Abstract
Non-small cell lung cancer (NSCLC) is a deadly and highly prevalent malignancy. Targeting activated-EGFR mutations in NSCLC via EGFR tyrosine kinase inhibitor (EGFR-TKI) initially achieves a profound therapeutic response, but resistance frequently evolves, reducing treatment options. Here, we present a small-molecule compound D6 which selectively inhibits tumor cell growth and migration in NSCLC cells with EGFR-TKI-resistant T790M-EGFR-activated mutations (T790M-EGFR-AM), e.g., L858R/T790M, 19Del/T790M and L858R/T790M/C797S. D6 mimics a natural product isolated from the roots of Codonopsis pilosula and selectively competes with T790M-EGFR-AM to bind to HSP90, thus facilitating the ubiquitination dependent proteasomal degradation of T790M-EGFR-AM. By contrast, D6 has little impact on typical HSP90 chaperone activity, suggesting low systemic toxicity. Promisingly, D6 combined with erlotinib or osimertinib shows efficacy in overcoming the EGFR-TKIs-resistance in NSCLCs. Our study raises an alternative strategy to overcome T790M-mediated EGFR-TKI resistance in NSCLC via targeting the protein–protein interaction of HSP90 and T790M-EGFR by intervention with D6.
Subject terms: Non-small-cell lung cancer, Target validation
Tang et al. propose the use of a small molecule compound D6 to treat EGFRTKI resistant tumors. The authors demonstrate potent and selective inhibition of NSCLC with T790M-mediated EGFR-TKI resistance through inhibiting HSP90-mediated protein–protein interaction.
Introduction
Lung cancer is the worldwide leading cause of cancer-related deaths1. Non-small cell lung cancer (NSCLC) constitutes almost 90% of lung cancer patients and has a poor prognosis with a 5-year survival of ∼15%2,3. Epidermal growth factor receptor (EGFR), also known as class I receptor tyrosine kinase, utilizes the tyrosine kinase (TK) domain to transduce extracellular signals upon ligand binding, e.g., EGF4. Activated EGFR signaling plays critical roles in driving NSCLC tumorigenesis and progression by activating two major downstream pathways in a kinase-dependent manner, i.e., the Ras/ERK1/2 and the PI3K/AKT4,5. EGFR typically exists in an autoinhibited conformation, and somatic mutations which destabilize this conformation can result in a ligand-independent activation: these mutations are widely observed and account for about 20% of advanced NSCLCs6. Most EGFR-activated mutations display as in-frame deletions in exon 19 E746_A750 (19Del-EGFR) or an L858R point mutation (L858R-EGFR)6–8.
NSCLCs harboring EGFR-activated mutations are more sensitive to EGFR tyrosine kinase inhibitors (EGFR-TKIs). The first generation of TKIs (erlotinib and gefitinib) are widely applied for 19Del- or L858R-EGFR-positive patients9. While initially achieving tumor regression, most patients develop acquired resistance 9–13 months post therapy10,11. Approximately 60% acquired resistance is attributed to an additional T790M mutation—the methionine substitution generates a bulky amino acid side chain which conducts a steric hindrance and thus interferes the binding of gefitinib or erlotinib to the EGFR12–14. To overcome the T790M-EGFR resistance, osimertinib, a representative third generation of EGFR-TKIs, which acts irreversibly and selectively for EGFR-activating mutations including T790M mutation, has been widely applied in clinic15,16. However, after approximately 10 months of exposure, tumor cells develop resistance, e.g., the C797 and L792 residues of T790M-EGFR are mutated with a portion of ∼26%15,17–19. Exploring new strategies to overcome the continuous acquisition of EGFR-TKI resistance is thus clinically urgent.
One avenue being pursued to overcome EGFR-TKI resistance is HSP90 inhibition. HSP90 belongs to the heat-shock protein family and assists protein folding or refolding20 and oncogenic proteins levels, maturation, and continued dysregulated activity are regulated by HSP90 chaperone activity in many cancers21. TKI-resistant T790M-EGFR NSCLC are more sensitive than wild-type (WT) cells in response to an HSP90 inhibitor which promotes fast degradation of T790M-EGFR to prevent tumor progression22–24. Thus far, most HSP90 inhibitors are designed to overcome the EGFR-TKI resistance by targeting either N- or C-terminus25, e.g., 17-AAG (tanespimycin), 17-DMAG (alvespimycin) and IPI-504 (retaspimycin hydrochloride), which elicit anti-neoplastic activity in NSCLC26–28. However, at present there are few approved HSP90 inhibitors because the severe toxicity is observed from clinic trials, such as induction of heat-shock response, retinopathy and gastrointestinal tract toxicity29. Development of HSP90 inhibitors with acceptable toxicity may provide an alternative strategy to overcome the EGFR-TKI resistance in NSCLCs.
The root of Codonopsis pilosula (Dangshen in Chinese) is a traditional Chinese medicine which is usually applied as an adjuvant to improve immune function and ameliorate therapy-induced fatigue for cancer patients30,31. Compounds isolated from C. pilosula have potentials to inhibit cancer cell growth or migration32,33. Here, we identified a small-molecule compound D6, a synthetic derivative of natural compound from C. pilosula, which possesses selective activity on NSCLC cells harboring T790M-EGFR mutation by targeting HSP90. In combination with erlotinib or osimertinib, D6 has potential to prevent the acquired EGFR-TKI resistance in NSCLC.
Results
Chemical structure of D6
We initially obtained a group of natural small molecules from the root of C. pilosula (n-D5, n-D6 and n-D7) and tested their anti-tumor activity in NSCLC cells. Interestingly, n-D6 (methyl (Z)-4-oxo-4-(9H-pyrido [3, 4-b] indol-1-yl) but-2-enoate) (hereafter code: D6-1, Supplementary Figs. 1–6) had promising anti-tumor activity. For further large-scale biological analysis, an analogue of D6-1, i.e., D6, was synthesized by a natural product derivative tryptamine as described in Supplementary methods (Scheme 1) and Supplementary Fig. 7. As shown, D6 is an HPLC-grade pure small-molecular compound (Supplementary Fig. 8). Its planar structure was further identified by spectroscopic methods including 1H, 13C NMR, 1H-1H COSY, HSQC, HMBC, and HRESIMS spectrum (Supplementary Figs. 9–17). The geometry of double bond was assigned as cis form by the coupling J = 12.2 Hz. Interestingly, the cis-D6 almost completely transformed to trans form in the cell culture medium (Supplementary Figs. 18 and 19), which indicates that the effect of D6 in our following performance was indeed elicited by the trans-D6.
NSCLC cells expressing L858/T790M-EGFR are sensitive to D6
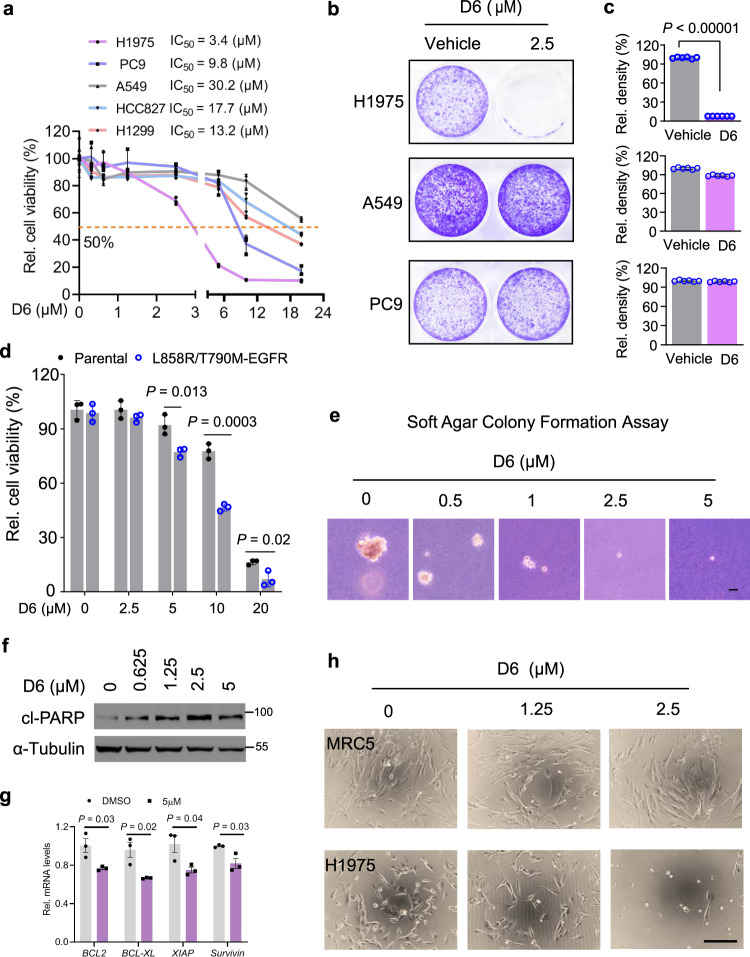
We next asked whether D6 had the similar anti-tumor activity as the natural product n-D6 for NSCLCs. Initially, three well-characterized human NSCLC cell lines (A549, PC9, and NCI-H1975) were treated with serial doses of D6 for 72 h and then cell viability was detected by CCK-8 assay. D6 reduced the cell viability of all NSCLC cells, showing IC50 values of 30.2 μM for A459, 9.8 μM for PC9, and 3.4 μM for NCI-H1975 (Fig. 1a). Notably, NCI-H1975 cells were more sensitive to D6.
