Abstract
Polycystic ovary syndrome (PCOS) is a common endocrinopathy in women. PCOS is characterized by anovulation, hyperandrogenism, polycystic ovaries, insulin resistance, and obesity. Despite the finding that the genetic origin of PCOS is well demonstrated in previous twin and familial clustering studies, genes and factors that can exactly explain the PCOS pathophysiology are not known. Objective(s). In this review, we attempted to identify genes related to secretion and signaling of insulin aspects of PCOS and their physiological functions in order to explain the pathways that are regulated by these genes which can be a prominent function in PCOS predisposition. Materials and Methods. For this purpose, published articles and reviews dealing with genetic evaluation of PCOS in women from peer-reviewed journals in PubMed and Google Scholar databases were included in this review. Results. The genomic investigations in women of different populations identified many candidate genes and loci that are associated with PCOS. The most important of them are INSR, IRS1-2, MTNR1A, MTNR1B, THADA, PPAR-γ2, ADIPOQ, and CAPN10. These are mainly associated with metabolic aspects of PCOS. Conclusions. In this review, we proposed that each of these genes may interrupt specific physiological pathways by affecting them and contribute to PCOS initiation. It is clear that the role of genes involved in insulin secretion and signaling is more critical than other pathways.
1. Introduction
The multigenic multifactorial endocrinopathy that affects about 5–10% of women of the world in their reproductive age is polycystic ovary syndrome (PCOS) [1]. The most important heterogeneous features of PCOS is infertility that originates from anovulation, hyperandrogenism, polycystic ovaries, insulin resistance, obesity, and cardiovascular diseases [2]. But, due to the heterogeneity of PCOS, the exact pathophysiological pathway that initiates the syndrome has not been known yet. Metabolic disorders such as insulin resistance, glucose intolerance, and type 2 diabetes are also observed in PCOS patients [3]. The role of different factors such as genetic, environmental, and developmental origin was explained in PCOS etiology [4]. In many cases, the genes involved in the pathology of these metabolic abnormalities were associated with PCOS and likely with one or more physiological routes interrupted by alteration of these genes. Therefore, the evaluation of these physiological pathways is valuable to clear the etiology of PCOS.
In retrospect to the fact that the etiology of PCOS is not known yet, we should consider two hypotheses hyperinsulinemia and intrauterine environment changes that have been well documented in PCOS formation by animal model studies. They can also be mediated by the genetic background of the individual [5]. Because the reproductive and metabolic implications are mainly observed in the first-class relatives, PCOS is considered as a genetic disorder [6]. Furthermore, familial aggregation studies have confirmed the genetic basis of PCOS hyperandrogenemia (hypersecretion of androgens in PCOS condition) and identified their related susceptible genetic variant in PCOS [4]. Based on twin studies, the heritability of PCOS is approximately 70% [7]. In spite of the effects of the susceptible genetic variants on PCOS that may be influenced by environmental factors, it seems that PCOS develops as a result of a combination of both genetic and environmental agents [4]. As noted, PCOS has heterogeneous characteristics due to involvement of either genetic or environmental factors, although the role of genetic factors is more severe [8]. The strong roles of inheritance and genetic background in PCOS development were confirmed based on twin and familial clustering studies [9, 10]. Unlike the increasing documents proving the heritability of PCOS and the effects of developmental origins of insulin resistance on PCOS development, the exact pathophysiological pathways in etiology of PCOS are not clear [11]. According to genome-wide association studies (GWAS) that identified risk loci for PCOS predisposition, researchers believed that the PCOS inheritance model is more likely to be oligogenic/polygenic than the autosomal dominant [10]. GWAS as a new approach presented a way for unbiased identification of genes without considering the probable role of causative variants [11]. The aim of the present review is brief description of susceptible genes contributed to PCOS development that are related to metabolic pathways such as insulin secretion and signaling. Therefore, according to the gene functions, the involved physiological routes affecting PCOS etiology are explained and novel hypotheses are categorized.
2. Materials and Methods
2.1. Focused Question
This review was done to answer this question: “What are the roles of insulin secretion and signaling-related genes in pathophysiologic mechanisms of polycystic ovary syndrome?”
2.2. Search and Study Selection
Key words and subject terms included (“PCOS” AND “insulin”) OR (“PCOS” AND “insulin” AND “gene”) OR (“PCOS” AND “insulin” AND “signaling”) OR (“PCOS” AND “diabetes”) OR (“PCOS” AND “diabetes” AND “gene”) OR (“PCOS” AND “diabetes” AND “signaling”). The search strategy was applied to PubMed, Elsevier, and Google Scholar databases, focused on the patient-related studies. English language research papers were considered. The review, abstracts without full manuscripts, the manuscripts related to the animal models, or in vitro studies were excluded. Data were collected from the full text of the articles as follows: (i) insulin resistance and diabetes mellitus type 2 or (ii) insulin secretion and signaling and (iii) the obtained results.
On the basis of physiological roles, identified genes related to PCOS can be classified into six groups including the following: a, gonadotropin secretion and actions; b, steroid hormones biosynthesis and functions; c, insulin resistance and type 2 diabetes mellitus; d, insulin secretion and signaling; e, obesity and dyslipidemia; f, chronic inflammatory reactions. In our previous review paper, the roles of genes involved in a, b, d, and f categories were explained in detail [2, 12]. In this review paper, we aimed to clarify the role of effective genes in the insulin secretion and signaling pathway.
3. Results and Discussion
3.1. Insulin Resistance and Diabetes Mellitus Type 2
Since insulin resistance is one of the underscored phenotypic features of PCOS that can have a genetic source, genetic variants of insulin resistance are also associated with PCOS [13]. Insulin resistance and hyperinsulinemia in adolescents are seen in the early stages of PCOS [14], and adolescents girls with PCOS are exposed to the increased risk of impaired glucose tolerance and diabetes mellitus type 2 [15]. Intrauterine growth retardation (IUGR) leads to alteration in the development of adipose tissue during fetal life [16], while adipose tissue has an effective role in the expansion of insulin resistance in adulthood [17]. Thus, insulin resistance resulting from IUGR can be a source of developmental and preprogramming changes that lead to some abnormalities in adulthood when the growth environment of the fetus is impaired [4]. The point is that PCOS and metabolic syndrome contain some common features such as having an intrinsic origin or due to being out of chronic adult illnesses, they are still originated from developmental age [18].
Obesity is another effective main factor of insulin resistance in PCOS. It is well known that the abdominal phenotype of obesity affects insulin resistance and subsequent compensatory hyperinsulinemia [19]. Obese and nonobese PCOS women have had insulin resistance and pancreatic beta-cell dysfunction, but this situation was not related to glucose intolerance in all PCOS participants [20].
The internal alterations of insulin function and hormonal environment may contribute to the development of PCOS insulin resistance. For instance, the defect of receptor auto-phosphorylation, stimulated by insulin, was exclusively observed in PCOS women and not in other metabolic abnormalities such as obesity, insulin resistance and non-insulin-dependent diabetes mellitus [21]. In some PCOS cases, the auto-phosphorylation of receptors is normal and there is a defect in postbinding receptor events of insulin signaling pathway which lead to insulin resistance [21].
Hyperandrogenism can alter the sensitivity of peripheral tissues to insulin, directly or indirectly, by increasing visceral fat and reducing the secretion of adiponectin. Adiponectin is a main insulin-sensitizing adipokine which contributes to PCOS insulin resistance [6]. Furthermore, there is evidence about heritability of hyperandrogenism and hyperinsulinemia among sisters of PCOS women [22].
Insulin sensitivity is affected by three factors: insulin receptor, peroxisome proliferator-activated receptor-gamma (PPAR-γ), and vitamin D [23]. Apa1 polymorphism of vitamin D receptor in Iranian PCOS women is highly associated with this syndrome [24]. A set of pathways leading to insulin resistance are described in Figure 1.
Figure 1.

Insulin resistance, producing factors and effective gene in each pathway, and ultimately insulin resistance by the formation of hyperinsulinemia and hyperandrogenism lead to PCOS. Insulin resistance can have different genetic, epigenetic (alteration during intrauterine development), and environmental origins or products derived from the adipose tissue. But, in PCOS condition, insulin resistance is mainly derived from postbinding receptor defects.
