Highlights
-
•
Comprehensive technical and applicative review concerning metaproteomics of the human gut.
-
•
Software and databases are essential for handling of big data amounts.
-
•
Presentation of the contributions of metaproteomics within integrative studies.
-
•
State of the art of metaproteomic research on the human gut in the clinical field.
Abbreviations: CD, Crohn's disease; BLAST, Basic Local Alignment Search Tool; DDA/DIA, Data Dependent Acquisition / Data Independent Acquisition; FASP, Filter Aided Sample Preparation; FDR, False Discovery Rate; IBD, Inflammatory Bowel Disease; IMS/MS, Ion-Mobility Spectrometry–Mass Spectrometry; LCA, Lowest Common Ancestor; MALDI/ESI, Matrix-Assisted Laser Desorption Ionisation/Electrospray Ionization; NGS, Next Generation Sequencing; (U)HPLC, (Ultra) High Performance Liquid Chromatography; RP/SCX, Reverse Phase /Strong Cation Exchange; SDS-PAGE, Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis; TOF, Time-Of-Flight
Keywords: Metaproteomics, Human gut microbiota, OMICS, Liquid chromatography, Mass spectrometry, Unipept
Abstract
Our digestive tract hosts more than a billion microorganisms comprising non-pathogenic bacteria, viruses, fungi and parasites. Understanding and characterizing the human gut microbiota has become a fundamental common theme to establish a link between its dysbiosis and certain pathologies, especially autoimmune and inflammatory diseases. Meta-Omics studies have, so far, provided great progress in this field. Genomics is conventionally used to determine the composition of the microbiota and, subsequently, metatranscriptomics lists the transcribed genes. However, to better understand the relationship between microbiota and health, protein-based studies are being applied. Proteomics enables the functional study of proteins as they are expressed by microbial communities. Metaproteomics exploits the power of mass spectrometry to identify broad protein profiles in complex samples, such as gut microbiota. The lastest technological advances in the field of mass spectrometry have opened the field of large-scale characterization of microbial proteins. Despite these hardware improvements, bioinformatics analysis remains a primary challenge. Herein, we describe the state-of-the-art concerning specific sample preparation and powerful shotgun analysis techniques. We also review several scientific studies of the human gut microbiota. Moreover, we discuss the advantages and limitations encountered in this research area, concerning new methods of sample preparation and innovative bioinformatic tools. Finally, prospects are addressed regarding the application of metaproteomic in the field of clinical microbiology and its integration with other meta-Omics.
1. Introduction
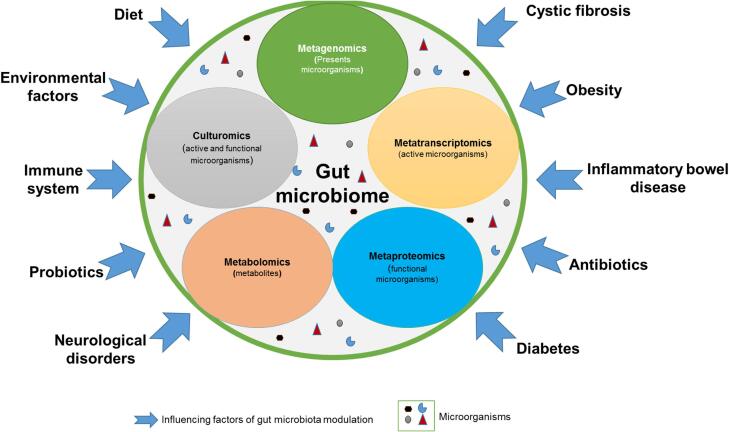
The human gut microbiota harbors complex communities of billions of microorganisms. These microorganisms, bacteria, viruses, archaea, yeasts and protozoa, are ten times more numerous than human cells [1], [2]. Under normal conditions, this complex population lives in mutual coexistence with the body and plays several fundamental roles that have a considerable impact on human health and physiology [3], [4]. Most of the microorganisms in the human microbiome are beneficial and play major metabolic and physiological roles. For example, the commensal microflora of the gut participate in the digestion of food [5], are involved in gut-brain intercommunication [6], and play an interactive role with immune system [3]. However, many factors can disturb the intestinal microbiota composition, known as dysbiosis. This microbial imbalance disrupts the microbiota composition and can lead to intestinal permeability. Alterations of the microbial ecosystem can occur due to several factors, such as environment, aging, diet and the immune system. As a result, changes in the bacterial composition of the gut microbiota have been associated with dysfunction of the digestive system, such as inflammatory bowel diseases, but also with obesity, metabolic, immune and neurological diseases and cancers [7], [8], [9], [10] (Fig. 1).
Fig. 1.
Understanding human gut microbiome: different omics that are involved and the factors influencing microbiota.
The different OMICs approaches have led to important advances in the study of the intestinal microbiome, the host and the intestinal environment. As well, next-generation sequencing (NGS) has allowed the use of genomic approaches to better understand the complex microbial environment from different biological samples. Mainly, metagenomics provides a comprehensive overview of the taxonomy and functional potential of microbial ecosystems [11], [12]. However, despite these advances, metagenomics cannot address all biological questions. The different NGS platforms used in laboratories, or the choice of bioinformatics tools, remain the main limitations [13]. Moreover, the least abundant microorganisms are statistically less likely to be detected, constituting a depth bias for high-throughput sequencing methods. In this respect, the metatranscriptomic (RNAseq) provides access to the metatranscriptome of the microbiome, allowing whole-genome profiling of the active microbial community and expressed biological signatures in the human microbiome [14]. However, bioinformatics tools for metatranscriptome data analysis are similar to those of metagenomics. Culturomics is also a culture-based omics approach that uses multiple culture conditions, MALDI-TOF mass spectrometry and 16S rRNA sequencing for the rapid identification of bacterial species [15]. Proteomics, initially defined by microbiologists as the study of all proteins expressed by a single organism, is in full emergence thanks to its application to complex bacterial communities. As a result, the analysis of the protein content of the microbial communities, such as gut microbiota is now named “metaproteomics” [16]. A metaproteomic analysis typically comprises 4 steps: 1) extraction and purification of proteins, 2) enzymatic digestion of proteins into peptides, 3) separation of peptides, usually by chromatography, followed by mass spectrometric analysis and 4) protein identification by database sequence comparison [17], [18], [19]. Metaproteomics is a rising technique but has some disadvantages related to the complexity of the sample, including both the complexity of the matrix as well as the microbial community itself. First, metaproteomes includes up to more than one thousand different species, each containing several hundred proteins, generating a myriad of peptides after digestion [20]. In addition, many peptides are common to many bacterial species or similar protein sequences, making data processing even more complex with a resultant high false-positive rate. Second, mass spectrometry generates hundreds of thousands of spectra, but the data analysis requires considerable bioinformatic effort to develop algorithms that will allow a reduction in the computational time needed. Third, one of the main elements of a successful metaproteomic study is the availability of a relevant database in order to match sequences with mass spectra. Moreover, a drawback of metaproteomics is its potential to generate numerous false positives from the use of large databases. In addition, data interpretation is recognized as a major limitation for metaproteomic analysis because huge amounts of data often result in high False Discovery Rates (FDR). Solutions are required to validate protein identifications across different MS and database search algorithms. Furthermore, metagenomics, metatranscriptomics, metabolomics and culturomics data can be integrated with metaproteomic to provide insight into the functioning of bacterial communities in the gut.
In light of these considerations, this review presents the current status of shotgun metaproteomic (bottom-up) studies applied to the human gut microbiota and highlights experimental and bioinformatics approaches, providing several examples. Finally, we address the prospects of gut metaproteomic analysis and future directions for clinical microbiology research.
2. Metaproteomics methodologies
2.1. Stool sample preparation
The study of the metaproteome of gut microbiota is primarily performed using faecal samples. However, stool comprises a complicated environmental matrix that can interfere with protein characterization studies [18]. Several challenges should be expected, such as: 1) a complex microbial composition, as faeces consist of a mix of gram-positive and gram-negative cells with various envelopes structures, 2) an abundance of host proteins, 3) the presence of proteins derived from consumed and undigested foods, 4) various physico-chemical properties of proteins involved in their solubility. Metaproteomic analysis can be altered by large inter- and intra-donor variabilities. Microbial species abundance in the gut can also vary >10 orders of magnitude across samples. The more complex and diverse the microbial community, the fewer proteins that can be identified for each taxa [21]. The performance of the metaproteomic analysis from human gut microbiota is also dependent on sample preparation [22]. Storage conditions can affect the sample, hence, strict protocol for stool storage following defecation is required, typically at −80 °C [23]. To date, numerous metaproteomic studies have achieved separation of microbial cells from feces by differential centrifugation, where insoluble material and large particles are separated at low speed, followed by pelleting microorganisms at higher centrifuge forces [24]. For example, Tanca et al. showed that stool samples previously treated by differential centrifugation revealed more proteins/peptides with a significantly higher microbial diversity than a direct conventional protein extraction step [25]. Additionally, Xiong et al developed a metaproteomic sample preparation strategy based on a double filtering (DF) differential separation step that selectively depletes human cells and proteins while enriching microbial biomass in the fecal sample [128]. The DF process constituted (1) a 20 μm vacuum filter unit to remove larger fibrous material and intact human cells, and (2) a 0.22 μm vacuum filter unit that permitted human proteins to be washed through while microbial cells were captured on the filter. This method resulted in greater than a 2-fold increase in microbial proteins that were identified and quantitated compared to the direct method whose protein extraction was performed using high speed centrifugation. Cell lysis should be adapted for gut microbiota. For instance, Gram positive bacteria, such as Firmicutes and Actinobacteria, which are two major phyla of the intestinal tract, have a thick peptidoglycan layer that is difficult to break down. Hence, a wide range of physical, mechanical and chemical methods are generally used in metaproteomic studies to disrupt cells, such as: heating, bead beating and ultrasonication with lysis detergent such as Sodium Dodecyl Sulfate (SDS) and chaotropic agents such as urea [20], [25], [26], [27], [28], [29], [30]. Many studies have reported that the use of SDS combined with mechanical disruption methods, such as bead beating or ultrasonication, provided better cell lysis yields than other buffers, in the case of gut microbial protein extraction [20], [30], [31].
Extracted proteins are then usually enzymatically digested into peptides, before or after the pre-fractionation step. The most frequently used enzyme is trypsin, because it generates many peptides, has great cleavage specificity and is easy to handle. The resulting peptides mostly have a molecular mass between 700 and 1500 Daltons, which is amenable to mass spectrometric analysis [32]. Nevertheless, other enzymes can be used alone or in combination with trypsin to enhance desired protein digestion effects [33].
After protein extraction, additional pre-concentration steps, such as filter-aided sample preparation (FASP) are often performed to obtain more concentrated peptides samples. This step allows a deeper coverage of metaproteomes [26]. Detergents and salt, which are commonly used during protein extraction, can interfere with mass spectrometric analysis, should be removed to the greatest extent possible during this step to increase analysis sensitivity [34].
