Abstract
Recessive Dystrophic Epidermolysis Bullosa (RDEB), is a rare genodermatosis caused by mutations in the gene coding for type VII collagen (COL7A1) (Hovnanian et al., 1994). More than 800 different pathogenic mutations in COL7A1 have been described to date (Mittapalli et al., 2019), however, the ancestral origins of many of these mutations have not been precisely identified. In this study, thirty-two RDEB patient samples from the Southwestern United States, Mexico, Chile, and Colombia carrying common mutations in the COL7A1 gene were investigated to determine the origins of these mutations and the extent to which shared ancestry contributes to disease prevalence. The results demonstrate both shared European and American origins of RDEB mutations in distinct populations in the Americas and suggest the influence of Sephardic ancestry in at least some RDEB mutations of European origins. Knowledge of ancestry and relatedness amongst RDEB patient populations will be crucial for the development of future clinical trials and the advancement of novel therapeutics.
Introduction
Recessive Dystrophic Epidermolysis Bullosa (RDEB), is a rare genodermatosis characterized by severe skin fragility and blistering resulting in chronic wounds with progressive fibrosis (Mittapalli et al., 2019), caused by mutations in gene coding for type VII collagen (COL7A1) (Hovnanian et al., 1994). More than 800 different pathogenic mutations in COL7A1 have been described to date (Stenson et al., 2017), however, the ancestral origins of these mutations have not been precisely identified. Furthermore, little knowledge exists of the descendants of isolated Sephardic communities on the Iberian Peninsula, populations that characteristically propagate recessive diseases such as RDEB (I. Nogueiro, J. Teixeira, A. Amorim, L. Gusmao, & L. Alvarez, 2015a; I. Nogueiro, J. C. Teixeira, A. Amorim, L. Gusmao, & L. Alvarez, 2015b). The successful identification of common origins in distinct RDEB populations will provide an important resource as new treatments involving gene editing approaches for RDEB and other severe genetic skin diseases become viable for early clinical trials. In this study, thirty-two Hispanic RDEB patient samples carrying common COL7A1 gene mutations from the Southwestern United States, Mexico, Chile, and Colombia were genotyped to investigate the ancestral origins for COL7A1 mutations and determine the extent to which shared ancestry contributes to RDEB in these populations.
Results
The thirty-two Hispanic RDEB patients included in this study from the Southwestern United States, Mexico, Chile, and Colombia (see Table 1) carry pathogenic COL7A1 gene mutations that were genotyped with different sequencing techniques and confirmed by Sanger Sequencing.
Table 1.
COL7A1 mutations in Hispanic RDEB patients
| FTHNA Kit No. | Mutation 1 | Mutation 2 | Sephardic | Test Site |
|---|---|---|---|---|
| MK40784 | c.6527_6528insC | c.8329C>T | 21% | Chile |
| 730451 | c.2470insG | c.2470insG | 18% | Mexico |
| MK40790 | c.6527_6528insC | c.5856+1G>T | 15% | Chile |
| MK40779 | c.7485+1G>A | c.4635+5G>A | 10% | Colombia |
| MK42684 | c.2470insG | c.2470insG | 9% | Mexico |
| 730456 | c.2470insG | c.2470insG | 7% | Mexico |
