Abstract
Background
Nonhuman primates (NHPs) play a significant role in zoonotic spill-overs, serving as either reservoirs, or amplifiers, of multiple neglected tropical diseases, including tick-borne infections. Anaplasma phagocytophilum are obligate intracellular bacteria of the family Anaplasmatacae, transmitted by Ixodid ticks and cause granulocytic anaplasmosis (formerly known as Human Granulocytic Ehrlichiosis (HGE)) in a wide range of wild and domestic mammals and humans too. The aim of this study was to determine whether Anaplasma phagocytophilum was circulating in olive baboons and vervet monkeys in Laikipia County, Kenya.
Results
Some 146 blood samples collected from olive baboons and 18 from vervet monkeys from Mpala Research Center and Ol jogi Conservancy in Laikipia County were screened for the presence of Anaplasma species using conventional Polymerase Chain Reaction (PCR), and then A. phagocytophilum was confirmed by sequencing using conventional PCR targeting 16S rRNA. This study found an overall prevalence of 18.3% for Anaplasma species. DNA sequences confirmed Anaplasma phagocytophilum in olive baboons for the first time in Kenya.
Conclusion
This study provides valuable information on the endemicity of A. phagocytophilum bacteria in olive baboons in Kenya. Future research is needed to establish the prevalence and public health implications of zoonotic A. phagocytophilum isolates and the role of nonhuman primates as reservoirs in the region.
Keywords: Zoonosis, Olive baboons, Vervet monkeys, Kenya, Anaplasma phagocytophilum
Background
There has been a rise in the frequency of emerging infectious diseases (EIDs), among which zoonotic tick-borne infections—especially rickettsial diseases such as anaplasmosis—are implicated [1]. What most of the recent pandemics have proven is that emerging infectious human diseases are mainly zoonosis of animal origin, particularly wildlife [2]. A complex series of interactions among wildlife, livestock and human populations and environmental factors contribute to their emergence [3].
Among wildlife species, non-human primates (NHPs) are often proprietors to many different microbial agents, some which have zoonotic potential. Primates are closely related to humans phylogenetically and ecologically [4], and they can indirectly transmit infectious agents to humans through intermediate hosts, arthropod vectors or directly through contact with or bites from the NHP, or through the consumption of NHP bush meat [5]. Certain factors such as forested tropical regions experiencing land-use changes and encroachment, as well as those with a high wildlife biodiversity, facilitate the spread of these diseases to livestock and man [6]. Others include adoption of new technology in farms, destruction of native forest habitats, climate change, global travel and human encroachment into new habitats [2].
This study focused on Anaplasma phagocytophium, a tick-borne pathogenic bacterium of zoonotic potential often harbored by wildlife, then passed into livestock and man [7]. Anaplasma belong to the family of Anaplasmataceae, order of Rickettsiales, class Alphaproteobacteria and genus Anaplasma [8]. This bacterium has a wide host range including domestic animals, wildlife and humans [9]. In the latter, the disease called human granulocytic anaplasmosis (HGA) (formerly known as human granulocytic ehrlichiosis [10] often presents with influenza-like symptoms which include fever, anorexia, diarrhea, leukopenia and thrombocytopenia [7]. Ixodid ticks are important in their maintenance as vectors [11]. The emergence of Anaplasmataceae as human pathogens has gained the attention of scientific community. Recent surveys have shown human infections of Anaplasma species in humans including A. platys in two women in Venezuela [12] and A. phagocytophilum exposure in dog owners in Morocco [13].
Recent reports on A. phagocytophilum infection in several animal species are available from France, Massachusetts, Brazil, Zambia, Ethiopia, and Kenya in both domestic and wild animals [14–19]. While Anaplasma in NHPs has been reported in some countries [17], its importance in NHPs in Kenya is not yet known which has therefore led us to investigate the occurrence of this bacteria in NHPs in Laikipia, Kenya.
