Abstract
Non-keratinizing nasopharyngeal carcinoma, the major subtype of nasopharyngeal carcinoma, is characterized by low differentiation and a close relation to Epstein-Barr virus infection, which indicates a link between Epstein-Barr virus oncogenesis and loss of differentiation, and raises our interest in investigating the involvement of Epstein-Barr virus in nasopharyngeal carcinoma dedifferentiation. Our previous study showed abundant expression of an Epstein-Barr virus-encoded microRNA, BART10-3p, in nasopharyngeal carcinoma tissues, but the association between BART10-3p and nasopharyngeal carcinoma differentiation remains unknown. Here, we examined the expression and prognostic value of BART10-3p, and undertook bioinformatics analysis and functional assays to investigate the influence of BART10-3p on nasopharyngeal carcinoma differentiation and proliferation and the underpinning mechanism. Microarray analysis identified BART10-3p as the most significantly upregulated Epstein-Barr virus-encoded microRNA in nasopharyngeal carcinoma tissues and the upregulation was confirmed in two public datasets. The expression of BART10-3p was an independent unfavorable prognosticator in nasopharyngeal carcinoma and its integration with the clinical stage showed improved prognosis predictive performance. Bioinformatics analysis suggested a potential role of BART10-3p in tumor differentiation and progression. Functional assays demonstrated that BART10-3p could promote nasopharyngeal carcinoma cell dedifferentiation, epithelial-mesenchymal transition, and proliferation in vitro, and tumorigenicity in vivo. Mechanistically, BART10-3p directly targeted the 3′UTR of ALK7 and suppressed its expression. Reconstitution of ALK7 rescued BART10-3p-induced malignant phenotypes. Overall, our study demonstrates that BART10-3p promotes dedifferentiation and proliferation of nasopharyngeal carcinoma by targeting ALK7, suggesting a promising therapeutic opportunity to reverse the malignant phenotypes of nasopharyngeal carcinoma.
Keywords: Nasopharyngeal carcinoma, Epstein-Barr virus, BART10-3p, dedifferentiation, proliferation, ALK7
Impact statement
This study demonstrated that the NPC-specific EBV miRNA, BART10-3p, could promote dedifferentiation and proliferation of NPC via direct suppression of ALK7. These data for the first time unravel the role and mechanism of BART10-3p in NPC dedifferentiation and proliferation, as well as its oncogenic effect in vivo, advancing our knowledge of EBV miRNA and ALK7 in NPC pathogenesis. Furthermore, the expression of BART10-3p was an independent unfavorable prognosticator in NPC and its integration with the clinical stage showed improved predictive performance. Therefore, BART10-3p represents a promising prognostic marker and therapeutic target that may allow an opportunity to reverse the malignant phenotypes of NPC.
Introduction
Nasopharyngeal carcinoma (NPC) is an epithelial neoplasm particularly prevalent in southern China, where around 98% of cases are histologically non-keratinizing carcinoma. 1 This particular subtype with low differentiation has a high propensity for systemic metastasis, posing a threat to long-term survival. 2 It has been proposed that loss of differentiation, also known as dedifferentiation, is a hallmark of cancer, for it endows cells with high tumorigenicity and metastatic capacity.3,4 Dissecting the molecules and mechanisms contributing to NPC dedifferentiation would provide novel intervention targets for the treatment of NPC. Another characteristic of non-keratinizing NPC is the prominent relationship with Epstein-Barr virus (EBV) infection, 5 which suggests a link between EBV oncogenesis and loss of differentiation, and leads us to investigate the involvement of EBV in NPC dedifferentiation.
EBV has been implicated in a set of human tumors, including lymphocytic malignancies, gastric carcinoma, lymphoepithelioma-like carcinoma, as well as NPC. In EBV-related tumors, only restricted viral products are expressed, and the viral gene expression varies with different types of tumors. 6 EBV-infected NPC is characterized by abundant expression of microRNAs (miRNAs), despite the absence of oncogenic proteins commonly detectable in EBV-transformed B lymphoblastoid cells. 7 The non-immunogenic feature of EBV-encoded miRNAs enables the virus to exert its oncogenic functions without inflaming the host immune system. 8 Thus, EBV-encoded miRNAs are potentially crucial in the pathogenesis of NPC.
Since the first report in 2004, more than 40 mature EBV-encoded miRNAs have been identified, mapping to two regions of the EBV genome: BamHI fragment A rightward transcript (BART) and BamHI fragment H rightward open reading frame 1 (BHRF1). 9 BART miRNAs are the predominantly expressed EBV miRNAs in NPC samples. 10 It has been documented that BART miRNAs can interfere with cancer cell survival, metastasis, immune evasion, resistance to treatment, and viral latency maintenance.11–15 Our previous microarray analysis identified BART10-3p as the most significantly upregulated EBV miRNA in NPC tissues. 16 Although BART10-3p has been shown to promote NPC cell metastasis, 17 its role in NPC differentiation remains unknown.
In this study, we demonstrate the abundant expression of BART10-3p in NPC tissues. BART10-3p promotes NPC dedifferentiation, epithelial-mesenchymal transition (EMT), and proliferation by directly targeting ALK7. Restored ALK7 expression rescues BART10-3p-driven malignant phenotypes. Moreover, we present evidence that BART10-3p expression predicts poor prognosis, and its incorporation with the clinical stage yields improved predictive value. Our findings may provide novel insights into the role of BART10-3p in NPC development and reveal a promising opportunity to reverse the malignant phenotypes of NPC.
Materials and methods
miRNA microarray analysis
Our previous miRNA microarray data (GEO accession number: GSE32960) 16 containing 312 NPC and 18 non-cancer tissues were exacted and reanalyzed using GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/), with the focus on EBV-encoded miRNAs. Those with absolute fold change > 2 and adjusted P < 0.05 were deemed as differentially expressed miRNAs. The work flow chart is shown in Figure 1.
Figure 1.
Flow chart of the study. EBV: Epstein-Barr virus; FC: fold change; GO: gene ontology; GSEA: gene set enrichment analysis; miRNA: microRNA; NPC: nasopharyngeal carcinoma.
Cell culture and transfection
The NPC cell lines HONE-1 and SUNE-1 (EBV-negative),18,19 as well as C666-1 (EBV-positive) 20 were gifted by Professor Mu-Sheng Zeng (Sun Yat-sen University Cancer Center [SYSUCC], China). The cells were authenticated by short-tandem repeat profiling and were free from mycoplasma contamination. All cell lines were cultured in RPMI-1640 (Gibco, Life Technologies, USA) with 10% fetal bovine serum (Gibco, Life Technologies, USA) as previously described. 21
For cell transfection, BART10-3p mimic (sequence: 5′-UACAUAACCAUGGAGUUGGCUGU-3′), BART10-3p inhibitor (sequence: 5′-ACAGCCAACUCCAUGGUUAUGUA-3′), and their corresponding negative controls (NC and NC inhibitor) were purchased from GenePharma (China). The coding sequence of the human ALK7 gene was cloned into the pHAGE-6tag-puro vector (Addgene, USA) for construction of ALK7 overexpression plasmid. NPC cells were transiently transfected with oligonucleotides (50 nM) or plasmids (2 μg) with the use of Lipofectamine 3000 reagent (Invitrogen, Life Technologies, USA), and collected for further assays at 48 h post transfection.
