Highlights
-
•
Relevant bacterial resistance markers are automatically detected by MALDI-TOF MS subtyping.
-
•
Subtyping detects KPC-producing enterobacteria and methicillin-resistant S. aureus.
-
•
Subtyping differentiates carbapenemase-producing Bacteroides fragilis.
-
•
Subtyping proved to have an excellent specificity.
-
•
Resistant strains can be detected in real-time during routine species identification.
Keywords: MALDI-TOF, Mass spectrometry, KPC, MRSA, Bacteroides fragilis, cfiA, Subtyping, Antibiotic resistance
Abstract
The spread of bacterial resistance has been continuously increasing in the recent decade. Multi-drug resistant (MDR) bacteria now represent one of the most worrisome public health issues, as they seriously complicate the treatment of infections, often leaving few therapeutic options.
Enterobacteria and Staphylococcus aureus are among the most common bacterial pathogens, while Bacteroides fragilis is the most frequent anaerobic pathogen. All of these species can cause severe and life-threatening infections, and represent the most frequent causes of antibiotic-resistant healthcare-associated infections worldwide, as they frequently exhibit resistance to various classes of antibiotics. Resistance to carbapenems, the last resort beta-lactam agent, is a particularly threatening problem. Achieved by different mechanisms, leads to total inefficacy of any beta-lactam agent.
During the recent years, MALDI-TOF mass spectrometry has become established as the reference method for bacterial identification in routine practice. It has proven to be a reliable and robust method to detect specific peaks in bacterial mass spectra, corresponding to specific resistance markers, enabling the instant detection of resistant isolates in real time during the standard routine identification process. Here, we investigated the performance of the subtyping module of the MALDI Biotyper system (Bruker Daltonik, GmbH) for the instant identification of KPC-producing Klebsiella pneumoniae, methicillin-resistant Staphylococcus aureus, and carbapenemase-producing Bacteroides fragilis during the identification workflow. We evaluated accuracy and potential impact on turnaround time. Furthermore, we investigated the possibility to extend the subtyping for detection of the KPC-specific marker to bacterial species other than K. pneumoniae.
1. Introduction
Bacterial resistance to antibiotics has been continuously increasing in the recent decade. Multi-drug resistant (MDR) bacteria now represent a concerning public health and economic threat [1], [2], recently indicated by the WHO as one of the most serious public health issues of our time [3]. Moreover, few new antibiotics are under development [4], hence, images of a return to the pre-antibiotic era can be easily conjured.
Previously confined to hospitals, MDR bacteria are now also found in the community, environment and animals [5], [6]. MDR bacteria can significantly complicate the treatment of infections, especially in critically ill patients, leaving clinicians with few and problematic treatment options [7], [8], [9].
Enterobacterales and Staphylococcus aureus, normally found as associated flora, are among the most clinically relevant bacterial pathogens, involved both in hospital-acquired and community-acquired infections. They play a major role among MDR organisms, exhibiting broad spectrum resistance to the various classes of antibiotics, and representing the most frequent causes of antibiotic-resistant healthcare-associated infections worldwide [10], [11]. Resistance to beta-lactam antibiotics, especially carbapenems, is a particularly threatening problem. It is caused by different mechanisms in the two groups of bacteria, which are leading to total inefficacy of any beta-lactam agent.
Resistance to carbapenems in Enterobacterales has been dramatically increasing worldwide over the last decade [8], [12], [13], [14]. This resistance involves various genera and species, and can be attributed to different molecular mechanisms [15]. Among them, bacteria producing carbapenem-hydrolyzing enzymes (carbapenemases) are the most worrisome, as they can spread easily and contaminate healthcare settings. The Klebsiella pneumoniae Carbapenemase (KPC) is the most clinically significant [12]. It is found predominantly in K. pneumoniae, and its global dissemination is a matter of great concern [16], given the high morbidity and mortality rates associated with invasive infection caused by these strains [12], [13], [17], [18].
S. aureus resistance to carbapenems is exhibited by methicillin-resistant strains (MRSA), which have developed resistance to methicillin, and to all other beta-lactam agents, except for new anti-MRSA cephalosporins (ceftaroline and ceftobiprole), by horizontal transfer and natural selection of genes that code for a mutant penicillin-binding protein (PBP2a). PBP2a has a low affinity for beta-lactam molecules, and thus prevents the activity of beta-lactam drugs. Despite a declining frequency of MRSA in the last few years (ECDC 2016), it remains a major cause of healthcare-associate infection. Moreover, the transfer and spread of healthcare-associated MRSA clones into the community have been occurring [19].