Fig. 1. D6 inhibits the viability of NCI-H1975 cells.
a PC9, H1299, A549, HCC827, and NCI-H1975 cells were treated with a range of concentrations of D6 for 72 h. Cell viability was detected by CCK8 and shown as relative viability compared to the control. b, c Representative images showed the cell proliferation of NCI-H1975, A549, and PC9 cells exposed to D6 (2.5 µM) and solvent DMSO. The bars of (c) represented the relative cell density. d HCC827 parental cells or the isogenic cells expressing L858R/T790M-EGFR were treated with the indicated concentrations of D6 for 72 h. Cell viability (%) was shown after normalized to the control group. e Representative images showed the cell growth of NCI-H1975 cells cultured in soft agar with the indicated concentrations of D6. Scale bar, 200 µm. f Immunoblotting analysis showing the levels of cleaved-PARP (cl-PARP) in NCI-H1975 cells treated with indicated doses of D6. g Quantitative PCR analysis showing the levels of anti-apoptosis genes in NCI-H1975 cells in the presence of D6 or not. h Representative images showing the effect of D6 on NCI-H1975 cells and normal lung fibroblast MRC5 cells. Scale bar, 200 µm. Data are presented as mean ± SEM (a, c, and g), n = 3 (a and g) or 6 (c) biologically independent samples. P values were calculated by two-tailed Student’s t-test.
One factor that may explain the differing responses observed between NSCLC cells is their EGFR status. Verified by genomic DNA sequencing, as shown, A549 cells have WT EGFR; PC9 cells express 19 Del (E746_A750) EGFR (19Del-EGFR); and NCI-H1975 cells possess L858R/T790M-EGFR3 (Supplementary Fig. 20a). Consistent with the intrinsic EGFR status, PC9 cells were most sensitive to erlotinib treatment, while A549 and NCI-H1975 cells showed apparent non-response or resistance (Supplementary Fig. 20b). Since NCI-H1975 cells expressing EGF-TKIs-resistant L858R/T790M-EGFR were more sensitive to D6, we investigated if D6 selectively targets the L858R/T790M-EGFR mutated NSCLCs. To reduce the impact of cell-context-dependent effects, we employed another two human NSCLC cell lines, HCC827 and H1299, which express 19Del-EGFR and WT EGFR, respectively. Both HCC827 and H1299 cells displayed much higher IC50 does of D6 than NCI-H1975 cells (Fig. 1a). Consistently, cell proliferation assays confirmed that D6 at the dose of 2.5 μM achieved marked cell growth arrest for NCI-H1975 cells but had marginal effect on either A549 or PC9 cells (Fig. 1b, c). To test the possibility that D6 preferentially inhibits L858/T790M-EGFR addicted NSCLC cells, we generated an isogenic cell line from HCC827 by stably expressing L858R/T790M-EGFR and then selected by erlotinib, to mimic the erlotinib resistance. D6 consistently displayed increased capacity to kill L858R/T790M-EGFR-expressing cells compared to their parental cells (Fig. 1d), indicating that the effect was not cell-type dependent.
Given that targeting the T790M-EGFR NSCLCs has clinical relevance, further analyses were performed in NCI-H1975 cells. D6 treatment at the dose 0.5 μM (lower than IC50 (3.4 μM)) was sufficient to suppress growth of NCI-H1975 cells in soft agar, which mimics tumor progress in vivo34 (Fig. 1e). Moreover, 48 h post exposure to D6, we observed a dose-dependent increase of cleaved PARP, a hallmark of apoptosis35, in NCI-H1975 cells, suggesting that D6 treatment eventually elicits cell apoptosis (Fig. 1f). Similarly, a significant downregulation of anti-apoptotic genes was observed after D6 treatment (Fig. 1g). Importantly, noncancerous cells, such as MRC5 (normal lung fibroblasts), HEK293, and LO2, were less sensitive to D6 in comparison with NCI-H1975 cells, suggesting a lower toxicity (Fig. 1h and Supplementary Fig. 20c). Together, these results suggest that D6 potently targets the NSCLC cells harboring L858R/T790M-EGFR.
D6 inhibits NCI-H1975 cell migration and invasion
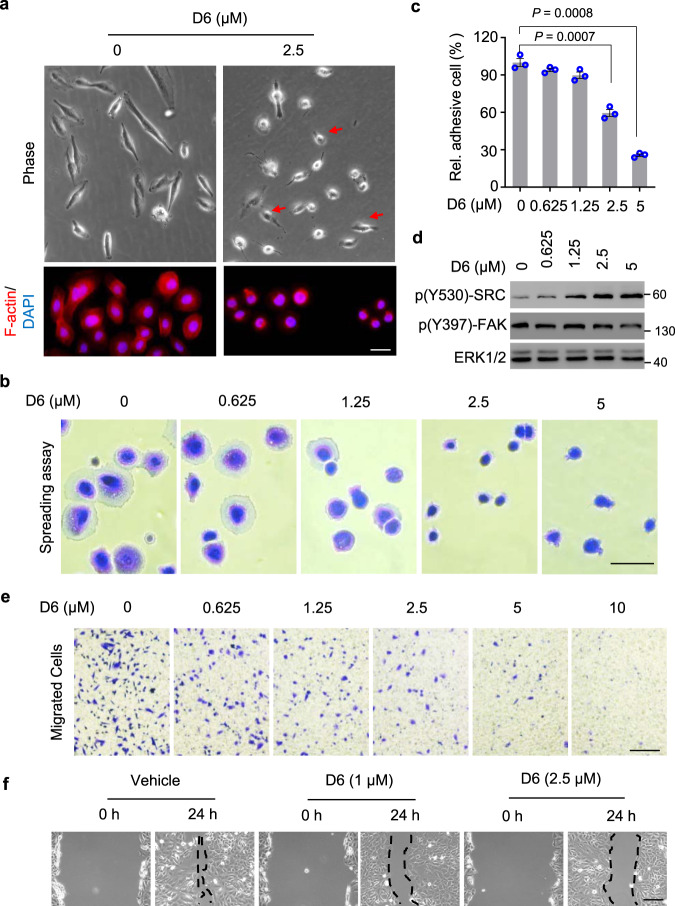
Approximately 30–40% of NSCLC patients have metastatic tumors after diagnosis, which is the main cause of death36. Tumor metastasis is a complex process largely dependent on enhanced cell motility37,38. Interestingly, with D6 treatment, NCI-H1975 cells shifted from spindle morphology to rounded morphology, which indicates that D6 might contribute to regulation of cell adhesion and spreading (Fig. 2a). Assessed by a cell spreading assay, D6 suppressed the adhesion of NCI-H1975 cells in a dose-dependent manner (Fig. 2b, c), noting that cell viability was marginally affected (Supplementary Fig. 21a). Cell adhesion and spreading are crucially regulated by SRC and FAK, two focal adhesion kinases39. Phosphorylation of SRC at Y530 (p-Y530) impairs its kinase activity and leads to downregulate (p-Y397) FAK, which disrupts the focal adhesion turnover and impairs cell migration40,41. D6 treatment increased (p-Y530) SRC but decreased (p-Y397) FAK in the NCI-H1975 cells (Fig. 2d). Cell migration also relies on efficient focal adhesion turnover42. We examined the effect of D6 on cell migration and showed that D6 treatment suppressed NCI-H1975 cells evading through transwell in a dose-dependent manner (Fig. 2e). Similarly, D6 treatment inhibited the migration of NCI-H1975 cells as determined by wound-healing assays (Fig. 2f). Of note, we did not observe apparent decrease of cell viability by D6 treatment in those assays (Supplementary Fig. 21b, c). Together, the results indicate that D6 has the potential to combat metastasis in NSCLCs.
Fig. 2. D6 inhibits NCI-H1975 cell migration.
a Representative images showing the cell morphology of NCI-H1975 cells post D6 exposure. Scale bar, 100 µm. b, c Representative images (b) showing NCI-H1975 cells spreading on cell culture plate with D6 incubation or not. The adhesive cells were counted and normalized to the solvent control (c). Scale bar, 50 µm. d Immunoblotting analysis of cell lysates prepared from NCI-H1975 cells exposed to different concentrations of D6. e NCI-H1975 cells with D6 pretreatment were subjected to transwell assay. The migrated cells were stained by 0.1% crystal violet solution. Scale bar, 200 µm. f Representative images showing the wound-healing assay for NCI-H1975 cells followed with D6 treatments. The dash line indicated the edge of wound closure. Scale bar, 200 µm. Data represent mean ± SEM (c), n = 3 biologically independent samples. P values were calculated by two-tailed Student’s t-test.
