Abstract
Drosophila dachshund is necessary and sufficient for compound eye development and is required for normal leg and brain development. A mouse homologue of dachshund, Dach1, is expressed in the developing retina and limbs, suggesting functional conservation of this gene. We have generated a loss-of-function mutation in Dach1 that results in the abrogation of the wild-type RNA and protein expression pattern in embryos. Homozygous mutants survive to birth but exhibit postnatal lethality associated with a failure to suckle, cyanosis, and respiratory distress. The heart, lungs, kidneys, liver, and skeleton were examined to identify factors involved in postnatal lethality, but these organs appeared to be normal. In addition, blood chemistry tests failed to reveal differences that might explain the lethal phenotype. Gross examination and histological analyses of newborn eyes, limbs, and brains revealed no detectable abnormalities. Since Dach1 mutants die shortly after birth, it remains possible that Dach1 is required for postnatal development of these structures. Alternatively, an additional Dach homologue may functionally compensate for Dach1 loss of function.
The control of retinal determination (RD) in Drosophila requires eyeless, sine oculis, eyes absent, and dachshund, which function together as components of a network (1, 6, 30, 33). These genes are expressed prior to photoreceptor differentiation and are required for retinal morphogenesis (2, 7, 25, 31). Moreover, eyeless, eyes absent, and dachshund are sufficient for inducing retinal fates, as targeted misexpression results in ectopic eye formation from nonneuronal tissues. eyeless is a member of the Pax gene family of transcription factors (9). sine oculis encodes a protein possessing a homeodomain motif with DNA-binding activity (17). eyes absent and dachshund encode novel nuclear proteins lacking DNA-binding motifs but have been proposed to act as transcription cofactors (2, 6, 25). The relationships between the RD genes cannot be summarized completely by a simple, linear pathway. Although Eyeless activates transcription of eyes absent and sine oculis, which in turn appear to activate dachshund, there is evidence of extensive cross-regulation between the RD genes (1, 6, 14, 30, 33). For example, misexpression of Dachshund induces ectopic eyeless, eyes absent, and sine oculis expression (6, 33). Together these findings suggest that the Drosophila RD genes cooperate as components of a network to control cell fate decisions by regulating target gene expression.
Vertebrate homologues of eyeless (Pax6), eyes absent (Eya1 to Eya3), sine oculis (Six3 and Six6), and dachshund (Dach) that are expressed in the developing eye have been identified (3, 4, 10, 11, 15, 20, 21, 29, 31, 37, 41). Evidence of a functional role for some of these genes during eye development has also been demonstrated. Mutations in Pax6 and PAX6 result in the Small eye phenotype in mice and ANIRIDIA in humans, respectively (12, 13, 19, 31, 35, 36). Furthermore, misexpression of Pax6 results in ectopic eye formation in Xenopus (8). Similarly, Six3 misexpression leads to ectopic lens and retina formation in the teleost Medaka (24, 28). Finally, reminiscent of the regulatory relationship between eyes absent and eyeless in Drosophila (14), expression of mouse Eya1 and Eya2 in the lens placode is dependent on Pax6 (41). These data suggest that the Drosophila RD network has been conserved in vertebrates. However, it is not known whether all vertebrate homologues of the Drosophila RD genes function during eye development and if these genes operate in the same or parallel pathways.
A mouse homologue of dachshund, Dach (hereafter referred to as Dach1), has been cloned, and its embryonic expression pattern has been characterized (4, 11, 15, 21, 25). Comparison of Dachshund and Dach1 sequences reveals the presence of two highly conserved sequences, Dachshund domain 1 (DD1) and DD2 (11). The Dachshund N terminus, including DD1, autonomously activates transcription in yeast, and DD2 has been shown to physically interact with Eyes absent, suggesting that Dachshund regulates gene expression through protein-protein interactions facilitated by DD1 and DD2 (6). Expression analysis of Dach1 demonstrates that it is expressed in the developing retina, limbs, and nervous system, which is analogous to the expression pattern of Drosophila dachshund (4, 11, 15, 25). As dachshund mutant flies lack eyes, have truncated limbs, and display defects in brain development, the structural and expression similarities shared between Dach1 and dachshund suggest a similar role for Dach1 during vertebrate development (22, 25, 26).
Here we present the construction of a Dach1 knockout allele and characterize the effect of this mutation on Dach1 expression and mouse development. Despite the abrogation of the wild-type embryonic expression pattern, gross and histological analyses of the homozygous mutant eyes, limbs, and brains revealed no detectable malformations. At the time of birth, the frequency of the homozygous mutant and wild-type alleles are roughly Mendelian. However, this mutation is associated with postnatal lethality, a failure to suckle, and respiratory distress, although additional analyses failed to reveal a cause for this phenotype.
MATERIALS AND METHODS
Generation of a Dach1 knockout allele.
