Ras proteins are now well recognized for their essential function in transducing extracellular signals that regulate cell growth, survival, and differentiation. Although ras genes were originally identified in the mid-1960s as the transforming elements of the Harvey and Kirsten strains of rat sarcoma viruses, investigation of the biological properties of their protein products did not gain momentum until the early 1980s, when mutated alleles of cellular ras genes were identified as dominant oncogenes in various types of human tumors. The mammalian ras gene family consists of three members: H-ras, K-ras, and N-ras, which are located on different chromosomes. The H-ras and K-ras genes are the cellular counterparts of the viral Harvey and Kirsten genes, respectively, and the N-ras gene is derived from a human neuroblastoma cell line. Are the different ras genes functionally redundant or does each ras gene have a specific role? This review briefly recounts results from biochemical and genetic studies supporting the unanticipated possibility that the answer to both questions may be yes.
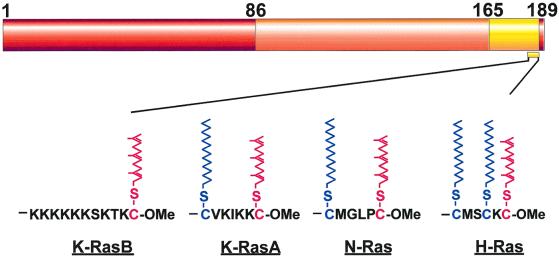
The basic anatomy of Ras proteins.
The three mammalian ras genes encode four highly related GTPases of 188 (K-RasB) or 189 (H-Ras, K-RasA, and N-Ras) amino acids in length. The A and B forms of K-Ras are generated by an alternative splicing of the fourth exon of this gene (2), and the abundance of K-RasB transcripts is higher in comparison to that of K-RasA transcripts (18). All of the critical domains for GTPase function (including sequence motifs important for nucleotide binding and GTP hydrolysis) are present within the N-terminal 165 amino acids of Ras proteins (18). Based on primary sequence comparisons, Ras proteins can be viewed as consisting of three contiguous regions (Fig. 1). The first region encompasses the N-terminal 86 amino acids, which are 100% identical among the different Ras proteins. Within this region lies the Ras effector binding domain (amino acids 32 to 40), which is the critical interaction site with all known downstream targets of Ras. The next 80 amino acids define a second region where mammalian Ras proteins diverge only slightly from each other, exhibiting an 85% homology between any protein pair. The remaining C-terminal sequence, known as the hypervariable region, starts at amino acid 165 and shows no sequence similarity among Ras proteins except for a conserved CAAX motif (C, cysteine; A, aliphatic amino acid; X, methionine or serine) at the very C-terminal end, which is present in all Ras proteins and directs posttranslational processing (7). Thus, if a unique role for each Ras protein is to be determined by sequence divergence, it would most likely be specified by the hypervariable region. While some of the functional studies on Ras proteins support this prediction, others indicate that this cannot be the entire story.
FIG. 1.
Sequence conservation between Ras proteins. Sequence identity from 0 to 100% is presented as a color gradient from yellow to orange to red. The sequences corresponding to the membrane anchor region of each human Ras protein are shown with their respective posttranslational modifications. The farnesylated cysteine residue which is conserved in all Ras proteins is shown in red. Palmitoylated cysteine residues in N- and H-Ras proteins are shown in blue.
Ras expression: lessons from molecular genetics.
Early quests for the physiological reasons underlying the existence of multiple forms of Ras proteins have focused on the analysis of expression patterns of ras genes in different cell lineages. The information that has emerged from these analyses indeed demonstrates significant variations in the levels of expression of the three ras genes between tissues as well as during development. For example, in mice, the level of H-ras transcripts is highest in brain, muscle, and skin and is lowest in liver, K-ras transcripts are most abundant in gut, lung, and thymus and are rare in skin and skeletal muscle, and N-ras transcripts are most prevalent in testis and thymus (17). Differential expression of the three ras genes has also been observed during mouse prenatal development, with N-ras expression being highest at day 10 of gestation and K-ras expression being lowest toward the end of gestation (17, 19). The simplest interpretation of these findings is that the three forms of Ras perform distinct cellular functions, with each function being biologically relevant in a particular tissue or cell type. However, this conclusion remains tentative for the following reasons. Firstly, the extent to which the diversified pattern of expression of ras genes correlates with the levels of their gene products has not been established. Immunohistochemical analysis of human tissue revealed considerable heterogeneity in the levels of Ras proteins between different tissues (9). However, because this analysis relied on an anti-Ras antibody that recognizes the products of all three ras genes, the relative amount of each Ras protein could not be discerned. Secondly, despite the variations in relative levels of expression, all three ras genes are concurrently expressed in most mouse and human tissues, suggesting that their protein products might possess overlapping functions (5, 9, 17).