Insulin resistance leads to compensatory increment of insulin secretion from beta cells resulting in hyperinsulinemia which in turn causes hyperandrogenism. However, when the beta cell is unable to compensate for insulin resistance, hyperglycemia occurs and is followed by glucose intolerance and type 2 diabetes [8]. The reasons of insulin resistance in PCOS are unknown, and it may be the result of postreceptor insulin signaling defects [8]. In addition, several factors, secreted by adipose tissue such as leptin, free fatty acids, interleukin-6, and tumor necrosis factor α (TNFα), promote insulin resistance and are considered as candidate PCOS genes [25]. The mistake of binding insulin to the receptor or alteration of insulin signal transduction is a forcible mechanism of insulin resistance [26]. The pathophysiological pathway through which hyperinsulinemia leads to hyperandrogenism is explained in Figure 2. Generally, abnormalities of insulin secretion and sensitivity are related to genes involved in insulin signaling and metabolism regulators which are explained in the following.
Figure 2.
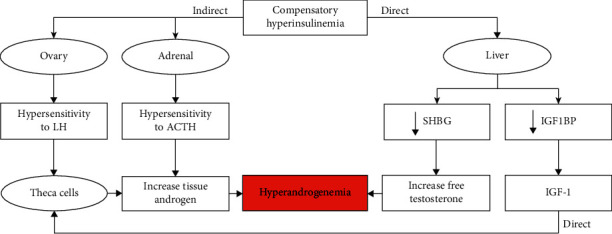
Mechanisms of direct and indirect effects of hyperinsulinemia on hyperandrogenemia. Insulin via increasing the sensitivity of theca cells to LH and adrenal cortex to ACTH elevated the synthesis of androgens in these tissues. Also, insulin via direct effect on the liver and by suppression of production of SHBG and IGF1BP leads to the increased serum level of androgens and eventually hyperandrogenemia.
3.2. ADIPOQ
Adiponectin is a protein that is specifically and abundantly expressed in adipocytes. Adiponectin gene polymorphisms affect the levels of this protein, obesity, insulin resistance, and type 2 diabetes [27]. Adiponectin has a special role in modulating insulin sensitivity [28]. Insulin sensitivity is controlled by several genes and interaction of gene products such as adiponectin and resistin (RETN). In a study on PCOS Japanese women, this syndrome was associated with RETN polymorphisms but did not show any association with ADIPOQ gene polymorphisms [29]. Adiponectin gene polymorphisms are more common in PCOS and had a significant correlation with glucose/insulin ratio [27]. In addition, the SNP rs1501299 polymorphism in adiponectin gene, specially based on its role in development of obesity caused by PCOS, was associated with PCOS risk in Chinese Han population [30]. Furthermore, G allele of rs1501299 increased the risk of PCOS in Jordanian population [31]. On the other hand, in a study of Polish women with PCOS, the SNP rs1501299 in the gene ADIPOQ was associated with a reduced risk of disease [32]. A meta-analysis by Liu et al. [33] demonstrated that rs1501299 polymorphisms are significantly associated with PCOS risk in East Asians. But, a meta-analysis of Asian population showed that women with the G276T polymorphism have decreased susceptibility to PCOS [34]. The strong association between 45T/G, +456G15G (T/G), +276 (G/T), 11391G > A, and G276T variants of ADIPOQ and the metabolic features of PCOS, such as insulin resistance, central obesity, dyslipidemia, hypertension, and hyperglycemia, was reported in different populations suggesting that ADIPOQ variants can be considered as the risk factors for PCOS development (Table 1).
Table 1.
Candidate genes involved in etiology of polycystic ovary syndrome related to insulin resistance and type 2 diabetes mellitus.
| Gene | Genetic marker(s) | Type of study | Physiologic function | Studied population | Type of polymorphism | References |
|---|---|---|---|---|---|---|
| ADIPOQ, RETN | SNPs at position −420 of the RETN and/or −11377 of the ADIPOQ | Case-control | Insulin resistance and obesity | Japanese | RETN−420G/G | [29] |
| ADIPOQ | rs1501299, rs2241766, and rs266729 | Case-control | Insulin resistance | Jordanian | G allele | [31] |
| ADIPOQ | rs17300539, rs266729, rs822395, rs822396, rs2241766, rs1501299, rs2241767, rs3774261, and rs17366743 | Case-control | Insulin resistance | Saudi Arabian | rs2241766, rs1501299, rs2241767, rs3774261, and rs17366743 | [35] |
| ADIPOQ | SNPs | Case-control | Modulating insulin sensitivity | Minia | Higher genotyping distributions of TG, GG, and TT | [27] |
| ADIPOQ | T45G and G276T | Meta-analysis | Insulin resistance, obesity, and T2DM | Asian | G276T | [34] |
| ADIPOQ | rs1501299 | Case-control | Lipid profile | Polish | GG | [32] |
| CAPN10 | UCSNP-44, UCSNP-43, UCSNP-19, and UCSNP-63 | Case-control | Ca-mediated intracellular signaling, and insulin secretion | Spanish | UCSNP-44 | [36] |
| CAPN10 | CAPN10 haplotypes | Haplotype-phenotype correlation | Ca-mediated intracellular signaling, and insulin secretion | Spanish | UCSNP-44 | [37] |
| CAPN10 | SNPs | Meta-analysis, meta-regression | Ca-mediated intracellular signaling, and insulin secretion | Asian | UCSNP-19, UCSNP-63, and UCSNP-45 | [38] |
| CAPN10 | UCSNP-43, UCSNP-44, UCSNP-19, and UCSNP-63 | Cross-sectional population-based | Ca-mediated intracellular signaling, and insulin secretion | Spanish | UCSNP-44, UCSNP-43, and UCSNP-19 | [39] |
| CAPN10 | UCSNP-43, UCSNP-19, and UCSNP-63 | Case-control | Ca-mediated intracellular signaling, and insulin secretion | Chilean | UCSNP-43 | [40] |
| CAPN10 | UCSNP-43, UCSNP-19, and UCSNP-63 | Cross-sectional | Ca-mediated intracellular signaling, and insulin secretion | Brazilian | UCSNP-43 | [41] |
| CAPN10 | UCSNP-44, UCSNP-43, UCSNP-56, UCSNP-19, and UCSNP-63 | Case-control | Ca-mediated intracellular signaling, and insulin secretion | Indian | UCSNP-44 | [42] |
| CAPN10 | UCSNP-43 and rs3792267 | Case-control | Ca-mediated intracellular signaling, and insulin secretion | Greek | UCSNP-43 | [43] |
| CAPN10 | UCSNP-43 and rs3792267 | Case-control | Ca-mediated intracellular signaling, and insulin secretion | Indian | ND | [44] |
| CAPN10 | UCSNP-19, UCSNP-63, UCSNP-44, and UCSNP-43 | Meta-analysis | Ca-mediated intracellular signaling, and insulin secretion | Different populations | UCSNP-19 and UCSNP-63 | [45] |
| PPAR-γ | Gly482Ser, PPAR-α Leu162Val, PPAR-δ rs2267668A/G, PPAR-δ−87T/C, PPAR-γ2 Pro12Ala, and PPAR-γ2-−681C/G | Case-control, meta-analysis | Glucose homeostasis, lipid metabolism, transport, and storage | Caucasian | Gly482Ser and Pro12Ala | [46] |
| PPAR-γ | Pro12Ala | Meta-analyses | Glucose homeostasis, lipid metabolism, transport, and storage | Different population | Pro12Ala | [47] |
| PPAR-γ2 | Pro12Ala | Case-control | Glucose homeostasis, lipid metabolism, transport, and storage | Chinese | ND | [48] |
| PPAR-γ2 | Pro12Ala | Case-control | Glucose homoeostasis, lipid metabolism, and adipocyte differentiation | South Indian | Pro12Ala | [49] |
| PPAR-γ | Pro12Ala (exon 2) and His447His (exon 6) | Case-control | Insulin resistance and adiposity | Southern Mediterranean | Pro12Ala (exon 2) | [50] |
Abbreviations: ADIPOQ, adiponectin; CAPN10, calpain 10; PPAR-γ, peroxisome proliferator-activated receptor-gamma; ND, no data.
3.3. CAPN10
Calpain protein is a cysteine protease that plays a role in pro-insulin processing and insulin secretion and action [51]. Women with PCOS are at the risk of impaired glucose tolerance (IGT) or a 2–7-fold increase in type 2 diabetes incidence. Therefore, all genes associated with type 2 diabetes mellitus can play an important role in the pathogenesis of PCOS [8]. CAPN10 gene was the first gene that was identified as type 2 diabetes risk gene [52]. The CAPN10 gene has multiple SNPs. In a meta-analysis study, the association of UCSNP-63 and UCSNP-19 polymorphisms with PCOS was proved [45]. In another meta-analysis study, the role of UCSNP-45 as well as UCSNP-63 and UCSNP-19 polymorphisms was confirmed as the risk factors for PCOS, especially in Asian women [38]. In many of the case-control studies, the association between various polymorphisms of CAPN10 and metabolic traits of PCOS was demonstrated and the diversity of populations was the only difference between studies (Table 1). Accordingly, CAPN10 which plays a role in insulin secretion and pathology of type 2 diabetes can also be an important susceptibility gene for PCOS.