A 600-bp SmaI-EcoRI fragment of human DACH cDNA clone 381801RG (Research Genetics) was used to screen a 129/SvEv genomic phage library. Three Dach1 genomic clones were isolated and mapped relative to each other and Dach1 cDNA sequences. The genomic clones contained the first coding exon: 218 bp of 5′ untranslated region, the putative translational start codon, 827 bp of open reading frame, and a splice site that is conserved between mouse Dach1 and Drosophila dachshund (11). These sequences encode amino acids 1 to 276, a region which includes most (98 of 107 amino acids) of the conserved DD1 domain. Dach1 genomic fragments flanking exon 1 were subcloned into pUSEFUL for assembly of a targeting vector (Allan Bradley, Baylor College of Medicine). Specifically, a 5′ recombination arm (3.7-kb SalI-SacI fragment) and a 3′ recombination arm (5.0-kb KpnI-SalI fragment) were subcloned into pUSEFUL flanking PGK-Hprt sequences (see Fig. 1A). The targeting vector was linearized with NotI and electroporated into the embryonic stem (ES) cell line AB2.1. Since homologous replacement abolishes a SacI site upstream of exon 1 (see Fig. 1A), SacI-digested Hprt+/herpes simplex virus thymidine kinase-negative AB2.1 colony DNA was screened by Southern analysis using 5′ and 3′ probes (flanking the recombination arms and not included in the targeting vector). As a consequence of recombination, PGK-Hprt sequences replace 319 bp upstream of exon 1, Dach1 exon 1, and approximately 2 kb of intron 1 (see Fig. 2A). From separate electroporations, two independent replacement events were isolated (ES cell lines B and G). In addition to the expected replacement, additional sequences derived from the targeting vector were inserted at the Dach1 locus (see Fig. 1A). A PCR-based assay was also developed for detecting the Dach1 mutant allele. Primers pl (5′-ACATGCACATACGCACACTTT) and p3 (5′-AAGAGGTCAAGACAGGAACATCA) amplify a 265-bp product from the wild-type allele, and primers p3 and p2 (5′-AGGCCACTTGTGTAGCGCCAA) produce a 381-bp amplicon from the mutant allele. The positions of these primers are shown in Fig. 1A.
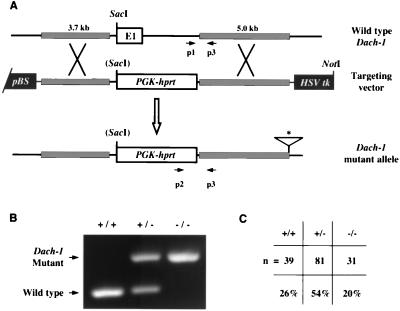
FIG. 1.
Dach1 knockout strategy and genotype analysis. (A) Dach1 targeting strategy. The top line shows the wild-type 5′ Dach1 region with exon 1 (white box) and intron 1 sequences (thin line) flanked by 5′ and 3′ recombination arms (grey boxes). The middle line shows the Dach1 knockout construct designed for homologous replacement of a SacI site, exon 1, and a 5′ portion of intron 1 with PGK-Hprt sequences. The resultant Dach1 mutant allele is illustrated on the bottom line. Positions of genotyping primers p1, p2, and p3 are shown as arrows below the wild-type and mutant allele diagrams. The line below the asterisk represents additional sequences, derived from the targeting vector, inserted at the Dach1 locus which were associated with two independently isolated homologous replacement events (Materials and Methods). pBS, pBluescript; HSV tk, herpes simplex virus thymidine kinase gene. (B) PCR genotype analysis of tail DNA. An ethidium bromide-stained agarose gel containing amplification products from wild-type (+/+), heterozygote (+/−), and homozygote (−/−) tail DNA is shown. Primer combination p1-p3 detects wild-type alleles, while primers p2 and p3 detect mutant alleles (Materials and Methods). (C) Genotype analysis of newborn mice. Tail DNA was collected from 151 newborn mice (21 intercross litters) and analyzed by PCR. The numbers and corresponding percentages of wild-type (+/+), heterozygote (+/−), and homozygote (−/−) mutant animals are shown. Tails from these mice were taken less than 2 h after birth. Homozygotes may be slightly underrepresented since they may be eaten prior to detection.
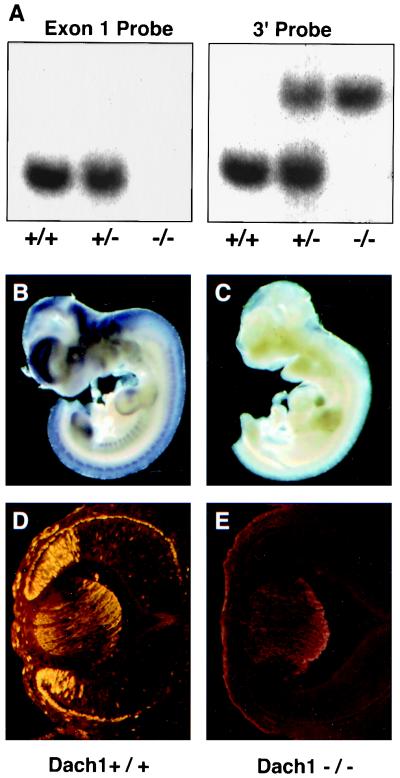
FIG. 2.
Targeted disruption of Dach1 exon1 is associated with abrogated RNA and protein expression. (A) Southern analysis of the Dach1 locus. Wild-type (+/+), heterozygote (+/−), and homozygote (−/−) newborn tail DNA was digested with SacI, subjected to gel electrophoresis, and transferred to a nylon membrane. The filter was hybridized to a Dach1 exon 1 probe (Fig. 1A), washed, and exposed to film. Subsequently, the filter was stripped and hybridized to a 3′ Dach1 probe (Materials and Methods). (B and C) Whole-mount in situ hybridization of E10 wild-type and homozygous mutant embryos. A Dach1 antisense riboprobe hybridizes to transcripts in the eye, limbs, neural tube, and brain in wild-type embryos (B), while this pattern is not detected in homozygous mutants (C) (11). This riboprobe corresponds to coding sequences located downstream of exon 1 and therefore is not deleted in the mutant allele. Tails were removed from the animals for PCR genotyping after color development. (D and E) Immunohistochemical analysis of E12.5 wild-type and homozygous mutant eye sections. A Dach1 antibody demonstrates protein expression within the peripheral retina, retinal pigmented epithelium, and developing cornea in wild-type eyes (D), while Dach1 staining is reduced to background levels in the homozygous mutant eyes (E). Lens and surface ectoderm staining is due to background staining of the secondary antibody.
In situ hybridization.
