Abstract
Autophagy is essential to cell function, as it enables the recycling of intracellular constituents during starvation and in addition functions as a quality control mechanism by eliminating spent organelles and proteins that could cause cellular damage if not properly removed. Recently, we reported on Wdfy3’s role in mitophagy, a clinically relevant macroautophagic scaffold protein that is linked to intellectual disability, neurodevelopmental delay, and autism spectrum disorder. In this study, we confirm our previous report that Wdfy3 haploinsufficiency in mice results in decreased mitophagy with accumulation of mitochondria with altered morphology, but expanding on that observation, we also note decreased mitochondrial localization at synaptic terminals and decreased synaptic density, which may contribute to altered synaptic plasticity. These changes are accompanied by defective elimination of glycogen particles and a shift to increased glycogen synthesis over glycogenolysis and glycophagy. This imbalance leads to an age-dependent higher incidence of brain glycogen deposits with cerebellar hypoplasia. Our results support and further extend Wdfy3’s role in modulating both brain bioenergetics and synaptic plasticity by including glycogen as a target of macroautophagic degradation.
Keywords: Glycogen, brain, mitochondria, synapses, electron microscopy
Introduction
Wdfy3 encodes an adaptor molecule centrally required for selective macroautophagy, the starvation-independent, discriminatory recruitment of cellular constituents for autophagic degradation. 1 Homozygous Wdfy3 mutation in mice leads to perinatal lethality, megalencephaly, and global long-range connectivity defects.2,3 Allele-dependent, heterozygous mutation leads to milder neurodevelopmental abnormalities including megalencephaly and diminished long-range connectivity. Human pathogenic WDFY3 variants have been associated with increased risk for intellectual disability/developmental delay, macrocephaly, microcephaly, and neuropsychiatric disorders including autism spectrum disorder (ASD).4–9 While neurodevelopmental defects associated with Wdfy3 loss are well-established, the functional consequences in adulthood remain more elusive. However, suggestions of important roles in this context come from work in Drosophila, where loss of the Wdfy3 homolog bchs, results in shorter lifespan, brain neurodegeneration, and altered endolysosomal transport, comparable to human neurodegenerative disorders, such as Alzheimer’s disease, amyotrophic lateral sclerosis, Wallerian neurodegeneration, and spastic paraplegia. Recent work in modeling Huntington’s disease (HD) in mice further underline the relevance of Wdfy3 function in maintaining brain health, as it apparently acts as a modifier whose depletion will accelerate the course of HD pathogenesis. 10
Our previous studies in Wdfy3+/lacZ mice, revealed persistent Wdfy3 expression in adult brain, motor deficits, and a critical requirement for Wdfy3 in mitophagy, the selective clearance of damaged mitochondria, mitochondrial transport, and axonogenesis.2,7,11 This requirement appears to be critical for brain function, considering that mitophagy is essential in sustaining brain plasticity by enabling mitochondrial trafficking.12,13 Although clearance of damaged mitochondria in Wdfy3+/lacZ mice was partly abrogated by the formation of mitochondria-derived vesicles targeted for lysosomal degradation in a process named micromitophagy, the accumulation of defective mitochondria likely compromised ATP supply, thereby playing a critical role in synaptic plasticity.
Recently, mitochondria have been identified as key organelles modulating the neuronal activity set point for homeostatic plasticity. This is accomplished by different processes, including buffering presynaptic calcium levels, 14 contributing to neurotransmitter synthesis and release in axons and during dendritic development and maintenance. 15 In addition, mitochondria provide local ATP to support protein synthesis required for cytoskeletal rearrangements during neuronal maturation and plasticity,16,17 axonal regeneration through mitochondrial transport, 18 and axonal development via mitochondrial docking and presynaptic regulation.19,20 The above-mentioned synaptic plasticity events along with neural circuits rely heavily on mitochondria-derived ATP; however, other pathways may contribute to sustain neuronal energy, including neuronal glycolysis especially during stress or high activity demands.21–23 However, the balance between energy production and demand may be altered under conditions in which both accumulation of damaged mitochondria and hampered glycogenolysis/glycophagy are evident. Even modest changes in energy availability may result in insufficient synaptic vesicle recycling, ensuing in defective synaptic transmission.
Based on the above concepts, we show here that Wdfy3 loss in Wdfy3+/lacZ mice dually affects brain bioenergetics by not only increasing the accumulation of defective mitochondria, but also increasing the number of glycophagosomes along with an age-dependent accelerated accumulation of brain glycogen. Furthermore, Wdfy3 mutation leads to degenerative processes specific to the adult cerebellum suggesting brain area specific effects of Wdfy3-mediated metabolic dysregulations.
Materials and methods
Animal breeding and husbandry
Wdfy3lacZ (Wdfy3tm1a(KOMP)Mbp) mice were generated and genotyped as previously described 2 and maintained on C57BL/6NJ background as a mixed wild type (WT)/heterozygous mutant colony in facilities approved by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) International. Animals were housed in Plexiglas cages (2–4 animals per cage; 55 x 33 x 19) and maintained under standard laboratory conditions (21 ± 2 °C; 55 ± 5% humidity) on a 12 h light/dark cycle, with ad libitum access to both water and food. The mice were fed with a standard rodent chow. All animals were handled in accordance with protocols approved by the University of California at Davis Institutional Animal Care and Use Committee (protocol #20512) overseen by the AAALAC International accreditation program (latest accreditation in February 14th, 2020) and in compliance with the ARRIVE 24 and NIH guidelines. 25 Animals were processed blindly of sex or genotype by the receiver until all outcomes were collected. While the Wdfy3lacZ allele is a hypomorph, as several lines of evidence suggest, we commonly refer to heterozygous mutants as haploinsufficient (HI). All mice were genotyped for Nnt as described before. 26
Chemicals and biochemicals
EDTA, EGTA, sodium succinate, mannitol, sucrose and HEPES were all purchased from Sigma (St. Louis, MO). Tris-HCl, glycine, sodium chloride, and potassium chloride were purchased from Fisher (Pittsburg, PA). Bovine serum albumin (fatty-acid free) was obtained from MP Biomedicals. All other reagents were of analytical or higher grade.
Untargeted proteomics and integrated bioinformatics to construct functional maps
Cortices from 3-m old female WT and Wdfy3+/lacZ mice (7 each) were used to obtain post-mitochondrial (PM) fractions. Sample preparation has been carried out as described before. 26 Briefly, after homogenization in MSHE buffer (0.22 M mannitol, 70 mM sucrose, 0.1% fatty acid-free BSA, 0.5 mM EGTA, 2 mM HEPES, pH 7.4), large cell debris and nuclei were pelleted by centrifugation at 600 × g for 5 min. Mitochondria were precipitated by centrifuging the supernatant for 10 min at 10,300 × g. The supernatant was called the PM fraction. Untargeted proteomics was performed at the UCD core facility as described in detail before, 27 followed by functional analysis27–30 to determine glucose metabolic pathways in cortex from each genotype allowing to construct a “functional fingerprinting” of intermediary metabolism. Integration of pathways and metabolic outcomes was performed by using bioinformatics approaches as described before.27,31,32 Identified proteins were compared with the latest computational infrastructure that takes advantage of public tools pathway databases (DAVID, Innate, PathVisio, STRING). The resulting differentially expressed proteins were used to perform pathway analyses to determine those altered or different in each of the genotypes vs. WT (NetworkAnalyst). The calculation of power in omics is complex. For example, the number of observed metabolites is unknown a priori and is highly dependent on the analytical platform and the dynamic range of metabolite concentrations. Furthermore, there is strong multicolinearity between variables, arising both from technical sources and possible inter-metabolite associations comprising common metabolic networks. Thus, there is no widely accepted method to determine power, and many times studies are driven by sample availability, based on pilot or literature data. 33 Nevertheless, despite the above limitations, several of our studies have showed a strong discrimination between probands and controls when using ∼20 subjects per diagnostic group.31,32,34
Biochemical evaluation of glycogen
Freshly isolated cortex and cerebellum of WT (n = 3) and Wdfy3+/lacZ (n = 5) 3 m old females was quickly dissected (∼5 min per brain), weighted, adjusted to a concentration of 10 mg tissue/200 µl ice-cold ddiH2O, and homogenized for 10 min on ice. Subsequently, samples were subjected to either sonication (3 strokes of 30 s each for a total of 90 s on ice with a Fisher Scientific Sonic Dismembrator 550) or no sonication. Homogenates were then boiled for 10 min to inactivate enzymes, centrifuged at 18,000 rpm for 10 min and supernatants were collected for glycogen levels analysis. Biochemical quantification of glycogen was performed by a commercial glycogen colorimetric assay kit (#169558, Abcam) following the manufacturer’s recommendations. Briefly, 50 µl of supernatant and glycogen standards were transferred to a 96 well plate, followed by incubation with 2 µl of hydrolysis enzyme mix for 30 min. Subsequently, the samples were incubated with 50 µl reaction mix for 30 min at room temperature. Absorption was followed with a Tecan Infinite M200 microplate reader at 450 nm. Glycogen concentration was calculated from the calibration curve obtained with glycogen standards. The final glycogen concentration was computed by subtracting the background value (the signal without hydrolysis enzyme mix).
Electron microscopy studies
Young adults (2–3 m old; 2 males per genotype) were anesthetized by isoflurane and tissues fixed by transcardiac perfusion with 25 ml phosphate-buffered saline (PBS) for 10 min, followed by 50 ml of fixative (4% paraformaldehyde in PBS) for 15 min. Tissues were rinsed twice in 0.1 M NaH2PO4 for a total of 30 min and placed in 1% osmium tetroxide, 0.1 M NaH2PO4 for 45 min. Tissues were then rinsed again in 0.1 M NaH2PO4, dehydrated in increasing concentrations of ethanol (from 50%, 75%, 95% and 100%). Propylene oxide was used as transitional solvent. Tissues were then pre-infiltrated overnight in a 50:50 ratio propylene oxide:resin. The following day, tissues were infiltrated with 100% resin for 5 h, and subsequently embedded in fresh resin. The embedded tissues were sectioned with an ultramicrotome at a thickness of 90 nm and collected on copper mesh grids. The sections were mounted on collodion-coated copper grids and stained with 4% uranyl acetate for 30 min and for 2 min in 0.2% lead citrate in 0.1 N NaOH. Images were taken with FEI Talos L120C TEM microscope.
