Abstract
Eukaryotic translation initiation factor 6 (eIF6), a monomeric protein of about 26 kDa, can bind to the 60S ribosomal subunit and prevent its association with the 40S ribosomal subunit. In Saccharomyces cerevisiae, eIF6 is encoded by a single-copy essential gene. To understand the function of eIF6 in yeast cells, we constructed a conditional mutant haploid yeast strain in which a functional but a rapidly degradable form of eIF6 fusion protein was synthesized from a repressible GAL10 promoter. Depletion of eIF6 from yeast cells resulted in a selective reduction in the level of 60S ribosomal subunits, causing a stoichiometric imbalance in 60S-to-40S subunit ratio and inhibition of the rate of in vivo protein synthesis. Further analysis indicated that eIF6 is not required for the stability of 60S ribosomal subunits. Rather, eIF6-depleted cells showed defective pre-rRNA processing, resulting in accumulation of 35S pre-rRNA precursor, formation of a 23S aberrant pre-rRNA, decreased 20S pre-rRNA levels, and accumulation of 27SB pre-rRNA. The defect in the processing of 27S pre-rRNA resulted in the reduced formation of mature 25S and 5.8S rRNAs relative to 18S rRNA, which may account for the selective deficit of 60S ribosomal subunits in these cells. Cell fractionation as well as indirect immunofluorescence studies showed that c-Myc or hemagglutinin epitope-tagged eIF6 was distributed throughout the cytoplasm and the nuclei of yeast cells.
Eukaryotic translation initiation factor 6 (eIF6), a monomeric protein of about 26 kDa, was originally isolated and characterized from both wheat germ (22, 23) and mammalian cell extracts (17, 34) based on an in vitro assay that measured the ability of the protein to bind specifically to the 60S ribosomal subunit and to prevent its association with the 40S ribosomal subunit to form 80S ribosomes. Because of this ribosomal subunit antiassociation property, eIF6 was thought to play a direct role in the provision of free ribosomal subunits required for initiation of protein synthesis. The protein was therefore classified as a eukaryotic translation initiation factor (13), although its role in translation of mRNAs was not defined in these original studies. More recently, to facilitate further characterization of eIF6 and to understand the function of this protein in translation, we cloned and then expressed in Escherichia coli both a human cDNA (28) and the yeast Saccharomyces cerevisiae gene (29) encoding functionally active eIF6, each of 245 amino acids (calculated Mr, 26,558 for human eIF6 and 25,550 for yeast eIF6). The two proteins are 72% identical. Detailed characterization of the yeast gene encoding eIF6, designated TIF6, showed that TIF6 is a single-copy gene that maps on chromosome XVI (as YPR016C) and is essential for cell growth and viability. These properties of TIF6 were used to construct a conditional null allele by placing its expression under the control of the regulated GAL10 promoter (29). We observed that depletion of eIF6 from this yeast strain resulted in inhibition of both cell growth and rate of in vivo protein synthesis (29). However, analysis of the polysome profiles of eIF6-depleted cells showed a reduction not only in the amounts of polysomes but also in the amounts of both 80S ribosomes and free 60S ribosomal subunits and accumulation of half-mer polysomes. Finally, analysis of total ribosomal subunit content in eIF6-depleted cells showed that there was a selective reduction of total 60S with respect to total 40S ribosomal subunits, causing a stoichiometric imbalance in the 60S/40S subunit ratio resulting in the formation of half-mer polysomes. Similar observations were also reported by Sanvito et al. (26), who identified eIF6 from mammalian cells as a β4 integrin-interacting protein p27 (3) and designated the yeast homologue p27BBP/eIF6 (26).
The polysome-ribosome profiles observed in eIF6-depleted cells are not characteristic of cells containing an inactive translation initiation factor. If eIF6 plays an essential role in the initiation phase of protein synthesis, its depletion in yeast cells would have caused not only a reduction in polysome content but also a simultaneous increase (not decrease) in the pool of free 40S, 60S, and 80S ribosomes. These results along with our observation that lysates of yeast cells lacking eIF6 remained active in translation of mRNAs in vitro (29) led us to conclude that eIF6 does not function as a translation initiation factor for global protein synthesis. Rather, the inhibition of translation observed in eIF6-depleted cells is due to selective reduction of 60S ribosomal subunits in these cells. Thus, if eIF6 is not a translation initiation factor, what essential cellular function does the protein perform? More specifically, how does eIF6 maintain the steady-state level of 60S ribosomal subunits in yeast cells?
In recent years, a number of yeast genes whose mutations cause selective reduction in the amount of 60S subunits with respect to 40S subunits and concomitant accumulation of half-mer polysomes have been identified. These genes encode several ribosomal proteins (5, 7, 14, 15, 20) and nonribosomal proteins like Nip7p (39), Dbp6p (11), Nmd3p (9), and Sqt1p (6). Removal of these proteins from yeast cells has been shown to impair the processing of rRNA precursors (pre-rRNAs) to mature 25S and 5.8S rRNAs, which are the constituents of the 60S ribosomal subunit. Failure to produce mature 25S and 5.8S rRNAs may account for the inhibition of synthesis of 60S ribosomal subunits. In light of these observations, the possibility exists that eIF6 itself may be involved in the processing of pre-rRNAs to mature 25S and 5.8S rRNAs and consequently in the biosynthesis of 60S ribosomal subunits.
In this report, we describe a functional analysis of the S. cerevisiae TIF6 gene. We show that the product eIF6 localizes both in the nucleus and in the cytoplasm. Depletion of eIF6 in yeast cells, which results in the selective reduction of 60S ribosomal subunits, does not affect the stability of preexisting 60S ribosomal particles. Rather, in vivo depletion of eIF6 causes impaired processing of 35S and 27S pre-rRNA precursors, resulting in decreased formation of mature 25S and 5.8S rRNAs that are the constituents of 60S ribosomal subunits. These observations are consistent with a role for eIF6 in 60S ribosomal subunit biogenesis.
MATERIALS AND METHODS
Strains, media, and genetic methods.
The S. cerevisiae haploid strains used in this work were derived from the diploid strain W303a/α. The genotypes of this strain and other yeast strains used in this study are listed in Table 1. Strain KSY603 (MATαtif6::HIS3 p[URA3 GAL10::Ub-TIF6]), constructed as described previously (29), contains a conditional eIF6 expression system in which N-terminally fused ubiquitinated eIF6 fusion protein was expressed from a transcription unit consisting of a ubiquitin gene cassette (Ub-R-lacI-TIF6, where R is the codon for arginine) (16) which acts as a protein-destabilizing genetic element fused to the NH2 terminus of the open reading frame (ORF) of the TIF6 gene under the transcriptional control of the GAL10 promoter. The haploid yeast strain KSY606, containing the chromosomal copy of the TIF6 gene inactivated by insertion of a HIS3 marker gene and harboring a centromeric (CEN) expression plasmid that expresses yeast eIF6 from its own natural promoter, was constructed as follows. A 1.752-kb yeast genomic fragment containing the entire TIF6 gene was amplified from yeast genomic DNA by PCR using a 5′-end primer, Yf6G5 (29), and a 3′-end primer, Yf6G3 (29). The PCR product was digested with BamHI and EcoRI and cloned into the same sites of CEN plasmid pRS316 (URA3 based) (30) to yield plasmid pRS316-TIF6. In this plasmid, the expression of eIF6 is under the control of its natural promoter present in the inserted 1.752-kb fragment. This plasmid was then used to transform the diploid strain KSY601 (MATa/MATα tif6::HIS3/TIF6), and transformants were sporulated at 30°C; the resulting tetrads were dissected, and spores were germinated on YPD plates followed by selection on SD−His−Ura plates. Only spores containing the tif6::HIS3 gene and harboring plasmid pRS316-TIF6 will germinate to yield His+ Ura+ colonies. This haploid yeast strain was named KSY606 (Table 1). Yeast strains were grown at 30°C either in rich YPD medium (1% [wt/vol] yeast extract, 2% [wt/vol] Bacto Peptone, 2% [wt/vol] dextrose) or in YPGal medium, where 2% (wt/vol) galactose replaced dextrose as the carbon source. Where indicated, haploid yeast cells were also grown in synthetic complete medium (0.67% Bacto Yeast Nitrogen Base without amino acids, 0.2% amino acid mixture supplemented with nutrients required for auxotrophic deficiencies) containing either 2% galactose (SGal) or 2% dextrose (SD) as the carbon source. For in vivo [35S]methionine incorporation, the 0.2% amino acid mixture in either the SGal or the SD medium did not contain methionine. The methionine-lacking media were designated SGal−Met and SD−Met, respectively. Yeast genetic methods were performed following standard protocols (19).