| 662380 | c.7485+5G>A | c.7485+5G>A | 8% | Colorado |
| MK40771 | c.8200G>A | c.4965C>T | 7% | Colombia |
| MK40789 | c.7708delG | c.7876–1G>A | 5% | Chile |
| 662381 | c.7485+5G>A | c.7485+5G>A | 4% | Colorado |
| MK40776 | c.7651G>C | c.4012G>A | 4% | Colombia |
| MK40782 | c.6781C>T | c.2044C>T | <2% | Colombia |
| MK40777 | c.1584G>T | c.1584G>T | - | Colombia |
| MK40787 | c.2005C>T | c.4342–2A>G | - | Chile |
| MK42706 | c.5108G>A | c.IVS23–1G>A | - | Mexico |
| 730454 | c.2470insG | c.2470insG | - | Mexico |
| MK40794 | c.2992+2T>G | c.6527_6528insC | - | Chile |
| MK40795 | c.3264_5293del | c.5532+1G>T | - | Chile |
| MK40785 | c.3759+2T>G | c.3759+2T>G | - | Chile |
| MK40772 | c.425A>G | c.8833T>C | - | Colombia |
| MK40775 | c.4510G>T | c.4510G>T | - | Colombia |
| MK40778 | c.4678G>A | c.4678G>A | - | Colombia |
| MK40793 | c.5532+1G>T | c.8245G>A | - | Chile |
| MK40792 | c.5932C>T | c.5932C>T | - | Chile |
| MK40780 | c.6091G>A | c.6091G>A | - | Colombia |
| MK40783 | c.6527_6528insC | c.6527_6528insC | - | Chile |
| MK40788 | c.6527_6528insC | c.8528–1G>A | - | Chile |
| MK40773 | c.6527_6528insC | c.5604+1G>A | - | Colombia |
| MK40791 | c.7708delG | c.7708delG | - | Chile |
| MK40786 | c.7708delG | c.8393T>G | - | Chile |
| MK40774 | c.8046+6G>A | c.5047C>T | - | Colombia |
| MK42683 | c.8709del11 | c.G2899del11 | - | Mexico |
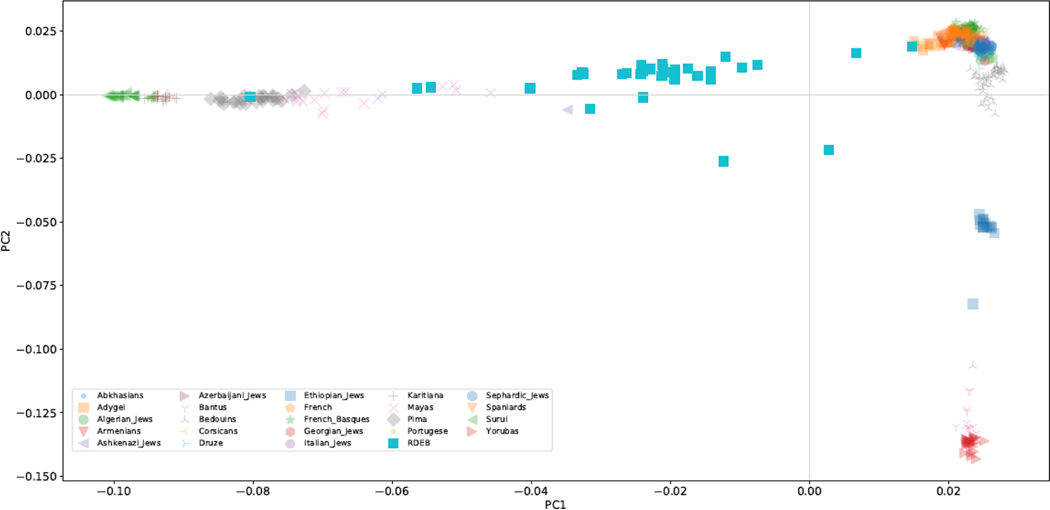
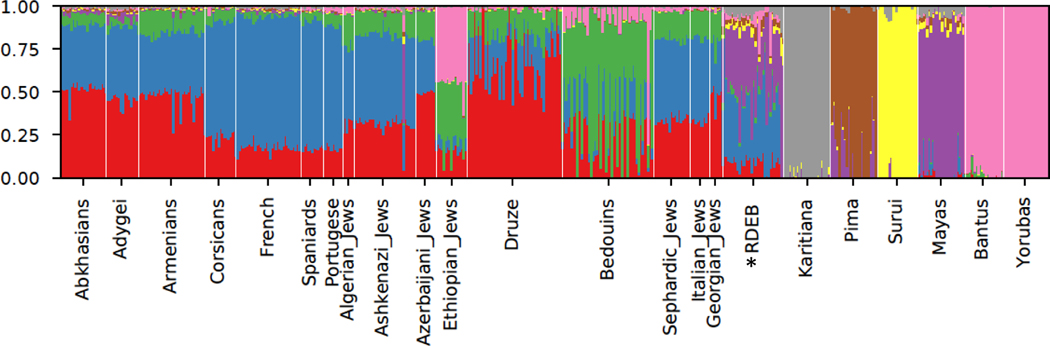
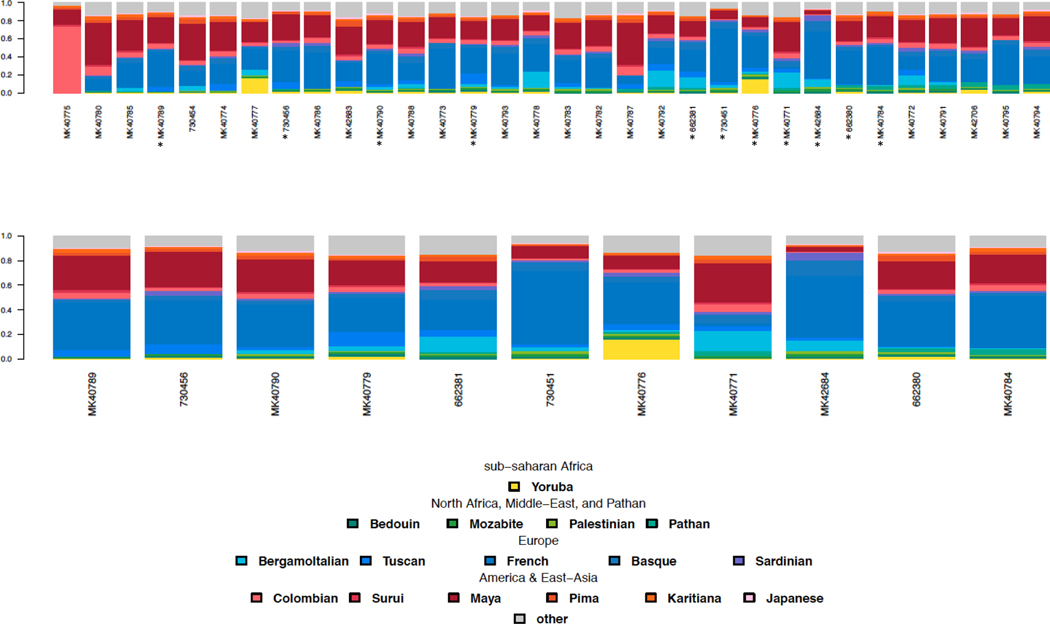
To further investigate ancestral patterns in the Hispanic RDEB population, a Principal Component Analysis (PCA) and Admixture Analysis were performed (Alexander, Novembre, & Lange, 2009) on genomewide genotyping data alongside a reference panel consisting of 500 individuals from African, Middle Eastern, and European ancestry (Chang et al., 2015). In the PCA, the majority of the individuals are scattered between European and Native American populations (Figure 1). For Admixture, we identified three different European components, evident in Figure 2 and Supplementary Figure 1, modal in Western Europe (blue), Middle East and the Caucasus (red) and the Arabian peninsula (green), supporting the presence of Sephardic ancestry. For the American component, we identified at K=8, a high variation in ancestral components, with an almost complete overlap between ancestry and population, possibly reflecting isolation and inbreeding dynamics, well-known in Jewish populations. Furthermore, Admixture analysis shows similar European and Native American ancestral components in the majority of the RDEB patients, with a minor, but consistent contribution from African groups (pink in Figure 2). A Non-Negative Least Squares (NNLS) haplotype-based method also supported overriding European and Native American origins, yet the majority (84%) of individuals harbor a small component (>2%) of North African and Middle-Eastern ancestry (Figure 3, shown in green), which may result from interactions between North Africa or Jewish populations and Iberian populations. The evidence of North African and Middle-Eastern lineage substantiates, at least in part, the influence of Sephardic Jews in the Hispanic RDEB population.