In interpreting the EM images, a synaptosome was defined as a clearly membrane-bound body containing 3 or more vesicles of 40-60 nm diameter (i.e. the typical diameter of synaptic vesicles). Synaptosome-like structures without intact plasma membrane were not considered as synaptosomes. Myelin was identified by its multilamellar structure. Myelin was measured as the length of transect line between the two widest points of intersection of a profile. Mitochondria were identified by the presence of a double membrane and cristae and were measured from outer membrane to outer membrane. Coated vesicles were identified by their size, typically 50-80 nm, and the characteristic electron-dense material adherent to their outer aspect. Unidentified material included all other profiles present, whether discretely membrane-bound or not.
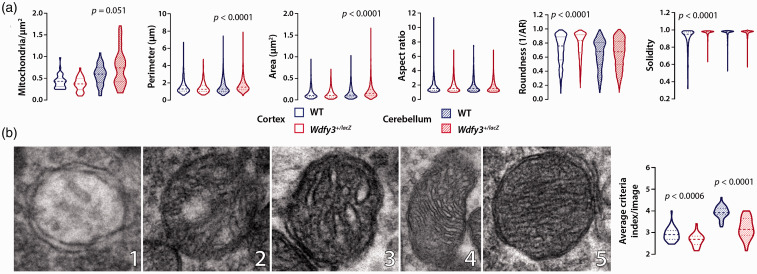
Using ImageJ software, 35 images from both brain regions and both genotypes were examined and analyzed. In total, we analyzed 855 mitochondria from 36 images of the WT mice and 2055 mitochondria from 46 images of the Wdfy3 mutant mice for cerebellum and 452 mitochondria in 38 images from two Wdfy3 mutant mice and 505 mitochondria in 39 images of cortices from WT mice. We focused on several key parameters, the first of which, size, which was quantified by area and perimeter of each mitochondrion. To quantify the images, the components (mitochondria and synapses) had to be identified by ImageJ, then visualized and (if needed) retraced by hand for morphological analysis. Mitochondria were identified as electron dense, roughly tubular structures with a visible double membrane and distinguishable cristae, identifiable through ImageJ. From the traced mitochondria, parameters of mitochondrial morphology, obtained with the ImageJ software, were the following: area, perimeter, aspect ratio (AR = major/minor axis), roundness (1/AR) and solidity (the fraction of pixels contained with a convex polygon fitted around a mitochondrion). Aspect ratio was calculated automatically by ImageJ by fitting an ellipse to the traced shape and determining the ratio between the major and minor axes and by utilizing the formula; [major axis]/[minor axis]. This formula calculates the ratio of the major axis to the minor axis of the fit ellipse (i.e., the smallest ellipse that circumscribes the selected mitochondrion). A value of 1 indicates a perfect circle. Values increase indefinitely as mitochondria become increasingly elongated. Roundness (inverse of aspect ratio) is reported as index of circularity. Circularity was also measured automatically by ImageJ with the formula (4π x area)/(perimeter2). On the circularity scale, a score of 1.0 is representative of a perfect circle, while values tend toward zero as mitochondria become increasingly elongated. Solidity [area/(convex hull area)] calculates the ratio of a mitochondrion’s area to the area of the smallest convex shape that can encircle the mitochondrion. A value of 1 indicates no concavity. Values tend toward 0 as mitochondria become more concave. In other words, it represents the shape of individual mitochondria, with values closer to 0 indicating more regular and less branched mitochondria. Solidity values closer to zero describe highly branched mitochondria, whereas higher solidity values (closer to 1) tend to describe more uniformly shaped mitochondria with low branching. 36 Likewise, synapses were characterized as the electron-dense junction between two cells with an abundance of synaptic vesicles carrying neurotransmitters. Additional measurements taken were regarding the relative densities of mitochondria and of synapses between genotypes. Mitochondria per synapse were also assessed by counting the number of mitochondria per frame and dividing by the number of synapses in the same area. This value did not mean to describe the number of mitochondria directly surrounding each synapse but rather the overall ratio of mitochondria to synapses. The median value and quartiles of the data sets were found and compared. Analysis of mitochondrial morphology was assessed by evaluating criteria for the five categories based on 37 and modified as it follows: 5—Intact, sharply defined, numerous and/or regular cristae; 4—Occasional swollen cristae, slightly irregular packing, round mitochondrion; 3—Fragmented and/or swollen cristae, irregularly packed; 2—Severely fragmented and/or swollen cristae, warped membranes; 1—Delamination of inner and outer mitochondrial membranes, absent cristae.
PAS staining
Brain slices from mice (cortex and cerebellum from 2 of each genotype, all male aged 3-4 m or 4 of each genotype aged 7-8 m) were stained with periodic acid–Schiff (PAS) as described. 38 Briefly, 60 µm-sectioned slices were oxidized by a treatment with 0.5% periodic acid for 10 min, followed by incubation in saturated dimedone aqueous solution at 60˚C for 30 min. Reaction with Schiff’s reagent was conducted for 5 min at room temperature. This analysis was performed at the UCD CPL Facility.
Statistical analyses
Type of data distribution was tested for all the outcomes with D’Agostino & Pearson normality test. In instances of normally distributed data and for comparisons of two groups, the Student’s unpaired t-test was used, whereas for multiple group comparisons, one-way ANOVA followed by the Bonferroni’s post-hoc test. In instances of non-normally distributed data, the non-parametric Mann-Whitney test was applied. Significance was set at p < 0.05. Outliers were computed with the ROUT (Robust Regression and Outlier removal) method. Statistical analysis was computed by using GraphPad Prism (version 8.1.2).
Results
Brain carbohydrate metabolism is altered in Wdfy3+/lacZ mice
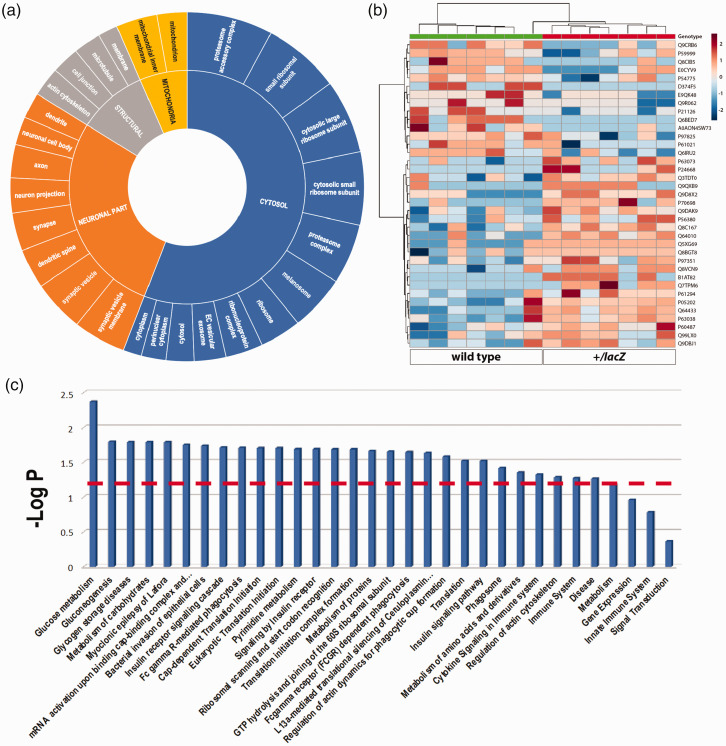
To assess whether Wdfy3 loss impairs brain carbohydrate metabolism and, consequently, brain bioenergetics, we performed untargeted proteomics on cytosolic fractions from cerebral cortex of each genotype. This approach yielded 1,531 differentially expressed proteins that according to the gene ontology cellular compartment enrichment analysis were, as expected, associated with the following subcellular compartments: cytosol, ribosomes, synapses, axons, dendrites, cytoskeleton, and mitochondria associated with ER (Figure 1(a)). To visualize inter- as well as intra-group variabilities in an unsupervised manner and present differential expression levels of the proteomes associated with either genotype, we opted for a heat map display (Figure 1(b)). The known cellular roles of identified proteins and their relative contents were assessed by pathway analysis utilizing the Reactome and KEGG databases. While this approach identified differentially expressed proteins associated with a multitude of pathways, we recognized a notable overrepresentation of pathways associated with carbohydrate metabolism (glucose metabolism, glycogen storage diseases, metabolism of carbohydrates, myoclonic epilepsy of Lafora, and insulin signaling) (Figure 1(c)). Indeed, the top association was with glucose metabolism suggesting a critical involvement of Wdfy3 in glucose processing and storage. Further, enrichment analysis of differentially expressed proteins that took significantly coordinated pathway shifts into account, indicated that pathways related to carbohydrate metabolism (including glycogen processing) were predominantly downregulated (Table 1).
Figure 1.
Cellular location and pathway overrepresentation analyses. Subcellular localization analysis (a) identified by untargeted proteomics performed on cerebral cortical cytosolic fractions of WT and Wdfy3+/lacZ mice. The identified 1,531 proteins were enriched (only the top quartile is shown after performing enrichment analysis with the GO:CC feature in g:Profiler 114 ) in the indicated cellular subcomponent. A heat map representation (b) was chosen to show individual protein levels selected by setting the p-value threshold at 0.05 for the Student’s t test. Pathway overrepresentation analysis (c) obtained by using as input proteins with significantly differential expression between genotypes suggested a critical involvement of Wdfy3 in glucose processing and storage. Data were filtered by the interquartile range (IQR) and normalized for each individual sum. Analysis was performed by using MetaboAnalyst, setting the -LOG (p-value) < 1.3. Pathways were ranked form left to right by most to least dysregulated.
Table 1.
