Abstract
Context
Serum insulin-like growth factor 1 (IGF-1) levels are relatively constant in somatropin-treated children with growth hormone deficiency (GHD), and guide dose adjustments for clinical efficacy and long-term safety. IGF-1 levels following treatment with long-acting growth hormones such as lonapegsomatropin (lonapegsomatropin-tcgd, TransCon hGH), a once-weekly somatropin prodrug, exhibit a characteristic profile over the dosing interval.
Objective
This study aimed to develop a method to predict average IGF-1 in lonapegsomatropin-treated GHD children to interpret IGF-1 data based on a single sample obtained any time at steady state.
Methods
A population nonlinear mixed-effect pharmacodynamic model for IGF-1 was developed based on 2 randomized, open-label trials of TransCon hGh in GHD children and used to develop a linear mixed model with Taylor series to fit simulated IGF-1 profiles of lonapegsomatropin-treated children.
A total of
49 896 IGF-1 sample data simulated from 105 lonapegsomatropin-treated GHD children were utilized for the final prediction model. The dosage range of TransCon hGh was 0.14 to 0.30 hGH mg/kg/week, and weekly average IGF-1 was calculated using IGF-1 profiles simulated from the nonlinear pharmacodynamic model. Predicted average IGF-1 was obtained by linear mixed model with Taylor series.
Results
The nonlinear mixed-effect model provided satisfactory model fit. The linear mixed model with Taylor series fit simulated IGF-1 data well, with a relatively straightforward prediction formula. IGF-1 values sampled at ~4.5 days post-dose coincided with weekly average IGF-1 at steady state.
Conclusion
A formula to predict average IGF-1 from a single sample of IGF-1 at steady state was established to aid clinicians in interpreting IGF-1 levels in GHD children administered lonapegsomatropin.
Keywords: growth hormone, long-acting growth hormone, IGF-1, prediction, prodrug, growth hormone deficiency
For more than 30 years, children with growth hormone deficiency (GHD) have been treated with daily injections of somatropin (recombinant human GH, rhGH) [1]. Both children and their caregivers find the daily injection frequency burdensome, leading to nonadherence rates of 5% to 82% [1-3]. In 2015, the Growth Hormone Research Society recognized the value of a long-acting growth hormone (LAGH) and agreed that by decreasing injection frequency and offering different pharmacokinetic properties, a LAGH would potentially increase adherence and improve outcomes [4]. Physicians report that they monitor insulin-like growth factor 1 (IGF-1) levels in patients treated with daily somatropin to guide dose adjustment for long-term safety and efficacy. In patients treated with daily somatropin, IGF-1 levels are relatively constant during the day and from day to day [5]. The different hGH exposure profiles of weekly growth hormones result in a pharmacodynamic (PD) response, as measured by serum IGF-1 levels, which follows a time-specific profile [6, 7]. Thus, a random IGF-1 concentration obtained at any time post-dose may not be representative of the overall or average IGF-1 exposure in the case of hGH administered weekly.
Lonapegsomatropin (lonapegsomatropin-tcgd, TransCon hGH; Ascendis Pharma A/S) is a long-acting prodrug in development for once-weekly administration to children and adults with GHD consisting of the parent drug, somatropin, an inert methyoxy polyethelene glycol carrier, and a TransCon linker [8-10]. Lonapegsomatropin was recently approved in the United States for once-weekly administration in children aged 1 year and older who weigh at least 11.5 kg and have growth failure due to inadequate secretion of endogenous growth hormone [11]. The apparent half-life of somatropin released from lonapegsomatropin is approximately 25 hours, as established in clinical phase 1 trials in adults [11].
We describe here the IGF-1 PD response to the weekly subcutaneous administration of lonapegsomatropin in GH-deficient children and provide a model that demonstrates how a single IGF-1 level obtained at steady state any time after an injection, but before the next, may be interpreted to predict an average level, which could aid clinicians to assess an individual patient’s response to therapy. A population nonlinear mixed-effect PD model for IGF-1 was developed based on 2 randomized, open-label trials evaluating TransCon hGh: CT-004, a 26-week phase 2 trial [8], and CT-301 (heiGHt Trial), a pivotal 52-week phase 3 trial [10]. Model-simulated IGF-1 profiles for all post-baseline samples from lonapegsomatropin subjects were generated for the calculation of average IGF-1 level in CT-301. The relationship between individual IGF-1 levels and the average IGF-1 values (weekly) was evaluated statistically based on simulated data from the nonlinear population PD model. As a result of this statistical evaluation, a linear mixed model was created to predict the average IGF-1 level from a single IGF-1 sample obtained at steady state at any time after dosing of weekly lonapegsomatropin treatment.