3.4. PPAR-γ2
PPAR-γ is a very important transcription factor which plays a role in regulating glucose homeostasis, lipid metabolism, and ovarian steroidogenesis [53]. The activation of the PPAR-γ by the thiazolidinedione drug, used to treat type 2 diabetes, induces differentiation of adipocytes and also increases insulin sensitivity [8]. The PPAR-γ gene contains a common SNP Pro12Ala [54]. This gene is expressed primarily in adipose tissue and stimulates the differentiation of preadipocytes into adipocytes, and also, it belongs to the family of nuclear hormone receptors [55]. Pro12Ala polymorphism of PPAR-γ gene was proposed in the women of South India as a PCOS susceptibility gene [49]. The meta-analysis has showed that Pro12Ala polymorphism in the PPAR-γ has the potential to reduce the risk of polycyclic ovarian syndrome in European patients, which was not observed in the Asians [47]. The Pro12Ala (exon 2) polymorphism of PPAR-γ had a protective effect on insulin resistance and beta-cell function in a population of Southern Mediterranean women with PCOS [50]. Therefore, the association between PCOS and Pro12Ala variant of PPAR-γ has a racial difference and is mainly related to metabolic abnormalities of PCOS.
3.5. Insulin Secretion and Signaling
Hyperinsulinemia is a result of insulin hypersecretion which is caused by the resistance of peripheral tissues to insulin. Also, insulin resistance is mainly due to impaired insulin signaling postbinding receptor pathway [6]. In addition, the glucose homeostasis abnormalities are common in PCOS patients; therefore, in PCOS condition, there is a defect in insulin secretion as well as insulin signaling pathway dysfunction [56]. On the one hand, human epidemiologic studies have demonstrated a correlation between low birth weight and metabolic diseases. On the other hand, IUGR leads to low birth weight which in turn promotes the fetuses into adults with metabolic diseases [57].
In PCOS, the initial defect in insulin secretion may indicate the dysfunction of pancreatic beta cells which is related to the occurrence of type 2 diabetes mellitus [58]. There is a basic overlapping link between type 2 diabetes and PCOS [59]. Type 2 diabetes is more likely due to secretion of impaired insulin from pancreatic beta cells. This pathogenic pathway for type 2 diabetes is well known [60], but in previous studies, there is a controversy about the role of beta-cellular impairment in PCOS [59]. The beta-cell dysfunction can affect PCOS development in two ways; firstly, the reduction of beta-cell activity can subsequently cause the impairment of glucose tolerance and hyperglycemia, and secondly, the elevation of beta-cell activity results in hyperinsulinemia, which is followed by adverse effects on peripheral tissues either alone or in combination, and affects the pathogenesis of PCOS [23] (Figure 3).
Figure 3.
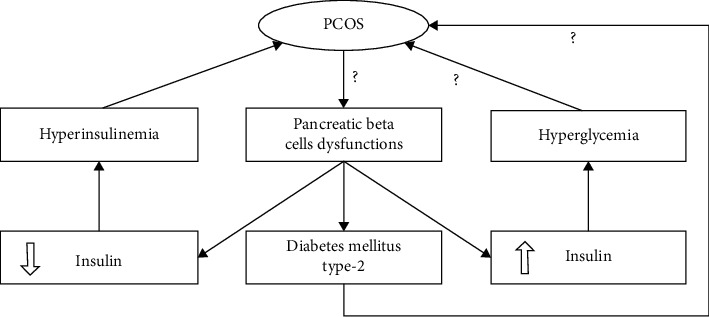
The effect of pancreatic beta-cell dysfunctions on PCOS pathogenesis. Beta-cell dysfunctions lead to type 2 diabetes mellitus and hyperinsulinemia that lead to PCOS by creating hyperandrogenism. However, it is not clear that the PCOS is an introduction to beta-cell abnormalities and then resulted in insulin secretion dysregulation or vice versa.
Women with PCOS have higher levels of fasting insulin and glucose-stimulated insulin, as well as less insulin sensitivity than healthy women who are matched according to age and body mass index [61]. Although the etiology of hyperinsulinemia has not been distinguished yet, clinical and molecular studies have believed that defects in insulin signaling and postbinding receptor, likely due to increase in insulin receptors and insulin receptor substrate-1 phosphorylation, affect metabolic pathways and lead to insulin sensitivity and secretion abnormalities [6]. In addition to pancreatic beta-cell dysfunctions, genes affecting the insulin secretion and signaling also play a role in insulin resistance. Genetic alterations and expression of these genes were investigated and explained in the following sections.
3.6. INS
The variable number of tandem repeat (VNTR) polymorphisms in the promoter region of the insulin gene affects its expression [62]. The results of the genetic evaluations of the insulin gene in relation to PCOS are highly controversial. It is mainly due to differences in diagnostic criteria for the identification of affected patients, VNTR genotyping methods, and the racial and geographic background of the participants [8]. Nevertheless, due to the impact of insulin resistance and hyperinsulinemia on anovulation, there may be an association between insulin-related genes and ovulation. While in a meta-analysis study, no association between INS VNTR gene and PCOS was observed [63], INS VNTR class III allele is correlated with increased HOMA-IR and BMI in Kashmiri women with PCOS [64]. A set of previous studies have shown that the variable number of tandem repeat of INS gene is not likely to be dependent on PCOS in different populations (Table 2).
Table 2.
Candidate genes involved in the etiology of polycystic ovary syndrome related to insulin secretion and signaling.
| Gene | Genetic marker(s) | Type of study | Physiologic function | Studied population | Type of polymorphism | References |
|---|---|---|---|---|---|---|
| INS | INS VNTR | Case-control | Insulin secretion | Czech | ND | [65] |
| INS | INS VNTR | Case-control, family-based association, quantitative trait analyses | Insulin secretion | British/Irish | ND | [66] |
| INS | −23/Hph I | Case-control | Insulin secretion | Korean | ND | [67] |
| INS | INS VNTR | Meta-analysis | Insulin secretion | Different | ND | [63] |
| INS | INS VNTR | Case-control | HOMA-IR | Kashmiri | ND | [64] |
| INSR | C/T SNP | Case-control | Insulin signaling | American | Exon 17C/T SNP | [68] |
| INSR | D19S884 | Case-control | Insulin signaling | Caucasian | D19S884 | [69] |
| INSR | T/C SNP | Case-control | Insulin signaling | Chinese | T/C SNP | [70] |
| INSR | Nine SNPs | Case-control | Insulin resistance | Korean | +176477C > T | [13] |
| INSR | Exon 17 C/T | Case-control | Insulin signaling | Turkish | ND | [71] |
| INSR | C/T SNP at exon 17 | Case-control | Insulin signaling | Chinese | C/T SNP at exon 17 | [72] |
| INSR | C/T polymorphism | Case-control | Insulin signaling | Indian | C/T polymorphism | [73] |
| INSR | rs1799817, rs2059807, rs8108622, and rs10500204 | Family association study | Insulin signaling | Chinese Han | ND | [74] |
| INSR | rs3786681, rs17253937, and rs2252673 | Family-based analysis | Insulin signaling | Chinese Han | rs2252673 | [75] |
| INSR | Susceptibility loci | Case-control | Insulin signaling | Europeans | INSR | [76] |
| INSR | Genotype and allele frequencies | Case-control | Insulin signaling | Indonesian | ND | [77] |
| INSR | rs1799817 | Case-control | Insulin signaling | Saudi Arabian | Allele T | [78] |
| INSR | rs2059807 and rs1799817 | Case-control | Insulin signaling | Indian | rs2059807 and rs1799817 | [79] |
| INSR | rs2059807 | GWAS | Metabolic syndrome and insulin resistance | Han Chinese | rs2059807 | [80] |
| INSR | INSR mutation | Case report | Insulin signaling | Jamaican | p.His1157Gln | [3] |
| ΙNSR, IRS-1, and IRS-2 | Gly972Arg (G972R) | Meta-analysis | Insulin signaling | Different | Gly972Arg (G972R) variant in IRSs | [81] |
| IRS-1 and IRS-2 | Gly972Arg and Gly1057Asp | Meta-analysis | Insulin signaling | Different | Gly972Arg 1 | [82] |
| IRS-2 | 295 SNPs | Case-control | Insulin signaling | Caucasian | Three SNPs | [83] |
| THADA | rs13429458 | Case-control | Regulation of energy homeostasis | Chinese Hui | ND | [76] |
| THADA | 2p21 chr | Case-control | Regulation of energy homeostasis | European | THADA | [83] |
| THADA | Susceptibility loci | Case-control | Regulation of energy homeostasis | Europeans | THADA | [76] |
| THADA | rs13429458, rs12478601, rs13405728, rs10818854, and rs2479106 | Family-based analysis | Regulation of energy homeostasis | Chinese Han | rs13429458 | [84] |
| THADA | rs12478601 | Case-control | Pancreatic beta-cell function | Iraqi | ND | [85] |
| THADA | rs13429458 | GWAS | Insulin resistance | Han Chinese | rs13429458 | [80] |
| THADA | rs13429458 | GWAS | Glucose metabolism | Indian | ND | [86] |
| THADA | rs13429458 | Meta-analysis | Regulation of energy homeostasis | Asian | Minor allele (C) | [87] |
| THADA | rsl3429458 | Case-control | Glucose metabolism | Xinjiang Uygur | Minor allele (T) | [88] |
| THADA | rs13429458 | Meta-analysis | Glucose metabolism | Chinese | rs13429458 | [89] |
| THADA | rs13429458 | Case-control | Glucose metabolism | Indian | rs13429458 | [90] |
| MTNR1B | rs10830963 and rs10830962 | Case-control | Regulator of circadian rhythms and reproductive processes | Chinese Han | rs10830963 | [91] |
| MTNR1A | rs2119882 | Case-control | Regulator of circadian rhythms and reproductive processes | Chinese Han | rs2119882 | [92] |
| MTNR | rs2119882 and rs10830963 | Family association study | Regulator of circadian rhythms and reproductive processes | Chinese Han | rs2119882 | [93] |
| MTNR1A MTNR1B | rs2119882 and rs10830963 | GWAS | Glycolipid metabolism | Chinese | MTNR1A rs2119882 and MTNR1B rs10830963 | [94] |
| MTNR1A MTNR1B | rs2119882 and rs10830963 | Meta-analysis | Insulin resistance | Different populations | MTNR1B rs10830963 and MTNR1B rs2119882 | [95] |
Abbreviations: GWAS, genome-wide association study; HOMA-IR, homeostatic model assessment for insulin resistance; INS, insulin gene; INSR, insulin receptor; IRS, insulin receptor substrate; MTNR1A, melatonin receptor 1A; THADA, thyroid adenoma associated; ND, no data.