TABLE 1.
Yeast strains used
| Strain | Genotype | Reference or source |
|---|---|---|
| W303a/α | MATa/MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100 | 21 |
| W303α | MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100 | 21 |
| KSY601 | MATa/MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100 tif6::HIS3/TIF6 | 29 |
| KSY603 | MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100 tif6::HIS3 p[URA3 GAL10::Ub-TIF6] | 29 |
| KSY605 | MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100 tif6::HIS3 p[TRP1 TIF6-myc] | This work |
| KSY606 | MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100 tif6::HIS3 p[URA3 TIF6] | This work |
| KSY607 | MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100 tif6::HIS3 p[LEU2 TIF6-HA] | This work |
Construction of a haploid yeast strain for expression of a C-terminally Myc-tagged or hemagglutinin (HA)-tagged eIF6 fusion protein.
To express the C-terminally Myc-tagged eIF6 fusion protein from its cognate promoter in yeast cells, two primers were designed: 5′-end primer Yf6G5 (5′CGGGATCCAAGGTGCAAGATCAGAC3′), which is complementary to a sequence 500 nucleotides upstream of the TIF6 ORF (BamHI site underlined); and 3′-end primer Yf6Myc (5′GGAATTCATGCATCTAGAGGTCTTCTTCGGAAATCAACTTCTGTTCTGAGTAGGTTTCAA3′), containing a Myc tag (in italics; the EcoRI site is underlined). A 1.2-kb S. cerevisiae genomic fragment containing the entire TIF6 gene was amplified by PCR using Pyrococcus DNA polymerase and the above two end primers. The PCR product was digested with BamHI and EcoRI and cloned into the same sites of the TRP1-based yeast CEN plasmid pRS314 (30) to yield pRS314-TIF6-Myc. This plasmid was then used to transform the haploid yeast strain KSY603 (MATα tif6::HIS3 p[URA3 GAL10::Ub-TIF6]), and Trp+ Ura+ transformants were selected on SGal plates. The transformants were then replica plated onto another SGal plate which contained uracil and 5-fluoroorotic acid (5-FOA) to promote loss of the URA3 plasmid. The yeast strain recovered from the 5-FOA plate, designated KSY605, had the disrupted chromosomal copy of TIF6 but harbored pRS314-TIF6-Myc as the complementing plasmid. These cells expressed, from its natural promoter, functional eIF6 with a Myc tag at the C-terminal end of the protein, at approximately wild-type levels.
To express C-terminally HA-tagged eIF6 in yeast cells, 3′-end primer yf63GF (5′CGAATTCGCGGCCGCCGCCTGAGTAGGTTTCAATCAA3′) (NotI site underlined) and 5′-end primer yf6G5 (see above) were used to amplify yeast genomic DNA by PCR, and the PCR product was digested with HindIII and NotI. The resulting HindIII/NotI fragment was used in a three-fragment ligation with the NotI/BamHI fragment derived from plasmid pGEXYP1 and LEU2-based CEN plasmid pRS315 that was digested with HindIII and BamHI to yield the new recombinant plasmid pRS315-TIF6-HA. This plasmid was then used to transform KSY606 (MATα tif6::HIS3 p[URA3-TIF6]) cells, and Leu+ Ura+ transformants were selected on appropriate SD plates. The transformants were then replica plated onto SD−Leu plates containing uracil and 5-FOA to promote loss of the URA3 plasmid. The yeast strain recovered from the 5-FOA plate, designated KSY607 (Table 1), had the disrupted chromosomal copy of TIF6 but harbored pRS315-TIF6-HA as the complementing plasmid. These cells expressed functional HA-tagged eIF6 from its natural promoter at approximately wild-type levels.
Subcellular fractionation of yeast cells and Western blotting.
Exponentially growing KSY605 yeast cells expressing c-Myc-tagged eIF6 were fractionated into nuclear and cytosolic fractions by an adaptation of the procedure of Aris and Blobel (1). Proteins of the subcellular fractions were precipitated with 10% trichloroacetic acid and analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis followed by immunoblotting using anti-c-Myc antibodies as probes to detect c-Myc-tagged-eIF6. Rabbit anti-Nop1p (a kind gift from T. Meier of this institution) and anti-yeast eIF5 (4) antibodies were used to detect nuclear and cytoplasmic protein markers, respectively.
Indirect immunofluorescence.
Cells (∼3 A600 units) from an exponentially growing culture of strain KSY607 (tif6::HIS3 p[LEU2 TIF6-HA]) in YPD medium were harvested by centrifugation. The harvested cells were prepared for immunofluorescence as described by Wang and Chang (37). DAPI (4′,6-diamidino-2-phenylindole dihydrochloride; Fluka) was used to stain chromosomal DNA. To detect eIF6-HA in permeabilized spheroplasts, cells were incubated overnight with monoclonal mouse anti-HA (BAbCo, Berkeley, Calif.) antibody primary at a dilution of 1:1,000, while goat anti-mouse Cy3-conjugated antibody (Jackson ImmunoResearch Laboratories) was used as the secondary antibody at a dilution of 1:1,000. Fluorescence-labeled cells were examined in an Olympus high-resolution IX70 microscope attached to a cooled charge-coupled device camera in the Analytical Imaging Facility of this institution. The figures were arranged using Adobe Photoshop.
Pulse-chase labeling of ribosomal subunits and analysis of their stability.
The haploid yeast strains W303α[TIF6] and KSY603[GAL10::Ub-TIF6] were each grown in 60 ml of SGal−Met medium to early logarithmic phase. Each culture was then treated with 200 μCi of [35S]methionine and shaken for 30 min at 30°C to label the ribosomal proteins. The cells from each culture were quickly harvested, resuspended in 60 ml of prewarmed SD-Met medium containing unlabeled methionine (500 μg/ml), and grown at 30°C. At the indicated time, 15 ml of each culture was harvested, and cells were washed with cold water and quick-frozen. Labeled ribosomal subunits were isolated from frozen cells as follows. Cells were thawed and washed with 1 ml of cold buffer R (20 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 50 mM KCl, 1 mM dithiothreitol, 0.2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 0.1% Triton X-100). The washed cells were then resuspended in 200 μl of buffer R and lysed by vortexing with an equal volume of glass beads. The cell lysates were clarified by centrifugation first at 12,000 × g for 15 min at 4°C, followed by recentrifugation of the supernatant at 17,000 × g for another 10 min. The clear supernatant was layered over 1.6 ml of 10% sucrose in buffer R and centrifuged in a Beckman TL100.3 rotor at 55,000 rpm for 2.5 h. Each ribosomal pellet was suspended in 100 μl of ice-cold buffer R by gentle shaking for 2 h and then treated with 2 mM puromycin at 30°C for 15 min. The suspension was then treated with 100 μl of 2× buffer S (1× buffer S is 50 mM Tris-HCl [pH 7.5], 800 mM KCl, 10 mM MgCl2, 10 mM dithiothreitol and 1 mM phenylmethylsulfonyl fluoride) and rotated slowly at 4°C for about 2 h to dissociate ribosomes into 40S and 60S ribosomal subunits. Equivalent amounts of A254-absorbing materials (approximately 4.0 A254 units in 200 μl) were layered on 11 ml of 15 to 35% (wt/vol) linear sucrose gradients in buffer S and centrifuged at 40,000 rpm for 8 h at 4°C in a Beckman SW41 rotor. The gradients were fractionated in an ISCO density gradient fractionator, and the absorbance profile at 254 nm was analyzed in an ISCO UA-5 absorbance monitor. An aliquot (100 μl) of each fraction was counted in Aquasol for 35S radioactivity in a liquid scintillation spectrometer. The stability of the labeled ribosomal subunits synthesized during growth of cells in SGal medium prior to transfer to SD medium was then calculated as total counts per minute of 35S radioactivity recovered in the 40S and 60S ribosomal subunit fractions.
