Abstract
Compromised autophagy and defective lysosomal clearance significantly contribute to impaired neuronal proteostasis, which represents a hallmark of Alzheimer's disease (AD) and other age-related neurodegenerative disorders. Growing evidence has implicated that modulating autophagic flux, instead of inducing autophagosome formation alone, would be more reliable to rescue neuronal proteostasis. Concurrently, selectively enhancing drug concentrations in the leision areas, instead of the whole brain, will maximize therapeutic efficacy while reduing non-selective autophagy induction. Herein, we design a ROS-responsive targeted micelle system (TT-NM/Rapa) to enhance the delivery efficiency of rapamycin to neurons in AD lesions guided by the fusion peptide TPL, and facilitate its intracellular release via ROS-mediated disassembly of micelles, thereby maximizing autophagic flux modulating efficacy of rapamycin in neurons. Consequently, it promotes the efficient clearance of intracellular neurotoxic proteins, β-amyloid and hyperphosphorylated tau proteins, and ameliorates memory defects and neuronal damage in 3 × Tg-AD transgenic mice. Our studies demonstrate a promising strategy to restore autophagic flux and improve neuronal proteostasis by rationally-engineered nano-systems for delaying the progression of AD.
Keywords: Autophagic flux, ROS-Responsive micelle system, Brain-neuron targeting, Rapamycin, Alzheimer's disease
Graphical abstract
Highlights
-
•
Modulating autophagic flux to restore neuronal proteostasis was proved to be effective in delaying the progression of AD.
-
•
We designed a novel ROS-responsive targeted micelle with superior targetability and desirable cargo release in AD neurons.
-
•
Our designed TPL peptide with high preferentiality to AD lesions showed great promise for developing AD-targeted therapeutics.
-
•
Systematic evaluation of TT-NM/Rapa would provide a rationale for applying rapamycin in neurodegenerative disease treatment.
1. Introduction
Alzheimer's disease (AD) is a progressive neurodegenerative disorder with rising prevalence yet poor therapeutic outcomes [1]. A cardinal pathological feature of AD is the anomalous accumulation of neurotoxic protein aggregates such as β-amyloid (Aβ) and hyperphosphorylated tau (p-tau), which further contributes to impaired axonal transport and neuronal dysfunction, and ultimately leads to neuronal apoptosis and loss [[2], [3], [4]]. Within eukaryotic cells, intracellular clearance of toxic aggregates is predominately managed by autophagy, a preserved cytoplasmic self-degradation process where autophagic substrates are sequestered within an autophagosome organelle and delivered to lysosomes for digestion [5]. In particular, autophagy is critical for the protein homeostasis and survival of terminally differentiated cells, such as neurons [6]. Several reports have revealed that neuronal autophagy is dysregulated in AD [[7], [8], [9]]. Hence, stimulating neuronal autophagy provides a feasible therapeutic approach for AD treatment. However, emerging evidence has highlighted that, compared with enhancing autophagosome formation alone, modulating autophagic flux (namely the entire dynamic process of autophagosome formation and autophagic degradation) may potentially be a more reliable strategy for the maintenance of neuronal proteostasis [7]. For example, partially increasing autophagosome formation that exceeds lysosomal proteolytic capacity can lead to abnormal accumulation of autophagosomes in axons, which in turn induces the overproduction of Aβ, and consequently, triggers a cascade of deleterious events [8]. Therefore, restoration of autophagic flux homeostasis in neurons holds great promise in the prevention and treatment of AD.
Rapamycin, a powerful and safe autophagy inducer, has been shown to provide therapeutic benefits in experimental models of AD [10]. Rapamycin can not only induce autophagosome assembly via inhibition of mammalian target of rapamycin complex 1 (mTORC1), but also promote autophagosome-lysosome fusion and lysosomal biogenesis via activation of transcription factor EB (TFEB) pathway, thereby improving autophagic flux [11]. Nevertheless, clinical application of rapamycin in AD is limited because of its extremely poor solubility of 2.6 μg/mL in water [12], and low blood-brain barrier (BBB) permeability with a brain-blood ratio (Cbrain/Cblood) of only 0.0057 [13]. Besides, non-selective autophagy induction in vivo may lead to potential side effects [14].
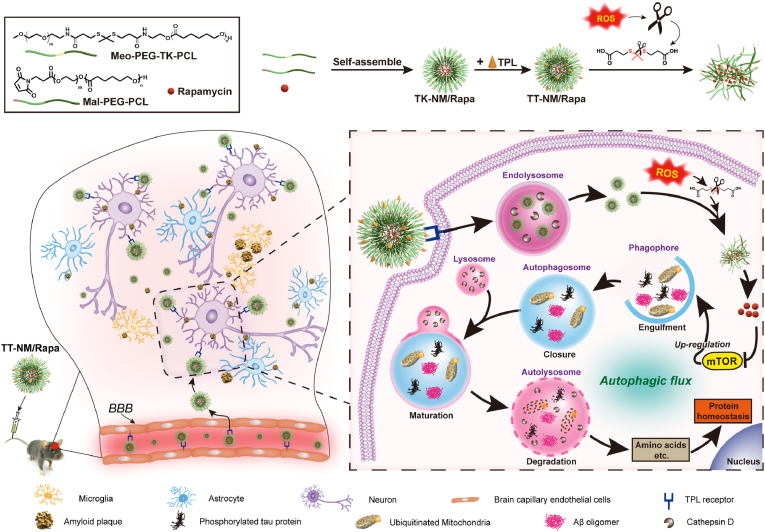
To overcome these disadvantages, we developed a brain-neuron targeting micelle system. Generally, nanocarriers are internalized by neurons mainly via clathrin-mediated endocytosis (Fig. S1), that is to say, the endosome-lysosome pathway plays a critical role in nanocarriers degradation and intraneuronal drug release [15]. Given the progressive failure of neuronal lysosomes in AD brains [16], administration of the conventional nano-system may lead to incomplete intracellular drug release and increased lysosomal hydrolysis burden, thus reducing the therapeutic effect. Therefore, exploiting the high level of reactive oxygen species (ROS) in AD neurons as well as the oxidative environment within endosomes, which contain abundant nicotinamide adenine dinucleotide phosphate (NADPH) oxidase membrane-bound enzymic complex, an important source of ROS in AD brains [[17], [18], [19]], we designed an intracellular ROS triggered micelle system to achieve endo/lysosome escape and efficient intracellular drug release. Inspired by that thioketal (TK) bond is stable in acidic and basic solution but undergoes cleavage in response to ROS [20], it has been introduced into the design of nanoparticles capable of swelling or disassembly to release drug quickly upon exposure to elevated ROS levels [[21], [22], [23]]. For this purpose, environment-sensitive micelle material Meo-PEG-TK-PCL containing ROS-cleavable TK moiety and biocompatible methoxy-polyethylene glycol-polycaprolactone (Meo-PEG-PCL) copolymer was synthesized as the polymer backbone for micelle preparation. After internalization in neurons, the micelle system exhibited fast, enhanced rapamycin release as a result of its instability under high levels of ROS, which greatly facilitated rapamycin to interact with targeted protein in the cytoplasm (Scheme 1). In addition, in order to increase the targetability of this nanocarrier, a dual-functional fusion peptide TPL was modified on the surface of the micelle system. TPL is composed of a BBB-penetrating peptide TGN and a GT1b ganglioside receptor-binding peptide Tet1 via a flexible four-glycine linker (sequence: l-TGNYKALHPHNG-GGGG-HLNILSTLWKYRC). In our previous study, we have demonstrated that TPL peptide showed improved blood stability and enhanced structural flexibility due to the inclusion of four-glycine linker, which resulted in higher binding affinity to either GT1b or brain capillary endothelial bEnd.3 cells than the mono-ligand peptide. Consequently, TPL significantly improved the targetability of nanocarriers to central neurons [24]. Hence, TPL peptide decoration was particularly employed in the study to facilitate efficient and specific delivery of rapamycin to neurons in lesions for AD therapy.
Scheme 1.
ROS-responsive targeted micellar system (TT-NM/Rapa) for improving neuronal proteostasis by upregulation of autophagic flux. TT-NM/Rapa can effectively penetrate through BBB and further target neurons in AD lesions mediated by selective GT1b binding of TPL. Upon endocytosis, the designed micelles consisting of ROS-sensitive thioketal moiety exhibit enhanced release of rapamycin to exert autophagy-modulating efficacy. TT-NM/Rapa efficiently regulate the entire autophagy-lysosomal pathway (initiation, elongation, and lysosomal fusion and degradation), thereby accelerating neurotoxic protein clearance and displaying superior neuroprotective effects for AD treatment.
Our results demonstrated the superior brain-neuron targetability and desirable ROS-responsiveness of targeted micelles (TT-NM). Moreover, the improved neuronal autophagic flux by rapamycin-loaded TT-NM (TT-NM/Rapa) resulted in better therapeutic effects on 3 × Tg-AD transgenic mice by reduction of neurotoxic Aβ and p-tau, and amelioration of memory defects as well. For the first time, the rationally designed nano-system improves the regulation of rapamycin on autophagic flux, achieving recovery of neuronal proteostasis for Alzheimer's treatment.
2. Results and discussion
Synthesis and characterization of ROS-sensitive polymer material. The ROS-responsive polymer material Meo-PEG-TK-PCL was synthesized through condensation of TK-containing diacid and amine (NH2)-terminated polymers referring to the previous studies (Fig. S2) [21,25]. The presence of the –CH3 proton peaks at ∼1.65 ppm and –CH2- proton peaks at both 2.88 ppm and 2.47–2.61 ppm corresponding to a thioketal linker in the 1H Nuclear Magnetic Resonance (1H NMR) spectrum verified the successful preparation of Meo-PEG-TK-PCL polymer (Fig. S3A). The number-average molar weight (Mn) of the resulting Meo-PEG-TK-PCL copolymer was 5537 g/mol with a relatively low polydispersity (Mw/Mn = 1.23) according to its gel permeation chromatography (GPC) profile (Fig. S4A). Its critical micelle concentration determined by pyrene fluorescence probe was 2.159 mg/L (Fig. S4B), indicating that micelle could assemble at extremely low concentrations.
Micelle preparation and characterization. The micelles containing ROS-sensitive Meo-PEG-TK-PCL backbone and TPL peptide modification (TT-NM) were fabricated in this study. In brief, ROS responsive micelles without ligand modification (TK-NM/Rapa) were first prepared by the solvent diffusion method with Meo-PEG-TK-PCL, maleimide-PEG-PCL (Mal-PEG-PCL) and rapamycin at an optimized mass ratio of 14:6:1 (w/w/w), followed by the preparation of TPL-conjugated TK-NM/Rapa (TT-NM/Rapa) via thiol-Mal Michael addition reactions (Figs. S3B–C). Meanwhile, we prepared a TPL-modified non-ROS-responsive control micelles (TP-NM/Rapa), which contains Meo-PEG-PCL and Mal-PEG-PCL at the same mass ratio of TT-NM/Rapa. To achieve the optimal central neuron targeting capability, the modification density of TPL peptide was subsequently screened through the cell uptake and mouse ex vivo imaging. TT-NM with TPL modification density of 10% or 15% exhibited the highest uptake in bEnd.3 cells (mouse brain capillary endothelial cells) and HT22 cells (mouse hippocampal neuronal cells) (Figs. S5A–D). Especially, 10% TT-NM displayed remarkably enhanced BBB penetration and decreased liver distribution among all the groups. (Figs. S5E–F). Consequently, TT-NM with 10% TPL modification density was chosen for further study.
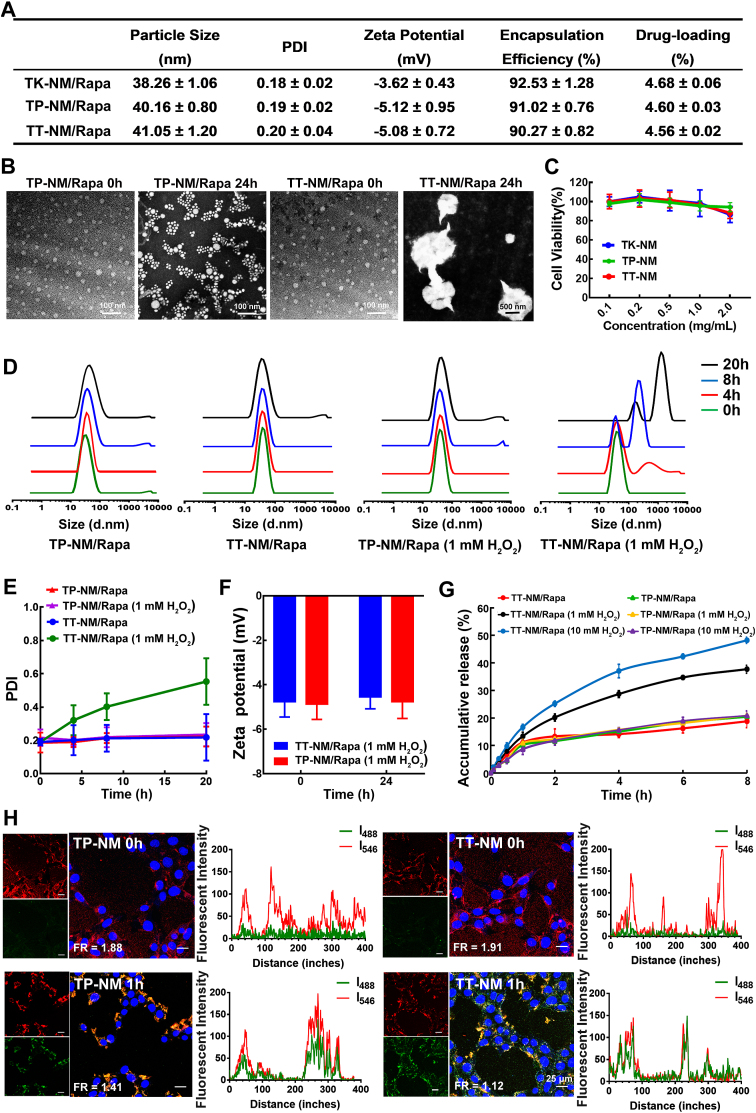
The hydrodynamic diameter of the optimized TT-NM/Rapa was 41.05 ± 1.20 nm with a relatively low polydispersity index of 0.20 ± 0.04 (Fig. 1A). The spherical morphology and narrow size distribution of micelles were also confirmed by transmission electron microscopy (TEM) imaging (Fig. 1B). Zeta potential of TT-NM/Rapa slightly decreased to −5.08 ± 0.72 mV after surface conjugation with TPL peptide. Its encapsulation efficiency and loading capacity was 90.27 ± 0.82% and 4.56 ± 0.02%, respectively. The drug concentration of micelle could reach 153 μg/mL, about 59-fold higher than the original water solubility of rapamycin (2.6 μg/mL). After being diluted 50 times with PBS and 10% fetal bovine serum (FBS), the micelles remained stable at 37 °C for 24 h (Fig. S6). Besides, blank TT-NM displayed favorable biocompatibility, which is essential for the long-term treatment of AD. There was no significant change in viability of HT22 cells after 48 h of incubation with TT-NM at concentrations up to 2 mg/mL (Fig. 1C).