Another line of investigation often considered as providing evidence in support of the differential function of the three ras genes concerns the analysis of ras mutation in human tumors. It is well documented that oncogenic forms of H-, K-, and N-ras are preferentially detected in certain tumor types. For example, more than 80% of pancreatic adenocarcinomas harbor a mutated K-ras gene, whereas in myeloid leukemia, the N-ras gene is most frequently mutated (3). However, in many tumor types, there is no absolute specificity for a mutated ras gene, and mutated forms of the three ras genes produce the same phenotype in in vitro transformation assays (3). Thus, differences in the function of Ras proteins, if they exist, cannot be the sole explanation for the bias in favor of particular ras mutations in certain tumor types. Taken together, while the conclusions drawn from gene expression studies have not answered definitively the question of functional divergence between ras genes, they certainly provide an interesting template for future work in this research area.
Ras localization: lessons from biochemistry.
In order to be biologically active, Ras proteins must be localized to the inner face of the plasma membrane, where they can effectively interact with their upstream activators and downstream targets. The biochemical process ensuring the association of Ras proteins with the plasma membrane involves a series of posttranslational modifications at their C termini. The first and obligatory step in this series is the farnesylation of the cysteine residue in the CAAX motif (4). Activated Ras proteins with substitutions in this cysteine residue fail to interact with the plasma membrane and are transformation defective (26). Subsequent to farnesylation, the AAX residues of the CAAX motif are proteolytically removed, generating a C-terminal cysteine which is then carboxyl methylated (10). While these CAAX modifications increase the hydrophobicity of the C-terminal end, they would not lead to the stable binding of Ras to the plasma membrane in the absence of a cooperating membrane targeting signal provided by the adjacent hypervariable region (Fig. 1). In H-Ras, N-Ras, and K-RasA, this signal involves one or two palmitoylated cysteine residues within the hypervariable region (12). In K-RasB, the signal is comprised of a polybasic domain containing multiple lysine residues (13).
The realization that each form of Ras protein interacts with the membrane via a different anchor fueled the notion that they might be localized to distinct membrane domains. In direct support of this concept are the findings that interfering with the function of caveolin, an integral membrane protein that binds cholesterol, impairs the signaling activity of H-Ras but not K-RasB (22). This suggests that H-Ras is preferentially localized to cholestrol-rich microdomains within the plasma membrane. The association of each Ras form with different membrane domains might be specified not only by their unique membrane-anchoring motifs but also by distinct targeting mechanisms, as suggested by recent experiments on the cellular trafficking of Ras proteins. Whereas the palmitoylated H- and N-Ras proteins traffic to the plasma membrane along the secretory pathway via the Golgi complex, K-RasB does not enter the conventional secretory pathway but instead is routed by virtue of its polybasic domain directly from the endoplasmic reticulum to the cell surface (1, 6).
In principle, there are two basic models for how the different mechanisms of membrane anchoring of Ras proteins could confer functional specificity. The first model postulates that the site of interaction of Ras proteins with the membrane would impact the availability of regulators and effectors because these molecules themselves might be localized to distinct subdomains within the plasma membrane. Several signaling molecules involved in Ras signaling, including receptor tyrosine kinases and members of the mitogen-activated protein kinase cascade, have been shown to be enriched in specialized cholesterol-rich membrane domains known as caveolae (21). Consistent with the idea that this compartmentalization could lead to differences in signaling activity between the various Ras proteins, it has been demonstrated that H-Ras, N-Ras, and K-RasB vary with respect to both their ability to engage the downstream targets Raf and phosphatidylinositol 3-kinase (11, 25, 27) and the extent of their activation by guanine nucleotide exchange factors (15). These variations are specified by the membrane-targeting C-terminal domain of Ras proteins, as determined by sequence shuffling experiments (15, 27).
The second model for how functional specificity among Ras proteins could be determined by membrane attachment postulates that the biochemical nature of the membrane anchor could dictate the efficiency of interaction with regulatory molecules. As described above, the association of H-Ras, N-Ras, and K-RasA with the membrane is mediated by a farnesyl-palmitoyl anchor, whereas K-RasB utilizes farnesylation and a polybasic domain to bind to the plasma membrane. Palmitate contributes to membrane interaction by virtue of its ability to insert itself deep into the lipid bilayer, whereas the polybasic domain facilities membrane association through ionic interactions with negatively charged phospholipid head groups (20, 23). In addition, the membrane-anchoring domain of K-RasB spans residues 175 to 186, whereas in H-Ras, N-Ras, and K-RasA, this domain consists of residues 180 to 186 (Fig. 1). These features could lead to differences in the positioning of the various Ras isoforms at the inner face of the plasma membrane, thereby affecting accessibility to effectors or activators. This model gains support from studies showing that, by eliminating the palmitoylation site on H-Ras or by modifying the length of the membrane anchor of H-Ras, it is possible to induce quantitative changes in the interaction of H-Ras with the exchanger Ras-GRF or with Raf, respectively (15, 27). Irrespective of whether one or both of the models turn out to be correct, it is clear that there are differences between the biochemical activities of each Ras protein. The extent to which these differences have physiological consequences has yet to be determined.