Pulse-chase labeling of pre-rRNA and analysis by gel electrophoresis.
rRNAs were labeled with [methyl-3H]methionine by an adaptation of the procedure of Kressler et al. (11). Briefly, the haploid strains W303α[TIF6] and KSY603[GAL10::Ub-TIF6] were grown in SGal−Met medium to an A600 of about 0.5. Cells were harvested by centrifugation and resuspended in SD-Met medium prewarmed to 30°C to an A600 of about 0.2. After about 1.5 h of growth at 30°C for eIF6 depletion in KSY603 cells, cells (∼40 A600 units) were suspended in 750 μl of SD−Met medium, prewarmed to 30°C and labeled with 250 μCi of [methyl-3H]methionine (70 to 85 Ci/mmol; Amersham) for 2 min at 30°C. The chase was initiated by adding 7 ml of SD medium containing unlabeled methionine (500 μg/ml). At 0, 1.5, 3.0, and 6.0 min, 2-ml samples were withdrawn and centrifuged for 10 s, and cells were quickly frozen in a dry-ice–ethanol bath until used for RNA isolation. For labeling RNA with [3H]uracil, exponentially growing yeast cells in SGal medium were harvested, resuspended in SD medium supplemented with uracil (20 μg/ml), and grown for 1.5 h at 30°C. Cells were harvested by centrifugation, resuspended in fresh SD medium lacking uracil, and pulse-labeled for 3 min at 30°C with 100 μCi of [5, 6-3H]uracil. The chase was initiated by the addition of unlabeled uracil to a final concentration of 300 μg/ml. At various times, samples were taken and quickly frozen in a dry-ice–ethanol bath. RNA was isolated from both [methyl-3H]methionine- and [3H]uracil-labeled cells by the hot-phenol method as described elsewhere (25).
The 3H-labeled small RNAs were analyzed by electrophoresis on 7% polyacrylamide gels containing 8 M urea, transferred to a nylon membrane, incubated in En3Hance (Dupont-NEN), and subjected to autoradiography. Large RNA molecules were resolved on 1.2% agarose–6% formaldehyde gels and transferred to Hybond-N+ nylon membranes (Amersham); the membranes were baked for 2 h at 80°C, sprayed with En3Hance (Dupont-NEN), dried, and subjected to autoradiography at −70°C with an intensifying screen.
Northern analysis.
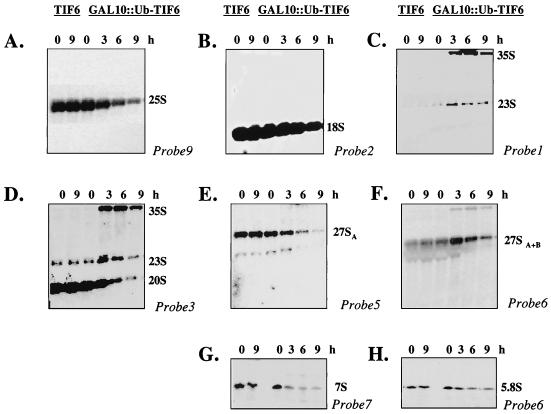
Steady-state levels of pre-rRNA and mature rRNA were determined by Northern blot analysis using 32P-labeled deoxyoligonucleotide probes (numbered 1 to 9 in Fig. 3). The sequences of these probes, which are complementary to various regions of 35S pre-rRNA, were 5′GGTCTCTCTGCTGCCGG3′ (probe 1), 5′CATGGCTTAATCTTTGAGAC3′ (probe 2), 5′CGGTTTTAATTGTCCTA3′ (probe 3), 5′TGTTACCTCTGGGCCC3′ (probe 4), 5′AATTTCCAGTTACGAAAATTCTTG3′ (probe 5), 5′TTTCGCTGCGTTCTTCATC3′ (probe 6), 5′GGCCAGCAATTTCAAGTTA3′ (probe 7), and 5′CTCCGCTTATTGATATGC3′ (probe 9). Approximately 100 ng of each oligonucleotide probe was end labeled with 32P by using 50 μCi of [γ-32P]ATP (6,000 Ci/mmol) and T4 polynucleotide kinase. Exponentially growing cultures of W303α[TIF6] and KSY603[GAL10::Ub-TIF6] were transferred to YPD medium. At various times following the shift to YPD, cells (10 A600 units) were withdrawn and total RNA was isolated. Approximately 5 μg of RNA was resolved on 1.2% agarose–6% formaldehyde gels or 7% polyacrylamide–8 M urea gels, transferred to Hybond-N+ nylon membranes (Amersham) as described by Sambrook et al. (25), and then cross-linked to the paper in a Stratagene UV cross-linker according to the manufacturer's specifications. The membrane blots were prehybridized in a buffer containing 1% bovine serum albumin, 1 mM EDTA, 0.5 M NaPO4 (pH 7.2), and 7% SDS at 65°C for 1 h and then hybridized for 12 h in the same buffer containing approximately 106 cpm of 32P-labeled oligonucleotide probes. The membrane blots were washed several times with 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) containing 0.5% SDS followed by 0.1× SSC–0.5% SDS at room temperature. The dried blots were analyzed by autoradiography.
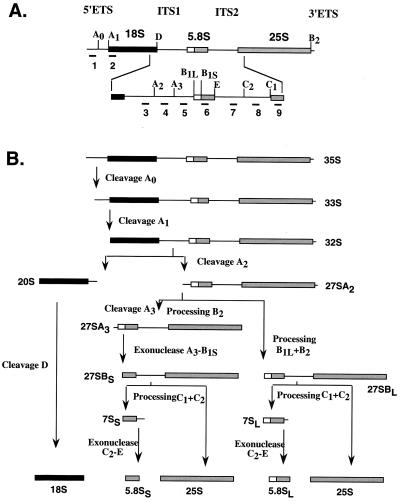
FIG. 3.
Schematic diagram of pre-rRNA processing in the yeast S. cerevisiae. (A) Organization and processing sites of 35S pre-rRNA. The 35S precursor contains the sequences for the mature 18S, 25S, and 5.8S rRNAs, which are separated by the internal transcribed spacers ITS1 and ITS2 and two external transcribed spacers, 5′ETS and 3′ETS, at the 5′ and 3′ ends, respectively. The oligonucleotides corresponding to different regions of 35S rRNA that were used as probes in Northern analysis are indicated by numbers 1 through 9. (B) Pre-rRNA processing steps and different processing intermediates. The major cleavage sites, endonucleolytic and exonucleolytic processing steps, processing intermediates, and pathways leading to mature rRNA are indicated (adapted from reference 11).
Other methods.
The procedure used for preparation of yeast cell lysates for immunoblot analysis was adapted from that described by Sachs and Davis (24). The rate of protein synthesis was measured by pulse-labeling 1.0 A600 unit of yeast cells with [35S]methionine for 5 min and measuring the incorporation of radioactivity into total cellular proteins as described previously (29).
RESULTS
Kinetic analysis of eIF6 disappearance, 60S ribosomal subunit reduction, and inhibition of protein synthesis in yeast cells.
We have previously constructed and used (29) the haploid yeast strain KSY603, in which the chromosomal copy of the TIF6 gene was inactivated by insertion of a HIS3 marker gene and the essential eIF6 function was provided by maintenance of a CEN plasmid harboring a conditional eIF6 expression system. In this expression system, eIF6 was expressed from a transcription unit containing a protein-destabilizing ubiquitin gene cassette consisting of GAL-UAS-UBI4-R-lacI-HA (where R is the codon for arginine) (16) fused to the NH2 terminus of the TIF6 ORF under the transcriptional control of the GAL10 promoter. When an exponentially growing culture of KSY603 in SGal medium was transferred to SD medium to repress synthesis of eIF6, the preexisting eIF6 protein was rapidly removed from yeast cells (29). Analysis of various parameters of protein synthesis in eIF6-depleted cells showed that there was a selective reduction in the level of 60S ribosomal subunits in these cells, resulting in inhibition of protein synthesis and eventually of cell growth (29).
