PURPOSE
Patients with locally advanced rectal cancer (LARC) are recommended to receive preoperative chemoradiotherapy (PCRT) followed by surgery. Response to PCRT varies widely: 60%-70% of patients with LARC do not derive therapeutic benefit from PCRT, whereas 15%-20% of patients achieve pathologic complete response (pCR). We sought to develop a liquid biopsy assay for identifying response to PCRT in patients with LARC.
MATERIALS AND METHODS
We analyzed two genome-wide microRNA (miRNA) expression profiling data sets from tumor tissue samples for in silico discovery (GSE68204) and validation (GSE29298). We prioritized biomarkers in pretreatment plasma specimens from clinical training (n = 41; 15 responders and 26 nonresponders) and validation (n = 65; 29 responders and 36 nonresponders) cohorts of patients with LARC. We developed an integrated miRNA panel and established a risk assessment model, which was combined with the miRNA panel and carcinoembryonic antigen levels.
RESULTS
Our comprehensive discovery effort identified an 8-miRNA panel that robustly predicted response to PCRT, with an excellent accuracy in the discovery (area under the curve [AUC] = 0.95) and validation (AUC = 0.92) cohorts. We successfully established a circulating miRNA panel with remarkable diagnostic accuracy in the clinical training (AUC = 0.82) and validation (AUC = 0.81) cohorts. Moreover, the predictive accuracy of the panel was significantly superior to conventional clinical factors in both cohorts (P < .01) and the risk assessment model was superior (AUC = 0.83). Finally, we applied our model to detect patients with pathologic complete response and showed that it was dramatically superior to currently used pathologic features (AUC = 0.92).
CONCLUSION
Our novel risk assessment signature for predicting response to PCRT has a potential for clinical translation as a liquid biopsy assay in patients with LARC.
INTRODUCTION
Colorectal cancer (CRC) is the third most common cancer worldwide and the second leading cause of cancer-related deaths.1,2 Among these, more than 100,000 people are diagnosed specifically with rectal cancer each year worldwide; 60%-70% of these patients have locally advanced rectal cancer (LARC).3 Preoperative (neoadjuvant) chemoradiotherapy (PCRT) followed by total mesorectal excision and adjuvant chemotherapy is the current standard treatment for patients with LARC.4-6 PCRT treatment has received increasing attention in the past decade and has been successfully used to achieve tumor regression and decrease the risk for local recurrence.7-9 The National Comprehensive Cancer Network guidelines suggest the use of PCRT such as fluorouracil-based chemotherapy plus long-course radiation therapy (RT) in patients with LARC.10 Because PCRT is associated with significantly worse postoperative intestinal and genitourinary functions, European Society for Medical Oncology guidelines suggest that chemotherapy plus short- or long-course RT can be used to balance the risk of tumor recurrence and likelihood of patient toxicity.11 Unfortunately, however, 60%-70% of patients with LARC do not respond to neoadjuvant therapy. These nonresponders can be defined as those who have no tumor regression changes, on the basis of resected specimens after neoadjuvant therapy.12,13
CONTEXT
Key Objective
Preoperative chemoradiotherapy (PCRT) followed by total mesorectal excision and adjuvant chemotherapy remains the preferred treatment choice in patients with locally advanced rectal cancer (LARC). However, the response to PCRT varies widely and 60%-70% of patients with LARC do not derive therapeutic benefit from PCRT, highlighting the need to develop predictive biomarkers of response to this treatment modality. In this study, we sought to develop a liquid biopsy assay for identifying response to PCRT in patients with LARC.
Knowledge Generated
We successfully established a circulating 8-miRNA panel that robustly predicts response to PCRT and established a risk assessment model that is superior to currently used pathologic features in predicting treatment response in LARC.
Relevance
Our noninvasive liquid biopsy assay offers a promising risk assessment tool for the identification of patients with LARC that will likely benefit from neoadjuvant PCRT and spare others from the toxicity and expense associated with such treatments.
Several studies have reported that more than 20% of patients experienced grade 3-4 toxic effects from PCRT, such as fever, diarrhea, nausea, and hematologic infections.14-16 Despite receiving limited or no benefit from neoadjuvant therapy, nonresponders still experience the associated toxicity from these treatments. More importantly, instead of a therapeutic benefit, tumor progression can occur in some patients during such treatment. This highlights the clinical challenge that a more robust prediction of nonresponders before PCRT is important for devising individualized treatment plans that avoid the toxicity and allow selection of alternative treatments in a timely manner. However, there is currently no robust method to accurately stratify patients into responder and nonresponder groups, other than by pathologic assessment after completion of neoadjuvant therapy. Therefore, using pretreatment specimens to identify potential responders and nonresponders may help to predict treatment outcomes and inform treatment choices.
In sharp contrast to nonresponse, pathologic complete response (pCR) is characterized by the complete elimination of malignant cells in resected specimens.17,18 Approximately 15%-20% of patients with LARC who undergo PCRT achieve pCR.19-22 Good pathologic response is associated with prolonged disease-free survival and lower local and distant recurrence rates.23-28 Some studies suggest that such patients could be closely followed without surgery, ie, a watch-and-wait strategy, if pCR can be accurately predicted.29-31 This would avoid a potential reduction in quality of life brought on by radical surgery, which can impair normal intestinal and genitourinary function.32
Previous studies reported that low levels of carcinoembryonic antigen (CEA), high pretreatment hemoglobin levels, early clinical T stage, lymph node metastasis–negative, small tumor size, and a long interval between RT and surgery are related to probability of a good response to PCRT.33-37 However, there are few established models or nomograms to predict this and even fewer are used clinically to predict good pathologic response after PCRT for patients with LARC. Therefore, developing more robust models to predict pathologic response has great clinical significance and remains a great challenge. However, an ideal clinical application of these biomarkers would be to use them to diagnose patients with high-risk LARC before surgery, before such tissue specimens are readily available. Therefore, translating these biomarkers into a liquid biopsy assay is attractive, as this would allow a noninvasive, facile, and inexpensive diagnostic assay for response prediction in patients with LARC. Accumulating evidence indicates that the expression pattern of microRNAs (miRNAs) reflects the physiologic and pathologic status of patients with cancer. Studies have shown that expression of specific miRNAs is directly involved in CRC pathogenesis and have emphasized their potential as circulating biomarkers for CRC.38-40 Our group has also previously reported a blood-based miRNA signature that allowed robust detection of recurrence and metastasis.41-43
In the present study, by analyzing pretreatment parameters that can be easily used in clinical decision making in patients with LARC before PCRT, we established an accurate model and developed a nomogram to predict the probability of good downstaging. Our final risk prediction model robustly identified patients who respond to PCRT.