Fig. 1.
Micelle characterization and ROS-responsive drug release. (A) Characterization of TK-NM/Rapa, TP-NM/Rapa and TT-NM/Rapa. TK-NM/Rapa, ROS responsive micelles without ligand modification; TP-NM/Rapa, TPL-modified non-ROS-responsive micelles; TT-NM/Rapa, ROS-responsive targeted micelles. Data are expressed as mean ± SD (n = 3). (B) Transmission electron micrograph of TT-NM/Rapa and TP-NM/Rapa before or after dispersed in PBS buffer (pH 7.4) containing 1 mM H2O2 at 37 °C for 24 h. (C) The viability of HT22 cells measured by the MTT assay following incubation with blank TK-NM, TP-NM and TT-NM at various concentrations at 37 °C for 48 h. The cells without treatment were used as the control. Data are expressed as mean ± SD (n = 6). (D) Size distribution of TT-NM/Rapa and TP-NM/Rapa before or after dispersed in PBS buffer (pH 7.4) containing 1 mM H2O2 at 37 °C for 4, 8 or 20 h. (E) PDI data and (F) zeta potential data. Data are expressed as mean ± SD (n = 3). (G) In vitro release of rapamycin from TT-NM and TP-NM in PBS buffer (pH 7.4), and PBS buffer (pH 7.4) containing 1 mM or 10 mM H2O2 at 37 °C for 8 h. Data are expressed as mean ± SD (n = 3). (H) Representative fluorescence images and quantitative analysis of fluorescent intensity of H2O2 treated HT22 cells following incubation with DiO/DiI loaded TT-NM and TP-NM at 37 °C. The FRET efficiency (FR) was calculated as I546/I488 by MATLAB (I546, fluorescence intensity of acceptor channel with an emission filter of 555–655 nm; I488, fluorescence intensity of donor channel with an emission filter of 500–530 nm; n = 6 images/time point).
ROS-triggered drug release. After incubation with 1 mM H2O2 for 20 h (ROS concentration in cells up to 1 mM during oxidative stress [26]), the particle size of TT-NM changed dramatically from 40 nm to > 1 μm, reflected by the disappearance of size peak at 40 nm and emergence of multi-dispersed distribution at 100 nm–1 μm (Fig. 1D and E). TEM images also substantiated the ROS-triggered disassembly of TT-NM, as a large number of irregular and swollen aggregates appeared after 24 h of incubation with 1 mM H2O2. However, only slight size variations were observed in the TP-NM group (Fig. 1B and D). There was little change in zeta potential values of TT-NM upon exposure to 1 mM H2O2, probably because the corresponding oxidation products (thiols and ketones) were barely charged (Fig. 1F). Meanwhile, in vitro release experiment showed that TT-NM underwent a fast release upon exposure to H2O2, whereas there was little change in the release rate of TP-NM/Rapa. The cumulative release percentage of TT-NM/Rapa was 1.91- and 2.30-fold higher than that of the control after 8 h of incubation with 1 mM and 10 mM H2O2, respectively, indicating that the release from TT-NM was H2O2-responsive through ROS-triggered destruction of micelles (Fig. 1G).
Next, fluorescence resonance energy transfer (FRET) analysis was conducted to investigate the intracellular ROS-triggered release from TT-NM. The micelles loaded with DiO (donor) and DiI (acceptor), a FRET pair of lipophilic probes, were incubated with H2O2-treated HT22 cells. At 0 h, only strong DiI signals (red) were observed in both TT-NM group and TP-NM groups, suggesting that it was intact DiO/DiI dual-loaded micelles that were internalized by HT22 cells. Thus the extreme proximity of core-loaded DiO and DiI resulted in the presence of FRET effect (quenched DiO, activated DiI). However, there was an observable decline in DiI signals and an increase in DiO signals (green) for the TT-NM group within 1 h, and the sharply decreased FRET signals were owing to a fast release of encapsulated probes to the cytoplasm (FRET efficiency value (FR) 1.91 → 1.12). Nevertheless, TP-NM group still exhibited a bright FRET signal during 1 h of period (FR = 1.41), suggesting only a small amount of drug release (Fig. 1H). These results confirmed that TT-NM could efficiently release drug into the cytoplasm, which may be credited to the disassembly of ROS-responsive TT-NM in highly oxidative endo/phagosomal vesicles and its rapid endo/lysosome escape (Fig. S7).
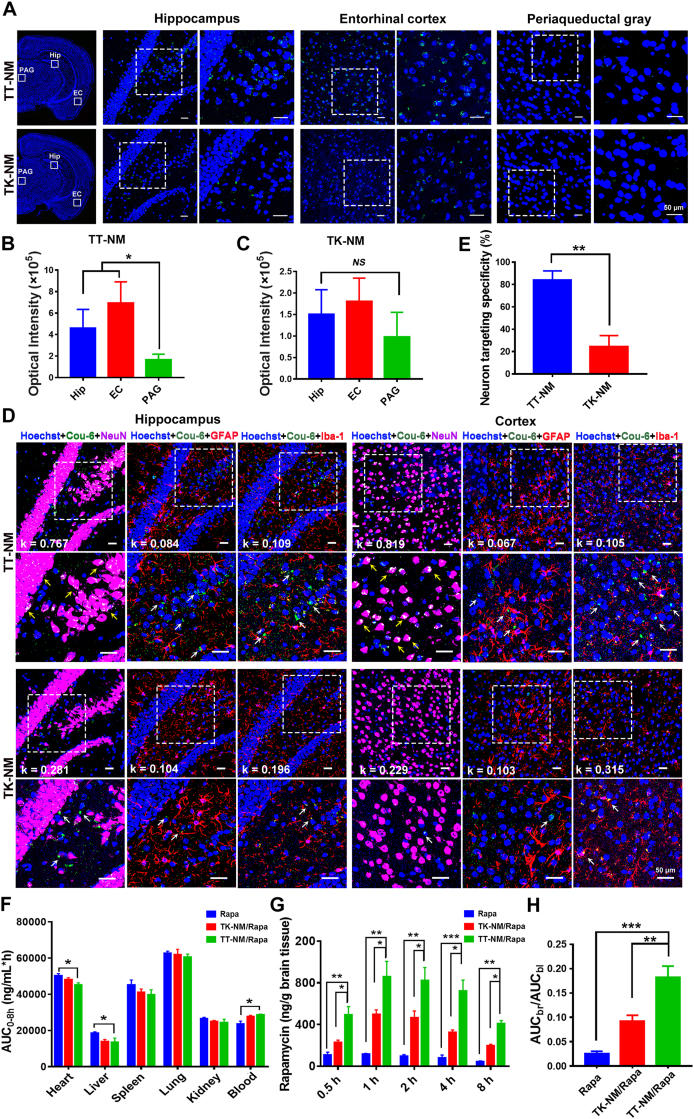
Targeting capability to central neurons and biodistribution of micelles. Following intravenous administration of micelles containing coumarin-6 (Cou-6, a hydrophobic fluorescent probe widely used for in vivo nanosystem tracking [27,28]) in 9-month-old 3 × Tg-AD mice, more intense fluorescence signals were observed in the brain of TT-NM group than that of unmodified micelle group (Fig. 2A). Noteworthily, the distribution of TT-NM was more concentrated in the hippocampus and entorhinal cortex, where Aβ deposition and neurofibrillary tangles were most frequently found [29], with 2.30-fold and 2.65-fold higher accumulation of TT-NM than that in the periaqueductal gray (a site less affected by AD) [30], respectively, whereas TK-NM did not exhibit any preferential accumulation in specific brain regions (Fig. 2B and C). This phenomenon can be attributed to a predominately high expression of trisialoganglioside GT1b in the cerebral cortex and hippocampal formation [31], leading to a differential distribution of micelles in the brain mediated by selective GT1b binding of TPL [24]. Furthermore, we evaluated the neuron-targeting specificity of TT-NM. Neurons, microglia and astrocytes in the cerebral cortex and hippocampus were visualized separately with NeuN staining in purplish red and Iba-1 or GFAP staining in red. As shown in Fig. 2D and E, the green fluorescence of TT-NM/Cou-6 was mainly co-localized with neurons but scarcely concentrated in microglia or astrocytes, with a 3.42-fold enhancement of the neuron targeting specificity (84%) compared with the TK-NM/Cou-6 group (24.6%). Correspondingly, our previous study has demonstrated that the inclusion of four-glycine linker enhanced the structural flexibility of TPL and facilitated its hydrogen bond interactions with GT1b receptors overexpressed on neurons [24,32]. Together, these results suggested that the modification of TPL facilitated the nanocarriers to penetrate BBB and preferentially accumulated in neurons in AD lesions.
Fig. 2.
Targeting capability to central neurons and biodistribution of TT-NM. (A) Representative confocal images of fluorescence distribution of TT-NM/Cou-6 or TK-NM/Cou-6 in different brain regions of 3 × Tg-AD mice at 1 h post injection (0.4 mg Cou-6/kg). The magnified insets of the white boxes of their respective images were shown on the right side respectively. Hip, hippocampus; EC, entorhinal cortex; PAG, periaqueductal gray. TT-NM/Cou-6 or TK-NM/Cou-6, green; Nuclei stained with Hoechst 33,342, blue. (B–C) Quantitative analysis of fluorescence intensity of TT-NM/Cou-6 (B) or TK-NM/Cou-6 (C) in the indicated mouse brain regions of Fig. 2A by Image J. 6 images fields were chosen. (D) Representative confocal images of fluorescence distribution of TT-NM/Cou-6 or TK-NM/Cou-6 in the hippocampus and cortex of 3 × Tg-AD mice at 1 h post injection. Lower row, the magnified insets of the white boxes of their respective images in the upper row. TT-NM/Cou-6 or TK-NM/Cou-6, green (white arrows); Nuclei stained with Hoechst 33,342, blue; Neurons stained with NeuN antibody, purplish red; Microglia stained with Iba-1 antibody, red; Astrocytes stained with GFAP antibody, red. Cell co-localized with Cou-6-loaded micelles were denoted by yellow arrows. The Manders' coefficients k indicating the degree of overlap of Cou-6-loaded micelles and different cellular markers were calculated by Image J. (E) Neuron targeting specificity was calculated by the fluorescence area ratio of Cou-6 co-localized with neurons and total Cou-6 signals. 6–8 images fields were chosen. (F) Area under the curve (AUC0–8h) of rapamycin in blood and main organs after intravenous injection of rapamycin solution (Rapa), TK-NM/Rapa and TT-NM/Rapa, respectively (n = 3). (G) Concentration-time profiles of rapamycin in brain after intravenous injection of three rapamycin preparations, respectively. (n = 3 mice/time point). (H) AUC ratios of brain to blood. (B–C) and (E–H) Data are presented as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, between the compared groups. NS, no significant difference.
We further investigated in vivo biodistribution of rapamycin preparations after intravenous administration by detecting the amounts of rapamycin in whole blood and tissues. TK-NM/Rapa and TT-NM/Rapa exhibited a slightly prolonged blood circulation compared to free rapamycin, and their area under the blood concentration-time curve (AUCblood) were 1.17- and 1.21-fold higher than that of free rapamycin, respectively (Fig. S8 and Table S1). At 8 h post-injection, there was a 9.9% and 27.4% reduction of AUCheart and AUCliver of TT-NM/Rapa relative to free rapamycin, respectively, indicating that TT-NM/Rapa were less prone to accumulate in the main organs (Fig. 2F). It was found that free rapamycin was poorly penetrated into brain, which was mainly restricted by P-glycoprotein efflux transport as previously reported [13]. TT-NM/Rapa presented the highest brain accumulation of rapamycin (1.83% ID/g brain at 1 h post-injection), and the corresponding brain/plasma ratio (AUCbrain/AUCblood) reached 0.182, about 1.98- and 7.11-fold higher than TK-NM/Rapa (0.092) and free rapamycin (0.0256), respectively (Fig. 2G and H). Hence, it was proved that TT-NM/Rapa possessed an enhanced potential for efficient drug delivery to brain.
