Abstract
Serum- and glucocorticoid-inducible kinases (SGKs) form a novel family of serine/threonine kinases that are activated in response to a variety of extracellular stimuli. SGKs are related to Akt (also called PKB), a serine/threonine kinase that plays a crucial role in promoting cell survival. Like Akt, SGKs are activated by the phosphoinositide-3 kinase (PI3K) and translocate to the nucleus upon growth factor stimulation. However the physiological substrates and cellular functions of SGKs remained to be identified. We hypothesized that SGKs regulate cellular functions in concert with Akt by phosphorylating common targets within the nucleus. The best-characterized nuclear substrates of Akt are transcription factors of the Forkhead family. Akt phosphorylates Forkhead transcription factors such as FKHRL1, leading to FKHRL1's exit from the nucleus and the consequent shutoff of FKHRL1 target genes. We show here that SGK1, like Akt, promotes cell survival and that it does so in part by phosphorylating and inactivating FKHRL1. However, SGK and Akt display differences with respect to the efficacy with which they phosphorylate the three regulatory sites on FKHRL1. While both kinases can phosphorylate Thr-32, SGK displays a marked preference for Ser-315 whereas Akt favors Ser-253. These findings suggest that SGK and Akt may coordinately regulate the function of FKHRL1 by phosphorylating this transcription factor at distinct sites. The efficient phosphorylation of these three sites on FKHRL1 by SGK and Akt appears to be critical to the ability of growth factors to suppress FKHRL1-dependent transcription, thereby preventing FKHRL1 from inducing cell cycle arrest and apoptosis. These findings indicate that SGK acts in concert with Akt to propagate the effects of PI3K activation within the nucleus and to mediate the biological outputs of PI3K signaling, including cell survival and cell cycle progression.
Serum- and glucocorticoid-induced kinases (SGKs) belong to a new family of serine/threonine kinases that are regulated at both the transcriptional and posttranslational levels by external stimuli. The mRNA encoding SGK1, the best-studied member of the SGK family, is rapidly induced in response to a variety of stimuli, including growth factors (51, 52), steroid and peptide hormones (3, 51, 52), cytokines (15, 50), changes in cell volume (49), and brain injury (24).
The SGK gene is conserved from yeast to human, and the SGK protein is expressed in a variety of tissues and cell lines in mammals (10, 51, 52). Although it has been proposed that SGK may play a role in cell cycle progression (8) or sodium homeostasis control (4, 12), the cellular functions of SGK are largely uncharacterized, and to date no in vivo SGK substrates have been identified.
Within the protein kinase superfamily, SGK is closely related to Akt (also called PKB), another serine/threonine kinase that is activated in response to growth and survival factors and plays a critical role in promoting cell survival (16, 20). Several recent reports have shown that growth and survival factors also trigger SGK activation (28, 41). The activation of SGK is dependent upon phosphoinositide 3-kinase (PI3K) activity and requires the phosphorylation of two regulatory sites, Thr-256 and Ser-422, that lie in the activation loop and the C-terminal domain of SGK, respectively (28, 41). The protein kinase PDK1, which had previously been identified as a PI3K-dependent serine/threonine kinase that phosphorylates and activates Akt, is also likely to be responsible for the phosphorylation of SGK at Thr-256 (28, 41). The kinase that phosphorylates Ser-422 has yet to be identified.
Upon activation by growth factors, endogenous SGK translocates rapidly into the nucleus, where it may encounter nuclear substrates (8). SGK and Akt are likely to phosphorylate related substrates, as they share a similar consensus phosphorylation site (RXRXXS/T) (1, 28, 41).
Despite their similarity, SGK and Akt display unique features. First, unlike Akt, whose expression appears not to be regulated by extracellular stimuli, SGK protein expression is induced upon treatment of cells with extracellular stimuli, including growth factors. Second, in contrast to Akt, SGK does not have a pleckstrin homology domain and appears not to be recruited to the plasma membrane prior to its activation. Third, the consensus sequence that is phosphorylated by SGK is not identical to the site phosphorylated by Akt. For example, SGK is more effective than Akt at phosphorylating a consensus peptide substrate in which the serine phosphoacceptor site is replaced with a threonine. Moreover, SGK, in contrast to Akt, is capable of phosphorylating peptide substrates that do not have a bulky hydrophobic amino acid immediately C terminal to the phosphoacceptor site (28). These differences between SGK and Akt suggested that these two kinases might have complementary rather than redundant functions.
The observation that SGK translocates to the nucleus following its activation by PI3K led us to hypothesize that SGK may act in concert with Akt to phosphorylate critical targets within the nucleus. The best-characterized nuclear substrates of Akt are transcription factors of the Forkhead family: FKHR, FKHRL1, and AFX (for a review, see reference 30). According to the new nomenclature for Forkhead transcription factors, these three proteins have been assigned to the FOXO (named for “Forkhead box, group O”) subfamily of Forkhead transcription factors (26).
When Akt is inactive, FOXO family members are dephosphorylated and localized in the nucleus, where they activate transcription. Upon growth factor stimulation, Akt is activated and FOXOs are phosphorylated at three key regulatory phosphorylation sites (Thr-32, Ser-253, and Ser-315 for FKHRL1). The phosphorylation of FOXOs promotes their exit from the nucleus, resulting in inhibition of FOXO-dependent transcription (6, 7, 23, 31, 38, 43, 45, 46). Akt has been reported to phosphorylate the second regulatory site of FOXOs (Ser-253 for FKHRL1) with greater affinity than the first and third sites (37, 43). However, it is not clear whether protein kinases in addition to Akt phosphorylate the first and third FOXO regulatory sites with high affinity or what the respective contribution of each site to the control of FOXO-dependent gene expression and FOXO biological functions is.
We report here that the protein kinase SGK mediates the biological effects of PI3K in parallel with Akt. Activated SGK is capable of promoting cell survival in part by phosphorylating and inactivating the Forkhead transcription factor FKHRL1 (also called FOXO3a). SGK, like Akt, phosphorylates FKHRL1, thereby leading to FKHLR1 translocation from the nucleus to the cytoplasm and to the inhibition of FKHRL1-dependent transcription. However, SGK and Akt, when expressed at physiological levels, display differences with respect to the efficacy with which they phosphorylate the regulatory sites on FKHRL1: Thr-32 is phosphorylated by both protein kinases, but SGK prefers Ser-315 whereas Akt favors Ser-253. The efficient phosphorylation of all three regulatory sites of FKHRL1 appears to be required for the ability of growth factors to completely repress FKHRL1-dependent transcription, thereby preventing this transcription factor from inducing apoptosis and/or cell cycle arrest. As mediators of PI3K signaling within the nucleus, SGK and Akt are likely to be important regulators of a variety of PI3K-dependent cellular responses, including cell proliferation and survival.
MATERIALS AND METHODS
Material.
Insulin-like growth factor I (IGF-I) and insulin were purchased, respectively, from R&D Systems and Roche Biochemicals. LY 294002 (LY) was obtained from Calbiochem. The antihemagglutinin (anti-HA) antibody (12CA5) was purchased from Roche Biochemicals, and the anti-M2 antibody was purchased from Sigma. The antibody against cleaved poly(ADP-ribose) polymerase (PARP) was purchased from Cell Signaling Technology. Purified SGK was obtained from Upstate Biotechnology.
Constructs.
The constructs encoding wild-type (WT) SGK and mutants with mutations of the phosphorylation sites were described previously (41). The constructs encoding WT Akt, the constitutively active (CA) mutant (myristylated Akt lacking the pleckstrin homology domain), and the kinase dead mutant (Akt with a K197M mutation) have been described previously (17). The constructs encoding glutathione S-transferase (GST)–FKHRL1, HA-FKHRL1, M2-FKHRL1, and the Forkhead responsive element (FHRE)-driven luciferase reporter were described previously (7). The constructs encoding the catalytic subunit of PKA (35), WT RSK2 (53), and the active forms of protein kinase C (PKC) ζ and λ (14, 47) have been described previously.
Antibodies.
The anti-FKHRL1, anti-phospho-T32, and anti-phospho-S253 antibodies were described previously (7). For the anti-phospho-S315 antibody, a phosphopeptide with the sequence CFRSRTNpSNATVS was synthesized (Tufts Synthesis Facility, Tufts Medical School, Boston, Mass.) and coupled to keyhole limpet hemocyanin (KLH) (Pierce). The KLH-coupled peptide was injected into New Zealand White rabbits (Covance Research Products, Denver, Pa.). Sera obtained from immunized rabbits were purified by passage over a protein A-Sepharose column (Pharmacia), followed by elution of the bound phosphoantibodies with 100 mM glycine (pH 2.5). The eluate was then passed over an agarose-iodoacetyl column to which was coupled the nonphosphorylated form of the peptide antigen. The flowthrough from the column contained the antiphosphopeptide antibodies used for the analysis of FKHRL1 phosphorylation.
For the anti-SGK1 antibody, a peptide with the sequence CVLHKQPYDRTVDWW was synthesized (Tufts Synthesis Facility, Tufts Medical School) and coupled to KLH (Pierce). The KLH-coupled peptide was injected into New Zealand White rabbits (Covance Research Products). Sera obtained from immunized rabbits were purified by passage over an agarose-iodoacetyl column to which was coupled the peptide antigen, followed by elution of the bound phosphoantibodies with 100 mM glycine (pH 2.5).
Cell culture.
Cells of the human embryonic kidney cell line 293 (HEK 293), the derivative HEK 293T, and the Chinese hamster lung fibroblast cell line CCL39 (American Type Culture Collection) were cultured in Dulbecco's modified Eagle medium (DMEM) (Gibco) supplemented with 10% fetal calf serum (FCS) (Gibco) and antibiotics (50 U of penicillin and 50 μg of streptomycin/ml) at 37°C in an atmosphere of 95% air–5% CO2. Cerebellar granule cells were obtained from P6 Long Evans rats and cultured in BME (Sigma) supplemented with 25 mM KCl, 10% heat-inactivated calf serum (HyClone), and antibiotics as previously described (21).
In vitro kinase assay.