3.7. INSR
Insulin receptor gene encodes insulin receptor that plays a pivotal role in insulin signaling pathway, and single nucleotide polymorphisms (SNPs) of this gene are likely to have an effect on PCOS metabolic disorders such as insulin resistance and obesity [13]. In various studies conducted in different populations, there is a strong association between the different varieties of INSR gene and PCOS indicating that the INSR, regardless of ethnicity and race, could be a good genetic marker for PCOS (Table 2). The reality is that a C/T polymorphism in the tyrosine kinase domain of INSR gene can be a susceptible variant for PCOS (Table 2). Furthermore, the rs2059807 and rs1799817 in INSR gene were significantly associated with IR in PCOS women in different populations [74, 78–80]. In fact, INSR mediates the effect of insulin resistance on PCOS. But, we should consider findings of studies of insulin resistance in PCOS condition demonstrating that only the metabolic tissues such as liver, skeletal muscle, and fibroblasts are insulin-resistant, whereas the ovary and pituitary tissues remain sensitive to insulin functions [96].
3.8. IRS-1 and IRS-2
Recent studies have shown that activation of phosphatidylinositol 3-kinase, being carried out by insulin receptor substrate-1 (IRS-1) and IRS-2 mediators, has an important role in the regulation of insulin-mediated glucose transfer and carbohydrate metabolism [82]. In PCOS women, there is an insulin receptor signaling defect, being accompanied with a decrease in IRS protein, and is related to phosphatidylinositol 3-kinase activity [97]. On the one hand, the Gly972Arg variant of IRS-1 gene was associated with low SHBG levels in adolescent girls with the history of premature pubarche [98]. On the other hand, the relationship between PCOS and insulin resistance is correlated with reduced SHBG-circulating levels leading to increased blood testosterone levels [99]. It is thought that decreasing the tyrosine phosphorylation of IRS-1 and increasing the phosphorylation of IRS-2 Ser312 in PCOS may be initial defects or possible molecular mechanisms in insulin resistance in PCOS [100]. In a meta-analysis, Arg972 polymorphism in IRS-1 has been shown as a PCOS susceptibility allele and it mediates its pathogenesis via an increased level of fasting glucose [81]. In another meta-analysis, the IRS-1 Gly972Arg polymorphism was found to be a risk factor for PCOS susceptibility [82]. The mRNA levels of IRS-1 and IRS-2 were significantly increased as the result of hyperandrogenic environment in PCOS women [101]. However, the value of IRS-1 and IRS-2 polymorphisms in association with PCOS is not as the value of INSR gene polymorphisms in PCOS etiology.
3.9. THADA
The thyroid adenoma-associated (THADA) gene has been initially identified in chromosomal defects of this genomic region in benign adenoma of thyroid glands, and its intron region was interconnected with peroxisome proliferator-activated receptor-gamma (PPAR-γ) [102]. Overtransmission of SNP rs13429458 in THADA suggested that this gene has the capacity to be a new candidate for PCOS [84]. Polymorphisms of THADA may be involved in pathogenesis of both diabetes mellitus type 2 and PCOS [84]. An SNP of THADA, being associated with type 2 diabetes mellitus, indicates that the THADA has the main role in insulin secretion [103]. Thus, further functional genetic studies are required to clarify the exact role of THADA in pathogenesis of both PCOS and diabetes mellitus type 2. However, the SNP rs13429458 of THDAD gene may be a genetic risk factor for PCOS in different populations [76, 87–90].
3.10. MTNR1A and MTNR1B
The action of melatonin is mediated by melatonin receptors (MTNRs) which include MTNR1A and MTNR1B, both of which belong to the G-protein coupled-receptors superfamily [92]. MTNR1A is mainly expressed in alpha cells and MTNR1B in beta cells of the pancreas [104]. The MTNR1B gene is a new candidate gene for type 2 diabetes [92], upregulation of which in the pancreatic islets of diabetic patients is a document for the main role of MTNR1B in T2DM pathogenesis [105]. The association of MTNR1B polymorphisms with PCOS has been documented [91, 106]. The rs10830963 SNP MTNR1B was associated with higher insulin resistance and plasma glucose levels and lower beta-cell function in Chinese PCOS women [91]. In another study, it has been demonstrated that the rs2119882 polymorphism of MTNR1A is also associated with metabolic properties of PCOS and could have a causal role in pathogenesis of PCOS [92]. Generally, launching the MTNR1B signaling pathway in the pancreatic beta cells reduces insulin secretion that resulted in elevated fasting glucose levels in PCOS individuals [107]. The MTNR1B rs10830963 and MTNR1B rs2119882 have been involved in the pathophysiology of insulin resistance in the Chinese PCOS women [94] as well as in the meta-analysis of different populations with PCOS [95], which indicates their involvement in the metabolic aspect of PCOS. Therefore, due to having an effective role in the pathology of diabetes, MTNR1A and MTNR1B may be a predisposition factor for metabolic disorders of PCOS.
4. Conclusions
The genetic aspect of PCOS is highly supported by different twin and investigations of familial aggregation. According to most of the studies, the critical genes for PCOS development were not reported yet, but scholars are in agreement with INSR, IRS1-2, MTNR1A, MTNR1B, THADA, PPAR-γ2, ADIPOQ, and CAPN10 as more susceptible genes in PCOS incidence. The significant point is that these genes were mostly associated with metabolic abnormalities of PCOS. For instance, the role of MTNR1A and MTNR1B, THADA, CAPN10, and PPAR-γ2 in pathology of type 2 diabetes and obesity has been confirmed. The animal transgenic model for genes involved in diabetes and insulin resistance can better interpret the physiological pathways involved in the onset of PCOS. New research studies can find the downstream and upstream agents regulating gene transcription and expression by genetic and bioinformatics studies. Then, they can identify most genetic markers which are related to PCOS. Generally, we proposed that after hyperandrogenism, the role of insulin resistance in pathology of PCOS is much more probable. In conclusion, in spite of complexity in finding the root cause, we can claim that PCOS heterogeneity has opened the way for many new research studies.
Acknowledgments
This work was financially supported by Shiraz University through grant no. 97gcu3m148075 for the PhD students.
Contributor Information
Mohammad Reza Jafarzadeh Shirazi, Email: jafarzd@shirazu.ac.ir.
Amin Tamadon, Email: amintamaddon@yahoo.com.
Data Availability
No data were used to support this study.
Ethical Approval
Not applicable.
Consent
Not applicable.
