Abstract
Antigenic characterization of emerging and re-emerging viruses is necessary for the prevention of and response to outbreaks, evaluation of infection mechanisms, understanding of virus evolution, and selection of strains for vaccine development. Primary analytic methods, including enzyme-linked immunosorbent/lectin assays, hemagglutination inhibition, neuraminidase inhibition, micro-neutralization assays, and antigenic cartography, have been widely used in the field of influenza research. These techniques have been improved upon over time for increased analytical capacity, and some have been mobilized for the rapid characterization of the SARS-CoV-2 virus as well as its variants, facilitating the development of highly effective vaccines within 1 year of the initially reported outbreak. While great strides have been made for evaluating the antigenic properties of these viruses, multiple challenges prevent efficient vaccine strain selection and accurate assessment. For influenza, these barriers include the requirement for a large virus quantity to perform the assays, more than what can typically be provided by the clinical samples alone, cell- or egg-adapted mutations that can cause antigenic mismatch between the vaccine strain and circulating viruses, and up to a 6-month duration of vaccine development after vaccine strain selection, which allows viruses to continue evolving with potential for antigenic drift and, thus, antigenic mismatch between the vaccine strain and the emerging epidemic strain. SARS-CoV-2 characterization has faced similar challenges with the additional barrier of the need for facilities with high biosafety levels due to its infectious nature. In this study, we review the primary analytic methods used for antigenic characterization of influenza and SARS-CoV-2 and discuss the barriers of these methods and current developments for addressing these challenges.
Keywords: Influenza, SARS-CoV-2, Antigenic analysis, Antigenic drift, Antigenic characterization, Vaccine strain selection
Influenza
Introduction to influenza viruses
The influenza virus is a recurring threat to public health. Seasonal influenza infections are associated with ~290,000–650,000 deaths annually worldwide [1], which includes ~12,000–61,000 deaths each year in the United States (US) alone [1, 2]. Unpredictably, but less frequently, global influenza pandemics occur, infecting 20–40% of the population in a single year [3–6] and dramatically raising death rates above normal levels. Influenza viruses belong to the Orthomyxoviridae family and are classified into four genera including type A, B, C, and the emerging type D [7, 8] based on their antigenic differences in the nucleoprotein and matrix 1 protein. Influenza viruses contain segmented, negative-sense, single-stranded RNA genomes. Influenza A viruses (IAVs) and influenza B viruses (IBVs) contain 8 viral RNA (vRNA) gene segments, whereas influenza C viruses (ICVs) and influenza D viruses (IDVs) contain 7 vRNA gene segments. Segments 1 (PB2), 2 (PB1), 3 (PA), 4 (HA), 5 (NP), 6 (NA), 7 (MP), and 8 (NS) of IAVs and IBVs encode polymerase basic protein 2 (PB2), polymerase basic protein 1 (PB1), polymerase acidic protein (PA), hemagglutinin (HA), nucleoprotein (NP), neuraminidase (NA), matrix proteins (M1 and M2), and nonstructural proteins (NS1 and NS2), respectively, which will be described in the subsequent sections. In addition, several novel accessory proteins of IAVs were identified that modulate viral pathogenicity, such as PB1-F2 [9] and PB1-N40 [10] encoded by the PB1 gene and PA-X [11], PA-N155, and PA-N182 [12] by the PA gene.
Evolution and antigenic variations of influenza viruses
The low-fidelity, “error-prone” RNA-dependent RNA polymerase (RdRp) of IAVs lacks the 3’ to 5’ exonuclease proofreading capability, leading to a rapid mutation rate that ranges from 0.4 × 10−3 to 2.0 × 10−6 mutations per nucleotide per year, depending on virus strain and gene [13–17]. Although the outcome of most random mutations is detrimental or lethal, non-deleterious mutations may be preserved and subsequently amplified in the population if they confer a fitness advantage [18]. High mutation frequencies and within-host selective pressures create quasi-species [19–22], defined as a proliferating population of non-identical but closely related viral genomes as seen with most RNA viruses, including influenza viruses [23, 24]. Some mutations can be positively selected in order for a virus to escape from host antibody neutralization or to replicate more efficiently, leading to virus variants becoming predominant in the population [25]. Population-level fitness has also been shown to be increased by cooperative interactions between variants within a quasi-species [26–30].
However, the overall mutation (at the nucleotide sequence level) and amino acid substitution (at the protein sequence level; from nonsynonymous mutations) frequencies are a complex association of factors that are genus-, strain-, and gene-specific and are even environmentally influenced (i.e., temperature or pH). These within- and between-host immune selection pressures result in variable evolutionary rates [13]. The phenomenon that amino acid substitutions accumulating on surface glycoproteins of influenza viruses gradually alter their antigenicity is referred to as “antigenic drift,” which allows influenza viruses to evade immune pressures from their hosts and is responsible for seasonal influenza epidemics that necessitate annual vaccine reformulations.
Unlike mutations, reassortment results in genome restructuring. Reassortment occurs when two strains from a shared genus infect the same host cell and produce a novel viral genotype, i.e., an assembly of segments from each “parental” strain. As is the case of random mutations, most reassortant events are deleterious, usually due to segment incompatibility [31]. When reassortment leads to the introduction of a novel HA and/or NA gene into a naive population (a population without existing immunity), it is commonly referred to as “antigenic shift” [32]. Antigenic shift, in combination with sustained human-to-human transmission, is a requirement for the emergence of an influenza pandemic strain. Reassortments have led to the emergence of the 1957, 1968, and 2009 IAV pandemics [33–36], contributed to the severe epidemics of 1947, 1951, and 2003, and facilitated the rise in antiviral drug resistance [37].
The influenza vaccine is the most viable option for counteracting and reducing the impact of influenza outbreaks [38]. Since the antigenicity of influenza viruses is continuously changing, new strains can potentially escape the immunity of previously exposed hosts [39, 40]. To account for antigenic drift, the vaccine composition of the influenza vaccine is revised almost yearly and separately in both the Northern and Southern Hemispheres and incorporates contemporary representatives of circulating viruses identified by continuous global monitoring and surveillance led by the World Health Organization’s (WHO) Global Influenza Surveillance and Response System (GISRS) [41].
Antigenic epitopes of HA and NA
HA and NA, which present in a ratio ranging from 4:1 to 10:1, are critical for viral attachment and release, respectively [42]. HA is a homo-trimeric glycoprotein containing a globular head subdomain spanning the primary HA sequences, and a stalk subdomain containing N and C termini of HA1 and most HA2 sequences (Fig. 1A). As the major target for immune responses, the HA globular head has high plasticity and is highly tolerant of amino acid substitutions and glycosylation alternations. Under selective pressure from the host immune system, the HA globular head subdomain has the highest amino acid substitution rate among all influenza virus proteins [43]. In comparison, the HA stalk subdomain is much less accessible to antibodies and is more conserved than the globular head subdomain [44].
Fig. 1.
Hemagglutinin (HA) structure and antigenic binding sites. (A) Structure of HA protein. (B) The five antigenic sites (i.e., Sa, Sb, Ca1, Ca2, and Cb) of H1 (A/California/04/2009; PDB 3UBE). (C) The five antigenic sites (i.e., A–E) of H3 (A/Aichi/2/1968; PDB 2YPG). (D) The four antigenic sites of influenza B viruses (B/Hong Kong/8/1973; PDB 3BT6)
Five major antibody binding sites (ABSs) are located in the HA1 globular head subdomain and are termed sites Sa, Sb, Ca1, Ca2, and Cb for subtype H1 IAVs (Fig. 1B) [45, 46] and as sites A, B, C, D, and E for subtype H3 IAVs [47, 48] (Fig. 1C). The corresponding five ABSs are reported in other subtype IAVs, particularly in subtype H5 viruses, which have been associated with rapid antigenic drift [49]. Three ABSs overlap with HA receptor binding sites (RBSs), including the residues in the 130-loop (i.e., 135, 136, 137, 138, 153) with ABS site A, the 190-helix (i.e., 186, 190, 194, 195) with site B, and the 220-loop (i.e., 226 and 228) with site D [50]. Compared to IAVs, IBVs undergo less antigenic drift. The ABSs of IBV HA are located in the 120-loop, 150-loop, 160-loop, and 190-helix (Fig. 1D) [51].
In contrast, the NA is a homo-tetrameric glycoprotein that is responsible for removing terminal sialic acids and releasing virions from infected cells, mucous, or other substances. The enzyme active site, including catalytic sites (i.e., R118, D151, R152, R224, E276, R292, R371, and Y406) (in N2 numbering) that directly interact with the substrate and framework sites (i.e., E119, R156, W178, S179, D/N198, I222, E227, H274, E277, N294, and E425), are conserved in all NA subtypes [52, 53]. However, the antigenic sites of NA are not fully understood. Partially antigenic epitopes have been mapped by sequence changes in escape mutant selection with monoclonal antibodies and by NA-antibody complex crystallography (Table 1). The positions of amino acid substitutions surrounding the enzyme’s active site have the ability to escape NA antibody binding and inhibit NA activity [68]. Recently, positions located laterally to the enzyme active site such as positions 329 and 390 have also been associated with antigenic drift [69, 70].
Table 1.
Amino acids essential for mAb binding to NA
| Amino acida | NA subtype and the antigen (virus) used as immunogenb | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N1 | N2 | N8 | N9 | ||||||||
| BR/07 | CA/09 | RI/57 | TOK/67 | MEM/98 | MN/10 | SGP/16 | Dk/UK/63 | Tern/AU/75 | SH/13 | AH/13 | |
| 95 (95) | +c | ||||||||||
| 150 | + | + | |||||||||
| 198 | + | ||||||||||
| 199 | + | + | |||||||||
| 220 | + | + | |||||||||
| 221 | + | + | + | ||||||||
| 244 | + | ||||||||||
| 245 | + | ||||||||||
| 247 (248) | + | + | + | + | |||||||
| 248 (249) | + | + | |||||||||
| 249 (250) | + | ||||||||||
| 250 | + | ||||||||||
| 251 | + | ||||||||||
| 253 | + | + | |||||||||
| 272 (273) | + | + | |||||||||
| 284 | + | ||||||||||
| 307 (308) | + | + | |||||||||
| 310 (311) | + | ||||||||||
| 329 | + | + | |||||||||
| 334 | + | + | |||||||||
| 338 | + | ||||||||||
| 340 (338) | + | + | + | ||||||||
| 342 (339) | + | + | + | + | + | + | |||||
| 344 (341) | + | ||||||||||
| 345 | + | ||||||||||
| 346 (343) | + | ||||||||||
| 346 | + | ||||||||||
| 367 (364) | + | + | + | ||||||||
| 368 | + | + | + | + | |||||||
| 369 | + | ||||||||||
| 370 | + | + | + | ||||||||
| 371 | + | ||||||||||
| 372 (369) | + | + | + | ||||||||
| 373 | + | ||||||||||
| 399 (396) | + | ||||||||||
| 400 (397) | + | + | + | + | + | ||||||
| 403 | + | ||||||||||
| 429 | + | + | |||||||||
| 431 | + | ||||||||||
| 434 | + | ||||||||||
| 450 (449) | + | ||||||||||
| 452 (451) | + | ||||||||||
| 457 (456) | + | ||||||||||
| 468 | + | ||||||||||
This table is an update of previously reported NA residues [54]
aAmino acid residues follow N2 numbering with N1 numbering shown in parentheses
bThe critical amino acid positions for antibody binding were originally reported for BR/07 [55], CA/09 [56–59], RI/57 [60], TOK/67 [61], MEM/98 [62], Dk/UK/63 [63], and Tern/AU/75 [64, 65], SH/13 [66], and AH/13 [67]
c“+” indicates the amino acid position is essential for monoclonal antibody binding
Antigenic drift of seasonal influenza viruses
Contemporary seasonal influenza viruses comprise of four co-circulating and antigenically distinct viruses. These include two IAVs (H1N1 and H3N2 subtypes) and two IBVs (B/Victoria/2/87-like [Victoria] and B/Yamagata/16/88-like [Yamagata] lineages) (Fig. 2A). The H3N2 IAV has been co-circulating in the human population since 1968 causing the 1968 pandemic. The 2009 H1N1 IAV [A(H1N1)pdm09] has been circulating in the human population since the 2009 pandemic, replacing the H1N1 seasonal virus that had been circulating in humans for 32 years from 1977 to 2009 [A(H1N1)season1977] [73]. Additionally, IBVs have been detected in the human population since their first isolation in 1940 [74], and the contemporary Victoria and Yamagata lineages of IBVs have been co-circulating in human population at least since 1983 [75].
Fig. 2.
Antigenic evolution of seasonal influenza viruses. (A) Timeline of influenza A and B virus pandemics and circulation in humans since 1918. There have been 4 pandemics during this period, which are indicated with arrows. The 1918 pandemic was caused by the H1N1pdm1918 virus, which circulated in humans until the 1957 H2N2 pandemic. The circulating H2N2 virus was then replaced by the 1968 H3N2 pandemic virus. In 1977, H1N1 reemerged was replaced by a reassortment H1N1pdm09 strain. H3N2 has been co-circulating with H1N1 since 1977. Influenza B viruses were first isolated in 1940 and have also been co-circulating in humans at least since 1987 as two antigenically distinct lineages, Victoria and Yamagata. (B) Antigenic cartography of 39,370 seasonal influenza A(H3N2) viruses (1968–2016) (adapted from Han et al. [71]). A total of 16 antigenic clusters were identified during this time period. Antigenic cartography of 13,591 human, swine, and avian influenza A(H1N1) viruses (adapted from Li et al. [72])
Over the past 50 years, H3N2 viruses have undergone frequent antigenic drift, and at least 18 antigenic variants have been reported (Fig. 2B and Table 2) [71]. The amino acid substitutions associated with antigenic variations are located on the five ABSs (A to E), but primarily in sites A and B, in which 7 residues are neighboring the RBS (145 at site A and 155, 156, 158, 159, 189, and 193 at site B) [82]. Additionally, a single amino acid substitution in HA can sufficiently alter the antigenicity of viruses, whereas most antigenic drift events have involved amino acid substitutions across multiple positions at the same site or two different sites [82]. In addition to those in HA, the amino acid substitutions in NA can also cause antigenic drift. For example, amino acid substitutions at sites 329 and 390 of N1 led to the antigenic drift of A/Solomon Island/3/2006 (H1N1) and A/Yokohama/94/2015 (H1N1) viruses, respectively [69, 70]. The acquisition of N-linked glycosylation at positions 245 and 247 (S245N/S247T) of N2 was associated with the poor vaccine performance against H3N2 viruses during the 2017–2018 influenza season [83] .
Table 2.
Antigenic drift events and associated amino acid substitutions in H3N2 (1968–2015) and H1N1 (1977–2009) seasonal influenza A viruses
| Subtype | Antigenic drift eventa | Predominant amino acid substitution(s) in HA (antibody binding sites)b |
| H3N2 [71, 76–80] | HK68→EN72 | G144D (A) |
| EN72→VI75 | S145N(A), S193D (B) | |
| VI75→TX77 | D53K(C), E82K(E) | |
| TX77→BK79 | K156E(B) | |
| BK79→SI87 | Y155H(B), K189R(B) | |
| SI87→BE89 | G135E(A), N145K(A), N193S(B) | |
| BE89→BE92 | E135K(A), K145N(A), E156K(B) | |
| BE92→WU95 | N145K(A), G172D(D) | |
| WU95→SY97 | K62E(E), K156Q(B), E158K(B) | |
| SY97→FU02 | Q156H(B) | |
| FU02→CA04 | K145N(A), Y159F(B) | |
| CA04→BR07 | S193F(B), D225N (near D) | |
| BR07→PE09 | K158N(B), N189K(B) | |
| PE09→TX12 | N278K(C), S45N(C) | |
| TX12→SW13 | N145S(A), N225D(near D), A138S(A), F159S(B) | |
| TX12→HK14 | N145S(A), N225D(near D), N144S(A), F159Y(B), Q311H(C), K160T(B) | |
| H1N1 [81] | CH83→SI86 | K197T(Sb), K192R(Sb) |
| SI86→TE91 | K82E(Cb) | |
| TE91→BE95 | T132V(Near Sa), S273P(Pa) | |
| BE95→NE99 | E156G(Sa), S77L(Cb) | |
| NE99→BR07 | T197K(Sb), R192M(Sb), K144E(Ca2), S46N(Pb) | |
| NE99→SO06 | E276K(Pa), R192K(Sb), K144E(Ca2), Y101H(near Cb) |
aFor H3N2 viruses: HK68, A/Hong Kong/1/1968; EN72, A/England/42/1972; VI75, A/Victoria/3/1975; TX77, A/Texas/1/77; BK79, A/Bangkok/01/1979; SI87, A/Sichuan/2/1987; BE89, A/Beijing/352/1989; BE92, A/Beijing/32/1992; WU95, A/Wuhan/359/1995; SY97, A/Sydney/5/1997; FU02, A/Fujian/411/2002; CA04, A/California/07/2004; BR07, A/Brisbane/59/2007; PE09, A/Perth/16/2009; TX12, A/Texas/50/2012; SW13, A/Switzerland/9715293/2013; HK14, A/Hong Kong/4801/2014. For H1N1 viruses: CH83, A/Chile/1/83; SI86, A/Singapore/6/1986; TE91, A/Texas/36/91 BE95, A/Beijing/262/95; NE99, A/New Caledonia/20/1999; BR07, A/Brisbane/59/2007; SO06, A/Solomon Islands/3/2006
bAll positions are under H3 numbering
Compared to H3N2 viruses, H1N1 viruses, including A(H1N1)season1977 (1977–2009) and A(H1N1)pdm09 (2009–present), undergo relatively less frequent antigenic drift events [84]. Between 1977 and 2009, at least six antigenic events were documented starting with the A(H1N1)season1977 epidemic (compared with 10 antigenic events for H3N2 virus during the same time period), and positions 156 at Sa, 192, 197, and 214 at Sb, 144 at Ca2, 77, 81, 82, and 94 at Cb, 273 and 276 at Pa, and 46 at Pb were shown to cause each of these events (Table 2) [72, 81]. Of note, Pa is located at the edge of head and stem regions (i.e., positions 53, 55–59, 61, 271–280, 285, 297, 304, under H3 numbering) and Pb is located at stem region (i.e., positions 23–25, 38–41, 45–48, 288–292, under H3 numbering) [85]. For H3N2 viruses, the HA protein mutates at a rate of 3.6 amino acid substitutions per year, compared to 2.5 amino acid substitutions per year for H1N1 viruses [76, 84]. Similar to those documented in H3N2 viruses, sites associated with antigenic variants are primarily located in ABSs within the protruding loops and helices of the HA molecule [47]. Of note, A(H1N1)pdm09 and A(H1N1)season1977 are antigenically distinct, and residues in the 153–157 regions and N-linked glycosylation sites of HA are shown to affect antigenic variations between these two viruses [86, 87]. Both A(H1N1)pdm09 and A(H1N1)season1977 are descendants of the 1918 H1N1 pandemic viruses (A[H1N1]pdm1918), and amino acid substitutions at the HA protein and N-linked glycosylation are shown to affect antigenic variations among three viruses [87]. Further attempts at characterizing antigenicity have led to the development and application of the Multi-Task Learning Sparse Group Lasso (MTL-SGL) machine learning method (discussed in a later section) by Li et al. [72] to identify key residues associated with antigenic changes for A(H1N1)season1977, A(H1N1)pdm09, and A(H1N1) swine influenza viruses. Of those 176 residues with amino acid substitutions, five at the N-glycosylation sites and 78 additional residues have been associated with H1N1 antigenicity, of which 46 residues were located at ABSs [72].
Although infrequent, antigenic drift events have also been documented in both Victoria and Yamagata lineages of IBVs. IBVs evolve more slowly than IAVs at 2.0×10−3 substitutions/site/year [88], compared to H3N2, which evolves at 5.5×10−3 substitutions/site/year, and H1N1 at 4.0×10−3 substitutions/site/year [89]. Between the two IBV lineages, the Yamagata lineage is more conserved, whereas the Victoria lineage is under greater positive selective pressure and experiences greater antigenic drift [88]. Antigenic cartography showed antigenic variations of the Victoria lineage during the period of 2002–2013, and the amino acid substitutions at the RBS, such as residues 142, 161, 218, 223, 224, and 272, were often found to be associated with the antigenic changes of IBVs [88]. Although occasionally, deletions have been detected in H3N2 and H1N1 IAVs during virus infection [90], and nucleotide deletions and/or insertions in HA and/or NA have been more frequently observed in epidemic IBVs [91, 92]. For example, among multiple genetic clades reported between the 1940s and the 1980s, multiple deletions were observed in HA proteins: compared with clade III (the precursor virus for the Victoria lineage), the viruses in clade I, which includes B/Lee/1940, the prototype strain of IBV, and the viruses in clade II (the precursor viruses for the Yamagata lineage) possessed a single amino acid deletion at position 163 on the HA protein [91]. Along with a deletion at 163, the precursor viruses of the Yamagata lineage that circulated before 1988 harbored double (positions 163 and 165) or triple (positions 162–164) deletions on HA, whereas no deletions were observed in the precursor Victoria viruses [91]. Of interests, since the period of 2011–2015, HA variants with double (positions 162–163) or triple (positions 162–164) deletions have been detected in the Victoria lineage IBVs [93].
As an RNA virus, influenza genetic variants continually emerge in the human population, and it is not uncommon to see multiple genetic variants co-circulating simultaneously. In both IAVs and IBVs, a genetic variant may become predominate in the population. Antigenic variants of seasonal IAVs have emerged from these genetic variants and co-circulate in human populations for a short period of time until one antigenic variant gradually predominates the viral population and replaces the others. For example, during the 2014–2015 influenza season, multiple genetic clades (i.e., Clades 3C.2a and 3C.3a) emerged from A/Texas/50/2012 (H3N2) (TX/12)-like viruses and co-circulated. Among them, two subclades (Clade 3C.3a, A/Switzerland/9715293/2013 [SWZ/13]-like, and Clade 3C.2a, A/Hong Kong/4801/2014 [HK/14]-like A/Hong Kong/2014) are antigenically distinct from TX/12-like viruses. Amino acid substitutions N145S-N225D-A138S-F159S and N145S-N225D-N144S-F159Y-Q311H were shown to drive the emergence of SWZ/13 and HK/14 from TX/12, and all of these amino acid substitutions are located in ABSs A or B. Thus, viruses with N145S-N225D likely served as intermediate precursors for SWZ/13 and HK/14 viruses [71]. In the following 2015–2016 season, the majority of epidemic strains were HK/14-like viruses with SWZ/13-like viruses co-circulating at a lower proportion [94].
Intra-lineage reassortments can occur among co-circulating antigenic variants for both IAVs and IBVs. Intra-lineage reassortments were likely responsible for the 1947 and 1951 A(H1N1) epidemics with the epidemic virus composition comprising of genome segments from separate phylogenetic histories when inferring separate phylogenetic trees for each gene segment [33]. Likewise, intra-lineage reassortment has been identified on the HA and NA surface glycoproteins of A(H3N2) [95]. Intra-lineage reassortment has also been identified in both Victoria and Yamagata lineages of influenza B, particularly in the HA, NA, and NP phylogenies [96, 97]. These intra-lineage reassortments of influenza viruses may contribute towards the antigenic evolution of IAVs and IBVs and cause epidemics. However, intra-lineage reassortment occurrences in H3N2 are undergoing overall negative selection, suggesting that this phenomenon may be rare [95].
Antigenic drift of avian influenza viruses (AIVs)
IAVs have been recovered from more than 105 wild bird species [98]. Migratory waterfowl, including birds in the orders Anseriformes (e.g., ducks, geese, swans) and Charadriiformes (e.g., gulls, terns, and waders), are considered to be the major natural reservoirs of IAVs [99]. Sixteen IAV HA (H1–H16) and nine NA (N1–N9) subtypes have been recovered from migratory waterfowl. Among wild birds, the prevalence of IAV infections can be up to 30% [99], and transmission typically occurs when they are exposed to viruses in the feces of infected animals [100, 101]. The antigenic evolution in migratory waterfowl may be static [102], as supported by recent studies showing minimal antigenic diversity among H3 and H7 IAVs in North American migratory waterfowl [103, 104].
Domestic poultry can be infected with non-pathogenic, lowly pathogenic, and highly pathogenic IAVs. There are a variety of HA subtypes that are less virulent and do not cause death from experimental infection. However, H5 and H7 subtypes can be highly virulent (although most H5 or H7 viruses are lowly pathogenic for poultry) and can cause a 100% death rate in experimental infections [105]. One example of a virulent H5 is the A/goose/Guangdong/1/1996 (H5N1) outbreak in 1996 [106]. The avian A/goose/Guangdong/1/1996(H5N1) was isolated from farmed geese and is the precursor for the H5N1 highly pathogenic avian influenza viruses (HPAIVs) that are currently in circulation and have spread globally to animals [106] with sporadic spillover cases to humans [107, 108].
The emergence of antigenic variants in domestic poultry has been shown to occur after escaping from immune responses generated by vaccination [109], particularly in H5 and H7 IAVs. Limited studies showed that avian influenza vaccinations led to antigenic drift of the H7N3 virus in Italy [110], H5N2 virus in Mexico [109], and H9N2 virus in Korea [111], and that H5N1 viruses rapidly evolved with increased selection in H5N1-vaccinated populations [112]. Since 2002, at least 15 countries have implemented avian influenza vaccinations for preventive and/or emergency programs [113]. Between 2002 and 2010, more than 113 billion vaccine doses were used, and China alone used over 90.9% of the total doses. For A/goose/Guangdong/1/1996(H5N1)-like H5 viruses, at least 20 antigenically different genetic clades or subclades of viruses have been reported [114, 115] and new antigenic variants are still emerging [116]. In China, similar observations, including both antigenic and genetic changes, were seen in enzootic H9N2 viruses [117–127]. However, whether avian influenza vaccinations facilitate the emergence of novel genetic and antigenic variants is not fully understood.
Similar to human seasonal influenza viruses, antigenic changes in AIVs are associated with amino acid substitutions in the ABSs or near the RBS of the HA protein. For example, in Italy, five amino acid substitutions (G133E, A151T, G177V, Q201H, and T112A) in H7N3 viruses during 2002–2004 were identified in the antigenic variant and parent strains, and four of them reside in ABSs [128]. Of interest, for H7N9 viruses, although there was a greater than 4-fold reduction in HI titers between the viruses in the fifth wave and those in the first wave [129], the in vitro and in vivo studies both showed that the heterologous antisera can neutralize virus infection [130]. Further analyses showed that the reduction of the HI titers is predominantly driven by the increased receptor-binding avidity determined by the amino acid substitution L226Q to red blood cells [131].
Antigenic drift of swine influenza viruses (SIVs)
Swine may play also a role in the reassortment and adaptation of novel zoonotic viruses. It is possible for a novel virus with pandemic potential to cause a spill-over event into the human population [51, 132, 133]. Influenza surveillance in swine is important for pandemic prevention and the protection of public health. SIVs not only cause outbreaks among swine, but can also be transmitted to humans, causing sporadic infections and even pandemic outbreaks in addition to the human seasonal IAVs. Domestic swine are thought to be the intermediate host for avian-origin IAV adaptations to human infections [134–137]. Both avian-like H1N1 and H3N2 viruses have been isolated from domestic swine [137–139]. Avian-origin IAVs of subtypes H1 to H13 have been shown to infect and replicate in swine under experimental conditions [137] as well as direct H2N3, H3N1, and H4N6 avian-to-swine transmission in nature [140–149].
Genetically diverse H1N1, H1N2, and H3N2 IAVs were detected during influenza surveillance studies in domestic swine from 2009 to 2012 [144–147]. The HA gene of H1 swine viruses were grouped into three genetic lineages: 1A (classical swine H1N1 (cH1N1) lineage), 1B (human seasonal lineage), and 1C (Eurasian avian lineage) (reviewed in [150]). The 1A lineage, which was derived from the 1918 pandemic H1N1 viruses, are detected globally and further grouped into α, β, or γ clades. The γ clade includes the A(H1N1)pdm09 virus. Of note, since the 2009 pandemic, A(H1N1)pdm09 has been repeatedly reintroduced into the swine populations and reassort with the enzootic H1 and H3 swine viruses. The 1B lineage includes the viruses spilt from humans into swine in North America, and this lineage evolved into 1B.2.2 and 1B.2.1 (formerly named δ1 and δ2 subclades, respectively). In the past decade, there were several reports of either H1N1 or H1N2 viruses in South America and Vietnam with an HA gene from the 1B lineage viruses [151–154]. The 1C lineage viruses were derived from an avian H1N1 virus, which was first detected in Europe and then Asia and has remained a predominant lineage across Eurasia [150].
In contrast to the diverse sources of swine H1N1 viruses, swine H3N2 viruses are associated with multiple introductions of seasonal influenza viruses from humans [150]. The contemporary H3N2 viruses in the European swine population (“Cluster-IV”, 1990.4 lineage) were suggested to be caused by a spillover event from humans in the 1970s [155, 156], and those in the North American swine population caused by spillover from humans during the 2010–2011 (2010.1 lineage) or 2016–2017 (2010.2 lineage) influenza seasons, which have not replaced Cluster-IV viruses in the US [150, 157, 158]. The H3N2 viruses are grouped into four major lineages, including two genetically and antigenically distinct lineages, C-IV H3 and 2010.1, which emerged and become enzootic in the past decade [158, 159]. Antigenic characterization studies demonstrate that these genetically diverse H1 and H3 viruses are antigenically distinct and show different extents of cross-reactivity in the serologic assays [160, 161].