RNA sequencing
Total RNA was extracted from SUNE-1 cells treated with BART10-3p mimic or NC using TRIzol reagent (Invitrogen, Life Technologies, USA). Agilent Bioanalyzer 2100 (Agilent technologies, USA) was used to qualify and quantify RNA, and RNeasy micro kit and RNase-Free DNase Set (QIAGEN, Germany) to purify RNA. TruSeq RNA Sample Prep kit (Illumina, USA) was employed for establishment of sequencing libraries. Briefly, mRNA was enriched from total RNA with oligo (dT) beads and fragmented at 94°C for 8 min. Then, double strand cDNA was synthesized and went through end repair, 3′-end adenylation, adapter ligation, and PCR amplification to generate the final cDNA libraries. The libraries were quantified by Qubit 2.0 Fluorometer (Life Technologies, USA), qualified by Agilent Bioanalyzer 2100 (Agilent technologies, USA), and subjected to cluster generation on the cBot system (Illumina, USA) and sequencing on the HiSeq 2500 system (Illumina, USA). Sequencing reads were mapped to the human genome (hg19) using Tophat 2.0.9. Cufflinks 2.1.1 was employed to assemble reads into transcripts and to evaluate differential expression. Genes with absolute fold change > 2 and P < 0.05 by Fisher’s exact test were defined as differentially expressed genes.
Bioinformatics analysis
The differentially expressed genes between BART10-3p-overexpressing and control cells were subjected to gene ontology (GO) analysis (http://www.pantherdb.org). GO terms with a Bonferroni-corrected P < 0.05 were deemed as significant. Gene set enrichment analysis (GSEA 4.1) was carried out with the use of gene sets downloaded from the molecular signatures database (MSigDB 7.2). Each gene set was assigned a normalized enrichment score (NES) and the significance was defined at false discovery rate < 0.05.
RNA extraction and quantitative RT-PCR
Total RNA was isolated from NPC cells with TRIzol reagent (Invitrogen, Life Technologies, USA) and transcribed into cDNA by reverse transcriptase (Promega, USA). Quantitative PCR reactions were monitored by the CFX96 Touch sequence detection system (Bio-Rad, USA) utilizing SYBR Green qPCR SuperMix-UDG reagents (Invitrogen, Life Technologies, USA). The relative gene expression was determined using the 2−ΔΔCT method 22 with GAPDH and U6 as endogenous controls for mRNA and miRNA, respectively. The primer sequences are listed in Supplementary Table 1.
Western blotting
Total protein was harvested from NPC cells with the use of RIPA buffer (Beyotime Biotechnology, China) that contained EDTA-free Protease Inhibitor Cocktail (Roche, Switzerland). An equal amount of extracted protein was subjected to 10–14% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrotransferred to polyvinylidene fluoride membranes (Merck Millipore, USA), after which the membranes were blocked for 1 h with 5% nonfat milk and incubated overnight at 4°C with primary antibodies targeting E-cadherin (1:1000, 24E10, Cell Signaling Technology, USA), MMP9 (1:2000, AP6214a, Abgent, USA), Vimentin (1:2000, 60330–1-Ig, Proteintech, China), CK13 (1:5000, 10164–2-AP, Proteintech, China), KLF4 (1:1000, 4038, Cell Signaling Technology, USA), ALK7 (1:1000, 12610–1-AP, Proteintech, China), and GAPDH (1:1000, 14C10, Cell Signaling Technology, USA). Horseradish peroxidase-conjugated antibodies (Proteintech, China) were used as secondary antibodies under room temperature for 1 h. The ECL detection system (Thermo Fisher Scientific, USA) was used for visualization of protein bands.
Immunofluorescence staining
NPC cells were seeded on coverslips for 24 h, fixed in 2% paraformaldehyde for 30 min, permeabilized in 0.5% Triton X-100 for 15 min, and blocked with 1% bovine serum albumin for 30 min. The coverslips were then incubated overnight at 4°C with primary antibodies against E-cadherin (1:50, 610181, BD Biosciences, USA) and Vimentin (1:500, 60330–1-Ig, Proteintech, China). Incubation with Alexa Fluor 488 or 594 donkey anti-mouse IgG secondary antibody (Life Technologies, USA) lasted for 1 h under room temperature. Cell nuclei were counterstained with Hoechst 33258 (Invitrogen, Life Technologies, USA) for 10 min and cell fluorescence images were acquired with a confocal laser-scanning microscope (Olympus FV1000, Olympus, Japan).
CCK-8 and colony formation assays
For the CCK-8 assay, 1000 transfected cells were cultured in 96-well plates. Each day on days 0–4 of incubation, the cells were treated with 10 μL of CCK-8 (Dojindo, Japan) and continued to be incubated at 37°C for 2 h, followed by measurement of absorbance at 450 nm using an ELX800 spectrophotometric plate reader (Bio-Tek, USA). Colony formation assay was done by culturing 400 transfected cells in 6-well plates for 7–12 days. The resulting colonies were fixed by methanol, stained by hematoxylin, and counted.
Dual-luciferase reporter assay
The ALK7 wild-type (wt) and mutant (mut) 3′UTR were generated and cloned into the psiCHECK-2 vector (Promega, USA), which comprises Renilla and firefly luciferase reporter genes. The wt or mut luciferase reporter plasmid (0.5 μg) was co-transfected with BART10-3p mimic or NC (50 nM), or with BART10-3p inhibitor or NC inhibitor (50 nM) using Lipofectamine 3000 (Invitrogen, Life Technologies, USA). Luciferase activity was determined using the Dual-Luciferase Reporter Assay System (Promega, USA) 24 h after transfection. The Renilla luciferase activity was normalized to firefly luciferase activity as a transfection control.
In vivo xenograft tumor growth model
Female BALB/c nude mice (four to five weeks) were purchased from the Medical Experimental Animal Center of Guangdong Province (China). The mice were randomly assigned to two groups (n = 5 per group) and injected with 1 × 10 6 SUNE-1 cells subcutaneously in the right axillary region to establish xenograft tumor growth models. After 12 days when the tumor volume reached 50–100 mm3, the two groups of mice received intratumoral injection of cholesterol-modified agomir-BART10-3p (5 nM) or agomir-NC (5 nM) (RiboBio, China) every three days. For evaluation of tumor size, the length (L) and width (W) were measured every three days, and tumor volume was calculated based on the formula: volume = (L × W2)/2. After 21 days of intratumoral injection, the mice were sacrificed and their tumors were dissected, weighted, fixed, and subjected to paraffin-embedded sectioning. The animal experiment was adhered to the Guide for the Care and Use of Laboratory Animals of Sun Yat-Sen University, and was approved by the Institutional Animal Care and Use Ethics Committee of SYSUCC (approval number: L102012019040G).