A Dach1 riboprobe was prepared as previously described (11). A Dachl EcoRI cDNA fragment (amino acid 365 to the stop codon and 386 bp of 3′ untranslated region) was cloned into the EcoRI site of pBluescript KS(+). These cDNA sequences are downstream of exon 1 and therefore are not deleted in the Dachl mutant allele. An antisense riboprobe was synthesized using T7 RNA polymerase. In situ hybridization of whole-mount embryos was performed as described elsewhere (32). Digoxigenin-labeled tissues were detected using antidigoxigenin antibody (FAb fragments; Boehringer Mannheim) coupled to alkaline phosphatase and nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate development (Boehringer Mannheim).
Immunohistochemistry.
Dach1 cDNA sequences encoding amino acids 461 to 538 (11) were subcloned into pGEX4T to create a glutathione S-transferase (GST)–Dach1 expression construct. GST-Dach1 fusion protein was purified from Escherichia coli using glutathione-agarose beads and injected into rabbits (Cocalico Biologicals, Inc.). The serum isolated from rabbits was then used for immunohistochemistry. Embryos were fixed in 4% paraformaldehyde–phosphate-buffered saline (PBS) for 3 h at 4°C. After several washes in PBS, the embryos were equilibrated in 30% sucrose–PBS and transferred to OCT embedding medium. Tissue sections (8 to 10 μM) were incubated in blocking solution (1% goat serum, 0.1% Triton X-100, PBS) for at least 30 min, washed briefly in PBS, and then incubated with 1/500 dilution of anti-Dach1 antiserum in blocking solution overnight at 4°C. The sections were then washed several times in PBS and incubated with Cy3 secondary antibody in blocking solution for 1 h at room temperature. Sections were then washed, mounted on slides, and visualized.
Histology.
Embryos and tissues were dissected, washed in cold PBS, and fixed in fresh 4% paraformaldehyde–PBS overnight at 4°C with gentle agitation. For central nervous system histology, newborn heads and necks were fixed in 10% formalin followed by decalcification in 10% formic acid. Tissues were washed twice in PBS for 30 min and then dehydrated with a PBS–distilled dH2O-ethanol (EtOH) series. In preparation for paraffin embedding, dehydrated tissues were washed once in 1:1 xylene-EtOH for 15 min and once in xylene for 30 min. Tissues were immediately washed in a 1:1 mixture of xylene and molten (59°C) paraffin (Paraplast tissue embedding medium; Oxford Labware) for 15 min, washed three times in molten paraffin for 2 h and then embedded in paraffin using Biopsy Uni-Cassettes (Tissue Tek). Embedded tissues were sectioned using a Leica RM2165 microtome, stained with hematoxylin and eosin (H & E), and mounted.
Skeleton preparations.
Skin and internal organs were carefully removed from euthanized newborn mice. Carcasses were fixed overnight in 95% EtOH and then stained with 0.015% alcian blue–20% acetic acid (Sigma) overnight. The samples were washed in 95% EtOH for at least 3 h and transferred to 2% KOH for 24 h, and any remaining soft tissue was removed carefully with forceps. Skeletons were stained overnight in 0.005% alizarin red sodium sulfate–1% KOH (Sigma) and then cleared in 1% KOH–20% glycerol for at least 2 days. Skeletons were subsequently stored in a 1:1 mixture of glycerol and 95% EtOH and photographed.
Blood analysis.
Serum was isolated from decapitated newborn mice using Microtainer serum separator tubes (Becton Dickinson). These samples were taken from pups with no obvious milk in their stomachs. Samples were processed and analyzed using a Vitros 950 chemistry analyzer (Ortho-Clinical Diagnostics, Johnson & Johnson), except for the analysis of blood glucose levels (see below). Averages and standard deviations are shown in Table 1. Measurements of sodium and potassium were performed on individual samples from three separate homozygous mutant and heterozygous newborns. To make these measurements, serum (8 to 10 μl) was diluted 1/11 in urine diluent. In control experiments, measurements of serial dilutions demonstrated values that were linearly proportional to the dilution factor. Measurements of alanine aminotransferase, alkaline phosphatase, blood urea nitrogen, calcium, and triglycerides were performed on two different pooled samples from a total of 15 homozygous mutant and 19 heterozygous newborns diluted 1/2.5 in 0.9% NaCl. Measurements of creatine and phosphate were performed on two pools of four homozygous mutant and three pools for four heterozygous newborns. Measurements of glucose levels were performed on five individual homozygous mutant and 15 heterozygous newborns within 10 min of birth, using an Accu-Chek glucometer (Roche). Abdomens of newborn pups (less than 12 h old) were examined for the presence of milk. Approximately twofold higher levels of triglycerides were present in heterozygotes with milk than without milk.
TABLE 1.
Analysis of heterozygote and Dach1 homozygote physiological parameters, determined as described in Materials and Methods
| Mouse group | Avg concn ± SD
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Potassium (meq/liter) | Sodium (meq/liter) | Alanine aminotransferase (U/liter) | Alkaline phosphatase (U/liter) | Blood urea nitrogen (mg/dl) | Calcium (mg/dl) | Triglycerides (mg/dl) | Creatinine (mg/dl) | Phosphate (mg/dl) | Glucose (mg/dl) | No. with milk present/no. with no milk | Mean body wt (g) ± SD | |
| Heterozygote | 10.6 ± 0.5 | 216 ± 11 | 103 ± 8.5 | 1,921 ± 496 | 22.5 ± 2.5 | 6.0 ± 0.5 | 85 ± 10 | 1.1 ± 0.1 | 11.7 ± 1.6 | 36.2 ± 9.7 | 36/11 | 1.32 ± 0.13 |
| Homozygote | 10.3 ± 0.5 | 191 ± 10 | 125 ± 9.0 | 2,034 ± 542 | 24.5 ± 2.5 | 5.4 ± 0.4 | 73 ± 21 | 1.3 ± 0.0 | 12.4 ± 0.6 | 37.2 ± 6.5 | 1/18 | 1.29 ± 0.12 |
RESULTS AND DISCUSSION
Generation of Dach1 mutant mice.
