Summary
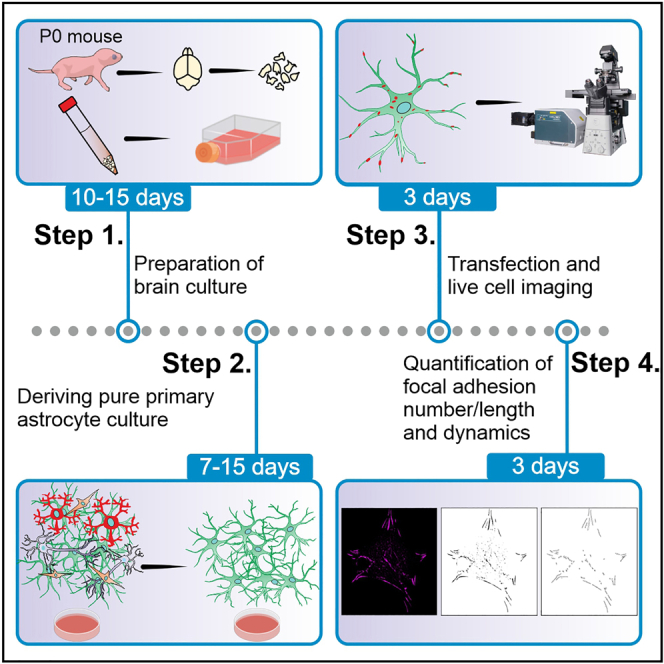
Primary astrocytes have gained attention as an important model for in vitro biological and biochemical research in the last decades. In this protocol, we describe a fast and cost-effective technique for isolating, culturing, and maintaining primary mouse astrocytes at ∼ 80% purity levels, which can be used in in vitro studies for migration and focal adhesion dynamics. In addition, we present an optimized transfection and manual quantification approach for focal adhesion analysis in fixed and living cells.
For complete details on the use and execution of this protocol, please refer to Kusuluri et al. (2021).
Subject areas: Cell Biology, Cell culture, Cell isolation, Microscopy, Neuroscience
Graphical abstract
Highlights
-
•
High purity of primary mouse astrocyte isolation without commercial kits
-
•
Isolated mouse primary astrocytes are functional for downstream applications
-
•
Quantitative analysis of focal adhesion properties in fixed and living astrocytes
Primary astrocytes have gained attention as an important model for in vitro biological and biochemical research in the last decades. In this protocol, we describe a fast and cost-effective technique for isolating, culturing and maintaining primary mouse astrocytes at ∼ 80% purity levels, which can be used in in vitro studies for migration and focal adhesion dynamics. In addition, we present an optimized transfection and manual quantification approach for focal adhesion analysis in fixed and living cells.
Before you begin
Dissection of mice and preparation of brain culture should be performed under sterile conditions. All equipment (Biosafety cabinet, forceps, scissors and plates, etc.) should be sterile to prevent any contamination. In this protocol, the isolation of primary astrocytes cells depends on the differential binding method which separates primary astrocytes, oligodendrocyte progenitor cells (OPCs) and microglia cells based on their different affinities for bacterial grade plastic dishes. Do not use plates that have been treated for animal cell culture because all cell types will attach to these plates with equal affinities. Primary astrocytes were isolated from brains of P0 C57BL/6J (wild type). This protocol was also successfully applied for two mouse models for Usher syndrome 2C namely Vlgr1/del7TM (McMillan and White, 2004) and Drum B mutated mice (Potter et al., 2016), bred on a C57BL/6J background and we did not observe any difference in protocol efficiency. All experiments described herein were performed in accordance with the guidelines provided by Association for Research in Vision and Ophthalmology.
Before starting the experiment several chemicals and surgical instruments should be prepared (For detailed information please see key resources table and materials and equipment section).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-GFAP (Glial fibrillary acidic protein), working dilution 1 in 1000 | DAKO-Agilent | Cat#ZO334 |
| Mouse monoclonal anti-GFAP (Glial fibrillary acidic protein), working dilution 1 in 1000 | Sigma-Aldrich | Cat#G3839 |
| Guineapig monoclonal anti-MAP2 (Microtubule associated protein), working dilution 1 in 1000 | Synaptic systems | Cat#188004 |
| Mouse monoclonal anti-Vinculin, working dilution 1 in 100 | Sigma-Aldrich | Cat#V9131 |
| Rabbit polyclonal anti- PDGFR-α, working dilution 1 in 200 | Santa Cruz Biotechnology, Inc. | Cat#sc-338 |
| Rat monoclonal anti-CD68, working dilution 1 in 200 | Bio-Rad Laboratories | Cat#MCA1957 |
| Chemicals, peptides, and recombinant proteins | ||
| Alexa Fluor 488-conjugated goat anti-rabbit IgG, working dilution 1 in 400 | Molecular Probes | Cat#A-11034 |
| Alexa Fluor 568-conjugated goat anti-rat IgG, working dilution 1 in 400 | Biotrend | Cat#20092-1 |
| Alexa Fluor 640-conjugated goat anti-mouse IgG, working dilution 1 in 200 | Biotrend | Cat#20177 |
| Phalloidin-TRITC, working dilution 1 in 400 | Sigma-Aldrich | Cat#P1951 |
| Dulbecco`s Modified Eagle Medium (DMEM) | GibcoTM | Cat#31966-021 |
| Fetal bovine serum (FBS) | GibcoTM | Cat#16000044 |
| Penicillin-streptomycin | GibcoTM | Cat#15140122 |
| 10× Hanks’ Balanced Salt Solution (HBSS) | GibcoTM | Cat#14065-056 |
| 1 M HEPES solution | GibcoTM | Cat#15630106 |
| DNase I, recombinant, Grade II, from bovine pancreas | Merck | Cat#10104159001 |
| 0.01% Poly-L-Lysine solution | Sigma-Aldrich | Cat#25988-63-0 |
| 0.05% Trypsin-EDTA (1×) | GibcoTM | Cat#25300-054 |
| 1×PBS | GibcoTM | Cat#14190-094 |
| Opti-MEM™ I Reduced Serum Medium | Thermo Fisher Scientific | Cat#31985062 |
| GeneJuice® Transfection Reagent | Merck | Cat#70967-6 |
| Fibronectin bovine plasma | Merck | Cat#F4759-5MG |
| Albumin crude from chicken egg = Ovalbumin | PanReac AppliChem | Cat#A4344 |
| Gelatin From Cold Water Fish Skin, 40–50% in Water | Sigma-Aldrich | Cat#G7765-250ML |
| Deposited data | ||
| Raw and analyzed data | This paper and Kusuluri et al. (2021) | N/A |
| Experimental models: organisms/strains | ||
| Mouse: C57BL/6J (P0 age) | The Jackson Laboratory | Stock no: 000664 |
| Mouse: Vlgr1/del7TM (P0 age) | Breed on a C57BL/6 background | McMillan and White (2004) |
| Mouse: Drum B (P0 age) | Breed on a C57BL/6 background | Potter et al. (2016) and Kusuluri et al. (2021) |
| Recombinant DNA | ||
| RFP-Paxillin | Rudolf E. Leube, Rick Horwitz | Rudolf E. Leube, Rick Horwitz |
| Software and algorithms | ||
| ImageJ software | National Institutes of Health (NIH) | http://imagej.nih.gov/ij/download.html |
| Focal adhesion analysis server | https://faas.bme.unc.edu | Berginski and Gomez (2013) |
| Other | ||
| Stereomicroscope | Leica Microsystems | Leica WILD M3B |
| Cold light source | Leica Microsystems | Schott KL750 |
| Inverted microscope | Nikon Instruments Inc. | Nikon Eclipse Ti2-E |
| Spinning disc unit | Yokogawa Electric Corporation | CSU-W1 |
| Leica confocal microscope | Leica Microsystems | DM6000B |
| Centrifuge | Eppendorf | Centrifuge 5430 R |
| Medium-sized scissors | A. Dumont & Fils | Cat#T5074 |
| Dumont #3 curved forceps | A. Dumont & Fils | Cat#T504 |
| Dumont #4 standard tip forceps | A. Dumont & Fils | Cat#T505 |
| Dumont #7 standard tip forceps | A. Dumont & Fils | Cat#T508 |
| Vannas spring scissors | A. Dumont & Fils | Cat#T5322 |
| 100 mm petri dish (cell culture) | Greiner Bio-One | Cat#664160 |
| 100 mm bacterial grade petri dish | SARSTEDT AG & Co. KG | Cat#82.1472 |
| T-75 culture flasks | Greiner Bio-One | Cat#658170 |
| 6-well culture plates | Greiner Bio-One | Cat#657160 |
| 15 mL Centrifuge Tubes | SARSTEDT AG & Co. KG | Cat#62.554.502 |
| 10 mm glass coverslips | Carl Roth GmbH +Co. KG | Cat#YX02.1 |
| 0.22 μm filter | Carl Roth GmbH +Co. KG | Cat#KH541 |
| 0.45 μm filter | Carl Roth GmbH +Co. KG | Cat#CCX9.1 |
| μ-Slide 4 Well chamber | Ibidi GmbH | Cat#80426 |
Materials and equipment
Growth medium
| Reagent | Final concentration | Amount |
|---|---|---|
| Dulbecco‘s Modified Eagle Medium (DMEM) | n/a | 450 mL |
| Fetal bovine serum (FBS) | 10% | 50 mL |
| Penicillin-streptomycin | 2% | 11 mL |
| Total | n/a | 511 mL |
CRITICAL: Final antibiotic concentration may be adjusted, since different antibiotic concentrations can affect cell proliferation and differentiation.