Ras gene targeting: the ultimate answer?
Given the inherent complexities in interpreting ectopic expression data, it is not surprising that targeted gene disruption in the mouse has been perceived as holding much promise for providing insights into the functional assignments of each ras gene. This approach was first applied to disrupt the murine N-ras gene, with the resulting phenotype indicating that the function of N-Ras is dispensable for normal mouse development, growth, and fertility (24). Next, the murine K-ras gene was disrupted by two groups independently (14, 16). Both studies have reported that K-ras-deficient mice die progressively, from embryonic day 12.5 until the term of gestation is complete, due to defects in hematopoiesis (14), myocardial cell proliferation, and neuronal cell survival (16). Esteban et al. (8) have described the phenotypes of mice that are homozygous null for either H-ras alone or H-ras and N-ras. A striking outcome of their analysis is that the double null mutant animals exhibit no detectable developmental and postnatal abnormalities (8). At first glance, these new findings, together with the earlier ras gene deletion studies, are most consistent with the interpretation that K-Ras possesses a unique function which is not shared by H-Ras or N-Ras and is necessary and sufficient for mouse development. However, an equally plausible explanation is that the selective requirement for K-Ras during development reflects its expression in a specific cell type during a critical developmental stage. Since the embryonic phenotypes associated with the loss of viability of the K-Ras null mutant mice have not been linked to specific biochemical defects, both possibilities remain formally valid at this time. In addition, the lack of observable phenotype in the H-Ras/N-Ras double mutant mice does not strictly imply functional overlap. Rather, it might be indicative of differential activities that are not amenable to scoring by gene disruption approaches. Thus, in the accompanying paper by Esteban et al. (8), a nearly 2-decade-old question has resurfaced: what's in a name?
ACKNOWLEDGMENTS
I thank members of my laboratory for helpful discussions.
REFERENCES
- 1.Apolloni A, Prior I A, Lindsay M, Parton R G, Hancock J F. H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol Cell Biol. 2000;20:2475–2487. doi: 10.1128/mcb.20.7.2475-2487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 3.Bos J L. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 4.Casey P J, Solski P A, Der C J, Buss J E. p21ras is modified by a farnesyl isoprenoid. Proc Natl Acad Sci USA. 1989;86:8323–8327. doi: 10.1073/pnas.86.21.8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chesa P G, Rettig W J, Melamed M R, Old L J, Niman H L. Expression of p21ras in normal and malignant human tissues: lack of association with proliferation and malignancy. Proc Natl Acad Sci USA. 1987;84:3234–3238. doi: 10.1073/pnas.84.10.3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choy E, Chiu V K, Silletti J, Feoktistov M, Morimoto T, Michaelson D, Ivanov I E, Philips M R. Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell. 1999;98:69–80. doi: 10.1016/S0092-8674(00)80607-8. [DOI] [PubMed] [Google Scholar]
- 7.Clarke S. Protein isoprenylation and methylation at carboxyl-terminal cysteine residues. Annu Rev Biochem. 1992;61:355–386. doi: 10.1146/annurev.bi.61.070192.002035. [DOI] [PubMed] [Google Scholar]
- 8.Esteban L M, Vicario-Abejon C, Fernandez-Salguero P, Fernandez-Medarde A, Swaminathan N, Yienger K, Lopez E, McKay R, Ward J M, Pellicar A, Santos E. Targeted genomic disruption of H-ras and N-ras, individually or in combination, reveals the dispensability of both loci for mouse growth and development. Mol Cell Biol. 2001;21:1444–1452. doi: 10.1128/MCB.21.5.1444-1452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furth M E, Aldrich T H, Cordon-Cardo C. Expression of ras proto-oncogene proteins in normal human tissues. Oncogene. 1987;1:47–58. [PubMed] [Google Scholar]