In summary, differential centrifugation, enzymatic digestion (trypsin) and removal of detergents and salts play an important role during stool sample preparation for the metaproteomic characterization of the human gut microbiota. Table 1 summarizes the key metaproteomics studies and their sample preparation protocols. Despite the heterogeneity of these different sample preparation and proteins extraction protocols, many groups use SDS and ultrasonication as a general procedure.
Table 1.
Metaproteomics studies of the human gut (classified in a chronological order).
| Samples (Feces) | Sample preparation | Pre- fractionation | Mass spectrometry | Database | Search engines | Results of analysis Number of validated proteins (>n peptides) | References |
|---|---|---|---|---|---|---|---|
| Childs N = 2 | Chemical and mechanical lysis Tryptic digestion | 2D PAGE, | MALDI-TOF-MS/MS | NCBI Swissprot Uniprot KB | PDQuest | >200 spots 1 protein identified | [36] |
| Healthy monozygotic twin N = 2 | Differential centrifugation Chemical and mechanical lysis Trypsin digestion | Nano2D-LC MudPIT RPC18-SCX-RPC18 | LTQ Orbitrap; DDA | In-house database: db1 and metadb | SEQUEST | 600–900 proteins per sample and replicate (db1) 970–1340 proteins per sample and replicate (metadb) | [24] |
| Healthy adult N = 2 | Chemical lysis (urea, thiourea) Tryptic digestion | SDS-PAGE, nanoLC-RPC18 | LTQ Orbitrap; DDA | In-house database from genomics | OMSSA | 2331 and 1870 peptides 1120 and 922 peptides | [131] |
| Healthy human N = 2 | Differential centrifugation, Direct extraction Chemical and mechanical lysisTrypsin digestion | SDS-PAGE, Nano2D-LC MudPIT RPC18-SCX-RPC18 | LTQ Orbitrap; DDA | In-house database | SEQUEST PepNovo + PEAKS | 5233 proteins (2 peptides) 6186 proteins (≥1 peptide) 3706 proteins (≥2 peptides) | [44] |
| Healthy adult N = 3 | Mechanical lysis: PBS + zirconium-silica beads Tryptic digestion | SDS-PAGE Nano2D-LC RPC18 | LTQ Orbitrap; DDA | In-house databases | OMSSA | 1790 microbial proteins (>2 peptides) | [86] |
| Adolescents N = 2 1 lean (female) and 1 obese (male) | Differential Centrifugation Mechanical lysis (sonication) Tryptic digestion | SDS-PAGE nanoUPLC-C18 | LTQ Orbitrap; DDA | Matched metagenomes unmatched metagenomes | Maxquant | 613 proteins (>2 peptides) | [8] |
| Patients with CD N = 6 (4 women and 2 men) Healthy Controls N = 6 | Ultracentrifugation Chemical and mechanical lysis Tryptic digestion | 2D-DIGE; nanoLC-C18 | LTQ Orbitrap; DDA LTQ-Linear Ion Trap; DDA | MetaHit database, Human SwissProt, In-house contaminant database | X!Tandem; | 141 proteins spots 89 bacterial proteins spots | [126] |
| Child N = 1 | Chemical and mechanical lysis Tryptic digestion | nano2D-LC: SCX; RPC18 | LTQ Orbitrap; DDA | in-house database from genomics | SEQUEST | and 4031 proteins (>1 peptides) | [85] |
| Healthy volunteer N = 1 | Direct extraction(DE) Differential Centrifugation (DC) Chemical and mechanical lysis Tryptic digestion | nanoLC: RPC18 | LTQ Orbitrap; DDA | UniProtKB, SwissProt, customized host-microbiome Unipept | SEQUEST | - DE:, 3911 proteins - DC: 4587 proteins | [25] |
| Premature infants N = 2 | Direct extraction Differential filtering Chemical and mechanical lysis Tryptic digestion | nano2D-LC SCX; RPC18 | LTQ Orbitrap; DDA | Customized database | Myrimatch | 807 proteins groups (DE) 1264 proteins groups (DF) (1 unique peptide per proteins) | [128] |
| Healthy individuals N = 29 (9 normal, 4 overweigh and 16 obese) | Mechanical lysis (bead beating) Tryptic digestion | SDS-PAGE; nanoHPLC: C18 | Q-Orbitrap; DDA | In-house human intestinal metaproteome database (HIMPdb) Unipept | OMSSA X!Tandem | 91.86% human and microbial proteins 73.90% assigned to Bacteroidetes phylum (obese) | [129] |
| Children with cystic fibrosis their unaffected siblings N = 30 | Differential centrifugation Chemical lysis Tryptic digestion | SDS-PAGE nanoLC-C18 | Linear Ion trap-FTICR; DDA | NCBI Unipept | Mascot X!Tandem | 1676 proteins 495 unique to patients 793 unique to siblings (≥4 peptides) | [93] |
| Mucosal lavage from distal colon of different adolescent N = 5 | Differential centrifugation; Chemical lysis Tryptic digestion | SDS-PAGE; LC-RPC18 | LTQ Orbitrap; DDA | In-house Database (HIMPD);Target-decoy database Unipept | X!Tandem | 4 014 protein groups (≥2 unique peptides) | [130] |
| Healthy adults N = 16 (8 probiotic and 8 placebo) | Mechanical lysis (bead beating) Tryptic digestion | SDS-PAGE Nano2D-LC:C18 | LTQ Orbitrap; DDA | In-house metaproteome database | Mascot | 66% identified peptides with LCA: 80,9% bacteria 1% Archeae 13.8% Eukaryotic 5.3% could not be assigned | [131] |
| Children with IBD N = 4 | Differential centrifugation; Chemical and mechanical lysis Tryptic digestion | nanoLC-C18 | Q-Orbitrap; DDA | Human gut gene catalog; human proteomeIn-house database Unipept | Maxquant, | : 20 558 protein groups (>2 peptides) | [88] |
| Mucosal lavage from healthy subjects N = 38 (205 lavage samples) | Differential centrifugation (DC), Mechanical lysis (magnetic beads) Tryptic digestion | nano2D-LC: RPC18 | LTQ Orbitrap; DDA | SwissProt (human and bacteria) | SEQUEST | 117 unique proteins: 63% human proteins 30% bacterial proteins 7% others | [132] |
| 56 patients with with Gut Colonization by Multidrug-Resistant Enterobacteriaceae (N = 212 stool samples) | Centrifugation Chemical and mechanical lysis Tryptic digestion | SDS-PAGE 2D- UHPLC RPC18 | Hybrid quadrupole Orbitrap; DDA | -Genome Reference Catalog – SWISS-PROT bacteria and human - Metagenomes | -Maxquant -Unipept | −60% of the identified peptides to a taxonomy level-80% of the peptides mapped to at least one Gene Ontolgy term | [102] |
2.2. Pre-fractionation and mass spectrometry
2.2.1. Pre-fractionation
In the field of metaproteomics, it is advisable to analyse sample fractions containing fewer proteins and peptides in order to increase the sensitivity of low abundance peptides and to increase the proteomic depth of the analysis. However, analysis of complex and multispecies samples, such as those from the human gut, where the total number of microbial genes may vastly exceed the number of human genes, is complicated for a variety of reasons, including the wide dynamic range of microbial proteins present and the high levels of protein sequence homology. The separation of proteins can be obtained by gel electrophoresis, while peptides are generally separated using liquid chromatography. Gel electrophoresis can separate proteins along one or two dimensions. The gel containing the resulting protein bands or spots can then be cut and subjected to enzymatic digestion. Two-dimensional gel electrophoresis is predominantly dedicated to the study of highly expressed proteins. The first published work in the field of metaproteomics was performed with two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) protein separation followed by matrix assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) analysis [35]. Subsequently, the first metaproteomics study on human fecal microbiota was carried out using the same technique [36]. However, the disadvantages of this technique include significant sample handling and limited reproducibility. In addition, some proteins are difficult to separate, such as those in low abundance, with high or low molecular weight, and particularly those of high hydrophobicity (e.g., membrane proteins) [37].
More recently, high performance liquid chromatography (HPLC) was used to separate tens of thousands of peptides from the enzymatic digestion of thousands of proteins. The main purpose of HPLC is to separate peptides so that fewer of them are entering the ionization source of the mass spectrometer at any one time. HPLC separates the compounds according to their affinity with a stationary phase and a mobile phase. In the field of metaproteomics, reverse phase (RP) liquid chromatography (LC) is the most commonly used technique for peptide separation due to its excellent resolving power, stability and ease of use [38], [39]. Analytical columns are composed of a C18-grafted silica stationary phase (apolar) and of a mobile phase generally composed of two solvents (water and acetonitrile). Both solvents are combined to adjust the hydrophobicity of the mobile phase and, thus, separate the peptides according to their interaction with the column and their affinity with this mobile phase. The emergence of nano, or capillary, liquid chromatography columns and adapted systems is also interesting because of their ability to separate very small quantities of peptides without a decrease in sensitivity. Nevertheless, nano chromatography is challenging and has numerous drawbacks (e.g., less stable LC systems, low column capacity, tricky maintenance) [40]. Recently, micro LC has become increasingly favoured because of its ease of use compared to nano chromatography and the sensitivity gain over conventional HPLC [41]. Ultra-High Performance Liquid Chromatography (UHPLC) has become a standard hardware update necessary to achieve greater chromatographic selectivity [42]. Furthermore, liquid chromatography can also combine two or three orthogonal separation dimensions (2D-LC or 2/3-phase MudPIT). The most frequently used configurations for the separation of peptides combines a strong cation exchange (SCX) column, usually in the first dimension, and a RP (C18) column in the last dimension [25], [43], [44]. Another interesting 2D configuration combining two columns in reverse phase with opposite pH values allows for increased identification when fractions are concatenated [45], [46]. Thus, it enables a straightforward depth screening of the metaproteomes analysed. In summary, the interest of sample pre-fractionation is to be able to analyse less complex mixtures and, thus, to detect more peptides. However, pre-fractionation increases the number of analysis steps for a single sample, which can significantly extend the overall analysis time, as well as greatly increase the cost [38], [47].