Note: Growth medium should be kept at +4°C for a maximum of 1 month. If there is a pH level change during storage which can be distinguished by a color change, new medium should be prepared.
1× HBSS for brain tissue storage during dissection
| Reagent | Final concentration | Amount |
|---|---|---|
| 10× HBSS | 1× | 100 mL |
| 1 M HEPES | 0.01 M | 10 mL |
| ddH2O | n/a | 890 mL |
| Total | n/a | 1000 mL |
CRITICAL: Adjust pH 7.0 of buffer using 1 N HCl or 1 N NaOH and sterile medium by passing through a 0.22 μm filter.
Note: Medium should be stored at +4°C for a maximum of 1 month. If there is a pH level change during storage which can be distinguished by the color change in medium, the new medium should be prepared.
DNase I solution
| Reagent | Final concentration | Amount |
|---|---|---|
| DNase I | 0.05% | 100 mg |
| 1× HBSS | n/a | 200 mL |
| Total | n/a | 200 mL |
CRITICAL: Do not dissolve the DNase I by using vortex as mechanical force may denaturate the enzyme which can cause decrease in the activity. The solution should be prepared by pipetting and sterilized by passing through a 0.22 μm filter. To prevent any freeze/thaw cycles which may reduce activity, 1–5 mL aliquots can be prepared and kept at –20°C or –80°C for a maximum of 6 months.
Blocking solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Ovalbumin | 0.1% | 0.1 g |
| 40–50% Fish gelatin | 0.5% | 1.1 g |
| 1× PBS | n/a | 100 mL |
| Total | n/a | 100 mL |
Note: Mix the Ovalbumin and 40%–50% Fish gelatin in 100 mL PBS for 12–16 h. The day after, bring the temperature of the solution to 50°C. Centrifuge for 10 min at 3000 RCF to remove undissolved particles. Sterile medium by passing through a 0.45 μm filter. In order to prevent freeze/thaw cycles, prepare 1–2 mL aliquots and keep at −20°C for a maximum of 12 months.
Step-by-step method details
Preparation of primary brain culture
Timing: 10–15 days
Before starting dissection of mice pups, place 10 mL of 1× HBSS (in a 15 mL Falcon tube), 1 mL of 0.05% DNase I and 1 mL of 0.05% trypsin on ice and arrange dissection tools, microscope, a cold light source and 70% ethanol under the dissection hood. This step includes dissection and preparation of primary brain cultures. To obtain a confluent layer of primary astrocytes cells, collect 3–4 pups’ brains in a 15 mL tube containing 1× HBSS solution.
Preparation of brain tissue
Timing: 15–30 min
-
1.
Before starting the dissection, gently hold the mice pup and spray the neck with 70% of ethanol to prevent contamination.
-
2.
Decapitation of mice can be done by using sharp scissors in one cut.
CRITICAL: Brain isolation can take time due to inexperience. In order to achieve high efficiency after the procedure and healthy brain culture, only one decapitation should be applied at a time and the remaining decapitation procedures should be carried out after step 14 is finished from the first mouse.
-
3.
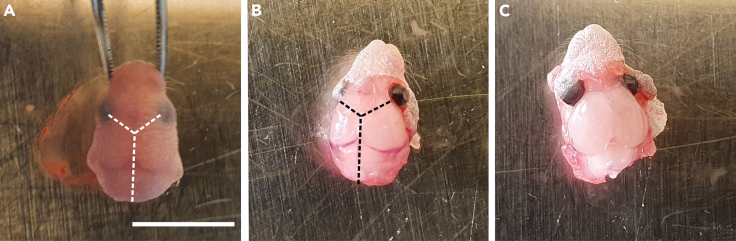
To keep the head fixed and stabile use anchoring forceps at orbital cavities (Figure 1A).
-
4.
Make an incision of the skin at the hindbrain and follow along the midline to eye level where transverse cuts to the eye cavity will be made (Figure 1B).
-
5.
Grasp the skin from both sides by using Dumont #5 forceps and pull it aside to reveal the skull (Figure 1C).
-
6.
Carefully cut the cranium using a small scissors. The cut should start from the neck where the vertebral foramen is located and extended anteriorly to the nose. Make additional transverse cuts to the eye cavity.
CRITICAL: While cutting the cranium be sure that no excess pressure is applied to the brain. This may cause deformation in brain tissue and bleeding, which can lower culture purity because of contamination with red blood cells and endothelial cells.
-
7.
Carefully pull apart the cranium using forceps to reveal the brain.
-
8.
Take out the brain with the help of a small spatula. First, place the spatula under the cerebellum and gently push it towards the olfactory bulbs and scoop out by lifting. This will disconnect the brain from the skull base.
-
9.
Transfer the brain to a 10 cm petri dish containing ice-cold 1× HBSS under sterile conditions. The brain should submerge completely in the liquid.
-
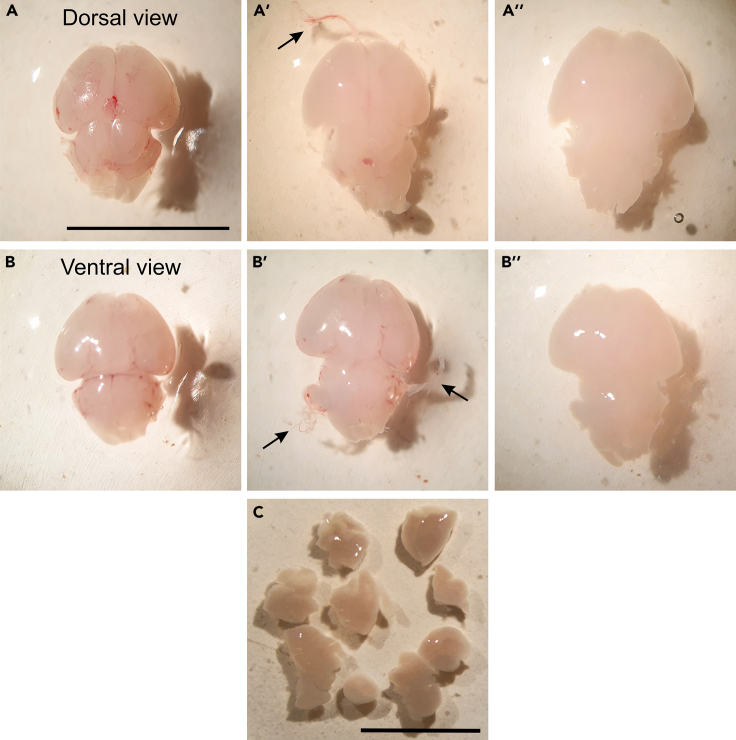
10.
Remove olfactory bulbs in order to free the meninges. Meninges can be distinguished by their reddish color resulting from blood vessels (Figures 2A and 2B).
-
11.
Reversing the brain and starting from the bottom side can help to remove meninges easily. Dissect the meninges from the surface of the cortical layer by using fine forceps.