Immunohistochemistry
Sections derived from xenograft mice tissues were deparaffinized and rehydrated, followed by endogenous peroxidase activity blocking and citrate-mediated high-temperature antigen retrieval. After blocking with 1% bovine serum albumin, the sections were incubated with anti-ALK7 antibody (1:100, 12610–1-AP, Proteintech, China) at 4°C overnight, with antibody diluent used as a negative control. The antigen-antibody reaction was visualized with the Dako REAL EnVision Detection system (Dako, Denmark) in compliance with the manufacturer’s protocol.
Statistical processing
All results are presented as mean ± standard error of mean (SEM) from at least three independent experiments. The Student’s t-test and Chi-square/Fisher’s exact test were applied in comparison of two groups. ANOVA analysis was utilized in comparison of multiple groups.
The cutoff to determine low and high BART10-3p expression was optimized to achieve the highest Chi-square value on Kaplan–Meier analysis and log-rank test using the X-tile 3.6.1 software (Yale University, USA). 23 Survival rates were estimated with the Kaplan–Meier method and comparisons were undertaken via the log-rank test. Estimates of hazard ratio were determined by fitting Cox regression models. To evaluate the added value of BART10-3p in overall survival prediction, an integrative model including BART10-3p expression and the clinical stage was built 16 and compared to the clinical stage-alone model in terms of area under the curve (AUC) and Akaike information criterion (AIC). A higher AUC and a lower AIC value indicate better predictive performance. All statistical analyses were exerted using R 3.6.0 (http://www.R-project.org/) and GraphPad Prism 7. Two-tailed P < 0.05 was considered significant. The authenticity of this article has been validated by uploading the key raw data onto the Research Data Deposit platform (http://www.researchdata.org.cn, approval number: RDDB2021001649).
Results
BART10-3p is upregulated in NPC tissues and predicts unfavorable prognosis
To identify NPC-specific EBV-encoded miRNAs, we examined the miRNA expression profile of 312 NPC and 18 non-cancer tissues based on our previous microarray. The analysis revealed that 11 out of 44 EBV-encoded miRNAs were upregulated, which were exclusively BART miRNAs (absolute fold change > 2, adjusted P < 0.05; Figure 2(a) and Supplementary Table 2). Notably, BART10-3p was the top upregulated EBV-encoded miRNA, and also among the most significantly upregulated human and viral miRNAs (Figure 2(b)). In addition, elevation of BART10-3p in NPC tissues was validated by two independent microarray datasets, GSE36682 (n = 68; Figure 2(c)) and GSE70970 (n = 263; Figure 2(d)).
Figure 2.
BART10-3p is increased in NPC tissues and correlates with unfavorable prognosis. (a) Heatmap of differentially expressed EBV-encoded miRNAs identified from the microarray dataset GSE32960 which contains 18 non-cancer tissues and 312 NPC tissues. Threshold was set at absolute fold change > 2 and adjusted P < 0.05. Columns: individual samples; rows: EBV-encoded miRNAs; blue: low expression; yellow: high expression. (b) Volcano plot for visualization of evidently differentially expressed miRNAs in the microarray dataset GSE32960. (c) Relative BART10-3p expression determined in the microarray dataset GSE36682 comprising 6 non-cancer tissues and 62 NPC tissues. (d) Relative BART10-3p expression determined in the microarray dataset GSE70970 which included 17 normal nasopharyngeal epithelial tissues and 246 NPC tissues. (e) Kaplan–Meier analysis of overall survival (OS), disease-free survival (DFS), and distant metastasis-free survival (DMFS) in patients stratified by BART10-3p expression. The log-rank test was used to calculate the P values. **P < 0.01.
We then explored whether the expression of BART10-3p has prognostic value in NPC. NPC tissues were classified into high (n = 60) and low (n = 252) BART10-3p groups based on the X-tile derived cutoff value. No significant correlation was noted between BART10-3p expression and clinical parameters (Table 1). In univariable analysis, high BART10-3p expression was related to inferior overall, disease-free, and distant metastasis-free survival compared with low expression, although with marginal significance (P = 0.050, 0.076, and 0.066, respectively; Figure 2(e)). Multivariable analysis revealed the independent prognostic significance of BART10-3p expression in all endpoints (all P < 0.05; Table 2). Further analysis showed that the model consisting of BART10-3p expression and the clinical stage had better predictive performance for overall survival relative to the clinical stage alone (AUC: 0.67 vs. 0.62, P = 0.032; AIC: 795.7 vs. 801.4).
Table 1.
Association between BART10-3p expression and clinicopathological features of nasopharyngeal carcinoma patients.
| Characteristic | No. of patients |
BART10-3p expression |
P valuea | |
|---|---|---|---|---|
| Low expression, n = 252 | High expression, n = 60 | |||
| Sex | 0.469 | |||
| Male | 233 | 186 (73.8) | 47 (78.3) | |
| Female | 79 | 66 (26.2) | 13 (21.7) | |
| Age | 0.833 | |||
| <45 y | 139 | 113 (44.8) | 26 (43.3) | |
| ≥45 y | 173 | 139 (55.2) | 34 (56.7) | |
| VCA-IgA | 0.494 | |||
| <1:80 | 46 | 40 (15.9) | 6 (10.0) | |
| 1:80–1:320 | 178 | 141 (56.0) | 37 (61.7) | |
| ≥1:640 | 88 | 71 (28.2) | 17 (28.3) | |
| EA-IgA | 0.502 | |||
| <1:10 | 76 | 64 (25.4) | 12 (20.0) | |
| 1:10–1:20 | 100 | 82 (32.5) | 18 (30.0) | |
| ≥1:40 | 136 | 106 (42.1) | 30 (50.0) | |
| T categoryb | 0.467 | |||
| T1 | 66 | 55 (21.8) | 11 (18.3) | |
| T2 | 89 | 69 (27.4) | 20 (33.3) | |
| T3 | 71 | 61 (24.2) | 10 (16.7) | |
| T4 | 86 | 67 (26.6) | 19 (31.7) | |
| N categoryb | 0.165 | |||
| N0 | 44 | 32 (12.7) | 12 (20.0) | |
| N1 | 148 | 116 (46.0) | 32 (53.3) | |
| N2 | 72 | 62 (24.6) | 10 (16.7) | |
| N3 | 48 | 42 (16.7) | 6 (10.0) | |
| Clinical stageb | 0.520 | |||
| I | 12 | 11 (4.4) | 1 (1.7) | |
| II | 86 | 66 (26.2) | 20 (33.3) | |
| III | 91 | 76 (30.2) | 15 (25.0) | |
| IV | 123 | 99 (39.3) | 24 (40.0) | |
| Chemotherapy | 0.824 | |||
| No | 268 | 217 (86.1) | 51 (85.0) | |
| Yes | 44 | 35 (13.9) | 9 (15.0) | |
aP value was calculated using the Chi-square test (or Fisher’s exact test if indicated).
bAccording to the 7th edition American Joint Committee on Cancer staging manual.