Enrichment analysis results performed on proteomics of brain cortex.
|
Downregulated |
Upregulated |
||||
|---|---|---|---|---|---|
| Pathway | Enrichment | -LOGP | Pathway | Enrichment | -LOGP |
| Tight junction | 15.24 | 3.56 | Translation | 13.82 | 4.38 |
| Amyotrophic lateral sclerosis (ALS) | 5.36 | 3.00 | SRP-dependent co-translational protein targeting to membrane | 14.05 | 3.81 |
| NRF2 pathway | 3.17 | 3.00 | Lysosome | 16.11 | 3.73 |
| Carbohydrate digestion and absorption | 20.00 | 2.78 | Eukaryotic translation elongation | 13.66 | 3.14 |
| SLC-mediated transmembrane transport | 15.17 | 2.53 | Formation of a pool of free 40S subunits | 13.49 | 3.02 |
| Platelet calcium homeostasis | 20.00 | 2.47 | Metabolism of proteins | 12.17 | 3.01 |
| Protein digestion and absorption | 18.18 | 2.31 | Peptide chain elongation | 13.59 | 2.91 |
| Proximal tubule bicarbonate reclamation | 17.14 | 2.27 | L13a-mediated translational silencing of ceruloplasmin expression | 13.19 | 2.81 |
| Amyotrophic lateral sclerosis | 15.45 | 2.18 | Cap-dependent translation initiation | 13.13 | 2.76 |
| Phosphatidylinositol signaling system | 15.45 | 2.18 | Eukaryotic translation initiation | 13.13 | 2.76 |
| Reduction of cytosolic Ca2+ levels | 20.00 | 2.16 | Cytoplasmic ribosomal proteins | 11.69 | 2.70 |
| Serotonin and anxiety | 3.64 | 2.15 | Eukaryotic translation termination | 13.42 | 2.63 |
| Ion transport by P-type ATPases | 16.25 | 2.06 | NMD independent of the exon junction complex (EJC) | 13.33 | 2.58 |
| Transport of inorganic cations/anions and amino acids/oligopeptides | 16.25 | 2.06 | Lipid metabolism pathway | 4.65 | 2.52 |
| Metabolism of xenobiotics by cytochrome P450 | 18.00 | 2.03 | GTP hydrolysis and joining of the 60S ribosomal subunit | 12.98 | 2.51 |
| Co-stimulation by the CD28 family | 16.92 | 2.03 | Ribosome | 13.09 | 2.35 |
| Glycerolipid metabolism | 16.92 | 2.03 | Gene expression | 11.70 | 2.27 |
| Pathways in cancer | 13.33 | 2.00 | PPAR alpha pathway | 3.57 | 2.15 |
| GABA receptor signaling | 4.36 | 1.88 | NMD enhanced by the exon junction complex | 12.84 | 2.07 |
| Effects of PIP2 hydrolysis | 20.00 | 1.85 | NMD | 12.84 | 2.07 |
| Huntington's disease | 12.13 | 1.77 | Transcription factor CREB and its extracellular signals | 16.92 | 1.90 |
| Glycogen metabolism | 6.38 | 1.77 | TGF-epithelial-mesenchymal transition | 4.00 | 1.89 |
| Robo4 and VEGF signaling pathways crosstalk | 5.71 | 1.77 | Activation of cAMP-dependent PKA | 20.00 | 1.77 |
| Endocytosis | 12.50 | 1.69 | Activation of CSK by cAMP-dependent protein kinase inhibits signaling through the T cell receptor | 20.00 | 1.77 |
| Transmembrane transport of small molecules | 11.71 | 1.59 | Glucagon signaling in metabolic regulation | 15.56 | 1.76 |
| Glycolysis/gluconeogenesis | 13.55 | 1.56 | Signaling events mediated by HDAC Class II | 16.67 | 1.67 |
| Attenuation phase | 20.00 | 1.54 | Eicosanoid synthesis | 1.78 | 1.55 |
| Glycogen breakdown | 20.00 | 1.54 | Centrosome maturation | 14.55 | 1.53 |
| SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion | 20.00 | 1.54 | Recruitment of mitotic centrosome proteins and complexes | 14.55 | 1.53 |
| Vitamin C metabolism | 20.00 | 1.54 | Collagen degradation | 20.00 | 1.48 |
| Folding of actin by CCT/TriC | 17.50 | 1.50 | DNA Damage/telomere stress induced senescence | 20.00 | 1.48 |
| Sema3A PAK dependent Axon repulsion | 17.50 | 1.50 | Senescence-associated secretory phenotype | 20.00 | 1.48 |
| Orphan transporters | 14.29 | 1.49 | Canonical NF-kB pathway | 20.00 | 1.48 |
| Bile secretion | 15.00 | 1.49 | Gata3 participate in activating the th2 cytokine genes expression | 20.00 | 1.48 |
| GABA synthesis, release, reuptake and degradation | 15.00 | 1.49 | Mechanism of protein import into the nucleus | 20.00 | 1.48 |
| Neuroactive ligand-receptor interaction | 15.00 | 1.49 | Repression of pain sensation by the transcriptional regulator dream | 20.00 | 1.48 |
| Oxidation by cytochrome P450 | 1.07 | 1.47 | Validated transcriptional targets of deltaNp63 isoforms | 20.00 | 1.48 |
| DNA Damage Response (only ATM dependent) | 2.88 | 1.46 | Loss of Nlp from mitotic centrosomes | 14.74 | 1.45 |
| Neurotransmitter metabolism | 2.61 | 1.46 | Loss of proteins required for interphase microtubule organization from the centrosome | 14.74 | 1.45 |
| Caloric restriction and aging | 3.08 | 1.40 | Degradation of the extracellular matrix | 16.36 | 1.44 |
| The citric acid cycle and respiratory electron transport | 11.67 | 1.39 | Amyloids | 17.50 | 1.42 |
| VEGF signaling pathway | 14.44 | 1.39 | Retinoid metabolism and transport | 17.50 | 1.42 |
| Polyol pathway | 3.08 | 1.35 | ChreBP regulation by carbohydrates and cAMP | 17.50 | 1.42 |
| Extracellular vesicle-mediated signaling in recipient cells | 1.94 | 1.33 | Regulation of BAD phosphorylation | 17.50 | 1.42 |
| Netrin-1 signaling | 16.00 | 1.31 | Glucagon-like peptide-1 regulates insulin secretion | 14.29 | 1.35 |
| Somatroph axis and its relationship to dietary restriction and aging | 3.33 | 1.30 | G-protein beta:gamma signaling | 15.38 | 1.29 |
| Ion channel transport | 13.33 | 1.30 | Phagosome | 12.61 | 1.22 |
| Dopaminergic Neurogenesis | 1.88 | 1.29 | |||
| NAD+ metabolism | 1.76 | 1.29 | |||
Note: The differential proteome was analyzed against the following pathway databases: KEGG, Reactome, Wikipathways, NETPath, PID Biocarta and PID NCI by utilizing InnateDB. 115 The table shows the combined results organized by enrichment and negative log adjusted p-value.
Notably, following the same trend as glycogen metabolism, pathways associated with neurotransmission were also downregulated further supporting the link between mitochondria- and glycogen-derived ATP and neurotransmission.39–41 Our proteomic analysis indicated a downregulation of mainly gamma aminobutyric acid (GABAergic) neurotransmission (Table 1). Several proteins involved in pre- and postsynaptic GABAergic (GABA-T, VGAT, PKA) as well as glutamatergic (GRK, PKA, EAAT, HOMER, ERK1/2, GIGO, IP3R) neurotransmission were found to be downregulated in Wdfy3+/lacZ cortex (Figure S1). The levels of critical enzymes involved in glucose metabolism including the tricarboxylic acid (TCA) cycle (Figure S1; Table 1) were also downregulated, as energy production and neurotransmitter synthesis and recycling are bidirectionally interconnected processes (e.g., GABA shunt; Figure S1). Among the upregulated pathways, lysosome and phagosome formation, translation, as well as protein and lipid metabolism appeared notable (Table 1; Figure S2).
Shift to glycogen synthesis vs. catabolism in Wdfy3 HI independent of lysosome content
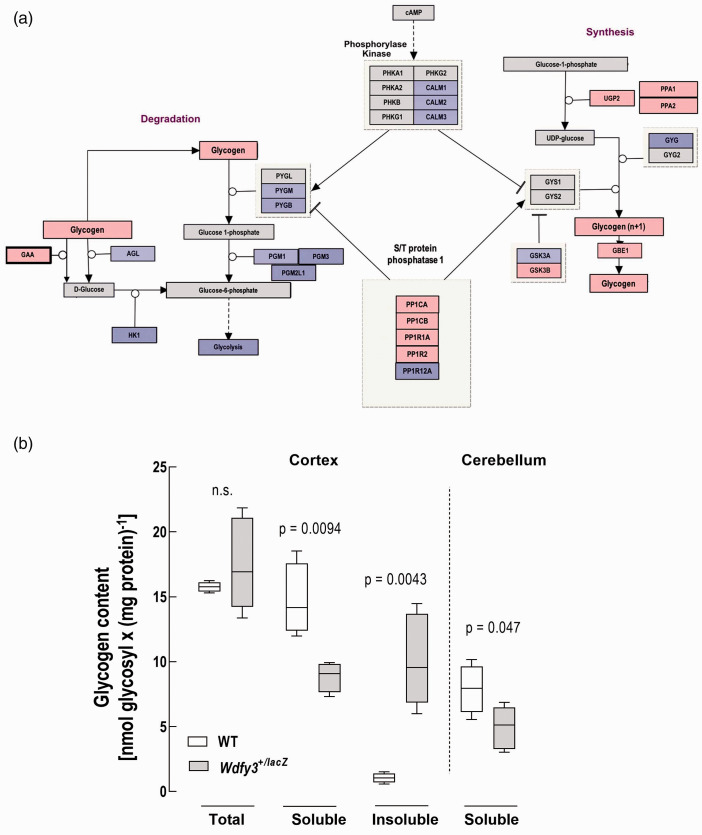
A more detailed analysis of carbohydrate metabolism indicated that Wdfy3 haploinsufficiency (HI) was associated with a shift towards glycogen synthesis at the expense of glycogen catabolism (Figure 2(a)). Particularly, overexpression of UTP-glucose-1-phosphate uridylyltransferase (Ugp2), 1,4-alpha-glucan-branching enzyme (Gbe1), and Ser/Thr-protein phosphatase PP1 (Ppp1) complex catalytic subunits alpha (Ppp1ca) and beta (Ppp1cb) in combination with lower content of its inhibitors Ppp1r1a and Ppp1r2 were indicative of increased glycogen synthesis, further confirmed by decreased levels of Gsk3a. However, decreased hexokinase 1 (Hk1) levels, required to form glucose-6-phosphate from glucose, and glycogenin, which serves as a starting point for glycogen synthesis, were also noted (Figure 2(a)), suggesting a combination of fewer glycogen granules with higher glycosyl units. Down-regulation of glycogen catabolism in Wdfy3+/lacZ mice was supported by decreased expression of glycogen phosphorylase (Pyg), phosphoglucomutase (Pgm), and debranching enzyme (Agl; Figure 2(a)). With respect to Lafora disease, a condition characterized by the accumulation of polyglucosans, Wdfy3+/lacZ mice showed overexpression of two of the five proteins encoded by Lafora disease-causing genes42–46 namely the laforin interacting proteins Epm2ap1 and the mitochondrial iron-sulfur cluster scaffold protein Hirip/Nfu1 were overexpressed in Wdfy3+/lacZ mice (log2 FC = 2.18 and 2.13, respectively).
Figure 2.