Influenza vaccination
In humans, vaccination is the primary option for counteracting and reducing the effects of influenza outbreaks [38]. However, national influenza vaccination policies differ from country to country. For example, in the US, the annual influenza vaccination has been recommended for persons ≥6 months of age since 2010 [162], especially for the elderly, very young children, pregnant women, and those with certain chronic medical conditions [163–165]. China [166], Canada [167], Australia [168], Estonia, and Poland follow the same national immunization guidelines as those of the US, whereas other European countries have vaccination recommendations for more specific populations or do not recommend influenza vaccinations for healthy children (reviewed in ref. [169]). Vaccine coverage during the past few years has reached ~60% in some regions (e.g., the US, Europe, Australia, and Japan). However, in other regions (e.g., Africa), influenza vaccine coverage has been non-existent or minimal. In the US, during the 2009–2010 through 2015–2016 influenza seasons, vaccination rates varied from 43.7 to 59.3% among children and from 38.8 to 43.6% among adults [170]; in France, during the 2012–2013 through 2014–2015 influenza seasons, vaccination rates were approximately 32% in the population that is targeted for vaccinations (ages 65 years and above, pregnant women, individuals with chronic respiratory diseases, and those with other comorbidities considered to place individuals at risk for severe influenza illness), varying from 42.7 to 44.2% in those ≥ 65 years of age [171]. Low vaccination rates and additional factors have led to the wide variations of influenza vaccine performance between seasons, and reduced vaccine effectiveness against some viruses has been observed (Table 3) [176, 182].
Table 3.
The adjusted vaccine effectiveness in the US from 2010–2011 to the 2019–2020 influenza seasons
| Influenza season | Epidemic virus(es) | % vaccine effectiveness (95% CI) for all age | Reference | ||||
| Overall influenza | A(H1N1)pdm09 | A/H3N2 | B/Yamagata | B/Victoria | |||
| 2010–2011 | A(H1N1)pdm09, A(H3N2) | 60 (53–66) | 66 (56–74) | 54 (42–64) | 60 (48–69)a | [172] | |
| 2011–2012 | A(H3N2) | 47 (35–65) | 65 (44–79) | 39 (23–52) | 66 (38–81) | 52 (8–75) | [173] |
| 2012–2013 | A(H3N2), IBVsb | 49 (43–55) | n.a.c | 39 (29–47) | 66 (58–73) | 51 (36–63) | [174] |
| 2013–2014 | A(H1N1)pdm09 | 52 (44–59) | 54 (46–61) | n.a.c | n.a.c | n.a.c | [175], CDCd |
| 2014–2015 | A(H3N2) | 19 (10–27) | n.a.c | 6 (−5–17) | 55 (43–65) | n.a.c | [176] |
| 2015–2016 | A(H1N1)pdm09, IBVs | 48 (41–55) | 45 (34–53) | 43 (4–66) | 57 (42–67) | 49 (30–64) | [177] |
| 2016–2017 | A(H3N2) | 40 (32–64) | n.a.c | 33 (23–41) | 52 (42–61) | 56 (23–57) | [178] |
| 2017–2018 | A(H3N2) | 38 (31–43) | 62 (50–71) | 22 (12–31) | 48 (39–55) | 76 (45–89) | [179] |
| 2018–2019 | A(H1N1)pdm09, A(H3N2) | 29 (21–35) | 44 (37–51) | 9 (−4–20) | 34 (−12–62)a | [180] | |
| 2019–2020e | B/Victoria, A(H1N1)pdm09 | 45 (36–53) | 37 (19–52) | n.a.c | n.a.c | 50 (39–59) | [181] |
aVaccine effectiveness against of B/Yamagata and B/Victoria lineage viruses
bIBVs influenza B viruses including B/Yamagata and B/Victoria lineages
cNot applied, there was no circulation of the type and/or subtype of virus during the corresponding season in the US
dCDC US Centers for Disease Control and Prevention: https://www.cdc.gov/flu/vaccines-work/effectiveness-studies.htm
eInterim estimates, data from October 23, 2019–January 25, 2020
The performance of vaccines is assessed by vaccine effectiveness, which is defined as reduction of influenza risk due to the influenza vaccine compared to unvaccinated individuals after adjusting for confounding factors. In the past decade, vaccines showed suboptimal effectiveness (<50%) for predominant circulating viruses in 8 of 10 seasons for a variety of reasons with a peak of 60% vaccine effectiveness (VE) during the 2010–2011 season and 52% VE during the 2013–2014 seasons to as low as 19% VE during the 2014–2015 season [183]. The low VE of the 2014–2015 influenza season was attributed to a mismatch between the circulating and vaccine A(H3N2) strains, but the vaccine strain was effective against influenza B [176]. As for the remaining seasons, during the 2018–2019 season, the overall VE was 29% with 44% VE against A(H1N1)pdm09 but only minor protection against A(H3N2), which had become the predominant influenza virus towards the latter part of the season [180]. Similarly, during the 2017–2018 (VE=38%) seasons and 2016–2017 (VE=40%), the VE against A(H3N2) viruses was less than A(H1N1)pdm09 and B viruses, which was speculated to be in part due to egg-adaptive amino acid substitutions in the HA protein resulting in poor inhibition of the circulating A(H3N2) viruses [178, 179, 184]. The 2015–2016 season (VE=48%) saw reduced VE of the live attenuated vaccine among children [177]. The 2012–2013 season (VE=49%) also had reduced VE against A(H3N2) among older adults, also speculated to be due to egg-adapted mutations, immunosenescence, or prior influenza exposures [174]. Finally, the 2011–2012 season (VE=47%) similarly saw lower VE against A(H3N2), potentially due to antigenic drift or incongruent influenza exposure history [173].
Although only available after vaccination implementation, the data for VE provide a retrospective, yet critical role in vaccine strain selection. A low VE indicating antigenic mismatch provides an opportunity to update the mismatched component(s) for the following influenza season.
Vaccine effectiveness has been shown to be affected by host factors including age, sex, comorbidities, and pre-existing immunity [185, 186], antigenic match [187–189], and the type of vaccine [188, 190]. Among these factors, identification of vaccine strains antigenically matching the epidemic strain is the key to a successful vaccination program [14]. The quadrivalent vaccine comprises all four co-circulating antigenically distinct strains whereas the trivalent vaccine contains A(H1N1), A(H3N2), and only one of two circulating IBVs. However, studies have shown that the inactivated quadrivalent influenza vaccines may provide limited overall benefit against influenza B illness when the IBV lineage in the trivalent vaccination matches the circulating B lineage [191, 192]. The global influenza surveillance network, which is coordinated by the WHO [193], characterizes antigenic and genetic properties of the epidemic influenza viruses as well as the prevalence patterns of these viruses, and then predicts which strains will be predominant during the upcoming influenza season [194, 195]. After the vaccine strains are determined, high yield strains of influenza viruses are often needed to be generated and optimized for vaccine production, although occasionally, wild-type strains (e.g., IBV) can be directly used for vaccine production [196, 197].
In domestic poultry, influenza vaccines are not only used for the prevention of disease and death, but also for the prevention of infection or reduction of viral replication in respiratory and digestive tracts to limit viral spread to uninfected flocks [198–201]. Currently, vaccines against H5 or H7 are available for a wider range of species including chickens, turkeys, ducks, geese, and zoo birds [200]. A bivalent H5 vaccine targeting clades 2.3.4.4 and 2.3.2.1 was replaced by a bivalent H5/H7 vaccine in 2017 [202] with a 97.9% coverage rate in Guangdong, China, and varying degrees of coverage in other Chinese provinces for a variety of bird species [203]. While there was no difference in protection between the prior H5 vaccine and the H5/H7 vaccine against H5 positive rates in humans, H7 positivity reduced by 98% [203]. Additionally, the H5/H7 vaccine has been shown to reduce virus shedding and mortality in chickens [202]. Challenges in poultry vaccination against H5 and H7 influenza viruses will encompass antigenic drift and potential cross-species transmission.
In domestic swine, vaccination is commonly used in the US. Swine vaccines are typically available as licensed commercial products consisting of culture-derived virions in crude allantoic fluid from specific-pathogen-free (SPF) chicken eggs, which are then chemically inactivated and formulated into a mineral oil emulsion vaccine [204]. Current swine influenza vaccines are strain-specific, consisting of two or more H1 and H3 isolates, and fail to induce cross-protection against genetic and antigenic virus variants [148]. Unlike human vaccines, this process avoids costly purification steps for the enrichment of the surface glycoproteins HA and NA [205], but booster vaccinations are necessary in order to achieve and maintain protective levels of systemic hemagglutination-inhibiting antibodies [206].
Selection of vaccine strains
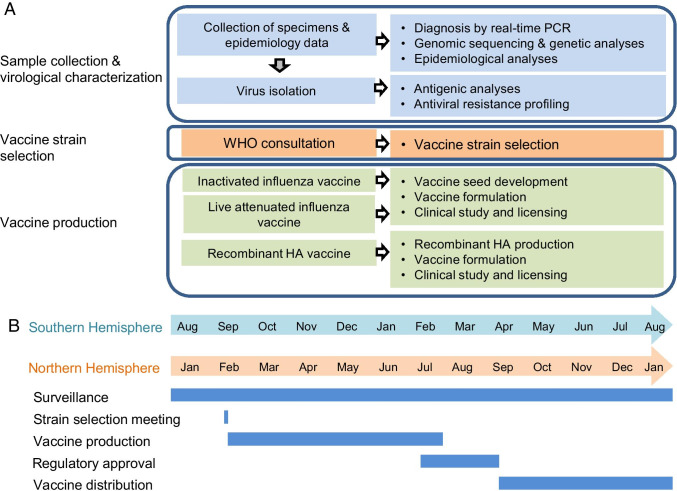
The GISRS was established to monitor the evolution and spread of influenza viruses and is composed of 144 national influenza centers (NICs) from 123 member states, six WHO collaborative centers (WHOCC), and numerous vaccine companies [207] (Fig. 3). In the US, more than 70 participating labs in the National Respiratory and Enteric Virus Surveillance System are involved. Tens of thousands of samples are collected from patients with influenza-like illnesses year-round and sent to one of the NICs. NICs identify the type and subtype of the samples and/or isolate them, and then send representative isolates and/or the original swab samples to one of the WHOCCs. WHOCCs perform genome sequencing and antigenic analyses on these virus samples and isolates [208, 209]. Based on the epidemiological and antigenic data, the GISRS committee meets in February (Northern hemisphere) and September (Southern hemisphere) and recommends vaccine strains to be used in the next influenza season [210].
Fig. 3.
Schematic of the influenza surveillance, vaccine strain-selection, and vaccine production process. (A) The process by the Global Influenza Surveillance and Response System (GISRS) was detailed in [208]. (B) Timeline of influenza vaccine production. The GISRS vaccine strain selection, production, approval process, and distribution in both the Northern and Southern Hemispheres are shown with the corresponding time of year that each procedure is performed
After the selection of vaccine components, WHOCC generates the virus seeds of each vaccine component and issue them to vaccine manufacturers. For inactivated vaccines, IAVs require the recombination of HA and NA genes from the candidate vaccine viruses and six internal genes from the A/Puerto Rico/8/34 strain by using reverse genetics to improve growth in eggs. For live attenuated vaccines, influenza A and B viruses require reassortment between the HA and NA genes from the candidate vaccine viruses and six internal genes from a cold-adapted master donor virus(A/Ann Arbor/6/60 [H2N2) or A/Leningrad/134/17/57[H2N2] for IAVs; B/Ann Arbor/1/66 or B/USSR/60/69 for IBVs) [211]. After the growth and antigenic characteristics are qualified, the corresponding vaccine virus seeds are applied to produce vaccines. The production of the vaccine can last more than 6 months. Vaccination starts in September for the Northern Hemisphere and in April for the Southern Hemisphere.
During these meetings, zoonotic influenza viruses, especially those with pandemic risks, are also discussed to update the vaccine strains used in pandemic preparedness. The procedure is similar to those for seasonal influenza viruses, and the current viruses include subtype H5, H7, and H9 AIVs and subtype H1N1 and H3N2 SIVs, all of which are enzootic in animals and have caused sporadic spillovers to humans [212]. The US Centers for Disease Control and Prevention (CDC) and WHO developed the Influenza Risk Assessment Tool (IRAT) and Tool for Influenza Pandemic Risk Assessment (TIPRA) to assess the emergence risk and public health impact risk of a novel (i.e., new in humans) IAV [213, 214]. The IRAT scores the risk by using 10 criteria based on biologic domain knowledge (e.g., virus properties, such as changes with known molecular signatures, receptor binding, transmission potential in laboratory animals, and drug susceptibility/resistance); population attributes (i.e., existing immunity, susceptibility to infection, severity of illness, and antigenic relationship to vaccine candidates); and virus ecology and epidemiology (i.e., global distribution, infections in animals, and infections in humans). IRAT then prioritizes the risks for enzootic IAVs by using laboratory-accumulated data. Similar to the IRAT, TIPRA evaluates the pandemic risk by assessing the likelihood for a virus to cause sustained human-to-human transmission based on virus properties, attributes (i.e., population immunity) in the human population, and virus ecology and epidemiology in animals [213–216].
However, selection of an influenza vaccine is not trivial. The influenza virus has a high mutation rate, and antigenic variants continually emerge in human populations. During the 6-month window between vaccine strain determination and subsequent vaccine development, antigenic drift can occur, leading to mismatch of the recommended vaccine strains. In addition, virus isolation and antigenic characterization are labor-intensive, and it is not possible to collect all potential influenza samples and include them into the analyses. Thus, even an antigenic variant that emerges before the GISRS committee meeting, especially those in the early stages of an epidemic, could be missed because of low prevalence or limited samples included in the laboratory analyses. To overcome sampling challenges, the CDC and Association of Public Health Laboratories (APHL) developed an Influenza Virologic Surveillance Right Size Roadmap, which provides tools to optimize sampling methods, disease surveillance, response and control efforts, and policy decisions [217]. An additional challenge is that the viruses can gain adaptive nonsynonymous mutations during isolation and/or propagation [77, 218], and these resulting amino acid substitutions can complicate antigenic analyses of epidemic strains in humans.
In addition to antigenic mismatch, a selected strain could produce low yields during vaccine production. A low yield vaccine strain will require additional laboratory work such as mutagenesis or egg passage to increase their growth rate. This could significantly delay vaccine production and delivery. For example, the 2009 H1N1 seed strain was a low yield strain (available on May 27, 2009) and required five WHO labs over 2 months to prepare the strain for production (August 6, 2009) [219]. Because of this delay, vaccine-derived immunity arose only after the second wave of the pandemic. Without such a delay, the vaccine may have reduced the impact of the second wave, and it is likely that many fewer infections and deaths would have occurred. Since different strains with similar antigenic profiles could have different growth ability in eggs, selection of a high yield strain with antigenic match is an important goal of vaccine strain selection.
An ideal method for antigenic variant identification includes comprehensive virus sampling, use of clinical specimens or limited virus isolation and passaging to avoid egg adaptations, and decreased time between strain selection and vaccine production. The following section will review the conventional and recent methods for antigenic characterization of influenza viruses with a discussion of their challenges.
Antigenic characterization of influenza viruses
Serological methods commonly used in antigenic characterization
Serological tests, such as hemagglutination inhibition (HI), neuraminidase inhibition (NI), and neutralization assays and the enzyme-linked lectin assay (ELLA), are routinely used to determine antigenic properties and identify antigenic variants during influenza surveillance. However, these conventional methods require a high quantity of viruses, which are more than what are typically available in the clinical samples, and, thus, a live testing virus will typically need to be isolated and propagated in either cells (e.g., Madin-Darby canine kidney, MDCK) or embryonated chicken eggs. Table 4 compares the principles of the common serological assays used in antigenic characterization.
Table 4.
Comparison between serological assays used in antigenic characterization of influenza virusesa
| Hemagglutination inhibition (HI) | Neutralization assays | Neuraminidase inhibition | Polyclonal serum–based proximity ligation assay (PolyPLA) | |||
|---|---|---|---|---|---|---|
| Micro-neutralization (MN) | Focus reduction neutralization test | Micro-neuraminidase inhibition | Enzyme-linked lectin assay (ELLA) | |||
| Principle | Antibodies inhibiting binding between virus and erythrocytes | Antibodies inhibiting viral cell entry and viral replication | Antibodies inhibiting the NA activity | Direct binding between virus and antibody | ||
| Primary epitopes | HA heads, particularly those close to RBD | HA and NA | NA | HA and NA | ||
| Standardization before assays | Yes, to determine 4 HAU | Yes, to determine 100 TCID50 | Yes, to determine 20–85% ICP | Yes, to determine an OD549 of 0.45 to 0.85 | Yes, to determine 90% maximum OD490 | Not needed |
| Sera/reaction | 25 uL | 50 uL | 50 uL | 50 uL | 50 uL | 0.5–1 uL |
| Virus/reaction | 4 HAU, and 1 HAU represents ~104 virion/25 uL [220] | 100 TCID50/50 uL | 20–85% ICP/50 uL | OD549 of 0.45 to 0.85/50 uL | 90% of maximum OD490/50 uL | ~103 TCID/mL in a volume of 0.5 uL |
| Terminal titer determination | Agglutination | ≥50% absorbance readout reduction compared to virus control | ≥80% ICP reduction compared to virus control | 50% inhibition concentration | PolyPLA units derived from normalized ΔCt | |
aHAU hemagglutinating unit; TCID50 50% tissue culture infectious dose; ICP infected cell population; OD optical density; ΔCt the change between threshold cycle of negative control and that of sample
HI assay
The HA of influenza viruses binds to sialic acid glycan receptors [221] of red blood cells (erythrocytes) and can agglutinate erythrocytes. Such agglutination ability of influenza viruses can be used to quantify viruses by determining the maximal dilution of viruses, which can achieve visible erythrocyte agglutination. Antibodies can inhibit erythrocyte agglutination by binding to HA ABSs at the RBS. The HI assay measures how a test influenza antigen and a reference antigen (e.g., a serum for a current vaccine strain) match through the immunological reaction between the test antigen and the reference antiserum (Fig. 4A). With its simplicity and moderate throughput, HI has been widely applied (and is expected to continue being a useful tool) in antigenic analyses of influenza viruses during surveillance and vaccine strain selection.
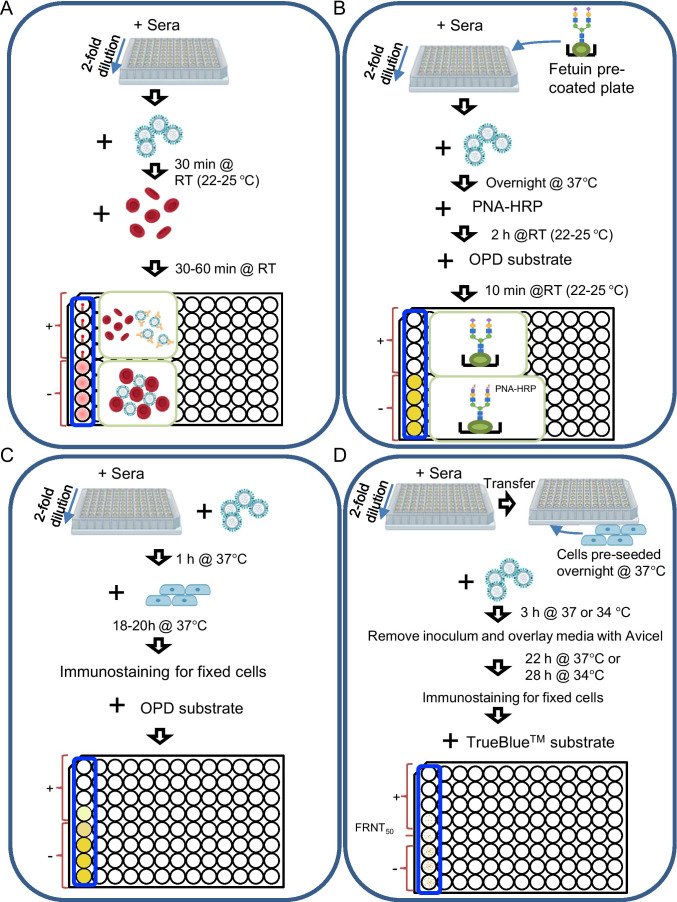
Fig. 4.
Conventional of serological assays used in influenza antigenic analyses. (A) Hemagglutination inhibition (HI) assay. After mixing 4 hemagglutination (HA) units of virus and 2-fold serially diluted reference sera, red blood cells (RBCs) are added to the reaction. If the binding of viruses to RBCs is not inhibited by the antibody, the RBCs will agglutinate in the micro-titrate. Otherwise, the RBCs are not agglutinated, forming (1) a button or a halo when using avian RBCs or mammalian RBCs or (2) a floating pellet (when the plate is tilted) using avian RBCs. The pattern of non-agglutinated chicken and turkey RBCs are shown in the top four wells to the left column and that of agglutinated chicken and turkey RBCs are shown in the bottom four wells. (B) Enzyme-linked lectin assay (ELLA). The mixture of a predetermined amount of virus and serially diluted reference sera is added to 96-well plates coated with fetuin, a liver protein with sialic acid and galactose at the glycan terminal, and then incubated overnight at 37°C. Peanut agglutinin conjugated to peroxidase (PNA-HRP) is then added, and the PNA-HRP binds to the exposed galactose due to the removal of sialic acid by neuraminidase activity. Otherwise, the neuraminidase is inhibited by the reference antibody, and the PNA-HRP does not bind to the fetuin. Finally, the signal is detected by adding o-phenylenediamine dihydrochloride (OPD) substrate. (C) Micro-neutralization (MN) assay. Reference sera are diluted by twofold, and then mixed with viruses in a titer of 100 median tissue culture infectious dose (TCID50) per well. The mixtures are used to infect cells (e.g., MDCK cells or MDCK-SIAT cells). After 1 day of incubation at 37°C, an ELISA can be performed to detect the fixed cells using anti-NP antibodies [222]. (D) Focus reduction neutralization test (FRNT). Reference sera are diluted by twofold and added to MDCK or MDCK-SIAT cell pre-seeded plates, and viruses caused 20–85% infected cell population (ICP) are then added. After 3 h incubation at 37°C (influenza A virus) or 34°C (influenza B virus), the inoculum is removed, and the monolayers are overlaid with the culture medium containing 1.2% (w/v) Avicel (FMC BioPolymer) and 2 μg/mL TPCK-trypsin. After 22-h incubation at 37°C (influenza A) or 28-h incubation at 34°C (influenza B), an immunostaining is performed to detect the fixed cells using NP-specific antibodies, a peroxidase-conjugated secondary antibody and TrueBlue substrate. The infected cell population is imaged by flatbed scanner. The neutralization titer is expressed as the reciprocal of the antiserum dilution that reduces ICP by 80% [223]
The success of the HI assay depends on the agglutination ability of erythrocytes and thus is affected by the influenza virus–associated glycan receptor distribution on the erythrocytes used in the assay and the receptor binding properties of the testing virus. Previous studies showed that turkey and chicken erythrocytes express α2,3- and α2,6-linked sialic acids, horse erythrocytes almost exclusively express α2,3-linked sialic acids [224], and guinea pig erythrocytes disproportionately express more α2,6- than 2,3-linked sialic acids [225]. Of these, turkey erythrocytes are commonly used and are still effective in antigenic analyses of human seasonal influenza viruses, IBVs and A(H1N1) (i.e., both A(H1N1)season1977 and A(H1N1)pdm09). However, for antigenic analyses of seasonal A(H3N2) viruses, chicken erythrocytes were used until mid-1990s [226]; turkey erythrocytes were then used to replace chicken erythrocytes until the 2004–2005 influenza season [34, 227, 228]; guinea pig erythrocytes were used from the 2004–2005 through the 2013–2014 influenza seasons [229]; after the 2010–2011 influenza season, the NA inhibitor is suggested to be added when using guinea pig erythrocytes [230, 231]. Multiple amino acid substitutions of the HA RBS [40] caused A(H1N1)season1977 [232, 233] and seasonal H3N2 IAVs [234] to lose their ability to bind to different species of erythrocytes that are typically used in influenza surveillance. For example, the amino acid substitutions at residues 193, 196, 197, and 225 in A(H1N1)season1977 in 1988 or later resulted in the loss of their abilities to agglutinate chicken erythrocytes [235]. For H3N2 viruses, the amino acid substitutions at positions 190, 226, and 194 of HA were associated with the loss of the ability for the A(H3N2) viruses to agglutinate chicken erythrocytes [234, 236–239]. Since the 2004–2005 influenza season, A(H3N2) viruses acquired Asp225Asn in HA, which caused the loss of their binding abilities to turkey erythrocytes [227, 230]; this may be attributed to the inability of viral binding to short oligosaccharides terminated with sialic acids (i.e., those having only one or two N-acetyl-lactosamine repeating units), which are mainly expressed by chicken or turkey erythrocytes [240]. As an extreme example, a large portion of A(H3N2) viruses, especially clade 3C.2a viruses, have lost the capacity to agglutinate chicken, turkey, and guinea pig erythrocytes [229, 241]; thus, they cannot be antigenically characterized by HI assays. In addition, the 3C.2a A(H3N2) viruses showed different adaptation patterns when using different types of MDCK cell lines: when passaged in conventional MDCK cell lines (CCL-34; ATCC), the 3C.2a viruses may acquire amino acid changes on HA and/or NA and regain the ability to agglutinate turkey and/or guinea pig erythrocytes, which likely affects antigenic analyses, but such changes were not observed with the MDCK-SIAT1 cell line [229]. The MDCK-SIAT1 cells, which are stably transfected with human CMP-N-acetylneuraminate:β-galactoside α-2,6-sialyltransferase, with increased SA2,6Gal but decreased SA2,3Gal expression [242], have been recommended to be used for virus isolation and propagation of current H3N2 viruses.
Of interest, NA activities were then found to affect viral erythrocyte agglutination in H3N2 viruses (e.g., those between 2005 and 2009) and can affect HI results [231]. Amino acid substitutions T148I and D151G were found to be artifacts of virus isolation and propagation in MDCK cells and associated with the NA-mediated viral erythrocyte-binding [231]. Subsequent studies reported that substitutions H150R [243] and G147R [244] on the NA protein were also associated with the phenomenon. Another study suggested that the HI assay measures the effect of antibodies against NA rather than HA for A(H3N2) viruses [243]. Thus, over time, to reduce the non-antigenic effects of virus variations, the protocols for HI assays have been updated by using neuraminidase inhibitors (e.g., oseltamivir) in HI assays using guinea pig erythrocytes [231].
In addition to the animal species, the sources and even batches of erythrocytes may impact the results from HI assays in practice, causing such data to be notoriously noisy and difficult to interpret, integrate, and reproduce between laboratories [245–247]. In addition, implementation of the HI protocol can generate additional noise in HI analyses. In the past decades, various mechanical practices have been used to improve the accuracy and throughput of HI assays, such as Cypher One developed by Wilson et al. in 2017 [248] and High-Throughput Imaging and Visualization Equipment (HIVE) T670 by Nguyen et al. in 2016 [249] to automate image analyses for interpretation of HI assays and to remove inconsistencies caused by plate tilting. Sanchez-Cano et al. [250] recently developed synthetic erythrocytes, called synthrocytes, which can be used for HI assays instead of animal erythrocytes. They created these synthetic erythrocytes by first identifying commercial beads that express distinct sedimentation patterns based on agglutination (Sicastar-blue beads), and then, based on the study by Suzuki et al. [221] that influenza viruses differ in their recognition of sialic acid–galactose linkages, developed sialylated beads as bioreceptors to bind influenza. Tested against conventional assays, synthrocytes offered improved speed and stability for A(H1N1) and IBVs and can easily be mass-produced [250]. While the sensitivity of synthrocytes remains less than fresh erythrocytes, the improved specificity, stability, and assay time suggest that synthetic erythrocytes offer great potential for improved serological analyses of influenza viruses. Nevertheless, these synthetic erythrocytes have not been used as a substitute of animal erythrocytes in the HI assays.
NI assay
While HA mediates receptor binding, NA cleaves N-acetyl neuraminic acid from the virus, which allows the virus to be released and infect additional cells [251]. Aminoff et al. [252] developed the original NI assay in 1961 as a macro-assay. This macro-assay was then modified by Van Deusen et al. in 1983 as a micro-neuraminidase-inhibition (micro-NI) assay performed on micro-titer plates to accommodate smaller quantities of reagents and allow for increased sensitivity compared to macro-NI and increased capacity to evaluate multiple isolates at a time [253]. NA can be subtyped using the antibody specificity of NA to influenza, and the susceptibility of influenza viruses to antiviral drugs can be determined using NI assays, which work by quantifying the amount of sialic acids cleaved by NA [222, 251]. Despite the development of the micro-assay, NI assays are still limited by the vast resource requirements including the need for individual glass tubes and use of toxic chemicals including arsenite and 2-thiobarbituric acid [222, 254]. Additionally, traditional NI assays may not adequately detect NA antigenicity when antigenic drift occurs at the lateral surface of the NA head and prevents the binding of anti-NA monoclonal antibodies [70]. These challenges are the reason that NI assays are not a commonly used method for assessing influenza antigenicity.
