ABSTRACT
In cancer patients, the clinical response to checkpoint-based immunotherapy is associated with the composition and functional quality of the host microbiome. While the relevance of the gut microbiome for checkpoint immunotherapy outcome has been addressed intensively, data on the role of the local tumor microbiome are missing. Here, we set out to molecularly characterize the local non-small cell lung cancer microbiome using 16S rRNA gene amplicon sequencing of bronchoscopic tumor biopsies from patients treated with PD-1/PD-L1-targeted checkpoint inhibitors. Our analyses showed significant diversity of the tumor microbiome with high proportions of Firmicutes, Bacteroidetes and Proteobacteria. Correlations with clinical data revealed that high microbial diversity was associated with improved patient survival irrespective of radiology-based treatment response. Moreover, we found that the presence of Gammaproteobacteria correlated with low PD-L1 expression and poor response to checkpoint-based immunotherapy, translating into poor survival. Our study suggests novel microbiome-specific/derived biomarkers for checkpoint immunotherapy response prediction and prognosis in lung cancer. In a broader sense, our data draw attention to the local tumor microbial habitat as an important addition to the spatially separated microbiome of the gut compartment.
KEYWORDS: Non-small cell lung cancer, microbiome, microbial diversity, Gammaproteobacteria, checkpoint inhibition
Background
Immune checkpoint inhibitor (ICI) therapy, either alone or in combination with classical chemotherapeutic drugs, constitutes the mainstay of treatment for advanced non-small cell lung cancer (NSCLC), in the absence of targetable molecular driver alterations.1,2 ICIs are powerful therapeutic agents that harness the natural specificity and adaptability of the immune system to reinvigorate anticancer immunity and tackle tumor heterogeneity3,4 and cancer-associated immune suppression.5 Across tumor entities, biomarkers for ICI response prediction include target antigen expression (e.g., PD-1/PD-L1),6 tumor mutational burden (TMB),7,8 DNA mismatch repair deficiency/microsatellite instability,9 and tumor T cell infiltration at baseline;10 however, ICI responsiveness also relies on the composition and functional quality of the host microbiome.11–13 Microbes are particularly abundant in the gastrointestinal (GI) tract,14 and certain species of this compartment (mostly bacterial commensals) have been shown to be associated with ICI treatment efficacy through various immunological and metabolic mechanisms.15,16 In the case of lung cancer, the GI microbiome is anatomically distant despite a postulated functional interaction via the gut-lung axis,17,18 which theoretically compromises the relevance of gut microbes in relation to more proximal habitats.19,20 Therefore, profiling of the local NSCLC tumor microbiome, as a reflection of the overarching pulmonary microbial landscape,18 is important to (i) get a better understanding of local tumor–microbiome interactions and (ii) potentially establish novel biomarkers with superior predictive value in ICI-treated NSCLC.
In this proof-of-concept study, we aimed at analyzing bacterial species directly in NSCLC tissue, using mostly bronchoscopic tumor biopsies. Our study reports novel microbiome-related NSCLC biomarkers in the tumor environment and emphasizes the importance of the local tumor microbial habitat for ICI responsiveness and patient outcome.
Methods
NSCLC biopsies and patient characteristics
Bronchoscopic (n = 35) and surgical (n = 3) biopsies from 38 patients with advanced-stage NSCLC treated with ICIs were obtained from the St. Gallen Lung Biopsy Biobank, a dedicated biobank for pulmonary samples maintained and operated by the Lung Center of the Cantonal Hospital St. Gallen. Healthy adjacent lung tissue macroscopically free of tumor was obtained from NSCLC patients undergoing curative-intent surgical treatment and was used for control purpose (n = 10). Tumor biopsies were from stage III–IV NSCLC patients who received a PD-1/PD-L1-targeting ICI (pembrolizumab, nivolumab, atezolizumab, durvalumab, or spartalizumab) in various therapy lines (first, second, or third) with or without a chemotherapy doublet (carboplatin-pemetrexed, cisplatin-pemetrexed, or carboplatin-paclitaxel) or an investigational treatment (MK-1308/anti-CTLA-4, NIS793/anti-TGF-β, NIR178/adenosine A2a receptor antagonist). Importantly, biopsies were sampled before the start of treatment. The patient population comprised 80% males and the main NSCLC histological subtypes were well-balanced (43% adenocarcinoma (ADC) and 53% squamous cell carcinoma (SCC)). PD-L1 expression status was available for 77% of the cases (12 patients with 0–20% of the cells positive, 11 patients with >20% of the cells positive) and was determined using the VENTANA PD-L1 (SP263) Assay from Roche Diagnostics (Rotkreuz, Switzerland). 30% of the patients had a known mutation in one of the following proto-oncogenes: KRAS, EGFR, ERBB2, PIK3CA, MET. Of note, the MET alteration in one patient concerned a missense mutation in exon 14 (T992I) that may have influenced immunotherapy outcome.21 In addition, the EGFR alteration found in another patient (a single nucleotide variant in exon 18) may also have impacted immunotherapy outcome.22 Median progression-free survival (PFS) and median overall survival (OS) were 3.1 and 15.3 months, respectively, and the overall response rate to ICI therapy was 27%. Patient characteristics are specified in Table 1.
Table 1.