2.2.2. Mass spectrometry
Regarding peptide detection, tandem mass spectrometry is currently the preferred technique. It monitors the mass of the peptides and their induced fragments. Generally, three main elements constitute a mass spectrometer: (i) an ion source, (ii) a mass analyzer and (iii) an ion current detector. The combination of two analyzers allows one to perform tandem spectrometry (MS/MS). This makes it possible to obtain structural information by fragmenting the parent ions and by measuring the fragment masses. The parent and fragment ions are detected by the detector and a mass spectrum is assembled. While there are several ionization techniques, the best for the analysis of biomolecules, such as proteins and peptides, are the soft ionization techniques: electrospray ionization (ESI) [133] and MALDI ionization (Matrix Assisted Laser Desorption Ionisation) [48]. Both techniques allow the ionization and transfer of intact biomolecules from the gas phase into the mass analyzer. Electrospray is an ionization technique that operates at atmospheric pressure, which makes it easy to couple with liquid chromatography. MALDI requires an overlay and co-crystallization the sample with an organic matrix and irradiation of the analyte-matrix crystal with a pulsed laser beam under vacuum. In the field of metaproteomics concerning the human gut, the mass analyzers most frequently used are the tandem hybrid mass analysers that enable high resolution analysis: Quadrupole Time-Of-Flight (Q-TOF) [49], Linear Ion Trap/Fourier Transform Ion Cyclotron Resonance (LIT/FTICR) [50] and especially Linear Trapping Quadrupole-Orbitrap (LTQ-Orbitrap) [51] or Quadrupole-Orbitrap (Q-Orbitrap). Each instrument offers a different mass accuracy, mass resolution, sensitivity or dynamic range. In any case, the purpose of mass spectrometry is to obtain sufficient selectivity and sensitivity to distinguish as many peptides as possible in complex samples [52]. In fact, sensitivity is important for the analysis of samples with limited quantities of peptide in order to increase the depth of metaproteome analysis at taxonomic and functional levels.
Currently, Ion mobility spectrometry (IMS) has been incorporated into a few instruments. Ion mobility is based on the separation of the molecular ions according to their mobility in a gas under the action of an electric field. The incorporation of ion mobility into MS/MS workflows allows an increase in selectivity. This enhanced selectivity can facilitate depth analysis for complex samples [53], [54]. Until now, the application of IMS for human gut metaproteomics has not yet been reported.
The MS/MS ion survey comprises three steps: i) selection of the peptide ion, ii) induced dissociation of the selected ion by collisions with an inert gas, iii) detection of the resulting ions. Two distinct MS/MS acquisition methods are used to collect peptide MS information: data dependent acquisition (DDA) or data independent acquisition (DIA). DDA mode is the most commonly used method in the field of shotgun proteomics due to its speed and sensitivity. In DDA, the precursors, usually the top 10–20 peptides per cycle, are sequentially selected from a full mass MS scan for fragmentation and acquisition in MS/MS mode. The selection parameters are pre-defined by the user. DDA mode generates fewer false positive rates because only the most intense peptides are fragmented. Nevertheless, it is a mode that often presents a loss of information especially in the case of weak peptide signals. Unlike the DDA mode, fragmentation of peptides is performed without pre-selection of the precursor during DIA [55]. DIA has recently been selected for a metaproteomic study on host-microbial interactions [56], but has not yet been used for the study of the human gut microbiota. Using this approach, all peptide ions are fragmented in the collision cell and all the resulting fragment ions are then recorded with alternate scans. This acquisition mode allows the recording of MS/MS data of all peptide signals, which greatly reduces information loss [57]. However, with this acquisition mode, many fragments are non-informative. Generally, the high number of fragment ions generated complicates the analysis in a classical database search strategy. However, this problem can be solved by the use of a reference spectral library, previously generated by a thorough analysis of the same / similar samples by the DDA. Currently, DIA mode is preferentially used with Q-TOF mass spectrometers.
In brief, research teams working in the field of metaproteomics, as applied to stool, have analysed their samples using LC-MS/MS systems. Table 1 summarizes the key metaproteomic studies, their pre-concentrations and MS methods. Most of the metaproteomic studies mentioned above use an Orbitrap as the mass analyzer with a DDA acquisition mode.
2.3. Metaproteomics data computation
2.3.1. Conventional sequence database search
The human gut microbiota is a complex environment and can be associated with a high number of protein sequences. On the other hand, mass spectrometry generates hundreds of thousands of peptide spectra that need to be compared with protein sequences. The number of identified proteins, as well as the identified taxonomies and functional annotations result from protein database selection. Large databases searches (>106 sequences), such as NCBI or Uniprot/Trembl are a challenge for metaproteomics studies in terms of computation times and the large number of peptides sequences matches (PSM) [58], [59]. Another important limitation with large databases concerns the evaluation of FDR, which may lead to the rejection of true protein identifications [60]. The assessment of FDR is performed by “Target-Decoy” methods [61], [62], [63]. FDR can then be calculated based on the matching scores. The sequence identifications are filtered according to the matching score to get an FDR lower than the defined threshold (generally <1% on the peptides). However, strict filtering based on the FDR to avoid false‐positive matches and the use of a restricted database would compromise the identification of microbial proteins by an increase of false negatives, thus limiting the number of peptide matches.
Protein databanks based on metagenomic data tend to get closer to the real protein content of samples. However, this approach may not provide a complete coverage of the protein content in the sample because there can be many different species in a single sample, for most of which a full genome is not available. Indeed, sequencing, assembly and annotation of the genome still generates incomplete or false metagenomic data [64]. Over the last few years, software has been developed in order to facilitate the automated analysis of high-throughput mass spectrometry-based proteomic data. Specialized algorithms have been implemented in research software to meet the requirements of mass spectrometry data. Generally, search engines assign a score to the peptide identifications [65]. The computation of this score differs between search algorithm. The most commonly used software programs in the field of proteomics are: Mascot [66], OMSSA [67], Sequest, X!Tandem [68], and ProteinLynx Global Server [69]. Table 1 describes the software used in the case of metaproteomics of the human gut. Despite the availability of software dedicated to metaproteomics, and the advances in DNA and RNA sequencing, metagenomes of the human gut usually contains hundreds of organisms with more than 106 proteins sequences. The determination of peptide sequence matches by searching against such databases could lead to an increased risk of false positives, but also a number of false-negative PSM. To address this challenge, metaproteomic studies based on iterative methods, where matches are derived from a primary search against a large database in order to create a smaller subset database, are increasing. The latter is called an in-house or customized database.
2.3.2. Customized iterative database approach
To meet computational challenges, customized databases are increasingly being used for protein identification. Taxonomic assignment is essential before any sequence search. Indeed, when searching for sequences, peptide sequences were often not proteotypic [70], [71]. It may, therefore, be difficult to distinguish certain organisms from a single peptide. In fact, shared peptides can be identified (potentially between homologous proteins between species). Thereby, bioinformatic pipelines were developed to assign peptides to their lowest unambiguous taxonomic rank, using an implementation of the Lowest Common Ancestor (LCA) algorithm generating taxonomic profiles at different levels. These pipelines are generally divided into two distinct steps: a first step where the peptides are matched to a reference database containing complete bacterial genome sequences assembled from NCBI or a reference protein database such as Uniprot or from metagenomes; a second step where peptides that passed the script of the first step are assigned to taxonomic ranks in an interactive tree in which a given tryptic peptide occurs [72]. These pipelines allow the generation of complete taxonomic profiles and lists of species-unique peptides (i.e., discriminative peptides). Metagenomic taxonomy-guided research strategies are increasingly being used in metaproteomics to improve the construction of protein databases. These strategies interactively explore the taxonomical content of the data using an algorithm based on the LCA peptides in order to assign each peptide to a taxon [73]. Unipept is an open source web application using the LCA algorithm to determine the taxonomic specificity of peptides [72]. For example, Tanca et al used the Unipept taxonomic assignment to generate a customized “host-microbiome” database containing sequences from specific microbial taxa and the host [25]. Thereby, iterative workflows can be used to build specific databases of biological samples. The study conducted by Xiao et al showed that a metagenomic taxonomy-guided database search strategy allows the construction of databases able to provide high sensitivity and precision in peptide identification in metaproteomic studies [74]. This strategy merges both taxonomy-guided reference protein sequences from public databases and metagenome assembly. Zhang et al. have also developed a universal workflow (MetaPro-IQ) to expand the sensitivity of peptide identification and greatly increase proteins identified for each sample [30]. A similar pipeline was used in the metaproteomics of saliva [75]. In brief, the implementation of Unipept algorithm became essential in the field of metaproteomics to prepare custom databases and simplify data processing pipelines.
2.3.3. De novo sequencing search
Otherwise, de novo peptide sequencing has become an alternative and complementary option for the assignment of peptide sequences to MS/MS spectra [76]. Peptide de novo sequencing in the analytical process derives a peptide’s amino acid sequence from its tandem mass spectrum without the assistance of sequence database. A clear advantage of de novo sequencing is that it works for both database and novel peptides. For example, a study combining protein databases search and peptide de novo sequencing, showed, respectively, the identification of 421 theoretical sequences and 333 new non-redundant proteins from faecal samples. The new peptides could not be mapped to the metagenomic sequence data [44]. As such, search engines for taxonomic and functional analysis are challenged by the vast amount of unannotated sequences [77]. De novo sequencing is often used to provide new unidentified sequences into databanks. This is possible thanks to a wide range of software tools. The most commonly used de novo peptide sequencing software programs are: PEAKS [78], PepNovo [79], Novor [80], NovoHMM [81], UniNovo [82] and MSNovo [83]. De novo peptide sequences are searched against databases using the Basic Local Alignment Search Tool Protein (BLAST p) algorithm [84]. However, during the process of de novo sequencing, some factors can cause difficulties including: incorrect assignment of ions, absence of ion fragments, existence of noise peaks in the spectrum, and post-translational modifications can contribute to the mass ambiguity and complicate the peptide fragmentation pattern. Moreover, the short length of tryptic peptides can impede MS-BLAST identification.
In summary, the use of customized databases with an iterative workflow should be encouraged in order to gain computational efficiency and focus on the protein content. This approach helps to reduce the rate of false identification associated with large databases and provides appropriate information. However, the processing of metagenomic data must be carried out carefully to ensure the best quality of the resulting metaproteomic databases. Besides, since databases generally do not cover all metaproteomes, de novo sequencing is highly useful for the detection of unknown peptide sequences directly from MS / MS spectra.
3. Metaproteomics of the human gut microbiota
3.1. Exploration of the gut metaproteome
A pioneering study of the human gut microbiota was conducted on two infants to investigate the functional role of gut microbiota during early growth [36]. However, despite the relatively simple faecal protein profile, the analysis was limited in depth due to the absence of an appropriate reference database. A few years later, with the development of analytical techniques and the availability of protein data from metagenomes, a study on the fecal microbiota of a preterm infant was performed. It revealed a much more detailed profile of the intestinal metaproteome and host microbiota interactions [85]. It will be interesting and valuable to collect more proteomic data that will allow a comparative study of microbial community functions between healthy preterm infants and those who develop diseases, such as neonatal necrotizing enterocolitis.