ELLA
ELLA is an alternative method for measuring NI titers that overcomes the resource limitations of the NI assays, allowing for increased scalability and safety [254]. Rather than individual glass tubes, ELLA is performed on 96-well plates that are coated with fetuin as the NA substrate [254, 255]. Heat treatment is first implemented to remove non-specific NA inhibitors, followed by careful virus titration to optimize assay sensitivity [254]. ELLA reactivity is measured on the cell population level, and thus is more accurate when the cell population is more homogenous, but will only report the average surface a-D-galactopyranasyl groups in a heterologous cell population (Fig. 4B) [256].
Neutralization assays
Neutralization assays quantify the ability of a testing antibody to neutralize viral infection. Typically, to evaluate antigenic properties of a testing virus, a series of reference sera dilutant is first incubated with the virus for antibody-virus binding and then the mixture is inoculated in cells or chicken embryonated eggs. Viruses with similar antigenic properties are expected to have similar results in neutralization assays against the same set of reference sera. Classical neutralization assays utilized embryonated chicken eggs or primary cells to detect the reduction of virus plaque formation or virus growth inhibition [257]. Many efforts have been made to further increase the throughput and optimize neutralization assays.
To increase the throughput and shorten the detection time, Okuno et al. [223] developed a focus reduction neutralization test (FRNT) based on micro-titration plates. In this assay, residual virus infectivity was obtained by counting foci stained by specific antibodies. Micro-neutralization (MN) assays increase the throughput in quantifying neutralizing antibodies. Multiple methods have been used in determining terminal titers, such as cytopathic effect (CPE) formation in cell culture, colorimetric assays to detect the cell viability or cytotoxicity, hemagglutination assays for detection of released virus, or usage of the enzyme-linked immunosorbent assay (ELISA) to identify infected cells [257, 258]. Lin et al. [259] established an optimized FRNT to mimic the plaque reduction of neutralization assays based on the infected cell population (ICP) in a 96-well-plate format by using an imaging method for more efficient quantification. Fig. 4C shows the ELISA-based MN assay described by the WHO manual for laboratory diagnosis and virological surveillance of influenza viruses [222], and Fig. 4D shows the optimized FRNT which has been used by WHOCCs in characterizing the antigenicity of seasonal H3N2 viruses to date. Based on FRNT, Baalen et al. [260] and Jorquera et al. [78] further improved the method parameters and developed the ViroSpot MN assay and high content imaging-based neutralization test (HINT), respectively. The HINT was performed at a single infection cycle by using a low multiplicity of infection (MOI) in order to reduce antigenic mischaracterization due to viral host-cell adaptation. To further minimize the impact of host-cell adaptation, the reference ferret antisera were generated directly against viruses in the human respiratory specimens instead of the propagated viruses. The cell immunostaining images are analyzed by a high-content imaging micro-plate reader to determine the HINT titer by calculating a 50% ICP.
Different from HI assays, neutralization assays determine neutralization ability by measuring the effects of virus propagation ability and can reflect antigenic properties of not only the global head subdomain of HA but also the stalk subdomain of HA and NA. Neutralization assays can overcome the challenges of HI assays, such as receptor-binding avidity variants [261]. However, neutralization assays require the virus to propagate efficiently in cells or chicken embryonated eggs. The poor growth ability of recent H3N2 viruses has generated challenges not only in viral isolation but also the implementation of MN assays (see the section of Challenges in conventional serological assays). Although MN assays generally showed a strong correlation with HI assays [262], there are often exceptions which result in difficulty integrating HI and MN data [247]. Additionally, these assays are lower-throughput and more much elaborate and time-consuming than HI assays [261].
ELISA
Indirect ELISA was routinely used in antigenic characterization. This assay typically requires virus or a recombinant protein (e.g., HA or NA) to be adsorbed to a micro-titer plate followed by the addition of sera samples, an enzyme-conjugated secondary antibody HA, and lastly the substrate, which is used for colorimetric quantification [257]. The binding strengths of antibodies can be quantified and compared across antigens and sera. For example, Li et al. [263] coated 15 HA units of purified viruses to micro-titer plates and characterized the antigenicity of the mutant viruses with an amino acid substitution N145K. Chambers were coated with virus-like particles expressing HA proteins in the micro-titer plates, suggesting that antigenic change was caused by the F159S substitution [79]. In both of these studies, monoclonal antibodies recognizing the conserved epitopes of HA proteins were used to calibrate the viruses before running the assays so that equal amounts of viruses are used across samples in the assays. One major limitation for ELISA is that this assay could suffer from high background due to substrate contamination, poor washing steps, or cross reactivity thus can lead to false positive or false negative results. Nevertheless, ELISA can measure different isotypes and subclasses of immunoglobulins (Ig), such as IgM, IgA, and IgG [257], which are valuable for dissecting humoral immune responses. However, it has not been reported that the antigenicity of influenza viruses is associated with the Igs other than IgG.
Mass spectrometry immunoassays
To overcome the shortcomings of not providing viral molecular detail in HI assays, a mass spectrometry (MS) immunoassay was developed to evaluate the antigenicity of influenza viruses utilizing monoclonal antibodies [264]. This assay is based on a comparison of matrix-assisted laser desorption ionization (MALDI) mass spectra obtained from proteolytic digestion of the whole virus [264] or an separated antigen [265, 266] either with or without antibody treatment. The mass maps provided not only the antigenicity characterization of a specific epitope but also the primary structure of the viral antigen. The basis of MS makes it possible to not immobilize either the antigen or antibody and avoid the antigenic alteration during immobilization. The antigenic characterization of this assay showed high consistency with that of HI assays for A(H1N1)season77 viruses [267]. The challenge of this assay is that the antigenicity can be only analyzed on the epitopes targeted by the given monoclonal antibodies.
Selection of reference sera in serological analyses
The selection of reference sera is one of the keys to unbiased serological analyses. Ideally, homologous antibodies are included in antigenic analyses for all testing strains. However, this is not possible during seasonal influenza surveillance because it is not practical to generate reference sera against all testing viruses as hundreds to thousands of viruses are assessed each year. It is critical to include homologous sera for those with potential antigenic variants, especially those with amino acid substitutions at known ABSs. Thus, serological analyses in influenza surveillance involve a dynamic update of the reference sera panel.
Due to their sensitivities to both seasonal influenza A and B viruses, influenza seronegative ferrets are widely used to generate the ferret sera used in seasonal influenza vaccine strain selection [41, 268–273]. One of the key criteria for WHO to make vaccine strain update is that variant viruses are identified to have ≥8-fold reduction of cross-reactivity to the ferret sera as compared to that of the homologous vaccine virus [274]. Vaccines have also been evaluated with human post-vaccination sera for cross-reactivity with circulating variants; a reduction of ≥50% in geometric mean titers significantly indicates low vaccine efficiency [261].
However, antigenic properties derived from ferret serological data may not reflect those in the human population, which contain varying degrees and compositions of pre-existing immunity. There are dramatic differences in specificity between human and ferret antibodies, and these differences are due in part to pre-exposure history [269]. This process, termed “original antigenic sin,” or immune imprinting, was observed in humans infected with the A(H1N1)pdm09 during the 2013–2014 season [275]. The human immune system has a preference for generating antibodies with cross-reactivities of previously exposed strains at the apparent expense of generating new antibodies that specifically recognize newer strains [276]. Although the circulating A(H1N1)pdm09 viruses showed antigenically neutral amino acid substitutions when evaluated using ferret sera, they acquired a genuine antigenic substitution located on HA that escaped antibody recognition and elicited infections in a large number of middle-aged humans [275]. Using representative A(H3N2) strains circulating during 2007–2014, we conducted head-to-head comparisons of the antigenic maps derived from human and ferret serologic data and illustrated their differences in antigenic characterizations [274, 277]. In general, data from ferret sera are more comparable with post-vaccination sera from children but not with those of adults with pre-existing immunity [277].
Antigenic cartography
Antigenic cartography, a computational approach to visualizing antigenic diversity, is often used in conjunction with serological assays. Antigenic cartography was initially proposed by Smith et al. [76] as a method for visualizing the antigenic diversity of IAVs based on antigenic distances. Antigenic distances in antigenic cartography are calculated using local pairwise distances between viruses that are in close temporal proximity; typically, each horizontal or vertical gridline on the map represents one antigenic unit distance corresponding to a 2-fold difference in serologic (e.g., HI) titers, and either the horizontal or the vertical axis can be used for this calculation.
Because challenges arise due to low reactors and potential biases from missing data, we developed AntigenMap, a publicly available, novel, low-rank matrix completion-based method (http://sysbio.missouri.edu/AntigenMap) [278, 279]. AntigenMap functions by first reconstructing serologic data (e.g., HI matrices with viruses and antibodies) by using low-rank matrix completion to account for missing data and, then, using multidimensional scaling, generates a 2- or 3-dimensional antigenic cartography [278, 279]. Antigenic cartography provides an intuitive visualization and interpretation of serological data and has been used routinely in antigenic analyses and vaccine strain selection for seasonal influenza viruses, pandemic preparedness, and basic research. Two-dimensional antigenic cartography may generate biases in antigenic distance during multidimensional scaling, especially in high dimensional serological data. However, such biases can be mitigated by using a three-dimensional antigenic map [278–280].
Three methods have been proposed for calculating antigenic distances for influenza vaccine selection, which were evaluated by Cai et al. [280]. These methods include average antigenic distance (A-distance), which measures the average difference between antigen and antisera interaction effects of two antigens, mutual antigenic distance (M-distance), which measures the distance between two clusters of antisera, and largest antigenic distance (L-distance), which uses the maximum difference between antigen and antisera interaction effects between antigens. These measurements were combined with a metric multidimensional scaling (MDS) method, which indirectly calculates antigenic distance as a Euclidean distance to minimize error and projects the antigens onto a graph. Cai et al. [280] determined that the most robust method for measuring antigenic distances for influenza strain selection is the M-distance.
Because it is derived from serological data, antigenic cartography is dependent on the input data. Thus, selection of the reference sera is extremely important. Biases in the reference sera can skew coordinates of antigens in the map and lead to misinterpretations of the antigenic relationships among the testing antigens. In addition, as mentioned above, potential biases in antigenic cartography may arise when used with high dimensional serological data. Thus, although antigenic cartography is a very useful and convenient tool, cautious interpretation on antigenic cartography should be used, often with an examination of the original serological data.
Challenges in conventional serological assays
Overall, these conventional serological assays described above have multiple barriers that include limited facility resources, culture-adapted mutations, and poor virus viability, leading to the inability to timely detect amino acid substitutions associated with antigenic drift [269]. As mentioned earlier, these assays require virus isolates from clinic samples. Compared to the seasonal surveillance of influenza viruses, antigenic characterization for pandemic preparedness of emerging pathogens presents even more challenges. Propagation of these viruses often requires facilities with biosafety level (BSL)-3 or BSL-4, which are difficult to obtain. Thus, most specimens must be shipped to laboratories with appropriate biosafety containment resources, which are often accompanied by administrative barriers, especially when shipping between countries.
Once the samples have reached an adequately equipped facility, the virus isolation process is not only time-consuming, but also vulnerable to emerging culture-adaptive mutations during viral propagation in either cells or embryonated eggs [281–285]. Since 1993, antigenic and biochemical differences in the HA proteins of influenza viruses have been noted after passage in both MDCK and eggs [232]. Growth in cell culture spiked with non-immune horse serum has produced altered patterns of agglutination for influenza viruses that are propagated in comparison to the original isolate [232, 286–294]. Additionally, sequence analyses have shown that changes in the H3 HA residues occur in the antigenic sites: residue 137 (antigenic site A), 156 (B), 186 (B), 248 (D), 276 (C) [287]. Both monoclonal and polyclonal antisera can detect a difference in antigenicity between HAs before and after passage in eggs [232]. The altered viruses negate the goal of choosing vaccine viruses which are likely to be circulating during the impending influenza seasons. For example, vaccine effectiveness in the 2012–2013 influenza season was only 49% in part due to egg adaptations in the A/Victoria/361/2011 vaccine strain (IVR-165) [174, 270].
Recently, viruses have also evolved to alter its receptor binding properties towards the various sialic acid receptor types of erythrocytes [239, 295–297], which includes losing the binding ability of human viruses to chicken erythrocytes [234, 236]. This has resulted in diminished effects of NA enzymatic activities in virus HI [235, 298, 299], further making HI assays not ideal for antigenic analyses. The limited growth ability of the influenza viruses in MDCKs [300] and/or embryonated chicken eggs without culture-adaptions [301], particularly the H3N2 and H1N1 human seasonal IAVs, two cells conventionally used in virus isolation, further challenges the usefulness of conventional serological assays. Thus, a clinical sample-based assay is needed. These barriers of high resource requirements, culture-adaptations, and poor virus growth often prevent timely and accurate surveillance and response to influenza outbreaks [302].
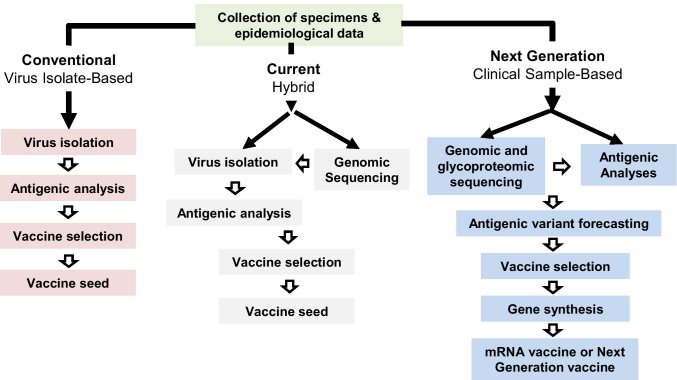
Antigenic characterization using clinical samples
A typical human patient with an acute seasonal IAV infection will have a detectable viral load of ~106 TCID50/mL in their nasopharyngeal swab samples [303], while viral loads in the swabs from those with asymptomatic infections can be much less [304]. Two potential approaches can be used to determine influenza antigenicity using clinical samples: (1) antibody-based assays, which are sensitive and require a low amount of influenza viruses equal to or less than the amount collected in the clinical specimen; and (2) sequence-based assays. Since the 1990s, sequencing of influenza virus genes, particularly HA, NA, and MP, has been routinely used in influenza surveillance. Recent advances in sequencing technology allow us to rapidly capture the genetic changes in influenza genomes [207, 268]. Genomic sequencing of influenza viruses requires as little as 49,350 copies of viruses in the sample (with a 95% probability of whole-genome recovery) [305] and are well documented to succeed for clinical specimens from both acute and asymptomatic infections [306].
Polyclonal serum–based proximity ligation assay (polyPLA)
We recently developed a novel quantitative PCR (qPCR)–based antigenic characterization method using a polyclonal antibody-based proximity ligation assay (polyPLA) [302, 307]. PolyPLA was developed based on a proximity ligation assay (PLA), which detects antigen–antibody interactions using monoclonal antibodies [308]. Specifically, the PLA incubates oligonucleotide-linked monoclonal antibodies with the analyte in question, and if the oligonucleotides are in close proximity, they can be ligated; the presence of analyte is then shown by the amplification of ligated products with qRT-PCR. The assay reporter signal depends on the proximity and dual recognition of each target analyte, which allows for high specificity [309].
Building on the PLA, PolyPLA is developed to quantify the antibody-antigen binding avidity by using the amplification signals in qPCR from the pairs of primers attached to a reference polyclonal antiserum (Fig. 5). Since the NP protein is antigenically conserved among influenza viruses, the ΔCt value based on the anti-NP monoclonal antibody was used to normalize the amount of intra-type viruses across the testing samples as well as those reference viruses used in the analyses. Therefore, the polyPLA units derived from polyclonal serum after normalization (i.e., NP monoclonal-based ΔCt value) are determined as the antigenic properties for the equal amounts of viruses and thus are comparable across the testing samples.
Fig. 5.
A diagram illustrates polyPLA. PolyPLA quantifies antibody-antigen binding avidity. Reference polyclonal antiserum (for antigenic analyses) or anti-NP monoclonal antibody (mAb) (for normalization) is biotinylated and then labeled using sodium azide-linked oligonucleotide probes. The labeled polyclonal antiserum or monoclonal antibody is incubated using a reference (virus) or testing antigen and ligated with the two oligonucleotides linked to the antibodies are ligated followed by qPCR, which is used to determine the amplification signals and quantify antibody-antigen binding avidity. The resulting cycle threshold (Ct) values of the polyclonal antisera and antigens are normalized by those by anti-NP monoclonal antibody and antigens and then analyzed for antigenic differences, and the normalization will ensure the equal amount of antigens in antigenic analyses. This figure was adapted from Martin et al. [302]
PolyPLA can detect a low virus titer of <1000 median tissue culture infectious dose (TCID50)/mL, distinguish between different IAV HA subtypes, and effectively identify antigenic variations within the same IAV HA subtype [302, 307]. Antigenic profiles determined by polyPLA have been validated to be consistent with those from HI and neutralization assays [302, 307]. In addition, antigenic maps derived from HI assays and polyPLA using the same set of viruses and sera were concordant [307]. Fig. 6 shows the correlation between the polyPLA and HI data obtained for 19 A(H3N2) clinical samples and 3 reference viruses by using 3 ferret reference sera. Correlation coefficients determined by linear regression showed that the titers between polyPLA units and log2(HI) had a coefficient of R = 0.8196 (p<0.0001). None of the 5 testing sera reacted with A/California/04/2009(H1N1) in a HI assay or a polyPLA. An eightfold increment in HI titer is correlated with a 3.26-fold increment in polyPLA units [307].
Fig. 6.
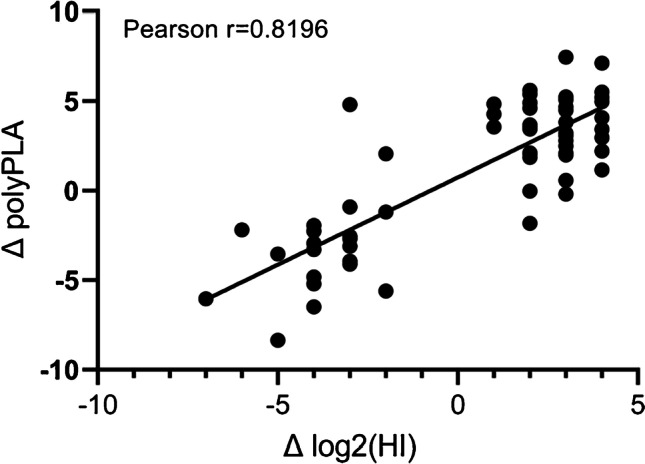
Correlation between polyPLA and HI titers. The Pearson correlation coefficient analysis was performed on paired polyPLA and HI titers for 19 H3N2 viruses against three reference sera, which were adapted from Martin et al. [302]
The polyPLA can address those aforementioned challenges in HI assays (dependence on types of erythrocytes and high noise) and in neutralization assays (labor intensiveness). Different from HI assays, the polyPLA detects antigenic variations in both HA and NA proteins. The polyPLA only requires a small volume of clinical samples (e.g., 2 uL), and this enables the feasibility to include replicates and test multiple reference sera for the same clinical samples. Because it is based on a common qRCR platform, polyPLA can be implemented in large-scale analyses with the same biosafety requirement as those typically used in diagnosis, e.g., BSL-2. Similar to most of those conventional serological assays, the polyPLA will work the best after narrowing down the virus subtype for the analyses, and such a challenge can be overcome by combining the analyses of the virus subtype(s) in the clinical samples through genomic sequences.
Sequence-based antigenic analyses
In the past three decades, sequence-based analyses, particularly those on the HA, NA, and MP genes, have been used routinely as an important component of influenza vaccine strain selection (Fig. 3). The sites affecting influenza antigenicity are primarily located in the head structures of the HA protein (Fig. 1) [45, 310, 311], and one or several of these antigenicity-associated sites frequently change during antigenic drift events [76, 271, 312–314]. Thus, these analyses have primarily focused on amino acid substitutions at the reported epitopes in the HA protein and drug resistance markers in these three proteins. These substitutions are typically mapped into phylogenetic trees to understand their evolutionary relationships and prevalence trends in the viral population. However, a comparison of antigenic and genetic maps showed that the antigenic impact of genetic changes varies. Thus, not all amino acid substitutions in HA sequences, including those at ABSs, equally affect antigenic properties [315, 316]. An ideal sequence-based strategy for vaccine strain selection is to quantify antigenic distances directly using protein sequences (Fig. 7).
Fig. 7.
Evolving analytic platform for vaccine strain selection and vaccine development. The conventional platform involves isolating viruses from clinical samples, and the viruses are used in antigenic analyses. Due to the labor intensiveness in laboratory efforts, typically only a small set of samples can be analyzed. In the past decade, the advances in genomic sequencing can allow us to quickly sequence viruses using clinical samples. To determine viral antigenicity, virus isolates are still needed. These sequences can be used to guide selection of samples in virus isolation and virological analyses. As a next-generation platform, we would sequence protein and glycans from clinical samples, and antigenicity would be determined by these sequences. The next-generation platform is expected to be higher throughput and can minimize sampling biases. The computational tools correlating antigenicity (and other vaccine strain required phenotypes) and these sequences would be available. Ideally, in this platform, the big data and artificial intelligence-based tool would be able to forecast antigenic evolution
A few attempts at simple machine learning methods have been implemented to identify influenza antigenicity–associated sites using a small set of serological data. For instance, Lee and Chen [317] developed a simple correlation method between HI titers and the number of amino acid changes between test viral HA and reference viral HA. Liao et al. [318] applied multiple logistic regressions between amino acid substitutions and HI values. Most recently, Huang et al. [319] developed a decision tree algorithm to predict variant drift by deriving association rules from HI data based on information theory. However, these earlier attempts did not consider the challenges of missing values and low reactors, which are commonly seen in serological data [278].
To overcome these challenges, we developed a set of sparse learning methods to identify antigenicity-associated residues by using serologic data and a quantitative function to identify antigenic distances using HA and NA proteins. Sparse learning methods have the advantages of efficiency and generalizability when using a small number of non-zero elements [320]. When data is limited, promoting sparsity has been shown to produce robust models that generalize well to extrapolated data [321]. Thus, sparse learning is suitable for this problem, which suffers from a relatively small data size and high noise levels. We formulated this as an optimization problem that measures the correlation between the antigenic distance changes in HI and NI data and the antigenic profiling by using a scoring function to characterize the number of amino acid substitutions in protein sequences. Structural and physiochemical features were also integrated into the scoring systems. This sparse learning algorithm effectively identifies antigenicity-associated residues in H5N1 [322] and H3N2 [323, 324] viruses. We then integrated antigenic mapping and machine learning via bootstrapped ridge selection using Antigen-Bridges [80]. This model identified 39 antigenicity-associated amino acid positions and, as a first, quantified antigenic distances on the basis of genetic distance of influenza A(H3N2) HA1 sequences from 1968 to 2009 [80].
After noticing the contribution of co-evolution in antigenic drift, we further developed AntigenCo to identify and quantify both single and co-evolutionary amino acid substitutions driving antigenic drift with higher accuracy [324]. To further identify the synergistic effects of multiple amino acid substitutions on antigenic changes, we further developed a generalized hierarchical square model (GHSM) [325]. GHSM enforces sparsity under hereditary structures in the interactions among the covariates and effectively incorporates the feature space up to the 5th-order interactions. We also developed a multi-task sparse learning algorithm and showed that a multi-task formulation can help solve challenges in integrating data from different platforms [71]. Based on this multi-task platform, we developed another algorithm, MTL-SGL [72], which can integrate different groups of features (e.g., amino acids and N-glycosylation). MTL-SGL enables us to identify key intra- and intergroup features by utilizing the l1 regularization term on the feature weights to force sparsity regularization on intragroup features and by utilizing the group structured l2 regularization to force sparsity regularization on the group of the feature weights. Antigenic maps for seasonal H1 and H3 viruses are developed for these viruses (Fig. 2B and C). Using sequence and N-glycosylation feature types, genetic determinants that affect influenza antigenicity were identified based on the weights of each feature. Compared with two other conventional single-task (LASSO and SGL) and multi-task models (ℓ1, 2 MTL and ℓ1,∞ MTL), MTL-SGL achieved the best performance with the lowest root mean squared error, highest accuracy, and highest sensitivity.
A few other attempts have been made to integrate antigenic analyses with phylogenetic analyses. Neher et al. [326] described sparse learning models based on the phylogenetic tree structure or amino acid substitutions to infer their antigenic properties, which also considers changes in avidity and serum potency, and demonstrated that the two models showed similar prediction accuracy and could be applied to all circulating subtypes. In addition to sparse learning algorithms, regression and Bayesian models have been introduced by treating the amino acid substitutions as features and the serological data or antigenic distances between sequences as responses [86, 316, 318, 327–332]. Steinbruck and McHardy [316] described a computational “antigenic tree” method by using nonnegative least-squares optimization to map pairwise antigenic distances onto the branches of a phylogenetic tree. Bedford et al. [332] implemented a Bayesian approach to combine antigenic cartography and phylogenetic information about the seasonal influenza viruses.
Harvey et al. [86] integrated sequences on the non-conserved and exposed HA surface and corresponding HI data for seasonal A(H1N1) viruses before 2009 using regression models. These models identified and quantified the impact of 18 determining substitutions and allowed for the prediction of antigenicity by HA sequence data with improved accuracy [86]. Cui et al. [328] combined multiple linear regression and physicochemical changes of 18 key amino acid positions to infer antigenic variants of A(H3N2) viruses with a low false-positive rate. By comparison, Rahman et al. [329] showed that combining a non-linear regression model with a scoring method based on the biochemical properties of amino acids for the specific antigenic areas on HA resulted in the best prediction.
To extend the multiple subtype prediction, Zhou et al. [333] proposed a Context-Free Encoding Scheme method to predict cross-subtype antigenic patterns by integrating them with a random forest classifier and to predict the antigenicity of different subtypes by using transfer learning. Yao et al. [334] emphasized that substitution metrics reflecting variant properties of amino acids are critical for improving the prediction performance of models and combined top substitution metrics into the random forest algorithm, which they named Joint Random Forest Regression (JRFR), and this method showed an improved prediction of antigenic variants of A(H3N2) HA1. Yin et al. [335] constructed a stacking model combined with residue-based, regional band-based, and epitope region–based feature extraction methods to predict the antigenic variants of both pandemic and epidemic A(H1N1) viruses. This model showed the capability of determining antigenic variants with an accuracy of 0.908 [335].
To improve predictions of H3N2 antigenic evolution, Suzuki et al. [327] evaluated de novo mutations by integrating HI titers with volume, isoelectric point, solvent accessibility, and distances from the RBS and N-linked glycosylation sites to quantify antigenic distances and while improved, prediction accuracy was still low. Du et al. [336] built the PREDict Antigenic Cluster (PREDAC) machine learning model integrated with network clustering to infer antigenic clusters of strains from HA sequences at the population level with an accuracy of 89.24–89.70%. HA antigenic similarity was predicted with a naïve Bayes classifier from 12 structural and physicochemical features of HA sequences, and 17 dominant A(H3N2) antigenic clusters were identified in mainland China between 1968 and 2010 [336]. This method was subsequently expanded to A(H1N1), A(H5N1), and all influenza subtype viruses [337–339]. Qiu et al. [340] designed a structure-based antigenicity scoring model by integrating a position-specific scoring matrix (PSSM) profile with local environmental change which fully incorporated the structural context of HA proteins to calculate the antigenicity. This model showed an accuracy of 0.875 for the antigenic inference of A(H3N2) [340]. To further predict the antigenicity of different pathogens, they established a new model, Conformational Epitope (CE)-BLAST, by comparing the conformational epitopes directly to suggest the relative antigenic distance between antigens. This model was validated using influenza and dengue experimental data [341].
In summary, antigenic characterization with direct utilization of clinical samples can overcome the challenges of culture adaptations during virus isolation and propagation. Both polyPLA and sequence-based analyses are high throughput but complementary. Genomic sequences can at least provide initial subtypes of viruses in the samples and help narrow down the targets for the polyPLA assays, and the polyPLA assays can help develop and refine those sequence-based computational models.
Predicting antigenic evolution of influenza viruses
Predicting antigenic evolution of influenza viruses using neuralization assays
The quasi-species nature of influenza viruses can lead to mutants that have escaped immune pressure to become predominant in the population after passages of viruses have been incubated with immune or convalescent sera in vitro or in previously immunized animals in vivo. A body of research utilized this classical approach to select virus escape variants and to aid the prediction of virus evolution [297, 342–345]. The classical approach is low throughput and constricted to individual mutants, and thus new high-throughput methods have been applied to this field.
