Abstract
Mammalian cells harbor three highly homologous and widely expressed members of the ras family (H-ras, N-ras, and K-ras), but it remains unclear whether they play specific or overlapping cellular roles. To gain insight into such functional roles, here we generated and analyzed H-ras null mutant mice, which were then also bred with N-ras knockout animals to ascertain the viability and properties of potential double null mutations in both loci. Mating among heterozygous H-ras+/− mice produced H-ras−/− offspring with a normal Mendelian pattern of inheritance, indicating that the loss of H-ras did not interfere with embryonic and fetal viability in the uterus. Homozygous mutant H-ras−/− mice reached sexual maturity at the same age as their littermates, and both males and females were fertile. Characterization of lymphocyte subsets in the spleen and thymus showed no significant differences between wild-type and H-ras−/− mice. Analysis of neuronal markers in the brains of knockout and wild-type H-ras mice showed that disruption of this locus did not impair or alter neuronal development. Breeding between our H-ras mutant animals and previously available N-ras null mutants gave rise to viable double knockout (H-ras−/−/N-ras−/−) offspring expressing only K-ras genes which grew normally, were fertile, and did not show any obvious phenotype. Interestingly, however, lower-than-expected numbers of adult, double knockout animals were consistently obtained in Mendelian crosses between heterozygous N-ras/H-ras mice. Our results indicate that, as for N-ras, H-ras gene function is dispensable for normal mouse development, growth, fertility, and neuronal development. Additionally, of the three ras genes, K-ras appears to be not only essential but also sufficient for normal mouse development.
In eukaryotes, Ras proteins are highly conserved from yeast to humans. These proteins include several subfamilies (Rho, Rab, Ras, Ran) of small GTP-binding proteins acting as a biological switches for various cellular processes. In mammals, the Ras subfamily includes three highly homologous H-, N-, and K-Ras proteins, as well as other structurally and functionally related proteins, such as Ral, Rap, R-Ras, and TC21 (14, 27, 29).
Ras proteins are essential signaling intermediates in eukaryotic cells. The Ras-signaling pathway begins with upstream activation at the cell surface via tyrosine kinase or cytokine receptors, or βγ subunits of heterotrimeric G proteins (8, 38). Subsequent formation of an active Ras-GTP complex triggers downstream signaling cascades resulting in modulation of DNA transcription at the cell nucleus (9, 16, 21, 27, 29, 32, 36). Although this pathway has been mostly depicted as a single, linear path linking the cell surface to nuclear responses, it is increasingly evident that Ras proteins are part of more versatile, branched signaling networks.
The mammalian H-, N-, and K-ras genes are expressed ubiquitously (7, 12, 26), raising questions about functional specificity or redundancy for each of these ras family members. Studies of yeast and mice indicate that ras gene function is partially dispensable for normal development and cell survival. Yeasts lacking one of their two ras genes are viable (20), while N-ras homozygous mutant mice grow normally (47). On the other hand, K-ras is essential for normal mouse development (18, 24). Homozygous K-ras−/− embryos die progressively between embryonic day 12.5 and term of gestation, with fetal liver defects and anemia (18). At day 11.5, there is increased cell death of motoneurons in the medulla and the cervical spinal chord, and at day 15.5 of gestation, ventricular walls are very thin (24).
Additional evidence for unique roles of H-, K-, and N-ras are as follows: (i) many tumors are associated with mutations in one specific ras family member (3), and (ii) although it is ubiquitous, the levels of ras mRNA in mice appear to be regulated both temporally and spatially, with certain tissues expressing one or more members of the family preferentially (26). N-ras and K-ras are highly expressed during early development, but levels decrease around postnatal day 10, while H-ras is highly expressed throughout development, with abundant expression in the adult brain. Furthermore, in the juvenile rat brain, H-ras is highly expressed in the neocortex, hippocampus, entorhinal cortex, striatum, thalamus, and cerebellum while the overall levels of expression of N- and K-ras are significantly lower (44, 49, 50).
Due to the ubiquitous expression of the three ras genes in mammalian tissues, it is difficult to determine specificity, if any, of each of the ras gene products regarding tissue or function or activation by specific guanine nucleotide exchange factors. Ras proteins and the neuronal Ras guanine nucleotide exchange factor Ras-GRF (also known as CDC25Mm) may play an important role in neurotransmission and plasticity in vivo (4, 6, 22). A recent in vitro report suggested that H-Ras alone is specifically activated by Ras-GRF (19), a finding consistent with the similar pattern of expression of H-ras and Ras-GRF observed in the rat brain (49). It has also been reported that H-ras, but not K-ras, traffics to the plasma membrane through the exocytic pathway (1).
Gene targeting experiments have indicated that N-ras is dispensable for mouse development or survival and that K-ras plays an important role in embryogenesis (18, 24, 47). In the present study, we targeted the H-ras gene in mice to determine the role of this gene in embryonic and adult mouse development, with emphasis on its potential role in neuronal differentiation. Furthermore, we also bred the H-ras knockout animals with previously available N-ras null mutant mice in order to ascertain the potential effects of the resultant double mutation in both ras loci.
MATERIALS AND METHODS
H-ras targeting vector and chimeric mouse production.
Two lambda genomic DNA clones corresponding to the murine H-ras gene were isolated from a 129SvJ mouse-derived library (Stratagene, La Jolla, Calif.), using the complete cDNA of m-H-ras as a probe (37). The fragments from these two genomic clones were subcloned into pBluescript II (Stratagene). Mapping and partial sequencing demonstrated that all coding exons of H-ras were contained in both λ phage clones. Plasmids pPNT (46), containing pgk-neo, and pMC1-TkpA (10), containing thymidine kinase selectable markers, were used to construct the H-ras targeting vector pLM102 (Fig. 1A). A 4.8-kb PvuII-PvuII fragment containing exon 0 (noncoding) was used as the 5′ arm of the construct, and a PvuII-PvuII fragment of 2.4 kb containing exon IV (last coding exon of H-ras) was used as the 3′ arm. The pPNT Neo cassette (XhoI-BamHI) was used as a positive marker and replaced a 1.63-kb fragment that contained exons I, II, and III (which code for amino acids 1 to 150, more than 75% of the protein). The negative marker (herpes tk) was placed 5′ to the regions of H-ras homology. The targeting vector, pLM102, was linearized with SalI, and 10 to 15 μg of DNA was electroporated (250 V, 250 μF; Gene Pulser; Bio-Rad) into RW-4 embryonic stem (ES cells) (Genome Systems, St. Louis, Mo.). After electroporation 2 × 106 cells were plated in 100-mm-diameter tissue culture dishes containing a monolayer of G418-resistant embryonic fibroblasts. Colonies resistant to double selection (350 μg of G418 [Gibco-BRL, Gaithersburg, Md.]/ml) and 5 μM ganciclovir (Syntex, Palo Alto, Calif.) were isolated and expanded. Southern blotting analysis showed that 8 out of 750 G418- and gancyclovir-resistant clones had targeted disruption of one H-ras locus by homologous recombination.