Glycogen pathway and content in cerebellum and cortex of WT and Wdfy3+/lacZ mice. Wikipathways generated visualization of the glycogen pathway (a) indicate proteins with more than LOG2 fold change in the proteomic analysis and used as input values for Pathvisio (red overexpressed and blue, underexpressed). Glycogen levels (b) are shown as box plots (median with IQR) showing total, soluble (cortex and cerebellum) and insoluble (cortex) glycogen levels assessed in WT and Wdfy3+/lacZ mice. Insoluble, membrane-bound glycogen content was calculated as the difference between total glycogen (obtained upon sample sonication) and soluble glycogen (nonsonicated samples). Statistical analysis of the data shown was carried out by Student’s t-test.
Glycophagy comprises the breakdown of intralysosomal glycogen mediated by acid α-glucosidase (Gaa47,48; In Wdfy3+/lacZ mice Gaa was overexpressed (log2 FC = 2.07), indicating that this enzyme was not the limiting step in glycophagy impairment of Wdfy3+/lacZ mice. To discern whether Gaa overexpression was an isolated phenomenon rather than a generalized increase in total lysosomal content, we analyzed the expression of proteins associated with the gene ontology term “lysosome” (Figure S2(a)). Wdy3 HI was associated with increased expression of constitutive lysosomal proteins (including proteases, glycosylases, lipases, ceramidase, subunits of the vacuolar ATPase or ATPV, Lamp1, among others), as well as other proteins associated with lysosomal biogenesis (Ap1/Ap3, Mpr). This finding suggested a generalized upregulation of lysosomal biogenesis (Table 1; Figure S2(a)) possibly as a compensatory mechanism to Wdfy3-mediated deficits in selective macroautophagy.
Glycophagy requires not only functional lysosomes but also active autophagy.49,50 Consequently, using the gene ontology term “phagosome” in the KEGG pathway database in conjunction with Pathview, 51 we sought to identify possible dysregulations in the expression of proteins associated with phagosome formation (Figure S2(b)). While components required for autophagosomal membrane nucleation and lysosomal fusion were overrepresented in Wdfy3+/lacZ mice compared with WT (Figure S2(b), in red; Table 1), factors needed for the phagophore complex (Atgs, Wif1, and Rab33b) were underrepresented (Figure S2(b), in blue; Table 1). These results were consistent with Wdfy3’s established role in phagosome formation by association with the Atg16l complex as we reported before. 11
Defective brain glycophagy in Wdfy3+/lacZ mice
To shed light into whether accumulated glycogen was readily accessible in its cytosolic form or sequestered in phagolysosomes, we evaluated the glycogen content in sonicated and nonsonicated samples from cortex and cerebellum obtained from WT and Wdfy3+/lacZ mice (Figure 2(b)). Values of sonicated samples were considered to reflect total glycogen, whereas values of naive samples were considered as accessible or soluble cytosolic glycogen. The difference between these two sets of values was representative of insoluble glycogen, sequestered within membrane-bound structures. Irrespective of sonication, samples containing glycogen were treated with amyloglucosidase (releases glucose from the hydrolysis of 1,4-, 1,6- and 1,3-α-D-glucosidic bonds) to ascertain the nature of the bonds within glycosydic residues. The free, soluble amount of glycogen was significantly lower in cortex of Wdfy3+/lacZ mice (53%; Figure 2(b)) with a concomitant increase in insoluble, but not total, glycogen (Figure 2(b)). A similar, albeit non-significant, trend was observed for soluble glycogen in cerebellum of Wdfy3+/lacZ mice, suggesting that other brain regions showed to a lesser extent this imbalance (Figure 2(b)). No significant difference was recorded between total and soluble cortical glycogen in WT mice (Figure 2(b)), suggesting that most glycogen (∼88%) is readily accessible in its soluble form. Of note, and as expected, total cortical and cerebellar glycogen contents in WT mice were respectively one- and two-orders of magnitude lower than that of the glycogen-rich organs skeletal muscle and liver 52 and consistent with several other studies,53–56 but lower than the highest reported values 57 (Table S1).
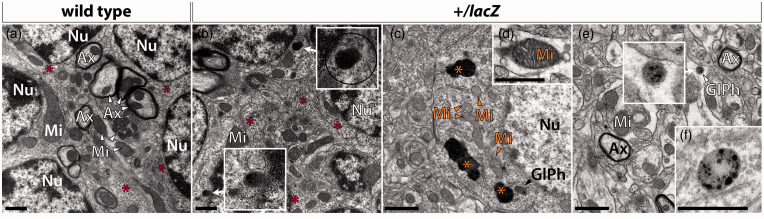
As the above results implied an accumulation of glycophagosomes in Wdfy3+/lacZ mice, we next sought to visualize glycogen distribution in cortex and cerebellum by utilizing electron microscopy. We identified electron opaque particles exhibiting ultrastructural features generally attributed to β-type glycogen58,59 that were distinguishable from other similarly sized particles by selectively enhancing electron density utilizing lead citrate staining. 60 In our preparations, other particulate structures - primarily ribosomes - exhibited about the same density as those in osmium tetroxide and uranyl acetate-stained preparations. Glycogen particles in WT cerebellum and cortex were abundant, appeared predominantly as a single particle (β-type) of 20-40 nm in diameter, and more seldom as compound particles (α-type), opposite to those noted in Wdfy3+/lacZ cerebellum (Figure 3(a) and (b)). Glycogen was associated with some profiles of the endoplasmic reticulum and occasionally in secondary lysosomes (Figure 3(c)). The electron microscopy analysis further revealed that Wdfy3 HI was associated with lipofuscin deposits (Figure 3(c)) in both cerebellum and cortex. These deposits appeared as highly electron-opaque, non-membrane bound, cytoplasmic aggregates consistent with the appearance of lipofuscin. While lipofuscin deposits appeared more numerous in cerebellum and cortex of Wdfy3+/lacZ mice, their highly irregular distribution and uncertain association with individual cells made their precise quantification impossible. We also noted in the mutants a buildup of mitochondria with distorted morphology, vacuolization, faded outer membranes, and formation of mitochondria-derived vesicles (Figure 3(c) and (d)). In addition, in Wdfy3+/lacZ mice the incidence of glycophagosomes was two-fold higher than in WT and typically presented as membrane-bound larger structures with dense matrix and/or accumulation of punctate material (Figure 3(e) and (f)).
Figure 3.
Aberrant subcellular glycogen deposits, glycophagosomes, and mitochondria in Wdfy3+/lacZ cerebellum and cortex. Representative TEM images (x 11,000) of WT (a) and Wdfy3+/lacZ cerebellum (b) and cortex (c–f). Red asterisks indicate glycogen particles that are dispersed in the cytosol. Glycogen particles included into secondary lysosomes are shown in the insets in (b). These secondary lysosomes appear as highly electron-opaque, non-membrane bound, cytoplasmic lipofuscin deposits. Orange arrowheads point to mitochondria with distorted morphology, vacuolization (d), faded outer membranes, and formation of mitochondria-derived vesicles. Glycophagosomes (GlPh) were noted in Wdfy3+/lacZ cortex (c), as well as highly electron-opaque lipofuscin deposits (orange asterisks in c). All scale bars are 1 μm.
Ax: axon; Mi: mitochondrion; Nu: nucleus.
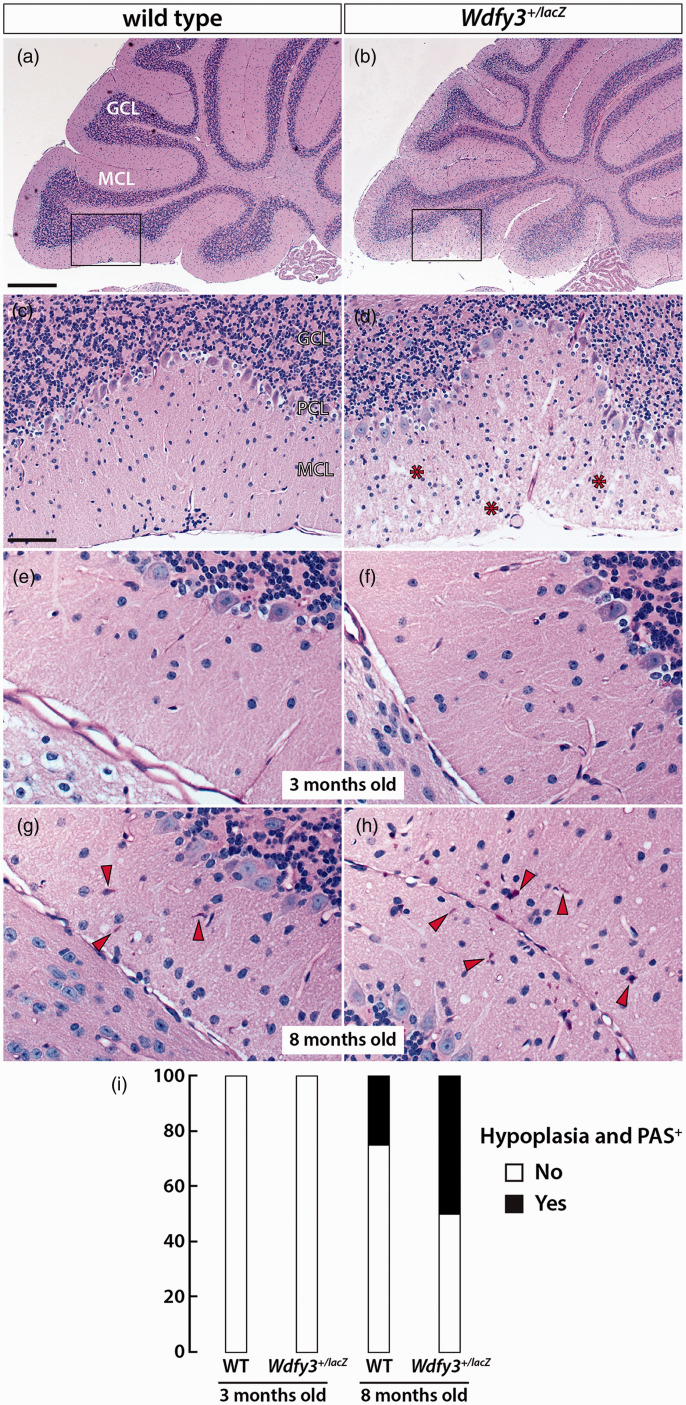
These results were comparable to those observed in Pompe disease. This disorder presents with a characteristic longitudinal trajectory of ever increasing severity, 61 accompanied by a decline of patchy glycogen with increases in high-intensity PAS positive clots (named polyglucosan bodies), 62 lipofuscin, as well as lysosomal and autophagy defects.63–65 Taking these observations into account, we wanted to test the effects of older age on the formation of brain glycogen deposits in Wdfy3+/lacZ mice. Histological analysis of H&E (Figure 4(a) to (d)) and periodic acid–Schiff (PAS) stained brain slices (Figure 4(e) to (h)) revealed cerebellar hypoplasia and accumulation of PAS+ material with disorganization of the granule and Purkinje cell layers in 7-8 m old mice (Figure 4(g) and (h)). None of these neuropathological features were observed in either WT or Wdfy3+/lacZ mice at 3-5 m of age (Figure 4(e) and (f)). Although these changes were evident in both genotypes with age, the incidence of the PAS+ material was almost 2-fold higher in Wdfy3+/lacZ mice compared to age-matched WT mice (Figure 4(i)).