Patient characteristics (n = 30)
| Parameter | Median (range) | |
|---|---|---|
| Age (years) | 67 (23–79) | |
| Progression-free survival (months) | 3.1 (.7–28.7) | |
| Overall survival (months) | 15.3 (2.6–50.9) | |
| Case number (n) | Fraction (%) | |
| Sex | ||
| female | 6 | 20.0 |
| male | 24 | 80.0 |
| NSCLC histological subtype | ||
| ADC | 13 | 43.3 |
| SCC | 16 | 53.3 |
| unknown | 1 | 3.3 |
| Tumor stage | ||
| III | 7 | 23.3 |
| IVA | 7 | 23.3 |
| IVB | 14 | 46.7 |
| unknown | 2 | 6.7 |
| ICI therapy line | ||
| first | 9 | 30.0 |
| second | 16 | 53.3 |
| third | 5 | 16.7 |
| ICI used | ||
| Pembrolizumab (anti-PD-1) | 12 | 40.0 |
| Nivolumab (anti-PD-1) | 12 | 40.0 |
| Atezolizumab (anti-PD-L1) | 1 | 3.3 |
| Durvalumab (anti-PD-L1) | 3 | 10.0 |
| Spartalizumab (anti-PD-1) | 2 | 6.7 |
| Combination anticancer therapy | ||
| yes (chemotherapy) | 5 | 16.7 |
| yes (investigational medicine) | 3 | 10.0 |
| no (ICI monotherapy) | 22 | 73.3 |
| ICI treatment response | ||
| CR | 0 | .0 |
| PR | 8 | 26.7 |
| SD | 6 | 20.0 |
| PD | 14 | 46.7 |
| unknown | 2 | 6.7 |
| PD-L1 expression status | ||
| 0–20% of cells positive | 12 | 40.0 |
| >20% of cells positive | 11 | 36.7 |
| unknown | 7 | 23.3 |
| Known mutation in a proto-oncogene | ||
| no | 21 | 70.0 |
| yes | 9 | 30.0 |
| KRAS G12C | 1 | 3.3 |
| KRAS G13C | 1 | 3.3 |
| KRAS G12F | 1 | 3.3 |
| KRAS G12D | 1 | 3.3 |
| KRAS G12A | 1 | 3.3 |
| EGFR SNV exon 18 | 1 | 3.3 |
| ERBB2 G776delinsVC | 1 | 3.3 |
| PIK3CA SNV exon 9 | 1 | 3.3 |
| MET T992I | 1 | 3.3 |
| Number of packyears | ||
| 0–40 | 13 | 43.3 |
| >40 | 13 | 43.3 |
| unknown | 4 | 13.3 |
ADC, adenocarcinoma; CR, complete response; ICI, immune checkpoint inhibitor; NSCLC, non-small cell lung cancer; PD, progressive disease; PR, partial response; SCC, squamous cell carcinoma; SD, stable disease; SNV, single nucleotide variant.
16S rRNA gene amplicon sequencing
Bronchoscopic (n = 35) and surgical (n = 3) NSCLC biopsies as well as healthy lung control tissues (n = 10) were subjected to 16S rRNA sequencing using Illumina MiSeq technology, a platform for targeted resequencing, expression profiling and metagenomics. To this end, PCR amplification protocols were optimized, and two-step NextEra PCR libraries were generated using 341 F (5′- CCT ACG GGN GGC WGC AG −3′) and 802 R (5′- GAC TAC HVG GGT ATC TAA TCC −3′) primers specific for the V3 and V4 regions of the bacterial 16S rRNA gene. The amount of total DNA (including host and other non-bacterial DNA) was determined using PicoGreen (Thermo Fisher Scientific, Waltham, MA) and 140.2 ± 33.7 ng of DNA (mean ± SEM) were used as input for PCR amplification emanating from similarly sized biopsies. Libraries were sequenced on the Illumina MiSeq platform employing a 500 cycles MiSeq Reagent Kit v2. Paired-end reads passing Illumina quality filtering were demultiplexed and trimmed of adaptor residuals using MiSeq reporter built-in data analysis software v2.6. The read quality was further analyzed using FastQC v0.11.8 and sequencing reads were trimmed of primers using cutadapt v2.8. Forward and reverse reads were merged to reproduce the sequenced molecule using USEARCH v11.0.667 and a minimum overlap of 15 bases. Further quality filtering allowed one expected error per merged read and discarded reads with ambiguous bases or an outlier amplicon size. Samples with a minimum of 5000 merged reads were denoised using the UNOISE module of the USEARCH package, and operational taxonomic units (OTUs) were defined. OTU abundances were filtered for possible bleed-in contaminations making use of the UNCROSS algorithm. Sequences from the RDP 16S rRNA database served as reference and taxonomies were predicted using the SINTAX module included in the USEARCH package, setting the minimum confidence threshold to .5. Ultimately, 5 NSCLC and 5 healthy lung samples did not pass the quality controls and were excluded from subsequent analysis; thus, a total of 30 NSCLC biopsies (28 bronchoscopic, 2 surgical) and 5 healthy lung samples were finally analyzed. Three non-template controls (NTCs) were run. Signals detected in the NTCs were subtracted from the whole analysis to correct for potential reagent contamination by trace amounts of bacterial DNA as reported.23–25 16S rRNA gene amplicon sequencing as described in this section was performed by Microsynth AG (Balgach, Switzerland). All samples were processed in parallel, starting from DNA isolation, PCR amplification, library preparation and sequencing, thus eliminating potential batch effects.