Disclosure
Zahra Shaaban and Arezoo Khoradmehr are the co-first authors.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
Authors' Contributions
Z. S., A. K., and A. T. conceived and designed the format of the manuscript. Z. S., A. K., F. N., and A. T. drafted and edited the manuscript. A. A. Y., M. R. J. S., and A. T. reviewed the manuscript. All authors contributed to the critical reading and discussion of the manuscript. All authors have read and agreed to the published version of the manuscript. Zahra Shaaban and Arezoo Khoradmehr were contributed equally to this work.
References
- 1.Azziz R., Carmina E., Chen Z., et al. Polycystic ovary syndrome. Nature Reviews Disease Primers . 2016;2(1) doi: 10.1038/nrdp.2016.57.16057 [DOI] [PubMed] [Google Scholar]
- 2.Shaaban Z., Khoradmehr A., Amiri-Yekta A., Jafarzadeh Shirazi M. R., Tamadon A. Pathophysiologic mechanisms of obesity-and chronic inflammation-related genes in etiology of polycystic ovary syndrome. Iranian Journal of Basic Medical Sciences . 2019;22(12):1378–1386. doi: 10.22038/IJBMS.2019.14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aghababaie A., Ford-Adams M., Buchanan C. R., Arya V., Hattersley A., Colclough K. Type A insulin resistance syndrome due to an INSR mutation presenting with diabetes mellitus evolving to hyperandrogenism and PCOS. Endocrine abstracts. Endocrine Abstracts . 2018;58 doi: 10.1530/endoabs.58.OC7.6. [DOI] [Google Scholar]
- 4.Fenichel P., Rougier C., Hieronimus S., Chevalier N. Which origin for polycystic ovaries syndrome: genetic, environmental or both? Annales d’Endocrinologie . 2017;78(3):176–185. doi: 10.1016/j.ando.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 5.Abbott D. H., Barnett D. K., Bruns C. M., Dumesic D. A. Androgen excess fetal programming of female reproduction: a developmental aetiology for polycystic ovary syndrome? Human Reproduction Update . 2005;11(4):357–374. doi: 10.1093/humupd/dmi013. [DOI] [PubMed] [Google Scholar]
- 6.Diamanti-Kandarakis E., Dunaif A. Insulin resistance and the polycystic ovary syndrome revisited: an update on mechanisms and implications. Endocrine Reviews . 2012;33(6):981–1030. doi: 10.1210/er.2011-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vink J. M., Sadrzadeh S., Lambalk C. B., Boomsma D. I. Heritability of polycystic ovary syndrome in a Dutch twin-family study. The Journal of Clinical Endocrinology & Metabolism . 2006;91(6):2100–2104. doi: 10.1210/jc.2005-1494. [DOI] [PubMed] [Google Scholar]
- 8.Roldán B., San Millán J. L., Escobar-Morreale H. F. Genetic basis of metabolic abnormalities in polycystic ovary syndrome. American Journal of PharmacoGenomics . 2004;4(2):93–107. doi: 10.2165/00129785-200404020-00004. [DOI] [PubMed] [Google Scholar]
- 9.Jones M. R., Goodarzi M. O. Genetic determinants of polycystic ovary syndrome: progress and future directions. Fertility and Sterility . 2016;106(1):25–32. doi: 10.1016/j.fertnstert.2016.04.040. [DOI] [PubMed] [Google Scholar]
- 10.McAllister J. M., Legro R. S., Modi B. P., Strauss J. F. Functional genomics of PCOS: from GWAS to molecular mechanisms. Trends in Endocrinology & Metabolism . 2015;26(3):118–124. doi: 10.1016/j.tem.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan S., Scherag A., Janssen O. E., et al. Large effects on body mass index and insulin resistance of fat mass and obesity associated gene (FTO) variants in patients with polycystic ovary syndrome (PCOS) BMC Medical Genetics . 2010;11(1):p. 12. doi: 10.1186/1471-2350-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaaban Z., Khoradmehr A., Jafarzadeh Shirazi M. R., Tamadon A. Pathophysiological mechanisms of gonadotropins–and steroid hormones–related genes in etiology of polycystic ovary syndrome. Iranian Journal of Basic Medical Sciences . 2019;22(1):3–16. doi: 10.22038/ijbms.2018.31776.7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee E.-J., Oh B., Lee J.-Y., Kimm K., Lee S.-H., Baek K.-H. A novel single nucleotide polymorphism of INSR gene for polycystic ovary syndrome. Fertility and Sterility . 2008;89(5):1213–1220. doi: 10.1016/j.fertnstert.2007.05.026. [DOI] [PubMed] [Google Scholar]
- 14.Lewy V. D., Danadian K., Witchel S. F., Arslanian S. Early metabolic abnormalities in adolescent girls with polycystic ovarian syndrome. The Journal of Pediatrics . 2001;138(1):38–44. doi: 10.1067/mpd.2001.109603. [DOI] [PubMed] [Google Scholar]
- 15.Palmert M. R., Gordon C. M., Kartashov A. I., Legro R. S., Emans S. J., Dunaif A. Screening for abnormal glucose tolerance in adolescents with polycystic ovary syndrome. The Journal of Clinical Endocrinology & Metabolism . 2002;87(3):1017–1023. doi: 10.1210/jcem.87.3.8305. [DOI] [PubMed] [Google Scholar]
- 16.Lapillonne A., Braillon P., Claris O., Chatelain P., Delmas P., Salle B. Body composition in appropriate and in small for gestational age infants. Acta Paediatrica . 1997;86(2):196–200. doi: 10.1111/j.1651-2227.1997.tb08868.x. [DOI] [PubMed] [Google Scholar]
- 17.McGarry J. D. Disordered metabolism in diabetes: have we underemphasized the fat component? Journal of Cellular Biochemistry . 1994;55(S1994A):29–38. doi: 10.1002/jcb.240550005. [DOI] [PubMed] [Google Scholar]
- 18.Barker D. J. P. The origins of the developmental origins theory. Journal of Internal Medicine . 2007;261(5):412–417. doi: 10.1111/j.1365-2796.2007.01809.x. [DOI] [PubMed] [Google Scholar]
- 19.Pasquali R., Gambineri A. Androgen Excess Disorders in Women . Berlin, Germany: Springer; 2006. The endocrine impact of obesity and body habitus in the polycystic ovary syndrome; pp. 283–291. [Google Scholar]
- 20.Dunaif A., Finegood D. T. Beta-cell dysfunction independent of obesity and glucose intolerance in the polycystic ovary syndrome. The Journal of Clinical Endocrinology & Metabolism . 1996;81(3):942–947. doi: 10.1210/jcem.81.3.8772555. [DOI] [PubMed] [Google Scholar]
- 21.Dunaif A. Hyperandrogenic anovulation (PCOS): a unique disorder of insulin action associated with an increased risk of non-insulin-dependent diabetes mellitus. The American Journal of Medicine . 1995;98(1):S33–S39. doi: 10.1016/s0002-9343(99)80057-6. [DOI] [PubMed] [Google Scholar]
- 22.Franks S., Webber L. J., Goh M., et al. Ovarian morphology is a marker of heritable biochemical traits in sisters with polycystic ovaries. Obstetrical & Gynecological Survey . 2009;64(3):166–167. doi: 10.1097/01.ogx.0000344392.60303.43. [DOI] [PubMed] [Google Scholar]
- 23.Barber T. M., Franks S. Genetics of polycystic ovary syndrome. Frontiers of Hormone Research . 2013;40:28–39. doi: 10.1159/000341682. [DOI] [PubMed] [Google Scholar]
- 24.Mahmoudi T. Genetic variation in the vitamin D receptor and polycystic ovary syndrome risk. Fertility and Sterility . 2009;92(4):1381–1383. doi: 10.1016/j.fertnstert.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Talbot J. A., Bicknell E. J., Rajkhowa M., Krook A., O’Rahilly S., Clayton R. N. Molecular scanning of the insulin receptor gene in women with polycystic ovarian syndrome. The Journal of Clinical Endocrinology & Metabolism . 1996;81(5):1979–1983. doi: 10.1210/jcem.81.5.8626868. [DOI] [PubMed] [Google Scholar]
- 26.Diamanti-Kandarakis E., Argyrakopoulou G., Economou F., Kandaraki E., Koutsilieris M. Defects in insulin signaling pathways in ovarian steroidogenesis and other tissues in polycystic ovary syndrome (PCOS) The Journal of Steroid Biochemistry and Molecular Biology . 2008;109(3-5):242–246. doi: 10.1016/j.jsbmb.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 27.Mahdi W. K., Mohammed M. S., Sanad A. S. Association of polycystic ovary syndrome and adiponectin gene polymorphisms. Archives of Clinical Microbiology . 2016;7(3):p. 20. [Google Scholar]