Utilizing a standard homologous knockout strategy, we generated a mutation in Dach1 by replacing exon 1 and flanking genomic sequences with PGK-Hprt (Fig. 1A). Southern blot analysis using a 3′ genomic probe was used to identify two independent replacement events in ES cell lines and follow the Dach1 mutant alleles during the establishment of mouse lines B and G (Fig. 2A, right). Consistent with a homologous replacement event, an exon 1-specific probe demonstrated that exon 1 is deleted in the homozygous mutant newborns by Southern analysis (Fig. 2A, left). Associated with the replacement event was the insertion of additional sequences from the targeting construct at the Dach1 locus in lines B and G (Fig. 1A). However, the inserted sequences are not associated with a dominant phenotype, as heterozygotes appear indistinguishable from wild-type littermates. We also developed a PCR assay to genotype embryos, newborns, and adults (Fig. 1B). Genotype analysis of 151 2-h-old newborns from 21 intercrosses demonstrated that the frequency of the wild-type, heterozygote, and mutant homozygote genotypes were roughly Mendelian (Fig. 1C). These results demonstrate that there is no pronounced embryonic or fetal lethal phenotype associated with the Dach1 mutant allele.
Dach1 expression analysis in mutant embryos.
To determine the effect of the knockout mutation on Dach1 RNA expression, whole-mount in situ hybridization analysis was performed. Embryos were hybridized in batch to a Dach1 antisense riboprobe and processed together through the color development step. PCR analysis of embryo tail DNA was then performed to compare genotypes with hybridization patterns (Fig. 2B). Unlike wild-type embryos that demonstrated expression in the brain, retina, and developing limbs, the Dach1 expression pattern in the homozygous mutants was dramatically reduced to background levels. Since the Dach1 antisense riboprobe used in these experiments is derived from coding sequences downstream of exon 1, the absence of Dach1 transcripts reflects changes in gene expression, presumably due to premature termination of nascent transcripts within PGK-hprt or the deletion of necessary regulatory elements.
We next performed immunohistochemistry using Dach1 antiserum to determine the effect of the mutation on protein expression levels. Antiserum raised against peptides encoded by sequences not deleted by the mutation was incubated with embryonic day 12.5 (E12.5) wild-type and homozygous mutant eye sections. In wild-type samples, the antiserum stained nuclei within the peripheral retina, which has been reported for Dach1 transcripts at this stage (4). Sporadic nuclear staining detected in the developing cornea is consistent with the reported expression of Dach1 RNA in mesoderm surrounding the eye (4). Finally, the antiserum demonstrated staining of retinal pigmented epithelial nuclei. Previous RNA studies did not demonstrate expression in the retinal pigmented epithelium at E12.5, most likely due to the presence of cytoplasmic pigment granules that could obscure the detection of staining. In homozygous mutants, Dach1 staining in the retina, cornea, and retinal pigmented epithelium is reduced to background levels. Taken together, the data show that the wild-type Dach1 RNA and protein are not detectable in a variety of tissues, at different stages of embryogenesis in Dach1 mutant embryos, indicating that we have generated a severe loss-of-function allele of Dach1.
Analysis of newborn Dach1 mutant mice.
Observations of newborn litters demonstrated postnatal lethality associated with the Dach1 mutant allele (see below). To characterize phenotypes prior to death, we focused our analysis on progeny less than 2 h old, when lethality was infrequent. Although 20% of these newborns were homozygous mutant (Fig. 1C), the external anatomy of the newborns appeared to be indistinguishable from that of control littermates (see below). This was also observed in intercrosses between Dach1 mutant lines B and G in 129/SvEv and 129/SvEv/C57BL/6J backgrounds. In addition, comparison of the weights and lengths of wild-type, heterozygote, and homozygous mutant newborns revealed no differences, consistent with the absence of a pronounced developmental phenotype (Table 1 and data not shown, respectively).
Drosophila dachshund mutant flies lack eyes, have truncated limbs, and exhibit brain abnormalities (22, 25, 26). Given the structural and expression similarities between Drosophila dachshund and mouse Dach1, we hypothesized that Dach1 mutant mice would exhibit defects in eye, limb, and/or brain development. However, an examination of the external anatomy of the heads and limbs of homozygous mutant newborns failed to reveal any detectable malformations (data not shown). We next inspected homozygous mutant and control sibling tissues for more subtle differences. H&E staining of eye sections revealed the presence of a normal immature retina, retinal pigmented epithelium, lens, optic nerve, cornea, and ciliary margin in newborn homozygous mutants (Fig. 3A and B). Although the retina is not fully differentiated in newborns, wild-type and homozygous mutant retinas exhibited similar degrees of retinal thickness and cell layering (Fig. 3C and D). We also tested newborn wild-type and homozygous mutant retinas for the expression of Pax6 and found no detectable differences (data not shown).
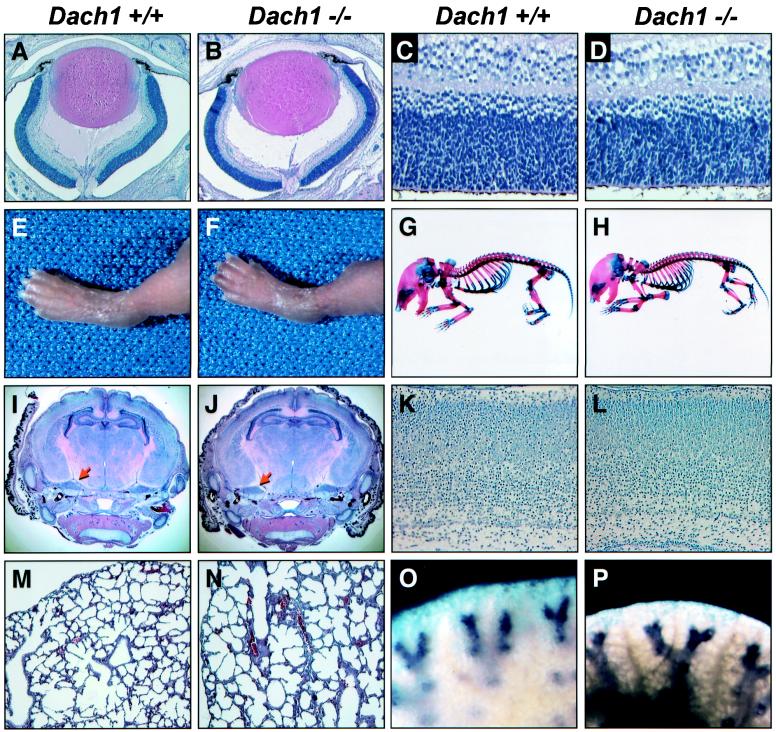
FIG. 3.