Bacteroides fragilis is the most frequent anaerobic pathogen, and can cause severe infections [20]. The species is split into two DNA homology groups, named Division I and Division II [21], [22]. Resistance to carbapenems is associated with the cfiA gene-encoded metallo-beta-lactamases, constitutively present in Division II of the species [23], and represents an emerging problem [24], [25].
The ability of clinical microbiology laboratories to detect resistant strains is crucial for diagnosis and treatment, as well as for epidemiological purposes. Rapid, reliable and cost-effective tests are required to improve patient outcome, and for the implementation of the adequate infection control practices [8], [26].
Various phenotypic and genotypic methods are currently available to detect carbapenemase-producing Enterobacterales and MRSA. Phenotypic methods are frequently inexpensive and easy-to-use, but are either slow, lack sensitivity and/or specificity [18], or are not suitable for epidemiological investigations, as many of them detect the presence of the enzymatic activity responsible for the antibiotic resistance, but don’t enable discrimination among the different enzymes types that can be involved. On the other hand, PCR-based methods are accurate and reliable, but expensive, and sometimes laborious.
Matrix-Assisted Laser Desorption-Ionization Time-of-Flight mass spectrometry (MALDI-TOF MS) has become the universal reference method for microbial species identification in clinical microbiology. Its introduction into laboratories has significantly reduced the time required for identification of bacteria and fungi (now minutes versus hours to days), both with lower cost and increased accuracy [27], [28].
Beyond identification, MALDI-TOF MS has further become a platform of investigation for antibiotic resistance. Recently, novel applications of this technology have facilitated the development of new methods for easy and rapid detection of bacterial antibiotic resistance. Specific peaks in the bacterial mass spectra were have been identified as antibiotic-resistance markers [29], [30]. The development of dedicated algorithms for their detection, implemented into the commercially available MALDI-TOF MS system, have enabled the automated instant identification of resistant strains, in real-time during the standard species identification process. KPC-producing K. pneumoniae strains can be identified by the detection of a peak at 11109 m/z, specifically associated with one of the most common KPC-carrying plasmids [29]. A subgroup of methicillin resistant S. aureus strains can be identified by the detection of a peak at 2412 m/z [30]. The carbapenemase-producing (Division II) B. fragilis subgroup can be differentiated from Division I by a predictable shift in the MALDI mass spectral pattern [31].
Here, we investigated the performance and the diagnostic value of the automated detection of KPC-producing Klebsiella pneumoniae, methicillin-resistant Staphylococcus aureus, and carbapenemase-producing Bacteroides fragilis using the subtyping functionality of the MALDI Biotyper system (Bruker Daltonik, GmbH) implemented as part of the routine workflow. The reliability and the robustness of the method have been evaluated in previous studies, investigating datasets of molecularly well-characterized isolates [31], [32], thus in this study we considered unnecessary any molecular confirmation of the results delivered by the MALDI approach.
2. Materials and methods
All routine clinical isolates of S. aureus and B. fragilis, and all surveillance and positive blood cultures isolates of K. pneumoniae collected in the laboratory of bacteriology of the University Hospital of Bologna Policlinico Sant’Orsola-Malpighi between 1st February – 15th May 2018 were included in this study. The strains were identified at species level by MALDI-TOF MS (MALDI Biotyper, Bruker Daltonik GmbH, Bremen, Germany), following the manufacturer’s instruction. Subtyping for the detection of resistance markers was performed simultaneously with species identification by the subtyping functionality implemented into the Biotyper MBT Compass software.
Identification results with low confidence level (log score < 2.0) were excluded.
S. aureus, B. fragilis and K. pneumoniae isolates derived from blood cultures were cultivated on Tryptone Soy Agar with 5% sheep blood (Meus S.r.l, Piove di Sacco, Italy) for 24–48 h. Surveillance K. pneumoniae isolates were cultivated on CHROMagar KPC agar or CHROMagar ORIENTATION Meus S.r.l., Piove di Sacco, Italy).