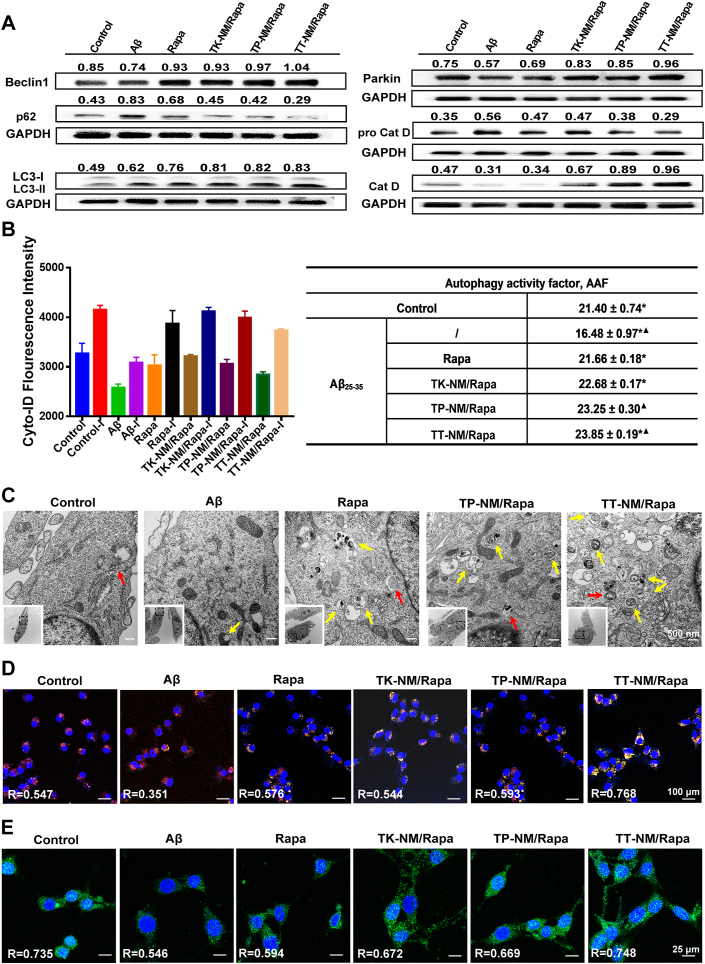
Upregulation of the autophagic flux by TT-NM/Rapa in vitro. To characterize autophagic flux, the expression of representative proteins involved in general autophagic-lysosomal pathway, including Beclin-1, Parkin, LC3-II, p62/SQSTM1, cathepsin D and its precursor, were monitored by Western blotting. Aβ25-35, a bioactive and neurotoxic fragment of full-length Aβ1-40 and Aβ1-42, was used for the establishment of AD model with autophagic dysregulation in vitro [[33], [34], [35]]. Exposure of HT22 cells to 50 μM Aβ25-35 for 48 h resulted in a decrease of Beclin-1 and Parkin proteins, suggesting a decline of autophagy induction. Besides, impaired clearance of autophagosomes might occur in Aβ25-35-treated cells, manifested by the accumulation of intracellular LC3-II and p62/SQSTM1 proteins. Furthermore, the overexpression of pro cathepsin D and a decline of its mature form also established the lysosomal hydrolysis defects in Aβ25-35-treated cells (Fig. 3A and Fig. S9). TT-NM/Rapa treatment at 100 nM of rapamycin (an optimized dose could achieve favorable therapeutic outcomes without neurotoxicity on HT22 cells, Fig. S10) strongly upregulated the expression of Beclin-1 and Parkin proteins (1.40- and 1.66-fold, respectively), much more effective than other rapamycin preparations. We also detected a significant elevation of LC3-II (1.34-fold) and a decrease of p62/SQSTM1 proteins (by 64.5%), implying that the assembly and subsequent clearance of autophagosomes were accelerated by TT-NM/Rapa. Moreover, TT-NM/Rapa could restore the lysosomal turnover of cathepsin D and consequently improve the lysosomal hydrolysis (Fig. 3A and Fig. S9), which is essential for the degradation of neurotoxic proteins Aβ and p-tau [36,37].
Fig. 3.
Upregulation of the autophagic flux by TT-NM/Rapa in vitro. (A) Western blot analysis of autophagic- and lysosomal-related proteins in neuronal cells. Aβ25-35-treated HT22 cells were incubated with Rapa, TK-NM/Rapa, TP-NM/Rapa and TT-M/Rapa (100 nM/Rapa) at 37 °C for 24 h. (B) Autophagy activity factors (AAF) of Aβ25-35-treated HT22 cells incubated with various rapamycin preparations either in presence or absence of chloroquine (lysosomal inhibitor, I) at 37 °C for 6 h. Autophagic flux analysis was conducted using flow cytometry for the measurement of Cyto-ID-stained autophagosomes, and then the AAF of each condition was calculated according to the formula in the Method section. Data were presented as mean ± SD (n = 3). *p < 0.05, compared with the TP-NM/Rapa; ▲p < 0.01, compared with the Rapa group. (C) Representative TEM images of autophagosomes or autolysosomes in HT22 cells without treatment (control), or HT22 cells exposed to 50 μM Aβ25-35 alone, or incubated with 50 μM Aβ25-35 plus Rapa, TP-NM/Rapa or TT-NM/Rapa (100 nM/Rapa). Red arrows, autophagosomes; yellow arrows, autolysosomes. (D) Representative confocal microscopy images of neuronal cells immunostaining with Anti-LAMP-2 (Red), Anti-LC3B (Green), and Hoechst 33,342 (Nuclear stain, blue). Aβ25-35-treated HT22 cells were incubated with various rapamycin preparations at 37 °C for 1 h. (E) Representative confocal microscopy images of neuronal cells immunostaining with Anti-TFEB (Green) and Hoechst 33,342 (nuclear stain, blue). Aβ25-35-treated HT22 cells were incubated with various rapamycin preparations at 37 °C for 1 h. The Pearson's correlation coefficient R represented the overlap ratio calculated by Image J. n = 5.
Due to the complexity of the autophagic flux that involves multistep processes from autophagosome formation to clearance, it is essential to check whether intracellular autophagosome accumulation results from the increased formation of autophagosomes or their impaired fusion with lysosomes [38]. Therefore, autophagic flux analysis was performed in the presence and absence of chloroquine, a classical lysosomal inhibitor, for the combinatorial assessment of individual sample that allowed us to quantify the total amount of autophagosome turnover during that period. The value of autophagic activity factors (AAF) in AD group decreased to only 16.48, which substantiated the compromise of autophagy in Aβ25-35-treated cells (Fig. 3B). Treatment with TT-NM/Rapa gave rise to a markedly enhanced autophagic activity with an AAF value of 23.85, the highest one among all four rapamycin formulations.
TEM analysis was further applied to examine the presence of autophagic structures within cells. As was shown in Fig. 3C, autophagosomes were scarcely observed in the untreated or Aβ25-35-treated controls, suggesting Aβ25-35 exposure did not cause increased formation of autophagosomes in HT22 cells. In contrast, incubation with rapamycin formulations led to a remarkable increase in the number of autophagosomes and autolysosomes, especially for the TT-NM/Rapa group. The robust activation of autophagic flux by TT-NM/Rapa were also supported by immunostaining detections of LC3 and LAMP2, markers of autophagic and lysosomal vesicles, respectively [39]. It was observed that the green fluorescence signals of LC3 were significantly enhanced and apparently co-localized with the red fluorescence signals of LAMP2 in the TT-NM/Rapa group (Pearson's correlation coefficient R = 0.768), suggesting that TT-NM/Rapa not only elevated the formation of autophagosomes but also promoted their fusion with lysosomes as well (Fig. 3D).
To explain the enhancement of lysosomal activities by TT-NM/Rapa, we also monitored the subcellular localization of TFEB upon cell treatment with various rapamycin preparations. TFEB is a key regulator of the Coordinated Lysosomal Expression and Regulation (CLEAR) network modulating lysosomal function and biogenesis, whose transcriptional activity is activated upon nuclear translocation [40,41]. As expected, TFEB mainly retained in the cytoplasm of Aβ25-35-treated cells. After incubation with rapamycin-loaded micelles, the intensity of green fluorescence was found to increase significantly accompanied by observable nuclear translocation. The Pearson's correlation coefficients obtained were 0.672, 0.669, and 0.748 for the TK-NM/Rapa, TP-NM/Rapa, and TT-NM/Rapa groups, respectively (Fig. 3E). These results hinted that TT-NM/Rapa promoted TFEB-mediated lysosomal activation most effectively, which corroborated with the results of Western blotting analysis as aforementioned.
Collectively, TT-NM/Rapa exhibited superior autophagy-inducing capability and ameliorated multiple stages of the autophagic flux.
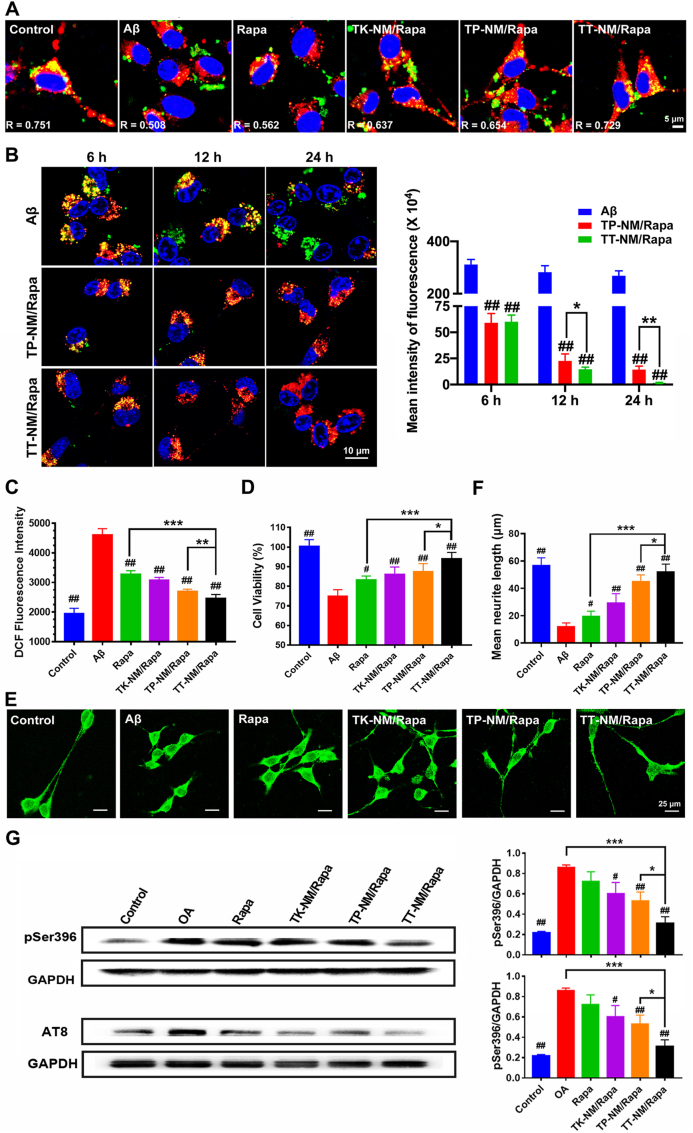
Neuroprotective effects of TT-NM/Rapa by accelerating the clearance of Aβ and p-tau in vitro. To investigate whether the upregulation of autophagic flux by TT-NM/Rapa could improve cellular clearance of intracellular Aβ, we monitored the phagocytosis and degradation of FAM-labeled Aβ1-42 (FAM-Aβ1-42) by lysosomes in HT22 cells. FAM-Aβ1-42 could be easily internalized and accumulated within the cytoplasm, but less co-localized with lysosomes in Aβ1-42 treated cells (R = 0.508). Moreover, there were still substantial amounts of Aβ1-42 within cells 24 h post-incubation, indicating the impaired clearance of intracellular Aβ (Fig. 4A and B). This is related to the strong toxicity of Aβ to lysosomes, as evidenced by the presence of a few lysosomes in Aβ1-42 treated cells after 24 h incubation. The reduction of the lysosomal phagocytosis and hydrolysis of Aβ1-42 was reversed by treatment with TT-NM/Rapa, as most of the internalized Aβ1-42 co-localized with the lysosomes in the HT22 cells (R = 0.729), and a gradual decrease in intracellular Aβ1-42 was observed during the prolonged incubation time. This result confirmed that TT-NM/Rapa facilitated lysosomal digestion of neurotoxic proteins through the activation of autophagic flux.
Fig. 4.
Neuroprotective effects of TT-NM/Rapa by accelerating the clearance of Aβ and p-tau in vitro. (A) Fluorescence monitoring of the intracellular distribution of FAM-Aβ1-42. HT22 cells were pre-incubated with 2.5 μM Aβ1-42 and various rapamycin formulations (100 nM/Rapa) at 37 °C for 24 h. The cells were further incubated with 2 μg/mL of FAM-labeled Aβ1-42 for 3 h. Healthy HT22 cells without any pretreatment served as the control. Green, FAM-Aβ1-42; Red, LysoTracker Red staining lysosomes; Blue, Hoechst 33,324 staining nuclei. The Pearson's correlation coefficient R demonstrated the overlap ratio of lysosomes and Aβ1-42. (B) Fluorescence monitoring of the intracellular clearance of FAM-Aβ1-42. HT22 cells were pre-incubated with 2.5 μM Aβ1-42 for 24 h in the presence or absence of TT-NM/Rap or TP-NM/Rapa (100 nM/Rapa). Following treated with FAM-Aβ1-42 for 3 h, the cells were further cultured in DMEM medium at 37 °C for 6, 12 and 24 h, respectively. Quantitative analysis of the fluorescence intensity of FAM-Aβ1-42 in cells was performed by Image J. (n = 6). Green, FAM-Aβ1-42; Red, lysosomes; Blue, nuclei. (C) TT-NM/Rapa inhibit Aβ25-35-induced oxidative stress. Aβ25-35-treated HT22 cells were incubated with various rapamycin formulations at 37 °C for 24 h. ROS level in neuronal cells staining with DCFH-DA probe was detected by flow cytometry. (n = 3). (D) Cell viability measured by MTT assay for HT22 cells exposed to Aβ25-35 alone or co-incubated with various rapamycin formulations at 37 °C for 24 h (n = 6). (E) TT-NM/Rapa promote neurite outgrowth. Representative confocal microscopy images of neuronal cells immunostaining with Anti-MAP-2 (Green) after exposure to Aβ25-35 alone or co-incubation with various rapamycin formulations at 37 °C for 24 h. (F) Quantitative analysis of the neurite length of the MAP2 immunostained HT22 cells in Fig. 4D by Image J. (n = 5). (G) TT-NM/Rapa promote the clearance of intercellular p-tau. Okadaic acid (OA)-treated HT22 cells were incubated with Rapa, TK-NM/Rapa, TP-NM/Rapa and TT-M/Rapa at 37 °C for 24 h. Intercellular p-tau (pSer 396 and AT8) levels were measured by Western blot analysis. (B–D) and (F–G) Data were presented as mean ± SD. #p < 0.05, ##p < 0.01, compared with Aβ or OA group; *p < 0.05, **p < 0.01, ***p < 0.001, compared with TT-NM/Rapa group.