Methods
Clinical Trial Design and Patient Population for Model Development
Trial CT-004
CT-004 was a phase 2, randomized, open-label trial of 3 different doses (0.14, 0.21, or 0.30 mg hGH/kg/week) of once-weekly ACP-001 (a bioequivalent predecessor of lonapegsomatropin [data on file]; TransCon hGH in this manuscript refers to both ACP-001 and lonapegsomatropin), compared to daily somatropin (Genotropin) over a period of 26 weeks in treatment-naïve children with GHD (N = 53). Full IGF-1 profile sampling occurred at Weeks 1 and 13 (pre-dose [at −0.5 h] and at 6, 8, 12, 18, 24, 48, 72, 96, 120, and 168 h post-dose) for all subjects [8].
Trial CT-301 (heiGHt Trial)
CT-301 was a phase 3, randomized, open-label trial of lonapegsomatropin compared with daily somatropin, over 52 weeks in 161 treatment-naïve children with GHD, of whom 105 were randomized to the lonapegsomatropin arm [10]. All randomized lonapegsomatropin subjects were medically eligible to participate in a subset for full IGF-1 profile sampling at Week 13 (pre-dose [at −0.5 hours] and at 8, 12, 16, 24, 36, 48, 72, 96, 120, and 168 hours post-dose). CT-301 sought to enroll 8 subjects into the subset, and ultimately enrolled 11 subjects across 6 sites and 2 countries. The 6 sites where subjects were enrolled into the subset had sufficient equipment, staff, and operating hours to conduct the rich sampling over the course of 1 week. Sparse sampling for the measurement of IGF-1 was performed for all subjects at Week 1 (baseline/pre-dose), Week 5 (pre-dose), Weeks 13, 26, 39 (48-72 hours post-dose), and Week 52 (164-172 hours post-dose) [10].
Statistical Analysis to Predict Average IGF-1 Level From a Single IGF-1 Level
Statistical methodology
A population nonlinear mixed-effect IGF-1 model was established for TransCon hGH based on all IGF-1 data collected from trials CT-004 and CT-301 [8, 10, 12]. Full weekly IGF-1 concentration profiles were simulated from the nonlinear mixed-effect model for all 105 lonapegsomatropin-treated subjects of CT-301 from time 0.0 to 7.0 days at Weeks 13, 26, 39, and 52. The actual dosing records for each patient were used in the simulation. All IGF-1 sampling timepoints in the CT-004 and CT-301 trials were included in the modeling. Of note, the trials enrolled children who were pre-pubertal; and a small number started the pubertal transition during the trials. A noncompartmental analysis (NCA) on the simulated profiles was performed to calculate area under the curve of the IGF-1 concentration from 0 to 7 days (AUC0-7). The average IGF-1 concentration over the weekly dosing interval was calculated as AUC0-7/7. This average IGF-1 concentration was then converted into an average IGF-1 standard deviation score (SDS) value using the age- and gender-specific reference intervals using the same assay reported by Bidlingmaier et al [13].
The simulated IGF-1 data for CT-301 were statistically analyzed to evaluate the relationship between the average IGF-1 level and a single IGF-1 sample. The goal of the statistical evaluation was to predict the average IGF-1 level from a single IGF-1 sample collected at any time during a dosing week at steady state. This goal was achievable without the need for other covariates, including dosage, because it was found that none of the covariates were significant in the nonlinear population PD model.
The data for a single IGF-1 sample can be accepted in units of SDS or concentration (e.g., ng/mL). The average IGF-1 level was calculated in the same units in which the single IGF-1 sample was expressed. Conversion between the IGF-1 SDS and concentration units can be calculated based on published literature [13] as was performed by the laboratories used for the 2 trials (CT-301 and CT-004). For an IGF-1 assay different from that used in the present study, the IGF-1 SDS calculation would be based on a formula applicable to that specific assay. The average IGF-1 SDS can be calculated as long as the IGF-1 SDS can be obtained for that particular IGF-1 assay.
Equation A
Let Delta(d) denote the difference between an IGF-1 value at d days from last dose and the average IGF-1 value of the week at steady state:
| (A) |
Here, Delta(d) is a function of d, where d ranges from 0.0 to 7.0 days and is the time from last dose of the weekly dosing interval in the units of days. The statistical goal is to predict Delta(d) based on the population nonlinear mixed PD model by analyzing model-simulated data for the phase 3 trial.