Figure 4.
Age- and Wdfy3-dependent cerebellar neurodegeneration and glycogen accumulation. H&E staining was performed on brain slices from 3 m old WT and 8 m old Wdfy3+/lacZ mice (a–d). White arrows indicate cerebellar hypoplasia. Asterisks indicate areas of cellular disarray. Brain glycogen was evidenced in brain slices from 3 (e, f) and 8 m (g, h) old WT and Wdfy3+/lacZ mice by staining with PAS. Red arrows indicate glycogen deposits which is visualized by a pink color. Incidence of brain hypoplasia and glycogen with age in WT and Wdfy3+/lacZ mice was calculated at 3–5 m and 7–8 m of age (i). Scale bars are 500 μm in A, B and 100 μm in (c–h).
GCL: granular cell layer; PCL: Purkinje cell layer; MCL: cellular layer.
Downregulation of synaptic neurotransmission pathways in cerebellum is reflected in decreased number of synapses and accumulation of aberrant synaptic mitochondria of Wdfy3+/lacZ mice
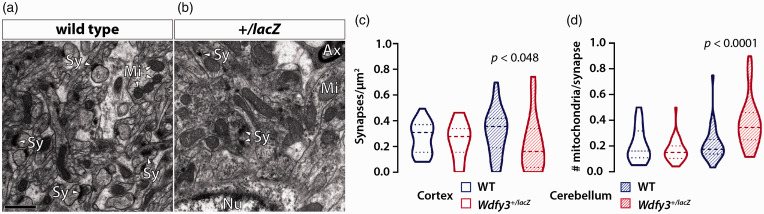
“Healthy” brain circuitry requires active glycogenolysis and functional mitochondria for adequate synaptic density, activity, and plasticity.12,13 We reasoned that deficits in selective macroautophagy may not only compromise fuel metabolism between glia and neurons, but also neurotransmission and synaptogenesis. To further explore this question and potentially identify ultrastructural morphological features that may explain the different effects of Wdfy3 loss on cortex compared to cerebellum, we performed transmission electron microscopy (TEM) to quantify mitochondria and their morphological features (area, perimeter, aspect ratio, roundness, and solidity), number of synapses, and analyze the expression of proteins involved in pre- and postsynaptic transmission. Our data confirmed in 2-3-months-old cerebellum, but not cortex, of Wdfy3+/lacZ mice, an increased number of enlarged mitochondria (Figure 5(a)). In cortex, the roundness and solidity of mitochondria were increased in Wdfy3+/lacZ compared with WT. Moreover, altered packing of cristae with fragmentation and delamination of inner and/or outer membrane was also noted in both brain regions based on a modified score system for evaluating mitochondrial morphology 37 (Figure 5(b)). Mitochondria with disrupted cristae and outer membrane (identified by lower scores) were evidenced in cortex (7%) and even more so in cerebellum (15%) of Wdfy3+/lacZ mice. Overall, the results indicated that defective mitochondrial clearance in Wdfy3+/lacZ resulted in the accumulation of damaged mitochondria with altered ultrastructural morphology. In cerebellum of Wdfy3+/lacZ mice, the number of synapses per µm2 was 30% lower than WT, but no significant changes were observed in cortex (Figure 6(a) to (c)). By combining both data sets (mitochondrial parameters and synapse numbers), the number of accumulated mitochondria with altered morphology per synapse was 1.8-fold higher in Wdfy3+/lacZ cerebellum compared with WT (p < 0.05; Figure 6(d)), but not significantly different in cortex.
Figure 5.
Altered mitochondrial morphology in Wdfy3+/lacZ brains. Mitochondrial parameters were evaluated in TEM images shown in Figure 3 with ImageJ software (a). Number of mitochondria is reported per µm2, and average mitochondrial area and perimeter respectively as µm2 and µm. Representative TEM images (b) show cristae and outer membrane degree of intactness (cerebellum), and quantification (cerebellum and cortex) of mitochondrial morphology. Numbers at bottom right of each micrograph indicates morphological score (from 1 to 5). All other details were included under Materials and Methods. Data are shown as violin plots (median + IQR). Statistical analysis performed with Mann-Whitney test for non-parametric data and significance is considered for p < 0.05.
Figure 6.
Changes in synaptic density and synaptic mitochondrial content in Wdfy3+/lacZ cerebellum. Representative images of cerebellar synapses (Sy; a, b)—defined as electron dense confined vesicular accumulations—and mitochondria in WT and Wdfy3+/lacZ mice. The number of synapse/µm2 and number of mitochondria/synapse are shown in in panels (c and d), respectively. Data are presented as violin plots, showing median and IQR. Statistical analysis was performed with Mann-Whitney test for non-parametric data. Scale bar is 1 μm.
Ax: axon; Mi: mitochondrion; Nu: nucleus.
As TEM did not reveal the contribution of pre- vs. postsynaptic processes to the cerebellar pathological alterations that we observed, we used orthogonal bioinformatic tools to identify biological pathways and cellular compartments relevant to synaptic processes in our differential cortical proteome dataset. We proceeded by testing our dataset for enrichment in proteins annotated to synaptic compartments and processes using the SynGO knowledgebase. 66 The analysis revealed significant enrichment of the input proteins in the presynaptic compartment (p = 3.35 × 10−8) including synaptic vesicles (p = 0.020), presynaptic membranes (p = 0.005), and their integral components (p = 0.022) (Figure S3). The postsynaptic compartment was also enriched (p = 1.3 × 10−5), albeit at a lower significance compared to the presynaptic compartment, mainly at the level of the postsynaptic cytoskeleton (p = 0.00025). In regards to biological function, enriched synaptic processes were constituted by synapse organization (p = 2.01 x 10−5), especially at the level of assembly (p = 0.001). At the presynaptic level (p = 0.0003), synaptic vesicle cycle (p = 0.0003) was over-represented, whereas regulation of postsynaptic membrane neurotransmitter receptor levels (p = 0.017) and chemical synaptic transmission (p = 0.006) were over-represented at the post-synaptic level (p = 0.017).
Taken together, these results indicated a relevant role for presynaptic events, mainly at the level of synaptic vesicle recycling, a process heavily supported by mitochondria-derived ATP in presynaptic terminals. 67
Discussion
The scaffold protein Wdfy3, a central component in selective macroautophagy, has been recognized as an important neurodevelopmental regulator. During prenatal development, Wdfy3 loss-of-function adversely impacts neural proliferation, as well as neuronal migration and connectivity.2,3 What remains much less explored are the consequences of Wdfy3 loss for adult brain function. Our previous work confirmed a requirement for Wdfy3 in regulating mitophagy, the targeted removal of functionally impaired mitochondria that is required for optimal bioenergetics and cell health, particularly so in energy-demanding neurons. 11 Intriguingly, the generation of cytosolic proteomic data and subsequent pathway analysis revealed that differentially expressed cortical proteins that were overrepresented in Wdfy3+/lacZ mice clustered within carbohydrate-associated pathways, namely glucose metabolism, glycogen storage diseases, carbohydrate metabolism, and myoclonic epilepsy of Lafora hinting at a possible role for Wdfy3 in glycogen degradation. Based on these observations, here we expand on Wdfy3’s mitophagic function and provide additional evidence that Wdfy3 mutation negatively affects glycophagy, synaptic density, and neurotransmission, processes connected to synaptic plasticity. Synaptic plasticity presents the dominant model underlying our understanding of how the brain stores information, i.e., how it forms new memories and recalls them, and if pathologically altered how it may affect subjects with autism and intellectual disabilities.68–72
Our results show that Wdfy3 HI decreases the number of synapses in cerebellum, but not cortex, suggesting that autophagic processing or some other Wdfy3-mediated mechanism is relevant to synaptic maintenance especially evident in tissues such as cerebellum with a higher content of neuron-to-glia ratios than cortex (∼10-fold 73 ). This result conforms to other recent findings that link autophagy in neural and non-neural cells (primarily microglia) in controlling dendritic spine pruning in mouse cortex.74,75 While loss of mTORC1-dependent macroautophagy was linked to defective synaptic pruning and altered social behaviors,74,76,77 to our knowledge no studies have implicated selective macroautophagy (i.e., mitophagy and glycophagy) as a critical effector in the same process and by extension brain plasticity. Several lines of evidence provided in this and our previous study support a role for Wdfy3 in modulating synaptic plasticity via coupling to selective macroautohagy. First, Wdfy3 is widely expressed in the postnatal brain, including hippocampal fields that undergo continuous synaptic remodeling. 11 Second, clearance of damaged mitochondria via mitophagy is essential to sustain normal mitochondrial trafficking and brain plasticity.12,13 Third, brain glycogen metabolism is relevant for memory processing78,79 and learning-dependent synaptic plasticity. 80 Fourth, as the balance between energy production and demand is altered when damaged mitochondria and hampered glycogenolysis/glycophagy are present, insufficient synaptic vesicle recycling can be expected resulting in defective synaptic transmission.