Data analysis and statistical considerations
Phylum, class and OTU abundances were analyzed in a descriptive manner. Bacterial α-diversity was estimated using the Shannon diversity index (SDI)26 as well as the number of unique OTUs observed. Correlation of the SDI with the number of unique OTUs was analyzed using Spearman’s rank correlation coefficient. Two-group data were tested for statistical significance using the Mann–Whitney U test, and three-group data were statistically analyzed using the Kruskal–Wallis test. A Cox proportional hazard model was used to analyze phylum and class abundance with respect to PFS and OS, and Wald test p-values are reported. Time to event data (patient survival) were analyzed using Kaplan–Meier estimates and log-rank tests (Χ2 statistics and associated p-values are reported). Where applicable, optimized cutoffs have been used for data analysis (e.g., to harmonize group sizes). Unless otherwise stated, quantitative data are shown as boxplots with whiskers and outliers. The nominal significance level was .05. Statistical analyses were performed using IBM SPSS Statistics 20 and the R statistical software (www.r-project.org).
Ethics statement
Patients donating their samples to the St. Gallen Lung Biopsy Biobank provided written informed consent for use of their biological material for research purpose (EKSG 11/044). The study was approved by the local Ethical Review Board (Ethikkommission Ostschweiz) under BASEC number 2019–02059.
Results
16S rRNA gene amplicon sequencing deciphers tumor microbial diversity
To characterize the tumor microbial habitat of NSCLC, bronchoscopic (n = 28) and surgical (n = 2) tumor biopsies as well as healthy lung control tissues (n = 5) were subjected to 16S rRNA gene amplicon sequencing.27,28 Bacterial species from various phyla were detectable in the samples, which could be allocated to 224 individual OTUs. OTU abundances ranged from <5000 reads to >30000 reads and showed high proportions of Firmicutes, Bacteroidetes and Proteobacteria in both NSCLC biopsies and healthy lung samples (Figure 1a). In NSCLC biopsies, the number of unique OTUs observed (ranging from 3 to 109) was highly correlated with the SDI, an established measure for α-diversity taking into account both the number of species and their abundance (r = .926, p < .001, Figure 1b). In summary, our results show that 16S rRNA gene amplicon sequencing represents a suitable method to analyze bacteria in bronchoscopic NSCLC biopsies and suggest significant diversity of the tumor microbial habitat along with marked interpatient variation.
Figure 1.
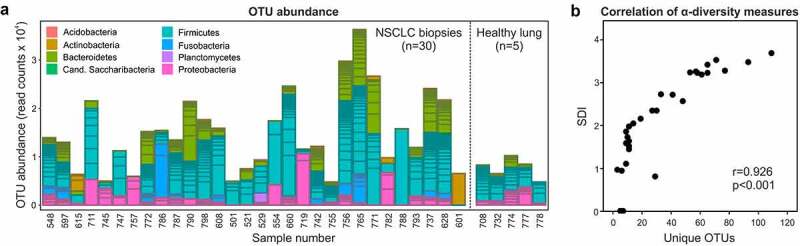
Microbial diversity of NSCLC tumors revealed through 16S rRNA gene amplicon sequencing. (a) OTU abundance in NSCLC and healthy lung samples and allocation to specific bacterial phyla. (b) Correlation of the SDI with the number of unique OTUs observed in NSCLC samples
NSCLC, non-small cell lung cancer; OTU, operational taxonomic unit; SDI, Shannon diversity index.
Survival benefit of patients with a higher tumor microbial diversity
Analyzing the SDI in stratified groups of patients did not reveal associations with sex (female vs. male, p = .494), the number of packyears (0–40 vs. >40, p = .650), tumor stage (III vs. IVA vs. IVB, p = .214), or the NSCLC histological subtype (ADC vs. SCC, p = 1.000) (Figure 2a-d). Along similar lines, the SDI was comparable in PD-L1low (0–20%) vs. PD-L1high (>20%) expressers (p = .786, Figure 2e). The SDI could not discriminate patients based on their observed radiological response to checkpoint-based immunotherapy (partial response (PR) vs. stable disease (SD) vs. progressive disease (PD)) (p = .161, figure 2f) and, accordingly, PFS was indifferent among SDIlow vs SDIhigh patients (χ2 = .152, p = .697, Figure 2g). However, patients with a high SDI showed significantly longer OS (χ2 = 4.036, p = .045, Figure 2h), which indicates benefit from a higher microbial diversity irrespective of checkpoint immunotherapy responsiveness. Collectively, the diversity of the tumor bacterial metagenome correlates with specific survival metrics in ICI-treated NSCLC.
Figure 2.