The investigation focused on the detection of A. phagocytophilum DNA in blood samples from olive baboons (Papio anubis) and Vervet monkeys (Chlorocebus pygerythrus) in Laikipia County, Kenya. Laikipia is part of Kenya’s rangelands mainly inhabited by trans-human pastoralists and has a large wildlife population including non-human primate species found close to human settlements. The complex interaction among wildlife, livestock and human populations contributes to the emergence of infectious diseases [3] alongside factors that facilitate disease spread [2, 6].
The objectives of this present study were to (i) establish the presence of A. phagocytophilum in olive baboons and vervet monkeys and (ii) establish whether the bacteria is genetically diverse.
Methods
Study area, sample population and sample size
The study area was based on a larger project (USAID Predict II) whose aim was to collect targeted information to support the interventions to mitigate spread of zoonotic viruses with pandemic potential. The focus was on highest risk locations and interfaces, where animals and people share changing landscapes. The study area, Laikipia County (Fig. 1) located in the Rift Valley of Kenya (005′N 36040′E), was picked as one of the locations. There is a diverse range of wildlife in the area, including eight NHPs found in this area.
Fig. 1.
Map of the NHPs sampling sites in Laikipia County, Kenya
This study focused on two NHP species: olive baboons and vervet monkeys, since they are closely related to humans phylogenetically and are implicated in most of the listed emerging human pathogens [20]. The sampling sites Mpala research Centre and Ol jogi Conservancy (Fig. 1) were conveniently chosen since large populations of these species inhabit these sites. Some of the whole blood samples from wild olive baboons and vervet monkeys that had been collected by the larger project from Laikipia County were included in this study constituting the study’s sample size of 146 olive baboon and 18 vervet monkey. The animals were not owned.
DNA extraction
Whole blood samples stored in TRIzol reagent at -80 °C were retrieved and allowed to thaw at room temperature. Extraction of genomic DNA was individually achieved from each whole blood samples using the DNeasy Blood & Tissue Kit (QIAgen Valencia, California, USA) following manufacturer’s instructions.
Targeted PCR
Of the extracted DNA, 2.0ul was used as template in the PCR reaction, targeting a 345 bp fragment of the Anaplasma 16S rRNA gene, using the EHR16SD/R primer set and previously defined conditions [21]. Amplicons were visualized on 1% agarose gels to validate amplicon size by comparison with a DNA molecular weight marker (1 Kb Plus DNA Ladder, Promega, Madison, USA).
DNA sequencing and data analysis
Ten selected positive Anaplasma spp. PCR products obtained with primers EHR16SD/R were purified with Thermo Scientific GeneJET PCR Purification Kit #K0701, #K0702 Protocol according to the manufacturer’s instructions. Purified DNA fragments were sequenced using an ABI PRISM 377 Genetic Analyzer (Applied Biosystems, USA), using the original PCR primers (Table 1). Sequence assembly of forward and reverse chromatograms was done using DNA Sequence Assembler v.4 (2013), Heracle BioSoft [22]. The sequences were matched to those deposited in the GenBank database using the BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple alignment of the sequences was done using BioEdit Sequence Alignment Editor (Hall, 1999). Construction of phylogenetic tree was done using Muscle 3.8 using the neighbor-joining method and visualization of the trees with FigTree v1.4.4 [23].
Table 1.
Primers used for detection and/or characterization of Anaplasma species in the present study
| Assay | Primer | Sequence 5′ to 3′ | Target gene | Amplicon size (bp) | References |
|---|---|---|---|---|---|
|
Conventional PCR |
EHR16SD EHR16SR |
GGTACCYACAGAAGAAGTCC TAGCACTCATCGTTTACAGC |
16S rRNA | 345 | [3] |
Results
Molecular survey of Anaplasma species
A total of 164 NHP blood samples—146 from olive baboons and 18 from vervet monkeys— were screened for the presence of Anaplasma spp through conventional PCR using primers targeting 345 bp of the 16S rRNA gene (Table 1). The overall prevalence for Anaplasma spp. was 18.3% (30/164) as estimated by EHR16SD/R PCR (Table 1), with 17.8 and 22.2% positivity rates in olive baboons and vervet monkeys respectively. Through sequencing, A. phagocytophilum was confirmed in three of ten gel positive samples, all from olive baboons from Mpala, Laikipia.