Note: We observed that keeping brain integrity during the process may reduce the risk of meningeal cells and fibroblast contamination and ensure full removal.
-
12.
Flip the brain and continue meninges removal.
-
13.
Carefully check whether there is any meninges residue and if not, cut the brain into 6–10 pieces using forceps or sharp blades in order to increase disassociation efficiency (Figure 2C).
-
14.
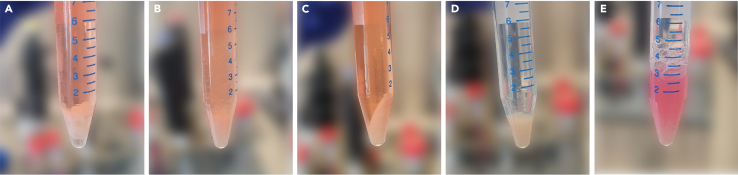
Place the brain pieces into a 15 mL tubes that contain 10 mL 1× HBSS and keep on ice (Figure 3A).
CRITICAL: Only 3–4 brains should be collected in 10 mL 1× HBSS. If more brain tissue is needed, use a 50 mL Falcon and increase the volume of 1× HBSS for 5 brains to 11 mL and for 12 brains 20 mL. If the brain pieces are not submerged completely, tap the bottom of the Falcon tube. This may help to submerge brain pieces that float on top.
Note: After obtaining meninges-free brain, repeat the steps 1–14 for the rest of the mice. Doing only one brain at a time will ensure healthy cultures.
-
15.
Under the sterile condition, add ice cold 0.05% DNase I into the tube and gently triturate brain tissue 10–15 times with a 10 mL and 2 mL pipette, respectively.
-
16.
Afterwards, add 0.05% trypsin, mix by using 10 mL pipette 20–30 times and incubate at room temperature (22°C–25°C) for 20 min.
CRITICAL: The amount of DNase I and trypsin can be adjusted depending on the number of brains in one tube. For 3–4 brains; 1 mL of DNase I and 1 mL of trypsin, for 5 brains; 1.5 mL of DNase I and 1.5 mL of trypsin, for 12 brains; 3 mL of DNase I and 3 mL of trypsin should be added.
-
17.
During incubation time, prepare one T-75 flask for 3–4 brains and coat with 3 mL of 0.01% Poly-L-Lysine (PLL) for 20 min. Place the T-75 flask in the incubator.
-
18.
20 min after incubation of brain pieces mix the Falcon tube thoroughly (Figure 3B).
-
19.
Centrifuge for 10 min at 150 RCF to the pellet brain tissue (Figure 3C).
-
20.
After centrifugation, carefully aspirate the supernatant using a Pasteur pipette connected to a pump (Figure 3D).
Note: If there is any meninges residue or blood vessel contamination, which can be distinguished by a reddish color on top of the pellet, also aspirate and discard this layer. Resuspend pellet with 2 mL of complete growth medium and repeat centrifugation step.
-
21.
Resuspend cell pellet with 2–4 mL of DMEM with a 10 mL pipette and mix thoroughly 10–20 times in order to obtain a single-cell suspension (Figure 3E).
-
22.
Aspirate PLL from the T-75 flask and rinse with 10 mL of DMEM to completely wash out any residue. Add 8–10 mL of pre-warmed 37°C DMEM supplemented with 10% FBS and 2% pen/strep.
-
23.
Plate the resuspended cell pellet and incubate at 37°C with 5% CO2 in an incubator.
-
24.
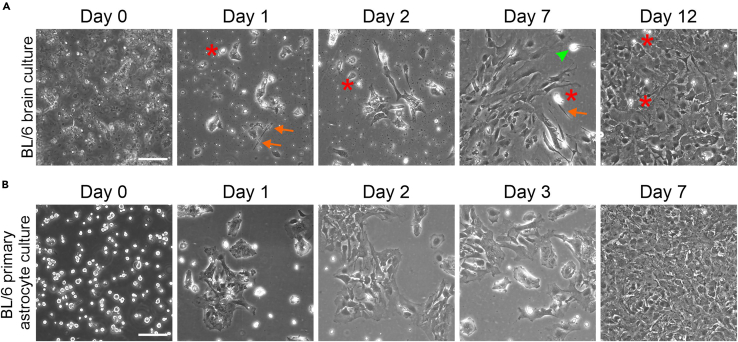
Change the medium on day 1, day 2 and day 7 after plating the primary cells (Figure 7A).
Note: In order to remove unattached and death cells wash the plate by using 1× PBS, before adding fresh complete growth medium.
Figure 1.
Dissection of the brain from PN0 mouse
(A and B) (A) Cut should be performed to remove the skin and (B) skull layers covering the brain. Dashed lines indicate the cuts using Vannas spring scissors. The cut starts from the neck where the vertebral foramen is located and extends anteriorly to the nose.
(C) Exposed brain. Scale bar: 5 mm.
Figure 2.
Dissection of mouse brain at P0
(A and B) Removing the meninges from mouse brain. (A) Dorsal view (B) and ventral view of isolated mouse brain. Meninges can be distinguished by reddish color in the cortex of the mouse brain. (A′ and B′) Meninges removal from the cortex of the mouse brain. Arrows indicate removed meninges from the cortex of the mouse brain. (A″ and B″) Dorsal and ventral view of completely meninges-free brain.
(C) Dissection of brain parts with a sharp razor blade or fine forceps. Scale bars: A and B: 10 mm; C: 5 mm.
Figure 3.
Dissociation of mouse brain tissue for primary cell cultivation
(A) Brain tissue dissected from wild type mice in 1× HBSS.
(B) Homogenized mouse brain tissue (MBT).
(C) MBT pellet after centrifugation.
(D) MBT pellet after removing supernatant.
(E) Resuspended MBT pellet in complete growth medium.
Figure 7.
Growth of BL/6 mouse primary astrocyte cells in mixed brain culture and pure culture
(A) Representative differential interference contrast (DIC) images show growth of different cell types during culturing days before applying the differential binding method.
(B) Primary astrocytes in culture after applying the differential binding method. After differential binding method primary astrocytes reached the confluency. Red asterisks indicate microglia cells, arrows indicate fibroblast cells and green arrowhead indicates a neuron. Scale bars: 100 μm.
Deriving pure primary mouse astrocyte culture
Timing: 7–15 days
This step will be used for the separation of primary astrocytes cells from oligodendrocyte progenitor cells (OPCs), microglia and neurons. In confluence, their tight arrangement at the bottom of the culture flask can distinguish primary astrocytes cells from other cell types. Widefield microcopy distinguishes microglia cells from other brain-derived cells revealing a bright and small rounded cell morphology.
Removal of oligodendrocyte progenitor cells (OPCs)
Timing: 10–20 min
-
25.
Mixed primary cell cultures which were obtained in steps 1 to 24 reach full confluency on days 10–15. When mixed cell culture reach confluency, gently tap the flask from both sides several times in order to detach the OPCs layer. After tapping the flask, check the cells under the microscope. Detach OPCs can be distinguished as floating cells in the plate (Figure 4).
Note: In case of incomplete detachment repeat tapping several times. Tapping should be done gently so as not detach the astrocyte layer.
-
26.
After the OPCs are released aspirate the medium completely to remove detached cells and rinse remaining cells carefully with 10 mL of complete growth medium.
Note: To avoid any loosening or loss of astrocyte layer, medium aspiration and rinsing should be done carefully. A 10 mL pipette should be placed at the corner of the flask and the medium should be slowly added against the wall of the flask and not directly onto the attached cells.
-
27.
After removing OPCs, add 10 mL of complete growth medium and ice cold 0.5 mL DNase I to loosen the remaining cell layer and incubate 5 min at 37°C with 5% CO2 in an incubator.
-
28.
After DNase I treatment, the remaining OPCs are released by horizontal shaking the T-75 Flask 20–30 times.
Note: While too gentle shaking may cause insufficient OPCs removal, too intense shaking can damage the astrocyte layer. Shaking should be performed at a steady speed by hand.
CRITICAL: Shaking can cause foaming; therefore, the shaken flask should be held vertically for 30 s to allow the foam to float on the top.