D6 decreases EGFR expression in NCI-H1975 cells
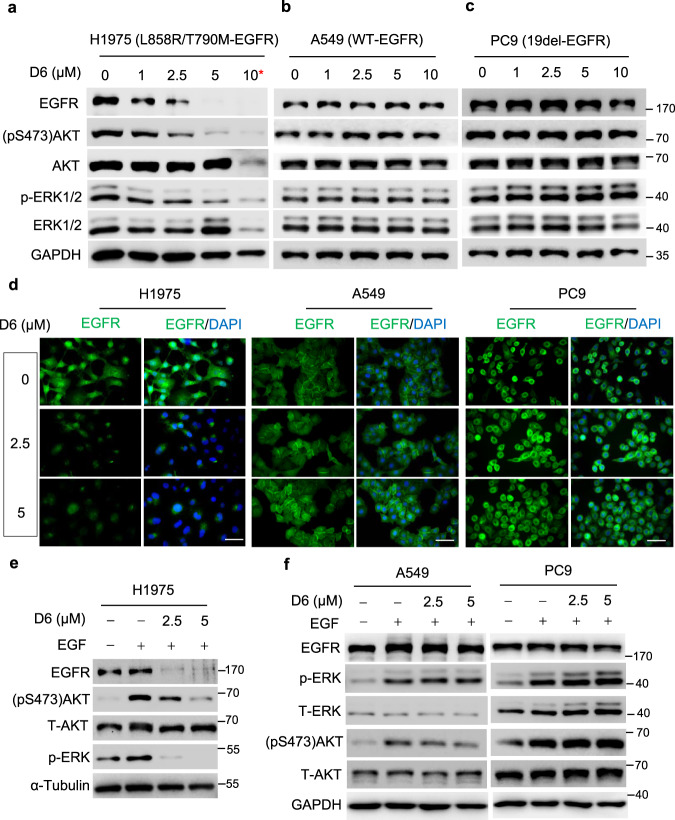
EGFR-mutant NSCLC cells are addicted to EGFR activity and require this activity to drive tumor growth, survival and metastasis43. D6 treatment leads to a dose-dependent decline of EGFR protein expression in NCI-H1975 cells, concurring with the inhibition of EGFR downstream effectors like AKT and ERK1/2 (Fig. 3a). By contrast, D6 treatment at the same dosages had a negligible effect on EGFR levels in both A549 and PC9 cells (Fig. 3b, c). Consistent results were observed by immunofluorescent staining of EGFR, where a prominent reduction of EGFR in NCI-H1975 cells but not in PC9 and A549 cells post D6 treatment was observed (Fig. 3d). D6 treatment markedly abrogated EGF-induced AKT and ERK activation in NCI-H1975 cells, but exerted no apparent effect on A549 and PC9 cells (Fig. 3e, f). These results indicate that D6 treatment elicits selective anti-tumor efficacy in L858R/T790M-EGFR NSCLC cells by downregulating EGFR expression.
Fig. 3. D6 suppressed EGFR expression in NCI-H1975 cells.
a−c NCI-H1975, A549, and PC9 cells were treated with indicated concentrations of D6 for 24 h. Cell lysates were prepared and subjected to immunoblotting analysis. *D6 resulted in cell death at the dose of 10 μM and led to total protein decrease. d Representative images showing the EGFR levels evaluated by immunofluorescent staining. NCI-H1975, A549, and PC9 cells were treated with D6 for 24 h and then fixed, permeabilized and incubated with anti-EGFR antibody. After incubation with an FITC-conjugated secondary antibody, nuclei were stained with DAPI. Scale bars, 50 µm. e, f Immunoblotting analysis of cell lysates derived from NCI-H1975, A549, and PC9 cells with or without D6 and EGF (10 ng/µl) treatment.
D6 promotes T790M-EGFR degradation through the ubiquitin-proteasome system
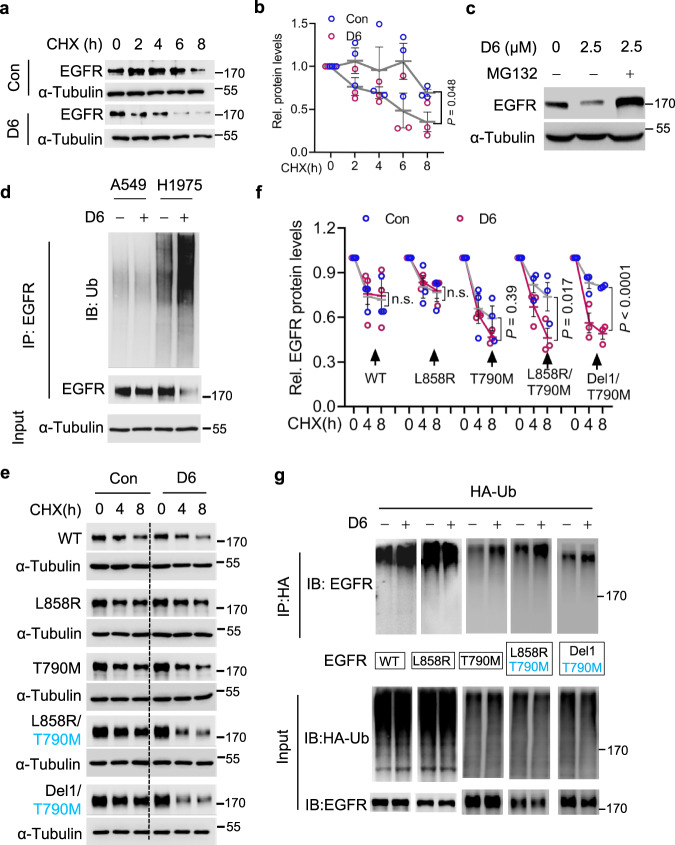
While decreasing EGFR protein expression, D6 treatment had little impact on the EGFR mRNA levels in NCI-H1975 cells (Supplementary Fig. 22), suggesting a potential effect on EGFR protein stability. We performed cycloheximide (CHX) chase assays, an experiment to measure protein stability44. D6 markedly facilitated EGFR protein turnover in NCI-H1975 cells (Fig. 4a, b) and this effect was potentially dependent on proteasome system as MG-132, a proteasome inhibitor45, nearly restored EGFR protein levels even with continued D6 treatment (Fig. 4c). This finding is consistent with a previous report that the degradation of mutated EGFR protein is at least partially dependent on the proteasome46. Supporting such notion, D6 treatment resulted in an obvious increase of ubiquitinated EGFR in NCI-H1975 cells. By contrast, D6 treatment had little effect on A549 cells which express the WT EGFR (Fig. 4d).
Fig. 4. D6 promotes L858R/T790M-EGFR degradation.
a, b Immunoblotting analysis (a) of cell lysates derived from NCI-H1975 cells treated by CHX (50 μg/ml) in the presence or absence of D6 (2.5 μM); protein quantification is presented in (b), n = 3 independent experiments. P values were calculated by two-way ANOVA analysis. c EGFR protein levels were detected by immunoblotting in NCI-H1975 cells treated by D6 (2.5 μM) with or without MG132 (10 μM). d NCI-H1975 or A549 cells transfected with HA-Ub were then treated with or without D6 (2.5 μM) for 6 h. Cell lysates were immunoprecipitated with anti-HA beads and beads elution was further analyzed by immunoblotting. e, f Immunoblotting analysis (e) of cell lysates derived from HEK293 cells transfected with different EGFR mutants and then were subjected to CHX (50 μg/ml) chase assay with or without D6 (2.5 μM) incubation. Protein degradation rate was shown as curves in (f), n = 3 independent experiments. P values were calculated by two-way ANOVA analysis. g Immunoblotting analysis of the elution of anti-HA beads prepared from HEK293 cells transfected with HA-Ub and EGFR mutant with or without D6 (2.5 μM) exposure. Data are presented as mean ± SEM (b and f). Representative results were analyzed from at least three independent experiments.
To confirm that D6 preferentially targets L858R/T790M-EGFR in NSCLC, we asked which architecture of EGFR is indispensable in response to D6, i.e., L858R, T790M alone or both; we also investigated if the observed effects are cell-type specific. We employed HEK293 cells which are entirely etiologically distinct to NCI-H1975 cells and performed CHX chase assay to evaluate the effect of D6 on several regularly mutated EGFRs. D6 treatment only marginally affected the stability of WT-EGFR, L858R-EGFR, and T790M-EGFR in HEK293 cells (Fig. 4e, f). However, L858R/T790M-EGFR and Del19/T790M-EGFR, two EGFR mutants that frequently emerge as TKI-resistant when patients are treated with gefitinib or erlotinib10, displayed prominent sensitivity to D6 exposure (Fig. 4e, f). Consistently, D6 increased the levels of ubiquitinated L858R/T790M-EGFR and Del19/T790M-EGFRs (Fig. 4g). These results suggest that D6 preferably targets EGFR which simultaneously harbors kinase activation (L858R or Del19) and erlotinib-resistant (T790M) mutations.