Our data point to an imbalance between glycogen synthesis and breakdown in Wdfy3+/lacZ mice, due to an impairment of glycophagy. This scenario is supported by our findings of equal total glycogen content in cortex and cerebellum between genotypes, but significant differences in distribution favoring insoluble glycogen in Wdfy3+/lacZ mice. A plausible explanation for this observation appears to be that routing of glycogen for lysosomal degradation via autophagosomes is diminished in Wdfy3+/lacZ brain due to the Wdfy3-dependent nature of these autophagosomes. This idea is supported by the higher content of lysosomes, but not autophagosomes, and the accumulation of glycophagosomes in the mutant. Although the molecular mechanism by which glycogen is transferred to the lysosome is still poorly understood, our findings suggest a direct requirement of Wdfy3 in this process. Currently, it remains unknown whether glycophagy provides a quantitatively different route of glycogen breakdown compared to phosphorylase-mediated glycogen catabolism. Plausible scenarios may include glycophagy-mediated glucose release in subcellular compartments with high-energy demand, such as synapses, or a different timescale of release to enable sustained or rapid availability. It is also conceivable that glycogen directed for glycophagy may be qualitatively different to that degraded in the cytosol, thus requiring a different route of degradation. For instance, abnormally branched, insoluble, and/or hyperphosphorylated glycogen may inhibit phosphorylase action and favor its recruitment to the glycophagosome. In a related example, loss-of-function of either the phosphatase laforin or the E3 ubiquitin ligase malin results in the accumulation of abnormally branched, hyperphosphorylated glycogen and polyglucosan inclusion bodies called Lafora bodies. 81 As expected, overexpression of laforin prevents stress-induced polyglucosan body formation in neurons, 82 but surprisingly also increases autophagy via the mTOR pathway, 83 providing a link between glycogen catabolism and autophagy. Notably, two of the five Lafora disease-causing genes, encoding the laforin interacting proteins Epm2aip1 44 and Hirip5/Nfu1, 45 showed higher expression in Wdfy3+/lacZ mice. While Epm2aip1 is yet of unknown function, it colocalizes with laforin in polyglucosan formations44,84 suggesting a role in glycogen quality control by preventing the formation of polyglucosans. 84 Relevant to mitochondria biology, the assembly protein Hirip5/Nfu145,85 is critical for the formation of mitochondrial iron-sulfur clusters.85,86
Historically, glycogen metabolism has been described primarily in glia87–91 with a defined role in behaviors associated with memory formation and consolidation 92 [see reviews92,93]. However, at a smaller scale neurons appear to actively metabolize glycogen as well, as they express both glycogen synthase and glycogen phosphorylase, 94 and accumulate some glycogen. 94 Neuronal glycogen has been associated with memory formation and synaptic plasticity, 95 and more recent studies in humans have shown accumulation of glycogen in neurons of the elderly in the form of abnormal glycogen deposits named polysaccharide-based aggregates, or alternatively corpora amylacea. 96 Similar deposits have been discovered in mouse and Drosophila brains, 97 as well as postmortem in frontal cortex of individuals with neurodegenerative disorders (Alzheimer’s and Pick’s diseases and Parkinson disease). 98 The inability to inhibit neuronal glycogen synthesis constitutes the basis of Lafora disease, 99 and impaired activity of glycogen branching enzyme has been reported in adult polyglucosan body disease. 100 Furthermore, targeted downregulation of Drosophila glycogen synthase in neurons improves neurological function with age and extends lifespan. 97 Consistent with these previous reports, we demonstrated that while cerebellar hypoplasia and accumulation of glycogen deposits increased with an animal’s age, their incidence, and likely their onset, was higher in Wdfy3+/lacZ mice suggesting a critical role for Wdfy3 in glycogen degradation and neurodegeneration, mirrored by an age-dependent decline in associative learning, cognitive, and memory-forming processes. Wdfy3 may act in this context as a modifier to disease progression as recently described in a mouse model of HD (BACHD, which expresses a full-length human mutant HTT gene). While Wdfy3 loss on its own would not initiate the accumulation of Htt aggregates, and BACHD mice will show only late-onset selective neuropathology, BACHD-Wdfy3 compound mutants revealed significant increases of Htt aggregates in cortex and striatum of 9 and 12 m old mice. 10 The accumulation of aggregates also correlated with an accelerated onset of HD symptoms in BACHD-Wdfy3 mice further supporting Wdfy3’s role as a disease modifier.
Additional associations exist between neuronal glycogen accumulation, autophagic flux, and HD. Specifically, glycogen deposits have been proposed as neuroprotective agents by enhancing the clearance of mutant Htt protein via activation of the autophagic machinery both in vitro and in a mouse model (R6/2). 98 The authors also showed that PAS+ glycogen deposits can be found in neurons of postmortem brain samples of individuals clinically diagnosed to have Alzheimer’s disease, Pick’s disease, or Parkinson’s disease suggesting a general link between neuronal glycogen and neurodegenerative disorders. However, as that study demonstrated, accumulation of glycogen in healthy neurons is detrimental even when autophagy is overactivated highlighting the delicate balance between glycogen homeostasis and brain function. A link between defective glucose metabolism and neuronal degeneration is also suggested by findings that hexokinase-II (HK-II), which catalyzes the first step of glycolysis, can induce apoptosis in primary neurons in response to glucose depletion. 101 Similarly, glucose deprivation results in dephosphorylation of the glucose metabolism modulator Bad protein (BCL-2-associated agonist of cell death) and Bad-dependent cell death. 102 Incidentally, in Bad mutant mouse lines reduced glucose metabolism increases the activity of metabolically sensitive neuronal K(ATP) channels and confers seizure resistance. 103
While our study did not differentiate between glial and neuronal glycogen, the fact that similar glycogen contents were observed in both cortex and cerebellum, areas with very different ratios of nonneuronal cells-to-neurons,73,104 supports the concept that observed changes also apply to neurons. Differences in glia-neuron ratios may also explain the perplexing differences in phenotypic severity between cortex and cerebellum. The dramatic accumulation of synaptic mitochondria with altered ultrastructural morphology and the lower number of synapses observed in mutant cerebellum compared with cortex may be explained by the relatively lower number of glycogen-containing glia in cerebellum and thus, diminished capacity to compensate for glycophagy impairment.
In summary and in line with other studies linking macroautophagy to synaptic pruning and aberrant behavior,74,76,77 here we suggest that Wdfy3-dependent selective macroautophagy may alter synaptic plasticity impacting neuronal circuits and brain health. The process may involve buffering glucose concentrations in the brain via rapid glycogenolysis as it offsets decreased glucose availability during periods of elevated activity followed by restoration of the glycogen pool during resting periods. 105 In addition, it is crucial for learning and memory processes where increased energy-demanding synaptic activity is required to elicit learning acquisition and storage under physiological conditions.106–109 The association between glucose availability and autophagy regulation has also been recognized in cardiomyocytes and other cells, were hexokinase-II (HK-II) downregulation diminished while overexpression increased glucose deprivation-induced autophagy via TORC1 inhibition. 110 Interestingly, several studies have shown that repression of the activity of glycogen synthase kinase 3 (GSK3), a multifunctional kinase involved in glycogen synthesis and a key modulator of synaptic plasticity, is associated with psychiatric, neurodegenerative and neurodevelopmental disorders,111–113 suggesting that defects in WDFY3 may contribute to the onset and/or morbidity of ASD and intellectual disability/developmental delay. This suggestion fits well with the larger context of Wdfy3-association with neuropsychiatric disorders as revealed by our in silico analysis (Figure S4) connecting several disorders including schizophrenia, global developmental delay, muscle hypotonia, seizures, epilepsy, intellectual disability, and bipolar disorder to Wdfy3 HI.
Electron microscopy images are publicly available at Dryad (doi:10.25338/B8PS6W).
Supplemental Material
Supplemental material, sj-pdf-1-jcb-10.1177_0271678X211027384 for Wdfy3 regulates glycophagy, mitophagy, and synaptic plasticity by Eleonora Napoli, Alexios A Panoutsopoulos, Patricia Kysar, Nathaniel Satriya, Kira Sterling, Bradley Shibata, Denise Imai, David N Ruskin, Konstantinos S Zarbalis and Cecilia Giulivi in Journal of Cerebral Blood Flow & Metabolism
Footnotes
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: KSZ is supported by Shriners Hospitals for Children and NIH grant R21MH115347. DNR is supported by NIH grant R15AT008742. EM analyses were conducted at Campus Research Core Facilities and funded by the UCD Pilot and Feasibility Program to CG. Ms. Sterling and Mr. Satriya performed their work as part of the Young Scholars Program at the University of California, Davis.
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors’ contributions: E.N. evaluated glycogen content, performed the EM studies, analyzed statistically most of the experiments, and contributed to the writing of the study; A. A. P. maintained Wdfy3lacZ mice, collected tissue for biochemical and histological examination; P.K. and B.S. performed tissue preparation for EM studies; N.S. and K.S. evaluated synapse numbers and mitochondrial morphology in EM images; D.I. performed the PAS-associated histology studies; D.N.R provided intellectual input and contributed to the writing; K.S.Z. maintained Wdfy3lacZ mice, collected tissue for biochemical and histological examination, and co-wrote the manuscript; C.G. conceived and design the study, wrote the manuscript and performed the interpretation and statistical analyses of the omics.
ORCID iD: Cecilia Giulivi https://orcid.org/0000-0003-1033-7435
Supplementary material: Supplemental material for this article is available online.