Figure 1.
Principal Component Analysis (PCA)
A Principal Component Analysis was carried out on a dataset composed of 516 individuals in total, including 32 RDEB samples. The RDEB samples are marked with a teal blue square ‘ ’ and represent substantial admixture with Native American and European individuals, including Sephardic Jews represented with light blue circles in the right upper corner ‘
’ and represent substantial admixture with Native American and European individuals, including Sephardic Jews represented with light blue circles in the right upper corner ‘ ’. The legend in the left lower corner denotes the color markers exhibited in the PCA analysis. The X and Y axis refer to Principal Component (PC) 1 and 2, respectively.
’. The legend in the left lower corner denotes the color markers exhibited in the PCA analysis. The X and Y axis refer to Principal Component (PC) 1 and 2, respectively.
Figure 2.
Admixture Analysis
Population structure of 32 RDEB samples inferred by Admixture analysis. Each individual is represented by a vertical (100%) stacked column of genetic components proportions shown in color for K8 (K=number of subpopulations). The RDEB population is marked with an asterisk.
Figure 3.
Non-Negative Least Squares (NNLS) Haplotype method
The NNLS Haplotype method exhibits a bar plot of the ancestral composition for a set of surrogate groups, as demonstrated in the legend. Implemented in ChromoPainter (CP) and merged with HGDP sequences, each individual (recipient) was painted as a combination of genomic fragments inherited by all the HGDP panel (donors). Thirty-two Hispanic RDEB patient samples carrying common mutations in the COL7A1 gene sorted according to Middle Eastern ancestry from the Southwestern United States, Mexico, Chile, and Colombia (Figure 3a). Asterisks in Figure 3a highlight the samples shown in Figure 3b. The analysis of RDEB samples with affinity to Sephardic origins sorted according to Middle Eastern ancestry demonstrate North African and Middle Eastern origins, highlighted in green, effectively supporting hidden genetic ancestry in heavily admixed individuals (Figure 3b).
To further explore the origin of these mutations, we estimated local ancestry across the COL7A1 locus in all 32 RDEB patients using a reference panel consisting of African, European, and Native American populations (see Methods and Materials). Pathogenic mutations at the locus were determined separately from the local ancestry and are thus not phased with the local ancestry estimates, however most patients (25/32) had just one ancestry at both copies of the locus, allowing us to gain insights into the ancestral source of the 20 mutations present on these haplotypes (Table 2). Seventeen of the mutations appear in patients with a single ancestry at both copies of COL7A1 (Table 2). Two additional mutations (c.7708delG and c.5532+1G>T) are present only in individuals with either a single ancestry at both copies of the locus or mixed ancestry at the locus, and may thus be attributed to a single ancestral population in our data. Notably, a single mutation, c.2470insG, appears to have originated independently in both ancestral European and Native American populations. In total, 10 of the mutations appear to have arisen on Native American haplotypes, 8 on European haplotypes, and 4 are observed only in individuals with mixed ancestry and cannot be determined.
Table 2.
Counts of local ancestry at COL7A in RDEB patients for each mutation
| Mutation | |||
|---|---|---|---|
| AMR | EUR | Mixed | |
| c.7485+5G>A | 4 | 0 | 0 |
| c.7708delG | 3 | 0 | 1 |
| c.2470insG | 2 | 6 | 0 |
| c.4510G>T | 2 | 0 | 0 |
| c.1584G>T | 2 | 0 | 0 |
| c.6091G>A | 2 | 0 | 0 |
| c.5108G>A | 1 | 0 | 0 |
| c.8200G>A | 1 | 0 | 0 |
| c.2005C>T | 1 | 0 | 0 |
| c.IVS23–1G>A | 1 | 0 | 0 |
| c.4965C>T | 1 | 0 | 0 |
| c.6527_6528insC | 0 | 6 | 1 |
| c.4678G>A | 0 | 2 | 0 |
| c.3759+2T>G | 0 | 2 | 0 |
| c.5932C>T | 0 | 2 | 0 |
| c.7651G>C | 0 | 1 | 0 |
| c.2992+2T>G | 0 | 1 | 0 |
| c.3264_5293del | 0 | 1 | 0 |
| c.5532+1G>T | 0 | 1 | 1 |
| c.8709del11 | 0 | 1 | 0 |
| c.425A>G | 0 | 0 | 1 |
| c.8046+6G>A | 0 | 0 | 1 |
| c.7485+1G>A | 0 | 0 | 1 |
| c.6781C>T | 0 | 0 | 1 |
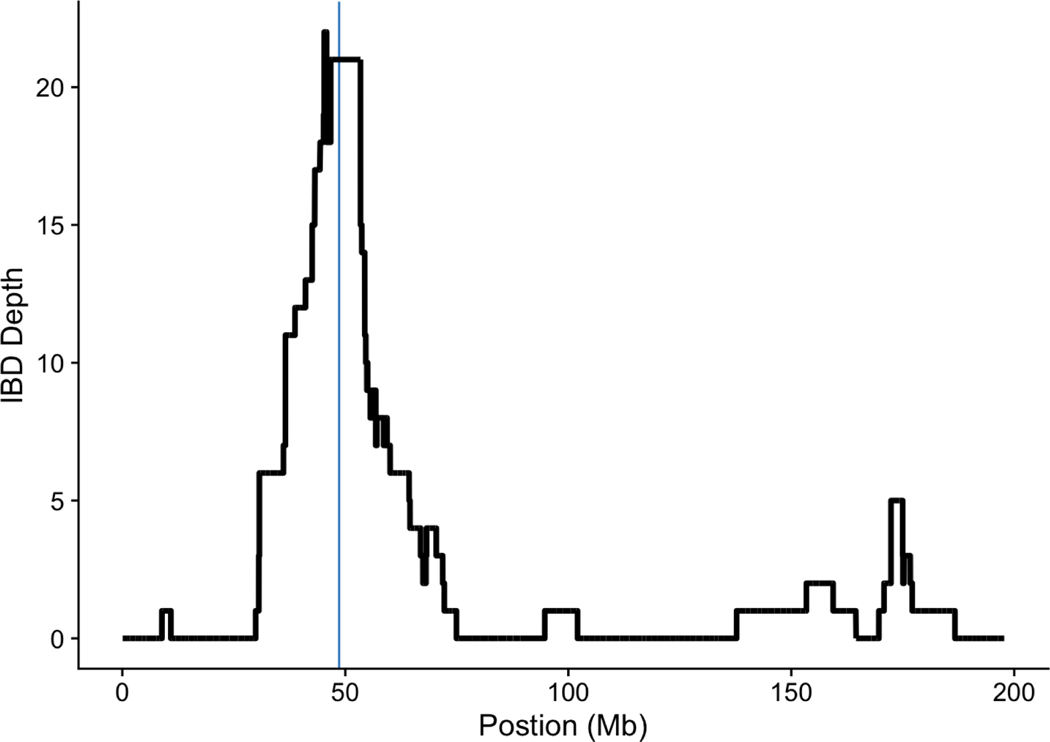
In order to evaluate the temporal origins of COL7A1 mutations in our dataset, we looked for shared haplotypes between patients and identified an enrichment of identity by descent (IBD) in the region surrounding the COL7A1 gene on chromosome 3 (Figure 4), supporting recent shared ancestry among at least some of the patients, even though none are known to be closely related. From among the 32 RDEB patients, we identified three different IBD clusters, each representing a single shared haplotype, indicating recent shared ancestry between these individuals (Table 3).