MATERIALS AND METHODS
Biomarker Discovery and Patient Cohorts
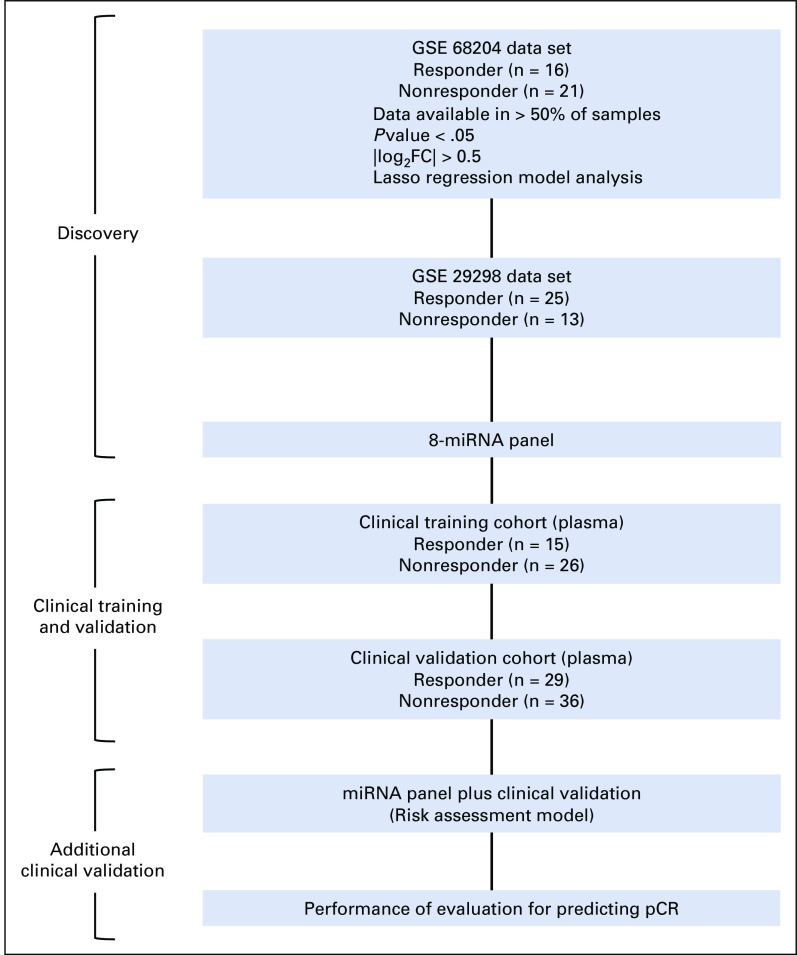
To perform a systematic and comprehensive biomarker discovery, we analyzed miRNA expression profiling results from two public data sets (GSE68204 and GSE29298) to identify and establish a miRNA panel for the prediction of response to PCRT in patients with LARC. In total, miRNA expression profiling data from 75 patients with LARC, which included miRNA microarray data from the GSE68204 cohort (n = 37; 16 responders and 21 nonresponders) and the GSE29298 cohort (n = 38; 25 responders and 13 nonresponders), were analyzed to identify a miRNA panel from tumor tissue samples in the discovery and internal validation phases, respectively (Appendix Fig A1).
We analyzed a total of 106 plasma specimens from patients with LARC, composed of two independent clinical cohorts: a training cohort (n = 41; 15 responders and 26 nonresponders) from Hospital Universitario de Donostia, Spain, and a validation cohort (n = 65; 29 responders and 36 nonresponders) from Asan Medical Center, Korea (Appendix Fig A1). The study was conducted in accordance with the Declaration of Helsinki. A written informed consent was obtained from all patients, and the study was approved by the institutional review boards of all participating institutions.
PCRT and Pathologic Assessment
The main neoadjuvant chemotherapy included a fluorouracil- or capecitabine-based regimen. The radiation was delivered using a three-dimensional conformal RT or intensity-modulated RT technique at a dose of 45-50.4 Gy in 25 or 28 fractions to the whole pelvis. Pathologic responses to PCRT were assessed according to the tumor regression grade (TRG) classification.44 On the basis of this classification, patients were divided into a responder group (TRG 1-2) and a nonresponder group (TRG 3-5). We defined TRG 1 as the complete pathologic response group.
RNA Extraction and Real-Time Quantitative Reverse Transcription Polymerase Chain Reaction From Plasma
We extracted total RNA by using 200 μL of plasma. Then, real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR) analysis was performed using a SensiFAST SYBR Lo-ROX Kit (Bioline, London, UK) on the Quantstudio 7 Flex Real-Time PCR System (Applied Biosystems, Foster City, CA), and expression levels were evaluated using the corresponding software system. The relative abundance of target transcripts was evaluated and normalized to the expression of miR-16-5p as an internal control, using the 2–ΔDCt method.45
Statistical Analysis
Clinicopathologic characteristics of the patient cohorts are shown as patient number and ratio except for age (median and range; Appendix Table A1). Binary logistic regression was used to train a classifier on the basis of the expression of miRNAs. Of note, once the model was trained in the training cohort, the same statistical model variables (weights and cutoff thresholds) were applied in the validation cohort. A P value < .05 was considered statistically significant. Detailed information is given in the Data Supplement.
RESULTS
Genome-Wide miRNA Expression Profiling Identifies a Novel 8-miRNA Panel for the Prediction of Response to PCRT in Patients With LARC
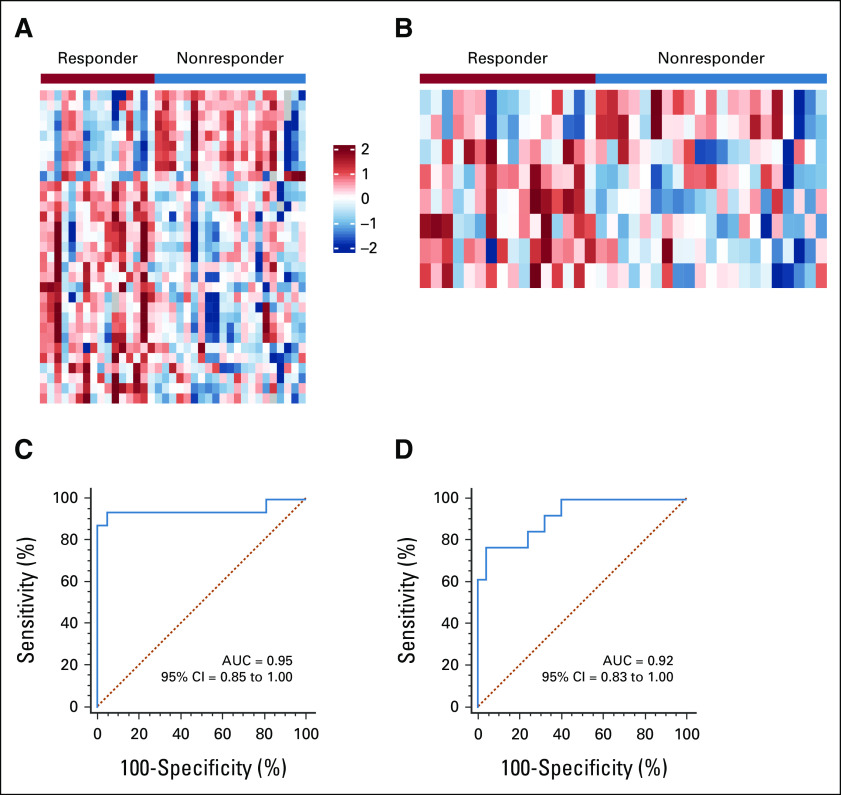
We performed a genome-wide, unbiased, comprehensive biomarker discovery analysis in two independent miRNA expression profiling data sets from tumor tissue samples (GSE68204 and GSE29298) to identify a miRNA panel for the prediction of response to PCRT in patients with LARC (Appendix Fig A1). We first compared miRNA expression profiles between patients with LARC defined as responders (TRG 1-2; n = 16) or nonresponders (TRG 3-5; n = 21) in the GSE68204 discovery cohort. These analyses identified differentially expressed (P < .05) candidate targets with data availability (no expression value for a given miRNA) in at least 50% of all cases and excluded highly correlated miRNAs (> 0.8) by using Pearson's correlation coefficient. The differential analysis was performed using LIMMA package, which fits a linear model and uses