Tumor microbial diversity associates with NSCLC patient survival. (a-d) Analysis of the SDI in stratified subgroups of NSCLC patients. (e-h) Analysis of the SDI in terms of PD-L1 expression, ICI treatment responsiveness, and patient survival. (a-f) Boxes indicate the median (highlighted in bold) and interquartile ranges. Whiskers indicate the minimum and maximum values except in the case of outliers (outliers are indicated by circles). (G + H) cutoffs used for SDI stratification: 2.73 for PFS and 2.65 for OS
ADC, adenocarcinoma; ICI, immune checkpoint inhibitor; NSCLC, non-small cell lung cancer; OS, overall survival; PD, progressive disease; PFS, progression-free survival; PR, partial response; SCC, squamous cell carcinoma; SD, stable disease; SDI, Shannon diversity index.
Gammaproteobacteria constitute a significant part of the NSCLC tumor microbiome
We next sought to investigate which types of bacteria would play a role in shaping or predicting the response to PD-1/PD-L1-targeted ICIs. As shown in Figure 3a, the phyla of Firmicutes, Bacteroidetes and Proteobacteria were largely dominating the scene, jointly accounting for almost 90% of the detected tumor microbiome. Other phyla still showing appreciable abundance included Fusobacteria and Actinobacteria (Figure 3a). A Cox proportional hazard model revealed that only the phylum of Proteobacteria was significantly associated with PFS (p < .001) and OS (p < .001) in our cohort and that the class of Gammaproteobacteria (accounting for almost 60% of Proteobacteria, Figure 3a), was most significantly correlated with survival (p < .001 for PFS and p = .011 for OS) (Figure 3b). In summary, Gammaproteobacteria account for roughly 7% of the total detected microbiome and are associated with survival in NSCLC patients receiving immunotherapy.
Figure 3.
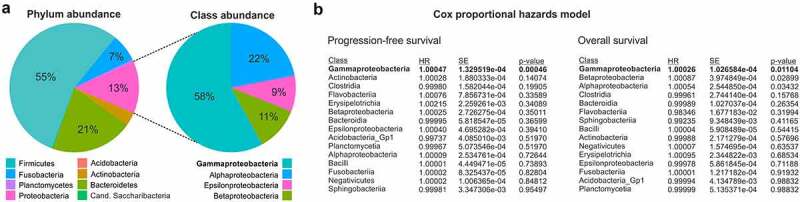
Gammaproteobacteria are abundant in NSCLC tumors and associate with patient survival. (a) Pie charts illustrating the relative abundance of particular bacterial phyla and classes within the total detected tumor microbiome. (b) Results of a Cox proportional hazards model for the analysis of possible associations of bacterial classes with patient survival ranked according to statistical significance
HR, hazard ratio; NSCLC, non-small cell lung cancer; SE, standard error.
Gammaproteobacteria correlate with low PD-L1 expression and predict poor checkpoint immunotherapy responsiveness
Based on the predictions from the Cox proportional hazard model (Figure 3b), we analyzed the abundance of Gammaproteobacteria in stratified patient subgroups and also comparatively investigated their levels in healthy lung vs. NSCLC tissues. Although not reaching statistical significance, Gammaproteobacteria levels appeared to be higher in lung cancer tissue as compared to healthy lung control tissue (p = .421, Figure 4a). Statistical significance was also not reached when stratifying according to sex (female vs. male, p = .900), the number of packyears (0–40 vs. >40, p = .153), tumor stage (III vs. IVA vs. IVB, p = .755), or the NSCLC histological subtype (ADC vs. SCC, p = .449), even though some trends were observed (Figure 4b-e). In contrast, high abundance of Gammaproteobacteria was significantly associated with low PD-L1 expression (p = .006, figure 4f), a finding that was also partly reflected in the response to ICI therapy (PR vs. SD vs. PD) (p = .275, Figure 4g). In line with these data, patients with a high abundance of Gammaproteobacteria in their tumors showed a significantly worse PFS (χ2 = 8.594, p = .003, Figure 4h), with OS revealing a similar trend without reaching statistical significance (χ2 = 1.739, p = .187, Figure 4i). Taken together, Gammaproteobacteria appear to be enriched in the cancerous lung and their abundance in the tumor surroundings correlates with low PD-L1 expression and poor PFS and a trend toward worse OS under ICI therapy.
Figure 4.

Gammaproteobacteria correlate with low PD-L1 expression and poor patient survival under ICI therapy. (a) Comparative analysis of Gammaproteobacteria abundance in healthy lung and NSCLC samples. (b-e) Analysis of Gammaproteobacteria abundance in stratified subgroups of NSCLC patients. (f-i) Analysis of Gammaproteobacteria abundance in terms of PD-L1 expression, ICI treatment responsiveness, and patient survival. (a-g) Boxes indicate the median (highlighted in bold) and interquartile ranges. Whiskers indicate the minimum and maximum values except in the case of outliers (outliers are indicated by circles and extreme outliers are indicated by stars). (H + I) cutoffs used for Gammaproteobacteria stratification: 480 for PFS and 811 for OS
ADC, adenocarcinoma; ICI, immune checkpoint inhibitor; NSCLC, non-small cell lung cancer; OS, overall survival; PD, progressive disease; PFS, progression-free survival; PR, partial response; SCC, squamous cell carcinoma; SD, stable disease.