D6 compromises the interaction of T790M/L858R-EGFR and HSP90
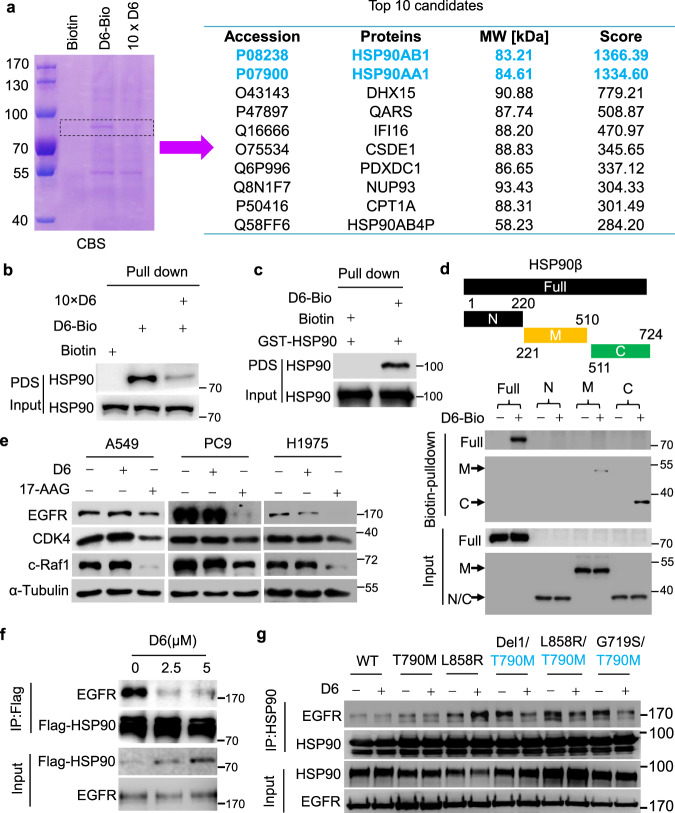
To explore the mechanism that how D6 impacts EGFR stability, we prepared biotin-labeled D6, i.e., D6-4, as shown in Supplementary methods (Scheme 2), and then performed biotin-based pulldown assay to identify D6 targeting proteins. Interestingly, D6-4 specifically captured some proteins with a molecular weight around ∼90 kDa (Fig. 5a, left), noting that 10 folded D6 successfully competed such bindings. Via protein mass spectrometry, we identified HSP90 as a D6 binding candidate (Fig. 5a, right). To confirm this, biotin-pulldown elution based on D6-4 derived from NCI-H1975 cell lysates was applied for immunoblotting. HSP90 was apparently trapped by D6-4, while D6 successfully antagonized the binding (Fig. 5b). The direct interaction of D6 and HSP90 was verified through the pulldown assay of D6-4 and purified HSP90 protein in vitro (Fig. 5c). HSP90 contains three major domains which rationally constitutes an ATP-dependent chaperone complex to regulate protein folding and degradation, i.e., the amino-terminal domain (NTD), the middle domain (MD) and the carboxy-terminal domain (CTD)47. To precisely define the D6 binding region, we performed domain-truncation analysis. Both MD and CTD were detected in the elution based on D6-4, but the CTD domain had the strongest affinity to D6 (Fig. 5d). These findings support that D6 directly targets HSP90.
Fig. 5. D6 attenuates the binding of T790M-EGFR mutants to HSP90.
a Coomassie blue staining (CBS) showing proteins trapped by biotin-labeled D6 (D6-4) from NCI-H1975 cell lysates. 10 × D6: using 10-fold D6 to saturate the target proteins and thus excluding non-specific binding. The protein gel indicated by dashed line was further analyzed by protein mass spectrometry and identified proteins are listed in the right panel. b, c Immunoblotting analysis of the protein elution based on biotin–avidin pulldown assays from NCI-H1975 cell lysates (b) or purified recombinant GST-HSP90 protein (c); PDS: pulldown solution. d Immunoblotting analysis showing the domains where D6 binds to HSP90. e Immunoblotting analysis of the cell lysates from A549, PC9, and NCI-H1975 cells treated with D6 (2.5 μM) or 17-AAG (2.5 μM), respectively. f Immunoblotting analysis of the anti-HSP90 immunoprecipitates prepared from cell lysates of NCI-H1975 cells with indicated concentrations of D6 treatment. g Immunoblotting analysis of anti-HSP90 immunoprecipitates derived from cell lysates of HEK293 cells transfected with HSP90 and indicated EGFR mutants with or without D6 treatment.
NSCLC cells harboring EGFR-activated mutations are much more sensitive to HSP90 inhibition than the wild type23,24. We then determined if D6 is a HSP90 inhibitor. We observed that 17-AAG, an HSP90 inhibitor targeting the NTD of ATP binding pocket, obviously downregulated expression of general clients, like EGFR, CDK4, and c-Raf1, in human NSCLC PC9, NCI-H1975, and A549 cells (Fig. 5e). However, D6 treatment had minor effect on these clients in all the tested cells with the exception of EGFR in NCI-H1975 (Fig. 5e), which suggests that D6 is unlikely to be a typical HSP90 inhibitor. Additionally, whilst D6 possessed a higher binding capacity to CTD (Fig. 5d) which is supposed to mediate HSP90 dimerization48, there was not a prominent decrease of HSP90 dimer formation in the presence of D6 (Supplementary Fig. 23a). Recently, targeting HSP90 protein–protein interactions (PPIs) is considered as an alternative strategy to avoid the system toxicity conferring by typical HSP90 inhibitors49. D6 might modulate HSP90-centered PPIs because D6 is biased to constrain the interaction between HSP90 and T790M/L858R-EGFRs. Supporting this notion, D6 was found to impair EGFR and HSP90 interaction in NCI-H1975 cells, noting that this interaction was unaffected in A549 and PC9 cells (Fig. 5f and Supplementary Fig. 23b, c). To exclude any cell-context-dependent effects, we recapitulated those experiments in the HEK293 cells. As shown, D6 weakened the interaction between HSP90 and EGFR carrying T790M mutation; notably, this is the case with L858R/T790M and Del19/T790M-EGFR mutants, both of which mainly evolve to resist the first generation of EGFR-TKIs (Fig. 5g and Supplementary Fig. 23d). These results indicate that D6 selectively promotes erlotinib-resistant EGFR degradation by jeopardizing HSP90 binding.
D6 inhibits the growth of erlotinib/gefitinib-resistant NSCLC xenografts
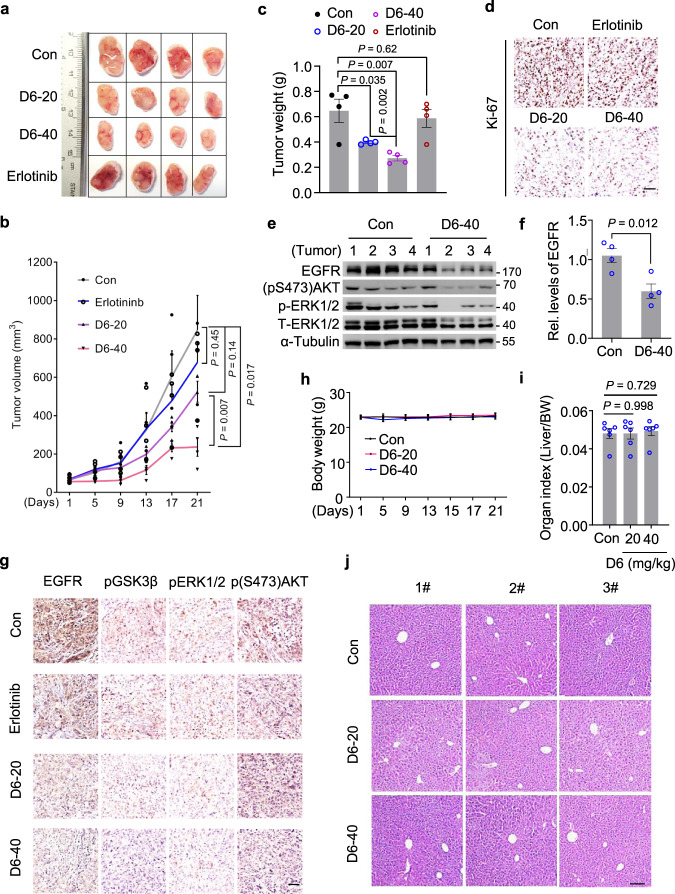
We next asked whether D6 could overcome T790M-EGFR-mediated TKI resistance. To investigate this, we generated isogenic erlotinib-resistant cells (Er-R) by stably expressing ectopic Del19/T790M-EGFR in the parental erlotinib-sensitive PC9 cells (Supplementary Fig. 24a). While Er-R cells showed a strong resistance to erlotinib (from 1000 to 5000 nM), D6 at lower dose (2.5 µM) was sufficient to overcome such resistance (Supplementary Fig. 24b). We further evaluated the in vivo anti-tumor effects of D6 on erlotinib-resistant NCI-H1975 cells expressing L858R/T790M EGFR. We generated xenografts by subcutaneous inoculation of NCI-H1975 cells in nude mice. After the tumors reached ∼100 mm3, mice were separated into four groups and treated (via i.p. injection) with D6 (20 mg/kg and 40 mg/kg), erlotinib (100 mg/kg) and solvent once every 2 days, respectively. The volume of xenografts showed reductions of 44.4% or 77.8% in mice treated with D6 at the dose of 20 or 40 mg/kg compared with solvent control; noting that erlotinib exerted no significant effect (Fig. 6a, b). D6 reduced tumor burden and elicited a marked decrease of Ki-67 expression (Fig. 6c, d). In accordance with the effect of D6 on tumor regression, D6 therapy suppressed the EGFR signaling activity: determined by immunoblotting analysis of isolated xenografts, D6 suppressed EGFR expression and relevant downstream effectors, e.g., (pS473)AKT and pERK1/2 (Fig. 6e, f). This effect was further confirmed by immunohistochemical (IHC) staining analysis, showing that tumors in mice exposed to D6 had a progressive decrease of EGFR protein, (pS473)AKT, phospho-ERK1/2 (pERK1/2), and (pS9)GSK3β (pGSK3β) (Fig. 6g). Notably, negligible toxicity of D6 was observed under such experimental conditions, as evidenced by little changes of the mouse body weight (Fig. 6h), the liver organ index (Fig. 6i), and the liver histological morphology (Fig. 6j). These results suggest the potential clinical significance of D6 by targeting T790M-EGFRs in erlotinib/gefitinib-resistant NSCLCs.