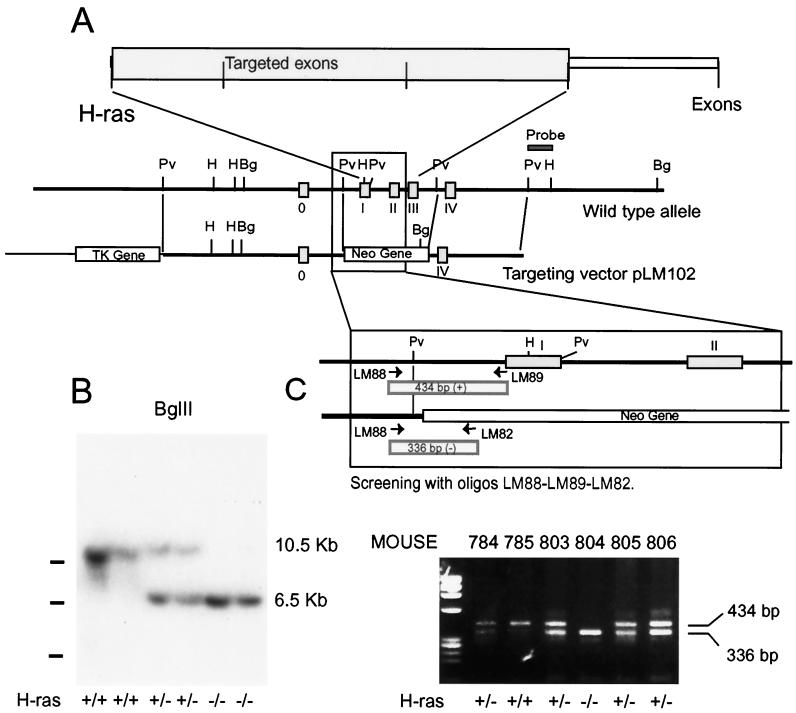
FIG. 1.
Targeted disruption of the murine H-ras gene in ES cells and mice. (A) Schematic representation of the H-ras locus and targeting vector. Boxes in the wild-type allele schematics represent the exons of the H-ras gene. The open boxes in the targeting vector schematics represent the pgk-neo and pgk-tk selectable marker genes. The position of the 3′ flanking probe used in Southern blotting is indicated. (B) Homologous recombination of the targeting vector in mice was verified by Southern blotting, digesting genomic DNA with BglII, and hybridizing with a 3′-flanking probe. The wild-type allele produced a 10.5-kb band, whereas the mutant allele yielded a 6.5-kb band due to the introduction of a new BglII site in the targeting vector. (C) Routine genotyping of mice was performed by PCR using the oligonucleotides indicated, whose sequences are given in Materials and Methods. The LM88 and LM89 primers are specific for the H-ras gene and amplified a 434-bp fragment. The LM82 primer is specific for the Neo-PGK promoter and amplified a 336-bp fragment with LM88. Pv, PvuII; H, HindIII; Bg, BglII.
Several of the recombinant ES cell lines exhibiting normal karyotypes were expanded and subsequently used to generate chimeras by injection into day 3.5 C57BL6/N blastocysts. The blastocysts were transferred to NIH-Swiss pseudopregnant foster mothers. Any chimeric offspring, identified by their agouti coat color, were mated with C57BL6/N females. Agouti offspring were then analyzed for H-ras disruption by Southern blotting and PCR analysis. Of the chimeras, two achieved germline transmission of the disrupted H-ras allele.
Genotyping of targeted ES cells, mice, and embryos.
Genomic DNA was extracted from cultured ES cells, mouse tail biopsies, or embryo yolk sacs as previously described (25). ES cells were incubated at 37°C while tail biopsies and embryo yolk sacs were incubated at 55°C in lysis buffer (100 mM Tris-HCl [pH 8.0] 5 mM EDTA, 0.2% sodium dodecyl sulfate [SDS], 200 mM NaCl, 200 μg of proteinase K/ml) for 4 to 5 h or overnight. DNA was precipitated using isopropanol, washed in 70% ethanol, and resuspended in 200 μl of Tris-EDTA buffer, pH 8.0. For Southern analysis, 20 μl of DNA was digested with HindIII, electrophoresed on 0.6% agarose gels, and transfered to GeneScreen Plus membranes (Dupont, Boston, Mass.). A probe flanking the 3′ end of the targeting vector sequence was radiolabeled using a random primer labeling kit (Stratagene) and was used in hybridizations. Wild-type and mutant alleles were identified by predicted restriction fragment size differences. Clones displaying homologous recombination were reassessed by restriction with BglII and hybridizing with the original 3′ probe (Fig. 1B). Another 5′ flanking probe was used to confirm proper homologous recombination (not shown). Digestion of ES cell DNA with enzymes that did not restrict within the targeting vector and Southern transfer and hybridization with a neo probe demonstrated a single band, confirming the presence of a single site of vector insertion in the targeted ES cell clones.
Routine genotyping of DNA isolated from mouse tail biopsies or embryo yolk sacs was performed by PCR. The primers for H-ras were LM88 (5′-ATAGTTGTAGGTTGCACCCACATGCCG-3′), LM89 (5′-ACCTGCCAATGAGAAGCACACTTAGCC-3′), and LM82 (5′-CTACCGGTGGATGTGGAATGTGTGCGA - 3′); LM88 and LM89 primers were specific for the H-ras gene (annealing to nucleotides 901 to 927 and 1308 to 1334 of the published genomic sequence of H-ras [5]) and amplified a fragment of 434 bp. LM82, specific for the Neo-PGK promoter (nucleotides 517 to 543; GenBank accession no. M18735), amplified a fragment of 336 bp with LM88. The primers for N-ras were LM164 (5′-CCAGGATTCTTACCGAAAGCAAGTGGTG-3′), LM205 (5′-GATGGCAAATACACAGAGGAACCCTTCG-3′), and LM166 (5′CAGAGCAGATTGTACTGAGAGTGCACC-3′). The LM164 and LM205 primers were specific for the N-ras gene (positions 4 to 31 and 121 to 148 on exon II; GenBank accession no. M12122) and amplified a fragment of 146 bp; LM166, specific for the cloning vector pUC19 (position 157 to 183), amplified a fragment of 315 bp with LM164. Oligonucleotides were used in a 50-μl reaction mixture with 1 to 2 μl of DNA and 1.25 U of Taq polymerase (Boehringer Mannheim, Indianapolis, Ind.). Cycling conditions were 94°C for 4 min followed by 30 cycles of 94°C for 1 min, 62°C for 1 min, and 72°C for 1 min, followed by an elongation cycle of 72°C for 10 min, using a Perkin-Elmer Thermal Cycler. Amplified products were analyzed by electrophoresis in 2.5% agarose gels (NuSieve 3:1).
RNA-PCR analysis.