- 28.Brochu-Gaudreau K., Rehfeldt C., Blouin R., Bordignon V., Murphy B. D., Palin M.-F. Adiponectin action from head to toe. Endocrine . 2010;37(1):11–32. doi: 10.1007/s12020-009-9278-8. [DOI] [PubMed] [Google Scholar]
- 29.Baba T., Endo T., Sata F., et al. The contributions of resistin and adiponectin gene single nucleotide polymorphisms to the genetic risk for polycystic ovary syndrome in a Japanese population. Gynecological Endocrinology . 2009;25(8):498–503. doi: 10.1080/09513590902972042. [DOI] [PubMed] [Google Scholar]
- 30.Zhang W., Wei D., Sun X., et al. Family-based analysis of adiponectin gene polymorphisms in Chinese Han polycystic ovary syndrome. Fertility and Sterility . 2014;101(5):1419–1423. doi: 10.1016/j.fertnstert.2014.01.035. e3. [DOI] [PubMed] [Google Scholar]
- 31.Alfaqih M. A., Khader Y. S., Al-Dwairi A. N., Alzoubi A., Othman A.-S., Hatim A. Lower levels of serum adiponectin and the T allele of rs1501299 of the ADIPOQ gene are protective against polycystic ovarian syndrome in Jordan. Korean Journal of Family Medicine . 2018;39(2):p. 108. doi: 10.4082/kjfm.2018.39.2.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Czeczuga-Semeniuk E., Galar M., Jarząbek K., Kozłowski P., Sarosiek N. A., Wołczyński S. The preliminary association study of ADIPOQ, RBP4, and BCMO1 variants with polycystic ovary syndrome and with biochemical characteristics in a cohort of Polish women. Advances in Medical Sciences . 2018;63(2):242–248. doi: 10.1016/j.advms.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 33.Liu Z., Wang Z., Hao C., Tian Y., Fu J. J. R. B. Endocrinology. Effects of ADIPOQ polymorphisms on PCOS risk: a meta-analysis. 2018;16(1):1–6. doi: 10.1186/s12958-018-0439-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tiongco R. E., Cabrera F. J., Clemente B., Flake C. C., Salunga M. A., Pineda-Cortel M. R., et al. G276T polymorphism in the ADIPOQ gene is associated with a reduced risk of polycystic ovarian syndrome: a meta-analysis of Asian population. Taiwanese Journal of Obstetrics and Gynecology . 2019;58(3):409–416. doi: 10.1016/j.tjog.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 35.Ezzidi I., Mtiraoui N., Mohmmed Ali M. E., Al Masoudi A., Abu Duhier F. Adiponectin (ADIPOQ) gene variants and haplotypes in Saudi Arabian women with polycystic ovary syndrome (PCOS): a case-control study. Gynecological Endocrinology . 2020;36(1):66–71. doi: 10.1080/09513590.2019.1632830. [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez A., Abril E., Roca A., et al. CAPN10 alleles are associated with polycystic ovary syndrome. The Journal of Clinical Endocrinology & Metabolism . 2002;87(8):3971–3976. doi: 10.1210/jcem.87.8.8793. [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez A., Abril E., Roca A., et al. Specific CAPN10 gene haplotypes influence the clinical profile of polycystic ovary patients. The Journal of Clinical Endocrinology & Metabolism . 2003;88(11):5529–5536. doi: 10.1210/jc.2003-030322. [DOI] [PubMed] [Google Scholar]
- 38.Shen W., Li T., Hu Y., Liu H., Song M. Calpain-10 genetic polymorphisms and polycystic ovary syndrome risk: a meta-analysis and meta-regression. Gene . 2013;531(2):426–434. doi: 10.1016/j.gene.2013.08.072. [DOI] [PubMed] [Google Scholar]
- 39.Sáez M. E., González-Sánchez J. L., Ramírez-Lorca R., et al. The CAPN10 gene is associated with insulin resistance phenotypes in the Spanish population. PLoS One . 2008;3(8) doi: 10.1371/journal.pone.0002953.e2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Márquez J. L., Pacheco A., Valdés P., Salazar L. A. Association between CAPN10 UCSNP-43 gene polymorphism and polycystic ovary syndrome in Chilean women. Clinica chimica acta; international journal of clinical chemistry . 2008;398(1-2):5–9. doi: 10.1016/j.cca.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 41.Wiltgen D., Furtado L., Kohek M. B. F., Spritzer P. M. CAPN10UCSNP-43, UCSNP-19 and UCSNP-63 polymorphisms and metabolic syndrome in polycystic ovary syndrome. Gynecological Endocrinology . 2007;23(3):173–178. doi: 10.1080/09513590701233661. [DOI] [PubMed] [Google Scholar]
- 42.Dasgupta S., Sirisha P. V. S., Neelaveni K., Anuradha K., Reddy B. M. Association of CAPN10 SNPs and haplotypes with polycystic ovary syndrome among South Indian Women. PLoS One . 2012;7(2) doi: 10.1371/journal.pone.0032192.e32192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anastasia K., Koika V., Roupas N. D., et al. Association of Calpain (CAPN) 10 (UCSNP-43, rs3792267) gene polymorphism with elevated serum androgens in young women with the most severe phenotype of polycystic ovary syndrome (PCOS) Gynecological Endocrinology . 2015;31(8):630–634. doi: 10.3109/09513590.2015.1032932. [DOI] [PubMed] [Google Scholar]
- 44.Thathapudi S., Erukkambattu J., Hasan Q., Addepally U., Kodati V. Association of calpain 10 gene UCSNP-43 polymorphism (rs3792267) with polycystic ovarian syndrome. International Journal of Reproduction, Contraception, Obstetrics and Gynecology . 2017;4(4):1185–1190. [Google Scholar]
- 45.Huang M., Xiao J., Zhao X., Liu C., Chen Q. Four polymorphisms of the CAPN 10 gene and their relationship to polycystic ovary syndrome susceptibility: a meta-analysis. Clinical Endocrinology . 2012;76(3):431–438. doi: 10.1111/j.1365-2265.2011.04218.x. [DOI] [PubMed] [Google Scholar]
- 46.San-Millán J. L., Escobar-Morreale H. F. The role of genetic variation in peroxisome proliferator-activated receptors in the polycystic ovary syndrome (PCOS): an original case-control study followed by systematic review and meta-analysis of existing evidence. Clinical Endocrinology . 2010;72(3):383–392. doi: 10.1111/j.1365-2265.2009.03679.x. [DOI] [PubMed] [Google Scholar]
- 47.Zhang H., Bi Y., Hu C., Lu W., Zhu D. Association between the Pro12Ala polymorphism of PPAR-γ gene and the polycystic ovary syndrome: a meta-analysis of case-control studies. Gene . 2012;503(1):12–17. doi: 10.1016/j.gene.2012.04.083. [DOI] [PubMed] [Google Scholar]
- 48.Yang J., Gong H., Liu W., Tao T. The association of Pro12Ala polymorphism in the peroxisome proliferator-activated receptor-gamma2 gene with the metabolic characteristics in Chinese women with polycystic ovary syndrome. International Journal of Clinical and Experimental Pathology . 2013;6(9):1894–902. [PMC free article] [PubMed] [Google Scholar]
- 49.Jacob R., Ramachandran C., Jude C., Venkatachalam U., Rao S. K. Peroxisome proliferator activated receptor gamma polymorphism Pro12Ala in polycystic ovary syndrome (PCOS) of South Indian Population. Asian Pacific Journal of Reproduction . 2016;5(3):210–213. doi: 10.1016/j.apjr.2016.04.002. [DOI] [Google Scholar]
- 50.Giandalia A., Pappalardo M. A., Russo G. T., et al. Influence of peroxisome proliferator-activated receptor-γ exon 2 and exon 6 and insulin receptor substrate (IRS)-1 Gly972Arg polymorphisms on insulin resistance and beta-cell function in southern mediterranean women with polycystic ovary syndrome. Journal of Clinical & Translational Endocrinology . 2018;13:1–8. doi: 10.1016/j.jcte.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sreenan S. K., Zhou Y.-P., Otani K., et al. Calpains play a role in insulin secretion and action. Diabetes . 2001;50(9):2013–2020. doi: 10.2337/diabetes.50.9.2013. [DOI] [PubMed] [Google Scholar]
- 52.Hanis C. L., Boerwinkle E., Chakraborty R., et al. A genome-wide search for human non-insulin-dependent (type 2) diabetes genes reveals a major susceptibility locus on chromosome 2. Nature Genetics . 1996;13(2):161–166. doi: 10.1038/ng0696-161. [DOI] [PubMed] [Google Scholar]