Equation (A) is applicable for data with an approximately normal distribution. It is applied to IGF-1 SDS data directly. For IGF-1 concentration data, natural log transformation is applied first since IGF-1 concentration is assumed to follow a lognormal distribution. The following description focuses on the method for IGF-1 SDS, while it is equally applicable to IGF-1 concentration with log-transformed data, except that the model outcomes with log-transformed data are required to be back transformed to their original concentration scale with exponential transformations.
Taylor series (Equation B)
A linear mixed model with Taylor series expansion was applied to predict Delta(d) based on the simulated weekly IGF-1 profile data, with 4 repetitions representing the 4 study visits at steady state (Weeks 13, 26, 39, and 52), from 0.0 to 7.0 days since last dose. The model has Delta(d) as the dependent variable, and time d with Taylor series expansion up to power k as independent variables:
| (B) |
where k is an integer to be determined from goodness of fit analysis, and ci, i = 1, …, k are coefficients for the fixed effects of the polynomial function of d. Subject is the random effect in the mixed model.
The precision of the prediction error was evaluated using the residual standard deviation (RSD) and the 90% prediction interval (PI) was calculated as ±1.645*RSD of the prediction. The model was fitted in SAS version 9.4 (SAS Institute Inc., Cary, NC, USA) with the proc mixed procedure. The simulated IGF-1 weekly profiles with dosing intervals less than 7.0 days of a dosing week were excluded for the final model, to remove the bias caused by earlier dosing, so that the prediction of the steady state profile includes data from full 7.0 days without the interruption of earlier dosing. Simple linear mixed models with
| (B’) |
for each d days since last dose was applied separately every half-day to examine the performance of the final mixed model in (B).
Results
Predicting Average IGF-1 Level
The simulation of all the data from the 105 children with GHD treated with lonapegsomatropin in CT-301 included a total of 416 weekly IGF-1 profiles across the 4 visits from Week 13 to 52. Each profile had a total of 168 simulated IGF-1 hourly samples to form a total of 69 888 simulated IGF-1 samples. Among those 416 weekly profiles, 297 profiles had durations of at least 7 days without earlier dosing interruption to form a total of 49 896 IGF-1 simulated samples for the final model. All 105 subjects had at least 1 IGF-1 profile with a full 7 days of duration without earlier dosing interruption.
Results for IGF-1 SDS
The IGF-1 weekly profile of Delta(d) was virtually identical across visits from Week 13 to Week 52 from the population perspective. Supplementary Fig. 1 includes boxplots of Delta(d) for every day of the 7-day dosing interval at each of the 4 visits (Weeks 13, 26, 39, and 52) with all data included [14]. The extreme outliers of high Delta(d) from Days 6 to 7 reflected some earlier dosing which randomly occurred due to the allowed flexible dosing schedule during the 7-day dosing interval.
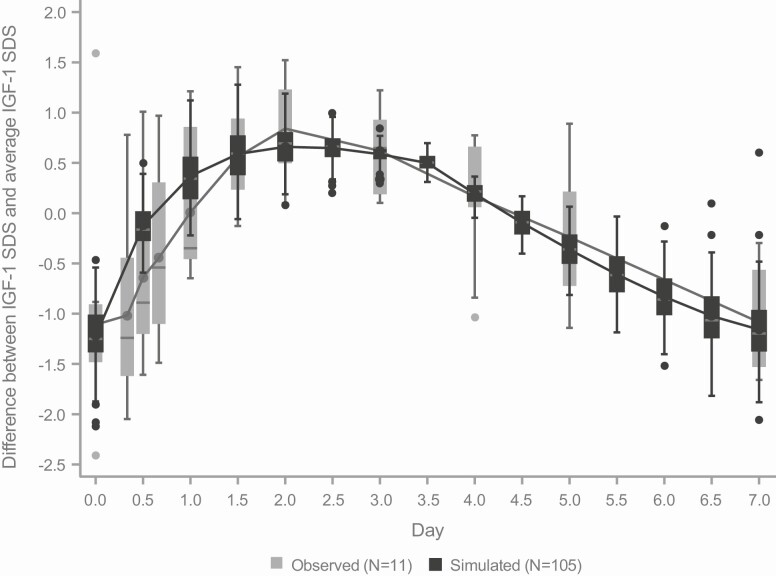
To examine whether the model-simulated weekly profiles agreed with the observed data, the weekly profile calculated from the 11 subjects from CT-301 (with intensive profile sampling at Week 13) was superimposed on the simulated profile every half-day at Week 13 for comparison (Fig. 1). The model-simulated IGF-1 boxplots and concentration curves overlapped with their corresponding observed boxplots and concentration curves based on this subset of 11 subjects (Fig. 1) with generally excellent agreement. The lack of smoothness of the profile based on the 11 subjects is not surprising due to the small sample size and limited sampling time points. Lonapegsomatropin had an average IGF-1 SDS of 0.312 at Week 13 to 0.717 at Week 52, an increase of 0.405 and daily somatropin had an average IGF-1 SDS of −0.596 at Week 13 to −0.024 at Week 52, an increase of 0.572. Therefore, the increases between the 2 treatment groups were similar after steady state, and lonapegsomatropin induced a numerically lower increase compared with somatropin. These data indicate the average IGF-1 level increased slowly from Week 13 to Week 52.