References
- 1.Filimonenko M, Isakson P, Finley KD, et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein ALFY. Mol Cell 2010; 38: 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orosco LA, Ross AP, Cates SL, et al. Loss of Wdfy3 in mice alters cerebral cortical neurogenesis reflecting aspects of the autism pathology. Nat Commun 2014; 5: 4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dragich JM, Kuwajima T, Hirose-Ikeda M, et al. Autophagy linked FYVE (ALFY/WDFY3) is required for establishing neuronal connectivity in the mammalian brain. Elife 2016; 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iossifov I, O'Roak BJ, Sanders SJ, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 2014; 515: 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iossifov I, Ronemus M, Levy D, et al. De novo gene disruptions in children on the autistic spectrum. Neuron 2012; 74: 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kadir R, Harel T, Markus B, et al. ALFY-Controlled DVL3 autophagy regulates Wnt signaling, determining human brain size. PLoS Genet 2016; 12: e1005919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Duc D, Giulivi C, Hiatt SM, et al. Pathogenic WDFY3 variants cause neurodevelopmental disorders and opposing effects on brain size. Brain 2019; 142: 2617–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stessman HA, Xiong B, Coe BP, et al. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat Genet 2017; 49: 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang T, Guo H, Xiong B, et al. De novo genic mutations among a Chinese autism spectrum disorder cohort. Nat Commun 2016; 7: 13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox LM, Kim K, Johnson CW, et al. Huntington's disease pathogenesis is modified in vivo by ALFY/Wdfy3 and selective macroautophagy. Neuron 2020; 105: 813–821 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Napoli E, Song G, Panoutsopoulos A, et al. Beyond autophagy: a novel role for autism-linked Wdfy3 in brain mitophagy. Sci Rep 2018; 8: 11348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacAskill AF, Atkin TA, Kittler JT. Mitochondrial trafficking and the provision of energy and calcium buffering at excitatory synapses. Eur J Neurosci 2010; 32: 231–240. [DOI] [PubMed] [Google Scholar]
- 13.Cai Q, Gerwin C, Sheng ZH. Syntabulin-mediated anterograde transport of mitochondria along neuronal processes. J Cell Biol 2005; 170: 959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Styr B, Gonen N, Zarhin D, et al. Mitochondrial regulation of the hippocampal firing rate set point and seizure susceptibility. Neuron 2019; 102: 1009–1024 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rangaraju V, Lewis TL, Jr, Hirabayashi Y, et al. Pleiotropic mitochondria: the influence of mitochondria on neuronal development and disease. J Neurosci 2019; 39: 8200–8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spillane M, Ketschek A, Merianda TT, et al. Mitochondria coordinate sites of axon branching through localized intra-axonal protein synthesis. Cell Rep 2013; 5: 1564–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rangaraju V, Lauterbach M, Schuman EM. Spatially stable mitochondrial compartments fuel local translation during plasticity. Cell 2019; 176: 73–84 e15. [DOI] [PubMed] [Google Scholar]
- 18.Cartoni R, Norsworthy MW, Bei F, et al. The Mammalian-specific protein Armcx1 regulates mitochondrial transport during axon regeneration. Neuron 2016; 92: 1294–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Courchet J, Lewis TL, Jr, Lee S, et al. Terminal axon branching is regulated by the LKB1-NUAK1 kinase pathway via presynaptic mitochondrial capture. Cell 2013; 153: 1510–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang C, Shin WS, Yeo D, et al. Data on the mode of binding between avenanthramides and IKKbeta domains in a docking model. Data Brief 2018; 17: 994–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashrafi G, Wu Z, Farrell RJ, et al. GLUT4 mobilization supports energetic demands of active synapses. Neuron 2017; 93: 606–615 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diaz-Garcia CM, Mongeon R, Lahmann C, et al. Neuronal stimulation triggers neuronal glycolysis and not lactate uptake. Cell Metab 2017; 26: 361–374 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jang S, Nelson JC, Bend EG, et al. Glycolytic enzymes localize to synapses under energy stress to support synaptic function. Neuron 2016; 90: 278–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Percie Du Sert N, Hurst V, Ahluwalia A, et al. The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. J Cereb Blood Flow Metab 2020; 40: 1769–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.National Research Council (U.S.). Committee for the update of the guide for the care and use of laboratory animals., Institute for Laboratory Animal Research (U.S.), National Academies Press (U.S.). Guide for the care and use of laboratory animals. 8th ed. Washington, D.C.: National Academies Press, 2011, xxv, 220 p. [Google Scholar]
- 26.Napoli E, Song G, Liu S, et al. Zdhhc13-dependent Drp1 S-palmitoylation impacts brain bioenergetics, anxiety, coordination and motor skills. Sci Rep 2017; 7: 12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Napoli E, Tassone F, Wong S, et al. Mitochondrial citrate transporter-dependent metabolic signature in the 22q11.2 deletion syndrome. J Biol Chem 2015; 290: 23240–23253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujisawa Y, Napoli E, Wong S, et al. Impact of a novel homozygous mutation in nicotinamide nucleotide transhydrogenase on mitochondrial DNA integrity in a case of familial glucocorticoid deficiency. BBA Clin 2015; 3: 70–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pon J, Napoli E, Luckhart S, et al. Mitochondrial NAD+-dependent malic enzyme from Anopheles stephensi: a possible novel target for malaria mosquito control. Malar J 2011; 10: 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vernau K, Napoli E, Wong S, et al. Thiamine deficiency-mediated brain mitochondrial pathology in Alaskan huskies with mutation in SLC19A3.1. Brain Pathol 2015; 25: 441–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giulivi C, Napoli E, Tassone F, et al. Plasma metabolic profile delineates roles for neurodegeneration, pro-inflammatory damage and mitochondrial dysfunction in the FMR1 premutation. Biochem J 2016; 473: 3871–3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giulivi C, Napoli E, Tassone F, et al. Plasma biomarkers for monitoring brain pathophysiology in FMR1 premutation carriers. Front Mol Neurosci 2016; 9: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blaise BJ, Correia G, Tin A, et al. Power analysis and sample size determination in metabolic phenotyping. Anal Chem 2016; 88: 5179–5188. [DOI] [PubMed] [Google Scholar]
- 34.Napoli E, Song G, Schneider A, et al. Warburg effect linked to cognitive-executive deficits in FMR1 premutation. Faseb J 2016; 30: 3334–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valente AJ, Maddalena LA, Robb EL, et al. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem 2017; 119: 315–326. [DOI] [PubMed] [Google Scholar]
- 36.Westrate LM, Drocco JA, Martin KR, et al. Mitochondrial morphological features are associated with fission and fusion events. PLoS One 2014; 9: e95265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coughlin L, Morrison RS, Horner PJ, et al. Mitochondrial morphology differences and mitophagy deficit in murine glaucomatous optic nerve. Invest Ophthalmol Vis Sci 2015; 56: 1437–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kong J, Shepel PN, Holden CP, et al. Brain glycogen decreases with increased periods of wakefulness: implications for homeostatic drive to sleep. J Neurosci 2002; 22: 5581–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goncalves CA, Rodrigues L, Bobermin LD, et al. Glycolysis-derived compounds from astrocytes that modulate synaptic communication. Front Neurosci 2018; 12: 1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dienel GA, Rothman DL. Glycogenolysis in cerebral cortex during sensory stimulation, acute hypoglycemia, and exercise: impact on astrocytic energetics, aerobic glycolysis, and astrocyte-neuron interactions. Adv Neurobiol 2019; 23: 209–267. [DOI] [PubMed] [Google Scholar]
- 41.Dienel GA. Does shuttling of glycogen-derived lactate from astrocytes to neurons take place during neurotransmission and memory consolidation? J Neurosci Res 2019; 97: 863–882. [DOI] [PubMed] [Google Scholar]
- 42.Minassian BA, Lee JR, Herbrick JA, et al. Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy. Nat Genet 1998; 20: 171–174. [DOI] [PubMed] [Google Scholar]
- 43.Chan EM, Young EJ, Ianzano L, et al. Mutations in NHLRC1 cause progressive myoclonus epilepsy. Nat Genet 2003; 35: 125–127. [DOI] [PubMed] [Google Scholar]
- 44.Ianzano L, Zhao XC, Minassian BA, et al. Identification of a novel protein interacting with laforin, the EPM2a progressive myoclonus epilepsy gene product. Genomics 2003; 81: 579–587. [DOI] [PubMed] [Google Scholar]
- 45.Ganesh S, Tsurutani N, Suzuki T, et al. The lafora disease gene product laforin interacts with HIRIP5, a phylogenetically conserved protein containing a NifU-like domain. Hum Mol Genet 2003; 12: 2359–2368. [DOI] [PubMed] [Google Scholar]
- 46.Fernandez-Sanchez ME, Criado-Garcia O, Heath KE, et al. Laforin, the dual-phosphatase responsible for lafora disease, interacts with R5 (PTG), a regulatory subunit of protein phosphatase-1 that enhances glycogen accumulation. Hum Mol Genet 2003; 12: 3161–3171. [DOI] [PubMed] [Google Scholar]
- 47.Kotoulas OB, Kalamidas SA, Kondomerkos DJ. Glycogen autophagy. Microsc Res Tech 2004; 64: 10–20. [DOI] [PubMed] [Google Scholar]
- 48.Kotoulas OB, Kalamidas SA, Kondomerkos DJ. Glycogen autophagy in glucose homeostasis. Pathol Res Pract 2006; 202: 631–638. [DOI] [PubMed] [Google Scholar]
- 49.Fukuda T, Roberts A, Plotz PH, et al. Acid alpha-glucosidase deficiency (pompe disease). Curr Neurol Neurosci Rep 2007; 7: 71–77. [DOI] [PubMed] [Google Scholar]
- 50.Raben N, Roberts A, Plotz PH. Role of autophagy in the pathogenesis of pompe disease. Acta Myol 2007; 26: 45–48. [PMC free article] [PubMed] [Google Scholar]
- 51.Luo W, Pant G, Bhavnasi YK, et al. Pathview web: user friendly pathway visualization and data integration. Nucleic Acids Res 2017; 45: W501–W8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tagliabracci VS, Girard JM, Segvich D, et al. Abnormal metabolism of glycogen phosphate as a cause for lafora disease. J Biol Chem 2008; 283: 33816–33825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sagar SM, Sharp FR, Swanson RA. The regional distribution of glycogen in rat brain fixed by microwave irradiation. Brain Res 1987; 417: 172–174. [DOI] [PubMed] [Google Scholar]
- 54.Cloix JF, Tahi Z, Boissonnet A, et al. Brain glycogen and neurotransmitter levels in fast and slow methionine sulfoximine-selected mice. Exp Neurol 2010; 225: 274–283. [DOI] [PubMed] [Google Scholar]
- 55.Picard M, Cloix JF, Hevor TK. Serotonergic neurotransmission plays a major role in the action of the glycogenic convulsant methionine sulfoximine. Neurosci Res 2011; 70: 313–320. [DOI] [PubMed] [Google Scholar]
- 56.Fan Z, Zhang Z, Zhao S, et al. Dynamic variations in brain glycogen are involved in modulating isoflurane anesthesia in mice. Neurosci Bull 2020; 36: 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsui T, Omuro H, Liu YF, et al. Astrocytic glycogen-derived lactate fuels the brain during exhaustive exercise to maintain endurance capacity. Proc Natl Acad Sci USA 2017; 114: 6358–6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Revel JP, Napolitano L, Fawcett DW. Identification of glycogen in electron micrographs of thin tissue sections. J Biophys Biochem Cytol 1960; 8: 575–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maxwell DS, Kruger L. The fine structure of astrocytes in the cerebral cortex and their response to focal injury produced by heavy ionizing particles. J Cell Biol 1965; 25: 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reynolds ES. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol 1963; 17: 208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hagemans ML, Winkel LP, Hop WC, et al. Disease severity in children and adults with pompe disease related to age and disease duration. Neurology 2005; 64: 2139–2141. [DOI] [PubMed] [Google Scholar]