Pioneering work done by Li et al. [346] involved the selection of antigenic variants from HA globular head random mutation libraries of A(H1N1)pdm and A(H3N2) viruses either in vitro under the pressure of human or ferret convalescent sera or in previously immunized mice. This proof-of-concept study identified the antigenic escape variants that showed similar antigenicity with variants that were observed in nature for A(H1N1)pdm and past A(H3N2) viruses. Furthermore, this study identified escape variants before they caused the epidemic in 2014-2015 [346]. On the other hand, the non-antigenic effects of amino acid substitutions, such as viral growth, also play a critical role in virus evolution.
Hence, Lee and colleagues [347] quantitatively characterized the effects on viral growth of all single amino acid substitutions to the HA of A(H3N2) by using deep mutational scanning. After deep sequencing, the amino acid preferences at each site were revealed and the results showed that beneficial amino acid substitutions generally tended to occur at higher frequencies in nature [347].
Predicting antigenic evolution in silico
Not all antigenic variants can succeed in fixing and spreading throughout a population (e.g., K158R) [348]. In many years, clades that are antigenically more distant from previously circulating viruses have died out [326]. Therefore, besides antigenicity, it is critical to include virus variants in vaccine strain selection that are predicted to become predominate in future influenza seasons using the surveillance and genetic data of the current season [349]. In other words, accurate forecasting would aid vaccine strain recommendations that must be determined one season in advance.
Prediction of influenza evolution requires the identification of viral clades/lineages coexisting in influenza population genetics and derivation of phylogenies from sequence data [208]. Then, the destiny of each clade can be predicted by estimating its fitness. In an earlier study, 18 HA1 codons under positive selection were identified based on the ratio of non-synonymous-to-synonymous mutations (dN/dS) on the trunk of a phylogenetic tree. The strain with the greatest number of substitutions in the positive selection sites determined the future dominant lineages of A(H3N2) [350]. However, the dN/dS ratio lacked sensitivity and was uninformative when applied to individual sites and for detecting selection pressures within a population [351].
Rather than using the dN/dS ratio, Steinbrück and McHardy described allele dynamics plots (AD plots) for visualizing the evolutionary dynamics of the different alleles of a gene within the population over time and identifying the alleles that might be associated with a selective advantage. With the application of this model in the HA of A(H3N2) isolates between 1998 and 2008, AD plots allowed for the correct identification of the alleles and their associated viral strains that subsequently became predominant in the viral population in four out of five test seasons [351]. To take the antigenic impact of the selected allele into account, they further combined AD plots with an antigenic tree [316] to estimate whether antigenically distinct HA alleles and the associated viral strains would become predominant within one season. This method predicted the predominant HA allele over nine influenza seasons with 78% accuracy and is currently used for recommending candidate vaccine viruses [352]. Klingen et al. [353] described Sweep Dynamics (SD) plots, an extension of AD plots, which combines phylogenetic algorithms with statistical techniques to detect the statistical significance of allele dynamics and can better dissect the influence of individual changes. The SD plots identified sweep-related changes in antigenic sites of A(H3N2) HAs that allowed for the timely prediction of antigenic variants [353].
Luksza and Lassig [349] described a fitness method by combining a susceptible-infectious-recovered (SIR) model with a mutational load model based on epidemiological data and HA sequences, respectively. Strain fitness is determined by similarity to past and presently circulating strains in epitope and non-epitope sites. This method can successfully anticipate the frequency of existing clades of A(H3N2) and other subtype viruses in the next year and can estimate how vaccination affects the course of influenza evolution [349]. In contrast, Neher et al. [354] developed a lineage fitness model based on local tree shape without using molecular data. This model used the simple assumption that an internal node with high fitness will be the root of a descendent lineage with high fitness in the genealogical tree. The fitness of different lineages was estimated with a growth rate measure derived from the branching patterns of the HA genealogy. This model was validated to predict the progenitor lineage of subsequent seasons using simulated and historical A(H3N2) data [354].
Castro et al. [355] identified early indicators of predicting cluster evolution and quantified fundamental trade-offs in prediction ability by using a phylodynamic model of influenza transmission. Their statistical logistic regression models can predict whether the emerging cluster at low frequencies will eventually rise to dominance using simulated data. However, the method achieved only 56% sensitivity when applied to 12 years of empirical influenza surveillance data [355].
In summary, both the experimental and computational prediction approaches shed insights into the understanding of the evolution patterns of influenza viruses. However, these approaches are challenged by our incomplete understanding of the complexity in influenza virus-host interactions at the individual, community, and population levels, particularly with the diversity and complexity of human immune histories and immune responses. Nevertheless, these prediction models have the potential to inform or improve the vaccine strain selection, and the accuracy and prediction range of these models to improve the predictability of influenza evolution remain to be improved.
Continued challenges in influenza antigenic variant analyses and vaccine strain selection
Worldwide epidemiological surveillance is necessary to monitor the continuing evolution of influenza viruses. The emergence and spread of antigenic variants will impair influenza vaccine effectiveness. Early detection of influenza antigenic variants is a key to a successful influenza vaccination program. Remarkable advancements have been made for influenza antigenic characterization techniques in the past decades, and these techniques were summarized in prior sections. However, the ongoing challenges discussed for influenza antigenic variant analyses and vaccine strain selection remain to be resolved.
These continued challenges can be summarized as the following: (1) The time required for the current process of vaccine production creates a window of opportunity for new virus variants to emerge and occasionally cause decreased vaccine effectiveness. The use of computationally predictive models, genomic and proteomic sequencing, and new vaccine platforms such as the mRNA vaccine (see later section) may help decrease the time from strain selection to vaccine production. (2) Culture-adapted mutations during virus isolation and propagation continue to present a challenge in antigenic characterization, seed preparation, and manufacturing. Methods that directly use clinical samples, including both assay- and sequence-based methods, are expected to play more important roles. (3) Selection of reference sera in antigenic characterization continues to be a challenge to reflect the heterologous and dynamic pre-existing immunity in the human population. (4) The majority of methods for antigenic characterization have been limited to protein sequences but not glycan sequences, and the conventional vaccine design does not consider site-specific glycosylation occupancy and glycan heterogeneity [356], which can affect antigenic properties of influenza viruses [83]. Thus, to robustly analyze antigenicity, integrating changes in N-glycosylation sites of influenza glycoproteins is necessary. Recent advances in mass spectrometry technologies allow us to use glycoproteomics and bioinformatics approaches to determine the glycosylation profiles of HA proteins and include site-specific glycosylation occupancy and glycan heterogeneity at each site [357–363], and such analytic method needs to be refined to be more sensitive and quantitative [364]. In addition, a robust tool is needed to predict influenza-specific N-glycosylation sites, associated glycan heterogeneity, and their impacts on influenza antigenicity for vaccine strain selection and vaccine preparation. (5) We are still unable to predict antigenic evolution or antigenic variants ahead of influenza seasons. Robust computational models, such as those using Big Data, AI, and machine learning models, remain to be developed. An ideal model will be able to predict the emergence and spread of an antigenic variant and its potential to become an epidemic in human populations at least one influenza seasonal ahead.
It is important to highlight that, despite these continued challenges, the advancements in influenza antigenic characterization techniques are effective and have proven to be invaluable in the rapid characterization and vaccine development during the coronavirus disease of 2019 (COVID-19), which is discussed in the following section.
SARS-CoV-2
Introduction to coronaviruses and SARS-CoV-2
In contrast to influenza viruses, coronaviruses belong to order Nidovirales, family Coronaviridae, and subfamily Coronaviridae, which is further subdivided into four genera: alpha, beta, gamma, and delta coronaviruses (CoVs) [365, 366]. Alpha and betacoronaviruses often infect mammalian species, while gamma and deltacoronaviruses primarily infect avian species [367]. Coronaviruses are non-segmented, enveloped, positive-sense, and single-stranded RNA viruses that contain the largest genomes of RNA viruses spanning between 28 and 32kb [365, 366]. The coronavirus genome is flanked by a 5’ cap and 3’ poly-A tail and 5’ and 3’ untranslated regions (UTRs). The 5’ region is comprised of a 5’ replicase gene, which synthesizes nonstructural proteins (NSPs) within open read frame (ORF)1ab [368], and the 3’ region is comprised of genes that encode four main structural proteins. The remainder of the genome is comprised of genes encoding an additional six accessory proteins [369]. The most abundant structural protein is the membrane (M) protein, which is made of three transmembrane domains, providing the virus its shape and plays a role in the assembly of viral particles [366, 370]. The trimeric spike (S) protein is a fusion protein that mediates viral attachment and fusion to the host receptor, making it the primary target of therapeutics and neutralizing antibodies [369]. Finally, the envelope (E) protein facilitates viral assembly and release and is highly divergent among coronaviruses, and the nucleocapsid (N) protein allows for binding of viral RNA in vitro [366]. Lineage A viruses of the betacoronaviruses genus also contain hemagglutinin-esterase (HE), a structural protein that acts as hemagglutinin, binds to surface glycoproteins, and may enhance S protein-mediated cell entry [366].
The majority of coronaviruses infect animals (i.e., livestock, birds, and other mammals), which become intermediate host reservoirs. Animal coronaviruses often infect livestock and poultry and create disruptions in both industries [371]. In animals, coronaviruses generally result in enteritis or upper respiratory disease [366]. During antigenic shift events, coronaviruses can gain the ability to infect humans. There are seven endemic human coronaviruses (HCoVs) that have been identified. The HCoV-NL63 and HCoV-229E alphacoronaviruses likely originated from bat reservoirs, while the HCoV-OC43 and HCoV-HKU1 betacoronaviruses likely emerged from rodent-associated viruses [367]. Recently, HCoV-229E has been shown to be more likely transferred from dromedary camels rather than bats [367]. These four endemic coronaviruses typically cause only mild upper or lower respiratory or gastrointestinal symptoms and are responsible for the seasonal common cold [372].
In addition to the four seasonal coronaviruses, there have been three instances of betacoronaviruses that have caused severe human diseases. The first outbreak occurred during the 2002–2003 season in China, caused by the SARS-CoV (27.9kb), was called the severe acute respiratory syndrome (SARS) with a 9% mortality rate, particularly in the elderly, and was widely accepted to have emerged from bats in China [366, 373]. However, the SARS-CoV was primarily spread through direct contact with infected individuals, and, thus, transmission was relatively limited and could be controlled by quarantining infected individuals [366]. During the early stages of the pandemic, the SARS-CoV genome had greater diversity and higher rates of nonsynonymous mutations than the later isolates, suggesting adaptation to the human host towards the beginning of the pandemic [373]. The functional receptor for SARS-CoV is the angiotensin converting enzyme 2 (ACE2) receptor, which binds to the S1 domain of the S protein and allows for viral replication [374]. Downregulation of ACE2 has been associated with acute lung injury [375, 376]. The next epidemic emerged in the Middle East in 2012 due to the Middle East respiratory syndrome-CoV (MERS-CoV, 30.1kb), and had a mortality rate of 36% [366, 377]. MERS-CoV is believed to have originated from dromedary camels, function through the dipeptidyl peptidase 4 (DPP4) receptor [366], and transmit through nosocomial contact, limiting its transmission potential [376]. Finally, the SARS-CoV-2 (29.9kb) [378] emerged in China in 2019 and caused the coronavirus-2019 disease (COVID-19) global pandemic and over 4.3 million deaths worldwide as of August 13, 2021 [379]. Similar to SARS-CoV, SARS-CoV-2 is suspected to have originated from bats [380, 381] and functions through ACE2 receptors which allows for efficient human-to-human spread [382, 383], but is transmitted by respiratory secretions, contributing to its rapid transmission [384, 385].
SARS-CoV-2 and antigenic variations
Antigenic relationships and known antigenic changes among CoVs
The genome order for coronaviruses is commonly 5’ replicase, S, E, M, N, accessory genes, and 3’ polyA sequence [386]. While structural and nonstructural genes remain relatively conserved between coronaviruses, the accessory genes vary [368]. Among the three pandemic HCoVs, SARS-CoV and SARS-CoV-2 are more genetically similar than MERS-CoV. SARS-CoV is 29,727 nucleotides long with five NSPs and eight accessory proteins (ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8a, ORF8b, ORF9b), SARS-CoV-2 contains 29,903 nucleotides and 15 NSPs with six accessory proteins (ORF3, ORF6, ORF7a, ORF7b, ORF8, and ORF9), and MERS-CoV contains 30,119 nucleotides and 16 NSPs with five accessory proteins (ORF3, ORF4a, ORF4b, ORF5, and ORF8) [368]. These variations in the accessory genes have been suggested to cause the differences in pathogenicity between coronaviruses. For example, in SARS-CoV, NSP1, papain-like protease (PLpro), NSP7, NSP15, ORF3b, M, ORF6, and N proteins have been shown to antagonize interferon (IFN) response [387].
In contrast, several proteins in SARS-CoV-2 have been shown to antagonize various sites in the IFN-I production and signaling pathways. ORF6, ORF8, and N have been implicated in SARS-CoV-2 to inhibit type I interferon (IFN-α and -β) expression, the NF-kB-responsive promotor, and the activation of IFN-simulated genes [388]. NSP6 and NSP13 delays IFN-I responses by targeting IFN regulatory factor 3 (IRF3) and antagonizing IFN-β production. They also bind TANK binding kinase 1 (TBK1) and suppress STAT1/STAT2 phosphorylation, leading to decreased IFN-stimulated gene formation. Additionally, Xia et al. [387] showed that NSP1 and NSP6 of SARS-CoV-2 can more efficiently suppress the IFN-I signaling than SARS-CoV and MERS-CoV.
SARS-CoV-2 shares 83.9% similarity in the receptor binding domain (RBD) and 87.2% similarity in the spike glycoprotein with SARS-CoV [389]. Kumar et al. [389] also identified novel glycosylation sites (NGTK, NFTI, NLTT, NTSN) on the spike glycoprotein of SARS-CoV-2 and multiple novel cytotoxic T lymphocyte epitopes compared with SARS-CoV, changing its binding capacity to host receptors and infectivity into the host cell. These variations may have led to the elevated transmission rate and severity of COVID-19.
Antigenic epitopes of spike
The RBD of the spike protein is of particular interest due to its role in mediating virus attachment to host receptors [368] and its role in binding with the human ACE2 receptors in SARS-CoV. Attachment to host receptor allows the virus to fuse with the cell membrane and enter the host cell. Lan et al. [390] showed that the RBD in SARS-CoV-2 is almost identical to that of SARS-CoV but has improved receptor binding affinity [390]. Additionally, anti-RBD antibodies have been shown to be the primary neutralizing polyclonal antibody response [391] and compete with the viral RBD binding to ACE2 receptors [392]. Amino acid changes (i.e., substitutions, insertions, and deletions) of the RBD in SARS-CoV-2 can lead to immune escape. Greaney et al. [393] performed complete mapping of these functional amino acid substitutions to the RBD of SARS-CoV-2 for ten human monoclonal antibodies and found that of the 36 RBD sites, 14 of those sites contained at least five mutants that could escape at least one of the ten tested antibodies. Greaney et al. [393] noted that the frequency of these substitutions was low, and the escape mutations generally had little impact on ACE2 binding and RBD folding. In contrast, Wang et al. [394] demonstrated that residue substitutions on the SARS-CoV-2 C-terminal domain on S1 interact with the human ACE2 receptor rather than the RBD, which also strengthens its receptor binding affinity compared to SARS-CoV. Regardless, the substitutions causing increased binding affinity of SARS-CoV-2 to human ACE2 receptors have contributed towards the increased pathogenesis of COVID-19.
Emerging SARS-CoV-2 variants
In addition to immune escape mutations, whole-genome sequencing (WGS) has allowed for the quick and comprehensive study of the rapid SARS-CoV-2 evolution and its genetic variants. In particular, amino acid substitutions in the NSP2, NSP3, and S proteins contribute towards SARS-CoV-2 virulence and transmission [395]. As more variants become predominant worldwide, there are increasing concerns regarding the potential for variants to escape the effects of vaccinations or natural immunity, worsen pathogenesis, and increase transmission. Weisblum et al. [396] demonstrated in vitro that amino acid substitutions in the RBD and N-terminal domain of the S protein have the potential for antibody evasion, although the frequency of natural occurrence for these substitutions was low. However, increasing prevalence of antibody-resistant strains may limit currently available antibody and convalescent plasma therapies.
SARS-CoV-2 variants have been rapidly emerging over the past year. By March 2020, the D614G variant, located on the spike protein, had already become predominant globally [397, 398]. D614G has been shown to be associated with increased viral loads [397–399] and potentially increased transmission [400], but no association with increased infectivity [397] or worse clinical outcomes in humans [398, 399]. Shortly after the discovery of the D614G variant, a new, rapidly spreading variant from the UK was identified as the WHO α (B.1.1.7) lineage containing the N501Y and 69-70del in the S gene [401]. This substitution N501Y was estimated to have an increased infectivity of 52% [402] and transmission of 75% [403]. Similar to the α lineage, the β (B.1.351) lineage contains the N501Y in addition to two other substitutions at the RBD (E484K, K417N) and became the dominant lineage in South Africa by November 2020 [404]. Additionally, the frequency of the γ (P.1) lineage rose to predominance in Brazil between September 2020 and February 2021 containing E484K, N501Y, and K417T [405]. Chen et al. [406] showed that strains containing the E484K had greatly reduced the neutralizing potential of multiple class I antibodies and even worse reductions on antibody binding when combined with the N501Y in vitro. The fourth and most recent variant of concern is the δ (B.1.617.2) lineage that has been predominant in India and contains G142D and E154K (N-terminal domain), L452R and E484Q (RBD), and P681R (furin cleavage site) [407, 408], and its clinical impacts are still being studied. While not yet fully elucidated, these emerging variants also raise concerns for possibilities for reinfections, vaccine evasion, atypical symptoms, and increased potency in younger adults and children. Continued meticulous surveillance of SARS-CoV-2 variants will be essential for containing the pandemic.
Antigenic characterization of SARS-CoV-2
The analytic methods used in antigenic characterization of SARS-CoV-2 are similar to those used in influenza viruses. The rapid mutation rate of SARS-CoV-2 requires constant monitoring of vaccine efficacy, changes in epitopes, and modifications of virus function and infectivity. Although genetic analyses can quickly identify the genetic variants, their antigenic properties must be assessed using conventional laboratory assays mentioned above. As with influenza viruses, culture-adaptations may occur in SARS-CoV-2 [409]. More data are needed to evaluate these culture-adapted amino acid changes in SARS-CoV-2 and their impacts on antigenic analyses. Similar to that for influenza viruses, an ideal method will be to assess the antigenic properties of the viruses using clinical samples. These barriers will need to be addressed as SARS-CoV-2 research expands and variants continue to emerge.
Enzyme-linked immunosorbent assay (ELISA)
ELISAs are used to detect IgM and IgG in COVID-19-positive individuals, which allows for the determination of disease state, detection of antibody titers, and identification of isotype. Antibodies can also be tracked for seroconversion in immune responses to SARS-CoV-2 over time [410]. In addition to Igs, ELISAs can detect the presence of certain peptides and proteins, including the viral S protein and human ACE2 receptor [411]. The limitations of ELISA are discussed in the ELISA assays section for influenza viruses.
Neutralization assays
Several types of neutralization assays including authentic viruses and pseudoviruses have been utilized for SARS-CoV-2 [412–414]. Authentic virus-based plaque reduction neutralization tests (PRNTs) and micro-neutralization (MN) assays have been well-adapted to quantitatively assess the extent that antibodies inhibit SARS-CoV-2 virus entry and replication [412, 413]. The limitation of these methods is the requirement of BSL3 facilities and suitably trained staff for authentic viruses. Limitations of pseudoviruses are discussed separately below. Although it is a common assay used in many viruses, the SARS-CoV-2 plaque formation using PRNT is time-consuming and low-throughput due to the requirements of agarose overlay and 24-well plates for the plaque forming stage as well as the process of plaque counting and data analysis [412, 413].
Micro-neutralization assays
The micro-neutralization assays, considered as the gold-standard [414], offer advantages over the PRNT by increasing throughput and reducing operation time and operator workload by the use of 96-well plates and the increased automation of washing, staining, and reading stages [412]. Manenti et al. [414] presented two detection methods of micro-neutralization assays that include the classical readout which entails measuring the CPE and a colorimetric readout using a spectrophotometer. The subjective CPE-based readout is carried out by checking the cell monolayers under an optical microscope, whereas the colorimetric readout is obtained by staining the healthy cells with neutral red dye followed by measuring absorbance at 540 nm. The comparison between the two methods showed that a well-trained operator is able to achieve a CPE-based readout in consistency with a colorimetric readout [414]. To avoid the subjective visual inspection of CPE, Amanat et al. [413] adapted the well-established ELISA-based micro-neutralization assay for other viruses such as influenza viruses to SARS-CoV-2 virus. In this assay, infected cells are immuno-stained depending on the viral NP protein, and the absorbance of the colorimetric product obtained by the reaction between substrate and peroxidase is measured by an ELISA reader. Bewley and colleagues [412] adapted the FRNT to SARS-CoV-2 viruses, which is also based on the immunostaining of SARS-CoV-2-specific proteins. The FRNT uses primary antibodies targeting the RBD on the S protein, and its readout is the count of immuno-stained foci (spots) rather than absorbance as seen in ELISA-based micro-neutralization assays. The advantage of foci counting is that it is easier to catch mishandling such as microbial contamination or other errors causing cell monolayer damage by the automatic foci counter (e.g., ImmunoSpot analyzer). Of note, a strong correlation was observed between PRNT and FRNT [412].
Pseudovirus neutralization assays
Pseudovirus neutralization assays have been developed to safely study viral entry inhibitors and neutralizing antibodies against SARS-CoV-2 and virus-host interactions without risk of SARS-CoV-2 infection. Pseudoviruses also help with accessibility for laboratories that do not have access to BSL-3 or BSL-4 facilities [415]. Three packaging systems have been used as the backbone for SARS-CoV-2 pseudoviruses: HIV-based lentiviral particles, murine leukemia virus (MLV)-based particles [416], and vesicular stomatitis virus (VSV) [417]. Ou et al. [418] and Hu et al. [419] generated a luciferase (Luc)-expressing pseudovirus that contained the S protein within a HIV-1 system. They also established a pseudovirus inhibition assay for testing two protease inhibitors that target virus entry after transfection with ACE2 receptors into the HEK293T cell line. Similarly, Nie et al. [415] and Hoffmann et al. [420] developed a pseudotyped virus expressing the S protein on a VSV backbone to quantify SARS-CoV-2 neutralizing antibodies. Case et al. [421] compared the neutralization activity between VSV-SARS-CoV-2 (spike) and SARS-CoV-2 isolates grown on Vero E6 cells, and found a strong correlation between the two tests as well as concordance between their EC50 values. Various commercial SARS-CoV-2 pseudoviruses are also available for purchase from mybiosource.com, Takara, eEnzyme, and multiple other companies. Limitations of this platform include the need to create a separate assay to test different aspects of virus mechanisms or other proteins and potential variability between the protein functions of pseudotyped virus and wild type virus [422]. While some studies as in Case et al. [421] had determined strong correlations between pseudovirus results and wild type, few studies using pseudoviruses confirm findings with wild type virus. Thus, more data need to evaluate how accurately pseudoviruses reflect the antigenic properties of wild type SARS-CoV-2 viruses.
High-throughput neutralization assays
Several high-throughput neutralization assays have been developed to meet the detection demands for the tremendous quantities of COVID-19 specimens. Muruato et al. [423] developed a fluorescent-based high-throughput neutralization assay. In this assay, the reporter virus was modified to introduce the mNeonGreen gene into the ORF7 of the SARS-CoV-2 virus genome. After being incubated with the testing sera, the reporter virus is inoculated onto Vero cells and the quantities of neutralizing titers are determined by the fluorescence signals quantified by high-content imaging. A strong correlation between this assay and the PRNT was observed (R2 of 0.85), and the reporter virus was shown to have similar replication ability with wild type virus [423]. Compared with other neutralization assays, this reporter virus-based assay can shorten the assay turnaround time by eliminating the immunostaining step and can be scalable to 384- or even 1536-well plates. Instead of a reporter virus, most other high-throughput neutralization assays use a surrogate virus and thus are called surrogate virus neutralization test (sVNT) [424–427]. Similar to conventional pseudovirus neutralization assays, sVNTs detect antibodies that are only specific to the spike protein or the RBD [428]. For example, the sVNT by Fenwich et al. [429] is developed based on the competitive inhibition of ACE2 binding to the spike protein trimer coupled with Luminex beads, and the quantification is dependent on the measurement of ACE2 recruited by the beads, which are fluorescently tagged in recognition of the ACE2-Fc fusion protein on a Bio-Plex 200 system. Compared with those assays with live viruses, the pseudovirus neutralization assays, especially those based on fluorescent pseudoviruses, are highly scalable [430, 431]. Nevertheless, compared with the conventional VN assays, one common challenge for these high throughput neutralization assays is that both systems need to update the virus component in either a reporter virus or a surrogate virus to match those of the epidemic strains, which, as an RNA virus, evolves rapidly.
mRNA vaccine and antigenic characterization of influenza viruses and SARS-CoV-2
A number of challenges were described for influenza antigenic variant analyses and vaccine strain selection, which are associated with the conventional vaccine platforms involving virus culture. Hence, rapid and flexible virus culture-independent vaccine production platforms, such as recombinant HA vaccines and nucleic acid vaccines (DNA vaccines and mRNA vaccines), are important and can potentially overcome those challenges.
Recombinant HA vaccines against seasonal influenza viruses (Flublock) have been licensed by the US Food and Drug Administration since 2013 [432]. However, they require threefold higher HA content than inactivated vaccines to provide a similar level of protection [433]. As for DNA vaccines, the main challenges are poor immunogenicity in humans and larger animal models compared to mouse models [434], eliciting of unintended antibody production that may lead to autoimmune diseases [435, 436], low specificity for target cells, poor anti-vector immunity, concerns for genome integration [437], and the requirement of high doses and devices (e.g., electroporation, aerosol, polymer condensing agents) [434, 438–440].
In recent decades, mRNA vaccines have emerged as a new vaccine platform. This platform began with the successful translation of mRNA injected into mice by Wolff et al. in 1990 [441], and multiple mRNA vaccine platforms have since been shown to elicit immunity against the influenza virus [442, 443], Zika virus [444, 445], rabies virus [446], and even some cancer types [447–449] in animal models [450]. Among the many advantages of current mRNA vaccines include its safety profile which removes the risks of potential infection or genome integration, efficient delivery to the cytoplasm rather than transfection to the nucleus as in DNA vaccines [451], stability and highly translatable nature after delivery [452], and potential for inexpensive, rapid, and scalable production [453]. The benefits conferred from mRNA vaccines (e.g., BNT162b2 [Pfizer-BioNTech] and mRNA-1273 [Moderna]) for human use was demonstrated during the COVID-19 pandemic as the fastest vaccines developed, with the aid of the Emergency Use Authorization by the United States Food and Drug Administration, at an astonishing 95% efficacy with minimal adverse effects [417]. The success of these vaccines is attributed to the use of nucleoside-modified mRNA to enhance translation capacity and reduce the innate immunity response and the unique use of lipid nanoparticles (LNPs) to improve mRNA stability and cellular penetration [454, 455]. As with all vaccine platforms, mRNA vaccines also present challenges that will need to be addressed moving forward which include thermal instability at elevated temperatures, allergic reactions to LNPs, and the unknowns of long-term properties in humans including the effects of repeated administration [417, 456]. However, the unprecedented speed and efficacy of these mRNA vaccines for COVID-19 opens the world of nanoparticle delivery systems for other infectious diseases and may become a viable option for future influenza vaccinations.
The mRNA vaccine platform can help overcome challenges in vaccine seed preparation and in vaccine manufacturing. The remaining challenges for the mRNA vaccine platforms rely on vaccine strain selection and antigenic characterization. A rapid and robust tool for sequence-based antigenicity analyses is urgently needed for influenza and SARS-CoV-2 viruses as well other potential diseases, and such tools will enable the assessment of virus antigenic properties using clinical samples directly, including swabs or genomic sequences as describe earlier [71, 72].
Conclusions
In summary, the importance of antigenic characterization for outbreak response and prevention, prediction of viral evolution, and vaccine strain selection cannot be understated. The necessity of these continuously improving techniques for characterizing antigenic properties of infectious diseases has been prominently displayed throughout the history of influenza epidemic outbreaks and pandemics. These existing methods have been modified and have become invaluable in identifying the mechanisms of infection, differences in lineages, and the unprecedented rapidity of response against the deadly COVID-19 pandemic, allowing for >90% effective vaccines to be developed within 1 year.
Currently available analytic methods for antigenic characterization include hemagglutination inhibition, neuraminidase inhibition, neutralization, and micro-neutralization assays as well as enzyme-linked lectin assays and antigenic cartography for influenza. Likewise, multiple existing methods have been modified to characterize SARS-CoV-2. Enzyme-linked immunosorbent assay, pseudoviruses, and micro-neutralization assays have been instrumental towards the rapid characterization of SARS-CoV-2 and the pandemic response. These methods are routinely used to assess the antigenic properties of emerging and re-emerging viruses and to identify antigenic variants of these viruses.