moderated t-statistic.46 Next, we selected candidate miRNAs on the basis of the following criteria: absolute log2 fold change > 0.5, P value < .05, and expression in at least half of the samples; miRNAs from passenger strand were excluded (Fig 1A). To find the lasso tuning parameter lambda, we performed five-fold cross-validation by cv.glmnet function from R package glmnet. A lambda value of 0.0735 (e–2.61) was used to select the candidates. After performing LASSO regression model analysis, we identified a panel of eight candidate miRNAs: miR-30e-5p, miR-33a-5p, miR-130a-5p, miR-210-3p, miR-214-3p, miR-320a, miR-338-3p, and miR-1260a (Fig 1B). To validate the potential of cancer specificity, we compared the candidate miRNAs between the tumor and normal specimens in the GSE68204. Seven miRNAs of eight candidate miRNAs were significantly upregulated, and six miRNAs of eight candidate miRNAs had log fold changes > 1 (Appendix Table A2). Subsequent application of the 8-miRNA panel to the GSE68204 data resulted in an area under the curve (AUC) of 0.95 (Fig 1C). Furthermore, we successfully validated the diagnostic ability of this 8-miRNA panel in an independent validation cohort (GSE29298; 25 responders [TRG 1-2] and 13 nonresponders [TRG 3-5]: AUC = 0.92; Fig 1D). Our results demonstrate the diagnostic accuracy of this panel in the identification of response to PCRT in patients with LARC.
FIG 1.
Genome-wide discovery and validation of a novel miRNA panel to predict response to PCRT in patients with LARC. (A) A heatmap of the expression profiles between responder and nonresponder patients with LARC in the GSE68204 cohort. Expression values were under Z-score normalization after log2 transformation, and the color scale represents the Z-score. Negative values and positive values indicate whether the expression levels are lower or greater than the mean. The red one is high expression, and the blue one is low expression between responders and nonresponders. (B) A heatmap of the expression profiles of the final 8-miRNA panel between responders and nonresponders in the GSE68204 cohort. ROC curves show the diagnostic performance of the 8-miRNA panel in distinguishing patients who responded to PCRT in the (C) GSE68204 discovery (responder = 16, nonresponder = 21, and AUC = 0.95) and (D) GSE29298 validation cohorts (responder = 25, nonresponder = 13, and AUC = 0.92). AUC, area under the curve; LARC, locally advanced rectal cancer; miRNA, microRNA; PCRT, preoperative chemoradiotherapy; ROC, receiver operator characteristic.
Clinical Training Confirms the Potential of the Blood-Based miRNA Assay To Predict Response to PCRT in Patients With LARC
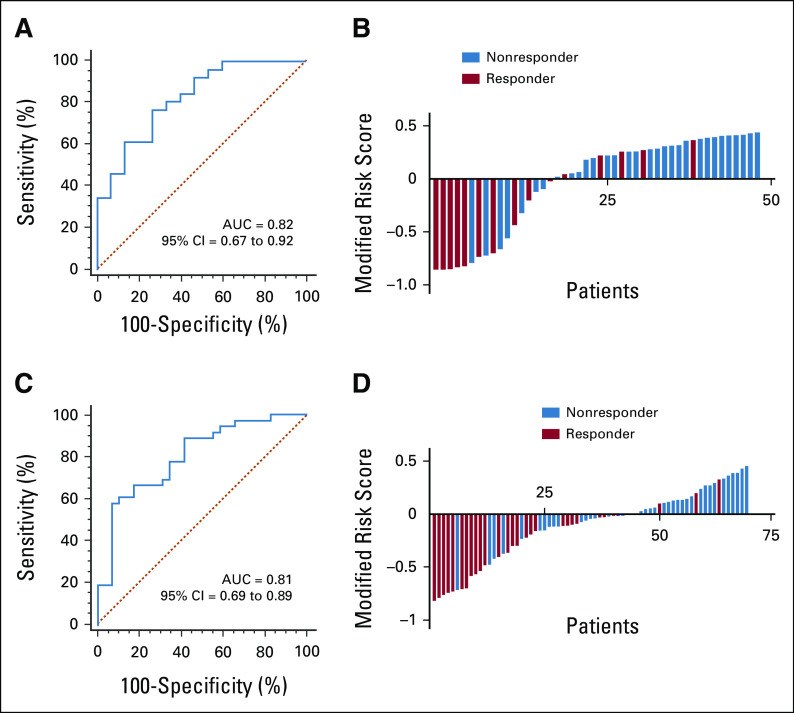
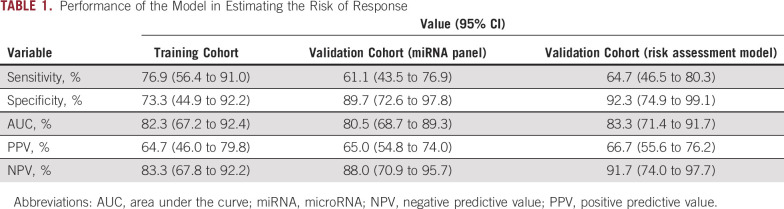
To evaluate the predictive capability of our miRNA panel as a blood-based liquid biopsy assay, we performed training and validation of the selected eight miRNAs using RT-qPCR assays in pretreatment blood specimens from two independent clinical cohorts. On the basis of the coefficients of individual miRNAs and a constant derived from analysis of the clinical training cohort (n = 41; 15 responders and 26 nonresponders), we developed a risk score: (–2.18799 × miR-30e-5p) + (1.24410 × miR-33a-5p) + (3.22741 × miR-130a-5p) + (2.16646 × miR-210-3p) + (0.37901 × miR-214-3p) + (0.33851 × miR-320a) + (0.73473 × miR-338-3p) + (–4.58404 × miR-1260a) + 11.96376. Our established miRNA panel demonstrated an excellent performance in predicting response, with an AUC value of 0.82 (95% CI, 0.67 to 0.92; Fig 2A) and a corresponding sensitivity of 0.77 and a specificity of 0.73 (Table 1). Subsequently, we dichotomized the patients into low- and high-risk groups on the basis of risk scores obtained from Youden's index–derived cutoff thresholds. According to the distribution of risk scores and response status, the risk scores of nonresponder patients were notably higher than those of responders (Fig 2B). These results demonstrate that we were able to successfully develop a blood-based pretreatment response prediction model in patients with LARC.