Correspondingly, we observed remarkable effects of TT-NM/Rapa against Aβ25-35-induced oxidative stress and neurotoxicity in vitro. Exposure of HT22 cells to 50 μM of Aβ25-35 for 48 h caused severe oxidative stress, evinced by a dramatic increase in intracellular ROS levels to 236% compared with that of the normal control. Besides, the viability also decreased to 74.5% in Aβ25-35-treated cells (Fig. 4C and D). TT-NM/Rapa treatment reversed the levels of ROS comparable to the normal control and enhanced the cell viability to 93.9%. Flow cytometric analysis of apoptosis also verified that the presence of TT-NM/Rapa could increase the cell survival to 90.2% as compared to 71.7% of survival for Aβ25-35-treated cells (Fig. S11). Aβ25-35 exposure resulted in severely impaired neurite outgrowth of HT22 cells, displaying a 79.2% decrease in neurite length (Fig. 4E and F). Rapamycin formulation treatment rescued neurite morphology and elongation to some extent. Particularly, TT-NM/Rapa exhibited the most remarkable effects in promoting neurite extension, and the recovery of neurite length was similar to that of the control. Taken together, these findings clearly showed that TT-NM/Rapa provided effective neuroprotection against Aβ25-35 in vitro by accelerating the elimination of toxic Aβ25-35.
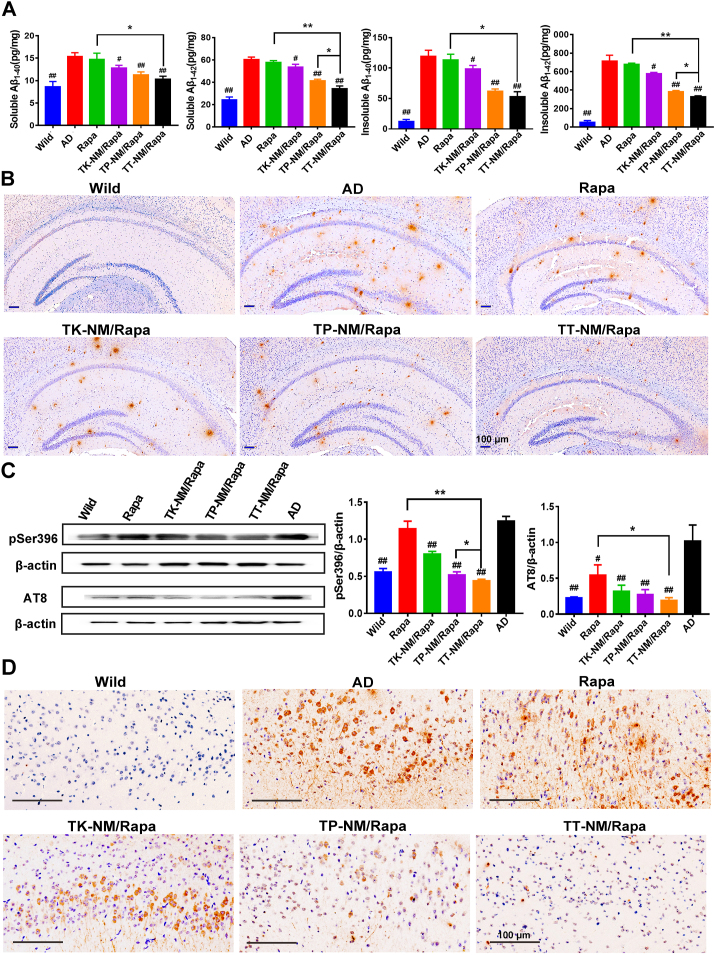
Rapamycin can promote autophagic clearance of p-tau and block its production via glycogen synthase kinase-3β (GSK-3β) inhibition, and hence decrease p-tau [42]. To test the effects of TT-NM/Rapa on tau hyperphosphorylation, we assessed the levels of p-tau in cells exposed to okadaic acid (OA) alone or subsequently treated with different rapamycin formulations. Western blotting analysis detected a significant increase in tau phosphorylation at Ser 386 and AT8 (Ser202/Thr205) site in the OA group compared to the normal control, respectively. Levels of p-tau were all lowered by rapamycin treatment to some extent. In particular, TT-NM/Rapa exhibited the strongest clearance on p-tau, demonstrated by 81.2% and 62.9% reduction of p-tau at Ser 396 and AT8 site compared with that of the OA group, respectively, establishing that TT-NM/Rapa inhibited accumulation of p-tau most significantly (Fig. 4G).
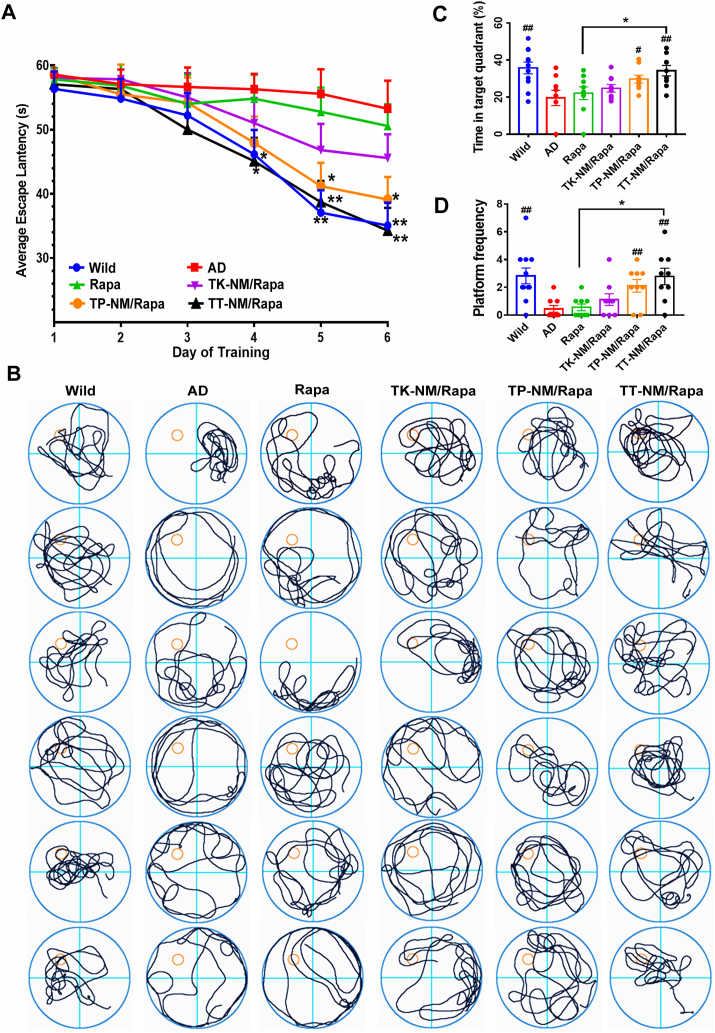
Rescue of the spatial cognitive deficits in triple-transgenic mice by TT-NM/Rapa. To investigate the effects of TT-NM/Rapa on cognition impairment of triple-transgenic (3 × Tg-AD) mice, a mouse model carries mutated human APP(Swe), tau (P301L) and PS1(M146V) transgenes that most closely characterizes the human AD pathology [43,44], the Morris water maze (MWM) tests were performed following daily administration with rapamycin solution or different rapamycin-loaded micelles for 4 consecutive weeks. Severe spatial learning disorder was established in 3 × Tg-AD mice aged 9–10-month-old [45], presented as no significant improvement regarding their escape latencies during the 6-day trials. After treatment with targeted micelles, 3 × Tg-AD mice exhibited improved spatial learning ability as they spent less time to find the submerged platforms. Specifically, the latencies of TT-NM/Rapa-treated mice were significantly shorter than the AD control since the 4th day of trial, comparable to that of the wild-type (WT) control (Fig. 5A). Similarly, in the probe trial where the platform was removed, the swimming path of TT-NM/Rapa group was preferentially towards the original platform area, whereas the untreated AD mice performed poorly concerning platform search. Moreover, the percentage of time that TT-NM/Rapa-treated mice spent in the targeted quadrant and their times of crossing of the previous platform area was significantly higher than that of the rapamycin solution and AD control group, demonstrating its prominent therapeutic effects to rescue spatial memory loss in 3 × Tg-AD mice (Fig. 5B–D).
Fig. 5.
Rescue of the spatial cognitive deficits in triple-transgenic mice by TT-NM/Rapa. 3 × Tg-AD mice at 9–10 months of age were i. v. Injected with saline or various rapamycin preparations (0.24 mg Rapa/kg/d) for 4 weeks consecutively, and Morris water maze experiment was carried out on the 23rd day after treatment. Wild-type mice treated with saline were used as the normal control (WT). (A) Mean escape latency in the Morris water maze experiment of the treated mice. *p < 0.05, **p < 0.01, compared with AD group. (B) Representative swimming path. (C) Percentage of time in targeted quadrant and (D) the times of crossing of the expected-platform area of the treated mice. #p < 0.05, ##p < 0.01, compared with AD group; *p < 0.05, compared with rapamycin solution. (A) and (C–D) Data were presented as mean ± SEM. (n = 9–11).
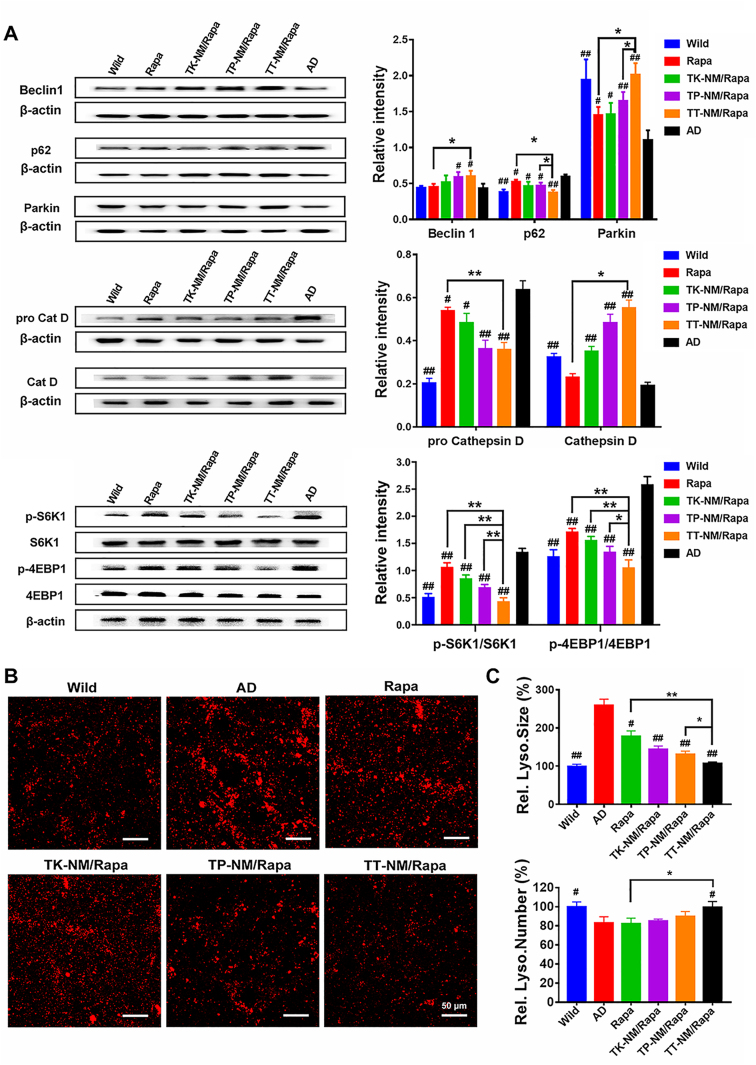
Restoration of autophagic flux in triple-transgenic mice by TT-NM/Rapa. All mice were immediately sacrificed and whole-brains were harvested following the behavioral tests. We examined the expression of representative proteins that control the different stages of the complete autophagic flux in the brains of 3 × Tg-AD and WT mice. We found that treatment with rapamycin solution alone slightly altered the expression of multiple autophagic- and lysosomal-related proteins in brains, whereas the administration of TT-NM/Rapa caused remarkable upregulation of Beclin-1, Parkin and cathepsin D proteins, and reduced p62/SQSTM1 proteins back to WT levels (Fig. 6A). TT-NM/Rapa boosted autophagy through downregulation of mTOR signaling pathways by rapamycin [46,47], as reflected by its most significant inhibition on phosphorylation of two main substrates of the mTORC1 complex, namely eukaryotic translation initiation factor (4 E-binding protein 1 4 E-BP1) and ribosomal protein S6 protein kinase 1 (S6K1) (Fig. 6A). In addition, LAMP2-immunostaining studies also showed that TT-NM/Rapa treatment successfully restored the abnormal enlargement of lysosomes to WT average size and increased the number of lysosomes to 120% relative to the AD control, which clearly illustrated that TT-NM/Rapa treatment achieved the most significant recovery of impaired lysosome reformation in 3 × Tg-AD mice (Fig. 6B and C).
Fig. 6.
Restoration of autophagic and lysosomal function in triple-transgenic mice by TT-NM/Rapa. (A) Western blot analysis of autophagic-lysosomal-related proteins as well as two downstream substrates of mTOR in the brain of 3 × Tg-AD mice treated with various rapamycin preparations or saline. (n = 3). (B) Representative confocal images of immunofluorescence staining of lysosomes in cortical region from 3 × Tg-AD mice treated with various rapamycin preparations or saline. Red, LAMP2-stained lysosomes. Blue, Hoechst-33342-stained nuclei. (C) Quantitative analysis of relative size and number of lysosomes per image field of Fig. 6B by Image J. (n = 6). (A) and (C) Data were presented as mean ± SD. #p < 0.05, ##p < 0.01, compared with AD group; *p < 0.05, **p < 0.01, between the compared groups.