Figure 1.
Comparison of observed and simulated Delta(d) IGF-1 SDS weekly profiles. Boxplots of Delta(d) for IGF-1 SDS based on simulated IGF-1 for subjects on lonapegsomatropin (N = 105; black boxes) and observed IGF-1 of subjects with profile sampling at Week 13 (N = 11; gray boxes).
The mixed model with Taylor series fits the best at k = 8 based on statistical evaluations of goodness of fit, including log likelihood ratio tests and other goodness of fit statistics. The coefficients of the linear model are included in Supplementary Table 1 [14].
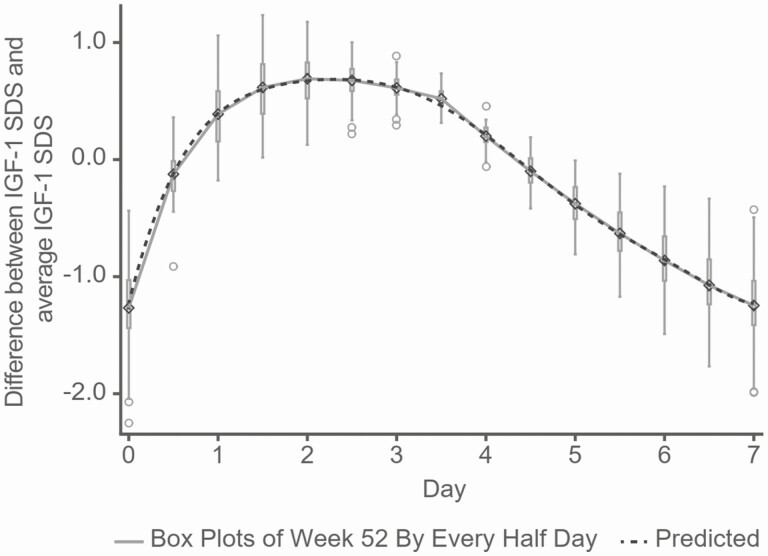
The mixed model with the Taylor series function fits the simulated profiles very well, illustrated by comparing to the boxplots of simulated IGF-1 SDS at Week 52 (Fig. 2). The Taylor series model provides a simple and smooth formula for the calculation of Delta(d) at any time during a 7-day dosing week.
Figure 2.
Comparing Delta(d) of IGF-1 SDS from the mixed model with Taylor series, and from boxplots of simulated data from the population nonlinear model every half-day at Week 52. Simulated IGF-1 SDS values are shown as boxplots and solid line at Week 52 and the model-predicted value is shown as a dotted line. Circles: outliers of boxplots. Diamonds: simulated data from the population nonlinear model.
The model predicted Delta(d) with 90% PI is displayed by every half-day in Table 1. Removing weekly profile data with less than 7 days of dosing interval improved the predicted profile so that IGF-1 levels at Day 0 and Day 7 were virtually identical, as expected at steady state.
Table 1.