HEK 293 cells were seeded at 106 cells per 10-cm dish and were transfected the following day by a modified calcium phosphate method with 2 μg of plasmid DNA. The transfection mixture was removed after 16 h of incubation and cells were serum starved for 24 h before stimulation for 15 min with 100 nM insulin. Cells were placed on ice and extracted with lysis buffer containing 50 mM Tris-HCl (pH 7.5), 1% Nonidet P-40 (vol/vol), 120 mM NaCl, 25 mM NaF, 40 mM β-glycerophosphate, 0.1 mM sodium orthopervanadate, 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, and 2 μM microcystin-LR. Lysates were centrifuged for 15 min at 12,000 × g, and the HA-conjugated SGK protein was immunoprecipitated from 400 μg of cell extracts with the anti-HA epitope 12CA5 monoclonal antibody coupled to protein A-Sepharose (Pharmacia Biotech). The immune complexes were washed once with lysis buffer containing 0.5 M NaCl, then with lysis buffer, and finally with kinase assay buffer (50 mM Tris-HCl [pH 7.5], 0.1% [vol/vol] 2-mercaptoethanol). In vitro kinase assays were performed for 60 min at 30°C in 50 μl of reaction mixture containing 30 μl of immunoprecipitate in kinase buffer, 5 μg of WT GST-FKHRL1 as a substrate, 10 mM MgCl2, 1 μM protein kinase A inhibitor peptide (Bachem), and 100 μM [γ-32P]ATP (Amersham; 1,000 to 2,000 cpm/pmol). All reactions were stopped by adding Laemmli sample buffer. HEK 293 cell extracts and immunoprecipitates were resolved by sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis (SDS–12% PAGE) and transferred to Immobilon P membranes (Millipore). The filters were blocked for 30 min in 1× phosphate-buffered saline (PBS) containing 5% skim milk, 0.5% Triton X-100, and 0.5% Tween 20, followed by a 2-h incubation with the anti-FKHRL1 antibody or the various anti-phospho-FKHRL1 antibodies diluted in the same blocking solution. The secondary antibody was alkaline phosphatase-conjugated anti-rabbit immunoglobulin G (IgG; Sigma) diluted 2,500-fold in the blocking buffer. The detection of proteins was carried out by using the alkaline phosphatase color development reagents from Bio-Rad.
Immunoblotting.
CCL39 fibroblasts were seeded in six-well plastic dishes at a density of 5 × 105/well. They were transfected by the calcium phosphate technique with 5 μg of the WT HA-FKHRL1 construct and 5 μg of the relevant constructs. Twenty-four hours after transfection, cells were incubated in serum-free DMEM for 20 h and treated with 10 μM LY for 1 h. Extracts were obtained by lysing the cells in lysis buffer (50 mM Tris-HCl [pH 8], 100 mM NaCl, 2 mM EGTA, 10 mM NaF, 40 mM β-glycerophosphate, 0.5% Triton X-100, 2 mM dithiothreitol, aprotinin, 1 mM phenylmethylsulfonyl fluoride). Proteins were resolved by SDS-PAGE (8 to 6%; 29:1, acrylamide/bisacrylamide ratio) and transferred to polyvinylidene difluoride or nitrocellulose membranes. The membranes were incubated with anti-HA, anti-FKHRL1, anti-phospho-Thr-32, anti-phospho-Ser-253, or anti-phospho-Ser-315 antibodies for 2 h at room temperature. The primary antibody was visualized using horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG secondary antibodies and enhanced chemiluminescence.
Immunolocalization.
CCL39 cells were plated onto glass coverslips at a density of 102/well in 12-well dishes. The cells were transfected by the calcium phosphate technique with 1 μg of M2-FKHRL1 and 3 μg of the relevant constructs. The day after transfection, cells were rendered quiescent by incubation in serum-free medium for 16 h and then treated with 20 μM LY for 1 h. The cells were then fixed for 15 min in 4% formaldehyde–2% sucrose at room temperature and permeabilized with 0.1% Triton X-100 for 10 min. Coverslips were washed with PBS, and nonspecific antibody binding sites were blocked by incubation with PBS containing 3% bovine serum albumin (BSA). The coverslips were then incubated with primary antibody diluted in PBS-BSA (anti-M2, 1/2,000) for 2 h and washed five times with PBS. Cells were then incubated for 1 h with a secondary antibody (goat anti-mouse IgG Cy3 conjugated: 1/500) diluted in PBS-BSA. After extensive washes in PBS, coverslips were mounted in Aquamount and examined under epifluorescent illumination. For quantification, 50 to 100 cells per coverslip were counted.
Luciferase assay.
CCL39 cells were seeded in 24-well plates at a density of 1.5 × 105/well and were cotransfected with 0.25 μg of empty vector (pECE) or WT HA-FKHRL1 or with 0.5 μg of the luciferase reporter gene, 0.75 μg of the EF-LacZ construct, 1 μg of carrier DNA (pBluescript), 0.25 to 0.5 μg of the SGK construct, and 0.05 to 0.01 μg of the Akt construct in order to achieve similar levels of expression between these two kinases. Two days after transfection, cells were lysed in 100 μl of lysis buffer and the luciferase activity of 1/5 of the samples was assayed according to the Promega protocol. β-Galactosidase activity was assayed as previously described (34).
RT-PCR analysis.
The expression of endogenous SGK was determined by reverse transcription (RT) of total RNA followed by PCR analysis. Total RNA was extracted using RNAzol (Telstat). Two micrograms of total RNA was reverse transcribed by extension of random hexamer primers using Superscript II reverse transcriptase (Life Technology) according to the manufacturer's protocol. PCR of the cDNA was performed using AdvanTaq (Life Technology) with the following pairs of primers: human SGK1-1 (hSGK1-1) (forward; GGAGCCTGAGCTTATGAATGCCAAC) and hSGK1-2 (reverse; TGCCACAGAAGGTGGATGTTGTGC); hSGK1-3 (forward; CCTTGTGGATATGCTGTGTGAACCG) and hSGK1-4 (reverse; TGGGGCATTGGTCCATAAAAACC). The PCR program used was as follows: 1 min at 95°C; 32 cycles of 30 s at 95°C, 1 min at 68°C, and 1 min at 72°C; and a 5-min extension at 72°C.
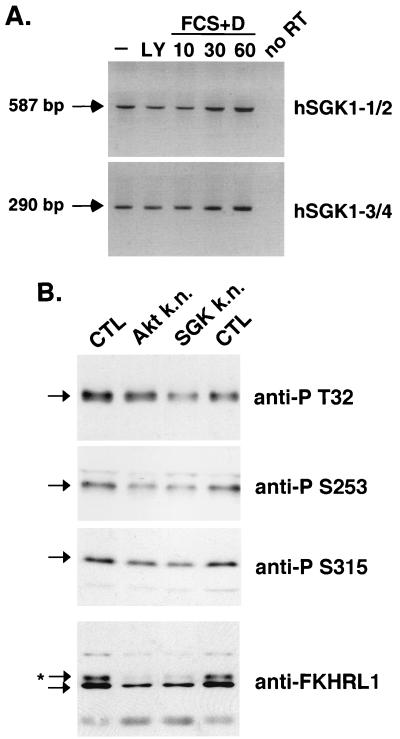
A PCR was also performed on total RNA that had not been reverse transcribed to control for the absence of genomic DNA in the RNA preparation (see Fig. 4A). The products of the PCRs were resolved on a 2% agarose gel.
FIG. 4.
Endogenous SGK is required for FKHRL1 phosphorylation induced by growth factors. (A) HEK 293T cells were starved for 6 h and then treated with 10 μM LY for 1 h or treated for the indicated periods of time (in minutes) with a combination of 20% FCS and 10−4 M dexamethasone (D) to induce SGK1 mRNA. RT-PCRs were performed on total RNA using two different pairs of primers that are specific for the human SGK1 isoform (hSGK1-1–hSGK1-2 and hSGK1-3–hSGK1-4). (B) HEK 293T cells were transfected with an empty vector (CTL) or constructs encoding the KN Akt (K197M) mutant (Akt k.n.) or the KN SGK (T256A/S422A) mutant (SGK k.n.). Cells were incubated in the presence of 10% FCS, and extracts were resolved by SDS-PAGE. Phosphorylation of endogenous FKHRL1 was detected by immunoblot analysis with antibodies directed against phospho-T32, phospho-S253, or phospho-S315 FKHRL1. Mobility shift of endogenous FKHRL1 was assessed by immunoblot analysis with the antibody directed against total FKHRL1. ∗, phosphorylated form of FKHRL1.
BrdU incorporation.
CCL39 fibroblasts were seeded onto glass coverslips in 12-well plates at a density of 65,000/well and were transfected with 4 μg of the construct of interest. At 8 h after transfection, cells were starved for 24 h in DMEM and then stimulated with 20% FCS in the presence of bromodeoxyuridine (BrdU) for 24 h. Cells were fixed in 100% methanol for 15 min at −20°C. Coverslips were incubated with 2 N HCl for 10 min at 37°C and washed extensively with PBS. Nonspecific antibody binding sites were blocked by incubation with PBS containing 8% BSA and 10% FCS. Coverslips were then incubated with primary antibodies (anti-FKHRL1, 1/2,000; antibromodeoxyuridine [anti-BrdU], 1/500) for 2 h and washed five times with PBS. Cells were then incubated for 1 h with a secondary antibody (goat anti-rabbit IgG, Cy3 conjugated, 1/500; goat anti-rat IgG, biotinylated, 1/300), followed by a 30-min incubation with Cy2-conjugated streptavidin (1/200). Coverslips were mounted in Aquamount and examined under epifluorescent illumination. For quantification, 50 to 100 cells per coverslip were counted.
Apoptosis assay.
Cerebellar granule neurons were seeded onto glass coverslips in 24-well plates at a density of 106 cells/well and at 5 or 6 days in vitro were cotransfected with 2 μg of the construct of interest and 0.5 μg of the plasmid encoding green fluorescent protein (GFP) under the control of the cytomegalovirus promoter, as described elsewhere (21). At 16 h after transfection, the medium was changed for BME containing IGF-I (50 ng/ml) for 24 h and then incubated in BME alone for an additional 8 h or not incubated. The transfected neurons were then fixed in 4% paraformaldehyde–2% sucrose. Nuclei were stained with Hoechst 33258 (2.5 mg/ml). Apoptotic nuclei of GFP-positive neurons were counted in a blinded manner.
RESULTS
SGK phosphorylates the Forkhead transcription factor FKHRL1 and displays a preference for Thr-32 and Ser-315 in vitro.
Within the protein kinase superfamily, the serine/threonine kinase SGK stands out as a good candidate for the phosphorylation of FOXO family members since (i) SGK is activated by growth and survival factors in a PI3K-dependent manner (28, 41), (ii) following growth factor stimulation, SGK translocates into the nucleus, where it might phosphorylate transcription factors (8), and (iii) the sequence of peptides that are selectively phosphorylated by SGK is compatible with the amino acid regions that surround the three known phosphorylation sites of FKHRL1 (28, 41).