One major barrier of these current influenza characterization methods include the need for a larger quantity of viruses than what is typically available from the original clinical samples, requiring isolation and amplification in cells or embryonated chicken eggs. This presents opportunities for culture-adapted mutations of the testing strain, which may generate modifications of the virus phenotype leading to antigenic mismatch during vaccine development. Additionally, vaccine composition must be determined almost 6 months prior to peak seasonal influenza activity, allowing time for antigenic drift to occur. This is another potential cause of vaccine mismatch with the circulating strain. SARS-CoV-2 characterization faces similar challenges. In addition to the requirement of high virus quantity and potential laboratory-acquired mutations, testing of each aspect of virus function requires a generation of a separate pseudovirus. The high mutations rates of both influenza and SARS-CoV-2 also present challenges for the relevancy of each test to the circulating strains.
Ideally, antigenic characterization would include comprehensive virus surveillance and sampling, use of original clinical specimens rather than virus isolates, genome-based methods, and increased efficiency of vaccine production from time of strain selection. Current methods in development to improve the techniques for antigenic characterization include mechanical and automated hemagglutination inhibition assays, synthetic erythrocytes, the enzyme-linked lectin assay, the combination of assays with imaging to increase quantification efficiency and throughput, 3D antigen cartography techniques for visualization and for mitigating bias introduced during multi-dimensional scaling, the novel polyclonal antibody-based proximity ligation assay which reduces the sample volume requirement while improving specificity, and advanced machine learning algorithms. These improved methods will help optimize sensitivity, specificity, and assay time. While antigenic characterization of influenza is complex, there are continuous advancements and innovation in the field, which have already served to rapidly address the COVID-19 pandemic. These advancements for both fields will continue to improve the accuracy, precision, and speed for characterizing and selecting vaccine strains in influenza and SARS-CoV-2.
Funding
This research was funded by NIH, grant numbers R01AI116744, R01AI147640, R01AI152521, and 5T32LM012410.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yang Wang and Cynthia Y. Tang contributed equally to this work.
References
- 1.Iuliano AD, Roguski KM, Chang HH, Muscatello DJ, Palekar R, Tempia S, et al. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet. 2018;391(10127):1285–1300. doi: 10.1016/S0140-6736(17)33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prevention CfDCa. Disease burden of influenza October 5, 2020 [Available from: https://www.cdc.gov/flu/about/burden/index.html.
- 3.Valleron A-J, Cori A, Valtat S, Meurisse S, Carrat F, Boëlle P-Y. Transmissibility and geographic spread of the 1889 influenza pandemic. Proc Natl Acad Sci U S A. 2010;107(19):8778–8781. doi: 10.1073/pnas.1000886107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mills CE, Robins JM, Lipsitch M. Transmissibility of 1918 pandemic influenza. Nature. 2004;432(7019):904–906. doi: 10.1038/nature03063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taubenberger JK, Morens DM. 1918 Influenza: the mother of all pandemics. Emerg Infect Dis. 2006;12(1):15–22. doi: 10.3201/eid1209.05-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly H, Peck HA, Laurie KL, Wu P, Nishiura H, Cowling BJ. The age-specific cumulative incidence of infection with pandemic influenza H1N1 2009 was similar in various countries prior to vaccination. PLoS One. 2011;6(8):e21828. doi: 10.1371/journal.pone.0021828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hause BM, Ducatez M, Collin EA, Ran Z, Liu R, Sheng Z, et al. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog. 2013;9(2):e1003176. doi: 10.1371/journal.ppat.1003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hause BM, Collin EA, Liu R, Huang B, Sheng Z, Lu W, et al. Characterization of a novel influenza virus in cattle and Swine: proposal for a new genus in the Orthomyxoviridae family. mBio. 2014;5(2):e00031–e00014. doi: 10.1128/mBio.00031-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, et al. A novel influenza A virus mitochondrial protein that induces cell death. Nat Med. 2001;7(12):1306–12. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- 10.Wise HM, Foeglein A, Sun JC, Dalton RM, Patel S, Howard W, et al. A complicated message: identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J Virol. 2009;83(16):8021–8031. doi: 10.1128/JVI.00826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, et al. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science. 2012;337(6091):199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muramoto Y, Noda T, Kawakami E, Akkina R, Kawaoka Y. Identification of novel influenza A virus proteins translated from PA mRNA. J Virol. 2013;87(5):2455–2462. doi: 10.1128/JVI.02656-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nobusawa E, Sato K. Comparison of the mutation rates of human influenza A and B viruses. J Virol. 2006;80(7):3675–3678. doi: 10.1128/JVI.80.7.3675-3678.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holland J, Spindler K, Horodyski F, Grabau E, Nichol S, VandePol S. Rapid evolution of RNA genomes. Science. 1982;215(4540):1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- 15.Steinhauer DA, Holland JJ. Rapid evolution of RNA viruses. Annu Rev Microbiol. 1987;41:409–433. doi: 10.1146/annurev.mi.41.100187.002205. [DOI] [PubMed] [Google Scholar]
- 16.Buonagurio DA, Nakada S, Parvin JD, Krystal M, Palese P, Fitch WM. Evolution of human influenza A viruses over 50 years: rapid, uniform rate of change in NS gene. Science. 1986;232(4753):980–982. doi: 10.1126/science.2939560. [DOI] [PubMed] [Google Scholar]
- 17.Martinez C, del Rio L, Portela A, Domingo E, Ortin J. Evolution of the influenza virus neuraminidase gene during drift of the N2 subtype. Virology. 1983;130(2):539–545. doi: 10.1016/0042-6822(83)90108-3. [DOI] [PubMed] [Google Scholar]
- 18.Cox NJ, Subbarao K. Global epidemiology of influenza: past and present. Annu Rev Med. 2000;51:407–421. doi: 10.1146/annurev.med.51.1.407. [DOI] [PubMed] [Google Scholar]
- 19.Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971;58(10):465–523. doi: 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- 20.Eigen M, Schuster P. The hypercycle. A principle of natural self-organization. Part A: Emergence of the hypercycle. Naturwissenschaften. 1977;64(11):541–565. doi: 10.1007/BF00450633. [DOI] [PubMed] [Google Scholar]
- 21.Schuster P. Quasispecies on Fitness Landscapes. Curr Top Microbiol Immunol. 2016;392:61–120. doi: 10.1007/82_2015_469. [DOI] [PubMed] [Google Scholar]
- 22.Wilke CO. Quasispecies theory in the context of population genetics. BMC Evol Biol. 2005;5:44. doi: 10.1186/1471-2148-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van den Hoecke S, Verhelst J, Vuylsteke M, Saelens X. Analysis of the genetic diversity of influenza A viruses using next-generation DNA sequencing. BMC Genomics. 2015;16:79. doi: 10.1186/s12864-015-1284-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rocha E, Cox NJ, Black RA, Harmon MW, Harrison CJ, Kendal AP. Antigenic and genetic variation in influenza A (H1N1) virus isolates recovered from a persistently infected immunodeficient child. J Virol. 1991;65(5):2340–2350. doi: 10.1128/jvi.65.5.2340-2350.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Air GM. Influenza virus antigenicity and broadly neutralizing epitopes. Curr Opin Virol. 2015;11:113–121. doi: 10.1016/j.coviro.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439(7074):344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ciota AT, Ehrbar DJ, Van Slyke GA, Willsey GG, Kramer LD. Cooperative interactions in the West Nile virus mutant swarm. BMC Evol Biol. 2012;12:58. doi: 10.1186/1471-2148-12-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shirogane Y, Watanabe S, Yanagi Y. Cooperation between different RNA virus genomes produces a new phenotype. Nat Commun. 2012;3:1235. doi: 10.1038/ncomms2252. [DOI] [PubMed] [Google Scholar]
- 29.Ke R, Aaskov J, Holmes EC, Lloyd-Smith JO. Phylodynamic analysis of the emergence and epidemiological impact of transmissible defective dengue viruses. PLoS Pathog. 2013;9(2):e1003193. doi: 10.1371/journal.ppat.1003193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borderia AV, Isakov O, Moratorio G, Henningsson R, Aguera-Gonzalez S, Organtini L, et al. Group selection and contribution of minority variants during virus adaptation determines virus fitness and phenotype. PLoS Pathog. 2015;11(5):e1004838. doi: 10.1371/journal.ppat.1004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White MC, Lowen AC. Implications of segment mismatch for influenza A virus evolution. J Gen Virol. 2018;99(1):3–16. doi: 10.1099/jgv.0.000989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Both GW, Sleigh MJ, Cox NJ, Kendal AP. Antigenic drift in influenza virus H3 hemagglutinin from 1968 to 1980: multiple evolutionary pathways and sequential amino acid changes at key antigenic sites. J Virol. 1983;48(1):52–60. doi: 10.1128/jvi.48.1.52-60.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson MI, Viboud C, Simonsen L, Bennett RT, Griesemer SB, St. George K, et al. Multiple reassortment events in the evolutionary history of H1N1 influenza A virus since 1918. PLoS Pathog. 2008;4(2):e1000012. doi: 10.1371/journal.ppat.1000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325(5937):197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vijaykrishna D, Poon LL, Zhu HC, Ma SK, Li OT, Cheung CL, et al. Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science. 2010;328(5985):1529. doi: 10.1126/science.1189132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith GJ, Bahl J, Vijaykrishna D, Zhang J, Poon LL, Chen H, et al. Dating the emergence of pandemic influenza viruses. Proc Natl Acad Sci U S A. 2009;106(28):11709–11712. doi: 10.1073/pnas.0904991106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tao H, Steel J, Lowen AC. Intrahost dynamics of influenza virus reassortment. J Virol. 2014;88(13):7485–7492. doi: 10.1128/JVI.00715-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harper SA, Fukuda K, Uyeki TM, Cox NJ, Bridges CB, Centers for Disease C et al. Prevention and control of influenza: recommendations of the Advisory Committee on Immunization Practices (ACIP) MMWR Recomm Rep. 2004;53(RR-6):1–40. [PubMed] [Google Scholar]
- 39.Webster RG, Laver WG, Air GM, Schild GC. Molecular mechanisms of variation in influenza viruses. Nature. 1982;296(5853):115–121. doi: 10.1038/296115a0. [DOI] [PubMed] [Google Scholar]
- 40.Nobusawa E, Nakajima K. Amino acid substitution at position 226 of the hemagglutinin molecule of influenza (H1N1) virus affects receptor binding activity but not fusion activity. Virology. 1988;167(1):8–14. doi: 10.1016/0042-6822(88)90048-7. [DOI] [PubMed] [Google Scholar]
- 41.Barr IG, McCauley J, Cox N, Daniels R, Engelhardt OG, Fukuda K, et al. Epidemiological, antigenic and genetic characteristics of seasonal influenza A(H1N1), A(H3N2) and B influenza viruses: basis for the WHO recommendation on the composition of influenza vaccines for use in the 2009-2010 northern hemisphere season. Vaccine. 2010;28(5):1156–1167. doi: 10.1016/j.vaccine.2009.11.043. [DOI] [PubMed] [Google Scholar]
- 42.Altman MO, Angeletti D, Yewdell JW. Antibody immunodominance: the key to understanding influenza virus antigenic drift. Viral Immunol. 2018;31(2):142–149. doi: 10.1089/vim.2017.0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhatt S, Holmes EC, Pybus OG. The genomic rate of molecular adaptation of the human influenza A virus. Mol Biol Evol. 2011;28(9):2443–2451. doi: 10.1093/molbev/msr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petrova VN, Russell CA. The evolution of seasonal influenza viruses. Nat Rev Microbiol. 2018;16(1):47–60. doi: 10.1038/nrmicro.2017.118. [DOI] [PubMed] [Google Scholar]
- 45.Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31(2 Pt 1):417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 46.Gerhard W, Yewdell J, Frankel ME, Webster R. Antigenic structure of influenza virus haemagglutinin defined by hybridoma antibodies. Nature. 1981;290(5808):713–717. doi: 10.1038/290713a0. [DOI] [PubMed] [Google Scholar]
- 47.Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289(5796):373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 48.Wiley DC, Skehel JJ. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- 49.Kaverin NV, Rudneva IA, Govorkova EA, Timofeeva TA, Shilov AA, Kochergin-Nikitsky KS, et al. Epitope mapping of the hemagglutinin molecule of a highly pathogenic H5N1 influenza virus by using monoclonal antibodies. J Virol. 2007;81(23):12911–12917. doi: 10.1128/JVI.01522-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ndifon W, Wingreen NS, Levin SA. Differential neutralization efficiency of hemagglutinin epitopes, antibody interference, and the design of influenza vaccines. Proc Natl Acad Sci U S A. 2009;106(21):8701–8706. doi: 10.1073/pnas.0903427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Q, Cheng F, Lu M, Tian X, Ma J. Crystal structure of unliganded influenza B virus hemagglutinin. J Virol. 2008;82(6):3011–3020. doi: 10.1128/JVI.02477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Colman PM, Varghese JN, Laver WG. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature. 1983;303(5912):41–44. doi: 10.1038/303041a0. [DOI] [PubMed] [Google Scholar]
- 53.Yen HL, Hoffmann E, Taylor G, Scholtissek C, Monto AS, Webster RG, et al. Importance of neuraminidase active-site residues to the neuraminidase inhibitor resistance of influenza viruses. J Virol. 2006;80(17):8787–8795. doi: 10.1128/JVI.00477-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eichelberger MC, Wan H. Influenza neuraminidase as a vaccine antigen. Curr Top Microbiol Immunol. 2015;386:275–299. doi: 10.1007/82_2014_398. [DOI] [PubMed] [Google Scholar]
- 55.Wan H, Gao J, Xu K, Chen H, Couzens LK, Rivers KH, et al. Molecular basis for broad neuraminidase immunity: conserved epitopes in seasonal and pandemic H1N1 as well as H5N1 influenza viruses. J Virol. 2013;87(16):9290–9300. doi: 10.1128/JVI.01203-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wan H, Yang H, Shore DA, Garten RJ, Couzens L, Gao J, et al. Structural characterization of a protective epitope spanning A(H1N1)pdm09 influenza virus neuraminidase monomers. Nat Commun. 2015;6:6114. doi: 10.1038/ncomms7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Job ER, Schotsaert M, Ibanez LI, Smet A, Ysenbaert T, Roose K, et al. Antibodies directed toward neuraminidase N1 control disease in a mouse model of influenza. J Virol. 2018;92(4). [DOI] [PMC free article] [PubMed]
- 58.Chen YQ, Wohlbold TJ, Zheng NY, Huang M, Huang Y, Neu KE, et al. Influenza infection in humans induces broadly cross-reactive and protective neuraminidase-reactive antibodies. Cell. 2018;173(2):417–29 e10. doi: 10.1016/j.cell.2018.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang L, Fantoni G, Couzens L, Gao J, Plant E, Ye Z, et al. Comparative efficacy of monoclonal antibodies that bind to different epitopes of the 2009 pandemic H1N1 influenza virus neuraminidase. J Virol. 2016;90(1):117–128. doi: 10.1128/JVI.01756-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Webster RG, Brown LE, Laver WG. Antigenic and biological characterization of influenza virus neuraminidase (N2) with monoclonal antibodies. Virology. 1984;135(1):30–42. doi: 10.1016/0042-6822(84)90114-4. [DOI] [PubMed] [Google Scholar]
- 61.Lentz MR, Air GM, Laver WG, Webster RG. Sequence of the neuraminidase gene of influenza virus A/Tokyo/3/67 and previously uncharacterized monoclonal variants. Virology. 1984;135(1):257–265. doi: 10.1016/0042-6822(84)90135-1. [DOI] [PubMed] [Google Scholar]
- 62.Gulati U, Hwang CC, Venkatramani L, Gulati S, Stray SJ, Lee JT, et al. Antibody epitopes on the neuraminidase of a recent H3N2 influenza virus (A/Memphis/31/98) J Virol. 2002;76(23):12274–12280. doi: 10.1128/JVI.76.23.12274-12280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saito T, Taylor G, Laver WG, Kawaoka Y, Webster RG. Antigenicity of the N8 influenza A virus neuraminidase: existence of an epitope at the subunit interface of the neuraminidase. J Virol. 1994;68(3):1790–1796. doi: 10.1128/jvi.68.3.1790-1796.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Air GM, Laver WG, Webster RG. Mechanism of antigenic variation in an individual epitope on influenza virus N9 neuraminidase. J Virol. 1990;64(12):5797–5803. doi: 10.1128/jvi.64.12.5797-5803.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tulip WR, Varghese JN, Laver WG, Webster RG, Colman PM. Refined crystal structure of the influenza virus N9 neuraminidase-NC41 Fab complex. J Mol Biol. 1992;227(1):122–148. doi: 10.1016/0022-2836(92)90687-F. [DOI] [PubMed] [Google Scholar]
- 66.Wilson JR, Guo Z, Reber A, Kamal RP, Music N, Gansebom S, et al. An influenza A virus (H7N9) anti-neuraminidase monoclonal antibody with prophylactic and therapeutic activity in vivo. Antivir Res. 2016;135:48–55. doi: 10.1016/j.antiviral.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wan H, Qi L, Gao J, Couzens LK, Jiang L, Gao Y, et al. Comparison of the efficacy of N9 neuraminidase-specific monoclonal antibodies against influenza A(H7N9) virus infection. J Virol. 2018;92(4). [DOI] [PMC free article] [PubMed]
- 68.Air GM. Influenza neuraminidase. Influenza Other Respir Viruses. 2012;6(4):245–256. doi: 10.1111/j.1750-2659.2011.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sandbulte MR, Westgeest KB, Gao J, Xu X, Klimov AI, Russell CA, et al. Discordant antigenic drift of neuraminidase and hemagglutinin in H1N1 and H3N2 influenza viruses. Proc Natl Acad Sci U S A. 2011;108(51):20748–20753. doi: 10.1073/pnas.1113801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yasuhara A, Yamayoshi S, Kiso M, Sakai-Tagawa Y, Koga M, Adachi E, et al. Antigenic drift originating from changes to the lateral surface of the neuraminidase head of influenza A virus. Nat Microbiol. 2019;4(6):1024–1034. doi: 10.1038/s41564-019-0401-1. [DOI] [PubMed] [Google Scholar]
- 71.Han L, Li L, Wen F, Zhong L, Zhang T, Wan XF. Graph-guided multi-task sparse learning model: a method for identifying antigenic variants of influenza A(H3N2) virus. Bioinformatics. 2019;35(1):77–87. doi: 10.1093/bioinformatics/bty457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li L, Chang D, Han L, Zhang X, Zaia J, Wan XF. Multi-task learning sparse group lasso: a method for quantifying antigenicity of influenza A(H1N1) virus using mutations and variations in glycosylation of Hemagglutinin. BMC Bioinformatics. 2020;21(1):182. doi: 10.1186/s12859-020-3527-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wentworth DE, McGregor MW, Macklin MD, Neumann V, Hinshaw VS. Transmission of swine influenza virus to humans after exposure to experimentally infected pigs. J Infect Dis. 1997;175(1):7–15. doi: 10.1093/infdis/175.1.7. [DOI] [PubMed] [Google Scholar]
- 74.Chakraverty P. Antigenic relationship between influenza B viruses. Bull World Health Organ. 1971;45(6):755–766. [PMC free article] [PubMed] [Google Scholar]
- 75.Rota PA, Wallis TR, Harmon MW, Rota JS, Kendal AP, Nerome K. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology. 1990;175(1):59–68. doi: 10.1016/0042-6822(90)90186-U. [DOI] [PubMed] [Google Scholar]
- 76.Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, et al. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305(5682):371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 77.Zost SJ, Parkhouse K, Gumina ME, Kim K, Diaz Perez S, Wilson PC, et al. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc Natl Acad Sci U S A. 2017;114(47):12578–12583. doi: 10.1073/pnas.1712377114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jorquera PA, Mishin VP, Chesnokov A, Nguyen HT, Mann B, Garten R, et al. Insights into the antigenic advancement of influenza A(H3N2) viruses, 2011-2018. Sci Rep. 2019;9(1):2676. doi: 10.1038/s41598-019-39276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chambers BS, Parkhouse K, Ross TM, Alby K, Hensley SE. Identification of hemagglutinin residues responsible for H3N2 antigenic drift during the 2014-2015 influenza season. Cell Rep. 2015;12(1):1–6. doi: 10.1016/j.celrep.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sun H, Yang J, Zhang T, Long LP, Jia K, Yang G, et al. Using sequence data to infer the antigenicity of influenza virus. mBio. 2013;4(4). [DOI] [PMC free article] [PubMed]
- 81.Quan LJ, Ji CY, Ding X, Peng YS, Liu M, Sun JY, et al. Cluster-transition determining sites underlying the antigenic evolution of seasonal influenza viruses. Mol Biol Evol. 2019;36(6):1172–1186. doi: 10.1093/molbev/msz050. [DOI] [PubMed] [Google Scholar]
- 82.Koel BF, Burke DF, Bestebroer TM, van der Vliet S, Zondag GC, Vervaet G, et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science. 2013;342(6161):976–979. doi: 10.1126/science.1244730. [DOI] [PubMed] [Google Scholar]
- 83.Wan H, Gao J, Yang H, Yang S, Harvey R, Chen YQ, et al. The neuraminidase of A(H3N2) influenza viruses circulating since 2016 is antigenically distinct from the A/Hong Kong/4801/2014 vaccine strain. Nat Microbiol. 2019;4(12):2216–2225. doi: 10.1038/s41564-019-0522-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Klein EY, Serohijos AWR, Choi J-M, Shakhnovich EI, Pekosz A. Influenza A H1N1 pandemic strain evolution – divergence and the potential for antigenic drift variants. PLoS One. 2014;9(4):e93632. doi: 10.1371/journal.pone.0093632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu A, Peng Y, Du X, Shu Y, Jiang T. Correlation of influenza virus excess mortality with antigenic variation: application to rapid estimation of influenza mortality burden. PLoS Comput Biol. 2010;6(8). [DOI] [PMC free article] [PubMed]
- 86.Harvey WT, Benton DJ, Gregory V, Hall JP, Daniels RS, Bedford T, et al. Identification of low- and high-impact hemagglutinin amino acid substitutions that drive antigenic drift of influenza A(H1N1) viruses. PLoS Pathog. 2016;12(4):e1005526. doi: 10.1371/journal.ppat.1005526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Medina RA, Stertz S, Manicassamy B, Zimmermann P, Sun X, Albrecht RA, et al. Glycosylations in the globular head of the hemagglutinin protein modulate the virulence and antigenic properties of the H1N1 influenza viruses. Sci Transl Med. 2013;5(187):187ra70-ra70. doi: 10.1126/scitranslmed.3005996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vijaykrishna D, Holmes EC, Joseph U, Fourment M, Su YC, Halpin R, et al. The contrasting phylodynamics of human influenza B viruses. eLife. 2015;4. [DOI] [PMC free article] [PubMed]
- 89.Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453(7195):615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cushing A, Kamali A, Winters M, Hopmans ES, Bell JM, Grimes SM, et al. Emergence of hemagglutinin mutations during the course of influenza infection. Sci Rep. 2015;5:16178. doi: 10.1038/srep16178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nerome R, Hiromoto Y, Sugita S, Tanabe N, Ishida M, Matsumoto M, et al. Evolutionary characteristics of influenza B virus since its first isolation in 1940: dynamic circulation of deletion and insertion mechanism. Arch Virol. 1998;143(8):1569–1583. doi: 10.1007/s007050050399. [DOI] [PubMed] [Google Scholar]
- 92.McCullers JA, Wang GC, He S, Webster RG. Reassortment and insertion-deletion are strategies for the evolution of influenza B viruses in nature. J Virol. 1999;73(9):7343–7348. doi: 10.1128/JVI.73.9.7343-7348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Virk RK, Jayakumar J, Mendenhall IH, Moorthy M, Lam P, Linster M, et al. Divergent evolutionary trajectories of influenza B viruses underlie their contemporaneous epidemic activity. Proc Natl Acad Sci U S A. 2020;117(1):619–628. doi: 10.1073/pnas.1916585116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Organization WH. Recommended composition of influenza virus vaccines for use in the 2015-2016 northern hemisphere influenza season 2015 [updated February 2015; cited 2021 May 1]. Available from: https://www.who.int/influenza/vaccines/virus/recommendations/201502_recommendation.pdf..
- 95.Villa M, Lässig M. Fitness cost of reassortment in human influenza. PLoS Pathog. 2017;13(11):e1006685-e. doi: 10.1371/journal.ppat.1006685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Oong XY, Ng KT, Lam TT-Y, Pang YK, Chan KG, Hanafi NS, et al. Epidemiological and evolutionary dynamics of influenza B viruses in Malaysia, 2012-2014. PloS one. 2015;10(8):e0136254-e. [DOI] [PMC free article] [PubMed]
- 97.Langat P, Raghwani J, Dudas G, Bowden TA, Edwards S, Gall A, et al. Genome-wide evolutionary dynamics of influenza B viruses on a global scale. PLoS Pathog 2017;13(12):e1006749-e. [DOI] [PMC free article] [PubMed]
- 98.Olsen B, Munster VJ, Wallensten A, Waldenstrom J, Osterhaus AD, Fouchier RA. Global patterns of influenza a virus in wild birds. Science. 2006;312(5772):384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- 99.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56(1):152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stallknecht DE, Kearney MT, Shane SM, Zwank PJ. Effects of pH, temperature, and salinity on persistence of avian influenza viruses in water. Avian Dis. 1990;34(2):412–418. doi: 10.2307/1591429. [DOI] [PubMed] [Google Scholar]
- 101.Ito T, Okazaki K, Kawaoka Y, Takada A, Webster RG, Kida H. Perpetuation of influenza A viruses in Alaskan waterfowl reservoirs. Arch Virol. 1995;140(7):1163–1172. doi: 10.1007/BF01322743. [DOI] [PubMed] [Google Scholar]
- 102.Sturm-Ramirez KM, Hulse-Post DJ, Govorkova EA, Humberd J, Seiler P, Puthavathana P, et al. Are ducks contributing to the endemicity of highly pathogenic H5N1 influenza virus in Asia? J Virol. 2005;79(17):11269–11279. doi: 10.1128/JVI.79.17.11269-11279.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bailey E, Long LP, Zhao N, Hall JS, Baroch JA, Nolting J, et al. Antigenic characterization of H3 subtypes of avian influenza A viruses from North America. Avian Dis. 2016;60(1 Suppl):346–353. doi: 10.1637/11086-041015-RegR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xu Y, Bailey E, Spackman E, Li T, Wang H, Long LP, et al. Limited antigenic diversity in contemporary H7 avian-origin influenza A viruses from North America. Sci Rep. 2016;6:20688. doi: 10.1038/srep20688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Alexander DJ. A review of avian influenza in different bird species. Vet Microbiol. 2000;74(1):3–13. doi: 10.1016/S0378-1135(00)00160-7. [DOI] [PubMed] [Google Scholar]
- 106.Wan XF. Lessons from emergence of A/goose/Guangdong/1996-like H5N1 highly pathogenic avian influenza viruses and recent influenza surveillance efforts in southern China. Zoonoses Public Health. 2012;59 Suppl 2(0 2):32-42. [DOI] [PMC free article] [PubMed]
- 107.Ungchusak K, Auewarakul P, Dowell SF, Kitphati R, Auwanit W, Puthavathana P, et al. Probable person-to-person transmission of avian influenza A (H5N1) N Engl J Med. 2005;352(4):333–340. doi: 10.1056/NEJMoa044021. [DOI] [PubMed] [Google Scholar]
- 108.Zhu X, Guo YH, Jiang T, Wang YD, Chan KH, Li XF, et al. A unique and conserved neutralization epitope in H5N1 influenza viruses identified by an antibody against the A/Goose/Guangdong/1/96 hemagglutinin. J Virol. 2013;87(23):12619–12635. doi: 10.1128/JVI.01577-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lee CW, Senne DA, Suarez DL. Effect of vaccine use in the evolution of Mexican lineage H5N2 avian influenza virus. J Virol. 2004;78(15):8372–8381. doi: 10.1128/JVI.78.15.8372-8381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Beato MS, Xu Y, Long LP, Capua I, Wan XF. Antigenic and genetic evolution of low pathogenic avian influenza viruses (H7N3) following heterologous vaccination. Clin Vacc Immunol. 2014;21(5):603–612. doi: 10.1128/CVI.00647-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lee DH, Fusaro A, Song CS, Suarez DL, Swayne DE. Poultry vaccination directed evolution of H9N2 low pathogenicity avian influenza viruses in Korea. Virology. 2015;488:225–231. doi: 10.1016/j.virol.2015.11.023. [DOI] [PubMed] [Google Scholar]
- 112.Cattoli G, Fusaro A, Monne I, Coven F, Joannis T, El-Hamid HS, et al. Evidence for differing evolutionary dynamics of A/H5N1 viruses among countries applying or not applying avian influenza vaccination in poultry. Vaccine. 2011;29(50):9368–9375. doi: 10.1016/j.vaccine.2011.09.127. [DOI] [PubMed] [Google Scholar]
- 113.Swayne DE. Impact of vaccines and vaccination on global control of avian influenza. Avian Dis. 2012;56(4 Suppl):818–828. doi: 10.1637/10183-041012-Review.1. [DOI] [PubMed] [Google Scholar]
- 114.WHO. Toward a unified nomenclature system for highly pathogenic avian influenza virus (H5N1). Emerg Infect Dis 2008;14(7):e1. [DOI] [PMC free article] [PubMed]
- 115.WHO Continuing progress towards a unified nomenclature for the highly pathogenic H5N1 avian influenza viruses: divergence of clade 2.2 viruses. Influenza Other Respir Viruses. 2009;3(2):59–62. doi: 10.1111/j.1750-2659.2009.00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Creanga A, Thi Nguyen D, Gerloff N, Thi Do H, Balish A, Dang Nguyen H, et al. Emergence of multiple clade 2.3.2.1 influenza A (H5N1) virus subgroups in Vietnam and detection of novel reassortants. Virology. 2013;444(1-2):12–20. doi: 10.1016/j.virol.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 117.Wei Y, Xu G, Zhang G, Wen C, Anwar F, Wang S, et al. Antigenic evolution of H9N2 chicken influenza viruses isolated in China during 2009-2013 and selection of a candidate vaccine strain with broad cross-reactivity. Vet Microbiol. 2016;182:1–7. doi: 10.1016/j.vetmic.2015.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu J, Okazaki K, Ozaki H, Sakoda Y, Wu Q, Chen F, et al. H9N2 influenza viruses prevalent in poultry in China are phylogenetically distinct from A/quail/Hong Kong/G1/97 presumed to be the donor of the internal protein genes of the H5N1 Hong Kong/97 virus. Avian Pathol. 2003;32(5):551–560. doi: 10.1080/0307-9450310001596728. [DOI] [PubMed] [Google Scholar]
- 119.Pu J, Wang S, Yin Y, Zhang G, Carter RA, Wang J, et al. Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus. Proc Natl Acad Sci U S A. 2015;112(2):548–553. doi: 10.1073/pnas.1422456112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li C, Yu K, Tian G, Yu D, Liu L, Jing B, et al. Evolution of H9N2 influenza viruses from domestic poultry in Mainland China. Virology. 2005;340(1):70–83. doi: 10.1016/j.virol.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 121.Li X, Shi J, Guo J, Deng G, Zhang Q, Wang J, et al. Genetics, receptor binding property, and transmissibility in mammals of naturally isolated H9N2 Avian Influenza viruses. PLoS Pathog. 2014;10(11):e1004508. doi: 10.1371/journal.ppat.1004508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cong YL, Pu J, Liu QF, Wang S, Zhang GZ, Zhang XL, et al. Antigenic and genetic characterization of H9N2 swine influenza viruses in China. J Gen Virol. 2007;88(Pt 7):2035–2041. doi: 10.1099/vir.0.82783-0. [DOI] [PubMed] [Google Scholar]
- 123.Xu KM, Li KS, Smith GJ, Li JW, Tai H, Zhang JX, et al. Evolution and molecular epidemiology of H9N2 influenza A viruses from quail in southern China, 2000 to 2005. J Virol. 2007;81(6):2635–2645. doi: 10.1128/JVI.02316-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gu M, Chen H, Li Q, Huang J, Zhao M, Gu X, et al. Enzootic genotype S of H9N2 avian influenza viruses donates internal genes to emerging zoonotic influenza viruses in China. Vet Microbiol. 2014;174(3-4):309–315. doi: 10.1016/j.vetmic.2014.09.029. [DOI] [PubMed] [Google Scholar]
- 125.Guan Y, Shortridge KF, Krauss S, Webster RG. Molecular characterization of H9N2 influenza viruses: were they the donors of the "internal" genes of H5N1 viruses in Hong Kong? Proc Natl Acad Sci U S A. 1999;96(16):9363–9367. doi: 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chu YC, Cheung CL, Hung Leung CY, Man Poon LL, Chen H, Peiris JS, et al. Continuing evolution of H9N2 influenza viruses endemic in poultry in southern China. Influenza Other Respir Viruses. 2011;5(Suppl 1):68–71. [PubMed] [Google Scholar]
- 127.Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333(6044):843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Beato MS, Xu Y, Long L-P, Capua I, Wan X-F. Antigenic and genetic evolution of low-pathogenicity avian influenza viruses of subtype H7N3 following heterologous vaccination. Clin Vacc Immunol. 2014;21(5):603–612. doi: 10.1128/CVI.00647-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Organization WH. Antigenic and genetic characteristics of zoonotic influenza viruses and development of candidate vaccine viruses for pandemic preparedness 2017 [updated March 2017; cited 2017. Available from: https://www.who.int/influenza/vaccines/virus/201703_zoonotic_vaccinevirusupdate.pdf?%20ua=1.