The predicted difference between observed IGF-1 SDS and average IGF-1 SDS by time with 90% prediction intervals (PI)
| Day | Mean | RSD | Lower limit of 90% PI | Upper limit of 90% PI |
|---|---|---|---|---|
| 0.0 | -1.22 | 0.35 | -1.80 | -0.65 |
| 0.5 | -0.12 | 0.20 | -0.46 | 0.22 |
| 1.0 | 0.39 | 0.28 | -0.07 | 0.86 |
| 1.5 | 0.60 | 0.27 | 0.16 | 1.04 |
| 2.0 | 0.67 | 0.21 | 0.32 | 1.02 |
| 2.5 | 0.68 | 0.15 | 0.43 | 0.93 |
| 3.0 | 0.62 | 0.10 | 0.45 | 0.78 |
| 3.5 | 0.46 | 0.08 | 0.33 | 0.59 |
| 4.0 | 0.21 | 0.09 | 0.06 | 0.36 |
| 4.5 | -0.09 | 0.14 | -0.31 | 0.13 |
| 5.0 | -0.39 | 0.18 | -0.68 | -0.09 |
| 5.5 | -0.64 | 0.23 | -1.01 | -0.26 |
| 6.0 | -0.85 | 0.27 | -1.29 | -0.42 |
| 6.5 | -1.07 | 0.30 | -1.57 | -0.57 |
| 7.0 | -1.22 | 0.33 | -1.77 | -0.68 |
Bold text indicates the day when sampling time for IGF-1 was coincident with the average IGF-1 SDS. Abbreviation: RSD, residual standard deviation.
The model prediction error was measured by model RSD. The prediction errors were generally low with RSD ≤ 0.35 from samples taken at any time during a dosing interval. The time range with an accurate prediction of RSD < 0.2 was between Days 2.5 and 5. The prediction errors were largest when the sampling times were close to Day 0 and Day 7. The sampling time for IGF-1 to be coincident with average IGF-1 level was around Day 4.5, and coincident with peak IGF-1 between Day 2.0 and Day 2.5 with the peak IGF-1 SDS about 0.7 above the average IGF-1 SDS.
Therefore, a simple formula was established to predict the average IGF-1 SDS based on a single sample collected at steady state at any given time of a dosing interval. The weekly IGF-1 profile with the mixed model using Taylor series provides a smoothed approximation of the population nonlinear mixed model. This smooth approximation might be more realistic compared with the original nonlinear PD model, which is affected by limited sampling time that makes it difficult to obtain a smooth weekly profile.
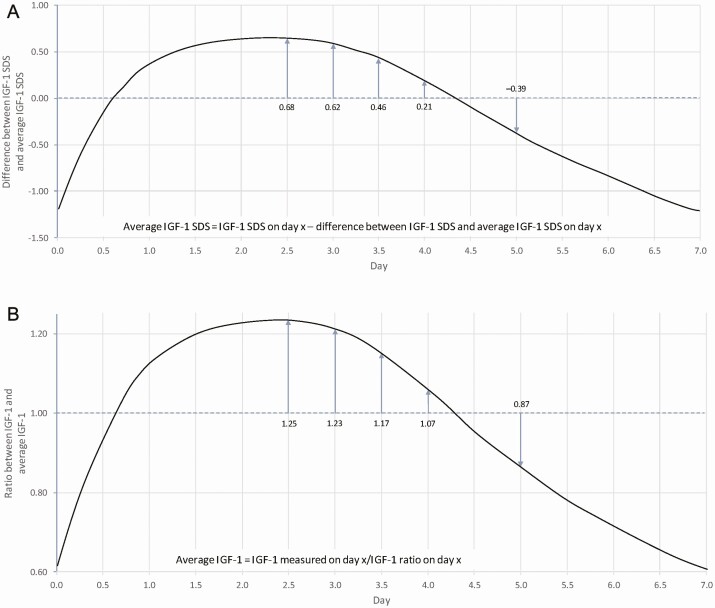
To interpret an IGF-1 SDS reading obtained at any given time since last dose, for example, Days 2.5 and 5.0, one would subtract 0.68 for the sample on Day 2.5, and add 0.39 for the sample on Day 5.0, to predict the average IGF-1 SDS (Fig. 3a, Table 1).
Figure 3.
Predicting average IGF-1 SDS and average IGF-1 concentrations with samples on Days 2.5 and 5.0. (a) The method to predict average IGF-1 SDS using an IGF-1 SDS reading at a time since the last dose was as follows: for an IGF-1 sample drawn 2.5 days post-dose, subtract 0.68 SDS from the measured value; for an IGF-1 sample drawn 5 days post-dose, add 0.39 to the measured value (b) The method to predict the average IGF-1 concentration using an IGF-1 concentration reading at a time since last dose is as follows: for an IGF-1 sample drawn 2.5 days post-dose, divide the sample concentration by the ratio of 1.25; for an IGF-1 sample drawn 5 days post-dose, divide the sample concentration by the ratio of 0.87.