EA: early antigen; IgA: immunoglobulin A; VCA: viral capsid antigen.
Table 2.
Multivariable Cox regression analysis in nasopharyngeal carcinoma patients.
| Variable |
OS |
DFS |
DMFS |
|||
|---|---|---|---|---|---|---|
| HR (95% CI) | P valuea | HR (95% CI) | P valuea | HR (95% CI) | P valuea | |
| Sex | ||||||
| Male | Reference | – | Reference | – | – | – |
| Female | 0.518 (0.272–0.988) | 0.046 | 0.571 (0.333–0.980) | 0.042 | – | – |
| BART10-3p | ||||||
| Low | Reference | – | Reference | – | Reference | – |
| High | 1.863 (1.102–3.150) | 0.020 | 1.733 (1.077–2.789) | 0.024 | 2.002 (1.149–3.488) | 0.014 |
| T category | ||||||
| T1-2 | Reference | – | Reference | – | Reference | – |
| T3-4 | 2.329 (1.430–3.792) | 0.001 | 1.813 (1.198–2.745) | 0.005 | 2.171 (1.301–3.624) | 0.003 |
| N category | ||||||
| N0-1 | Reference | – | Reference | – | Reference | – |
| N2-3 | 2.042 (1.279–3.260) | 0.003 | 1.892 (1.250–2.866) | 0.003 | 2.399 (1.460–3.942) | 0.001 |
aP value was calculated using multivariable Cox regression models with backward elimination; included covariates were: sex, age, VCA-IgA, EA-IgA, T and N category, and BART10-3p expression.
CI: confidence interval; DFS: disease-free survival; DMFS: distant metastasis-free survival; EA: early antigen; HR: hazard ratio; IgA: immunoglobulin A; OS: overall survival; VCA: viral capsid antigen.
BART10-3p overexpression is correlated with NPC differentiation and progression
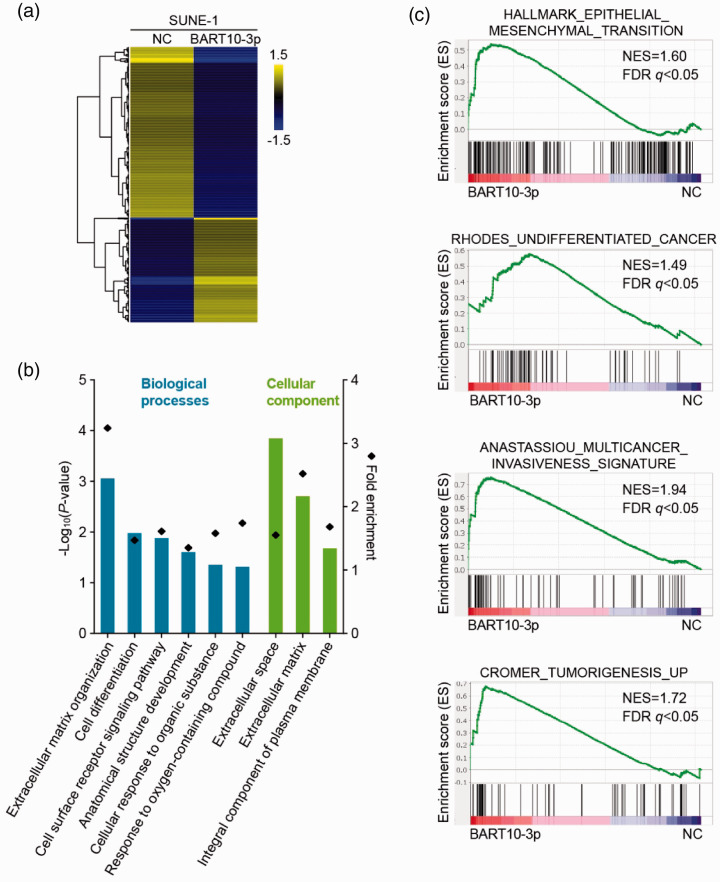
To elucidate the potential role of BART10-3p in NPC pathogenesis, we employed RNA sequencing of SUNE-1 cells with or without BART10-3p overexpression and compared their gene expression profiles. The comparison showed that 608 genes were differentially expressed (absolute fold change > 2, P < 0.05; Figure 3(a)), of which 231 were upregulated and 377 were downregulated. GO analysis revealed that the significantly altered genes were enriched for terms related to cell differentiation and extracellular matrix (Figure 3(b)). GSEA illustrated that increased expression of BART10-3p was positively correlated with gene signatures of EMT, undifferentiated cancer, and tumor development (Figure 3(c)). Taken together, BART10-3p may serve as a critical regulator in differentiation and progression of NPC.
Figure 3.
BART10-3p overexpression is related to NPC differentiation and progression. RNA sequencing was performed in SUNE-1 cells transfected with BART10-3p mimic or negative control. (a) Heatmap of differentially expressed genes identified by RNA sequencing in transfected SUNE-1 cells. Threshold was set at absolute fold change > 2 and P < 0.05. Columns: individual cells; rows: genes; blue: low expression; yellow: high expression. (b) Gene ontology (GO) analysis of differentially expressed genes between transfected SUNE-1 cells. (c) Gene set enrichment analysis (GSEA) plots showing enrichment of gene signatures of epithelial-mesenchymal transition, undifferentiated cancer, cancer invasiveness, and tumorigenesis in response to overexpression of BART10-3p. NES: normalized enrichment score; FDR: false discovery rate.
BART10-3p induces NPC cell dedifferentiation and EMT
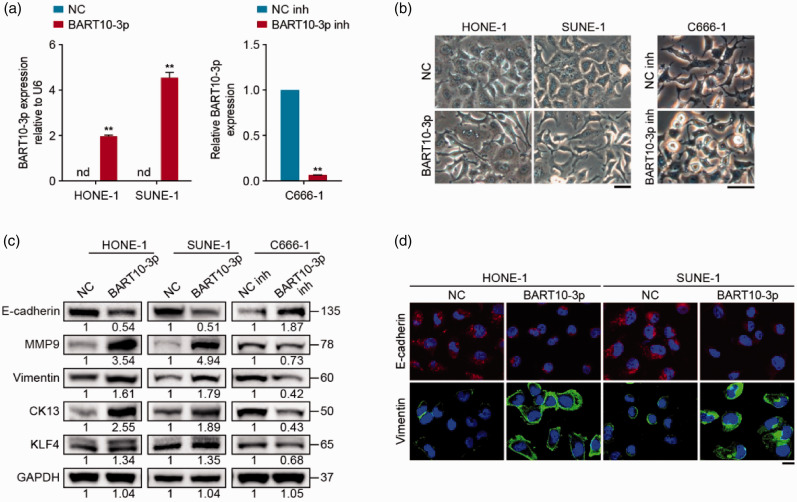
To verify the above data, NPC cell lines of different degrees of differentiation were used: poorly differentiated HONE-1 and SUNE-1 cells with cobblestone morphology typical of epithelial cells, and undifferentiated C666-1 cells with spindle morphology identical to mesenchymal cells. We transiently transfected HONE-1 and SUNE-1 cells with BART10-3p mimic or NC, and transfected C666-1 cells with BART10-3p inhibitor or NC inhibitor; the transfection efficiency was confirmed by quantitative RT-PCR (Figure 4(a)). Notably, BART10-3p overexpression induced HONE-1 and SUNE-1 cells to adopt a spindle-like mesenchymal phenotype, while BART10-3p inhibition induced mesenchymal-epithelial transition in C666-1 cells (Figure 4(b)). Moreover, Western blotting showed that introduction of BART10-3p significantly increased the expression of the mesenchymal markers MMP9 and Vimentin, and the low differentiation markers CK13 and KLF4, and decreased that of the epithelial marker E-cadherin, while downregulation of BART10-3p in C666-1 cells resulted in reversed changes in these molecules (Figure 4(c)). Immunofluorescence staining validated the altered expression of E-cadherin and Vimentin (Figure 4(d)). These data indicate that BART10-3p could drive dedifferentiation and EMT of NPC cells.