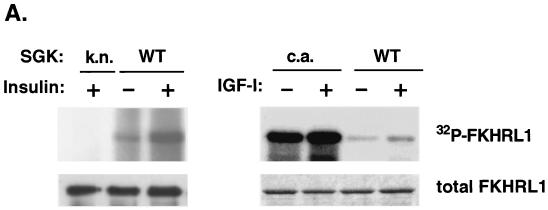
To determine if SGK can phosphorylate FKHRL1, a WT SGK, a kinase-inactive (KN) form of SGK (K127Q), or a CA form of SGK (S422D) (41) was expressed in HEK 293 cells and activated by exposure of the cells to insulin or IGF-I. In vitro kinase assays of immunoprecipitated SGK were then performed using bacterially purified FKHRL1 as a substrate. We found that in response to insulin or IGF-I, WT SGK, but not a KN mutant of SGK, phosphorylated FKHRL1 in vitro (Fig. 1A, left panel). The CA form of SGK efficiently phosphorylated FKHRL1 in vitro, in the absence or presence of IGF-I (Fig. 1A, right panel).
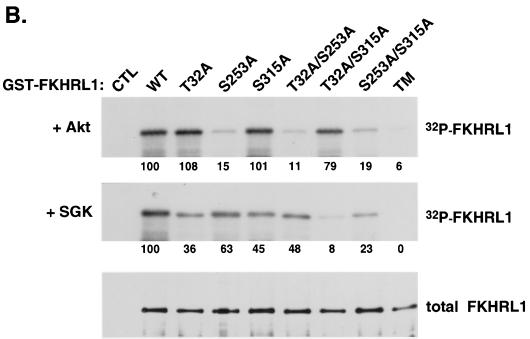
FIG. 1.
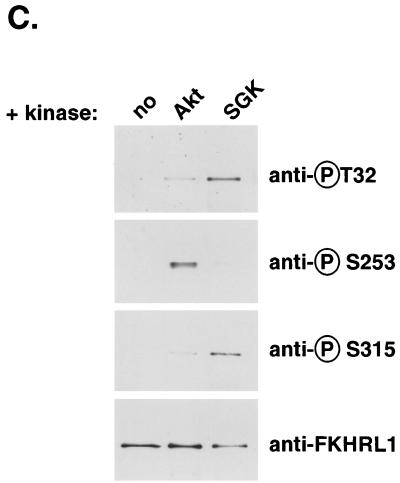
SGK phosphorylates FKHRL1 in vitro. (A) HEK 293 cells were transfected with WT SGK, a KN mutant of SGK (K127Q), or a CA mutant of SGK (S422D) and stimulated with insulin or IGF-I for 15 min. SGK was immunoprecipitated from cell lysates with the anti-HA antibody and incubated in the presence of [γ-32P]ATP with WT GST-FKHRL1 (upper panels). The total level of GST-FKHRL1 was analyzed by immunoblotting with an antibody directed against total FKHRL1 (lower panels). (B) HEK 293 cells were transfected with a CA mutant of Akt. Akt was immunoprecipitated from cell lysates with the anti-HA antibody and incubated in the presence of [γ-32P]ATP with WT GST-FKHRL1 or a series of phosphorylation mutants of FKHRL1 (upper panel). Purified CA SGK was obtained from Upstate Biotechnology and incubated in the presence of [γ-32P]ATP with WT GST-FKHRL1 or a series of phosphorylation mutants of FKHRL1 (middle panel). The total levels of GST-FKHRL1 were analyzed by immunoblotting with an antibody directed against total FKHRL1 (lower panel). CTL, control; TM, triple mutant of FKHRL1 (T32A/S253A/S315A). The numbers represent the percentage of phosphorylation of FKHRL1 mutants compared to wild-type FKHRL1. (C) Extracts obtained for panel B with WT-FKHRL1 were analyzed by immunoblotting with antibodies directed against phospho-T32, phospho-S253, and phospho-S315 or with the antibody directed against total FKHRL1.
To identify the regulatory sites of FKHRL1 that are phosphorylated by SGK, in vitro kinase assays were performed using as substrates WT FKHRL1 or a series of mutants in which the three FKHRL1 phosphorylation sites (Thr-32, Ser-253, and Ser-315) had been replaced by alanines either individually or in combination (Fig. 1B). We found that SGK no longer phosphorylated the T32A/S315A double mutant but still phosphorylated the S253A single mutant, indicating that Thr-32 and Ser-315 are the major FKHRL1 sites phosphorylated by SGK in vitro. In contrast, Akt still phosphorylated the FKHRL1 T32A/S315A double mutant but no longer phosphorylated the FKHRL1 S253A mutant. These experiments show that in contrast to Akt, which prefers Ser-253, SGK favors Thr-32 and Ser-315 in vitro.
To confirm directly the difference in the efficacy with which SGK and Akt phosphorylate the three regulatory sites of FKHRL1, we performed immunoblot analyses on the samples obtained by kinase assay, using antibodies that specifically recognize each of the three phosphorylation sites of FKHRL1 (Fig. 1C). Consistent with the results obtained by kinase assays, we found that SGK favored the phosphorylation of Thr-32 and Ser-315 of FKHRL1 whereas Akt markedly preferred Ser-253. These results are consistent with previous studies which showed that although Akt can phosphorylate all three FOXO regulatory sites when highly active, it has a preference for the second site of phosphorylation under conditions that are within the linear range of activity for this kinase (7, 37, 43).
These experiments show that SGK, like Akt, phosphorylates FKHRL1 in vitro but that SGK and Akt display a difference in the efficacy with which they phosphorylate the three FKHRL1 phosphorylation sites. Our results are consistent with the observation that the amino acid sequences surrounding Thr-32 and Ser-315 correspond more closely to the SGK consensus phosphorylation sequence than to the Akt consensus phosphorylation site (28). In contrast, Ser-253 is surrounded by amino acids that form a better consensus Akt phosphorylation site (1).
SGK phosphorylates the Forkhead transcription factor FKHRL1 and displays a preference for Ser-315 within cells.
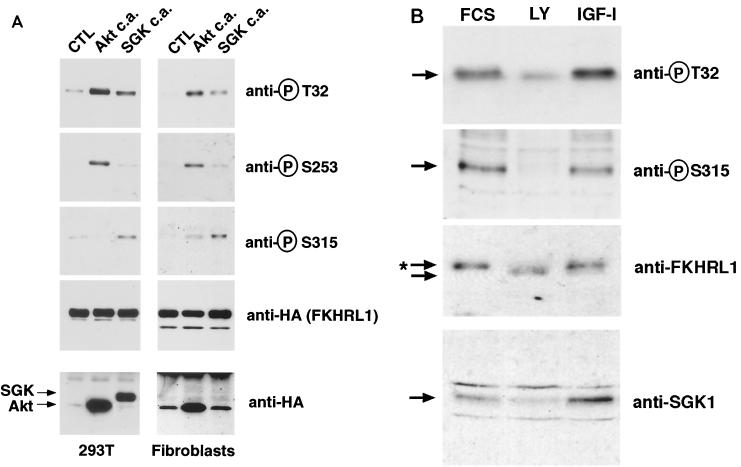
We next determined whether SGK also induces the phosphorylation of FKHRL1 within cells and whether the difference in the efficacy with which SGK and Akt phosphorylate FKHRL1 regulatory sites is also observed in vivo. To this end, we cotransfected fibroblasts or HEK 293T cells with constructs expressing CA SGK or Akt together with FKHRL1 and performed immunoblotting with phosphoantibodies to FKHRL1. Overexpression of CA versions of Akt or SGK within cells led to the phosphorylation of all three regulatory sites of FKHRL1 (data not shown). However, when the amount of the expression plasmids transfected into cells was reduced so that the levels of expressed SGK and Akt were lower and closer to the endogenous levels, we found that SGK was clearly more potent than Akt at inducing the phosphorylation of FKHRL1 at Ser-315 (Fig. 2A). In contrast, Akt was more efficient than SGK at phosphorylating FKHRL1 at Ser-253. Thr-32, although preferred in vitro by SGK, appeared to be phosphorylated by both Akt and SGK within cells. These findings corroborate the specificity of SGK and Akt for FKHRL1 observed in vitro. These results also indicate that although when overexpressed, Akt and SGK can phosphorylate all three phosphorylation sites of FKHRL1, when expressed at lower, potentially more physiological levels, these two kinases display a marked preference for particular regulatory sites of FKHRL1.
FIG. 2.
SGK induces FKHRL1 phosphorylation in vivo. (A) CCL39 fibroblasts or HEK 293T cells were cotransfected with a construct encoding WT HA-FKHRL1 and either a control empty vector (CTL) or constructs encoding CA mutants of Akt (Akt c.a.) or SGK (SGK c.a.). Small amounts of the Akt plasmid were transfected in order to obtain a level of expression of Akt that is close to the low level of expression of SGK. Cell lysates were analyzed by immunoblotting with antibodies directed against phospho-T32, phospho-S253, or phospho-S315 FKHRL1 or with the antibody directed against the HA epitope (for HA-FKHRL1, HA-Akt, and HA-SGK). In fibroblasts, the levels of SGK expression are too low to be detected by the anti-HA antibody. (B) Primary cultures of cerebellar granule neurons were incubated in the presence of 10% serum, treated with 10 μM LY for 1 h, or stimulated with 100 ng of IGF-I/ml for 20 min. Cell lysates were analyzed by immunoblotting with antibodies directed against SGK1, phospho-T32, or phospho-S315 or with the antibody directed against total FKHRL1. ∗, phosphorylated form of FKHRL1.
To determine if endogenous FKHRL1 is phosphorylated within cells at the preferred SGK phosphorylation sites in response to extracellular stimuli, we used primary cultures of cerebellar granule neurons, which express high levels of endogenous FKHRL1 (A. Brunet and M. E. Greenberg, unpublished data). The neurons were incubated in the presence of survival factors (FCS or IGF-I) or in the presence of a chemical inhibitor that blocks the PI3K pathway (LY). The levels of endogenous FKHRL1 protein remained constant in the absence or presence of extracellular stimulation (Fig. 2B). In contrast, immunoblots with phosphoantibodies to FKHRL1 showed that endogenous FKHRL1 was phosphorylated at the SGK-preferred sites Thr-32 and Ser-315 (Fig. 2B) in response to survival factors in cerebellar granule neurons and that the phosphorylation of endogenous FKHRL1 at these sites was blocked by the PI3K inhibitor LY. We found that FKHRL1 was also phosphorylated at the Akt-preferred site Ser-253 in cerebellar granule cells in response to growth factors (data not shown). Consistent with these findings, endogenous FKHRL1 underwent a retardation in its electrophoretic mobility upon growth factor stimulation (Fig. 2B), an event which had been previously described as being due to the phosphorylation of FKHRL1 at all three regulatory sites (7). We then generated an antibody directed against rat SGK1 and showed by immunoblot analyses that endogenous SGK is expressed in the cerebellar granule neurons (Fig. 2B). As reported previously for other cell types (8), endogenous SGK protein is expressed at low levels in the absence of growth factors but SGK's expression is up-regulated when neurons are incubated in the presence of IGF-I for 20 min. These findings show that the phosphorylation of endogenous FKHRL1 at SGK-preferred sites is an event that occurs in vivo in response to growth and survival factors and is correlated with the expression of endogenous SGK.