Results for IGF-1 Concentration
The results for the IGF-1 concentration (Supplementary Figs. 2 and 3) [14] were very similar to those for the IGF-1 SDS. As for the IGF-1 SDS, the mixed model with Taylor series fit the best at k = 8 based on statistical evaluations of goodness of fit, including log likelihood ratio tests and other goodness of fit statistics. The coefficients of the linear model are shown in Supplementary Table 2 [14]. The mixed model with the Taylor series function fit the simulated profiles very well when compared with the boxplots of the simulated IGF-1 concentration in log scale at Week 52 (Supplementary Fig. 4) [14]. The model predicted Delta(d) with 90% PI is displayed by every half-day in Supplementary Table 3 [14]. The model prediction error was measured by model RSD. The prediction errors were generally low with an RSD < 0.17 from samples taken at any time during a dosing week. The time range with an accurate prediction of RSD < 0.07 was between Days 2.5 and 5.0. The prediction errors were largest when the sampling times were close to Day 0.0 and Day 7.0. Table 2 shows similar results in the original concentration scale. The sampling time for IGF-1 to be coincident with average IGF-1 is around Day 4.5, and coincident with the peak IGF-1 between Day 2.0-2.5 with the peak IGF-1 concentration about 25% higher than the average IGF-1 concentration.
Table 2.
Predicted ratio between IGF-1 and average IGF-1 concentrations by time with 90% prediction interval (PI)
| Day | Ratio | Lower limit of 90% PI | Upper limit of 90% PI |
|---|---|---|---|
| 0.0 | 0.62 | 0.47 | 0.81 |
| 0.5 | 0.96 | 0.85 | 1.08 |
| 1.0 | 1.14 | 0.98 | 1.33 |
| 1.5 | 1.22 | 1.06 | 1.40 |
| 2.0 | 1.25 | 1.11 | 1.40 |
| 2.5 | 1.25 | 1.15 | 1.36 |
| 3.0 | 1.23 | 1.15 | 1.31 |
| 3.5 | 1.17 | 1.10 | 1.23 |
| 4.0 | 1.07 | 1.02 | 1.13 |
| 4.5 | 0.97 | 0.89 | 1.05 |
| 5.0 | 0.87 | 0.78 | 0.98 |
| 5.5 | 0.79 | 0.68 | 0.92 |
| 6.0 | 0.73 | 0.60 | 0.87 |
| 6.5 | 0.67 | 0.54 | 0.83 |
| 7.0 | 0.62 | 0.48 | 0.79 |
Bold text indicates the day when sampling time for IGF-1 was coincident with the average IGF-1 concentration.
A simple formula was established to predict the average IGF-1 concentration based on a single sample at steady state for any time after dosing. The weekly IGF-1 concentration profile with the mixed model using Taylor series provides a smoothed approximation of the population nonlinear mixed model, which might be more realistic compared with the original nonlinear PD model that is affected by limited sampling time.
To interpret an IGF-1 reading at a time since the last dose, for example, at Days 2.5 and 5.0, one would divide the ratio by 1.25 for the sample on Day 2.5 and divide the ratio by 0.87 for the sample on Day 5.0, to predict the average IGF-1 (Fig. 3b, Table 2).
Discussion
The pharmacokinetic (PK) and PD profiles of lonapegsomatropin differ from daily somatropin as well as from other LAGHs with different molecular structures and physical-chemical characteristics. In managing patients taking daily somatropin, many physicians monitor serum IGF-1 levels. Given the PK profile of GH released from lonapegsomatropin, the average IGF-1 level may be a more clinically relevant measure to represent overall exposure compared with peak or trough levels and may be useful to physicians [4]. With lonapegsomatropin therapy, IGF-1 is estimated to reach steady state by 5 weeks [12]. At steady state, the IGF-1 level sampled approximately 4.5 days after a dose administration best represented the average IGF-1 level over the weekly interval between doses. However, collecting blood samples for serum IGF-1 at a specific date and time may not be practical in a clinical setting. Here, we provide information to permit the prediction of the weekly average serum IGF-1 level at steady state as long as the timing relative to the last dose is known to the nearest half-day. The prediction has very good accuracy with a known error range from a single IGF-1 sample collected anytime during the dosing week, with the best precision for a sample from 2.5 to 5.0 days after the last dose.
Linear models to predict the average IGF-1 level for some LAGH molecules with unique profiles have been published [6, 7]. A novelty of the research presented here is that it creates a single linear function explaining the entire IGF-1 weekly profile at steady state, instead of relying on a unique formula at each time point; a single linear formula that provides a straightforward smooth profile can be used to help understand and interpret IGF-1 results. The method takes into account the lack of normality of IGF-1 concentration data by using the same approach to handle normalized data of either IGF-1 SDS or log-transformed IGF-1 data. Additionally, the analysis described here includes a direct comparison of model-simulated data and the observed IGF-1 levels with very good general agreement, as well as the ability to predict average IGF-1 in the SDS scale or concentration units.