Fig. 6. D6 inhibits the growth of xenografts developed from NCI-H1975 cells.
a Nude mice inoculated with NCI-H1975 cells were randomly divided into four groups (n = 4 mice per group). When tumor volume reached ~100 mm3, mice were administered (i.p.) with D6 (20 or 40 mg/kg) or erlotinib (100 mg/kg), respectively. The remaining mice injected with an equivalent volume of solvent were taken as control. After 21 days, mice were sacrificed and tumors were collected for analysis. b, c Tumor volume (b) and tumor weight (c) were analyzed in each group of mice. To calculate tumor volume, the tumor width and length were measured by a caliper. d Representative images showing the IHC staining of Ki-67 expression in tumors isolated from (c). Scale bar, 100 µm. e, f Immunoblotting analysis of protein lysates isolated from tumors in (c). EGFR expression levels were quantified (f), n = 4 mice for indicated groups. g Representative images showing the IHC staining of indicated proteins in tumors isolated from (c). Scale bar, 100 µm. h−j Mouse body weight (h), mouse liver organ index (i), and mouse liver histological morphology (j) were analyzed post-application with indicated dosages of D6. Con: solvent solution; D6-20 or D6-40: mice treated with D6 at the dose of 20 or 40 mg/kg. Data are presented as mean ± SEM, n = 4 mice (b, c, and f) or n = 6 mice (i). P values were calculated by two-way ANOVA analysis (b) or two-tailed Student’s t-test (c, f, and i).
D6 has the potential to overcome the osimertinib resistance in NSCLC
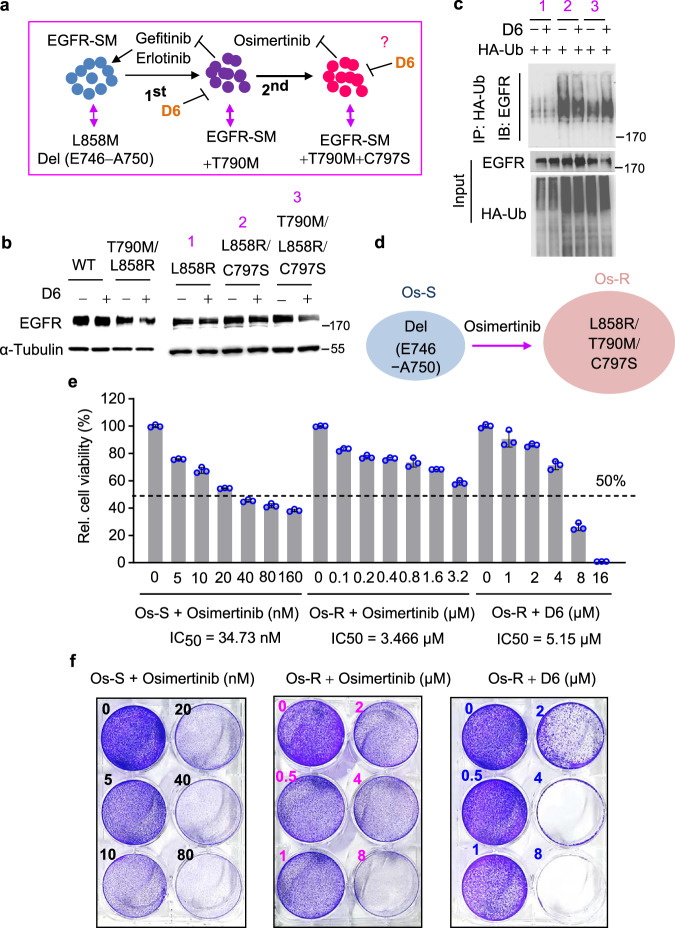
Osimertinib is a representative third-generation EGFR-TKI and has been approved to treat gefitinib or erlotinib-resistant T790M-positive NSCLCs15. However, osimertinib-resistance mutation, such as the frequently identified C797S, frequently emerge within the first year of treatment18,50. As shown in Fig. 7a, tumor cells in some patients might evolve with multiple mutations including T790M, C797S, plus the original EGFR-TKI-sensitive mutations (SM: sensitive mutation, e.g., L858R or Del19 E746_A750) (Fig. 7a). Under such circumstances, applying EGFR-TKIs including osimertinib is ineffective. As D6 destabilized T790M-EGFR mutants, we assessed if D6 would achieve similar efficacy in osimertinib-resistant NSCLCs. As shown, D6 downregulated the protein level of both L858R/T790M and L858R/T790M/C797S-EGFR but had little effect on L858R alone or L858R/C797S mutated forms (Fig. 7b). Consistently, D6 greatly facilitated the ubiqutination of L858R/T790M/C797S-EGFR (Fig. 7c). These results suggest that D6 has the potential to treat osimertinib-resistant NSCLCs. To support that notion, we generated an osimertinib-resistant PC-9 cell line (Os-R) which was transfected with L858R/T790M/C797S-EGFR and then selected by osimertinib (Fig. 7d). In comparison with the parental PC-9 cells (Os-S) which were sensitive to osimertinib (IC50 = 34.73 nM), Os-R cells displayed apparent resistance (IC50 = 3.466 µM, ~100 folds higher) (Fig. 7e, left and middle). Importantly, D6 achieved the potent inhibitory effect on Os-R cells at a low dose (IC50 = 5.15 µM) (Fig. 7e, right). As determined by cell proliferation assay, D6 was detrimental to Os-R cell growth at the dose of 4 µM (Fig. 7f, right). By contrast, Os-R cells showed resistance to osimertinib even at the dose of 8 µM (Fig. 7f, middle). Altogether, these results indicate that D6 may have the potential to treat the osimertinib-resistant NSCLCs.
Fig. 7. D6 overcomes osimertinib resistance in NSCLCs.
a Schematic diagram illustrating the development of EGFR-TKI resistance. b Immunoblotting analysis showing the expression of indicated EGFR mutants transfected into HEK293 cells and treated with or without D6 (2.5 µM). The numbers 1, 2, and 3 represented indicated EGFR mutants. c Cell lysates derived from HEK293 cells transfected with HA-Ub and indicated EGFR mutants with D6 (2.5 μM) treatment or not were incubated with anti-HA beads. The elution of anti-HA beads was then subjected to immunoblotting analysis. d Schematic diagram showing the generation of osimertinib-resistant PC9 cells (Os-R) from the osimertinib-sensitive parental cells (Os-S). e Os-S or Os-R cells were treated with the indicated concentrations of osimertinib or D6. Cell viability was detected by CCK8 and displayed after normalized to the control. f The representative images showing the cell proliferation of Os-S or Os-R cells exposed to the indicated concentrations of osimertinib or D6 for long-term culture in 6-well plate. Data are presented as mean ± SEM (e), n = 3 biologically independent samples.
Discussion
Most NSCLC patients harboring EGFR-activating mutations have a high response rate and achieve a survival benefit when treated with the first generation of EGFR-TKIs, gefitinib, or erlotinib4. However, acquired resistance elicited by the secondary EGFR T790M mutation typically emerges within 9–13 months10,13. Here, we demonstrate that D6 treatment reduces the viability of NSCLC cells harboring T790M-EGFR mutations that mediated TKI resistance in both in vitro and in vivo models. Particularly, D6 treatment does not widely affect noncancerous cells, suggesting its low toxicity. At the molecular level, D6 induces a fast degradation of TKI-resistant T790M-EGFR by jeopardizing its binding to HSP90 but has minor effect on the chaperone activity. Therefore, theoretically, D6 treatment may lead to less side-effects in comparison with canonical HSP90 inhibitors. Our results provide an alternative strategy to overcome the T790M-EGFR mutant in NSCLCs based on D6 treatment. Considering the further application of D6, it is crucial to analyze the systemic bioavailability in future studies.