Figure 4.
Identity By Descent (IBD)
Cumulative depth of identical by descent (IBD) segments from all pairwise sample comparisons across chromosome 3 (blue vertical line indicates the location of COL7A1).
Table 3.
Counts of RDEB mutations and local ancestry populations within identity by descent COL7A1 haplotypes
| IBD Cluster ID | Count of Patients in Cluster | Count of Patient Haplotype Zygosity for Cluster (#Homozygous/#Heterozygous) | Count of Patient Mutation Zygosity (#Homozygous/#Heterozygous) | Local Ancestry Counts at COL7A1 in Cluster | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| c.6527_6528insC | c.5604+1G>A | c.8528–1G>A | c.8709del11 | c.G2899del11 | c.2470insG | c.7708delG | c.7876–1G>A | AMR | EUR | Mixed | |||
| A | 4 | 0/4 | 1/2 | 0/1 | 0/1 | 0/1 | 0/1 | 0/0 | 0/0 | 0/0 | 0 | 3 | 1 |
| B | 2 | 2/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 2/0 | 0/0 | 0/0 | 0 | 2 | 0 |
| C | 2 | 1/1 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 1/1 | 0/1 | 2 | 0 | 0 |
The largest cluster is comprised of 4 individuals (2 from Chile, 1 from Colombia, and 1 from Mexico) (Table 3, Cluster A). The observation that ¾ of the patients carry at least one copy of the pathogenic RDEB mutation c.6527_6528insC mutation, with no other COL7A1 mutation being shared by all individuals in the cluster, suggests that the haplotype shared by individuals in the cluster carries the c.6527_6528insC mutation. It is possible that the pathogenic mutation of the patient who does not carry the c.6527_6528insC mutation was incompletely characterized, or that the shared haplotype does not encompass the mutation. Local ancestry indicates that the shared haplotype is of European origin.
A second IBD haplotype is shared by two RDEB patients from Mexico (Table 3, Cluster B). Both patients are homozygous for the c.2470insG mutation, and both are homozygous for the shared haplotype. Local ancestry across the COL7A1 locus on the shared haplotype indicates that it is likely of European origin.
The third IBD haplotype is shared by two RDEB patients from Chile carrying the c.7708delG mutation (Table 3, Cluster C), one of whom is homozygous for the shared haplotype and the mutation. Native American local ancestry across the COL7A1 locus in all patients suggests that this mutation arose in the Americas.
Multiple other mutations are observed multiple times in the RDEB patients without being part of an IBD cluster. We do not rule out the possibility that at least some of these mutations may have been inherited from a common ancestor, but recombination in intervening generations has interrupted the inherited haplotype to a size below detection thresholds. On the other hand, it is also reasonable to consider that at least some of these mutations may have arisen independently.
Discussion
Admixed populations present challenges in understanding the history of disease, particularly in regions where each population can have a different admixture history. The high frequency of shared haplotypes harboring pathogenic RDEB mutations among unrelated patients in our data suggests that founder or other demographic events may have increased the prevalence of these pathogenic haplotypes in Hispanic populations in the Americas. We also present evidence that RDEB mutations in multiple Hispanic populations have ancestral origins from both European and Native American ancestral populations, with at least one mutation (c.2470insG) having origins from both ancestral populations. We note that because the number of observations of each mutation in our dataset is limited by our patient sampling, it is possible that other mutations presented here also have other origins that simply are not sampled in our patients. Thus, we cannot exclude the possibility that these other mutations have independent origins in other ancestral populations.