FIG 2.
Clinical training and validation of the miRNA panel as a blood-based assay for identifying response to PCRT in patients with LARC. (A) ROC curve shows the diagnostic performance of the panel in the clinical training cohort (responder = 15, nonresponder = 26, and AUC = 0.82). (B) Risk score distribution plot in the training cohort. Modified risk score was obtained by subtracting individual risk scores from Youden's index value of the risk model. (C) ROC curve shows the diagnostic performance of the panel in the clinical validation cohort (responder = 29, nonresponder = 36, and AUC = 0.81). (D) Risk score distribution plot in the validation cohort. AUC, area under the curve; LARC, locally advanced rectal cancer; miRNA, microRNA; PCRT, preoperative chemoradiotherapy; ROC, receiver operator characteristic.
TABLE 1.
Performance of the Model in Estimating the Risk of Response
Validation of the miRNA Panel in an Independent Cohort Confirms Its Translational Potential
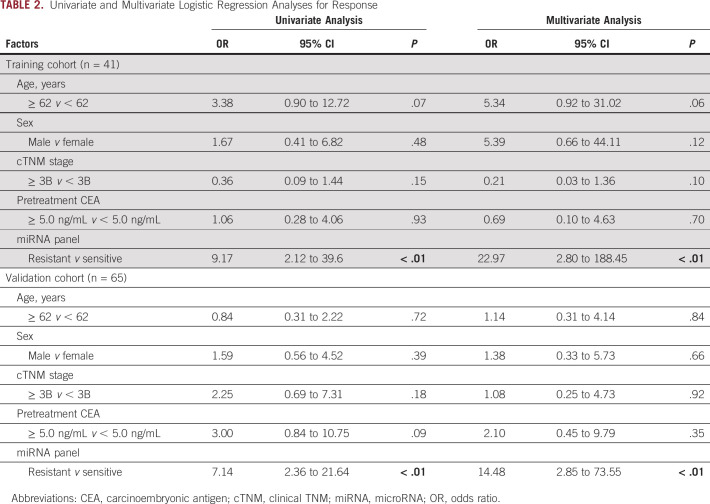
To evaluate the translational potential of our miRNA panel for identifying high-risk patients, we deliberately examined its performance in an independent clinical validation cohort (n = 65; 29 responders and 36 nonresponders). To this end, we applied the miRNA panel using the same statistical model, coefficients, and cutoff values derived from the training cohort to the independent validation cohort. We confirmed the robustness of our biomarker panel in predicting response, with an AUC value of 0.81 (95% CI, 0.69 to 0.89; Figs 2C and 2D), a sensitivity of 0.61, and a specificity of 0.90 (Table 1). We next categorized all patients into high- and low-risk groups using the cutoff thresholds derived using Youden's index from this miRNA panel and subsequently performed logistic regression for univariate and multivariate analyses. The multivariate analysis revealed that our newly established panel emerged as an independent predictor of response in both clinical cohorts (training cohort: odds ratio = 22.97, 95% CI, 2.80 to 188.45, P < .01; validation cohort: odds ratio = 14.48, 95% CI, 2.85 to 73.55, P < .01; Table 2).
TABLE 2.
Univariate and Multivariate Logistic Regression Analyses for Response
A Risk Assessment Model That Combines the miRNA Panel With Key Clinical Factors Demonstrates Greater Accuracy for Response Prediction in Patients With LARC
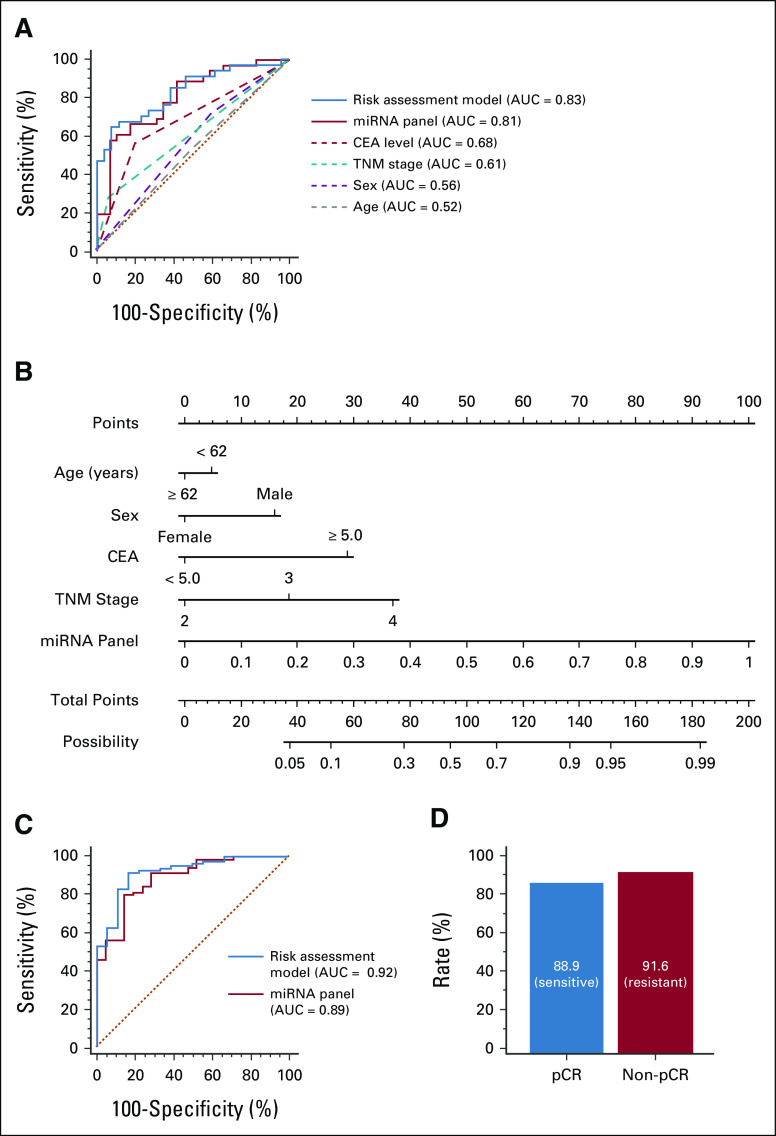
Given that CEA is an important biomarker for response to PCRT, we next determined the extent to which a model that combined our miRNA panel along with CEA expression levels would further improve its predictive accuracy. As five patients lacked sufficient clinical information, a total of 60 patients were included in this risk assessment model. We compared predictive accuracy between the miRNA panel with and without CEA, relative to other clinical risk factors (CEA levels, TNM stage, sex, and age). The risk assessment model that included CEA levels exhibited a notably improved performance in predicting patient response (AUC = 0.83; Fig 3A) compared with the miRNA panel alone. Furthermore, this model demonstrated dramatically superior predictive accuracy compared with other clinical risk factors. To further assess the ability of our biomarker signature to predict response when combined with key clinical features (ie, age, sex, CEA level, and TNM stage), we used the logistic regression model and established a nomogram incorporating these features. The cutoff threshold for the response risk score was chosen as 0.63, which was determined using Youden's index. We determined that although individual risk factors added some weight to the model, our miRNA panel had the highest weight in this model (Fig 3B).