- 130.Dong J, Chen P, Wang Y, Lv Y, Xiao J, Li Q, et al. Evaluation of the immune response of a H7N9 candidate vaccine virus derived from the fifth wave A/Guangdong/17SF003/2016. Antivir Res 2020:104776. [DOI] [PubMed]
- 131.Wang Y, Lv Y, Niu X, Dong J, Feng P, Li Q, et al. L226Q Mutation on influenza H7N9 virus hemagglutinin increases receptor-binding avidity and leads to biased antigenicity evaluation. J Virol. 2020;94(20). [DOI] [PMC free article] [PubMed]
- 132.Kuiken T, Holmes EC, McCauley J, Rimmelzwaan GF, Williams CS, Grenfell BT. Host species barriers to influenza virus infections. Science. 2006;312(5772):394–397. doi: 10.1126/science.1122818. [DOI] [PubMed] [Google Scholar]
- 133.Van Reeth K. Avian and swine influenza viruses: our current understanding of the zoonotic risk. Vet Res. 2007;38(2):243–260. doi: 10.1051/vetres:2006062. [DOI] [PubMed] [Google Scholar]
- 134.Yasuda J, Shortridge KF, Shimizu Y, Kida H. Molecular evidence for a role of domestic ducks in the introduction of avian H3 influenza viruses to pigs in southern China, where the A/Hong Kong/68 (H3N2) strain emerged. J Gen Virol. 1991;72(Pt 8):2007–2010. doi: 10.1099/0022-1317-72-8-2007. [DOI] [PubMed] [Google Scholar]
- 135.Webster RG, Campbell CH, Granoff A. The "in vivo" production of "new" influenza A viruses. I. Genetic recombination between avian and mammalian influenza viruses. Virology. 1971;44(2):317–328. doi: 10.1016/0042-6822(71)90263-7. [DOI] [PubMed] [Google Scholar]
- 136.Webster RG, Campbell CH, Granoff A. The "in vivo" production of "new" influenza viruses. 3. Isolation of recombinant influenza viruses under simulated conditions of natural transmission. Virology. 1973;51(1):149–162. doi: 10.1016/0042-6822(73)90375-9. [DOI] [PubMed] [Google Scholar]
- 137.Kida H, Shortridge KF, Webster RG. Origin of the hemagglutinin gene of H3N2 influenza viruses from pigs in China. Virology. 1988;162(1):160–166. doi: 10.1016/0042-6822(88)90405-9. [DOI] [PubMed] [Google Scholar]
- 138.Scholtissek C, Burger H, Bachmann PA, Hannoun C. Genetic relatedness of hemagglutinins of the H1 subtype of influenza A viruses isolated from swine and birds. Virology. 1983;129(2):521–523. doi: 10.1016/0042-6822(83)90194-0. [DOI] [PubMed] [Google Scholar]
- 139.Shortridge KF, Webster RG, Butterfield WK, Campbell CH. Persistence of Hong Kong influenza virus variants in pigs. Science. 1977;196(4297):1454–1455. doi: 10.1126/science.867041. [DOI] [PubMed] [Google Scholar]
- 140.Kida H, Ito T, Yasuda J, Shimizu Y, Itakura C, Shortridge KF, et al. Potential for transmission of avian influenza viruses to pigs. J Gen Virol. 1994;75(Pt 9):2183–2188. doi: 10.1099/0022-1317-75-9-2183. [DOI] [PubMed] [Google Scholar]
- 141.Pensaert M, Ottis K, Vandeputte J, Kaplan MM, Bachmann PA. Evidence for the natural transmission of influenza A virus from wild ducts to swine and its potential importance for man. Bull World Health Organ. 1981;59(1):75–78. [PMC free article] [PubMed] [Google Scholar]
- 142.Guan Y, Shortridge KF, Krauss S, Li PH, Kawaoka Y, Webster RG. Emergence of avian H1N1 influenza viruses in pigs in China. J Virol. 1996;70(11):8041–8046. doi: 10.1128/jvi.70.11.8041-8046.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Peiris JS, Guan Y, Markwell D, Ghose P, Webster RG, Shortridge KF. Cocirculation of avian H9N2 and contemporary "human" H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J Virol. 2001;75(20):9679–9686. doi: 10.1128/JVI.75.20.9679-9686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Xu C, Fan W, Wei R, Zhao H. Isolation and identification of swine influenza recombinant A/Swine/Shandong/1/2003(H9N2) virus. Microbes Infect. 2004;6(10):919–925. doi: 10.1016/j.micinf.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 145.Choi YK, Nguyen TD, Ozaki H, Webby RJ, Puthavathana P, Buranathal C, et al. Studies of H5N1 influenza virus infection of pigs by using viruses isolated in Vietnam and Thailand in 2004. J Virol. 2005;79(16):10821–10825. doi: 10.1128/JVI.79.16.10821-10825.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Karasin AI, Brown IH, Carman S, Olsen CW. Isolation and characterization of H4N6 avian influenza viruses from pigs with pneumonia in Canada. J Virol. 2000;74(19):9322–9327. doi: 10.1128/JVI.74.19.9322-9327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Karasin AI, West K, Carman S, Olsen CW. Characterization of avian H3N3 and H1N1 influenza A viruses isolated from pigs in Canada. J Clin Microbiol. 2004;42(9):4349–4354. doi: 10.1128/JCM.42.9.4349-4354.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Vincent AL, Ma W, Lager KM, Janke BH, Richt JA. Swine influenza viruses a North American perspective. Adv Virus Res. 2008;72:127–154. doi: 10.1016/S0065-3527(08)00403-X. [DOI] [PubMed] [Google Scholar]
- 149.Kaplan BS, DeBeauchamp J, Stigger-Rosser E, Franks J, Crumpton JC, Turner J, et al. Influenza virus surveillance in coordinated swine production systems, United States. Emerg Infect Dis. 2015;21(10):1834–1836. doi: 10.3201/eid2110.140633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Anderson TK, Chang J, Arendsee ZW, Venkatesh D, Souza CK, Kimble JB, et al. Swine influenza A viruses and the tangled relationship with humans. Cold Spring Harb Perspect Med. 2021;11(3). [DOI] [PMC free article] [PubMed]
- 151.Pereda A, Rimondi A, Cappuccio J, Sanguinetti R, Angel M, Ye J, et al. Evidence of reassortment of pandemic H1N1 influenza virus in swine in Argentina: are we facing the expansion of potential epicenters of influenza emergence? Influenza Other Respir Viruses. 2011;5(6):409–412. doi: 10.1111/j.1750-2659.2011.00246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Biondo N, Schaefer R, Gava D, Cantao ME, Silveira S, Mores MA, et al. Genomic analysis of influenza A virus from captive wild boars in Brazil reveals a human-like H1N2 influenza virus. Vet Microbiol. 2014;168(1):34–40. doi: 10.1016/j.vetmic.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 153.Nelson M, Culhane MR, Rovira A, Torremorell M, Guerrero P, Norambuena J. Novel human-like influenza A viruses circulate in swine in Mexico and Chile. PLoS Curr. 2015;7. [DOI] [PMC free article] [PubMed]
- 154.Nelson MI, Schaefer R, Gava D, Cantao ME, Ciacci-Zanella JR. Influenza A viruses of human origin in swine, Brazil. Emerg Infect Dis. 2015;21(8):1339–1347. doi: 10.3201/eid2108.141891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Brown IH. History and epidemiology of swine influenza in Europe. In: Richt JA, Webby RJ, editors. Swine Influenza. Berlin, Heidelberg: Springer Berlin Heidelberg; 2013. pp. 133–146. [Google Scholar]
- 156.Zell R, Scholtissek C, Ludwig S. Genetics, evolution, and the zoonotic capacity of European swine influenza viruses. In: Richt JA, Webby RJ, editors. Swine Influenza. Berlin, Heidelberg: Springer Berlin Heidelberg; 2013. pp. 29–55. [DOI] [PubMed] [Google Scholar]
- 157.Rajao DS, Gauger PC, Anderson TK, Lewis NS, Abente EJ, Killian ML, et al. Novel reassortant human-like H3N2 and H3N1 influenza A viruses detected in pigs are virulent and antigenically distinct from swine viruses endemic to the United States. J Virol. 2015;89(22):11213–22. doi: 10.1128/JVI.01675-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zeller MA, Li G, Harmon KM, Zhang J, Vincent AL, Anderson TK, et al. Complete genome sequences of two novel human-like H3N2 influenza a viruses, A/swine/Oklahoma/65980/2017 (H3N2) and A/Swine/Oklahoma/65260/2017 (H3N2), Detected in Swine in the United States. Microbiol Resour Announc. 2018;7(20). [DOI] [PMC free article] [PubMed]
- 159.Zeller MA, Anderson TK, Walia RW, Vincent AL, Gauger PC. ISU FLUture: a veterinary diagnostic laboratory web-based platform to monitor the temporal genetic patterns of Influenza A virus in swine. BMC Bioinformatics. 2018;19(1):397. doi: 10.1186/s12859-018-2408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Feng Z, Baroch JA, Long LP, Xu Y, Cunningham FL, Pedersen K, et al. Influenza A subtype H3 viruses in feral swine, United States, 2011-2012. Emerg Infect Dis. 2014;20(5):843–846. doi: 10.3201/eid2005.131578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lewis NS, Russell CA, Langat P, Anderson TK, Berger K, Bielejec F, et al. The global antigenic diversity of swine influenza A viruses. Elife. 2016;5:e12217. doi: 10.7554/eLife.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fiore AE, Uyeki TM, Broder K, Finelli L, Euler GL, Singleton JA, et al. Prevention and control of influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2010. MMWR Recomm Rep. 2010;59(RR-8):1-62. [PubMed]
- 163.Sakala IG, Honda-Okubo Y, Fung J, Petrovsky N. Influenza immunization during pregnancy: benefits for mother and infant. Hum Vaccines Immunother. 2016;12(12):3065–3071. doi: 10.1080/21645515.2016.1215392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Grohskopf LA, Sokolow LZ, Broder KR, Walter EB, Bresee JS, Fry AM, et al. Prevention and control of seasonal influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices - United States, 2017-18 Influenza Season. MMWR Recomm Rep. 2017;66(2):1–20. doi: 10.15585/mmwr.rr6602a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.American Academy of Pediatrics Committee on Infectious D. Recommendations for influenza immunization of children. Pediatrics. 2004;113(5):1441–7. [DOI] [PubMed]
- 166.National Immunization Advisory Committee Technical Working G. [Technical guidelines for seasonal influenza vaccination in China (2020-2021)]. Zhonghua Yu Fang Yi Xue Za Zhi. 2020;54(10):1035–59. [DOI] [PubMed]
- 167.Canada Go. Canadian Immunization Guide Chapter on Influenza and Statement on Seasonal Influenza Vaccine for 2020–2021 2020 [Available from: https://www.canada.ca/en/public-health/services/publications/vaccines-immunization/canadian-immunization-guide-statement-seasonal-influenza-vaccine-2020-2021.html. Accessed 15 Oct 2021.
- 168.Health AGDo. Australian Immuisation Handbook 2021 [Available from: https://immunisationhandbook.health.gov.au/vaccine-preventable-diseases/influenza-flu.
- 169.Principi N, Camilloni B, Esposito S, Group EVS Influenza immunization policies: which could be the main reasons for differences among countries? Human Vacc Immunother. 2018;14(3):684–692. doi: 10.1080/21645515.2017.1405188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.CDC. Flu vaccination coverage, United States, 2015-16 Influenza Season 2016 [Available from: https://www.cdc.gov/flu/pdf/fluvaxview/2015-16/nfid-coverage-2015-16-final.pdf. Accessed 3 Mar 2020.
- 171.Robert J, Detournay B, Levant MC, Uhart M, Gourmelen J, Cohen JM. Flu vaccine coverage for recommended populations in France. Med Mal Infect. 2020;50(8):670–675. doi: 10.1016/j.medmal.2019.12.004. [DOI] [PubMed] [Google Scholar]
- 172.Treanor JJ, Talbot HK, Ohmit SE, Coleman LA, Thompson MG, Cheng PY, et al. Effectiveness of seasonal influenza vaccines in the United States during a season with circulation of all three vaccine strains. Clin Infect Dis. 2012;55(7):951–959. doi: 10.1093/cid/cis574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ohmit SE, Thompson MG, Petrie JG, Thaker SN, Jackson ML, Belongia EA, et al. Influenza vaccine effectiveness in the 2011-2012 season: protection against each circulating virus and the effect of prior vaccination on estimates. Clin Infect Dis. 2014;58(3):319–327. doi: 10.1093/cid/cit736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.McLean HQ, Thompson MG, Sundaram ME, Kieke BA, Gaglani M, Murthy K, et al. Influenza vaccine effectiveness in the United States during 2012-2013: variable protection by age and virus type. J Infect Dis. 2015;211(10):1529–1540. doi: 10.1093/infdis/jiu647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gaglani M, Pruszynski J, Murthy K, Clipper L, Robertson A, Reis M, et al. Influenza vaccine effectiveness against 2009 pandemic influenza A(H1N1) virus differed by vaccine type during 2013-2014 in the United States. J Infect Dis. 2016;213(10):1546–1556. doi: 10.1093/infdis/jiv577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zimmerman RK, Nowalk MP, Chung J, Jackson ML, Jackson LA, Petrie JG, et al. 2014-2015 influenza vaccine effectiveness in the United States by vaccine type. Clin Infect Dis. 2016;63(12):1564–1573. doi: 10.1093/cid/ciw635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Jackson ML, Chung JR, Jackson LA, Phillips CH, Benoit J, Monto AS, et al. Influenza vaccine effectiveness in the United States during the 2015-2016 Season. N Engl J Med. 2017;377(6):534–543. doi: 10.1056/NEJMoa1700153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Flannery B, Chung JR, Thaker SN, Monto AS, Martin ET, Belongia EA, et al. Interim estimates of 2016-17 seasonal influenza vaccine effectiveness - United States, February 2017. MMWR Morb Mortal Wkly Rep. 2017;66(6):167–171. doi: 10.15585/mmwr.mm6606a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Rolfes MA, Flannery B, Chung JR, O'Halloran A, Garg S, Belongia EA, et al. Effects of influenza vaccination in the United States during the 2017-2018 influenza season. Clin Infect Dis. 2019;69(11):1845–1853. doi: 10.1093/cid/ciz075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Flannery B, Kondor RJG, Chung JR, Gaglani M, Reis M, Zimmerman RK, et al. Spread of antigenically drifted influenza A(H3N2) viruses and vaccine effectiveness in the United States during the 2018–2019 season. J Infect Dis. 2020;221(1):8–15. doi: 10.1093/infdis/jiz543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Dawood FS, Chung JR, Kim SS, Zimmerman RK, Nowalk MP, Jackson ML, et al. Interim estimates of 2019-20 seasonal influenza vaccine effectiveness - United States, February 2020. MMWR Morb Mortal Wkly Rep. 2020;69(7):177–182. doi: 10.15585/mmwr.mm6907a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Belongia EA, Simpson MD, King JP, Sundaram ME, Kelley NS, Osterholm MT, et al. Variable influenza vaccine effectiveness by subtype: a systematic review and meta-analysis of test-negative design studies. Lancet Infect Dis. 2016;16(8):942–951. doi: 10.1016/S1473-3099(16)00129-8. [DOI] [PubMed] [Google Scholar]
- 183.Prevention CfDCa. Past Seasons Vaccine Effectiveness Estimates January 29, 2020 [Available from: https://www.cdc.gov/flu/vaccines-work/past-seasons-estimates.html. Accessed 3 Mar 2020.
- 184.Xu X, Blanton L, Elal AIA, Alabi N, Barnes J, Biggerstaff M, et al. Update: influenza activity in the United States during the 2018-19 season and composition of the 2019-20 influenza vaccine. MMWR Morb Mortal Wkly Rep. 2019;68(24):544–551. doi: 10.15585/mmwr.mm6824a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Dhakal S, Klein SL. Host factors impact vaccine efficacy: implications for seasonal and universal influenza vaccine programs. J Virol. 2019;93(21):e00797–e00719. doi: 10.1128/JVI.00797-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Castrucci MR. Factors affecting immune responses to the influenza vaccine. Hum Vacc Immunother. 2018;14(3):637–646. doi: 10.1080/21645515.2017.1338547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Tenforde MW, Kondor RJG, Chung JR, Zimmerman RK, Nowalk MP, Jackson ML, et al. Effect of antigenic drift on influenza vaccine effectiveness in the United States—2019–2020. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed]
- 188.Tricco AC, Chit A, Soobiah C, Hallett D, Meier G, Chen MH, et al. Comparing influenza vaccine efficacy against mismatched and matched strains: a systematic review and meta-analysis. BMC Med. 2013;11:153. doi: 10.1186/1741-7015-11-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Belongia EA, Kieke BA, Donahue JG, Greenlee RT, Balish A, Foust A, et al. Effectiveness of inactivated influenza vaccines varied substantially with antigenic match from the 2004-2005 season to the 2006-2007 season. J Infect Dis. 2009;199(2):159–167. doi: 10.1086/595861. [DOI] [PubMed] [Google Scholar]
- 190.Chung JR, Flannery B, Ambrose CS, Bégué RE, Caspard H, DeMarcus L, et al. Live attenuated and inactivated influenza vaccine effectiveness. Pediatrics. 2019;143(2):e20182094. doi: 10.1542/peds.2018-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Gaglani M, Vasudevan A, Raiyani C, Murthy K, Chen W, Reis M, et al. Effectiveness of trivalent and quadrivalent inactivated vaccines against influenza B in the United States, 2011–2012 to 2016–2017. Clin Infect Dis. 2021;72(7):1147–1157. doi: 10.1093/cid/ciaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Milne GJ, Halder N, Kelso JK, Barr IG, Moyes J, Kahn K, et al. Trivalent and quadrivalent influenza vaccination effectiveness in Australia and South Africa: results from a modelling study. Influenza Other Respir Viruses. 2016;10(4):324–332. doi: 10.1111/irv.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.WHO. Global epidemiological surveillance standards for influenza2013. Available from: http://www.who.int/influenza/resources/documents/WHO_Epidemiological_Influenza_Surveillance_Standards_2014.pdf?ua=1. Accessed 5 Mar 2020.
- 194.Layne SP, Beugelsdijk TJ, Patel CK, Taubenberger JK, Cox NJ, Gust ID, et al. A global lab against influenza. Science. 2001;293(5536):1729. doi: 10.1126/science.293.5536.1729. [DOI] [PubMed] [Google Scholar]
- 195.Kitler ME, Gavinio P, Lavanchy D. Influenza and the work of the World Health Organization. Vaccine. 2002;20(Suppl 2):S5–14. doi: 10.1016/S0264-410X(02)00121-4. [DOI] [PubMed] [Google Scholar]
- 196.Palese P. Making better influenza virus vaccines? Emerg Infect Dis. 2006;12(1):61–65. doi: 10.3201/eid1201.051043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Influenza B Victoria lineage candidate vaccine viruses for development and production of vaccines for use in the northern hemisphere 2015-2016 influenza season. World Health Organization; September 14, 2015.
- 198.Swayne DE. Vaccines for List A poultry diseases: emphasis on avian influenza. Dev Biol (Basel) 2003;114:201–212. [PubMed] [Google Scholar]
- 199.Swayne DE, Beck JR, Garcia M, Stone HD. Influence of virus strain and antigen mass on efficacy of H5 avian influenza inactivated vaccines. Avian Pathol. 1999;28(3):245–255. doi: 10.1080/03079459994731. [DOI] [PubMed] [Google Scholar]
- 200.Swayne DEaK, D. R. Avian influenza. In: Swayne DE, editor. Vaccines, vaccination, and immunology for avian influenza viruses in poultry: John Wiley & Sons, Inc.; 2008.
- 201.Capua I, Terregino C, Cattoli G, Toffan A. Increased resistance of vaccinated turkeys to experimental infection with an H7N3 low-pathogenicity avian influenza virus. Avian Pathol. 2004;33(2):158–163. doi: 10.1080/03079450310001652077. [DOI] [PubMed] [Google Scholar]
- 202.Peng C, Hou G, Li J, Wang S, Wang Y, Cheng S, et al. Protective efficacy of an inactivated chimeric H7/H5 avian influenza vaccine against highly pathogenic avian influenza H7N9 and clade 2.3.4.4 H5 viruses. Vet Microbiol. 2018;223:21–26. doi: 10.1016/j.vetmic.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 203.Wu J, Ke C, Lau EHY, Song Y, Cheng KL, Zou L, et al. Influenza H5/H7 virus vaccination in poultry and reduction of zoonotic infections, Guangdong Province, China, 2017-18. Emerg Infect Dis. 2019;25(1):116–118. doi: 10.3201/eid2501.181259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Van Reeth K, Ma W. Swine influenza virus vaccines: to change or not to change-that's the question. Curr Top Microbiol Immunol. 2013;370:173–200. doi: 10.1007/82_2012_266. [DOI] [PubMed] [Google Scholar]
- 205.Bruschke CJ, Pittman M, Laddomada A. International regulations and standards for avian influenza, including the vaccine standards of the World Organisation for Animal Health. Rev Sci Tech. 2009;28(1):379–389. doi: 10.20506/rst.28.1.1852. [DOI] [PubMed] [Google Scholar]
- 206.Rudolf M, Poppel M, Frohlich A, Breithaupt A, Teifke J, Blohm U, et al. Longitudinal 2 years field study of conventional vaccination against highly pathogenic avian influenza H5N1 in layer hens. Vaccine. 2010;28(42):6832–6840. doi: 10.1016/j.vaccine.2010.08.038. [DOI] [PubMed] [Google Scholar]
- 207.Ampofo WK, Baylor N, Cobey S, Cox NJ, Daves S, Edwards S, et al. Improving influenza vaccine virus selectionReport of a WHO informal consultation held at WHO headquarters, Geneva, Switzerland, 14-16 June 2010. Influenza Other Respir Viruses. 2011. [DOI] [PMC free article] [PubMed]
- 208.Morris DH, Gostic KM, Pompei S, Bedford T, Luksza M, Neher RA, et al. Predictive modeling of influenza shows the promise of applied evolutionary biology. Trends Microbiol. 2018;26(2):102–118. doi: 10.1016/j.tim.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Klingen TR, Reimering S, Guzman CA, McHardy AC. In silico vaccine strain prediction for human influenza viruses. Trends Microbiol. 2018;26(2):119–131. doi: 10.1016/j.tim.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 210.Ampofo WK, Azziz-Baumgartner E, Bashir U, Cox NJ, Fasce R, Giovanni M, et al. Strengthening the influenza vaccine virus selection and development process: report of the 3rd WHO Informal Consultation for Improving Influenza Vaccine Virus Selection held at WHO headquarters, Geneva, Switzerland, 1-3 April 2014. Vaccine. 2015;33(36):4368–4382. doi: 10.1016/j.vaccine.2015.06.090. [DOI] [PubMed] [Google Scholar]
- 211.Krutikova EV, Stepanova EA, Wong PF, Kiseleva IV, Grigor’eva EP, Rudenko LG. Genetic basis of attenuation of cold-adapted influenza strain B/Leningrad/14/17/55 — backup master donor virus for influenza type B live attenuated vaccines. Bull Exp Biol Med. 2020;168(5):669–672. doi: 10.1007/s10517-020-04777-x. [DOI] [PubMed] [Google Scholar]
- 212.Antigenic and genetic characteristics of zoonotic influenza A viruses and development of candidate vaccine viruses for pandemic preparedness March 2021.
- 213.Trock SC, Burke SA, Cox NJ. Development of an influenza virologic risk assessment tool. Avian Dis. 2012;56(4 Suppl):1058–1061. doi: 10.1637/10204-041412-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 214.WHO. Tool for influenza pandemic risk assessment (TIPRA)2016.
- 215.Organization WH. Tool for influenza pandemic risk assessment (TIPRA) 2020 [cited 2021 September 13]. Available from: https://www.who.int/publications/m/item/tool-for-influenza-pandemic-risk-assessment-(tipra). Accessed 15 Oct 2021.
- 216.Burke SA, Trock SC. Use of influenza risk assessment tool for prepandemic preparedness. Emerg Infect Dis. 2018;24(3):471–477. doi: 10.3201/eid2403.171852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Influenza Virologic Surveillance Right Size Roadmap. July 2013.