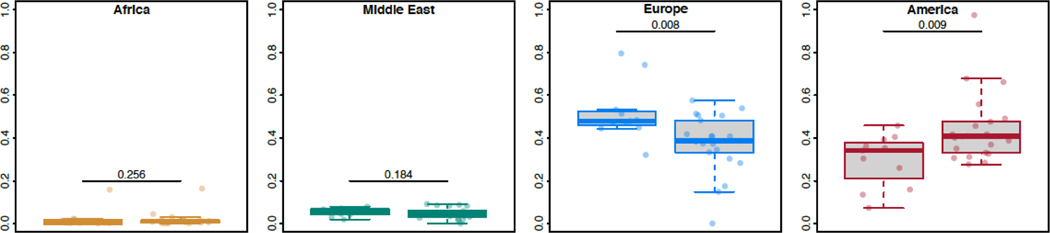
Eleven of the RDEB patients presented here had substantial amounts (>4%) of Sephardic ancestry using FamilyTreeDNA ancestry testing, a finding confirmed by a significantly higher proportion of European and American ancestry in these patients compared to the remainder of the RDEB patients (Figure 5). The finding is not entirely unexpected as it is known that Sephardic individuals arrived in the Americas in the late 15th century, and Sephardic ancestry has been previously detected in some Hispanic populations, with some Sephardic mutations being observed in former Spanish colonies such as the Southwestern US, Mexico and El Salvador (Ellis et al., 1998; Ostrer, 2016; Shahrabani-Gargir et al., 1998; Struewing et al., 1995; Velez et al., 2012). Notably, significant admixture also occurred between North African non-Jewish populations and Sephardic Jews following their expulsion from Spain during the Inquisition (Aksentijevich et al., 1993; Campbell et al., 2012), which may explain the ancestral components in the NNLS related to African and Middle Eastern populations (Figures 2 and 3).
Figure 5.
Ancestries Proportions
This figure represents the ancestry proportions for Africa, the Middle East, Europe and America. Within each panel, the left hand side bar graph represents RDEB samples with Sephardic ancestry and the right hand side bar graph represents RDEB samples without Sephardic ancestry. The 11 RDEB samples with Sephardic ancestry clearly demonstrate significantly greater European ancestry and less American ancestry than RDEB samples without Sephardic ancestry. There are no significant differences noted amongst the African or Middle Eastern ancestry.
The recognition that the European haplotype shared by individuals carrying the c.6527_6528insC mutation represents patients from three separate populations could suggest that the arrival of the mutation on the haplotype predates European settlement. This knowledge would be consistent with estimates of the first occurrence of the c.6527_6528insC mutation more than 3000 years ago when pre-Roman communities settled in the Iberian Peninsula (Sanchez-Jimeno et al., 2013). Interestingly, this region was also home to a closed endogamous community of Sephardic Jews during a time period more than a millennium ago, coinciding with the estimated origin of the c.6527_6528insC founder mutation (Adams et al., 2008). The c.6527_6528insC mutation remains frequent on the Iberian Peninsula of Spain and it is possible that at least some Hispanic populations inherited this mutation through Hispanic or Sephardic migration. Data from the original source of Sephardic Jews on the Iberian Peninsula is limited and the question of Sephardic origins is challenging as Sephardic and Spanish ancestry signals are likely to be largely overlapping due to centuries of cohabitation (Álvarez-Álvarez et al., 2018; Nogueiro et al., 2015b).
From the analyses presented here, the observation that at least some RDEB mutations appear to have European origins lends some support to the hypothesis that some of the pathogenic RDEB mutations presented here are of Sephardic origin. Population frequencies represented in gnomAD indicate slight enrichment with European populations in eight Hispanic RDEB mutations (Supplementary Table 1), providing rationale for further investigation of Sephardic ancestry in these variants (Collins et al., 2020; Karczewski et al., 2020; Landrum et al., 2020). Interestingly, there are a few rare variants demonstrating enrichment with African populations, which may represent the Sephardic populations of North Africa (Gonçalves et al., 2005). Establishing more confident and specific inferences of the ancestral origins of the mutations present in the Hispanic RDEB patients will benefit from additional sampling from ancient and modern-day Sephardic populations as well as from the populations from which the RDEB samples were collected.
Gene-editing treatments for RDEB patients using guide RNAs specific to pathogenic mutations are anticipated in the near future (Bonafont et al., 2019; Mencía et al., 2018). As regulatory agencies may consider each guide RNA as a separate drug, separate clinical trials for each RNA would be required. Patient populations who share the same founder mutations therefore represent an increasingly important resource to facilitate early clinical trials and advance novel treatments.
Materials and Methods
Thirty-two RDEB homozygous and compound heterozygous patient samples from the Southwestern United States, Mexico, Chile, and Colombia with common mutations in COL7A1 included in this study were previously identified using different sequencing technologies and subsequently confirmed by Sanger sequencing (Table 1). Informed written consent was obtained from all patients in concordance with Institutional Review Board approval from the USA: Colorado Multiple Institutional Review Board (COMIRB no: 09–0192), Mexico: Universidad de Monterrey (132012-CE), Chile: Comité Ético Científico, Facultad de Medicina, Clinica Alemana - Universidad del Desarrollo (Project number 2013–145), and Colombia: Universidad del Rosario (CIE-UR DVO005 1149-CV1192).
Illumina Bead Chip Array and FTDNA Family Finder Analysis
DNA samples were genotyped by GeneByGene, Inc. utilizing the Illumina Human OmniExpress BeadChip array and analyzed with the FTDNA Family Finder autosomal DNA test. The Family Finder test returns results for about 690,000 pairs of single nucleotide polymorphisms (SNPs) on the 22 pairs of autosomal chromosomes. Autosomal SNPs are clustered into sets about 50 to 100 SNPs long that are predefined based on the reliability, variability, average centiMorgans (cM) and density of the SNPs. The Family Finder software then evaluates SNP sets for matching as half identical or a non-match based on an autosomal DNA algorithm demonstrating shared DNA segments. Adjacent SNP sets are also analyzed to see if they qualify as identical by descent (IBD) segments. A segment is considered a candidate autosomal match if it contains at least 500 SNPs, and it is at least 1 cM long. Then, proprietary rules based on total shared cM and longest segment cM are applied to infer whether the match is valid.