2.1. KPC-producing K. pneumoniae
N = 684 clinical and surveillance isolates of K. pneumoniae were subtyped by the detection of a specific peak at m/z 11,109 related to the pKpQIL blaKPC harbouring plasmid [29].
As the reliability and the robustness of the method has been previously assessed [32], when the KPC-related peak was detected, no further routine investigations were performed, besides the standard susceptibility testing (NM-EN51 panel, Microscan WalkAway, Beckman Coulter Inc., Brea, CA, USA).
Strains that did not show the KPC-related peak, but exhibited a reduced susceptibility to carbapenems (defined as MIC for at least one of the carbapenems higher than the epidemiological cut off) with routine susceptibility testing underwent verification of carbapenemase production by the reference method (disc-diffusion synergy test – KPC + MBL Confirm ID Pack, ROSCO Diagnostika, Taastrup, Denmark), to evaluate whether a carbapenemase other than the pKpQIL plasmid-related KPC was present.
The impact of this new method in terms of time-to-response for KPC-producing isolates was evaluated by comparing the reporting time of these samples with the reporting time of an equal number of routine samples randomly selected from the period of time prior to implementation of the Biotyper subtyping module. The former workflow included, as the first step, the Carba-NP test (Neo-Rapid CARB-Kit, Rosco Diagnostics, Taastrup, Denmark), in case of negativity, followed by a disc-diffusion synergy test).
2.2. Methicillin resistant S. aureus (MRSA)
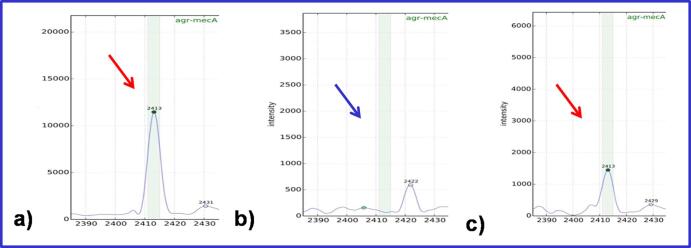
S. aureus clinical isolates (n = 593) were subtyped by the Biotyper software through detection of a specific peak at m/z 2412. This peak corresponds to a PSM-mec peptide, related to one of the mechanisms that cause methicillin-resistance [30] – Fig. 1.
Fig. 1.
Automated detection of the specific PSM peak at m/z 2411–2419. a) S. aureus ATCC 33591 (PSM-mec positive); b) clinical isolate of MRSA (PSM-mec negative); c) clinical isolate of MRSA (PSM-mec positive).
Classification into methicillin-resistant/susceptible was performed on the basis of susceptibility to oxacillin and to cefoxitin, as specific screening beta-lactam antibiotics, included into the routine susceptibility test (PM STA-36 panel, Microscan WalkAway, Beckman Coulter).
2.3. Carbapenemase-producing B. fragilis
B. fragilis clinical isolates (n = 35) were subtyped into Division I/II (cfiA-negative/positive) by their specific pattern in the mass spectra, as previously described [31]. Susceptibility to carbapenems was investigated as part of a routine diagnostic workflow to determine the Minimum Inhibitory Concentration (MIC) of one of the carbapenems (meropenem) by the MIC gradient strip methodology (M.I.C. Evaluator, Oxoid, Basingstoke, UK) according to EUCAST guidelines, version 8.1 (www.eucast.org).
2.4. KPC subtyping of enterobacteria other than K. pneumoniae
The KPC-related peak was searched in other Enterobacterales species (all species for which at least one KPC-producing isolate was found in the Bacteriology laboratory of the University Hospital of Bologna since the appearance of the first KPC-producing isolate in 2010).
MALDI-TOF mass spectra of 8801 clinical and surveillance isolates of clinically relevant Enterobacterales species, other than K. pneumoniae, were investigated for the presence of the KPC-related peak at 11109 m/z described in KPC-producing K. pneumoniae (see Section 2.2). The spectra were collected in the Bacteriology laboratory of the MVZ Lab in Dortmund (n = 7694) and in Bologna (n = 1107), and exhibited different susceptibility patterns to carbapenems (Table 1).
Table 1.