Total RNA was extracted and purified from frozen mouse tissues (strain C57BL6/N × 129SvJ) using TRIzol reagent (Gibco-BRL, Grand Island, N.Y.). First-strand cDNA was generated using SuperScript II RNase H− reverse transcriptase (RT) (Gibco-BRL) and oligo(dT) as described by the manufacturer. PCR was performed using primers for the various ras genes: for H-ras, LM99 (5′-AAGCTTGTGGTGGTGGGCGCTAAAGGC-3′) and LM111 (5′-CTTTCACCCGCTTGATCTGCTCCCTGTACT-3′), corresponding to positions 13 to 39 and 284 to 313 of the coding sequence (GenBank accession no. M10035); for N-ras, oligonucleotides LM164 (5′-CCAGGATTCTTACCGAAAGCAAGTGGTG-3′) and LM165 (5′-CCTGTAGAGGTTAATATCTGCAAATG-3′), corresponding to positions 4 to 31 and 162 to 187 on exon II; GenBank accession no. M12122); and for K-ras, LM209 (5′-AGTACGACCCTACGATAGAGGACTCCT-3′), bp 92 to 118, LM210 (5′-CAATCTGTACTGTCGGATCTCTCTCACC-3′), specific for K-ras4A bp 477 to 504, and LM211 (5′-CTAATGTATAGAAGGCATCGTCAACACCC-3′), specific for K-ras4B, bp 450 to 478, of their respective coding sequences. The conditions for PCR were as described above. Amplified products were analyzed directly in 2.5% agarose NuSieve (3:1) gels.
Histopathological analysis.
Necropsies were performed on embryos and young adult H-ras−/− and H-ras+/+ mice. Tissues were fixed in formalin and embedded in paraffin. Sections were stained with hematoxylin and eosin stain.
Western blot analysis.
Protein extracts were obtained from snap-frozen mouse tissues. Tissues were homogenized in radioimmunoprecipitation buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton X-100, 1% sodium desoxycholate, 0.1% SDS) and centrifuged in a Sorvall S1256 at 30,000 × g for 30 min. Supernatant was recovered and proteins were quantified. Lysates (50 to 70 μg/lane) were loaded onto SDS-polyacrylamide gels, and the proteins were transferred to polyvinylidene difluoride membranes (Millipore Immobilon-P) by electroblotting. Membranes blocked in TTBS (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.05% Tween 20 plus 1% bovine serum albumin) were incubated, as appropriate, with 1:1,000 dilutions of commercial antibodies from Santa Cruz Biotechnology, Santa Cruz, Calif. Antibodies used included polyclonal anti-H-ras antibodies (C-20, sc-520), polyclonal anti-N-ras antibodies (C-20, sc-519), and monoclonal anti-K-ras antibodies (F234, sc-30). Western blottings were developed using ProtoBlot Western blot AP (Promega) following procedures recommended by the supplier.
Hippocampal cultures.
Reagents for tissue culture were purchased from Gibco-BRL, Sigma (St. Louis, Mo.), and Intergen (Purchase, N.Y.). Fetal bovine serum (FBS) was inactivated during a 30-min incubation at 56°C prior to use. Cultures were prepared from wild-type and homozygous mutant mouse embryonic hippocampus on day 16 of gestation (E16). The hippocampi were minced and trypsinized. Cells suspended in Dulbecco's minimum Eagle medium (DMEM)-F12-N2 and 10% FBS were plated on glass coverslips coated with 15 μg of polyornithine per ml and 1 μg of fibronectin per ml at a density of 190,000 cells/cm2. After 6 days in culture, a third part of the medium was replaced by DMEM-N2 and 10% FBS, and 5 μM 1-β-d-arabinofuranosylcytosine (Ara-C) was added in order to halt glial proliferation (48).
Immunostaining of cultured cells.
Cells grown in culture for 14 to 18 days were fixed with 4% paraformaldehyde–0.1 M phosphate buffer (pH 7.4) for 30 min. After treatment with 0.1% Triton X-100–10% normal serum–phosphate-buffered saline, cells were incubated overnight at 4°C with the primary antibodies against microtubule-associated protein 2ab (MAP-2ab; mouse monoclonal antibody at 1:200; Sigma); GABA (rabbit polyclonal antibody at 1:1,000; Sigma); calretinin (rabbit polyclonal antibody at 1:1,500; Swant, Bellinzona, Switzerland); synapsin-I (rabbit polyclonal antibody at 1:1,000; from M. Kennedy); Ca2+/calmodulin-dependent protein kinase IIα (CaMKIIα; mouse monoclonal antibody at 1:100; Boehringer Mannheim); phosphorylated CaMKIIα (PCaMKIIα; mouse monoclonal antibody at 1:100; Affinity Bioreagents, Golden, Colo.). The cells were then incubated with the corresponding fluorescein- and/or rhodamine-conjugated secondary antibodies (1:100) (Jackson Immuno Research, West Grove, Pa., or Cappel, Durham, N.C.) or with a biotinylated secondary antibody (1:200) followed by an avidin-biotin-horseradish peroxidase complex (Vectastain ABC kit; Vector, Burlingame, Calif.) and developed using diaminobenzidine (DAB). Coverslips were mounted in 1,4-diazabicyclo[2.2.2]octane (DABCO)-glycerol.
Fluorescence-activated cell sorter (FACS) analyses.
Single-cell suspensions from thymus and spleen of wild-type and mutant mice were prepared in sorter medium (phenol red-free Hanks balanced salt solution containing 0.1% sodium azide, 0.2% bovine serum albumin, 10 mM EDTA, and 4 mM sodium bicarbonate). Cells were resuspended at 2 × 107 cells/ml, and 50-μl (106 cells) aliquots were stained with the following reagents: fluorescein isothiocyanate (FITC)-conjugated rat anti-mouse CD45, clone 30F11.1; phycoerythrin (PE)-rat anti-mouse CD45(R)/B220; FITC-rat anti-mouse ThB, clone 49-H4; PE-rat anti-mouse CD43, clone S7; FITC-hamster anti-mouse T-cell receptor α/β (TCRα/β, clone H57-597; PE-rat anti-mouse CD5, clone 53-7.3; PE-rat anti-mouse CD11b/Mac-1, clone M1/70; FITC-anti-mouse-Thy1.2 (Becton Dickinson, Mansfield, Mass.); PE-rat anti-mouse CD19, clone 1D3; PE-rat anti-mouse CD4, clone H129.19; or FITC-rat anti-mouse CD8, clone 53-6.7. To prevent FcR-mediated binding of labeled antibodies, unlabeled monoclonal antibody 2.4G2, which is specific for mouse FcRII, was added prior to the addition of labeled reagents. Once stained, cells were washed twice in sorter medium and resuspended at 2.5 × 106 cells/ml for analysis on a Becton Dickinson FACScan. Nonviable cells were excluded by forward-angle scatter and uptake of propidium iodide. Unless otherwise noted, all antibodies were obtained from Pharmingen, San Diego, Calif.