The RDEB DNA samples were then subset to approximately 245,000 unlinked SNPs and run through ADMIXTURE using FamilyTreeDNA’s proprietary global reference panel called myOrigins2. The twenty-four global population clusters represent modern human genetic variation and results reflect admixture between historical gene pools. They include Sephardic, Ashkenazi, North and Central America, South America, British Isles, Scandinavia, Finland, West and Central Europe, Southeast Europe, East Europe, Iberia, West Middle East, East Middle East, Asia Minor, North Africa, East Central Africa, South Central Africa, West Africa, Central Asia, South Central Asia, Siberia, Northeast Asia, Southeast Asia, and Oceania. Affinity to Sephardic origins is determined significant by at least a 4% match to the Sephardic cluster (Chacon-Duque et al., 2018).
Identity by Descent Analysis
We merged the genotyping data from RDEB patients with genotypes from a combined reference dataset consisting individuals from the Human Genome Diversity Project (HGDP) (Cann et al., 2002) and 1000 Genomes (Genomes Project et al., 2010; Genomes Project et al., 2012; Genomes Project et al., 2015), and phased the entire merged dataset using EAGLE2 (Loh et al., 2016) with default parameters. Phased haplotypes were then fed through iLASH to detect identical by descent (IBD) regions greater than or equal to 3 cM between pairs of RDEB patients. Cumulative IBD depth at each marker was calculated using a custom python script that counts the total number of times each locus is part of an IBD segment shared between two individuals. We used DASH (Gusev et al., 2011) with a minimum cluster size of 3 and a minimum haplotype length of 3 cM to identify clusters of patients that share the same IBD haplotypes across the entirety of COL7A1.
Local Ancestry Analysis
We used phased haplotypes (see ‘Identity by Descent Analysis’) from RDEB patients in RFMix (Maples, Gravel, Kenny, & Bustamante, 2013) (version 1.5.4) with a reference panel consisting of European and non-admixed African populations (ESN, GWD, MSL, LWK, YRI) from 1000 Genomes and 107 indigenous American individuals from HGDP to train the random forests. The estimates for local ancestry presented in the manuscript are generated from the RFMix Viterbi output.
Principal Component Analysis (PCA) and Admixture Analysis
In order to explore the ancestry of the analyzed individuals, an explorative analysis was carried out using a Principal Component Analysis (PCA) and Admixture approach (Alexander et al., 2009). We merged the genotype data with publicly available genome-wide datasets (Behar et al., 2013; Behar et al., 2010; Kushniarevich et al., 2015; Ongaro et al., 2019; Tambets et al., 2018; Tamm et al., 2019; Yunusbayev et al., 2012; Yunusbayev et al., 2015) using PLINK 1.9, a widely used program for research in population genetics (Chang et al., 2015). After merging, SNPs and individuals characterized by less than 3% and 5% of missing data were retained for a total of 87,181 SNPs and 516 individuals. A PCA was carried out using the PCA flag in PLINK1.9 (Chang et al., 2015).
The Admixture analysis was carried out on the dataset with random seed and cross-validation based on data resampling. Although the most supported configuration is K=9, we are showing K=8, for which European and the Middle East ancestries show a reasonable degree of differentiation (Supplementary Figure 1).
ChromoPainter (CP) and Non-Negative Least Square-NNLS (NNLS)
The ancestry composition of the analyzed individuals was explored utilizing haplotype method implemented in ChromoPainter (Lawson, Hellenthal, Myers, & Falush, 2012). The analyzed samples were merged with the HGDP sequences in order to increase the density of the data (Bergström et al., 2019). First, the sequence data SNPs coordinates were lifted from Ghch38 to hg19 using Picard Tools (Picard, 2020). The merged dataset was subsequently phased using shapeit ver 2. using default settings (Delaneau, Marchini, & Zagury, 2011). Second, each individual (recipient) was painted as a combination of genomic fragments inherited by all the HGDP panel (donors), an effective way to unlock the hidden genetic ancestry in heavily admixed individuals (Montinaro et al., 2015; Ongaro et al., 2019). The nuisance parameters were set as M=0.0018 and n=409, estimated based on Expectation Maximization run in a subset of 5 individuals from each population.
The resulting copying vectors were subsequently normalized and each RDEB patient individual ancestry was reconstructed as a combination of copying vectors from all the populations in the HGDP dataset, using a modified version of the NNLS function of R software implemented in GlobeTrotter (Hellenthal et al., 2014; Leslie et al., 2015). The results for the RDEB patients are summarized in Figure 4. Populations contributing on average less than 1% were labelled as others.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgements
We thank all the patients and their families for helping us to carry out this study. This study was supported by the Avotaynu Foundation, Epidermolysis Bullosa Research Partnership, Epidermolysis Bullosa Medical Research Foundation, Cure EB, National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) of the National Institute of Health (NIH) (R01AR059947 and U01AR075932), the Department of Defense (DOD) (W81XWH-18-1-0706), Dystrophic Epidermolysis Bullosa Research Association (DEBRA) International, and the Gates Frontiers Fund. We also want to give special thanks to Stephen Berman, MD, the Founding Director of the Epidermolysis Bullosa Center of Excellence at Children’s Hospital Colorado, who has cared for Hispanic RDEB patients in Colorado for over 3 decades and originally suggested to us that these patients may be of Converso ancestry.
Footnotes
Conflict of Interest
The authors state no conflict of interest.