Molecular characterization of Anaplasma phagocytophilum. 16S rRNA genotypes
Nine of the ten PCR products were successfully sequenced on both DNA strands and generated nucleotide sequences with primers EHR16SD/R targeting 345 bp of the 16S rRNA gene of Anaplasma species. Multiple alignment of Anaplasma nucleotide sequences of three A. phagocytophilum isolates revealed that most sequences were conserved expect the following: for our isolates, nucleotides at the first 3 positions and at position 174 differ with the rest of the sequences. At position 304, 39A differs with the rest and at the following positions 378, 381–384,386–388 and 391, 39A and 41A differ with rest (Fig. 2). They all shared 99 to 100% nucleotide similarity (Table 2).
Fig. 2.
Multiple sequence alignment of 16S rRNA gene for A. phagocytophilum isolates. The conserved regions are represented by the dots (.) while the variable regions are indicated by the letters representing the nucleotide A, C, G and T
Table 2.
BLASTn analysis results using 16S rRNA sequences of isolates from olive baboons
| Sample number | Animal species | Pathogen identity | E-values | Identity (%) |
|---|---|---|---|---|
| 28A | Olive baboon | A. phagocytophilum | 4 | 100 |
| 41A | Olive baboon | A. phagocytophilum | 4 | 99.39 |
| 39A | Olive baboon | A. phagocytophilum | 3 | 99.39 |
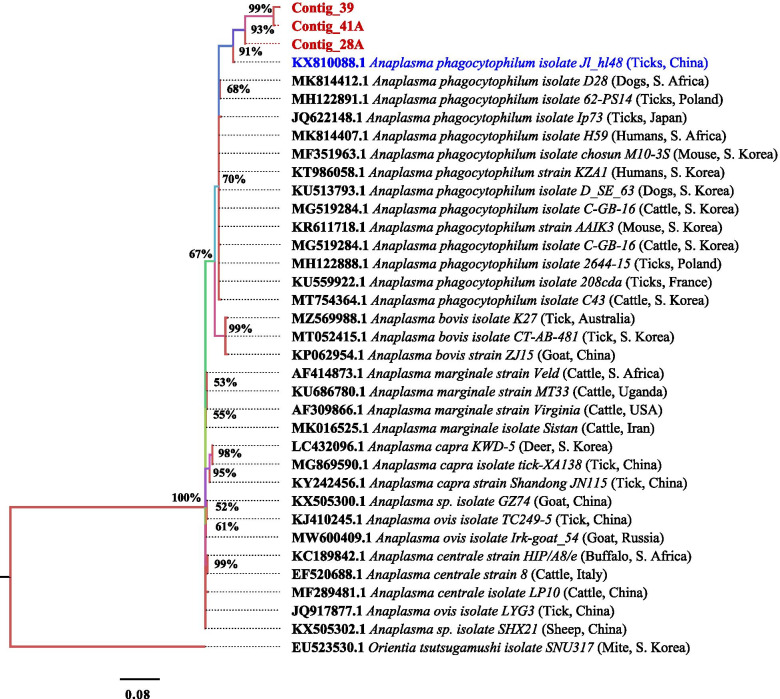
The sequences of the A. phagocytophilum isolates from Kenya were identical to those from Japan, South Korea, France, China, South Africa and Denmark. The species isolates were from human, cattle, ticks, dogs and mice. The accession numbers JQ622148, MF351963, KU559922, MG519284, KX810088, MH122888, MK814411, MK814407, MF582329, MK814412, MK271308, MH122891, AY776165, KT986058, KU513793, and KR611718 were all of A. phagocytophilum isolates (Fig. 2). Phylogenetic analysis revealed that the isolates from Japan, South Korea, France, China, South Africa, Denmark and Poland belonged to clade I but have recent common ancestor with the Kenyan isolates clustered into one clade II (Fig. 3).
Fig. 3.