RESULTS
Generation of homozygous mutant mice for H-ras gene.
The targeting vector pLM102 was constructed by substituting a Neo cassette for exons I, II, and III of H-ras (Fig. 1A). However, the vector still contained significant homologous regions 5′ and 3′ to the coding exons to facilitate homologous recombination. The linearized targeting vector was electroporated into RW-4 murine ES cells, which were grown in the presence of G418-ganciclovir selection. A total of 750 clones were screened by Southern hybridization, using a probe 3′ of the short arm of homology (data not shown). We identified eight clones positive for homologous recombination. After karyotyping to eliminate chromosomal abnormalities, five cell lines were microinjected into blastocysts to produce chimeric mice.
Chimeric animals carrying the targeted H-ras gene were mated to generate heterozygous mice carrying one normal and one mutant H-ras allele. No obvious phenotype was apparent in heterozygous mutant mice, and when these were inbred, wild-type, heterozygous, and homozygous mutant mice were produced in the expected Mendelian ratios (data not shown). Furthermore, homozygous mutant males and females were both fertile. Mutant genotypes were initially confirmed by Southern hybridization (Fig. 1B), and further routine genotyping was carried out by PCR, using oligonucleotides which hybridized within the H-ras gene and on the neo gene (see Materials and Methods) (Fig. 1C).
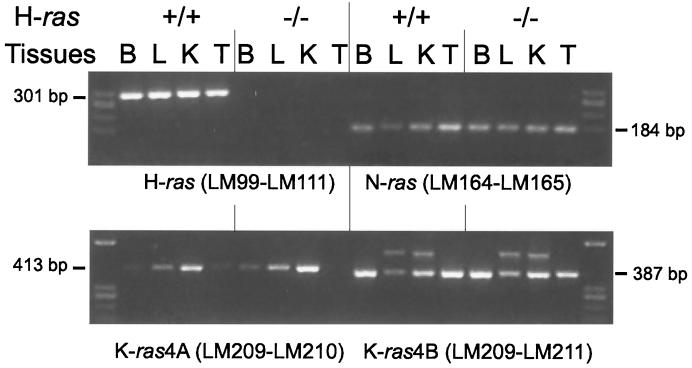
To ensure that the modification of the H-ras gene resulted in a null mutation, we used RT-PCR to examine ras mRNA expression in tissues from wild-type and mutant H-ras mice. Figure 2 depicts the failure of oligonucleotides specific for H-ras gene to amplify cDNA from homozygous mutant −/− animals. However, RT-PCR of the other ubiquitously expressed ras genes (N-ras, K-ras4A, and K-ras4B) showed normal levels of expression in homozygous mutant mice, in comparison with wild-type mice. Therefore, a null mutation in the H-ras gene in vivo was successfully achieved in this study, and there was no modification in the level of expression of the rest of the ras genes due to the absence of H-ras. In the case of K-ras4A, the levels of expression looked quite variable depending on the tissues analyzed (Fig. 2, lower panel), in agreement with a previous report (35). On the other hand, the levels of K-ras4B and N-ras were more or less similar in all tissues checked.
FIG. 2.
Detection by RT-PCR of H-ras, N-ras, K-ras4A, and K-ras4B in total RNA from tissues of animals with wild-type and mutant H-ras. Oligonucleotides specific for detecting each of the different ras gene transcripts were used as indicated. Their sequences are detailed in Materials and Methods. (Above) LM99 and LM111 amplified H-ras, and LM164 and LM165 amplified N-ras. (Below) LM209 and LM210 amplified K-ras4A, and LM209 and LM211 were specific for K-ras4B. Tissues used were brain (B), liver (L), kidney (K), and testis (T) from mice with wild-type (+/+) and null (−/−) H-ras genes. The levels of expression of N-ras and both K-ras4A and K-ras4B molecules in tissues were not affected by the absence or presence of H-ras gene products. As previously described, K-ras4A shows different expression levels depending on the tissues studied. The oligonucleotides used for K-ras4B occasionally lighted up an alternative band in liver and kidney whose intensity was not affected by either the presence or the absence of H-ras.
Viability and fertility of mice lacking H-ras.
Breeding among H-ras+/− mice produced offspring with the expected Mendelian ratios for the wild-type, heterozygous, and homozygous mutant genotypes. The Mendelian ratios for the genotypes were maintained among male and female offspring. Thus, the absence of H-Ras protein in mice did not compromise the development of either gender. The sizes of the litters were similar to those of wild-type and mutant animals (average of seven or eight pups per litter). To confirm that the mutation did not disrupt embryonic development, we performed histopathological analysis of embryos on day 12 or 13 of gestation. No differences were observed between wild-type and mutant embryos (data not shown). Furthermore, histopathological analysis of adult H-ras−/− animals was also carried out using standard procedures. Detailed analysis of necropsies and histological sections of brain, heart, liver, testis, thymus, lung, spleen, kidney, pancreas, and parotid of H-ras−/− animals did not show any gross or histological abnormality compared with these organs in wild-type mice. Therefore, a functional H-ras gene appears not to be required for normal organ development in mice.
Neuronal development in H-ras knockout mice.