Discussion
The host microbiome serves important functions in health and disease and is particularly abundant in the GI tract,14 the respiratory tract,18 and on the skin.29 Commensal microbes are essential gatekeepers for epithelial barrier integrity30 and transient (e.g., antibiotics and infection) or chronic (e.g., long-term medication and underlying conditions) dysbiosis can prime to a variety of immune-related and metabolic disorders including inflammatory bowel disease, obesity, and type 2 diabetes.31–35 In contrast, ‘gain-of-function’ dysbiosis can lead to the overgrowth of particular microbial strains in specific anatomical compartments, thus causing local opportunistic infections.36,37 A high diversity of the host microbiome with a well-balanced and time-stable composition of bacteria, viruses and fungi is therefore essential to prevent disease and maintain health until old age.38–40 Interactions of the microbiome with host cells are mediated mostly by immunological and metabolic means, and can be local, distant, or systemic.18
While, with the notable exception of colorectal cancer,41 the significance of the microbiome for cancer development remains elusive, accumulating evidence suggests a pivotal role of host microbes in shaping the response to cancer therapy. Specifically, the use of antibiotics curtails ICI treatment efficacy42–44 and also has a negative impact on the performance of classical cytotoxic drugs such as platinum and cyclophosphamide.45 Importantly, unresponsiveness of germ-free or antibiotic-treated mice to checkpoint inhibition can be rescued through oral supplementation with certain bacteria.11,12 In ICI-treated melanoma, the fecal microbiome is significantly different between responding and non-responding patients,13 and fecal microbiome transplantation (FMT) from responding patients is clinically evaluated as an ICI re-sensitizing intervention in the subgroup of treatment-refractory patients.46 While these data clearly bear witness to a key role of host commensal microbes in cancer immunotherapy outcome, most studies have prioritized the GI microbiome irrespective of the tumor site of origin. However, the host microbiome in its totality is much wider, and more proximal tumor ecosystems may be equally important, if not dominant, in governing disease progression and treatment responsiveness.19,20,47–51 Here, we have pictured the local tumor microbiome of NSCLC using 16S rRNA gene amplicon sequencing of tumor biopsies from patients treated with PD-1/PD-L1-targeted ICIs.
Results from this study converge on three key messages as follows: (i) 16S rRNA gene amplicon sequencing is technically feasible in bronchoscopic tumor biopsies that typically provide limited sample input and are further enriched for host cells rather than metagenomic signals (very low microbial biomass). (ii) A higher diversity of the tumor microbiome represents a prognostic factor, as it correlates with improved OS of ICI-treated NSCLC patients irrespective of the clinical response to ICI treatment or PFS. (iii) A high abundance of Gammaproteobacteria, a diverse class of gram-negative bacteria,52 predicts low PD-L1 expression and poor response to PD-1/PD-L1-targeted checkpoint blockade, resulting in unfavorable patient survival. Importantly, the associations of SDI and Gammaproteobacteria with survival were cross-validated using random permutation of both strata based on the log-rank test Χ2 statistics (data not shown).
While shotgun sequencing represents the gold standard for metagenomics in stool samples and additionally covers viruses, fungi and protozoa,28 16S rRNA sequencing is a valuable approach especially when microbial density is low such that sample composition is biased toward host cells as is the case for lung tissue53 and fetal organs.54 Here, we have characterized the local NSCLC microbiome as a potential mirror of the overarching pulmonary metagenomic landscape18 using the latter approach. Our data suggest high tumoral abundances especially of Firmicutes, Bacteroidetes, and Proteobacteria. Our observation that a higher α-diversity predicts improved OS independently from the clinical response to ICI treatment is interesting and may indicate that a more diverse microbiome is selected for patients with a better health status or other factors associated with favorable outcome (e.g., less disease burden or more indolent disease). In addition, a higher diversity of the tumor microbiome may increase the chances for protective immunological cross-reactivity between microbial-derived peptide products and current or future arising tumor neoantigens,15 indicating a more causal role of the microbiome as a mediator of a better tumor response to ICI therapy. It is challenging to uncouple prognostic vs. predictive features of α-diversity in the current study, and it cannot be inferred that a higher microbial diversity would also correlate with longer OS in untreated patients or patients not treated with ICIs but with other systemic agents. In comparison, a higher gut microbial diversity clearly represents a predictive biomarker in anti-PD-1-treated melanoma.13 Our finding that Gammaproteobacteria correlate with low PD-L1 expression and poor ICI-related survival suggests a new potential biomarker for ICI response prediction in NSCLC while leaving unanswered questions about the underlying mechanisms. Certainly, Gammaproteobacteria represent a highly diverse class of bacteria with numerous metabolic pathways involved, and member species can act as both facultative (e.g., E. coli) and bona fide pathogens (P. aeruginosa, S. typhimurium, H. influenzae, L. pneumophila).52 Whether the totality of Gammaproteobacteria downregulates PD-L1 expression and/or curtails anticancer immunity through metabolic or immunological rewiring55 remains to be shown in future mechanistic studies.