There is a clear clinical need to overcome T790M-mediated resistance to EGFR-TKIs, and avenues to overcome this resistance are important to pursue. One approach is the development of the second generation of EGFR-TKIs that covalently modify EGFR, such as afatinib12,14. However, due to irreversible covalent binding, they potently suppress the kinase activity of both WT and mutated EGFR and lead to dose-limiting toxicities, which limits the therapeutic window. Alternatively, osimertinib is an oral and irreversible third generation of EGFR-TKI with potent and selective inhibition of T790M EGFR mutant over WT, which has been widely used for EGFR-T790M-positive NSCLC patients15. Unfortunately, resistance mutations also emerge over the course of treatment, most frequently observed as C797S15–17. In this study, we unravel that D6 treatment displays a promising efficacy to eliminate the L858R/T790M/C797S-EGFR-expressing NSCLC cells which are resistant to osimertinib. Additionally, a complicated issue not being deeply explored in current study is the efficacy of D6 when applied to NSCLC patients harboring C797S mutation in cis or trans with T790M-EGFRs. Firstly, we have demonstrated that the modified PC9 cells expressing C797S-EGFR in cis with L858R/T790M were feasible to be targeted by D6 (Fig. 7). Furthermore, D6 had little effect on L858R/C797S-EGFR protein expression (Fig. 7b). These results suggest that D6 most likely hits EGFR mutants harboring T790M in cis with C797S but less likely manipulates the form in trans. Critically, considering that C797S-EGFR is still sensitive to quinazoline-based EGFR-TKIs, patients harboring T790M mutation in trans with C797S are expected to respond well to the traditional first generation of EGFR-TKI in combination with osimertinib16. Here, we highlight the clinical significance of D6 targeting the EGFR mutants of C797S in cis with T790M, as which displays resistance to almost all EGFR-TKIs applied alone or in combination. In future study, it is worthwhile to explore the efficacy of D6 combining with various EGFR-TKIs to prevent the acquisition of TKI resistance.
Cancer cells are dependent on chaperone proteins to survive in acute stress environments, e.g., hypoxic and nutrient-starved microenvironments, which makes most solid cancers sensitive to HSP90 inhibition47. Indeed, it is reported that mutant T790M-EGFRs in NSCLCs are more sensitive to HSP90 inhibitors than the WT cells and HSP90 inhibitors have the potential to overcome EGFR-TKI resistance22–24. Several HSP90 inhibitors have been evaluated in early clinical trial phase26,29,49,51,52. Unfortunately, most HSP90 inhibitors unselectively block chaperone activity and thus have severe side-effects, e.g., hepatotoxicity, and are abandoned for further applications53. In our study, we discover a small-molecule D6 targets HSP90 and damages HSP90-mediated stabilizing T790M-EGFRs without affecting other canonical clients like CDK4 and c-Raf. Mechanically, D6 preferably disrupts the interaction of HSP90 and T790M-EGFRs, whilst showing minimal effect on WT EGFR or first-generation TKI-sensitive EGFR mutants. Therefore, compared with the canonical HSP90 inhibitors, D6 might have less toxicity. Future study to analyze these effects in vivo is necessary to understand how these findings can be translated into clinical therapy.
How D6 exactly manipulates the interaction of HSP90 and T790M-EGFRs is unclear without detailed structural analysis. Indeed, HSP90-mediated chaperone system is complicated as different HSP90 clients are proposed to have co-chaperones or effectors to facilitate correct folding47,54. We assume that T790M-EGFR, comparing to WT or other EGFR mutants, might have evolved some distinctive mechanisms to well cooperate with HSP90 and paired cofactors. For instance, the association of T790M-EGFR with HSP90 is more evident in comparison with non-T790M mutations23 and T790M-EGFR-expressing NSCLC cells are much more sensitive to HSP90 inhibitors22,24. Thus, there may exist a HSP90-centred PPI network which accounts for T790M-EGFR stabilization. Another clue to support this notion is found in cases where erlotinib-resistant NSCLC cells have developed a mysterious HSP90-associated chaperone system to stabilize L858R/T790M-EGFR: it requires the help of cytosolic PKM255. Like D6 treatment, PKM2 has little effect on the HSP90 chaperone activity over other clients but a strong impact on T790M-EGFR. Recently, targeting HSP90-PPI is being proposed as an alternative strategy to precisely suppress HSP90’s partial function, and is expected to have lower toxicity than conventional HSP90 inhibitors56. Several teams have successfully developed molecules which are applied to disrupt the PPI of HSP90-CDC37, HSP90-P23, HSP90-AHA1, and HSP90-HOP49. Herein, this study highlights the D6 targeting the PPIs of HSP90-T790M-EGFR, which may be beneficial to overcome the clinically vital issue of EGFR-TKI resistance.
In summary, we characterize D6 as a novel small-molecule compound that shows potent and selective inhibition of NSCLC cells with T790M-mediated EGFR-TKI resistance. Particularly, we emphasize that D6 differs from current typical HSP90 inhibitors as it specifically disrupts the interaction of HSP90 and T790M-EGFR without affecting the chaperone activity, which may lead to reduced toxicities. D6 may impact the T790M-EGFR-positive NSCLCs by targeting the PPIs of HSP90/T790M-EGFRs, and these findings pave the way for further research into how D6 treatment may treat EGFR-TKI resistance in clinical setting.
Methods
Cell culture
HCC827, PC9, A549, H1299, LO2, and HEK293 cells were obtained from ATCC (American Type Culture Collection). NCI-H1975 and MRC5 cells were purchase from the cell bank of the Chinese Academy of Sciences (Shanghai, China). NCI-H1975, HCC827, and PC9 cells were maintained in RPMI 1640 (Gibco, USA) and A549, MRC5, LO2, and HEK293 in DMEM (Corning, USA) supplemented with 10% FBS (PAN-Biotech, Germany) and antibiotics (100 U/ml Penicillin-Streptomycin, Gibco) at 37 °C in an atmosphere of 5% CO2 humidified environment. All cell lines are regularly authenticated by checking cell morphology and growth rates. If mycoplasma is contamination free is verified by PCR analysis (TaKaRa, Cat. #6601, Japan).
Reagents and antibodies
Erlotinib (HY-50896), 17-AAG (HY-10211), MG132 (HY-13259), Osimertinib (HY-15772) and Recombinant Human Epidermal Growth Factor (rHuEGF, HY-P7109) were purchased from MedChemExpress (MCE), USA. Cycloheximide (No.S7418) was from Selleckchem, USA. Antibodies against cleaved PARP (#9548), ERK1/2 (#4695), phospho-ERK1/2 (Thr202/Tyr204) (#9101), AKT (#4691), and phospho-AKT (Ser473) (#4060) were purchased from Cell Signaling Technology, USA. Antibodies against β-actin, GAPDH, and α-Tubulin were from Beyotime Biotechnology, China. Secondary antibody coupled to Alexa Fluor 488 dye was purchased from Thermo Fisher, USA. HRP-conjugated secondary antibodies used for western blotting were from Jackson Immuno Research Lab., USA. Full antibody details are available in Supplementary Table 1.
Cell viability
Cells were seeded onto 96-well plates at a density of 5000 cells per well and were allowed to adhere for 24 h. Cells with different treatments were subjected to viability detection by using the Cell Counting Kit-8 (CCK-8, MCE) according to the manufacturer’s notifications. In brief, 10 μl CCK-8 working solution was added to each well. After incubation at 37 °C for 1–2 h, the absorbance was measured at 450 nm using a Microplate Reader (PerkinElmer, USA).
Transwell migration assay
In all, 5 × 104 cells pretreated with D6 for 1 h were suspended in 200 µl FBS-free medium and placed in the upper chamber (8 µm pore size, Corning). The insert was then incubated in a 24-well plate with 500 µl 10% FBS-supplemented medium as chemoattractant. Then, 4–6 h later, the top membrane was swiped with cotton swabs to remove non-migrated cells, and the cells on the other side membrane were stained by 0.1% crystal violet solution. Lastly, cell number was counted and normalized to evaluate the relative migration ability.
Cell proliferation assay
In all, 2 × 103 cells were seeded onto a 6-well plate and then treated by indicated concentrations of D6 or equal volume of DMSO solvent. After 7 days, cells were fixed by 4% PFA and stained by 0.1% crystal violet solution. For quantification, the dye was eluted by 33% acetic acid and measured at 570 nm by using a microplate reader57. Relevant cell proliferation rate is presented as relative cell density by normalizing to control.
Cell wound-healing assay
Cells were seeded onto a 12-well plate and allowed to grow to full confluency in complete medium. After D6 treatment for 4 h, cell monolayer was scratched with a 10 µl-pipette tip to generate the wound. Cell migratory ability was measured and analyzed by recording the cell closure at an indicated time point.
RNA isolation and qRT-PCR
Cells were lysed in Trizol reagent RNAiso Plus (Takara, Japan) and the total RNA was isolated by standard protocol and then transcribed into cDNA using 5× Primescript® RT Master Mix (Takara, Japan) according to the manufacturer’s instructions. Gene expression was determined by qRT-PCR analysis by using 2× SYBR® Green Mix (Takara, Japan) on a Bio-Rad detection system (Bio-Rad). The primers used in this study are listed below. hEGFR-F: 5′- GGCACTTTTGAAGATCATTTTCTC-3′, hEGFR-R: 5′-CTGTGTTGAGGGCAATGAG-3′; hXIAP-F: 5′-AGTGGTAGTCCTGTTTCAGCATCA-3′, hXIAP-R: 5′-CCGCACGGTATCTCCTTCA-3′; hBCL-2-F: 5′-CATGCTGGGGCCGTACAG-3′, hBCL-2-R: 5′-GAACCGGCACCTGCACAC-3′; hBCL-XL-F:5′-TGCATTGTTCCCATAGAGTTCCA-3′, hBCL-XL-R: 5′-CCTGAATGACCACCTAGAGCCTT-3′; hSurvivin-F:5′-TGCCTGGCAGCCCTTTC-3′, hSurvivin-R: 5′-CCTCCAAGAAGGGCCAGTTC-3′.