The first comprehensive intestinal metaproteome from a human adult was extracted from two healthy monozygotic twins [24]. This study explained an asymmetric and distinctive, but relatively stable distribution of proteins for each individual. The study also highlighted discrepancies between predicted protein levels from the metagenome and actual results. This confirms the importance of metaproteomics in the understanding of proteins expression because several unknown proteins represented previously undescribed microbial pathways. Another study showed a highly comparable clustering of the metaproteomic and phylogenetic profiles at the phylum level. The study showed differences in the relative share of Actinobacteria [86]. Soon after, a comparative study was performed between one lean and one obese adolescent. Their fecal samples showed subject-specific metaproteome differences that correlated with compositional differences of the microbiota [8]. In the lean subject, proteins classified as Bacteroidetes were in high representation (81%), while according to metagenomics, this phylum represents only about 20% of the microbial community. In the obese subject, the total microbiota was more abundant in the phylum Firmicutes (94%) and protein expression was predominantly attributed (56%). These previous studies show that metaproteomes provide complementary information about potentially active and functional bacteria in the gut microbial community. This study should be supported by further studies dealing with large cohorts of different unrelated individuals and alternative integrated omics approaches, such as metatranscriptomic and metabolomics in order to determine the metabolic links between obesity and gut microbiota.
In another context, metaproteomics could play an important role in the characterization of the gut microbial community in health and disease [87], [88], [89], [90]. The number of taxa in fecal samples is estimated at >21,000 with >63,000,000 unique proteins [91]. Studies concerning bacterial phylotypes and their identification in relation to the host, therefore, remain a vast expanse to be explored. The composition of microbial communities has been studied by different methodologies, such as culture, microscopy and especially, metagenomics. At present, metaproteomics might have a considerable contribution to explore the diversity of the gut microbiota. It also provides new information, such as the description of new functional genes. Zhang et al, recently combined an efficient sample preparation technique, high-resolution mass spectrometry and bioinformatics tools for the ultra-deep metaproteomic characterization of the human gut microbiome [92]. They reported the deepest analysis of the microbiome to date with an average of 20,558 protein groups identified per analyzed sample. Using an LCA approach with the Unipept tool, the taxonomic characterization of peptides pointed to 155 different microbe species with at least 3 distinctive peptides. This work also revealed variations in the microbiome from different individuals. However, because of the relatively long MS time for deep metaproteomics, this application for clinical samples analyses is limited.
In comparison to metagenomics, the study of the metaproteome for the characterization of microbial communities still has a long way to go. Nevertheless, in the field of microbial ecology, metaproteomics delivers a great amount of valuable data for in-depth analysis of microbiomes in response to human and microbial changes [93]. It appears as though a complementary approach to metagenomics, and a tool for large-scale taxonomic characterization of proteins in microbial ecosystems [94], could respond to diverse biologic questions concerning the host biology in health and disease. Rapid technical advances are expected and should focus on detection methods for protein modifications, which should reduce analysis cost and time. The integration of other omics platforms, such as metatranscriptomics metabolomics and culturomics could also allow in-depth study of diverse microbial communities at different pathological states.
3.2. Gut microbiota in health and diseases
A few years later, in addition to characterizing the microbial intestinal metaproteome of healthy subjects, comparative studies have increased in number. These studies were carried out to determine the expression of microbial proteins in case of gut dysbiosis. It has been suggested that an imbalance of the microbiota plays a central role in the chronic inflammation associated with the disease commonly named Inflammatory Bowel Disease (IBD). The first study compared healthy and unhealthy adults and was based on Crohn's disease (CD) [95]. In this study, Erikson et al. combined shotgun metagenomics and metaproteomics to identify potential functional signatures of CD. Stool samples were collected from six twins, either healthy or affected by CD in the ileum or colon. The study revealed several genes of the microbial community, as well as microbial and human proteins, that differentiated CD from healthy subjects, including depletion of many proteins in CD in the ileum. Another study focused on host–microbe relationships in Inflammatory Bowel Disease and was performed through bacterial characterisation and metaproteomics analysis. It reported that the examination of relationships between the bacteria and metaproteomes allowed identification of a high frequency of 14 bacterial phylotypes that significantly differentiate human subjects by disease type, namely Crohn’s disease and ulcerative colitis [96]. Furthermore, gut microbiota dysbiosis was reported in patients with cystic fibrosis. Fecal metaproteomics allows the analysis of host and microbial proteins to elucidate the functional changes resulting from this dysbiosis. For example, Debyser et al demonstrated that fecal protein from patients with cystic fibrosis were dominated by host proteins involved in inflammation and mucus formation [97]. Taxonomic analysis of the microbial proteins, based on LCA, confirmed significant differences in the gut microbial diversity with a strong reduction of butyrate reducers, such as Faecalibacterium prausnitzii and an increase of Enterobacteriaceae, Ruminococcus gnavus and Clostridia species. This study also highlights a list of host and microbial proteins that could be potential biomarkers for cystic fibrosis. So, metaproteomics enhances the understanding of the microbial world and establishes a link between microbial communities to its function. The functional distribution of COGs (clusters of orthologous groups) allows identification of responsible bacterial members of health status under altered physiological conditions revealing differential protein profiles. For example, the extraction of the metaproteome allows functional classification of bacterial proteins from a classification of COG [86]. The shotgun metaproteomics approach has identified several COG categories that are more highly represented in the microbial metaproteome, compared to the average metagenome, in fecal samples from a female twin pair [24]. In this study, 50% of total proteins detected in the metaproteome were involved in translation, carbohydrate metabolism, or energy production. The other categories of COG were underrepresented in the metaproteomes, relative to metagenomes, including proteins involved in inorganic ion metabolism, cell wall and membrane biogenesis, cell division, and biosynthesis of secondary metabolites. Moreover, the best understanding of the study of gut microbiota function is to associate metaproteomics with COGs classification [120].
The bidirectional communication between the host and its microbiota is complex [98]. It involves a third partner, which is the immune system of the host, via innate immunity receptors. The immune system protects us against the constant aggressions of our environment and the gut microbiota plays an essential role in maintaining immunity. Influencing factors, such as stress, inappropriate diet, repetitive consumption of drugs and toxic substances can cause an imbalance of the microbiota or intestinal permeability. An important aspect to keep in mind is that the human microbiome is overly exposed to antibiotics that can rapidly alter its composition with potential immediate effects on health. Gut microbiota alterations induced by antibiotics can also indirectly affect health on long-term basis [99], [100]. The effects of antibiotic-induced microbiota alterations have an impact on the immune system and, therefore, cause an increased susceptibility to infections, inducing metabolic deregulation of the host. For example, a metaproteomic analysis study showed important changes in the protein profiles of the gut microbiota responses following β-lactam therapy [101]. The authors demonstrated that antibiotics targeting specific pathogenic infections and diseases may alter gut microbial ecology. Metaproteome results suggest the restoration of the microbiota indicating that the initial profile was recovered at the end of the treatment. To date, one of the largest clinical metaproteomic studies on the human gut microbiota was conducted on acute leukaemia patients with multidrug-resistant Enterobacteriaceae gut colonization [102]. This study allowed the authors to describe the taxonomic composition and functional process of patients during the Enterobacteriaceae gut colonization. The analysis showed that public metagenome databases are incomplete and that sample-specific metagenomes improve results. This supports the idea that large database sizes come with several issues.
Despite the symbiosis between the host and the gut microbiota, major changes can affect the functionality of the microbiome. From this dysbiosis, an inappropriate immune response of the host may result. It is now suggested that pathologies related to disorders of immunity or metabolism can be triggered or aggravated by the bacteria that we host. For instance, alteration of the gut microbiota has been implicated in metabolic diseases, such as obesity and diabetes [103]. A metaproteomic and genomic study of the gut microbiota showed that microbial taxa associated with host proteins involved in the function of the mucus barrier and microvilli adhesion were depleted in patients with new-onset type 1 diabetes [104]. Recently, a study of the gut microbiota was correlated with liver cirrhosis. The authors used metaproteomics to detect proteome changes in the case of affected patients [105]. They found that the abundances of 14 proteins were increased in the fecal microbiota from liver cirrhosis patients. Seven proteins, such as ketol-acid reductoisomerase, phosphoglycerate kinase, ribose-phosphate pyrophosphokinase, and probable thiol peroxidase were more highly expressed in patient’s intestinal microbiota compared with normal. These specific proteins can serve as potential biomarkers and therapeutic targets for the development of treatments. Furthermore, metaproteomic analysis of the gut microbiota has been increasingly applied to the identification of specific proteins as targets for treatment. Several pathologies or functional disorders have been linked with gut microbiota dysbiosis, such as Alzheimer's disease, cardiovascular diseases, Parkinson's disease, depression and anxiety [98], [106], [107]. The gut-metaproteome is a key element in maintaining the relationship between the host and the microbiota. Consequently, advances need to focus on the identification of human gut biomarkers. This could lead to the implementation of new clinical diagnostic tests and treatments to heal microbiota-related diseases.
4. Metaproteomics combined with other omics
The human gut microbiota has been conceptualized as a dynamic ecological community consisting of several taxa, potentially interacting with each other, the host and the environment [108]. The fundamental objectives of human microbiome research focus on the various changes in the abundance and composition of the microbiota in relation to health and disease. Four key omics technologies are used to study the functions of cells: genomics for DNA, transcriptomics for RNA, proteomics for proteins, and metabolomics for small molecules/metabolites. To explore the dynamics of the microbial community, meta-omics approaches have been used to analyze large-scale gene or protein expressions and metabolite compositions [90]. Therefore, metagenomics, metatranscriptomics, metaproteomics and metabolomics are closely linked and metaproteomics plays a central role to more effectively decipher the composition and functions of microbial communities. Indeed, recent technological progress in the field of mass spectrometry and computational informatics has allowed metaproteomics to become a significant approach for the characterization of the human gut microbiome. The application of metaproteomics, combined with metagenomic analysis, has shown that the gut microbiome contains distinctive sets of active microorganisms between individuals [95]. However, the study of the relationship between taxonomic alterations and functional repercussions linked to the disease remains difficult. To resolve the taxonomic and functional attributes of gastrointestinal microbiota, Heintz-Buschart et al. combined data from genomics, metagenomics, metatranscriptomics and metaproteomics, and showed that the associated microbial functional signatures were linked to metabolic traits in distinct taxa [109]. The use of multi-omics approaches would also identify small molecules and bacterial peptides affecting the physiology of the host, such as gastrointestinal motility induced by metabolites (e.g., CH4, H2, H2S, SFCA) from the microbiota, or deregulation of the microbiota-gut-brain in neurodegenerative diseases [110]. Another multi-omics study provided novel insights into metabolic changes caused by antibiotic disturbance [101]. In this study, the integrative analysis showed an oscillatory imbalance between Gram-negative and Gram-positive bacteria after initiation of the β-lactam therapy. During this process, metabolic disorders associated with the different stages of the therapy were noted, such as an overall attenuation of energetic metabolism of gut bacteria and their capacity to transport and metabolize bile acid, cholesterol, hormones and vitamins.