Dataset of enterobacteria other than K. pneumoniae, and sensitivity of KPC-production by MALDI Biotyper subtyping for the different species.
| Species | Tot. | Carbapenem-S |
carbapenem-R |
KPC-producing | KPC-peak detected | |
|---|---|---|---|---|---|---|
| Italy | Germany | Italy | ||||
| E. coli | 3502 | 398 | 2735 | 369 | 146 | 126 (86.3%) |
| K. aerogenes | 414 | 18 | 362 | 34 | 11 | 9 (81.8%) |
| E. cloacae complex | 2249 | 90 | 2085 | 74 | 5 | 5 (100%) |
| K. oxytoca | 1460 | 31 | 1413 | 16 | 5 | 3 (60%) |
| C. freundii | 639 | 30 | 587 | 22 | 9 | 8 (88.9%) |
| S. marcescens | 537 | 19 | 512 | 6 | 6 | 4 (66.7%) |
| 8801 | 586 | 7694 | 521 | |||
All strains had undergone routine susceptibility testing by Vitek2 (bioMerieux, Marcy L’Etoile, France), and confirmation of carbapenemase-production by a disk-diffusion synergy test (KPC + MBL Confirm ID Pack, Rosco Diagnostics) in case of reduced susceptibility to carbapenems.
In detail, E. coli (n = 3502), E. cloacae (n = 2249) complex, K. aerogenes (n = 414), K. oxytoca (n = 1460), Citrobacter spp. (n = 639) and S. marcescens (n = 537) clinical and surveillance isolates were included (Table 1).
As this functionality was not part for the MALDI Biotyper software at the time of this study, for automated detection of the KPC-related peak in non-K. pneumoniae enterobacteria, a specific software algorithm was developed for each species (E. coli, E. cloacae, E. kobei, E. asburiae, E. ludwigii, K. aerogenes, K. oxytoca, C. freundii, S. marcescens). The peak detection was based on results of visual analysis of spectra performed with the flexAnalysis® software version 3.4 (Bruker Daltonik, Bremen, Germany) – Fig. 2. Spectra were normalized and smoothed with standard settings. For precise detection, the spectra were screened for species-specific potential peaks for an internal recalibration, as described earlier [33]. After internal recalibration, intensities 3-times higher than the surrounding noise were counted as peaks. If a peak was detected in a window ±5 m/z around the previously described mass of 11109 m/z, the algorithm accepted the detection of the KPC-related peak. Examples of the automated detection results are shown in Fig. 3.
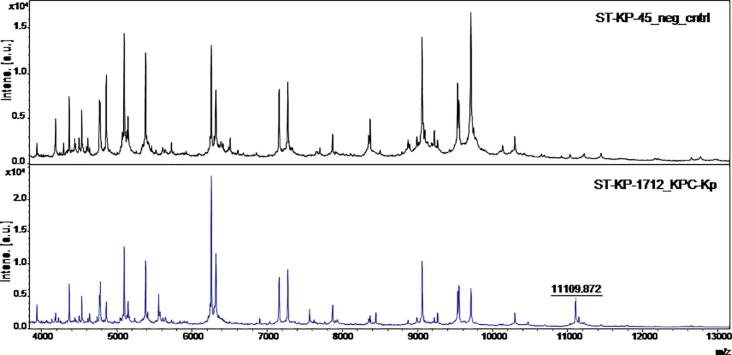
Fig. 2.
The KPC-related peak at 11.109 m/z in the bacterial MALDI-TOF MS mass spectra of a KPC-producing isolate (lower spectrum), in comparison with a non-KPC producing isolate (upper spectrum).
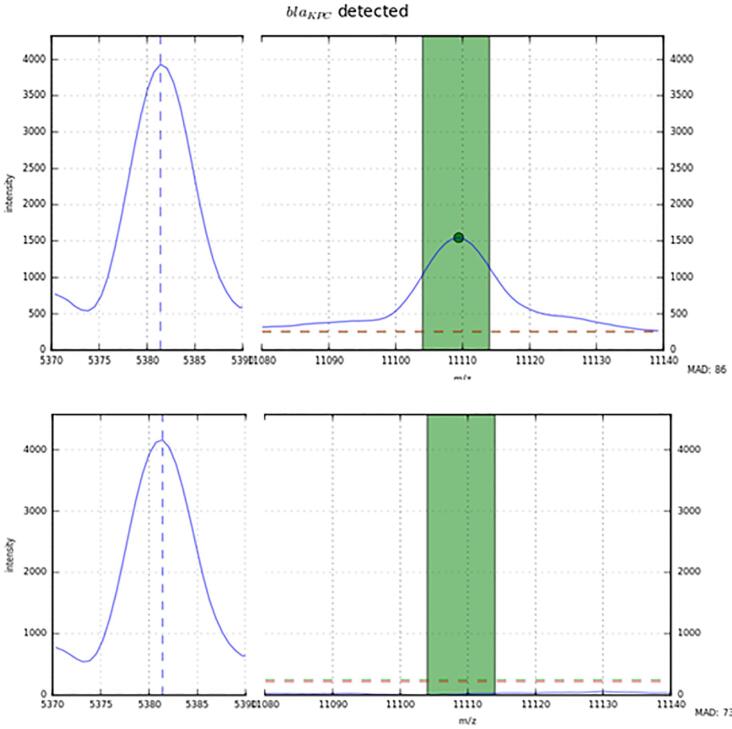
Fig. 3.