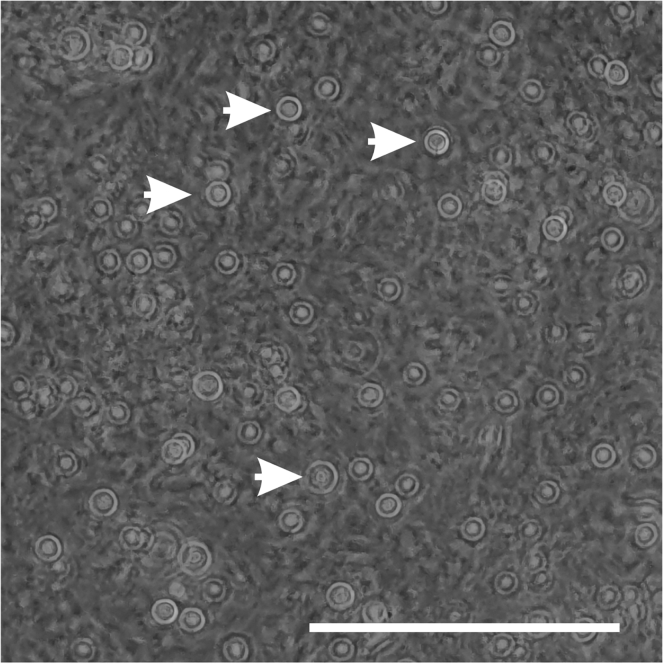
Figure 4.
Detached oligodendrocyte progenitor cells after tapping the flask
OPCs are distinguished as floating cells above the monolayer of astrocytes and microglia. Arrow heads indicate round shaped floating OPCs. Scale bars: 100 μm.
Removal of microglia
Timing: 2–2.30 h
-
29.
Aspirate the medium which may still contains OPCs, and wash the astrocytes and microglia layer with 3–4 mL PBS.
-
30.
Add 3–4 mL of 0.05% trypsin and incubate 3–5 min at 37°C with 5% CO2 in an incubator.
CRITICAL: Cells should not be incubated with trypsin for more than 5 min. Longer incubation time may harm primary astrocytes and results in an unhealthy culture.
-
31.
After incubation, check the cells under the microscope. If there is incomplete detachment, tap the slide of the flask several times and check under the microscope again.
-
32.
After trypsinization, add 6–7 mL of complete growth medium and resuspend the cells.
-
33.
Transfer the suspension to a new 15 mL Falcon tube and pellet detached cells by centrifugation for 10 min at 150 rcf.
-
34.
During centrifugation, prepare 4 sterile 10-cm bacterial grade plates. Add 2 mL of complete growth medium in each of 4 Petri dishes.
-
35.
Aspirate the supernatant which you obtain after step 33 and resuspend cells with 4 mL of complete growth medium.
-
36.
Add 1 mL of the cell suspension to each petri dish (Figure 5A) and incubate at 37°C with 5% CO2 in the incubator for 20 min (Figure 5B).
Note: In this step, remaining microglia cells should adhere to the bacterial grade plate while primary astrocyte cells should not.
-
37.
After incubation, collect all supernatants containing astrocytes of all 4 Petri dishes and pool them to a new 10-cm bacterial grade plate (Figure 5C).
CRITICAL: The transfer of supernatant should be performed carefully as shaking of the plate may result in loosening bound microglia. Gently tilt the plate and collect the supernatant from the side of the plate.
-
38.
Incubate the supernatant (obtained in step 37) contains ∼10–12 mL medium at 37°C with 5% CO2 in the incubator for 90 min (Figure 5D).
Note: After incubation, check the cells under the microscope. Microglia cells should completely adhere after this step and primary astrocytes can be distinguished as floating cells in the culture medium.
-
39.
Transfer the supernatant containing primary astrocytes into a new T-75 flask.
Note: According to volume of the obtained supernatant, add additional amount of pre-warmed complete growth medium to the T-75 flask to fill up to 20 mL.
-
40.
Incubate the primary astrocytes for 7–15 days at 37°C with 5% CO2 in the incubator. Primary astrocytes are reaching confluency in between day 7–15 after isolation. After culture reach confluency, primary astrocytes can be used for downstream experiments (Figure 7B).
CRITICAL: Change the complete growth medium by aspirating half of the old medium (approx. 10 mL) and replacing it with a fresh, pre-warmed complete growth medium (approx. 10 mL) every 4 days during cultivation. This will provide you with a conditioned medium that will help maintain a healthy culture.
Figure 5.
Differential surface binding of primary astrocytes from mouse brain
(A) Whole brain cell suspension which obtained in step 35.
(B) Red arrow heads indicate settled microglia after 20 min of incubation.
(C) Transferring the supernatant which obtained in step 37 to a new bacterial grade plate.
(D) Red arrows indicate settled microglia after 90 min of incubation. Scale bar: 50 μm.
Transfection of isolated primary astrocytes and live-cell imaging
Timing: 3 days
In these steps, we describe how to transfect primary mouse astrocytes with a focal adhesion (FA) construct namely RFP-Paxillin and how to perform live imaging to investigate FA dynamics. For live-cell imaging, we use a μ-Slide 4 Well chamber (Ibidi, Munich, Germany) and for transfection, we use GeneJuice® transfection reagent (Merck, Darmstadt, Germany).
-
41.
First, coat the μ-Slide 4 Well chamber with 300 μL of fibronectin 5 μg/mL dissolved in dH2O for 1 h at 37°C.
-
42.
Next, remove the fibronectin from the wells and wash the wells once with 500 μL of sterile 1× PBS or water. After the washing, place the chamber in the incubator in order to dry it.
Note: Fibronectin solution can be used up to 3 times, therefore discarded fibronectin can be store at −20°C for further usage.
CRITICAL: If the purpose of the experiment is to study migration or focal adhesion dynamics, we strongly recommend the usage of fibronectin instead of poly-L-Lysine coating, since fibronectin may enhance cell adhesion and spreading (Blau 2013).
-
43.
Wash the primary astrocytes when reach confluency which are cultured in T-75 flask with 1× PBS and add 3 mL of 0.05% trypsin for 5 min at 37°C to detach cells.
-
44.
Collect the detached cells in a 15 mL tube and add 1–5 mL of growth media for preparing cell suspension. Count the number of cells and prepare 5 × 104 cell/mL suspension.
-
45.
Add 700 μL (∼35,000 cells) of cell suspension into each well and incubate 1 day at 37°C with 5% CO2 in the incubator.
Note: Cell number can be adjusted based on chamber size used. Cells should be at least 50% confluent before transfection.
-
46.
The day after incubation, change the medium to fresh 700 μL of complete growth medium.
Note: Transfection reagent volumes and DNA amount can be scaled up if more than 4 wells are needed.
-
47.
For the single transfection of focal adhesion construct, prepare 1 Eppendorf tube and add 100 μL of Opti-MEMTM and add 6 μL of GeneJuice®. Vortex the solution thoroughly to mix it and incubate at room temperature for 5 min.
-
48.
Add 2 μL of focal adhesion construct at a 1 μg/mL concentration into the serum-free/ GeneJuice® mixture and mix it by tapping the tubes and incubate at room temperature for 15 min.
CRITICAL: Do not vortex the solution after adding the DNA construct, vigorous mixing can cause low transfection efficiency.
-
49.
Drop-wisely add 27 μL of reagent/DNA mixture to the cells and rock the chamber to homogenize the solution in the complete growth medium.
-
50.
After 24 h of transfection remove the old medium which contains transfection reagent and add pre-warmed complete growth medium and incubate the cells for additional 24 h at 37°C with 5% CO2 in the incubator.
-
51.
Before starting live-cell imaging, turn on the temperature and CO2 controller of Nikon Eclipse Ti2-E/Yokogawa CSU-W1 Spinning disk microscope and wait till the condition stabilizes for usage.
-
52.
Acquire the movies at 1 frame/3–5 min with 20 z-stacks at 0.5 μm step size for 1.25 h to 3-h time course (Methods video S1).
Note: Time interval between frames and z-stacks size and steps can be adjusted based on your experimental setup. After determining the parameter, all movies should be acquired using the same settings in order to provide experimental consistence.
-
53.
Apply maximum projections to the movies and analyze focal adhesion assembly/disassembly rates by using Focal Adhesion Analysis Server (FAAS) (Figure 10) (Berginski and Gomez 2013).