In fact, the study of the function of the gut microbiota is better understood by combining metaproteomics with other OMICS approaches. Among these combined analyses, it is possible to identify potential genes, proteins and metabolic pathways that can be associated with a healthy condition. For example, the study of the functionality of the gut microbiota by combining multi-omics has shown a considerable divergence between potential functions and active expression in the gut microbiota of a healthy human cohort [111]. The authors found an overlap between the metagenome and the metaproteome regarding the most abundant phyla and genera. Nevertheless, they found considerable differences that highlight a divergence of microbial functions, especially with a carbohydrate metabolism.
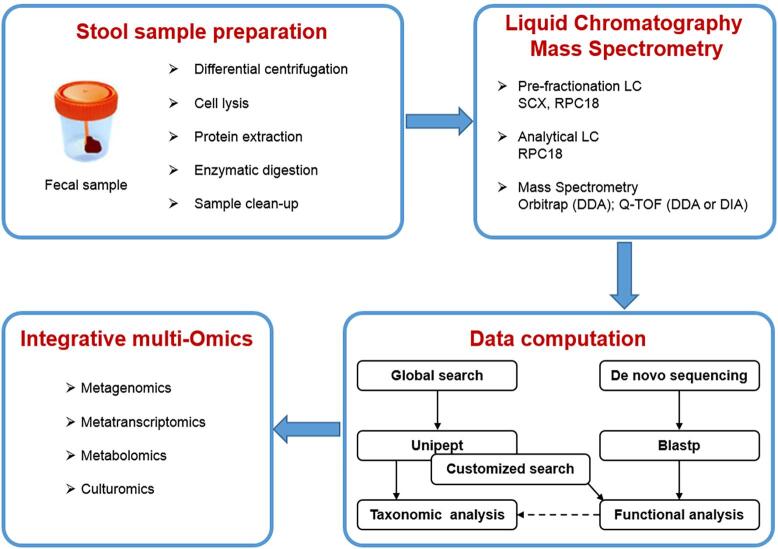
Overall, analyses exploring the interactions between the intestinal microbiota and its functions towards humans would be more relevant if metaproteomics was included with other OMICs (Fig. 2). The latest of the omics approaches is culturomics, developed at our institute [15]. This in-depth study tested 212 culture conditions to select 18 best conditions for the isolation of prokaryotes. This appraoch was effectively combined to rapid identification of bacterial colonies by MALDI TOF MS. Indeed, metaproteomics provides distinct and complementary microbial functional information to metagenomics and other approaches [112]. Each “omics” dataset can be studied separately, but relevant information can certainly be extracted from a joint analysis of several of them. The power of these integrative tools on the descriptive level allows today a deep and large-scale characterization of biological systems. However, these technologies face some explanatory limitations. Among the most frequently mentioned difficulties, these must be emphasized: data management requires powerful bioinformatics tools, formulation of objective hypotheses is required to comprehend biological systems; large sets of data obtained under different experimental conditions are difficult to compare; relationship between a molecular signature and the biological interpretation of biomarkers is not always obvious.
Fig. 2.
Representation of a typical workflow in a metaproteomic analysis of fecal sample.
5. Future directions for human gut metaproteomics research
5.1. Application challenges
Despite the great potential of metaproteomics for characterizing microbial ecosystems and their various roles within the human host, many challenges remain. Metaproteomics should enable measurement of proteome expression for the entire microbial community at a specific moment in the gut microbial ecosystem. Beyond the difficulties encountered during protein extraction, analytical platforms also show limitations for a sensitivity analysis of protein samples in such a complex dynamic range. Thanks to the recent emerging mass spectrometry technologies, the depth of metaproteome analysis can be improved with a data independent acquisition mode [113]. The DIA mode usually results in an increased sensitivity and enables a significant improvement concerning reproducibility and quantification of proteins in complex samples, as compared to DDA [114]. Nevertheless, it is not widely used as a routine method because of technological drawbacks, such as the high number of clinical samples. These numerous samples require efficient tools, such as software that can handle very large amounts of data with a computational time that does not exceed 24 h. In addition to this analytical aspect, identifying the proteins of the complex microbial consortium, comprising hundreds or thousands of species, has also proved a difficult task. The absence of complete genomic sequences, particularly of poorly characterized and uncultivated species, is a major challenge for researchers. The availability of a relevant database is one of the essential elements of metaproteomics for the complete analysis of the gut microbiota samples. Some researchers use protein databases from completed genomes, while others use sequences from different metagenomes, complete or not, and from diverse origins [115]. A solution could be to use standardized metaproteomic databases from non-redundant and complete metagenomes for each type of microbial community [116]. All of this is intended to simplify and speed up computational analysis. Moreover, future improvements in software and algorithms will significantly contribute to the development of advanced meta-proteomic analyses [30], [67], [78], [79], [117].
5.2. Clinical diagnosis
Advances in scientific research have shown the important role of intestinal flora in the regulation of many functions of the body, including the immune system. Changes in the composition of the intestinal microbiota (dysbiosis) are often due to environmental or dietary factors that can lead to chronic diseases (e.g., metabolic, inflammatory, cardiac). Therefore, it is important to consider the role of the gut microbiome when selecting therapy. Gut microbiota represents a variable factor and in case of a dysbiosis, the solution to prevent any pathogenesis of chronic diseases seems to be probiotics, prebiotics and diet. Furthermore, fecal transplantation (from healthy individuals) is used as a community replacement approach to restore the composition of intestinal flora (in particular for Clostridium difficile infection). The gut microbiota is, therefore, used as a tool for diagnosis and personalized treatment strategies [127].
In this context, metaproteomics research has already led to some remarkable discoveries about the functional and taxonomic characteristics of gut microbiota. Considered as a tool to observe the consequences of the modulation of the intestinal flora, metaproteomics could help adapt a personalized treatment in cases of dysbiosis, since it is a powerful tool for observing modulation of the intestinal flora. However, this is still an emerging area where an increase in the number of studies involving complex microbial communities is expected. The number of samples in clinical microbiology labs can reach hundreds or even thousands per day. Consequently, metaproteomics might be an application tool for the routine diagnosis of fecal samples, such as MALDI-TOF-MS, which became a standard tool in clinical microbiology laboratories [118]. Indeed, metaproteomics could help identify markers for clinical diagnosis and provide an overview of antigens, functions and taxa. However, not all the conditions are set at this time to allow routine metaproteomic analyses. Such a quantity of samples would require qualified personnel, standardization of sample preparation with a short processing time at low cost. One of the main challenges would also be the implementation of powerful and automated software. Moreover, software and the databases should conform to high-quality standards and specific privacy regulations for the medical applications.
5.3. Multi-omics contribution
The study of metaproteomes helps to better understand the molecular interactions of the bacterial communities with the host [21]. Apart from protein identification, metaproteomics can determine the main microbial actors contributing to the gut metabolic functions [119], [120]; this is not possible with metagenomics based on the 16S RNA. For instance, the advent of “culturomics”, allowed culturing of many human microbial species that were not previously culturable [15], [121], [122]. The combination of culturomics and metagenomics, showed that both approaches are complementary, each providing data/results confirming the other approach, however also providing unique information [123]. For example, Li et al showed that the combination of culturomics with metaproteomics allowed a systemic understanding of the human microbiome thanks to the evaluation of the nutritional composition of the culture medium [124]. They demonstrated that the metaproteomic profile changed with the nutrional components of the culture medium. Therefore, metaproteomics has become a complementary approach to metagenomic [125] data and other omics approaches. In summary, the combination of OMICS approaches allows an exhaustive understanding of the intestinal microbiota thanks to the complementarities of the results.
As for metagenomics, rapid technical progress is needed in the field of metaproteomics in order to facilitate integration with other OMICS.
6. Conclusion
This review on the metaproteomics of the human gut microbiome shows a recent and powerful approach that can be used to characterize and better understand the human intestinal environment. Given the complexity of samples, metaproteomics of the human gut still faces several challenges, such as sample preparation, limitations of analytical tools and data interpretation. To date, major improvements and developments have made it possible to rigorously validate metaproteomic analyses, thanks to (i) optimised extraction, lysis and cell purification procedures, (ii) improved separation methods by liquid chromatography, and (iii) broader analysis of metaproteomes by rapid, accurate and sensitive mass spectrometry. Moreover, the availability of tailored sequence databases from high-quality metagenomics and development of bioinformatics tools, as well as efficient workflow pipelines, have improved the number of proteins identified.
Furthermore, the contribution of metaproteomic data to other meta-omics datasets provides an exhaustive and complementary view of the functional state of the intestinal microbiome. Altogether, metaproteomics is the cornerstone in the study of microbial ecosystems. It has great potential to become a valuable tool for routine diagnosis in clinical microbiology laboratories. However, the multifaceted, diverse and complex metaproteomics approaches should be standardized to enable a more conclusive understanding of the function of the microbial communities in the human gut.
Declaration of Competing Interest
The authors declare that there is no conflict of interest regarding the publication of this article.