Attenuation of Aβ burden and p-tau in triple-transgenic mice by TT-NM/Rapa. Senile plaques composed of misfolded Aβ and neurofibrillary tangles formed by p-tau represent two main neuropathological hallmarks of AD, strongly indicating that the proteostasis defects play a critical role in the onset and progression of AD [2]. We measured the levels of both soluble and insoluble Aβ1-40 and Aβ1-42 in whole-brain fractions of 3 × Tg-AD mice by ELISA kits. As anticipated, both soluble and insoluble Aβ levels were markedly increased in the 3 × Tg-AD brains, about 1.8–2.5 times and 9.8–13.7 times higher than that of the age-matched counterparts. It was shown that both soluble and insoluble Aβ levels declined to some extent after treatment with different rapamycin formulations. Especially, we observed a 43.2% reduction in soluble Aβ1-42 levels, a neurotoxic form of Aβ species, in the TT-NM/Rapa group, coordinated with a 54.0% decrease in its insoluble Aβ1-42 levels (Fig. 7A). We further calculated the Aβ1-42/Aβ1-40 protein ratio, a key indicator whose increase was identified as a more accurate precursor of deleterious events leading to AD, rather than the changes in the absolute level of Aβ1-42 [48]. Among all rapamycin preparations, TT-NM/Rapa treatment resulted in the lowest ratio of Aβ1-42 to Aβ1-40, which was also significantly lower than that of AD group (Fig. S12A), suggesting that it effectively reduced amyloid pathology in AD brains. Besides, the results of the immunohistochemical staining of Aβ deposition also coincided with ELISA data. Mice treated with TT-NM/Rapa exhibited apparently decreased numbers of plaques in the brains (Fig. 7B), and the area percentage of Aβ plaques in the hippocampal region of 3 × Tg-AD mice was reduced by 69.2% in the TT-NM/Rapa group, while the corresponding value in the TP-NM/Rapa group was still 1.94-fold higher than TT-NM/Rapa group, underlining the importance of complete rapamycin release from TT-NM nanocarriers for efficient autophagic flux regulation and subsequent clearance of neurotoxic proteins (Fig. S12B).
Fig. 7.
Attenuation of Aβ burden and tau hyperphosphorylation in triple-transgenic mice by TT-NM/Rapa. (A) Quantification of the levels of soluble and insoluble Aβ1-40 and Aβ1-42 in 3 × Tg-AD mice treated with various rapamycin preparations or saline by ELISA kits. (n = 5). (B) Representative immunohistochemical images of amyloid plaque deposit in the brain of 3 × Tg-AD mice treated with various rapamycin preparations or saline. Aβ plaque was stained with anti-β-Amyloid1-42 antibody. (C) Western blot analysis of p-tau proteins in the brain of 3 × Tg-AD mice treated with various rapamycin preparations or saline. (n = 3). (D) Representative immunohistochemical images of neurofibrillary tangles in the cortex of 3 × Tg-AD mice treated with various rapamycin preparations or saline. Neurofibrillary tangles were stained with anti-Phospho-tau (Ser202, Thr205) antibody. (A) and (C) Data were presented as mean ± SD. #p < 0.05, ##p < 0.01, compared with AD group; *p < 0.05, **p < 0.01, between the compared groups.
In 3 × Tg-AD mice aged 9–10 months, the levels of p-tau at Ser 386 and AT8 site were upregulated (Fig. 7C). The targeted micelles loaded with rapamycin significantly diminished the levels of p-tau, especially TT-NM/Rapa treatment exhibited the strongest inhibition effect. The marked reduction of p-tau by TT-NM/Rapa was further confirmed by immunohistochemical analysis. Hyperphosphorylation of tau at8 is commonly recognized as a marker for neurofibrillary tangles and neuropil threads, thus serves as an indicator of p-tau lesions [49]. Among all four rapamycin formulations, TT-NM/Rapa most remarkably downregulated the AT8-immunoreactivity in the cortex of the treated mice, where few neurofibrillary lesions could be found (Fig. 7D).
Overall, these results indicated that the activation of neuronal autophagic flux by TT-NM/Rapa could lead to enhanced clearance of Aβ and p-tau, thereby effectively inhibiting the generation of Aβ deposition and neurofibrillary tangles in 3 × Tg-AD brains.
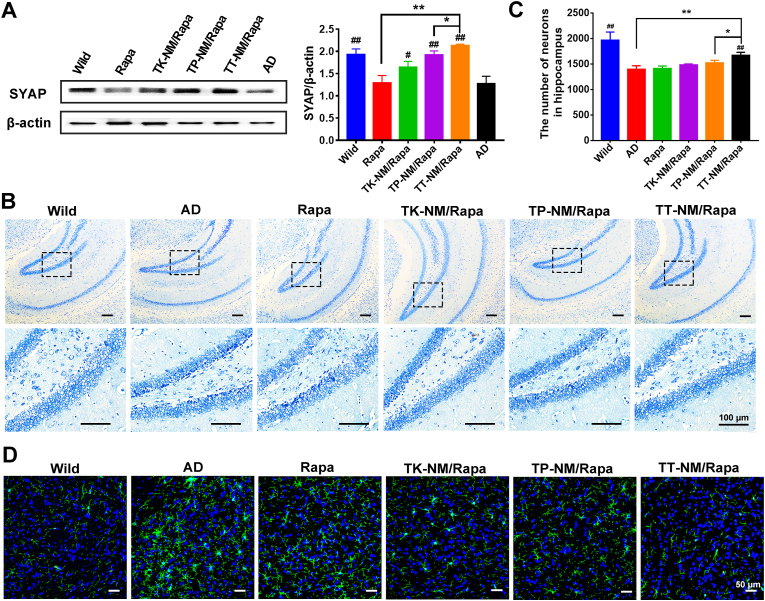
Upregulation of synaptophysin and alleviation of hippocampal damage by TT-NM/Rapa. Synaptophysin, a major glycoprotein of the synaptic membrane, is critically involved in synapse formation and the assembly of synaptic vesicles. Therefore, it is recognized as a reliable marker of synaptic plasticity and function [50]. The expression of synaptophysin was markedly downregulated in 3 × Tg-AD mice at the age of 9–10 months old (Fig. 8A), which closely correlated with the cognitive decline as described above. Treatment with various rapamycin micelles achieved different degrees of increase in synaptophysin level, remarkably, the synaptophysin level increased to 110.3% relative to the WT control in the TT-NM/Rapa group. As reported, Aβ1-42 can weaken neurotransmission and impair synaptic plasticity via GABAA receptor inhibition [51]. We could therefore infer that TT-NM/Rapa protects against synaptic damage by promoting the clearance of intraneuronal Aβ. Accordingly, the results of Nissl staining also indicated that TT-NM/Rapa therapy resulted in a prominent inhibition of neuronal loss in 3 × Tg-AD mice, presented as abundant Nissl bodies and a notable increase in the number of neurons in the hippocampus region. In contrast, obvious cell death and shrinkage still existed in the TP-NM/Rapa group after treating with the same rapamycin dose for 28 days (Fig. 8B and C). Taken together, above results strongly supported the superior neuroprotective ability of TT-NM/Rapa to other rapamycin preparations.
Fig. 8.
Alleviation of hippocampal neuron damage and inhibition of abnormal microglia activation by TT-NM/Rapa. (A) Western blot analysis of synaptophysin (SYAP) protein in the brain of 3 × Tg-AD mice treated with various rapamycin formulations or saline. (n = 3). (B) Representative images of Nissl staining of hippocampal region from 3 × Tg-AD mice treated with various rapamycin formulations or saline. Image in lower row of each group were the enlarged regions of the black boxes in the upper images. (C) Quantitative analysis of the number of neurons in the entire hippocampal region of Fig. 8B by Image J. (n = 3). (D) Representative confocal images of immunofluorescence staining of Iba1 in cortical region from 3 × Tg-AD mice treated with various rapamycin preparations or saline. Green, Iba1-stained activated microglia; Blue, nuclei stained by Hoechst 33,342. (A) and (C) Data were presented as mean ± SD. #p < 0.05, ##p < 0.01, compared with AD group; *p < 0.05, **p < 0.01, between the compared groups.
Inhibition of abnormal microglia activation by TT-NM/Rapa. Growing evidence has shown that the continuous accumulation of Aβ or neuronal damage can stimulate microglia to be over-activated, leading to neuroinflammatory cascade and furthering AD progression [52]. As shown in Fig. 8D, activated amoeboid microglia appeared to dominate the cortical region in 9- to 10-month-old 3 × Tg-AD mice. Rapamycin solution, TK-NM/Rapa, or TP-NM/Rapa treatment attenuated the microgliosis to some extent, demonstrated by the decreased number of hypertrophic amoeboid microglia in the brains. Noteworthily, the morphologies of most microglia transferred from amoeboid to less ramified shape in the TT-NM/Rapa group, whose fluorescence intensity also declined to the level comparable to the WT control. The results indicated that TT-NM/Rapa mitigated microglial hyperactivities of 3 × Tg-AD mice, as a result of that TT-NM/Rapa alleviated Aβ burden and protected neurons by restoring autophagic flux.
In vivo biocompatibility. As good biocompatibility of administered formulations is important and favorable for the long-term management of AD, we further examined the biocompatibility profiles of TT-NM/Rapa. There was no obvious alteration of mouse serum hematological indicators of liver and kidney function after TT-NM/Rapa treatment for 28 days, which were comparable to that of the WT control group. In addition, no histopathological changes were found in major organs, indicating that the micelles did not cause undesirable side effects in vivo (Fig. S13).
3. Conclusion
In this study, we developed a ROS-responsive targeted micelle (TT-NM/Rapa) to precisely deliver rapamycin to neurons in AD lesions and achieve efficient release in response to elevated intracellular levels of ROS, which greatly strengthened the regulation of rapamycin on autophagic flux for AD therapy. TT-NM/Rapa restored autophagic clearance of neurotoxic Aβ and p-tau protein through inducing autophagy, facilitating autophagosome-lysosome fusion, and enhancing lysosomal activities. Hence, it greatly alleviated the microgliosis, rescued neuronal loss and hippocampal damage, and ameliorated cognitive defects in 3 × Tg-AD transgenic mice. Our results present the great potential of rapamycin-loaded nano-system to better maintain neuronal proteostasis via autophagic flux restoration for AD therapy.
4. Materials and methods
Materials. Rapamycin (Rapa), coumarin-6 (Cou-6) and thiazolyl blue tetrazolium bromide (MTT) were purchased from Sigma-Aldrich Inc. (St.Louis, MO, USA). TPL (l-TGNYKALHPHNGGGGG-HLNILSTLWKYRC), Aβ25-35, Aβ1-42 and FAM-Aβ1-42 were synthesized by the Chinese Peptide Co., Ltd. (Hangzhou, China). 1,1′-Dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI), 3,3′-dioctadecyloxacarbocyanine perchlorate (DiO), Total ROS assay kit and Cell Counting Kit-8 (CCK-8) were purchased from Beyotime Biotechnology Co., Ltd. (Shanghai, China). CYTO-ID Autophagy Detection Kit and BCA Protein Assay Kit were attained from Enzo Life Sciences, Inc. (Farmingdale, NY, USA) and BD Biosciences, Inc. (Franklin Lakes, NJ, USA) respectively. Amyloid-β1-40 Human ELISA Kit and Amyloid-β1-42 Human ELISA Kit were purchased from Elabscience Biotechnology Co., Ltd. (Shanghai, China). Lysotracker Red DND-99, Gibco Dulbecco's Modified Eagle medium (DMEM), fetal bovine serum (FBS), penicillin-streptomycin solution and trypsin-EDTA 0.25% solution were purchased from Thermo Fisher Scientific, Inc. (Eugene, OR, USA). The mouse brain microvascular endothelial cells (bEnd.3) and mouse hippocampal neuronal cells (HT22) were obtained from the Chinese Academy of Sciences Cell Bank (Shanghai, China). All chemicals used were analytical or reagent grade.
Animals. ICR mice (18–22 g, male) were obtained from Sino-British Sippr/BK Lab Animal Ltd. (Shanghai, China). Triple-transgenic AD (3 × Tg-AD) mice (B6; 129-Tg (APPSwe, tauP301L)1Lfa Psen 1tm1Mpm/Mmjax, male and female in half) were obtained from the Jackson Laboratory (Bar Harbor, ME, USA). Age- and gender-matched wild-type (WT) mice were purchased from Shanghai Model Organisms Center (Shanghai, China). All the mice were housed under specific pathogen-free conditions with an alternating 12 h light/dark cycle, controlled temperature at 23 ± 1 °C, relative humidity at 50 ± 5% and ad libitum feeding to food and water. The animal studies were performed under the approval of institutional animal care and use committee (IACUC), School of Pharmacy, Fudan University.
Preparation of micelles. Solvent diffusion method was applied to prepare ROS responsive micelles (TK-NM/Rapa) according to our pre-experiments. In brief, 14 mg Meo-PEG-TK-PCL, 6 mg maleimide-PEG-PCL (Mal-PEG-PCL) and 1.0 mg rapamycin (5% mass ratio of polymeric materials) were dissolved in 4 mL of acetone under water-bath sonicating, followed by the rapid injected into 10 mL of stirring distilled water and stirring last for additional 1 h at 400 r/min. Then the acetone was completely removed by rotary evaporation to obtain TK-NM/Rapa, and the resulting micelles were separated from unentrapped drug by sepharose CL-4B gel filtration. Fluorescent-probe-loaded micelles (Cou-6 or DiO/DiI) were prepared with almost the same preparation method by replacing the rapamycin with corresponding fluorescent-probe (0.04 mg, 0.2% mass ratio of polymeric materials).
ROS responsive targeted micelles (TT-NM/Rapa) were prepared with the same mass ratio of Meo-PEG-TK-PCL, Mal-PEG-PCL and rapamycin as aforementioned. TPL peptide was conjugated on the surface of micelle by maleimide-thiol coupling reaction under nitrogen at room temperature for 4 h. Therefore, micelles with different modification density (5%, 10%, 15%) could be obtained by varying the molar ratio of TPL peptide to maleimide (1:6, 1:3, 1:2), termed as 5%TT-NM, 10%TT-NM and 15%TT-NM, respectively. Besides, TPL-modified non-responsive micelles (TP-NM/Rapa) composed of Meo-PEG-PCL and Mal-PEG-PCL were prepared by the above method as the control group.
Characterization of micelles. The particle size, polydispersity index (PDI) and surface charge of micelles in PBS containing 1 mM H2O2 or not were measured at 25 °C using Zetasizer Nano ZS (Malvern, UK). The critical micelle concentration (CMC) of TK-NM was determined by the change of vibrational band intensity of pyrene monomer as previously described [53]. The morphology of micelles was characterized by JEM-1230 transmission electron microscope (TEM, JEOL, Japan). In addition, HPLC-UV assay was applied for the examination of encapsulation efficiency (EE%) and drug-loading capacity (DLC%) according to the previous report [54].