Note: Microtubule induced focal adhesion disassembly experiment can be used as a control experiment in order to establish the protocol (Ezratty et al., 2005).
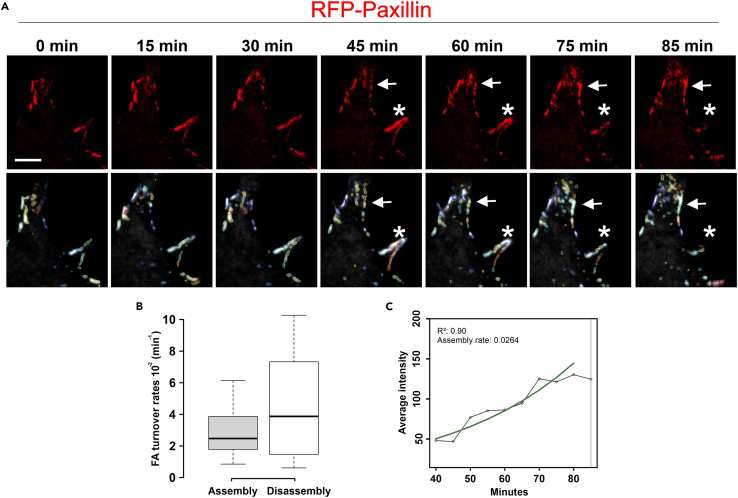
Figure 10.
Analysis of assembly and disassembly of focal adhesions by life cell imaging in RFP-Paxillin transfected BL/6 mouse primary astrocytes
(A) Tracking of RFP-Paxillin molecules in focal adhesions (FAs) by live cell imaging. Images were acquired for 85 min with 3 min time intervals. Arrows point to an individual FA which assembles and asterisk indicated a FA which disassembles during the time course of imaging. Lower panel shows color coded individual FAs identified and analyzed via the Focal adhesion analysis server (FAAS) (Berginski and Gomez 2013).
(B) Assembly and disassembly rates of FAs in a single BL/6 primary mouse astrocyte.
(C) Automated intensity analysis of RFP-paxillin in an individual FAs during assembly by FAAS. Green line in the intensity plot indicates the trend line of assembling FA. Scale bar: 10 μm.
Images were acquired for 85 min with 3 min time intervals. Live-cell imaging performed under 5% CO2 and at 37°C using Nikon Eclipse Ti2-E equipped with a spinning disc microscope. Scale bar: 20 μm.
Immunostaining of focal adhesion molecules in fixed primary astrocytes
Timing: 4 days
We used a Leica DM6000B microscope (Leica, Bensheim, Germany) for imaging of specimens.
-
54.
Prepare 10 mm glass coverslips in 6-well plates and coat with fibronectin as described in step 41–42.
-
55.
Seed 1–1.5 × 105 cells in each well and incubate the primary astrocytes cells for 48 h at 37°C with 5% CO2 in the incubator.
-
56.
After the 48 h incubation, carefully remove the growth medium and wash one time rapidly with 1× PBS and do second time washing using 1× PBS for 10 min in room temperature.
-
57.
Prepare a parafilm sheet and place it a wet chamber for keeping humidity during incubation times. Take the coverslips out from wells, place onto parafilm sheet and fix the cells by using 50 μL of 2% of PFA in 1× PBS for each coverslip for 10 min at room temperature.
-
58.
Wash the cells 2 times for 10 min with 1× PBS to remove any remaining PFA.
-
59.
Permeabilize the cells with 50 μL of 0.2% Triton-X-100 in PBS solution.
-
60.
Remove the permeabilization solution and wash cells with 50 μL of 50 mM NH4Cl in PBS for 5 min in room temperature.
-
61.
Incubate the cells with 50 μL of blocking solution (0.1% Ovalbumin and 0.5% Fish gelatin in 1× PBS) for 60 min in room temperature.
-
62.
After incubation, treat the cells with 50 μL of primary antibodies 12–16 h at 4°C. We used rabbit anti-GFAP (1:400 dilution) and mouse anti-vinculin (1:200 dilution) prepared in blocking solution.
Note: Primary antibodies can be incubated at room temperature for 2–3 h.
-
63.
After primary antibody incubation, wash the cells with 1× PBS one time rapidly and two additional times for 10 min.
-
64.
Apply the 50 μL of secondary antibodies for each coverslip. Alexa Flour 488-conjugated goat anti-rabbit 488 IgG antibody (1:400 diluted in blocking solution), Alexa Fluor 640-conjugated goat anti-mouse IgG antibody (1:400 diluted in blocking solution) and the dyes TRITC-phalloidin (1:400 diluted in blocking solution) for labeling actin filaments (F-actin) and 4′,6-Diamidino-2-Phenylindole (DAPI) (1:400 diluted in blocking solution) for counterstaining of nucleus were prepared in a 1.5 mL tube. Incubate the cells with antibodies at room temperature for 60–75 min in room temperature.
-
65.
Wash the cells with 1× PBS one time rapidly and three times for 10 min.
-
66.
Mount the coverslips with Mowiol.
CRITICAL: Step 63–66 should be conducted in subdued lighting to prevent bleaching of the fluorescent probes.
Quantification of number and length of focal adhesion molecules in the primary astrocyte cells
Timing: 1–2 days
Images were acquired on Leica DM6000B microscope (Leica, Bensheim, Germany) with a 63× objective and 20 cells for each condition were used for quantification. The number and length of focal adhesion molecules were quantified by using Fiji (Figure 6). For this, we modified a protocol which previously described (Horzum et al., 2014).
-
67.
Open the images using ImageJ software.
-
68.If Z-projection is not created
-
a.Go to Image-Stacks-Z Project-Maximum Projection.
-
b.Save the processed image in the folder.
-
a.
-
69.
Pick an exported and processed image from the images folder.
-
70.To set the scale: First, draw a straight line on the image scale bar.
-
a.Go to Analyze-Set scale
-
b.And mark as global.
-
a.
-
71.
Verify the scale by drawing a line on the scale bar.
-
72.
Make a duplicate image named DUP1 for the RGB image (for comparison).
-
73.Draw a scale bar in the image near to the cell to be cropped.
-
a.Analyze-Tools-Scale bar
-
a.
-
74.
Now, select the Rectangle box symbol from the imageJ toolbar and draw around the cell.
-
75.Then right-click within the drawn box and press duplicate.
-
a.Go to Image-Adjust-Brightness and contrast for adjustments and then save the image in the folder.
-
a.
-
76.
Again, go to 1st duplicate RGB image and again right-click within yellow box for the FFT image.
-
77.Separate the merged imaged
-
a.Go to Image-Color-Split channels
-
b.And pick the channels which contain focal adhesion staining
-
a.
-
78.To acquire the FFT image, change the RGB image to an 8-bit image.
-
a.Image-Type 8-bit
-
a.
-
79.In order to subtract background
-
a.Process-FFT Bandpass filter
-
b.Set pixel size for large structure to 20 pixels and small 1 pixel.
-
c.Adjust tolerance of direction to 5%
-
d.Select Autoscale after filtering
-
e.Select Saturate image when autoscaling
-
f.And press OK
-
a.
-
80.Go to Image-Adjust-Brightness
-
a.Adjust brightness till getting black background and save the Min/Max values for applying to other images.
-
b.Adjust maximum & then minimum.
-
a.
-
81.Save the FFT filtered image.
-
a.Duplicate image.
-
b.Image-Type-RGB color as in this step you cannot save 8-bit image.
-
c.Close RGB image.
-
a.
-
82.Go to Image-Adjust-Threshold
-
a.Select Dark Background.
-
b.In first drop-down box select-Huang.
-
c.In the second drop-down box select-B&W.
-
d.Then apply.
-
a.
-
83.Run analyze particles.
-
a.Set the parameter for size: 40-Infinity and for circularity 0.00–0.99.
-
b.Select pixel units and show the outlines option.
-
c.Select display results, clear results, summarize & include holes.
-
a.
-
84.Quantification of focal adhesion numbers per cell area
-
a.Open the image with F-actin staining.
-
b.Draw a line around the cell body using the freehand tool in imageJ tool bar.
-
c.Press control+M, this will give the area of the cell body.
-
d.Save the results.
-
e.Transfer the data to an Excel file.
-
f.Count the number of focal adhesions and divide it by the total cell area.