Other PI3K-activated protein kinases do not play a significant role in phosphorylating FKHRL1.
We have shown that SGK and Akt, two protein kinases activated by growth and survival factors in a PI3K-dependent manner, are capable of phosphorylating FKHRL1 in vitro and in vivo. However, an increasing number of protein kinases have recently been reported to be regulated by PI3K, including protein kinase A (13), p70S6 kinase (2, 42), p90RSK (25, 44), and the conventional and atypical isoforms of PKC (22, 32). Most of these serine/threonine kinases are structurally related to Akt and known to phosphorylate a consensus sequence with an arginine three amino acids N-terminal to the phosphoacceptor site, which corresponds to the amino acid sequence that surrounds the three FKHRL1 phosphorylation sites.
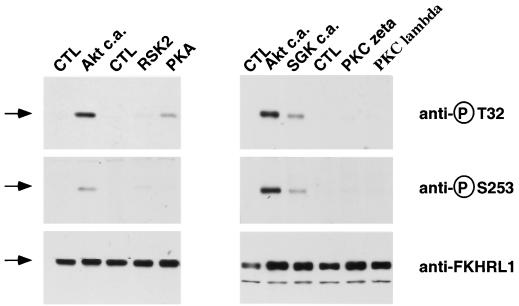
We therefore determined if these other PI3K-activated protein kinases were capable of inducing FKHRL1 phosphorylation. In contrast to our findings with SGK and Akt, immunoblot experiments using anti-FKHRL1 phosphospecific antibodies showed that overexpression of CA forms of the atypical PKC ζ or λ did not lead to an enhancement of FKHRL1 phosphorylation (Fig. 3). Likewise, expression of PKA or p90RSK only weakly induced the phosphorylation of FKHRL1 at Thr-32, Ser-253 (Fig. 3), and Ser-315 (data not shown).
FIG. 3.
Effects of various protein kinases on FKHRL1 phosphorylation. CCL39 fibroblasts were cotransfected with a construct encoding WT HA-FKHRL1 and constructs encoding CA forms of various protein kinases. Immunoblot analyses were performed as described for Fig. 2.
These results show that overexpression of protein kinases related to Akt does not necessarily lead to FKHRL1 phosphorylation and support the conclusion that SGK phosphorylation of FKHRL1 is an event that likely occurs in vivo. We conclude that SGK and Akt are unique among the known PI3K-regulated protein kinases in their ability to efficiently phosphorylate FKHRL1.
Endogenous SGK participates in FKHRL1 phosphorylation in response to growth factor signaling.
To determine if SGK activity is required for FKHRL1 phosphorylation in response to growth factors, we interfered with the activity of endogenous SGK in cells by expressing dominant-negative SGK alleles. To this end, we first verified that endogenous SGK was expressed in HEK 293T cells. Since available antibodies do not recognize the endogenous SGK protein in these cells, we performed RT-PCR analysis of mRNA extracted from HEK 293T cells with pairs of primers that are specific for the SGK1 isoform. We found that the SGK1 mRNA is transcribed in HEK 293T cells, indicating that SGK1 protein is likely expressed in these cells (Fig. 4A). Consistent with the previous finding that SGK expression is induced by serum and glucocorticoids (8), we found that the levels of SGK mRNA expression were slightly up-regulated in HEK 293T cells that were exposed to serum and dexamethasone for 1 h (Fig. 4A).
We then transfected HEK 293T cells with a KN mutant of Akt (K197M) or SGK (T256A/S422A) and assessed by immunoblot analyses the level of phosphorylation of endogenous FKHRL1 after growth factor addition. We found that expression of KN Akt partially inhibited phosphorylation of endogenous FKHRL1, especially at Ser-253. Likewise, expression of KN SGK partially inhibited phosphorylation of endogenous FKHRL1 at Thr-32, Ser-253, and Ser-315 (Fig. 4B). The findings obtained with the phosphospecific FKHRL1 antibodies were corroborated by an analysis of the effect the KN Akt or SGK mutants had on the electrophoretic mobility of endogenous FKHRL1. In the presence of growth factors, endogenous FKHRL1 migrates as two forms: a slowly migrating form that is phosphorylated at Thr-32, Ser-253, and Ser-315 and a more rapidly migrating form that is unphosphorylated at these sites (7). As shown in Fig. 4B, the expression of KN SGK or Akt led to the disappearance of the slowly migrating phosphorylated form of FKHRL1, suggesting that these mutants block the ability of endogenous SGK or Akt to phosphorylate FKHRL1. However, it is possible that these dominant-negative forms of SGK or Akt each inhibit the common activator of SGK and Akt, PDK1. If this is true, then expression of dominant-negative Akt or SGK mutants alone may be sufficient to inhibit the endogenous forms of both Akt and SGK. Thus, in the absence of genetic disruption of the SGK and Akt family members or the availability of specific chemical inhibitors of Akt and SGK, it is not possible to show conclusively which family of kinases is required for the phosphorylation of endogenous FKHRL1 at specific sites.
SGK activity is important for promoting FKHRL1 relocalization to the cytoplasm upon growth factor stimulation.
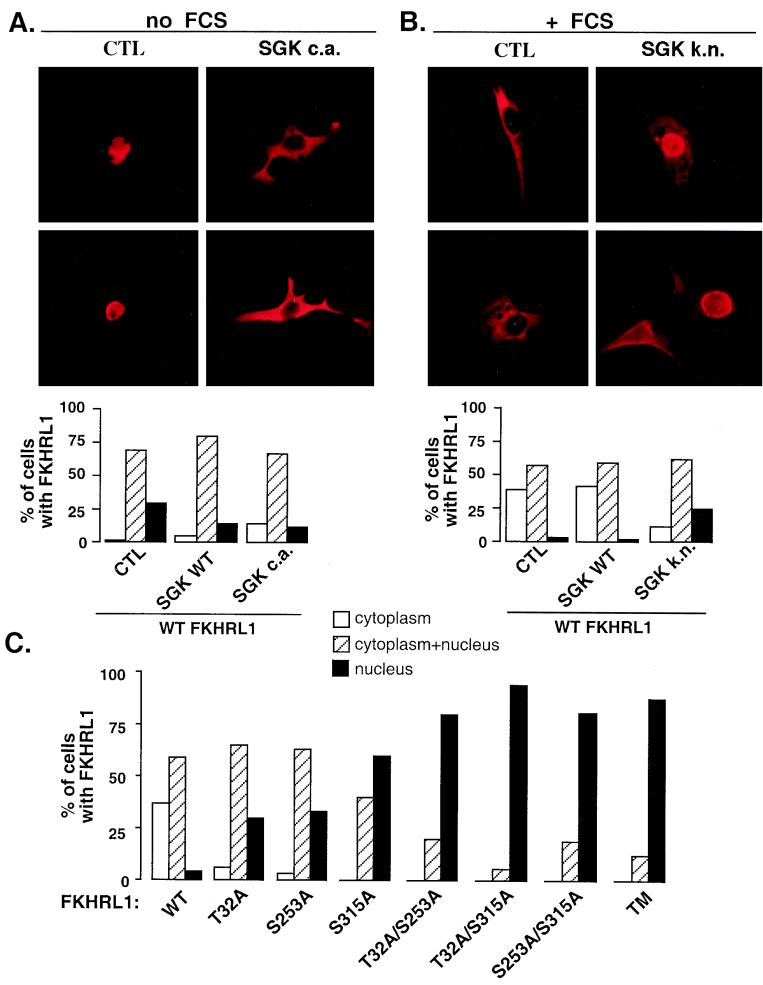
To determine if SGK, by phosphorylating FKHRL1, promotes the sequestration of FKHRL1 within the cytoplasm, we first coexpressed WT SGK, the CA SGK mutant (S422D), or the KN SGK mutant (T256A/S422A) together with FKHRL1 and examined the effect on the subcellular localization of FKHRL1. In the absence of growth factors, FKHRL1 was found to be predominantly in the nucleus (Fig. 5A). However, when FKHRL1 was coexpressed with the CA S422D mutant of SGK, a significant fraction of FKHRL1 was present in the cytoplasm (Fig. 5A). This suggests that SGK phosphorylation of FKHRL1 leads to the translocation of FKHRL1 from the nucleus to the cytoplasm.
FIG. 5.
SGK plays a critical role in promoting FKHRL1 exclusion from the nucleus in response to growth factors. (A) CCL39 fibroblasts were cotransfected with a WT M2-tagged FKHRL1 construct and either an empty vector (CTL) or plasmids encoding WT SGK or CA SGK (S422D) (SGK c.a.); then they were serum starved for 8 h and incubated in the presence of 20 μM LY for the last hour. FKHRL1 was detected by immunolocalization with an anti-M2 antibody. Quantification of a representative experiment is shown. (B) CCL39 fibroblasts were cotransfected with a WT M2-tagged FKHRL1 construct and either an empty vector (CTL) or plasmids encoding WT SGK or KN SGK (T256A/S422A) and were incubated with 10% FCS. FKHRL1 was detected by immunolocalization with the anti-M2 antibody. Quantification of a representative experiment is shown. (C) CCL39 fibroblasts were transiently transfected with an HA-tagged FKHRL1 construct (WT or the different phosphorylation site mutants) and were incubated in the presence of 10% FCS. FKHRL1 was detected by immunolocalization with the anti-HA antibody. Quantification of a representative experiment is shown. TM, triple mutant of FKHRL1 (T32A/S253A/S315A).
In contrast to the effects of the CA SGK, the expression of the KN SGK T256A/S422A mutant prevented growth factor induction of FKHRL1 translocation from the nucleus to the cytoplasm (Fig. 5B). This result supports the conclusion that the KN T256A/S422A mutant of SGK functions as a dominant-interfering form of SGK that, when expressed in cells, blocks growth factor activation of endogenous SGK. This result further suggests that activation of endogenous SGK is necessary for growth factor-induced translocation of FKHRL1 from the nucleus to the cytoplasm.