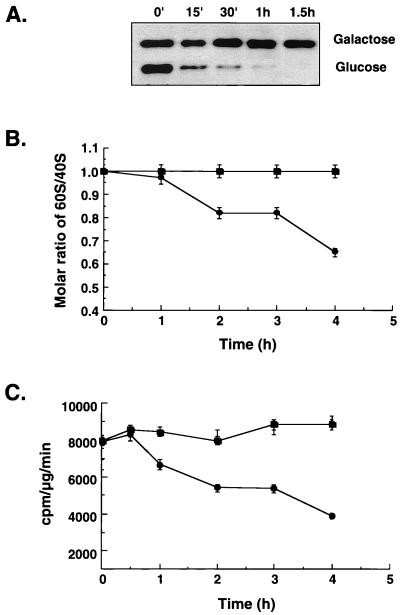
In this work, we first carried out a more detailed kinetic analysis of the decline in the level of 60S ribosomal subunits in eIF6-depleted cells. For this purpose, we transferred exponentially growing cultures of wild-type W303α and KSY603 from galactose- to glucose-containing medium and then monitored the rates of disappearance of eIF6 and the level 60S ribosomal subunits by measuring (i) the levels of eIF6 in cell lysates by immunoblot analysis, (ii) the relative amounts of total 40S and 60S ribosomal subunits, and (iii) the relative rates of protein synthesis in KSY603 cell lysates (Fig. 1). As shown in Fig. 1A, there was a rapid decline of eIF6 fusion protein in KSY603 cells after transfer to glucose medium. Within an hour, the eIF6 fusion protein was barely detectable in these cell lysates. In contrast, the level of eIF6 in KSY603 cell lysates continuing to grow in SGal medium remained fairly constant during a 6-h period (only data up to the 1.5-h time point are shown for comparison). However, significant reduction in the molar ratio of 60S to 40S ribosomal subunits occurred only after near-complete disappearance of eIF6 from yeast cells, and this reduction occurred gradually over a period of several hours (Fig. 1B). The kinetics of inhibition of the rate of protein synthesis was similar to that observed for the reduction of the 60S/40S molar ratio (Fig. 1C), indicating that inhibition of protein synthesis in eIF6-depleted cells was probably due to deficit of 60S ribosomal subunits in these cells. As expected, in control wild-type W303α[TIF6] cells grown in SD medium or KSY603 cells grown in galactose-containing medium, both the molar ratio of 60S to 40S subunits as well as the rate of protein synthesis remained unchanged (in Fig. 1, only data for W303α are shown). These results clearly indicate that reduction in the steady-state levels of 60S ribosomal subunits begins only after near-complete disappearance of eIF6 from yeast cells, and this reduction occurs gradually over a period of several hours. It should also be noted that while eIF6 was undetectable in about 1.5 h following transfer of KSY603 cells from galactose- to glucose-containing medium, the cell growth rate did not begin to decrease until after about 3 h. (There was only about a 10% decrease in growth rate at 4 h.) From this time point, there was a progressive increase in the doubling time until about 15 h, when cell growth ceased (data not shown) (29).
FIG. 1.
Kinetics of disappearance of 60S ribosomal subunits and inhibition of protein synthesis in eIF6-depleted cells. (A) Exponentially growing cultures of KSY603[GAL10::Ub-HA-TIF6] cells in SGal−Met medium were transferred to SD−Met medium and allowed to grow. At the indicated times, cell lysates were prepared from 1 A600 unit of cells, and approximately 100 μg of protein from each cell lysate was analyzed by Western blotting using monoclonal anti-HA antibodies as probes. (B) Total ribosomes were isolated from KSY603 (●) and control wild-type (■) cells at indicated times following a shift of each culture from SGal-Met to SD-Met medium. Ribosomes were dissociated into subunits and sedimented through 15 to 40% sucrose gradients as described in Materials and Methods and in reference 29. (C) Rates of protein synthesis. At the indicated times following shift of cells from SGal-Met to SD-Met medium, 1 A600 unit of cells from each culture was harvested and suspended in 300 μl of SD-Met medium containing 0.5 μCi of [35S]methionine (1,175 Ci/mmol), and cells were shaken for 5 min at 30°C. The pulse was then terminated by the addition of a stop buffer containing unlabeled methionine (1.8 mg/ml) and cycloheximide (50 μg/ml). The rate of protein synthesis at each time point was calculated as described previously (29). The data presented in panels B and C are averages of two independent experiments.
eIF6 is not required for the stability of 60S ribosomal subunits in the cytoplasm.
The selective reduction in the steady-state level of 60S ribosomal subunits due to removal of eIF6 from yeast cells suggested that eIF6 may be required either for the stability of 60S particles in the cytoplasm or for the biogenesis of 60S ribosomal subunits.
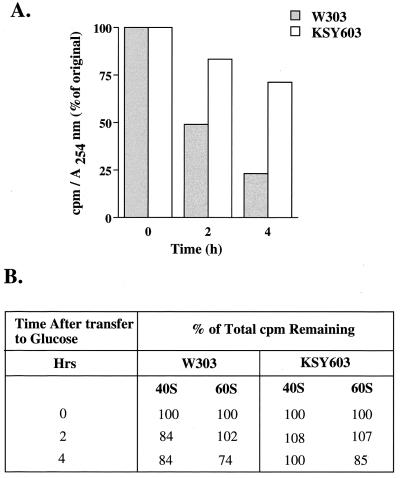
The effect of eIF6 depletion on the stability of 60S ribosomal subunits in yeast cells was first investigated. An exponentially growing culture of KSY603[GAL10::Ub-TIF6] as well as the wild-type W303α[TIF6] cells were labeled with [35S]methionine for 30 min. Cells were then transferred to glucose-containing medium containing an excess of unlabeled methionine, and the stability of [35S]methionine-labeled 60S ribosomal subunits synthesized during growth in galactose medium was monitored over the growth period in glucose medium when rapid eIF6 depletion occurred. We isolated both 40S and 60S ribosomal subunits from each cell lysate by sucrose gradient centrifugation and determined the total 35S radioactivity in each ribosomal subunit pool. If eIF6 is required for the stability of 60S ribosomal subunits, removal of eIF6 from yeast cells should result in a progressive decline in the amount of 35S radioactivity in the 60S ribosomal subunit pool. Figure 2B shows that in both wild-type W303α[TIF6] and KSY603 cells, most of the 35S radioactivity incorporated into 60S and 40S subunits during growth in SGal medium was recovered following growth in SD medium. These results suggest that eIF6 depletion did not significantly reduce the stability of preexisting 60S ribosomal subunits. However, we observed that the specific radioactivity of 60S subunits decreased in wild-type W303α[TIF6] cell lysates exponentially with time during growth in SD medium. This is expected because of the synthesis of new unlabeled ribosomal subunits during the growth period in SD medium. The specific radioactivities of 60S subunits decreased nearly 50 and 25% during growth of W303α in SD medium for 2 and 4 h, respectively. These results correlate well with the doubling time of about 2 h for W303α cells in SD medium. In contrast, the specific radioactivity of 60S ribosomal subunits in KSY603 cells did not decrease significantly during growth in SD medium after eIF6 depletion (Fig. 2A). The small (approximately 15%) decrease in specific radioactivity of 60S subunits in KSY603 cells at 2 h following transfer of yeast cells from SGal to SD medium can be explained by our observation that eIF6 does not disappear completely immediately following the transfer of KSY603 cells from galactose- to glucose-containing medium. Rather, there was a progressive decline in eIF6 levels during the first hour of growth in SD medium (Fig. 1A). Presumably, during this period new 60S subunits were synthesized at a reduced level in KSY603 cells, thus decreasing slightly the specific radioactivity of 60S subunits. At any rate, since removal of eIF6 from yeast cells did not significantly reduce the stability of preexisting 60S ribosomal subunits (Fig. 2B), this result suggests that eIF6 depletion inhibits the synthesis of new 60S ribosomal subunits. Otherwise, the specific radioactivity of 60S ribosomes would have decreased exponentially as was observed for wild-type W303 cells (Fig. 2A). It should be noted that there was no significant reduction in growth rate of KSY603 cells in SD medium during the first 2 h of growth. After 4 h, there was only about a 10% reduction in the growth rate (data not shown).
FIG. 2.
Synthesis but not stability of 60S ribosomes is affected by eIF6 depletion. Exponentially growing cultures of W303α[TIF6] or KSY603[GAL10::Ub-TIF6] in SGal−Met medium (approximately 50 A600 units of cells) were labeled with 200 μCi of [35S]methionine for 30 min at 30°C. Cells were then transferred to 100 ml of SD-Met medium containing unlabeled methionine (500 μg/ml). At the indicated times, cells (10 A600 units) were harvested, washed, and lysed as described in Materials and Methods. Each lysate was subjected to 15 to 40% (wt/vol) sucrose gradient centrifugation to separate the ribosomal subunits and fractionated in an ISCO density gradient fractionator attached to a UA-5 absorbance monitor. The amounts of 60S subunits were quantified from absorbance at 254 nm, while 35S radioactivity content of each fraction was determined by counting in Aquasol in a liquid scintillation spectrometer. The total amount of 35S radioactivity present in either the 40S or the 60S ribosomal subunit pool under each condition following transfer to SD medium was calculated. These values are shown in panel B as the percentage of total counts per minute originally incorporated into 60S or 40S ribosomal subunits prior to transfer to SD medium. The specific radioactivity of 60S ribosomal subunits during growth in SD medium was calculated as counts per minute of 35S radioactivity per 1 A254 unit of 60S ribosomal subunits.
eIF6 is required for pre-rRNA processing and consequently for the biogenesis of 60S ribosomes.