- 53.Panda P. K., Rane R., Ravichandran R., Singh S., Panchal H. Genetics of PCOS: a systematic bioinformatics approach to unveil the proteins responsible for PCOS. Genomics data . 2016;8:52–60. doi: 10.1016/j.gdata.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ek J., Urhammer S. A., Sørensen T. I. A., Andersen T., Auwerx J., Pedersen O. Homozygosity of the Pro12Ala variant of the peroxisome proliferation-activated receptor-γ2 (PPAR-γ2): divergent modulating effects on body mass index in obese and lean Caucasian men. Diabetologia . 1999;42(7):892–895. doi: 10.1007/s001250051243. [DOI] [PubMed] [Google Scholar]
- 55.Michalik L., Auwerx J., Berger J. P., et al. International union of pharmacology. LXI. Peroxisome proliferator-activated receptors. Pharmacological Reviews . 2006;58(4):726–741. doi: 10.1124/pr.58.4.5. [DOI] [PubMed] [Google Scholar]
- 56.Bergman R. N., Finegood D. T., Kahn S. E. The evolution of β-cell dysfunction and insulin resistance in type 2 diabetes. European Journal of Clinical Investigation . 2002;32:35–45. doi: 10.1046/j.1365-2362.32.s3.5.x. [DOI] [PubMed] [Google Scholar]
- 57.Cresswell J., Barker D., Osmond C., Egger P., Phillips D., Fraser R. Fetal growth, length of gestation, and polycystic ovaries in adult life. The Lancet . 1997;350(9085):1131–1135. doi: 10.1016/s0140-6736(97)06062-5. [DOI] [PubMed] [Google Scholar]
- 58.Colilla S., Cox N. J., Ehrmann D. A. Heritability of insulin secretion and insulin action in women with polycystic ovary syndrome and their first degree relatives. Journal of Clinical Endocrinology & Metabolism . 2001;86(5):2027–2031. doi: 10.1210/jc.86.5.2027. [DOI] [PubMed] [Google Scholar]
- 59.Barber T. M., McCarthy M. I., Wass J. A. H., Franks S. Obesity and polycystic ovary syndrome. Clinical Endocrinology . 2006;65(2):137–145. doi: 10.1111/j.1365-2265.2006.02587.x. [DOI] [PubMed] [Google Scholar]
- 60.Rutter G. A., Parton L. E. The &Bgr;-Cell in type 2 diabetes and in obesity. Obesity and Metabolism . 2008;36:118–134. doi: 10.1159/000115360. [DOI] [PubMed] [Google Scholar]
- 61.Diamanti-Kandarakis E., Piperi C., Argyrakopoulou G., et al. Polycystic ovary syndrome: the influence of environmental and genetic factors. Hormones . 2006;5(1):17–34. doi: 10.14310/horm.2002.11165. [DOI] [PubMed] [Google Scholar]
- 62.Bennett S. T., Lucassen A. M., Gough S. C. L., et al. Susceptibility to human type 1 diabetes at IDDM2 is determined by tandem repeat variation at the insulin gene minisatellite locus. Nature Genetics . 1995;9(3):284–292. doi: 10.1038/ng0395-284. [DOI] [PubMed] [Google Scholar]
- 63.Song L.-y., Luo J.-r., Peng Q.-l., et al. Lack of association of INS VNTR polymorphism with polycystic ovary syndrome: a meta-analysis. Journal of Assisted Reproduction and Genetics . 2014;31(6):675–681. doi: 10.1007/s10815-014-0211-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rasool S. U. A., Ashraf S., Nabi M., et al. Insulin gene VNTR class III allele is a risk factor for insulin resistance in Kashmiri women with polycystic ovary syndrome. Meta Gene . 2019;21 doi: 10.1016/j.mgene.2019.100597.100597 [DOI] [Google Scholar]
- 65.Vanková M., Vrbíková J., Hill M., Cinek O., Bendlová B. Association of insulin gene VNTR polymorphism with polycystic ovary syndrome. Annals of the New York Academy of Sciences . 2002;967(1):558–565. doi: 10.1111/j.1749-6632.2002.tb04317.x. [DOI] [PubMed] [Google Scholar]
- 66.Powell B. L., Haddad L., Bennett A., et al. Analysis of multiple data sets reveals no association between the insulin gene variable number tandem repeat element and polycystic ovary syndrome or related traits. The Journal of Clinical Endocrinology & Metabolism . 2005;90(5):2988–2993. doi: 10.1210/jc.2004-2485. [DOI] [PubMed] [Google Scholar]
- 67.Yun J.-H., Gu B.-H., Kang Y.-B., Choi B.-C., Song S., Baek K.-H. Association betweenINS-VNTRpolymorphism and polycystic ovary syndrome in a Korean population. Gynecological Endocrinology . 2012;28(7):525–528. doi: 10.3109/09513590.2011.650658. [DOI] [PubMed] [Google Scholar]
- 68.Siegel S., Futterweit W., Davies T. F., et al. A C/T single nucleotide polymorphism at the tyrosine kinase domain of the insulin receptor gene is associated with polycystic ovary syndrome. Fertility and Sterility . 2002;78(6):1240–1243. doi: 10.1016/s0015-0282(02)04241-3. [DOI] [PubMed] [Google Scholar]
- 69.Tucci S., Futterweit W., Concepcion E. S., et al. Evidence for association of polycystic ovary syndrome in caucasian women with a marker at the insulin receptor gene locus. The Journal of Clinical Endocrinology & Metabolism . 2001;86(1):446–449. doi: 10.1210/jcem.86.1.7274. [DOI] [PubMed] [Google Scholar]
- 70.Jin L., Zhu X.-M., Luo Q., Qian Y., Jin F., Huang H.-F. A novel SNP at exon 17 of INSR is associated with decreased insulin sensitivity in Chinese women with PCOS. MHR: Basic science of reproductive medicine . 2006;12(3):151–155. doi: 10.1093/molehr/gal022. [DOI] [PubMed] [Google Scholar]
- 71.Unsal T., Konac E., Yesilkaya E., et al. Genetic polymorphisms of FSHR, CYP17, CYP1A1, CAPN10, INSR, SERPINE1 genes in adolescent girls with polycystic ovary syndrome. Journal of Assisted Reproduction and Genetics . 2009;26(4):205–216. doi: 10.1007/s10815-009-9308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen Z., Shi Y., Zhao Y., Li Y., Tang R., Zhao L. Correlation between single nucleotide polymorphism of insulin receptor gene with polycystic ovary syndrome. Zhonghua Fu Chan Ke Za Zhi . 2004;39(9):582–585. [PubMed] [Google Scholar]
- 73.Mukherjee S., Shaikh N., Khavale S., et al. Genetic variation in exon 17 of INSR is associated with insulin resistance and hyperandrogenemia among lean Indian women with polycystic ovary syndrome. European Journal of Endocrinology . 2009;160(5):855–862. doi: 10.1530/eje-08-0932. [DOI] [PubMed] [Google Scholar]
- 74.Xu X., Zhao H., Shi Y., et al. Family association study between INSR gene polymorphisms and PCOS in Han Chinese. Reproductive Biology and Endocrinology . 2011;9(1):p. 76. doi: 10.1186/1477-7827-9-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Du J., Wang J., Sun X., et al. Family-based analysis of INSR polymorphisms in Chinese PCOS. Reproductive Biomedicine Online . 2014;29(2):239–244. doi: 10.1016/j.rbmo.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 76.Brower M. A., Jones M. R., Rotter J. I., et al. Further investigation in europeans of susceptibility variants for polycystic ovary syndrome discovered in genome-wide association studies of Chinese individuals. The Journal of Clinical Endocrinology & Metabolism . 2015;100(1):E182–E186. doi: 10.1210/jc.2014-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Suhron M., ZJSRiP Z. How were stress family and INSR (insulin receptor) expression in polycystic ovary syndrome (PCOS) insulin resistant in madurese tribe? Indonesia . 2021;12(1):170–175. [Google Scholar]
- 78.MHJJoMB D. Rs1799817 in INSR associates with susceptibility to polycystic ovary syndrome. 2020;39(3):p. 149. doi: 10.2478/jomb-2019-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dakshinamoorthy J., Jain P. R., Ramamoorthy T., Ayyappan R., Balasundaram U. Association of GWAS identified INSR variants (rs2059807 & rs1799817) with polycystic ovarian syndrome in Indian women. International Journal of Biological Macromolecules . 2020;144:663–670. doi: 10.1016/j.ijbiomac.2019.10.235. [DOI] [PubMed] [Google Scholar]