Phylogenetic tree of the 16S rRNA gene from A.phagocytophilum isolates
Discussion
This study is the first to investigate the presence of A. phagocytophilum DNA in olive baboons and vervet monkeys in Kenya and it provides molecular evidence of its occurrence in olive baboons within Laikipia County, a wild habitat in close proximity to livestock and humans.
With the rise in reported cases of HGA in United states [24] and exposure of humans to the bacteria [13], detection of this bacteria in primates is a valuable finding since very little information is available in Africa. Anaplasma phagocytophilum has been previously reported to infect a wide range of animal hosts in various parts of the world [14–16], including Africa, [17, 18, 25] but from only one study in Kenya [26].
This study found a prevalence of Anaplasma spp. of 18.3% (30/164) in olive baboons (17.8%) and vervet monkeys (22.2%) sampled in Laikipia county, Kenya. However, due to limited funding from the project, the current study was limited to sequencing only a few of the gel positive samples. Ten of thirty gel positive samples from olive baboons and vervet monkeys were sequenced which confirmed the presence of A. phagocytophilum DNA in three samples from olive baboons in Mpala, Laikipia county, Kenya. This result suggests that if all the 30 samples been sequenced, more A. phagocytophilum DNA might have been confirmed even in samples from vervet monkeys and the prevalence of A. phagocytophilum could have been determined. Nonetheless, we only report the presence of A. phagocytophilum in olive baboons in Kenya.
In this study, primers targeting the 16S rRNA genes of A. phagocytophilum were identified from a previous study and used for the detection of A. phagocytophilum [21]. The 16S rRNA is a ribosomal RNA gene which are conserved greatly among all bacterial species [27] and are shown to be the most sensitive assay in detection DNA of A. phagocytophilum [28]. This makes them very useful in molecular diagnostics. However, 16S rRNA sequences evolve slowly, and while this region is perfect to diagnostic assays, it lacks the population-level variation required when distinguishing genotypes of A. phagocytophilum [29, 30]. In this study, the 16S rRNA gene yielded positive bands and therefore reinforced that it is a good marker for detection of Anaplasma species as reported by other studies [13, 17, 18]. However, for future studies a longer fragment of 16S rRNA gene should be amplified and obtained with additional primer set for in depth molecular characterization of A. phagocytophilum.
Sequencing analysis revealed that the sequences of the 16S rRNA gene were very conserved not only between African isolates but also between the other isolates of world-wide origin agreeably with the only available study on A. phagocytophilum in baboons in Zambia [17]. This proved to be a limiting factor to the current study as there is no sufficient data to draw comparison from. The sequences of the A. phagocytophilum isolates from Kenya were identical to those from Japan, South Korea, France, South Africa and Denmark and most especially from China (KX810088) in ticks. The species isolates were from cattle, ticks, dogs, rodents and human (Fig. 3). This find was consistent with the study in Zambia [17]. Previous studies have reported genetic relatedness between both A. phagocytophilum infecting animals and humans [12, 13].
Multiple alignment of Anaplasma nucleotide sequences of the A. phagocytophilum isolates revealed that all the sequences of samples from baboons were conserved (Fig. 2). The isolates from Japan, South Korea, France, China, South Africa and Denmark belonged to clade I but have recent common ancestor with the Kenyan isolates clustered into one clade II (Fig. 3). All our isolates were similar to that of ticks from China (KX810088). Amongst them, 39A and 4IA were shown to be evolving much faster than 28A which is because of sequence differences as a result of mutations. Additionally, they are new isolates as suggested by the posterior probability (50%) (Fig. 3).
The presence of A. phagocytophilum DNA in baboons in close proximity to humans (wildlife conservancies, ranches) raises the question: to what extent is this bacterial presence in baboon blood a concern for humans? The study localities where the samples were collected is a research center and conservancy with researchers, tourists and local community members who either work or grazed their livestock within. Therefore, this study shows that there is a need for an expansive bio-surveillance of these pathogens with particular attention to these groups of people and their livestock to evaluate the risk of disease transmission in such communities.