Detection of the KPC-specific peak by the automated algorithm. For each species, the dedicated specific algorithm recognized an internal calibration peak, related to the species, and the KPC-related peak at 11,109 m/z, if present.
3. Results
3.1. KPC-producing K. pneumoniae
371 K. pneumoniae strains were subtyped as “presumptive KPC”.
Among the 313 strains subtyped as “non-KPC”, 24 strains resulted positive for the production of a class A carbapenemase to synergy test (KPC). Therefore, sensitivity of KPC detection was calculated as 94% (371/395) for the tested sample collective (Table 2).
Table 2.
Sensitivity and specificity of detection of KPC-production in K. pneumoniae by MALDI Biotyper subtyping.
| K. pneumoniae | All isolates (n = 684) |
|
|---|---|---|
| KPC-peak detected | KPC-peak NOT detected | |
| KPC+ (n = 395) | 371 (93.9%) | 24 |
| KPC- (n = 289) | – | 289 (100%) |
The method in this study enabled us to report the presence of KPC-producing isolates earlier compared to the former routine approach. The time saved ranged from 1.5 to 24 h, depending on which of the methods included in the multistep routine workflow detected the positive result (Carba NP or disc-diffusion synergy test – KPC, respectively).
3.2. Methicillin resistant S. aureus (MRSA)
28 S. aureus strains were subtyped as “presumptive PSM-positive MRSA”, corresponding to 15.7% (28/178) of the total number of MRSA detected by standard susceptibility testing. None of the 415 methicillin-susceptible strains were subtyped as “presumptive PSM-positive MRSA”.
The “presumptive PSM-positive MRSA” warning provided by the instrument during routine practice allowed a reduction in the time to reporting by one day.
3.3. Carbapenemase-producing B. fragilis
32 (91.4%) B. fragilis strains were subtyped as Division I, and 3 (8.6%) as Division II. The result of routine susceptibility testing for meropenem was coherent with the Biotyper classification. The MALDI subtyping into Division I/II (carbapenem-susceptible/resistant, respectively) enabled a reduction in the time-to-response, regarding susceptibility to carbapenems, by 24–48 h in comparison with the MIC determination.
3.4. KPC subtyping of other enterobacteria
The same KPC-related peak at 11109 m/z described in K. pneumoniae was detected in 126/146 (86.3%) E. coli, 5/5 (100%) E. cloacae complex, 9/11 (81.8%) K. aerogenes, 3/5 (60%) K. oxytoca, 8/9 (88.9%) C. freundii, and 4/6 (66.7%) S. marcescens KPC-producers (Table 1).
4. Discussion
In this study, we investigated the performance and the diagnostic value of the automated detection of KPC-producing Klebsiella pneumoniae, methicillin-resistant Staphylococcus aureus, and carbapenemase-producing Bacteroides fragilis using the subtyping functionality of the MALDI Biotyper system (Bruker Daltonik, GmbH) and its suitability to be implemented into the routine workflow. K. pneumoniae isolates were subtyped for the presence of the blaKPC pKpQIL carrying a plasmid-related peak present only in KPC-producing strains. S. aureus isolates were subtyped for the presence of the PSM peak, related to a subgroup of methicillin-resistant strains. B. fragilis isolates were subtyped into Division I and II by the detection of a specific mass spectral pattern.
MALDI-TOF MS subtyping proved to be a reliable and promising method to recognize resistant strains simultaneously with identification at species level, in a short time, and without requiring any further tasks besides classical routine identification procedures. The implementation of KPC subtyping further simplified and shortened the laboratory workflow, as it reduced the handling time and the number of samples that required further analytical steps.