References
- 1.Grice Elizabeth A., Segre Julia A. The human microbiome: our second genome. Annu. Rev. Genom. Hum. Genet. 2012;13(1):151–170. doi: 10.1146/annurev-genom-090711-163814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macpherson A.J., Harris N.L. Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 4.Turnbaugh P.J., Ley R.E., Hamady M., Fraser-Liggett C., Knight R., Gordon J.I. The human microbiome project: exploring the microbial part of ourselves in a changing world. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snelson M., Coughlan M.T. Dietary advanced glycation end products: digestion, metabolism and modulation of gut microbial ecology. Nutrients. 2019;11(2):E215. doi: 10.3390/nu11020215. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Candeias E.M., Sebastião I.C., Cardoso S.M., Correia S.C., Carvalho C.I., Plácido A.I., Santos M.S., Oliveira C.R., Moreira P.I., Duarte A.I. Gut-brain connection: the neuroprotective effects of the anti-diabetic drug liraglutide. World J. Diabetes. 2015;6(6):807–827. doi: 10.4239/wjd.v6.i6.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Vos W.M., de Vos E.A.J. Role of the intestinal microbiome in health and disease: from correlation to causation. Nutr Rev. 2012;70(Suppl 1):S45–S56. doi: 10.1111/j.1753-4887.2012.00505.x. Review. [DOI] [PubMed] [Google Scholar]
- 8.Ferrer M., Ruiz A., Lanza F., Haange S.-B., Oberbach A., Till H., et al. Microbiota from the distal guts of lean and obese adolescents exhibit partial functional redundancy besides clear differences in community structure. Environ. Microbiol. 2013 Jan;15(1):211–226. doi: 10.1111/j.1462-2920.2012.02845. [DOI] [PubMed] [Google Scholar]
- 9.Dicks L.M.T., Geldenhuys J., Mikkelsen L.S., Brandsborg E., Marcotte H. Our gut microbiota: a long walk to homeostasis. Benef Microbes. 2018;9(1):3–20. doi: 10.3920/BM2017.0066. [DOI] [PubMed] [Google Scholar]
- 10.Hansen T.H., Gøbel R.J., Hansen T., Pedersen O. The gut microbiome incardio-metabolic health. Genome Med. 2015 Mar 31;7(1):33. doi: 10.1186/s13073-015-0157-z. eCollection 2015. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckburg P.B., Bik E.M., Bernstein C.N., Purdom E., Dethlefsen L., Sargent M., et al. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandal Rahul Shubhra, Saha Sudipto, Das Santasabuj. Metagenomic surveys of gut microbiota. Genom. Proteom. Bioinform. 2015;13(3):148–158. doi: 10.1016/j.gpb.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jovel J., Patterson J., Wang W., Hotte N., O’Keefe S., Mitchel T., et al. Characterization of the gut microbiome using 16S or shotgun metagenomics. Front. Microbiol. 2016;20(7):459. doi: 10.3389/fmicb.2016.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gosalbes M.J., Durbán A., Pignatelli M., Abellan J.J., Jiménez-Hernández N., Pérez-Cobas A.E., Latorre A., Moya A. Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS One. 2011;6(3) doi: 10.1371/journal.pone.0017447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lagier J.-C., Khelaifia S., Alou M.T., Ndongo S., Dione N., Hugon P., et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 2016 Nov;7(1):16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PubMed] [Google Scholar]
- 16.Schneider T., Riedel K. Environmental proteomics: analysis of structure and function of microbial communities. Proteomics. 2010;10(4):785–798. doi: 10.1002/pmic.200900450. Review. [DOI] [PubMed] [Google Scholar]
- 17.Petriz B.A., Franco O.L. Metaproteomics as a complementary approach to gut microbiota in health and disease. Front. Chem. 2017;26(5):4. doi: 10.3389/fchem.2017.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolmeder C.A., de Vos W.M. Metaproteomics of our microbiome – developing insight in function and activity in man and model systems. J. Proteomics. 2014;31(97):3–16. doi: 10.1016/j.jprot.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 19.Lee P.Y., Chin S.-F., Neoh H.-M., Jamal R. Metaproteomic analysis of human gutmicrobiota: where are we heading? J. Biomed. Sci. 2017;24(1):36. doi: 10.1186/s12929-017-0342-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X., Li L., Mayne J., Ning Z., Stintzi A., Figeys D. Assessing the impact ofprotein extraction methods for human gut metaproteomics. J. Proteom. 2018;30(180):120–127. doi: 10.1016/j.jprot.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 21.Haange S.B., Jehmlich N. Proteomic interrogation of the gut microbiota: potential clinical impact. Expert. Rev. Proteom. 2016;13(6):535–537. doi: 10.1080/14789450.2016.1190652. [DOI] [PubMed] [Google Scholar]
- 22.Cañas B., Piñeiro C., Calvo E., López-Ferrer D., Gallardo J.M. Trends in sample preparation for classical and second generation proteomics. J. Chromatogr. A. 2007;1153:235–258. doi: 10.1016/j.chroma.2007.01.045. [DOI] [PubMed] [Google Scholar]
- 23.Morris L.S., Marchesi J.R. Assessing the impact of long term frozen storage of faecal samples on protein concentration and protease activity. J. Microbiol. Methods. 2016;123:31–38. doi: 10.1016/j.mimet.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verberkmoes N.C., Russell A.L., Shah M., Godzik A., Rosenquist M., Halfvarson J., et al. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2009;3(2):179–189. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- 25.Tanca A., Palomba A., Pisanu S., Addis M.F., Uzzau S. Enrichment or depletion? The impact of stool pretreatment on metaproteomic characterization of the human gutmicrobiota. Proteomics. 2015;15(20):3474–3485. doi: 10.1002/pmic.201400573. [DOI] [PubMed] [Google Scholar]
- 26.Wiśniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparationmethod for proteome analysis. Nat Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 27.Deusch S., Seifert J. Catching the tip of the iceberg – evaluation of samplepreparation protocols for metaproteomic studies of the rumen microbiota. Proteomics. 2015;15(20):3590–3595. doi: 10.1002/pmic.201400556. [DOI] [PubMed] [Google Scholar]
- 28.Chourey K., Jansson J., VerBerkmoes N., Shah M., Chavarria K.L., Tom L.M., et al. Direct cellular lysis/protein extraction protocol for soil metaproteomics. J. Proteome Res. 2010;9(12):6615–6622. doi: 10.1021/pr100787q. [DOI] [PubMed] [Google Scholar]
- 29.Tanca A., Palomba A., Pisanu S., Deligios M., Fraumene C., Manghina V., et al. A straightforward and efficient analytical pipeline formetaproteome characterization. Microbiome. 2014;2(1):49. doi: 10.1186/s40168-014-0049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X., Ning Z., Mayne J., Moore J.I., Li J., Butcher J., et al. MetaPro-IQ: a universal metaproteomicapproach to studying human and mouse gut microbiota. Microbiome. 2016;4(1):31. doi: 10.1186/s40168-016-0176-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaiyarit S., Thongboonkerd V. Comparative analyses of cell disruption methods for mitochondrial isolation in high-throughput proteomics study. Anal Biochem. 2009;394(2):249–258. doi: 10.1016/j.ab.2009.07.026. [DOI] [PubMed] [Google Scholar]
- 32.Burkhart J.M., Schumbrutzki C., Wortelkamp S., Sickmann A., Zahedi R.P. Systematic and quantitative comparison of digest efficiency and specificity reveals the impact of trypsin quality on MS-based proteomics. J. Proteom. 2012;75(4):1454–1462. doi: 10.1016/j.jprot.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 33.Giansanti P., Tsiatsiani L., Low T.Y., Heck A.J.R. Six alternative proteases for mass spectrometry-based proteomics beyond trypsin. Nat. Protoc. 2016;11(5):993–1006. doi: 10.1038/nprot.2016.057. [DOI] [PubMed] [Google Scholar]
- 34.Antharavally B.S. Removal of detergents from proteins and peptides in a spin-column format Chapter 6: Unit 6.12. Curr. Protoc. Protein Sci. 2012 doi: 10.1002/0471140864. ps0612s69. [DOI] [PubMed] [Google Scholar]
- 35.Wilmes P., Bond P.L. The application of two-dimensional polyacrylamide gel electrophoresis and downstream analyses to a mixed community of prokaryotic microorganisms. Environ. Microbiol. 2004;6:911–920. doi: 10.1111/j.1462-2920.2004.00687.x. [DOI] [PubMed] [Google Scholar]
- 36.Klaassens E.S., de Vos W.M., Vaughan E.E. Metaproteomics approach to study the functionality of the microbiota in the human infant gastrointestinal tract. Appl. Environ. Microbiol. 2007;73:1388–1392. doi: 10.1128/AEM.01921-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gygi S.P., Corthals G.L., Zhang Y., Rochon Y., Aebersold R. Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc. Natl. Acad. Sci. USA. 2000;97:9390–9395. doi: 10.1073/pnas.160270797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bandhakavi S., Markowski T.W., Xie H., Griffin T.J. Three-dimensional peptide fractionation for highly sensitive nanoscale LC-based shotgun proteomic analysis of complex protein mixtures. Methods Mol. Biol. 2011;790:47–56. doi: 10.1007/978-1-61779-319-6_4. [DOI] [PubMed] [Google Scholar]
- 39.Josic D., Kovac S. Reversed-phase high performance liquid chromatography of proteins. Curr. Protoc. Protein Sci. 2010 doi: 10.1002/0471140864.ps0807s61. [DOI] [PubMed] [Google Scholar]
- 40.Gaspari M., Cuda G. Nano LC-MS/MS: a robust setup for proteomic analysis. Methods Mol. Biol. 2011;790:115–126. doi: 10.1007/978-1-61779-319-6_9. [DOI] [PubMed] [Google Scholar]
- 41.Arnold D.W., Needham S.R. Micro-LC-MS/MS: the future of bioanalysis. Bioanalysis. 2013 Jun;5(11):1329–1331. doi: 10.4155/bio.13.31. [DOI] [PubMed] [Google Scholar]
- 42.Walter T.H., Andrews R.W. Recent innovations in UHPLC columns and instrumentation. TrAC Trends Anal. Chem. 2014 doi: 10.1016/j.trac.2014.07.016. [DOI] [Google Scholar]
- 43.Nägele E., Vollmer M., Hörth P., Vad C. 2D-LC/MS techniques for the identification of proteins in highly complex mixtures. Expert Rev. Proteom. 2004;1:37–46. doi: 10.1586/14789450.1.1.37. [DOI] [PubMed] [Google Scholar]
- 44.Cantarel B.L., Erickson A.R., VerBerkmoes N.C., Erickson B.K., Carey P.A., Pan C., et al. Strategies for metagenomic-guided whole-community proteomics of complex microbial environments. PLoS One. 2011;6(11) doi: 10.1371/journal.pone.0027173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang F., Shen Y., Camp D.G., Smith R.D. Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics. 2011;11(10):2019–2026. doi: 10.1002/pmic.201000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilar M., Olivova P., Daly A.E., Gebler J.C. Orthogonality of separation in two-dimensional liquid chromatography. Anal. Chem. 2005;77:6426–6434. doi: 10.1021/ac050923i. [DOI] [PubMed] [Google Scholar]
- 47.Righetti P.G., Castagna A., Antonioli P., Boschetti E. Prefractionation techniques in proteome analysis: the mining tools of the third millennium. Electrophoresis. 2005;26:297–319. doi: 10.1002/elps.200406189. [DOI] [PubMed] [Google Scholar]