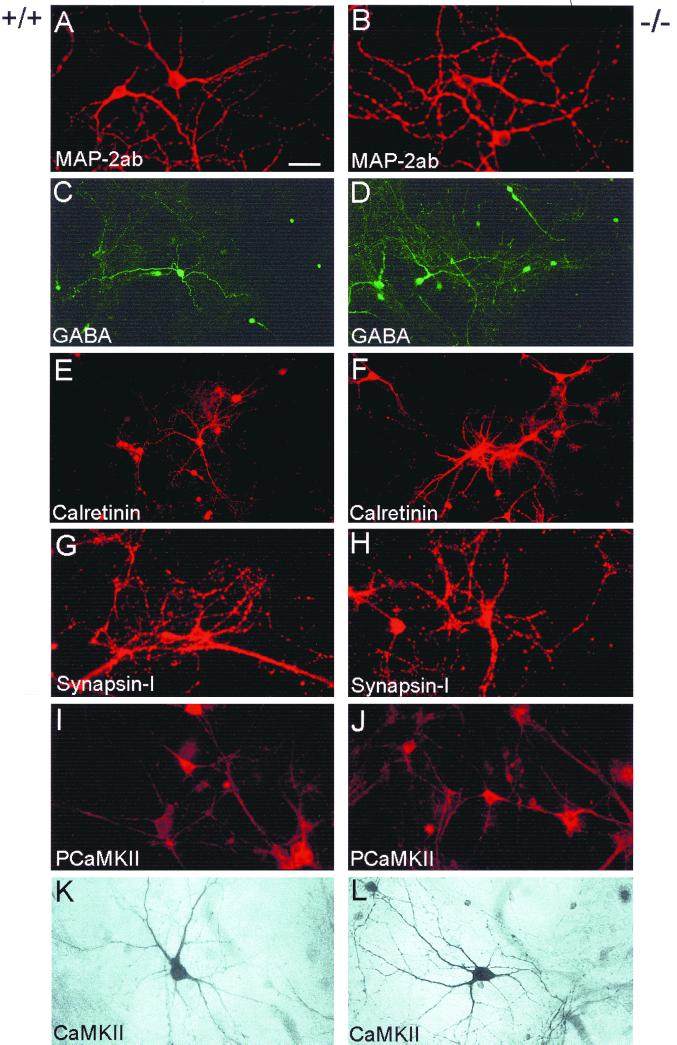
It is thought that the Ras-MAP kinase pathway plays an important role during neuronal survival and differentiation (2, 23, 40). To test whether the absence of the H-Ras protein could affect neuronal differentiation, we prepared cultures of hippocampal neurons and stained them with specific antibodies against MAP-2ab, a general marker for neurons (33), CaMKIIα, an abundant and functionally important dendritic protein expressed at high levels in hippocampal glutamatergic neurons (41, 42, 43), and phosphorylated CaMKIIα (34). An interaction between CaMKIIα and Ras proteins at excitatory synapses has been described recently (6, 22). Staining was also performed to detect GABA, calretinin (a calcium-binding protein labeling a subpopulation of GABAergic hippocampal neurons [13]), and the synaptic-vesicle-associated protein synapsin I (11). As shown in Fig. 3, hippocampal neurons prepared from H-ras−/− knockout mice differentiated in culture in a manner similar to their wild-type counterparts. Most neurons in the cultures possessed a pyramid-like morphology, bearing multiple dendrites and expressing phosphorylated CaMKIIα and CaMKIIα (Fig. 3A and B, I to L), suggesting that they were glutamatergic. Some neurons expressed GABA and calretinin, indicating that they were inhibitory (Fig. 3C to F). We found no appreciable differences in the numbers of glutamatergic and GABAergic cells between H-ras−/− and wild-type neurons (data not shown). As reported for mature hippocampal neurons in culture (11, 48), expression of synapsin I became concentrated in puncta, a fact that was independent of the presence or absence of H-ras in the cells (Fig. 3G and H).
FIG. 3.
Neuronal differentiation is not altered in neurons lacking H-ras. Cultured hippocampal neurons from wild-type mice (A, C, E, G, I, K) and H-ras−/− mice (B, D, F, H, J, L) were stained with specific antibodies against MAP-2ab, GABA, calretinin, synapsin I, phosphorylated CaMKIIα (PCaMKIIα), or CaMKIIα. Scale bar: A, B, and G to J, 35 μm; C to F, 55 μm; K and L, 25 μm.
Analysis of hematopoietic cells in H-ras knockout mice.
Finally, to determine the effects of the H-ras null mutation on components of the immune system, cells from thymus and spleen were analyzed by flow cytometry. The cells were stained using fluorescent antibodies specific for markers of macrophage, B-cell, and T-cell lineages. No significant differences were observed in the lymphoid or myeloid component of the spleen or in the T cells populating the thymus (data not shown). Therefore, the H-ras null mutation does not prevent the maturation of effector cells of the immune system.
Double null mutant mice deficient for H-ras and N-ras are viable.
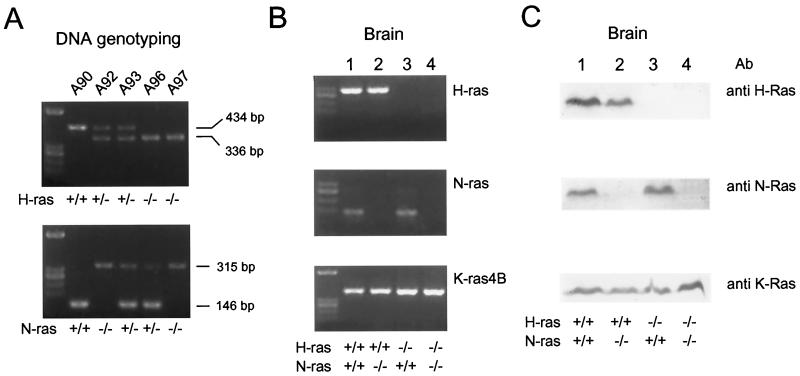
As previously shown for N-ras (47), the mice deficient for H-ras had no apparent phenotypic defects, and we wished to mate these mutant ras strains to try and generate potential double mutant H-ras/N-ras mice. As shown in Fig. 4, breeding between H-ras and N-ras mutant mice gave rise to viable, adult offspring whose genomes carried disrupted versions of both ras loci (for example, Fig. 4A, mouse A97). Furthermore, we confirmed the absence of expression of both genes, as we did for H-ras alone, by RT-PCR and by Western blot analysis. Figure 4B shows that there was no expression of H-ras or N-ras mRNA in the null double H-ras/N-ras mutants (Fig. 4B, lane 4), while there were normal levels of both RNAs in the wild type (Fig. 4B, lane 1) or heterozygous combinations of the two null mutations (Fig. 4B, lanes 2 and 3) in both loci. It is interesting that the levels of K-ras expression were roughly similar in all wild-type and mutant animals, suggesting that normal levels of K-ras are sufficient to sustain viability of adult animals and that there is no need of an overexpression of the K-ras locus to compensate for the absence of expression of the other two mammalian ras loci. Analysis of Ras protein expression using Western immunoblottings with antibodies specific for each of the H-Ras, N-Ras, and K-Ras protein products (Fig. 4C) completely paralleled the observations made with RNA expression. FACS analyses of cells from the thymus and spleen of adult animals did not show any significant differences among the B- and T-cell lineages of the N-ras–H-ras double knockouts and the wild-type animals (not shown). These results confirm the viability of animals lacking expression of H-Ras and N-Ras proteins in all of their tissues and the sufficiency of normal levels of K-Ras to sustain such viability.
FIG. 4.
Analysis of double mutant mice deficient for H-ras and/or N-ras. (A) Genotyping of representative animals resulting from crossing of H-ras- and N-ras-disrupted mice. Oligonucleotides used were as described in Materials and Methods: LM82, LM88, and LM89 were used for the H-ras gene, and LM164, LM205, and LM166 were used for the N-ras gene. Note that mouse A97 was deficient in both gene loci. (B) RT-PCR detection of expression of the different ras genes. Procedures were exactly as those used for Fig. 2. For K-ras, only K-ras4B expression is shown since the levels of K-ras4A were rather low in the brain and were similar for all four genotypes studied here (data not shown). (C) Western immunoblotting of H-Ras, N-Ras, and K-Ras proteins in double and single mutation mice. A 50-μg sample of total protein from brain was electrophoresed on SDS–14% polyacrylamide gels. The antibodies used were polyclonal anti-H-Ras and anti-N-Ras antibodies and monoclonal anti-K-Ras antibodies (recognizing both K-Ras4A and K-Ras4B proteins). (B and C) Representative examples showing only expression of RNA and protein from brain. Other tissues analyzed yielded similar results, with no increase in the expression of K-Ras in the double mutation mice compared with mice with single mutations of H-Ras or N-Ras or with wild-type mice.