-
a.
-
85.
Use Mann-Whitney-Wilcoxon Test for the significant test.
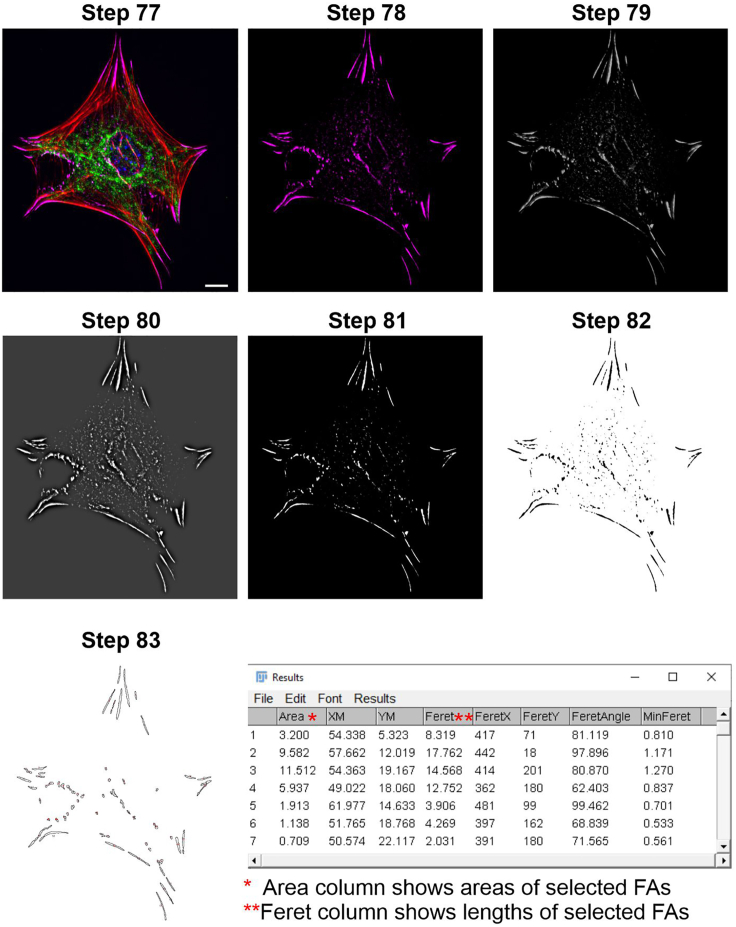
Figure 6.
Step by step quantification of focal adhesion (FA) numbers and lengths
Step 77: Representative image shows merged image of primary mouse astrocytes which co-labeled with vinculin marker (magenta), actin filaments (red), GFAP (green) and for nuclear staining DAPI (blue). Step 78: After splitting channels, only vinculin staining is shown. Step 79: 8-bit image. Step 80: FFT Bandpass filter applied image. Step 81–83: Subtracting background and detecting FAs in the cell. Scale bar: 10 μm.
Expected outcomes
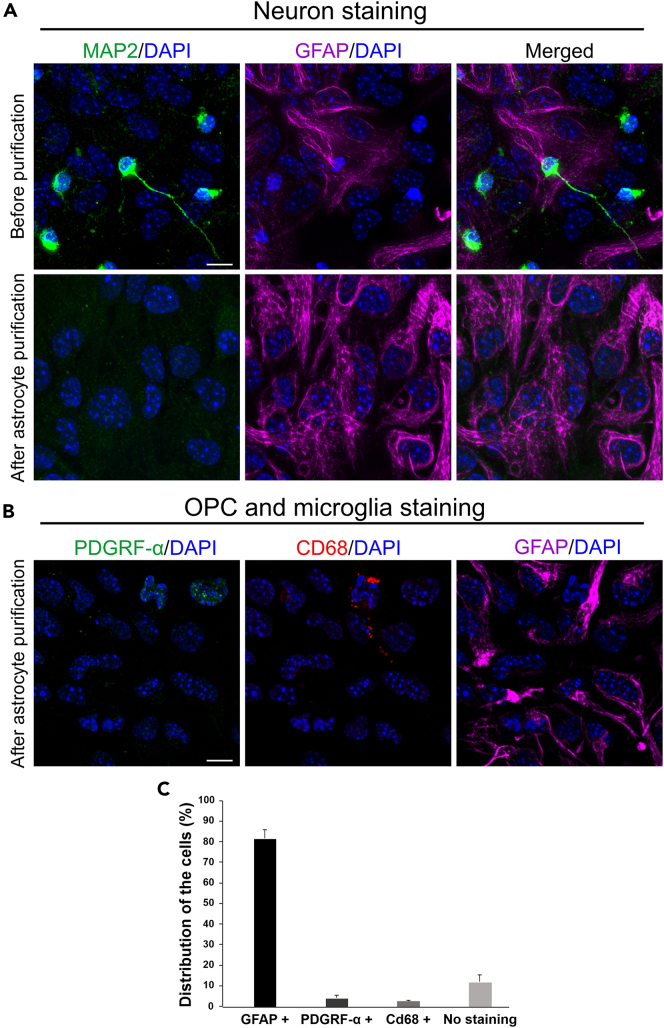
In this protocol, we provide a cost-efficient, reproducible isolation of primary astrocytes from P0 BL/6 mice without the requirement of commercial kits or special equipment. Applying our protocol, a healthy and easy to maintain primary astrocyte culture can be obtained from wild type or mutant mice strains (Figure 7). Furthermore, and most importantly, the purity of the primary astrocyte cell culture is suitable for studying several astrocytes dependent or related physiological processes. Isolated primary astrocytes can be identified by indirect immunofluorescence using antibodies against the glial fibrillary acidic protein (GFAP). In addition, we applied immunofluorescence staining of microtubule-associated protein (MAP2), PDGRF-alpha and CD68 as marker, respectively, for the identification primary neurons, OPC and microglia, to confirm the purity of astrocyte culture (Figure 8).
Figure 8.
Representative indirect immunostaining of BL/6 mouse brain primary cell cultures
(A) Indirect immunofluorescence double staining of MAP2 (green) and primary astrocyte marker GFAP (magenta), as common markers for primary neurons and primary astrocytes, counterstained by DAPI (blue) for nuclear DNA, before and after the purification of astrocytes.
(B) Immunofluorescence staining of a primary astrocyte culture after primary astrocyte purification. OPC, microglia and primary astrocytes cells were stained by antibodies against PDGRF-α, CD68 and GFAP, respectively.
(C) Quantification of the number of GFAP, PDGRF-α and CD68-stained positive cells after astrocyte purifications demonstrates that primary astrocytes are the most prominent cell type (∼80%). N = 360–490 cells per condition, 3 independent experiments. Scale bar: 10 μm.