The following limitations are applicable to the study, which lower the generalizability of the results: (i) The study is purely correlative and case numbers are limited. (ii) The study population was retrospectively selected and heterogeneous in terms of clinical parameters. (iii) Bronchoscopic biopsies provide a locoregional snapshot of the tumor and may not be representative of the tumor as a whole. (iv) The sampling and handling of bronchoscopic and surgical biopsies in non-sterile environments may have led to some degree of cross-contamination, a potential pitfall that might be more relevant to the smaller biopsy specimens. (v) No systematic data on the use of antibiotics immediately before or during checkpoint immunotherapy were available for the study population, thus prohibiting corresponding investigations. (vi) The study did not investigate non-bacterial microbes such as viruses and fungi. (vii) The bacterial signal in bronchoscopic tumor biopsies was low such that PCR amplification may have introduced some bias. In addition, results need to be interpreted in awareness of the fact that microbial signals detected in NTCs were subtracted from the test samples to account for reagent contamination as reported.23–25 (viii) Many bacterial species were detectable in a few samples only such that species-level analyses could not be performed for statistical reasons.
We here show the technical feasibility of 16S rRNA gene amplicon sequencing-based bacterial identification in bronchoscopic tumor biopsies and report the identification of novel microbiome-specific biomarkers for prognosis and checkpoint immunotherapy response prediction in NSCLC. Our proof-of-concept study sets the stage for larger validation trials and gives impetus for endeavors to mechanistically dissect the underlying mechanisms and define therapeutic leads. In the long run, it is hoped that basic knowledge about the role of the local tumor microbiome can inspire rational combination therapies to re-sensitize to – or boost – checkpoint blockade,56 e.g., through oral or inhaled supplementation with commensal microbes (re-installation of diversity) or the informed use of narrow-spectrum antibiotic/antiviral/antifungal agents (elimination of detrimental microbes).
Acknowledgments
We would like to thank Dr. Sebastian Strempel and Dr. Bruno Müller from Microsynth AG for providing excellent support during data analysis and interpretation.
Funding Statement
This work was kindly supported by the Lungenliga St. Gallen-Appenzell based in St. Gallen, Switzerland (grant to MB, no grant number available), and the Stiftung Propter Homines based in Vaduz, Principality of Liechtenstein (grant to MHB, no grant number available).
Specific abbreviations used
ADC: Adenocarcinoma
CR: Complete response
FMT: Fecal microbiome transplantation
GI: Gastrointestinal
HR: Hazard ratio
ICI: Immune checkpoint inhibitor
NSCLCNon-small cell lung cancer
NTC: Non-template control
OS: Overall survival
OTU: Operational taxonomic unit
PD: Progressive disease
PFS: Progression-free survival
PRPartial response
SCC: Squamous cell carcinoma
SD: Stable disease
SDI: Shannon diversity index
SE: Standard error
SEM: Standard error of the mean
SNV: Single nucleotide variant
TMB: Tumor mutational burden
Disclosure statement
The author(s) have no conflicts of interest relevant to this study to declare. No medical writer was involved in the preparation of the manuscript.
Data availability
Sequencing data are available from the corresponding author upon reasonable request.
Authors’ contributions
Conception and design: MB, MHB
Data collection: MB, FB, RR
Data analysis and interpretation: MB, FB, WCA, LF, RR, SIR, MJ, MF, MHB
Statistics: FB
Wrote the first draft of the paper: MB, MHB
Wrote the final version of the paper: MB, FB, WCA, LF, RR, SIR, MJ, MF, MHB
Approved the paper for publication: MB, FB, WCA, LF, RR, SIR, MJ, MF, MHB
References
- 1.Paz-Ares L, Luft A, Vicente D, Tafreshi A, Gumus M, Mazieres J, Hermes B, Çay Şenler F, Csőszi T, Fülöp A, et al. Pembrolizumab plus chemotherapy for squamous non–small-cell lung cancer. N Engl J Med. 2018;379(21):2040–9. doi: 10.1056/NEJMoa1810865. [DOI] [PubMed] [Google Scholar]
- 2.Gandhi L, Rodriguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F, Domine M, Clingan P, Hochmair MJ, Powell SF, et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med. 2018;378(22):2078–2092. doi: 10.1056/NEJMoa1801005. [DOI] [PubMed] [Google Scholar]
- 3.Boesch M, Zeimet AG, Reimer D, Schmidt S, Gastl G, Parson W, Spoeck F, Hatina J, Wolf D, Sopper S, et al. The side population of ovarian cancer cells defines a heterogeneous compartment exhibiting stem cell characteristics. Oncotarget. 2014;5(16):7027–7039. doi: 10.18632/oncotarget.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boesch M, Sopper S, Zeimet AG, Reimer D, Gastl G, Ludewig B, Dominik Wolf. Heterogeneity of cancer stem cells: rationale for targeting the stem cell Niche. Biochim Biophys Acta. 2016;1866(2):276–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolf D, Fiegl H, Zeimet AG, Wieser V, Marth C, Sprung S, Sopper S, Hartmann G, Reimer D, Boesch M, et al. High RIG-I expression in ovarian cancer associates with an immune-escape signature and poor clinical outcome. Int J Cancer. 2020;146(7):2007–2018. doi: 10.1002/ijc.32818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel SP, Kurzrock R.. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther. 2015;14(4):847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 7.Hellmann MD, Ciuleanu TE, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C, Minenza E, Linardou H, Burgers S, Salman P, et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med. 2018;378(22):2093–2104. doi: 10.1056/NEJMoa1801946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, Barron DA, Zehir A, Jordan EJ, Omuro A, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51(2):202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, Morse MA, Van Cutsem E, McDermott R, Hill A, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol. 2018;36(8):773–779. doi: 10.1200/JCO.2017.76.9901. [DOI] [PubMed] [Google Scholar]