In vitro cytotoxicity study. HT22 cells were seeded into a collagen-coated 96-well plate at a density of 1.0 × 104 cells/well and incubated with DMEM medium supplemented with 10% (w/v) FBS and 1% (w/v) penicillin-streptomycin (hereinafter referred to as regular DMEM) overnight. Then cells were treated with fresh media containing TK-NM or TT-NM at a range of concentrations (0.1, 0.2, 0.5, 1, 2 mg/mL) for 48 h, followed by the additional incubation with 10 μL of MTT (5 mg/mL) for 4 h. The culture media were replaced with 200 μL of DMSO to fully dissolve the generated formazan crystals afterwards. With UV-absorbance at 570 nm recorded by Multiskan Mk3 microplate reader (Thermo Fisher, USA), cell viability was expressed as the percentage of control.
ROS-mediated release of Rapa. TT-NM/Rapa and TP-NM/Rapa (containing 40 μg of Rapa) were transferred to dialysis bags (MWCO = 3500Da). The dialysis bags were completely submerged in 40 mL of PBS buffer containing 0.5% SDS as well as 0, 1 or 10 mM H2O2. The samples were incubated at 37 °C by continuous shaking at 100 r/min for 8 h. At the defined intervals, 400 μL of samples were withdrawn and replenished with the same volume of pre-warmed fresh medium. After 8 h, samples were also collected for calibration of the original drug loading after destroying the dialysis bags and vortex mixing. The concentration of samples with three replicates was determined by HPLC-UV assay, and the cumulative drug release from TT-NM/Rapa and TP-NM/Rapa was further calculated as described before [54].
FRET-based imaging of intracellular ROS-responsive drug release. HT22 cells were plated on collagen-coated confocal dishes (1.0 × 105/dish). As soon as cells grew to 70% confluency, cells were incubated with regular DMEM containing 100 μM H2O2 for 4 h to obtain a high-ROS neuronal model as previously reported [55]. After several washes with PBS buffer, cells were treated with TT-NM/DiO-DiI and TP-NM/DiO-DiI solution (DiO = 50 ng/mL) and incubated at 37 °C for 30 min. Then cells were gently washed with PBS and incubated with fresh regular DMEM for additional 1 h, followed by the 4% paraformaldehyde fixation and Hoechst 33,324 staining. Images at predetermined intervals were acquired by LSM 710 Confocal Microscope (Carl Zeiss, Germany) equipped with Argon (488 nm) and HeNe (546 nm) lasers as well as emission filter sets of 500–530 nm for green emission (DiO) and 555–655 nm for red emission (DiI or FRET). The fluorescence intensity of signals was quantified and the FRET efficiency (FR) was further calculated as I546/I488 by MATLAB.
In vivo evaluation of central neuron targeting. TK-NM/Cou-6 and TT-NM/Cou-6 were intravenously injected into 9-month-old 3 × Tg-AD mice at the dosage of 0.5 mg/kg Cou-6. One hour after administration, the mice were anesthetized, perfused with saline and following 4% paraformaldehyde. The brains were harvested and fixed in 4% paraformaldehyde for 48 h, followed by the gradient dehydration with sucrose solution and embedment with Tissue-Tek O·C.T compound (Sakura Finetek, USA). After being frozen at −80 °C overnight, the brain samples were cut into 8-μm sections and pasted onto slides. Later, the sections were incubated with blocking buffer containing 10% of normal goat serum and 0.2% of Triton X-100 at 37 °C for 1 h, and subsequentially incubated with anti-NeuN antibody (1:100 dilution, ab190565, Abcam), anti-Iba-1 antibody (1:100 dilution, ab178846, Abcam) or anti-GFAP antibody (1:200 dilution, ab68428, Abcam) for 1 h, respectively. Then the sections were rinsed extensively with PBS buffer and incubated with diluted secondary antibody (goat anti-mouse IgG H&L Alexa Fluor 594, 1:300 dilution, ab150116, Abcam; goat anti-rabbit IgG H&L Alexa Fluor 594, 1:300 dilution, ab150080, Abcam) at 37 °C for 1 h. After nuclei staining with Hoechst 33,324, the sections were examined under TCS-SP5 confocal microscope (Leica, Germany). Quantitative analysis of fluorescence intensity of Cou-6 signals in the mouse brain regions and Manders’ coefficients k of Cou-6-loaded micelles and different cellular markers was performed by Image J. Then neuron targeting specificity was calculated by the fluorescence area ratio of Cou-6 co-localized with neurons and total Cou-6 signals [24]. 6–8 images fields were chosen.
Biodistribution studies in mice. ICR mice were randomly divided into three groups (n = 15/group) and intravenously administered with TK-NM/Rapa, TT-NM/Rapa or free rapamycin at the same dose (1 mg/kg rapamycin). At predetermined time points (30 min, 1, 2, 4 and 8 h post-dose), three mice of each group were anesthetized, exsanguinated via cardiac puncture for blood sample collection. After transcardiac perfusion with physiological saline, the brain and main organs of mice were withdrawn, washed twice with saline, blotted to remove excess fluid, weighed and homogenized using SKSI homogenizer (Biheng Biotechnology, China, tissue: saline = 1:3, w/w). 10 μL of internal standard solution (tacrolimus, 20 ng/mL) was spiked with every aliquot of 100 μL blood sample or tissue homogenate. Following steps, including protein precipitation, rapamycin extraction and quantification analysis with API4000 Q-Trap LC-MS/MS (AB SCIEX, USA) were analogous to previous report [56]. The optimized mass spectrometry conditions were ESI-positive mode, 5.5 kV of ionspray voltage, 25 L/h of curtain gas flow and 500 °C of desolvation temperature. The rapamycin scan was selected at m/z 931.7 as parent ion and m/z 864.5 as daughter ion, while tacrolimus scan was set at m/z 821.6 and 768.5. Spectrometric data were processed by Analyst software, and corresponding pharmacokinetic parameters were calculated by PKSolver program using noncompartmental data analysis mode.
TEM characterization of autophagosomes and autolysosomes. HT22 cells were grown in 25 cm2 cell culture flasks to 70% confluency, followed by incubation with 50 μM Aβ25-35 for 48 h. Then cells were separately treated with rapamycin solution, TP-NM/Rapa or TT-NM/Rapa (100 nM/Rapa) for 1 h. Afterwards, the cultures were replaced with regular DMEM for another 2 h. The cells were subsequently washed with cold PBS for three times, and fixed with 2.5% glutaraldehyde at 4 °C in the dark. After 2 h of fixation, the cells were carefully scraped and collected for TEM sample preparation. The effect of different rapamycin preparations on autophagic activity was observed under Tecnai G2 F20 S-TWIN Transmission Electron Microscopy (FEI, USA).
Autophagic flux analysis. HT22 cells were seeded into a 24-well plate covered with rat-tail collagen and allowed to adhere. HT22 cells at 70–80% confluency were then exposed to 50 μM Aβ25-35 for 42 h. After being thoroughly washed with PBS, cells were treated with various rapamycin formulations either in presence or absence of chloroquine at 37 °C for 6 h. Meanwhile, untreated cells and cells solely treated with chloroquine served as the control [57]. The final concentrations of Rapa and chloroquine were fixed at 100 nM and 10 μM, respectively. After that, cells were stained with 1 μM CYTO-ID Green autophagy dye for 15 min. Then cells were gently washed with serum-free medium, harvested by trypsinization, and resuspended in 200 μL of PBS buffer before performing flow cytometry analysis (Ex = 480 nm, Em = 530 nm). The autophagy activity factor (AAF) of each condition was calculated according to the mean fluorescence intensity (MFI) of each sample and corresponding chloroquine-treated sample [58]:
Autophagy induction, TFEB nuclear translocation, intracellular Aβ1-42clearance and neurite outgrowth detected by immunofluorescence assay. To confirm the autophagy-inducing ability of rapamycin preparations in HT22 cells, autophagy was examined by immunostaining of LC3 autophagosome marker. With a density of 1.0 × 105 cells cultured in per sterile confocal dish at 37 °C overnight, HT22 cells were first treated with 50 μM of Aβ25‐35 for 48 h. After incubated with Rapa, TK-NM/Rapa, TP-NM/Rapa, TT-NM/Rapa (100 nM/Rapa) or regular DMEM for 1 h, respectively, cells were gently washed by serum-free medium and continued incubation with regular DMEM for another 2 h. Following being washed twice with cold PBS and fixed with 4% paraformaldehyde for 20 min, cells were permeabilized with 0.1% of Triton X-100 for 20 min and then blocked with 5% bovine serum albumin in PBS at 37 °C for 1 h. Subsequently, cells were immunostained with anti-LC3 antibody (1:200 dilution, ab192890, abcam) and anti-LAMP-2 antibody (1:200 dilution, 15,665, Cell Signaling) at 37 °C for 1 h, washed with PBS again, and then incubated with goat anti-mouse IgG H&L Alexa Fluor 488 (1:300 dilution, ab150113, Abcam) and goat anti-rabbit IgG H&L Alexa Fluor 594 (1:200 dilution, ab150080, Abcam) at 37 °C for another 1 h followed by nuclei staining with Hoechst 33,324. Images were captured by LSM 710 Confocal Microscope (Carl Zeiss, Germany) with excitation/emission spectra of 488/519 nm, 594/617 nm, and 350/461 nm for AF488, AF594 and Hoechst 33,324, respectively. The corresponding Pearson's correlation coefficient (R) of LC-3 and LAMP-2 immunofluorescence signals were analyzed by Image J software.
For assessment of nuclear translocation of TFEB stimulated by rapamycin formulations, HT22 cells were incubated with various rapamycin preparations (100 nM/Rapa) for 12 h after the indicated treatment with Aβ25-35 for 48 h. After fixation, permeabilization and blocking of non-specific staining, cells were incubated with anti-TFEB antibody (1:100 dilution, P19484, Bethyl Laboratories) and then probed with goat anti-mouse IgG H&L Alexa Fluor-488 (1:300 dilution, ab150113, Abcam) at 37 °C for 1 h. Following nuclei-stained with Hoechst 33,324, the TFEB subcellular distribution of HT22 cells was examined under LSM 710 Confocal Microscope. The corresponding Pearson's correlation coefficient (R) of TFEB signals and nucleic dye were analyzed by Image J software.
To evaluate the intracellular Aβ1-42 clearance, HT22 cells were pre-incubated with 2.5 μM Aβ1-42 and various rapamycin formulations (100 nM/Rapa) at 37 °C for 24 h. Then the cells were washed with PBS buffer three times and then exposed to 2 μg/mL of FAM-Aβ1-42 for another 3 h (evaluation of lysosomal phagocytosis). For the evaluation of lysosomal degradation, the culture medium containing FAM-Aβ1-42 was subseqnently discarded and replaced by fresh DMEM medium and further incubated for 6, 12 and 24 h, respectively. After removal of the medium and being washed three times, the cells were treated with Lysotracker Red DND-99 for lysosome staining and Hoechst 33,342 for nuclei staining successively. Images were captured using LSM 710 Confocal Microscope. The Pearson's correlation coefficient (R) of FAM-Aβ1-42 and lysosomes as well as the fluorescence intensity of FAM-Aβ1-42 in cells were analyzed by Image J software.
To characterize the morphology changes of neuronal cells, HT22 cells were treated with 50 μM of Aβ25‐35 for 24 h, followed by co-incubation with different rapamycin formulations (100 nM/Rapa) for another 24 h. Cells were fixed, permeabilized and immunostained following the abovementioned protocols, using anti-MAP-2 antibody (1:100 dilution, ab32454, Abcam) as the primary antibody and goat anti-rabbit IgG H&L Alexa Fluor-488 (1:200 dilution, ab150077, Abcam) as the secondary antibody, respectively. After nuclei staining with Hoechst 33,324, cells were visualized by LSM 710 Confocal Microscope, and the mean neurite length of each group was further quantified by Image J software.
Aβ25-35-induced cytotoxicity analysis. HT22 cells were cultured in a collagen-coated 96-well plate (1.0 × 105 cells/well) and treated with 50 μM Aβ25-35 at 37 °C for 24 h. Afterwards, cells were exposed to 50 μM Aβ25-35 or co-incubated with various rapamycin preparations (100 nM/Rapa) for an additional 24 h. Besides, the untreated cells were prepared parallel as the control. Following cells were added with 10 μL CCK solution per well and continued incubation at 37 °C for 45 min, the absorbance at 450 nm was measured by Multiskan Mk3 microplate reader (Thermo Fisher, USA) and the viability of HT22 cells was presented as a percentage of the control.
Intracellular ROS detection. HT22 cells were exposed to 50 μM Aβ25-35 for 24 h followed by the co-incubation with various rapamycin formulations (100 nM/Rapa) for additional 24 h as aforementioned. Then cells were thoroughly washed with PBS buffer, and subsequently incubated with 10 μM of DCFH-DA at 37 °C for 20 min. After that, cells were gently washed with serum-free medium to remove extracellular DCFH-DA probes, followed by appropriately trypsinized and transferred into a 96-well black microplate. The DCF fluorescence intensity was assessed by Infinite M1000 microplate reader (Tecan, Switzerland, Ex = 488 nm, Em = 525 nm).
Animal treatment. 3 × Tg-AD mice were randomly divided into five groups (n = 9–11/group, 9–10-month-old) with wild-type counterparts serving as the normal control group, among which there was no significant difference regarding age, body-weight or gender distribution. For rapamycin preparations groups (rapamycin solution, TK-NM/Rapa, TP-NM/Rapa, and TT-NM/Rapa), the mice were given 0.24 mg/kg/d Rapa. For the WT group and AD control group, the mice only received the same volume of saline as rapamycin preparations groups (200 μL/day). All the mice were administered intravenously for 4 weeks consecutively, and Morris water maze experiment was carried out on the 23rd day after treatment.
Morris water maze (MWM) experiment. The ability of TT-NM/Rapa to reverse behavioral deficits was examined by MWM following the previously described methodology [24]. Concisely, the spatial navigation tasks lasted for 6 consecutive days. Each mouse needed to receive training for four times randomly which started in different quadrants every day. The mice could find the underwater platform guided by markers on the maze's wall and the cut-off time of a trial was 60 s. In case the mouse failed to locate the concealed submerged platform within 60 s or stay on it for 10 s, it would be led to the platform and stayed 30 s for an additional memory training, and then the escape latency was recorded as 60 s.