FIG 3.
Additional clinical validation of the miRNA signature for identifying response to PCRT in patients with LARC. (A) ROC curves show the diagnostic performance of the new risk assessment model (combined miRNA panel and CEA) relative to indicated individual clinical factors in the clinical validation cohort (AUC = 0.83). (B) Nomogram indicates the possibility of responding to PCRT among patients with LARC. For clinical use, the scores of each covariate are added and the total score is located on the total score points axis. (C) ROC curves for predicting pCR show the diagnostic performance of the miRNA panel (AUC = 0.89) and the risk assessment model (AUC = 0.92) in total participants in the clinical cohorts (pCR = 21 and non-pCR = 85). (D) pCR and non-pCR rate in PCRT-sensitive and PCRT-resistant patients by using the risk assessment model. AUC, area under the curve; CEA, carcinoembryonic antigen; LARC, locally advanced rectal cancer; miRNA, microRNA; pCR, pathologic complete response; PCRT, preoperative chemoradiotherapy; ROC, receiver operator characteristic.
The Risk Assessment Model Has Robust Potential in Predicting pCR in Clinical Cohorts
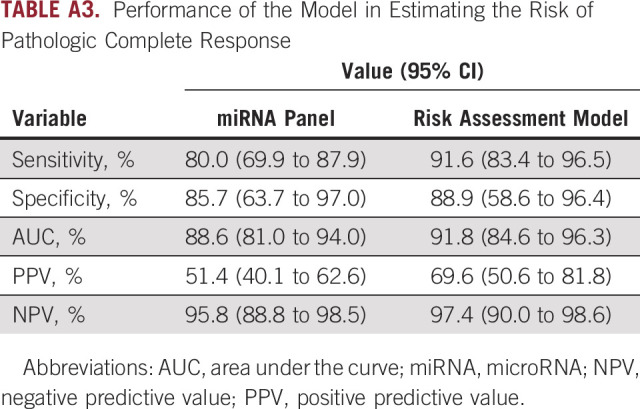
Since it is important to predict pCR for treatment decision making after PCRT, we categorized all participants from both clinical cohorts into a pCR group (n = 21) and a non-pCR group (n = 85) to evaluate the performance of the risk assessment model, which was combined with the miRNA panel and CEA levels for predicting pCR. These analyses demonstrated that LARC patients with pCR were 19.8% (21 of 106). We then predicted PCRT-sensitive (panel score: low) and PCRT-resistant (panel score: high) groups using the cutoff thresholds derived using Youden's index. Generating a receiver operator characteristic curve revealed that our miRNA panel predicted pCR with an AUC value of 0.89 (95% CI, 0.81 to 0.94; Fig 3C), a sensitivity of 0.80, and a specificity of 0.86 (Appendix Table A3).
We next evaluated the ability of the miRNA signature to stratify known pCR and non-pCR groups into predicted PCRT-sensitive and PCRT-resistant groups. The model stratified 85.7% of pCR patients (18 of 21) into the sensitive group, and 80.0% (68 of 85) of non-pCR patients into the resistant group (Table 3). In other words, of the 35 total patients that the panel classified as PCRT-sensitive, 18 (51.4%) had experienced pCR after PCRT, which represents a notably higher rate of accuracy for predicting pCR compared with other clinical factors. Next, we evaluated the risk assessment model, which demonstrated a remarkably higher accuracy in predicting pCR, with an AUC value of 0.92 (95% CI, 0.85 to 0.96; Fig 3C), a sensitivity of 0.92, and a specificity of 0.83 (Appendix Table A2). Moreover, our risk model was even more accurate than the miRNA panel alone, as it stratified 88.9% of pCR patients (16 of 18) into the sensitive group and 91.6% (76 of 83) of non-pCR patients into the resistant group (Fig 3D). Similarly, of the 23 patients who were classified as PCRT-sensitive, 16 patients (69.6%) had experienced pCR and of the 78 total patients that the model classified as PCRT-resistant, only 2 (2.6%) had experienced pCR, indicating that the remaining 97.4% did not experience pCR, which represents a notably higher rate of accuracy in predicting pCR and non-pCR. These results suggest that our established risk assessment model has a predictive ability not only for PCRT sensitivity but also for pCR. Moreover, the demonstration that PCRT was successfully applied to achieve pCR in many of the PCRT-sensitive patients classified by our model underscores the potential for a more effective strategy for screening patients with LARC.
TABLE 3.
pCR Rate in Training and Validation Cohorts
DISCUSSION
The importance of multimodality treatment is well-recognized, and there is a growing interest in developing novel, multimodal strategies in patients with LARC. Patients who respond well to PCRT have an excellent long-term prognosis.23,47,48 Importantly, recent studies showed that a regimen combining mFOLFOX6 and RT achieved a higher pCR rate of 38% in a clinical trial.49-52 Accurately predicting an excellent pathologic response to PCRT is critical for making appropriate treatment decisions. Patients who are not expected to respond to normal management can choose more aggressive regimens before PCRT or palliative surgery depending on their performance status. However, radical surgery may be associated with a high rate of temporary or permanent stoma, defecatory disorders, urinary and sexual dysfunction, and unnecessary mortality, resulting in an unfavorable quality of life.53,54 For patients who are expected to respond, noninvasive treatment strategies, such as the watch-and-wait strategy, are gaining popularity because pCR after PCRT has shown a good long-term prognosis regardless of the treatment strategy.55,56 Thus, it is crucial to understand the factors that predict a pathologic response to PCRT before radical surgery. Our study highlights the inadequacy of the clinicopathologic features currently used to identify patients as potential responders and offers an improved alternative that may ultimately lead to individualized treatment strategies.
In the present study, we established an accurate model and evaluated the ability of our 8-miRNA signature to predict response in pretreatment plasma samples. More importantly, our newly established model demonstrated significantly superior diagnostic accuracy for pCR in all cohorts (pCR rate = 69.6%), compared with other clinical risk factors (approximately 30%). Although all patients received radical surgery, patients who demonstrated pCR composed of 19.8% of patients, but 88.9% of the patients were selected as responders by using our new model. This demonstrates the potential clinical significance of our risk assessment model for reducing the number of unnecessary surgeries and considering watch-and-wait treatment in these patients. Also, our model revealed that 97.4% of patients classified as PCRT-resistant did not experience pCR, indicating that these patients require more intensive treatment and organ-preserving approach by omitting unnecessary treatment. Our model can be used to support individualized therapy as follows. For patients who are expected to show poor pathologic response to the standard PCRT regimen, an enhanced mFOLFOX6-RT regimen can be considered, so that toxicity caused by PCRT in patients with poor performance status can be avoided. For patients who are expected to have good pathologic response from the standard regimen, radical surgery can be chosen after PCRT. Especially for patients with a high probability of pCR after PCRT, local resection or a watch-and-wait strategy can be used to avoid surgical complications.