Sensitivity of this approach was different for the three groups of bacteria investigated (∼95% for KPC-K. pneumoniae, ∼20% for methicillin-resistant S. aureus, and ∼100% for carbapenemase-producing B. fragilis). This difference can be easily explained with some epidemiological considerations. In fact, subtyping for S. aureus and K. pneumoniae depends on the epidemiological situation, as it relies on the detection of one of the possible mechanisms for that given resistance, and thus, it was shown to be related only to the prevalence of the resistance type detectable by MALDI among all the circulating resistant strains (prevalence of the KPC-producing strains harbouring the pKpQIL plasmid among all the KPC-producing K. pneumoniae circulating clones, and prevalence of the subgroup of methicillin-resistant S. aureus harbouring the mecA cassette containing the gene which encodes the psm-peak related small protein).
In contrast, for B. fragilis, subtyping relies on the detection of strains that belong to one of the two DNA-homology groups in which the species is divided. Division II harbors the only mechanism for resistance to carbapenems currently known for B. fragilis. Hence, sensitivity overlaps with prevalence of the Division II subfamily among all the B. fragilis isolates and was found to be 100%.
For the three groups investigated, specificity was found to be excellent (100%), as the specific peaks/pattern were not found in any of the susceptible isolates.
5. Conclusions
The MALDI subtyping approach enabled a significantly faster time to report in comparison to the routine procedure, saving between 1 and 24 h for KPC-producing K. pneumoniae, 24 h for methicillin-resistant S. aureus, and 24–48 h for carbapenem-resistant B. fragilis. Thus, the method could enable an earlier adoption of proper surveillance measures. Moreover, the shortened time to report in KPC-producing strains from blood cultures might have the potential to significantly reduce the time to therapeutic escalation.
Furthermore, this study showed that KPC subtyping can be successfully extended to further species of enterobacteria. The same KPC-related peak described in K. pneumoniae was detected in the majority of the KPC-producing strains of all the other species of Enterobacterales investigated (providing interesting hints for further investigations regarding the transfer of this genetic determinant among the different species). The prevalence of the KPC-specific peak in KPC-bearing strains of these other species was similar to the one in KPC K. pneumoniae.
These findings on one hand prove that MALDI subtyping for detection of antibiotic resistance markers is a valid and useful method, mature enough to be implemented into the routine practice. On the other hand, this approach could have a huge potential to be expanded to other combinations of bacteria/resistances, as soon as specific resistance markers are identified.
This research did not receive any specific grant from funding agencies in the public, commercial or not-for-profit sectors.
Declaration of Competing Interest
MC, ABP and SA have any conflicts of interest to disclose.
Authors with financial interests or relationships to disclose are listed with their details below: MK is employee of Bruker Daltonik GmbH, manufacturer of the MALDI Biotyper system.
Corresponding author confirms here the conflict of interest statements provided during the initial submission of the manuscript.
Glossary
- MDR
Multi-Drug Resistant
- KPC
Klebsiella pneumoniae Carbapenemase
- MRSA
Methicillin-resistant Staphylococcus aureus
References
- 1.Carlet J., Pulcini C., Piddock L.J. Antibiotic resistance: a geopolitical issue. Clin. Microbiol. Infect. 2014;20(10):949–953. doi: 10.1111/1469-0691.12767. [DOI] [PubMed] [Google Scholar]
- 2.Shriber D.E., Baris E., Marquez P.V., et al. Final report. The World Bank. 2017;2017:1–172. http://documents.worldbank.org/curated/en/323311493396993758/pdf/114679-REVISED-v2-Drug-Resistant-Infections-Final-Report.pdf [Google Scholar]
- 3.WHO. Antimicrobial Resistance: Global Report on Surveillance. (2014) http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf?ua=1).