- 62.Oe Y, Baba O, Ashida H, et al. Glycogen distribution in the microwave-fixed mouse brain reveals heterogeneous astrocytic patterns. Glia 2016; 64: 1532–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grune T, Jung T, Merker K, et al. Decreased proteolysis caused by protein aggregates, inclusion bodies, plaques, lipofuscin, ceroid, and ‘aggresomes' during oxidative stress, aging, and disease. Int J Biochem Cell Biol 2004; 36: 2519–2530. [DOI] [PubMed] [Google Scholar]
- 64.Yang DS, Stavrides P, Saito M, et al. Defective macroautophagic turnover of brain lipids in the TgCRND8 alzheimer mouse model: prevention by correcting lysosomal proteolytic deficits. Brain 2014; 137: 3300–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brandenstein L, Schweizer M, Sedlacik J, et al. Lysosomal dysfunction and impaired autophagy in a novel mouse model deficient for the lysosomal membrane protein Cln7. Hum Mol Genet 2016; 25: 777–791. [DOI] [PubMed] [Google Scholar]
- 66.Koopmans F, van Nierop P, Andres-Alonso M, et al. SynGO: an evidence-based, expert-curated knowledge base for the synapse. Neuron 2019; 103: 217–234 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pathak D, Shields LY, Mendelsohn BA, et al. The role of mitochondrially derived ATP in synaptic vesicle recycling. J Biol Chem 2015; 290: 22325–22336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bhakar AL, Dolen G, Bear MF. The pathophysiology of fragile X (and what it teaches us about synapses). Annu Rev Neurosci 2012; 35: 417–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bernardinelli Y, Nikonenko I, Muller D. Structural plasticity: mechanisms and contribution to developmental psychiatric disorders. Front Neuroanat 2014; 8: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Phillips M, Pozzo-Miller L. Dendritic spine dysgenesis in autism related disorders. Neurosci Lett 2015; 601: 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nishiyama J. Plasticity of dendritic spines: molecular function and dysfunction in neurodevelopmental disorders. Psychiatry Clin Neurosci 2019; 73: 541–550. [DOI] [PubMed] [Google Scholar]
- 72.Banerjee A, Miller MT, Li K, et al. Towards a better diagnosis and treatment of Rett syndrome: a model synaptic disorder. Brain 2019; 142: 239–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bandeira F, Lent R, Herculano-Houzel S. Changing numbers of neuronal and non-neuronal cells underlie postnatal brain growth in the rat. Proc Natl Acad Sci USA 2009; 106: 14108–14113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim HJ, Cho MH, Shim WH, et al. Deficient autophagy in microglia impairs synaptic pruning and causes social behavioral defects. Mol Psychiatry 2017; 22: 1576–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lieberman OJ, McGuirt AF, Tang G, et al. Roles for neuronal and glial autophagy in synaptic pruning during development. Neurobiol Dis 2019; 122: 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bowling H, Klann E. Shaping dendritic spines in autism spectrum disorder: mTORC1-dependent macroautophagy. Neuron 2014; 83: 994–996. [DOI] [PubMed] [Google Scholar]
- 77.Tang G, Gudsnuk K, Kuo SH, et al. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron 2014; 83: 1131–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gibbs ME, Anderson DG, Hertz L. Inhibition of glycogenolysis in astrocytes interrupts memory consolidation in young chickens. Glia 2006; 54: 214–222. [DOI] [PubMed] [Google Scholar]
- 79.Suzuki A, Stern SA, Bozdagi O, et al. Astrocyte-neuron lactate transport is required for long-term memory formation. Cell 2011; 144: 810–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Duran J, Saez I, Gruart A, et al. Impairment in long-term memory formation and learning-dependent synaptic plasticity in mice lacking glycogen synthase in the brain. J Cereb Blood Flow Metab 2013; 33: 550–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Turnbull J, Wang P, Girard JM, et al. Glycogen hyperphosphorylation underlies lafora body formation. Ann Neurol 2010; 68: 925–933. [DOI] [PubMed] [Google Scholar]
- 82.Wang Y, Ma K, Wang P, et al. Laforin prevents stress-induced polyglucosan body formation and lafora disease progression in neurons. Mol Neurobiol 2013; 48: 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aguado C, Sarkar S, Korolchuk VI, et al. Laforin, the most common protein mutated in lafora disease, regulates autophagy. Hum Mol Genet 2010; 19: 2867–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chan EM, Ackerley CA, Lohi H, et al. Laforin preferentially binds the neurotoxic starch-like polyglucosans, which form in its absence in progressive myoclonus epilepsy. Hum Mol Genet 2004; 13: 1117–1129. [DOI] [PubMed] [Google Scholar]
- 85.Olive JA, Cowan JA. Role of the HSPA9/HSC20 chaperone pair in promoting directional human iron-sulfur cluster exchange involving monothiol glutaredoxin 5. J Inorg Biochem 2018; 184: 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Melber A, Winge DR. Steps toward understanding mitochondrial Fe/S cluster biogenesis. Methods Enzymol 2018; 599: 265–292. [DOI] [PubMed] [Google Scholar]
- 87.Brown AM. Brain glycogen re-awakened. J Neurochem 2004; 89: 537–552. [DOI] [PubMed] [Google Scholar]
- 88.Cataldo AM, Broadwell RD. Cytochemical identification of cerebral glycogen and glucose-6-phosphatase activity under normal and experimental conditions. II. Choroid plexus and ependymal epithelia, endothelia and pericytes. J Neurocytol 1986; 15: 511–524. [DOI] [PubMed] [Google Scholar]
- 89.Hertz L, Xu J, Song D, et al. Astrocytic glycogenolysis: mechanisms and functions. Metab Brain Dis 2015; 30: 317–333. [DOI] [PubMed] [Google Scholar]
- 90.Quach TT, Duchemin AM, Rose C, et al. 3[H]glycogenolysis in brain slices mediated by beta-adrenoceptors: comparison of physiological response and [3H]dihydroalprenolol binding parameters. Neuropharmacology 1988; 27: 629–635. [DOI] [PubMed] [Google Scholar]
- 91.Xu J, Song D, Bai Q, et al. Basic mechanism leading to stimulation of glycogenolysis by isoproterenol, EGF, elevated extracellular K+ concentrations, or GABA. Neurochem Res 2014; 39: 661–667. [DOI] [PubMed] [Google Scholar]
- 92.Obel LF, Muller MS, Walls AB, et al. Brain glycogen – new perspectives on its metabolic function and regulation at the subcellular level. Front Neuroenergetics 2012; 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kreft M, Bak LK, Waagepetersen HS, et al. Aspects of astrocyte energy metabolism, amino acid neurotransmitter homoeostasis and metabolic compartmentation. ASN Neuro 2012; 4: AN20120007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Saez I, Duran J, Sinadinos C, et al. Neurons have an active glycogen metabolism that contributes to tolerance to hypoxia. J Cereb Blood Flow Metab 2014; 34: 945–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Duran J, Gruart A, Varea O, et al. Lack of neuronal glycogen impairs memory formation and learning-dependent synaptic plasticity in mice. Front Cell Neurosci 2019; 13: 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cavanagh JB. Corpora-amylacea and the family of polyglucosan diseases. Brain Res Brain Res Rev 1999; 29: 265–295. [DOI] [PubMed] [Google Scholar]
- 97.Sinadinos C, Valles-Ortega J, Boulan L, et al. Neuronal glycogen synthesis contributes to physiological aging. Aging Cell 2014; 13: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rai A, Singh PK, Singh V, et al. Glycogen synthase protects neurons from cytotoxicity of mutant huntingtin by enhancing the autophagy flux. Cell Death Dis 2018; 9: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Minassian BA. Lafora's disease: towards a clinical, pathologic, and molecular synthesis. Pediatr Neurol 2001; 25: 21–29. [DOI] [PubMed] [Google Scholar]
- 100.Robitaille Y, Carpenter S, Karpati G, et al. A distinct form of adult polyglucosan body disease with massive involvement of central and peripheral neuronal processes and astrocytes: a report of four cases and a review of the occurrence of polyglucosan bodies in other conditions such as lafora's disease and normal ageing. Brain 1980; 103: 315–336. [DOI] [PubMed] [Google Scholar]
- 101.Mergenthaler P, Kahl A, Kamitz A, et al. Mitochondrial hexokinase II (HKII) and phosphoprotein enriched in astrocytes (PEA15) form a molecular switch governing cellular fate depending on the metabolic state. Proc Natl Acad Sci USA 2012; 109: 1518–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Danial NN, Gramm CF, Scorrano L, et al. BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature 2003; 424: 952–956. [DOI] [PubMed] [Google Scholar]
- 103.Gimenez-Cassina A, Martinez-Francois JR, Fisher JK, et al. BAD-dependent regulation of fuel metabolism and K(ATP) channel activity confers resistance to epileptic seizures. Neuron 2012; 74: 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Herculano-Houzel S, Mota B, Lent R. Cellular scaling rules for rodent brains. Proc Natl Acad Sci USA 2006; 103: 12138–12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shulman RG, Hyder F, Rothman DL. Cerebral energetics and the glycogen shunt: neurochemical basis of functional imaging. Proc Natl Acad Sci USA 2001; 98: 6417–6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Brown AM, Tekkok SB, Ransom BR. Glycogen regulation and functional role in mouse white matter. J Physiol 2003; 549: 501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dienel GA, Ball KK, Cruz NF. A glycogen phosphorylase inhibitor selectively enhances local rates of glucose utilization in brain during sensory stimulation of conscious rats: implications for glycogen turnover. J Neurochem 2007; 102: 466–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sickmann HM, Walls AB, Schousboe A, et al. Functional significance of brain glycogen in sustaining glutamatergic neurotransmission. J Neurochem 2009; 109 Suppl 1: 80–86. [DOI] [PubMed] [Google Scholar]
- 109.Swanson RA. Physiologic coupling of glial glycogen metabolism to neuronal activity in brain. Can J Physiol Pharmacol 1992; 70 Suppl: S138–44. [DOI] [PubMed] [Google Scholar]
- 110.Roberts DJ, Tan-Sah VP, Ding EY, et al. Hexokinase-II positively regulates glucose starvation-induced autophagy through TORC1 inhibition. Mol Cell 2014; 53: 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Beaulieu JM. Not only lithium: regulation of glycogen synthase kinase-3 by antipsychotics and serotonergic drugs. Int J Neuropsychopharmacol 2007; 10: 3–6. [DOI] [PubMed] [Google Scholar]
- 112.Caracci MO, Avila ME, De Ferrari GV. Synaptic Wnt/GSK3beta signaling hub in autism. Neural Plast 2016; 2016: 9603751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hur EM, Zhou FQ. GSK3 signalling in neural development. Nat Rev Neurosci 2010; 11: 539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Raudvere U, Kolberg L, Kuzmin I, et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 2019; 47: W191–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Breuer K, Foroushani AK, Laird MR, et al. InnateDB: systems biology of innate immunity and beyond – recent updates and continuing curation. Nucleic Acids Res 2013; 41: D1228–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-jcb-10.1177_0271678X211027384 for Wdfy3 regulates glycophagy, mitophagy, and synaptic plasticity by Eleonora Napoli, Alexios A Panoutsopoulos, Patricia Kysar, Nathaniel Satriya, Kira Sterling, Bradley Shibata, Denise Imai, David N Ruskin, Konstantinos S Zarbalis and Cecilia Giulivi in Journal of Cerebral Blood Flow & Metabolism