scholar]
- 54.Parmar DR, Soni JY, Guduru R, et al. . Discovery of new anticancer thiourea-azetidine hybrids: design, synthesis, in vitro antiproliferative, SAR, in silico molecular docking against VEGFR-2, ADMET, toxicity, and DFT studies. Bioorg Chem 2021;115:105206. [DOI] [PubMed] [Google Scholar]
- 55.El-Demerdash A, Metwaly AM, Hassan A, et al. . Comprehensive virtual screening of the antiviral potentialities of marine polycyclic guanidine alkaloids against SARS-CoV-2 (COVID-19). Biomolecules 2021;11:460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eissa IH, Khalifa MM, Elkaeed EB, et al. . In silico exploration of potential natural inhibitors against SARS-Cov-2 nsp10. Molecules 2021;26:6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.