Protein extraction and immunoblotting
Cell lysate was prepared following RIPA buffer and then protein extracts resolved in 5× Laemmli sample buffer (100 mM Tris-HCl [pH6.8], 4% SDS, 20% glycerol and 1 mM DTT) were separated by SDS-PAGE and transferred to a PVDF membrane (Millipore, USA). After being blocked with 5% non-fat milk and incubated with the respective primary antibodies and HRP-conjugated secondary antibodies, immunoblotting images were collected on a Bio-Rad system (Bio-Rad, USA). The detailed information about antibodies is shown in Supplementary Table 1.
Immunoprecipitation
Cells with indicated treatments were lysed in IP lysis buffer (200 mM NaCl, 20 mM Tris-HCl (pH 7.9), 5 mM MgCl2, 10% glycerol, 0.2 mM EDTA and 0.1% NP-40) supplemented with protease inhibitors (Roche cOmplete Protease Inhibitor, Swiss). After sonication and centrifugation, the clear supernatant was collected and incubated with respective antibodies at 4 °C for 4 h or overnight. Immunoprecipitates were washed with IP lysis buffer and further resolved in 1.5× Laemmli sample buffer for further western blotting analysis.
Immunofluorescence staining
Cells seeded onto coverslips were firstly treated with D6 for 12 h and then fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100 and blocked with 1% BSA. The anti-EGFR antibody was incubated overnight and recognized by the secondary antibody conjugated with Alex-488 dyes. After nuclei stained with DAPI, cells were observed and photographed under a fluorescent microscope (Zeiss, Germany).
Animals
Nude mice (BALB/c nu/nu) (5–6 weeks old) were purchased from the Shanghai Laboratory Animal Center (SLAC) and maintained under regular conditions (22 °C, 40–60% humidity, a 12 h light/12 h dark light cycle). For xenograft implantation, NCI-H1975 cells (1 × 106 cells/0.1 ml per mouse) were injected into the right front axilla of mice (s.c.) and then were randomly separated into four groups. When tumor volume reached ~100 mm3, mice of each group were applied to D6 (20 or 40 mg/kg), erlotinib (100 mg/kg) or solvent by i.p. injection. Tumor volume was measured with clipper ruler every other day. At the end of the experiment, tumor-bearing mice were anaesthetized, and tumors were removed and weighed. Experimental animals were housed and handled in accordance with protocols approved by the Committee on the Use of Live Animals in Teaching and Research of Shenzhen University.
Immunohistochemical staining
Tumors were fixed in 4% paraformaldehyde for 48 h. Following embedding in paraffin, 5 μm sections were coated on slides, deparaffinized, and rehydrated. After quenching endogenous peroxidase activity and blocking non-specific binding sites, slides were incubated at 4 °C overnight with indicated primary antibodies and then followed with a proper secondary antibody incubating for 1 h. Sections were observed and imaged under optical microscope (ZEISS, Germany). Detailed information about used antibodies is shown in Supplementary Table 1.
Statistics and reproducibility
The results are based on at least three independent experiments or biologically independent samples. Data are presented as means ± SEM or SD as indicated in the figure legend. The statistical P values were determined by Student’s t-test or two-way ANOVA analysis as indicated in this article.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files.
Acknowledgements
This study was supported by grants from the National Key R&D Program of China (2017YFA0503900 to B.L. and Y.C.), the National Natural Science Foundation of China (32090030 to X.T. and Z.L., 81972602, 81702909 to X.T., 91849208 to B.L., 81525026 to Y.C., 81871114, 82022025, 91949124 to M.Q., and 82071578 to Z.L.), the Shenzhen Municipal Commission of Science and Technology Innovation (ZDSYS20190902093401689, KQJSCX20180328093403969 and JCYJ20180507182044945 to B.L., JCYJ20210324120213038 to Y.Y. and X.T.) and SZU Top Ranking Project (86000000210). The authors would like to thank Dr. Jessica Tamanini (Shenzhen University and ETediting) for editing the manuscript prior to submission.
Author contributions
B.L., Y.C., and X.T. conceived and supervised the study; X.T., L.C., G.L., F.S., and S.Z. performed experiments, collected data and analyzed the results; Y.Y. and D.H. carried out the chemical synthesis; M.Q., Z.L., and J.L. helped to discuss the results; B.L., Y.C., and X.T. organized data and wrote the paper.
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary handling editor: Eve Rogers.
Data availability
All the data that support this study are included in this published article, Supplementary information file, and Source Data file. All source data are included in Supplementary Data 1 and Supplementary Fig. 25.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xiaolong Tang, Lizhi Cheng, Guo Li.
Contributor Information
Xiaolong Tang, Email: tangxiaolong05@126.com.
Yong-Xian Cheng, Email: yxcheng@szu.edu.cn.
Baohua Liu, Email: ppliew@szu.edu.cn.
Supplementary information
The online version contains Supplementary material available at 10.1038/s42003-021-02906-4.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Altorki NK, et al. The lung microenvironment: an important regulator of tumour growth and metastasis. Nat. Rev. Cancer. 2019;19:9–31. doi: 10.1038/s41568-018-0081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hong Z, et al. Luteolin is effective in the non-small cell lung cancer model with L858R/T790M EGF receptor mutation and erlotinib resistance. Br. J. Pharm. 2014;171:2842–2853. doi: 10.1111/bph.12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur. J. Cancer. 2001;37:S9–15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 5.Hirsch FR, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J. Clin. Oncol. 2003;21:3798–3807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 6.Cataldo VD, Gibbons DL, Perez-Soler R, Quintas-Cardama A. Treatment of non-small-cell lung cancer with erlotinib or gefitinib. N. Engl. J. Med. 2011;364:947–955. doi: 10.1056/NEJMct0807960. [DOI] [PubMed] [Google Scholar]
- 7.Balius TE, Rizzo RC. Quantitative prediction of fold resistance for inhibitors of EGFR. Biochemistry. 2009;48:8435–8448. doi: 10.1021/bi900729a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yun CH, et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark GM, et al. Clinical utility of epidermal growth factor receptor expression for selecting patients with advanced non-small cell lung cancer for treatment with erlotinib. J. Thorac. Oncol. 2006;1:837–846. [PubMed] [Google Scholar]
- 10.Suda K, Onozato R, Yatabe Y, Mitsudomi T. EGFR T790M mutation: a double role in lung cancer cell survival? J. Thorac. Oncol. 2009;4:1–4. doi: 10.1097/JTO.0b013e3181913c9f. [DOI] [PubMed] [Google Scholar]
- 11.Sundaresan TK, et al. Detection of T790M, the acquired resistance EGFR mutation, by tumor biopsy versus noninvasive blood-based analyses. Clin. Cancer Res. 2016;22:1103–1110. doi: 10.1158/1078-0432.CCR-15-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwak EL, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc. Natl Acad. Sci. USA. 2005;102:7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yun CH, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc. Natl Acad. Sci. USA. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;28:S24–31. doi: 10.1038/onc.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Remon J, Steuer CE, Ramalingam SS, Felip E. Osimertinib and other third-generation EGFR TKI in EGFR-mutant NSCLC patients. Ann. Oncol. 2018;29:i20–i27. doi: 10.1093/annonc/mdx704. [DOI] [PubMed] [Google Scholar]
- 16.Leonetti A, et al. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br. J. Cancer. 2019;121:725–737. doi: 10.1038/s41416-019-0573-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang ZH, Lu JJ. Osimertinib resistance in non-small cell lung cancer: mechanisms and therapeutic strategies. Cancer Lett. 2018;420:242–246. doi: 10.1016/j.canlet.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 18.Le X, et al. Landscape of EGFR-dependent and -independent resistance mechanisms to osimertinib and continuation therapy beyond progression in EGFR-mutant NSCLC. Clin. Cancer Res. 2018;24:6195–6203. doi: 10.1158/1078-0432.CCR-18-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Z, et al. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor osimertinib in non-small cell lung cancer patients. Clin. Cancer Res. 2018;24:3097–3107. doi: 10.1158/1078-0432.CCR-17-2310. [DOI] [PubMed] [Google Scholar]
- 20.Pearl LH, Prodromou C. Structure and in vivo function of Hsp90. Curr. Opin. Struct. Biol. 2000;10:46–51. doi: 10.1016/s0959-440x(99)00047-0. [DOI] [PubMed] [Google Scholar]