Potential limitations of our study include our retrospective study design, which may have potential selection bias. Thus, a prospective clinical trial is required to further confirm the diagnostic accuracy of our risk model. Second, our nomogram was based on the experience of a cohort of patients from a single institution. Thus, additional studies in a group of independent external institutions are required to answer clinical questions regarding which patients need an aggressive regimen, which need radical surgery, which can undergo local excision, and which can be managed with a watch-and-wait strategy after achieving a complete response. Third, as we selected the candidate miRNAs from tissue specimens in the discovery effort, we consider that these miRNAs originated from rectal tumors. However, we could not evaluate the expression of these miRNAs in the clinical cohort. Nonetheless, accumulating reports show that miR-320a was identified to significantly correlate with sensitivity to chemoradiation of CRC.57 Similarly, miR-338-3p was identified as one of the miRNAs with the rectal cancer specificity.58 Previous reports showed that using pre- and post-treatment magnetic resonance imaging data, the researchers developed a radiomics model with excellent performance for individualized, noninvasive prediction of pCR and the multiparametric magnetic resonance imaging radiomic features have potential for predicting nonresponse to neoadjuvant therapy in patients with LARC.3,59 Since accumulating reports have shown that circulating tumor DNA (ctDNA) was used to predict the response of chemoradiotherapy (CRT) and postoperative recurrence in patients with CRC, the ctDNA analysis could potentially be useful to guide patient selection for surgery, surveillance, and adjuvant chemotherapy after CRT.60-63 However, detection of ctDNA suffers from poor sensitivity because of its limited abundance in circulation, and hence, the ctDNA levels are generally classified as detectable (ctDNA-positive) or undetectable (ctDNA-negative) on the basis of the mutation frequencies. However, since cell-free RNA changes are dynamic yet more abundant and do not suffer from sensitivity issues, a combination of our established signature and ctDNA classifier could be more accurate for predicting the response to CRT treatment. So, future clinical studies could aim to design prospective multicenter studies, which recruit patients with LARC and collect serial blood specimens (pre- and post-CRT and postoperation) to predict the response for CRT and survival outcomes. These studies can then evaluate the expression of cell-free RNA and ctDNA markers individually and in combination to assess their performance in improving patient outcomes in this malignancy.
In conclusion, we have identified and validated that a novel risk assessment model, which was combined with the miRNA panel and CEA levels, allows prediction of response to PCRT in a liquid biopsy assay. Furthermore, the final risk prediction model robustly identified patients who completely respond to PCRT. Our findings highlight the potential clinical impact of our model for improved selection and management of patients with this malignancy.
ACKNOWLEDGMENT
We would like to thank Drs Tatsuhiko Kakisaka, Satoshi Nishiwada, Yasuyuki Okada, Huanlin Wang, Divya Sahu, Geeta Sharma, and In-Seob Lee for their thoughtful discussions and advice during the course of this project. We would also like to extend our thanks to Dr Sarah Wilkinson for her significant editing and useful suggestions for improving the quality of our article.
APPENDIX
FIG A1.
Overview of the study.
TABLE A1.
Clinicopathologic Characteristics of Clinical Cohorts
TABLE A2.
Differential Analysis Between Normal and Tumor Specimens for Cancer Specificity of the Eight Selected Markers
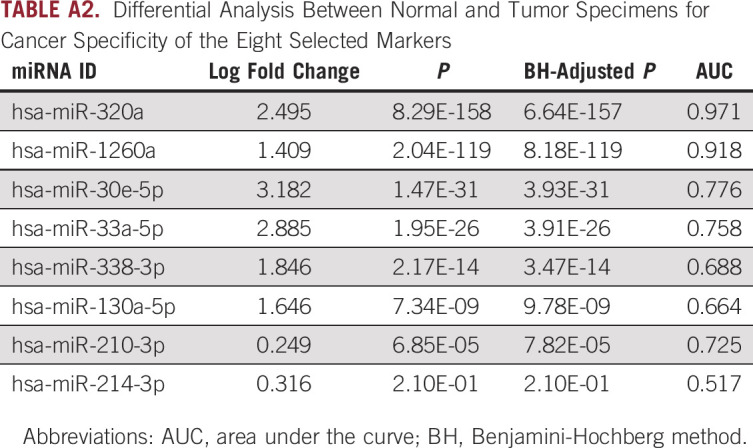
TABLE A3.
Performance of the Model in Estimating the Risk of Pathologic Complete Response
Mitsuo Shimada
Research Funding: Taiho Pharmaceutical (Inst), Chugai Pharma (Inst), TSUMURA & CO (Inst), Ono Pharmaceutical Co, Ltd (Inst), Takeda (Inst), AbbVie (Inst), EA Pharma (Inst), Eisai (Inst), Covidien Japan (Inst), Novartis (Inst)¸ Bayer Yakuhin (Inst), Astellas Pharma (Inst)
No other potential conflicts of interest were reported.
SUPPORT
Supported by CA72851, CA181572, CA184792, CA202797, and CA227602 grants from the National Cancer Institute, National Institutes of Health.
AUTHOR CONTRIBUTIONS
Conception and design: Yuma Wada, Yuji Morine, In Ja Park, Ajay Goel
Financial support: Xin Wang, Ajay Goel
Administrative support: Ajay Goel
Provision of study materials or patients: Tetsuya Ikemoto, Ane Etxart, Luis Bujanda
Collection and assembly of data: Mitsuo Shimada, Tetsuya Ikemoto, Yu Saito, Ane Etxart, Luis Bujanda, In Ja Park
Data analysis and interpretation: Zhongxu Zhu, Xin Wang, Yangsoon Park, In Ja Park, Ajay Goel
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I =Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Mitsuo Shimada
Research Funding: Taiho Pharmaceutical (Inst), Chugai Pharma (Inst), TSUMURA & CO (Inst), Ono Pharmaceutical Co, Ltd (Inst), Takeda (Inst), AbbVie (Inst), EA Pharma (Inst), Eisai (Inst), Covidien Japan (Inst), Novartis (Inst)¸ Bayer Yakuhin (Inst), Astellas Pharma (Inst)
No other potential conflicts of interest were reported.