- 10.Gutierrez L, Magee A I, Marshall C J, Hancock J F. Post-translational processing of p21ras is two-step and involves carboxyl-methylation and carboxy-terminal proteolysis. EMBO J. 1989;8:1093–1098. doi: 10.1002/j.1460-2075.1989.tb03478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamilton M, Wolfman A. Ha-ras and N-ras regulate MAPK activity by distinct mechanisms in vivo. Oncogene. 1998;16:1417–1428. doi: 10.1038/sj.onc.1201653. [DOI] [PubMed] [Google Scholar]
- 12.Hancock J F, Magee A I, Childs J E, Marshall C J. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell. 1989;57:1167–1177. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- 13.Hancock J F, Paterson H, Marshall C J. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- 14.Johnson L, Greenbaum D, Cichowski K, Mercer K, Murphy E, Schmitt E, Bronson R T, Umanoff H, Edelmann W, Kucherlapati R, Jacks T. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 1997;11:2468–2481. doi: 10.1101/gad.11.19.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones M K, Jackson J H. Ras-GRF activates Ha-Ras, but not N-Ras or K-Ras 4B, protein in vivo. J Biol Chem. 1998;273:1782–1787. doi: 10.1074/jbc.273.3.1782. [DOI] [PubMed] [Google Scholar]
- 16.Koera K, Nakamura K, Nakao K, Miyoshi J, Toyoshima K, Hatta T, Otani H, Aiba A, Katsuki M. K-ras is essential for the development of the mouse embryo. Oncogene. 1997;15:1151–1159. doi: 10.1038/sj.onc.1201284. [DOI] [PubMed] [Google Scholar]
- 17.Leon J, Guerrero I, Pellicer A. Differential expression of the ras gene family in mice. Mol Cell Biol. 1987;7:1535–1540. doi: 10.1128/mcb.7.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowy D R, Willumsen B M. Function and regulation of ras. Annu Rev Biochem. 1993;62:851–891. doi: 10.1146/annurev.bi.62.070193.004223. [DOI] [PubMed] [Google Scholar]
- 19.Muller R, Slamon D J, Adamson E D, Tremblay J M, Muller D, Cline M J, Verma I M. Transcription of c-onc genes c-rasKi and c-fms during mouse development. Mol Cell Biol. 1983;3:1062–1069. doi: 10.1128/mcb.3.6.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray D, Ben-Tal N, Honig B, McLaughlin S. Electrostatic interaction of myristoylated proteins with membranes: simple physics, complicated biology. Structure. 1997;5:985–989. doi: 10.1016/s0969-2126(97)00251-7. [DOI] [PubMed] [Google Scholar]
- 21.Okamoto T, Schlegel A, Scherer P E, Lisanti M P. Caveolins, a family of scaffolding proteins for organizing “preassembled signaling complexes” at the plasma membrane. J Biol Chem. 1998;273:5419–5422. doi: 10.1074/jbc.273.10.5419. [DOI] [PubMed] [Google Scholar]
- 22.Roy S, Luetterforst R, Harding A, Apolloni A, Etheridge M, Stang E, Rolls B, Hancock J F, Parton R G. Dominant-negative caveolin inhibits H-Ras function by disrupting cholesterol-rich plasma membrane domains. Nat Cell Biol. 1999;1:98–105. doi: 10.1038/10067. [DOI] [PubMed] [Google Scholar]
- 23.Silvius J R, l'Heureux F. Fluorimetric evaluation of the affinities of isoprenylated peptides for lipid bilayers. Biochemistry. 1994;33:3014–3022. doi: 10.1021/bi00176a034. [DOI] [PubMed] [Google Scholar]
- 24.Umanoff H, Edelmann W, Pellicer A, Kucherlapati R. The murine N-ras gene is not essential for growth and development. Proc Natl Acad Sci USA. 1995;92:1709–1713. doi: 10.1073/pnas.92.5.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voice J K, Klemke R L, Le A, Jackson J H. Four human ras homologs differ in their abilities to activate Raf-1, induce transformation, and stimulate cell motility. J Biol Chem. 1999;274:17164–17170. doi: 10.1074/jbc.274.24.17164. [DOI] [PubMed] [Google Scholar]
- 26.Willumsen B M, Norris K, Papageorge A G, Hubbert N L, Lowy D R. Harvey murine sarcoma virus p21 ras protein: biological and biochemical significance of the cysteine nearest the carboxy terminus. EMBO J. 1984;3:2581–2585. doi: 10.1002/j.1460-2075.1984.tb02177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan J, Roy S, Apolloni A, Lane A, Hancock J F. Ras isoforms vary in their ability to activate Raf-1 and phosphoinositide 3-kinase. J Biol Chem. 1998;273:24052–24056. doi: 10.1074/jbc.273.37.24052. [DOI] [PubMed] [Google Scholar]