Experiments presented above clearly suggested that eIF6 depletion affected the biogenesis of 60S ribosomal subunits.
Ribosome biogenesis in eukaryotic cells is a complex process that occurs mostly in the nucleolus, where four rRNAs are formed, modified, and processed during assembly with 78 (in yeast) and 79 (in mammals) ribosomal proteins into mature 40S and 60S ribosomal subunits. In the yeast S. cerevisiae where the process has been best characterized both biochemically and genetically, the 18S rRNA of 40S subunits and the 25S and 5.8S rRNAs of 60S subunits are transcribed by RNA polymerase I as a single 35S pre-rRNA, whereas the fourth rRNA, 5S, a constituent of the 60S subunit, is transcribed independently from 5S DNA by RNA polymerase III. The 35S pre-rRNA then undergoes a sequence of ordered endo- and exonucleolytic cleavage and trimming reactions requiring the participation of small nucleolar RNAs and several trans-acting protein factors to generate mature 18S, 25S, and 5.8S rRNAs (Fig. 3) (for reviews, see references 12, 32, 33, and 35).
Decreases in the levels of ribosomal subunits due to mutations in or depletion of a nonribosomal protein are usually the result of defects in pre-rRNA processing and/or assembly of ribosomal subunits. Mutation or depletion of a number of yeast proteins causes a defect in pre-rRNA processing (see references 12 and 35 for reviews). In some cases, the stability of mature rRNAs is also affected (9). Presumably, these proteins associate with pre-rRNA at one or more steps during the processing reactions.
To study the role of eIF6 in 60S subunit biogenesis, we examined the effects of eIF6 depletion on the synthesis and processing of pre-rRNA as measured by [methyl-3H]methionine pulse-chase labeling experiments. Since 25S and 18S rRNAs are highly and specifically methylated during processing, processing of pre-rRNA is easily monitored by labeling yeast RNA with [methyl-3H]methionine (33, 36). For this purpose, exponentially growing cultures of yeast strain KSY603[GAL10::Ub-TIF6] and the wild-type strain W303α[TIF6] in SGal medium were transferred to SD−Met medium, and cells were allowed to grow for 90 min to remove eIF6 from KSY603 cells. Cells were then pulsed-labeled with [methyl-3H]methionine for 2 min and chased for 1.5, 3.0, and 6.0 min with an excess of unlabeled methionine.
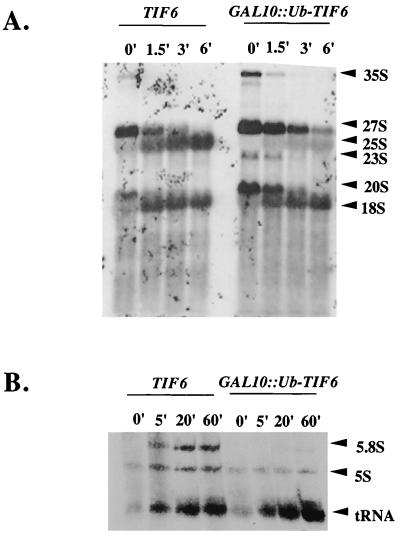
In wild-type W303α cells, 35S pre-rRNA and 27S and 20S processing intermediates were rapidly chased into mature 25S and 18S rRNAs, as expected (Fig. 4A). In contrast, in eIF6-depleted KSY603 cells, processing of all of the pre-rRNA intermediates was slightly slower. This was true of 35S to 27S and 20S species, of the processing of 20S to 18S mature rRNA, and of the disappearance of 27S pre-rRNA. The striking aspect is that in eIF6-depleted cells, the processing of 27S pre-rRNA leads to complete loss of the RNA rather than production of the 25S mature species. In addition, there was a transient appearance of a weakly labeled unusual 23S species that was not observed in wild-type cells. Taken together, these results suggest that the primary defect due to eIF6 depletion is in the formation of 60S-specific 25S rRNA from 27S pre-rRNA. The processing of 35S pre-RNA was also slightly affected, resulting in the reduced formation of all precursor and mature rRNAs.
FIG. 4.
Depletion of eIF6 leads to reduced synthesis 25S and 5.8S rRNAs. (A) Exponentially growing cultures of wild-type strain W303α[TIF6] and strain KSY603[GAL10::Ub-TIF6] in SGal−Met medium were shifted to SD−Met medium and grown for 90 min to remove eIF6 from KSY603 cells. Cells (40 A600 units) were pulse-labeled with 250 μCi of [methyl-3H]methionine for 2 min at 30°C and then chased with an excess of unlabeled methionine (500 μg/ml) for the indicated times. Total RNA was isolated from each batch of cells; for each time point, an RNA sample containing about 20,000 cpm of 3H radioactivity was analyzed in a 1.2% formaldehyde-agarose gel and subjected to fluorography as described in Materials and Methods. The positions of mature 18S and 25S rRNAs and pre-rRNAs are indicated. (B) Exponentially growing cultures of KSY603[GAL10::Ub-TIF6] and control KSY606[TIF6][URA3] cells in SGal-Ura medium (A600 = 0.5) were transferred to SD-Ura medium and allowed to grow for 90 min, pulsed with 200 μCi of [5,6-3H]uracil for 3 min, and then chased with an excess of unlabeled uracil (1 mg/ml) for the indicated times. Total cellular RNA from each sample was isolated, and approximately 60,000 cpm of each [3H]RNA sample was subjected to 7% polyacrylamide–8 M urea gel electrophoresis, transferred to a Hybond-N+ membrane, and analyzed by fluorography as described in Materials and Methods. The positions of 5.8S and 5S RNAs and 4S tRNA are indicated.
To rule out the possibility that altered or defective methylation was responsible for impaired formation of 25S rRNA, as well as to monitor the processing and formation of low-molecular-weight rRNAs, a similar experiment was carried out with [3H]uracil. A pulse-label with [5,6-3H]uracil for 3 min was chased for 5, 20, and 60 min with an excess of unlabeled uracil. (It should be noted that much shorter periods of labeling and chase time were used in experiments with [3H]methionine than in those with [3H]uracil. This was because the endogenous pool of S-adenosylmethionine is known to be rapidly saturated and chased [36]). In contrast, [3H]uracil is utilized with much slower kinetics.) Analysis of total [3H]RNA samples by agarose-formaldehyde gel electrophoresis under conditions similar to those used for Fig. 4A yielded results similar to those obtained with [methyl-3H]labeled RNA (data not shown). More important, analysis of low-molecular-weight (5.8S and 5S) rRNAs from the [3H]uracil labeling experiments by polyacrylamide-urea gel electrophoresis showed that almost no 5.8S rRNA was formed in eIF6-depleted cells (Fig. 4B). The lack of formation of 5.8S rRNA is consistent with lack of formation of 25S rRNA since both mature rRNAs are formed by processing of 27S pre-rRNA. The formation of 5S RNA and 4S tRNA was, however, readily observed, although eIF6-depleted KSY603 cells showed a lower overall incorporation of [3H]uracil into 5S RNA than eIF6-containing wild-type cells (Fig. 4B), possibly due to turnover of 5S RNA that is not incorporated into 60S particles.