- 10.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CPM, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350(6264):1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359(6371):91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 13.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359(6371):97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cani PD. Human gut microbiome: hopes, threats and promises. Gut. 2018;67(9):1716–1725. doi: 10.1136/gutjnl-2018-316723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boesch M, Baty F, Rothschild SI, Tamm M, Joerger M, Fruh M, Brutsche MH. Tumour neoantigen mimicry by microbial species in cancer immunotherapy. Br J Cancer. 2021;125(3):313–323. doi: 10.1038/s41416-021-01365-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Man Lei Y, Jabri B, Alegre M-L, et al. Commensal bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350(6264):1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dang AT, Marsland BJ. Microbes, metabolites, and the gut-lung axis. Mucosal Immunol. 2019;12(4):843–850. doi: 10.1038/s41385-019-0160-6. [DOI] [PubMed] [Google Scholar]
- 18.Wypych TP, Wickramasinghe LC, Marsland BJ. The influence of the microbiome on respiratory health. Nat Immunol. 2019;20(10):1279–1290. doi: 10.1038/s41590-019-0451-9. [DOI] [PubMed] [Google Scholar]
- 19.Tsay JJ, Wu BG, Sulaiman I, Gershner K, Schluger R, Li Y, Yie T-A, Meyn P, Olsen E, Perez L, et al. Lower airway dysbiosis affects lung cancer progression. Cancer Discov. 2021;11(2):293–307. doi: 10.1158/2159-8290.CD-20-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong Q, Chen ES, Zhao C, Jin C. Host-microbiome interaction in lung cancer. Front Immunol. 2021;12:679829. doi: 10.3389/fimmu.2021.679829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kron A, Scheffler M, Heydt C, Ruge L, Schaepers C, Eisert AK, Merkelbach-Bruse S, Riedel R, Nogova L, Fischer RN, et al. Genetic heterogeneity of MET-aberrant NSCLC and its impact on the outcome of immunotherapy. J Thorac Oncol. 2021;16(4):572–582. doi: 10.1016/j.jtho.2020.11.017. [DOI] [PubMed] [Google Scholar]
- 22.Lee CK, Man J, Lord S, Links M, Gebski V, Mok T, Yang JCH. Checkpoint inhibitors in metastatic EGFR-mutated non-small cell lung cancer-a meta-analysis. J Thorac Oncol. 2017;12(2):403–407. doi: 10.1016/j.jtho.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 23.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12(1):87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marsh RL, Nelson MT, Pope CE, Leach AJ, Hoffman LR, Chang AB, Smith-Vaughan HC. How low can we go? The implications of low bacterial load in respiratory microbiota studies. Pneumonia (Nathan). 2018;10(1):7. doi: 10.1186/s41479-018-0051-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Goffau MC, Lager S, Salter SJ, Wagner J, Kronbichler A, Charnock-Jones DS, Peacock SJ, Smith GCS, Parkhill J. Recognizing the reagent microbiome. Nat Microbiol. 2018;3(8):851–853. doi: 10.1038/s41564-018-0202-y. [DOI] [PubMed] [Google Scholar]
- 26.Reese AT, Dunn RR, McFall-Ngai MJ. Drivers of microbiome biodiversity: a review of general rules, feces, and ignorance. mBio. 2018;9(4). doi: 10.1128/mBio.01294-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanschagrin S, Yergeau E. Next-generation sequencing of 16S ribosomal RNA gene amplicons. J Vis Exp. 2014;90. doi: 10.3791/51709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ranjan R, Rani A, Metwally A, McGee HS, Perkins DL. Analysis of the microbiome: advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem Biophys Res Commun. 2016;469(4):967–977. doi: 10.1016/j.bbrc.2015.12.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol. 2018;16(3):143–155. doi: 10.1038/nrmicro.2017.157. [DOI] [PubMed] [Google Scholar]
- 30.Kayama H, Okumura R, Takeda K. Interaction between the microbiota, epithelia, and immune cells in the intestine. Annu Rev Immunol. 2020;38(1):23–48. doi: 10.1146/annurev-immunol-070119-115104. [DOI] [PubMed] [Google Scholar]
- 31.Duvallet C, Gibbons SM, Gurry T, Irizarry RA, Alm EJ. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat Commun. 2017;8(1):1784. doi: 10.1038/s41467-017-01973-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amabebe E, Robert FO, Agbalalah T, Orubu ESF. Microbial dysbiosis-induced obesity: role of gut microbiota in homoeostasis of energy metabolism. Br J Nutr. 2020;123(10):1127–1137. doi: 10.1017/S0007114520000380. [DOI] [PubMed] [Google Scholar]
- 33.Gurung M, Li Z, You H, Rodrigues R, Jump DB, Morgun A, Shulzhenko N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine. 2020;51:102590. doi: 10.1016/j.ebiom.2019.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. 2021;19(1):55–71. doi: 10.1038/s41579-020-0433-9. [DOI] [PubMed] [Google Scholar]