- 4.Nambiar S., Laessig K., Toerner J., Farley J., Cox E. Antibacterial drug development: challenges, recent developments, and future considerations. Clin. Pharmacol. Ther. 2014;96(2):147–149. doi: 10.1038/clpt.2014.116. [DOI] [PubMed] [Google Scholar]
- 5.Kelly A.M., Mathema B., Larson E.L. Carbapenem-resistant Enterobacteriaceae in the community: a scoping review. Int. J. Antimicrob. Agents. 2017;50:127–134. doi: 10.1016/j.ijantimicag.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abraham S., Wong H.S., Turnidge J., Johnson J.R., Trott D.J. Carbapenemase-producing bacteria in companion animals: a public health concern on the horizon. J. Antimicrob. Chemother. 2014;69:1155–1157. doi: 10.1093/jac/dkt518. [DOI] [PubMed] [Google Scholar]
- 7.Livermore D.M. Fourteen years in resistance. Int. J. Antimicrob. Agents. 2012;39:283–294. doi: 10.1016/j.ijantimicag.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 8.Nordmann P., Poirel L. The difficult-to-control spread of carbapenemase producers in Enterobacteriaceae worldwide. Clin. Microbiol. Infect. 2014;20:821–830. doi: 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 9.Falagas M., Lourida P., Puolikakos P., Rafailidis P.I., Tansarli G.S. Antibiotic treatment of infections due to carbapenem-resistant Enterobacteriaceae: a systematic evaluation of the available evidence. Antimicrob. Agents Chemother. 2014;58(2):654–663. doi: 10.1128/AAC.01222-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lowy F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998;339(8):520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 11.Ko W.C., Paterson D.L., Sagnimeni A.J., Hansen D.S., Von Gottberg A., Mohapatra S., Casellas J.M., Goossens H., Mulazimoglu L., Trenholme G., Klugman K.P., McCormack J.G., Yu V.L. Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg. Infect. Dis. 2002;8:160–166. doi: 10.3201/eid0802.010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munoz-Price L.S., Poirel L., Bonomo R.A., Schwaber M.J., Daikos G.L., Cormican M., Cornaglia G., Garau J., Gniadkowski M., Hayden M.K., Kumarasamy K., Livermore D.M., Maya J.J., Nordmann P., Patel J.B., Paterson D.L., Pitout J., Villegas M.V., Wang H., Woodford N., Quinn J.P. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 2013;13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tangden T., Giske C.G. Global dissemination of extensively drug-resistant carbapenemase-producing Enterobacteriaceae: clinical perspectives on detection, treatment and infection control. J. Intern. Med. 2015;277:501–512. doi: 10.1111/joim.12342. [DOI] [PubMed] [Google Scholar]
- 14.Patel G., Bonomo R.A. “Stormy waters ahead”: global emergence of carbapenemases. Front. Microbiol. 2013;14(4):48. doi: 10.3389/fmicb.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nordmann P., Dortet L., Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol. Med. 2012;18:263–272. doi: 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 16.van Duin D., Doi Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence. 2017;8(4):460-469. doi: 10.1080/21505594.2016.1222343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esterly J.S., Wagner J., McLaughlin M.M., Postelnick M.J., Qi C., Scheetz M.H. Evaluation of clinical outcomes in patients with bloodstream infections due to Gram-negative bacteria according to carbapenem MIC stratification. Antimicrob. Agents Chemother. 2012;56:4885–4890. doi: 10.1128/AAC.06365-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamma P.D., Goodman K.E., Harris A.D., Tekle T., Robert A., Taiwo A., Simner P.J. Comparing the outcomes of patients with carbapenemase-producing and non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae bacteremia. Clin. Infect. Dis. 2017;64:257–264. doi: 10.1093/cid/ciw741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grundmann H., Schouls L.M., Aanensen D.M., Pluister G.N., Tami A., Chlebowicz M., Glasner C., Sabat A.J., Weist K., Heuer O., Friedrich A.W., ESCMID Study Group on Molecular Epidemiological Markers, European Staphylococcal Reference Laboratory Working Group The dynamic changes of dominant clones of Staphylococcus aureus causing bloodstream infections in the European region: results of a second structured survey. EuroSurveill. 2014;19(49) doi: 10.2807/1560-7917.es2014.19.49.20987. [DOI] [PubMed] [Google Scholar]