Figure 4.
BART10-3p induces NPC cell dedifferentiation and EMT. NPC cells were transfected with BART10-3p mimic, inhibitor, or their negative controls. (a) Quantitative RT-PCR analysis of BART10-3p expression in transfected cells. U6 was used as an internal control. (b) Morphology of transfected cells under inverted microscope. Scale bar, 40 μm. (c) Western blotting analysis of E-cadherin, MMP9, Vimentin, CK13, and KLF4 expression in transfected cells. GAPDH was used as an internal control. (d) Immunofluorescence images for E-cadherin and Vimentin expression in transfected cells. Scale bar, 20 μm. Data are presented as mean (n = 3) ± SEM. **P < 0.01. nd: not detected.
BART10-3p promotes NPC cell proliferation
To explore whether BART10-3p affects NPC cell proliferation, we adopted CCK-8 and colony formation assays. CCK-8 assay revealed that introduction of BART10-3p significantly facilitated proliferation of NPC cells, whereas BART10-3p inhibition suppressed cell proliferation (Figure 5(a)). Colony formation assay showed that BART10-3p-overexpressing cells formed more colonies than control cells (Figure 5(b)). Collectively, these findings suggest that BART10-3p can significantly promote NPC cell proliferation.
Figure 5.
BART10-3p promotes NPC cell proliferation. NPC cells were transfected with BART10-3p mimic, inhibitor, or their negative controls. (a) CCK-8 assay of transfected cells. (b) Representative images (left) and quantification (right) of the colony formation assay in transfected cells. Data are presented as mean (n = 3) ± SEM. **P < 0.01.
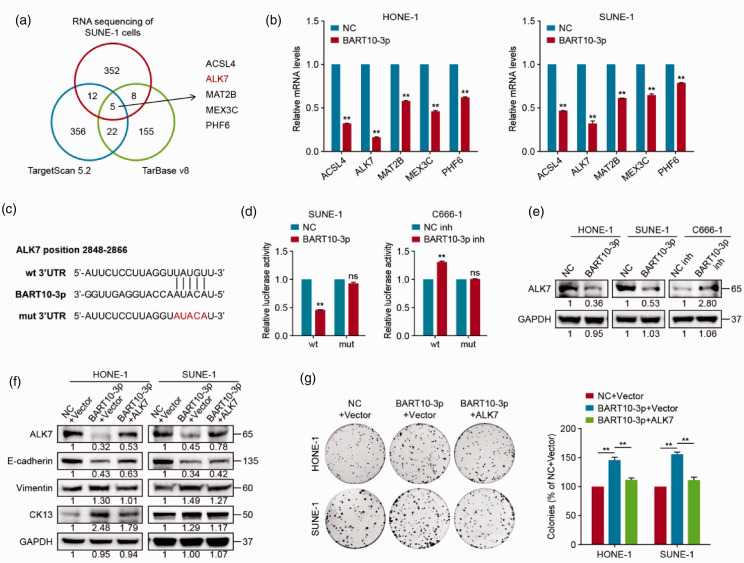
ALK7 is a direct target of BART10-3p in NPC cells
We subsequently investigated the mechanism by which BART10-3p induces dedifferentiation and proliferation of NPC. Putative BART10-3p target genes predicted by TargetScan 5.2 and DIANA-TarBase v8 were overlapped with RNA-sequencing identified genes downregulated by more than 2-fold, resulting in five candidate genes: ACSL4, ALK7, MAT2B, MEX3C, and PHF6 (Figure 6(a)). Among these, ALK7 was validated as the most significantly altered gene after overexpression of BART10-3p (Figure 6(b)). Then, we carried out dual-luciferase reporter assay to verify the regulatory effect of BART10-3p on ALK7. As expected, the luciferase activity of the ALK7 wt group was significantly reduced or enhanced upon overexpression or attenuation of BART10-3p, whereas that of the ALK7 mut group was not influenced (Figure 6(c) and (d)). Additionally, BART10-3p overexpression evidently suppressed the protein level of ALK7 (Figure 6(e)). These findings elucidate that ALK7 is a direct target of BART10-3p in NPC cells.
Figure 6.
ALK7 is a direct target and functional mediator of BART10-3p in NPC cells. (a) Venn diagram displaying the number of predicted BART10-3p target genes based on TargetScan 5.2 and DIANA-TarBase v8 programs and RNA sequencing data. The overlapping genes are shown to the right. (b) Quantitative RT-PCR analysis of the expression of the five predicted target genes (ACSL4, ALK7, MAT2B, MEX3C, and PHF6) in NPC cells transfected with BART10-3p mimic or negative control. GAPDH was used as an internal control. (c) Predicted binding site of BART10-3p on ALK7 wild-type 3′UTR. Mutant 3′UTR was generated as indicated. (d) Luciferase reported assay after co-transfection of wild-type or mutant luciferase reporter plasmids with BART10-3p mimic, inhibitor, or their negative controls. (e) Western blotting analysis of ALK7 expression in NPC cells transfected with BART10-3p mimic, inhibitor, or their negative controls. GAPDH was used as an internal control. (f, g) HONE-1 and SUNE-1 cells were co-transfected with BART10-3p mimic and ALK7 overexpression plasmid or empty vector, and the control group was co-transfected with BART10-3p negative control and empty vector. (f) Western blotting analysis of ALK7, E-cadherin, Vimentin, and CK13 expression in transfected cells. GAPDH was used as an internal control. (g) Representative images (left) and quantification (right) of the colony formation assay in transfected cells. Data are presented as mean (n = 3) ± SEM. **P < 0.01. ns: not significant.