REFERENCES
- 1.Siegel RL, Miller KD, Goding Sauer A, et al. : Colorectal cancer statistics, 2020. CA Cancer J Clin 70:145-164, 2020 [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A: Cancer statistics, 2020. CA Cancer J Clin 70:7-30, 2020 [DOI] [PubMed] [Google Scholar]
- 3.Liu Z, Zhang XY, Shi YJ, et al. : Radiomics analysis for evaluation of pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Clin Cancer Res 23:7253-7262, 2017 [DOI] [PubMed] [Google Scholar]
- 4.Gérard JP, Conroy T, Bonnetain F, et al. : Preoperative radiotherapy with or without concurrent fluorouracil and leucovorin in T3-4 rectal cancers: Results of FFCD 9203. J Clin Oncol 24:4620-4625, 2006 [DOI] [PubMed] [Google Scholar]
- 5.Kalyan A, Rozelle S, Benson A III: Neoadjuvant treatment of rectal cancer: Where are we now? Gastroenterol Rep (Oxf) 4:206-209, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Gijn W, Marijnen CA, Nagtegaal ID, et al. : Preoperative radiotherapy combined with total mesorectal excision for resectable rectal cancer: 12-year follow-up of the multicentre, randomised controlled TME trial. Lancet Oncol 12:575-582, 2011 [DOI] [PubMed] [Google Scholar]
- 7.Sauer R, Becker H, Hohenberger W, et al. : Preoperative versus postoperative chemoradiotherapy for rectal cancer. N Engl J Med 351:1731-1740, 2004 [DOI] [PubMed] [Google Scholar]
- 8.Sauer R, Liersch T, Merkel S, et al. : Preoperative versus postoperative chemoradiotherapy for locally advanced rectal cancer: Results of the German CAO/ARO/AIO-94 randomized phase III trial after a median follow-up of 11 years. J Clin Oncol 30:1926-1933, 2012 [DOI] [PubMed] [Google Scholar]
- 9.Zhao L, Bai C, Shao Y, et al. : A phase II study of neoadjuvant chemoradiotherapy with oxaliplatin and capecitabine for rectal cancer. Cancer Lett 310:134-139, 2011 [DOI] [PubMed] [Google Scholar]
- 10.Benson AB, Venook AP, Al-Hawary MM, et al. : Rectal cancer, version 2.2018, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 16:874-901, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glynne-Jones R, Wyrwicz L, Tiret E, et al. : Rectal cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol 28:iv22-iv40, 2017 [DOI] [PubMed] [Google Scholar]
- 12.Hofheinz RD, Wenz F, Post S, et al. : Chemoradiotherapy with capecitabine versus fluorouracil for locally advanced rectal cancer: A randomised, multicentre, non-inferiority, phase 3 trial. Lancet Oncol 13:579-588, 2012 [DOI] [PubMed] [Google Scholar]
- 13.Schmoll HJ, Tabernero J, Maroun J, et al. : Capecitabine plus oxaliplatin compared with fluorouracil/folinic acid as adjuvant therapy for stage III colon cancer: Final results of the NO16968 randomized controlled phase III trial. J Clin Oncol 33:3733-3740, 2015 [DOI] [PubMed] [Google Scholar]
- 14.Bosset JF, Collette L, Calais G, et al. : Chemotherapy with preoperative radiotherapy in rectal cancer. N Engl J Med 355:1114-1123, 2006 [DOI] [PubMed] [Google Scholar]
- 15.Rödel C, Liersch T, Becker H, et al. : Preoperative chemoradiotherapy and postoperative chemotherapy with fluorouracil and oxaliplatin versus fluorouracil alone in locally advanced rectal cancer: Initial results of the German CAO/ARO/AIO-04 randomised phase 3 trial. Lancet Oncol 13:679-687, 2012 [DOI] [PubMed] [Google Scholar]
- 16.Roh MS, Colangelo LH, O'Connell MJ, et al. : Preoperative multimodality therapy improves disease-free survival in patients with carcinoma of the rectum: NSABP R-03. J Clin Oncol 27:5124-5130, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Campos-Lobato LF, Stocchi L, da Luz Moreira A, et al. : Pathologic complete response after neoadjuvant treatment for rectal cancer decreases distant recurrence and could eradicate local recurrence. Ann Surg Oncol 18:1590-1598, 2011 [DOI] [PubMed] [Google Scholar]
- 18.Martin ST, Heneghan HM, Winter DC: Systematic review and meta-analysis of outcomes following pathological complete response to neoadjuvant chemoradiotherapy for rectal cancer. Br J Surg 99:918-928, 2012 [DOI] [PubMed] [Google Scholar]
- 19.Duldulao MP, Lee W, Streja L, et al. : Distribution of residual cancer cells in the bowel wall after neoadjuvant chemoradiation in patients with rectal cancer. Dis Colon Rectum 56:142-149, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Aguilar J, Smith DD, Avila K, et al. : Optimal timing of surgery after chemoradiation for advanced rectal cancer: Preliminary results of a multicenter, nonrandomized phase II prospective trial. Ann Surg 254:97-102, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maas M, Nelemans PJ, Valentini V, et al. : Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: A pooled analysis of individual patient data. Lancet Oncol 11:835-844, 2010 [DOI] [PubMed] [Google Scholar]
- 22.Tulchinsky H, Shmueli E, Figer A, et al. : An interval >7 weeks between neoadjuvant therapy and surgery improves pathologic complete response and disease-free survival in patients with locally advanced rectal cancer. Ann Surg Oncol 15:2661-2667, 2008 [DOI] [PubMed] [Google Scholar]
- 23.Fokas E, Liersch T, Fietkau R, et al. : Tumor regression grading after preoperative chemoradiotherapy for locally advanced rectal carcinoma revisited: Updated results of the CAO/ARO/AIO-94 trial. J Clin Oncol 32:1554-1562, 2014 [DOI] [PubMed] [Google Scholar]
- 24.Jalilian M, Davis S, Mohebbi M, et al. : Pathologic response to neoadjuvant treatment in locally advanced rectal cancer and impact on outcome. J Gastrointest Oncol 7:603-608, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reggiani Bonetti L, Lionti S, Domati F, et al. : Do pathological variables have prognostic significance in rectal adenocarcinoma treated with neoadjuvant chemoradiotherapy and surgery? World J Gastroenterol 23:1412-1423, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rödel C, Martus P, Papadoupolos T, et al. : Prognostic significance of tumor regression after preoperative chemoradiotherapy for rectal cancer. J Clin Oncol 23:8688-8696, 2005 [DOI] [PubMed] [Google Scholar]
- 27.Valentini V, Coco C, Picciocchi A, et al. : Does downstaging predict improved outcome after preoperative chemoradiation for extraperitoneal locally advanced rectal cancer? A long-term analysis of 165 patients. Int J Radiat Oncol Biol Phys 53:664-674, 2002 [DOI] [PubMed] [Google Scholar]
- 28.Vecchio FM, Valentini V, Minsky BD, et al. : The relationship of pathologic tumor regression grade (TRG) and outcomes after preoperative therapy in rectal cancer. Int J Radiat Oncol Biol Phys 62:752-760, 2005 [DOI] [PubMed] [Google Scholar]
- 29.Appelt AL, Pløen J, Harling H, et al. : High-dose chemoradiotherapy and watchful waiting for distal rectal cancer: A prospective observational study. Lancet Oncol 16:919-927, 2015 [DOI] [PubMed] [Google Scholar]
- 30.Habr-Gama A, Sabbaga J, Gama-Rodrigues J, et al. : Watch and wait approach following extended neoadjuvant chemoradiation for distal rectal cancer: Are we getting closer to anal cancer management? Dis Colon Rectum 56:1109-1117, 2013 [DOI] [PubMed] [Google Scholar]