Finally, despite the lack of any observable phenotype of adult, double null mutant H-ras–N-ras mice, it is interesting that less-than-expected numbers of these double knockout adult animals were consistently obtained in Mendelian crosses between heterozygous N-ras–H-ras animals (Table 1). In any event, the viability and fertility of the double mutant mice indicate that K-Ras alone is not only necessary (as previously shown [18, 24]) but also sufficient to support development of mice to the adult stage.
TABLE 1.
Analysis of offspring resulting from crossings between mice carrying null mutations of H-ras and N-rasa
| Parameter | Offspring of mice carrying null mutations (♂H-ras+/−/N-ras−/− × ♀H-ras−/−/N-ras+/−)
|
|||
|---|---|---|---|---|
| H-ras+/−/N-ras−/− | H-ras+/−/N-ras+/− | H-ras−/−/N-ras+/− | H-ras−/−/N-ras−/− | |
| Total no. of animals | 8 | 10 | 14 | 2 |
| Expected Mendelian ratio | 1/4 | 1/4 | 1/4 | 1/4 |
Representative genotypic analysis of all newborn animals from six litters obtained by breeding a male H-ras+/−/N-ras−/− animal with two female H-ras−/−N-ras+/− animals. Similar percentage distributions of offspring genotypes were obtained in crosses in which parental genotype and sex were inverted (♀H-ras+/−/N-ras−/− × ♂H-ras−/−/N-ras+/−). Genotypes were determined for weaned animals by following procedures described in Materials and Methods.
DISCUSSION
The present study provides a detailed analysis of mice carrying a null mutation in the H-ras gene, although the viability of H-ras−/− animals was mentioned briefly in reports of K-ras knockout mice (18, 24). In addition, we describe preliminary analyses of mice with a double null mutation in two of the three ras genes (H-ras and N-ras). Using gene targeting and microinjection techniques, offspring with a heterozygous H-ras gene were obtained. Mating these animals resulted in mice with a homozygous null mutation of H-ras that were phenotypically indistinguishable from their wild-type littermates. The successful ablation of the H-ras gene occurred because the neomycin cassette used in the targeting vector lacked the first three exons of murine H-ras. The deleted exons contained the initiation codon and greater than 75% of the coding sequence. This was confirmed by the lack of expression of H-ras mRNA and protein, or truncated derivatives thereof, in tissues from mice deficient in genomic H-ras.
H-, N-, and K-ras genes are expressed ubiquitously, but tissue-specific and developmental stage-specific quantitative differences in mRNA expression of the three ras genes have been described (12, 26, 28). This, together with the observation that different ras genes are predominantly activated in different tumors, suggests specific roles for the members of the ras gene family. However, we successfully generated H-ras homozygous mutant animals in the expected Mendelian ratios. A breeding colony of adult H-ras−/− animals has been maintained in our laboratory for more than 1 year. The animals appear healthy and normal with no signs of any apparent associated lesions. The growth rates of H-ras−/− animals were indistinguishable from those of wild-type animals, and mutant mice reproduced normally. Histological analysis of embryos and various adult tissues failed to reveal any differences between wild-type and H-ras-deficient mice. Therefore, we conclude that there is no absolute requirement for H-ras function in embryonic or adult mouse development or in sexual maturation.
In a previous study, H-ras was implicated in playing a role in thymocyte development (44). However, analysis of immune composition showed the thymocyte composition to be nearly identical in wild-type and mutant mice, comprised mainly of the double positive (CD4+ CD8+) T cells. Additionally, statistically similar proportions of mature CD4+ and CD8+ T cells were present in the spleens of wild-type and H-ras mutant mice. The proportions of T cells with rearranged TCRα/β in the spleen and thymus were not significantly different between H-ras+/+ and H-ras−/− animals. Thus, loss of H-ras gene function does not affect peripheral immune system components in mice.
The preferential expression of H-ras over N- and K-ras in the brain suggests an important role for H-ras in brain development and neurotransmission. This is supported by almost exclusive expression of Ras-GRF1, a guanine exchange factor for H-Ras in the brain (19, 44, 49, 50). However, in the present study, analysis of neuronal development in wild-type and H-ras−/− mice showed no significant differences between the two groups. We did not find differences in the number of neurons or in the expression of synapsin I, CaMKIIα, or phosphorylated CaMKIIα, suggesting that H-Ras function was dispensable for the survival and differentiation of hippocampal neurons. However, these results do not rule out the possibility that the absence of H-Ras could affect synaptic plasticity. Brambilla et al. (4) have shown that deletion of the gene coding for Ras-GRF (also known as CDC25Mm) disrupted synaptic plasticity in the basolateral amygdala and memory consolidation. While this report was in preparation, two publications on H-ras knockout mice were released, documenting a possible regulatory role of the product of this gene in the regulation of long-term potentiation through regulation of NMDA receptor phosphorylation (31) or in tumor formation in skin carcinogenesis (17).
Taken together, our results indicate that H-ras gene expression is dispensable for mouse development, growth, and fertility. These observations are similar to those for N-ras (47), wherein, using gene targeting, it was determined that N-ras is also dispensable for mouse development. Of the three ras genes, only K-ras appears to be critical for normal mouse development based on the observation that K-ras deficiency results in embryonic lethality (18, 24). Furthermore, it appears that K-ras is not only necessary but also sufficient for mouse development, since we also observed that double null mutant mice deficient for H-ras and N-ras are still viable, have normal development, and are fertile. Interestingly, however, the number of adult double knockout animals resulting from crosses between heterozygous N-ras/H-ras animals was lower than expected according to Mendelian ratios, suggesting a lower viability of embryos carrying double null mutations in the H-ras and N-ras loci. The reason for this lower number remains to be determined in future studies.
It could be speculated that the differences in C-terminal amino acid sequence and processing exhibited by the K-Ras proteins in relation to both the H-Ras and N-Ras proteins (15, 30, 39) may account, at least in part, for the lethality of the K-ras knockout mutations and the viability of the H-ras and N-ras knockout mutants. It should also be noted that the reports describing K-ras knockout mice so far have dealt only with mutations resulting in an absence of expression of both the K-ras4A and K-ras4B alternative exons. Generation of individual null mutations of each of these alternative forms of K-ras is required to distinguish any functional differences between the two K-ras isoforms in mammalian development.
Bearing in mind the limitation that mouse development under the controlled conditions of a pathogen-free animal facility does not reproduce the conditions that mice find in nature, we conclude the dispensability of H-ras and N-ras gene function, singly or in combination, for mouse growth and development. Although dispensability has been clearly established for H- and N-Ras proteins in the laboratory environment, their particular function(s) in vivo has yet to be established. It is possible that H-Ras and N-Ras function in targeted mice is taken over by K-Ras proteins or by the other structurally and functionally related members of the Ras subfamily (Ral, Rap, R-Ras, and TC21). Generation of mice with these mutant genes may provide further insight into the possible specificity of function of the various ras gene products.