To characterize the steps in pre-RNA processing pathway that are defective in the absence of eIF6, the steady-state levels of pre-rRNAs and mature rRNAs were determined by Northern blot analysis of total RNA isolated from eIF6-depleted and control wild-type cells, using deoxyoligonucleotide probes complementary to defined regions of 35S pre-rRNA as shown in Fig. 3. RNA was isolated from wild-type W303α[TIF6] and KSY603[GAL10::Ub-TIF6] cells that were initially grown in YPGal medium and then shifted to YPD medium and allowed to grow for various times. Figure 5 shows that eIF6 depletion caused a marked decrease in 25S rRNA but only a slight decrease in 18S rRNA steady-state levels (Fig. 5A and B). Hybridization with oligonucleotide probe 1, which is complementary to sequences 5′ to site A0 (Fig. 3) and detects the precursor 35S rRNA, showed that eIF6-depleted KSY603 cells accumulated 35S pre-rRNA, indicating a processing defect at site A0 (Fig. 5C). In addition, an aberrant intermediate processing product of 23S accumulated in these cells (Fig. 5C). This 23S pre-rRNA was also detected with oligonucleotide probe 3 (Fig. 5D) and probe 4 (not shown) but not with probe 5 (Fig. 5E), indicating that the 23S RNA may correspond to the region of the 35S pre-rRNA that extends from the 5′ externally transcribed spacer (5′ETS) to site A3. Thus, the aberrant 23S pre-rRNA probably originated from a premature cleavage of 35S pre-rRNA at site A3. It should be noted that other investigators observed accumulation of the 23S rRNA intermediate in yeast cells that were defective in the 35S pre-rRNA processing pathway (11, 39; see also references 12 and 35). Hybridization with oligonucleotide probe 3 (Fig. 5D) also showed that concomitant with eIF6 depletion, the amount of 20S pre-rRNA diminished significantly. The decrease in the level of 20S pre-rRNA observed in eIF6-depleted cells was probably due to defects in the processing of 35S pre-rRNA and accumulation of the aberrant 23S pre-rRNA. In view of our observation that synthesis of 18S rRNA was only weakly impaired in eIF6-depleted cells (Fig. 5B), some of the mature 18S rRNA could have also been derived by processing of 23S rRNA. When oligonucleotide 5 was used as a hybridization probe (Fig. 5E), we observed that levels of 27SA2 and 27SA3 pre-rRNAs were much lower in eIF6-depleted cells than in wild-type cells. However, probing with oligonucleotide 6 (Fig. 5F), which should detect both 27SA2, and 27SA3 pre-RNAs as well as 27SB pre-RNA, we observed that eIF6 depletion caused initial accumulation of 27S pre-rRNA. Similar results were obtained with probe 7 (data not shown). Based on the results in Fig. 5E and F, we therefore conclude that depletion of eIF6 in yeast cells caused accumulation of 27SB pre-rRNA, indicating a block in the processing of 27SB pre-rRNA to 25S rRNA and 7S pre-rRNA. This was confirmed by determining the steady-state levels of 7S pre-rRNA and 5.8S mature rRNA in eIF6-depleted KSY603 cells relative to wild-type W303α cells using oligonucleotide probes 7 (Fig. 5G) and 6 (Fig. 5H), respectively. Concomitant with gradual depletion of eIF6 from KSY603 cells, the steady-state levels of 7S pre-rRNA and mature 5.8S rRNA were decreased. Taken together, the Northern hybridization data suggest that removal of eIF6 from yeast cells results in some inhibition of A0-A2 cleavage, leading to increased accumulation of 35S pre-rRNA, the appearance of a 23S aberrant rRNA by premature cleavage of 35S pre-rRNA at site A3, reduced steady-state levels of 20S, 27SA2, and 27SA3 pre-rRNAs, and increased accumulation of 27SB pre-rRNA, with further processing blocked at 27SB pre-rRNA. Inhibition of the processing of 27SB pre-rRNA results in the decreased formation of mature 25S rRNA and of 7S pre-rRNA.
FIG. 5.
Steady-state levels of mature rRNAs and rRNA precursors in eIF6-depleted cells. Haploid yeast strains W303α[TIF6] and KSY603[GAL10::Ub-TIF6] were grown in YPGal medium and shifted to YPD medium. At the indicated times, cells were harvested, and total RNA was extracted and subjected to Northern analysis using 32P-labeled oligonucleotide probes complementary to different regions of 35S pre-rRNA as described in Materials and Methods and Fig. 3. (A) Probe 9, sequences within the mature 25S rRNA; (B) probe 2, sequences within the mature 18S rRNA; (C) probe 1, sequences upstream of site A0 in the 5′ETS; (D) probe 3, sequences in ITS1 between sites D and A2; (E) probe 5, sequences in ITS1 between sites A3 and B1L; (F) probe 6, sequences within mature 5.8S rRNA; (G) probe 7, sequences in ITS2 between E and C2; (H) probe 6, for detection of small 5.8S rRNA. The positions of mature 25S and 18S rRNAs and different precursor rRNAs are indicated. The blots were also analyzed using a probe corresponding to U1 small nuclear RNA, which served as an internal loading control (not shown).
eIF6 is present in both nucleus and cytoplasm.
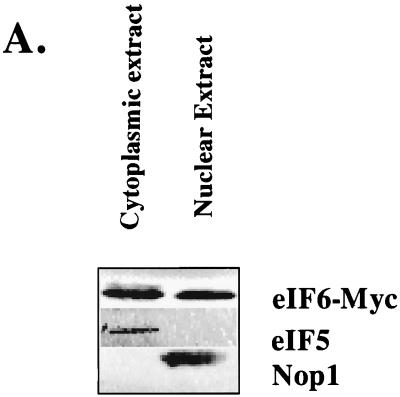
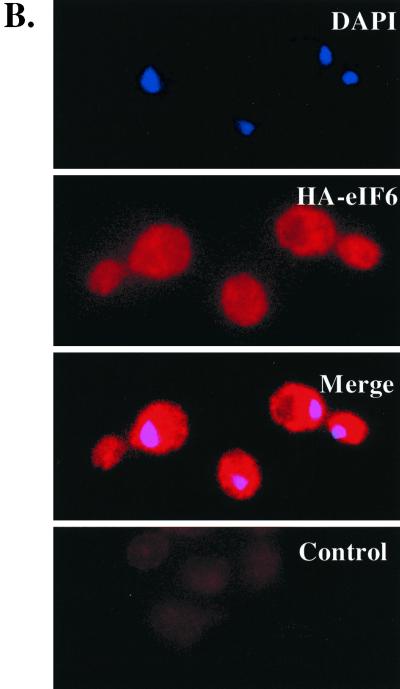
eIF6 was originally isolated from the cytoplasm of cell extracts of higher eukaryotes (17, 22, 23, 34), including extracts of rabbit reticulocytes, that lack a nucleus (17, 28). In the yeast strain KSY603, HA-eIF6 expressed from a GAL10 promoter was found to be associated with free cytoplasmic 60S ribosomal subunits (29). However, our observation in this work that eIF6 is required for processing of pre-rRNAs to mature rRNAs, a process that is known to occur in the nucleolus, suggests that eIF6 should also be present in the nuclei of yeast cells. We investigated the subcellular localization of eIF6 by constructing a yeast strain (KSY605) in which functional eIF6 was expressed from its own natural promoter as a C-terminally c-Myc-tagged fusion protein. Yeast cells expressing c-Myc-tagged eIF6 fusion protein were then fractionated into nuclear and cytoplasmic fractions, and the nuclei were further purified as described in Materials and Methods. The proteins in the nuclear and cytoplasmic fractions were analyzed by Western blotting using anti-c-Myc antibodies. Figure 6A shows that eIF6-Myc was present in both nuclear and postnuclear (cytoplasmic) fractions. The validity of the fractionation was verified using antibodies to eIF5 (a marker for cytoplasmic proteins) and Nop1p (a marker for nucleolar proteins). To visualize directly the presence of eIF6 in subcellular fractions, we constructed the haploid yeast strain KSY607, in which eIF6 was expressed as a C-terminally-tagged HA fusion protein from its cognate promoter. This eIF6-HA fusion protein could functionally complement a disruption in the genomic copy of TIF6, indicating that the presence of HA at the C terminus of eIF6 does not alter its function in yeast cells (data not shown). Exponentially growing cells of KSY607 expressing eIF6-HA were examined by indirect immunofluorescence using anti-HA antibodies followed by decoration with goat anti-mouse Cy3-conjugated antibodies. The nuclei were stained with DAPI. The fluorescence photographs showed that HA-eIF6 was distributed evenly in the nucleus and the cytoplasm and fully apparent in the bud lacking any nucleus (Fig. 6B). No signal was obtained with the combination of anti-HA and goat anti-mouse Cy3-conjugated antibodies when cells of KSY606 expressing untagged eIF6 were analyzed by indirect immunofluorescence (Fig. 6B, Control). It should be noted that calculation of the relative distribution of total eIF6 in nuclear and cytosolic fractions in the experiment presented in Fig. 6A showed that >90% of the total cellular eIF6 is present in the cytosolic fraction, while only about 5 to 8% is present in the nuclear fraction (data not shown).
FIG. 6.