- 21.Pratt WB. The hsp90-based chaperone system: involvement in signal transduction from a variety of hormone and growth factor receptors. Proc. Soc. Exp. Biol. Med. 1998;217:420–434. doi: 10.3181/00379727-217-44252. [DOI] [PubMed] [Google Scholar]
- 22.Sawai A, et al. Inhibition of Hsp90 down-regulates mutant epidermal growth factor receptor (EGFR) expression and sensitizes EGFR mutant tumors to paclitaxel. Cancer Res. 2008;68:589–596. doi: 10.1158/0008-5472.CAN-07-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimamura T, Lowell AM, Engelman JA, Shapiro GI. Epidermal growth factor receptors harboring kinase domain mutations associate with the heat shock protein 90 chaperone and are destabilized following exposure to geldanamycins. Cancer Res. 2005;65:6401–6408. doi: 10.1158/0008-5472.CAN-05-0933. [DOI] [PubMed] [Google Scholar]
- 24.Shimamura T, et al. Hsp90 inhibition suppresses mutant EGFR-T790M signaling and overcomes kinase inhibitor resistance. Cancer Res. 2008;68:5827–5838. doi: 10.1158/0008-5472.CAN-07-5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim YS, et al. Update on Hsp90 inhibitors in clinical trial. Curr. Top. Med. Chem. 2009;9:1479–1492. doi: 10.2174/156802609789895728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Egorin MJ, et al. Pharmacokinetics, tissue distribution, and metabolism of 17-(dimethylaminoethylamino)-17-demethoxygeldanamycin (NSC 707545) in CD2F1 mice and Fischer 344 rats. Cancer Chemother. Pharmacol. 2002;49:7–19. doi: 10.1007/s00280-001-0380-8. [DOI] [PubMed] [Google Scholar]
- 27.Sydor JR, et al. Development of 17-allylamino-17-demethoxygeldanamycin hydroquinone hydrochloride (IPI-504), an anti-cancer agent directed against Hsp90. Proc. Natl Acad. Sci. USA. 2006;103:17408–17413. doi: 10.1073/pnas.0608372103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian ZQ, et al. Synthesis and biological activities of novel 17-aminogeldanamycin derivatives. Bioorg. Med. Chem. 2004;12:5317–5329. doi: 10.1016/j.bmc.2004.07.053. [DOI] [PubMed] [Google Scholar]
- 29.Garg G, Khandelwal A, Blagg BS. Anticancer Inhibitors of Hsp90 function: beyond the usual suspects. Adv. Cancer Res. 2016;129:51–88. doi: 10.1016/bs.acr.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zneg XL, Li XA, Zhang BY. [Immunological and hematopoietic effect of Codonopsis pilosula on cancer patients during radiotherapy] Zhongguo Zhong Xi Yi Jie He Za Zhi. 1992;12:581. [PubMed] [Google Scholar]
- 31.Sun YX. Immunological adjuvant effect of a water-soluble polysaccharide, CPP, from the roots of Codonopsis pilosula on the immune responses to ovalbumin in mice. Chem. Biodivers. 2009;6:890–896. doi: 10.1002/cbdv.200800154. [DOI] [PubMed] [Google Scholar]
- 32.Chen M, et al. Exopolysaccharides from a Codonopsis pilosula endophyte activate macrophages and inhibit cancer cell proliferation and migration. Thorac. Cancer. 2018;9:630–639. doi: 10.1111/1759-7714.12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu J, et al. The natural compound codonolactone attenuates TGF-beta1-mediated epithelial-to-mesenchymal transition and motility of breast cancer cells. Oncol. Rep. 2016;35:117–126. doi: 10.3892/or.2015.4394. [DOI] [PubMed] [Google Scholar]
- 34.Horibata, S., Vo, T. V., Subramanian, V., Thompson, P. R. & Coonrod, S. A. Utilization of the soft agar colony formation assay to identify inhibitors of tumorigenicity in breast cancer cells. J. Vis. Exp.99, e52727 (2015). [DOI] [PMC free article] [PubMed]
- 35.Chaitanya GV, Steven AJ, Babu PP. PARP-1 cleavage fragments: signatures of cell-death proteases in neurodegeneration. Cell Commun. Signal. 2010;8:31. doi: 10.1186/1478-811X-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamura T, et al. Specific organ metastases and survival in metastatic non-small-cell lung cancer. Mol. Clin. Oncol. 2015;3:217–221. doi: 10.3892/mco.2014.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao X, Guan JL. Focal adhesion kinase and its signaling pathways in cell migration and angiogenesis. Adv. Drug Deliv. Rev. 2011;63:610–615. doi: 10.1016/j.addr.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cooper J, Giancotti FG. Integrin signaling in cancer: mechanotransduction, stemness, epithelial plasticity, and therapeutic resistance. Cancer Cell. 2019;35:347–367. doi: 10.1016/j.ccell.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nat. Rev. Mol. Cell Biol. 2005;6:56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- 40.Elsberger B, Tan BA, Mallon EA, Brunton VG, Edwards J. Is there an association with phosphorylation and dephosphorylation of Src kinase at tyrosine 530 and breast cancer patient disease-specific survival. Br. J. Cancer. 2010;103:1831–1834. doi: 10.1038/sj.bjc.6605913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamadi A, et al. Regulation of focal adhesion dynamics and disassembly by phosphorylation of FAK at tyrosine 397. J. Cell Sci. 2005;118:4415–4425. doi: 10.1242/jcs.02565. [DOI] [PubMed] [Google Scholar]
- 42.Huttenlocher A, Sandborg RR, Horwitz AF. Adhesion in cell migration. Curr. Opin. Cell Biol. 1995;7:697–706. doi: 10.1016/0955-0674(95)80112-x. [DOI] [PubMed] [Google Scholar]
- 43.Politi K, et al. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev. 2006;20:1496–1510. doi: 10.1101/gad.1417406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kao, S. H. et al. Analysis of protein stability by the cycloheximide chase assay. Bio. Protoc.5, e1374 (2015). [DOI] [PMC free article] [PubMed]
- 45.Tsubuki S, Saito Y, Tomioka M, Ito H, Kawashima S. Differential inhibition of calpain and proteasome activities by peptidyl aldehydes of di-leucine and tri-leucine. J. Biochem. 1996;119:572–576. doi: 10.1093/oxfordjournals.jbchem.a021280. [DOI] [PubMed] [Google Scholar]
- 46.Longva KE, et al. Ubiquitination and proteasomal activity is required for transport of the EGF receptor to inner membranes of multivesicular bodies. J. Cell Biol. 2002;156:843–854. doi: 10.1083/jcb.200106056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat. Rev. Cancer. 2010;10:537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biebl, M. M. & Buchner, J. Structure, function, and regulation of the Hsp90 machinery. Cold Spring Harb. Perspect. Biol. 11, a034017 (2019). [DOI] [PMC free article] [PubMed]
- 49.Dutta Gupta S, Bommaka MK, Banerjee A. Inhibiting protein–protein interactions of Hsp90 as a novel approach for targeting cancer. Eur. J. Med. Chem. 2019;178:48–63. doi: 10.1016/j.ejmech.2019.05.073. [DOI] [PubMed] [Google Scholar]
- 50.Oxnard GR, et al. Assessment of resistance mechanisms and clinical implications in patients with EGFR T790M-positive lung cancer and acquired resistance to osimertinib. JAMA Oncol. 2018;4:1527–1534. doi: 10.1001/jamaoncol.2018.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chatterjee S, Bhattacharya S, Socinski MA, Burns TF. HSP90 inhibitors in lung cancer: promise still unfulfilled. Clin. Adv. Hematol. Oncol. 2016;14:346–356. [PubMed] [Google Scholar]
- 52.Johnson ML, et al. Phase I/II study of HSP90 inhibitor AUY922 and erlotinib for EGFR-mutant lung cancer with acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors. J. Clin. Oncol. 2015;33:1666–1673. doi: 10.1200/JCO.2014.59.7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saif MW, et al. Open-label, dose-escalation, safety, pharmacokinetic, and pharmacodynamic study of intravenously administered CNF1010 (17-(allylamino)-17-demethoxygeldanamycin [17-AAG]) in patients with solid tumors. Cancer Chemother. Pharmacol. 2013;71:1345–1355. doi: 10.1007/s00280-013-2134-9. [DOI] [PubMed] [Google Scholar]
- 54.Karagoz GE, Rudiger SG. Hsp90 interaction with clients. Trends Biochem. Sci. 2015;40:117–125. doi: 10.1016/j.tibs.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 55.Yang YC, et al. Cytosolic PKM2 stabilizes mutant EGFR protein expression through regulating HSP90-EGFR association. Oncogene. 2016;35:3387–3398. doi: 10.1038/onc.2015.397. [DOI] [PubMed] [Google Scholar]
- 56.Petta I, Lievens S, Libert C, Tavernier J, De Bosscher K. Modulation of protein–protein interactions for the development of novel therapeutics. Mol. Ther. 2016;24:707–718. doi: 10.1038/mt.2015.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang JT, et al. Propyl gallate exerts an antimigration effect on temozolomide-treated malignant glioma cells through inhibition of ROS and the NF-κB pathway. J. Immunol. Res. 2017;2017:9489383. doi: 10.1155/2017/9489383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files.
Data Availability Statement
All the data that support this study are included in this published article, Supplementary information file, and Source Data file. All source data are included in Supplementary Data 1 and Supplementary Fig. 25.