The observation that SGK expression, like Akt expression, leads to FKHRL1 translocation from the nucleus to the cytoplasm taken together with the observation that these two kinases phosphorylate distinct regulatory sites on FKHRL1 raised the possibility that SGK and Akt might cooperate in promoting an efficient exclusion of FKHRL1 from the nucleus. To test this hypothesis, we analyzed the subcellular localization of FKHRL1 mutants in which the three regulatory phosphorylation sites were replaced by alanines, either individually or in combination (Fig. 5C). In the presence of growth factors, WT FKHRL1 was mainly localized in the cytoplasm of transfected cells. In contrast, the number of cells in which FKHRL1 single mutants were found in the nucleus was significantly higher than the number of cells expressing WT FKHRL1 in the nucleus (Fig. 5C). Moreover, the number of cells expressing FKHRL1 double or triple phosphorylation site mutants in the nucleus was even greater than the number of cells displaying each single FKHRL1 mutant in the nucleus. These results indicate that the phosphorylation of FKHRL1 at Thr-32, Ser-253, and Ser-315 is necessary to promote the efficient translocation of FKHRL1 from the nucleus to the cytoplasm.
SGK suppresses FKHRL1 transcriptional activity.
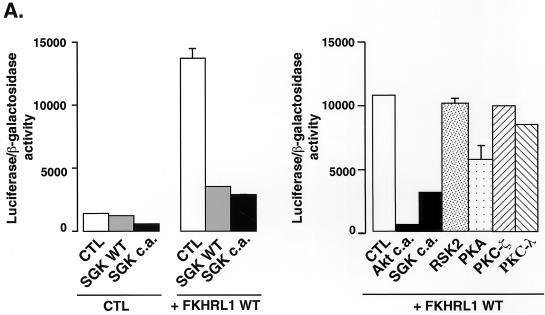
Since SGK phosphorylation of FKHRL1 promotes the exit of FKHRL1 from the nucleus, we expected that SGK phosphorylation of FKHRL1 might lead to a reduction in FKHRL1-dependent transcription. We tested whether SGK inhibits FKHRL1-dependent transcription by performing luciferase assays using a FHRE-driven luciferase reporter construct. We found that WT SGK and the CA mutants of SGK effectively blocked FKHRL1-dependent transcription (Fig. 6A, left panel). In contrast to the expression of SGK or Akt, the expression of PKA, p90RSK, PKC ζ, or PKC λ, which does not lead to FKHRL1 efficient phosphorylation (Fig. 3), did not have a significant effect on FKHRL1-dependent transcription (Fig. 6A, right panel).
FIG. 6.
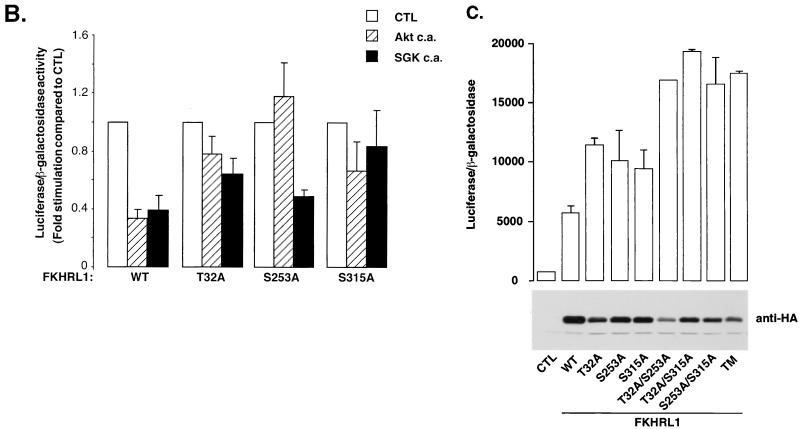
SGK inhibits FKHRL1-dependent transcription. (A) CCL39 fibroblasts were transiently cotransfected with an empty vector (CTL) or a vector encoding WT FKHRL1 together with WT SGK, CA SGK (S422D) (SGK c.a.), or CA mutants of various protein kinases and the FHRE-Luc reporter construct. The day after transfection, cells were starved for 24 h and luciferase activity was assayed. Data are the means and variances for two independent experiments conducted in duplicate. (B) CCL39 fibroblasts were transiently cotransfected with an empty vector (CTL), a vector encoding CA SGK (S422D) (SGK c.a.), or a vector encoding CA Akt (Akt c.a.), together with vectors encoding WT FKHRL1 or the different FKHRL1 phosphorylation site mutants and the FHRE-Luc reporter construct. Luciferase assays were performed as described for panel A. Data are the means and variances for two independent experiments conducted in duplicate. (C) CCL39 fibroblasts were transiently cotransfected with an empty vector (CTL) or vectors encoding WT FKHRL1 or the different FKHRL1 phosphorylation site mutants and the FHRE-Luc reporter construct. The day after transfection, cells were incubated in the presence of IGF-I for 24 h and luciferase activity was assayed. Data are the means and variances of two independent experiments conducted in duplicate. Expression of the different mutants of FKHRL1 was monitored by immunoblotting with the anti-HA antibody. TM, triple mutant of FKHRL1 (T32A/S253A/S315A) .
Since SGK phosphorylates FKHRL1 in vivo mainly at Thr-32 and Ser-315 but poorly at Ser-253, we predicted that SGK would no longer block FKHRL1-dependent transcription when FKHRL1 was mutated at Thr-32 or Ser-315. To test this prediction, we performed luciferase assays using the FHRE-luciferase reporter gene and either WT FKHRL1 or a FKHRL1 T32A, S253A, or S315A mutant in the absence or presence of CA SGK or Akt (Fig. 6B). We found that SGK was less capable of inhibiting the transcriptional activity of FKHRL1 T32A and S315A mutants but still efficiently repressed the transcriptional activity of the FKHRL1 S253A mutant. By contrast, Akt, which favors the phosphorylation of Ser-253 and Thr-32 in vivo, no longer effectively inhibited transcription mediated by FKHRL1 S253A or T32A mutants but still inhibited to some extent transcription mediated by FKHRL1 S315A mutants. The partial rather than complete inhibition of the transcriptional activity of the FKHRL1 S315A mutant by Akt is probably due to the fact that Akt, especially when overexpressed, can lead to an increase in Ser-315 phosphorylation. These results indicate that SGK and Akt promote the repression of FKHRL1-dependent transcription by phosphorylating distinct FKHRL1 phosphorylation sites.
To further investigate whether the SGK- or Akt-favored phosphorylation sites cooperate to achieve the efficient repression of FKHRL1-dependent transcription, we tested the transcriptional activity of various combinations of FKHRL1 phosphorylation site mutants in the presence of survival factors (Fig. 6C). We found that replacement of each phosphorylation site of FKHRL1 individually by an alanine led to an increase in FKHRL1-dependent transcription, indicating that the phosphorylation of each site plays a role in the repression of FKHRL1-dependent transcription. In addition, replacement of two or three of the FKHRL1 phosphorylation sites by alanine led to an even further enhancement of FKHRL1-dependent transcription. These results indicate that the phosphorylation of different phosphorylation sites of FKHRL1, catalyzed by SGK and Akt, is critical for the effective repression of FKHRL1-dependent transcription.
FKHRL1 induces cell cycle arrest and apoptosis when mutated at SGK or Akt phosphorylation sites.
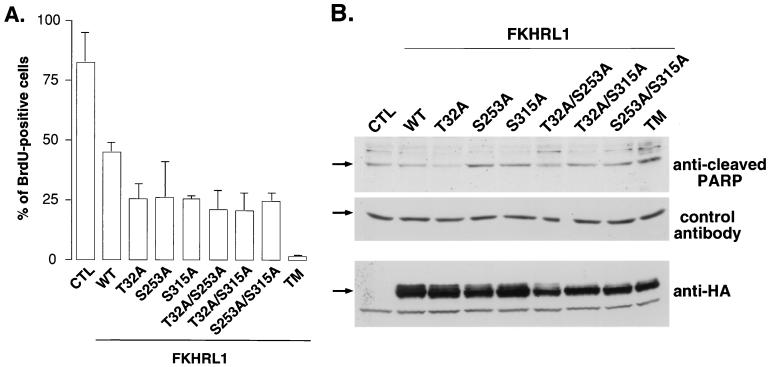
A recently described transcriptional target of FKHRL1 is the cell cycle inhibitor gene p27KIP1, and indeed, FKHRL1 and the other FOXOs have been shown to induce cell cycle arrest when present in the nucleus (36). Since the phosphorylation of each regulatory site of FKHRL1 contributes to the exclusion of FKHRL1 from the nucleus and the repression of FKHRL1-dependent transcription, we asked if the efficient phosphorylation of the different regulatory sites of FKHRL1 by Akt and SGK was required to block FKHRL1-dependent cell cycle arrest. To this end, we transfected the various FKHRL1 phosphorylation site mutants into fibroblasts and measured reentry into the S phase of the cell cycle by BrdU incorporation after growth factor stimulation (Fig. 7A). We found that FKHRL1 single and double phosphorylation site mutants were more efficient at blocking cell cycle reentry than WT FKHRL1. The mutant of FKHRL1 in which all three phosphorylation sites were replaced by an alanine was the most effective at blocking cell cycle reentry. These results indicate that the phosphorylation of the three regulatory sites of FKHRL1 is important to prevent FKHRL1 from inducing cell cycle arrest.
FIG. 7.
The phosphorylation of FKHRL1 at the three regulatory sites is required to prevent FKHRL1-induced cell cycle arrest and apoptosis. (A) CCL39 fibroblasts were transfected with the various phosphorylation site mutants of FKHRL1, starved for 24 h, and stimulated for 24 h with 20% FCS in the presence of BrdU. Transfected cells and the incorporation of BrdU were identified by immunofluorescence with an anti-FKHRL1 antibody and an anti-BrdU antibody, respectively. Data are the means and variances of two independent experiments. CTL, control; TM, triple mutant of FKHRL1 (T32A/S253A/S315A). (B) HEK 293T cells were transfected with the various FKHRL1 phosphorylation site mutants and stimulated for 24 h with IGF-I. Immunoblot analyses were performed with an antibody directed against the cleaved product of PARP, a control antibody to a protein whose levels remain constant (MKK1), and an antibody to HA. TM, triple mutant of FKHRL1 (T32A/S253A/S315A) .