ALK7 is required for BART10-3p-driven malignant phenotypes
We next explored whether ALK7 is involved in BART10-3p-driven biological effects by transfecting the ALK7 overexpression plasmid or empty vector into HONE-1 and SUNE-1 cells with BART10-3p mimic. Notably, ALK7 overexpression reversed BART10-3p-mediated downregulation of E-cadherin and upregulation of Vimentin and CK13 in protein level (Figure 6(f)). In addition, the stimulatory effect of BART10-3p on cell proliferation was attenuated by ALK7 reconstitution (Figure 6(g)). Overall, these results illustrate that BART10-3p enhances NPC cell dedifferentiation and proliferation by inhibiting ALK7.
BART10-3p promotes NPC tumorigenicity in vivo
To further investigate the function of BART10-3p in NPC in vivo, we established xenograft tumor models (n = 5 per group) and increased BART10-3p expression via intratumoral injection of agomir-BART10-3p. Introduction of agomir-BART10-3p significantly increased tumor volume and weight as well as tumor growth rate, in comparison to the agomir-NC group (Figure 7(a) to (c)). Additionally, immunohistochemistry of the dissected specimens showed that the expression of ALK7 was markedly decreased in the agomir-BART10-3p group relative to the agomir-NC group (Figure 7(d)). These results indicate that BART10-3p can significantly promote tumor growth in vivo.
Figure 7.
BART10-3p facilitates NPC xenograft tumor growth in vivo. SUNE-1 cells were subcutaneously injected into the right axillary region of nude mice. After the tumor volume reached 50–100 mm3, intratumoral injection of 5 nM agomir-BART10-3p or agomir-NC (n = 5 per group) was performed. (a) Photograph of xenograft mice. (b) Photograph and weights of xenograft tumors. (c) Tumor growth curves of xenograft mice. The arrows indicate intratumoral injection of 5 nM agomir-BART10-3p or agomir-NC every three days. (d) Immunohistochemical staining for ALK7 expression in xenograft tumors. Scale bar, 100 μm. Data are presented as mean (n = 5) ± SEM. *P < 0.05, **P < 0.01.
Discussion
The current study highlights the crucial role of BART10-3p in NPC dedifferentiation and proliferation. We found that BART10-3p was dramatically upregulated in NPC tissues and associated with inferior outcomes. Overexpression of BART10-3p promoted NPC cell dedifferentiation, EMT, and proliferation in vitro and tumorigenicity in vivo. Further, we identified ALK7 as a direct target and functional mediator of BART10-3p. Collectively, our study demonstrated the oncogenic role of BART10-3p and unveiled a novel mechanism underlying differentiation and proliferation of NPC.
As an oncovirus, EBV has evolved diverse strategies to persist and propagate in malignant cells. One of the most important mechanisms is through generation of viral miRNAs, which can manipulate both viral and host gene expression. Studies of systematic profiling revealed that EBV-encoded BART miRNAs are abundantly expressed in NPC samples both in vitro and in vivo, amounting to over 10% of the total detectable miRNAs.24,25 In the present study, we identified 11 significantly upregulated BART miRNAs in NPC tissues. Among these, BART10-3p showed the highest expression comparable to that of the top upregulated human miRNA. Although BART10-3p expression has shown potential in distinguishing recurrent NPC patients from control, 26 its prognostic value has not been sufficiently elucidated. In our cohort of over 300 NPC patients, high BART10-3p expression was associated with unfavorable prognosis, and its combination with the clinical stage resulted in significantly improved predictive performance. Therefore, BART10-3p represents a promising prognostic biomarker of NPC.
Dedifferentiation, a biological process whereby cells regress to a less differentiated state, is considered a hallmark of cancer. 3 This process is seemingly important for the maintenance of EBV latent infection and malignant transformation in NPC because various EBV latent genes serve this purpose. For example, LMP2A and LMP1 are EBV-encoded proteins that can inhibit differentiation of NPC.27,28 Moreover, a few EBV-derived miRNAs have been reported to contribute to NPC dedifferentiation. Cluster 2 BART miRNAs were found to collectively prevent terminal differentiation of NPC by downregulating an epithelial differentiation marker NDRG1. 29 BART7 and BART22 were shown to promote EMT and stemness of NPC by targeting SMAD7 and MAP2K4, respectively.14,30 In our study, bioinformatics analysis indicated that BART10-3p may participate in differentiation and EMT of NPC. Subsequent assays confirmed that BART10-3p induced NPC cell dedifferentiation as demonstrated by its influence on cell morphology and differentiation-related makers (MMP9, Vimentin, CK13, KLF4, and E-cadherin). Furthermore, the oncogenic role of BART10-3p is supported by previous studies in NPC and EBV-associated gastric carcinoma.17,31 Collectively, BART10-3p acts as an oncogene in NPC dedifferentiation and proliferation.
ALK7 is a serine/threonine kinase receptor for several members of transforming growth factor beta (TGFβ) superfamily, including nodal, activin B, activin AB, and GDFs.32–34 Prior studies have implicated ALK7 in human tissue morphogenesis, metabolic disorders, and tumorigenesis. 35–41 Similar to the bidirectional functions of ALK7 in stem cell differentiation under different conditions, ALK7 has contrasting roles in different tumors. The ALK7 signaling is found to impair metastasis in breast and pancreatic neuroendocrine tumors, 37 suppress proliferation and chemoresistance in ovarian cancer, 38 and induce apoptosis in hepatoma. 39 Conversely, ALK7 exerts tumor-promoting effects in bladder cancer and retinoblastoma.40,41 However, little is documented concerning the role of ALK7 in NPC. Our results suggested that ALK7 may serve as a tumor suppressor in NPC, given that it was identified as a direct target of the tumor-promoting gene BART10-3p both in vitro and in vivo and that ectopic expression of ALK7 could reverse the oncogenic effects of BART10-3p. Our research enables new insights into the regulation of ALK7 signaling by BART10-3p in NPC.
Experience from the management of hematological malignancies suggests that differentiation-based therapy which reverses tumor dedifferentiation has the potential to deprive tumor cells of malignant traits. Nevertheless, its application in NPC and other solid tumors is still at an early stage. 42 Yan et al. 43 documented that restored expression of IKKα via retinoic acid treatment could activate differentiation and reduce tumorigenicity of poorly differentiated NPC cells. The present study found that BART10-3p played a crucial role in dedifferentiation and proliferation of NPC, providing additional insights into the mechanisms underlying development of NPC. Although miRNA-based therapies have not reached trials of large cohorts, our findings provide important implications for inducing differentiation of NPC through interfering the BART10-3p/ALK7 axis.
Our study has several limitations. First, the role of BART10-3p in NPC differentiation was not explored in vivo. Besides, although we identified ALK7 as a functional mediator of BART10-3p, we did not provide direct evidence concerning the function of ALK7 in NPC. These aspects warrant investigation in future work. In addition, the expression and prognostic value of BART10-3p in NPC were assessed only on microarray data. More reliable and accurate tests, such as quantitative RT-PCR, need to be conducted on clinical samples for confirmation. Despite the limitations, this study represents the first to examine the role and mechanism of BART10-3p in NPC differentiation and proliferation.