The age- and gender-specific reference intervals of IGF-1 SDS are Z-scores representing the number of standard deviations above (if positive) or below (if negative) the median of the general population. The IGF-1 SDS scores are usually provided by the laboratory since derivation depends on the assay and supporting database utilized by the laboratory, while some common assays have published algorithms [13]. Therefore, although the reported absolute concentration levels may be assay dependent, the pharmacodynamic pattern should be similar across different assays.
The analyses in this manuscript focused on the average IGF-1, as this best represents the total exposure to IGF-1. However, it was observed that the peak IGF-1 SDS can also be predicted as about 0.7 above average IGF-1 SDS, or 25% higher than the average IGF-1 concentration, and it is reached at approximately Day 2.0 to 2.5.
There are theoretical concerns that peak IGF-1 levels above the normal range may increase the risk of long-term adverse events although no adverse events have been associated with IGF-1 levels above +2 SD in children receiving daily growth hormone therapy [15]. However, due to the short duration of the IGF-1 peak during lonapegsomatropin treatment, the average IGF-1 level is more likely to correlate with the efficacy and long-term safety of the therapy. In addition, a specific IGF-1 level has not been identified above which there is a documented increase in the risk of any known adverse event of rhGH therapy [16].
It is recognized that there are some limitations of this method. The development of the analysis model was retrospective without a prospective study for confirmation. The population studied was limited to enroll children with GHD in the dose range of 0.14 to 0.30 mg/kg/week. Additional data may be needed for children treated outside of the dose range. Finally, an accurate IGF-1 assay is required.
Conclusion
A simple linear mixed model was established to predict average IGF-1 for children with GHD receiving lonapegsomatropin based on a single sample at steady state in the scale of SDS or concentration units to assist the clinician in evaluating the safety and efficacy of the product in an individual patient. The average IGF-1 may be predicted from a sample collected at any time following the lonapegsomatropin injection as long as the interval between the injection and sample collection is known to the nearest half-day. The sampling time that is coincident with average IGF-1 following lonapegsomatropin is approximately 4.5 days after dosing. IGF-1 levels peak at Day 2.0 to 2.5 and are back to baseline at Day 7.0. The average IGF-1 predicted by this model may potentially be used by clinicians to guide dose adjustments of lonapegsomatropin. Average IGF-1 values predicted by this model are likely to be important real-world tools in monitoring safety and efficacy during long-term lonapegsomatropin therapy.
Acknowledgments
We thank the study participants and their families, the trial investigators, and study site staff. We thank Yuhua Su for statistical support. We also wish to acknowledge members of the trial’s independent safety committee and Andrew Occiano and J. Ludovic Croxford for writing assistance.
Financial Support: This study was sponsored by Ascendis Pharma Endocrinology Division A/S.
Glossary
Abbreviations
- GH
growth hormone
- GHD
growth hormone deficiency
- IGF-1
insulin-like growth factor 1
- LAGH
long-acting growth hormone
- PD
pharmacodynamic
- PI
prediction interval
- PK
pharmacokinetic
- rhGH
recombinant human growth hormone
- RSD
residual standard deviation
- SDS
standard deviation score
Additional Information
Disclosures: A.D.R. is a consultant for Antares Pharma, Ascendis Pharma A/S, Clarus Therapeutics, Lumos Pharma, United States Anti-doping Agency (USADA), and Ultragenyx Pharmaceutical. He has not received honoraria or reimbursement for this manuscript. B.S.M. is a consultant for Abbvie, Ascendis Pharma A/S, BioMarin, EMD Serono, Novo Nordisk, Orchard, Pfizer, Sandoz, Sanofi Genzyme, Tolmar, and Vertice and has received research support from Alexion, Abbvie, Amgen, Ascendis, Lumos, Novo Nordisk, Opko, Pfizer, Sandoz, and Tolmar. He has not received honoraria or reimbursement for this manuscript. Z.L., A.D.S., and M.B. are employees of Ascendis Pharma, Inc.
Data Availability
The datasets generated during and/or analyzed during the current trial are not publicly available but are available from the corresponding author on reasonable request.
Current Affiliation: Current affiliation for M.B. is ShouTi, Inc., South San Francisco, CA 94080, USA
Reprint Requests: Reprint requests should be addressed to the corresponding author.