In addition to inducing cell cycle arrest in fibroblasts, another function of the FOXO subfamily of Forkhead transcription factors is to trigger apoptosis (7, 36, 45, 46). To assess whether the phosphorylation of FKHRL1 at the different regulatory sites is required to prevent this transcription factor from inducing apoptosis, we transfected the various FKHRL1 phosphorylation site mutants into HEK 293T cells in the presence of the survival factor IGF-I. We assessed apoptosis of the transfected cells by performing immunoblot analyses with an antibody that specifically binds the cleaved product of PARP (Fig. 7B). In living cells, PARP is intact and thus not recognized by the antibody, whereas in cells undergoing apoptosis, caspases are activated and cleave PARP, one of whose cleavage products is recognized by the antibody. We found that while WT FKHRL1 did not induce PARP cleavage, the mutants of FKHRL1 with single and especially double and triple phosphorylation site mutations triggered PARP cleavage. These results indicate that the efficient phosphorylation of FKHRL1 at all three regulatory sites is critical to prevent FKHRL1 from inducing cell death.
SGK plays a role in the promotion of growth factor-induced cell survival.
Our observation that SGK suppresses FKHRL1-dependent transcription and that the phosphorylation of FKHRL1 at all three regulatory sites is important for the prevention of FKHRL1-induced cell cycle arrest and apoptosis suggested that SGK may regulate these two biological functions. Indeed, a role for SGK in cell cycle progression had been suggested in a recent study (8).
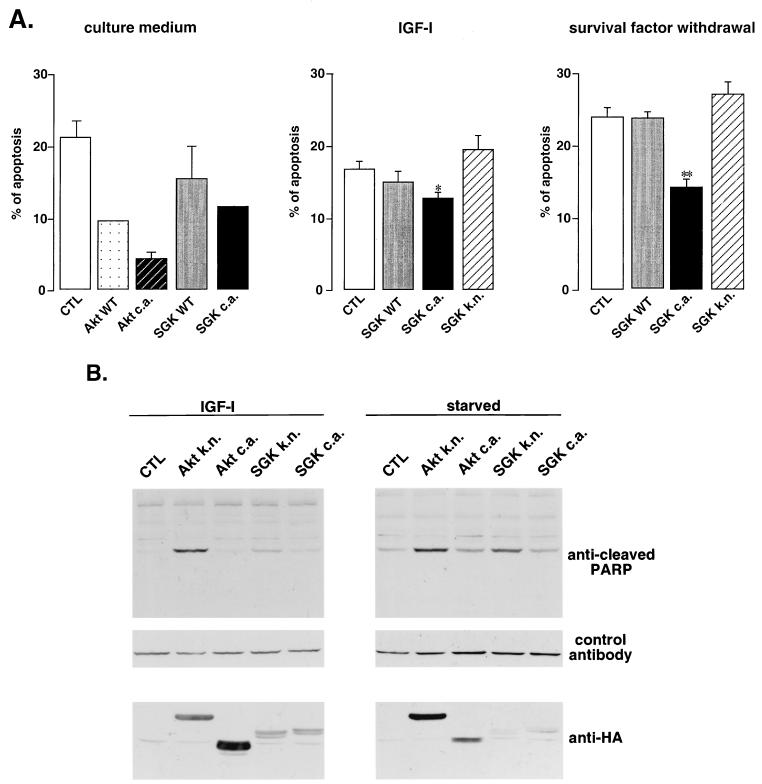
To determine whether SGK plays a critical role in regulating cell survival, we first employed primary cultures of cerebellar granule neurons, a population of neurons that is dependent both on PI3K and on the repression of FKHRL1-dependent transcription for cell survival (7, 21). As shown in Fig. 2B, these neurons express an endogenous form of SGK as well as an endogenous form of FKHRL1 that is phosphorylated in response to survival factors. After transfecting cerebellar granule cells with various forms of SGK and counting the number of apoptotic and surviving cells among the transfected cells, we then asked whether SGK could promote the survival of neurons. The expression of a CA form of SGK, as with Akt, leads to a significant reduction of apoptotic neurons whether the cerebellar neurons are grown in the presence or absence of survival factors (with a P value of <0.01 according to analysis of variance) (Fig. 8A). In contrast, the expression of KN SGK (T256A/S422A) in cerebellar neurons led to a small increase in the number of cells undergoing apoptosis (Fig. 8A), which is consistent with ability of this SGK mutant to act in a dominant-interfering manner to prevent endogenous SGK from phosphorylating FKHRL1 and relocalizing it to the cytoplasm.
FIG. 8.
SGK plays a role in cell survival regulation. (A) Cerebellar granule cells were transiently cotransfected with a vector encoding GFP and either an empty vector (CTL), vectors encoding WT or CA Akt (Akt c.a.), or vectors encoding WT SGK, CA SGK (S422D) (SGK c.a.), or KN SGK (T256A/S422A) (SGK k.n.). Cells were either left in culture medium, incubated with IGF-I for 24 h, or deprived of all survival factors for 8 h. Transfected cells were followed by detection of GFP fluorescence, and DNA was stained with the Hoechst compound. The experiments were performed in duplicate, and 100 to 200 cells per coverslip were counted in a blinded fashion. Data in the left panel are the means and variances of two independent experiments conducted in duplicate; data in the middle and right panels are the means and standard errors of the means of three independent experiments conducted in duplicate. The results comparing the effects of the KN and CA mutants of SGK are statistically significant according to analysis of variance, with a P value of <0.05 (∗) or <0.01 (∗∗). (B) HEK 293T cells were transiently transfected with a vector alone (CTL), a vector encoding CA Akt or SGK (S422D), or a vector encoding KN Akt (K197M) or SGK (T256A/S422A). At 24 h after transfection, cells were either starved or incubated in the presence of 50 ng of IGF-I/ml for 24 h. Similar amounts of total proteins were resolved by SDS-PAGE. Immunoblot analyses were performed using an antibody directed against the cleaved product of PARP, an antibody to a control protein (MKK1), and an antibody to HA.
To further determine if SGK activity is critical for mediating cell survival, we expressed KN mutants of SGK or Akt in HEK 293T cells in the presence or absence of IGF-I and monitored apoptosis by the appearance of the cleaved form of PARP (Fig. 8B). We found that the KN forms of SGK and Akt in HEK 293T cells induced the cleavage of PARP, while the CA forms did not.
It has to be noted that in cerebellar neurons as well as in HEK 293T cells, the effects of Akt mutants on cell survival were more pronounced than the effects of SGK mutants. However this difference could be due to a difference in the expression or activity of the mutants of Akt or SGK, making it difficult to determine which kinase, Akt or SGK, plays a more critical role in mediating cell survival.
Taken together, these findings indicate that SGK, when expressed and activated, may together with Akt mediate survival effects of PI3K, in part by phosphorylating FKHRL1 and repressing its transcriptional activity. The inhibition of SGK and Akt activity may trigger the dephosphorylation of FKHRL1, the translocation of FKHRL1 from the cytoplasm to the nucleus, and the activation of FKHRL1-regulated death genes, thereby inducing apoptosis.
DISCUSSION
In this report we describe experiments demonstrating that the PI3K-regulated kinase SGK1 phosphorylates the FOXO transcription factor FKHRL1, thereby inducing the exit of FKHRL1 from the nucleus and the repression of FKHRL1-dependent transcription. Our experiments identify FKHRL1 as a physiological substrate of SGK and suggest a biological role for this protein kinase in the regulation of cell survival and cell cycle progression. Our findings suggest that SGK functions as a critical link between growth factor activation of the PI3K and the regulation of gene expression events that are critical for biological responses such as cell survival and cell cycle reentry.
SGK and Akt may cooperate in promoting a variety of biological responses by phosphorylating common substrates at overlapping but different regulatory sites. SGK and Akt differ with respect to the efficacy with which they phosphorylate specific sites on FKHRL1, since SGK preferentially phosphorylates Ser-315 whereas Akt favors Ser-253. The coordinated phosphorylation of FKHRL1 at three sites by SGK and Akt could involve the sequential phosphorylation of the different sites of FKHRL1. In a recent study of the FKHRL1 family member FKHR, the site equivalent to FKHRL1 Ser-253 was suggested to act as a “gatekeeper,” since the phosphorylation of Ser-253 was found to be required to release a negative constraint on the FKHR molecule so that FKHR could then be phosphorylated at additional sites (37). Nakae and colleagues suggested that while Akt is responsible for phosphorylating FKHR at Ser-253, other unidentified protein kinases that are distinct from Akt may phosphorylate the two other sites on FKHR (37). Given the findings in the present study, it is tempting to speculate that Akt may first phosphorylate FKHRL1 at Ser-253 and that this phosphorylation event may induce a conformational change in FKHRL1 that allows SGK to phosphorylate FKHRL1 at Ser-315. Alternatively, the phosphorylation of FKHRL1 at Ser-253 by Akt may allow the binding to FKHRL1 of 14-3-3 proteins, which then act as cofactors that promote the phosphorylation of FKHRL1 by SGK. A role for 14-3-3 proteins as cofactors that promote sequential protein phosphorylation was recently demonstrated for another Akt target, the Bcl-2 family member BAD (18).
The efficient phosphorylation of the three regulatory sites of FKHRL1 appears to be required to ensure the effective translocation of FKHRL1 from the nucleus to the cytoplasm and the effective repression of FKHRL1-dependent transcription. The phosphorylation of FKHRL1 at each of the regulatory sites may involve different mechanisms that cooperate to promote the efficient exclusion of FKHRL1 from the nucleus. FKHRL1 Ser-315, which is the phosphorylation site favored by SGK, is located close to the nuclear export signal (NES) of FOXOs (6). One possibility is that SGK phosphorylation of Ser-315 unmasks the NES of FKHRL1, thereby rendering the NES accessible to the nuclear export machinery. Thus, SGK phosphorylation of Ser-315 may play a primary role in exporting nuclear FKHRL1 to the cytoplasm upon activation of the PI3K-SGK signaling pathway. The phosphorylation of the other two sites on FKHRL1, Thr-32 and Ser-253, is catalyzed more effectively by Akt and is required for FKHRL1 binding to 14-3-3 proteins (7). When bound to FKHRL1, 14-3-3 proteins may participate in the nuclear export of FKHRL1, since the 14-3-3 proteins have been shown to contain a NES and to be involved in the coexport of their binding partners (33). 14-3-3 protein binding to FKHRL1 may also ensure the retention of FKHRL1 in the cytoplasm. As FKHRL1 Ser-253 is located within the nuclear localization signal of FKHRL1, Akt phosphorylation of Ser-253 may also inhibit the function of the nuclear localization signal of FKHRL1, thereby preventing the reentry of cytoplasmic FKHRL1 into the nucleus.