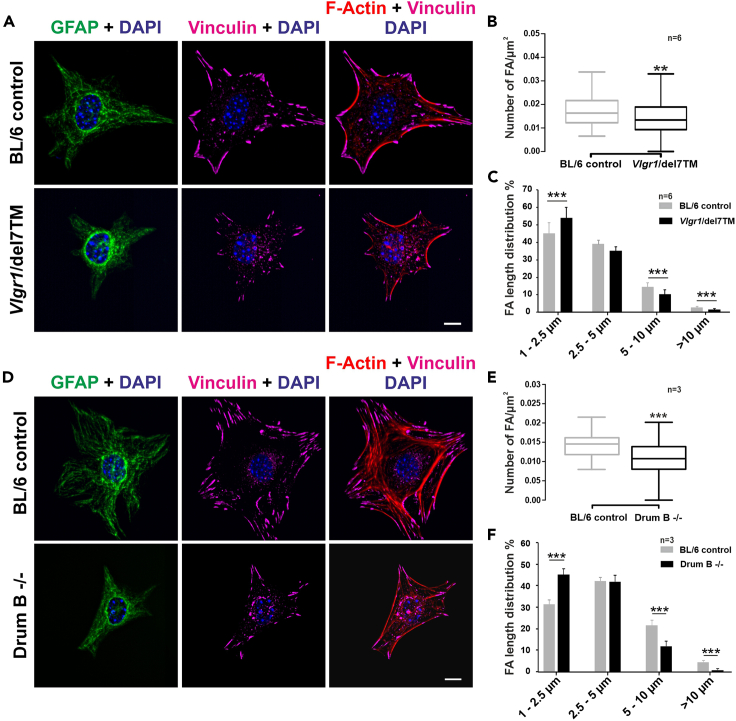
We applied our isolation protocol described above for the analysis of focal adhesion dynamics in murine isolated primary astrocytes (Figure 9). Focal adhesions (FAs) are highly dynamic molecules which serve as bi-directional signaling hub in the cell by sensing environmental (outside-in) and intracellular cues (inside-out) (Shen et al., 2012; Sun et al., 2016). With this unique bi-directional signaling feature FA molecules play essential roles in the migration and spreading (Kim et al., 2012; Kim and Wirtz 2013). Namely, size and turnover of FAs are highly important for the migration capacity of the cell. We recently showed that the adhesion G protein coupled receptor (GPCR) ADGRV1/VLGR1 (very large G protein coupled-receptor-1) is a component of FAs and mutations in VLGR1 cause defects in FA growth and maturation processes (Kusuluri et al., 2021). In the latter study, we used isolated primary BL/6 wild-type astrocytes, Vlgr1/del7TM mice lacking the seven transmembrane domains (7TM) and the intracellular domain (ICD) of Vlgr1 (McMillan and White, 2004), and mutant Drum B mice in which only a small portion of the N-terminal fragment is present and the entire C-terminal fragment is missing (Potter et al., 2016; Kusuluri et al., 2021). A significant decrease in the number and length of FAs was observed in both Vlgr1 mutant mice compared with primary BL/6 wild-type astrocytes (Figure 10) (Kusuluri et al., 2021). For the quantifications, we applied the ImageJ protocol described above in step 67 to 85. Moreover, our study demonstrated that VLGR1 is metabotropic mechanosensor at FAs and provided novel insights into the pathomechanisms underlying VLGR1-associated diseases, such as human Usher syndrome (Reiners and Wolfrum 2006) and epilepsy (Wang et al., 2015; Myers et al., 2018).
Figure 9.
VLGR1 deficiency alters FA morphology
Representative images of FAs in VLGR1-deficient mouse primary astrocytes.
(A and D) FAs were stained for vinculin (magenta), F-actin with TRITC-phalloidin (red), and nuclei with DAPI (blue); GFAP (green) was used as an astrocyte marker. FA number and length were reduced after VLGR1-depletion in primary astrocytes of both Vlgr1 mutant mouse lines.
(B and C) (B) 20 cells per condition, (C) ∼1300–1700 FAs per category for control and ∼200–1000 FAs for mutant astrocytes.
(E and F) (E) 20 cells per condition, (F) ∼950–1250 FAs per condition for control and ∼410–510 FAs for mutant astrocytes. Scale bars: (A and D) 10 μm. Data are represented in B and E are shown as box plot and statistical analyses were done using two-tailed Mann-Whitney U test. Data in C and F are represented as mean±SD. Statistical analyses were done using Sidak’s multiple comparison test; ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001.
(A–F). Figure is acquired and adapted from Figures 3D–3H in Kusuluri et al. (2021) with the permission of Elsevier.
Limitations
This protocol is designed for isolation of primary astrocytes from P0 mice and all isolation steps are optimized for this age. Nevertheless, our protocol can be adjusted for different aged mice brain culture. It might be taken under consideration that precursor and adult primary astrocyte can show different transcriptional profile which may affect experimental outcome (Zhang et al., 2016; Clarke et al., 2018; Bronzuoli et al., 2019). In addition, there might be species-specific differences between species that primary astrocyte derived may show differences in transcriptomic and protein interaction level. In human and mice astrocytes, it has been reported that metabolism and mitochondria related protein network was higher in mice astrocyte whereas defensive response and extracellular space related proteins were higher in human astrocytes (Li et al., 2021). In rat and mouse astrocytes, significant differences in GFAP expression level, cell proliferation and morphology has been also shown (Puschmann et al., 2010). We observed that primary astrocytes can only be sub-passaged up to 3 times. As more than 3 sub passaging cause astrocyte marker (GFAP) expression loss and unhealthy cell phenotype such as poor cell growth, reduce in cell migration capacity and changes in cell morphology, we highly recommend that experiments should be planned in advance and passage 1 and passage 2 should be used. Astrocytes are one of the main parts of active information transport in central nervous system by interacting with neurons. Neuron-astrocyte interaction through messenger and signaling molecules such as Ca2+ and glutamate, is highly important in the CNS for function, development and pathology of the brain (Wilhelm et al., 2004; Benarroch 2005; Nimmerjahn 2009). Therefore, it should be taken under consideration that pure primary astrocyte culture may not always reflect in-vivo physiology in the absence of different cell types.
Troubleshooting
Problem 1
Contamination of fibroblast and endothelial cells in the primary cultures (step 10).
Potential solution
Fibroblast and endothelial cells are the two major cell types present in meninges. Poor handling in meningeal removing increases the contamination level in culture. These two cell types can reach confluency very rapidly which may cause a reduced number of astrocytes and purity of cultures. Meninges should peel away using fine forceps. Be sure that meninges are removed in one piece, if not there can be patches of meninges left which may cause contamination.
Problem 2
Insufficient dissociation of brain tissue (step 21).
Potential solution
When performing dissociation of brain tissue, be sure to use freshly thawed ice-cold DNase I solution as DNase I is enzymatically helping dissociation of brain tissue. Due to environmental and mechanical stress during the dissociation process DNA can leak as a result of cell damage. Released DNA is often the cause of cell clumps that affects efficiency of dissociation. Start the resuspension of cell pellet with a 10 mL pipette and continue with 5 mL and 2 mL pipettes, respectively. Using a smaller pipette size can increase dissociation efficiency and reduce aggregation levels.
Problem 3
Cell viability is not sufficient after plating (step 24).
Potential solution
This problem may occur because of excessive Poly-L-Lysine (PLL) residue in flask. Therefore, wash the plates coated with PLL thoroughly before plating single-cell suspension. After plating single-cell suspension, cellular growth should be observed on day 1, day 2, day 7 and day 14. Differential binding should be performed when cultures reach full confluency, as insufficient cell numbers may affect the cell viability.
Problem 4
OPCs contamination in isolated astrocyte cultures (step 25).
Potential solution
After 7–15 days of culturing, primary astrocytes reach confluency. It is important to keep the astrocyte layer attached to the flask surface during the OPCs. While too gentle shaking of flask may cause lower yield of OPCs removal, too intense shanking may damage the astrocyte layer. Therefore, shaking should be done carefully.
Problem 5
Microglia contamination after differential binding (step 29).
Potential solution
After trypsinization, transfer medium containing astrocytes and potential microglia cells to the bacterial grade plates. Microglia cells are attached to the surface of bacterial grade plates whereas primary astrocytes do not. After 20 min of transferring to bacterial grade plate, check the cells, primary astrocytes can be distinguished easily as they float in the medium. Collect the medium containing primary astrocytes and repeat the step carefully for another 90 min. When collecting medium, be careful not to shake the plate or the microglia may become detached.
Problem 6
Low transfection rate (step 41).
Potential solution
An efficient transfection rate is very important for successful experiment. We have observed that transfection efficiency in primary astrocyte was the range of 15%–30%. To optimize transfection efficiency for your gene/protein of interest, you may try different transfection methods, such as transfection by electroporation or transfection by liposomes. Transfection efficiency may depend on several factors, such as DNA:transfection reagent ratio, purity and composition of the DNA, the degree of confluency of the culture and transfection method. Therefore, to achieve sufficient number of transfected cells and transfection protocol must be optimized. Plasmid DNA should be prepared by using Endotoxin-free isolation kit, as residue of bacterial toxin may lower the efficiency. After obtaining endotoxin-free DNA, different transfection reagent:DNA ratios should be tested to optimize transfection.
Problem 7
Poor migration speed during live-cell imaging (step 52).