Subcellular distribution of eIF6. (A) Cell fractionation. Lysates of the haploid yeast strain KSY605 expressing a c-Myc-tagged eIF6 from its natural promoter were fractionated into postnuclear supernatant (cytosolic fraction) and nuclear pellet as described in Materials and Methods. Approximately 25 μg of protein from each fraction was subjected to Western blot analysis using rabbit polyclonal anti-c-Myc, anti-yeast eIF5, and anti-Nop1p antibodies as probes. (B) Localization of eIF6-HA fusion protein. Indirect immunofluorescence staining was performed with KSY607 cells expressing eIF6-HA from the TIF6 promoter. Chromatin DNA of yeast cells was stained with DAPI. eIF6-HA was detected by immunofluorescence staining with monoclonal mouse anti-HA primary antibody and Cy3-conjugated secondary antibody (second panel). In the third panel, the photographs of the top two panels were merged. Note that in this merged photograph, the nuclear region assumes a magenta color. In the control panel, indirect immunofluorescence was performed with untagged eIF6 expressed from the TIF6 promoter in KSY606 cells.
DISCUSSION
A molecular genetic approach using the yeast S. cerevisiae was previously used in this laboratory (29) to investigate whether eIF6, originally isolated and characterized on the basis of its in vitro ability to bind to 60S ribosomal subunits (17, 22, 23, 34) and prevent its association with the 40S ribosomal subunits, indeed functions as a translation initiation factor in vivo in yeast cells. Using yeast strain KSY603, which contains a conditional eIF6 expression system, we were able to remove eIF6 rapidly from yeast cells and to measure various parameters of protein synthesis in vivo and in vitro (29). We observed that removal of eIF6 from yeast cells caused a progressive inhibition in the rate of protein synthesis in yeast cells. However, this inhibition of protein synthesis was due not to a defect in translation initiation but rather to selective reduction of the level of 60S ribosomal subunits in yeast cells, causing a stoichiometric imbalance between 40S and 60S ribosomal subunits with consequent formation of half-mer polysomes (29). Similar observations were reported by Sanvito et al. (26), who identified eIF6 from mammalian cells as a β4 integrin-interacting protein (3). The question, however, remained: how does eIF6 function to maintain the steady-state level of 60S ribosomal subunits in yeast cells?
Experiments presented here show that the stability of mature 60S particles synthesized in KSY603 cells in the presence of eIF6 was not significantly affected following removal of eIF6 from these cells. Rather, our evidence clearly indicates that in eIF6-depleted cells, the reduction of 60S subunit levels is due to a primary defect in pre-rRNA processing. A pulse-label shortly after eIF6 depletion led to relatively normal incorporation into rRNA molecules and the expected, if slightly delayed, processing to 27S and 20S pre-rRNA species. However, while the 20S precursor RNA was processed to 18S rRNA efficiently, most of the 27S pre-rRNA species was degraded without forming 25S and 5.8S mature rRNAs.
These results were confirmed by Northern blot analysis. In eIF6-depleted cells, there is an increase in the steady-state level of 35S and 27SB precursor rRNAs and a marked decrease in the level of mature 25S and 5.8S rRNAs as well as of 7S pre-rRNA. This result reflects a defect in the processing of 27SB species to mature 25S rRNA and 7S pre-rRNA. Whereas depletion of eIF6 leads to a marked reduction in the steady-state level of 25S rRNA, presumably due to dilution, there was only a slight decrease in the amount of 18S rRNA during this time period. This observation points to a rather specific and immediate block in the synthesis of 60S-specific rRNAs in eIF6-depleted cells. Taken together, these results suggest that eIF6 is necessary for the formation of 60S subunits because it is necessary in the formation of 25S and 5.8S rRNAs. Perhaps the absence of eIF6 induces the processing machinery to make an error that results in immediate degradation of the 27S RNA. Alternatively, eIF6 may be needed to signal to the degradation machinery that the 60S particle is functional.
We do not know whether accumulation of 35S and 23S pre-rRNAs in eIF6-depleted cells results directly from eIF6 depletion or is a consequence of the defective 27SB pre-rRNA processing which causes some form of feedback inhibition or delayed processing of 35S pre-rRNA. Inhibition of processing of the 35S pre-rRNA, accumulation of the aberrant 23S pre-rRNA by cleavage of 35S RNA at site A3, and reduced steady-state levels of 20S pre-rRNA have all been reported for many mutants that affect biogenesis of 60S ribosomal subunits (2, 10, 12, 18, 31, 38, 39; see also reference 35). These observations suggest that delayed processing at sites A0 to A2 may be a general feature of mutations that inhibit the formation of mature 25S and 5.8S rRNAs. It should also be noted that the pre-rRNA processing defects observed in eIF6-depleted cells closely resemble those previously reported for yeast cells lacking Nip7p, Nop2p, and Dbp3p (10, 18, 35, 39). Depletion of each of Nip7p and Nop2p in yeast cells caused a block in the processing of 27SB pre-rRNA to 25S rRNA and 7S pre-rRNA, while cells lacking Dbp3p were shown to be defective in the processing of 27SA2 to 27SA3 pre-rRNA, resulting in the inhibition of synthesis of mature 25S and 5.8S rRNAs; these cells also accumulated 35S and 23S pre-rRNAs and reduced levels of 20S pre-rRNA. Reduction in the level of 20S pre-rRNA, in turn, causes some inhibition in the formation of 18S rRNA. It has been postulated (35) that the requirement for 25S and 5.8S processing proteins for 18S rRNA synthesis is indirect. It is likely that the pre-rRNA processing machinery is a single large complex formed well before the specific steps in 25S and 5.8S rRNA formation. Thus, defects in any processing protein might inhibit early steps including eventually 18S rRNA synthesis.
It is now well established that the synthesis of ribosomal proteins and assembly of ribosomes are tightly coupled to processing and modification of pre-rRNA. Mutation or depletion of proteins involved in ribosome biogenesis usually leads to defects at multiple steps in the pathway of synthesis of ribosomes (12, 35). Thus, our experiments do not rule out the possibility that eIF6, in addition to being required for efficient processing of pre-rRNA, is required for the synthesis of ribosomal proteins and/or their assembly into mature ribosomes. It should be noted that measurement of the rate of transcription of the rRNA genes using the nuclear run-on assays described by Elion and Warner (8) did not show any significant difference in the synthesis of 35S rRNA between the wild-type and eIF6-depleted cell extracts (data not shown).
In this work, we have also studied the subcellular localization of eIF6 in yeast cells. Although eIF6 is associated with free cytoplasmic 60S subunits both in mammalian cells (17, 22, 23, 28, 34) and in the yeast S. cerevisiae (29), indirect immunofluorescence studies using eIF6-HA fusion protein as well as cell fractionation studies clearly show that the protein is localized both in the cytoplasm and in the nucleus. These localization data are consistent with the role of eIF6 in pre-rRNA processing, mainly a nucleolar event. However, the predominant cytoplasmic localization of eIF6 suggests that eIF6 may have a function in the cytoplasm. It is tempting to speculate that eIF6 is exported from the nucleus in association with 60S ribosomal subunits and is released from the 60S subunit before or after it binds to the 40S initiation complex to form the 80S initiation complex. This is in accord with our observation that HA-tagged eIF6 expressed in yeast cells associates with free cytosolic 60S subunits and is not part of 80S monosomes or polyribosomes (29). Association of eIF6 with free cytoplasmic 60S subunits suggests that eIF6 may act as a chaperone in transporting 60S subunits from the nucleus to the cytoplasm. The possibility also exists that although eIF6 does not directly function as a general translation initiation factor, its binding to 60S ribosomal subunits may serve as a checkpoint of the subunit joining step during initiation of protein synthesis. We found some time ago (34) that 60S ribosomal subunits containing bound mammalian eIF6 are incapable of joining the 40S initiation complex to form the 80S initiation complex. These observations suggest that a mechanism must exist for the release of eIF6 from the 60S subunit either prior to or concomitant with the joining of the 60S subunit to the 40S initiation complex. Interestingly, Nip7p (39) and Nmd3p (9) have also been identified as proteins required for 60S biogenesis that also associate specifically with free 60S subunits in the cytoplasm of yeast cells. It remains to be seen whether these proteins possess a ribosomal subunit antiassociation activity similar to that observed for eIF6 (29). On the other hand, the cytoplasmic localization and association of eIF6 with 60S ribosomal subunits might have no functional consequence or might be involved in a yet unknown function of eIF6.
eIF6 is an evolutionarily conserved protein. Proteins structurally homologous to eIF6 have also been found in phylogenetically distant species including archeaons and plants but not in eubacteria (3, 28). The high degree of conservation in amino acid sequence among phylogenetically distant species indicates that eIF6 plays an important role in an essential process in the cell.