Gross and histological analyses of newborn wild-type (+/+) and Dach1 homozygous mutant (−/−) mice. (A and B) H&E staining of eye sections. The retina, optic nerve, lens, retinal pigmented epithelium, cornea, and ciliary margin are present and appear to be normal in newborn Dach1 homozygous mutant eyes. (C and D) High magnification of H&E-stained retinas. Dach1 mutant retinas exhibit a similar degree of layering and cell numbers as newborn wild-type retinas. (E and F) The external dimensions and morphology of wild-type and Dach1 mutant legs and digits are similar. (G and H) Alcian blue-alizarin red staining of wild-type and homozygous mutant skeletons does not reveal any detectable differences in either bone or cartilage morphology. The homozygote right forelimb was detached to reveal the structure of the limb. (I and J) H&E-stained coronal sections of newborn brains showing cortical layers, hippocampi, and trigeminal ganglia (arrow) in both wild-type and homozygous mutant animals. (K and L) High magnification of H&E-stained neocortex from newborn mice. The marginal zone, cortical plate, subplate, and intermediate zone of the cortex can be seen in newborn Dach1 mutant brains. (M and N) Lungs of wild-type and homozygous mutant newborn animals do not show any detectable differences in airway branching, alveolar septation, spacing, and cellular composition. (O and P) Whole-mount in situ hybridization of newborn lungs using a Clara cell-specific riboprobe, CC10. Epithelial cells lining the terminal bronchioles with similar branching patterns are found in both wild-type and homozygous mutant lungs.
Analysis of limbs also revealed that the external dimensions of wild-type and Dach1 mutant limbs and digits appeared to be indistinguishable (Fig. 3E and F). In particular, the anterior-posterior, dorsal-ventral, and proximal-distal axes of the homozygous mutant limbs were normal. Similarly, the limb skeletal anatomy was not detected to be abnormal in newborn homozygous mutants (Fig. 3G and H). Since Dach1 is expressed in a perichondral pattern in the developing hand plate (11), we examined H&E-stained newborn paws and found no evidence of soft tissue malformations (data not shown).
As Dach1 is expressed in the nervous system, we performed histological analysis of serial coronal or axial sections through the cerebra and brainstem. Comparison of cerebral cortex, hippocampi, subcortical gray matter, brainstem cranial nerve nuclei, cerebellum, and trigeminal ganglia between homozygous mutant and wild-type controls did not reveal any abnormalities (Fig. 3I-L). In addition, terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling staining showed no differences in numbers of apoptotic cells in the central nervous system and cranial nerve ganglion between wild-type and homozygous mutants (data not shown).
Despite the absence of Dach1 expression during early eye, limb, and brain development, these tissues are established and patterned normally. There are several explanations for a lack of a developmental phenotype in Dach1 mutant mice. First, because Dach1 mutants die after birth, we cannot rule out that Dach1 is required for postnatal development. For example, since differentiation of retinal progenitors continues for about 2 weeks after birth (5), Dach1 may be required for maturation rather than the establishment of the neuroretina. Similarly, Dach1 may be required for postnatal neurogenesis in the brain (16). The creation of tissue-specific knockouts could lead to the rescue of the Dach1 lethal phenotype and thereby the determination of a role for Dach1 in the maturation of the neuroretina and/or the brain.
A second possibility is that an additional Dach homologue compensates for Dach1 loss of function. Indeed, we have isolated mouse cDNA sequences encoding a protein showing greater amino acid identity with chick Dach2 than with chick Dach1 (G. Mardon, unpublished data) (18). Perhaps by making Dach1/Dach2 double-mutant mice, an eye-, limb-, or brain-specific function can be attributed to the vertebrate homologues of dachshund in the future. A similar situation exists for determining the role of vertebrate eyes absent (Eya1 to Eya3) genes during eye development. Although Eya1 knockout mice exhibit kidney and ear developmental abnormalities, no gross eye abnormalities were reported (40). Eya2 and Eya3 are expressed during eye development making it possible that either of these genes compensate for the loss of Eya1 function (41).
Another reason for the absence of developmental phenotypes in the Dach1 mutants may be that we have not created a null allele. This is a formal possibility, as we have not deleted the entire coding region. However, deletion of exon 1 results in the abrogation of the wild-type Dach1 RNA and protein expression patterns. Furthermore, this deletion removes most (98 of 107 amino acids) of the DD1 domain. Since DD1 demonstrates a high level of conservation (78% identity) between insects and vertebrates and is the most conserved domain in the entire protein, the possibility that DD1 is dispensable for Dach1 function in multiple tissues seems unlikely (11).
The Dach1 mutant allele is associated with postnatal lethality.
Genotype analysis of intercross progeny revealed that Dach1 mutant homozygotes did not survive to weaning. Postnatal lethality was observed in intercrosses between Dach1 mutant lines B and G in 129/SvEv and 129/SvEv/C57BL/6J backgrounds. Analysis of 2 h-old litters demonstrated that 93% (29 of 31) of homozygous mutants were alive (Fig. 1C). Thus, most Dach1 mutants die between this time and 3 weeks of age.