In summary, this study distinguishes the oncogenic role of BART10-3p in dedifferentiation and proliferation of NPC via inhibition of ALK7. We also demonstrate the prognostic value of BART10-3p expression. These findings may contribute to further insights into the mechanisms underlying differentiation and proliferation of NPC and provide evidence for targeting the BART10-3p/ALK7 axis as a therapeutic strategy.
Supplemental Material
Supplemental material, sj-pdf-1-ebm-10.1177_15353702211037261 for Epstein-Barr virus microRNA BART10-3p promotes dedifferentiation and proliferation of nasopharyngeal carcinoma by targeting ALK7 by Wei-Jie Luo, Shi-Wei He, Wen-Qing Zou, Yin Zhao, Qing-Mei He, Xiao-Jing Yang, Rui Guo and Yan-Ping Mao in Experimental Biology and Medicine
Footnotes
AUTHORS’ CONTRIBUTIONS: All authors made a significant contribution to the work reported, whether that is in the study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING: This study was supported by the National Natural Science Foundation of China (81672988, 81930072, 81803052); the Natural Science Foundation of Guangdong Province (2017A030312003); and the Sun Yat-Sen University Clinical Research 5010 Program (2017-FXY-114).
ORCID iD: Yan-Ping Mao https://orcid.org/0000-0003-1027-071X
SUPPLEMENTAL MATERIAL: Supplementary material for this article is available online.
References
- 1.Wei WI, Sham JST. Nasopharyngeal carcinoma. Lancet 2005; 365:2041–54 [DOI] [PubMed] [Google Scholar]
- 2.Reddy SP, Raslan WF, Gooneratne S, Kathuria S, Marks JE. Prognostic significance of keratinization in nasopharyngeal carcinoma. Am J Otolaryngol 1995; 16:103–8 [DOI] [PubMed] [Google Scholar]
- 3.Sell S, Nicolini A, Ferrari P, Biava PM. Cancer: a problem of developmental biology; scientific evidence for reprogramming and differentiation therapy. Curr Drug Targets 2016; 17:1103–10 [DOI] [PubMed] [Google Scholar]
- 4.Yamada Y, Haga H, Yamada Y. Concise review: dedifferentiation meets cancer development: proof of concept for epigenetic cancer. Stem Cells Transl Med 2014; 3:1182–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hadar T, Rahima M, Kahan E, Sidi J, Rakowsky E, Sarov B, Sarov I. Significance of specific Epstein-Barr virus IgA and elevated IgG antibodies to viral capsid antigens in nasopharyngeal carcinoma patients. J Med Virol 1986; 20:329–39 [DOI] [PubMed] [Google Scholar]
- 6.Farrell PJ. Epstein-Barr virus and cancer. Annu Rev Pathol 2019; 14:29–53 [DOI] [PubMed] [Google Scholar]
- 7.Kieff E, Rickinson AB. Epstein-Barr virus and its replication. Fields virology. 4th ed. Philadelphia, PA, 2001, pp. 2511–73
- 8.Vojtechova Z, Tachezy R. The role of miRNAs in virus-mediated oncogenesis. Int J Mol Sci 2018; 19:1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. Identification of virus-encoded microRNAs. Science 2004; 304:734–6 [DOI] [PubMed] [Google Scholar]
- 10.Lo AK, Dawson CW, Jin DY, Lo KW. The pathological roles of BART miRNAs in nasopharyngeal carcinoma. J Pathol 2012; 227:392–403 [DOI] [PubMed] [Google Scholar]
- 11.Bernhardt K, Haar J, Tsai MH, Poirey R, Feederle R, Delecluse HJ. A viral microRNA cluster regulates the expression of PTEN, p27 and of a bcl-2 homolog. PLoS Pathog 2016; 12:e1005405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin C, Zong J, Lin W, Wang M, Xu Y, Zhou R, Lin S, Guo Q, Chen H, Ye Y, Zhang B, Pan J. EBV-miR-BART8-3p induces epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma cells through activating NF-kappaB and Erk1/2 pathways. J Exp Clin Cancer Res 2018; 37:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murer A, Rühl J, Zbinden A, Capaul R, Hammerschmidt W, Chijioke O, Münz C. MicroRNAs of Epstein-Barr virus attenuate T-cell-mediated immune control in vivo. mBio 2019; 10:e01941-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Jiang Q, Liu X, Lin X, Tang Z, Liu C, Zhou J, Zhao M, Li X, Cheng Z, Li L, Xie Y, Liu Z, Fang W. Cinobufotalin powerfully reversed EBV-miR-BART22-induced cisplatin resistance via stimulating MAP2K4 to antagonize non-muscle myosin heavy chain IIA/glycogen synthase 3β/β-catenin signaling pathway. EBioMedicine 2019; 48:386–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiu J, Thorley-Lawson DA. EBV microRNA BART 18-5p targets MAP3K2 to facilitate persistence in vivo by inhibiting viral replication in B cells. Proc Natl Acad Sci U S A 2014; 111:11157–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu N, Chen NY, Cui RX, Li WF, Li Y, Wei RR, Zhang MY, Sun Y, Huang BJ, Chen M, He QM, Jiang N, Chen L, Cho WC, Yun JP, Zeng J, Liu LZ, Li L, Guo Y, Wang HY, Ma J. Prognostic value of a microRNA signature in nasopharyngeal carcinoma: a microRNA expression analysis. Lancet Oncol 2012; 13:633–41 [DOI] [PubMed] [Google Scholar]
- 17.Yan Q, Zeng Z, Gong Z, Zhang W, Li X, He B, Song Y, Li Q, Zeng Y, Liao Q, Chen P, Shi L, Fan S, Xiang B, Ma J, Zhou M, Li X, Yang J, Xiong W, Li G. EBV-miR-BART10-3p facilitates epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma by targeting BTRC. Oncotarget 2015; 6:41766–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, Chen J, Zeng M, Li M, Jian S, Pan W, Zhang L, Wu Y. The study of Epstein-Barr virus genome form in nasopharyngeal carcinoma. Chin J Cancer 1997; 16:85–9 [Google Scholar]
- 19.Glaser R, Zhang HY, Yao KT, Zhu HC, Wang FX, Li GY, Wen DS, Li YP. Two epithelial tumor cell lines (HNE-1 and HONE-1) latently infected with Epstein-Barr virus that were derived from nasopharyngeal carcinomas. Proc Natl Acad Sci U S A 1989; 86:9524–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheung ST, Huang DP, Hui AB, Lo KW, Ko CW, Tsang YS, Wong N, Whitney BM, Lee JC. Nasopharyngeal carcinoma cell line (C666-1) consistently harbouring Epstein-Barr virus. Int J Cancer 1999; 83:121–6 [DOI] [PubMed] [Google Scholar]