References
- Adams SM, Bosch E, Balaresque PL, Ballereau SJ, Lee AC, Arroyo E, . . . Jobling MA. (2008). The genetic legacy of religious diversity and intolerance: paternal lineages of Christians, Jews, and Muslims in the Iberian Peninsula. American Journal of Human Genetics, 83(6), 725–736. doi: 10.1016/j.ajhg.2008.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksentijevich I, Pras E, Gruberg L, Shen Y, Holman K, Helling S, . . . et al. (1993). Familial Mediterranean fever (FMF) in Moroccan Jews: demonstration of a founder effect by extended haplotype analysis. American Journal of Human Genetics, 53(3), 644–651. [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, & Lange K. (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Research, 19(9), 1655–1664. doi: 10.1101/gr.094052.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Álvarez MM, Risch N, Gignoux CR, Huntsman S, Ziv E, Fejerman L, . . . Athanasiadis G. (2018). Genetic analysis of Sephardic ancestry in the Iberian Peninsula. bioRxiv, 325779. doi: 10.1101/325779 [DOI] [Google Scholar]
- Behar DM, Metspalu M, Baran Y, Kopelman NM, Yunusbayev B, Gladstein A, . . . Rosenberg NA. (2013). No evidence from genome-wide data of a Khazar origin for the Ashkenazi Jews. Human Biology, 85(6), 859–900. doi: 10.3378/027.085.0604 [DOI] [PubMed] [Google Scholar]
- Behar DM, Yunusbayev B, Metspalu M, Metspalu E, Rosset S, Parik J, . . . Villems R. (2010). The genome-wide structure of the Jewish people. Nature, 466(7303), 238–242. doi: 10.1038/nature09103 [DOI] [PubMed] [Google Scholar]
- Bergström A, McCarthy SA, Hui R, Almarri MA, Ayub Q, Danecek P, . . . Tyler-Smith C. (2019). Insights into human genetic variation and population history from 929 diverse genomes. bioRxiv, 674986. doi: 10.1101/674986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonafont J, Mencia A, Garcia M, Torres R, Rodriguez S, Carretero M, . . . Larcher F. (2019). Clinically Relevant Correction of Recessive Dystrophic Epidermolysis Bullosa by Dual sgRNA CRISPR/Cas9-Mediated Gene Editing. Molecular Therapy, 27(5), 986–998. doi: 10.1016/j.ymthe.2019.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CL, Palamara PF, Dubrovsky M, Botigué LR, Fellous M, Atzmon G, . . . Ostrer H. (2012). North African Jewish and non-Jewish populations form distinctive, orthogonal clusters. Proceedings of the National Academy of Sciences of the United States of America, 109(34), 13865–13870. doi: 10.1073/pnas.1204840109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, . . . Cavalli-Sforza LL. (2002). A human genome diversity cell line panel. Science, 296(5566), 261–262. doi: 10.1126/science.296.5566.261b [DOI] [PubMed] [Google Scholar]
- Chacon-Duque JC, Adhikari K, Fuentes-Guajardo M, Mendoza-Revilla J, Acuna-Alonzo V, Barquera R, . . . Ruiz-Linares A. (2018). Latin Americans show wide-spread Converso ancestry and imprint of local Native ancestry on physical appearance. Nat Commun, 9(1), 5388. doi: 10.1038/s41467-018-07748-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, & Lee JJ (2015). Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience, 4, 7. doi: 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RL, Brand H, Karczewski KJ, Zhao X, Alföldi J, Francioli LC, . . . Talkowski ME. (2020). A structural variation reference for medical and population genetics. Nature, 581(7809), 444–451. doi: 10.1038/s41586-020-2287-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Marchini J, & Zagury JF (2011). A linear complexity phasing method for thousands of genomes. Nature Methods, 9(2), 179–181. doi: 10.1038/nmeth.1785 [DOI] [PubMed] [Google Scholar]
- Ellis NA, Ciocci S, Proytcheva M, Lennon D, Groden J, & German J. (1998). The Ashkenazic Jewish Bloom syndrome mutation blmAsh is present in non-Jewish Americans of Spanish ancestry. American Journal of Human Genetics, 63(6), 1685–1693. doi: 10.1086/302167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project C, Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, . . . McVean GA. (2010). A map of human genome variation from population-scale sequencing. Nature, 467(7319), 1061–1073. doi: 10.1038/nature09534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, . . . McVean GA. (2012). An integrated map of genetic variation from 1,092 human genomes. Nature, 491(7422), 56–65. doi: 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, . . . Abecasis GR. (2015). A global reference for human genetic variation. Nature, 526(7571), 68–74. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves R, Freitas A, Branco M, Rosa A, Fernandes AT, Zhivotovsky LA, . . . Brehm A. (2005). Y-chromosome lineages from Portugal, Madeira and Açores record elements of Sephardim and Berber ancestry. Annals of Human Genetics, 69(Pt 4), 443–454. doi: 10.1111/j.1529-8817.2005.00161.x [DOI] [PubMed] [Google Scholar]
- Gusev A, Kenny EE, Lowe JK, Salit J, Saxena R, Kathiresan S, . . . Pe’er I. (2011). DASH: a method for identical-by-descent haplotype mapping uncovers association with recent variation. American Journal of Human Genetics, 88(6), 706–717. doi: 10.1016/j.ajhg.2011.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellenthal G, Busby GBJ, Band G, Wilson JF, Capelli C, Falush D, & Myers S. (2014). A genetic atlas of human admixture history. Science, 343(6172), 747–751. doi: 10.1126/science.1243518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovnanian A, Hilal L, Blanchet-Bardon C, de Prost Y, Christiano AM, Uitto J, & Goossens M. (1994). Recurrent nonsense mutations within the type VII collagen gene in patients with severe recessive dystrophic epidermolysis bullosa. American Journal of Human Genetics, 55(2), 289–296. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/8037207 [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, . . . Genome Aggregation Database, C. (2020). The mutational constraint spectrum quantified from variation in 141,456 humans. Nature, 581(7809), 434–443. doi: 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushniarevich A, Utevska O, Chuhryaeva M, Agdzhoyan A, Dibirova K, Uktveryte I, . . . Balanovsky O. (2015). Genetic Heritage of the Balto-Slavic Speaking Populations: A Synthesis of Autosomal, Mitochondrial and Y-Chromosomal Data. PloS One, 10(9), e0135820. doi: 10.1371/journal.pone.0135820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Chitipiralla S, Brown GR, Chen C, Gu B, Hart J, . . . Kattman BL. (2020). ClinVar: improvements to accessing data. Nucleic Acids Research, 48(D1), D835–d844. doi: 10.1093/nar/gkz972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson DJ, Hellenthal G, Myers S, & Falush D. (2012). Inference of population structure using dense haplotype data. Plos Genetics, 8(1), e1002453. doi: 10.1371/journal.pgen.1002453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie S, Winney B, Hellenthal G, Davison D, Boumertit A, Day T, . . . Bodmer W. (2015). The fine-scale genetic structure of the British population. Nature, 519(7543), 309–314. doi: 10.1038/nature14230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh PR, Danecek P, Palamara PF, Fuchsberger C, Y AR, H KF, . . . A LP. (2016). Reference-based phasing using the Haplotype Reference Consortium panel. Nature Genetics, 48(11), 1443–1448. doi: 10.1038/ng.3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maples BK, Gravel S, Kenny EE, & Bustamante CD (2013). RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. American Journal of Human Genetics, 93(2), 278–288. doi: 10.1016/j.ajhg.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencía Á, Chamorro C, Bonafont J, Duarte B, Holguin A, Illera N, . . . Murillas R. (2018). Deletion of a Pathogenic Mutation-Containing Exon of COL7A1 Allows Clonal Gene Editing Correction of RDEB Patient Epidermal Stem Cells. Mol Ther Nucleic Acids, 11, 68–78. doi: 10.1016/j.omtn.2018.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittapalli VR, Kuhl T, Kuzet SE, Gretzmeier C, Kiritsi D, Gaggioli C, . . . Nystrom A. (2019). STAT3 targeting in dystrophic epidermolysis bullosa. British Journal of Dermatology. doi: 10.1111/bjd.18639 [DOI] [PubMed] [Google Scholar]
- Montinaro F, Busby GB, Pascali VL, Myers S, Hellenthal G, & Capelli C. (2015). Unravelling the hidden ancestry of American admixed populations. Nat Commun, 6, 6596. doi: 10.1038/ncomms7596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueiro I, Teixeira J, Amorim A, Gusmao L, & Alvarez L. (2015a). Echoes from Sepharad: signatures on the maternal gene pool of crypto-Jewish descendants. European Journal of Human Genetics, 23(5), 693–699. doi: 10.1038/ejhg.2014.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueiro I, Teixeira JC, Amorim A, Gusmao L, & Alvarez L. (2015b). Portuguese crypto-Jews: the genetic heritage of a complex history. Front Genet, 6, 12. doi: 10.3389/fgene.2015.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongaro L, Scliar MO, Flores R, Raveane A, Marnetto D, Sarno S, . . . Montinaro F. (2019). The Genomic Impact of European Colonization of the Americas. Current Biology, 29(23), 3974–3986 e3974. doi: 10.1016/j.cub.2019.09.076 [DOI] [PubMed] [Google Scholar]
- Ostrer H. (2016). The origin of the p.E180 growth hormone receptor gene mutation. Growth Hormone and IGF Research, 28, 51–52. doi: 10.1016/j.ghir.2015.08.003 [DOI] [PubMed] [Google Scholar]
- Picard. (2020). Retrieved from http://broadinstitute.github.io/picard/ [Google Scholar]
- Sanchez-Jimeno C, Cuadrado-Corrales N, Aller E, Garcia M, Escamez MJ, Illera N, . . . Del Rio M. (2013). Recessive dystrophic epidermolysis bullosa: the origin of the c.6527insC mutation in the Spanish population. British Journal of Dermatology, 168(1), 226–229. doi: 10.1111/j.1365-2133.2012.11128.x [DOI] [PubMed] [Google Scholar]
- Shahrabani-Gargir L, Shomrat R, Yaron Y, Orr-Urtreger A, Groden J, & Legum C. (1998). High frequency of a common Bloom syndrome Ashkenazi mutation among Jews of Polish origin. Genet Test, 2(4), 293–296. doi: 10.1089/gte.1998.2.293 [DOI] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Evans K, Hayden M, Heywood S, . . . Cooper DN. (2017). The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Human Genetics, 136(6), 665–677. doi: 10.1007/s00439-017-1779-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struewing JP, Abeliovich D, Peretz T, Avishai N, Kaback MM, Collins FS, & Brody LC (1995). The carrier frequency of the BRCA1 185delAG mutation is approximately 1 percent in Ashkenazi Jewish individuals. Nature Genetics, 11(2), 198–200. doi: 10.1038/ng1095-198 [DOI] [PubMed] [Google Scholar]
- Tambets K, Yunusbayev B, Hudjashov G, Ilumae AM, Rootsi S, Honkola T, . . . Metspalu M. (2018). Genes reveal traces of common recent demographic history for most of the Uralic-speaking populations. Genome Biology, 19(1), 139. doi: 10.1186/s13059-018-1522-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm E, Di Cristofaro J, Mazieres S, Pennarun E, Kushniarevich A, Raveane A, . . . Montinaro F. (2019). Publisher Correction: Genome-wide analysis of Corsican population reveals a close affinity with Northern and Central Italy. Scientific Reports, 9(1), 18827. doi: 10.1038/s41598-019-55185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velez C, Palamara PF, Guevara-Aguirre J, Hao L, Karafet T, Guevara-Aguirre M, . . . Ostrer H. (2012). The impact of Converso Jews on the genomes of modern Latin Americans. Human Genetics, 131(2), 251–263. doi: 10.1007/s00439-011-1072-z [DOI] [PubMed] [Google Scholar]
- Yunusbayev B, Metspalu M, Jarve M, Kutuev I, Rootsi S, Metspalu E, . . . Villems R. (2012). The Caucasus as an asymmetric semipermeable barrier to ancient human migrations. Mol Biol Evol, 29(1), 359–365. doi: 10.1093/molbev/msr221 [DOI] [PubMed] [Google Scholar]
- Yunusbayev B, Metspalu M, Metspalu E, Valeev A, Litvinov S, Valiev R, . . . Villems R. (2015). The genetic legacy of the expansion of Turkic-speaking nomads across Eurasia. Plos Genetics, 11(4), e1005068. doi: 10.1371/journal.pgen.1005068 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.