To better estimate the age of lethality, cages were checked for deliveries and deaths five times a day, the time of either event was noted, and genotyping was performed on dead and weaned animals. From the progeny of 11 intercrosses, we genotyped 12 wild-type, 42 heterozygotic, and 14 homozygous mutant newborns. Among the dead animals, 1 was wild type, 3 were heterozygotes, and 14 were homozygotes. Nine of the 14 mutants demonstrated a minimum age range of 4 to 16 h and maximum age range of 16 to 32 h; the remaining 5 were dead when the litters were first discovered. These litters were, at most, 8 h old. Thus, the Dach1 mutant allele is associated with postnatal lethality, and homozygous mutants fail to survive beyond the first day.
In Dach1 intercross litters with at least one suckling, we found a strong correlation between the homozygous mutant genotype and the absence of milk in the stomach (Table 1). This phenotype is consistent with a failure to suckle and in other knockout mice has been associated with postnatal death due to malnutrition (23). Since body weights of homozygous mutant and heterozygous newborn pups are similar, the maternal supply of nutrients and fetal metabolism is likely to be normal for homozygous mutants (Table 1). Similarly, blood glucose and triglyceride levels determined after birth were similar for homozygous mutants and heterozygotes (Table 1). These data suggest that the physiological state of homozygotes may be normal before and just after birth, but a failure to suckle may contribute to the lethal phenotype.
A second characteristic associated with the homozygous mutant genotype was cyanosis and respiratory distress. We observed that 10% (3 of 31) of 2-h-old homozygotes were cyanotic (Fig. 1C). Cyanosis was accompanied by respiratory distress in one of these mutants. In older mutants, we have observed the two phenomena occurring together, although the frequencies of these phenotypes have not been determined (http://www.bcm.tmc.edu/pathology/db/Mardon/). The respiratory abnormality is characterized by a gaping mouth and exaggerated diaphragmatic contractions, which in some animals continued for several hours before the pup expired. It is unclear if all homozygotes demonstrate cyanosis and respiratory distress.
Abnormal lung development may explain a predilection for a cyanosis/respiratory distress phenotype. For example, epidermal growth factor receptor-deficient mice were found to demonstrate a variable incidence of cyanosis and breathing difficulties associated with abnormal branching and alveolar septation of the lungs (27). In further support of this hypothesis, Dach1 expression has been detected in embryonic lung mesoderm (Mardon, unpublished). However, gross and histological analyses of the lungs showed no abnormalities in lobe formation, airway branching, alveolar septation, or cellular architecture (Fig. 3M and N). In addition, whole-mount in situ hybridization of wild-type and homozygous mutant lungs revealed no detectable differences in staining of surfactant protein B and CC10, type II alveolar cell and Clara cell markers, respectively (34, 38) (data not shown and Fig. 3O and P). Gross examination of homozygous mutants diaphragms revealed no structural defects that might serve as a precondition for the respiratory behavior (data not shown). Although a malformed skeleton may restrict breathing, we did not identify any skeletal abnormalities that would explain the respiratory distress phenotype (Fig. 3G and H).
To identify any other anatomical or functional abnormalities, we examined major organs for malformations and measured several blood chemistry parameters in noncyanotic newborns. Examination of the Dach1 homozygous mutant hearts and major vessels by gross and histological analyses revealed no apparent defects in structure (data not shown). Similarly, histological analysis of livers and liver-related functional tests, alanine aminotransferase and alkaline phosphatase, did not reveal any significant differences between wild-type and homozygous mutants (data not shown and Table 1). Dach1 expression has been detected in the kidneys, suggesting that the lethal/cyanotic phenotype may be related to an inability to regulate electrolyte concentrations and/or eliminate metabolic waste products (21). Histological analysis of the kidney did not reveal any structural defects between wild-type and mutant animals (data not shown). Furthermore, analysis of serum blood- urea- nitrogen, calcium, creatinine, potassium, and phosphate also did not reveal any significant differences (Table 1). We observed a small difference in sodium levels, although it is not clear if this is sufficient to explain the Dach1 lethal phenotype. Finally, as Dach1 is expressed in the nervous system, a neurologic defect may explain lethality, a failure to suckle, cyanosis, and respiratory distress (11). Histological analysis of brain sections revealed no differences in the cerebral cortex, hippocampi, subcortical gray matter, brainstem cranial nerve nuclei, cerebellum, and trigeminal ganglia between homozygous mutant and wild-type controls (Fig. 3I to L).
In this work, we focused our histological and blood analyses on animals that were noncyanotic and did not display respiratory distress. A single factor that would satisfactorily serve as a precondition for the homozygous lethal phenotype has not been identified. Our data indicate that homozygous mutants die within the first 24 h and fail to suckle. Brn-3a and NMDA e2 receptor knockout mice exhibit similar phenotypes (23, 39). However, impaired suckling in these mutants was also associated with both a decreased number of neurons in the trigeminal ganglion and the absence of a suckling response. This contrasts with the Dach1 phenotype, where homozygote mutant trigeminal ganglia appear to be normal and the newborns do have a suckling response (data not shown). Other possible causes include rejection of the pup by the mother, defects in a pheromone response to the mother, an inability to compete for food, mechanical problems with swallowing, and a failure of the stomach to accommodate milk.
Although homozygous mutants fail to suckle, starvation may not be the only mechanism of death. In particular, some mutant pups have been observed to die when their littermates have not suckled. Since mutants have glucose and triglyceride levels similar to those of unfed littermates, it is unlikely that starvation is responsible for lethality soon after birth. Furthermore, cyanosis or respiratory distress is not an expected outcome of starvation. There may be a single or multiple underlying defects that interact with a rapidly changing newborn physiology to produce variation in the timing and mechanism of death. In the future, generation of tissue-specific conditional mutations may provide the means to determine which tissue is responsible for postnatal lethality. This would also serve as a way to rescue the lethal phenotype and to determine if Dach1 is required for developmental processes beyond birth.