ACKNOWLEDGMENTS
This work was supported by FEDER grant 1FD1997-1735 from Ministerio de Ciencia y Tecnología, Spain.
We thank Alberto Orfao and Marta Ayuso for help with FACS analysis. M. Kennedy (Caltech) is gratefully acknowledged for anti-synapsin I antibody.
REFERENCES
- 1.Apolloni A, Prior I A, Lindsay M, Parton R G, Hancock J F. H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol Cell Biol. 2000;20:2475–2487. doi: 10.1128/mcb.20.7.2475-2487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonni A, Brunet A, West A E, Datta S R, Takasu M A, Greenberg M E. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science. 1999;286:1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- 3.Bos J L. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 4.Brambilla R, Gnesutta N, Minichiello L, White G, Roylance A J, Herron C E, Ramsey M, Wolfer D P, Cestari V, Rossi-Arnaud C, Grant S G, Chapman P F, Lipp H P, Sturani E, Klein R. A role for the Ras signalling pathway in synaptic transmission and long-term memory. Nature. 1997;390:281–286. doi: 10.1038/36849. [DOI] [PubMed] [Google Scholar]
- 5.Brown K, Bailleul B, Ramsden M, Fee F, Krumlauf R, Balmain A. Isolation and characterization of the 5′ flanking region of the mouse c-Harvey-ras gene. Mol Carcinog. 1988;1:161–170. doi: 10.1002/mc.2940010304. [DOI] [PubMed] [Google Scholar]
- 6.Chen H J, Rojas-Soto M, Oguni A, Kennedy M B. A synaptic Ras-GTPase activating protein (p135 SynGAP) inhibited by CaM kinase II. Neuron. 1998;20:895–904. doi: 10.1016/s0896-6273(00)80471-7. [DOI] [PubMed] [Google Scholar]
- 7.Chesa P G, Rettig W J, Melamed M R, Old L J, Niman H L. Expression of p21ras in normal and malignant human tissues: lack of association with proliferation and malignancy. Proc Natl Acad Sci USA. 1987;84:3234–3238. doi: 10.1073/pnas.84.10.3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crespo P, Xu N, Simonds W F, Gutkind J S. Ras-dependent activation of MAP kinase pathway mediated by G-protein beta gamma subunits. Nature. 1994;369:418–420. doi: 10.1038/369418a0. [DOI] [PubMed] [Google Scholar]
- 9.Egan S E, Weinberg R W. The pathway to signal achievement. Nature. 1993;365:781–783. doi: 10.1038/365781a0. [DOI] [PubMed] [Google Scholar]
- 10.Fernández-Salguero P, Pineau T, Hilber D M, McPhail T, Lee S S, Kimura S, Nebert D W, Rudikoff S, Ward J M, Gonzalez F J. Immune system impairment and hepatic fibrosis in mice lacking the dioxin-binding Ah receptor. Science. 1995;268:722–726. doi: 10.1126/science.7732381. [DOI] [PubMed] [Google Scholar]
- 11.Fletcher T L, Cameron P, De Camilli P, Banker G. The distribution of synapsin I and synaptophysin in hippocampal neurons developing in culture. J Neurosci. 1991;11:1617–1626. doi: 10.1523/JNEUROSCI.11-06-01617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furth M E, Aldrich T H, Cordon-Cardo C. Expression of ras proto-oncogene proteins in normal human tissues. Oncogene. 1987;1:47–58. [PubMed] [Google Scholar]
- 13.Gulyás A I, Miettinen R, Jacobowitz D M, Freund T F. Calretinin is present in non-pyramidal cells of the rat hippocampus. I. A new type of neuron specifically associated with the mossy fiber system. Neuroscience. 1992;48:1–27. doi: 10.1016/0306-4522(92)90334-x. [DOI] [PubMed] [Google Scholar]
- 14.Hall A. Small GTP-binding proteins and the regulation of the actin cytoskeleton. Annu Rev Cell Biol. 1994;10:31–54. doi: 10.1146/annurev.cb.10.110194.000335. [DOI] [PubMed] [Google Scholar]
- 15.Hancock J F, Magee A I, Childs J E, Marshall C J. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell. 1989;57:1167–1177. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- 16.Hunter T. Oncoprotein networks. Cell. 1997;88:333–346. doi: 10.1016/s0092-8674(00)81872-3. [DOI] [PubMed] [Google Scholar]
- 17.Ise K, Nakamura K, Nakao K, Shimizu S, Harada H, Ichise T, Miyoshi J, Gondo Y, Ishikawa T, Aiba A, Katsuki M. Targeted deletion of the H-ras gene decreases tumor formation in mouse skin carcinogenesis. Oncogene. 2000;19:2951–2956. doi: 10.1038/sj.onc.1203600. [DOI] [PubMed] [Google Scholar]
- 18.Johnson L, Greenbaum D, Cichowski K, Mercer K, Murphy E, Schmitt E, Bronson R T, Umanoff H, Edelmann W, Kucherlapati R, Jacks T. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 1997;11:2468–2481. doi: 10.1101/gad.11.19.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones M K, Jackson J H. Ras-GRF activates Ha-Ras, but not N-Ras or K-Ras 4B, protein in vivo. J Biol Chem. 1998;273:1782–1787. doi: 10.1074/jbc.273.3.1782. [DOI] [PubMed] [Google Scholar]
- 20.Kataoka T, Powers S, McGill C, Fasano O, Strathern J, Broach J, Wigler M. Genetic analysis of yeast RAS1 and RAS2 genes. Cell. 1984;37:437–445. doi: 10.1016/0092-8674(84)90374-x. [DOI] [PubMed] [Google Scholar]
- 21.Katz M E, McCormick F. Signal transduction from multiple Ras effectors. Curr Opin Genet Dev. 1997;7:75–79. doi: 10.1016/s0959-437x(97)80112-8. [DOI] [PubMed] [Google Scholar]
- 22.Kim J H, Liao D, Lau L F, Huganir R L. SynGAP: a synaptic RasGAP that associates with the PSD-95/SAP90 protein family. Neuron. 1998;20:683–691. doi: 10.1016/s0896-6273(00)81008-9. [DOI] [PubMed] [Google Scholar]
- 23.Klesse L J, Parada L F. Trks: signal transduction and intracellular pathways. Microsc Res Tech. 1999;45:210–216. doi: 10.1002/(SICI)1097-0029(19990515/01)45:4/5<210::AID-JEMT4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 24.Koera K, Nakamura K, Nakao K, Miyoshi J, Toyoshima K, Hatta T, Otani H, Aiba A, Katsuki M. K-ras is essential for the development of the mouse embryo. Oncogene. 1997;15:1151–1159. doi: 10.1038/sj.onc.1201284. [DOI] [PubMed] [Google Scholar]