Non-human primates have been shown to host different pathogens, including several Anaplasma species [17, 31]. We assume that they could serve as a good indicator of bacteria circulation in ecosystem and explain the persistence of anaplasmosis in domestic animals despite consistent control. Therefore, epidemiological surveillance of NHPs’ pathogens is important in generating information that can generate actions developing strategies on prevention and control of emerging and re-emerging zoonotic diseases [17].
Conclusions
This paper reports the presence of Anaplasma phagocytophilum in olive baboons in Laikipia County, Kenya for the first time. This finding open concerns on the public health implications of potentially zoonotic A. phagocytophilum infections in the Laikipia community. It proves the need to investigate whether these NHP species are infected with other Anaplasma species, raises concerns on the role they play in the maintenance of Anaplasma species and therefore, the implications in disease control and prevention.
Acknowledgements
This study was made possible by the generous support of the American people through the United States Agency for International Development (USAID) Emerging Pandemic Threats PREDICT project (cooperative agreement number GHN-A-OO-09-00010-00) and especially through provision of the samples included in this study. We thank the staff at the Molecular Laboratory at the Institute of Primate Research, Kenya who assisted with laboratory analysis of the work. The authors would like to thank Dr. Mercy Akinyi, Dr. Lilian Waiboci and Pheris Namakwa for their invaluable contribution in the work.
Abbreviations
- NHPs
Nonhuman primates
- USAID
United States AID
- PCR
Polymerase chain reaction
- DNA
Deoxyribonucleic acid
- RNA
Ribonucleic acid
- rRNA
Ribosomal Ribonucleic acid
- EIDs
Emerging infectious diseases
- HGA
Human granulocytic anaplasmosis
- DNTPs
Deoxyribonucleotide triphosphate
- BLAST
Basic Local Alignment Search Tool
- USA
United States of America
Authors’ contributions
SJM conducted the laboratory work, participated in the study design, conducted the data analysis and prepared the first draft of the manuscript. GMM, JMK, TAO and DZ were involved in the conception of the study, study design and the writing and review of this manuscript. SM was involved in the laboratory work. All authors have read and approved the final manuscript.
Funding
The work was supported by the funding from the USAID Predict II project, 2018.
Availability of data and materials
Data and materials are available upon reasonable request from the corresponding author.
Declarations
Ethics approval and consent to participate
Approval for this study was given by the Institute of Primate research Institutional Scientific and Ethics Review Committee (ISERC): Ethical approval number 1SERC/T01/18, 16th March 2018. Since the information collected for this study was not of a sensitive nature and non-invasive as banked blood samples were used, the ethical approval and guidelines received from this committee was deemed sufficient to conduct the study. The study followed laid biosafety guidelines on handling and processing of biological samples.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Heymann DL, Chen L, Takemi K, Fidler DP, Tappero JW, Thomas MJ, Kenyon TA, Frieden TR, Yach D, Nishtar S, Kalache A, Olliaro PL, Horby P, Torreele E, Gostin LO, Ndomondo-Sigonda M, Carpenter D, Rushton S, Lillywhite L, Devkota B, Rannan-Eliya RP. Global health security: the wider lessons from the west African Ebola virus disease epidemic. Lancet (London, England) 2015;385(9980):1884–1901. doi: 10.1016/S0140-6736(15)60858-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones BA, Grace D, Kock R, Alonso S, Rushton J, Said MY, McKeever D, Mutua F, Young J, McDermott J, Pfeiffer DU. Zoonosis emergence linked to