ACKNOWLEDGMENTS
We thank John M. Hicks for help in the histological analysis of the Dach1 mutants, Jeffery Whitsett for supplying the CC10 riboprobe construct, and Maricella Ortiz for technical assistance during generation of the Dach1 knockout lines.
A.L.B. was supported by Mutants for Cell Adhesion Molecules Merit Award R37 AI32177. R.J.D. and Y.I.S. were supported by NIH grants from the National Eye Institute. This work was supported by funds awarded to G.M. from the National Eye Institute, Baylor Mental Retardation Research Center, Retina Research Foundation, and Moran Foundation.
REFERENCES
- 1.Bonini N M, Bui Q T, Grayboard G L, Warrick J M. The Drosophila eyes absent gene directs ectopic eye formation in a pathway conserved between flies and vertebrates. Development. 1997;124:4819–4826. doi: 10.1242/dev.124.23.4819. [DOI] [PubMed] [Google Scholar]
- 2.Bonini N M, Leiserson W M, Benzer S. The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell. 1993;72:379–395. doi: 10.1016/0092-8674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- 3.Bovolenta P, Mallamaci A, Puelles L, Boncinelli E. Expression pattern of cSix3, a member of the Six/Sine oculis family of transcription factors. Mech Dev. 1998;70:201–203. doi: 10.1016/s0925-4773(97)00183-4. [DOI] [PubMed] [Google Scholar]
- 4.Caubit X, Thangarajah R, Theil T, Wirth J, Nothwang H G, Ruther U, Krauss S. Mouse Dac, a novel nuclear factor with homology to Drosophila dachshund shows a dynamic expression in the neural crest, the eye, the neocortex, and the limb bud. Dev Dyn. 1999;214:66–80. doi: 10.1002/(SICI)1097-0177(199901)214:1<66::AID-DVDY7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 5.Cepko C L, Austin C P, Yang X J, Alexiades M. Cell fate determination in the vertebrate retina. Proc Natl Acad Sci USA. 1996;93:589–595. doi: 10.1073/pnas.93.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen R, Amoui M, Zhang Z H, Mardon G. Dachshund and Eyes Absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell. 1997;91:893–903. doi: 10.1016/s0092-8674(00)80481-x. [DOI] [PubMed] [Google Scholar]
- 7.Cheyette B N, Green P J, Martin K, Garren H, Hartenstein V, Zipursky S L. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- 8.Chow R L, Altmann C R, Lang R A, Hemmati-Brivanlou A. Pax6 induces ectopic eyes in a vertebrate. Development. 1999;126:4213–4222. doi: 10.1242/dev.126.19.4213. [DOI] [PubMed] [Google Scholar]
- 9.Dahl E, Koseki H, Balling R. Pax genes and organogenesis. Bioessays. 1997;19:755–765. doi: 10.1002/bies.950190905. [DOI] [PubMed] [Google Scholar]
- 10.Davis J A, Reed R R. Role of Olf-1 and Pax-6 transcription factors in neurodevelopment. J Neurosci. 1996;16:5082–5094. doi: 10.1523/JNEUROSCI.16-16-05082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis R J, Shen W, Heanue T A, Mardon G. Mouse Dach, a homologue of Drosophila dachshund, is expressed in the developing retina, brain and limbs. Dev Genes Evol. 1999;209:526–536. doi: 10.1007/s004270050285. [DOI] [PubMed] [Google Scholar]
- 12.Glaser T, Walton D S, Maas R L. Genomic structure, evolutionary conservation and Aniridia mutations in the human PAX6 gene. Nat Genet. 1992;2:232–239. doi: 10.1038/ng1192-232. [DOI] [PubMed] [Google Scholar]
- 13.Grindley J C, Davidson D R, Hill R E. The role of Pax-6 in eye and nasal development. Development. 1995;121:1433–1442. doi: 10.1242/dev.121.5.1433. [DOI] [PubMed] [Google Scholar]
- 14.Halder G, Callaerts P, Flister S, Walldorf U, Kloter U, Gehring W J. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development. 1998;125:2181–2191. doi: 10.1242/dev.125.12.2181. [DOI] [PubMed] [Google Scholar]
- 15.Hammond K L, Hanson I M, Brown A G, Lettice L A, Hill R E. Mammalian and Drosophila dachshund genes are related to the Ski proto-oncogene and are expressed in eye and limb. Mech Dev. 1998;74:121–131. doi: 10.1016/s0925-4773(98)00071-9. [DOI] [PubMed] [Google Scholar]
- 16.Hatten M E. Central nervous system neuronal migration. Annu Rev Neurosci. 1999;22:511–539. doi: 10.1146/annurev.neuro.22.1.511. [DOI] [PubMed] [Google Scholar]
- 17.Hazbun T R, Stahura F L, Mossing M C. Site-specific recognition by an isolated DNA-binding domain of the Sine oculis protein. Biochemistry. 1997;36:3680–3686. doi: 10.1021/bi9625206. [DOI] [PubMed] [Google Scholar]
- 18.Heanue T A, Reshef R, Davis R J, Mardon G, Oliver G, Tomarev S, Lassar A B, Tabin C J. Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes Dev. 1999;13:3231–3243. doi: 10.1101/gad.13.24.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill R E, Favor J, Hogan B L, Ton C C, Saunders G F, Hanson I M, Prosser J, Jordan T, Hastie N D, van Heyningen V. Mouse Small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. . (Erratum, Nature 355:750, 1992.) [DOI] [PubMed] [Google Scholar]
- 20.Jean D, Bernier G, Gruss P. Six6 (Optx2) is a novel murine Six3-related homeobox gene that demarcates the presumptive pituitary/hypothalamic axis and the ventral optic stalk. Mech Dev. 1999;84:31–40. doi: 10.1016/s0925-4773(99)00068-4. [DOI] [PubMed] [Google Scholar]