References
- 1. Guyda HJ. Four decades of growth hormone therapy for short children: what have we achieved? J Clin Endocrinol Metab. 1999;84(12):4307-4316. [DOI] [PubMed] [Google Scholar]
- 2. Fisher BG, Acerini CL. Understanding the growth hormone therapy adherence paradigm: a systematic review. Horm Res Paediatr. 2013;79(4):189-196. [DOI] [PubMed] [Google Scholar]
- 3. Cutfield WS, Derraik JG, Gunn AJ, et al. Non-compliance with growth hormone treatment in children is common and impairs linear growth. Plos One. 2011;6(1):e16223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Christiansen JS, Backeljauw PF, Bidlingmaier M, et al. Growth Hormone Research Society perspective on the development of long-acting growth hormone preparations. Eur J Endocrinol. 2016;174(6):C1-C8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jørgensen JO, Møller N, Lauritzen T, Alberti KG, Orskov H, Christiansen JS. Evening versus morning injections of growth hormone (GH) in GH-deficient patients: effects on 24-hour patterns of circulating hormones and metabolites. J Clin Endocrinol Metab. 1990;70(1):207-214. [DOI] [PubMed] [Google Scholar]
- 6. Fisher DM, Rosenfeld RG, Jaron-Mendelson M, Amitzi L, Koren R, Hart G. Pharmacokinetic and Pharmacodynamic Modeling of MOD-4023, a Long-Acting Human Growth Hormone, in Growth Hormone Deficiency Children. Horm Res Paediatr. 2017;87(5):324-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Juul Kildemoes R, Højby Rasmussen M, Agersø H, Overgaard RV. Optimal Monitoring of Weekly IGF-I Levels During Growth Hormone Therapy With Once-Weekly Somapacitan. J Clin Endocrinol Metab. 2021;106(2):567-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chatelain P, Malievskiy O, Radziuk K, et al. ; TransCon GH Working Group. A Randomized Phase 2 Study of Long-Acting TransCon GH vs Daily GH in Childhood GH Deficiency. J Clin Endocrinol Metab. 2017;102(5):1673-1682. [DOI] [PubMed] [Google Scholar]
- 9. Sprogøe K, Mortensen E, Karpf DB, Leff JA. The rationale and design of TransCon Growth Hormone for the treatment of growth hormone deficiency. Endocr Connect. 2017;6(8):R171-R181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thornton PS, Maniatis AK, Aghajanova E, et al. Weekly lonapegsomatropin in treatment-naïve children with growth hormone deficiency: the phase 3 heiGHt trial. J Clin Endocrinol Metab. 2021;106(11):3184-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. SKYTROFA (lonapegsomatropin) [package insert]. Palo Alto, CA: Ascendis Pharma, Inc; Revised August 2021. [Google Scholar]
- 12. Lin Z, Su Y, Chessler S, Komirenko A, Christoffersen ED, Shu AD. Estimating the weekly average IGF-1 from a single IGF-1 sample in children with growth hormone deficiency (GHD) treated with lonapegsomatropin. In: Horm Res Paediatr. 2021;94(suppl 2):76. Accessed November 30, 2021. https://www.karger.com/Article/Pdf/519117 [Google Scholar]
- 13. Bidlingmaier M, Friedrich N, Emeny RT, et al. Reference intervals for insulin-like growth factor-1 (igf-i) from birth to senescence: results from a multicenter study using a new automated chemiluminescence IGF-I immunoassay conforming to recent international recommendations. J Clin Endocrinol Metab. 2014;99(5):1712-1721. [DOI] [PubMed] [Google Scholar]
- 14. Lin Z, Shu AD, Bach M, Miller BS, Rogol AD. Data from Average IGF-1 Prediction for Once-Weekly Lonapegsomatropin in Children with Growth Hormone Deficiency. figshare. 2021. Deposited June 24, 2021. 10.6084/m9.figshare.14838465 [DOI] [PMC free article] [PubMed]
- 15. Miller BS. rhGH safety and efficacy update. Adv Pediatr. 2011;58(1):207-241. [DOI] [PubMed] [Google Scholar]
- 16. Allen DB, Backeljauw P, Bidlingmaier M, et al. GH safety workshop position paper: a critical appraisal of recombinant human GH therapy in children and adults. Eur J Endocrinol. 2016;174(2):P1-P9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current trial are not publicly available but are available from the corresponding author on reasonable request.
Current Affiliation: Current affiliation for M.B. is ShouTi, Inc., South San Francisco, CA 94080, USA
Reprint Requests: Reprint requests should be addressed to the corresponding author.