agricultural intensification and environmental change. Proc Natl Acad Sci U S A. 2013;110(21):8399–8404. doi: 10.1073/pnas.1208059110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brierley L, Vonhof MJ, Olival KJ, Daszak P, Jones KE. Quantifying global drivers of zoonotic bat viruses: a process-based perspective. Am Nat. 2016;187(2):E53–E64. doi: 10.1086/684391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Locatelli S, Peeters M. Cross-species transmission of simian retroviruses: how and why they could lead to the emergence of new diseases in the human population. AIDS (London, England) 2012;26(6):659–673. doi: 10.1097/QAD.0b013e328350fb68. [DOI] [PubMed] [Google Scholar]
- 5.Jones-Engel L, May CC, Engel GA, Steinkraus KA, Schillaci MA, Fuentes A, Rompis A, Chalise MK, Aggimarangsee N, Feeroz MM, Grant R, Allan JS, Putra A, Wandia IN, Watanabe R, Kuller L, Thongsawat S, Chaiwarith R, Kyes RC, Linial ML. Diverse contexts of zoonotic transmission of simian foamy viruses in Asia. Emerg Infect Dis. 2008;14(8):1200–1208. doi: 10.3201/eid1408.071430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen T, Murray KA, Zambrana-Torrelio C, et al. Global hotspots and correlates of emerging zoonotic diseases. Nat Commun. 2017;8:1124. doi: 10.1038/s41467-017-00923-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ismail N, Bloch KC, McBride JW. Human ehrlichiosis and anaplasmosis. Clin Lab Med. 2010;30(1):261–292. doi: 10.1016/j.cll.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dumler JS, Barbet AF, Bekker CPJ, Dasch GA, Palmer GH, Ray SC, Rurangirwa FR. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and “HGE agent” as subjective synonyms of Ehrlichia phagocytophila. Int J Syst Evol Microbiol. 2001;51(6):2145–2165. doi: 10.1099/00207713-51-6-2145. [DOI] [PubMed] [Google Scholar]
- 9.Kocan KM, de la Fuente J, Cabezas-Cruz A. The genus Anaplasma: new challenges after reclassification. Revue scientifique et technique (International Office of Epizootics) 2015;34(2):577–586. doi: 10.20506/rst.34.2.2381. [DOI] [PubMed] [Google Scholar]
- 10.Bakken JS, Dumler JS, Chen SM, Eckman MR, Van Etta LL, Walker DH. Human granulocytic ehrlichiosis in the upper Midwest United States. A new species emerging? JAMA. 1994;272(3):212–218. doi: 10.1001/jama.1994.03520030054028. [DOI] [PubMed] [Google Scholar]
- 11.Zaid T, Ereqat S, Nasereddin A, Al-Jawabreh A, Abdelkader A, Abdeen Z. Molecular characterization of Anaplasma and Ehrlichia in ixodid ticks and reservoir hosts from Palestine: a pilot survey. Vet Med Sci. 2019. 10.1002/vms3.150. [DOI] [PMC free article] [PubMed]
- 12.Arraga-Alvarado CM, Qurollo BA, Parra OC, Berrueta MA, Hegarty BC, Breitschwerdt EB. Case report: molecular evidence of Anaplasma platys infection in two women from Venezuela. Am J Trop Med Hyg. 2014;91(6):1161–1165. doi: 10.4269/ajtmh.14-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elhamiani Khatat S, Daminet S, Kachani M, et al. Anaplasma spp. in dogs and owners in North-Western Morocco. Parasit Vectors. 2017;10:202. doi: 10.1186/s13071-017-2148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Afonso E, Goydadin A-C. Molecular detection of Anaplasma phagocytophilum DNA in the lesser horseshoe bat (Rhinolophus hipposideros) guano. Epidemiol Infect. 2018;146:1253–1258. doi: 10.1017/S0950268818001279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galemore ER, Labato MA, O'Neil E. Prevalence of Anaplasma phagocytophilum infection in feral cats in Massachusetts. JFMS Open Rep. 2018;4(1):2055116917753804. doi: 10.1177/2055116917753804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silveira JAG, Rabelo EML, Ribeiro MFB. Molecular detection of tick-borne pathogens of the family Anaplasmataceae in Brazilian Brown brocket deer (Mazama gouazoubira, Fischer, 1814) and marsh deer (Blastocerus dichotomus, Illiger, 1815) Transbound Emerg Dis. 2011;59(4):353–360. doi: 10.1111/j.1865-1682.2011.01278.x. [DOI] [PubMed] [Google Scholar]