The probe trail tests were conducted in the final day of training 2 h later than the spatial navigation tasks. In the probe trails, the hidden platform was withdrawn from the maze. The mice were placed in the quadrant diametrically opposite the original platform and swam freely for 60 s. Their swimming paths were tracked and recorded by Water maze video-track system (Coulbourn, USA).
Enzyme-linked immunosorbent assay (ELSA) of Aβ1-40and Aβ1-42. First, frozen hippocampal tissue from transgenic and WT control mice was isolated and homogenized in RIPA buffer containing a cocktail of protease inhibitors (1.04 mM AEBSF, 0.8 μM aprotinin, 40 μM bestatin, 14 μM E−64, 20 μM leupeptin, and 15 μM pepstatin A). After 2 h of lysis under constant agitation at 4 °C, the homogenized mixtures were briefly sonicated followed by centrifugation at 100,000×g for 30 min at 4 °C, and the supernatant was collected as the soluble fractions. Then the pellet was further sonicated in 70% (w/v) formic acid followed by centrifugation at 100,000×g for 30 min at 4 °C again, and the supernatant was retrieved as the insoluble fractions. Levels of Aβ1-40 and Aβ1-42 in both soluble and insoluble fractions were sequentially assessed following the operating instructions of Amyloid1-40 Human ELISA Kit and Amyloid-β1-42 Human ELISA Kit (Elabscience, China), respectively. For soluble fractions, proteins were loaded directly onto ELISA immunoplates, whereas insoluble fractions were neutralized by 1 M Tris buffer and diluted to 1:20 prior to sample loading. Besides, protein concentration of individual sample was determined by a BCA protein assay kit (Thermo Fisher, USA) for normalization of protein levels.
Western blotting. The alterations in the expression of autophagic- and lysosomal-related proteins (Beclin-1, LC-3, p62/SQSTM1, Parkin and Cathepsin D), the downstream substrates of mTOR (S6K1, 4EBP1, and their phosphorylated forms), hyperphosphorylated tau proteins (p-Ser396, p-Ser202/Thr205) and synaptophysin (SYAP) were detected by Western immunoblotting analysis as mentioned elsewhere [42]. In brief, the concentrations of extracted protein samples were detected by BCA Protein Assay Kit for normalization of protein concentrations prior to mixture with SDS-PAGE Sample Loading Buffer (Beyotime, China) and denaturation. Next, samples of equal total protein were separated by 10% of SDS-polyacrylamide gel (15% for LC3 protein) electrophoresis, and then transferred to a pre-activated PVDF membrane (0.22 μm, Merck Millipore, USA). After being blocked with TBS containing 5% (w/v) nonfat milk for 1.5 h, specific protein bands were probed with corresponding primary antibodies and appropriate HRP-conjugated secondary antibodies following the routine protocols. Subsequently, the immunoblotting signals were visualized by Immobilon Western chemiluminescent HRP reagents (Millipore, USA) with FluorChem-M Western imaging system (ProteinSimple, USA). GAPDH or β-actin staining served for standardization of the samples. Antibodies and their working dilutions used in this study were as followed: anti-Beclin-1 antibody (1:1000 dilution, ab207612, Abcam), anti-LC3B antibody (1:3000 dilution, ab192890, Abcam), anti-SQSTM1/p62 antibody (1: 5000 dilution, ab109012, Abcam), anti-Parkin antibody (1: 1000 dilution, 4211, Cell Signaling), anti-Cathepsin D antibody (1: 2000 dilution, ab75852, Abcam), Anti-S6K1 antibody (1: 2500 dilution, ab32529, Abcam), anti-p-S6K1(Thr389) antibody (1:1000 dilution, 9234, CST), anti-4EBP1 (1:1000 dilution, ab32024, Abcam), anti-p-4EBP1(Thr37/46) (1:1000 dilution, 2855, CST), Phospho-tau (Ser396) antibody (1:1000 dilution, 710,298, Thermo Fisher), Phospho-tau (Ser202, Thr205) antibody (1:500 dilution, MN1020, Thermo Fisher), anti-SYAP-antibody (1:1000 dilution, ab203045, Abcam), anti-GAPDH antibody (1: 1000 dilution, ab59164, Abcam), anti-β-actin antibody (1:1000 dilution, ab8227, Abcam), HRP-conjugated Goat Anti-Rabbit IgG H&L (1: 3000 dilution, ab6721, Abcam) and HRP-conjugated Goat Anti-Mouse IgG H&L (1:4000 dilution, ab6789, Abcam).
Nissl staining. The brain tissue was immersion-fixed in 4% paraformaldehyde at 4 °C overnight, gradient dehydrated by sequential ethanol series and xylene, and then embedded in paraffin to facilitate cutting into 10-μm slices. After deparaffinization in xylene and hydration by graded ethanol and distilled water, the brain sections were incubated with Nissl staining solution (C0117, Beoyotime, China) at 54 °C for 20 min, followed by rinsing with distilled water for several times. Then the brain sections were dehydrated by ethanol gradients and cleared in xylene. After being enclosed in Permount™ mounting medium (G8590, Solarbio, China), the sections were observed under DMI4000B fluorescent microscope (Leica, Germany), and the number of neurons in entire hippocampal areas was quantified by Image J software.
Immunofluorescent staining. Frozen brain sections of the individual sample were processed basically as aforementioned. Then the slides were incubated with anti-LAMP-2 antibody (1:200 dilution, 15,665, Cell Signaling) or anti-Iba1 antibody (1:100 dilution, ab178846, Abcam) at 37 °C for 1 h, followed by several washes with PBS buffer and subsequent incubation with goat anti-rabbit IgG H&L Alexa Fluor-594 (1:200 dilution, ab150080, Abcam) or goat anti-rabbit IgG H&L Alexa Fluor-488 (1:200 dilution, ab150077, Abcam) for another 1 h. After nuclei staining by Hoechst 33,324, the brain sections were viewed under TCS-SP5 confocal microscope (Leica, Germany) with excitation/emission spectra of 594/617 nm and 350/461 nm, respectively. For morphometric analysis of lysosomes, the relative size and relative number of LAMP2-positive vesicles (presenting lysosomes) were measured by Image J software.
Immunohistochemical assay. Paraffin-embedded brain sections were prepared following the procedures described above. After deparaffinization and hydration, the slides were first treated with 3% (w/v) of H2O2 for 5 min, rinsed with PBS, and then blocked with QuickBloc blocking buffer for immunol staining (P0260, Beoyotime, China) at 37 °C for 1 h. Afterwards, the sections were incubated with anti-β-Amyloid1-42 antibody (1:50 dilution, ab10148, Abcam) or anti-Phospho-tau (Ser202, Thr205) antibody (1:100 dilution, MN1020, Thermo Fisher) at 4 °C overnight, following several washes with PBS and incubation with HRP-conjugated Goat Anti-mouse IgG (1: 200 dilution, ab6789, Abcam) at 37 °C for 1 h. Subsequently, the immunolabeled sections were visualized by Pierce™ DAB substrate kit (34,002, Thermo Fisher, USA) and then counterstained with Mayer’ hematoxylin solution (G1080, Solarbio, China). After air-dried, dehydrated by ethanol gradients and coverslipped with mounting medium, the sections were examined by DMI4000B fluorescent microscope, and the percentage area of amyloid plaque deposit in the hippocampal CA1 region was quantitively analyzed by Image J software.
Statistical analysis. All the data were presented as mean ± SD except for specific notification. Unpaired two-tailed student's t-test was utilized for between-group comparisons, whereas one-way ANOVA test combined with the Dunnett's post hoc test was used for multi-group analysis. P < 0.05 was considered as a statistical significance.
CRediT authorship contribution statement
Shuting Xu: Conceptualization, Methodology, Investigation, Visualization, Software, Formal analysis, Data curation, Writing – original draft, Writing – review & editing. Peng Yang: Investigation, Visualization, Validation, Software, Formal analysis, Data curation. Kang Qian: Investigation, Visualization, Validation. Yixian Li: Investigation, Validation. Qian Guo: Investigation. Pengzhen Wang: Investigation. Ran Meng: Writing – review & editing. Jing Wu: Investigation. Jinxu Cao: Investigation. Yunlong Cheng: Investigation. Minjun Xu: Investigation. Qizhi Zhang: Conceptualization, Supervision, Project administration, Writing – review & editing, Resources, Funding acquisition.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 82073780 and 81690263), and Shanghai Municipal Natural Science Foundation (No. 19ZR140620).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2021.09.017.
Author declaration
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
We confirm that the manuscript has been read and approved by all named authors and that there are no other persons who satisfied the criteria for authorship but are not listed.
We further confirm that the order of authors listed in the manuscript has been approved by all of us.
We confirm that we have given due consideration to the protection of intellectual property associated with this work and that there are no impediments to publication, including the timing of publication, with respect to intellectual property. In so doing we confirm that we have followed the regulations of our institutions concerning intellectual property.
We further confirm that any aspect of the work covered in this manuscript that has involved experimental animals has been conducted with the ethical approval of all relevant bodies and that such approvals are acknowledged within the manuscript.
We understand that the Corresponding Author is the sole contact for the Editorial process (including Editorial Manager and direct communications with the office). She is responsible for communicating with the other authors about progress, submissions of revisions and final approval of proofs. We confirm that we have provided a current, correct email address which is accessible by the Corresponding Author.
Author list:
Shuting Xu1, Peng Yang 1, Kang Qian, Yixian Li, Qian Guo, Pengzhen Wang, Ran Meng, Jing Wu, Jinxu Cao, Yunlong Cheng, Minjun Xu, Qizhi Zhang*
*Corresponding author, Prof. Q. Zhang. E-mail address: qzzhang@fudan.edu.cn.
1 These authors made equal contributions to this work Date: April 24, 2021.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Boland B., Yu W.H., Corti O., Mollereau B., Henriques A., Bezard E., Pastores G.M., Rubinsztein D.C., Nixon R.A., Duchen M.R., Mallucci G.R., Kroemer G., Levine B., Eskelinen E.L., Mochel F., Spedding M., Louis C., Martin O.R., Millan M.J. Promoting the clearance of neurotoxic proteins in neurodegenerative disorders of ageing. Nat. Rev. Drug Discov. 2018;17(9):660–688. doi: 10.1038/nrd.2018.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reddy P.H., Oliver D.M. Amyloid beta and phosphorylated tau-induced defective autophagy and mitophagy in alzheimer's disease. Cells. 2019;8(5) doi: 10.3390/cells8050488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caccamo A., Majumder S., Richardson A., Strong R., Oddo S. Molecular interplay between mammalian target of rapamycin (mTOR), amyloid-beta, and Tau: effects on cognitive impairments. J. Biol. Chem. 2010;285(17):13107–13120. doi: 10.1074/jbc.M110.100420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kulkarni A., Chen J., Maday S. Neuronal autophagy and intercellular regulation of homeostasis in the brain. Curr. Opin. Neurobiol. 2018;51:29–36. doi: 10.1016/j.conb.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Zhu X.C., Yu J.T., Jiang T., Tan L. Autophagy modulation for Alzheimer's disease therapy. Mol. Neurobiol. 2013;48(3):702–714. doi: 10.1007/s12035-013-8457-z. [DOI] [PubMed] [Google Scholar]
- 6.Perluigi M., Di Domenico F., Butterfield D.A. mTOR signaling in aging and neurodegeneration: at the crossroad between metabolism dysfunction and impairment of autophagy. Neurobiol. Dis. 2015;84:39–49. doi: 10.1016/j.nbd.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 7.Menzies F.M., Fleming A., Caricasole A., Bento C.F., Andrews S.P., Ashkenazi A., Fullgrabe J., Jackson A., Jimenez Sanchez M., Karabiyik C., Licitra F., Lopez Ramirez A., Pavel M., Puri C., Renna M., Ricketts T., Schlotawa L., Vicinanza M., Won H., Zhu Y., Skidmore J., Rubinsztein D.C. Autophagy and neurodegeneration: pathogenic mechanisms and therapeutic opportunities. Neuron. 2017;93(5):1015–1034. doi: 10.1016/j.neuron.2017.01.022. [DOI] [PubMed] [Google Scholar]
- 8.Di Meco A., Curtis M.E., Lauretti E., Pratico D. Autophagy dysfunction in alzheimer's disease: mechanistic insights and new therapeutic opportunities. Biol. Psychiatr. 2020;87(9):797–807. doi: 10.1016/j.biopsych.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Son J.H., Shim J.H., Kim K.H., Ha J.Y., Han J.Y. Neuronal autophagy and neurodegenerative diseases. Exp. Mol. Med. 2012;44(2):89–98. doi: 10.3858/emm.2012.44.2.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bove J., Martinez-Vicente M., Vila M. Fighting neurodegeneration with rapamycin: mechanistic insights. Nat. Rev. Neurosci. 2011;12(8):437–452. doi: 10.1038/nrn3068. [DOI] [PubMed] [Google Scholar]
- 11.Martini-Stoica H., Xu Y., Ballabio A., Zheng H. The autophagy-lysosomal pathway in neurodegeneration: a TFEB perspective. Trends Neurosci. 2016;39(4):221–234. doi: 10.1016/j.tins.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao J., Wang H., Xie Y., Fu Y., Li Y., Liu P., Du H., Zhu J., Dong L., Hussain M., Li Y., Zhang L., Zhu J., Tao J. Transdermal delivery of rapamycin with poor water-solubility by dissolving polymeric microneedles for anti-angiogenesis. J. Mater. Chem. B. 2020;8(5):928–934. doi: 10.1039/c9tb00912d. [DOI] [PubMed] [Google Scholar]