- 21.Kozmik Z, Pfeffer P, Kralova J, Paces J, Paces V, Kalousova A, Cvekl A. Molecular cloning and expression of the human and mouse homologues of the Drosophila dachshund gene. Dev Genes Evol. 1999;209:537–545. doi: 10.1007/s004270050286. [DOI] [PubMed] [Google Scholar]
- 22.Kurusu M, Nagao T, Walldorf U, Flister S, Gehring W J, Furukubo-Tokunaga K. Genetic control of development of the mushroom bodies, the associative learning centers in the Drosophila brain, by the eyeless, twin of eyeless, and dachshund genes. Proc Natl Acad Sci USA. 2000;97:2140–2144. doi: 10.1073/pnas.040564497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kutsuwada T, Sakimura K, Manabe T, Takayama C, Katakura N, Kushiya E, Natsume R, Watanabe M, Inoue Y, Yagi T, Aizawa S, Arakawa M, Takahashi T, Nakamura Y, Mori H, Mishina M. Impairment of suckling response, trigeminal neuronal pattern formation, and hippocampal LTD in NMDA receptor epsilon 2 subunit mutant mice. Neuron. 1996;16:333–344. doi: 10.1016/s0896-6273(00)80051-3. [DOI] [PubMed] [Google Scholar]
- 24.Loosli F, Winkler S, Wittbrodt J. Six3 overexpression initiates the formation of ectopic retina. Genes Dev. 1999;13:649–654. doi: 10.1101/gad.13.6.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mardon G, Solomon N M, Rubin G M. dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development. 1994;120:3473–3486. doi: 10.1242/dev.120.12.3473. [DOI] [PubMed] [Google Scholar]
- 26.Martini S R, Roman G, Meuser S, Mardon G, Davis R L. The retinal determination gene, dachshund, is required for mushroom body cell differentiation. Development. 2000;127:2663–2672. doi: 10.1242/dev.127.12.2663. [DOI] [PubMed] [Google Scholar]
- 27.Miettinen P J, Warburton D, Bu D, Zhao J S, Berger J E, Minoo P, Koivisto T, Allen L, Dobbs L, Werb Z, Derynck R. Impaired lung branching morphogenesis in the absence of functional EGF receptor. Dev Biol. 1997;186:224–236. doi: 10.1006/dbio.1997.8593. [DOI] [PubMed] [Google Scholar]
- 28.Oliver G, Loosli F, Koster R, Wittbrodt J, Gruss P. Ectopic lens induction in fish in response to the murine homeobox gene Six3. Mech Dev. 1996;60:233–239. doi: 10.1016/s0925-4773(96)00632-6. [DOI] [PubMed] [Google Scholar]
- 29.Oliver G, Mailhos A, Wehr R, Copeland N G, Jenkins N A, Gruss P. Six3, a murine homologue of the sine oculis gene, demarcates the most anterior border of the developing neural plate and is expressed during eye development. Development. 1995;121:4045–4055. doi: 10.1242/dev.121.12.4045. [DOI] [PubMed] [Google Scholar]
- 30.Pignoni F, Hu B, Zavitz K H, Xiao J, Garrity P A, Zipursky S L. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell. 1997;91:881–891. doi: 10.1016/s0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- 31.Quiring R, Walldorf U, Kloter U, Gehring W J. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- 32.Riddle R D, Johnson R L, Laufer E, Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75:1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- 33.Shen W, Mardon G. Ectopic eye development in Drosophila induced by directed dachshund expression. Development. 1997;124:45–52. doi: 10.1242/dev.124.1.45. [DOI] [PubMed] [Google Scholar]
- 34.Stripp B R, Sawaya P L, Luse D S, Wikenheiser K A, Wert S E, Huffman J A, Lattier D L, Singh G, Katyal S L, Whitsett J A. cis-acting elements that confer lung epithelial cell expression of the CC10 gene. J Biol Chem. 1992;267:14703–14712. [PubMed] [Google Scholar]
- 35.Ton C C, Hirvonen H, Miwa H, Weil M M, Monaghan P, Jordan T, van Heyningen V, Hastie N D, Meijers-Heijboer H, Drechsler M, et al. Positional cloning and characterization of a paired box- and homeobox-containing gene from the Aniridia region. Cell. 1991;67:1059–1074. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- 36.Ton C C, Miwa H, Saunders G F. Small eye (Sey): cloning and characterization of the murine homolog of the human Aniridia gene. Genomics. 1992;13:251–256. doi: 10.1016/0888-7543(92)90239-o. [DOI] [PubMed] [Google Scholar]
- 37.Walther C, Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development. 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- 38.Weaver T E, Sarin V K, Sawtell N, Hull W M, Whitsett J A. Identification of surfactant proteolipid SP-B in human surfactant and fetal lung. J Appl Physiol. 1988;65:982–987. doi: 10.1152/jappl.1988.65.2.982. [DOI] [PubMed] [Google Scholar]
- 39.Xiang M, Gan L, Zhou L, Klein W H, Nathans J. Targeted deletion of the mouse POU domain gene Brn-3a causes selective loss of neurons in the brainstem and trigeminal ganglion, uncoordinated limb movement, and impaired suckling. Proc Nat Acad Sci USA. 1996;93:11950–11955. doi: 10.1073/pnas.93.21.11950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu P X, Adams J, Peters H, Brown M C, Heaney S, Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet. 1999;23:113–117. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- 41.Xu P X, Woo I, Her H, Beier D R, Maas R L. Mouse Eya homologues of the Drosophila eyes absent gene require Pax6 for expression in lens and nasal placode. Development. 1997;124:219–231. doi: 10.1242/dev.124.1.219. [DOI] [PubMed] [Google Scholar]