- 17.Nakayima J, Hayashida K, Nakao R, Ishii A, Ogawa H, Nakamura I, Moonga L, Hang'ombe BM, Mweene AS, Thomas Y, Orba Y, Sawa H, Sugimoto C. Detection and characterization of zoonotic pathogens of free-ranging non-human primates from Zambia. Parasit Vectors. 2014;7:490. doi: 10.1186/s13071-014-0490-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teshale S, Geysen D, Ameni G, et al. Survey of Anaplasma phagocytophilum and Anaplasma sp. ‘Omatjenne’ infection in cattle in Africa with special reference to Ethiopia. Parasit Vectors. 2018;11:162. doi: 10.1186/s13071-018-2633-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peter SG, Gakuya DW, Maingi N, Mulei CM. Prevalence and risk factors associated with Ehrlichia infections in smallholder dairy cattle in Nairobi City county, Kenya. Vet World. 2019;12(10):1599–1607. doi: 10.14202/vetworld.2019.1599-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Locatelli S, Peeters M. Non-human Primates, retroviruses, and zoonotic infection risks in the human population. Nat Educ Knowledge. 2012;3(10):62. [Google Scholar]
- 21.Parola P, Roux V, Camicas J, Baradji I, Brouqui P, Raoult D. Detection of Ehrlichiae in African ticks by polymerase chain reaction. Trans R Soc Trop Med Hyg. 2000;94:707–708. doi: 10.1016/S0035-9203(00)90243-8. [DOI] [PubMed] [Google Scholar]
- 22.DNA Baser Database [https://www.dnabaser.com/download/DNA-Baser-sequence-assembler/index.html] Assessed 13 June 2020.
- 23.Molecular Evolution, Phylogenetics and Epidemiology. [http://tree.bio.ed.ac.uk/]. Assessed 20 June 2020.
- 24.Center for Disease Control and Prevention (CDC). 2019. Anaplasmosis. Epidemiology and statistics. Retrieved from https://www.cdc.gov/anaplasmosis/stats/index.html. Accessed 20 June 2020.
- 25.Mtshali K, Nakao R, Sugimoto C, Thekisoe O. Occurrence of Coxiella burnetii, Ehrlichia canis, Rickettsia species and Anaplasma phagocytophilum-like bacterium in ticks collected from dogs and cats in South Africa. J S Afr Vet Assoc. 2017;88(1):1–6. doi: 10.4102/jsava.v88i0.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mwamuye MM, Kariuki E, Omondi D, Kabii J, Odongo D, Masiga D, Villinger J. Novel Rickettsia and emergent tick-borne pathogens: a molecular survey of ticks and tick-borne pathogens in Shimba Hills National Reserve, Kenya. Ticks Tick Borne Dis. 2017;8(2):208–218. doi: 10.1016/j.ttbdis.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Massung RF, Priestley RA, Miller NJ, Mather TN, Levin ML. Inability of a variant strain of Anaplasma phagocytophilum to infect mice. J Infect Dis. 2003;188(11):1757–1763. doi: 10.1086/379725. [DOI] [PubMed] [Google Scholar]
- 28.Yang J, Liu Z, Qingli N, Liu J, Xie J, Chen Q, et al. Evaluation of different nested PCRs for detection of Anaplasma phagocytophilum in ruminants and ticks. BMC Vet Res. 2016;12. 10.1186/s12917-016-0663-2. [DOI] [PMC free article] [PubMed]
- 29.Bown KJ, Lambin X, Ogden NH, Petrovec M, Shaw SE, Woldehiwet Z, Birtles RJ. High-resolution genetic fingerprinting of European strains of Anaplasma phagocytophilum by use of multilocus variable-number tandem-repeat analysis. J Clin Microbiol. 2007;45(6):1771–1776. doi: 10.1128/JCM.00365-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bown KJ, Lambin X, Ogden NH, Begon M, Telford G, Woldehiwet Z, Birtles RJ. Delineating Anaplasma phagocytophilum ecotypes in coexisting, discrete enzootic cycles. Emerg Infect Dis. 2009;15(12):1948–1954. doi: 10.3201/eid1512.090178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dumler JS. Anaplasma and Ehrlichia infection. Ann N Y Acad Sci. 2005;1063:361–373. doi: 10.1196/annals.1355.069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data and materials are available upon reasonable request from the corresponding author.