The regulation of FKHRL1 subcellular localization by phosphorylation is reminiscent of the regulation of the yeast transcription factor Pho4 (27, 29, 40). Pho4 is phosphorylated at several regulatory sites by different kinases and the multiple phosphorylation events cooperate to promote the nuclear export of Pho4 and to prevent the nuclear import of Pho4, thus resulting in the exclusion of this transcription factor from the nucleus (29). The sequential phosphorylation of transcription factors, such as Pho4 or FKHRL1, at several sites that have overlapping but distinct roles in regulating nuclear export may therefore provide a general and efficient mechanism that ensures the effective translocation of these transcription factors from the nucleus to the cytoplasm.
The primary consequence of FKHRL1 phosphorylation at its regulatory sites appears to be the translocation of FKHRL1 from the nucleus to the cytoplasm and the inhibition of FKHRL1-dependent transcription. However, a question that remains to be answered is that of whether there are kinase-dependent mechanisms in addition to the regulation of the subcellular localization of FKHRL1 that contribute to the control of FKHRL1 function. Given that Ser-315, the amino acid residue that is preferentially phosphorylated by SGK, is located within the transactivation domain of FKHRL1, it is possible that the phosphorylation of Ser-315 may also regulate the transactivation function of FKHLR1. In addition, phosphorylation of FKHRL1 at different phosphorylation sites may facilitate FKHRL1 binding to distinct protein partners that have yet to be identified but could prove to be important for FKHRL1 function.
It is not yet clear why there are two families of PI3K-regulated kinases that phosphorylate FKHRL1. One possibility is that when expressed at physiological levels, SGK and Akt display a strict preference for one FKHRL1 site over the others, so that SGK exclusively phosphorylates FKHRL1 at Ser-315 while Akt selectively phosphorylates FKHRL1 at Ser-253. This is consistent with the observation that when expressed at low levels, SGK selectively phosphorylated Ser-315 while Akt was more effective at phosphorylating Ser-253. Another possibility is suggested by the observation that Akt and SGK are differentially expressed in cells in response to environmental stimuli. Whereas Akt is activated within seconds upon growth factor addition and may be involved in the phosphorylation of FKHRL1 at early times after growth factor stimulation, SGK protein is present only at very low levels in cells in the absence of growth factor stimulation (8). The expression of SGK is induced within hours of exposure of the cells to growth factors (8). Thus, active SGK could phosphorylate FKHRL1 at later times after growth factor stimulation and may prolong the effects of growth factor stimulation on gene expression.
The overexpression of KN mutants of SGK or Akt in cells reduces the phosphorylation of FKHRL1 and inhibits FKHRL1 cytoplasmic relocalization in response to growth factors, suggesting that endogenous SGK and Akt may both be required for the phosphorylation of FKHRL1 and for FKHRL1 cytoplasmic retention. In addition, overexpression of KN mutants of SGK or Akt in cells promotes apoptosis, raising the possibility that endogenous SGK and Akt may both play an important role in mediating PI3K-dependent cell survival. However, a potential limitation to the use of the dominant-interfering SGK and Akt mutants to selectively inhibit their endogenous counterparts is that SGK and Akt are both activated by the same upstream kinase, PDK1 (28, 41). Thus, the expression of high levels of dominant-interfering mutants of SGK may lead to inhibition of PDK1 and consequently may prevent the activation of both SGK and Akt (48). Conversely, it is possible that the overexpression of dominant-negative mutants of Akt may interfere with endogenous SGK function. Therefore, many of the data that had been generated in the past few years using dominant-negative mutants of Akt to block endogenous Akt function may have to be reexamined, as the results obtained may reflect the inhibition of both endogenous Akt and SGK. The identification of chemical inhibitors that specifically block SGK or Akt will be particularly useful for resolving this issue.
In our present study, we have defined a new function for SGK in mediating survival in response to survival factors. It will be important to determine whether the survival function of SGK can be generalized to cells that have been challenged with a range of apoptotic stimuli, such as DNA damage, matrix detachment, or cell cycle discordance. The transcription factor FKHRL1 is also likely just one of a number of SGK substrates that mediate the survival function of SGK. Other previously characterized targets of Akt that had been implicated in the regulation of cell survival, such as BAD and caspase 9 (9, 17, 19), may also be found to be phosphorylated and regulated by SGK.
The physiological roles of SGK are not likely to be restricted to the promotion of cell survival. Previous studies have indicated that SGK is present in the nucleus during the S phase of the cell cycle, suggesting that SGK plays an active role in cell cycle progression (8). Importantly, FOXO transcription factors have recently been shown to promote cell cycle arrest by inducing the expression of the cell cycle inhibitor p27KIP1 (36). Consistent with a role for SGK phosphorylation of FKHRL1 in cell cycle progression, we find that mutating any one of the three phosphorylation sites of FKHRL1 generates a form of FKHRL1 that promotes cell cycle arrest. These results suggest that SGK and Akt may together promote cell cycle progression by phosphorylating and inactivating FOXOs, thereby decreasing p27KIP1 levels.
Given the importance of SGK and Akt for cell survival and proliferation, mutations in these kinases are likely to have an impact on cancer development in vertebrates. CA versions of Akt and PI3K are transduced by oncogenic retroviruses and can trigger cell transformation (5, 11). It will be interesting to determine whether the CA versions of SGK will also be found to be tumorigenic. The ability of CA forms of Akt and SGK to induce cell transformation may be a consequence of the inappropriate suppression of FOXO-dependent transcription, due to SGK and/or Akt phosphorylation of FOXO transcription factors. One attractive model is that when cells are exposed to deleterious conditions that inhibit PI3K signaling, FOXOs will translocate to the nucleus and induce the expression of cell death or cell cycle arrest genes. CA SGK and Akt may shut off the expression of these cell death and arrest genes by excluding FOXOs from the nucleus, thereby triggering abnormal proliferation and survival and ultimately cell transformation.
Members of the PI3K pathway are conserved throughout evolution. In particular, SGK homologues are present in yeast, nematodes, flies, and mammals. In the nematode Caenorhabditis elegans, the PI3K-Akt pathway plays an important role in the regulation of insulin metabolism and organismal aging by suppressing the activity of a Forkhead transcription factor termed Daf16 (39). Since in addition to Akt, there exists an SGK counterpart in the nematode, it will be interesting to determine whether the nematode SGK contributes to the suppression of Daf16 function and participates in the control of insulin metabolism and organismal aging.
In summary, the protein kinase SGK appears to integrate the effects of different extracellular stimuli and to regulate a variety of biological functions by phosphorylating key substrates including the FOXO transcription factor FKHRL1. The identification of the array of biological functions and substrates of SGK will be of critical importance in understanding how the PI3K signaling pathway orchestrates cellular responses to various extracellular cues.
ACKNOWLEDGMENTS
We thank G. Firestone, A. Toker, and F. Uberall for kindly providing reagents. We thank S. R. Datta, M. Z. Lin, K. F. Tolias, and J. Zieg for critical reading of the manuscript. We also thank S. R. Datta for stimulating discussion.
This research was supported by an NIH grant (CA43855), a Mental Retardation Research Center grant (HD 18655), and a grant from Daiichi Pharmaceuticals to M.E.G. A.B. is supported by a Human Frontier Long Term Fellowship.
ADDENDUM IN PROOF
While this ipaper was in review, Sonyang and colleagues (Curr. Biol. 10:1233–1236, 2000) demonstrated that SGK-3 (also known as CISK) can both promote cell survival and directly phosphorylate and inactivate the proapoptotic proteins FKHR and BAD.
REFERENCES
- 1.Alessi D R, Caudwell F B, Andjelkovic M, Hemmings B A, Cohen P. Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett. 1996;399:333–338. doi: 10.1016/s0014-5793(96)01370-1. [DOI] [PubMed] [Google Scholar]
- 2.Alessi D R, Kozlowski M T, Weng Q P, Morrice N, Avruch J. 3-Phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6 kinase in vivo and in vitro. Curr Biol. 1998;8:69–81. doi: 10.1016/s0960-9822(98)70037-5. [DOI] [PubMed] [Google Scholar]
- 3.Alliston T N, Maiyar A C, Buse P, Firestone G L, Richards J S. Follicle stimulating hormone-regulated expression of serum/glucocorticoid-inducible kinase in rat ovarian granulosa cells: a functional role for the Sp1 family in promoter activity. Mol Endocrinol. 1997;11:1934–1949. doi: 10.1210/mend.11.13.0033. [DOI] [PubMed] [Google Scholar]
- 4.Alvarez de la Rosa D, Zhang P, Naray-Fejes-Toth A, Fejes-Toth G, Canessa C M. The serum and glucocorticoid kinase sgk increases the abundance of epithelial sodium channels in the plasma membrane of Xenopus oocytes. J Biol Chem. 1999;274:37834–37839. doi: 10.1074/jbc.274.53.37834. [DOI] [PubMed] [Google Scholar]
- 5.Bellacosa A, Testa J R, Staal S P, Tsichlis P N. A retroviral oncogene, akt, encoding a serine threonine kinase containing a SH2-like region. Science. 1991;254:274–277. doi: 10.1126/science.254.5029.274. [DOI] [PubMed] [Google Scholar]
- 6.Biggs W H I, Meisenhelder J, Hunter T, Cavenee W K, Arden K C. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc Natl Acad Sci USA. 1999;96:7421–7426. doi: 10.1073/pnas.96.13.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunet A, Bonni A, Zigmond M J, Lin M Z, Juo P, Hu L S, Anderson M J, Arden K C, Blenis J, Greenberg M E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 8.Buse P, Tran S H, Luther E, Phu P T, Aponte G W, Firestone G L. Cell cycle and hormonal control of nuclear-cytoplasmic localization of the serum- and glucocorticoid-inducible protein kinase, Sgk, in mammary tumor cells. A novel convergence point of anti-proliferative and proliferative cell signaling pathways. J Biol Chem. 1999;274:7253–7263. doi: 10.1074/jbc.274.11.7253. [DOI] [PubMed] [Google Scholar]