- 218.Chambers BS, Li Y, Hodinka RL, Hensley SE. Recent H3N2 influenza virus clinical isolates rapidly acquire hemagglutinin or neuraminidase mutations when propagated for antigenic analyses. J Virol. 2014;88(18):10986–10989. doi: 10.1128/JVI.01077-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Robertson JS, Nicolson C, Harvey R, Johnson R, Major D, Guilfoyle K, et al. The development of vaccine viruses against pandemic A(H1N1) influenza. Vaccine. 2011;29(9):1836–1843. doi: 10.1016/j.vaccine.2010.12.044. [DOI] [PubMed] [Google Scholar]
- 220.Ryu W-S. Chapter. 4 - Diagnosis and methods. In: Ryu W-S, editor. Molecular virology of human pathogenic viruses. Academic Press; 2017. pp. 47–62. [Google Scholar]
- 221.Suzuki Y, Ito T, Suzuki T, Holland RE, Chambers TM, Kiso M, et al. Sialic acid species as a determinant of the host range of influenza A viruses. J Virol. 2000;74(24):11825. doi: 10.1128/JVI.74.24.11825-11831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Organization WH. Manual for the laboratory diagnosis and virological surveillance of influenza2011.
- 223.Okuno Y, Tanaka K, Baba K, Maeda A, Kunita N, Ueda S. Rapid focus reduction neutralization test of influenza A and B viruses in microtiter system. J Clin Microbiol. 1990;28(6):1308–1313. doi: 10.1128/jcm.28.6.1308-1313.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Ito T, Suzuki Y, Mitnaul L, Vines A, Kida H, Kawaoka Y. Receptor specificity of influenza A viruses correlates with the agglutination of erythrocytes from different animal species. Virology. 1997;227(2):493–499. doi: 10.1006/viro.1996.8323. [DOI] [PubMed] [Google Scholar]
- 225.Ovsyannikova IG, White SJ, Albrecht RA, Garcia-Sastre A, Poland GA. Turkey versus guinea pig red blood cells: hemagglutination differences alter hemagglutination inhibition responses against influenza A/H1N1. Viral Immunol. 2014;27(4):174–178. doi: 10.1089/vim.2013.0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Grassauer A, Egorov AY, Ferko B, Romanova I, Katinger H, Muster T. A host restriction-based selection system for influenza haemagglutinin transfectant viruses. J Gen Virol. 1998;79(Pt 6):1405–1409. doi: 10.1099/0022-1317-79-6-1405. [DOI] [PubMed] [Google Scholar]
- 227.Lin YP, Xiong X, Wharton SA, Martin SR, Coombs PJ, Vachieri SG, et al. Evolution of the receptor binding properties of the influenza A(H3N2) hemagglutinin. Proc Natl Acad Sci U S A. 2012;109(52):21474–21479. doi: 10.1073/pnas.1218841110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ndifon W, Dushoff J, Levin SA. On the use of hemagglutination-inhibition for influenza surveillance: surveillance data are predictive of influenza vaccine effectiveness. Vaccine. 2009;27(18):2447–2452. doi: 10.1016/j.vaccine.2009.02.047. [DOI] [PubMed] [Google Scholar]
- 229.Lin Y, Wharton SA, Whittaker L, Dai M, Ermetal B, Lo J, et al. The characteristics and antigenic properties of recently emerged subclade 3C.3a and 3C.2a human influenza A(H3N2) viruses passaged in MDCK cells. Influenza Other Respir Viruses. 2017;11(3):263–274. doi: 10.1111/irv.12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Oh DY, Barr IG, Mosse JA, Laurie KL. MDCK-SIAT1 cells show improved isolation rates for recent human influenza viruses compared to conventional MDCK cells. J Clin Microbiol. 2008;46(7):2189–2194. doi: 10.1128/JCM.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Lin YP, Gregory V, Collins P, Kloess J, Wharton S, Cattle N, et al. Neuraminidase receptor binding variants of human influenza A(H3N2) viruses resulting from substitution of aspartic acid 151 in the catalytic site: a role in virus attachment? J Virol. 2010;84(13):6769–6781. doi: 10.1128/JVI.00458-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Azzi A, Bartolomei-Corsi O, Zakrzewska K, Corcoran T, Newman R, Robertson JS, et al. The haemagglutinins of influenza A (H1N1) viruses in the 'O' or 'D' phases exhibit biological and antigenic differences. Epidemiol Infect 1993;111(1):135-142. [DOI] [PMC free article] [PubMed]
- 233.Morishita T, Kobayashi S, Miyake T, Ishihara Y, Nakajima S, Nakajima K. Host-specific hemagglutination of influenza A (H1N1) virus. Microbiol Immunol. 1993;37(8):661–665. doi: 10.1111/j.1348-0421.1993.tb01689.x. [DOI] [PubMed] [Google Scholar]
- 234.Nobusawa E, Ishihara H, Morishita T, Sato K, Nakajima K. Change in receptor-binding specificity of recent human influenza A viruses (H3N2): a single amino acid change in hemagglutinin altered its recognition of sialyloligosaccharides. Virology. 2000;278(2):587–596. doi: 10.1006/viro.2000.0679. [DOI] [PubMed] [Google Scholar]
- 235.Morishita T, Nobusawa E, Nakajima K, Nakajima S. Studies on the molecular basis for loss of the ability of recent influenza A (H1N1) virus strains to agglutinate chicken erythrocytes. J Gen Virol. 1996;77(Pt 10):2499–2506. doi: 10.1099/0022-1317-77-10-2499. [DOI] [PubMed] [Google Scholar]
- 236.Medeiros R, Escriou N, Naffakh N, Manuguerra JC, van der Werf S. Hemagglutinin residues of recent human A(H3N2) influenza viruses that contribute to the inability to agglutinate chicken erythrocytes. Virology. 2001;289(1):74–85. doi: 10.1006/viro.2001.1121. [DOI] [PubMed] [Google Scholar]
- 237.Fitch WM, Bush RM, Bender CA, Cox NJ. Long term trends in the evolution of H(3) HA1 human influenza type A. Proc Natl Acad Sci U S A. 1997;94(15):7712–7718. doi: 10.1073/pnas.94.15.7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Cox NJ, Bender CA. The molecular epidemiology of influenza viruses. Semin Virol. 1995;6(6):359–370. doi: 10.1016/S1044-5773(05)80013-7. [DOI] [Google Scholar]
- 239.Lindstrom S, Sugita S, Endo A, Ishida M, Huang P, Xi SH, et al. Evolutionary characterization of recent human H3N2 influenza A isolates from Japan and China: novel changes in the receptor binding domain. Arch Virol. 1996;141(7):1349–1355. doi: 10.1007/BF01718836. [DOI] [PubMed] [Google Scholar]
- 240.Broszeit F, van Beek RJ, Unione L, Bestebroer TM, Chapla D, Yang JY, et al. Glycan remodeled erythrocytes facilitate antigenic characterization of recent A/H3N2 influenza viruses. Nat Commun. 2021;12(1):5449. doi: 10.1038/s41467-021-25713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Skowronski DM, Sabaiduc S, Chambers C, Eshaghi A, Gubbay JB, Krajden M, et al. Mutations acquired during cell culture isolation may affect antigenic characterisation of influenza A(H3N2) clade 3C.2a viruses. Euro surveillance : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2016;21(3):30112. [DOI] [PubMed]
- 242.Matrosovich M, Matrosovich T, Carr J, Roberts NA, Klenk HD. Overexpression of the alpha-2,6-sialyltransferase in MDCK cells increases influenza virus sensitivity to neuraminidase inhibitors. J Virol. 2003;77(15):8418–8425. doi: 10.1128/JVI.77.15.8418-8425.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Mögling R, Richard MJ, Svd V, Rv B, Schrauwen EJA, Spronken MI, et al. Neuraminidase-mediated haemagglutination of recent human influenza A(H3N2) viruses is determined by arginine 150 flanking the neuraminidase catalytic site. J Gen Virol. 2017;98(6):1274–1281. doi: 10.1099/jgv.0.000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Hooper KA, Bloom JD. A mutant influenza virus that uses an N1 neuraminidase as the receptor-binding protein. J Virol. 2013;87(23):12531–12540. doi: 10.1128/JVI.01889-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Martin BE, Bowman AS, Li L, Nolting JM, Smith DR, Hanson LA, et al. Detection of antigenic variants of subtype H3 swine influenza A viruses from clinical samples. J Clin Microbiol. 2017;55(4):1037–1045. doi: 10.1128/JCM.02049-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Yuan XT, Zhang T, Wan XF. A joint matrix completion and filtering model for influenza serological data integration. PLoS One. 2013;8(7):e69842. doi: 10.1371/journal.pone.0069842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Stephenson I, Heath A, Major D, Newman RW, Hoschler K, Junzi W, et al. Reproducibility of serologic assays for influenza virus A (H5N1) Emerg Infect Dis. 2009;15(8):1252–1259. doi: 10.3201/eid1508.081754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Wilson G, Ye Z, Xie H, Vahl S, Dawson E, Rowlen K. Automated interpretation of influenza hemagglutination inhibition (HAI) assays: is plate tilting necessary? PLoS One. 2017;12(6):e0179939. doi: 10.1371/journal.pone.0179939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Nguyen M, Fries K, Khoury R, Zheng L, Hu B, Hildreth SW, et al. Automated imaging and analysis of the hemagglutination inhibition assay. J Lab Autom. 2016;21(2):287–296. doi: 10.1177/2211068215610061. [DOI] [PubMed] [Google Scholar]
- 250.Sánchez-Cano A, Andrés C, Herance JR, Pumarola T, Antón A, Baldrich E. Detection of viruses and virus-neutralizing antibodies using synthetic erythrocytes: toward a tuneable tool for virus surveillance. ACS Sensors. 2021;6(1):83–90. doi: 10.1021/acssensors.0c01830. [DOI] [PubMed] [Google Scholar]
- 251.Pedersen JC. Neuraminidase-inhibition assay for the identification of influenza A virus neuraminidase subtype or neuraminidase antibody specificity. Methods Mol Biol (Clifton, NJ) 2008;436:67–75. doi: 10.1007/978-1-59745-279-3_9. [DOI] [PubMed] [Google Scholar]
- 252.Aminoff D. Methods for the quantitative estimation of N-acetylneuraminic acid and their application to hydrolysates of sialomucoids. Biochem J. 1961;81(2):384–392. doi: 10.1042/bj0810384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Van Deusen RA, Hinshaw VS, Senne DA, Pellacani D. Micro neuraminidase-inhibition assay for classification of influenza A virus neuraminidases. Avian Dis. 1983;27(3):745–750. doi: 10.2307/1590317. [DOI] [PubMed] [Google Scholar]
- 254.Gao J, Couzens L, Eichelberger MC. Measuring influenza neuraminidase inhibition antibody titers by enzyme-linked lectin assay. J Vis Exp. 2016;115:54573. doi: 10.3791/54573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Lambré CR, Terzidis H, Greffard A, Webster RG. Measurement of anti-influenza neuraminidase antibody using a peroxidase-linked lectin and microtitre plates coated with natural substrates. J Immunol Methods. 1990;135(1-2):49–57. doi: 10.1016/0022-1759(90)90255-T. [DOI] [PubMed] [Google Scholar]
- 256.McCoy JP, Varani J, Goldstein IJ. Enzyme-linked lectin assay (ELLA): II. Detection of carbohydrate groups on the surface of unfixed cells. Exp Cell Res. 1984;151(1):96–103. doi: 10.1016/0014-4827(84)90359-8. [DOI] [PubMed] [Google Scholar]
- 257.Katz JM, Hancock K, Xu X. Serologic assays for influenza surveillance, diagnosis and vaccine evaluation. Expert Rev Anti-Infect Ther. 2011;9(6):669–683. doi: 10.1586/eri.11.51. [DOI] [PubMed] [Google Scholar]
- 258.Frank AL, Puck J, Hughes BJ, Cate TR. Microneutralization test for influenza A and B and parainfluenza 1 and 2 viruses that uses continuous cell lines and fresh serum enhancement. J Clin Microbiol. 1980;12(3):426–432. doi: 10.1128/jcm.12.3.426-432.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Lin Y, Gu Y, Wharton SA, Whittaker L, Gregory V, Li X, et al. Optimisation of a micro-neutralisation assay and its application in antigenic characterisation of influenza viruses. Influenza Other Respir Viruses. 2015;9(6):331–340. doi: 10.1111/irv.12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.van Baalen CA, Jeeninga RE, Penders GH, van Gent B, van Beek R, Koopmans MP, et al. ViroSpot microneutralization assay for antigenic characterization of human influenza viruses. Vaccine. 2017;35(1):46–52. doi: 10.1016/j.vaccine.2016.11.060. [DOI] [PubMed] [Google Scholar]
- 261.Ampofo WK, Al Busaidy S, Cox NJ, Giovanni M, Hay A, Huang S, et al. Strengthening the influenza vaccine virus selection and development process: outcome of the 2nd WHO Informal Consultation for Improving Influenza Vaccine Virus Selection held at the Centre International de Conferences (CICG) Geneva, Switzerland, 7 to 9 December 2011. Vaccine. 2013;31(32):3209–3221. doi: 10.1016/j.vaccine.2013.05.049. [DOI] [PubMed] [Google Scholar]
- 262.Veguilla V, Hancock K, Schiffer J, Gargiullo P, Lu X, Aranio D, et al. Sensitivity and specificity of serologic assays for detection of human infection with 2009 pandemic H1N1 virus in U.S. populations. J Clin Microbiol. 2011;49(6):2210–2215. doi: 10.1128/JCM.00229-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Li Y, Bostick DL, Sullivan CB, Myers JL, Griesemer SB, Stgeorge K, et al. Single hemagglutinin mutations that alter both antigenicity and receptor binding avidity influence influenza virus antigenic clustering. J Virol. 2013;87(17):9904–9910. doi: 10.1128/JVI.01023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Kiselar JG, Downard KM. Antigenic surveillance of the influenza virus by mass spectrometry. Biochemistry. 1999;38(43):14185–14191. doi: 10.1021/bi991609j. [DOI] [PubMed] [Google Scholar]
- 265.Morrissey B, Streamer M, Downard KM. Antigenic characterisation of H3N2 subtypes of the influenza virus by mass spectrometry. J Virol Methods. 2007;145(2):106–114. doi: 10.1016/j.jviromet.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 266.Morrissey B, Downard KM. A proteomics approach to survey the antigenicity of the influenza virus by mass spectrometry. Proteomics. 2006;6(7):2034–2041. doi: 10.1002/pmic.200500642. [DOI] [PubMed] [Google Scholar]
- 267.Schwahn AB, Downard KM. Antigenicity of a type A influenza virus through comparison of hemagglutination inhibition and mass spectrometry immunoassays. J Immunoass Immunochem. 2009;30(3):245–261. doi: 10.1080/15321810903084350. [DOI] [PubMed] [Google Scholar]
- 268.Russell CA, Jones TC, Barr IG, Cox NJ, Garten RJ, Gregory V, et al. Influenza vaccine strain selection and recent studies on the global migration of seasonal influenza viruses. Vaccine. 2008;26(Suppl 4):D31–D34. doi: 10.1016/j.vaccine.2008.07.078. [DOI] [PubMed] [Google Scholar]
- 269.Hensley SE. Challenges of selecting seasonal influenza vaccine strains for humans with diverse pre-exposure histories. Curr Opin Virol. 2014;8:85–89. doi: 10.1016/j.coviro.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Skowronski DM, Janjua NZ, De Serres G, Sabaiduc S, Eshaghi A, Dickinson JA, et al. Low 2012-13 influenza vaccine effectiveness associated with mutation in the egg-adapted H3N2 vaccine strain not antigenic drift in circulating viruses. PLoS One. 2014;9(3):e92153. doi: 10.1371/journal.pone.0092153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Jin H, Zhou H, Liu H, Chan W, Adhikary L, Mahmood K, et al. Two residues in the hemagglutinin of A/Fujian/411/02-like influenza viruses are responsible for antigenic drift from A/Panama/2007/99. Virology. 2005;336(1):113–119. doi: 10.1016/j.virol.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 272.Belser JA, Katz JM, Tumpey TM. The ferret as a model organism to study influenza A virus infection. Dis Model Mech. 2011;4(5):575–579. doi: 10.1242/dmm.007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Fonville JM, Wilks SH, James SL, Fox A, Ventresca M, Aban M, et al. Antibody landscapes after influenza virus infection or vaccination. Science. 2014;346(6212):996–1000. doi: 10.1126/science.1256427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Xie H, Wan X-F, Ye Z, Plant EP, Zhao Y, Xu Y, et al. H3N2 mismatch of 2014-15 northern hemisphere influenza vaccines and head-to-head comparison between human and ferret antisera derived antigenic maps. Sci Rep. 2015;5:1527. doi: 10.1038/srep15279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Linderman SL, Chambers BS, Zost SJ, Parkhouse K, Li Y, Herrmann C, et al. Potential antigenic explanation for atypical H1N1 infections among middle-aged adults during the 2013-2014 influenza season. Proc Natl Acad Sci U S A. 2014;111(44):15798–15803. doi: 10.1073/pnas.1409171111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Davenport FM, Hennessy AV, Francis T., Jr Epidemiologic and immunologic significance of age distribution of antibody to antigenic variants of influenza virus. J Exp Med. 1953;98(6):641–656. doi: 10.1084/jem.98.6.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Xie H, Li L, Ye Z, Li X, Plant EP, Zoueva O, et al. Differential effects of prior influenza exposures on H3N2 cross-reactivity of human postvaccination sera. Clin Infect Dis. 2017;65(2):259–267. doi: 10.1093/cid/cix269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Cai Z, Zhang T, Wan XF. A computational framework for influenza antigenic cartography. PLoS Comput Biol. 2010;6(10):e1000949. doi: 10.1371/journal.pcbi.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Barnett JL, Yang J, Cai Z, Zhang T, Wan XF. AntigenMap 3D: an online antigenic cartography resource. Bioinformatics. 2012;28(9):1292–1293. doi: 10.1093/bioinformatics/bts105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Cai Z, Zhang T, Wan XF. Antigenic distance measurements for seasonal influenza vaccine selection. Vaccine. 2012;30(2):448–453. doi: 10.1016/j.vaccine.2011.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Gambaryan AS, Robertson JS, Matrosovich MN. Effects of egg-adaptation on the receptor-binding properties of human influenza A and B viruses. Virology. 1999;258(2):232–239. doi: 10.1006/viro.1999.9732. [DOI] [PubMed] [Google Scholar]
- 282.Gambaryan AS, Tuzikov AB, Piskarev VE, Yamnikova SS, Lvov DK, Robertson JS, et al. Specification of receptor-binding phenotypes of influenza virus isolates from different hosts using synthetic sialylglycopolymers: non-egg-adapted human H1 and H3 influenza A and influenza B viruses share a common high binding affinity for 6'-sialyl(N-acetyllactosamine) Virology. 1997;232(2):345–350. doi: 10.1006/viro.1997.8572. [DOI] [PubMed] [Google Scholar]
- 283.Gambaryan AS, Marinina VP, Tuzikov AB, Bovin NV, Rudneva IA, Sinitsyn BV, et al. Effects of host-dependent glycosylation of hemagglutinin on receptor-binding properties on H1N1 human influenza A virus grown in MDCK cells and in embryonated eggs. Virology. 1998;247(2):170–177. doi: 10.1006/viro.1998.9224. [DOI] [PubMed] [Google Scholar]
- 284.Govorkova EA, Matrosovich MN, Tuzikov AB, Bovin NV, Gerdil C, Fanget B, et al. Selection of receptor-binding variants of human influenza A and B viruses in baby hamster kidney cells. Virology. 1999;262(1):31–38. doi: 10.1006/viro.1999.9892. [DOI] [PubMed] [Google Scholar]
- 285.Tang CY, Segovia K, McElroy JA, Li T, Guan M, Zhang X, et al. Cell-adapted mutations and antigenic diversity of influenza B viruses in Missouri, 2019-2020 Season. Viruses. 2021;13(10). [DOI] [PMC free article] [PubMed]
- 286.Katz JM, Naeve CW, Webster RG. Host cell-mediated variation in H3N2 influenza viruses. Virology. 1987;156(2):386–395. doi: 10.1016/0042-6822(87)90418-1. [DOI] [PubMed] [Google Scholar]
- 287.Rocha EP, Xu X, Hall HE, Allen JR, Regnery HL, Cox NJ. Comparison of 10 influenza A (H1N1 and H3N2) haemagglutinin sequences obtained directly from clinical specimens to those of MDCK cell- and egg-grown viruses. J Gen Virol. 1993;74(Pt 11):2513–2518. doi: 10.1099/0022-1317-74-11-2513. [DOI] [PubMed] [Google Scholar]
- 288.Rogers GN, Paulson JC, Daniels RS, Skehel JJ, Wilson IA, Wiley DC. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983;304(5921):76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 289.Williams SP, Robertson JS. Analysis of the restriction to the growth of nonegg-adapted human influenza virus in eggs. Virology. 1993;196(2):660–665. doi: 10.1006/viro.1993.1522. [DOI] [PubMed] [Google Scholar]
- 290.Robertson JS, Bootman JS, Newman R, Oxford JS, Daniels RS, Webster RG, et al. Structural changes in the haemagglutinin which accompany egg adaptation of an influenza A(H1N1) virus. Virology. 1987;160(1):31–37. doi: 10.1016/0042-6822(87)90040-7. [DOI] [PubMed] [Google Scholar]
- 291.Rogers GN, Pritchett TJ, Lane JL, Paulson JC. Differential sensitivity of human, avian, and equine influenza A viruses to a glycoprotein inhibitor of infection: selection of receptor specific variants. Virology. 1983;131(2):394–408. doi: 10.1016/0042-6822(83)90507-X. [DOI] [PubMed] [Google Scholar]
- 292.Rogers GN, Daniels RS, Skehel JJ, Wiley DC, Wang XF, Higa HH, et al. Host-mediated selection of influenza virus receptor variants. Sialic acid-alpha 2,6Gal-specific clones of A/duck/Ukraine/1/63 revert to sialic acid-alpha 2,3Gal-specific wild type in ovo. J Biol Chem. 1985;260(12):7362–7367. doi: 10.1016/S0021-9258(17)39617-5. [DOI] [PubMed] [Google Scholar]
- 293.Choppin PW, Tamm I. Two kinds of particles with contrasting properties in influenza A virus strains from the 1957 pandemic. Virology. 1959;8:539–542. doi: 10.1016/0042-6822(59)90059-5. [DOI] [PubMed] [Google Scholar]
- 294.Choppin PW, Tamm I. Studies of two kinds of virus particles which comprise influenza A2 virus strains : I. Characterization of stable homogeneous substrains in reactions with specific antibody, mucoprotein inhibitors, and erythrocytes. J Exp Med. 1960;112(5):895–920. doi: 10.1084/jem.112.5.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Rogers GN, Paulson JC. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology. 1983;127(2):361–373. doi: 10.1016/0042-6822(83)90150-2. [DOI] [PubMed] [Google Scholar]
- 296.Thompson AJ, Paulson JC. Adaptation of influenza viruses to human airway receptors. J Biol Chem. 2020;296:100017. doi: 10.1074/jbc.REV120.013309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Hensley SE, Das SR, Bailey AL, Schmidt LM, Hickman HD, Jayaraman A, et al. Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science. 2009;326(5953):734–736. doi: 10.1126/science.1178258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Allen JD, Ross TM. H3N2 influenza viruses in humans: viral mechanisms, evolution, and evaluation. Hum Vacc Immunother. 2018;14(8):1840–1847. doi: 10.1080/21645515.2018.1462639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Zhu X, McBride R, Nycholat CM, Yu W, Paulson JC, Wilson IA. Influenza virus neuraminidases with reduced enzymatic activity that avidly bind sialic acid receptors. J Virol. 2012;86(24):13371–13383. doi: 10.1128/JVI.01426-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Asaoka N, Tanaka Y, Sakai T, Fujii Y, Ohuchi R, Ohuchi M. Low growth ability of recent influenza clinical isolates in MDCK cells is due to their low receptor binding affinities. Microbes Infect. 2006;8(2):511–519. doi: 10.1016/j.micinf.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 301.Wu NC, Zost SJ, Thompson AJ, Oyen D, Nycholat CM, McBride R, et al. A structural explanation for the low effectiveness of the seasonal influenza H3N2 vaccine. PLoS Pathog. 2017;13(10):e1006682. doi: 10.1371/journal.ppat.1006682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Martin BE, Jia K, Sun H, Ye J, Hall C, Ware D, et al. Detection of influenza antigenic variants directly from clinical samples using polyclonal antibody based proximity ligation assays. Virology. 2015;476:151–158. doi: 10.1016/j.virol.2014.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Ip DK, Lau LL, Leung NH, Fang VJ, Chan KH, Chu DK, et al. Viral shedding and transmission potential of asymptomatic and paucisymptomatic influenza virus infections in the community. Clin Infect Dis. 2017;64(6):736–742. doi: 10.1093/cid/ciw841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Hancioglu B, Swigon D, Clermont G. A dynamical model of human immune response to influenza A virus infection. J Theor Biol. 2007;246(1):70–86. doi: 10.1016/j.jtbi.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 305.Hopken MW, Piaggio AJ, Pabilonia KL, Pierce J, Anderson T, Abdo Z. Predicting whole genome sequencing success for archived avian influenza virus (Orthomyxoviridae) samples using real-time and droplet PCRs. J Virol Methods. 2020;276:113777. doi: 10.1016/j.jviromet.2019.113777. [DOI] [PubMed] [Google Scholar]
- 306.Houlihan CF, Frampton D, Ferns RB, Raffle J, Grant P, Reidy M, et al. Use of whole-genome sequencing in the investigation of a nosocomial influenza virus outbreak. J Infect Dis. 2018;218(9):1485–1489. doi: 10.1093/infdis/jiy335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Martin BE, Bowman AS, Li L, Nolting JM, Smith DR, Hanson LA, et al. Detection of antigenic variants of swine subtype H3 influenza A viruses from clinical samples. J Clin Microbiol. 2017. [DOI] [PMC free article] [PubMed]
- 308.Schlingemann J, Leijon M, Yacoub A, Schlingemann H, Zohari S, Matyi-Toth A, et al. Novel means of viral antigen identification: improved detection of avian influenza viruses by proximity ligation. J Virol Methods. 2010;163(1):116–122. doi: 10.1016/j.jviromet.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 309.Fredriksson S, Dixon W, Ji H, Koong AC, Mindrinos M, Davis RW. Multiplexed protein detection by proximity ligation for cancer biomarker validation. Nat Methods. 2007;4(4):327–329. doi: 10.1038/nmeth1020. [DOI] [PubMed] [Google Scholar]
- 310.Wilson IA, Cox NJ. Structural basis of immune recognition of influenza virus hemagglutinin. Annu Rev Immunol. 1990;8:737–771. doi: 10.1146/annurev.iy.08.040190.003513. [DOI] [PubMed] [Google Scholar]
- 311.Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE, Jr, Wilson IA. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328(5976):357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Shih AC, Hsiao TC, Ho MS, Li WH. Simultaneous amino acid substitutions at antigenic sites drive influenza A hemagglutinin evolution. Proc Natl Acad Sci U S A. 2007;104(15):6283–6288. doi: 10.1073/pnas.0701396104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Zhou R, Das P, Royyuru AK. Single mutation induced H3N2 hemagglutinin antibody neutralization: a free energy perturbation study. J Phys Chem B. 2008;112(49):15813–15820. doi: 10.1021/jp805529z. [DOI] [PubMed] [Google Scholar]
- 314.Ansaldi F, Icardi G, Gasparini R, Campello C, Puzelli S, Bella A, et al. New A/H3N2 influenza variant: a small genetic evolution but a heavy burden on the Italian population during the 2004-2005 season. J Clin Microbiol. 2005;43(6):3027–3029. doi: 10.1128/JCM.43.6.3027-3029.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Durviaux S, Treanor J, Beran J, Duval X, Esen M, Feldman G, et al. Genetic and antigenic typing of seasonal influenza virus breakthrough cases from a 2008-2009 vaccine efficacy trial. Clin Vacc Immunol. 2014;21(3):271–279. doi: 10.1128/CVI.00544-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Steinbruck L, McHardy AC. Inference of genotype-phenotype relationships in the antigenic evolution of human influenza A (H3N2) viruses. PLoS Comput Biol. 2012;8(4):e1002492. doi: 10.1371/journal.pcbi.1002492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Lee MS, Chen JS. Predicting antigenic variants of influenza A/H3N2 viruses. Emerg Infect Dis. 2004;10(8):1385–1390. doi: 10.3201/eid1008.040107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Liao YC, Lee MS, Ko CY, Hsiung CA. Bioinformatics models for predicting antigenic variants of influenza A/H3N2 virus. Bioinformatics. 2008;24(4):505–512. doi: 10.1093/bioinformatics/btm638. [DOI] [PubMed] [Google Scholar]
- 319.Huang JW, King CC, Yang JM. Co-evolution positions and rules for antigenic variants of human influenza A/H3N2 viruses. BMC Bioinformatics. 2009;10(Suppl 1):S41. doi: 10.1186/1471-2105-10-S1-S41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Zhang T. Adaptive forward-backward greedy algorithm for sparse learning with linear models. Proceedings of the Twenty-Second Annual Conference on Neural Information Processing Systems (NIPS), Vancouver, British Columbia, Canada, December 8-11, 20082008. p. 1921-8.