- 48.Hillenkamp F., Karas M. Mass spectrometry of peptides and proteins by matrix-assisted ultraviolet laser desorption/ionization. Methods Enzymol. 1990;193:280–295. doi: 10.1016/0076-6879(90)93420-P. [DOI] [PubMed] [Google Scholar]
- 49.Rietschel B., Baeumlisberger D., Arrey T.N., Bornemann S., Rohmer M., Schuerken M., et al. The benefit of combining nLC-MALDI-Orbitrap MS data withnLC-MALDI-TOF/TOF data for proteomic analyses employing elastase. J. Proteome Res. 2009;8(11):5317–5324. doi: 10.1021/pr900557k. [DOI] [PubMed] [Google Scholar]
- 50.Makarov A., Denisov E., Kholomeev A., Balschun W., Lange O., Strupat K., et al. Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal. Chem. 2006;78:2113–2120. doi: 10.1021/ac0518811. [DOI] [PubMed] [Google Scholar]
- 51.Hu Q., Noll R.J., Li H., Makarov A., Hardman M., Graham Cooks R. The Orbitrap: a new mass spectrometer. J. Mass Spectrom. 2005;40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- 52.Graham C., McMullan G., Graham R.L.J. Proteomics in the microbial sciences. Bioeng. Bugs. 2011;2:17–30. doi: 10.4161/bbug.2.1.14413. [DOI] [PubMed] [Google Scholar]
- 53.Sans M., Feider C.L., Eberlin L.S. Advances in mass spectrometry imaging coupled to ion mobility spectrometry for enhanced imaging of biological tissues. Curr. Opin. Chem. Biol. 2018;42:138–146. doi: 10.1016/j.cbpa.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meier F., Brunner A.-D., Koch S., Koch H., Lubeck M., Krause M., et al. Online parallel accumulation-serial fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol. Cell Proteom. 2018;17(12):2534–2545. doi: 10.1074/mcp.TIR118.000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gillet L.C., Navarro P., Tate S., Röst H., Selevsek N., Reiter L., et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell Proteom. 2012;11(6) doi: 10.1074/mcp.O111.016717. O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Starr Amanda E., Deeke Shelley A., Li Leyuan, Zhang Xu, Daoud Rachid, Ryan James, Ning Zhibin, Cheng Kai, Nguyen Linh V.H., Abou-Samra Elias, Lavallée-Adam Mathieu, Figeys Daniel. Proteomic and metaproteomic approaches to understand host–microbe interactions. Anal. Chem. 2018;90(1):86–109. doi: 10.1021/acs.analchem.7b04340. [DOI] [PubMed] [Google Scholar]
- 57.Geromanos S.J., Vissers J.P.C., Silva J.C., Dorschel C.A., Li G.-Z., Gorenstein M.V., et al. The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics. 2009;9(6):1683–1695. doi: 10.1002/pmic.200800562. [DOI] [PubMed] [Google Scholar]
- 58.Jagtap P., Goslinga J., Kooren J.A., McGowan T., Wroblewski M.S., Seymour S.L., Griffin T.J. A two-step database search method improves sensitivity in peptide sequence matches for metaproteomics and proteogenomics studies. Proteomics. 2013;13(8):1352–1357. doi: 10.1002/pmic.201200352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chatterjee S., Stupp G.S., Park S.K., Ducom J.C., Yates J.R., 3rd, Su A.I., Wolan D.W. A comprehensive and scalable database search system for metaproteomics. BMC Genom. 2016;17(1):642. doi: 10.1186/s12864-016-2855-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cargile B.J., Bundy J.L., Stephenson J.L. Potential for false positive identifications from large databases through tandem mass spectrometry. J. Proteome Res. 2004;3:1082–1085. doi: 10.1021/pr049946o. [DOI] [PubMed] [Google Scholar]
- 61.Wang G., Wu W.W., Zhang Z., Masilamani S., Shen R.-F. Decoy methods for assessingfalse positives and false discovery rates in shotgun proteomics. Anal. Chem. 2009;81(1):146–159. doi: 10.1021/ac801664q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elias J.E., Gygi S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 63.Elias J.E., Gygi S.P. Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol. Biol. 2010;604:55–71. doi: 10.1007/978-1-60761-444-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wooley J.C., Godzik A., Friedberg I. A primer on metagenomics. PLoS Comput. Biol. 2010;6 doi: 10.1371/journal.pcbi.1000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chepanoske C.L., Richardson B.E., von Rechenberg M., Peltier J.M. Average peptide score: a useful parameter for identification of proteins derived from database searches of liquid chromatography/tandem mass spectrometry data. Rapid Commun. Mass Spectr. 2005;19:9–14. doi: 10.1002/rcm.1741. [DOI] [PubMed] [Google Scholar]
- 66.Perkins D.N., Pappin D.J., Creasy D.M., Cottrell J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 67.Geer L.Y., Markey S.P., Kowalak J.A., Wagner L., Xu M., Maynard D.M., et al. Open mass spectrometry search algorithm. J. Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 68.Craig R., Beavis R.C. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 69.Prieto G., Aloria K., Osinalde N., Fullaondo A., Arizmendi J.M., Matthiesen R. PAnalyzer: a software tool for protein inference in shotgun proteomics. BMC Bioinform. 2012;5(13):288. doi: 10.1186/1471-2105-13-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuster B., Schirle M., Mallick P., Aebersold R. Scoring proteomes with proteotypic peptide probes. Nat. Rev. Mole. Cell Biol. 2005;6:577–583. doi: 10.1038/nrm1683. [DOI] [PubMed] [Google Scholar]
- 71.Boulund F., Karlsson R., Gonzales-Siles L., Johnning A., Karami N., Al-Bayati O., Åhrén C., Moore E.R.B., Kristiansson E. Typing and characterization of bacteria using bottom-up tandem mass spectrometry proteomics. Mol. Cell Proteom. 2017;16(6):1052–1063. doi: 10.1074/mcp.M116.061721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mesuere B., Willems T., Van der Jeugt F., Devreese B., Vandamme P., Dawyndt P. Unipept web services for metaproteomics analysis. Bioinformatics. 2016;32(11):1746–1748. doi: 10.1093/bioinformatics/btw039. [DOI] [PubMed] [Google Scholar]
- 73.Huson D.H., Auch A.F., Qi J., Schuster S.C. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xiao J., Tanca A., Jia B., Yang R., Wang B., Zhang Y., et al. Metagenomic taxonomy-guided database-searching strategy for improving metaproteomic analysis. J. Proteome Res. 2018;17(4):1596–1605. doi: 10.1021/acs.jproteome. [DOI] [PubMed] [Google Scholar]
- 75.Jagtap P., McGowan T., Bandhakavi S., Tu Z.J., Seymour S., Griffin T., et al. Deep metaproteomic analysis of human salivary supernatant. Proteomics. 2012;12(7):992–1001. doi: 10.1002/pmic.201100503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ma B., De Johnson R. novo sequencing and homology searching. Mol. Cell Proteom. 2012;11(2) doi: 10.1074/mcp.O111.014902. O111.014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Muth T., Renard B.Y., Martens L. Metaproteomic data analysis at a glance: advances in computational microbial community proteomics. Expert Rev. Proteom. 2016;13(8):757–769. doi: 10.1080/14789450.2016.1209418. [DOI] [PubMed] [Google Scholar]
- 78.Ma B., Zhang K., Hendrie C., Liang C., Li M., Doherty-Kirby A., et al. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003;17:2337–2342. doi: 10.1002/rcm.1196. [DOI] [PubMed] [Google Scholar]
- 79.Frank A., Pevzner P. PepNovo: de novo peptide sequencing via probabilistic network modeling. Anal. Chem. 2005;77:964–973. doi: 10.1021/ac048788h. [DOI] [PubMed] [Google Scholar]
- 80.Ma B. Novor: real-time peptide de novo sequencing software. J. Am. Soc. Mass Spectrom. 2015;26(11):1885–1894. doi: 10.1007/s13361-015-1204-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fischer B., Roth V., Roos F., Grossmann J., Baginsky S., Widmayer P., et al. NovoHMM: a hidden markov model for de novo peptide sequencing. Anal. Chem. 2005;77:7265–7273. doi: 10.1021/ac0508853. [DOI] [PubMed] [Google Scholar]
- 82.Jeong K., Kim S., Pevzner P.A. UniNovo: a universal tool for de novo peptide sequencing. Bioinformatics. 2013;29(16):1953–1962. doi: 10.1093/bioinformatics/btt338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mo L., Dutta D., Wan Y., Chen T. MSNovo: a dynamic programming algorithm for de novo peptide sequencing via tandem mass spectrometry. Anal. Chem. 2007;79:4870–4878. doi: 10.1021/ac070039n. [DOI] [PubMed] [Google Scholar]
- 84.Shevchenko A., Sunyaev S., Loboda A., Shevchenko A., Bork P., Ens W., et al. Charting the proteomes of organisms with unsequenced genomes by MALDI-quadrupole time-of-flight mass spectrometry and BLAST homology searching. Anal. Chem. 2001;73:1917–1926. doi: 10.1021/ac0013709. [DOI] [PubMed] [Google Scholar]
- 85.Young J.C., Pan C., Adams R., Brooks B., Banfield J.F., Morowitz M.J., et al. Metaproteomics reveals functional shifts in microbial and human proteins during a preterm infant gut colonization case. Proteomics. 2015;15(20):3463–3473. doi: 10.1002/pmic.201400563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kolmeder C.A., de Been M., Nikkilä J., Ritamo I., Mättö J., Valmu L., et al. Comparative metaproteomics and diversity analysis of human intestinal microbiota testifies for its temporal stability and expression of core functions. PLoS One. 2012;7(1) doi: 10.1371/journal.pone.0029913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mulle J.G., Sharp W.G., Cubells J.F. The gut microbiome: a new frontier in autism research. Curr. Psychiatry Rep. 2013;15(2):337. doi: 10.1007/s11920-012-0337-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mao L., Franke J. Symbiosis, dysbiosis, and rebiosis-the value of metaproteomics in human microbiome monitoring. Proteomics. 2015;15(5–6):1142–1151. doi: 10.1002/pmic.201400329. [DOI] [PubMed] [Google Scholar]
- 89.Baothman O.A., Zamzami M.A., Taher I., Abubaker J., Abu-Farha M. The role of gut microbiota in the development of obesity and diabetes. Lipids Health Dis. 2016;18(15):108. doi: 10.1186/s12944-016-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xiao M., Yang J., Feng Y., Zhu Y., Chai X., Wang Y. Metaproteomic strategies and applications for gut microbial research. Appl. Microbiol. Biotechnol. 2017;101(8):3077–3088. doi: 10.1007/s00253-017-8215-7. [DOI] [PubMed] [Google Scholar]
- 91.Wilmes P., Heintz-Buschart A., Bond P.L. Microbiota from the distal guts of lean and obese adolescents exhibit partial functional redundancy besides clear differences in community structure. Environ. Microbiol. 2013;15(1):211–226. doi: 10.1111/j.1462-2920.2012.02845.x. [DOI] [PubMed] [Google Scholar]
- 92.Zhang X., Chen W., Ning Z., Mayne J., Mack D., Stintzi A., et al. Deep metaproteomics approach for the study of human microbiomes. Anal. Chem. 2017;89(17):9407–9415. doi: 10.1021/acs.analchem.7b02224. [DOI] [PubMed] [Google Scholar]
- 93.Armengaud J. Microbiology and proteomics, getting the best of both worlds. Environ. Microbiol. 2013;15(1):12–23. doi: 10.1111/j.1462-2920.2012.02811. x.Epub 2012 Jun 19. [DOI] [PubMed] [Google Scholar]