- 9.Cardone H M, Roy N, Stennicke H R, Salvesen G S, Franke T F, Stanbridge E, Frisch S, Reed J C. Regulation of cell death protease caspase-9 by phosphorylation. Science. 1998;282:1318–1321. doi: 10.1126/science.282.5392.1318. [DOI] [PubMed] [Google Scholar]
- 10.Casamayor A, Torrance P D, Kobayashi T, Thorner J, Alessi D R. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr Biol. 1999;9:186–197. doi: 10.1016/s0960-9822(99)80088-8. [DOI] [PubMed] [Google Scholar]
- 11.Chang H W, Aoki M, Fruman D, Auger K R, Bellacosa A, Tsichlis P N, Cantley L C, Roberts T M, Vogt P K. Transformation of chicken cells by the gene encoding the catalytic subunit of PI 3-kinase. Science. 1997;276:1848–1850. doi: 10.1126/science.276.5320.1848. [DOI] [PubMed] [Google Scholar]
- 12.Chen S Y, Bhargava A, Mastroberardino L, Meijer O C, Wang J, Buse P, Firestone G L, Verrey F, Pearce D. Epithelial sodium channel regulated by aldosterone-induced protein sgk. Proc Natl Acad Sci USA. 1999;96:2514–2519. doi: 10.1073/pnas.96.5.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng X, Ma Y, Moore M, Hemmings B A, Taylor S S. Phosphorylation and activation of cAMP-dependent protein kinase by phosphoinositide-dependent protein kinase. Proc Natl Acad Sci USA. 1998;95:9849–9854. doi: 10.1073/pnas.95.17.9849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou M M, Hou W, Johnson J, Graham L K, Lee M H, Chen C S, Newton A C, Schaffhausen B S, Toker A. Regulation of protein kinase C zeta by PI 3-kinase and PDK-1. Curr Biol. 1998;8:1069–1077. doi: 10.1016/s0960-9822(98)70444-0. [DOI] [PubMed] [Google Scholar]
- 15.Cowling R T, Birnboim H C. Expression of serum- and glucocorticoid-regulated kinase (sgk) mRNA is up-regulated by GM-CSF and other proinflammatory mediators in human granulocytes. J Leukoc Biol. 2000;67:240–248. doi: 10.1002/jlb.67.2.240. [DOI] [PubMed] [Google Scholar]
- 16.Datta S R, Brunet A, Greenberg M E. Cellular survival: a play in three Akts. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 17.Datta S R, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg M E. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 18.Datta S R, Katsov A, Hu L, Petros A, Fesik S W, Yaffe M B, Greenberg M E. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol Cell. 2000;6:41–51. [PubMed] [Google Scholar]
- 19.del Peso L, Gonzalez-Garcia M, Page C, Herrera R, Nunez G. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 20.Downward J. Mechanisms and consequences of activation of protein kinase B/Akt. Curr Opin Cell Biol. 1998;10:262–267. doi: 10.1016/s0955-0674(98)80149-x. [DOI] [PubMed] [Google Scholar]
- 21.Dudek H, Datta S R, Franke T F, Birnbaum M, Yao R, Cooper G M, Segal R A, Kaplan D R, Greenberg M E. Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science. 1997;275:661–665. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- 22.Dutil E M, Toker A, Newton A C. Regulation of conventional protein kinase C isozymes by phosphoinositide-dependent kinase 1 (PDK1) Curr Biol. 1998;8:1366–1375. doi: 10.1016/s0960-9822(98)00017-7. [DOI] [PubMed] [Google Scholar]
- 23.Guo S, Rena G, Cichy S, He X, Cohen P, Unterman T. Phosphorylation of serine 256 by protein kinase B disrupts transactivation by FKHR and mediates effects of insulin on IGF binding protein-1 promoter activity through a conserved insulin response sequence. J Biol Chem. 1999;274:17184–17192. doi: 10.1074/jbc.274.24.17184. [DOI] [PubMed] [Google Scholar]
- 24.Imaizumi K, Tsuda M, Wanaka A, Tohyama M, Takagi T. Differential expression of sgk mRNA, a member of the Ser/Thr protein kinase gene family, in rat brain after CNS injury. Brain Res Mol Brain Res. 1994;26:189–196. doi: 10.1016/0169-328x(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 25.Jensen C J, Buch M B, Krag T O, Hemmings B A, Gammeltoft S, Frodin M. 90-kDa ribosomal S6 kinase is phosphorylated and activated by 3-phosphoinositide-dependent protein kinase-1. J Biol Chem. 1999;274:27168–27176. doi: 10.1074/jbc.274.38.27168. [DOI] [PubMed] [Google Scholar]
- 26.Kaestner K H, Knochel W, Martinez D E. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–146. [PubMed] [Google Scholar]
- 27.Kaffman A, Rank N M, O'Shea E K. Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev. 1998;12:2673–2683. doi: 10.1101/gad.12.17.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi T, Cohen P. Activation of serum- and glucocorticoid-regulated protein kinase by agonists that activate phosphatidylinositide 3-kinase is mediated by 3-phosphoinositide-dependent protein kinase-1 (PDK1) and PDK2. Biochem J. 1999;339:319–328. [PMC free article] [PubMed] [Google Scholar]
- 29.Komeili A, O'Shea E K. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science. 1999;284:977–980. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- 30.Kops G J, Burgering B M. Forkhead transcription factors: new insights into protein kinase B (c-akt) signaling. J Mol Med. 1999;77:656–665. doi: 10.1007/s001099900050. [DOI] [PubMed] [Google Scholar]
- 31.Kops G J, de Ruiter N D, De Vries-Smits A M, Powell D R, Bos J L, Burgering B M. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature. 1999;398:630–634. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- 32.Le Good J A, Ziegler W H, Parekh D B, Alessi D R, Cohen P, Parker P J. Protein kinase C isotypes controlled by phosphoinositide 3-kinase through the protein kinase PDK1. Science. 1998;281:2042–2045. doi: 10.1126/science.281.5385.2042. [DOI] [PubMed] [Google Scholar]
- 33.Lopez-Girona A, Furnari B, Mondesert O, Russell P. Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature. 1999;397:172–175. doi: 10.1038/16488. [DOI] [PubMed] [Google Scholar]
- 34.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Maurer R A. Both isoforms of the cAMP-dependent protein kinase catalytic subunit can activate transcription of the prolactin gene. J Biol Chem. 1989;264:6870–6873. [PubMed] [Google Scholar]
- 36.Medema R H, Kops G J, Bos J L, Burgering B M. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature. 2000;404:782–787. doi: 10.1038/35008115. [DOI] [PubMed] [Google Scholar]
- 37.Nakae J, Barr V, Accili D. Differential regulation of gene expression by insulin and IGF-1 receptors correlates with phosphorylation of a single amino acid residue in the forkhead transcription factor FKHR. EMBO J. 2000;19:989–996. doi: 10.1093/emboj/19.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakae J, Park B C, Accili D. Insulin stimulates phosphorylation of the forkhead transcription factor FKHR on serine 253 through a wortmannin-sensitive pathway. J Biol Chem. 1999;274:15982–15985. doi: 10.1074/jbc.274.23.15982. [DOI] [PubMed] [Google Scholar]
- 39.Ogg S, Paradis S, Gottlieb S, Patterson G I, Lee L, Tissenbaum H A, Ruvkun G. The Forkhead transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 40.O'Neill E M, Kaffman A, Jolly E R, O'Shea E K. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- 41.Park J, Leong M L, Buse P, Maiyar A C, Firestone G L, Hemmings B A. Serum and glucocorticoid-inducible kinase (SGK) is a target of the PI 3-kinase-stimulated signaling pathway. EMBO J. 1999;18:3024–3033. doi: 10.1093/emboj/18.11.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pullen N, Dennis P B, Andjelkovic M, Dufner A, Kozma S C, Hemmings B A, Thomas G. Phosphorylation and activation of p70s6k by PDK1. Science. 1998;279:707–710. doi: 10.1126/science.279.5351.707. [DOI] [PubMed] [Google Scholar]
- 43.Rena G, Guo S, Cichy S, Unterman T G, Cohen P. Phosphorylation of the transcription factor forkhead family member FKHR by protein kinase B. J Biol Chem. 1999;274:17179–17183. doi: 10.1074/jbc.274.24.17179. [DOI] [PubMed] [Google Scholar]
- 44.Richards S A, Fu J, Romanelli A, Shimamura A, Blenis J. Ribosomal S6 kinase 1 (RSK1) activation requires signals dependent on and independent of the MAP kinase ERK. Curr Biol. 1999;9:810–820. doi: 10.1016/s0960-9822(99)80364-9. [DOI] [PubMed] [Google Scholar]
- 45.Takaishi H, Konishi H, Matsuzaki H, Ono Y, Shirai Y, Saito N, Kitamura T, Ogawa W, Kasuga M, Kikkawa U, Nishizuka Y. Regulation of nuclear translocation of forkhead transcription factor AFX by protein kinase B. Proc Natl Acad Sci USA. 1999;96:11836–11841. doi: 10.1073/pnas.96.21.11836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang E D, Nunez G, Barr F G, Guan K-L. Negative regulation of the Forkhead transcription factor FKHR by Akt. J Biol Chem. 1999;274:16741–16746. doi: 10.1074/jbc.274.24.16741. [DOI] [PubMed] [Google Scholar]
- 47.Uberall F, Hellbert K, Kampfer S, Maly K, Villunger A, Spitaler M, Mwanjewe J, Baier-Bitterlich G, Baier G, Grunicke H H. Evidence that atypical protein kinase C-lambda and atypical protein kinase C-zeta participate in Ras-mediated reorganization of the F-actin cytoskeleton. J Cell Biol. 1999;144:413–425. doi: 10.1083/jcb.144.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vanhaesebroeck B, Alessi D R. The PI3K-PDK1 connection: more than just a road to PKB. Biochem J. 2000;346:561–576. [PMC free article] [PubMed] [Google Scholar]
- 49.Waldegger S, Barth P, Raber G, Lang F. Cloning and characterization of a putative human serine/threonine protein kinase transcriptionally modified during anisotonic and isotonic alterations of cell volume. Proc Natl Acad Sci USA. 1997;94:4440–4445. doi: 10.1073/pnas.94.9.4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waldegger S, Klingel K, Barth P, Sauter M, Rfer M L, Kandolf R, Lang F. h-sgk serine-threonine protein kinase gene as transcriptional target of transforming growth factor beta in human intestine. Gastroenterology. 1999;116:1081–1088. doi: 10.1016/s0016-5085(99)70011-9. [DOI] [PubMed] [Google Scholar]
- 51.Webster K M, Goya L, Firestone G L. Immediate-early transcriptional regulation and rapid mRNA turnover of a putative serine/threonine protein kinase. J Biol Chem. 1993;268:11482–11485. [PubMed] [Google Scholar]
- 52.Webster K M, Goya L, Ge Y, Maiyar A C, Firestone G L. Characterization of sgk, a novel member of the serine/threonine protein kinase gene family which is transcriptionally induced by glucocorticoids and serum. Mol Cell Biol. 1993;13:2031–2040. doi: 10.1128/mcb.13.4.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xing J, Ginty D D, Greenberg M E. Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science. 1996;273:959–963. doi: 10.1126/science.273.5277.959. [DOI] [PubMed] [Google Scholar]