- 20.Ng L.S., Kwang L.L., Rao S., Tan T.Y. Anaerobic bacteremia revisited: species and susceptibilities. Ann. Acad. Med. Singapore. 2015;44(1):13–18. [PubMed] [Google Scholar]
- 21.Podglajen I., Breuil J., Casin I., Collatz E. Genotypic identification of two groups within the species Bacteroides fragilis by ribotyping and by analysis of PCR-generated fragment patterns and insertion sequence content. J. Bacteriol. 1995;177(18):5270–5275. doi: 10.1128/jb.177.18.5270-5275.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutacker M., Valsangiacomo C., Piffaretti J.C. Identification of two genetic groups in Bacteroides fragilis by multilocus enzyme electrophoresis: distribution of antibiotic resistance (cfiA, cepA) and enterotoxin (bft) encoding genes. Microbiology. 2000;146:1241–1254. doi: 10.1099/00221287-146-5-1241. [DOI] [PubMed] [Google Scholar]
- 23.Cuchural G.J., Jr, Malamy M.H., Tally F.P. b-Lactamase-mediated imipenem resistance in Bacteroides fragilis. Antimicrob. Agents Chemother. 1986;30(5):645–648. doi: 10.1128/aac.30.5.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagy E., Urbán E., Nord C.E., ESCMID Study Group on Antimicrobial Resistance in Anaerobic Bacteria Antimicrobial susceptibility of Bacteroides fragilis group isolates in Europe: 20 years of experience. Clin. Microbiol. Infect. 2011;17(3):371–379. doi: 10.1111/j.1469-0691.2010.03256.x. [DOI] [PubMed] [Google Scholar]
- 25.Hartmeyer G.N., Sóki J., Nagy E., Justesen U.S. Multidrug-resistant Bacteroides fragilis group on the rise in Europe? J. Med. Microbiol. 2012;61:1784–1788. doi: 10.1099/jmm.0.049825-0. [DOI] [PubMed] [Google Scholar]
- 26.Banerjee R., Humpries R. Clinical and laboratory considerations for the rapid detection of carbapenem-resistant Enterobacteriaceae. Virulence. 2017;8:427–439. doi: 10.1080/21505594.2016.1185577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel R. MALDI-TO MS for the diagnosis of infectious diseases. Clin. Chem. 2015;61:100–111. doi: 10.1373/clinchem.2014.221770. [DOI] [PubMed] [Google Scholar]
- 28.Dingle T.C., Butler-Wu S.M. MALDI-TOF mass spectrometry for microorganism identification. Clin. Lab Med. 2013;33:589–609. doi: 10.1016/j.cll.2013.03.001. http://dx.doi.org/101016/j.cll.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Lau A.F., Wang H., Weingarten R.A., Drake S.K., Suffredini A.F., Garfield M.K., Chen Y., Gucek M., Youn J.H., Stock F., Tso H., DeLeo J., Cimino J.J., Frank K.M., Dekker J.P. A rapid matrix-assisted laser desorption ionization-time of flight mass spectrometry-based method for single-plasmid tracking in an outbreak of carbapenem-resistant Enterobacteriaceae. J. Clin. Microbiol. 2014;52(8):2804–2812. doi: 10.1128/JCM.00694-14. Epub 2014 May 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Josten M., Dischinger J., Szekat C., Reif M., Al-Sabti N., Sahl H.G., Parcina M., Bekeredjian-Ding I., Bierbaum G. Identification of agr-positive methicillin-resistant Staphylococcus aureus harbouring the class A mec complex by MALDI-TOF mass spectrometry. Int. J. Med. Microbiol. 2014;304(8):1018–1023. doi: 10.1016/j.ijmm.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 31.Nagy E., Becker S., Sóki J., Urbán E., Kostrzewa M. Differentiation of division I (cfiA-negative) and division II (cfiA-positive) Bacteroides fragilis strains by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Med. Microbiol. 2011;60(Pt. 11):1584–1590. doi: 10.1099/jmm.0.031336-0. [DOI] [PubMed] [Google Scholar]
- 32.Cordovana M., Kostrzewa M., Glandorf J., Bienia M., Ambretti S., Pranada A.B. A full MALDI-based approach to detect plasmid-encoded KPC-producing Klebsiella pneumoniae. Front. Microbiol. 2018;23(9):2854. doi: 10.3389/fmicb.2018.02854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pranada A.B., Witt E., Bienia M., Kostrzewa M., Timke M. Accurate differentiation of Mycobacterium chimaera from Mycobacterium intracellulare by MALDI-TOF MS analysis. J. Med. Microbiol. 2017;66(5):670–677. doi: 10.1099/jmm.0.000469. [DOI] [PubMed] [Google Scholar]