Potential solution
Extracellular matrix is vital for migration of cells in vitro and in vivo. Primary astrocytes produce and secrete extracellular matrix (ECM) proteins such as laminin in vitro condition to provide growth and spreading condition (Chiu et al., 1991). However, in low cell density such as single cell migration study, migration speed can reduce dramatically due to lack of sufficient ECM. A potential solution to this problem is to coating the surface using fibronectin at a 5 μg/mL. Since, different ECM concentrations have an effect on migration speed and migrating cell numbers (Desban and Duband 1997; Millon-Fremillon et al., 2008), effective fibronectin concentration from 1 μg/mL to 10 μg/mL may be tested before starting the experiment. Also, providing conditioned astrocyte medium can enhance the migration speed of primary astrocyte cells as it contains ECM molecules.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Uwe Wolfrum (wolfrum@uni-mainz.de).
Materials availability
Reagents and resources used in this study are commercially available with the exception of RFP-paxillin plasmid which was kindly provided by Drs. Leube and Hortwitz. Nevertheless, requests for resources and reagents can be directed to and will be fulfilled by the lead contact.
Acknowledgments
This work was supported by the German Research Council DFG FOR 2149 “Elucidation of Adhesion-GPCR signaling” WO 548/8 (UW). We thank Drs Rudolf E. Leube and Rick Horwitz for kindly sharing reagents and Dr Karl R. Fath for critical reading and language corrections.
Author contributions
Conceptualization, B.E.G. and U.W.; investigation, B.E.G. and J.K.; writing – original draft, B.E.G. and U.W.; writing – review & editing, B.E.G., J.K. and U.W.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2021.100954.
Data and code availability
This study did not generate new data sets.
References
- Benarroch E.E. Neuron-astrocyte interactions: partnership for normal function and disease in the central nervous system. Mayo Clin. Proc. 2005;80:1326–1338. doi: 10.4065/80.10.1326. [DOI] [PubMed] [Google Scholar]
- Berginski M.E., Gomez S.M. The Focal Adhesion Analysis Server: a web tool for analyzing focal adhesion dynamics. F1000Res. 2013;2:68. doi: 10.12688/f1000research.2-68.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau A. Cell adhesion promotion strategies for signal transduction enhancement in microelectrode array in vitro electrophysiology: an introductory overview and critical discussion. Curr. Opin. Colloid Interface Sci. 2013;18:481–492. [Google Scholar]
- Bronzuoli M.R., Facchinetti R., Valenza M., Cassano T., Steardo L., Scuderi C. Astrocyte function is affected by aging and not Alzheimer's disease: a preliminary investigation in hippocampi of 3xTg-AD mice. Front. Pharmacol. 2019;10:644. doi: 10.3389/fphar.2019.00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu A.Y., Espinosa De Los Monteros A., Cole R.A., Loera S., De Vellis J. Laminin and s-laminin are produced and released by astrocytes, Schwann cells, and schwannomas in culture. Glia. 1991;4:11–24. doi: 10.1002/glia.440040103. [DOI] [PubMed] [Google Scholar]
- Clarke L.E., Liddelow S.A., Chakraborty C., Munch A.E., Heiman M., Barres B.A. Normal aging induces A1-like astrocyte reactivity. Proc. Natl. Acad. Sci. U S A. 2018;115:E1896–E1905. doi: 10.1073/pnas.1800165115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desban N., Duband J.L. Avian neural crest cell migration on laminin: interaction of the alpha1beta1 integrin with distinct laminin-1 domains mediates different adhesive responses. J. Cell Sci. 1997;110:2729–2744. doi: 10.1242/jcs.110.21.2729. [DOI] [PubMed] [Google Scholar]
- Ezratty E.J., Partridge M.A., Gundersen G.G. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat. Cell Biol. 2005;7:581–590. doi: 10.1038/ncb1262. [DOI] [PubMed] [Google Scholar]
- Horzum U., Ozdil B., Pesen-Okvur D. Step-by-step quantitative analysis of focal adhesions. MethodsX. 2014;1:56–59. doi: 10.1016/j.mex.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Wirtz D. Focal adhesion size uniquely predicts cell migration. FASEB J. 2013;27:1351–1361. doi: 10.1096/fj.12-220160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.C., Kim C., Wood L., Neal D., Kamm R.D., Asada H.H. Integrating focal adhesion dynamics, cytoskeleton remodeling, and actin motor activity for predicting cell migration on 3D curved surfaces of the extracellular matrix. Integr. Biol. 2012;4:1386–1397. doi: 10.1039/c2ib20159c. [DOI] [PubMed] [Google Scholar]
- Kusuluri D.K., Guler B.E., Knapp B., Horn N., Boldt K., Ueffing M., Aust G., Wolfrum U. Adhesion G protein-coupled receptor VLGR1/ADGRV1 regulates cell spreading and migration by mechanosensing at focal adhesions. iScience. 2021;24:102283. doi: 10.1016/j.isci.2021.102283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Pan L., Pembroke W.G., Rexach J.E., Godoy M.I., Condro M.C., Alvarado A.G., Harteni M., Chen Y.W., Stiles L., et al. Conservation and divergence of vulnerability and responses to stressors between human and mouse astrocytes. Nat. Commun. 2021;12:3958. doi: 10.1038/s41467-021-24232-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan D.R., White P.C. Loss of the transmembrane and cytoplasmic domains of the very large G-protein-coupled receptor-1 (VLGR1 or Mass1) causes audiogenic seizures in mice. Mol. Cell Neurosci. 2004;26:322–329. doi: 10.1016/j.mcn.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Millon-Fremillon A., Bouvard D., Grichine A., Manet-Dupe S., Block M.R., Albiges-Rizo C. Cell adaptive response to extracellular matrix density is controlled by ICAP-1-dependent beta1-integrin affinity. J. Cell Biol. 2008;180:427–441. doi: 10.1083/jcb.200707142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers K.A., Nasioulas S., Boys A., Mcmahon J.M., Slater H., Lockhart P., Sart D.D., Scheffer I.E. ADGRV1 is implicated in myoclonic epilepsy. Epilepsia. 2018;59:381–388. doi: 10.1111/epi.13980. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn A. Astrocytes going live: advances and challenges. J. Physiol. 2009;587:1639–1647. doi: 10.1113/jphysiol.2008.167171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter P.K., Bowl M.R., Jeyarajan P., Wisby L., Blease A., Goldsworthy M.E., Simon M.M., Greenaway S., Michel V., Barnard A., et al. Novel gene function revealed by mouse mutagenesis screens for models of age-related disease. Nat. Commun. 2016;7:12444. doi: 10.1038/ncomms12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puschmann T.B., Dixon K.J., Turnley A.M. Species differences in reactivity of mouse and rat astrocytes in vitro. Neurosignals. 2010;18:152–163. doi: 10.1159/000321494. [DOI] [PubMed] [Google Scholar]
- Reiners J., Wolfrum U. Molecular analysis of the supramolecular usher protein complex in the retina. Harmonin as the key protein of the Usher syndrome. Adv. Exp. Med. Biol. 2006;572:349–353. doi: 10.1007/0-387-32442-9_49. [DOI] [PubMed] [Google Scholar]
- Shen B., Delaney M.K., Du X. Inside-out, outside-in, and inside-outside-in: G protein signaling in integrin-mediated cell adhesion, spreading, and retraction. Curr. Opin. Cell Biol. 2012;24:600–606. doi: 10.1016/j.ceb.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Guo S.S., Fassler R. Integrin-mediated mechanotransduction. J. Cell Biol. 2016;215:445–456. doi: 10.1083/jcb.201609037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Fan X., Zhang W., Zhang C., Wang J., Jiang T., Wang L. Deficiency of very large G-protein-coupled receptor-1 is a risk factor of tumor-related epilepsy: a whole transcriptome sequencing analysis. J. Neurooncol. 2015;121:609–616. doi: 10.1007/s11060-014-1674-0. [DOI] [PubMed] [Google Scholar]
- Wilhelm A., Volknandt W., Langer D., Nolte C., Kettenmann H., Zimmermann H. Localization of SNARE proteins and secretory organelle proteins in astrocytes in vitro and in situ. Neurosci. Res. 2004;48:249–257. doi: 10.1016/j.neures.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Sloan S.A., Clarke L.E., Caneda C., Plaza C.A., Blumenthal P.D., Vogel H., Steinberg G.K., Edwards M.S., Li G., et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Images were acquired for 85 min with 3 min time intervals. Live-cell imaging performed under 5% CO2 and at 37°C using Nikon Eclipse Ti2-E equipped with a spinning disc microscope. Scale bar: 20 μm.
Data Availability Statement
This study did not generate new data sets.