ACKNOWLEDGMENTS
U. Basu and K. Si contributed equally to this work.
This work was supported by grants GM15399 to U.M. and GM25532 to J.R.W. from the National Institutes of Health and by Cancer Core Support Grant P30CA13330 from the National Cancer Institute.
We are indebted to Amy Chang of this institution for considerable help in studies on indirect immunofluorescence of eIF6.
REFERENCES
- 1.Aris J P, Blobel G. Isolation of yeast nuclei. Methods Enzymol. 1991;194:735–749. doi: 10.1016/0076-6879(91)94056-i. [DOI] [PubMed] [Google Scholar]
- 2.Berges T, Petfalski E, Tollervey D, Hurt E C. Synthetic lethality with fibrillarin identifies NOP77p, a nucleolar protein required for pre rRNA processing and modification. EMBO J. 1994;13:3136–3148. doi: 10.1002/j.1460-2075.1994.tb06612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biffo S, Sanvito F, Costa S, Preve L, Pignatelli R, Spinardi L, Marchisio P C. Isolation of a novel beta 4 integrin-binding protein (p27BBP) highly expressed in epithelial cells. J Biol Chem. 1997;272:30314–30321. doi: 10.1074/jbc.272.48.30314. [DOI] [PubMed] [Google Scholar]
- 4.Chakravarti D, Maiti T, Maitra U. Isolation and immunochemical characterization of eukaryotic translation initiation factor 5 from Saccharomyces cerevisiae. J Biol Chem. 1993;268:5754–5762. [PubMed] [Google Scholar]
- 5.Deshmukh M, Tsay Y-F, Paulovich A G, Woolford J L., Jr Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits. Mol Cell Biol. 1993;13:2835–2845. doi: 10.1128/mcb.13.5.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisinger D P, Dick F A, Denke E, Trumpower B L. SQT1, which encodes an essential WD domain protein of Saccharomyces cerevisiae, suppresses dominant-negative mutations of the ribosomal protein gene QSR1. Mol Cell Biol. 1997;17:5146–5155. doi: 10.1128/mcb.17.9.5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eisinger D P, Dick F A, Trumpower B L. Qsr1p, a 60S ribosomal subunit protein, is required for joining of 40S and 60S subunits. Mol Cell Biol. 1997;17:5136–5145. doi: 10.1128/mcb.17.9.5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elion E A, Warner J R. The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell. 1984;39:663–673. doi: 10.1016/0092-8674(84)90473-2. [DOI] [PubMed] [Google Scholar]
- 9.Ho J H-N, Johnson A W. NMD3 encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2389–2399. doi: 10.1128/mcb.19.3.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong B, Brockenbrough J S, Wu P, Aris J P. Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol Cell Biol. 1997;17:378–388. doi: 10.1128/mcb.17.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kressler D, Cruz J D-L, Rojo M, Linder P. Dbp6p is an essential putative ATP-dependent RNA helicase required for 60S ribosomal-subunit assembly in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:1855–1865. doi: 10.1128/mcb.18.4.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kressler D, Linder P, Cruz J D-L. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7897–7912. doi: 10.1128/mcb.19.12.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merrick W C, Hershey J W B. The pathway and mechanism of eukaryotic protein synthesis. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 31–69. [Google Scholar]
- 14.Moritz M, Pulaski B A, Woolford J L., Jr Assembly of 60S ribosomal subunits is perturbed in temperature-sensitive yeast mutants defective in ribosomal protein L16. Mol Cell Biol. 1991;11:5681–5692. doi: 10.1128/mcb.11.11.5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moritz M, Paulovich A G, Tsay Y F, Woolford J L., Jr Depletion of yeast ribosomal proteins L16 or rp59 disrupts ribosome assembly. J Cell Biol. 1990;111:2261–2274. doi: 10.1083/jcb.111.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park E-C, Finley D, Szostak J W. A strategy for the generation of conditional mutations by protein destabilization. Proc Natl Acad Sci USA. 1992;89:1249–1252. doi: 10.1073/pnas.89.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raychaudhuri P, Stringer E A, Valenzuela D M, Maitra U. Ribosomal subunit antiassociation activity in rabbit reticulocyte lysates. Evidence for a low molecular weight ribosomal subunit antiassociation protein factor (Mr = 25,000) J Biol Chem. 1984;259:11930–11935. [PubMed] [Google Scholar]
- 18.Ripmaster T L, Vaughn G P, Woolford J L., Jr A putative ATP-dependent RNA helicase involved in Saccharomyces cerevisiae ribosome assembly. Proc Natl Acad Sci USA. 1992;89:11131–11135. doi: 10.1073/pnas.89.23.11131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rose M D, Winston F, Hieter P. Methods in yeast genetics, a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 20.Rotenberg M O, Moritz M, Woolford J L., Jr Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes Dev. 1988;2:160–172. doi: 10.1101/gad.2.2.160. [DOI] [PubMed] [Google Scholar]
- 21.Rothstein R J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 22.Russell D W, Spremulli L L. Purification and characterization of a ribosome dissociation factor (eukaryotic initiation factor 6) from wheat germ. J Biol Chem. 1979;254:8796–8800. [PubMed] [Google Scholar]
- 23.Russell D W, Spremulli L L. Mechanism of action of the wheat germ ribosome dissociation factor: interaction with the 60 S subunit. Arch Biochem Biophys. 1980;201:518–526. doi: 10.1016/0003-9861(80)90540-8. [DOI] [PubMed] [Google Scholar]
- 24.Sachs A B, Davis R W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 26.Sanvito F, Piatti S, Villa A, Bossi M, Lucchini G, Marchisio P C, Biffo S. The β4 integrin interactor p27 (BBP/eIF6) is an essential nuclear matrix protein involved in 60S ribosomal subunit assembly. J Cell Biol. 1999;144:823–837. doi: 10.1083/jcb.144.5.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitt M E, Brown T A, Trumpower B L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Si K, Chaudhuri J, Chevesich J, Maitra U. Molecular cloning and functional expression of a human cDNA encoding translation initiation factor 6. Proc Natl Acad Sci USA. 1997;94:14285–14290. doi: 10.1073/pnas.94.26.14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Si K, Maitra U. The Saccharomyces cerevisiae homologue of mammalian translation initiation factor 6 does not function as a translation initiation factor. Mol Cell Biol. 1999;19:1416–1426. doi: 10.1128/mcb.19.2.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun C, Woolford J L., Jr The yeast NOP4 gene is an essential nucleolar protein required for pre-rRNA processing and accumulation of 60S ribosomal subunits. EMBO J. 1994;13:3127–3135. doi: 10.1002/j.1460-2075.1994.tb06611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tollervey D. Trans-acting factors in ribosome synthesis. Exp Cell Res. 1996;229:226–232. doi: 10.1006/excr.1996.0364. [DOI] [PubMed] [Google Scholar]
- 33.Udem S, Warner J R. Ribosomal RNA synthesis in Saccharomyces cerevisiae. J Mol Biol. 1972;65:227–242. doi: 10.1016/0022-2836(72)90279-3. [DOI] [PubMed] [Google Scholar]
- 34.Valenzuela D M, Chaudhuri A, Maitra U. Eukaryotic ribosomal subunit anti-association activity of calf liver is contained in a single polypeptide chain protein of Mr = 25,500 (eukaryotic initiation factor 6) J Biol Chem. 1982;257:7712–7719. [PubMed] [Google Scholar]
- 35.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 36.Warner J R. Labeling of RNA and phosphoproteins in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:423–427. doi: 10.1016/0076-6879(91)94033-9. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Chang A. Eps1, a novel PDI-related protein involved in ER quality control in yeast. EMBO J. 1999;18:5972–5982. doi: 10.1093/emboj/18.21.5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weaver P L, Sun C, Chang T-H. Dbp3p, a putative RNA helicase in Saccharomyces cerevisiae, is required for efficient pre-rRNA processing predominantly at site A3. Mol Cell Biol. 1997;17:1354–1365. doi: 10.1128/mcb.17.3.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zanchin N I T, Roberts P, DeSilva A, Sherman F, Goldfarb D S. Saccharomyces cerevisiae Nip7p is required for efficient 60S ribosome subunit biogenesis. Mol Cell Biol. 1997;17:5001–5015. doi: 10.1128/mcb.17.9.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]