- 94.Guirro M., Costa A., Gual-Grau A., Mayneris-Perxachs J., Torrell H., Herrero P., et al. Multi-omics approach to elucidate the gut microbiota activity: metaproteomics and metagenomics connection. Electrophoresis. 2018;39(13):1692–1701. doi: 10.1002/elps.201700476. [DOI] [PubMed] [Google Scholar]
- 95.Erickson A.R., Cantarel B.L., Lamendella R., Darzi Y., Mongodin E.F., Pan C., et al. Integrated metagenomics/metaproteomics reveals human host-microbiota signatures of Crohn's disease. PLoS One. 2012;7(11) doi: 10.1371/journal.pone.0049138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Presley L.L., Ye J., Li X., Leblanc J., Zhang Z., Ruegger P.M., et al. Host-microbe relationships in inflammatory bowel disease detected by bacterial and metaproteomic analysis of the mucosal-luminal interface. Inflamm. Bowel Dis. 2012;18(3):409–417. doi: 10.1002/ibd.21793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Debyser G., Mesuere B., Clement L., Van de Weygaert J., Van Hecke P., Duytschaever G., et al. Faecal proteomics: A tool to investigate dysbiosis and inflammation in patients with cystic fibrosis. J. Cyst Fibros. 2016;15(2):242–250. doi: 10.1016/j.jcf.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 98.Valles-Colomer M., Falony G., Darzi Y., Tigchelaar E.F., Wang J., Tito R.Y., et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 2019 doi: 10.1038/s41564-018-0337-x. [DOI] [PubMed] [Google Scholar]
- 99.Francino M.P. Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front. Microbiol. 2016;12(6):1543. doi: 10.3389/fmicb.2015.01543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jernberg C., Löfmark S., Edlund C., Jansson J.K. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007;1:56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 101.Pérez-Cobas A.E., Gosalbes M.J., Friedrichs A., Knecht H., Artacho A., Eismann K., et al. Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut. 2013;62(11):1591–1601. doi: 10.1136/gutjnl-2012-303184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rechenberger J., Samaras P., Jarzab A., Behr J., Frejno M., Djukovic A., Sanz J., González-Barberá E.M., Salavert M., López-Hontangas J.L., Xavier K.B., Debrauwer L., Rolain J.M., Sanz M., Garcia-Garcera M., Wilhelm M., Ubeda C., Kuster B. Challenges in clinical metaproteomics highlighted by the analysis of acute leukemia patients with gut colonization by multidrug-resistant enterobacteriaceae. Proteomes. 2019;7(1):E2. doi: 10.3390/proteomes7010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Meijnikman A.S., Gerdes V.E., Nieuwdorp M., Herrema H. Evaluating causality of gut microbiota in obesity and diabetes in humans. Endocr. Rev. 2018;39(2):133–153. doi: 10.1210/er.2017-00192. [DOI] [PubMed] [Google Scholar]
- 104.Gavin P.G., Mullaney J.A., Loo D., Cao K.-A.L., Gottlieb P.A., Hill M.M., et al. Intestinal metaproteomics reveals host-microbiota interactions in subjects at risk for Type 1 diabetes. Diabetes Care. 2018;41(10):2178–2186. doi: 10.2337/dc18-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wei Xiao, Jiang Shan, Chen Yuye, Zhao Xiangna, Li Huan, Lin Weishi, Li Boxing, Wang Xuesong, Yuan Jing, Sun Yansong. Cirrhosis related functionality characteristic of the fecal microbiota as revealed by a metaproteomic approach. BMC Gastroenterol. 2016;16(1) doi: 10.1186/s12876-016-0534-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mancuso C., Santangelo R. The long way between preclinical studies and clinical evidence. Pharmacol. Res. 2018;129:329–336. doi: 10.1016/j.phrs.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 107.Yoshida N., Yamashita T., Hirata K. Gut microbiome and cardiovascular diseases. Diseases. 2018;6(3):E56. doi: 10.3390/diseases6030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Foxman B., Goldberg D., Murdock C., Xi C., Gilsdorf J.R. Conceptualizing human microbiota: from multicelled organ to ecological community. Interdiscip Perspect Infect. Dis. 2008;2008 doi: 10.1155/2008/613979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Heintz-Buschart A., May P., Laczny C.C., Lebrun L.A., Bellora C., Krishna A., et al. Integrated multi-omics of the human gut microbiome in a case study of familial type 1 diabetes. Nat. Microbiol. 2016;10(2):16180. doi: 10.1038/nmicrobiol.2016.180. [DOI] [PubMed] [Google Scholar]
- 110.Reigstad C.S., Kashyap P.C. Beyond phylotyping: understanding the impact of gutmicrobiota on host biology. Neurogastroenterol. Motil. 2013;25(5):358–372. doi: 10.1111/nmo.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tanca A., Abbondio M., Palomba A., Fraumene C., Manghina V., Cucca F., et al. Potential and active functions in the gut microbiota of a healthy human cohort. Microbiome. 2017;5(1):79. doi: 10.1186/s40168-017-0293-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Del Chierico F., Gnani D., Vernocchi P., Petrucca A., Alisi A., Dallapiccola B., et al. Meta-omic platforms to assist in the understanding of NAFLD gut microbiota alterations: tools and applications. Int. J. Mol. Sci. 2014;15(1):684–711. doi: 10.3390/ijms15010684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Heyer R., Schallert K., Zoun R., Becher B., Saake G., Benndorf D. Challenges and perspectives of metaproteomic data analysis. J. Biotechnol. 2017;10(261):24–36. doi: 10.1016/j.jbiotec.2017.06.1201. [DOI] [PubMed] [Google Scholar]
- 114.Sajic T., Liu Y., Aebersold R. Using data-independent, high-resolution mass spectrometry in protein biomarker research: perspectives and clinical applications. Proteom. Clin. Appl. 2015;9(3–4):307–321. doi: 10.1002/prca.201400117. [DOI] [PubMed] [Google Scholar]
- 115.Pible O., Armengaud J. Improving the quality of genome, protein sequence, and taxonomy databases: a prerequisite for microbiome meta-omics 2.0. Proteomics. 2015;15:3418–3423. doi: 10.1002/pmic.201500104. https://doi-org.lama.univ-amu.fr/10.1002/pmic.201500104. [DOI] [PubMed] [Google Scholar]
- 116.Tanca A., Palomba A., Fraumene C., Pagnozzi D., Manghina V., Deligios M., et al. The impact of sequence database choice on metaproteomic results in gut microbiota studies. Microbiome. 2016;4:51. doi: 10.1186/s40168-016-0196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mesuere B., Van der Jeugt F., Devreese B., Vandamme P., Dawyndt P. The unique peptidome: Taxon-specific tryptic peptides as biomarkers for targeted metaproteomics. Proteomics. 2016;16(17):2313–2318. doi: 10.1002/pmic.201600023. [DOI] [PubMed] [Google Scholar]
- 118.Seng P., Rolain J.-M., Fournier P.E., La Scola B., Drancourt M., Raoult D. MALDI-TOF-mass spectrometry applications in clinical microbiology. Future Microbiol. 2010 Nov;5(11):1733–1754. doi: 10.2217/fmb.10.127. [DOI] [PubMed] [Google Scholar]
- 119.Mayers M.D., Moon C., Stupp G.S., Su A.I., Wolan D.W. Quantitative metaproteomics and activity-based probe enrichment reveals significant alterations in protein expression from a mouse model of inflammatory bowel disease. J. Proteome Res. 2017;16(2):1014–1026. doi: 10.1021/acs.jproteome.6b00938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hagen Live H., Frank Jeremy A., Zamanzadeh Mirzaman, Eijsink Vincent G.H., Pope Phillip B., Horn Svein J., Arntzen Magnus Ø., Liu Shuang-Jiang. Quantitative metaproteomics highlight the metabolic contributions of uncultured phylotypes in a thermophilic anaerobic digester. Appl. Environ. Microbiol. 2017;83(2) doi: 10.1128/AEM.01955-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lagier J.-C., Armougom F., Million M., Hugon P., Pagnier I., Robert C., et al. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin. Microbiol. Infect. 2012;18(12):1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 122.Lagier J.-C., Hugon P., Khelaifia S., Fournier P.-E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin. Microbiol. Rev. 2015;28(1):237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lagier J.-C., Dubourg G., Million M., Cadoret F., Bilen M., Fenollar F., et al. Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 2018;1:540–550. doi: 10.1038/s41579-018-0041-0. [DOI] [PubMed] [Google Scholar]
- 124.Li L., Zhang X., Ning Z., Mayne J., Moore J.I., Butcher J., et al. Evaluating in vitro culture medium of gut microbiome with orthogonal experimental design and a metaproteomics approach. J. Proteome Res. 2018;17(1):154–163. doi: 10.1021/acs.jproteome.7b00461. [DOI] [PubMed] [Google Scholar]
- 125.Rooijers K., Kolmeder C., Juste C., Doré J., de Been M., Boeren S., et al. An iterative workflow for mining the human intestinal metaproteome. BMC Genom. 2011;12(6) doi: 10.1186/1471-2164-12-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Juste C., Kreil D.P., Beauvallet C., Guillot A., Vaca S., Carapito C., et al. Bacterial protein signals are associated with Crohn's disease. Gut. 2014;63(10):1566–1577. doi: 10.1136/gutjnl-2012-303786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kashyap P.C., Chia N., Nelson H., Segal E., Elinav E. Microbiome at the frontier of personalized medicine. Mayo Clin. Proc. 2017;92(12):1855–1864. doi: 10.1016/j.mayocp.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Xiong W., Giannone R.J., Morowitz M.J., Banfield J.F., Hettich R.L. Development of an enhanced metaproteomic approach for deepening the microbiome characterization of the human infant gut. J. Proteome Res. 2015;14(1):133–141. doi: 10.1021/pr500936p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kolmeder C.A., Ritari J., Verdam F.J., Muth T., Keskitalo S., Varjosalo M., et al. Colonic metaproteomic signatures of active bacteria and the host in obesity. Proteomics. 2015;15(20):3544–3552. doi: 10.1002/pmic.201500049. [DOI] [PubMed] [Google Scholar]
- 130.Zhang X., Ning Z., Mayne J., Deeke S.A., Li J., Starr A.E., et al. In vitro metabolic labeling of intestinal microbiota for quantitative metaproteomics. Anal. Chem. 2016;88(12):6120–6125. doi: 10.1021/acs.analchem.6b01412. [DOI] [PubMed] [Google Scholar]
- 131.Kolmeder C.A., Salojärvi J., Ritari J., de Been M., Raes J., Falony G., et al. Faecal metaproteomic analysis reveals a personalized and stable functional microbiome and limited effects of a probiotic intervention in adults. PLoS One. 2016;11(4) doi: 10.1371/journal.pone.0153294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li X., LeBlanc J., Truong A., Vuthoori R., Chen S.S., Lustgarten J.L., et al. A metaproteomic approach to study human-microbial ecosystems at the mucosal luminal interface. PLoS One. 2011;6(11) doi: 10.1371/journal.pone.0026542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fenn J., Mann M., Meng C., Wong S., Whitehouse C. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246(4926):64–71. doi: 10.1126/science:2675315. [DOI] [PubMed] [Google Scholar]