- 80.Tian Y., Li J., Su S., et al. PCOS-GWAS susceptibility variants in THADA, INSR, TOX3, and DENND1A are associated with metabolic syndrome or insulin resistance in women with PCOS. Frontiers in Endocrinology . 2020;11:p. 274. doi: 10.3389/fendo.2020.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ioannidis A., Ikonomi E., Dimou N. L., Douma L., Bagos P. G. Polymorphisms of the insulin receptor and the insulin receptor substrates genes in polycystic ovary syndrome: A Mendelian randomization meta-analysis. Molecular Genetics and Metabolism . 2010;99(2):174–183. doi: 10.1016/j.ymgme.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 82.Ruan Y., Ma J., Xie X. Association of IRS-1 and IRS-2 genes polymorphisms with polycystic ovary syndrome: a meta-analysis. Endocrine Journal . 2012;59(7):601–609. doi: 10.1507/endocrj.ej11-0387. [DOI] [PubMed] [Google Scholar]
- 83.Goodarzi M. O., Louwers Y. V., Taylor K. D., et al. Replication of association of a novel insulin receptor gene polymorphism with polycystic ovary syndrome. Fertility and Sterility . 2011;95(5):1736–1741. doi: 10.1016/j.fertnstert.2011.01.015. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao H., Xu X., Xing X., et al. Family-based analysis of susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Human Reproduction . 2011;27(1):294–298. doi: 10.1093/humrep/der379. [DOI] [PubMed] [Google Scholar]
- 85.Alizzi F. J., Kokaz H. T., Al-Mayah Q. S. J. I. J. O. W. S. H., SCIENCES R. DENND1A and THADA gene polymorphism among Iraqi women with polycystic ovary syndrome. International Journal of Women’s Health and Reproduction Sciences . 2020;8(3):265–271. [Google Scholar]
- 86.Dadachanji R., Sawant D., Patil A., Mukherjee S. J. G. E. Replication study of THADA rs13429458 variant with PCOS susceptibility and its related traits in Indian women. Gynecological Endocrinology . 2021;37(8):1–5. doi: 10.1080/09513590.2021.1906854. [DOI] [PubMed] [Google Scholar]
- 87.Park S., Liu M., Zhang T., Research M. THADA_rs13429458 minor allele increases the risk of polycystic ovary syndrome in Asian, but not in caucasian women: a systematic review and meta-analysis. Hormone and Metabolic Research . 2019;51(10):661–670. doi: 10.1055/a-0969-1872. [DOI] [PubMed] [Google Scholar]
- 88.Li X., Huang Y.-H., Tian H.-Q., Zhang M., La X.-L. J. R., Medicine D. Association study between polycystic ovary syndrome and THADA gene polymorphisms in xinjiang uygur women. 2017;1(2):p. 80. [Google Scholar]
- 89.Wan P., Meng L., Huang C., Dai B., Jin Y., Chai L. Replication study and meta-analysis of selected genetic variants and polycystic ovary syndrome susceptibility in Asian population. Journal of Assisted Reproduction and Genetics . 2021;38(10):1–9. doi: 10.1007/s10815-021-02291-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vishnubotla D. S., Shek A. P., Madireddi S. Pooled genetic analysis identifies variants that confer enhanced susceptibility to PCOS in Indian ethnicity. Gene . 2020;752 doi: 10.1016/j.gene.2020.144760.144760 [DOI] [PubMed] [Google Scholar]
- 91.Li C., Shi Y., You L., Wang L., Chen Z.-J. Association of rs10830963 and rs10830962 SNPs in the melatonin receptor (MTNR1B) gene among Han Chinese women with polycystic ovary syndrome. Molecular Human Reproduction . 2010;17(3):193–198. doi: 10.1093/molehr/gaq087. [DOI] [PubMed] [Google Scholar]
- 92.Li C., Shi Y., You L., Wang L., Chen Z.-J. Melatonin receptor 1A gene polymorphism associated with polycystic ovary syndrome. Gynecologic and Obstetric Investigation . 2011;72(2):130–134. doi: 10.1159/000323542. [DOI] [PubMed] [Google Scholar]
- 93.Song X., Sun X., Ma G., et al. Family association study between melatonin receptor gene polymorphisms and polycystic ovary syndrome in Han Chinese. European Journal of Obstetrics & Gynecology and Reproductive Biology . 2015;195:108–112. doi: 10.1016/j.ejogrb.2015.09.043. [DOI] [PubMed] [Google Scholar]
- 94.Xu X. H., Kou L. C., Wang H. M., Bo C. M., Song X. C. Genetic polymorphisms of melatonin receptors 1A and 1B may result in disordered lipid metabolism in obese patients with polycystic ovary syndrome. Molecular Medicine Reports . 2019;19(3):2220–2230. doi: 10.3892/mmr.2019.9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yi S., Xu J., Shi H., Li W., Li Q., Sun Y. P. Association between melatonin receptor gene polymorphisms and polycystic ovarian syndrome: a systematic review and meta-analysis. Bioscience Reports . 2020;40(6) doi: 10.1042/BSR20200824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nestler J. E., Jakubowicz D. J., Falcon de Vargas A., Brik C., Quintero N., Medina F. Insulin stimulates testosterone biosynthesis by human thecal cells from women with polycystic ovary syndrome by activating its own receptor and using inositolglycan mediators as the signal transduction System1. The Journal of Clinical Endocrinology & Metabolism . 1998;83(6):2001–2005. doi: 10.1210/jcem.83.6.4886. [DOI] [PubMed] [Google Scholar]
- 97.Dunaif A., Wu X., Lee A., Diamanti-Kandarakis E. Defects in insulin receptor signaling in vivo in the polycystic ovary syndrome (PCOS) American Journal of Physiology. Endocrinology and Metabolism . 2001;281(2):E392–E399. doi: 10.1152/ajpendo.2001.281.2.e392. [DOI] [PubMed] [Google Scholar]
- 98.Ibáñez L., Potau N., Zampolli M., et al. Source localization of androgen excess in adolescent girls. The Journal of Clinical Endocrinology & Metabolism . 1994;79(6):1778–1784. doi: 10.1210/jcem.79.6.7989484. [DOI] [PubMed] [Google Scholar]
- 99.Al-Rubiay T. S. J. Investigation mutations of sex hormones binding globulin (SHBG) gene and androgens levels in Iraqi women effected with polycystic ovary syndrome (PCOS) A thesis . Baghdad, Iraq: University of Baghdad; 2014. [Google Scholar]
- 100.Corbould A., Zhao H., Mirzoeva S., Aird F., Dunaif A. Enhanced mitogenic signaling in skeletal muscle of women with polycystic ovary syndrome. Diabetes . 2006;55(3):751–759. doi: 10.2337/diabetes.55.03.06.db05-0453. [DOI] [PubMed] [Google Scholar]
- 101.Lee M. H., Yoon J. A., Kim H. R., et al. Hyperandrogenic milieu dysregulates the expression of insulin signaling factors and glucose transporters in the endometrium of patients with polycystic ovary syndrome. Reproductive Sciences (Thousand Oaks, Calif.) . 2019 doi: 10.1177/1933719119833487. [DOI] [PubMed] [Google Scholar]
- 102.Drieschner N., Belge G., Rippe V., Meiboom M., Loeschke S., Bullerdiek J. Evidence for a 3p25 breakpoint hot spot region in thyroid tumors of follicular origin. Thyroid . 2006;16(11):1091–1096. doi: 10.1089/thy.2006.16.1091. [DOI] [PubMed] [Google Scholar]
- 103.Goodarzi M. O., Jones M. R., Li X., et al. Replication of association of DENND1A and THADA variants with polycystic ovary syndrome in European cohorts. Journal of Medical Genetics . 2012;49(2):90–95. doi: 10.1136/jmedgenet-2011-100427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Staiger H., Machicao F., Schäfer S. A., et al. Polymorphisms within the novel type 2 diabetes risk locus MTNR1B determine β-cell function. PLoS One . 2008;3(12) doi: 10.1371/journal.pone.0003962.e3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Peschke E., Stumpf I., Bazwinsky I., Litvak L., Dralle H., Mühlbauer E. Melatonin and type 2 diabetes ? a possible link? Journal of Pineal Research . 2007;42(4):350–358. doi: 10.1111/j.1600-079x.2007.00426.x. [DOI] [PubMed] [Google Scholar]
- 106.Wang L., Wang Y., Zhang X., et al. Common genetic variation in MTNR1B is associated with serum testosterone, glucose tolerance, and insulin secretion in polycystic ovary syndrome patients. Fertility and Sterility . 2010;94(6):2486–2489. doi: 10.1016/j.fertnstert.2010.01.059. [DOI] [PubMed] [Google Scholar]
- 107.Pandiperumal S., Trakht I., Srinivasan V., et al. Physiological effects of melatonin: role of melatonin receptors and signal transduction pathways. Progress in Neurobiology . 2008;85(3):335–353. doi: 10.1016/j.pneurobio.2008.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data were used to support this study.