- 321.Dempster AP. Covariance selection. Biometrics. 1972;28:157–175. doi: 10.2307/2528966. [DOI] [Google Scholar]
- 322.Cai Z, Ducatez MF, Yang J, Zhang T, Long LP, Boon AC, et al. Identifying antigenicity-associated sites in highly pathogenic H5N1 influenza virus hemagglutinin by using sparse learning. J Mol Biol. 2012;422(1):145–155. doi: 10.1016/j.jmb.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Sun H, Yang J, Zhang T, Long LP, Jia K, Yang G, et al. Inferring influenza virus antigenicity using sequence data. mBio. 2013;4:4. doi: 10.1128/mBio.00230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Yang J, Zhang T, Wan XF. Sequence-based antigenic change prediction by a sparse learning method incorporating co-evolutionary information. PLoS One. 2014;9(9):e106660. doi: 10.1371/journal.pone.0106660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Han L, Zhang Y, Wan X-F, Zhang T, editors. Generalized hierarchical sparse model for arbitrary-order interactive antigenic sites identification in flu virus data. Proceedings of the 22nd ACM SIGKDD Conference on Knowledge Discovery and Data Mining (KDD); 2016; San Francisco. [DOI] [PMC free article] [PubMed]
- 326.Neher RA, Bedford T, Daniels RS, Russell CA, Shraiman BI. Prediction, dynamics, and visualization of antigenic phenotypes of seasonal influenza viruses. Proc Natl Acad Sci U S A. 2016;113(12):E1701–E1709. doi: 10.1073/pnas.1525578113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Suzuki Y. Predictability of antigenic evolution for H3N2 human influenza A virus. Genes Genet Syst. 2013;88(4):225–232. doi: 10.1266/ggs.88.225. [DOI] [PubMed] [Google Scholar]
- 328.Cui H, Wei X, Huang Y, Hu B, Fang Y, Wang J. Using multiple linear regression and physicochemical changes of amino acid mutations to predict antigenic variants of influenza A/H3N2 viruses. Biomed Mater Eng. 2014;24(6):3729–3735. doi: 10.3233/BME-141201. [DOI] [PubMed] [Google Scholar]
- 329.Rahman T, Mahapatra M, Laing E, Jin Y. Evolutionary non-linear modelling for selecting vaccines against antigenically variable viruses. Bioinformatics. 2015;31(6):834–840. doi: 10.1093/bioinformatics/btu768. [DOI] [PubMed] [Google Scholar]
- 330.Ren X, Li Y, Liu X, Shen X, Gao W, Li J. Computational identification of antigenicity-associated sites in the hemagglutinin protein of A/H1N1 seasonal influenza virus. PLoS One. 2015;10(5):e0126742. doi: 10.1371/journal.pone.0126742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Davies V, Harvey WT, Reeve R, Husmeier D. Improving the identification of antigenic sites in the H1N1 influenza virus through accounting for the experimental structure in a sparse hierarchical Bayesian model. J R Stat Soc: Ser C: Appl Stat. 2019;68(4):859–885. doi: 10.1111/rssc.12338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Bedford T, Suchard MA, Lemey P, Dudas G, Gregory V, Hay AJ, et al. Integrating influenza antigenic dynamics with molecular evolution. eLife. 2014;3:e01914. doi: 10.7554/eLife.01914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Zhou X, Yin R, Kwoh CK, Zheng J. A context-free encoding scheme of protein sequences for predicting antigenicity of diverse influenza A viruses. BMC Genomics. 2018;19(Suppl 10):936. doi: 10.1186/s12864-018-5282-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Yao Y, Li X, Liao B, Huang L, He P, Wang F, et al. Predicting influenza antigenicity from hemagglutintin sequence data based on a joint random forest method. Sci Rep. 2017;7(1):1545. doi: 10.1038/s41598-017-01699-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Yin R, Tran VH, Zhou X, Zheng J, Kwoh CK. Predicting antigenic variants of H1N1 influenza virus based on epidemics and pandemics using a stacking model. PLoS One. 2018;13(12):e0207777. doi: 10.1371/journal.pone.0207777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Du X, Dong L, Lan Y, Peng Y, Wu A, Zhang Y, et al. Mapping of H3N2 influenza antigenic evolution in China reveals a strategy for vaccine strain recommendation. Nat Commun. 2012;3:709. doi: 10.1038/ncomms1710. [DOI] [PubMed] [Google Scholar]
- 337.Peng Y, Wang D, Wang J, Li K, Tan Z, Shu Y, et al. A universal computational model for predicting antigenic variants of influenza A virus based on conserved antigenic structures. Sci Rep. 2017;7:42051. doi: 10.1038/srep42051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Li H, Peng Y, Zou Y, Huang Z, Wu A, Li K, et al. PREDAC-H5: a user-friendly tool for the automated surveillance of antigenic variants for the HPAI H5N1 virus. Infect Genet Evol. 2014;28:62–63. doi: 10.1016/j.meegid.2014.08.030. [DOI] [PubMed] [Google Scholar]
- 339.Liu M, Zhao X, Hua S, Du X, Peng Y, Li X, et al. Antigenic patterns and evolution of the human influenza A (H1N1) virus. Sci Rep. 2015;5:14171. doi: 10.1038/srep14171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Qiu J, Qiu T, Yang Y, Wu D, Cao Z. Incorporating structure context of HA protein to improve antigenicity calculation for influenza virus A/H3N2. Sci Rep. 2016;6:31156. doi: 10.1038/srep31156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Qiu T, Yang Y, Qiu J, Huang Y, Xu T, Xiao H, et al. CE-BLAST makes it possible to compute antigenic similarity for newly emerging pathogens. Nat Commun. 2018;9(1):1772. doi: 10.1038/s41467-018-04171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Chang P, Sealy JE, Sadeyen JR, Iqbal M. Amino acid residue 217 in the hemagglutinin glycoprotein is a key mediator of avian influenza H7N9 virus antigenicity. J Virol. 2018;93(1):e01627–e01618. doi: 10.1128/JVI.01627-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Long J, Bushnell RV, Tobin JK, Pan K, Deem MW, Nara PL, et al. Evolution of H3N2 influenza virus in a guinea pig model. PLoS One. 2011;6(7):e20130. doi: 10.1371/journal.pone.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.DeDiego ML, Anderson CS, Yang H, Holden-Wiltse J, Fitzgerald T, Treanor JJ, et al. Directed selection of influenza virus produces antigenic variants that match circulating human virus isolates and escape from vaccine-mediated immune protection. Immunology. 2016;148(2):160–173. doi: 10.1111/imm.12594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Hoper D, Kalthoff D, Hoffmann B, Beer M. Highly pathogenic avian influenza virus subtype H5N1 escaping neutralization: more than HA variation. J Virol. 2012;86(3):1394–1404. doi: 10.1128/JVI.00797-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Li C, Hatta M, Burke DF, Ping J, Zhang Y, Ozawa M, et al. Selection of antigenically advanced variants of seasonal influenza viruses. Nat Microbiol. 2016;1(6):16058. doi: 10.1038/nmicrobiol.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Lee JM, Huddleston J, Doud MB, Hooper KA, Wu NC, Bedford T, et al. Deep mutational scanning of hemagglutinin helps predict evolutionary fates of human H3N2 influenza variants. Proc Natl Acad Sci U S A. 2018;115(35):E8276–E8E85. doi: 10.1073/pnas.1806133115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Dinis JM, Florek NW, Fatola OO, Moncla LH, Mutschler JP, Charlier OK, et al. Deep sequencing reveals potential antigenic variants at low frequencies in influenza A virus-infected humans. J Virol. 2016;90(7):3355–3365. doi: 10.1128/JVI.03248-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Luksza M, Lassig M. A predictive fitness model for influenza. Nature. 2014;507(7490):57–61. doi: 10.1038/nature13087. [DOI] [PubMed] [Google Scholar]
- 350.Bush RM, Bender CA, Subbarao K, Cox NJ, Fitch WM. Predicting the evolution of human influenza A. Science. 1999;286(5446):1921–1925. doi: 10.1126/science.286.5446.1921. [DOI] [PubMed] [Google Scholar]
- 351.Steinbruck L, McHardy AC. Allele dynamics plots for the study of evolutionary dynamics in viral populations. Nucleic Acids Res. 2011;39(1):e4. doi: 10.1093/nar/gkq909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Steinbruck L, Klingen TR, McHardy AC. Computational prediction of vaccine strains for human influenza A (H3N2) viruses. J Virol. 2014;88(20):12123–12132. doi: 10.1128/JVI.01861-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Klingen TR, Reimering S, Loers J, Mooren K, Klawonn F, Krey T, et al. Sweep Dynamics (SD) plots: computational identification of selective sweeps to monitor the adaptation of influenza A viruses. Sci Rep. 2018;8(1):373. doi: 10.1038/s41598-017-18791-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Neher RA, Russell CA, Shraiman BI. Predicting evolution from the shape of genealogical trees. eLife. 2014;3. [DOI] [PMC free article] [PubMed]
- 355.Castro LA, Bedford T, Ancel ML. Early prediction of antigenic transitions for influenza A/H3N2. PLoS Comput Biol. 2020;16(2):e1007683. doi: 10.1371/journal.pcbi.1007683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Chang D, Zaia J. Why glycosylation matters in building a better flu vaccine. Mol Cell Proteomics. 2019;18(12):2348–2358. doi: 10.1074/mcp.R119.001491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Cruz E, Cain J, Crossett B, Kayser V. Title: site-specific glycosylation profile of influenza A (H1N1) hemagglutinin through tandem mass spectrometry. Hum Vacc Immunother 2017:0. [DOI] [PMC free article] [PubMed]
- 358.She YM, Farnsworth A, Li X, Cyr TD. Topological N-glycosylation and site-specific N-glycan sulfation of influenza proteins in the highly expressed H1N1 candidate vaccines. Sci Rep. 2017;7(1):10232. doi: 10.1038/s41598-017-10714-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.An Y, Rininger JA, Jarvis DL, Jing X, Ye Z, Aumiller JJ, et al. Comparative glycomics analysis of influenza hemagglutinin (H5N1) produced in vaccine relevant cell platforms. J Proteome Res. 2013;12(8):3707–3720. doi: 10.1021/pr400329k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Xie H, Doneanu C, Chen W, Rininger J, Mazzeo JR. Characterization of a recombinant influenza vaccine candidate using complementary LC-MS methods. Curr Pharm Biotechnol. 2011;12(10):1568–1579. doi: 10.2174/138920111798357447. [DOI] [PubMed] [Google Scholar]
- 361.An Y, Cipollo JF. An unbiased approach for analysis of protein glycosylation and application to influenza vaccine hemagglutinin. Anal Biochem. 2011;415(1):67–80. doi: 10.1016/j.ab.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 362.Blake TA, Williams TL, Pirkle JL, Barr JR. Targeted N-linked glycosylation analysis of H5N1 influenza hemagglutinin by selective sample preparation and liquid chromatography/tandem mass spectrometry. Anal Chem. 2009;81(8):3109–3118. doi: 10.1021/ac900095h. [DOI] [PubMed] [Google Scholar]
- 363.Schwarzer J, Rapp E, Reichl U. N-glycan analysis by CGE-LIF: profiling influenza A virus hemagglutinin N-glycosylation during vaccine production. Electrophoresis. 2008;29(20):4203–4214. doi: 10.1002/elps.200800042. [DOI] [PubMed] [Google Scholar]
- 364.Chang D, Hackett WE, Zhong L, Wan XF, Zaia J. Measuring site-specific glycosylation similarity between influenza A virus variants with statistical certainty. Mol Cell Proteomics. 2020;19(9):1533–1545. doi: 10.1074/mcp.RA120.002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.González JM, Gomez-Puertas P, Cavanagh D, Gorbalenya AE, Enjuanes L. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch Virol. 2003;148(11):2207–2235. doi: 10.1007/s00705-003-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. New York: Springer; 2015. pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Corman VM, Muth D, Niemeyer D, Drosten C. Hosts and sources of endemic human coronaviruses. Elsevier. 2018:163–88. [DOI] [PMC free article] [PubMed]
- 368.Asrani P, Hasan GM, Sohal SS, Hassan MI. Molecular basis of pathogenesis of coronaviruses: a comparative genomics approach to planetary health to prevent zoonotic outbreaks in the 21st Century. OMICS: A Journal of. Integr Biol. 2020;24(11):634–644. doi: 10.1089/omi.2020.0131. [DOI] [PubMed] [Google Scholar]
- 369.Saksena N, Bonam SR, Miranda-Saksena M. Epigenetic lens to visualize the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection in COVID-19 pandemic. Front Genet. 2021;12. [DOI] [PMC free article] [PubMed]
- 370.Triggle CR, Bansal D, Ding H, Islam MM, Farag EABA, Hadi HA, et al. A comprehensive review of viral characteristics, transmission, pathophysiology, immune response, and management of SARS-CoV-2 and COVID-19 as a basis for controlling the pandemic. Front Immunol. 2021;12. [DOI] [PMC free article] [PubMed]
- 371.Saif LJ. Animal coronaviruses: what can they teach us about the severe acute respiratory syndrome? Rev Sci Tech. (2). [DOI] [PubMed]
- 372.Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Peiris JSM, Guan Y, Yuen KY. Severe acute respiratory syndrome. Nat Med. 2004;10(S12):S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Imai Y, Kuba K, Rao S, Huan Y, Guo F, Guan B, et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436(7047):112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.De Wit E, Van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Gussow AB, Auslander N, Faure G, Wolf YI, Zhang F, Koonin EV. Genomic determinants of pathogenicity in SARS-CoV-2 and other human coronaviruses. Proc Natl Acad Sci. 2020;117(26):15193–15199. doi: 10.1073/pnas.2008176117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Khailany RA, Safdar M, Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Reports. 2020;19:100682. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Organization WH. WHO Coronavirus (COVID-19) Dashboard [updated June 4, 2021. Available from: https://covid19.who.int/.
- 380.Phan T. Novel coronavirus: from discovery to clinical diagnostics. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2020;79:104211-. [DOI] [PMC free article] [PubMed]
- 381.Liu P, Jiang JZ, Wan XF, Hua Y, Li L, Zhou J, et al. Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog. 2020;16(5):e1008421. doi: 10.1371/journal.ppat.1008421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Xu X, Chen P, Wang J, Feng J, Zhou H, Li X, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63(3):457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–92.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Yu IT, Li Y, Wong TW, Tam W, Chan AT, Lee JH, et al. Evidence of airborne transmission of the severe acute respiratory syndrome virus. N Engl J Med. 2004;350(17):1731–1739. doi: 10.1056/NEJMoa032867. [DOI] [PubMed] [Google Scholar]
- 385.Harrison AG, Lin T, Wang P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 2020;41(12):1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Brian DA, Baric RS. Coronavirus genome structure and replication. Berlin Heidelberg: Springer; 2005. pp. 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Xia H, Cao Z, Xie X, Zhang X, Chen JY-C, Wang H, et al. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33(1):108234. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Li J-Y, Liao C-H, Wang Q, Tan Y-J, Luo R, Qiu Y, et al. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286:198074. doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Kumar S, Maurya VK, Prasad AK, Bhatt MLB, Saxena SK. Structural, glycosylation and antigenic variation between 2019 novel coronavirus (2019-nCoV) and SARS coronavirus (SARS-CoV) Virusdisease. 2020;31(1):13–21. doi: 10.1007/s13337-020-00571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 391.Zost SJ, Gilchuk P, Chen RE, Case JB, Reidy JX, Trivette A, et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat Med. 2020;26(9):1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Wu Y, Wang F, Shen C, Peng W, Li D, Zhao C, et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science (New York, NY) 2020;368(6496):1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29(1):44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4):894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Angeletti S, Benvenuto D, Bianchi M, Giovanetti M, Pascarella S, Ciccozzi M. COVID-2019: the role of the nsp2 and nsp3 in its pathogenesis. J Med Virol. 2020;92(6):584–588. doi: 10.1002/jmv.25719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JC, et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9. [DOI] [PMC free article] [PubMed]
- 397.Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592(7852):116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–27.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Müller NF, Wagner C, Frazar CD, Roychoudhury P, Lee J, Moncla LH, et al. Viral genomes reveal patterns of the SARS-CoV-2 outbreak in Washington State. Sci Transl Med. 2021. [DOI] [PMC free article] [PubMed]
- 400.Zhou R, Li F, Chen F, Liu H, Zheng J, Lei C, et al. Viral dynamics in asymptomatic patients with COVID-19. Int J Infect Dis. 2020;96:288–290. doi: 10.1016/j.ijid.2020.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Tang JW, Tambyah PA, Hui DS. Emergence of a new SARS-CoV-2 variant in the UK. J Inf Secur. 2021;82(4):e27–ee8. doi: 10.1016/j.jinf.2020.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Zhao S, Lou J, Cao L, Zheng H, Chong MKC, Chen Z, et al. Quantifying the transmission advantage associated with N501Y substitution of SARS-CoV-2 in the UK: an early data-driven analysis. J Travel Med. 2021;28(2). [DOI] [PMC free article] [PubMed]
- 403.Leung K, Shum MH, Leung GM, Lam TT, Wu JT. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro surveillance : bulletin Europeen sur les maladies transmissibles = Eur Commun Dis Bull. 2021;26(1). [DOI] [PMC free article] [PubMed]
- 404.Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020:2020.12.21.20248640.
- 405.Dos Santos CA, Bezerra GVB, Azevedo Marinho ARRA, Alves JC, Tanajura DM, Martins-Filho PR. SARS-CoV-2 genomic surveillance in Northeast Brazil: timing of emergence of the Brazilian variant of concern P1. J Travel Med 2021:taab066. [DOI] [PMC free article] [PubMed]
- 406.Chen RE, Zhang X, Case JB, Winkler ES, Liu Y, VanBlargan LA, et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat Med. 2021;27(4):717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Edara V-V, Lai L, Sahoo MK, Floyd K, Sibai M, Solis D, et al. Infection and vaccine-induced neutralizing antibody responses to the SARS-CoV-2 B.1.617.1 variant. 2021:2021.05.09.443299. [DOI] [PMC free article] [PubMed]
- 408.Yadav PD, Sapkal GN, Abraham P, Ella R, Deshpande G, Patil DY, et al. Neutralization of variant under investigation B.1.617 with sera of BBV152 vaccinees. 2021:2021.04.23.441101. [DOI] [PubMed]
- 409.Lau SY, Wang P, Mok BW, Zhang AJ, Chu H, Lee AC, et al. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg Microbes Infect. 2020;9(1):837–842. doi: 10.1080/22221751.2020.1756700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Espejo AP, Akgun Y, Al Mana AF, Tjendra Y, Millan NC, Gomez-Fernandez C, et al. Review of current advances in serologic testing for COVID-19. Am J Clin Pathol. 2020;154(3):293–304. doi: 10.1093/ajcp/aqaa112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Zhang L, Guo H. Biomarkers of COVID-19 and technologies to combat SARS-CoV-2. Adv Biomark Sci Technol. 2020;2:1–23. doi: 10.1016/j.abst.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Bewley KR, Coombes NS, Gagnon L, McInroy L, Baker N, Shaik I, et al. Quantification of SARS-CoV-2 neutralizing antibody by wild-type plaque reduction neutralization, microneutralization and pseudotyped virus neutralization assays. Nat Protoc. 2021;16(6):3114–3140. doi: 10.1038/s41596-021-00536-y. [DOI] [PubMed] [Google Scholar]
- 413.Amanat F, White KM, Miorin L, Strohmeier S, McMahon M, Meade P, et al. An in vitro microneutralization assay for SARS-CoV-2 serology and drug screening. Curr Protoc Microbiol. 2020;58(1):e108. doi: 10.1002/cpmc.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Manenti A, Maggetti M, Casa E, Martinuzzi D, Torelli A, Trombetta CM, et al. Evaluation of SARS-CoV-2 neutralizing antibodies using a CPE-based colorimetric live virus micro-neutralization assay in human serum samples. J Med Virol. 2020;92(10):2096–2104. doi: 10.1002/jmv.25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Nie J, Li Q, Wu J, Zhao C, Hao H, Liu H, et al. Quantification of SARS-CoV-2 neutralizing antibody by a pseudotyped virus-based assay. Nat Protoc. 2020;15(11):3699–3715. doi: 10.1038/s41596-020-0394-5. [DOI] [PubMed] [Google Scholar]
- 416.Pinto D, Park Y-J, Beltramello M, Walls AC, Tortorici MA, Bianchi S, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583(7815):290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 417.Khurana A, Allawadhi P, Khurana I, Allwadhi S, Weiskirchen R, Banothu AK, et al. Role of nanotechnology behind the success of mRNA vaccines for COVID-19. Nano Today. 2021;38:101142. doi: 10.1016/j.nantod.2021.101142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11(1):1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Hu J, Gao Q, He C, Huang A, Tang N, Wang K. Development of cell-based pseudovirus entry assay to identify potential viral entry inhibitors and neutralizing antibodies against SARS-CoV-2. Genes Dis. 2020;7(4):551–557. doi: 10.1016/j.gendis.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–80.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Case JB, Rothlauf PW, Chen RE, Liu Z, Zhao H, Kim AS, et al. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. bioRxiv : the preprint server for biology. 2020. [DOI] [PMC free article] [PubMed]
- 422.Wan XF, Tang CY, Ritter D, Wang Y, Li T, Segovia K, et al. SARS-CoV-2 show no infectivity at later stages in a prolonged COVID-19 patient despite positivity in RNA testing. J Med Virol. 2021;93(7):4570–4575. doi: 10.1002/jmv.27001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Muruato AE, Fontes-Garfias CR, Ren P, Garcia-Blanco MA, Menachery VD, Xie X, et al. A high-throughput neutralizing antibody assay for COVID-19 diagnosis and vaccine evaluation. Nat Commun. 2020;11(1):4059. doi: 10.1038/s41467-020-17892-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Abe KT, Li Z, Samson R, Samavarchi-Tehrani P, Valcourt EJ, Wood H, et al. A simple protein-based surrogate neutralization assay for SARS-CoV-2. JCI Insight. 2020;5(19). [DOI] [PMC free article] [PubMed]
- 425.Tan CW, Chia WN, Qin X, Liu P, Chen MI, Tiu C, et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat Biotechnol. 2020;38(9):1073–1078. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- 426.Byrnes JR, Zhou XX, Lui I, Elledge SK, Glasgow JE, Lim SA, et al. Competitive SARS-CoV-2 serology reveals most antibodies targeting the spike receptor-binding domain compete for ACE2 binding. mSphere. 2020;5(5). [DOI] [PMC free article] [PubMed]
- 427.Bosnjak B, Stein SC, Willenzon S, Cordes AK, Puppe W, Bernhardt G, et al. Low serum neutralizing anti-SARS-CoV-2 S antibody levels in mildly affected COVID-19 convalescent patients revealed by two different detection methods. Cell Mol Immunol. 2021;18(4):936–944. doi: 10.1038/s41423-020-00573-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Lu Y, Wang J, Li Q, Hu H, Lu J, Chen ZJSJoI. Advances in neutralization assays for SARS-CoV-2. 2021;94(3):e13088.
- 429.Fenwick C, Turelli P, Pellaton C, Farina A, Campos J, Raclot C, et al. A high-throughput cell- and virus-free assay shows reduced neutralization of SARS-CoV-2 variants by COVID-19 convalescent plasma. Sci Transl Med. 2021;13(605). [DOI] [PMC free article] [PubMed]
- 430.Oguntuyo KY, Stevens CS, Hung CT, Ikegame S, Acklin JA, Kowdle SS, et al. Quantifying absolute neutralization titers against SARS-CoV-2 by a standardized virus neutralization assay allows for cross-cohort comparisons of COVID-19 sera. mBio. 2021;12(1). [DOI] [PMC free article] [PubMed]
- 431.Tsai WY, Ching LL, Hsieh SC, Melish ME, Nerurkar VR, Wang WK. A real-time and high-throughput neutralization test based on SARS-CoV-2 pseudovirus containing monomeric infrared fluorescent protein as reporter. Emerg Microbes Infect. 2021;10(1):894–904. doi: 10.1080/22221751.2021.1925163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.(CDC) CfDCaP. Recombinant influenza (flu) vaccine May 6, 2021 [Available from: https://www.cdc.gov/flu/prevent/qa_flublok-vaccine.htm.
- 433.Cox MM, Izikson R, Post P, Dunkle L. Safety, efficacy, and immunogenicity of Flublok in the prevention of seasonal influenza in adults. Ther Adv Vacc. 2015;3(4):97–108. doi: 10.1177/2051013615595595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Klinman DM, Klaschik S, Tross D, Shirota H, Steinhagen F. FDA guidance on prophylactic DNA vaccines: analysis and recommendations. Vaccine. 2010;28(16):2801–2805. doi: 10.1016/j.vaccine.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Gilkeson GS, Grudier JP, Karounos DG, Pisetsky DS. Induction of anti-double stranded DNA antibodies in normal mice by immunization with bacterial DNA. J Immunol (Baltimore, Md : 1950). 1989;142(5):1482-6. [PubMed]
- 436.Zafrir Y, Agmon-Levin N, Paz Z, Shilton T, Shoenfeld Y. Autoimmunity following hepatitis B vaccine as part of the spectrum of 'autoimmune (auto-inflammatory) syndrome induced by adjuvants' (ASIA): analysis of 93 cases. Lupus. 2012;21(2):146–152. doi: 10.1177/0961203311429318. [DOI] [PubMed] [Google Scholar]
- 437.Würtele H, Little KC, Chartrand P. Illegitimate DNA integration in mammalian cells. Gene Ther. 2003;10(21):1791–1799. doi: 10.1038/sj.gt.3302074. [DOI] [PubMed] [Google Scholar]
- 438.Forde GM. Rapid-response vaccines—does DNA offer a solution? Nat Biotechnol. 2005;23(9):1059–1062. doi: 10.1038/nbt0905-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Lee J, Arun Kumar S, Jhan YY, Bishop CJ. Engineering DNA vaccines against infectious diseases. Acta Biomater. 2018;80:31–47. doi: 10.1016/j.actbio.2018.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Klinman DM, Takeno M, Ichino M, Gu M, Yamshchikov G, Mor G, et al. DNA vaccines: safety and efficacy issues. Springer Semin Immunopathol. 1997;19(2):245–256. doi: 10.1007/BF00870272. [DOI] [PubMed] [Google Scholar]
- 441.Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, et al. Direct gene transfer into mouse muscle in vivo. Science. 1990;247(4949):1465. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 442.Petsch B, Schnee M, Vogel AB, Lange E, Hoffmann B, Voss D, et al. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat Biotechnol. 2012;30(12):1210–1216. doi: 10.1038/nbt.2436. [DOI] [PubMed] [Google Scholar]
- 443.Bahl K, Senn JJ, Yuzhakov O, Bulychev A, Brito LA, Hassett KJ, et al. Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Mol Ther. 2017;25(6):1316–1327. doi: 10.1016/j.ymthe.2017.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Pardi N, Hogan MJ, Pelc RS, Muramatsu H, Andersen H, DeMaso CR, et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543(7644):248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Chahal JS, Fang T, Woodham AW, Khan OF, Ling J, Anderson DG, et al. An RNA nanoparticle vaccine against Zika virus elicits antibody and CD8+ T cell responses in a mouse model. Sci Rep. 2017;7(1):252. doi: 10.1038/s41598-017-00193-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Schnee M, Vogel AB, Voss D, Petsch B, Baumhof P, Kramps T, et al. An mRNA vaccine encoding rabies virus glycoprotein induces protection against lethal infection in mice and correlates of protection in adult and newborn pigs. PLoS Negl Trop Dis. 2016;10(6):e0004746-e. doi: 10.1371/journal.pntd.0004746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Vallazza B, Petri S, Poleganov MA, Eberle F, Kuhn AN, Sahin U. Recombinant messenger RNA technology and its application in cancer immunotherapy, transcript replacement therapies, pluripotent stem cell induction, and beyond. WIREs RNA. 2015;6(5):471–499. doi: 10.1002/wrna.1288. [DOI] [PubMed] [Google Scholar]
- 448.Conry RM, LoBuglio AF, Wright M, Sumerel L, Pike MJ, Johanning F, et al. Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res. 1995;55(7):1397–1400. [PubMed] [Google Scholar]
- 449.Van Lint S, Goyvaerts C, Maenhout S, Goethals L, Disy A, Benteyn D, et al. Preclinical evaluation of TriMix and antigen mRNA-based antitumor therapy. Cancer Res. 2012;72(7):1661–1671. doi: 10.1158/0008-5472.CAN-11-2957. [DOI] [PubMed] [Google Scholar]
- 450.Sahin U, Karikó K, Türeci Ö. mRNA-based therapeutics--developing a new class of drugs. Nat Rev Drug Discov. 2014;13(10):759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 451.Kauffman KJ, Webber MJ, Anderson DG. Materials for non-viral intracellular delivery of messenger RNA therapeutics. J Control Release. 2016;240:227–234. doi: 10.1016/j.jconrel.2015.12.032. [DOI] [PubMed] [Google Scholar]
- 452.Karikó K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther. 2008;16(11):1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines — a new era in vaccinology. Nat Rev Drug Discov. 2018;17(4):261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384(5):403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N Engl J Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Jackson NAC, Kester KE, Casimiro D, Gurunathan S, DeRosa F. The promise of mRNA vaccines: a biotech and industrial perspective. npj Vaccines. 2020;5(1):11. doi: 10.1038/s41541-020-0159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]