- 21.Liao WT, Song LB, Zhang HZ, Zhang X, Zhang L, Liu WL, Feng Y, Guo BH, Mai HQ, Cao SM, Li MZ, Qin HD, Zeng YX, Zeng MS. Centromere protein H is a novel prognostic marker for nasopharyngeal carcinoma progression and overall patient survival. Clin Cancer Res 2007; 13:508–14 [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001; 25:402–8 [DOI] [PubMed] [Google Scholar]
- 23.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 2004; 10:7252–9 [DOI] [PubMed] [Google Scholar]
- 24.Yang HJ, Huang TJ, Yang CF, Peng LX, Liu RY, Yang GD, Chu QQ, Huang JL, Liu N, Huang HB, Zhu ZY, Qian CN, Huang BJ. Comprehensive profiling of Epstein-Barr virus-encoded miRNA species associated with specific latency types in tumor cells. Virol J 2013; 10:314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong AM, Kong KL, Tsang JW, Kwong DL, Guan XY. Profiling of Epstein-Barr virus-encoded microRNAs in nasopharyngeal carcinoma reveals potential biomarkers and oncomirs. Cancer 2012; 118:698–710 [DOI] [PubMed] [Google Scholar]
- 26.Gao W, Wong TS, Lv KX, Zhang MJ, Tsang RK, Chan JY. Detection of Epstein-Barr virus (EBV)-encoded microRNAs in plasma of patients with nasopharyngeal carcinoma. Head Neck 2019; 41:780–92 [DOI] [PubMed] [Google Scholar]
- 27.Fotheringham JA, Mazzucca S, Raab-Traub N. Epstein-Barr virus latent membrane protein-2A-induced DeltaNp63alpha expression is associated with impaired epithelial-cell differentiation. Oncogene 2010; 29:4287–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang CF, Yang GD, Huang TJ, Li R, Chu QQ, Xu L, Wang MS, Cai MD, Zhong L, Wei HJ, Huang HB, Huang JL, Qian CN, Huang BJ. EB-virus latent membrane protein 1 potentiates the stemness of nasopharyngeal carcinoma via preferential activation of PI3K/AKT pathway by a positive feedback loop. Oncogene 2016; 35:3419–31 [DOI] [PubMed] [Google Scholar]
- 29.Kanda T, Miyata M, Kano M, Kondo S, Yoshizaki T, Iizasa H. Clustered microRNAs of the Epstein-Barr virus cooperatively downregulate an epithelial cell-specific metastasis suppressor. J Virol 2015; 89:2684–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai L, Long Y, Chong T, Cai W, Tsang CM, Zhou X, Lin Y, Ding T, Zhou W, Zhao H, Chen Y, Wang J, Lyu X, Cho WC, Li X. EBV-miR-BART7-3p imposes stemness in nasopharyngeal carcinoma cells by suppressing SMAD7. Front Genet 2019; 10:939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Min K, Lee SK. EBV miR-BART10-3p promotes cell proliferation and migration by targeting DKK1. Int J Biol Sci 2019; 15:657–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reissmann E, Jornvall H, Blokzijl A, Andersson O, Chang C, Minchiotti G, Persico MG, Ibanez CF, Brivanlou AH. The orphan receptor ALK7 and the activin receptor ALK4 mediate signaling by nodal proteins during vertebrate development. Genes Dev 2001; 15:2010–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuchida K, Nakatani M, Yamakawa N, Hashimoto O, Hasegawa Y, Sugino H. Activin isoforms signal through type I receptor serine/threonine kinase ALK7. Mol Cell Endocrinol 2004; 220:59–65 [DOI] [PubMed] [Google Scholar]
- 34.Andersson O, Korach-Andre M, Reissmann E, Ibanez CF, Bertolino P. Growth/differentiation factor 3 signals through ALK7 and regulates accumulation of adipose tissue and diet-induced obesity. Proc Natl Acad Sci U S A 2008; 105:7252–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pauklin S, Vallier L. Activin/nodal signalling in stem cells. Development 2015; 142:607–19 [DOI] [PubMed] [Google Scholar]
- 36.Carlsson LM, Jacobson P, Walley A, Froguel P, Sjostrom L, Svensson PA, Sjoholm K. ALK7 expression is specific for adipose tissue, reduced in obesity and correlates to factors implicated in metabolic disease. Biochem Biophys Res Commun 2009; 382:309–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michael IP, Saghafinia S, Tichet M, Zangger N, Marinoni I, Perren A, Hanahan D. ALK7 signaling manifests a homeostatic tissue barrier that is abrogated during tumorigenesis and metastasis. Dev Cell 2019; 49:409–24.e6 [DOI] [PubMed] [Google Scholar]
- 38.Ye G, Fu G, Cui S, Zhao S, Bernaudo S, Bai Y, Ding Y, Zhang Y, Yang BB, Peng C. MicroRNA 376c enhances ovarian cancer cell survival by targeting activin receptor-like kinase 7: implications for chemoresistance. J Cell Sci 2011; 124:359–68 [DOI] [PubMed] [Google Scholar]
- 39.Kim BC, van Gelder H, Kim TA, Lee HJ, Baik KG, Chun HH, Lee DA, Choi KS, Kim SJ. Activin receptor-like kinase-7 induces apoptosis through activation of MAPKs in a Smad3-dependent mechanism in hepatoma cells. J Biol Chem 2004; 279:28458–65 [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Zhong W, Zhu M, Hu S, Su X. Nodal regulates bladder cancer cell migration and invasion via the ALK/smad signaling pathway. Onco Targets Ther 2018; 11:6589–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asnaghi L, White DT, Key N, Choi J, Mahale A, Alkatan H, Edward DP, Elkhamary SM, Al-Mesfer S, Maktabi A, Hurtado CG, Lee GY, Carcaboso AM, Mumm JS, Safieh LA, Eberhart CG. ACVR1C/SMAD2 signaling promotes invasion and growth in retinoblastoma. Oncogene 2019; 38:2056–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu WP, Zhang X, Xie WF. Differentiation therapy for solid tumors. J Dig Dis 2014; 15:159–65 [DOI] [PubMed] [Google Scholar]
- 43.Yan M, Zhang Y, He B, Xiang J, Wang ZF, Zheng FM, Xu J, Chen MY, Zhu YL, Wen HJ, Wan XB, Yue CF, Yang N, Zhang W, Zhang JL, Wang J, Wang Y, Li LH, Zeng YX, Lam EW, Hung MC, Liu Q. IKKalpha restoration via EZH2 suppression induces nasopharyngeal carcinoma differentiation. Nat Commun 2014; 5:3661. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ebm-10.1177_15353702211037261 for Epstein-Barr virus microRNA BART10-3p promotes dedifferentiation and proliferation of nasopharyngeal carcinoma by targeting ALK7 by Wei-Jie Luo, Shi-Wei He, Wen-Qing Zou, Yin Zhao, Qing-Mei He, Xiao-Jing Yang, Rui Guo and Yan-Ping Mao in Experimental Biology and Medicine