- 35.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104(34):13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilkins LJ, Monga M, Miller AW. Defining dysbiosis for a cluster of chronic diseases. Sci Rep. 2019;9(1):12918. doi: 10.1038/s41598-019-49452-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galloway-Pena JR, Shi Y, Peterson CB, Sahasrabhojane P, Gopalakrishnan V, Brumlow CE, Daver NG, Alfayez M, Boddu PC, Khan MAW, et al. Gut microbiome signatures are predictive of infectious risk following induction therapy for acute myeloid leukemia. Clin Infect Dis. 2020;71(1):63–71. doi: 10.1093/cid/ciz777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larsen OFA, Claassen E. The mechanistic link between health and gut microbiota diversity. Sci Rep. 2018;8(1):2183. doi: 10.1038/s41598-018-20141-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng F, Li Y, Zhao J. The gut microbiome of healthy long-living people. Aging (Albany NY). 2019;11:289–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, Harris HMB, Coakley M, Lakshminarayanan B, O’Sullivan O, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488(7410):178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 41.Tilg H, Adolph TE, Gerner RR, Moschen AR. The intestinal microbiota in colorectal cancer. Cancer Cell. 2018;33(6):954–964. doi: 10.1016/j.ccell.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Derosa L, Hellmann MD, Spaziano M, Halpenny D, Fidelle M, Rizvi H, Long N, Plodkowski AJ, Arbour KC, Chaft JE, et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann Oncol. 2018;29(6):1437–1444. doi: 10.1093/annonc/mdy103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schett A, Rothschild SI, Curioni-Fontecedro A, Krahenbuhl S, Fruh M, Schmid S, Driessen C, Joerger M. Predictive impact of antibiotics in patients with advanced non small-cell lung cancer receiving immune checkpoint inhibitors: antibiotics immune checkpoint inhibitors in advanced NSCLC. Cancer Chemother Pharmacol. 2020;85(1):121–131. doi: 10.1007/s00280-019-03993-1. [DOI] [PubMed] [Google Scholar]
- 44.Huang XZ, Gao P, Song YX, Xu Y, Sun JX, Chen XW, Zhao J-H, Wang Z-N. Antibiotic use and the efficacy of immune checkpoint inhibitors in cancer patients: a pooled analysis of 2740 cancer patients. Oncoimmunology. 2019;8(12):e1665973. doi: 10.1080/2162402X.2019.1665973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pflug N, Kluth S, Vehreschild JJ, Bahlo J, Tacke D, Biehl L, Eichhorst B, Fischer K, Cramer P, Fink A-M, et al. Efficacy of antineoplastic treatment is associated with the use of antibiotics that modulate intestinal microbiota. Oncoimmunology. 2016;5(6):e1150399. doi: 10.1080/2162402X.2016.1150399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, Adler K, Dick-Necula D, Raskin S, Bloch N, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science. 2021;371(6529):602–609. doi: 10.1126/science.abb5920. [DOI] [PubMed] [Google Scholar]
- 47.Shi Y, Zheng W, Yang K, Harris KG, Ni K, Xue L, Lin W, Chang EB, Weichselbaum RR, Fu Y-X, et al. Intratumoral accumulation of gut microbiota facilitates CD47-based immunotherapy via STING signaling. J Exp Med. 2020;217(5). doi: 10.1084/jem.20192282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Livyatan I, Nejman D, Shental N, Straussman R. Characterization of the human tumor microbiome reveals tumor-type specific intra-cellular bacteria. Oncoimmunology. 2020;9(1):1800957. doi: 10.1080/2162402X.2020.1800957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, Rotter-Maskowitz A, Weiser R, Mallel G, Gigi E, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 2020;368(6494):973–980. doi: 10.1126/science.aay9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, Quesada P, Sahin I, Chandra V, San Lucas A, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. 2019;178(4):795–806 e12. doi: 10.1016/j.cell.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, Gavert N, Zwang Y, Cooper ZA, Shee K, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357(6356):1156–1160. doi: 10.1126/science.aah5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams KP, Gillespie JJ, Sobral BW, Nordberg EK, Snyder EE, Shallom JM, Dickerman AW. Phylogeny of gammaproteobacteria. J Bacteriol. 2010;192(9):2305–2314. doi: 10.1128/JB.01480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mathieu E, Escribano-Vazquez U, Descamps D, Cherbuy C, Langella P, Riffault S, Remot A, Thomas M. Paradigms of lung microbiota functions in health and disease, particularly, in asthma. Front Physiol. 2018;9:1168. doi: 10.3389/fphys.2018.01168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mishra A, Lai GC, Yao LJ, Aung TT, Shental N, Rotter-Maskowitz A, Shepherdson E, Singh GSN, Pai R, Shanti A, et al. Microbial exposure during early human development primes fetal immune cells. Cell. 2021;184(13):3394–3409.e20. doi: 10.1016/j.cell.2021.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity. 2018;48(3):434–452. doi: 10.1016/j.immuni.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Finlay BB, Goldszmid R, Honda K, Trinchieri G, Wargo J, Zitvogel L. Can we harness the microbiota to enhance the efficacy of cancer immunotherapy? Nat Rev Immunol. 2020;20(9):522–528. doi: 10.1038/s41577-020-0374-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequencing data are available from the corresponding author upon reasonable request.