- 25.Laird P W, Zijderveld A, Linders K, Rudnicki M A, Jaenisch R, Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leon J, Guerrero I, Pellicer A. Differential expression of the ras gene family in mice. Mol Cell Biol. 1987;7:1535–1540. doi: 10.1128/mcb.7.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowy D R, Willumsen B M. Function and regulation of ras. Annu Rev Biochem. 1993;62:851–891. doi: 10.1146/annurev.bi.62.070193.004223. [DOI] [PubMed] [Google Scholar]
- 28.Lu X, Chou T B, Williams N G, Roberts T, Perrimon N. Control of cell fate determination by p21ras/Ras1, an essential component of torso signaling in Drosophila. Genes Dev. 1993;7:621–632. doi: 10.1101/gad.7.4.621. [DOI] [PubMed] [Google Scholar]
- 29.Macara I G, Lounsbury K M, Richards S A, McKiernan C, Bar-Sagi D. The Ras superfamily of GTPases. FASEB J. 1996;10:625–630. doi: 10.1096/fasebj.10.5.8621061. [DOI] [PubMed] [Google Scholar]
- 30.Magee T, Marshall C. New insights into the interaction of Ras with the plasma membrane. Cell. 1999;98:9–12. doi: 10.1016/S0092-8674(00)80601-7. [DOI] [PubMed] [Google Scholar]
- 31.Manabe T, Aiba A, Yamada A, Ichise T, Sakagami H, Kondo H, Katsuki M. Regulation of long-term potentiation by H-Ras through NMDA receptor phosphorylation. J Neurosci. 2000;20:2504–2511. doi: 10.1523/JNEUROSCI.20-07-02504.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marshall M S. Ras target proteins in eukaryotic cells. FASEB J. 1995;9:1311–1318. doi: 10.1096/fasebj.9.13.7557021. [DOI] [PubMed] [Google Scholar]
- 33.Matus A, Bernhardt R, Bodmer R, Alaimo D. Microtubule-associated protein 2 and tubulin are differently distributed in the dendrites of the developing neurons. Neuroscience. 1986;17:371–389. doi: 10.1016/0306-4522(86)90253-8. [DOI] [PubMed] [Google Scholar]
- 34.Ouyang Y, Kantor D, Harris K M, Schuman E M, Kennedy M B. Visualization of the distribution of autophosphorylated calcium/calmodulin-dependent protein kinase II after tetanic stimulation in the CA1 area of the hippocampus. J Neurosci. 1997;17:5416–5427. doi: 10.1523/JNEUROSCI.17-14-05416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pells S, Divjak M, Romanowski P, Impey H, Hawkins N J, Clarke A R, Hooper M L, Williamson D J. Developmentally-regulated expression of murine K-ras isoforms. Oncogene. 1997;15:1781–1786. doi: 10.1038/sj.onc.1201354. [DOI] [PubMed] [Google Scholar]
- 36.Santos E, Nebreda A R. Structural and functional properties of Ras proteins. FASEB J. 1989;3:2151–2163. doi: 10.1096/fasebj.3.10.2666231. [DOI] [PubMed] [Google Scholar]
- 37.Santos E, Tronick S R, Aaronson S A, Pulciani S, Barbacid M. T24 human bladder carcinoma oncogene is an activated form of the normal human homologue of BALB- and Harvey-MSV transforming genes. Nature. 1982;298:343–347. doi: 10.1038/298343a0. [DOI] [PubMed] [Google Scholar]
- 38.Satoh T, Nakafuku M, Kaziro Y. Function of Ras as a molecular switch in signal transduction. J Biol Chem. 1992;267:24149–24152. [PubMed] [Google Scholar]
- 39.Schafer W R, Rine J. Protein prenylation: genes, enzymes, targets, and functions. Annu Rev Genet. 1992;26:209–237. doi: 10.1146/annurev.ge.26.120192.001233. [DOI] [PubMed] [Google Scholar]
- 40.Segal R A, Greenberg M E. Intracellular signaling pathways activated by neurotrophic factors. Annu Rev Neurosci. 1996;19:463–489. doi: 10.1146/annurev.ne.19.030196.002335. [DOI] [PubMed] [Google Scholar]
- 41.Sík A, Hájos N, Gulácsi A, Mody I, Freund T F. The absence of a major Ca2+ signaling pathway in GABAergic neurons of the hippocampus. Proc Natl Acad Sci USA. 1998;95:3245–3250. doi: 10.1073/pnas.95.6.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silva A, Stevens C F, Tonegawa S, Wang Y. Deficient hippocampal long-term potentiation in α-calcium calmodulin kinase II mutant mice. Science. 1992;257:201–206. doi: 10.1126/science.1378648. [DOI] [PubMed] [Google Scholar]
- 43.Silva A, Paylor R, Wehner J M, Tonegawa S. Impaired spatial learning in α-calcium calmodulin kinase II mutant mice. Science. 1992;257:206–211. doi: 10.1126/science.1321493. [DOI] [PubMed] [Google Scholar]
- 44.Sturani E, Abbondio A, Branduardi P, Ferrari C, Zippel R, Martegani E, Vanoni M, Denis-Donini S. The Ras guanine nucleotide exchange factor CDC25Mm is present at the synaptic junction. Exp Cell Res. 1997;235:117–123. doi: 10.1006/excr.1997.3660. [DOI] [PubMed] [Google Scholar]
- 45.Swan K A, Alberola-Ila J, Gross J A, Appleby M W, Forbush K A, Thomas J F, Perlmutter R M. Involvement of p21ras distinguishes positive and negative selection in thymocytes. EMBO J. 1995;14:276–285. doi: 10.1002/j.1460-2075.1995.tb07001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tybulewicz V L J, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 47.Umanoff H, Edelmann W, Pellicer A, Kucherlapati R. The murine N-ras gene is not essential for growth and development. Proc Natl Acad Sci USA. 1995;92:1709–1713. doi: 10.1073/pnas.92.5.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vicario-Abejón C. Long-term culture of hippocampal neurons. In: Crawley J N, Gerfen C R, McKay R, Rogawsky M A, Sibley D R, Skolnick P, editors. Current protocols in neuroscience. New York, N.Y: John Wiley and Sons; 1997. pp. 3.2.1–3.2.12. [DOI] [PubMed] [Google Scholar]
- 49.Wei W, Schreiber S S, Baudry M, Tocco G, Broek D. Localization of the cellular expression pattern of cdc25NEF and ras in the juvenile rat brain. Mol Brain Res. 1993;19:339–344. doi: 10.1016/0169-328x(93)90136-d. [DOI] [PubMed] [Google Scholar]
- 50.Zippel R, Gnesutta N, Matus-Leibovitch N, Mancinelli E, Saya D, Vogel Z, Sturani E. Ras-GRF, the activator of Ras, is expressed preferentially in mature neurons of the central nervous system. Mol Brain Res. 1997;48:140–144. doi: 10.1016/s0169-328x(97)00120-4. [DOI] [PubMed] [Google Scholar]