- 31.Maas M, Beets-Tan RG, Lambregts DM, et al. : Wait-and-see policy for clinical complete responders after chemoradiation for rectal cancer. J Clin Oncol 29:4633-4640, 2011 [DOI] [PubMed] [Google Scholar]
- 32.Habr-Gama A, Perez RO, Nadalin W, et al. : Operative versus nonoperative treatment for stage 0 distal rectal cancer following chemoradiation therapy: Long-term results. Ann Surg 240:711-717, 2004; discussion 717-718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al-Sukhni E, Attwood K, Mattson DM, et al. : Predictors of pathologic complete response following neoadjuvant chemoradiotherapy for rectal cancer. Ann Surg Oncol 23:1177-1186, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalady MF, de Campos-Lobato LF, Stocchi L, et al. : Predictive factors of pathologic complete response after neoadjuvant chemoradiation for rectal cancer. Ann Surg 250:582-589, 2009 [DOI] [PubMed] [Google Scholar]
- 35.Lorimer PD, Motz BM, Kirks RC, et al. : Pathologic complete response rates after neoadjuvant treatment in rectal cancer: An analysis of the national cancer database. Ann Surg Oncol 24:2095-2103, 2017 [DOI] [PubMed] [Google Scholar]
- 36.Park YA, Sohn SK, Seong J, et al. : Serum CEA as a predictor for the response to preoperative chemoradiation in rectal cancer. J Surg Oncol 93:145-150, 2006 [DOI] [PubMed] [Google Scholar]
- 37.Wallin U, Rothenberger D, Lowry A, et al. : CEA - a predictor for pathologic complete response after neoadjuvant therapy for rectal cancer. Dis Colon Rectum 56:859-868, 2013 [DOI] [PubMed] [Google Scholar]
- 38.Chen X, Ba Y, Ma L, et al. : Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 18:997-1006, 2008 [DOI] [PubMed] [Google Scholar]
- 39.Kanaan Z, Rai SN, Eichenberger MR, et al. : Plasma miR-21: A potential diagnostic marker of colorectal cancer. Ann Surg 256:544-551, 2012 [DOI] [PubMed] [Google Scholar]
- 40.Ng EK, Chong WW, Jin H, et al. : Differential expression of microRNAs in plasma of patients with colorectal cancer: A potential marker for colorectal cancer screening. Gut 58:1375-1381, 2009 [DOI] [PubMed] [Google Scholar]
- 41.Hur K, Toiyama Y, Okugawa Y, et al. : Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut 66:654-665, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toiyama Y, Hur K, Tanaka K, et al. : Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann Surg 259:735-743, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toiyama Y, Takahashi M, Hur K, et al. : Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst 105:849-859, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mandard AM, Dalibard F, Mandard JC, et al. : Pathologic assessment of tumor regression after preoperative chemoradiotherapy of esophageal carcinoma. Clinicopathologic correlations. Cancer 73:2680-2686, 1994 [DOI] [PubMed] [Google Scholar]
- 45.Livak KJ, Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402-408, 2001 [DOI] [PubMed] [Google Scholar]
- 46.Smyth GK: Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:Article 3, 2004. [DOI] [PubMed] [Google Scholar]
- 47.Fokas E, Ströbel P, Fietkau R, et al. : Tumor regression grading after preoperative chemoradiotherapy as a prognostic factor and individual-level surrogate for disease-free survival in rectal cancer. J Natl Cancer Inst 109:djx095, 2017. [DOI] [PubMed] [Google Scholar]
- 48.Trakarnsanga A, Gönen M, Shia J, et al. : Comparison of tumor regression grade systems for locally advanced rectal cancer after multimodality treatment. J Natl Cancer Inst 106:dju248, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garcia-Aguilar J, Chow OS, Smith DD, et al. : Effect of adding mFOLFOX6 after neoadjuvant chemoradiation in locally advanced rectal cancer: A multicentre, phase 2 trial. Lancet Oncol 16:957-966, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huerta S, Hrom J: Oxaliplatin as a radiosensitizing agent in rectal cancer. Anticancer Drugs 22:317-323, 2011 [DOI] [PubMed] [Google Scholar]
- 51.Martin LK, Bekaii-Saab T: Optimizing neoadjuvant therapy for rectal cancer with oxaliplatin. J Natl Compr Canc Netw 11:298-307, 2013; quiz 307 [DOI] [PubMed] [Google Scholar]
- 52.Ren DL, Li J, Yu HC, et al. : Nomograms for predicting pathological response to neoadjuvant treatments in patients with rectal cancer. World J Gastroenterol 25:118-137, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bregendahl S, Emmertsen KJ, Lous J, et al. : Bowel dysfunction after low anterior resection with and without neoadjuvant therapy for rectal cancer: A population-based cross-sectional study. Colorectal Dis 15:1130-1139, 2013 [DOI] [PubMed] [Google Scholar]
- 54.Guren MG, Eriksen MT, Wiig JN, et al. : Quality of life and functional outcome following anterior or abdominoperineal resection for rectal cancer. Eur J Surg Oncol 31:735-742, 2005 [DOI] [PubMed] [Google Scholar]
- 55.Smith JD, Ruby JA, Goodman KA, et al. : Nonoperative management of rectal cancer with complete clinical response after neoadjuvant therapy. Ann Surg 256:965-972, 2012 [DOI] [PubMed] [Google Scholar]
- 56.Stijns RCH, Tromp MR, Hugen N, et al. : Advances in organ preserving strategies in rectal cancer patients. Eur J Surg Oncol 44:209-219, 2018 [DOI] [PubMed] [Google Scholar]
- 57.Salendo J, Spitzner M, Kramer F, et al. : Identification of a microRNA expression signature for chemoradiosensitivity of colorectal cancer cells, involving miRNAs-320a, -224, -132 and let7g. Radiother Oncol 108:451-457, 2013 [DOI] [PubMed] [Google Scholar]
- 58.Gaedcke J, Grade M, Camps J, et al. : The rectal cancer microRNAome–microRNA expression in rectal cancer and matched normal mucosa. Clin Cancer Res 18:4919-4930, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou X, Yi Y, Liu Z, et al. : Radiomics-based pretherapeutic prediction of non-response to neoadjuvant therapy in locally advanced rectal cancer. Ann Surg Oncol 26:1676-1684, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tie J, Cohen JD, Wang Y, et al. : Circulating tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol 5:1710-1717, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khakoo S, Carter PD, Brown G, et al. : MRI tumor regression grade and circulating tumor DNA as complementary tools to assess response and guide therapy adaptation in rectal cancer. Clin Cancer Res 26:183-192, 2020 [DOI] [PubMed] [Google Scholar]
- 62.Tie J, Cohen JD, Wang Y, et al. : Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: A prospective biomarker study. Gut 68:663-671, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murahashi S, Akiyoshi T, Sano T, et al. : Serial circulating tumour DNA analysis for locally advanced rectal cancer treated with preoperative therapy: Prediction of pathological response and postoperative recurrence. Br J Cancer 123:803-810, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]