- 13.Brandt C., Hillmann P., Noack A., Romermann K., Ohler L.A., Rageot D., Beaufils F., Melone A., Sele A.M., Wymann M.P., Fabbro D., Loscher W. The novel, catalytic mTORC1/2 inhibitor PQR620 and the PI3K/mTORC1/2 inhibitor PQR530 effectively cross the blood-brain barrier and increase seizure threshold in a mouse model of chronic epilepsy. Neuropharmacology. 2018;140:107–120. doi: 10.1016/j.neuropharm.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Tramutola A., Lanzillotta C., Di Domenico F. Targeting mTOR to reduce Alzheimer-related cognitive decline: from current hits to future therapies. Expert Rev. Neurother. 2017;17(1):33–45. doi: 10.1080/14737175.2017.1244482. [DOI] [PubMed] [Google Scholar]
- 15.Zhao F., Zhao Y., Liu Y., Chang X., Chen C., Zhao Y. Cellular uptake, intracellular trafficking, and cytotoxicity of nanomaterials. Small. 2011;7(10):1322–1337. doi: 10.1002/smll.201100001. [DOI] [PubMed] [Google Scholar]
- 16.Lee S., Sato Y., Nixon R.A. Primary lysosomal dysfunction causes cargo-specific deficits of axonal transport leading to Alzheimer-like neuritic dystrophy. Autophagy. 2011;7(12):1562–1563. doi: 10.4161/auto.7.12.17956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trushina E., McMurray C.T. Oxidative stress and mitochondrial dysfunction in neurodegenerative diseases. Neuroscience. 2007;145(4):1233–1248. doi: 10.1016/j.neuroscience.2006.10.056. [DOI] [PubMed] [Google Scholar]
- 18.Tejada-Simon M.V., Serrano F., Villasana L.E., Kanterewicz B.I., Wu G.-Y., Quinn M.T., Klann E. Synaptic localization of a functional NADPH oxidase in the mouse hippocampus. Mol. Cell. Neurosci. 2005;29(1):97–106. doi: 10.1016/j.mcn.2005.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ushio-Fukai M. Compartmentalization of redox signaling through NADPH oxidase–derived ROS. Antioxidants Redox Signal. 2009;11(6):1289–1299. doi: 10.1089/ars.2008.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ballance W.C., Qin E.C., Chung H.J., Gillette M.U., Kong H. Reactive oxygen species-responsive drug delivery systems for the treatment of neurodegenerative diseases. Biomaterials. 2019;217:119292. doi: 10.1016/j.biomaterials.2019.119292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Q., Wen Y., Wen J., Zhang Y.-P., Xu X.-D., Victorious A., Zavitz R., Xu X. A new biosafe reactive oxygen species (ROS)-responsive nanoplatform for drug delivery. RSC Adv. 2016;6(45):38984–38989. [Google Scholar]
- 22.Li Q., Liang J., You X., Liu X., Wu H., Gao W., Wen Y., Wu J., Xu X., Zhang F. A novel reactive oxygen species triggered polymeric nanoplatform for controlled drug delivery and cancer therapy. J. Biomed. Nanotechnol. 2017;13(5):513–521. [Google Scholar]
- 23.Sun C., Liang Y., Hao N., Xu L., Cheng F., Su T., Cao J., Gao W., Pu Y., He B. A ROS-responsive polymeric micelle with a π-conjugated thioketal moiety for enhanced drug loading and efficient drug delivery. Org. Biomol. Chem. 2017;15(43):9176–9185. doi: 10.1039/c7ob01975k. [DOI] [PubMed] [Google Scholar]
- 24.Guo Q., Xu S., Yang P., Wang P., Lu S., Sheng D., Qian K., Cao J., Lu W., Zhang Q. A dual-ligand fusion peptide improves the brain-neuron targeting of nanocarriers in Alzheimer's disease mice. J. Contr. Release. 2020;320:347–362. doi: 10.1016/j.jconrel.2020.01.039. [DOI] [PubMed] [Google Scholar]
- 25.Wang M., Zhai Y., Ye H., Lv Q., Sun B., Luo C., Jiang Q., Zhang H., Xu Y., Jing Y., Huang L., Sun J., He Z. High Co-loading capacity and stimuli-responsive release based on cascade reaction of self-destructive polymer for improved chemo-photodynamic therapy. ACS Nano. 2019;13(6):7010–7023. doi: 10.1021/acsnano.9b02096. [DOI] [PubMed] [Google Scholar]
- 26.Hu B., Dai F., Fan Z., Ma G., Tang Q., Zhang X. Nanotheranostics: Congo red/Rutin‐MNPs with enhanced magnetic resonance imaging and H2O2‐responsive therapy of Alzheimer's disease in APPswe/PS1dE9 transgenic mice. Adv. Mater. 2015;27(37):5499–5505. doi: 10.1002/adma.201502227. [DOI] [PubMed] [Google Scholar]
- 27.Hu K., Li J., Shen Y., Lu W., Gao X., Zhang Q., Jiang X. Lactoferrin-conjugated PEG–PLA nanoparticles with improved brain delivery: in vitro and in vivo evaluations. J. Contr. Release. 2009;134(1):55–61. doi: 10.1016/j.jconrel.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 28.Pang Z., Lu W., Gao H., Hu K., Chen J., Zhang C., Gao X., Jiang X., Zhu C. Preparation and brain delivery property of biodegradable polymersomes conjugated with OX26. J. Contr. Release. 2008;128(2):120–127. doi: 10.1016/j.jconrel.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 29.von Gunten A., Kovari E., Bussiere T., Rivara C.B., Gold G., Bouras C., Hof P.R., Giannakopoulos P. Cognitive impact of neuronal pathology in the entorhinal cortex and CA1 field in Alzheimer's disease. Neurobiol. Aging. 2006;27(2):270–277. doi: 10.1016/j.neurobiolaging.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 30.Whitesell J.D., Buckley A.R., Knox J.E., Kuan L., Graddis N., Pelos A., Mukora A., Wakeman W., Bohn P., Ho A., Hirokawa K.E., Harris J.A. Whole brain imaging reveals distinct spatial patterns of amyloid beta deposition in three mouse models of Alzheimer's disease. J. Comp. Neurol. 2019;527(13):2122–2145. doi: 10.1002/cne.24555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vajn K., Viljetic B., Degmecic I.V., Schnaar R.L., Heffer M. Differential distribution of major brain gangliosides in the adult mouse central nervous system. PloS One. 2013;8(9) doi: 10.1371/journal.pone.0075720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J.K., Teng Q., Garrity-Moses M., Federici T., Tanase D., Imperiale M.J., Boulis N.M. A novel peptide defined through phage display for therapeutic protein and vector neuronal targeting. Neurobiol. Dis. 2005;19(3):407–418. doi: 10.1016/j.nbd.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 33.Li L.S., Lu Y.L., Nie J., Xu Y.Y., Zhang W., Yang W.J., Gong Q.H., Lu Y.F., Lu Y., Shi J.S. Dendrobium nobile Lindl alkaloid, a novel autophagy inducer, protects against axonal degeneration induced by Aβ25‐35 in hippocampus neurons in vitro. CNS Neurosci. Ther. 2017;23(4):329–340. doi: 10.1111/cns.12678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh A.K., Bissoyi A., Kashyap M.P., Patra P.K., Rizvi S.I. Autophagy activation alleviates amyloid-β-induced oxidative stress, apoptosis and neurotoxicity in human neuroblastoma SH-SY5Y cells. Neurotox. Res. 2017;32(3):351–361. doi: 10.1007/s12640-017-9746-5. [DOI] [PubMed] [Google Scholar]
- 35.Frozza R.L., Horn A.P., Hoppe J.B., Simão F., Gerhardt D., Comiran R.A., Salbego C.G. J.N.r. A comparative study of β-amyloid peptides Aβ1-42 and Aβ25-35 toxicity in organotypic hippocampal slice cultures. Neurochem. Res. 2009;34(2):295–303. doi: 10.1007/s11064-008-9776-8. [DOI] [PubMed] [Google Scholar]
- 36.Vidoni C., Follo C., Savino M., Melone M.A., Isidoro C. The role of cathepsin D in the pathogenesis of human neurodegenerative disorders. Med. Res. Rev. 2016;36(5):845–870. doi: 10.1002/med.21394. [DOI] [PubMed] [Google Scholar]
- 37.Kenessey A., Nacharaju P., Ko L.W., Yen S.H. Degradation of tau by lysosomal enzyme cathepsin D: implication for Alzheimer neurofibrillary degeneration. J. Neurochem. 1997;69(5):2026–2038. doi: 10.1046/j.1471-4159.1997.69052026.x. [DOI] [PubMed] [Google Scholar]
- 38.Benito-Cuesta I., Diez H., Ordonez L., Wandosell F. Assessment of autophagy in neurons and brain tissue. Cells. 2017;6(3) doi: 10.3390/cells6030025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xue X., Wang L.R., Sato Y., Jiang Y., Berg M., Yang D.S., Nixon R.A., Liang X.J. Single-walled carbon nanotubes alleviate autophagic/lysosomal defects in primary glia from a mouse model of Alzheimer's disease. Nano Lett. 2014;14(9):5110–5117. doi: 10.1021/nl501839q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martina J.A., Chen Y., Gucek M., Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8(6):903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song W., Soo Lee S., Savini M., Popp L., Colvin V.L., Segatori L. Ceria nanoparticles stabilized by organic surface coatings activate the lysosome-autophagy system and enhance autophagic clearance. ACS Nano. 2014;8(10):10328–10342. doi: 10.1021/nn505073u. [DOI] [PubMed] [Google Scholar]
- 42.Ozcelik S., Fraser G., Castets P., Schaeffer V., Skachokova Z., Breu K., Clavaguera F., Sinnreich M., Kappos L., Goedert M., Tolnay M., Winkler D.T. Rapamycin attenuates the progression of tau pathology in P301S tau transgenic mice. PloS One. 2013;8(5) doi: 10.1371/journal.pone.0062459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oddo S., Caccamo A., Shepherd J.D., Murphy M.P., Golde T.E., Kayed R., Metherate R., Mattson M.P., Akbari Y., LaFerla F.M. Triple-transgenic model of Alzheimer's disease with plaques and tangles: intracellular Abeta and synaptic dysfunction. Neuron. 2003;39(3):409–421. doi: 10.1016/s0896-6273(03)00434-3. [DOI] [PubMed] [Google Scholar]
- 44.Gimenez-Llort L., Mate I., Manassra R., Vida C., De la Fuente M. Peripheral immune system and neuroimmune communication impairment in a mouse model of Alzheimer's disease. Ann. N. Y. Acad. Sci. 2012;1262:74–84. doi: 10.1111/j.1749-6632.2012.06639.x. [DOI] [PubMed] [Google Scholar]
- 45.Pietropaolo S., Feldon J., Yee B.K. Age-dependent phenotypic characteristics of a triple transgenic mouse model of Alzheimer disease. Behav. Neurosci. 2008;122(4):733–747. doi: 10.1037/a0012520. [DOI] [PubMed] [Google Scholar]
- 46.Alayev A., Holz M.K. mTOR signaling for biological control and cancer. J. Cell. Physiol. 2013;228(8):1658–1664. doi: 10.1002/jcp.24351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ravikumar B., Vacher C., Berger Z., Davies J.E., Luo S., Oroz L.G., Scaravilli F., Easton D.F., Duden R., O'Kane C.J. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004;36(6):585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 48.Mucke L., Masliah E., Yu G.Q., Mallory M., Rockenstein E.M., Tatsuno G., Hu K., Kholodenko D., Johnson-Wood K., McConlogue L. High-level neuronal expression of abeta 1-42 in wild-type human amyloid protein precursor transgenic mice: synaptotoxicity without plaque formation. J. Neurosci. 2000;20(11):4050–4058. doi: 10.1523/JNEUROSCI.20-11-04050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Attems J., Thomas A., Jellinger K. Correlations between cortical and subcortical tau pathology. Neuropathol. Appl. Neurobiol. 2012;38(6):582–590. doi: 10.1111/j.1365-2990.2011.01244.x. [DOI] [PubMed] [Google Scholar]
- 50.Gudi V., Gai L., Herder V., Tejedor L.S., Kipp M., Amor S., Suhs K.W., Hansmann F., Beineke A., Baumgartner W., Stangel M., Skripuletz T. Synaptophysin is a reliable marker for axonal damage. Journal of Neuropatholgy & Experimental Neurology. 2017;76(2):109–125. doi: 10.1093/jnen/nlw114. [DOI] [PubMed] [Google Scholar]
- 51.Russell C.L., Semerdjieva S., Empson R.M., Austen B.M., Beesley P.W., Alifragis P. Amyloid-beta acts as a regulator of neurotransmitter release disrupting the interaction between synaptophysin and VAMP2. PloS One. 2012;7(8) doi: 10.1371/journal.pone.0043201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krause D.L., Muller N. Neuroinflammation, microglia and implications for anti-inflammatory treatment in Alzheimer's disease. Int. J. Alzheimer's Dis. 2010;2010 doi: 10.4061/2010/732806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H., Hu D., Liang F., Huang X., Zhu Q. Influence factors on the critical micelle concentration determination using pyrene as a probe and a simple method of preparing samples. Royal Society open science. 2020;7(3):192092. doi: 10.1098/rsos.192092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Forrest M.L., Won C.Y., Malick A.W., Kwon G.S. In vitro release of the mTOR inhibitor rapamycin from poly(ethylene glycol)-b-poly(epsilon-caprolactone) micelles. J. Contr. Release. 2006;110(2):370–377. doi: 10.1016/j.jconrel.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 55.Ray B., Bisht S., Maitra A., Maitra A., Lahiri D.K. Neuroprotective and neurorescue effects of a novel polymeric nanoparticle formulation of curcumin (NanoCurc) in the neuronal cell culture and animal model: implications for Alzheimer's disease. J. Alzheim. Dis. 2011;23(1):61–77. doi: 10.3233/JAD-2010-101374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sallustio B.C., Noll B.D., Morris R.G. Comparison of blood sirolimus, tacrolimus and everolimus concentrations measured by LC-MS/MS, HPLC-UV and immunoassay methods. Clin. Biochem. 2011;44(2–3):231–236. doi: 10.1016/j.clinbiochem.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 57.Mauthe M., Orhon I., Rocchi C., Zhou X., Luhr M., Hijlkema K.J., Coppes R.P., Engedal N., Mari M., Reggiori F. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14(8):1435–1455. doi: 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chan L.L., Shen D., Wilkinson A.R., Patton W., Lai N., Chan E., Kuksin D., Lin B., Qiu J. A novel image-based cytometry method for autophagy detection in living cells. Autophagy. 2012;8(9):1371–1382. doi: 10.4161/auto.21028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.